Abstract
Purpose: Variability in response to warfarin is one of the main obstacles challenging its use in clinical practice. Vitamin K epoxide reductase complex (VKORC) is the target enzyme of warfarin, and variations in the form of single nucleotide polymorphisms (SNPs) in VKORC1, coding for this enzyme, are known to cause resistance to warfarin treatment. This study aimed to explore VKORC1 variants in Emirati patients receiving warfarin treatment and to correlate their genotypes at the studied SNPs to their maintenance warfarin dose.
Patients and methods: Sanger sequencing of the majority of the VKORC1 gene was applied to samples from 90 patients and 117 normal individuals recruited from Tawam Hospital, Al-Ain, UAE. Genotypes at the following variants were determined (rs9923231, rs188009042, rs61742245, rs17708472, rs9934438, rs8050894, rs2359612, rs7294). Statistical analysis was applied, including ANOVA, cross-tabulation, and multiple linear regression analysis, to determine the ability of nongenetic factors (age and gender) and genetic factors (VKORC1 genotypes) to explain variability in warfarin dose in patients.
Results: Different frequencies of minor alleles were detected in the selected SNPs. Significant variation among genotypes at six VKORC1 variants were identified (rs9923231, rs9934438, rs8050894, rs2359612, rs7294). The main predictors for warfarin dose were rs9923231, age, and rs61742245 with 50.7% of the average warfarin dose in our sample could be explained by a regression model built on these three factors.
Conclusion: This is the first report of the explanatory power of VKORC1 genotypes and nongenetic factors (age and gender) on warfarin dose among Emiratis. Also, this study highlighted the positive effect of considering rare pharmacogenomic variants on explaining warfarin dose variability.
Keywords: anticoagulants, pharmacogenomics, single nucleotide polymorphisms, United Arab Emirates
Introduction
For >50 years, warfarin continued to be the mainstay anticoagulant worldwide.1 It is commonly prescribed in the management and prophylaxis of thromboembolism in patients with atrial fibrillation, cardiac valve prosthesis, deep vein thrombosis, pulmonary embolism, and recurrent stroke among other indications.1 Although it proved to be highly effective in reducing thrombosis related morbidity and mortality, warfarin has a narrow therapeutic window and therefore maintaining the required anticoagulant effect is often challenging. Warfarin dose is usually titrated through monitoring its blood thinning effect defined by the international normalized ratio (INR).2,3 Sequelae of overdosing include bleeding and hospitalization which are among the most common adverse events in USA,4 and under-dosing puts patients at high risk of thrombotic events.2 New classes of anticoagulants were developed, and they are gradually replacing warfarin in many indications due to their superior safety profile.5 However, warfarin is still one of the 50 most commonly prescribed medications. It continues to be the most prescribed anti-coagulant worldwide especially in developing countries.1,3,6
The difficulty in reaching target INR with standard warfarin doses lies in the wide range of variability exhibited in individuals’ response. Warfarin is crucially affected by drug–drug interactions, alcohol consumption, and some food supplements.7 Vitamin K intake was found, in several studies, to correlate with dose. Moreover, several physical factors, such as age and weight have shown to affect warfarin dose requirement.8,9 The first clues of a genetic factor in this variability appeared in the 1990s.8 At present, genetic differences can explain around 50% of the interindividual variability in warfarin treatment.10
Initially, pharmacogenomics studies of warfarin traced its metabolic pathway looking for candidate genes that might be involved in interindividual variability. This candidate gene approach identified the metabolizing enzyme cytochrome P450 2C9 encoded by the CYP2C9 gene as a significant contributor. It was soon found that individuals with nonfunctional haplotypes that produce a metabolically impaired enzyme require lower doses of warfarin. More genes in the metabolic pathway were investigated with results of little significance. Despite strong indications on the presence of other genetic determinants, there were no significant advances in identifying them until 2004 when the molecular target of the drug, Vitamin K epoxide reductase, was identified.8,11
Vitamin K epoxide reductase complex subunit 1 protein is the target enzyme of warfarin and is encoded by the VKORC1 gene. Warfarin inhibits this reductase, limiting the conversion of vitamin K epoxide into vitamin K and consequently decreasing the formation of clotting factors.12,13 Single nucleotide polymorphisms (SNPs) occurring on VKORC1 mainly including rs9923231, rs17708472, rs9934438, and rs2359612 were found to affect the availability or activity of the reductase impacting the anticoagulant effect of drugs targeting this enzyme, like warfarin.14 Some of the variants were found to cause partial or complete resistance to warfarin without reaching the target INR.15 In general, VKORC1 variants can explain approximately 20–30% of the variation in warfarin dosing.16 Consequently, haplotype maps were established from the common VKORC1 SNPs. These efforts yielded two main ways to report VKORC1 haplotypes: the H system17 and the star system.18 Both systems were built in European populations. The H haplotypes (from H1 to H9) depend on the genotypes at 10 variants (rs7196161, rs17880887, rs17881535, rs9923231, rs2884737, rs17708472, rs9934438, rs8050894, rs2359612, rs7294). These haplotypes can be further grouped into two main groups: group A that includes H1 and H2 which are low-dose haplotypes, and group B that includes H7, H8, H9 which are high-dose haplotypes.17 The star system relies on the genotypes at five variants (rs9923231, rs17708472, rs9934438, rs2359612, rs7294). In the star system, *2 haplotype is associated with low-dose warfarin while *3 and *4 alleles are associated with a high dose of warfarin.18
Numerous studies have established the contribution of VKORC1 genomic variants on warfarin dose in patients from different populations. Some SNPs were found to be common in diverse populations, while others were found sporadically and were considered rare.11 The United Arab Emirates (UAE) indigenous citizens are considered an admixed population that is under-represented in pharmacogenomics studies. The current study aimed at evaluating the effect of the eight variants in VKORC1 on warfarin maintenance dose in a group of Emirati patients receiving a stabilized dose of warfarin. The chosen variants included those in the star system (five SNPs) and three more SNPs that were found to be actionable in some populations.19 Furthermore, we aimed at estimating the frequencies of haplotypes and analyzing their association with warfarin dose.
