Abstract
Background: Leptin (LEP) is a human analogous form of the mouse obese gene and plays a critical role in energy expenditure as well as the progression of carcinogenesis. Many studies exploring the relationship between the LEP rs7799039 (G2548A) polymorphism and cancer risk have observed controversial results. To extensively evaluate this potential association, we conducted this meta-analysis.
Methods: All eligible studies published up to August 2018 on the relationship between the LEP rs7799039 G>A polymorphism and cancer risk were obtained by searching PubMed, EMBASE, and the China Biology Medicine databases. The association of LEP rs7799039 G>A polymorphism with cancer risk was evaluated by crude ORs together with their 95% CI's.
Results: Thirty-one case–control studies involving 25,799 subjects were included for meta-analysis. We identify a significant correlation with an overall cancer risk when these eligible case–control studies were pooled for analysis: for AA vs GG: an OR = 1.22, 95% CI = 1.01–1.48, P = 0.042 and for AA/GA vs GG: an OR = 1.16, 95% CI = 1.02–1.33, P = 0.026. A significant association was also detected in Asians, prostate cancer, other cancers, and hematopoietic malignancy subgroups. Sensitivity analysis was conducted by deleting an individual study in turn and calculation of the pooled ORs and CIs of the remainders. The results of sensitivity analyses indicated that no eligible study influenced the pooled ORs and CIs materially. Begg’s and Egger’s tests revealed that there was no evidence of publication bias.
Conclusion: In conclusion, our study suggests that the LEP rs7799039 G>A polymorphism might contribute to the development of cancer. In order to further verify or refute our findings, large and well-designed epidemiological studies are needed.
Keywords: leptin, polymorphism, cancer, risk, energy, meta-analysis
Introduction
Cancer is one of the major public health burden with over 18.1 million new cancer cases and 9.6 million cancer deaths in 2018.1 According to assessments of World Health Organization in 2015, cancer is among the leading cause of death in most countries. The reasons may be very complex. Aging and growth of the population, as well as risk factors for cancer, might influence the development of cancer. Recently, accumulating evidence indicates that there is a connection between diabetes and obesity with cancer.2 Thus, any variation in diabetes and obesity-related genes may influence the risk of cancer.
LEP, a 16-kDa adipocyte-derived peptide hormone, is a mediator of obesity and homeostasis. LEP interacts with the LEP receptor and its function is mediated through this receptor. Previous studies have demonstrated that the LEP signal may be transmitted through several signaling pathways (eg JAK/STAT, MAPK, PI3K, Wnt/β-catenin, and ERK).3,4 It is also reported that LEP may affect angiogenesis, inflammation, thrombosis, and tumor growth, invasion, and metastasis.4–13 Hardwick et al reported that LEP was very important for phosphorylation of the p42/44 mitogen-activated protein kinase and for enhancing proliferation of colonic epithelial cells.14 It is well known that single-nucleotide polymorphisms (SNPs) in genes may be implicated in the pathogenesis of a number of cancers and can be used as an indicator of early screening, diagnostics, and prevention measures.15 The human LEP gene maps to chromosome 7 (location: 128241278–128257629, NCBI Build 38). The LEP gene is polymorphic. And LEP SNPs may influence the risk of cancer.16 The rs7799039 G>A (G2548A) polymorphism in the LEP gene is the most widely studied for its relationship between this locus and the risk of human diseases. Terrasi et al suggested that the occurrence of LEP rs7799039 G>A variants could promote LEP protein expression in breast cancer cells through a Sp1- and nucleolin-dependent pathway, resulting in the LEP overexpression in tumor tissue.17
Recently, many molecular epidemiological studies have been carried out to identify the relationship between the LEP rs7799039 G>A polymorphism and cancer risk, but the findings have been conflicting. Three meta-analyses have been performed to explore the relationship between this SNP and cancer risk.18–20 Results of these studies indicated that individuals carrying the LEP rs7799039 A allele might have an increased susceptibility of overall cancer. However, only a case–control study focusing on the association between the LEP rs7799039 G>A polymorphism and the risk of gastric cancer was included.21 The relationship of this polymorphism with cancer risk in Asians is unclear. Recently, several case–control studies conducted in Asians were carried out to explore the association between the LEP rs7799039 G>A polymorphism and cancer risk. To obtain a more precise assessment of the correlation of LEP rs7799039 G>A polymorphism with the risk of cancer, we performed an updated meta-analysis of all eligible studies focusing on the relationship of the rs7799039 G>A polymorphism to the susceptibility of developing cancer.
Materials and methods
Search strategy
In this meta-analysis, we carried out an electronic literature retrieval in PubMed, Embase, and the China Biology Medicine databases up to August 2018 using the following search strategies: (‘LEP” or “leptin”) and (“carcinoma” or “cancer” or ‘malignancy ‘ or “neoplasms”) and (“polymorphism” or “SNP” or “variation”). There was no restriction on language. The references in included studies and reviewers were carefully checked for other potential data. When a publication involved some subgroups, it was treated separately. This study was reported based on the Preferred Reporting Items for Meta-analyses (PRISMA) guideline (Table S1: PRISMA checklist).22
Selection and exclusion criteria
The major selection criteria were as follows: (1) designed as case–control study that assessed the relationship between LEP rs7799039 G>A variants and cancer risk; (2) presented sufficient data (eg genotype number or other available data) to calculate the pooled-estimating; and (3) genotype distribution in controls did not violate Hardy–Weinberg equilibrium (HWE).
The exclusion criteria were defined as follows: (1) the publication was not designed as a case–control study; (2) the genotype data was not presented or could not be calculated; (3) genotype distribution in controls violated HWE; and (4) review articles and letters.
Data extraction
Two authors (W. Tang and C. Liu) independently extracted the information from each eligible study. If the extracted information was different, they would review the publication again and reached consensus. If they could not get a consistent assessment, another author (H. Qiu) would be invited to resolve the dispute and a final decision was made. The following data were extracted from each study: the surname of the first author, year of publication, country, ethnicity, numbers of participants, source of control, genotype frequencies, and genotyping method.
