Abstract
Polydactyly is a common congenital anomaly of the hand and foot. Postaxial polydactyly (PAP) is characterized by one or more posterior or postaxial digits. In a Pakistani family with autosomal recessive nonsyndromic postaxial polydactyly type A (PAPA), we performed genomewide genotyping, linkage analysis, and exome and Sanger sequencing. Exome sequencing revealed a homozygous nonsense variant (c.478C>T, p.[Arg160*]) in the FAM92A gene within the mapped region on 8q21.13-q24.12 that segregated with the PAPA phenotype. We found that FAM92A is expressed in the developing mouse limb and E11.5 limb bud including the progress zone and the apical ectodermal ridge, where it strongly localizes at the cilia level, suggesting an important role in limb patterning. The identified variant leads to a loss of the FAM92A/Chibby1 complex that is crucial for ciliogenesis and impairs the recruitment and the colocalization of FAM92A with Chibby1 at the base of the cilia. In addition, we show that Fam92a−/− homozygous mice also exhibit an abnormal digit morphology, including metatarsal osteomas and polysyndactyly, in addition to distinct abnormalities on the deltoid tuberosity of their humeri. In conclusion, we present a new nonsyndromic PAPA ciliopathy due to a loss-of-function variant in FAM92A.
Keywords: CILIOPATHY, POSTAXIAL POLYDACTYLY, FAM92A, CHIBBY1, PAPA
Introduction
Postaxial polydactyly (PAP) is a frequent congenital digit malformation characterized by one or more extra posterior digits. Polydactyly results from defective patterning of the anterior-posterior axis of the developing limb and can manifest as an isolated malformation (nonsyndromic) or as part of a syndrome.(1) Syndromic cases encompass 14.6% of all polydactyly and 11.8% of postaxial polydactyly.(2) Nonsyndromic PAP can be broadly divided into two types: PAPA, with fully developed extra digits, and PAPB, showing incompletely developed extra digits (pedunculated postminimus).(3)
Seven nonsyndromic PAPA loci have been mapped through linkage analysis, PAPA1-PAPA4 (autosomal dominant)(4–7) and PAPA5-PAPA7 (autosomal recessive).(8,9) Only three causal genes, GLI3 (PAPA1),(10) ZNF141 (PAPA6),(9) and IQCE (PAPA7),(11) have been identified. Variants in GLI3 are associated with both PAPA and PAPB, even within the same family.(12) However, most identified PAP genes are involved in syndromic disease. There are >170 genes currently associated with syndromic PAP.(13) Genes associated with syndromic or nonsyndromic PAP can be classified under ciliopathies (ie, genes related to cilia biogenesis, structure, and functions) and nonciliopathies.(3) Known PAP syndromic ciliopathies include Meckel and Bardet-Biedl syndromes.(3) GLI3 and IQCE both regulate sonic hedgehog (SHH) signaling,(14,15) which localizes to or shuttles through the primary cilium.(15)
Herein, we describe the mapping of a new PAPA locus on chromosome 8q22.1 and identification of the gene underlying gene, FAM92A, which is also a new PAPA ciliopathy. In addition, we show that Fam92a−/− homozygous mice exhibit distinct bone and digit abnormalities, demonstrating the importance of this gene in both human and mouse bone metabolism.
Materials and Methods
Ascertainment
The study was approved by the Institutional Review Boards of the Quaid-i-Azam University, Pakistan, and Baylor College of Medicine (BCM) and Affiliated Hospitals, USA (protocols #IRB-QAU-155 and H-41049). Written informed consent was obtained from all study participants.
Family BD152 (Fig. 1A) with autosomal recessive nonsyndromic postaxial polydactyly type A was ascertained from a remote area in district Dera Ismail Khan of Khyber Pakhtunkhwa (KPK) province in Pakistan. Information on the pedigree structure and family history were obtained through interview with family members. All family members who participated in the study underwent a medical examination to document PAPA and to rule out that PAPA is part of a syndrome. X-rays were obtained from hands and feet. Patients were examined for features of ciliopathy, renal, and ocular anomalies. Blood samples were obtained from three affected (V:1, V:2, V:3) and three unaffected (III:3, IV:1, V:4) family members, and DNA was extracted using a standard phenol chloroform procedure.(16)
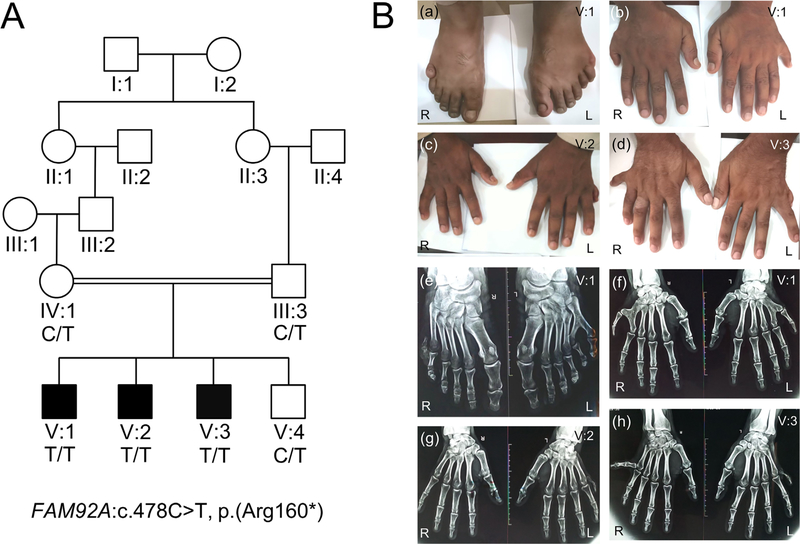
Fig. 1.
Pedigree and clinical features. (A) Pedigree drawing of family BD152 with autosomal recessive PAPA. Clear symbols represent unaffected individuals, whereas filled ones represent family members with PAPA. The individuals with a genotype had available DNA samples that underwent genomewide genotyping. The DNA sample from affected family member V:2 also underwent exome sequencing. (B) Patient V:1 showing bilateral PAPA in feet (a) and bilateral PAPA in hands (b). Patient V:2 showing postaxial extra finger in the left hand (c). Patient V:3 showing extra finger deviated to the ulnar side in the right hand and underdeveloped finger in the left hand (d). Radiograph of feet of patient V:1 showing 6th digit without metatarsal in right foot and varus deviation of the 5th and valgus deviation of the 6th toe with extra toe attached to the 5th metatarsal in left foot (e). Radiograph of hands of the same patient showing bilateral extra fingers attached to the 5th metacarpals in each hand (f). Radiograph of hands of affected individual V:2 displaying unilateral PAP in the left hand (g). Radiograph of hands of affected individual V:3 showing well-developed phalanges of 6th finger with valgus deviation in the right hand and underdeveloped phalanges of extra finger in the left hand (h). R=right; L=left.
