Abstract
Background: Although 80% of childhood acute lymphoblastic leukemia (ALL) cases are cured with current treatment protocols, exposure to chemotherapeutics or radiation therapy during a vulnerable period of child development has been associated with a high frequency of late adverse effects (LAE). Previous observations suggest important skeletal muscle size, density and function deficits in ALL survivors.
Purpose: Given that only a fraction of all patients will suffer from this particular complication, we investigated whether it could be predicted by genetic markers.
Patients and methods: We analysed associations between skeletal muscle force (Fmax) and power (Pmax) and germline genetic variants from 1039 genes derived through whole-exome sequencing. Top-ranking association signals retained after correction for multiple testing were confirmed through genotyping, and further analysed through stratified analyses and multivariate models.
Results: Our results show that skeletal muscle function deficit is associated with two common single nucleotide polymorphisms (SNPs) (rs2001616DUOX2, P=0.0002 (Pmax) and rs41270041ADAMTS4, P=0.02 (Fmax)) and two rare ones located in the ALOX15 gene (P=0.001 (Pmax)). These associations were further modulated by sex, body mass index and risk groups, which reflected glucocorticoid dose and radiation therapy (P≤0.02).
Conclusion: Occurrence of muscle function deficit in childhood ALL is thus strongly modulated by variations in the DUOX2, ADAMTS4 and ALOX15 genes, which could lead to personalized prevention strategies in childhood ALL survivors.
Keywords: acute lymphoblastic leukemia, late adverse effects, skeletal muscle deficit, genetic association study, whole exome sequencing
Introduction
Childhood acute lymphoblastic leukemia (ALL) accounts for nearly 25% of all paediatric malignancies.1,2 Optimization of multi-agent risk-adapted protocols has improved the 5-year survival rate, reaching by 2017 an estimated 90% in children less than 14 years old.3 This therapeutic success introduces new challenges, as 60% of childhood ALL survivors now experience a broad spectrum of late adverse effects (LAE) due to aggressive chemo- and radiation-therapy administered during critical child development stages.4,5 Childhood ALL survivors are more likely to develop a metabolic syndrome, cardiotoxicity, neuropsychological deficit and musculoskeletal morbidity — a cluster of interconnected factors that globally contribute to severe impairment of their quality of life.6–11
Clinical evaluation of childhood ALL survivors has revealed skeletal muscle size, density and function deficits as compared with healthy controls.12 Evidence suggests acute oxidative stress induced alteration of functional muscle mitochondria, motor neurons and satellite cells could contribute to these functional deficits.13 Muscle morbidity could lead to long-term mobility and autonomy deficits, such as impaired walking and gait or a diminished capacity to carry out daily activities.14–16 As lipid oxidation and glucose consumption is primarily performed in skeletal muscle, it is likely that treatment-induced muscle function deficit ultimately contributes to obesity, and cardiovascular or metabolic syndrome-related morbidity in ALL survivors.17
While molecular mechanisms involved in skeletal muscle atrophy remain unclear, sarcopenia as a response to glucocorticoid (GC) signalling, irrespective of cancer treatment, has been associated with various genetic markers and molecular pathways in the adult population. Evidence shows that GCs act as up-regulators of atrophy-related genes such as the Notch signalling pathway and constituents of the E3 ubiquitin ligase cascade.18 Likewise, mTOR (mechanistic target of rapamycin kinase) and AMPK (AMP-activated protein kinase) pathways are involved in GC-induced delays of muscle development in an animal model, while GCs increase adiposity in muscle tissue by affecting IL-4 (Interleukin 4) regulation of fibro/adipogenic progenitors.19,20
Genetic susceptibility to long-term skeletal muscle morbidity in childhood ALL survivors has yet to be documented. To address this issue, we present correlations between relevant clinical outcomes and genotypic profiling obtained through whole exome sequencing (WES) of ALL survivors.
Identification of genetic markers with predictive potential would allow further risk stratification of patients and personalisation of LAE prevention programs.
Methods
Study participants
The study group is composed of 242 survivors from the PETALE cohort, diagnosed with childhood ALL since 1987 at the Sainte-Justine University Hospital Center (SJUHC) in Montreal.10 Eligibility criteria include an age at diagnosis of 19 years or less, a 2-year adherence to a Dana-Farber Cancer Institute (DFCI)-ALL treatment protocol, Caucasian descent, and a post-remission delay of at least 5 years. Patients who relapsed, received hematopoietic stem cell transplants, suffered from concomitant syndromes, congenital bone disease, sickle-cell anemia or Legg-Calvé-Perthes disease, or were given osteotoxic drugs outside of their ALL treatment were considered non-eligible for the study. All patients fulfilling eligibility criteria and who consented to participate were included in this study.10 A written informed consent was obtained in accordance with the principles laid down by the Declaration of Helsinki. The SJUHC Institutional Review Board approved the research proposal.