Methods
Sample
The study cohort included 90 unrelated patients treated with warfarin and 107 normal controls. All the participants were Emiratis who were recruited from Tawam Hospital, Al-Ain, UAE. Participants had read the information sheet and signed informed consent forms. This study was conducted according to the declaration of Helsinki after the approval of Al-Ain Medical District Human Research Ethics Committee (Ethical Approval CRD 261-Protocol No. 13/38).
Patients data were collected retrospectively and included warfarin stabilized dose, target INR, indication for warfarin therapy, comorbidities, other prescribed medications, and demographic characteristics. About 5 mL of peripheral venous blood was collected and placed in an EDTA tube to be genotyped.
Genotyping
DNA was extracted from peripheral leukocytes using a whole-blood extraction kit (Flexigene DNA isolation kit; Qiagen, Germany). The isolated genomic DNA samples were kept in sterile plastic vials at 4°C until analysis or stored at −20°C. Genotype study was carried out using the PCR. The forward and reverse primers sequences for amplifying most of the VKORC1 gene and its promoter, and the PCR conditions are listed in Table 1.
Table 1.
Primers and PCR conditions used to amplify and sequence the vast majority of genomic sequence including the reported SNPs on VKORC1
| Forward primer | Reverse primer | Annealing temp (°C) |
Annealing time (min) | Product size (bp) |
|---|---|---|---|---|
| F1-GCCAGCAGGAGAGGGAAATA | R1-AGTTTGGACTACAGGTGCCT | 58 | 60 | 290 |
| F2-GCAGGCACCTGTAGTCCAA | R2-CGCTTACCCTATGCCAAGTC | 58 | 60 | 842 |
| F3-TGATCCGCTGGTCTCTAGGT | R3-GAGAGCACTAAGCCCGTCAG | 58 | 60 | 786 |
| F5-GTCTGGCTGTGCGTGATG | R5-GCCATAGCGCCCGATTAATT | 58 | 45 | 313 |
| F6-AGGCGTTAGCATAATGACGG | R6-GGGTGGAACCAGGTTAGGAC | 60 | 60 | 725 |
| F7-GTCCTAACCTGGTTCCACCC | R7-CCCTCCAAGGGACTGGTCT | 60 | 60 | 995 |
| F9-GAATACGTGCGTAAGCCACC | R9-GGTTCAGACTTGGCTGATTG | 60 | 45 | 659 |
The quality of PCR amplification was checked by separation on 0.6% agarose gel electrophoresis before proceeding to Sanger sequencing. For sequencing, the PCR products were purified using ExoSAP-IT® (Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s protocol. Cycle sequencing was performed using the BigDye Terminator kit v3.1 (Applied Biosystems) under standard conditions. Capillary electrophoresis was performed in a 3130xl Genetic Analyzer (Applied Biosystems).
Statistical analysis
Frequencies, means, and standard deviations, when applicable, were calculated. The frequencies of each allele were calculated from the genotypes of patients and controls. The retrieved genotypes and allele frequencies for each variant were compared between the patients and control groups by chi-square (χ2) and Fisher’s exact test. The same tests were used to compare the frequencies in all participants with frequencies from the global population of the 1000 Genomes Project. The minor alleles at each studied SNP were determined depending on Ensembl database19 and PharmGKB database.20 Hardy–Weinberg equilibrium was tested by χ2 test. Pairwise linkage disequilibrium (LD) between the selected SNPs was assessed by calculating D’ and r2.
ANOVA was used to assess the effect of individual VKORC1 polymorphisms on the maintenance dose of warfarin. Patients were assigned into three groups according to warfarin dose: group 1 included patients who used doses ≤3 mg/day (ie, denoted as the low-dose group). Group 2 included patients with doses between 3.1 and 5.9 mg/day (ie, intermediate-dose group), and group 3 includes patients who used doses ≥6 mg/day (ie, high-dose group). Cross-tabulation with χ2 statistics were performed to describe the relationship between genotypes and dose-dependent groups. Haplotypes were constructed based on selected SNPs using the SNPSstats21 and SNPAlyze ver 9.022 software.
Multivariant analysis using linear regression was used to determine the ability of nongenetic factors (ie, age and gender) and genetic factors (VKORC1 genotypes) to predict warfarin dose. Statistical analysis was performed using SPSS (version 24; SPSS Inc, an IBM Company, Chicago, IL, USA). The significance level of two-sided P-value was chosen to be ≤0.05.
