Abstract
Background: Bladder cancer is a common malignancy that affects the human urinary tract. Muscle-invasive bladder cancer (MIBC) is aggressive and has poor prognosis. Previous studies have reported that the tumor-infiltrating lymphocytes (TILs) were associated with MIBC outcome; however, inconsistency remains and mRNA level TIL markers’ prognostic significance in MIBC is unclear.
Materials and methods: In the present study, we reanalyzed data from four public datasets (the Cancer Genome Atlas for investigation; and CIT, GSE5287, and GSE31684 for validation) to examine the prognostic significance of CD3D, CD4, CD8A, CD3D/CD4 and CD3D/CD8A in MIBC.
Results: We found that the CD3D/CD4 ratio was a stable independent prognostic factor in MIBC (beta = −0.87, P = 0.025); high CD3D/CD4 ratio predicted better survival in MIBC, and the power of this association was much stronger in basal-squamous tumors (beta = −4.73, P = 2.67E-06). We also noted that the CD4 expression was significantly higher than CD3D (P < 0.05), indicating the presence of CD3−CD4+ cells which could be immune-suppressing.
Conclusion: The CD3D/CD4 ratio can be viewed as a prognostic marker and a rough measurement for the interaction between immune-effecting CD3+ TILs and immune-suppressing CD3−CD4+ cells in MIBC, and this interaction may play a particularly important role in anti-cancer immunity in basal-squamous tumors as it has a very strong association with survival in this subtype, and may be used to select potential responders to immunotherapy.
Keywords: bladder cancer, muscle-invasive, tumor-infiltrating lymphocytes, CD3, CD4, CD8, prognosis, immunotherapy, basal-squamous subtype
Introduction
Bladder cancer (BC) is the most common malignancy affecting the human urinary tract, with approximately 430,000 new cases diagnosed in 2012 and 165,000 annual deaths.1 Muscle-invasive bladder carcinoma (MIBC, defined as BC with pathologic T≥2) is a major clinical issue and displays unfavorable prognosis, with 5-year survival rate being 50%. Radical cystectomy in combination with neo-adjuvant and/or adjuvant chemoradiotherapy is currently recommended treatment option,2,3 and synchronous radiotherapy with chemotherapy4 or bladder-sparing trimodal strategies5 are also alternatives to cystectomy in properly selected patients for MIBC. However, no major improvement in MIBC patients’ survival has been achieved over the last 20 years.6 Quite recently, the emerging immune checkpoint inhibitors have yielded promising efficacy and improvement on overall survival (OS) in subjects with advanced and metastatic BC,7,8 but there is yet no established protocol to select these responders.
Theoretically, durable response induced by anti-cancer immunotherapy depends on the following elements: 1) tumor immunogenicity, mainly associated with tumor mutation burden (TMB) and neo-antigen load; 2) tumor microenvironment, in particular the abundance and composition of tumor-infiltrating lymphocytes (TILs); and 3) the effects of tumor-associated immune suppression.9,10 BC is known as the third top cancer in terms of TMB, just following melanoma and non-small-cell lung cancer (NSCLC);11,12 therefore, it is considered a possibly ideal target disease of immunotherapy.8,13,14 Meanwhile, identification of TIL markers able to predict prognosis will largely improve risk-evaluation and therapeutic decision-making for BC, especially MIBC patients. Previously, several articles reported that TILs and their subpopulation structure were associated with prognosis in BC patients,15–17 whereas others found negative prognostic value of TILs in a global pan-cancer analysis.18 Debates and uncertainty remain for this issue. Moreover, traditional investigations regarding the prognostic value of TILs were based on either immunohistochemistry or flow cytometry counting of infiltrated immune cells, but both are time-consuming and require considerable resources. Gene expression biomarkers with adequate predictability can be an alternative.
Recently, the increasing accessibility of cancer-omics datasets have largely boosted the discovery of cancer biomarkers. Refined data mining of bioinformatics datasets, especially those stored in open data repositories like the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), are becoming the very first step of the translational pipeline of identifying clinically useful biomarkers.
We noted that the association between mRNA expression of key TIL markers (CD3, CD4, and CD8) and clinical outcome is not clear in BC. In the present study, we aimed to investigate the prognostic significance of CD3D (total T cells), CD4 (helper T cells) and CD8A (cytotoxic T cells) gene expression level and the ratios of CD3D/CD4 and CD3D/CD8A expression in MIBC patients using public datasets (one investigation cohort from TCGA,19 and three validation cohorts from GEO).