Muscle force and power phenotypic data
Study participants underwent comprehensive clinical, biological and psychosocial investigations carried at the SJUHC and the Montreal Shriners Hospital for Children (MSHC), in the course of a day, between 2013-02-12 and 2016-05-19.
We used the Leonardo Mechanograph® Ground Reaction Force Plate (Novotec Medical GmbH, Pforzheim, Germany) as the measurement device for peak muscle force and power data acquisition. Data were analysed using the Leonardo Mechanography GRFP Research Edition® software, which measures body mass during quiet stance, and calculates the instantaneous vertical acceleration upon jumping, by recording the force as a function of time. The acceleration is time integrated to obtain the instantaneous vertical velocity. Peak power (Pmax) designates the highest value of force times velocity during the initial acceleration phase, while peak force (Fmax) refers to the maximum vertical force applied to the platform. Each participant performed a single two-legged jump and multiple one-legged hopping.21 Weight-adjusted Pmax and Fmax data were collected for each participant during a single two-leg jump and transformed into age- and gender-specific z-scores by comparison with a normal population.22 A z-score of −2 was used as a threshold to discriminate between affected and unaffected individuals.
Sequencing and candidate gene selection
A list of candidate genes was generated through a review of the literature. Based on the functional bone-muscle local unit theory, genes involved in both muscle and bone molecular pathways were targeted. Studied pathways include Wnt-β catenin signalling, osteoclastogenesis, osteoblast and osteocyte maturation, Vitamin D signalling, and pathways involved in glucocorticoid and methotrexate (MTX) action.23–30 Additional candidate genes were selected via a Gene Ontology Enrichment Analysis and Visualisation Tool (Gorilla).31 A total of 1039 candidate genes were chosen.
DNA was extracted from whole blood obtained during phenotyping evaluation of PETALE participants. Whole exomes were captured in solution with Agilent’s SureSelect Human All Exon 50Mb kits and sequenced on either Life Technologies SOLiD System 4.0 (mean coverage = 40X) or Illumina HiSeq 2500 platform (mean coverage = 113.1X) at SJUHC integrated clinical genomic centre in paediatrics. Reads were aligned to the hg19 reference genome using SOLiD LifeScope software for the SOLiD samples and BWA-MEM for the Illumina samples.32,33 PICARD was used to mark PCR duplicates and collect sequencing quality control metrics.34 Germline variant calling was performed using the Haplotype Caller and quality score recalibration was performed using Variant Recalibrator, both implemented in the Genome Analysis Tool Kit (GATK).35 Variants were selected based on the variant quality score (VQSR = PASS) and minimum depth of coverage (DP ≥ 10). WES data were available for 222 participants. Common and rare variants were defined respectively by their minor allele frequency (MAF) in the reference population as ≥5% and <5%, as based on 1000genome and ESP6500 (European American population) datasets.36 A functional annotation from ANNOVAR was used to apply filters to genotype data so that only variants with missense (at least potentially damaging, Polyphen2 scores ≥ 0.85 and Sift scores ≤ 0.1) or nonsense variations, or variations in splicing sites, were further considered.37
Of those, common variants either exceeding a missing rate of 20%, not in the Hardy-Weinberg Equilibrium (HWE, p<0.001), or having a pairwise linkage disequilibrium (LD, r2>0.8) were excluded, leaving 253 common SNPs located in 188 genes. The rare variants were analysed in an exploratory fashion using a collapsing approach whereby minor alleles of at least 2 potentially functional variants within one gene, each in HWE and with missing rate <20%, were analysed together, resulting in an analysis of 655 genes.
Statistical analysis
We used a whole genome data analysis toolset (PLINK©) to quantify associations between common SNPs and phenotypic datasets applying an allelic ratio test for dichotomous variables and a linear regression for continuous phenotypes. Quantitative datasets were tested for normality through an assessment of the distribution of residuals. For rare variant associations, the SKAT-O test (Optimal Sequence Kernel Association Test) in R Studio© software, which collapses variants within the same gene, was used.38 Resulting P-values were adjusted for multiple testing using a Benjamini-Hochberg correction method. A maximum false discovery rate (FDR) of 5% was chosen as a cut-off value for significance.
Selective genotyping of top-ranking SNPs (FDR<5%) following WES data analysis was carried out for all participants on the Sequenom platform at the McGill University and Génome Québec Innovation Centre. DNA samples from all 242 participants were available for genotyping. Depending on the studied SNP, genotyping was inconclusive for 3–14 participants. Concordance percentages between sequencing and genotyping results for top-ranking signals were recorded. Resulting genotypes and phenotypic data were analysed by IBM SPSS Statistics for Mac Version 24.0 (IBM Corp. Armonk, NY) with the chi-square or the Fisher exact tests, using appropriate genetic models. Rare SNPs were analysed through a collapsing approach with iterative exclusion of a single SNP per iteration, allowing weighting the SNP contribution to association signals.