Results
Patients characteristics
Warfarin stabilized dose was defined as the dose that was prescribed and maintained for the last three clinic visits at least to achieve the target INR. In the patients group (n=90) warfarin dose ranged from 0.5 to 15 mg (mean 4.7±2.48) for a target INR of 2–3 for these patients. Descriptive statistics of the patients are summarized in Table 2. Briefly, 45 males and 45 females were included in this study with an average age of 60.54±17.8 years. Atrial fibrillation was the most common indication for warfarin treatment in this study.
Table 2.
Descriptive statistics for 90 Emirati individuals treated with warfarin recruited for VKORC1 genotyping
| Variable | Value |
|---|---|
| Gender, n (%) Female Male |
45 (50) 45 (50) |
| Age in years, range Mean age (SD) |
21–98 60.54 (17.8) |
| Indication for warfarin therapy, n Atrial fibrillation Prosthetic heart valve Deep vein thrombosis Cardiomyopathy Hypercoagulable state Stroke Pulmonary embolism Left ventricular thrombosis Valve disease |
38 26 16 3 2 2 1 1 1 |
VKORC1 SNPs genotyping
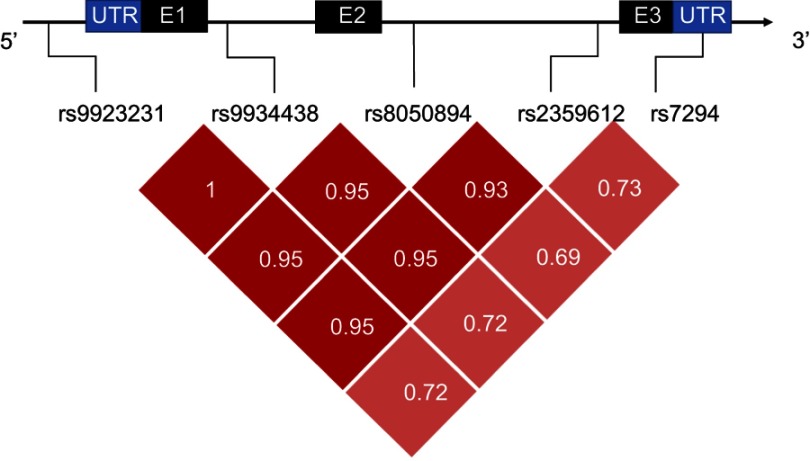
Genotypes at the studied SNPs were analyzed in both patients and controls. For LD analysis, only variants that had allele frequencies >0.1 were evaluated. It was found that SNPs rs9923231 and rs9934438 are in complete LD (pairwise r2=1). Figure 1 illustrates the pairwise LD statistics of selected markers.
Figure 1.
Linkage disequilibrium analysis: a diagram of VKORC1 with the location of selected SNPs (Transcript: VKORC1-205 ENST00000394975.2). Black and blue boxes represent exons (E) and untranslated regions (UTR), respectively. Dark red blocks represent stronger LD than the lighter ones. Values represent pairwise r2.
The allele frequencies at each variant were calculated and compared between patients and controls, and no statistically significant (P<0.05) differences between these frequencies was detected. Further on, the allele counts in the whole sample (patients and controls) were compared to the counts retrieved from the global population of the 1000 Genomes Project and statistically significant differences were found at all SNPs except rs17708472 (Table 3).
Table 3.
Frequencies of minor alleles at the studied SNPs in comparison to allele frequencies in different populations from the 1000 Genomes Projecta
| Variant | Minor allele | MAF in current study | MAF in ALL | P-valueb | AFR | AMR | EAS | EUR | SAS |
|---|---|---|---|---|---|---|---|---|---|
| rs9923231 | A | 0.52 | 0.356 | <0.001 | 0.054 | 0.411 | 0.885 | 0.388 | 0.145 |
| rs61742245 | T | 0.033 | ˂0.01 | <0.001 | 0.001 | – | – | 0.001 | |
| rs188009042 | G | 0.01 | 0.003 | a | – | 0.014 | – | – | |
| rs17708472 | T | 0.083 | 0.094 | 0.79 | 0.033 | 0.140 | 0.003 | 0.234 | 0.092 |
| rs9934438 | T | 0.52 | 0.356 | <0.001 | 0.054 | 0.412 | 0.885 | 0.388 | 0.145 |
| rs8050894 | C | 0.52 | 0.416 | 0.008 | 0.256 | 0.442 | 0.885 | 0.400 | 0.148 |
| rs2359612 | T | 0.53 | 0.610 | 0.0003 | 0.821 | 0.575 | 0.115 | 0.612 | 0.855 |
| rs7294 | A | 0.328 | 0.420 | 0.013 | 0.454 | 0.402 | 0.112 | 0.366 | 0.759 |
Notes: aFrequencies in different populations are adapted from www.ensembl.org; bP-values calculated between the current allele frequencies and allele frequency of global populations (1000 Genomes Project) using Fisher’s exact test.
Abbreviations: MAF, minor allele frequency; ALL, all individuals from phase 3 of 1000 Genomes Project; AFR, African; AMR, American; EAS, East Asian; EUR, European; SAS, South Asian.