Materials and methods
Datasets
The TCGA dataset (investigation dataset), including the transcriptome data by RNA-sequencing and clinical data of a cohort of 407 MIBC patients, was downloaded from the Genomic Data Commons data portal (https://gdc.cancer.gov). The information of these patients’ mRNA clustering subtype was obtained from the supplementary materials of Robertson et al19. The validation datasets (CIT, GSE5287, and GSE31684) were downloaded from the GEO data portal (https://www.ncbi.nlm.nih.gov/geo) and supplementary materials of corresponding publications. The characteristics of the included datasets were summarized in Table S1.
Table S1.
Basic characteristics of included datasets
| Investigation | Validation | |||||||
|---|---|---|---|---|---|---|---|---|
| TCGA (n=407) | CIT (n=73) | GSE5287 (n=30) | GSE31684 (n=78) | |||||
| n | Percentage | n | Percentage | n | Percentage | n | Percentage | |
| Age | ||||||||
| ≤65 | 160 | 39.3% | 32 | 43.8% | NA | NA | 25 | 32.1% |
| >65 | 246 | 60.4% | 41 | 56.2% | NA | NA | 53 | 67.9% |
| Unknown | 1 | 0.2% | 0 | 0.0% | NA | NA | 0 | 0.0% |
| Gender | ||||||||
| Male | 300 | 73.7% | 62 | 84.9% | NA | NA | 57 | 73.1% |
| Female | 107 | 26.3% | 11 | 15.1% | NA | NA | 21 | 26.9% |
| Tumor stage | ||||||||
| II | 132 | 32.4% | 25 | 34.2% | 0 | 0 | 17 | 21.8% |
| III/IV | 273 | 67.1% | 48 | 65.8% | 30 | 100.0% | 61 | 78.2% |
| Unknown | 2 | 0.5% | 0 | 0.0% | 0 | 0 | 0 | 0.0% |
| Molecular subtype | ||||||||
| Luminal Papillary | 136 | 33.4% | NA | NA | NA | NA | NA | NA |
| Luminal Infiltrated | 76 | 18.7% | NA | NA | NA | NA | NA | NA |
| Luminal | 43 | 10.6% | NA | NA | NA | NA | NA | NA |
| Basal-Squamous | 133 | 32.7% | NA | NA | NA | NA | NA | NA |
| Neuronal | 19 | 4.7% | NA | NA | NA | NA | NA | NA |
| Adjuvant chemotherapy | ||||||||
| Yes | 176 | 43.2% | NA | NA | 30 | 100.0% | 34 | 43.6% |
| No | 182 | 44.7% | NA | NA | 0 | 0.0% | 44 | 56.4% |
| Unknown | 149 | 36.6% | NA | NA | 0 | 0.0% | 0 | 0.0% |
| Adjuvant radiotherapy | ||||||||
| Yes | 10 | 2.5% | NA | NA | NA | NA | NA | NA |
| No | 263 | 64.6% | NA | NA | NA | NA | NA | NA |
| Unknown | 134 | 32.9% | NA | NA | NA | NA | NA | NA |
Abbreviation: NA, not available.
Statistical analysis
For the TCGA dataset, the RNA-Seq by Expectation-Maximization expression values of CD3D, CD4, and CD8A were subjected to log2 transformation. The association between CD3D, CD4, and CD8A expression, ratios of CD3D/CD4 and CD3D/CD8A, and clinical characteristics including age, gender, and tumor pathologic stage was investigated in the TCGA cohort using Pearson correlation analysis and analysis of variance (ANOVA), or their non-parametric counterpart (Spearman correlation and Kruskal–Wallis test). The association between CD3D, CD4, and CD8A expression, ratios of CD3D/CD4 and CD3D/CD8A, and OS was analyzed with univariate and multivariate (adjusted for age, gender, and pathologic stage) Cox proportional hazard model. For independent prognostic markers identified by multivariate analysis, Kaplan–Meier survival curves were drawn for the low vs high (cutoff by median and optimal threshold suggested by Cox regression tree analysis) group comparisons; and median survivals and P-values by log-rank test were also calculated. Subgroup survival analyses stratified by molecular subtypes based on transcriptome were conducted. The identified prognostic factors were further subjected to external validation and pooled analysis using the validation datasets (original gene expression values by microarray were log2 transformed before analysis). A P<0.05 was considered statistically significant. The R version 3.4.3 was used for all statistical analyses.
