Abstract
Background/aims: MiR-216b and forkhead box M1 (FOXM1) were demonstrated to exert their biological effects on the development and progression of tumors. This study aimed to investigate the expression and role of miR-216b and FOXM1 in tissues and cell lines of non-small cell lung cancer (NSCLC).
Methods: The expressions of miR-216b and FOXM1 in NSCLC tissues and cells were detected by qRT-PCR and Western blot analysis. Cell proliferation was measured by CCK-8 assay. Cell migration and invasion were confirmed by Transwell assay. Finally, the bioinformatics and dual-luciferase reporter assay were conducted to validate the relationship of miR-216b and FOXM1.
Results: Compared with normal tissues and cells, the expression of miR-216b was obviously decreased in NSCLC tissues and cells. However, the expressions of FOXM1 mRNA and protein were significantly increased, and negatively correlated with the expression of miR-216b. Multivariate Cox’s regression analysis suggested that miR-216b or FOXM1 expression was an independent prognostic factor for patients with NSCLC. MiR-216b overexpression remarkably repressed cell proliferation, migration, invasion, and epithelial–mesenchymal transition (EMT) of NSCLC cells. The bioinformatics and dual-luciferase reporter assay validated that the 3ʹ-untranslated region (3ʹ-UTR) of FOXM1 mRNA was indeed a direct target of FOXM1. In vitro, overexpression of FOXM1 partially eliminated inhibitory effects of miR-216b on cell proliferation, migration, and invasion, whereas inhibition of FOXM1 contributed to inhibitory effects mediated by miR-216b.
Conclusion: MiR-216b inhibits cell proliferation, migration, invasion, and EMT by targeting the expression of FOXM1 in human NSCLC. These findings suggested a potential therapeutic role of miR-216b in patients of NSCLC.
Keywords: MiR-216b,FOXM1,NSCLC,suppressor
Introduction
Non-small cell lung cancer (NSCLC), including squamous cell carcinoma, adenocarcinoma, adenosquamous cell carcinoma and large cell carcinoma, accounts for approximately 80% cases in the worldwide.1–3 In spite of advancements in the early detection, clinical diagnosis, and treatment of NSCLC, the prognosis of patients with NSCLC remains unsatisfactory.4 High mortality of patients with NSCLC is mainly attributed to tumor heterogeneity, metastasis, and resistance to targeted therapies.5 Therefore, it is essential to investigate the molecular mechanisms of NSCLC cells.
MicroRNAs (miRNAs or miRs) play important roles in the development and progression of various cancers, including cell differentiation, proliferation, cell cycle, and metastasis.6,7 Since it was found that miRNAs were involved in chronic lymphocytic leukemia, more and more miRNAs were reported to be deregulated and correlated with tumorigenesis. As reported, deregulated expressions of miRNAs are associated with various cancers, including osteosarcoma, colorectal cancer, and lung cancer,8–10 which influence cancer initiation, maintenance, and metastatic progression by regulating the expressions of multiple target genes. Recently, some miRNAs have been identified to be deregulated in NSCLC samples compared with normal samples.9,10 However, the molecular mechanisms of miR-216b in the pathogenesis of NSCLC are unclear.
Forkhead box M1 (FOXM1), a member of the FOX family of transcription factors, plays a vital important role in cell growth, cell cycle, and metabolism.11–13 Aberrant expression of FOXM1 correlates with tumorigenesis and progression in several kinds of malignancies, such as cholangiocarcinoma, breast cancer, hepatocellular carcinoma, and gliomas.14 However, the role of FOXM1 in NSCLC is still unclear. In this study, we found that miR-216b expression was decreased in NSCLC specimens and inhibited the proliferative, migratory, and invasive properties of NSCLC cells by regulating the protein expression of EMT biomarkers and targeting the 3’-UTR of FOXM1. We proved miR-216b might be a potential therapeutic strategy for patients of NSCLC.
Materials and methods
Ethics statement
This study was approved by the Ethical Review Committee of Shandong Provincial Third Hospital. All biological samples were obtained with patients’ written informed consent.
Clinical NSCLC tissues
Between 2013 and 2015, we collected a total of 30 primary NSCLC and adjacent non-cancerous tissues from the Department of Surgery, Shandong Provincial Third Hospital and Shandong Cancer Hospital. All patients provided informed consent before surgery. All tissue specimens were snap-frozen immediately in liquid nitrogen. The patients did not undergo chemotherapy or radiotherapy before surgery. Experienced pathologists histologically determined both cancerous and adjacent non-tumor tissues. All collected tissue samples were rapidly snap-frozen in liquid nitrogen and stored in liquid nitrogen at ‒80°C.
Cell culture
The human NSCLC cell lines A549, H460, and H1299 were obtained from the American Type Culture Collection (Manassas, VA, USA), and were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA, USA), and HEK-293T cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen) at 37°C in 5% CO2.
Cell transfection
MiR-216b mimics and negative control miRNAs (miR-NC) were obtained from GeneCopoeia (Guangzhou, China). FOXM1 expression vector and FOXM1-specific siRNA (si-FOXM1) and negative control siRNAs (si-control) were purchased from Ruibo Biotechnology Co. (Guangzhou, China). The transfection of these vectors into A549, H460, H1299, or HEK-293T cells was performed in 6-well plates with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) based on the manufacturer’s instructions.
Total RNA extraction and reverse transcription-quantitative polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissue samples and cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). TaqMan® microRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) was adopted to determine miR-216b expression, with U6 serving as an internal control. For quantitative analysis of FOXM1 mRNA, reverse transcription was carried out using PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan). The qPCR was performed using SYBR® Premix Ex Taq (Takara Bio, Inc., Otsu, Japan) on Applied Biosystems 7500 Real-time PCR System (Applied Biosystems, CA, USA), with GAPDH as an internal control. All reactions were performed in triplicate and the relative expression of miR-216b and FOXM1 mRNA was calculated using the 2−∆∆Ct method.