Statistical analysis
The strength of the correlation of the LEP rs7799039 G>A polymorphism with cancer susceptibility was determined by crude ORs with 95% CIs. The relationship between LEP rs7799039 G>A and cancer risk was evaluated using allele model (A vs G), homozygote model (AA vs GG), recessive model (AA vs GG/GA), and dominant model (AA/GA vs GG). We used the Q and I2 test to check the heterogeneity among the included studies. A P>0.1 and I2<50% indicated that there was low heterogeneity, and then the Mantel–Haenszel method (fixed-effects model) was used to calculate the ORs and CIs;23 otherwise, the DerSimonian and Laird method (random-effects model) was used to assess the association.24,25 The sources of heterogeneity were analyzed by subgroup analyses. Sensitivity analysis was analyzed by omitting an individual study in turn and re-calculating the ORs and CIs. Publication bias was checked by using Bgger’s and Egger’s test. An internet chi-square test was used to determine whether the distribution of the genotypes in controls conformed to HWE (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). All data were calculated and analyzed by using Stata 12.0 software (Stata Corp., College Station, TX). A P<0.05 (two-sided) was considered as statistical significance.
Results
Study characteristics
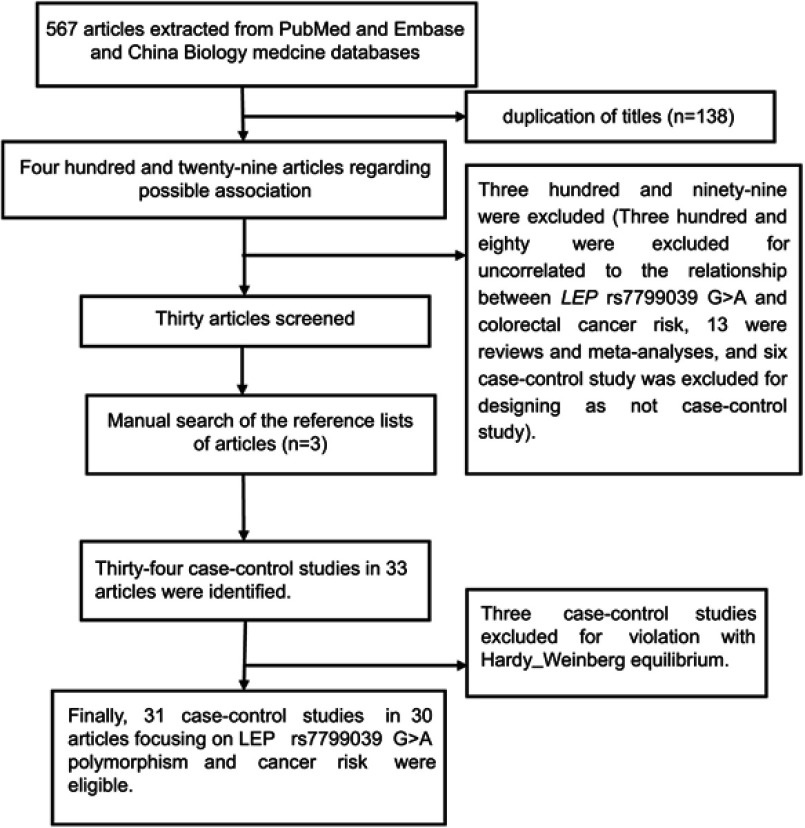
Based on the selection criteria, 30 publications focusing on the association of the LEP rs7799039 G>A polymorphism with cancer risk were included.21,26–53 One publication contained two independent case–control studies that we treated as two investigations.49 The detail selecting process is shown in Figure 1. A total of 31 case–control studies involving 25,799 subjects were included in this meta-analysis. Among them, 19 were conducted in Caucasians,26–44 eight performed in Asians,21,45–50,54 and four were in mixed populations.51–53 Nine were population-based,27,28,30,33,37,38,43,44,50 and 22 case–control were hospital-based studies.21,26,29,31,32,34–36,39–42,45–49,51–54 Of all the eligible studies, 11 focused on breast cancer,33,37–40,44,48,49,51,53 four focused on colorectal cancer (CRC),31,32,41,52 three focused on prostate cancer (PC),26,30,34 and 13 focused on other cancers.21,27–29,35,36,42,43,45–47,50,54 Other information includingd case–control studies in the pooled analysis is summarized in Table 1. The genotypes and alleles of LEP rs7799039 G>A polymorphism are shown in Table 2.
Figure 1.
Flow diagram of the meta-analysis of the association between LEP rs7799039 G>A polymorphism and overall cancer risk.
Table 1.
Characteristics of the studies in meta-analysis
| Study | Publication year | Country | Ethnicity | Cancer type | Sample size (case/control) | Source of control | Genotype method |
|---|---|---|---|---|---|---|---|
| Ribeiro et al.26 | 2004 | Portugal | Caucasians | Prostate cancer | 150/118 | HB | PCR-RFLP |
| Skibola et al.27 | 2004 | USA | Caucasians | Lymphoma | 376/805 | PB | TaqMan |
| Willett et al.28 | 2005 | UK | Caucasians | Lymphoma | 593/754 | PB | TaqMan |
| Snoussi et al.51 | 2006 | Tunisia | Mixed | Breast cancer | 308/222 | HB | PCR-RFLP |
| Slattery et al.52 | 2008 | USA | Mixed | Colorectal cancer | 1565/1965 | HB | TaqMan |
| Chovanec et al.29 | 2009 | Czech | Caucasians | Endometrial cancer | 66/543 | HB | PCR |
| Moore et al.30 | 2009 | Finland | Caucasians | Prostate cancer | 1053/1053 | PB | TaqMan |
| Pechlivanis et al.31 | 2009 | Czech | Caucasians | Colorectal cancer | 702/752 | HB | TaqMan |
| Vasku et al.32 | 2009 | Czech | Caucasians | Colorectal cancer | 102/101 | HB | PCR-sequencing |
| Cleveland et al.33 | 2010 | USA | Caucasians | Breast cancer | 1059/1101 | PB | PCR |
| Kim et al.21 | 2012 | Korea | Asians | Gastric cancer | 48/48 | HB | PCR-RFLP |
| Ribeiro et al.34 | 2012 | Portugal | Caucasians | Prostate cancer | 449/557 | HB | TaqMan |
| Tavil et al.35 | 2012 | Turkey | Caucasians | Leukemia | 72/70 | HB | PCR-RFLP |
| Garcia-Robles et al.53 | 2013 | Mexico | Mixed | Breast cancer | 130/189 | HB | PCR |
| Unsal et al.36 | 2014 | Turkey | Caucasians | Lung cancer | 162/130 | HB | PCR-RFLP |
| Zhang et al.45 | 2018 | China | Asians | Hepatocellular carcinoma | 584/923 | HB | SNPscan |
| Hussain et al.46 | 2015 | India | Asians | Oral cancer | 306/228 | HB | PCR-RFLP |
| Karakus et al.37 | 2015 | Turkey | Caucasians | Breast cancer | 199/185 | PB | PCR |
| Mahmoudi et al.38 | 2015 | Iran | Caucasians | Breast cancer | 45/41 | PB | PCR-RFLP |
| Mohammadzadeh et al.39 | 2015 | Iran | Caucasians | Breast cancer | 100/100 | HB | PCR-RFLP |
| Rostami et al.40 | 2015 | Iran | Caucasians | Breast cancer | 203/171 | HB | PCR-RFLP |
| Mahmoudi et al.41 | 2016 | Iran | Caucasians | Colorectal cancer | 261/339 | HB | PCR-RFLP |
| Amer et al.42 | 2017 | Egypt | Caucasians | Hepatocellular carcinoma | 150/100 | HB | PCR-RFLP |
| Ali et al.43 | 2017 | Pakistan | Caucasians | Bladder carcinoma | 200/200 | PB | PCR |
| Qiu et al.47 | 2017 | China | Asians | Esophageal cancer | 507/1496 | HB | SNPscan |
| Rodrigo et al.44 | 2017 | Sri Lanka | Caucasians | Breast cancer | 80/80 | PB | PCR |
| Cao et al.54 | 2015 | China | Asians | Lung cancer | 162/200 | HB | PCR-RFLP |
| Yuan et al.48 | 2017 | China | Asians | Breast cancer | 703/805 | HB | MALDI-TOF MS |
| Liu et al.49 | 2018 | China | Asians | Breast cancer | 434/440 | HB | MALDI-TOF MS |
| Liu et al.49 | 2018 | China | Asians | Breast cancer | 334/331 | HB | MALDI-TOF MS |
| Zhang et al.50 | 2014 | USA | Mixed | Pancreatic cancer | 173/476 | PB | TaqMan |
Abbreviations: PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; PCR, polymerase chain reaction; MALDI-TOF MS, Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry.