Results
Distribution of investigated markers in TCGA cohort
The distribution of CD3D and CD8A mRNA expression was similar with each other, and significantly lower than CD4 expression across the 407 TCGA MIBC patients (mean difference (MD) =−2.21, 95% CI =[−2.30, −2.12], between CD3D and CD4; MD =−1.82, 95% CI =[−1.91, −1.72], between CD8A and CD4; Figure 1A). The CD3D/CD8A ratio was significantly larger than CD3D/CD4 ratio (MD =0.31, 95% CI =[0.28, 0.33]; Figure 1B).
Figure 1.
Distribution of TIL markers and their ratios. (A) Distribution of CD3D, CD4 and CD8A; (B) distribution of CD3D/CD4 and CD3D/CD8A ratios.
Abbreviation: TIL, tumor-infiltrating lymphocyte.
Correlation between investigated markers and clinical parameters in TCGA cohort
According to Pearson correlation analysis, the age was not correlated with any of the investigated markers (CD3D: r=−0.04, P=0.43; CD4: r=−0.005, P=0.9171; CD8A: r=0.005, P=0.92; CD3D/CD4: r=−0.07, P=0.17; CD3D/CD8A: r=−0.07, P=0.15). A significant gender difference was detected only for CD4 (ANOVA, P=0.037) and CD3D/CD8A (Kruskal–Wallis test, P=0.0001). No significant association was found between pathologic stage (II, III, and IV) and any markers (CD3D, ANOVA, P=0.40; CD4, ANOVA, P=0.67; CD8A, ANOVA, P=0.054; CD3D/CD4, Kruskal–Wallis test, P=0.18; CD3D/CD8, Kruskal–Wallis test, P=0.070).
Prognostic significance of investigated markers in TCGA cohort
According to the findings of univariate Cox regression analysis, the CD3D and CD3D/CD4 ratio were significantly associated with OS (Table 1). Multivariate Cox regression adjusted for age, gender and pathologic stage (II vs III–IV) indicated that both the CD3D and CD3D/CD4 ratio were independent prognostic factors (CD3D, beta =−0.11, P=0.032; CD3D/CD4, beta =−0.87, P=0.025; Table 1). The CD3D/CD4 ratio had a better prognosis predictability than CD3 expression as measured by the concordance index in both univariate (0.586 vs 0.57) and multivariate models (0.657 vs 0.655).
Table 1.
Prognostic significance of TIL markers in MIBC (TCGA, n=406)
| Markersa | Univariate | Multivariateb | ||
|---|---|---|---|---|
| Beta | P | Beta | P | |
| CD3D | −0.11 | 0.025* | −0.11 | 0.032* |
| CD4 | −0.04 | 0.57 | −0.05 | 0.48 |
| CD8A | −0.09 | 0.065 | −0.09 | 0.06 |
| CD3D/CD4 | −0.99 | 0.013* | −0.87 | 0.025* |
| CD3D/CD8A | −0.19 | 0.46 | −0.13 | 0.59 |
Notes: aGene expression measured as log2 RSEM, or derived ratio, as continuous variable. bMultivariate Cox regression adjusted by age, gender and tumor stage. *Statistically significant.
Abbreviations: MIBC, muscle-invasive bladder cancer; TIL, tumor-infiltrating lymphocytes; TCGA, the Cancer Genome Atlas.
