Abstract
Purpose: Gliomas are the most common primary malignant neoplasms of the central nervous system. Secreted phospholipases A2 (sPLA2s) are known to play an important role in various physiological processes, including bioactive lipid production, defense mechanisms, and cell signaling. However, their roles and clinical importance in gliomas remain unclear.
Patients and methods: In this study, we analyzed the association between the expression of various sPLA2-encoding genes and the clinicopathology of gliomas, using the data of 1047 patients obtained from a public database. Immunohistochemical analysis of 82 glioma tissues was also carried out to assess the relationship between phospholipase A2 group V (PLA2G5) protein expression and the World Health Organization (WHO) glioma grades.
Results: We found that high PLA2G5 gene expression was associated with unfavorable prognosis in both low-grade and high-grade gliomas. The immunohistochemistry of the 82 glioma tissues further confirmed that PLA2G5 protein expression was dependent on the WHO glioma grade. In addition, we found a correlation between PLA2G5 gene expression and both epithelial–mesenchymal transition and the isocitrate dehydrogenase 1 mutation status in these tumors.
Conclusion: Our results indicate that PLA2G5 could be a potential biomarker for predicting poor prognosis in patients with gliomas.
Keywords: secretory phospholipases A2, glioma, PLA2G5, biomarker
Introduction
Gliomas, which are the most common primary malignant neoplasms in the central nervous system, can be assigned to World Health Organization (WHO) tumor grades according to their histopathological features, with grade IV representing high-grade gliomas (HGGs) and grades I–III representing low-grade gliomas (LGGs).1 This classification method is also used in the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). Patients with HGG frequently have a poor prognosis, and the median total survival time is only 14.6 months.2 Although considerable progress has been made in the treatment strategies of neurosurgery, chemotherapy, radiotherapy, and biotherapy, the survival rates associated with gliomas remain low, particularly with HGGs.3 Some well-known molecular markers, including O-6-methylguanine-DNA methyltransferase, isocitrate dehydrogenase 1 (IDH1), epidermal growth factor receptor, and p53, can be helpful to not only predict prognosis but also to provide an accurate comprehensive glioma diagnosis when combined with the classic WHO central nervous system tumor classification.4 Therefore, research on new prognostic markers would help us understand the molecular biology of gliomas and improve the prognosis of patients with this disease.
Secreted phospholipases A2 (sPLA2s) are involved in various physiological processes, including defense mechanisms, bioactive lipid production, and cell signaling.5–7 The members of this enzyme family catalyze the hydrolysis of membrane phospholipids to lysophospholipids, free fatty acids, and arachidonic acid, which can be metabolized to eicosanoids.6 Increasing lines of evidence show that sPLA2s are involved in various human tumors, and some members of this family of phospholipases have been proposed as potential therapeutic targets.8 For example, expression of the phospholipase A2 group III (sPLA2-III)-encoding gene PLA2G3 was significantly increased in adenocarcinomas of the left and right colon, and PLA2G3 was considered to be a novel biomarker for colon cancer.9 Studies by Sved et al showed that sPLA2-IIA (PLA2G2A) gene overexpression was associated with both the carcinogenesis of prostate cancer and poor prognosis.10,11 However, the roles and clinical importance of sPLA2s in gliomas remain unclear.
In this study, we analyzed the expression levels of the sPLA2-encoding genes and survival information of 1047 patients with gliomas, using data obtained from TCGA. The notable discoveries were that sPLA2-V (PLA2G5) was the only sPLA2 associated with a poor prognosis for both LGGs and HGGs, and that the PLA2G5 gene expression level was associated with the WHO tumor grade. Immunohistochemical analysis of 82 gliomas obtained from a hospital in China further confirmed the changes in the PLA2G5 protein levels in glioma tumors and its clinical significance.
Materials and methods
Patients and tissue samples
Clinical information and molecular data of 1047 patients with gliomas (524 with LGGs and 523 with LGGs) were obtained from TCGA. The gene expression of PLA2G5 in glioma tumors from 524 LGGs was obtained using the RNASeq (Illumina HiSeq) platform in TCGA. The gene expression of PLA2G5 in glioma tumors from 523 HGGs was obtained using the gene expression array (Affymetrix U133A) in TCGA as well as RNASeq (Illumina HiSeq) for 172 of the HGGs. The basic information of the patients is summarized in Tables 1 and 2. For the age factor, the patients with LGGs were roughly separated into two groups by the median age of 41 years (age <41 years, n=258; age ≥41 years, n=266), whereas the patients with HGG were separated into two groups according to the median age of 59 years (age <59 years, n=255; age ≥59 years, n=268) (Tables 1 and 2).
Table 1.
Clinical parameters of LGG patients in TCGA (n=524)
| Variable | Number of patients | Percentage of patients | |
|---|---|---|---|
| Gender | Male | 288 | 55.0% |
| Female | 236 | 45.0% | |
| Age (years) | <41 | 258 | 49.2% |
| ≥41 | 266 | 50.8% | |
| WHO Grade | II | 254 | 48.5% |
| III | 270 | 51.5% | |
| IDH1 mutation | Yes | 388 | 74.0% |
| No | 136 | 26.0% |
Abbreviations: IDH1, isocitrate dehydrogenase 1; LGG, low-grade gliomas; TCGA, the Cancer Genome Atlas.