Western blot
The proteins in the lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride membranes (Corning Incorporated). The proteins were first blocked using 5% non-fat dry milk for 1 hr at room temperature and then incubated with primary antibodies (Cell Signaling Technology, Beverly, MA, USA) at 4°C overnight. The membranes were washed with Tris-buffered saline Tween, and then incubated with secondary antibodies (Applygen) for 1 hr at room temperature. An electrochemiluminescence detection system was used to visualize the protein bands, and the protein expression levels were measured by Image-Pro Plus 6.0 software (Media Cybernetics, Rockville, MD, USA). GAPDH (Cell Signaling Technology, Beverly, MA, USA) was used as an internal control. The band densities were quantified by ImageQuant software (GE Healthcare, UK).
Cell proliferation assay
Cells were plated at 96-well plate with a density of 2×103 or 4×103 cells per well, CCK-8 was added and returned to incubation conditions for 2–4 hrs. Light absorbance at 450 nm was measured daily with a microplate reader. All experiments were triplicates.
Cell apoptosis
Cell apoptosis was examined using the Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (556547; BD Pharmingen, San Diego, CA, USA) according to the manufacturer’s instructions. Cells were re-suspended in 1X binding buffer solution (BD Pharmingen) with Annexin V-FITC and propidium iodide and incubated for 15 mins at room temperature in the dark. Apoptotic cells were analyzed using a BD Accuri C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) with BD Accuri C6 software 1.0 (BD Biosciences).
Wound healing assay
Cells were grown on plastic 6-well plates at the density of 5×105 cells per well and cultured for 12 hrs. Uniform wounds were scraped by a sterile pipette tip after transfection with miR-216b mimics or miR-NC for 24 hrs. The wound closure was observed by microscope and photographed at 0, and 24 hrs after scratching.
Transwell invasion assays
Invasion assays were conducted in Transwell chambers (Costar, Corning Inc., NY, USA) coated without or with Matrigel (BD Biosciences) on the upper surface of the 8-μm (pore size) membrane. Briefly, transfected cells were harvested, suspended in serum-free medium, and plated into the upper chamber for the migration or invasion assays, respectively, and media supplemented with 10% FBS were placed into the lower chamber. After 24-hr incubation, the cells that had migrated or invaded through the membrane to the lower surface were fixed, stained, and counted under an inverted microscope (Olympus, Tokyo, Japan).
Luciferase reporter assay
MiRwalk database (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) were used to screen for direct targets of miR-216b. The 1.8 kb full-length of FOXM1 3′-UTR was cloned into dual-luciferase reporter vector (Promega; Madison, WI, USA) and named wild-type (WT) 3′-UTR. Site-directed mutagenesis of the miR-216b binding site in the FOXM1 3′-UTR was carried out using the GeneTailor Site-Directed Mutagenesis System (Invitrogen) and named mutant (MUT) 3′-UTR. Cells were seeded in 96-well culture plates (Costar) and transfected with WT or MUT 3′-UTR construct along with miR-216b mimics in triplicate. Firefly and renilla luciferase activities were measured at 48 hrs after cotransfection with the Dual-Luciferase Reporter Assay kit (Promega, Madison, WI, USA).
Statistical analysis
All statistical analyses were performed using the SPSS 17.0 statistical software package (SPSS, Inc., Chicago, IL, USA). The means ±SD was calculated, and a two-tailed Student’s t-test was performed using the data analysis tools provided by the software. The overall survival was calculated as the period from surgery to death or last contact. A Kaplan–Meier survival curve was created and compared with the log-rank test results. The Cox proportional hazards regression model was used for univariate and multivariate analyses to explore the effects of clinicopathological variables, miR-216b, and FOXM1 on survival. In all cases, P ≤ 0.05 was considered to indicate significant difference.