Table 2.
Distribution of LEP rs7799039 G>A polymorphism genotype and allele
| Study | Publication year | Case AA | Case AG | Case GG | Control AA | Control AG | Control GG | Case A | Case G | Control A | Control G | HWE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ribeiro et al.26 | 2004 | 24 | 89 | 30 | 12 | 62 | 44 | 137 | 149 | 86 | 150 | Yes |
| Skibola et al.27 | 2004 | 91 | 167 | 118 | 167 | 376 | 259 | 349 | 403 | 710 | 894 | Yes |
| Willett et al.28 | 2005 | 127 | 294 | 170 | 145 | 348 | 260 | 548 | 634 | 638 | 868 | Yes |
| Snoussi et al.51 | 2006 | 37 | 152 | 119 | 11 | 99 | 112 | 226 | 390 | 121 | 323 | Yes |
| Slattery et al.52 | 2008 | 284 | 782 | 499 | 393 | 938 | 634 | 1350 | 1780 | 1724 | 2206 | Yes |
| Chovanec et al.29 | 2009 | 20 | 33 | 13 | 131 | 255 | 149 | 73 | 59 | 517 | 553 | Yes |
| Moore et al.30 | 2009 | 281 | 453 | 213 | 216 | 437 | 210 | 1015 | 879 | 869 | 857 | Yes |
| Pechlivanis et al.31 | 2009 | 120 | 309 | 230 | 150 | 334 | 227 | 549 | 769 | 634 | 788 | Yes |
| Vasku et al.32 | 2009 | 24 | 41 | 35 | 20 | 44 | 36 | 89 | 111 | 84 | 116 | Yes |
| Cleveland et al.33 | 2010 | 226 | 492 | 341 | 180 | 561 | 360 | 944 | 1174 | 921 | 1281 | Yes |
| Kim et al.21 | 2012 | 29 | 18 | 1 | 33 | 14 | 1 | 76 | 20 | 80 | 16 | Yes |
| Ribeiro et al.34 | 2012 | 73 | 212 | 164 | 84 | 268 | 203 | 358 | 540 | 436 | 674 | Yes |
| Tavil et al.35 | 2012 | 26 | 31 | 15 | 27 | 29 | 14 | 83 | 61 | 83 | 57 | Yes |
| Garcia-Robles et al.53 | 2013 | 22 | 71 | 37 | 46 | 95 | 48 | 115 | 145 | 187 | 191 | Yes |
| Unsal et al.36 | 2014 | 50 | 86 | 26 | 27 | 63 | 40 | 186 | 138 | 117 | 143 | Yes |
| Zhang et al.45 | 2014 | AA+AG:122 | - | 51 | AA+AG:318 | - | 158 | - | - | - | - | Yes |
| Hussain et al.46 | 2015 | 50 | 154 | 102 | 23 | 92 | 113 | 254 | 358 | 138 | 318 | Yes |
| Karakus et al.37 | 2015 | 49 | 105 | 45 | 47 | 98 | 40 | 203 | 195 | 192 | 178 | Yes |
| Mahmoudi et al.38 | 2015 | 27 | 11 | 7 | 17 | 19 | 5 | 65 | 25 | 53 | 29 | Yes |
| Mohammadzadeh et al.39 | 2015 | 36 | 55 | 9 | 52 | 45 | 3 | 127 | 73 | 149 | 51 | Yes |
| Rostami et al.40 | 2015 | 115 | 64 | 24 | 63 | 77 | 31 | 294 | 112 | 203 | 139 | Yes |
| Cao et al.54 | 2015 | 57 | 75 | 30 | 33 | 80 | 87 | 189 | 135 | 146 | 254 | Yes |
| Mahmoudi et al.41 | 2016 | 76 | 135 | 50 | 113 | 154 | 72 | 287 | 235 | 380 | 298 | Yes |
| Amer et al.42 | 2017 | 60 | 69 | 21 | 49 | 47 | 4 | 189 | 111 | 145 | 55 | Yes |
| Ali et al.43 | 2017 | 61 | 103 | 36 | 61 | 100 | 39 | 225 | 175 | 222 | 178 | Yes |
| Qiu et al.47 | 2017 | 291 | 184 | 29 | 797 | 591 | 101 | 766 | 242 | 2185 | 793 | Yes |
| Rodrigo et al.44 | 2017 | 32 | 43 | 5 | 53 | 24 | 3 | 107 | 53 | 130 | 30 | Yes |
| Yuan et al.48 | 2017 | 416 | GG+GA:276 | - | 426 | GG+GA:347 | - | - | - | - | - | Yes |
| Zhang et al.50 | 2018 | 295 | 221 | 59 | 505 | 360 | 56 | 811 | 339 | 1370 | 472 | Yes |
| Liu et al.49 | 2018 | 252 | GG+GA:182 | - | 236 | GG+GA:206 | - | - | - | - | - | Yes |
| Liu et al.49 | 2018 | 201 | GG+GA:133 | - | 190 | GG+GA:141 | - | - | - | - | - | Yes |
Abbreviation: HWE, Hardy–Weinberg equilibrium.