Genomewide genotyping
Genotyping was performed using the Infinium HumanCytoSNP-12 BeadChip (Illumina Inc., San Diego, CA, USA), which interrogates 299,140 single-nucleotide polymorphism (SNP) markers, following the manufacturer’s instructions. Data quality control included removing SNPs with possible genotyping errors that were detected through Mendelian inconsistences (PedCheck) and double recombination events over short genetic distances (Merlin).(17,18) Analysis of the genotype data was performed using parametric multipoint and two-point linkage analysis (Allegro 1.2) and homozygosity mapping (HomozygosityMapper).(19,20) For the linkage analysis, a fully penetrant autosomal recessive mode of inheritance was assumed with a disease frequency of 1.0×10−4. Marker allele frequencies were estimated from founders and reconstructed founders of the Pakistani families that were genotyped at the same time.
Exome sequencing
Genomic DNA of one affected family member (V:2) was selected for exome sequencing. Exome library preparation was performed using the Roche/Nimblegen SeqCap EZ v2.0 (~36.5 MB). Barcoded libraries were pooled and sequencing was performed on an Illumina HiSeq with an average on-target coverage of 54×. Reads were aligned to GRCh37/Hg19 using Burrows-Wheeler Aligner (BWA-MEM).(21) Duplicate removal, indel-realignment, quality recalibration, and variant detection and calling were performed using Picard and Genome Analysis Toolkit (GATK).(22)
Variants were annotated using ANNOVAR.(23) Variants were selected based on the following criteria: exonic and splicing variants (±12 bp); population-specific minor allele frequency (MAF) <5.0×10−3 in the Genome Aggregation Database (gnomAD) database;(24) and a scaled C-score of ≥10 in the Combined Annotation Dependent Depletion (CADD) database.(25) Variants located in the linkage/homozygosity region were given priority, but other genomic areas were also considered. Sanger sequencing was used to verify segregation of candidate variants within the family.
Overrepresentation analysis
To check commonly shared pathways in PAP, we downloaded all postaxial polydactyly-associated genes from HP:0100259 (n=172)(13) to run a PANTHER Overrepresentation Test based on GO Ontology database released 2017–09-26.(26)
RNA expression in the developing mouse limb
To evaluate expression of FAM92A and other polydactyly genes during early mouse limb development, expression data were downloaded and further analyzed from data sets GSE78345, GSE78372, GSE78405, GSE78465, and GSE82919 from the Gene Expression Omnibus (GEO) database.(27) These data contain gene expression data evaluated in total RNA extracted from developing limbs in C57BL/6 (B6NCrl substrain; males and females) mouse embryos at stages E11.5–E15.5. Data were generated as part of the Encyclopedia of DNA Elements (ENCODE) project, were analyzed by Bernstein and colleagues,(28) and nonstrand-specific RNA-seq data containing the TPM (transcripts per kilobase million) values were downloaded and further analyzed with R3.1.2.
cDNA constructs and antibodies
Human FAM92A1 cDNA (#SC100149, OriGene, Rockville, MD, USA) clone was subcloned into pEGFP-C2 and ptdTomato-C1 vectors. Green fluorescent protein (GFP)-FAM92A1R160* and tdTomato-FAM92A1R160* constructs were generated using site-directed mutagenesis protocol (Agilent, Santa Clara, CA, USA). The following primary antibodies were used: anti-FAM92A (1:200, #HPA034760, Atlas, Bromma, Sweden), anti-dsred/tdTomato (1:1000, Clontech, Mountain View, CA, USA), anti-GFP (1:1000, #A11122, Invitrogen, Carlsbad, CA, USA), anti-acetylated tubulin (1:200, #T7451, Sigma, St. Louis, MO, USA).
Immunostaining on cells
COS-7, HeLa, and LLC-PK1/CL4 cells were transfected with 2 μg of cDNA according to the Lipofectamine 2000 protocol and were immunostained and imaged as described previously.(29)
Immunostaining on cryosections
Limbs from E11.5 mouse and E21 rat embryos were harvested and fixed in 4% paraformaldehyde solution for 24 hours at 4°C. Forelimbs were then decalcified using 0.25 M EDTA for 24 hours and then incubated overnight with a 30% sucrose solution. Samples were then processed for optimal cutting temperature (OCT) compound embedding and stored at −80°C. OCT blocks were sectioned at 14 μm thickness. Sections were then processed for immunocytochemistry using anti-FAM92A antibodies and imaged with Zeiss 710 (Thornwood, NY, USA) confocal microscope using a 4× objective for low magnification ora 100×1.46 NAoil immersion objective. The study of animals was approved by the Institutional Animal Care and Use Committee (IACUC #0417014).
Induction of ciliogenesis
In HeLa cells, ciliogenesis was induced by serum starvation. Twenty-four hours after transfection, HeLa cells were maintained for another 24 hours in Dulbecco’s modified eagle medium (DMEM) medium without fetal bovine serum (FBS). Cells were washed and fixed with 4% paraformaldehyde (PFA) after the immunostaining protocol.
Nano SPD 2.0 assay
A Nano SPD 2.0 assay was performed as previously described.(30) Here, using a Lipofectamine 2000 protocol, cells were cotransfected with GFP-binding MYO10NANOTRAP, GFP-FAM92A, GFP-Chibby1, tdTomato-FAM92A, or tdTomato-FAM92AR160* constructs. The quantification of fluorescence intensities at the tips of the filopodia was performed as described previously.(31)
Co-immunoprecipitation assay
HEK 293 cells were maintained in DMEM supplemented with 10% FBS, glutamine, and penicillin-streptomycin at 37 °C in 5% CO2. Cells were grown in a 100-mm culture dish and transfected using 10 μg of each cDNA using polyethylenimine (Polysciences, Warrington, PA, USA). Twenty-four hours after transfection, cells were washed with 1× PBS and sonicated for 10 seconds in radioimmunoprecipitation assay (RIPA) buffer containing a protease inhibitor mixture. Protein A-Sepharose CL-4B beads were incubated for 4 hours with 4 μg of GFP antibody. Cell lysates were then incubated with the beads overnight at 4 °C. The following day, beads were centrifuged at 10,000 g for 3 minutes, washed three times with RIPA buffer, and boiled for 5 minutes in 2× SDS sample buffer. Samples were processed for Western blot using 4% to 20% Tris-Glycine gel (Novex, Thermo Fisher Scientific, Waltham, MA, USA).
3D modeling
The tertiary and quaternary structures of FAM92A homodimer were modeled using the cothreading of protein-protein complex structure server COTH (CO-THreader) (see Supplemental Web Resources). Only models with a Z-score >2.5 were analyzed. The tertiary structure of FAM92A was also modeled using Phyre2 and SWISS-MODEL (see Supplemental Web Resources) to confirm the tertiary structure obtained by COTH. The pdb (protein data bank) files generated by COTH server were analyzed using UCSF Chimera (see Supplemental Web Resources).