Results
MiR-216b expression is reduced in NSCLC tissues and cell lines
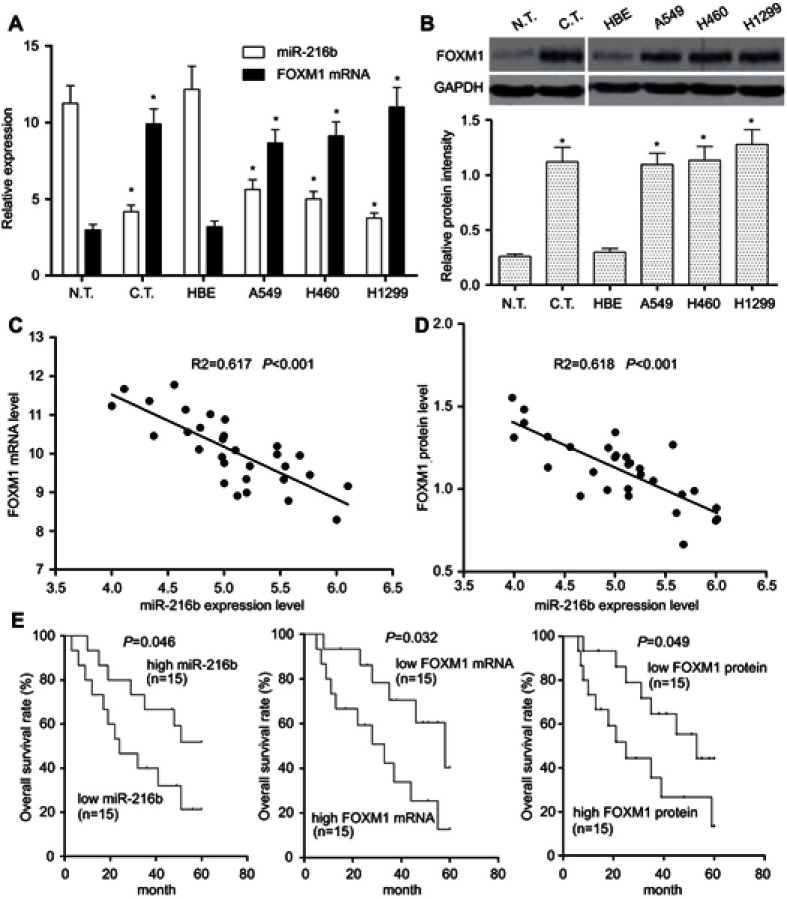
To confirm whether miR-216b was abnormally expressed in NSCLC tissues, 30 pairs of NSCLC tissues and adjacent normal tissues were collected to examine the relative expression of miR-216b by qRT-PCR. As shown in Figure 1A, compared with paired adjacent normal tissues, the expression of miR-216b was lower in human NSCLC tissues (P<0.01). Afterward, the expression of miR-216b was further examined in normal human bronchial epithelial (HBE) and NSCLC cells lines (A549, H460, and H1299) by qRT-PCR. As illustrated in Figure 1A, the expression of miR-216b was markedly decreased in NSCLC cell lines than that in HBE cells (P<0.01). These data indicated that miR-216b was reduced in NSCLC tissues and cell lines.
Figure 1.
Expression and association miR-216b and FOXM1 in NSCLC tissues and cells. (A) qRT-PCR was performed to detect the expressions of miR-216b and FOXM1 mRNA in 30 cases of NSCLC tissues and four kinds of cell lines (A549, H460, H1299, and normal human bronchial epithelial cell line HBE). (B) Western blot was used to detect FOXM1 protein level in NSCLC tissues and cell lines. Relative grayscale values of Western blotting were calculated to analyze the protein levels. U6 and GAPDH were used as internal controls, respectively. (C) Spearman’s correlation analysis was carried out to investigate the relationship between miR-216b and FOXM1 mRNA in 30 cases of NSCLC tissues. (D) Spearman’s correlation analysis was carried out to investigate the relationship between miR-216b and FOXM1 protein in 30 cases of NSCLC tissues. (E) Kaplan–Meier analysis of overall survival rates of patients with NSCLC according to miR-216b and FOXM1 expression status. Asterisks indicated statistically significant differences. Data were represented as means±SD. * P<0.01 vs control.
Abbreviations: FOXM1, forkhead box M1; NSCLC, non-small cell lung cancer
FOXM1 expression is increased in NSCLC tissues and cell lines
To investigate the role and mechanism of FOXM1 in NSCLC, the expressions of FOXM1 mRNA and protein in adjacent non-tumor tissues (n=30), and NSCLC tissues (n=30) were determined by qRT-PCR and Western blot, respectively. The results revealed that the expression levels of FOXM1 mRNA and protein were obviously increased in NSCLC tissues compared with those in the normal tissues (P<0.01; Figure 1A and B). Furthermore, we measured the expression levels of FOXM1 mRNA and protein in NSCLC cell lines, and identified that FOXM1 mRNA and protein were generally upregulated in A549, H460, and H1299 cell lines than those in normal lung epithelial cell HBE (P<0.01; Figure 1A and B).
MiR-216b negatively correlates with FOXM1 in NSCLC tissues
To further investigate the relationship between miR-216b and FOXM1 in NSCLC tissues, Spearman’s correlation analysis was carried out, and revealed that miR-216b expression level was negatively correlated with FOXM1 mRNA expression levels in NSCLC tissues (Figure 1C; R2=0.617, P<0.001). Likely, miR-216b expression level was negatively correlated with FOXM1 protein expression level in NSCLC tissues (Figure 1D; R2=0.618, P<0.001). These results suggested that downregulation of miR-216b may be associated with upregulation of FOXM1 in NSCLC.
Correlation of miR-216b or FOXM1 with the prognosis of patients with NSCLC
Here, we used the median value to divide samples into high and low expression group. Kaplan–Meier survival model showed that the overall survival duration of patients with NSCLC in the high miR-216b expression group was longer than those in the low miR-216b expression group (P=0.046; Figure 1E). Inversely, the overall survival duration of patients with low FOXM1 mRNA and protein expressions was longer than those with high FOXM1 mRNA and protein expressions (P=0.032, 0.049, respectively; Figure 1E). Using a Cox proportional hazard model, we analyzed the prognostic value of each variable on the overall survival of patients with NSCLC. Multivariate analysis validated that miR-216b or FOXM1 was an independent prognostic predictor of patients with NSCLC (Table 1).
Table 1.
Univariate and multivariate Cox regression analysis of overall survival
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Age (years, ≥60 vs <60) | 1.088 (0.546–1.877) | 0.565 | - | - |
| Gender (male vs female) | 1.281 (0.883–1.858) | 0.193 | - | - |
| Tumor size (cm, >3 vs ≤3 cm) | 1.495 (1.101–2.245) | 0.048* | - | - |
| Histologic subtype (squamous cell carcinoma vs adenocarcinoma) | 1.084 (0.764–1.539) | 0.650 | - | - |
| TNM stage (I–II vs III) | 0.399 (0.180–0.885) | 0.024* | 0.651 (0.437–0.972) | 0.036* |
| Lymph nodes metastasis (positive vs negative) | 2.259 (1.129–4.522) | 0.021* | 2.166 (1.289–3.639) | 0.004* |
| MiR-216b | 2.143 (1.069–4.297) | 0.032* | 2.561 (1.411–4.653) | 0.002* |
| FOXM1 mRNA | 2.066 (1.430–2.985) | <0.001* | 2.217 (1.494–3.291) | 0.013* |
| FOXM1 protein | 2.265 (0.907–5.658) | <0.001* | 2.298 (1.537–3.434) | 0.009* |
*Statistically significant result.