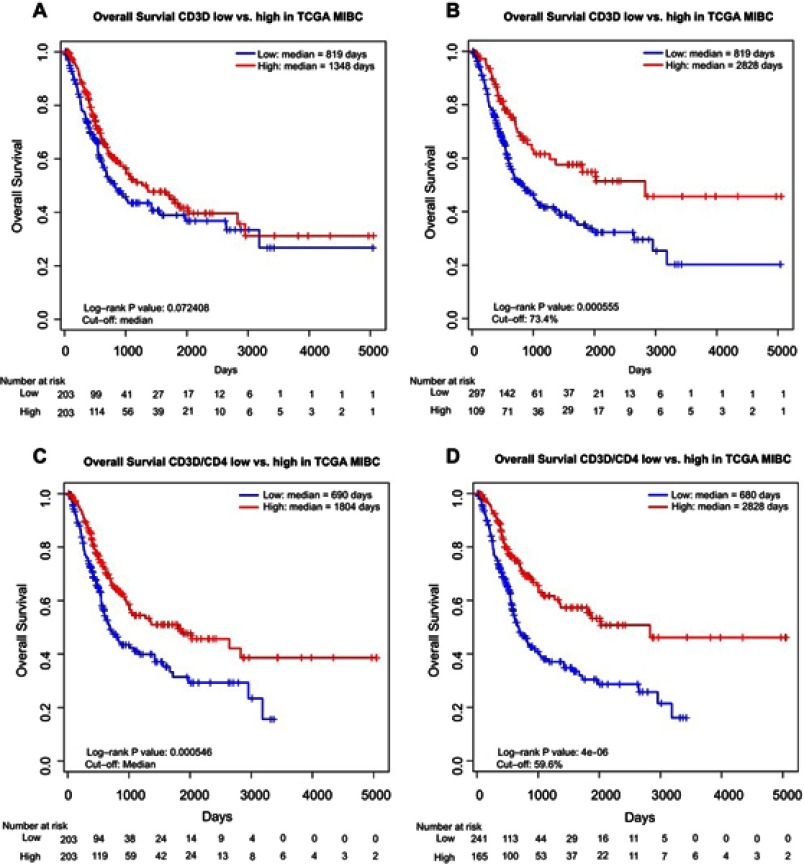
We then divided the subjects into low vs high groups according to the expression of CD3D and CD3D/CD4 ratio, using median and the optimal cutoffs identified by Cox regression tree analysis. The CD3D high group showed non-significantly better OS than the CD3D low group when using median cutoff (median OS: 1,348 days vs 819 days, log-rank test P=0.072; Figure 2A), but the difference became significant taking the optimal cutoff by 73.4% (median OS: 2,828 days vs 819 days, log-rank test P=0.00056; Figure 2B). In contrast, the CD3D/CD4 high group showed significantly better OS than the CD3D/CD4 low group with both the median (median OS: 1,804 days vs 690 days, log-rank test P=0.00055; Figure 2C) and the optimal cutoff by 59.6% (median OS: 2,828 days vs 680 days, log-rank test P=4E-06; Figure 2D).
Figure 2.
Kaplan–Meier curves showing difference in OS in MIBC patients between (A) low and high CD3D groups by median cutoff; (B) low and high CD3D groups by optimal cutoff; (C) low and high CD3D/CD4 groups by median cutoff; (D) low and high CD3D/CD4 groups by optimal cutoff.
Abbreviation: MIBC, muscle-invasive bladder cancer.
Subgroup analyses by molecular subtypes based on transcriptome for CD3D and CD3D/CD4 in TCGA cohort
It had been shown that some investigated TILs markers (precisely CD8A, CXCL9, and CXCL10) were unequally distributed among previously defined different TCGA molecular subtypes based on transcriptome.10,20 Therefore, the subgroup-specific prognostic significance of CD3D and CD3D/CD4 was further investigated by univariate and multivariate Cox regression analysis within these subtypes. As shown in Table 2, the CD3 expression and CD3D/CD4 ratio showed no significance in luminal-papillary, luminal and neuronal subtypes, and displayed limited prognostic predictability in luminal-infiltrated subtype. In contrast, the two markers demonstrated very strong independent prognostic predictability in patients with basal-squamous tumor (CD3D, beta =−0.37, adjusted P=0.00065; CD3D/CD4, beta =−4.8, adjusted P=2.7E-06). Besides, in basal-squamous subtype, the CD3D/CD4 ratio exhibited a better prognosis predictability than CD3 expression alone as measured by the concordance index in both univariate (0.67 vs 0.631) and multivariate models (0.686 vs 0.668).
Table 2.
Subgroup analyses by molecular subtype based on transcriptome for CD3D and CD3D/CD4 in MIBC (TCGA, n=406)
| Molecular subtype | CD3Da | CD3D/CD4a | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariateb | Univariate | Multivariateb | |||||
| Beta | P | Beta | P | Beta | P | Beta | P | |
| Luminal papillary (n=135) | −0.19 | 0.12 | −0.09 | 0.45 | −0.51 | 0.5 | 0.08 | 0.92 |
| Luminal infiltrated (n=76) | −0.33 | 0.035* | −0.24 | 0.14 | −2.25 | 0.09 | −1.54 | 0.29 |
| Luminal (n=43) | −0.21 | 0.37 | −0.31 | 0.25 | −1.64 | 0.27 | −2.75 | 0.15 |
| Basal squamous (n=133) | −0.37 | 0.00059* | −0.38 | 0.00065* | −4.8 | 6.09E-07* | −4.73 | 2.67E-06* |
| Neuronal (n=19) | −0.06 | 0.8 | −0.11 | 0.72 | 0.088 | 0.95 | −0.59 | 0.74 |
Notes: aGene expression measured as log2 RSEM, or derived ratio, as continuous variable. bMultivariate Cox regression adjusted by age, gender and tumor stage. *Statistically significant.