Table 2.
Clinical parameters of HGG patients in TCGA (n=523)
| Variable | Number of patients | Percentage of patients | |
|---|---|---|---|
| Gender | Male | 318 | 60.8% |
| Female | 205 | 39.2% | |
| Age (years) | <59 | 255 | 48.8% |
| ≥59 | 268 | 51.2% | |
| Chemotherapy | Yes | 516 | 98.7% |
| No | 7 | 1.3% | |
| Radiotherapy | Yes | 511 | 97.7% |
| No | 12 | 2.3% | |
| IDH1 mutation | Yes | 32 | 6.1% |
| No | 491 | 93.2% |
Abbreviations: IDH1, isocitrate dehydrogenase 1; HGG, high-grade glioma; TCGA, the Cancer Genome Atlas.
In addition, glioma tissues from 82 patients were selected from the Department of Neurosurgery, Xiangya Hospital, Hunan, China. This group comprised 23 grade II, 19 grade III, and 40 grade IV tumors. Eight normal brain tissue samples were taken from paracancerous edema tissues of HGGs. Written informed consent was provided by all the patients. The study, carried out in accordance with the Declaration of Helsinki, was approved by the Ethics Committee of Xiangya Hospital, Hunan, China. The tissue samples from the 82 patients were embedded in paraffin blocks and then sectioned into slices of 5-µm thickness. The basic information of these 82 patients is provided in Table S1.
Table S1.
Clinical parameters of 82 patients form the department of neurosurgery, Xiangya Hospital, Hunan, China
| Variable | Number of patients | Percentage of patients | |
|---|---|---|---|
| Gender | Male | 48 | 58.5% |
| Female | 34 | 41.5% | |
| Age (years) | <50 | 40 | 48.8% |
| ≥50 | 42 | 51.2% | |
| WHO Grade | II | 23 | 28.0% |
| III | 19 | 23.2% | |
| IV | 40 | 48.8% |
Immunohistochemistry
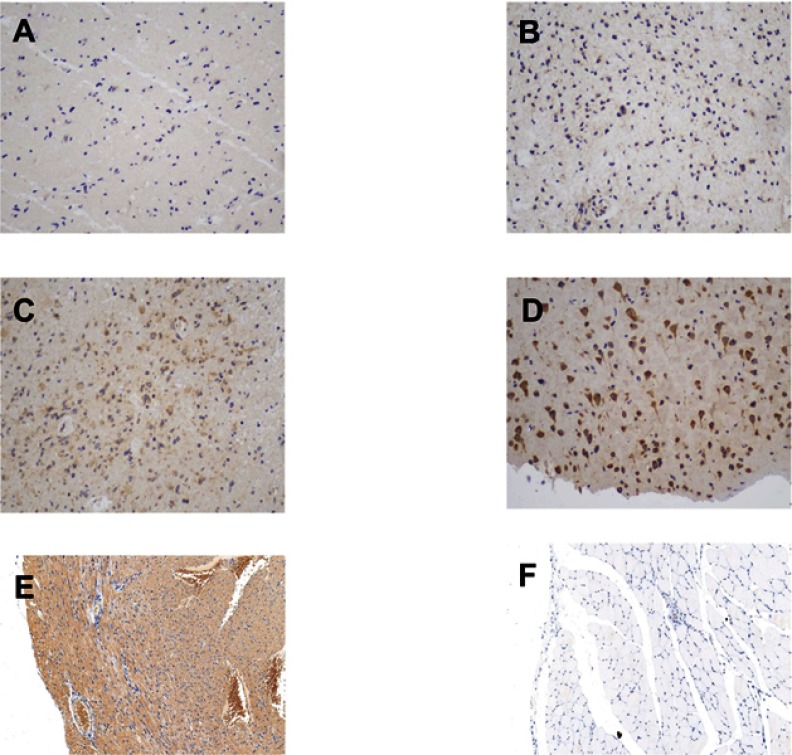
The tissue sections from the 82 patients were first deparaffinized and hydrated for antigen retrieval. They were then incubated with 0.3% hydrogen peroxide for 10 min at room temperature and washed twice with phosphate-buffered saline (PBS). After blocking with 5% goat serum for 10 min, the sections were washed with PBS and incubated overnight with a primary antibody against the PLA2G5 protein (1:100, Biorbyt, Cambridge, CA, USA) at 4 °C. A horseradish peroxidase-labeled secondary antibody (1:400, Abcam, Cambridge, MA, USA) was added dropwise to the sections and incubation was carried out at 37 °C for 30 min. After washing with PBS, the sections were developed using a DAB substrate kit (Sangon Biotech, Shanghai, China) and counterstained with hematoxylin (Sangon Biotech). Two pathologists, who were blinded to the sample identities, independently scored the PLA2G5 immunostaining results on the basis of the staining intensity and percentage of positive cells. The intensity scores were as follows: 0 (negative), 1 (weak positive), 2 (moderately positive), and 3 (strong positive). The associated percentages of PLA2G5-positive cells were 0% (for score 0), 1–25% (score 1), 26–50% (score 2), and >50% (score 3). The immunohistochemical PLA2G5 score was calculated by multiplying the intensity score by the positive percentage, up to a maximum of 9 points; illustrative figures are provided in Figure S1.
Figure S1.