Fam92a−/− mice
A Fam92A−/− mouse, Fam92atm11b(KOMP)Wtsi, was created and phenotyped as part of the International Mouse Phenotyping Consortium (IMPC) project.(32) In short, a lacZ-tagged knockout was developed on a C57BL/6N background using a “knockout-first” design. A critical exon (exon 2) was deleted to create a frameshift of FAM92A.(33) Fourteen homozygous mutants (7 males and 7 females) and >2900 controls were phenotyped at the Sanger Institute using the “Mouse Genetics Project (MGP) Select pipeline” for a variety of parameters.(32,34) All X-ray procedures were carried out at week 14 using a Faxitron X-Ray system. Phenotype data for the 14 mice and >2800 controls were evaluated at the Sanger Institute and bioinformatics analysis was performed using the Fisher’s exact test for categorical data and a linear, mixed model or Mann-Whitney U rank sum test for continuous data.(32) Because these data showed indications for specific defects in the limbs and digits of the Fam92a−/− mice, X-rays of the limbs of 14 Fam92a−/− mice and 14 controls with the same genetic background (7 males and 7 females) were reevaluated blindly and independently by the Mouse Phenotyping Core and Center for Statistical Genetics at BCM. A one-sided Fisher’s exact test was performed to evaluate the statistical significance of these defects in Fam92a−/− mice. The care and use of mice were in accordance with the UK Home Office regulations, UK Animals (Scientific Procedures) Act of 1986.
Results
Clinical evaluation
The expression of postaxial polydactyly phenotype was highly variable among affected individuals of the family as well as between the limbs in the same individual (Fig. 1). Bilateral postaxial polydactyly of the hands and feet was observed in affected individual V:1, with a slightly varied phenotype in the hands and feet (Fig. 1). Affected family member V:2 has postaxial polydactyly only in his left hand, while his right hand and both feet are normal (Fig. 1). Affected individual V:3 has postaxial polydactyly in both hands with the extra finger deviated to the ulnar side on the right hand and straight on the left hand (Fig. 1) and normal feet. Teeth, nails, sweating, and hearing were normal in all three affected individuals. Patients have normal heights, weights, and body mass index (BMI) with normal food intake. Additional blood tests (serum creatinine, GFR, and blood urea nitrogen) and urine tests (urinalysis, urine protein test, and creatinine clearance) were normal, and renal ultrasounds were normal. No microphthalmia or anaphthalmia was observed, and vision was normal. X-rays of the hands and feet did not show any tendon calcifications (Fig. 1); unfortunately, we were unable to exclude the presence of tendon calcifications in other areas. Neurological problems and facial dysmorphism were also not observed in any of the affected individuals. Heterozygous carriers in the family had normal hands and feet and were clinically indistinguishable from non-carrier individuals.
Linkage analysis and exome sequencing
Homozygosity mapping and linkage analysis localized the PAPA locus to a region on chromosome 8q21.13-q24.12, which is flanked by SNPs rs2053962 and rs2514582 (GRCh37/Hg19; chr8:80870910–119596377) (Supplemental Figs. S1 and S2). A multipoint LOD score of 2.82 was obtained with SNP markers in this region. Exome sequencing uncovered a homozygous nonsense variant (NM_001283034.1:c.478C>T, NP_001269963.1: p.[Arg160*]) in the FAM92A gene. This variant lies within the mapped region on 8q21.13-q24.12 (GRCh37/Hg19; chr8:94722038C>T). Sanger sequencing confirmed that this variant segregates with PAPA (Fig. 1A; Supplemental Fig. S3). The c.478C>T variant (rs368652620) was observed in the gnomAD database, which contains 123,136 exomes and 15,496 whole genomes of unrelated individuals, with a very low MAF in the exome data (all populations MAF=2.04×10−5, South East Asians MAF=6.55×10−5 and no homozygous individuals observed) and was also not observed in the genome data.(24) In addition, the variant is not present in the Greater Middle East (GME) Variome Project that contains 1111 unrelated individuals from the Greater Middle East, including 168 Persians and Pakistanis.(35) The variant was also not present in Sanger sequence data from 186 in-house control Pakistani DNAs. The c.478C>T variant has a CADD C-score of 37 and is conserved amongst species (PhastCons: 0.998; PhyloP: 2.447). More importantly, it is predicted to be targeted by the classical nonsense-mediated decay pathway as it is located more than 50 to 54 nucleotides 5′ to the final exon-exon junction.(36)
No rare splice or exonic variants were found in previous nonsyndromic postaxial polydactyly–associated genes and syndromic genes (n=172) from Human Phenotype Ontology database (HP:0100259; postaxial polydactyly(13)). Although several additional rare variants were identified in the exome data of affected individual V-1, they did not segregate with the PAPA phenotype (Supplemental Table S1).
To evaluate commonly shared pathways in PAP, we ran an overrepresentation test,(26) using 172 known postaxial polydactyly genes. This analysis demonstrated the GO biological processes cilium organization (GO:0044782; p=8.72×10−50), cilium assembly (GO:0060271; p=6.78×10−49) and plasma membrane bounded cell projection assembly (GO:0120031; p=2.27×10−44) are highly overrepresented underlying biological processes involved in PAP disorders. FAM92A is annotated to these three processes as well, indicating FAM92A is part of these shared underlying pathological processes involved in PAP.