SNPs variants association with dose requirement
A significant difference (P<0.05) in average doses of warfarin among different genotype groups was found at the following SNPs: rs9923231, rs9934438, rs8050894, rs2359612, and rs7294. A difference was also observed between genotype groups at rs61742245, but at lower significance. In contrast, no significant difference between genotype groups at the two variants (rs188009042, rs17708472) was detected (Table 4). Cross-tabulation showed that all the patients who were minor allele carriers at rs9923231, rs9934438, rs8050894, and rs2359612 required warfarin doses lower than 6 mg/day, ie, were in the low or intermediate dose group. Moreover, around 70% of those patients were found in the warfarin low-dose group (≤3 mg/day). In contrast, patients that carry the wild-type at the same variants clustered in the high-dose group (≥6 mg/day) and only a few of them were in the intermediate or low-dose groups. A similar significant association was reported at rs7294 but it was inverted; ie, minor allele carriers clustered in the high-dose group and most of the wild-type carriers were in the low-dose group (results of cross-tabulations are found in the supplementary material; Table S1).
Table 4.
Frequencies of genotypes at the studied SNPs and results of ANOVA
| VKORC1 SNP | Genotypes frequencies, n (%) | Average dose±SD | ANOVA | |
|---|---|---|---|---|
| F (2,87) | P-value | |||
| rs9923231 | 32.955 | 0.00* | ||
| GG | 22 (24.4%) | 7.05±2.89 | ||
| GA | 43 (47.8%) | 4.73±1.63 | ||
| AA | 25 (27.8%) | 2.56±1.00 | ||
| rs61742245 | ||||
| GG | 86 (95.6%) | 4.53±2.22 | 5.3.5 | 0.007* |
| GT | 2 (2.2%) | 7.50±2.12 | ||
| TT | 2 (2.2%) | 9.25±8.13 | ||
| rs188009042 | ||||
| AA | 89 (98.9%) | 4.66±2.47 | 1.80 | 0.18 |
| AG | 1 (1.0%) | 8.00 | ||
| GG | 0 | – | ||
| rs17708472 | ||||
| CC | 76 (84.4%) | 4.56±2.44 | 1.37 | 0.258 |
| CT | 13 (14.4%) | 5.27±2.60 | ||
| TT | 1 (1.1%) | 8.00 | ||
| rs9934438 | 32.955 | 0.00* | ||
| CC | 22 (24.4%) | 7.05±2.89 | ||
| CT | 43 (47.8%) | 4.73±1.63 | ||
| TT | 25 (27.8%) | 2.56±1.00 | ||
| rs8050894 | 29.541 | 0.00* | ||
| GG | 21 (26.7%) | 7.03±2.94 | ||
| GC | 45 (50.0%) | 4.73±1.71 | ||
| CC | 24 (23.3%) | 2.58±1.01 | ||
| rs2359612 | 28.37 | 0.00* | ||
| CC | 19 (21.1%) | 7.09±3.04 | ||
| CT | 47 (52.2%) | 4.81±1.76 | ||
| TT | 24 (26.7%) | 2.58±1.01 | ||
| rs7294 | 9.23 | 0.00* | ||
| GG | 39 (43.3%) | 3.60±2.48 | ||
| GA | 43 (47.8%) | 5.32±1.99 | ||
| AA | 8 (8.9%) | 6.71±2.63 | ||
Note: *Statistically significant.
Haplotype analysis and association with warfarin doses
Analyzing haplotypes in the patient’s group depending on the star system revealed that VKORC1*2 haplotype was the most frequent one with a 50% frequency. The second most frequent haplotype was VKORC1*3 with 32% frequency, followed by VKORC1*4 with 7% frequency, followed by the reference sequence VKORC1*1 with 6% frequency, and the remaining 5% were rare haplotypes. However, a global test for statistical difference among these haplotypes did not give a significant association with dose in linear regression analysis (P=0.62). Accordingly, we reconstructed haplotypes depending on all genotyped variants excluding the two variants, rs188009042 and rs17708472, that did not show significant variation within genotype or significant linkage to other variants in our sample. Haplotypes extracted from the six remaining variants (rs9923231, rs61742245, rs9934438, rs8050894, rs2359612, rs7294) have shown significant global haplotype association with dose (P=0.009). The most frequent haplotype (AGTCTG) was denoted by H1 and it had 50% frequency, followed by haplotype H2 (GGCGCA) with 32% frequency, followed by H3 (GGCGCG) with 11% frequency, then haplotype H4 (GTCGCG) with 2% frequency. The rare haplotypes which occurred in ≤2 individuals were combined in one group denoted as “rare”. Logistic regression was used to determine the association between haplotypes and warfarin dose. Table 5 lists the retrieved haplotypes, their frequencies, and association with dose.
Table 5.