MiR-216b inhibits NSCLC cell proliferation in vitro
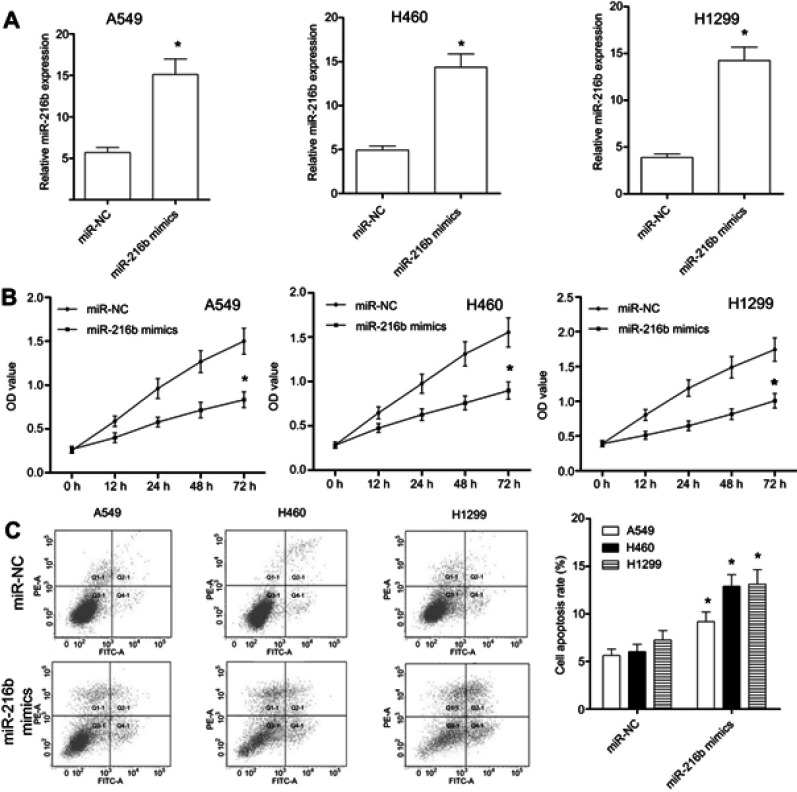
To determine the role of miR-216b in NSCLC cells, we overexpressed miR-216b in A549, H460, and H1299 cells by transfecting miR-216b mimics. At 48 hrs after transfection, miR-216b expression level in miR-216b mimics group was significantly increased as compared with miR-NC in A549, H460, and H1299 cells (Figure 2A). Then, CCK-8 assay was conducted to analyze the proliferation capacity of A549, H460, and H1299 cells at 24, 48, and 72 hrs post-transfection (Figure 2B). We found that overexpression of miR-216b obviously inhibited cell proliferation in A549, H460, and H1299 cells. To further investigate the effects of miR-216b on cell apoptosis, flow cytometry was applied. Our findings showed that, at 48 hrs after transfection with miR-216b mimics, miR-216b mimics significantly induced A549, H460, and H1299 cell apoptosis (early and late apoptosis) compared with miR-NC (Figure 2C).
Figure 2.
MiR-216b affects the proliferation and apoptosis of NSCLC cells. (A) qRT-PCR was performed to detect the expression level of miR-216b in A549, H460, and H1299 cells transfected with miR-216b mimics. (B) The proliferation of A549, H460, and H1299 cells was determined by CCK-8 assay. (C) The apoptosis of A549, H460, and H1299 cells was analyzed by flow cytometry. Q2 and Q3 indicated the early and late apoptosis, respectively. Asterisks indicated statistically significant differences. Data were represented as mean±SD. * P<0.01 vs control.
MiR-216b inhibits NSCLC cell migration, invasion, and EMT
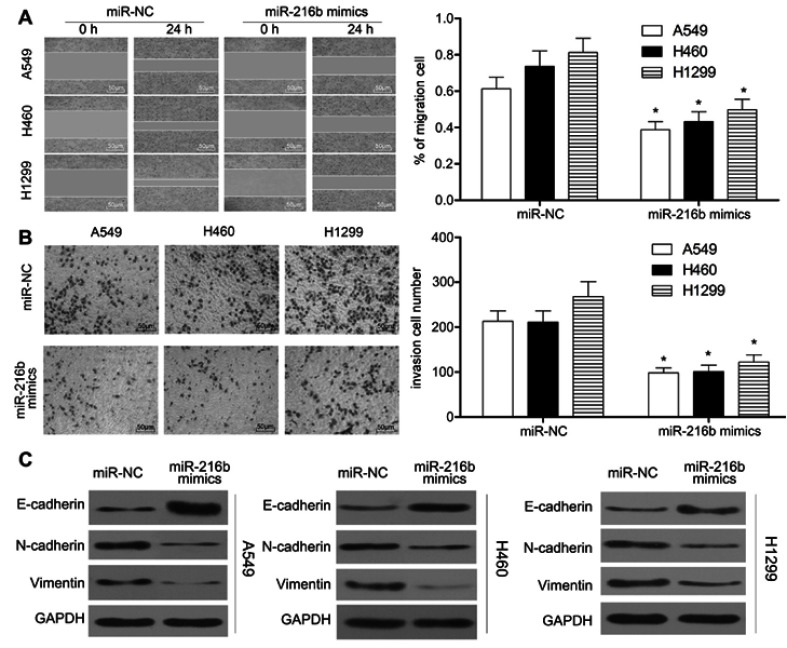
We further evaluated the effect of miR-216b on cell migration and invasion using wound healing and Transwell matrigel assay. Wound healing assay showed that A549, H460, and H1299 cells transfected with miR-216b mimics underwent slower scratch wound closure than miR-NC group (P<0.01, Figure 3A). Transwell matrigel assay demonstrated that miR-216b significantly dramatically inhibited the invasion capacity of A549, H460, and H1299 cells compared with miR-NC (P<0.01, Figure 3B). Furthermore, we investigated the relationship of miR-216b expression and EMT biomarkers in NSCLC cells using Western blot. The results revealed that overexpression of miR-216b increased the levels of epithelial marker E-cadherin, while reducing the levels of mesenchymal markers (N-cadherin and Vimentin) in A549, H460, and H1299 cells (P<0.05, Figure 3C). These results suggested that overexpression of miR-216b can regulate EMT of NSCLC cells.