Meta-analysis results
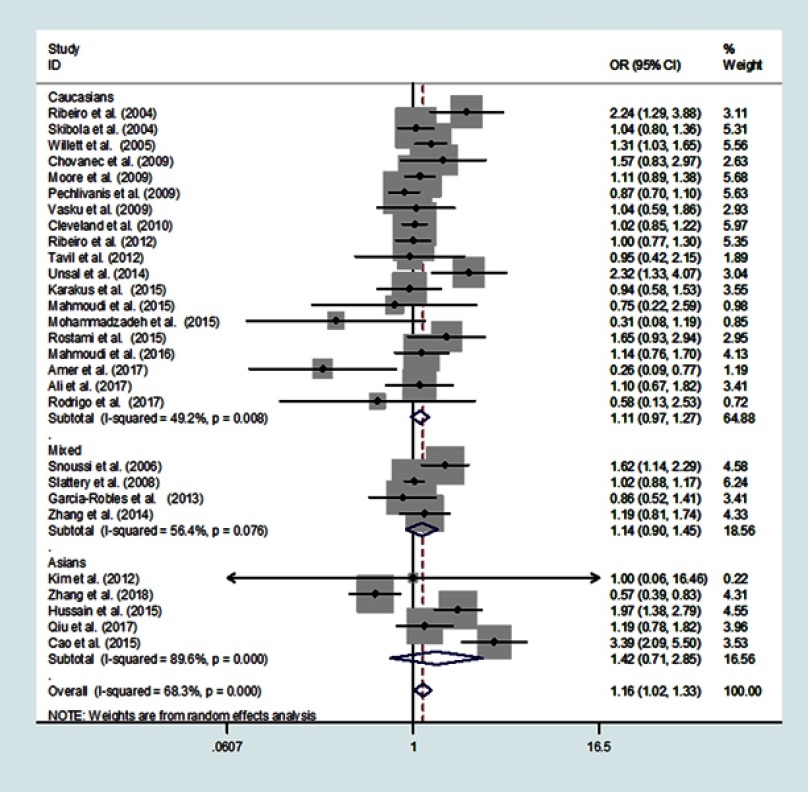
Table 3 summarizes the results of this meta-analysis. We found that the LEP rs7799039 G>A polymorphism was associated with overall cancer risk (Figure 2). The A vs G genetic model has an OR = 1.10 with a 95% CI = 1.00–1.21 and a P = 0.051. The AA vs GG genetic model has an OR = 1.22 with a 95% CI = 1.01–1.48, and a P = 0.042. When we compared AA/GA vs GG model, we found an OR = 1.16 with a 95% CI = 1.02–1.33 and P = 0.026. Comparing the AA genotype with GA/GG, we calculated an OR =1.12 with a 95% CI = 1.00–1.26 and a P = 0.059.
Table 3.
Results of the meta-analysis from different genetic models
| No. of cases/controls | A vs G | AA vs GG | AA+GA vs GG | AA vs.GA+GG | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | I2 | P (Q-test) | OR (95% CI) | P | I2 | P (Q-test) | OR (95% CI) | P | I2 | P (Q-test) | OR (95% CI) | P | I2 | P (Q-test) | ||
| Total | 11,276/14,523 | 1.10(0.1.00–1.21) | 0.051 | 79.1% | <0.001 | 1.22(1.01–1.48) | 0.042 | 74.9% | <0.001 | 1.16(1.02–1.33) | 0.026 | 68.3% | <0.001 | 1.12(1.00–1.26) | 0.059 | 69.6% | <0.001 |
| Ethnicity | |||||||||||||||||
| Caucasians | 6,022/7,200 | 1.07(0.96–1.18) | 0.216 | 69.5% | <0.001 | 1.18(0.98–1.41) | 0.088 | 58.5% | 0.001 | 1.11(0.97–1.27) | 0.122 | 49.2% | 0.008 | 1.09(0.92–1.28) | 0.314 | 67.2% | <0.001 |
| Mixed | 2,176/2,852 | 1.07(0.78–1.47) | 0.687 | 83.9% | 0.002 | 1.17(0.55–2.48) | 0.676 | 83.9% | 0.002 | 1.14(0.90–1.45) | 0.279 | 56.4% | 0.076 | 1.08(0.58–2.00) | 0.816 | 81.0% | 0.005 |
| Asians | 3,078/4,471 | 1.27(0.87–1.87) | 0.216 | 91.7% | <0.001 | 1.60(0.67–3.80) | 0.287 | 90.6% | <0.001 | 1.42(0.71–2.85) | 0.326 | 89.6% | <0.001 | 1.23(1.01–1.49) | 0.044 | 70.0% | 0.001 |
| Cancer type | |||||||||||||||||
| Prostate cancer | 1,652/1,728 | 1.17(0.97–1.40) | 0.098 | 59.5% | 0.085 | 1.36(0.94–1.97) | 0.106 | 56.8% | 0.099 | 1.24(0.89–1.72) | 0.208 | 70.7% | 0.033 | 1.24(1.04–1.47) | 0.014 | 0.0% | 0.470 |
| Breast cancer | 3,595/3,665 | 1.02(0.79–1.31) | 0.878 | 82.4% | <0.001 | 1.11(0.70–1.75) | 0.669 | 71.3% | 0.001 | 1.07(0.84–1.38) | 0.576 | 46.1% | 0.072 | 1.12(0.87–1.43) | 0.385 | 78.3% | <0.001 |
| Colorectal cancer | 2,630/3,157 | 0.95(0.88–1.03) | 0.203 | 0.0% | 0.671 | 0.90(0.77–1.05) | 0.169 | 0.0% | 0.675 | 0.99(0.88–1.11) | 0.849 | 0.0% | 0.615 | 0.88(0.77–1.00) | 0.046 | 0.0% | 0.699 |
| Others | 3,399/5,973 | 1.17(0.98–1.40) | 0.081 | 82.7% | <0.001 | 1.34(0.92–1.94) | 0.127 | 81.1% | <0.001 | 1.25(0.95–1.65) | 0.104 | 78.2% | <0.001 | 1.18(0.98–1.42) | 0.083 | 63.0% | 0.002 |
| System of cancer | |||||||||||||||||
| Reproductive and breast cancer | 5,313/5,936 | 1.10(0.95–1.29) | 0.207 | 75.9% | <0.001 | 1.28(0.98–1.68) | 0.074 | 63.0% | 0.002 | 1.15(0.97–1.37) | 0.105 | 47.9% | 0.032 | 1.17(0.97–1.40) | 0.096 | 70.7% | <0.001 |
| Hematopoietic malignancy | 1,041/1,629 | 1.13(1.01–1.26) | 0.038 | 0.0% | 0.595 | 1.25(1.00–1.55) | 0.049 | 0.0% | 0.680 | 1.17(0.99–1.39) | 0.068 | 0.0% | 0.399 | 1.15(0.95–1.39) | 0.140 | 0.0% | 0.730 |