Analysis of haplotypes and their association with dose
| Haplotype | rs9923231 | rs61742245 | rs9934438 | rs8050894 | rs2359612 | rs7294 | F (%) | Difference in mean (CI)* |
P |
|---|---|---|---|---|---|---|---|---|---|
| H1 | A | G | T | C | T | G | 50 | 0.00 | – |
| H2 | G | G | C | G | C | A | 32 | 2.29 (1.6–2.95) |
<0.0001** |
| H3 | G | G | C | G | C | G | 11 | 1.87 (0.93–2.81) |
0.00015** |
| H4 | G | T | C | G | C | G | 2 | 3.18 (1.9–4.46) |
<0.0001** |
| H5 | G | G | C | G | T | G | 1.2 | 1.02 (−1.61–3.65) |
0.45 |
| Rare | * | * | * | * | * | * | 2.8 | 2.36 (0.62–4.1) |
0.0089** |
Notes: *The most frequent haplotype is considered the reference and the difference in means measures the difference between the mean of dose among each haplotype and the reference. **Significant association.
Multilinear regression analysis
A stepwise multivariate regression that included all the selected variants at VKORC1 with age and gender revealed that the main predictors for warfarin dose were rs9923231, age, and rs61742245. The most potent indicator was rs9923231, which alone explained 0.424 of dose variability (indicated by adjusted r2). Adding age increased the prediction of the model to 0.482, then by adding rs61742245 r2 it reached 0.507, which suggests that 50.7% of the average warfarin dose in our sample could be explained by genotype at rs9923231 and rs61742245 and age (P<0.05). Table 6 lists predictors and logistic regression for this model.
Table 6.
Model summary and predictors of logistic regression
| Model | R | R square | Adjusted R square | Std error of the estimate | |||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.656a | 0.431 | 0.424 | 1.88 | |||||
| 2 | 0.702b | 0.493 | 0.482 | 1.79 | |||||
| 3 | 0.724c | 0.524 | 0.507 | 1.74 |
Notes: aPredictors (constant), rs9923231. bPredictors (constant), rs9923231, Age. cPredictors (constant), rs9923231. Age, rs61742245.
Abbreviation: Std, standard.
Discussion
The aim of the current study is to examine the effect of genotypes at some actionable variants of VKORC1 on warfarin dose. The main finding is the significant prediction power of some of these variants and age on warfarin dose in the Emirati population.
UAE is situated along the coast of the Arabian Gulf in a central area to the old-world that made it a vital nexus in the dispersal of modern human and historical migration waves.23 Indigenous UAE citizens, ie, Emiratis, who constitute <15% of the whole population (one out of nine million, UAE government census), include subgroups from different origins that demonstrate genetic isolates and semi-isolates that preferably practice consanguineous marriages.24 Bertrand et al found that Emiratis share 23.7% of their DNA with Southwest Asian population and, compared to other populations from Arabian Peninsula, they exhibit the highest contribution from Asia and the lowest from North Africa.23 In our data, statistically significant differences (P<0.05) were found between allele frequencies at the studied SNPs among 207 Emiratis (patients and controls) and frequencies reported in the 1000 Genomes Project. This finding is not surprising in an admixed population, that is composed of a rarely studied complex substructure. Genomic research that takes the population substructure into account would give deeper insights into the genomic similarities and differences with other populations.25
ANOVA in warfarin dosage between different genotypes revealed a significant difference at five variants. Mainly, rs9923231 was a leading contributor that remained a significant predictor of dose in our multiple regression generated model. The association between genotypes at this SNP and warfarin dose have repeatedly been reported in several populations and among different ethnicities.26 Moreover, rs9923231 was found to be a main determinant of warfarin dose among a Saudi Arabian population27 where it was reported in a frequency that was not statistically different from the current study (X2=3.4, P=0.6). This variant, which is commonly called in literature as −1639G>A, is known to occur in the promotor of VKORC1 affecting the transcription factor binding site and reducing the gene mRNA expression.17,28 The result of such reduction in activity was found to reduce vitamin K carboxylation rate and hence, warfarin requirement.18 Accordingly, clustering of >70% of minor alleles carriers at this SNP in the warfarin low-dose group (warfarin dose ≤3 mg/day) is justified. The opposite is true for wild-type carriers and both observations are consistent with previous reports.18,29
The intronic variant rs9934438 has repeatedly been found in near perfect concordance with rs9923231 (ie, perfect LD).17,26,30 A similar finding was reported in our population (LD pairwise r2=1). Accordingly, an identical clustering of genotypes in patients-dose groups has been seen in both variants. Moreover, the minor allele homozygous carriers of both SNPs (GG at rs9923231 and CC at rs9934438) required the highest doses of warfarin (7.05±2.89) compared to other genotype groups at the same variants. Variation in any of the two SNPs explained 18% and 5% of dose variability in Caucasians and African Americans, respectively.31 In our group, around 42.4% of warfarin dose variability could be explained by either of these two variants. This prediction power is also more than Chinese (31%),32 Iranian (20.3%),33 and close to Omani (45%) patients.34
Furthermore, the genotypes at the two SNPs, rs8050894 and rs2359612, were associated with a similar dose variance. These two SNPs, which are in intron 2, were associated with warfarin requirement in several populations.17,18 The mechanism underlying their effect is presumed to be through affecting transcription factor binding sites.17 In our population, none of the homozygote carriers of the minor alleles at these two variants required a high dose of warfarin and around 70% were in the warfarin low-dose group.