Abbrevaiton: TCGA, the Cancer Genome Atlas.
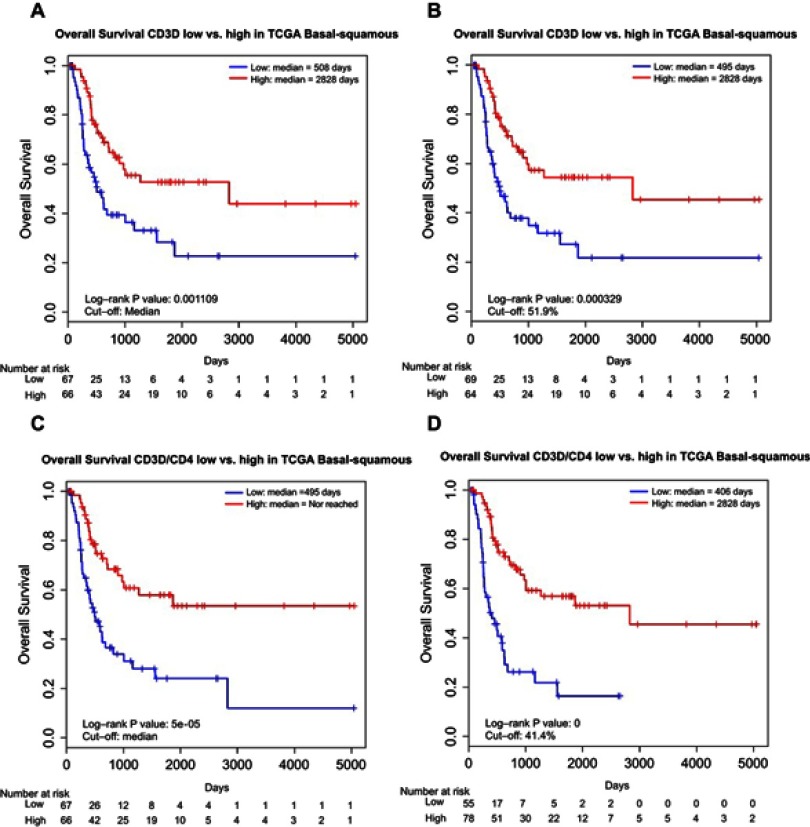
We also performed low vs high stratification of CD3D and CD3D/CD4 expression in patients with basal-squamous tumors using the same aforementioned cutoffs. We found a significantly better OS in the CD3D high group with both the median (median OS: 2,828 days vs 508 days, log-rank test P=0.0011; Figure 3A) and the optimal cutoff by 51.9% (median OS: 2,828 days vs 495 days, log-rank test P=0.00033; Figure 3B). Moreover, distinct and even stronger differences were revealed for the CD3D/CD4 high group with both the median (median OS: Not reached vs 495 days, log-rank test P=5E-05; Figure 3C) and optimal cutoff by 41.4% (median OS: 2,828 days vs 406 days, log-rank test P<1E-06; Figure 3D).
Figure 3.
Kaplan–Meier curves showing difference in OS in basal-squamous MIBC patients between (A) low and high CD3D groups by median cutoff; (B) low and high CD3D groups by optimal cutoff; (C) low and high CD3D/CD4 groups by median cutoff; (D) low and high CD3D/CD4 groups by optimal cutoff.
Abbreviation: MIBC, muscle-invasive bladder cancer.