The immunohistochemical staining score of PLA2G5 in glioma and positive/negative control (magnification ×200). (A) Percentage score = 1, intensity score = 1; (B) Percentage score = 2, intensity score = 1; (C) Percentage score = 3, intensity score = 2; (D) Percentage score = 3, intensity score = 3; (E) Positive control: cardiac muscle; (F) Negative control: striated muscle.
Statistical analysis
Differences between two groups were analyzed using the Student's t-test or Mann-Whitney test. Differences between groups were analyzed by analysis of variance. The overall-survival curves were plotted according to the Kaplan-Meier method and compared using a log-rank test. Descriptive statistical analysis was used to ensure normality. Cox proportional hazard regression analysis was used for the univariate and multivariate analyses, where the parameters used were the gender, age, WHO grade, chemoradiotherapy, IDH1 status, and PLA2G5 gene expression, which were potentially related to survival. Associations between the gene expression levels of PLA2G5 and epithelial–mesenchymal transition (EMT) markers were analyzed using Pearson’s Chi-square test. Statistical analysis was performed using GraphPad Prism 5 (LaJolla, CA, USA) and SPSS version 20.0 (IBM Corp., Armonk, NY, USA). A P-value of <0.05 was considered to be statistically significant.
Results
Prognostic value of the sPLA2 family in gliomas
We analyzed the gene expression data and prognosis of 1047 patients with gliomas, obtained from TCGA; namely, 255 grade II, 269 grade III, and 523 grade IV gliomas. We assigned the patients equally on the basis of the median expression levels of sPLA2-encoding genes. We found that patients with LGGs expressing high levels of PLA2G2A, PLA2G4A, PLA2G5, PLA2G6, PLA2G15 and PLA2G16, and those with HGGs expressing high levels of PLA2G1B, PLA2G2E, PLA2G3, PLA2G4D and PLA2G5, had poorer survival. This shows that PLA2G5 was the only sPLA2 associated with an unfavorable prognosis for both LGGs and HGGs (Table 3).
Table 3.
Relationship between sPLA2s and patient prognosis in LGGs and HGGs from TCGA database
| PLA2Gs | LGG | HGG | ||
|---|---|---|---|---|
| Cut-off value | P-value | Cut-off value | P-value | |
| PLA2G1B | 0.577 | 0.8824 | 0.585 | 0.0156 |
| PLA2G2A | 1.446 | 0.0068 | 6.908 | 0.3268 |
| PLA2G2D | 0.930 | 0.7239 | 1.324 | 0.7288 |
| PLA2G2E PLA2G2F PLA2G3 PLA2G4A |
NA NA NA 6.799 |
- - - 0.0017 |
3.851 3.918 4.571 8.267 |
0.0061 0.4204 0.0303 0.2306 |
| PLA2G4C | 9.100 | 0.1362 | 8.568 | 0.9348 |
| PLA2G4D | 0.477 | 0.4380 | 0.536 | 0.0469 |
| PLA2G5 | 1.338 | 0.0002 | 6.447 | 0.0020 |
| PLA2G6 | 9.622 | <0.0001 | 8.443 | 0.8509 |
| PLA2G7 | 6.167 | 0.3866 | 5.562 | 0.3538 |
| PLA2G10 | 0.520 | 0.2176 | NA | - |
| PLA2G12A | 9.805 | 0.1205 | 9.429 | 0.3966 |
| PLA2G12B PLA2G15 |
NA 9.114 |
- 0.0139 |
NA 9.575 |
- 0.2904 |
| PLA2G16 | 10.030 | 0.0043 | 10.26 | 0.3123 |
Abbreviations: HGG, high-grade glioma; LGG, low-grade gliomas; NA, not available; TCGA,the Cancer Genome Atlas.