The fifth significant genotype was seen at rs7294. This variant is located in the 3ʹ untranslated region of VKORC1.18 The A allele was associated with significantly higher doses of warfarin in previous studies,31,35 as well as in our group. More recently, rs7294 was found to have a significant effect on warfarin plasma concentration.36 In our patients, ANOVA and cross-tabulation have shown significant differences between average dosages among genotype groups and significant association between minor allele carriers and high-dose group, respectively, at this SNP. However, it was not a significant predictor in our dose prediction model like what was found in European populations,29,31,37 and in contrast to results from Sudanese patients.38
The G>T at rs61742245 leads to a substitution of tyrosine for aspartic acid at amino acid 36 (p.D36Y). It was found to be associated with the requirement of high doses of warfarin in sporadic reports.18 It is mainly identified among Ashkenazi Jews and Ethiopians with minor allele frequency (MAF) around 4% and 15%, respectively.39,40 Kurnik et al focused on the magnitude of the effect of this variant on warfarin dose and found that after accounting of genetic and nongenetic covariates, the carriers of a single (D36Y) allele require more than double the dose of warfarin in comparison to non-carriers.41 In our cohort, four individuals had the minor allele (T) at rs61742245, of which two were homozygous. Remarkably, one of the homozygous carriers of the deficient allele was stabilized on an intermediate dose of warfarin (3.5 mg) while the other homozygous and the two heterozygous cases required high doses of warfarin (15, 6, and 9 mg/day, respectively). The latter three individuals’ requirement of high warfarin dose is consistent with findings from other populations,40 while the lower than expected dosage of the first homozygous patient suggests a possible presence of another factor affecting warfarin dose that was not included in our study (CYP2C9 status for instance). Regardless of the few individuals present in each genotype group, rs61742245 was a significant predictor in the regression model. Our study is the first to examine this variant in Arabs. Moreover, due to its low MAF, few studies have ever evaluated its effect on warfarin dose in other populations.
It is noteworthy, that five SNPs showed strong LD (rs9923231, rs9934438, rs8050894, rs235961, rs7294) (Figure 1), which was strongest at the first four (r2≥0.93) and moderate at rs7294 (r2≈0.7). Similar findings were reported from some other populations.17,26 LD describes the nonrandom association of alleles at two or more loci, and statistical analysis of LD can be used to extract haplotype blocks.42 When SNPs are in a strong LD, alleles of some of these variants can be predicted from other alleles. Hence, testing all the alleles will give redundant information.43 Accordingly, in our population rs9923231 and perhaps rs7294 can be chosen from the five previous SNPs, besides the significant variant at rs61742245, in case a limited number of VKORC1 SNPs are to be assessed.
The two remaining SNPs (rs188009042, rs17708472) exhibited low genotype variability in our sample, which has affected assessing their association with dose. The highest population frequency of the minor allele of (rs188009042) approaches 0.03 that was reported mainly in Asian populations19 and is less than its rate in our group. Though, due to the small sample size, the frequency in the Emirati population cannot be extrapolated. A similar observation applies to rs17708472. It is noteworthy that only one patient who represented an outlier with warfarin dose equal to 8 mg/day, carried one minor allele at rs188009042 and was homozygous for the alternative allele at rs17708472. The same individual carried the wild-type alleles on all other VKORC1 SNPs. The effect of these two variants on dose requirement could not be confirmed in this case, though it is not excluded.
Haplotype analysis based on star system did not give a significant association with dose. In contrast, haplotypes relying on the six variants that have shown significant outcomes in our primary variance analysis gave a significant association with dose. A better haplotype/dose model would be constructed if more variants were included and variants from other genes were tested.
Regarding the nongenetic factors, gender and age were examined. While the gender did not show any effect on dose, age was the second most reliable dose predicting variable. It is already established that patients of older age have to be treated with more conservative regimens of warfarin. The higher risk of bleeding, comorbidities, and concomitant used medications all are among the leading reasons for starting with lower doses in geriatric patients.44
Our study has some limitations; a limited number of nongenetic factors was included in our analysis while body mass index and smoking status might be other significant contributors. Moreover, we did not genotype other genes known to affect warfarin dosing (eg, CYP2C9) for the same studied individuals, which would have explained the outlier group and added to the prediction power of our model.
Conclusions
This is the first report of the explanatory power of VKORC1 genotypes and nongenetic factors (age and gender) on warfarin dose in Emiratis. The common variant rs9923231 along with the rare variant at rs61742245 and age attributed to >50% of the variability in warfarin dose. Although VKORC1 genotype and age are well-known predictors of warfarin dose in different populations, a model based on these factors was remarkably a strong predictor of warfarin dose in our population. We highlighted here that under-represented populations exhibit different allele frequencies, haplotype structures, and might have some rare actionable pharmacogenomic variants.
Acknowledgments
We would like to thank the patients for taking part in this study. In addition, we thank the Department of Pathology and Molecular Genetics lab staff at the United Arab Emirates University for their assistance in this study. This project was funded by the United Arab Emirates University (grant 31R091).
Disclosure
The authors confirm that there are no potential conflicts of interest associated with this work.
Supplementary material
Table S1.