Fam92a is expressed in the embryonic mouse limb bud and rat forelimb
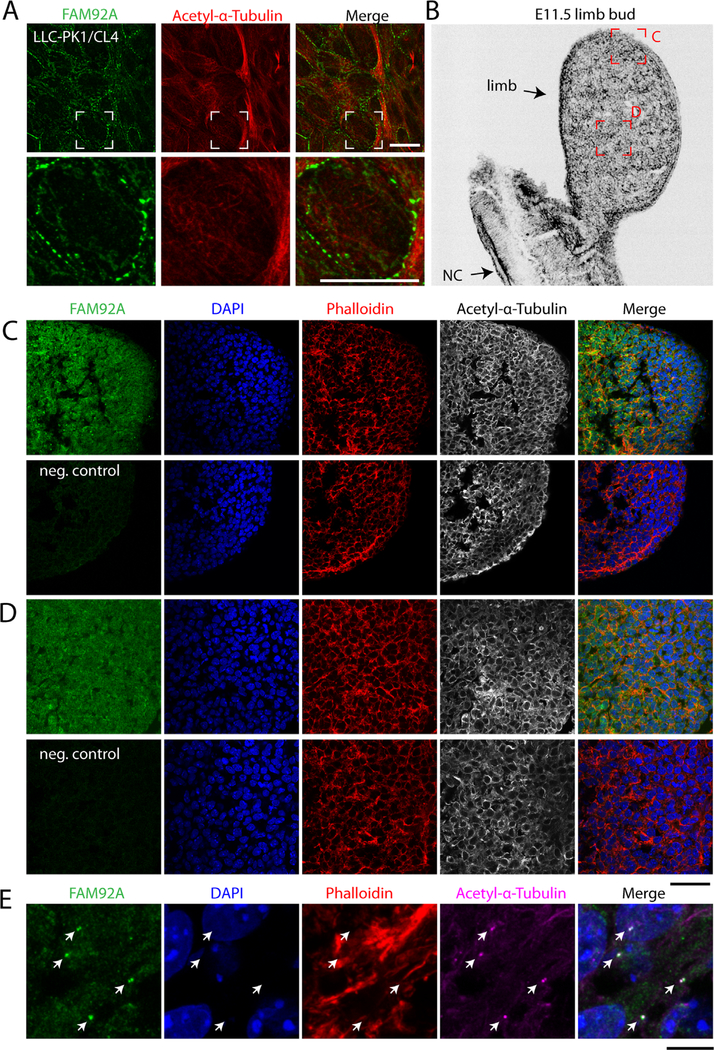
We first evaluated FAM92A protein expression in the rat forelimb. To validate the FAM92A antibody, LLC-PK1/CL4 cells were transfected with a human GFP-FAM92A construct and immunostained with an FAM92A antibody that recognized the C-terminal end of the protein (Supplemental Fig. S4A). The FAM92A antibody strongly recognized the GFP-FAM92A tagged protein (Supplemental Fig. S4B). The Anti-FAM92A antibody did not recognize the GFP-FAM92AR160* tagged protein (Supplemental Fig. S4B), suggesting that the antibody is specific to human FAM92A. Anti-FAM92A antibody was then used to detect endogenous FAM92A levels in LLC-PK1/CL4 cells (Fig. 2A). We detected strong FAM92A-positive clusters along the plasma membrane of the cells. Next, E11.5 embryonic mouse sections of the limb were immunostained with FAM92A antibody and showed that FAM92A is strongly expressed throughout the developing limb bud, including the progress zone and the apical ectodermal ridge (AER) compared with the negative control (Fig. 2B–D). High-magnification confocal images of cells taken from the AER of the limb bud are showing that Fam92a strongly localizes at the cilia level (Fig. 2E). Embryonic sections of E21 rat forelimbs were also immunostained with FAM92A antibody and revealed that FAM92A is strongly expressed in multiple layers of the skin, including the stratra corneum, granulosum, and basale when compared with the negative control sections (Supplemental Fig. S5). FAM92A is also strongly expressed in the chondrocytes where it forms clusters and in the mesenchymal tissue (Supplemental Fig. S5).
Fig. 2.
FAM92A is strongly expressed in the embryonic mouse limb bud. (A) LLC-PK1/CL4 cells were immunostained with FAM92A (green) and acetylated alpha-tubulin (red) antibodies. (B) Inverted grayscale image of an E11.5 mouse limb bud 14 μm cryosection counterstained with phalloidin. The red boxes are showing the areas imaged in C and D. NC=notochord. (C, D) Confocal images of E11.5 mouse limb bud 14 μm cryosections immunostained with FAM92A (green) and acetylated-α-tubulin (grayscale) antibodies and counterstained with rhodamin phalloidin (red) and 4′,6-Diamidine-2′-phenylindole dihydrochloride (DAPI) (blue). The merging of green, red, and blue channels are shown. Negative control samples (neg. control) were immunostained with the secondary antibody only. (E) High magnification of cells taken from the AERof the E11.5 limb bud immunostained with FAM92A (green) and acetylated-α-tubulin (magenta) antibodies and counterstained with rhodamine phalloidin (red) and DAPI (blue). Arrowheads are pointing to the cilia. Scale bars=10 μm (A), 50 μm (C, D), and 10 μm (E).
We also evaluated Fam92a mRNA gene expression in the developing limb from C57BL/6 mouse embryos at stages E11.5, E12.5, E13.5, E14.5, and E15.5 (Supplemental Fig. S6), which also revealed a high and steady expression during E11.5-E15.5.
The p.Arg160* truncated protein disrupts FAM92A homodimerization
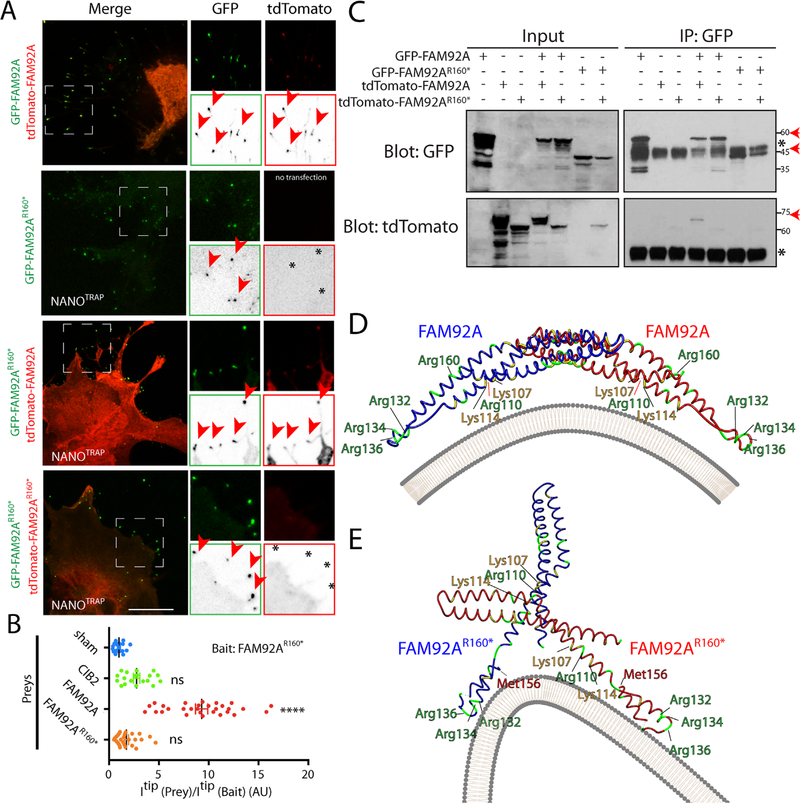
Because FAM92A can homodimerize,(37) we next investigated if the p.Arg160* truncated protein could disrupt FAM92A/FAM92A interaction. Cos-7 cells were cotransfected with GFP-FAM92A and tdTomato-FAM92A or tdTomato-FAM92AR160* constructs (Supplemental Fig. S7A, B). GFP-FAM92A and tdTomato-FAM92A form clusters within the cell cytoplasm as previously shown,(37) and the analysis of the expression profile and signal intensity on a line selection showed strong colocalization between both constructs, suggesting an interaction between both tagged proteins (Supplemental Fig. S7A). On the contrary, GFP-FAM92A and tdTomato-FAM92AR160* did not form any significant clusters within the cells. Scan line analysis revealed that the colocalization of the wild type and mutated protein were reduced (Supplemental Fig. S7B), suggesting a poor or weak association of both tagged proteins. The impact of the p.Arg160* variant on FAM92A homodimerization was further analyzed using nanoSPD 2.0 assays(30) (Fig. 3A, B; Supplemental Fig. S7C, D; and Supplemental Fig. S8). The nanoSPD 2.0 assay, using the Myo10nanotrap, GFP-FAM92A, and tdTomato-FAM92A constructs, revealed a strong accumulation of both proteins at the tip of filopodia in COS-7 cells compared with single transfected cells and to the negative control, suggesting a strong homodimerization (Supplemental Fig. S7C, D). Similarly, GFP-FAM92A and tdTomato-FAM92AR160* both accumulated to the tips of filopodia but in a less efficient way (Fig. 3A, B and Supplemental Fig. S7C, D), confirming that the FAM92A/FAM92AR160* complex is less stable compared with FAM92A homodimer. We next used GFP-FAM92AR160* as a bait. TdTomato-FAM92AR160* failed to accumulate at the tips of filopodia in the presence of GFP-FAM92AR160*, suggesting a complete disruption of FAM92A homodimerization induced by the p.Arg160* variant (Fig. 3A, B). To confirm these data and the impact of p.Arg160* variant on FAM92A homodimerization, we performed co-immunoprecipitation studies using both FAM92A and FAM92AR160* as baits. Similarly to the nanoSPD 2.0 assay, the p.Arg160* variant completely abolished the FAM92A homodimerization (Fig. 3C).
