Abstract
Background: Cancer metastasis is the leading cause of cancer-related death in breast cancer. However, our understanding of its mechanisms is still limited. At this study, the biological roles and clinical significance of NRF3 (NFE2L3, nuclear factor, Erythroid 2 Like 3) in breast cancer are evaluated for the first time.
Methods: NRF3 expression in breast cancer cell lines and clinical specimens was determined by western blot and immunohistochemistry, respectively. Cell proliferation, cell cycle distribution, cell migration, and invasion were detected by MTT, colony formation, flow cytometry, and transwell assays, respectively. All other proteins were measured by western blot. The clinical significance of NRF3 was analyzed using the data from tissue microarray.
Results: We found that NRF3 expression was obviously suppressed in breast cancer tissues, and negatively associated with the Lymph node metastasis status and tumor stages. Our data also indicated NRF3 expression was much higher in MCF-7 cells than that in MDA-MB-231 and SKBR3 cells which were more malignant. Silence of NRF3 in MCF-7 cells could significantly promote cell proliferation by reducing the cell number in the G0/G1 phase. Exogenous expression of NRF3 in SKBR3 and MDA-MB-231 cells effectively inhibited both cell growth and metastasis with epithelial–mesenchymal transition and MMPs expression suppressed. NRF3 overexpression also impaired the ID3 expression by inactivating the AKT signaling pathway. Exogenous expression of ID3 could not only effectively promote breast cancer cell invasion by inhibiting E-cadherin expression and upregulating MMP-2 expression, but also attenuated the inhibitory function of NRF3 on the breast cancer cell invasion.
Conclusion: Our findings suggested that NRF3 inhibited breast cancer cell proliferation and metastasis via inhibiting AKT/ID3 axis at least partially, and potentially to be a valuable clinic marker in breast cancer prognosis.
Keywords: human breast cancer, NRF3, metastasis, EMT, AKT/ID3, overall survival
Introduction
Breast cancer is the second most common tumors usually diagnosed in more and less developed regions, ranking as the fifth cause of cancer-related death. As so far, surgery combined with chemotherapy is still the preferable therapeutic plans for breast cancer treatment.1,2 Like the extensive research on finding more proteins for more accurate judgment about the breast cancer stage, increasing groups move to focus on finding the metastatic makers because the metastasis accounts for the overwhelming cause of death in patients with solid tumors.3 Although the detection of breast cancer metastasis at the earliest stage is extremely important for its management and prognosis, our understanding of its molecular and cellular determinants is still limited. Thus, urgent identification more and better new predictors to assess the ability of metastasis can help us to develop a more accurate prognosis and suitable treatment for breast cancer.
NRF3, which is also named NFE2L3, was first identified as a basic-region leucine zipper family of transcription factors over 10 years ago.4 NRF3 belongs to the Cap “n” Collar (CNC) proteins family, which include NFE2L1 (NRF1, Nuclear Factor, Erythroid 2 Like 1),5 NFE2L2 (NRF2, Nuclear Factor, Erythroid 2 Like 2),6 NFE2L3 (NRF3,Nuclear Factor, Erythroid 2 Like 3), as well as the distantly related BACH1 (BTB Domain And CNC Homolog 1) and BACH2 (BTB Domain And CNC Homolog 2) proteins.7 As a most recently identified member of this family, only extremely limited information for NRF3 was published to date. Previous studies showed NRF3 played an important role in placental development by regulating gene expression as a transcription factor.8,9 Different from the other members of this family extensively studied in multiple cancer types,10,11 NRF3 was only reported to exert a protective role against lymphomagenesis.12 More recently, researchers found NRF3 could translocate into nucleus and activate the gene expression of UHMK1 (the cell cycle regulator U2AF homology motif kinase 1) for controlling cell proliferation, and could also be degraded by β-TRCP (Beta-Transducin Repeat Containing E3 Ubiquitin Protein Ligase), an adaptor for the Skp1-Cul1-F-box protein (SCF) ubiquitin ligase.13 However, the biological functions and the underlying mechanisms of NRF3 in breast cancer are still remained to be established.
In this study, we found that NRF3 expression was suppressed in breast cancerous specimens compared with paracancerous tissues. The expression of NRF3 in breast cancer cell was negatively associated with cell proliferation. Moreover, our results also showed the migration and invasion ability of breast cancer cells were decreased responding to NRF3 overexpression by inhibiting EMT process and the expression of MMPs, which indicated NRF3 possessed a negative role on breast cancer metastasis. The repression of AKT/ID3 axis was found to might account for the antitumor functions of NRF3 in human breast cancer cells. Finally, the NRF3 expression level was negatively associated with the metastasis statue of the breast cancer cells in lymph nodes and positively related to the survival period of breast cancer patients.
Methods and material
Cell culture
The breast cancer cell lines MCF7, MDA-MB-231, and SKBR3 cells were purchased from ATCC (Manassas, VA, USA). All cells were grown in Dulbecco’s Modified Eagle’s medium (DMEM; Gibco, Life Technologies Corp., Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone, GE Healthcare Life Sciences), 2mM L-glutamine (Gibco), 1% penicillin (100 units/ml) and streptomycin (100 μg/mL) (Gibco) and incubated at 37°C, 5% CO2 in a humidified incubator and passaged at ≥80% confluence by trypsin (Gibco).
Immunohistochemistry (IHC)
The breast cancer tissues and the adjacent cancer tissues were harvested at the Central Hospital of Taizhou City when surgery for patients diagnosed with breast cancer and fixed in buffered 10% formalin, transferred to 70% ethanol, embedded in paraffin, sectioned into 5 μm sections and kept in −80℃. All patients provided written informed consent for the use of these clinical materials in research, and the project was approved by the Institutional Ethics Committee of the Central Hospital of Taizhou City and was conducted in accordance with the Declaration of Helsinki. Rabbit polyclonal antibody for NRF3 (HPA05588) was purchased from Sigma–Aldrich Company (Merck KGaA). IHC assay was performed as previously described.14 Stained tissues were imaged with an Olympus microscope.