Results of cross-tabulation
| Low dose | Intermediate dose | High dose | |
|---|---|---|---|
| GG | 2 | 4 | 16 |
| GA | 6 | 30 | 7 |
| AA | 20 | 5 | 0 |
| Total | 28 | 39 | 23 |
| GG | 2 | 4 | 15 |
| GC | 7 | 30 | 8 |
| CC | 19 | 5 | 0 |
| Total | 28 | 39 | 23 |
| CC | 2 | 4 | 16 |
| CT | 6 | 30 | 7 |
| TT | 20 | 5 | 0 |
| Total | 28 | 39 | 23 |
| CC | 2 | 3 | 14 |
| CT | 7 | 31 | 9 |
| TT | 19 | 5 | 0 |
| Total | 28 | 39 | 23 |
| GG | 22 | 11 | 6 |
| GA | 4 | 28 | 11 |
| AA | 2 | 0 | 6 |
| Total | 28 | 39 | 23 |
| AA | 28 | 39 | 22 |
| AG | 0 | 0 | 1 |
| GG | 0 | 0 | 0 |
| Total | 28 | 39 | 23 |
| GG | 28 | 38 | 20 |
| GT | 0 | 0 | 2 |
| TT | 0 | 1 | 1 |
| Total | 28 | 39 | 23 |
| CC | 26 | 33 | 17 |
| CT | 2 | 6 | 5 |
| TT | 0 | 0 | 1 |
| Total | 28 | 39 | 23 |
References
- 1.Johnson JA, Cavallari LH. Warfarin pharmacogenetics. Trends Cardiovasc Med. 2015;25:33–41. doi: 10.1016/j.tcm.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawai VK, Cunningham A, Vear SI, et al. Genotype and risk of major bleeding during warfarin treatment. Pharmacogenomics. 2014;15:1973–1983. doi: 10.2217/pgs.14.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes GD, Lucas E, Alexander GC, Goldberger ZD. National trends in ambulatory oral anticoagulant use. Am J Med. 2015;128:1300–1305.e1302. doi: 10.1016/j.amjmed.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90:625–629. doi: 10.1038/clpt.2011.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirley K, Qato DM, Kornfield R, Stafford RS, Alexander GC. National trends in oral anticoagulant use in the United States, 2007 to 2011. Circ Cardiovasc Qual Outcomes. 2012;5:615–621. doi: 10.1161/CIRCOUTCOMES.112.967299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bista D, Chalmers L, Bereznicki L, Peterson G. Potential use of NOACs in developing countries: pros and cons. Eur J Clin Pharmacol. 2014;70:817–828. doi: 10.1007/s00228-014-1693-y [DOI] [PubMed] [Google Scholar]
- 7.Duraes AR, Roriz PD, Bulhoes FV, et al. Dabigatran versus warfarin after bioprosthesis valve replacement for the management of atrial fibrillation postoperatively protocol. JMIR Res Protoc. 2014;3:e21. doi: 10.2196/resprot.3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee MT, Klein TE. Pharmacogenetics of warfarin. challenges and opportunities. J Hum Genet. 2013;58:334–338. doi: 10.1038/jhg.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Self TH, Wallace JL, Sakaan S, Sands CW. Effect of body weight on dose of Vitamin K antagonists. South Med J. 2015;108:637–643. doi: 10.14423/SMJ.0000000000000356 [DOI] [PubMed] [Google Scholar]
- 10.Johnson JA. Warfarin: an old drug but still interesting. Pharmacotherapy. 2008;28:1081–1083. doi: 10.1592/phco.28.9.1081 [DOI] [PubMed] [Google Scholar]
- 11.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254 [DOI] [PubMed] [Google Scholar]
- 12.Rost S, Fregin A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214 [DOI] [PubMed] [Google Scholar]
- 13.Johnson JA, Caudle KE, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing. 2017 update. Clin Pharmacol Ther. 2017;102:397–404. doi: 10.1002/cpt.668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin T, Miyata T. Warfarin dose and the pharmacogenomics of CYP2C9 and VKORC1 –rationale and perspectives. Thromb Res. 2007;120:1–10. doi: 10.1016/j.thromres.2006.10.021 [DOI] [PubMed] [Google Scholar]
- 15.Watzka M, Geisen C, Bevans CG, et al. Thirteen novel VKORC1 mutations associated with oral anticoagulant resistance: insights into improved patient diagnosis and treatment. J Thromb Haemost. 2011;9:109–118. doi: 10.1111/j.1538-7836.2010.04095.x [DOI] [PubMed] [Google Scholar]
- 16.Lee MT, Chen CH, Chuang HP, et al. VKORC1 haplotypes in five East-Asian populations and Indians. Pharmacogenomics. 2009;10:1609–1616. doi: 10.2217/pgs.09.80 [DOI] [PubMed] [Google Scholar]
- 17.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503 [DOI] [PubMed] [Google Scholar]
- 18.Geisen C, Watzka M, Sittinger K, et al. VKORC1 haplotypes and their impact on the inter-individual and inter-ethnical variability of oral anticoagulation. Thromb Haemost. 2005;94:773–779. doi: 10.1160/TH05-04-0290 [DOI] [PubMed] [Google Scholar]
- 19.Ensembl genome browser. Available at: www.Ensembl.org. Accessed September 10, 2018.
- 20.PharmGKB. Pharmacogenomics Database. Available at: www.Pharmgkb.org. Accessed September 10, 2018.
- 21.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268 [DOI] [PubMed] [Google Scholar]
- 22.dynacom. SNPAlyze Ver.9.0. Available from: https://www.dynacom.co.jp/english/snpalyze_e/e_sa_demo.html. Accessed August 8, 2018.