Fig. 3.
The p.Arg160* variant disrupts FAM92A dimerization. (A) COS-7 cells were cotransfected with nonfluorescent MYO10NANOTRAP construct and GFP-FAM92AR160* (baits, green) and/or tdTomato-FAM92A or tdTomato-FAM92AR160* (Preys, red) constructs. Single channels are also shown as inverted grayscale images. Accumulations at the tip of bait and prey are shown with an arrowhead. Stars indicate the absence of accumulation of prey at the filopodia tips. Scale bar=10 μm. (B) Quantification of nanoSPD 2.0 assays. CIB2 was used as a negative control. ****p ≤ 1.0×10−4; ns=not significant. AU=Arbitrary Unit. (C) Co-immunoprecipitation assay is showing the impairment of FAM92A homodimerization by p.Arg160* variant. The red arrows indicate the expected sizes of fusion proteins. *IgG heavy chain. (D, E) 3D modeling of the quaternary structure of FAM92A/FAM92A (D) and FAM92AR160* /FAM92AR160* (E) homodimers. Arg and Lys residues are shown in green and yellow, respectively. The plasma membrane is shown in gray.
We next modeled the tertiary and quaternary structure of FAM92A homodimer using the cothreading of protein-protein complex structure server COTH.(38) The tertiary BAR Domain-Containing FAM92A protein was successfully modeled (Fig. 3D) and the predicted model was confirmed using Phyre2 and SWISS-MODEL. The quaternary structure revealed a strong homodimerization of FAM92A (Z-score=2.52). The structure FAM92A homodimer is banana shaped, consistent with the quaternary structure of other crystalized BAR-containing protein complexes.(37) The concave face of FAM92A homodimer contains the Lys107, Lys114, Arg110, Arg132, Arg134, and Arg136, five positively charged residues that have been shown to be important for the association of FAM92A with curved membranes, for its function and for its ability to induce membrane remodeling in the presence of Chibby1 (Fig. 3D). In contrast, the Arg160 residue is localized on the convex face of the homodimer and resides within an alpha helix that appears to be important for the proper protein folding.
The quaternary structure of FAM92AR160* complex revealed impaired association between two proteins (Z-score=2.70) (Fig. 3E), with the concave face of one FAM92AR160* molecule not facing the plasma membrane. The recruitment of this complex, if existent, to curved membranes would be compromised because some of the key positively charged residues (Lys107, Lys114, Arg110) would be now exposed to the wrong side of the complex (Fig. 3E). If the FAM92AR160* mRNA is not degraded through the nonsense-mediated mRNA decay mechanism, a FAM92A/FAM92R160* dimer could potentially exist in human carriers. We tested this hypothesis and modeled the FAM92A/FAM92R160* dimer. Confidence of 3D modeling (Supplemental Fig. S7E) was poor (Z-score=2.173) and showed a poor association of both proteins.
The p.Arg160* truncated protein disrupts the FAM92A/Chibby1 complex
We next investigated if the p.Arg160* variant could disrupt the previously characterized FAM92A/Chibby1 complex.(37) Cos-7 cells were cotransfected with GFP-Chibby1 and tdTomato-FAM92A or tdTomato-FAM92AR160* constructs (Supplemental Fig. S9A, B). GFP-Chibby1 and tdTomato-FAM92A form clusters within the cell cytoplasm as previously shown(37) and the analysis of line scan showed strong colocalization between constructs, suggesting an interaction between both tagged proteins (Supplemental Fig. S9A). In contrast, GFP-Chibby1 and tdTomato-FAM92AR160* did not form any significant clusters within the cells and the colocalization was poor (Supplemental Fig. S9B), suggesting a disruption of the FAM92A/Chibby1 complex by the p.Arg160* variant.
The disruption of the FAM92A/Chibby1 complex by the p.Arg160* protein was further confirmed by a nanoSPD 2.0 assay (Fig. 4A and Supplemental Fig. S8). A NanoSPD 2.0 assay using the Myo10nanotrap, GFP-Chibby1, and tdTomato-FAM92A constructs revealed a strong accumulation of both proteins at the tips of filopodia in COS-7 cells showing a strong interaction of the two proteins (Fig. 4A, B and Supplemental Fig. S8). Similar to our negative control (tdTomato-CIB2), the tdTomato-FAM92AR160* construct did not accumulate to the tips of filopodia in the presence of GFP-Chibby1, suggesting a loss of interaction (Fig. 4A, B and Supplemental Fig. S8). To confirm these data and the impact of p.Arg160* variant on FAM92A/Chibby1 interaction, we performed co-immunoprecipitation studies using both GFP-Chibby1 as a bait. Similarly to the nanoSPD 2.0 assay, the p.Arg160* variant completely abolished the FAM92A/Chibby1 interaction (Fig. 4C).
Fig. 4.
The p.Arg160* variant disrupts the FAM92A/Chibby1 complex. (A) COS-7 cells were cotransfected with nonfluorescent MYO10NANOTRAP construct, GFP-Chibby1 (bait, green), and/or tdTomato-FAM92A or tdTomato-FAM92AR160* (Preys, red) constructs. Single channels are also shown as inverted grayscale images. Accumulations at the tip of bait and prey are shown with an arrowhead. Stars indicate the absence of accumulation of prey at the filopodia tip. Scale bar=10 μm. (B) Quantification of nanoSPD 2.0 assay. CIB2 was used as a negative control. ****p ≤ 0.0001; ns=nonsignificant; AU=Arbitrary Unit. (C) Co-immunoprecipitation of FAM92A variants with Chibby1 and FAM92A variants. The red arrows indicate the expected sizes of fusion proteins. *IgG heavy chain.