Western blot
Cultured cells were lysed in RIPA buffer containing 50 mM Tris-HCl at pH 8.0, 150 mM sodium chloride, 2 mM EDTA at pH 8.0, 0.1% Triton-X 100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate, 1% protease inhibitor (Sigma). Keep the supernatant after centrifugation at high speed and prepare the cell lyase with 2X SDS loading buffer. Gel electrophoresis was performed on an acrylamide gel and proteins were transferred onto a PVDF membrane (Bio-rad).
Antibodies for immunoblotting included anti-Vimentin (CST, 5741), anti-N-cadherin(CST, 13116), anti-E-cadherin (CST, 3195), anti-MMP-2 (CST, 40994), anti-MMP-9 (CST, 13667), anti-TIMP-1 (CST, 8946), anti-TIMP-2 (CST, 5738), anti-AKT (CST, 4691), anti-p-AKT (S473) (CST, 4060), anti-NF-κB p65 (CST, 8242), anti-p- NF-κB p65 (S536) (CST, 3033) and anti-ID3 (CST, 9837) diluted 1:1000, anti-β-actin (Sigma, A8481) diluted 1:5000. Protein bands were detected with an ECL chemiluminescence reaction kit (Thermo Scientific).
NRF3 knockdown and overexpression
The sequences for small interfering RNA (siRNA) against NRF3 were purchased from Genepharma (Shanghai, China) are 5ʹ-GCACGAAGCUGUGGAUAUTT-3ʹ (si-NRF3-1), 5ʹ-CCGCGUAGACCUAGAUCUUTT-3ʹ (si-NRF3-2) and 5ʹ-GAACU CACUUCAGCAGAAUTT-3ʹ (si-NRF3-3). The coding sequence of human NRF3 mRNA and ID3 mRNA were synthesized and subcloned into the pcDNA3.1 vector to construct the overexpression plasmids. The integrity of the respective plasmid constructs was confirmed by DNA sequencing. After cells were seeded in six well plates for overnight, 2 μg overexpression plasmid or empty vector was transfected with Lipofectamine 3000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Western blot was used to validate the expression efficiency after cells were transfected for 24 hrs.
RNA sequencing
The mRNA expression profiling in NRF3 overexpressing MDA-MB-231 cells was analyzed using RNA sequencing. Total RNA was extracted from MDA-MB-231 cells transfected with pcDNA3.1 or NRF3 overexpression vector using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. After the RNA purification, the remained RNAs were fragmented using Ambion Fragmentation Solution (Thermo Fisher Scientific, USA). Then, the mRNA fragments were used to synthesize cDNAs, followed by end repairing and adenine connection. RNA sequence analysis was performed by the HiSeq. 2000 sequencer (Illumina, USA). Three independent RNA samples for each cell were analyzed.
Cell proliferation assay
Cells were seeded in 96-well plate and incubated for the indicated time. After treatment, cells were incubated with 50 mL of 1 mg/mL 3-(4,5-dimethylthiazol-2-yl) −2,5- diphenyltetrazolium bromide (MTT, Sigma–Aldrich Chemical) in phosphate-buffered saline (PBS) for 3 hrs according to manufacturer’s instructions. The purple formazan was then solubilized by DMSO and absorbance at 570 nm was read by a microplate reader (Molecular Devices, Sunnyvale, CA).
Cell cycle analysis by flow cytometry
3×105 cells were transferred into a 6 cm dish and subjected to cell cycle arrest by incubating with 0.1% FBS for 24 hrs. Next, the synchronized cells were transfected relative siRNAs according to standard protocol. They were then released from arrest by replacing the medium with fresh DMEM +10% FBS. Cells were harvested after 24 hrs later, washed with PBS and fixed in 75% ethanol at −20°C. The fixed cells were washed with PBS, incubated for 30 mins at 37°C with RNase A (100 ug/mL) and 0.1% TritonX-100, stained with propidium iodide (30 μg/mL), and analyzed by flow cytometry (FACS Calibur, BD Biosciences). The percentages of cells at G1, S, and G2/M were calculated by the FlowJo software program (Flowjo, LLC) based on the DNA content.
Colony formation assay
The colony formation assay was performed according to a previously established protocol.15 Briefly, the six-well plate placed 800 breast cancer cells transiently overexpressed NRF3 per well. The cells were washed with PBS buffer after 14 days of culture, and the colonies were fixed with methanol and stained with crystal violet solution and the number of colonies (>50 cells) was counted.
Cell migration and invasion assays
The migration and invasion assays were performed as previously described.16 Briefly, Chambers (8 μm pore, BD Falcon, BD Biosciences) with or without matrigel (BD Biosciences) were used to investigate invasion and migration, respectively. After treatment, 3×104 cells in 100 μL serum-free medium were placed into the chambers without matrigel for migration assay. For invasion assay, 5×104 cells in 100 μL serum-free medium were placed into chambers which were coated with matrigel. And then these chambers were put in 24-well plates filled with 600 μL medium containing 20% FBS as an attractant. After incubation for 24 hrs, cells in the upper sides were removed and the migrated or invaded cells on the underside of the membrane were fixed with iced methanol and stained with 0.1% crystal violet for 30 min at 37°C and then washed twice with PBS. Stained cells were counted in three independent areas under microscope.
Quantitative RT-PCR analysis
Total RNA was extracted by lysing cells with TRizol reagent (Invitrogen, Carlsbad, CA). Total RNA was used to synthesize the first strand cDNA by M-MLV reverse transcriptase (Promega, Madison, WI). Real-time PCR amplification was performed in the ABI 7300 Real-Time PCR system (Applied Bioscience, Foster City, CA) according to the manufacturer’s procedure for relative quantification. The amplification reactions were carried out with 2X Power SYBR Green PCR Master Mix (Applied Biosystems). The standard temperature profile included initial denaturing at 95°C for 10 mins, followed by 40 cycles of denaturing at 95°C for 15 s, annealing and extension at 60°C for 1 min. A DNA dissociation curve was generated to confirm the specificity of amplification. Relative Standard Curve Method (2–ΔΔCt) was used to determine the relative mRNA expression using β-actin as a control. The sequences for all primers used in the present study are listed in Table 1.
Table 1.