- 23.Garcia-Bertrand R, Simms TM, Cadenas AM, Herrera RJ. United Arab Emirates: phylogenetic relationships and ancestral populations. Gene. 2014;533:411–419. doi: 10.1016/j.gene.2013.09.092 [DOI] [PubMed] [Google Scholar]
- 24.Al-Gazali LI. Attitudes toward genetic counseling in the United Arab Emirates. Community Genet. 2005;8:48–51. doi: 10.1159/000083339 [DOI] [PubMed] [Google Scholar]
- 25.John SE, Antony D, Eaaswarkanth M, et al. Genetic variants associated with warfarin dosage in Kuwaiti population. Phagmacogenomics. 2017;8:757–764. doi: 10.2217/pgs-2017-0020 [DOI] [PubMed] [Google Scholar]
- 26.Limdi NA, Wadelius M, Cavallari L, et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115:3827–3834. doi: 10.1182/blood-2009-12-255992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alzahrani AM, Ragia G, Hania H, Manolopoulos VG. Genotyping of CYP2C9 and VKORC1 in the Arabic Population of Al-Ahsa, Saudi Arabia. Biomed Res Int. 2013;2013:315890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owen RP, Gong L, Sagreiya H, Klein TE, Altman RB. VKORC1 pharmacogenomics summary. Pharmacogenet Genomics. 2010;20:642–644. doi: 10.1097/FPC.0b013e32833433b6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313 [DOI] [PubMed] [Google Scholar]
- 30.Jia L, Wang Z, Men J, Cai H, Wei M. Polymorphisms of VKORC1 and CYP2C9 are associated with warfarin sensitivity in Chinese population. Ther Clin Risk Manag. 2017;13:421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavallari LH, Langaee TY, Momary KM, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:459–464. doi: 10.1038/clpt.2009.223 [DOI] [PubMed] [Google Scholar]
- 32.Veenstra DL, You JH, Rieder MJ, et al. Association of Vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics. 2015;15:687–691. doi: 10.1097/01.fpc.0000174789.77614.68 [DOI] [PubMed] [Google Scholar]
- 33.Namazi S, Azarpira N, Hendijani F, Khorshid MB, Vessal G, Mehdipour AR. The impact of genetic polymorphisms and patient characteristics on warfarin dose requirements. A cross-sectional study in Iran. Clin Ther. 2010;32:1050–1060. doi: 10.1016/j.clinthera.2010.06.010 [DOI] [PubMed] [Google Scholar]
- 34.Pathare A, Al Khabori M, Alkindi S, et al. Warfarin pharmacogenetics. development of a dosing algorithm for Omani patients. J Hum Genet. 2012;57:665–669. doi: 10.1038/jhg.2012.94 [DOI] [PubMed] [Google Scholar]
- 35.D’Andrea G, D’Ambrosio RL, Di Perna P, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–649. doi: 10.1182/blood-2004-06-2111 [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Zhu J, Ding J. VKORC1 −1639G/A and 1173 C/T genetic polymorphisms influence individual differences in warfarin maintenance dose. Genet Test Mol Biomarkers. 2015;19:488–493. doi: 10.1089/gtmb.2015.0097 [DOI] [PubMed] [Google Scholar]
- 37.Perera MA, Gamazon E, Cavallari LH, et al. The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther. 2011;89:408–415. doi: 10.1038/clpt.2010.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shrif NE, Won HH, Lee ST, et al. Evaluation of the effects of VKORC1 polymorphisms and haplotypes, CYP2C9 genotypes, and clinical factors on warfarin response in Sudanese patients. Eur J Clin Pharmacol. 2011;67:1119–1130. doi: 10.1007/s00228-011-1060-1 [DOI] [PubMed] [Google Scholar]
- 39.Scott SA, Edelmann L, Kornreich R, Desnick RJ. Warfarin pharmacogenetics. CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazi and Sephardi Jewish populations. Am J Hum Genet. 2008;82:495–500. doi: 10.1016/j.ajhg.2007.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aklillu E, Leong C, Loebstein R, Halkin H, Gak E. VKORC1 Asp36Tyr warfarin resistance marker is common in Ethiopian individuals. Blood. 2008;111:3903–3904. doi: 10.1182/blood-2008-01-135863 [DOI] [PubMed] [Google Scholar]
- 41.Kurnik D, Qasim H, Sominsky S, et al. Effect of the VKORC1 D36Y variant on warfarin dose requirement and pharmacogenetic dose prediction. Thromb Haemost. 2012;108:781–788. doi: 10.1160/TH12-03-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slatkin M. Linkage disequilibrium – understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9:477–485. doi: 10.1038/nrg2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi F, Yanai K, Morii T, et al. Linkage disequilibrium grouping of Single Nucleotide Polymorphisms (SNPs) reflecting haplotype phylogeny for efficient selection of tag SNPs. Genetics. 2005;70:291–304. doi: 10.1534/genetics.104.038232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neidecker M, Patel AA, Nelson WW, Reardon G. Use of warfarin in long-term care. A systematic review. BMC Geriatr. 2012;12:14. doi: 10.1186/1471-2318-12-14 [DOI] [PMC free article] [PubMed] [Google Scholar]