External validation for CD3D and CD3D/CD4’s prognostic predictability
We validated the prognostic significance of CD3D and CD3D/CD4 in three public datasets. Based on the results (Table 3), high CD3D expression was associated with improved survival in all three datasets (CIT, GSE5287, and GSE31684), but the association was significant in only one dataset (GSE31684, P=0.012). High CD3D/CD4 ratio was associated with significantly improved survival in two of three cohorts (GSE31684, P=0.02; GSE5287, P=0.035) and marginally significant survival benefits in the other cohort (CIT, P=0.07). Furthermore, we investigated whether the prognostic predictability of CD3D and CD3D/CD4 was stronger among subjects possibly classified as basal-squamous tumors in these validation cohorts. We considered subjects whose average expression level of the 10 key basal-squamous markers reported in Robertson et al19 was within the top third as having basal-squamous tumors. However, no significant association with OS was observed (Table 3).
Table 3.
Results of validation analyses in three independent cohorts
| Dataset | Overall subjects | Subjects with high basal-squamous markers | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | CD3Da | CD3D/CD4a | n | CD3Da | CD3D/CD4a | |||||
| Beta | P | Beta | P | Beta | P | Beta | P | |||
| CIT | 73 | −0.024 | 0.9 | −3.77 | 0.07 | 22 | 0.18 | 0.61 | −4.79 | 0.37 |
| GSE31684 | 78 | −0.21 | 0.012* | −1.14 | 0.02* | 26 | −0.32 | 0.052 | −0.26 | 0.86 |
| GSE5287 | 30 | −0.26 | 0.28 | −6.04 | 0.035* | 10 | −0.4 | 0.36 | −12.9 | 0.07 |
Notes: aGene expression measured as log2 RSEM, or derived ratio, as continuous variable. *Statistically significant.
Pooled analysis for CD3D/CD4 ratio across TCGA and validation cohorts
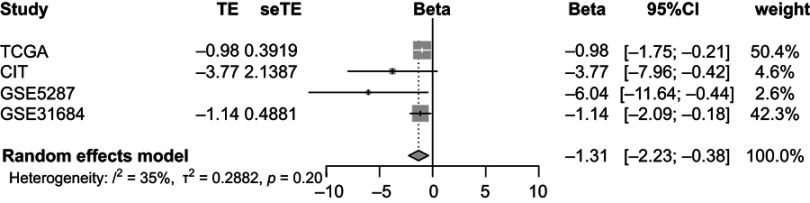
Random-effects meta-analysis was performed to combine the individual analysis results across TCGA and validation cohorts for CD3D/CD4 ratio. As shown in Figure 4, the pooled analysis indicated that considering all observations of included datasets, there was a significant association between CD3D/CD4 ratio and OS (beta =−1.31, 95% CI =[−2.23, −0.38]).
Figure 4.
Forest plots of random-effects meta-analysis for CD3D/CD4 across cohorts.
Abbrevaiton: TCGA, the Cancer Genome Atlas.
Discussion
BC is a common and aggressive cancer that represents significant disease burden. Treatment options regarding MIBC are still limited. Although emerging active immunotherapy (anti-PD1/PD-L1) has shown promising efficacy in BC, only a subset of patients respond well. Besides, it is still largely unknown the way to predict/identify these responders and the reason why some MIBC molecular subtypes have poor TILs. On the other hand, TIL therapy, a passive immunotherapy using infusion of ex vivo expanded self-TILs to trigger anti-tumor immunity, has achieved great success over the past 10–15 years9,21 and actively studied in clinical trials involving several cancer types including melanoma, NSCLC and cervical cancer (NCT02360579; NCT03419559 and NCT03108495). Regarding TMB, which provide a preferential niche for immunotherapy, BC ranked third just following melanoma and NSCLC. In view of this, TIL biomarkers with prognostic significance may also be used to help selecting patients who may respond to immunotherapy and improve the clinical management of MIBC.