The sequences for the primers used in the qRT-PCR assay
| Gene | Sequences |
|---|---|
| LGALS3BP | Forward: 5ʹ-CTATCAGTCCAGACGGGGG-3’ |
| Reverse: 5ʹ-GGCTTTGTTTTCGTAGGCAA-3ʹ | |
| ANXA2 | Forward: 5ʹ- GGCAAAGGGTAGAAGAGC-3’ |
| Reverse: 5ʹ-ATCAGCAAAATACAGGGG-3’ | |
| EIF4A1 | Forward:5ʹ-CTATGTGACTTGTATGAAACCC-3’ |
| Reverse: 5ʹ-GATGACTAAAGAAACCTGCTG-3’ | |
| ALDOA | Forward: 5ʹ-ACATCGCTCACCGCATCG-3’ |
| Reverse: 5ʹ-CGCCTTCTGGTAGAGTGTCTCA-3’ | |
| ID3 | Forward: 5ʹ-GGCTGCTACGAGGCGGTG-3’ |
| Reverse: 5ʹ-TGGATGGGAAGGTGGGGGC-3’ | |
| RPL18 | Forward: 5ʹ-ATGGGAGTGGACATCCGCCATAACA-3’ |
| Reverse: 5ʹ-TGTTATGGCGGATGTCCACTCCCAT-3’ | |
| RAB5A | Forward: 5ʹ-ACATTGTAATAGCTTTATCGG-3’ |
| Reverse: 5ʹ-GGTTGGTTGTGTGGGTTC-3’ | |
| KRT18 | Forward: 5ʹ-CACCTTCTCCACCAACTACCG-3’ |
| Reverse: 5ʹ-ATGCCTCCCATTCCTGCC-3’ | |
| SERPINH1 | Forward: 5ʹ-AGATGCAGAAGAAGGCTGTTGC-3’ |
| Reverse 5ʹ-GGGGTGGTCGGCGTAGAA-3’ | |
| SSR1 | Forward: 5ʹ-TTCACCGAGTGCAGATAC-3’ |
| Reverse 5ʹ-AGATTGATGACCAAACCA-3’ | |
| β-actin | Forward: 5ʹ-AGCACAGAGCCTCGCCTT TGC-3’ |
| Reverse 5ʹ-CTGTAGCCGCGCTCGGTGAG-3’ |
Abbreviations: LGALS3BP, Galectin 3 Binding Protein; ANXA2, Annexin A2; EIF4A1, Eukaryotic Translation Initiation Factor 4A1; ALDOA, Aldolase, Fructose-Bisphosphate A; ID3, Inhibitor of DNA Binding 3; RPL18, Ribosomal Protein L18; RAB5A, Member RAS Oncogene Family; KRT18, Keratin 18; SERPINH1, Serpin Family H Member 1; SSR1, Signal Sequence Receptor Subunit 1.
Statistical analysis
Comparisons were performed by two-tailed unpaired Student’s t-test . A p-value ≤0.05 was considered statistically significant. Statistical significance was classified as: *. p≤0.05. Survival curves based on the Kaplan–Meier method were compared using a log-rank test. The correlation between NRF3 expression and clinicopathological features was evaluated by the chi-square test. The software used to perform these analyses was SPSS version 18 (SPSS; Chicago, IL).
Ethics approval and informed consent
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Ethics Committee of the Taizhou Central Hospital (Zhejiang, China) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Results
NRF3 expression is suppressed in high malignant breast cancer cell lines, and its silence promotes cell proliferation
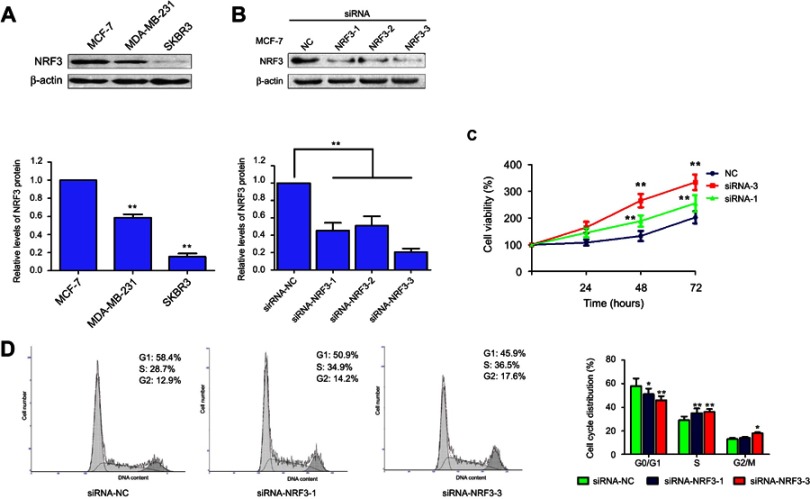
We first measured the NRF3 expression levels in three human breast cancer cell lines including MCF-7, MDA-MB-231, and SKBR3 using western blotting. As shown in Figure 1A, NRF3 was expressed in MCF-7 cells with the highest level and expressed in SKBR3 cells with the lowest level among them. As we knew, MCF-7 is a less malignant breast cancer cell line compared with MDA-MB-231 and SKBR3 cells, especially in migration and invasion capabilities. Our data suggest that NRF3 might act as a tumor suppressor in breast cancer cells.
Figure 1.
NRF3 is suppressed in high malignant breast cancer cells and inhibits cell proliferation. (A) Western blot was performed to detect the expression of NRF3 in breast cancer cell line MCF-7, MDA-MB-231, and SKBR3 (top panel). β-actin was used as loading control. The quantification of NRF3 protein level for the above three different breast cancer cell lines (bottom panel). Error bar indicated SD (**P<0.01). (B) Western blot was performed to detect the knock-down efficiency of NRF3 in MCF-7 cell line by three different siRNA (top panel). β-actin was used as loading control. The quantification of NRF3 protein level (bottom panel) for western blot. Error bar indicated SD (**P<0.01). (C) MTT assay to evaluate the proliferation difference between breast cell lines with NC siRNA (blue), siNRF3-1 (green), and siNRF3-3 (red). Error bar indicated SD (**P<0.01). (D) flow cytometry analysis (left panel) of the cell cycle with NRF3 knock-down or not. The quantification of the cell cycle distribution (right panel) in the G0/G1 phase, S phase and G2/M phase. Error bar indicated SD (*P<0.05 and **P<0.01). Three independent experiments were performed for all assays.