The p.Arg160* variant impairs the recruitment and the colocalization of FAM92A with Chibby1 at the base of cilia
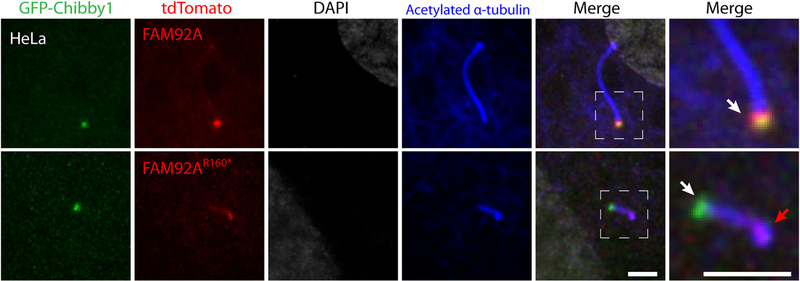
The centriolar and basal body localization of FAM92A depends largely on its interaction with Chibby1.(37) To test the effect of the p.(Arg160*) on FAM92A localization at the basal body, we cotransfected HeLa cells with the FAM92AR160* variant and Chibby1 constructs and we induced ciliogenesis by serum starvation for 24 hours (Fig. 5). As previously shown, GFP-Chibby1 and tdTomato-FAM92A strongly colocalized at the base of the cilia (Fig. 5, white arrow). Interestingly, this colocalization was impaired when cells were overexpressing the tdTomato-FAM92AR160* variant (Fig. 5).TdTomato-FAM92AR160* was accumulating at the tip rather than the base of cilia (Fig. 5, red arrow).
Fig. 5.
The p.Arg160* variant impairs the recruitment and the colocalization of FAM92A with Chibby1 at the base of cilia. HeLa cells were cotransfected with GFP-Chibby1 (green) and tdTomato-FAM92A or tdTomato-FAM92AR160* (red) constructs. Ciliogenesis was induced by 24-hour serum starvation. HeLa cells were immunostained with acetylated α-tubulin antibody (blue), a marker of cilia, and counterstained with DAPI (grayscale). Single channel and merge images are shown. Accumulations of Chibby1 and Fam92a variants are indicated with white and red arrows. Scale bars=2 μm.
Fam92a−/− mice show abnormal humeri and digit morphology
Seven homozygous male and 7 homozygous female Fam92atm11b(KOMP)Wtsi mice were phenotyped as part of the IMPC project.(32) The homozygous male and female mice were evaluated by X-rays and showed a significant and distinct humeri morphology compared with 2935 wild-type controls (7 mice; p=7.99×10−15) (Fig. 6A–C).(32) The abnormality was similar in all cases (Fig. 6A–C) and appears as an extra bone growth or exostosis on the deltoid tuberosity of the humerus. This is consistent with a tendon calcification. Upon detailed reevaluation of 14 Fam92a−/− mice and 14 wild-type controls of the same genetic background, we found 9 of 14 (64%; p=2.90×10−4) Fam92a−/− mice had this abnormality and none of the controls. Three mice had bilateral and 6 had unilateral abnormalities. In addition, two homozygous mice also showed abnormalities at the left stifle consistent with a tendon calcification (Fig. 6D,E). In total, 10 of 14 homozygous mice show abnormal bone growth consistent with tendon calcifications on their limbs (71%;p=7.63×10−5). Large-scale phenotyping also suggested that homozygous male and female mice also have an abnormal digit morphology (compared with 2935 controls). Although digit abnormalities are not uncommon and can also be observed in a small fraction of controls (2.7% of 2935 controls), this observation was enriched in the Fam92a−/− mice (28.6%; n=4; p=3.97×10−4). Four mice were identified either showing polysyndactyly at the phalanges (n=1) or an osteoma at one of the metatarsal digits (n=3; Fig. 6F–I). Upon detailed reevaluation of 14 Fam92a−/− mice and 14 wild-type controls of the same genetic background, the same digit integrity abnormalities were identified in the cases (n=4), and none were found in controls (p=0.049). Further detailed analysis of radiographs revealed no other significant abnormalities to the limbs or digits, tail, nor any other significant skeletal abnormalities.
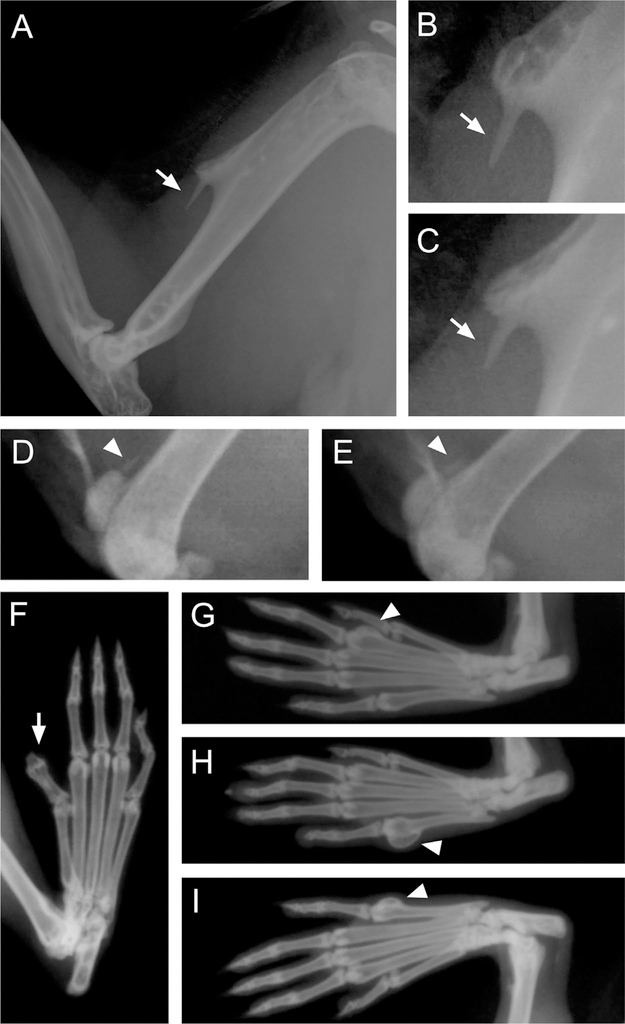
Fig. 6.
Abnormalities of the humeri and digits in homozygous Fam92a−/− mice shown by X-ray analysis. (A–C) Nine of 14 homozygous mice exhibit a similar unilateral or bilateral exostosis on the deltoid tuberosity of the humerus. This deltoid spur is compatible with a tendon calcification, particularly calcification of the m. deltoideus (scapular part). (D, E) Two Fam92a−/− mice also show tendon calcifications on their left stifle (quadriceps tendon). (F–H) Four homozygous mice showed an abnormal digit morphology. (F) Polysyndactyly of the right hind paw at phalanges. (G) Osteoma on digit two of left hind paw at the metatarsal. (H) Osteoma on digit five of the left hind paw at the metatarsal. (I) Osteoma on digit five of the right hind paw at the metatarsal. Arrows or arrowheads indicate the abnormality.