| Digestive system cancer | 4,398/6,428 | 0.99(0.86–1.13) | 0.838 | 73.4% | <0.001 | 0.94(0.70–1.25) | 0.658 | 70.4% | 0.001 | 1.01(0.81–1.27) | 0.928 | 71.7% | <0.001 | 0.95(0.82–1.09) | 0.454 | 47.5% | 0.055 |
| Others | 524/530 | 1.60(0.96–2.66) | 0.070 | 88.2% | <0.001 | 2.48(0.99–6.23) | 0.053 | 85.1% | 0.001 | 2.06(1.05–4.02) | 0.035 | 80.3% | 0.006 | 1.66(0.90–3.03) | 0.102 | 78.6% | 0.009 |
| Sample size | |||||||||||||||||
| <1000 | 3,685/4,312 | 1.13(0.94–1.37) | 0.200 | 81.6% | <0.001 | 1.32(0.92–1.90) | 0.129 | 74.8% | <0.001 | 1.29(1.03–1.62) | 0.027 | 64.3% | <0.001 | 1.15(0.92–1.43) | 0.224 | 73.3% | <0.001 |
| ≥1000 | 7,591/10,211 | 1.04(0.96–1.12) | 0.378 | 62.4% | 0.006 | 1.05(0.88–1.26) | 0.559 | 67.9% | 0.002 | 1.01(0.90–1.13) | 0.836 | 51.2% | 0.037 | 1.09(0.97–1.23) | 0.135 | 61.6% | 0.005 |
| Source of control | |||||||||||||||||
| Hospital-based | 7,498/9,828 | 1.13(0.98–1.30) | 0.087 | 83.6% | <0.001 | 1.26(0.94–1.68) | 0.118 | 81.3% | <0.001 | 1.21(0.98–1.49) | 0.073 | 77.7% | <0.001 | 1.13(0.98–1.31) | 0.100 | 71.1% | <0.001 |
| Population-based | 3,778/4,695 | 1.08(0.97–1.19) | 0.151 | 47.6% | 0.064 | 1.24(1.09–1.42) | 0.001 | 0.0% | 0.731 | 1.10(0.99–1.21) | 0.069 | 0.0% | 0.811 | 1.11(0.90–1.37) | 0.314 | 65.2% | 0.005 |
Figure 2.
Meta-analysis of the association between LEP rs7799039 G>A polymorphism and overall cancer risk (dominant model, random-effects model).
In a subgroup analysis by ethnicity, we found an association in Asian populations with AA/GA vs GG having an OR = 1.23, a 95% CI = 1.01–1.49 and a P = 0.044, Table 3.
In a subgroup analysis by cancer type, we found that the LEP rs7799039 G>A polymorphism moderately increased the risk of PC; AA vs GA/GG: OR =1.24, 95%CI = 1.04–1.470, P = 0.014. However, we found that this G>A polymorphism might actually confer a decreased the risk to CRC, AA vs GA/GG: OR =0.88, 95%CI = 0.77–1.00, P = 0.046. When we conducted a subgroup analysis by cancer system, we found that this G>A polymorphism might increase the susceptibility of hematopoietic cancer; A vs G: OR = 1.13, 95% CI = 1.01–1.26, P = 0.038; AA vs GG: OR = 1.25, 95% CI = 1.00–1.55, P = 0.049) and other system cancers (AA/GA vs GG: OR = 2.06, 95% CI = 1.05–4.02, P = 0.035.
Heterogeneity analysis
For this meta-analysis, we found that there was significant heterogeneity among the included case–control studies (Table 3). To identify the major sources of heterogeneity, we carried out subgroup analyses. The results indicated that Asians, small sample size studies (<1000), and hospital-based studies might lead to the major heterogeneity in this meta-analysis.
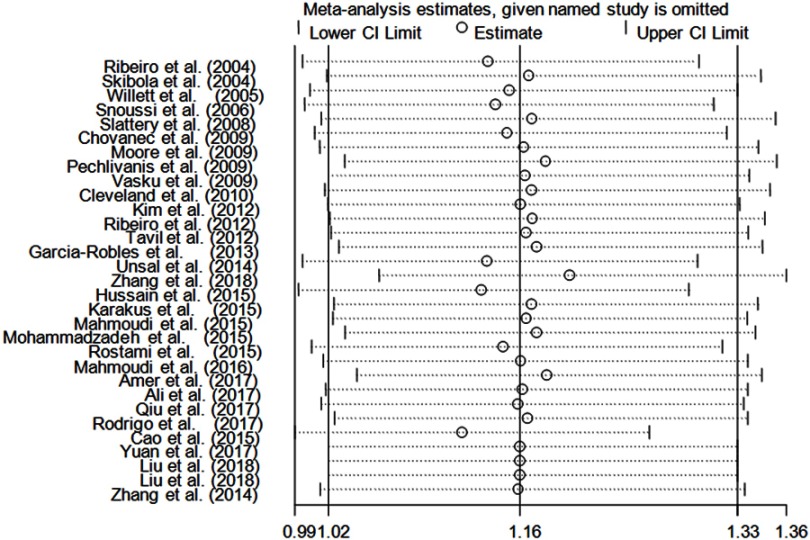
Sensitivity analysis
Sensitivity analysis was conducted by deleting an individual study in turn and calculating the pooled ORs and CIs of the remaining studies. For this SNP, the results under all genetic comparisons were not influenced by removing any eligible study (Figure 4).
Figure 4.
Sensitivity analysis of the influence of dominant model (random-effects estimates).