Figure 3.
MiR-216b inhibits the migration, invasion, and EMT of NSCLC cells. (A) Wound healing assay was performed to observe the migration capacity of A549, H460, and H1299 cells. Transwell assay was performed to observe the invasion capacity of A549, H460, and H1299 cells. (C) EMT biomarkers in A549, H460, and H1299 cells were detected by Western blot. Asterisks indicated statistically significant differences. Data were represented as mean±SD. * P<0.01 vs control.
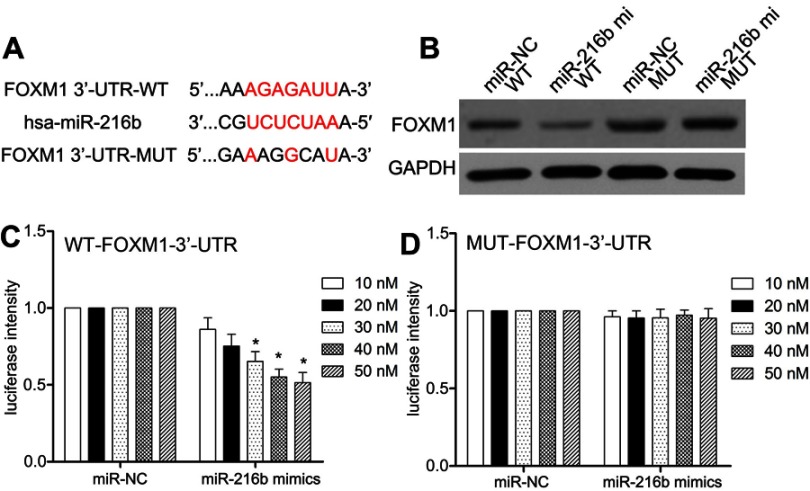
MiR-216b directly targets FOXM1 3′-UTR
Firstly, we predicted potential target genes by MicroRNA.org (http://www.microrna.org/) and found conserved putative miR-216b binding sites at the 3′-UTR of FOXM1 (Figure 4A). Next, we analyzed the FOXM1 protein expression by Western blot and semiquantitative analysis after transfecting HEK-293T cells with miR-216b mimics and pcDNA3.1-FOXM1 containing the WT or MUT 3′-UTR. Western blot analysis showed that the expression level of FOXM1 protein was downregulated in HEK-293T cells transfected with miR-216b mimics and pcDNA3.1-FOXM1 containing the WT 3′-UTR, while it was highly expressed in HEK-293T cells transfected with pcDNA3.1-FOXM1 lacking the 3′-UTR (P<0.01; Figure 4B). Subsequently, we performed dual-luciferase reporter assay to determine whether there was an interaction between FOXM1 mRNA and miR-216b. As shown in Figure 4C and D, miR-216b mimics decreased the luciferase activity of reporter containing the WT 3′-UTR of FOXM1, while the reporter containing the MUT 3′-UTR of FOXM1 did not have similar effects (P>0.05).
Figure 4.
MiR-216b directly targets FOXM1. (A) The schematic of the predicted base pairing between miR-216b and FOXM1 3ʹ-UTR. (B) The expression of FOXM1 protein in HEK-293T cells co-transfected with miR-216b mimics and FOXM1-3ʹ-UTR-WT/MUT was analyzed by Western blot. (C and D) The dual-luciferase activity assay was used to analyze the relative luciferase activities of FOXM1-3ʹ-UTR-WT and FOXM1-3ʹ-UTR-MUT in HEK-293T cells with miR-216b mimics or miR-NC. Each experiment was assayed in triplicate independently. Data were represented as mean±SD. * P<0.01 vs control.