Logistic regression models (in IBM SPSS) were used to assess an influence of genotypes on studied outcomes in the presence of covariables that significantly correlated with studied phenotypes and included sex, risk status either high (HR) or standard (SR), and body mass index (BMI). BMI was included as a cofactor as adipose tissue accumulation in obese individuals is thought to interfere with muscle function through aberrant adipokines production.39 A participant BMI was classified as normal (BMI ≤ 24.9 kg/m2), overweight (25 kg/m2 ≤ BMI < 30 kg/m2), or obese (BMI ≥ 30 kg/m2), in accordance with the adult classification scale.
Due to the DUOX2 gene role in thyroid hormone synthesis, the polymorphism rs2001616 located in DUOX2, for which significant associations with musculoskeletal deficit were found, was analysed for a correlation with TSH and T4 levels available for 235 and 231 PETALE participants, respectively.
To allow better characterisation of the impact of genotypes in relation to Fmax and Pmax, stratified analyses were also performed according to sex, risk group, and BMI at visit (obese vs non-obese).
Results
Cohort characteristics
Fmax measurements were available for 215 participants (Table 1), of whom 40% had a z-score below −1 while 13% had a z-score ≤ −2. As for Pmax, results were available for 226 participants: 55% and 27% with z-scores ≤ −1 and ≤−2, respectively. Thus, the PETALE cohort is more affected in terms of muscle morbidity than the normal population (P<10−4).
Table 1.
Cohort characteristics and distribution of study outcomes
| Category | Overall count (%) | |
|---|---|---|
| Sex | Female | 124 (51.2) |
| Male | 118 (48.8) | |
| Age at PETALE investigation visit (years) | Median (range) | 21.92 (9 - 41) |
| Age at Dx (years) | Median (range) | 4.00 (0-18) |
| Time since end of Tx (years) | Median (range) | 13.06 (4-26) |
| Risk level | Standard | 111 (45.9) |
| High | 131 (54.1) | |
| DFCI Protocol | DFCI 87-01 | 20 (8.3) |
| DFCI 91-01 | 48 (19.8) | |
| DFCI 95-01 | 71 (29.3) | |
| DFCI 2000-01 | 77 (31.8) | |
| DFCI 2005-01 | 26 (10.7) | |
| MTX Cumulative (mg/m2) | Median (range) | 6723 (1004-12999) |
| GC Cumulative dose (Pred. Eq. mg/m2) | Median (range) | 9025(4078-30210) |
| CRT | No | 99 (40.9) |
| Yes | 143 (59.1) | |
| BMI category at visita | Normal | 163 (67.4) |
| Overweight | 46 (19.0) | |
| Obese | 33 (13.6) | |
| Fmax | Average Z-score (range) | -0.86 (-5.64 - 2.03) |
| Z-score ≤ -1 SD | 96 (39.7) | |
| Z-score ≤ -2 SDs | 32 (13.2) | |
| Missing | 27 (11.2) | |
| Pmax | Average Z-score (range) | -1.45 (-7.30 - 1.36) |
| Z-score ≤ -1SD | 133 (55.0) | |
| Z-score ≤ -2 SDs | 65 (26.9) | |
| Missing | 16 (6.6) |
Notes: a Normal ≤ 24.9 kg/m2; 25 kg/m2 ≤ overweight <30 kg/m2; obese ≥ 30 kg/m2.
Abbreviations: Dx, diagnosis; Tx, treatment; DFCI, Dana-Farber Cancer Institute; CRT, cranial radiation therapy; BMI, body mass index; MTX, methotrexate; GC, glucocorticoids.
Association study with WES data
Association analyses were performed between functionally predicted common and rare variants derived from selected candidate genes obtained from WES data. Among common SNPs, rs41270041 located in the ADAMTS4 gene, was positively associated with Fmax z-scores ≤ −2 (P=10−4), whereas rs2001616 in the DUOX2 gene was significantly associated with Pmax, analysed both as a categorical (z-score ≤ −2) and continuous variable (P=10−5 and P=4.6×10−5, respectively. Table S1).
Table S1.