The cancer patients’ outcome is determined by the interaction between tumor and surrounding microenvironment. Previous studies have revealed that the microenvironment plays an important role in BC carcinogenesis and progression. Of note, it has been reported that TILs and their subpopulation structure are associated with prognosis in BC patients. Like MIBC tumors which are heterogeneous in terms of molecular profiles, the TILs are also a heterogeneous group of immune cells. The majority of TILs belong to the T cell population, which are mostly CD3+. Cytotoxic T cells (CTLs), with CD8+ as a specific marker, are the main mediator of the host’s anti-cancer immunity. The activity of CD8+ CTLs depends on a complicated interaction network involving other T cell subtypes including T-helper cells (CD4+) and regulatory T cells (Treg), and other immune cell populations such as tumor-associated macrophages (TAMs). One of the first investigations on the role of TILs in MIBC by Sharma and colleagues revealed that higher infiltration of CD8+ CTLs was associated with better survival.17 Sjodahl et al found that presence of CD3+ T cell infiltration predicted good prognosis in MIBC patients, which involved modulation by CD68+ TAMs.16 Horn et al reported ratio between FOXP3 and CD3D or CD8 positive cells were significantly associated with BC survival.15 However, others found lack of evidence for a prognostic value of TILs in BC through a global pan-cancer analysis with TCGA data.18 We supposed that this inconsistency could be a result of ignoring the heterogeneous immune-infiltration status across different MIBC molecular subtypes. According to Mariathasan and colleagues, the immune-infiltration markers were enriched in luminal-infiltrated and basal-squamous subtypes compared with other subtypes.10 This can be partially explained by the PPARγ/RXRα pathway, which is mainly activated in luminal-papillary and luminal tumors, and confers immune evasion through impairing CD8+ T-cell infiltration.22,23 Accordingly, it has been shown that tumors belong to the luminal-infiltrated or basal-squamous subtypes respond better to immunotherapy than other subtypes.7,10,20
Despite the aforementioned research efforts concerning the role of TILs in MIBC, the prognostic predictability of mRNA expression of major TIL markers (CD3D, for overall T cells; CD4, for T-helpers; and CD8A, for CTLs) and their ratios have not been reported. The immune infiltration status was usually measured by immunohistochemistry or flow cytometry counting of infiltrated immune cells. Without doubt, both of them were precise but time-consuming and technically complicated. Biomarkers based on gene expression are possible alternatives. Moreover, mRNA detection is more feasible to take advantage of liquid-based urine cytology, which represents the “gold standard” for surveillance of urothelial carcinoma diagnosis and prognosis.24
In the TCGA investigation cohort, we observed that the CD4 expression was significantly higher than CD3D expression. The CD3D/CD4 ratio was less than 1 in most MIBC subjects, and CD3D/CD8A less than 1 in more than half of the subjects. It seems that in many of the subjects the TILs had a large number of and/or were actively proliferating CD3− CD4+ (macrophages, monocytes, and dendric cells) and CD3−CD8+cells (NK cells). We thus suppose that the CD3D/CD4 ratio can be considered a rough measurement of interaction between T cells and CD3− CD4+ cells which could be immune-suppressing cells, which is in accordance with Sjodhal et al’s findings. In line with previous reports,15,17 we also found CD8A was an independent prognostic factor but only in basal-squamous subtypes (beta =−0.35, P=0.001). Interestingly, the overall T cell marker CD3D and the ratio of CD3D/CD4 were independent favorable prognostic factors in the TCGA cohort; furthermore, a stable association was observed in several validation cohorts for CD3D/CD4, strengthening the reliability of this predictor. Strikingly, both CD3D and the ratio of CD3D/CD4 exhibited largely stronger predictability in TCGA patients with basal-squamous tumor; although the validation in other cohorts failed to identify a stable significant association which could be due to lack of consensus classification among different cohorts and very limited sample size. This suggests that the interaction between immune-effecting CD3+ TILs and immune-suppressing CD3− CD4+ cells may play a particularly important role in the anti-tumor immunity in basal-squamous tumors. Following this clue, we speculate that basal-squamous MIBC patients with higher expression ratio of CD3D/CD4 (stronger effecting TILs and weaker suppressing cells) could be potential responders to cancer immunotherapy, which should be further investigated.
In conclusion, we have found that high CD3D/CD4 ratio predicts better survival in MIBC, and its association with OS may be stronger in basal-squamous tumors. A hypothesis is made that CD3D/CD4 may also predict response to immunotherapy in basal-squamous tumors, which needs further investigation.
Acknowledgment
This work has been supported by the Fundamental Research Funds for the Central Universities (2662018PY023).
Disclosure
The authors report no conflicts of interest in this work.