Restoration of FOXM1 reverses the inhibitory effects of miR-216b
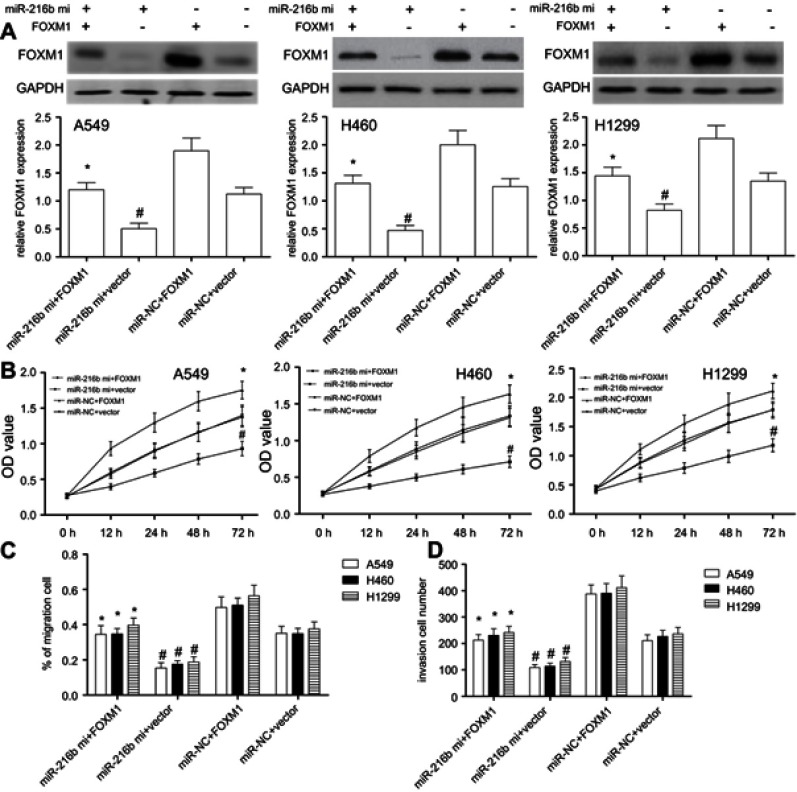
In view of the results above, we assumed that miR-216b suppressed cell proliferation, migration, and invasion via downregulating FOXM1. Due to the lower expression level of miR-216b, rescue experiments were performed by co-transfecting the miR-216b mimics with or without FOXM1 overexpression, followed by identification of cell proliferation, migration, and invasion of A549, H460, or H1299 cells. We found that FOXM1 protein was overexpressed in A549, H460, or H1299 cells with FOXM1 overexpression vector compared with their respective vector control (P<0.01; Figure 5A). In addition, A549, H460, or H1299 cells with miR-216b mimics and vector showed significantly decreased FOXM1 protein expression compared with miR-NC and vector group (P<0.01; Figure 5A), indicating that FOXM1 expression was affected by miR-216b. Then, CCK-8 assay showed overexpression of miR-216b alone greatly inhibited cell proliferation, while overexpression of FOXM1 alone notably promoted cell proliferation. Further, co-expression of FOXM1 with miR-216b rescued the inhibitory effects of miR-216b mimics on cell proliferation (P<0.01; Figure 5B). In addition, wound healing and invasion assay revealed overexpression of miR-216b alone greatly inhibited wound closure and invasion capacity, whereas overexpression of FOXM1 alone promoted cell migration and invasion; co-expression of FOXM1 with miR-216b rescued the inhibitory effects of miR-216b mimics on cell migration and invasion (P<0.01; Figure 5C and D).
Figure 5.
Restored FOXM1 expression reverses the inhibitory effects of miR-216b in NSCLC cells. A549, H460, and H1299 cell lines were co-transfected with miR-216b mimics and pcDNA3.1-FOXM1 without the 3′-UTR. FOXM1 expression was measured by Western blot for each group. (A) FOXM1 protein expression was analyzed by Western blot. (B) Cell proliferation was measured by CCK-8 assay. (C and D) The migration and invasion were analyzed by wound healing and Transwell invasion assay, respectively. Each experiment was assayed in triplicate independently. Data were represented as mean±SD. * P<0.01 vs miR-216b mimics+vector; # P<0.01 vs miR-NC+vector.
Silencing of FOXM1 enhances the inhibitory effects of miR-216b
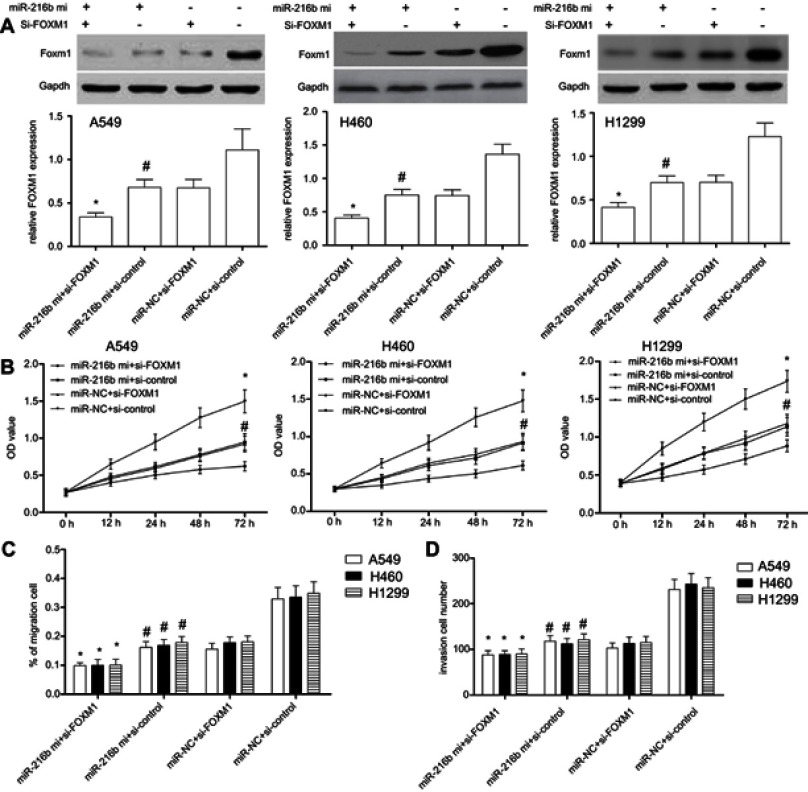
To finally determine whether FOXM1 was involved in miR-216b regulated cell proliferation, migration, and invasion, we used FOXM1 siRNAs to silence the expression of FOXM1. Western blot results showed that the expression of FOXM1 was significantly decreased following transfection with FOXM1 siRNAs in A549, H460, and H1299 cells (P<0.01; Figure 6A). In addition, A549, H460, or H1299 cells with miR-216b mimics and si-control showed significantly decreased FOXM1 protein expression compared with miR-NC and si-control group (P<0.01; Figure 6A), suggesting that FOXM1 expression was negatively regulated by miR-216b. Further, CCK-8 results showed that silencing of FOXM1 inhibited cell proliferation in A549, H460, and H1299 cells (P<0.01; Figure 6B). In addition, wound healing and invasion assay revealed co-transfection of miR-216b mimics and si-FOXM1 greatly enhanced the inhibitory effects of miR-216b mimics on cell migration and invasion compared with three other groups (P<0.01; Figure 6C and D).