Publication bias
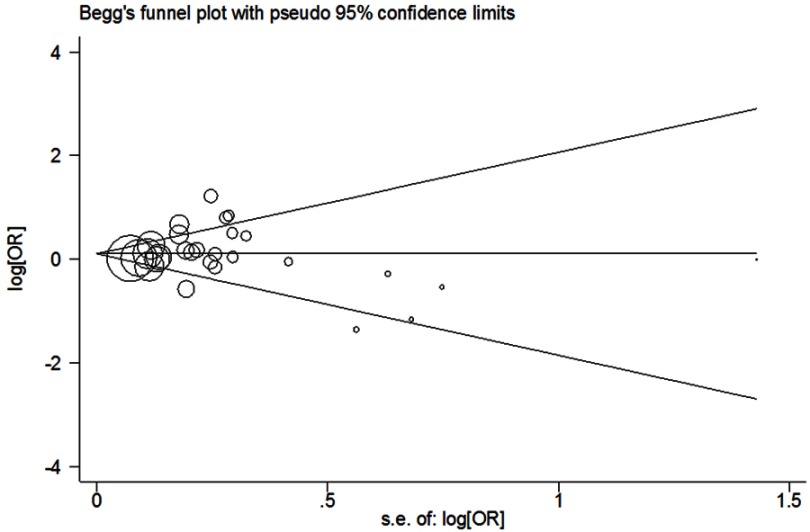
Begg’s test and Egger’s test were used to determine whether there was publication bias in genetic comparisons. The shapes of the Begg’s funnel plot revealed that they were symmetrical; A vs G had a P Begg’s= 0.588, AA vs GG had a P Begg’s= 0.802; AA/GA vs GG had a P Begg’s = 0.953; and AA vs GA/GG had a P Begg’s= 0.887 (Figure 3). The results of Egger’s test also highlighted that there was no evidence of publication bias (A vs G: PEgger’s = 0.559; AA vs GG: PEgger’s = 0.579; AA/GA vs GG: PEgger’s = 0.639 and AA vs GA/GG: PEgger’s = 0.660).
Figure 3.
Begg’s funnel plot of meta-analysis of the association between LEP rs7799039 G>A polymorphism and cancer risk (dominant genetic model, random-effects model).
Discussion
The adipocyte-derived peptide hormone LEP has a well-known influence on inflammation, tumor growth, and metastasis. Rs7799039 G>A is a common promoter SNP in the LEP gene, that may affect the transcriptional level and LEP expression.55 We therefore hypothesized that the LEP rs7799039 G>A polymorphism might be closely related to the susceptibility of cancer. Although a number of studies have focused on the relationship between the LEP rs7799039 G>A polymorphism and cancer risk, the observed results have been inconsistent. Three meta-analyses carried out by Liu et al18, He et al19, and Yang et al20, including 12, 15, and 15 eligible case–control studies, respectively, yielded conflicting results in some subgroups. Of late, some new data regarding the relationship of the LEP rs7799039 G>A polymorphism and cancer risk have been reported.36–50,53,54 Therefore, an updated meta-analysis is needed to address this issue. In our meta-analysis, data of 31 independent case–control studies including 11,276 cancer cases and 14,523 controls were pooled, which is more participants than were in the meta-analyses mentioned above. Thus, this updated analysis should be more comprehensive. To the best of our knowledge, the present study is the most convincing pooled analysis to explore the association between the LEP rs7799039 G>A polymorphism and cancer risk. Results of our meta-analysis did indicate that the LEP rs7799039 G>A polymorphism was associated with an increased risk of overall cancer, especially in Asians, PC, hematopoietic malignancy, and other system cancer subgroups.
With the accumulating evidence of genetic association investigations, it is urgent to synthesize all available data to obtain a robust result. According to the findings, the association of increased cancer risk with the LEP rs7799039 G>A polymorphism was found in overall populations. Race also could be a critical biological factor for the genetic comparison. In previous meta-analyses,18–20 most of the eligible studies contained only Caucasians. In the current study, more case–control studies included Asians.21,45–50,54 The results suggest that the LEP rs7799039 G>A polymorphism might increase the risk of cancer in Asians. We are the first to report the relationship between this SNP and cancer risk in this ethnicity.
An interesting phenomenon observed during stratified analysis was that the LEP rs7799039 G>A polymorphism decreased the risk of CRC, while this SNP increased the risk of PC, other cancers, and hematopoietic malignancy. One possible explanation is that there were insufficient sample sizes for subgroup analysis. Although our findings were stable by one-way sensitivity analysis, publication bias was not found.
Among the included studies, significant heterogeneity was found in four genetic models for overall analysis. Stratified analyses indicated that heterogeneity was significant in some subgroups (eg Asians, small sample sizes, and hospital-based studies). These factors may contribute to the major heterogeneity in this study.
Several limitations, in this meta-analysis, should be acknowledged. First, although the Begg’s funnel plot and Egger’s test suggested no significant publication bias, it is possible that certain unpublished data are yet to be included. Selection bias for this study might have existed. Second, for lack of detailed information in the included studies, only crude ORs and CIs were calculated. We did not carry out the analysis adjusted for other potential risk factors (eg smoking, alcohol consumption, body mass index, and vegetable intake). Finally, heterogeneity among the eligible case–control studies was statistically significant in multiple genetic models. These findings should be considered with caution.