Abbreviations: NRF3, Nuclear Factor, Erythroid 2 Like 3; NC, negative control.
Next, siRNA was used to knock down the NRF3 expression in MCF-7 cells to define its biological functions especially in cell proliferation. All three siRNAs (1–3) could effectively impair the NRF3 expression in MCF-7 cells (Figure 1B). The most effective siRNA-1 and 3 were chosen to perform the MTT assay to determine the influence of NRF3 silence on cell proliferation. As shown in Figure 1C, both siRNA-1and 3 significantly enhanced the cell proliferation of MCF-7 cells compared to the cells transfected with a negative control siRNA. Finally, the data from flow cytometry assay indicated that NRF3 silence significantly increased the percentage of cell number in S and G2/M phase, while decreased the percentage of cell number in G0/G1 phase (Figure 1D).
Exogenous expression of NRF3 significantly inhibits migration ability and cell proliferation of human breast cancer cells
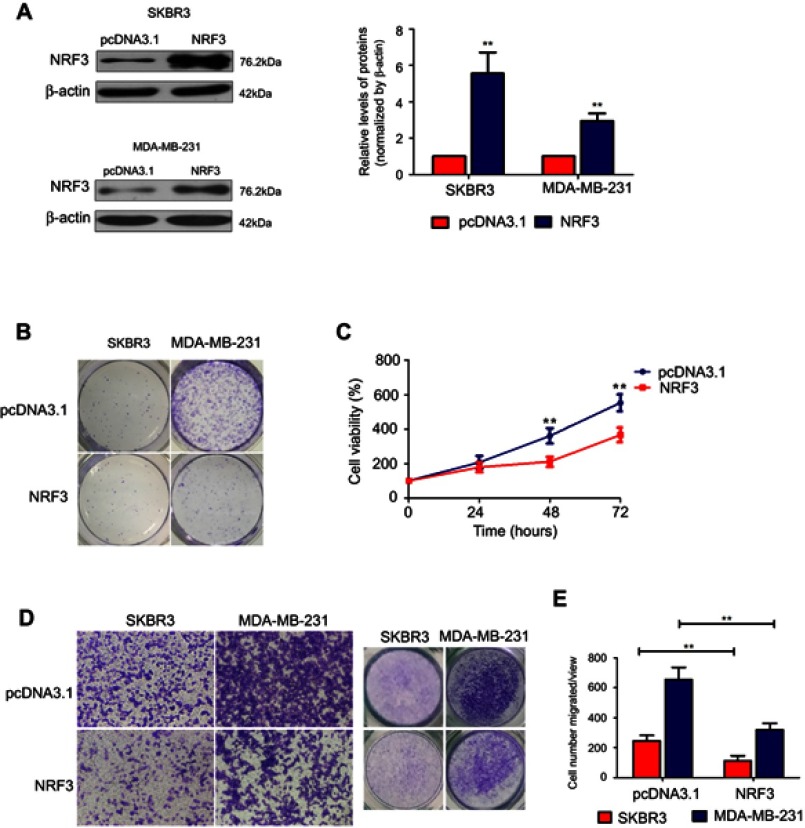
To further confirm the biological functions of NRF3 in human breast cancer, NRF3 overexpression plasmid was constructed and introduced into SKBR3 and MDA-MB-231 cells. The expression efficiency of the overexpression plasmid was validated by western blot assay. As shown in Figure 2A, NRF3 levels were effectively upregulated in both SKBR3 and MDA-MB-231 cells after transfecting the overexpression plasmid. The colony formation experiment was performed to determine the cell growth ability of the breast cancer cells with NRF3 overexpression. Our data revealed that the exogenous expression of NRF3 effectively inhibited the colony forming ability of both cell lines especially in MDA-MB-231 cells (Figure 2B). In contrast to the positive role of NRF3 silence on proliferation in MCF-7 cells, NRF3 overexpression significantly impaired the cell proliferation of MDA-MB-231 cells (Figure 2C). More interestingly, upregulation of NRF3 in both two breast cancer cell lines significantly inhibited their migration ability (Figure 2D and E).
Figure 2.
Ectopic expression of NRF3 suppresses breast cancer cell proliferation and migration. (A) Western blot was performed to determine the overexpression of NRF3 in SKBR3 and MDA-MB-231 cells by transfected ectopic NRF3-expressed plasmids. β-actin was used as loading control. (B) Colony form assay for SKBR3 and MDA-MB-231 cells transfected with NRF3-overexpressed plasmids or empty vector as 800 cells per well. The colonies were stained with crystal violet solution. (C) MTT assay was performed to evaluate the proliferation variation between MDA-MB-231 cells transfected with empty vector (blue) and NRF3-expressed plasmid (red). Error bar indicated SD (**P<0.01). (D) Cell migration assay was performed to detect the migration ability of SKBR3 and MDA-MB-231 cells with NRF3-overexpressed plasmids or empty vector by transwell. The cells were stained with crystal violet solution (left and middle panels). The quantification of the cell number migrated per view at 24 hours after transfected (right panel). pcDNA3.1 was used as negative control. Error bar indicated SD (**P<0.01). Three independent experiments were performed for all assays.
Abbreviations: NRF3, Nuclear Factor, Erythroid 2 Like 3.