Figure 6.
FOXM1 silencing enhances miR-216b-induced effects in NSCLC cells. (A) FOXM1 protein expression was analyzed by Western blot. (B) The proliferation of A549, H460, and H1299 cells co-transfected with FOXM1 siRNAs and/or miR-216b mimics was measured through CCK-8 assay. (C and D) The migration and invasion of A549, H460, and H1299 cells co-transfected with FOXM1 siRNAs and/or miR-216b mimics were analyzed by wound healing and Transwell invasion assay, respectively. Data were represented as mean±SD. * P < 0.01 vs miR-216b mimics+si-control; # P < 0.01 vs miR-NC+si-control.
Discussion
Emerging evidence revealed the function of miRNAs in the progression of NSCLC.15–18 For instance, miR-96 promotes invasion and metastasis by targeting GPC3 in NSCLC.15 miR-598 acts as a tumor suppressor, and negatively regulates DERL1 and EMT to suppress the invasion and migration in NSCLC.18 However, the role of miR-216b in NSCLC remains unclear. In previous studies, it has been reported that miR-216b functioned as a tumor suppressor gene by inhibiting proliferation and migration and invasion. In the present study, we found that miR-216b level was downregulated in NSCLC tissues and cell lines. Multivariate Cox’s regression analysis suggested that miR-216b expression was an independent prognostic factor for patients with NSCLC, suggesting that miR-216b was a potential biomarker and therapeutic target for patients with NSCLC.
Previous studies showed that miR-216b inhibited cell progression and promoted apoptosis by downregulating KRAS in pancreatic cancer and clear cell renal cell carcinoma,19,20 and JAK2/STAT3 signaling in colorectal cancer.21 MiR-216b also regulated proliferation and invasion of NSCLC by targeting SOX9.22 In this work, we found that FOXM1 level was upregulated in NSCLC tissues, and negatively correlated with the expression of miR-216b. According to current reports, Zheng et al found that miR-216b was downregulated in hepatocellular carcinoma and inhibited HepG2 cell growth by targeting FOXM1.23 Zhang et al demonstrated that miR-216b inhibited glioma cell migration and invasion through suppression of FOXM1.24 Here, we confirmed that FOXM1 expression was directly downregulated by miR-216b, and involved in the anti-oncogenicity of miR-216b. Further, restored FOXM1 expression level showed an earlier obvious rescue effect on cell viability, migration, and invasion, indicating a potential and more important role of FOXM1 on NSCLC cells.
FOXM1 belongs to the forkhead box superfamily of transcription factors, which is known as a regulator of cell proliferation, differentiation, apoptosis, and invasion.25,26 It has been revealed that high expression of FOXM1 positively correlated with tumor grade and poor survival of cancer patients.12 In colorectal cancer cells, dysregulation of miR-6868-5p/FOXM1 circuit contributes to colorectal cancer angiogenesis and promotes cancer cell invasion and migration by regulating heat shock protein family A member 5 transactivation.27 Therefore, our study indicated that miR-216b might form a complex regulation network via downregulating FOXM1 in NSCLC.
In summary, our data revealed that the expression level of miR-216b was reduced in NSCLC tissues and cell lines. MiR-216b functioned as a tumor suppressor, and inhibited cell proliferation migration, invasion, and EMT by directly downregulating FOXM1 in NSCLC. Therefore, regulation of the expressions of miR-216b and FOXM1 may be a promising therapeutic method for patients with NSCLC.