In conclusion, this study performed an extensive assessment based on a larger sample size than the previous pooled analysis. Our study indicates that the LEP rs7799039 G>A polymorphism may contribute to the development of cancer. In order to further verify or refute our findings, large well-designed epidemiological studies are warranted. As investigations among Asian populations are limited, further well-designed epidemiological studies involving a wider spectrum of subjects to explore the potential role of this SNP in Asians are needed.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Garg SK, Maurer H, Reed K, Selagamsetty R. Diabetes and cancer: two diseases with obesity as a common risk factor. Diabetes Obes Metab. 2014;16(2):97–110. doi: 10.1111/dom.2014.16.issue-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena NK, Sharma D, Ding X, et al. Concomitant activation of the JAK/STAT, PI3K/AKT, and ERK signaling is involved in leptin-mediated promotion of invasion and migration of hepatocellular carcinoma cells. Cancer Res. 2007;67(6):2497–2507. doi: 10.1158/0008-5472.CAN-06-3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang X, Wang S, Wang X, Zhang L, Zhao H, Zhang L. Leptin promotes the growth of breast cancer by upregulating the Wnt/beta-catenin pathway. Exp Ther Med. 2018;16(2):767–771. doi: 10.3892/etm.2018.6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nwadozi E, Ng A, Stromberg A, et al. Leptin is a physiological regulator of skeletal muscle angiogenesis and is locally produced by PDGFRalpha and PDGFRbeta expressing perivascular cells. Angiogenesis. 2019;22(1):103–115. doi:10.1007/s10456-018-9641-6 [DOI] [PubMed] [Google Scholar]
- 6.Sun X, Wei J, Tang Y, et al. Leptin-induced migration and angiogenesis in rheumatoid arthritis is mediated by reactive oxygen species. FEBS Open Bio. 2017;7(12):1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clements VK, Long T, Long R, Figley C, Smith DMC, Ostrand-Rosenberg S. Frontline science: high fat diet and leptin promote tumor progression by inducing myeloid-derived suppressor cells. J Leukoc Biol. 2018;103(3):395–407. doi: 10.1002/JLB.4HI0517-210R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray A, Cleary MP. The potential role of leptin in tumor invasion and metastasis. Cytokine Growth Factor Rev. 2017;38:80–97. doi: 10.1016/j.cytogfr.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye XL, Lu CF. Association of polymorphisms in the leptin and leptin receptor genes with inflammatory mediators in patients with osteoporosis. Endocrine. 2013;44(2):481–488. doi: 10.1007/s12020-013-9899-9 [DOI] [PubMed] [Google Scholar]
- 10.Patraca I, Martinez N, Busquets O, et al. Anti-inflammatory role of Leptin in glial cells through p38 MAPK pathway inhibition. Pharmacol Rep. 2017;69(3):409–418. doi: 10.1016/j.pharep.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 11.Santos CL, Bobermin LD, Souza DO, Quincozes-Santos A. Leptin stimulates the release of pro-inflammatory cytokines in hypothalamic astrocyte cultures from adult and aged rats. Metab Brain Dis. 2018. doi: 10.1007/s11011-018-0311-6 [DOI] [PubMed] [Google Scholar]
- 12.Lu W, Zhou S, Xue L, et al. High preoperative serum leptin level is an independent risk factor for deep vein thrombosis after total knee arthroplasty in osteoarthritis patients: a prospective and cross-sectional study. Medicine. 2018;97(21):e10884. doi: 10.1097/MD.0000000000010884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrini S, Neri T, Lombardi S, et al. Leptin induces the generation of procoagulant, tissue factor bearing microparticles by human peripheral blood mononuclear cells. Biochim Biophys Acta. 2016;1860(6):1354–1361. doi: 10.1016/j.bbagen.2016.03.029 [DOI] [PubMed] [Google Scholar]
- 14.Hardwick JC, Van Den Brink GR, Offerhaus GJ, Van Deventer SJ, Peppelenbosch MP. Leptin is a growth factor for colonic epithelial cells. Gastroenterology. 2001;121(1):79–90. [DOI] [PubMed] [Google Scholar]
- 15.Yuzhalin AE, Kutikhin AG. Integrative systems of genomic risk markers for cancer and other diseases: future of predictive medicine. Cancer Manag Res. 2012;4:131–135. doi: 10.2147/CMAR.S30855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Zhong Z, Tang W, Chen J. Leptin rs2167270 G > A (G19A) polymorphism may decrease the risk of cancer: A case-control study and meta-analysis involving 19 989 subjects. J Cell Biochem. 2019. doi: 10.1002/jcb.28378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terrasi M, Fiorio E, Mercanti A, et al. Functional analysis of the −2548G/A leptin gene polymorphism in breast cancer cells. Int J Cancer. 2009;125(5):1038–1044. doi: 10.1002/ijc.24372 [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Wu H, Zhu Y, Gao Y. Genetic association between leptin-2548G/A polymorphism and risk of cancer: a meta analysis. Int J Clin Exp Med. 2015;8(1):448–455. [PMC free article] [PubMed] [Google Scholar]
- 19.He J, Xi B, Ruiter R, et al. Association of LEP G2548A and LEPR Q223R polymorphisms with cancer susceptibility: evidence from a meta-analysis. PLoS One. 2013;8(10):e75135. doi: 10.1371/journal.pone.0075135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Liu P, Guo F, et al. Genetic G2548A polymorphism of leptin gene and risk of cancer: a meta-analysis of 6860 cases and 7956 controls. J BUON. 2014;19(4):1096–1104. [PubMed] [Google Scholar]
- 21.Kim EY, Chin HM, Park SM, et al. Susceptibility of gastric cancer according to leptin and leptin receptor gene polymorphisms in Korea. J Korean Surg Soc. 2012;83(1):7–13. doi: 10.4174/jkss.2012.83.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269, W264. [DOI] [PubMed] [Google Scholar]
- 23.Mantel NHW, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. [DOI] [PubMed] [Google Scholar]
- 26.Ribeiro R, Vasconcelos A, Costa S, et al. Overexpressing leptin genetic polymorphism (−2548 G/A) is associated with susceptibility to prostate cancer and risk of advanced disease. Prostate. 2004;59(3):268–274. doi: 10.1002/pros.20004 [DOI] [PubMed] [Google Scholar]
- 27.Skibola CF, Holly EA, Forrest MS, et al. Body mass index, leptin and leptin receptor polymorphisms, and non-hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2004;13(5):779–786. [PubMed] [Google Scholar]
- 28.Willett EV, Skibola CF, Adamson P, et al. Non-Hodgkin’s lymphoma, obesity and energy homeostasis polymorphisms. Br J Cancer. 2005;93(7):811–816. doi: 10.1038/sj.bjc.6602762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chovanec J, Bienertova-Vasku JA, Dostalova Z. Leptin–2548 g/A polymorphism in endometrial cancer. Klinicka. 2009;22(5):223–227. [PubMed] [Google Scholar]