Exogenous expression of NRF3 significantly impairs invasion potential of human breast cancer cells by inhibiting EMT and suppressing MMP2 expression
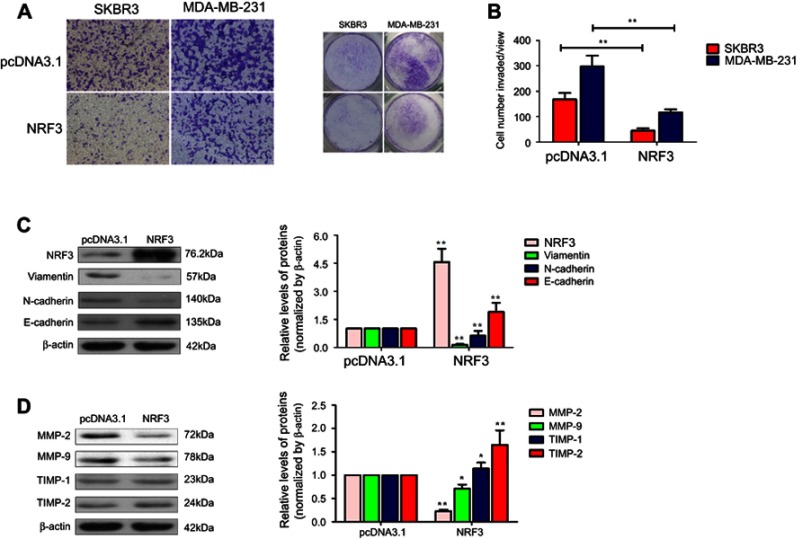
Next, the invasion capability of both SKBR3 and MDA-MB-231 cells with NRF3 overexpression was determined using the transwell assay. As expected, NRF3 overexpression could also significantly repress the invasion potential of both two cell lines (Figure 3A and B). As the EMT is the critical initial step for cancer cell migration,17 western blot assay was performed to confirm whether NRF3 could influence the EMT process. As shown in Figure 3C, exogenous expression of NRF3 could effectively upregulate the expression of E-cadherin, while downregulate the expression of N-cadherin and Vimentin. Furthermore, the degradation of extracellular matrix (ECM) is also an essential process for cancer cell invasion.18 Therefore, we also detected the expression patterns of MMPs and TRIMs under the NRF3 overexpression. As shown in Figure 3D, high level of NRF3 indeed effectively downregulated the expression of MMP-2 and MMP-9, while upregulated the expression of TIMP-2 with no obvious change on TIMP-1.
Figure 3.
NRF3 is required to suppress breast cancer cell invasion related to EMT and MMPs. (A and B) Cell invasion assays were performed to detect the invasion potential of SKBR3 and MDA-MB-231 cells with NRF3-overexpressed plasmids or empty vector by transwell coated with matrigel. Representative images at 40× (left panel), total view for whole well (right panel) and the quantification for cell number invaded per view (B). Error bar indicated SD (**P<0.01). (C) Western blot was performed to detect the expression of E-cadherin, N-cadherin, and Vimentin protein levels in MDA-MB-231 cells over-expressed with NRF3 or not (left panel). β-actin was as a loading control. The quantification for NRF3 (pink), Viamentin (green), N-cadherin (blue), and E-cadherin (red) protein levels according to western blotting. pcDNA3.1 was used as negative control. Error bar indicated SD (**P<0.01). (D) Western blot was performed to detect the expression of MMP-2, MMP-9, TIMP-1, and TIMP-2 protein levels in MDA-MB-231 cells overexpressed with NRF3 or not (left panel). β-actin was used as loading control. The quantification for MMP-2 (pink), MMP-9 (green), TIMP-1 (blue), and TIMP-2 (red) protein levels according to western blotting. pcDNA3.1 as a negative control. Error bar indicated SD (*P<0.05 and **P<0.01). Three independent experiments were performed for all assays.
Abbreviations: NRF3, Nuclear Factor, Erythroid 2 Like 3; EMT, epithelial–mesenchymal transition; MMP, matrix metallopeptidase; TIMP, TIMP metallopeptidase inhibitor.
AKT/ID3 axis might account for the anti-metastasis function of NRF3 in human breast cancer cells
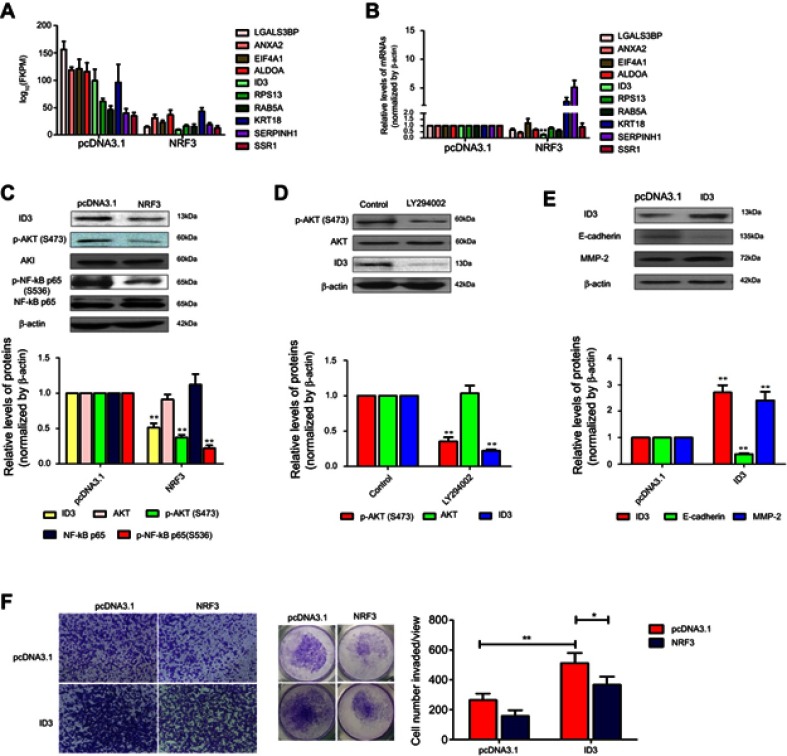
Subsequent to the phenotype experiments, high throughput sequencing was performed to compare the mRNA expression profiling between MDA-MB-231 cells transfected with pcDNA3.1 vector and NRF3 overexpression plasmid. As shown in Figure 4A, ten most downregulated genes under NRF3 overexpression were listed. qPCR experiment was then performed to validate the data of sequencing and indicated that the expression of ANXA2, ALDOA, and ID3 were indeed suppressed by exogenous NRF3, while the expression of KRT18 and SERPINH1 was upregulated which was inconsistent with the sequencing results (Figure 4B). Previous research has reported ID3 played a critical role in mediating the lung metastasis of breast cancer cell.19 Therefore, ID3 protein expression in NRF3 overexpressed MDA-MB-231 cells was detected using western blot assay. As shown in Figure 4C, exogenously expressed NRF3 could effectively impair the ID3 expression. We also observed that the AKT signaling pathway, an upstream regulator of ID3, was inactivated, which was also supported that the phosphorylation of NF-κB p65 was repressed (Figure 4C).