Acknowledgment
We are thankful to other members in our laboratory for their suggestions.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Watanabe T, Hosaka T, Ohmori-Matsuda K, et al. High preoperative plasma vasohibin-1 concentration predicts better prognosis in patients with non-small cell lung carcinoma. Health Sci Rep. 2018;1(6):e40. doi: 10.1002/hsr2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Li J, Wang X, et al. Promising clinical outcome with long term follow-up after body gamma knife stereotactic radiosurgery for patients with early stage non-small cell lung cancer. Front Oncol. 2018;8:618. doi: 10.3389/fonc.2018.00618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohan A, Harris K, Bowling MR, Brown C, Hohenforst-Schmidt W. Therapeutic bronchoscopy in the era of genotype directed lung cancer management. J Thorac Dis. 2018;10(11):6298–6309. doi: 10.21037/jtd.2018.08.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xi JJ, Yin JC, Wang L, Lu C-L, Wang Q, Jiang W. A surveillance method-oriented detection of post-operative spatial-temporal recurrence for non-small cell lung cancer. J Thorac Dis. 2018;10(11):6107–6117. doi: 10.21037/jtd.2018.10.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung YY, Shanmugam MK, Narula AS, et al. Oxymatrine attenuates tumor growth and deactivates STAT5 signaling in a lung cancer xenograft model. Cancers (Basel). 2019;11(1):pii: E49. doi: 10.3390/cancers11010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen KH, Hung JH, Chang CW, et al. Solasodine inhibits invasion of human lung cancer cell through downregulation of miR-21 and MMPs expression. Chem Biol Interact. 2017;268:129–135. doi: 10.1016/j.cbi.2017.03.005 [DOI] [PubMed] [Google Scholar]
- 8.Cai W, Xu Y, Zuo W, et al. MicroR-542-3p can mediate ILK and further inhibit cell proliferation, migration and invasion in osteosarcoma cells. Aging (Albany NY). 2019;11(1):18–32. doi: 10.18632/aging.101698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Wang J, Li N, et al. miR-34a increases the sensitivity of colorectal cancer cells to 5-fluorouracil in vitro and in vivo. Am J Cancer Res. 2018;8(2):280–290. [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez S, Risolino M, Mandia N, et al. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2015;34(25):3240–3250. doi: 10.1038/onc.2014.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang N, Pati D. Separase inhibitor sepin-1 inhibits FOXM1 expression and breast cancer cell growth. J Cancer Sci Ther. 2018;10(3):pii: 517. doi: 10.4172/1948-5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao X, Yu W, Qian J, et al. ADAM-17 is a poor prognostic indicator for patients with hilar cholangiocarcinoma and is regulated by FOXM1. BMC Cancer. 2018;18(1):570. doi: 10.1186/s12885-018-4242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogura S, Yoshida Y, Kurahashi T, et al. Targeting the mevalonate pathway is a novel therapeutic approach to inhibit oncogenic FOXM1 transcription factor in human hepatocellular carcinoma. Oncotarget. 2018;9(30):21022–21035. doi: 10.18632/oncotarget.24781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pal S, Kozono D, Yang X, et al. Dual HDAC and PI3K inhibition abrogates NFκB- and FOXM1-mediated DNA damage response to radiosensitize pediatric high-grade gliomas. Cancer Res. 2018;78(14):4007–4021. doi: 10.1158/0008-5472.CAN-17-3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fei X, Zhang J, Zhao Y, Sun M, Zhao H, Li S. miR-96 promotes invasion and metastasis by targeting GPC3 in non-small cell lung cancer cells. Oncol Lett. 2018;15(6):9081–9086. doi: 10.3892/ol.2018.8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao W, Sun Q, Yu Z, et al. MiR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer via PI3K/Akt pathway. Gene. 2018;670:31–37. doi: 10.1016/j.gene.2018.05.100 [DOI] [PubMed] [Google Scholar]
- 17.He J, Yu L, Wang CM, Zhou X-F. MiR-1275 promotes non-small cell lung cancer cell proliferation and metastasis by regulating LZTS3 expression. Eur Rev Med Pharmacol Sci. 2018;22(9):2680–2687. doi: 10.26355/eurrev_201805_14964 [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Wei K, Qin Z, et al. MiR-598 suppresses invasion and migration by negative regulation of derlin-1 and epithelial-mesenchymal transition in non-small cell lung cancer. Cell Physiol Biochem. 2018;47(1):245–256. doi: 10.1159/000489803 [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Chen W, Cai H, et al. MiR-216b inhibits pancreatic cancer cell progression and promotes apoptosis by down-regulating KRAS. Arch Med Sci. 2018;14(6):1321–1332. doi: 10.5114/aoms.2018.72564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Dong D, Jiang S, et al. miR-216b post-transcriptionally downregulates oncogene KRAS and inhibits cell proliferation and invasion in clear cell renal cell carcinoma. Cell Physiol Biochem. 2018;49(5):1755–1765. doi: 10.1159/000493621 [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Liu X, He B, et al. MiR-216b functions as a tumor suppressor by targeting HMGB1-mediated JAK2/STAT3 signaling way in colorectal cancer. Am J Cancer Res. 2017;7(10):2051–2069. [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Dong H, Dai H, Liu D, Wang Z. MicroRNA-216b regulated proliferation and invasion of non-small cell lung cancer by targeting SOX9. Oncol Lett. 2018;15(6):10077–10083. doi: 10.3892/ol.2018.8573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng WW, Zhou J, Zhang CH, Liu X-S. MicroRNA-216b is downregulated in hepatocellular carcinoma and inhibits HepG2 cell growth by targeting forkhead box protein M1. Eur Rev Med Pharmacol Sci. 2016;20(12):2541–2550. [PubMed] [Google Scholar]
- 24.Zhang T, Ma G, Zhang Y, Huo H, Zhao Y. miR-216b inhibits glioma cell migration and invasion through suppression of FOXM1. Oncol Rep. 2017;38(3):1751–1759. doi: 10.3892/or.2017.5824 [DOI] [PubMed] [Google Scholar]
- 25.Laissue P. The forkhead-box family of transcription factors: key molecular players in colorectal cancer pathogenesis. Mol Cancer. 2019;18(1):5. doi: 10.1186/s12943-019-0938-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao Y, Wang X, Jiang L, Shao X, Zhu X, He S. Prognostic and clinicopathological value of FOXM1 expression in colorectal cancer: a systematic review and meta-analysis. Medicine (Baltimore). 2018;97(52):e13899. doi: 10.1097/MD.0000000000013899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Wu M, Lei Z, et al. Dysregulation of miR-6868-5p/FOXM1 circuit contributes to colorectal cancer angiogenesis. J Exp Clin Cancer Res. 2018;37(1):292. doi: 10.1186/s13046-018-0970-5 [DOI] [PMC free article] [PubMed] [Google Scholar]