- 30.Moore SC, Leitzmann MF, Albanes D, et al. Adipokine genes and prostate cancer risk. Int J Cancer. 2009;124(4):869–876. doi: 10.1002/ijc.24043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pechlivanis S, Bermejo JL, Pardini B, et al. Genetic variation in adipokine genes and risk of colorectal cancer. Eur j endocrinol. 2009;160(6):933–940. doi: 10.1530/EJE-09-0039 [DOI] [PubMed] [Google Scholar]
- 32.Vasku A, Vokurka J, Bienertova-Vasku J. Obesity-related genes variability in Czech patients with sporadic colorectal cancer: preliminary results. Int J Colorectal Dis. 2009;24(3):289–294. doi: 10.1007/s00384-008-0553-6 [DOI] [PubMed] [Google Scholar]
- 33.Cleveland RJ, Gammon MD, Long CM, et al. Common genetic variations in the LEP and LEPR genes, obesity and breast cancer incidence and survival. Breast Cancer Res Treat. 2010;120(3):745–752. doi: 10.1007/s10549-009-0503-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribeiro RJ, Monteiro CP, Azevedo AS, et al. Performance of an adipokine pathway-based multilocus genetic risk score for prostate cancer risk prediction. PLoS One. 2012;7(6):e39236. doi: 10.1371/journal.pone.0039236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavil B, Balta G, Ergun EL, et al. Leptin promoter G-2548A genotypes and associated serum leptin levels in childhood acute leukemia at diagnosis and under high-dose steroid therapy. Leuk Lymphoma. 2012;53(4):648–653. doi: 10.3109/10428194.2011.626881 [DOI] [PubMed] [Google Scholar]
- 36.Unsal M, Kara N, Karakus N, Tural S, Elbistan M. Effects of leptin and leptin receptor gene polymorphisms on lung cancer. Tumour Biol. 2014;35(10):10231–10236. doi: 10.1007/s13277-014-2293-2 [DOI] [PubMed] [Google Scholar]
- 37.Karakus N, Kara N, Ulusoy AN, Ozaslan C, Tural S, Okan I. Evaluation of CYP17A1 and LEP gene polymorphisms in breast cancer. Oncol Res Treat. 2015;38(9):418–422. doi: 10.1159/000438940 [DOI] [PubMed] [Google Scholar]
- 38.Mahmoudi R, Noori Alavicheh B, Nazer Mozaffari MA, Fararouei M, Nikseresht M. Polymorphisms of leptin (−2548 G/A) and leptin receptor (Q223R) genes in Iranian women with breast cancer. Int J Genomics. 2015;2015:132720. doi: 10.1155/2015/132720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammadzadeh G, Ghaffari MA, Bafandeh A, Hosseini SM, Ahmadi B. The relationship between −2548 G/A leptin gene polymorphism and risk of breast cancer and serum leptin levels in Ahvazian women. Iran J Cancer Prev. 2015;8(2):100–108. [PMC free article] [PubMed] [Google Scholar]
- 40.Rostami S, Kohan L, Mohammadianpanah M. The LEP G-2548A gene polymorphism is associated with age at menarche and breast cancer susceptibility. Gene. 2015;557(2):154–157. doi: 10.1016/j.gene.2014.12.021 [DOI] [PubMed] [Google Scholar]
- 41.Mahmoudi T, Farahani H, Nobakht H, Dabiri R, Zali MR. Genetic variations in leptin and leptin receptor and susceptibility to colorectal cancer and obesity. Iran J Cancer Prev. 2016;9(3):e7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amer T, El-Baz R, Mokhtar AR, El-Shaer S, Elshazli R, Settin A. Genetic polymorphisms of IL-23R (rs7517847) and LEP (rs7799039) among Egyptian patients with hepatocellular carcinoma. Arch Physiol Biochem. 2017;123(5):279–285. doi: 10.1080/13813455.2017.1320680 [DOI] [PubMed] [Google Scholar]
- 43.Ali SHB, Bangash KS, Rauf A, et al. Identification of novel potential genetic predictors of urothelial bladder carcinoma susceptibility in Pakistani population. Fam Cancer. 2017;16(4):577–594. doi: 10.1007/s10689-017-9991-z [DOI] [PubMed] [Google Scholar]
- 44.Rodrigo C, Tennekoon KH, Karunanayake EH, De Silva K, Amarasinghe I, Wijayasiri A. Circulating leptin, soluble leptin receptor, free leptin index, visfatin and selected leptin and leptin receptor gene polymorphisms in sporadic breast cancer. Endocr J. 2017;64(4):393–401. doi: 10.1507/endocrj.EJ16-0448 [DOI] [PubMed] [Google Scholar]
- 45.Zhang S, Jiang J, Chen Z, et al. Investigation of LEP and LEPR polymorphisms with the risk of hepatocellular carcinoma: a case-control study in Eastern Chinese Han population. Onco Targets Ther. 2018;11:2083–2089. doi: 10.2147/OTT.S153931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hussain SR, Naqvi H, Gupta S, et al. A study on oncogenic role of leptin and leptin receptor in oral squamous cell. Tumour Biol. 2015;36(8):6515–6523. doi: 10.1007/s13277-015-3342-1 [DOI] [PubMed] [Google Scholar]
- 47.Qiu H, Lin X, Tang W, et al. Investigation of TCF7L2, LEP and LEPR polymorphisms with esophageal squamous cell carcinomas. Oncotarget. 2017;8(65):109107–109119. doi: 10.18632/oncotarget.22619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan XL, Xu ZP, Liu CR, et al. [Study of the association between polymorphism of persistent obesity, human leptin gene/leptin receptor gene and molecular subtypes of breast cancer]. Zhonghua Yu Fang Yi Xue Za Zhi. 2017;51(6):533–538. doi: 10.3760/cma.j.issn.0253-9624.2017.06.015 [DOI] [PubMed] [Google Scholar]
- 49.Liu CR, Li Q, Hou C, et al. Changes in body mass index, leptin, and leptin receptor polymorphisms and breast cancer risk. DNA Cell Biol. 2018;37(3):182–188. doi: 10.1089/dna.2017.4047 [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Dhakal IB, Zhang X, Prizment AE, Anderson KE. Genetic variability in energy balance and pancreatic cancer risk in a population-based case-control study in Minnesota. Pancreas. 2014;43(2):281–286. doi: 10.1097/MPA.0b013e3182a7c829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snoussi K, Strosberg AD, Bouaouina N, Ben Ahmed S, Helal AN, Chouchane L. Leptin and leptin receptor polymorphisms are associated with increased risk and poor prognosis of breast carcinoma. BMC Cancer. 2006;6:38. doi: 10.1186/1471-2407-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slattery ML, Wolff RK, Herrick J, Caan BJ, Potter JD. Leptin and leptin receptor genotypes and colon cancer: gene-gene and gene-lifestyle interactions. Int J Cancer. 2008;122(7):1611–1617. doi: 10.1002/ijc.23135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luan H, Zhang H, Li Y, et al. Association of two obesity-related gene polymorphisms LEPG2548A rs7799039 and LEPRQ223R rs1137101 with the risk of breast cancer. Oncotarget. 2017;8(35):59333–59344. doi: 10.18632/oncotarget.19580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao FJWG, Yu YD, Yu XJ, Die QS, Lei JH, Deng SH. Chen P association of leptin gene −2548 G/A polymorphism with the susceptibility and prognosis of non-small cell lung cancer. Chin J Exp Surg. 2015;32(3):621–623. [Google Scholar]
- 55.Hoffstedt J, Eriksson P, Mottagui-Tabar S, Arner P. A polymorphism in the leptin promoter region (−2548 G/A) influences gene expression and adipose tissue secretion of leptin. Horm Metab Res. 2002;34(7):355–359. doi: 10.1055/s-2002-33466 [DOI] [PubMed] [Google Scholar]