In addition to abnormal skeletal features, male mice also show some aberrations in food intake and a decreased white blood cell count. Additionally, some biochemical measurements such as glucose (in males only), glycerol, and alpha-amylase are different from normal levels.(32) The significance of these aberrations is unclear. Select phenotyping data and other current measurements are available on the IMPC website (see Supplemental Web Resources).(32)
Discussion
Via a linkage analysis and exome sequencing approach, we identified a homozygous nonsense variant in FAM92A (p.[Arg160*]), that segregates with PAPA in a consanguineous family from Pakistan. The variant is located in the mapped disease region on chr8q21.13-q24.12, and we provide evidence it leads to a loss-of-function (LOF) of FAM92A. Little is currently known about the function of FAM92A, also called FAM92A1 or Family with Sequence Similarity 92 Member A. A human paralog of FAM92A is FAM92B, which shares a similar BAR domain. BAR domain proteins function in endocytosis, vesicle fission and fusion, regulation of the actin cytoskeleton, and membrane remodeling.(37) Here we show that the FAM92AR160* truncated protein affects the tertiary structure of FAM92A and leads to the disruption of FAM92A homodimerization. According to 3D modeling data, this variant is also highly likely to affect the remodeling of plasma membrane of the cells. FAM92A is also an important factor in ciliogenesis. When FAM92A is knocked down in cells, an impaired ciliogenesis is observed.(37,39) FAM92A localization was detected at the transition zone of the primary cilium in IMCD3 cells.(39) In addition, FAM92A interacts with Cby1 (Chibby1) at the ciliary base to promote ciliogenesis via regulation of membrane-remodeling processes.(37) Cby1 knockout mice show several hallmarks of ciliary defects, including chronic upper airway infection, subfertility, and polycystic kidneys as well as polydactyly and hydrocephalus at low frequencies.(37)
Cilia are microtubule-based structures, which are involved in a wide range of essential biological processes and have been linked to several human skeletal and visceral abnormalities. Our data show a strong and steady expression of FAM92A in the mouse limb during development and a strong FAM92A expression in the embryonic rat forelimbs, particularly in the chondrocytes. We also show that FAM92A is expressed in the developing mouse limb bud, including the progress zone and the apical ectodermal ridge, and strongly localizes at the cilia level (Fig. 2), suggesting an important role in limb patterning. Ciliogenesis is known to be important for the chondrocyte function. For instance, variants in IFT52 or IFT81 lead to the loss of IFT-B cilia core complex, disrupt the ciliogenesis in chondrocytes, and lead to Short-Rib Polydactyly Syndrome.(40,41) Impaired ciliogenesis is often associated with polydactyly disorders, including PAP.(3) We illustrate this as well as cilium-associated processes are highly and significantly overrepresented in PAP in an overrepresentation analysis.(13) FAM92A is also involved in these shared underlying pathological processes.
Fam92A−/− mice (Fam92atm11b(KOMP)Wtsi) show a variety of skeletal limb abnormalities. Most significantly, 64% of homozygous mice show a uni- or bilateral abnormal bone growth or exostosis on the deltoid tuberosity of the humerus (Fig. 6A–C). This deltoid spur is compatible with a tendon calcification, particularly calcification of the m. deltoideus (scapular part). Two Fam92a−/− mice also showed abnormalities on their left stifle consistent with a tendon calcification (Fig. 6D, E). In addition, 4 of 14 homozygous mice also showed an abnormal digit morphology, including polysyndactyly and osteomas on the hind paw metatarsals (Fig. 6F–I). As mentioned above, polydactyly disorders are associated with impaired ciliogenesis.(3) However, the primary cilium has been implicated in the growth of neoplasms as well, such as osteomas, as it is crucial in the development, growth, patterning, and orientation of cells and tissues.(42) Defective cilia can lead to a disorganization of vital signaling during cell replication and the cell fate decisions that can contribute to the growth of neoplasms early in life.(42)
The premature stop variant we identified in individuals with PAPA is predicted to be targeted by the classical nonsense-mediated decay (NMD) pathway, a molecular mechanism whereby potentially defective messenger RNAs (mRNAs) are degraded to prevent translation and avoid yielding a truncated peptide,(36) though there are exceptions to this rule and alternate NMD mechanisms exist as well.(43) Triggering of NMD leads to a greatly reduced truncated protein expression to no detectable protein at all, which is often observed in recessive pediatric diseases.(44,45) Because patient cells are unavailable, we could not verify NMD and examine any truncated protein levels. However, we investigated the function of the truncated FAM92AR160* protein to evaluate the overall likelihood of loss-of-function of FAM92A due to the p.(Arg160*) variant. We show that the FAM92AR160* protein disrupts FAM92AR160* homodimerization and leads to the loss of FAM92A/Chibby1 complex (Fig. 4). This latter complex, as mentioned above, is important in ciliogenesis via regulation of membrane-remodeling processes.(37) A reduced or absent FAM92A protein expression due to NMD, expression of a truncated protein, or a combination of both due to the p.(Arg160*) nonsense variant will lead to loss of the FAM92A/Chibby1 complex and a ciliogenesis defect.(37) The FAM92A nonsense variant is therefore a LOF variant that will likely impact ciliogenesis, which is important for the development of the limb, where Fam92a endogenously localizes to the cilia (Fig. 2). In addition, we evaluated the impact of the FAM92AR160* variant on ciliogenesis and show that this variant is not recruited to the base of the cilia anymore but rather accumulates at the tip (Fig. 5). This also indicates that the chibby1/FAM92A interaction is important to recruit FAM92A at the base of the cilia.
Overall, we demonstrate that both mice and humans homozygous for FAM92A/Fam92a LOF variants show an abnormal digit morphology, although the presentation and expression is variable, even within the BD152 PAPA family, within Fam92a−/− mice, and even between limbs of the same patient/mouse. This phenotypic variability is not uncommon in developmental disorders and might be driven by genetic and environmental modifier influences during development. Although a percentage of the mice also present with tendon calcification, we did not observe this in the patients in the family included in this study; however, we could not exclude the presence of tendon calcifications in other areas than the hand and feet in the PAPA family.
In conclusion, we show for the first time to our knowledge that loss-of-function variants in FAM92A cause a skeletal phenotype in humans and mice, demonstrate a role of FAM92A in limb development, and present a new ciliopathy associated with PAPA.
Supplementary Material
Acknowledgments
The authors thank the family members that participated in this study. We also acknowledge the ENCODE Consortium and the Barbara J. Wold lab (California Institute of Technology) for generating and sharing mouse developing limb RNA-seq data. We thank the entire Sanger Institute mouse pipeline and phenotyping team for their contribution in generating and phenotyping the Fam92a−/− mouse and Dr Corey Reynolds and Taylor Vales for their assistance and insights in the X-ray analysis. GFP-hChibbyl was a gift from Ken-Ichi Takemaru (Addgene plasmid # 89472).