Supplementary material
References
- 1.Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96–108. doi: 10.1016/j.eururo.2016.06.010 [DOI] [PubMed] [Google Scholar]
- 2.Alfred Witjes J, Lebret T, Compérat EM, et al. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol. 2017;71(3):462–475. doi: 10.1016/j.eururo.2016.06.020 [DOI] [PubMed] [Google Scholar]
- 3.Sanli O, Dobruch J, Knowles MA, et al. Bladder cancer. Nat Rev Dis Prim. 2017;3(17022):121–127. doi: 10.1038/nrdp.2017.22 [DOI] [PubMed] [Google Scholar]
- 4.James ND, Hussain SA, Hall E, et al. Radiotherapy with or without chemotherapy in muscle-invasive bladder cancer. N Engl J Med. 2012;366(16):1477–1488. doi: 10.1056/NEJMoa1106106 [DOI] [PubMed] [Google Scholar]
- 5.Ploussard G, Daneshmand S, Efstathiou JA, et al. Critical analysis of bladder sparing with trimodal therapy in muscle-invasive bladder cancer: a systematic review. Eur Urol. 2014;66(1):120–137. doi: 10.1016/j.eururo.2014.02.038 [DOI] [PubMed] [Google Scholar]
- 6.Mahe M, Dufour F, Neyret-Kahn H, et al. An FGFR3/MYC positive feedback loop provides new opportunities for targeted therapies in bladder cancers. EMBO Mol Med. 2018:1–18. doi: 10.15252/emmm.201708163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellmunt J, de Wit R, Vaughn DJ, et al. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med. 2017;376(11):1015–1026. doi: 10.1056/NEJMoa1613683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balar AV, Galsky MD, Rosenberg JE, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389(10064):67–76. doi: 10.1016/S0140-6736(16)32455-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borst J, Ahrends T, Bąbała N, Melief CJM, Kastenmüller W. CD4+ T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18. doi: 10.1038/s41577-018-0044-0 [DOI] [PubMed] [Google Scholar]
- 10.Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell PJ, Getz G, Stuart JM, Korbel JO, Stein LD. Net – ICGC/TCGA pan-cancer analysis of whole genomes. Pan-cancer analysis of whole genomes. bioRxiv. 2017;3(10):162784. doi: 10.1101/162784 [DOI] [Google Scholar]
- 12.Henn BM, Botigué LR, Bustamante CD, Clark AG, Gravel S. Estimating the mutation load in human genomes. Nat Rev Genet. 2015;16(6):333–343. doi: 10.1038/nrg3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2016;348(6230):124. doi: 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horn T, Laus J, Seitz AK, et al. The prognostic effect of tumour-infiltrating lymphocytic subpopulations in bladder cancer. World J Urol. 2016;34(2):181–187. doi: 10.1007/s00345-015-1615-3 [DOI] [PubMed] [Google Scholar]
- 16.Sjödahl G, Lövgren K, Lauss M, et al. Infiltration of CD3+ and CD68+ cells in bladder cancer is subtype specific and affects the outcome of patients with muscle-invasive tumors. Urol Oncol Semin Orig Investig. 2014;32(6):791–797. doi: 10.1016/j.urolonc.2014.02.007 [DOI] [PubMed] [Google Scholar]
- 17.Sharma P, Shen Y, Wen S, et al. CD8 tumor-infiltrating lymphocytes are predictive of survival in muscle-invasive urothelial carcinoma. Proc Natl Acad Sci. 2007;104(10):3967–3972. doi: 10.1073/pnas.0611618104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danaher P, Warren S, Dennis L, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5(1):1–15. doi: 10.1186/s40425-017-0215-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(3):540.e25–556.e25. doi: 10.1016/j.cell.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–1920. doi: 10.1016/S0140-6736(16)00561-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S, Margolin K. Tumor-infiltrating lymphocytes in melanoma. Curr Oncol Rep. 2012;14(5):468–474. doi: 10.1007/s11912-012-0257-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korpal M, Puyang X, Wu ZJ, et al. Evasion of immunosurveillance by genomic alterations of PPARγ/RXRα in bladder cancer. Nat Commun. 2017;8(1):1–14. doi: 10.1038/s41467-017-00147-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rochel N, Krucker C, Coutos-Thévenot L, et al. Recurrent activating mutations of PPARγ associated with luminal bladder tumors. Nat Commun. 2019;10(1):253. doi: 10.1038/s41467-018-08157-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H, Kim K, Woo J, et al. Quantitative proteomic analysis identifies AHNAK (Neuroblast differentiation-associated protein AHNAK) as a novel candidate biomarker for bladder urothelial carcinoma diagnosis by liquid-based cytology. Mol Cell Proteomics. 2018;17(9):1788–1802. doi: 10.1074/mcp.RA118.000562 [DOI] [PMC free article] [PubMed] [Google Scholar]