Figure 4.
NRF3 impairs breast cancer cell invasion through the AKT/ID3 axis. (A) .The most ten downregulated genes under NRF3 overexpression obtained from the data of RNA seq, relative expression level, normalized by log10 (FPKM). (B) qRT-PCR was used to validate the RNA seq data, relative expression level, normalized by GADPH. (C) Western blot for ID3, AKT, p-AKT, NF-κB p65, and p-NF-κB p65 protein levels in MDA-MB-231 cells overexpressed with NRF3 or not. β-actin was as a loading control. (D) Western blot (left panel) for ID3, AKT and p-AKT protein levels in MDA-MB-231 cells treated with LY294002 or not. β-actin was as a loading control. (E) Western blot for ID3, E-cadherin and MMP-2 protein levels in MDA-MB-231 cells over-expressed with ID3 or not. β-actin was used as a loading control. (F) Cell invasion assays for MDA-MB-231 cells with overexpressed NRF3, ID3 alone or in combination by transwell with matrigel. Representative images at 40× (left panel), total view for whole well (middle panel) and the quantification for cell number invaded per view (right panel). Error bar indicated SD (*P<0.05 and **P<0.01). Three independent experiments were performed for all assays.
Abbreviations: NRF3, nuclear factor, Erythroid 2 Like 3; AKT, AKT serine/threonine Kinase; ID3, inhibitor of DNA binding 3; FPKM, Reads Per Kilobase of exon model per Million mapped reads; MMP, matrix metallopeptidase; TIMP, TIMP metallopeptidase inhibitor.
To further confirm AKT signaling pathway indeed involved in ID3 expression in MDA-MB-231 cells, an AKT signaling pathway antagonist, LY294002, was used to treat the MDA-MB-231 cells and the phosphorylation of AKT and ID3 expression were detected by western blot assay. As shown in Figure 4D, LY294002 not only effectively inhibited the phosphorylation of AKT, but also downregulated the expression of ID3 in MDA-MB-231 cells. Subsequently, the overexpression plasmid of ID3 was constructed and subjected to MDA-MB-231 cells to evaluate its role in breast cancer cell metastasis. As shown in Figure 4E, ID3 was successfully high expressed by its overexpression plasmid, while exogenous expression of ID3 effectively downregulated the expression of E-cadherin and upregulated the expression of MMP-2. However, we did not observe the obvious change in the expression of N-cadherin, Vimentin, MMP-9, TIMP-1, and TIMP-2 (data not shown). Finally and as expected, exogenous expression of ID3 not only excited the invasion potential of MDA-MB-231 cells, but also attenuated the inhibitory effect of NRF3 on the cell invasion (Figure 4F).
NRF3 is suppressed in breast cancer specimens and positively related to the survival of breast cancer patients
Considering that NRF3 protein expression was predicted to be downregulated in breast cancer tissue according to The Human Protein Atlas (http://www.proteinatlas.org/, and data not shown), we evaluate the difference of NRF3 expression between breast cancer tissues and the paired adjacent tissues of cancer by IHC. The result showed that the expression of NRF3 in breast cancer tissues was inhibited severely, while was very abundant in corresponding non-tumor normal tissue (Figure 5A).
Figure 5.
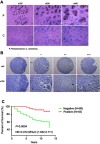
NRF3 was suppressed in breast cancer tissues and positively associated with the overall survival of breast cancer patients. (A) Immunohistochemistry (IHC) staining of NRF3 on breast cancer tissues and the paired adjacent tissues of cancer showing the difference of NRF3 expression between cancer tissues and normal tissues. The brown part indicated NRF3 positive. The representative data were shown from ten pairs of tissues. P: adjacent tissues of cancer. C: cancer tissues. (B) Representative images of immunohistochemistry staining of NRF3 for 144 patients’ breast cancer tissues microarray showing NRF3 expression low in cancer tissues. ‘-’ indicates the negative expression of NRF3. ‘+’ indicates the degree of the positive expression level of NRF3 (C) Kaplan–Meier survival analysis between NFR3-positive and NRF3-negative breast cancer patients. N=99 in NRF3-negative breast cancer patients group. N=45 in NRF3-positive breast cancer patients group. p=0.0004.
Abbreviations: NRF3, nuclear factor, Erythroid 2 Like 3; IHC, immunohistochemistry.
To further determine the clinical significance of NRF3 in breast cancer, we collected cancer tissues from 144 breast cancer patients to construct a tissue microarray for IHC. The result showed that 99 of them (68.7%) were negative, while only 45 of them (31.3%) were positive. As to those in the positive group, the expression of NRF3 was also extremely weak (Figure 5B). We next investigated whether the NRF3 could be used to predict patient prognosis. We examined the correlation between the patients’ survival period and NRF3 expression by comparing the lifespan for these patients in the above two groups after diagnosed. Interestingly, Kaplan–Meier survival analysis showed the percentage of 10 years over survival for NFR3-positive breast cancer patients was almost 90%, while only nearly 50% for those NRF3-negative (Figure 5C).
Baseline characteristics were also compared between NRF3-positive and -negative patients (as shown in Table 2). The NRF3-negative patients showed a higher rate of tumors with metastasis (p=0.0082) and higher stage (p=0.0365). However, NRF3 expression was not related to other clinical characteristics (as shown in Table 2). Taken together, all these findings suggested the significant positive relationship between the NRF3 expression level and the overall survival of breast cancer patients probably through influencing cancer metastasis.
Table 2.