This work was supported by funds from the Higher Education Commission, Islamabad, Pakistan (to WA). Genotyping and exome sequencing was provided by the University of Washington Center for Mendelian Genomics (UW-CMG) and was funded by the National Human Genome Research Institute and the National Heart, Lung and Blood Institute grant UM1 HG006493 (to DAN, MJB, and SML). Part of the studies at University of Maryland Baltimore was supported by the NIH grant R01DC016295 (to ZMA).This project was also supported by the Mouse Phenotyping Core at BCM with funding from the National Institute of Health (UM1HG006348) and NIH (RO1DK114356). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Disclosures
All authors state that they have no conflicts of interest.
References
- 1.Biesecker LG. Polydactyly: how many disorders and how many genes? 2010 update. Dev Dyn. 2011;240(5):931–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castilla EE, Lugarinho R, da Graça Dutra M, Salgado LJ. Associated anomalies in individuals with polydactyly. Am J Med Genet. 1998;80(5):459–65. [DOI] [PubMed] [Google Scholar]
- 3.Verma PK, El-Harouni AA. Review of literature: genes related to postaxial polydactyly. Front Pediatr. 2015;3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galjaard R-JH, Smits APT, Tuerlings JHAM, et al. A new locus for postaxial polydactyly type A/Bon chromosome 7q21-q34. Eur J Hum Genet. 2003;11(5):409–15. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H, Tian Y, Breedveld G, et al. Postaxial polydactyly type A/B (PAP-A/B) is linked to chromosome 19p13.1–13.2 in a Chinese kindred. Eur J Hum Genet. 2002;10(3):162–6. [DOI] [PubMed] [Google Scholar]
- 6.van der Zwaag PA, Dijkhuizen T, Gerssen-Schoorl KBJ, et al. An interstitial duplication of chromosome 13q31.3q32.1 further delineates the critical region for postaxial polydactyly type A2. Eur J Med Genet. 2010;53(1):45–9. [DOI] [PubMed] [Google Scholar]
- 7.Radhakrishna U, Blouin JL, Mehenni H, et al. Mapping one form of autosomal dominant postaxial polydactyly type A to chromosome 7p15-q11.23 by linkage analysis. Am J Hum Genet. 1997;60(3):597–604. [PMC free article] [PubMed] [Google Scholar]
- 8.Umm-e-Kalsoom, Basit S, Kamran-ul-Hassan Naqvi S, Ansar M, Ahmad W. Genetic mapping of an autosomal recessive postaxial polydactyly type A to chromosome 13q13.3-q21.2 and screening of the candidate genes. Hum Genet. 2012;131(3):415–22. [DOI] [PubMed] [Google Scholar]
- 9.Kalsoom U, Klopocki E, Wasif N, et al. Whole exome sequencing identified a novel zinc-finger gene ZNF141 associated with autosomal recessive postaxial polydactyly type A. J Med Genet. 2013;50(1):47–53. [DOI] [PubMed] [Google Scholar]
- 10.Furniss D, Critchley P, Giele H, Wilkie AOM. Nonsense-mediated decay and the molecular pathogenesis of mutations inSALL1 andGLI3. Am J Med Genet A. 2007;143A(24):3150–60. [DOI] [PubMed] [Google Scholar]
- 11.Umair M, Shah K, Alhaddad B, et al. Exome sequencing revealed a splice site variant in the IQCEgene underlying post-axial polydactyly type A restricted to lower limb. Eur J Hum Genet. 2017;25(8):960–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radhakrishna U, Bornholdt D, Scott HS, et al. The phenotypic spectrum of GLI3 morphopathies includes autosomal dominant preaxial polydactyly type-IV and postaxial polydactyly type-A/B; no phenotype prediction from the position of GLI3 mutations. Am J Hum Genet. 1999;65(3):645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pusapati GV, Hughes CE, Dorn KV, et al. EFCAB7 and IQCE regulate hedgehog signaling by tethering the EVC-EVC2 complex to the base of primary cilia. Dev Cell. 2014;28(5):483–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murdoch JN, Copp AJ. The relationship between sonic Hedgehog signaling, cilia, and neural tube defects. Birth Defects Res A Clin Mol Teratol. 2010;88(8):633–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc. 2006;2006(1):pdb.prot 4455. [DOI] [PubMed] [Google Scholar]
- 17.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. [DOI] [PubMed] [Google Scholar]
- 18.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25(1):12–3. [DOI] [PubMed] [Google Scholar]
- 20.Seelow D, Schuelke M. HomozygosityMapper2012—bridging the gap between homozygosity mapping and deep sequencing. Nucleic Acids Res. 2012;40:W516–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164–e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi H, Huang X, Muruganujan A,et al. PANTHER version 11:expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1): D183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed ZM, Jaworek TJ, Sarangdhar GN, et al. Inframe deletion of human ESPN is associated with deafness, vestibulopathy and vision impairment. J Med Genet. 2018;55(7):479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bird JE, Barzik M, Drummond MC, et al. Harnessing molecular motors for nanoscale pulldown in live cells. Mol Biol Cell. 2017;28(3):463–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yousaf R, Ahmed ZM, Giese APJ, et al. Modifier variant of METTL13 suppresses human GAB1-associated profound deafness. J Clin Invest. 2018;128(4):1509–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koscielny G, Yaikhom G, Iyer V, et al. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 2014;42(D1):D802–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skarnes WC, Rosen B, West AP, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474(7351):337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White JK, Gerdin A-K, Karp NA, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154(2):452–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott EM, Halees A, Itan Y, et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet. 2016;48(9):1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76(1):51–74. [DOI] [PubMed] [Google Scholar]
- 37.Li F-Q, Chen X, Fisher C, et al. BAR domain-containing FAM92 proteins interact with Chibby1 to facilitate ciliogenesis. Mol Cell Biol. 2016;36(21):2668–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee S, Zhang Y. Protein-protein complex structure predictions by multimeric threading and template recombination. Structure. 2011;19(7):955–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breslow DK, Hoogendoorn S, Kopp AR, et al. A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat Genet. 2018;50(3):460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duran I, Taylor SP, Zhang W, et al. Destabilization of the IFT-B cilia core complex due to mutations in IFT81 causes a Spectrum of Short-Rib Polydactyly Syndrome. Sci Rep. 2016;6(1):34232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Taylor SP, Nevarez L, et al. IFT52 mutations destabilize anterograde complex assembly, disrupt ciliogenesis and result in short rib polydactyly syndrome. Hum Mol Genet. 2016;25(18): 4012–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klasson T, Giles R. The role of the cilium in hereditary tumor predisposition syndromes. J Pediatr Genet. 2015;3(2):129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller JN, Pearce DA. Nonsense-mediated decay in genetic disease: friend or foe? Mutat Res Rev Mutat Res. 2014;762: 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinttala R, Sasarman F, Nishimura T, et al. An N-terminal formyl methionine on COX 1 is required for the assembly of cytochrome c oxidase. Hum Mol Genet. 2015;24(14):4103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14(10):1074–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.