Association of PLA2G5 gene expression with the clinicopathological features of patients with gliomas
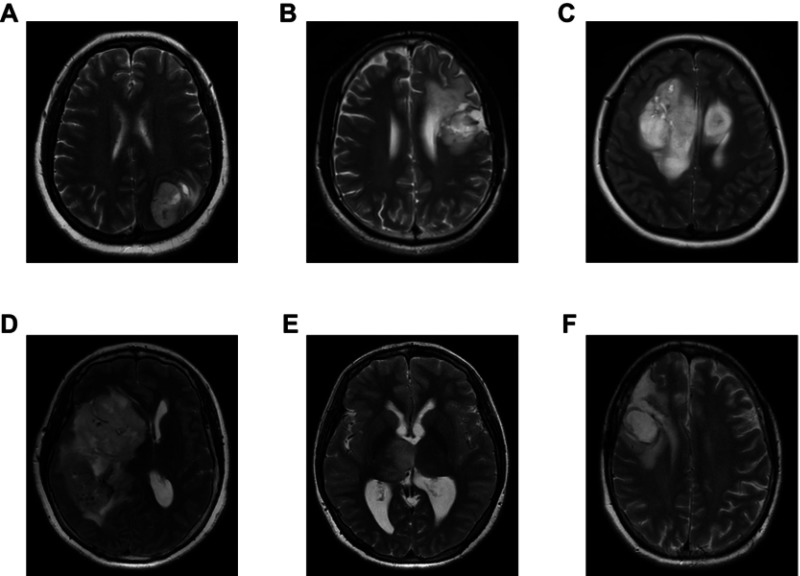
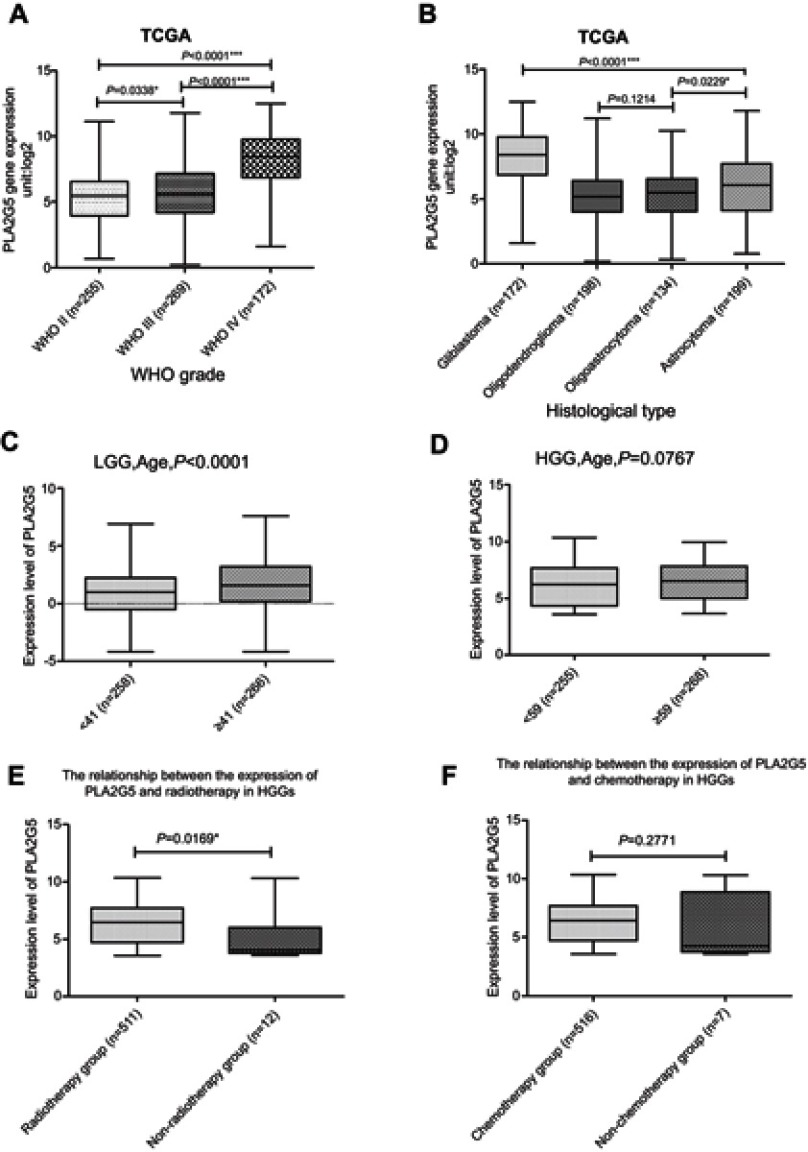
We performed a statistical evaluation of the associations between PLA2G5 gene expression and various clinicopathological features, and found that the expression of this gene was positively associated with the WHO grade of the gliomas in the patients (Figure 1A). PLA2G5 gene expression was also associated with the histological type of the glioma. It was clear that the PLA2G5 gene expression level was the highest in glioblastomas, whereas its expression in astrocytoma subtypes was higher than that in oligodendroglioma and oligoastrocytoma subtypes (Figure 1B). Representative images of various glioma subtypes are presented in Figure S2. When we grouped the patients according to age, PLA2G5 gene expression was positively correlated with patient age. For LGGs, the PLA2G5 gene expression levels in the elderly group (age ≥41 years) were significantly higher than those in the younger group (age <41 years) (P<0.0001, Figure 1C). However, for HGGs, the PLA2G5 expression levels in patients aged ≥59 years were not higher than in those aged <59 years (P=0.0767, Figure 1D). Most of the patients with HGGs had undergone radiotherapy and/or chemotherapy after surgery. We found that the association of PLA2G5 gene expression with radiotherapy for HGG was statistically significant, but not with chemotherapy (Figure 1E and F). However, because the sample size of the non-radiotherapy group was too small, we could not rule out a possible sampling bias. In addition, we did not obtain radiotherapy or chemotherapy data for the patients with LGGs in TCGA. Furthermore, there was no association between the PLA2G5 gene expression level and gender for both HGGs and LGGs (Figure S3A and B). The correlation between PLA2G5 gene expression and the IDH1 status is described below.
Figure S2.
Representative MRI images of various glioma subtypes. (A) Glioblastoma (WHO IV); (B) Astrocytoma (WHO III); (C) Astrocytoma (WHO II); (D) Oligodendroglioma (WHO III); (E) Oligodendroglioma (WHO II); (F) Oligoastrocytoma (WHO II).
Figure 1.
PLA2G5 gene expression was associated with the clinicopathological features of glioma patients(*P<0.05;***P<0.0001). (A) PLA2G5 gene expression was dependent on glioma WHO grade; (B) PLA2G5 gene expression was associated with the histological type of glioma; (C)-(D) PLA2G5 gene expression was positively correlated with patient age in LGGs (P<0.0001) but not in HGGs (P=0.0767); (E)-(F) PLA2G5 gene expression was associated with radiotherapy of HGGs (P=0.0169), but not with chemotherapy (P=0.2771).