Patient characteristics based on NRF3 expression
| NRF3 negative (N=99) | NRF3 positive (N=45) | P-value | |
|---|---|---|---|
| Age | 51.5±12.56 | 49.5±0.71 | 0.7448 |
| Tumor size, n (%) | 0.5124 | ||
| <3 cm | 34 (34%) | 18 (40%) | |
| ≥3 cm | 65 (66%) | 27(60%) | |
| Lymph node status, n (%) | 0.0082** | ||
| Metastasis | 67(68%) | 20(44%) | |
| No Metastasis | 32(32%) | 25(56%) | |
| Stage, n (%) | 0.0365* | ||
| I | 9 (9%) | 5(11%) | |
| II | 51(51%) | 32(71%) | |
| III | 39(40%) | 8(18%) | |
| Histologic grade, n (%) | 0.2235 | ||
| I | 4(4%) | 1(2%) | |
| II | 73(74%) | 39(87%) | |
| III | 22(22%) | 5(11%) | |
| ER, n (%) | 0.5602 | ||
| Negative | 38(38%) | 15(33%) | |
| Positive | 61(62%) | 30(67%) | |
| PR, n (%) | 0.1259 | ||
| Negative | 51(52%) | 17(38%) | |
| Positive | 48(48%) | 28(62%) | |
| HER2, n (%) | 0.7697 | ||
| Negative | 68(69%) | 32(71%) | |
| Positive | 31(31%) | 13(29%) | |
| Subtype, n (%) | 0.7745 | ||
| TN | 15(15%) | 6(13%) | |
| Non-TN | 84(85%) | 39(87%) |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor-2; TN, triple-negative.
Discussion
It is well known that there is a tight connection between oxidant stress and inflammation, and the abnormality of them could finally contribute to the carcinogenesis of many cancer types including breast cancer.20 The Cap “n” Collar (CNC) proteins family, which mainly include NRF1, NRF2, and NRF3, play a critical role in the regulation of oxidant stress and multiple cancers development and progress.21 As a most intensively studied member, hundreds of publications have well defined the functions of NRF2 in multiple cancer types.22 However, little is known about the roles of its homologous gene NRF3 in cancer especially in breast cancer. To our best knowledge, this is the first study to evaluate the roles of NRF3 in human breast cancer.
At first, we first compared the NRF3 expression in three human breast cancer cell lines. Our data suggested that NRF3 was highly expressed in MCF-7 cells, while was less expressed in SKBR3 and MDA-MB-231 cells (Figure 1A). These finding implied NRF3 might be a tumor suppressor in human breast cancer, which is consistent with that NRF3 exerts a protection role against lymphomagenesis.12 Subsequently, lose and gain function experiments revealed that NRF3 could inhibit the cell proliferation by reducing the cell number in the G0/G1 phase, while increase the cell number in S and G2/M phase in MCF-7 cells (Figure 1C and D).
Furthermore, our data indicated that exogenous expression of NRF3 could significantly impair both migration and invasion abilities in SKBR3 and MDA-MB-231 cells (Figure 2D and Figure 3A), which was consistent with the data that NRF3 is negatively associated the metastasis statue of breast cancer (Table 2). The invasive tumor cells must first alter cell-to-cell adhesion and the adhesion to the ECM.18 EMT, a process for cell migration initiation, starts with the disintegration of cell–cell adhesion by losing epithelial markers, such as E-cadherin, and expressing mesenchymal markers, such as vimentin, play an indispensable role in cancer metastasis.23 Therefore, we next tried to determine the role of NRF3 in the EMT of breast cancer cells. Interestingly, ectopic expression of NRF3 significantly inhibited the EMT of MDA-MB-231 cell by upregulating the expression of E-cadherin and downregulating the expression of vimentin and N-cadherin (Figure 3C). More importantly, exogenous expression of NRF3 also downregulated the expression of MMP-2 and MMP-9 and upregulated the expression of MMP inhibitor TIMP-2 in MDA-MB-231 cells (Figure 3D). All these findings suggested that NRF3 could cripple the migration and invasion abilities of breast cancer cells by inhibiting both the EMT process and the degradation of the ECM.
To look insight into the underlying mechanisms of anti-tumor effects of NRF3 in human breast cancer, RNA sequencing was performed to determine the mRNA expression profiling in MDA-MB-231 cell with NRF3 overexpressed. After validation using qPCR, we found ID3 (Inhibitor of DNA Binding 3, helix-loop-helix (HLH) Protein) may play an important role in NRF3 mediated anti-tumor functions (Figure 4A and B). ID3 is a HLH protein that can form heterodimers with other HLH proteins,24 and Id protein family plays pivotal roles in distinct aspects of normal and malignant breast biology.25 In human breast cancer, ID3 is a potential target as it takes part in regulating multiple malignant behaviors including cell proliferation, apoptosis, and metastasis.19,26,27 Several researches have implied that ID3 located downstream of AKT signaling pathway and could be activated by the later in other cancer types such as glioma.28,29 Here, we also tried to confirm whether AKT signaling pathway was suppressed by NRF3. Interestingly, exogenous expression of NRF3 not only impaired the phosphorylation of AKT but also inhibited the downstream NF-κB p65 activation (Figure 4C). This finding was further validated by the data that LY294002 could reduce the ID3 expression (Figure 4D). More importantly, the ectopic expression of ID3 effectively downregulated the expression of E-cadherin and upregulated the expression of MMP-2 (Figure 4E). As expected, ID3 not only promoted the invasion potential but also attenuated the inhibitory effect of NRF3 on the cell invasion in MDA-MB-231 cells (Figure 4F). All these results supported that NRF3 might play its anti-tumor effect by inhibiting AKT/ID3 axis in human breast cancer.
Finally, the NRF3 expression in breast cancer and paracancerous specimens was compared, and the association between NRF3 expression and survival of breast cancer patients was also determined. We observed that NRF3 expression level was much high paracancerous specimens, but extremely weak in breast cancer tissues (Figure 5A). Similarly, NRF3 immunoreactivity detected as positive was only 31.3% in 144 breast cancer patients, and the positive signal was very weak in our study (Figure 5B). This finding is highly consistent with the results from the dataset The Human Protein Atlas. Furthermore, we have shown the poor survival for these patients negative in NRF3 immunoreactivity according to clinic information from postoperative follow-up and management (Figure 5C).