Abbreviations: TCGA, the Cancer Genome Atlas; LGG, low-grade glioma; HGG, high-grade glioma; GBM, glioblastoma.
Figure S3.
There was no correlation between the PLA2G5 gene expression level and gender in both LGGs (A) and HGGs (B).
Abbreviations: TCGA, the Cancer Genome Atlas; LGG, low-grade glioma; HGG, high-grade glioma.
PLA2G5 protein expression in glioma tumors and its association with the WHO grade
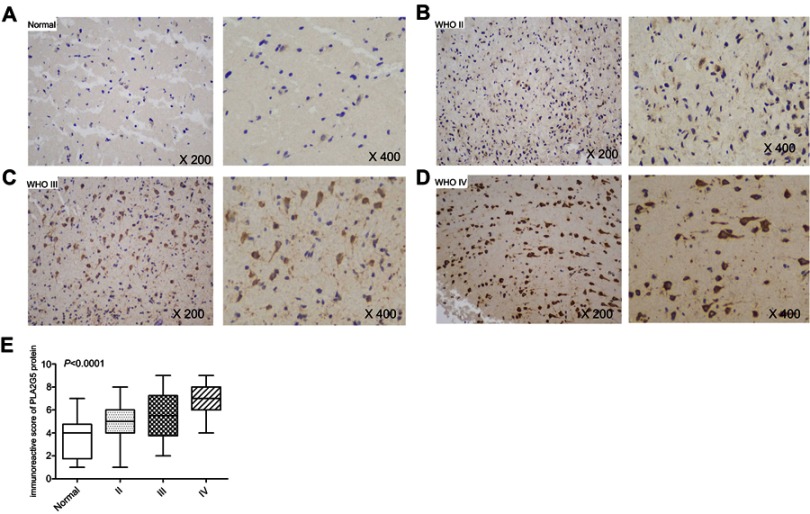
Using immunohistochemistry, we further evaluated the PLA2G5 protein expression levels in 82 glioma tissues obtained from Xiangya Hospital. We observed strong PLA2G5 immunostaining in the cytoplasm and/or nucleus of the tumor cells, but not in the normal brain tissues. Furthermore, the PLA2G5 protein levels were positively correlated with the WHO glioma grade (P<0.0001, Figure 2A–E). This was consistent with our results of PLA2G5 gene expression in data from TCGA.
Figure 2.
PLA2G5 protein expression in glioma examined by immunohistochemistry. (A)-(D) Immunohistochemical staining images of grade II, III and IV glioma tissues and non-tumor brain tissues (magnification ×200 and ×400); (E) The immunohistochemical score indicates that PLA2G5 protein expression was dependent on WHO grade.
Association of increased PLA2G5 gene expression with poor patient prognosis
As previously described, we assigned all patients to two groups on the basis of the median (low and high) PLA2G5 gene expression levels and analyzed the complete follow-up data for the 524 LGGs and 523 HGGs from TCGA. For the LGG cases, we found a significant statistical difference in overall survival between the two groups by Kaplan-Meier survival analysis. The median survival time of patients with high PLA2G5 gene expression levels was approximately 20.5 months, which was significantly shorter than the 26.2 months for patients with low PLA2G5 expression levels (log rank P=0.0002, Figure 3A). For the HGG cases, the median survival times of patients were not significantly different regardless of high or low PLA2G5 gene expression (11.9 and 12.6 months, respectively). However, patients with high PLA2G5 gene expression levels had lower long-term survival times than patients with low expression levels (P=0.002, Figure 3B).
Figure 3.

Kaplan-Meier survival analysis based on the median (low and high) PLA2G5 gene transcript expression in glioma. (A) High PLA2G5 gene expression indicated poor survival in LGGs (P=0.0002); (B) High PLA2G5 gene expression indicated poor survival in HGGs (P=0.002).
Abbreviations: TCGA, the Cancer Genome Atlas; LGG, low-grade glioma; HGG, high-grade glioma.
Verification of PLA2G5 gene expression as an independent factor affecting patient prognosis
Using univariate and multivariate Cox regression models, we analyzed the relationship between PLA2G5 gene expression and various clinicopathological factors and the prognosis of patients with gliomas. The age, WHO grade, IDH1 mutation status, and PLA2G5 gene expression were independent prognostic factors for patients with LGGs (Table 4). For patients with HGGs, the age, chemotherapy, radiotherapy, IDH1 mutation status, and PLA2G5 gene expression were the independent prognostic factors (Table 5). We concluded that PLA2G5 gene expression is an independent indicator of poor prognosis for both LGGs and HGGs (Tables 4 and 5).
Table 4.
Univariate and multivariate Cox regression analyses of PLA2G5 expression and clinicopathological features in affecting the OS of patients with LGG
| Variables | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Gender | 0.872 | 0.618–1.232 | 0.438 | 0.773 | 0.543–1.102 | 0.155 |
| Age | 2.914 | 2.001–4.245 | <0.001 | 2.774 | 1.857–4.142 | <0.001 |
| WHO Grade | 3.354 | 2.296–4.895 | <0.001 | 2.785 | 1.897–4.089 | <0.001 |
| IDH1 mutation | 0.294 | 0.209–0.414 | <0.001 | 0.362 | 0.252–0.520 | <0.001 |
| PLA2G5 expression | 1.936 | 1.363–2.749 | <0.001 | 1.572 | 1.088–2.274 | 0.016 |
Abbreviations: HR, hazard ratio; CI, confidence interval; LGG, low-grade glioma; IDH1, isocitrate dehydrogenase 1; OS, overall survival.
Table 5.
Univariate and multivariate Cox regression analyses of PLA2G5 expression and clinicopathological features in affecting the OS of patients with HGG
| Variables | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Gender | 0.851 | 0.700–1.034 | 0.105 | 0.895 | 0.735–1.091 | 0.272 |
| Age | 1.862 | 1.534–2.260 | <0.001 | 1.761 | 1.447–2.144 | <0.001 |
| Chemotherapy | 0.581 | 0.481–0.703 | <0.001 | 0.639 | 0.517–0.790 | <0.001 |
| Radiotherapy | 0.537 | 0.444–0.649 | <0.001 | 0.702 | 0.567–0.868 | 0.001 |
| IDH1 mutation | 0.308 | 0.164–0.577 | <0.001 | 0.345 | 0.182–0.653 | 0.001 |
| PLA2G5 expression | 1.360 | 1.122–1.649 | 0.002 | 1.258 | 1.032–1.533 | 0.023 |
Abbreviations: HR, hazard ratio; CI, confidence interval; HGG, high-grade glioma; IDH1, isocitrate dehydrogenase 1; OS, overall survival.
Possible mechanisms behind the poor prognosis in glioma patients with high PLA2G5 gene expression
We analyzed the PLA2G5 gene expression levels in gliomas with wild-type IDH1 and in those with mutated IDH1 and found that they were related. The PLA2G5 gene expression levels in LGGs carrying wild-type IDH1 were significantly higher than in those carrying mutated IDH1 (P<0.0001, Figure 4A), and the same results were found with the HGGs (P<0.0001, Figure 4B). These findings indicate that the poor prognosis associated with PLA2G5 gene expression in gliomas is possibly mediated by the IDH1 status. In addition, we analyzed the correlation of PLA2G5 gene expression with EMT marker gene expression (Table 6). In the HGG cohort, the expression levels of the PLA2G5 gene were significantly and positively correlated with those of the cadherin 2 (CDH2), vimentin (VIM), and fibronectin 1 (FN1) genes (P<0.0001, Figure 4C–E), whereas in the LGG cohort, they were positively correlated with the expression levels of the CDH2, VIM, Snail family transcriptional repressor 1 (SNAI1), SNAI2, Twist family BHLH transcription factor 1 (TWIST1), and TWIST2 genes (Table 6). Our findings suggest that PLA2G5 may play a role in the poor prognosis of gliomas by promoting EMT.
Figure 4.

The relationship between PLA2G5 gene expression and IDH1 status and EMT markers in glioma. (A)-(B) PLA2G5 expression in IDH1 wild-type cases was higher than that in IDH1 mutant cases in both LGGs (p<0.0001) and HGGs (p<0.0001); (C) The correlation between of PLA2G5 and FN1 in HGGs; (D) The correlation between PLA2G5 and VIM in HGGs; (E) The correlation between PLA2G5 and CDH2 in HGGs.
Abbreviations: TCGA, the Cancer Genome Atlas; LGG, low-grade glioma; HGG, high-grade glioma.
Table 6.
The correlation of PLA2G5 expression with the expression of EMT markers
| EMT markers | LGG | HGG | ||
|---|---|---|---|---|
| Pearson’s r | P-value | Pearson’s r | P-value | |
| CDH2 | 0.1933 | <0.0001 | 0.4545 | <0.0001(Figure4E) |
| VIM | 0.2577 | <0.0001 | 0.4052 | <0.0001(Figure4D) |
| FN1 | 0.07693 | 0.0785 | 0.2173 | <0.0001(Figure4C) |
| SNAI1 | 0.2903 | <0.0001 | −0.07391 | 0.0913 |
| SNAI2 | 0.1239 | 0.0045 | −0.02494 | 0.5693 |
| TWIST1 | 0.3166 | <0.0001 | −0.004144 | 0.9247 |
| TWIST2 | 0.2464 | <0.0001 | NA | - |
Abbreviation: EMT, epithelial-mesenchymal transition; LGG, low-grade glioma; HGG, high-grade glioma.
Discussion
Although there are reports of the expression of sPLA2 family proteins in various human cancers,9–12 their roles and clinical significance in gliomas remain unclear. In our study, we explored the relationship between gene expression of the sPLA2 family members and disease prognosis, using data of 1047 patients with gliomas obtained from TCGA. We demonstrated that PLA2G5 was the only sPLA2 associated with a poor prognosis in both LGGs and HGGs. Although some of the other sPLA2s (eg, PLA2G2A, PLA2G2E, PLA2G3, and PLA2G6) may also play an important role in LGGs or HGGs, further research is needed to elucidate what those roles are. In fact, the survival curves related to PLA2G5 gene expression suggest that its role may be more relevant in LGGs than in HGGs and at “early times.” In addition, we found that PLA2G5 gene expression depended on the tumor grade and was an independent prognostic indicator for gliomas. Using immunohistochemistry, we further explored the correlation between PLA2G5 protein expression and the clinicopathology of gliomas and confirmed that this sPLA2 member has clinical importance in these tumors.