Conclusion
In summary, NRF3 could inhibit the cell proliferation and metastasis of breast cancer cells through AKT/ID3 axis at least partially. Based on all our findings, we proposed that NRF3 was necessary for breast cancer metastasis suppression and potential to be a clinic marker in breast cancer prognosis.
Acknowledgments
This work was supported in part by Zhejiang Medical Association (No. 2013ZYC-A134), Technology Division of Taizhou (No. 1501KY19), Department of Education of Zhejiang province (No. 201738081) and Department of Science and Technology of Zhejiang province (No. 2016C33230).
Data Availability
All data for this study are presented in this published article.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Berry DA, Cronin KA, Plevritis SK, et al. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353(17):1784–1792. doi: 10.1056/NEJMoa050518 [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. Ca-Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi A, Ito E, Toki T, et al. Molecular cloning and functional characterization of a new Cap’n’ collar family transcription factor Nrf3. J Biol Chem. 1999;274(10):6443–6452. [DOI] [PubMed] [Google Scholar]
- 5.Chan JY, Han XL, Kan YW. Cloning of Nrf1, an Nf-E2-related transcription factor, by genetic selection in yeast. P Natl Acad Sci USA. 1993;90(23):11371–11375. doi: 10.1073/pnas.90.23.11371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of Nf-E2-related factor-2 (Nrf2), a Nf-E2-like basic leucine-zipper transcriptional activator that binds to the tandem Nf-E2/Ap1 repeat of the beta-globin locus-control region. P Natl Acad Sci USA. 1994;91(21):9926–9930. doi: 10.1073/pnas.91.21.9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oyake T, Itoh K, Motohashi H, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16(11):6083–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chenais B, Derjuga A, Massrieh W, et al. Functional and placental expression analysis of the human NRF3 transcription factor. Mol Endocrinol. 2005;19(1):125–137. doi: 10.1210/me.2003-0379 [DOI] [PubMed] [Google Scholar]
- 9.Braun S, Hanselmann C, Gassmann MG, et al. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol Cell Biol. 2002;22(15):5492–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohta T, Iijima K, Miyamoto M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68(5):1303–1309. doi: 10.1158/0008-5472.CAN-07-5003 [DOI] [PubMed] [Google Scholar]
- 11.Yamadori T, Ishii Y, Homma S, et al. Molecular mechanisms for the regulation of Nrf2-mediated cell proliferation in non-small-cell lung cancers. Oncogene. 2012;31(45):4768–4777. doi: 10.1038/onc.2011.628 [DOI] [PubMed] [Google Scholar]
- 12.Chevillard G, Paquet M, Blank V. Nfe2l3 (Nrf3) deficiency predisposes mice to T-cell lymphoblastic lymphoma. Blood. 2011;117(6):2005–2008. doi: 10.1182/blood-2010-02-271460 [DOI] [PubMed] [Google Scholar]
- 13.Chowdhury A, Katoh H, Hatanaka A, et al. Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Sci Rep. 2017;7(1):12494. doi: 10.1038/s41598-017-12675-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y, Xiao CH, Tan LD, Wang QS, Li XQ, Feng YM. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-beta signalling. Br J Cancer. 2014;110(3):724–732. doi: 10.1038/bjc.2013.768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franken NAP, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1(5):2315–2319. doi: 10.1038/nprot.2006.339 [DOI] [PubMed] [Google Scholar]
- 16.Justus CR, Leffler N, Ruiz-Echevarria M, Yang LV. In vitro cell migration and invasion assays. Jove-J Vis Exp Jun. 2014;88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heerboth S, Housman G, Leary M, et al. EMT and tumor metastasis. Clin Transl Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown GT, Murray GI. Current mechanistic insights into the roles of matrix metalloproteinases in tumour invasion and metastasis. J Pathol. 2015;237(3):273–281. doi: 10.1002/path.4586 [DOI] [PubMed] [Google Scholar]
- 19.Gupta GP, Perk J, Acharyya S, et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A. 2007;104(49):19506–19511. doi: 10.1073/pnas.0709185104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49(11):1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sykiotis GP, Bohmann D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci Signal. 2010;3(112):re3. doi: 10.1126/scisignal.3112re3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menegon S, Columbano A, Giordano S. The dual roles of NRF2 in cancer. Trends Mol Med. 2016;22(7):578–593. doi: 10.1016/j.molmed.2016.05.002 [DOI] [PubMed] [Google Scholar]
- 23.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellmeier W, Aguzzi A, Kleiner E, Kurzbauer R, Weith A. Mutually exclusive expression of a helix-loop-helix gene and N-myc in human neuroblastomas and in normal development. Embo J. 1992;11(7):2563–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Candia P, Benera R, Solit DB. A role for Id proteins in mammary gland physiology and tumorigenesis. Adv Cancer Res. 2004;92:81–94. doi: 10.1016/S0065-230X(04)92004-0 [DOI] [PubMed] [Google Scholar]
- 26.Mern DS, Hoppe-Seyler K, Hoppe-Seyler F, Hasskarl J, Burwinkel B. Targeting Id1 and Id3 by a specific peptide aptamer induces E-box promoter activity, cell cycle arrest, and apoptosis in breast cancer cells. Breast Cancer Res Treat. 2010;124(3):623–633. doi: 10.1007/s10549-010-0810-6 [DOI] [PubMed] [Google Scholar]
- 27.Chen YH, Wu ZQ, Zhao YL, Si YL, Guo MZ, Han WD. FHL2 inhibits the Id3-promoted proliferation and invasive growth of human MCF-7 breast cancer cells. Chin Med J. 2012;125(13):2329–2333. [PubMed] [Google Scholar]
- 28.Sato A, Mizobuchi Y, Nakajima K, et al. Blocking COX-2 induces apoptosis and inhibits cell proliferation via the Akt/survivin- and Akt/ID3 pathway in low-grade-glioma. J Neurooncol. 2017;132(2):231–238. doi: 10.1007/s11060-017-2380-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin X, Yin J, Kim SH, et al. EGFR-AKT-Smad signaling promotes formation of glioma stem-like cells and tumor angiogenesis by ID3-driven cytokine induction. Cancer Res. 2011;71(22):7125–7134. doi: 10.1158/0008-5472.CAN-11-1330 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data for this study are presented in this published article.