As is well established, sPLA2s play a critical role in the generation of bioactive lipids, defense mechanisms, and cell signaling,5–7 and some of these proteins have been identified as potential pathological biomarkers13–16 or therapeutic targets.8,17–19 These enzymes can generate bioactive lipid mediators, including lysophosphatidic acid, linoleic acid, and arachidonic acid.9 These lipids have been reported to regulate cell proliferation, migration, and angiogenesis by promoting mitosis.6,17 These biological properties have been confirmed in related studies of colorectal cancer.9 As a member of the sPLA2 family, PLA2G5 also has these biological properties. In addition, Sadaria et al showed that the metastatic potential of esophageal cancer could be inhibited using an sPLA2 inhibitor, which decreased the expression of intercellular adhesion molecule-1 and attenuated the activation of nuclear factor-kappa beta.20 The studies by Wang et al and Dong et al suggested PLA2G2A as a metastasis-promoting gene and a cancer biomarker in lung cancer.21,22 PLA2G2A was found to regulate the Src/ERK/Akt/mTOR/p70S6K/rS6 pathway to promote the growth and migration of human astrocytoma cells.23 These studies indicate that high PLA2G5 gene expression may play a role in the development and progression of gliomas by promoting their proliferation, migration, and angiogenesis.
EMT is known to play an important role in cancer cell metastasis and the development of chemoresistance.24 The EMT process is reportedly also present in gliomas, and thus the study of the mechanism involved would be of great significance for exploring new treatments for these tumors.25 In our study, we observed significant and positive correlations between the gene expression levels of PLA2G5 and those of several EMT markers, in both the LGGs and HGGs. These results suggest that PLA2G5 may affect the prognosis of patients with glioma through its involvement with the EMT process. One possible mechanism is that PLA2G5 promotes EMT by generating arachidonic acid. Studies have shown that arachidonic acid promotes the EMT process by inducing the increased expression of VIM, N-cadherin, and matrix metallopeptidase 9 secretion, and lowering the level of E-cadherin junctions.26 In addition, arachidonic acid accelerates the EMT process by regulating CDH1, VIM, and TWIST1 gene expression.27 These findings are in agreement with our results.
IDH1 plays a crucial role in cellular metabolism. Mutations of this protein were first reported in 2008 by Parsons et al, and were subsequently found to occur in most of the LGGs and secondary adult glioblastomas.28,29 The mutated IDH1 is now one of the most widely known glioma biomarkers. The prognosis of patients with gliomas carrying the mutated IDH1 is significantly better those carrying the wild-type IDH1,29 as was confirmed by our findings (Figure S4). In our results, we found a correlation between the wild-type IDH1 status and high PLA2G5 gene expression in both LGGs and HGGs. This may indicate that PLA2G5 affects the prognosis of patients by affecting the metabolism of gliomas through its interaction with IDH1. However, the exact mechanism of how PLA2G5 affects the prognosis of these patients remains to be further studied.
Figure S4.

Patients with IDH1 mutation survived longer than those with wild-type IDH1 in both LGGs (A) and HGGs (B).
Abbreviations: TCGA, the Cancer Genome Atlas; LGG, low-grade glioma; HGG, high-grade glioma.
Conclusion
In conclusion, the present work shows that PLA2G5 could be a novel biomarker of prognosis for patients with gliomas. PLA2G5 is associated with both the glioma grade and survival time of patients and may be involved in the initiation and progression of these central nervous system tumors.
Acknowledgments
This study was supported by the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2014BAI04B01).
Disclosure
The authors report no conflicts of interest in this work.
Supplementary materials
References
- 1.Ostrom QT, Gittleman H, Xu J, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol. 2016. doi: 10.1093/neuonc/nov189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das S, Marsden PA. Angiogenesis in glioblastoma. N Engl J Med. 2013;369:1561–1563. doi: 10.1056/NEJMcibr1309402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walid M. Prognostic factors for long-term survival after glioblastoma. Perm J. 2008;12. doi: 10.7812/TPP/08-027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis DN, Perry A, Reifenberger G, et al. The 2016 World health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 5.Dennis EA, Deems RA, Yu L. Extracellular phospholipase A2. Adv Exp Med Biol. 1992;318:35–39. [DOI] [PubMed] [Google Scholar]
- 6.Murakami M, Lambeau G. Emerging roles of secreted phospholipase A2 enzymes: an update. Biochimie. 2013. doi: 10.1016/j.biochi.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 7.Menschikowski M, Hagelgans A, Siegert G. Secretory phospholipase A2 of group IIA: is it an offensive or a defensive player during atherosclerosis and other inflammatory diseases? Prostaglandins Other Lipid Mediat. 2006;79:1–33. doi: 10.1016/j.prostaglandins.2005.10.005 [DOI] [PubMed] [Google Scholar]
- 8.Cummings BS. Phospholipase A2 as targets for anti-cancer drugs. Biochem Pharmacol. 2007;74:949–959. doi: 10.1016/j.bcp.2007.04.021 [DOI] [PubMed] [Google Scholar]
- 9.Mounier CM, Wendum D, Greenspan E, Fléjou JF, Rosenberg DW, Lambeau G. Distinct expression pattern of the full set of secreted phospholipases A2 in human colorectal adenocarcinomas: SPLA2-III as a biomarker candidate. Br J Cancer. 2008;98:587–595. doi: 10.1038/sj.bjc.6604184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sved P, Scott KF, McLeod D, et al. Oncogenic action of secreted phospholipase A2 in prostate cancer. Cancer Res. 2004;64:6934–6940. doi: 10.1158/0008-5472.CAN-03-3018 [DOI] [PubMed] [Google Scholar]
- 11.Graff JR, Konicek BW, Chedid M, et al. Expression of group IIa secretory phospholipase A2 increases with prostate tumor grade. Clin Cancer Res. 2001;7(12):3857–3861. [PubMed] [Google Scholar]
- 12.Menschikowski M, Hagelgans A, Nacke B, et al. Epigenetic control of group V phospholipase A2 expression in human malignant cells. Tumor Biol. 2016;37:8097–8105. doi: 10.1007/s13277-015-4670-x [DOI] [PubMed] [Google Scholar]
- 13.Murakami M, Masuda S, Shimbara S, Ishikawa Y, Ishii T, Kudo I. Cellular distribution, post-translational modification, and tumorigenic potential of human group III secreted phospholipase A2. J Biol Chem. 2005;280:24987–24998. doi: 10.1074/jbc.M502088200 [DOI] [PubMed] [Google Scholar]
- 14.Smith MW, Yue ZN, Geiss GK, et al. Identification of novel tumor markers in hepatitis C Virus-associated hepatocellular carcinoma. Cancer Res. 2003;63(4):859–864. [PubMed] [Google Scholar]
- 15.Mallat Z, Steg PG, Benessiano J, et al. Circulating secretory phospholipase A2 activity predicts recurrent events in patients with severe acute coronary syndromes. J Am Coll Cardiol. 2005;46:1249–1257. doi: 10.1016/j.jacc.2005.06.056 [DOI] [PubMed] [Google Scholar]
- 16.Wootton PTE, Drenos F, Cooper JA, et al. Tagging-SNP haplotype analysis of the secretory PLA2IIa gene PLA2G2A shows strong association with serum levels of sPLA2IIa: results from the UDACS study. Hum Mol Genet. 2006;15:355–361. doi: 10.1093/hmg/ddi453 [DOI] [PubMed] [Google Scholar]
- 17.Laye JP, Gill JH. Phospholipase A2 expression in tumours: a target for therapeutic intervention? Drug Discov Today. 2003;8:710–716. doi: 10.1016/S1359-6446(03)02754-5 [DOI] [PubMed] [Google Scholar]
- 18.Rodrigues RS, Izidoro LF, de Oliveira RJJ, Sampaio SV, Soares AM, Rodrigues VM. Snake venom phospholipases A2: a new class of antitumor agents. Protein Pept Lett. 2009;16:894–898. doi: 10.2174/092986609788923266 [DOI] [PubMed] [Google Scholar]
- 19.Henderson WR, Chi EY, Bollinger JG, et al. Importance of group X–secreted phospholipase A2 in allergen-induced airway inflammation and remodeling in a mouse asthma model. J Exp Med. 2007;204:865–877. doi: 10.1084/jem.20070029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadaria MR, Meng X, Fullerton DA, et al. Secretory phospholipase A2 inhibition attenuates intercellular adhesion molecule-1 expression in human esophageal adenocarcinoma cells. Ann Thorac Surg. 2011;91:1539–1545. doi: 10.1016/j.athoracsur.2011.01.017 [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Hao FY, Wang JG, Xiao W. Group IIa secretory phospholipase A2 (sPLA2IIa) and progression in patients with lung cancer. Eur Rev Med Pharmacol Sci. 2014;18(18):2648–2654. [PubMed] [Google Scholar]
- 22.Dong Z, Meller J, Succop P, et al. Secretory phospholipase A2-IIa upregulates HER/HER2-elicited signaling in lung cancer cells. Int J Oncol. 2014;45(3):978–984. doi: 10.3892/ijo.2014.2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martín R, Cordova C, Gutiérrez B, Hernández M, Nieto ML. A dangerous liaison: leptin and sPLA2-IIA join forces to induce proliferation and migration of astrocytoma cells. PLoS One. 2017. doi: 10.1371/journal.pone.0170675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwatsuki M, Mimori K, Yokobori T, et al. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010;101:293–299. doi: 10.1111/j.1349-7006.2009.01419.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahlert UD, Nikkhah G, Maciaczyk J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 2013;331:131–138. doi: 10.1016/j.canlet.2012.12.010 [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Orozco R, Navarro-Tito N, Soto-Guzman A, Castro-Sanchez L, Perez Salazar E. Arachidonic acid promotes epithelial-to-mesenchymal-like transition in mammary epithelial cells MCF10A. Eur J Cell Biol. 2010;89:476–488. doi: 10.1016/j.ejcb.2009.12.005 [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Liu Y, Weng J, et al. Decrease of lactogenic hormones induce epithelial-mesenchymal transition via TGFβ1 and arachidonic acid during mammary gland involution. J Reprod Dev. 2017;63(3):325–332. doi: 10.1262/jrd.2016-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]