Top-ranking common SNPs, WES data
| SNP | rs41270041 | rs2001616 | |
|---|---|---|---|
| Gene | ADAMTS4 | DUOX2 | |
| Locus | chr1: 161161284 | chr15: 45404066 | |
| Genomic Location | Exon | Exon | |
| Major allele > minor allele | G>C | A>G | |
| MAF (%) | 8.6 | 15.5 | |
| Role of protein connected with muscle metabolism | Degradation of aggrecan proteoglycan | Role in thyroid hormone synthesis | |
| Associated Phenotype | Fmax (Z-score ≤ −2 SDs) |
Pmax (Z-score ≤ −2 SDs) |
Pmax (Z-score) |
| P-value | 1.19×10−4 | 1.02×10−5 | 4.63×10−5 |
| Allelic Ratio | Affected:12/46 | Affected: 33/83 | |
| Unaffected:19/309 | Unaffected: 24/214 | ||
| Allelic OR (95%-CI) | 4.24 (1.93–9.31) |
3.55 (1.98–6.36) |
NA |
Abbreviations: MAF, minor-allele frequency; OR, Odds ratio; CI, confidence interval.
Rare-SNP analyses identified top-ranking ALOX15 (P=5.7×10−6) gene, associated with continuous Pmax z-scores. The signal originated from three collapsed rare variants in the ALOX15 gene (Table S2).
Table S2.
Top-ranking rare SNPs, WES data
| Gene | ALOX15 | ||
|---|---|---|---|
| SNPs | |||
| rs34210653 | rs11568142 | rs113604586 | |
| MAF (%) | 1.61 | 0.78 | 0.23 |
| Major allele > minor allele | G > A | C > G | T > C |
| Genomic location | Exon | ||
| Gene locus | 17p13.2 | ||
| Role of protein | Arachidonic acid metabolism pathway | ||
| Associated phenotype | Pmax (Z-score) | ||
| P -value | 5.7x10−6 | ||
|
Participants with > 1 minor allele |
9 (4.05%) | ||
Abbreviation: MAF; minor-allele frequency.
Confirmation by genotyping and association analyses of obtained genotypes
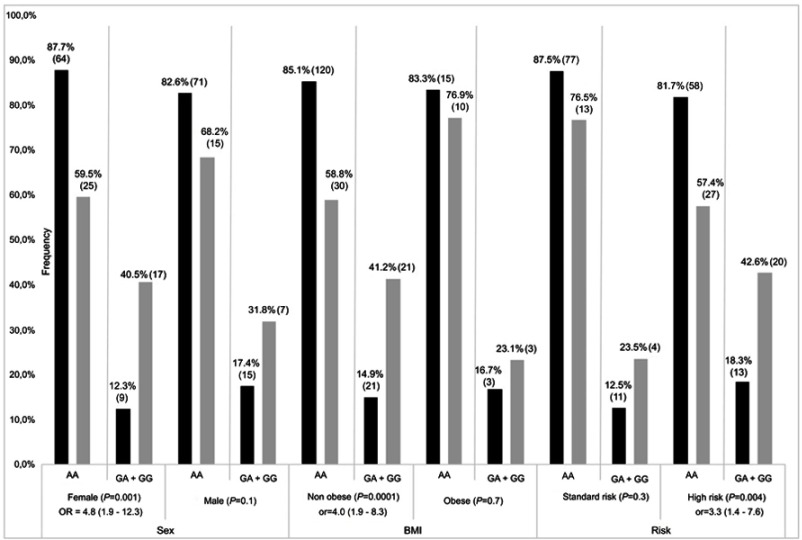
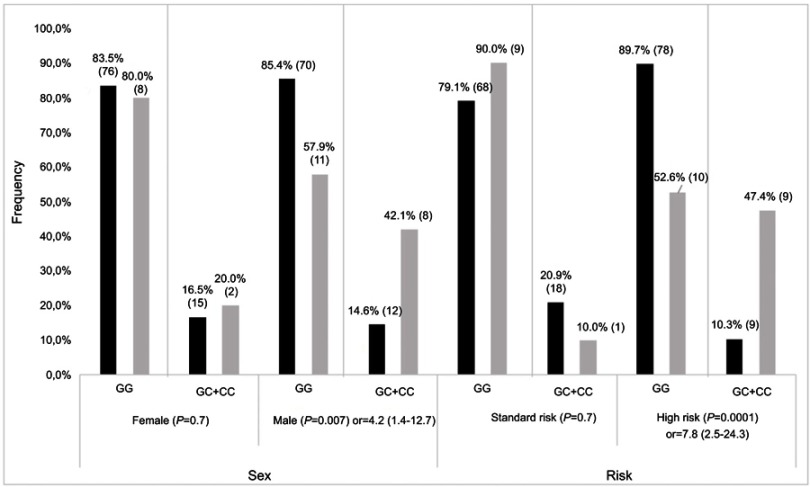
Participants were screened for top-ranking SNPs, including 2 common SNPs (rs41270041 in ADAMTS4 and rs2001616 in DUOX2) and 3 rare variants (rs113604586, rs34210653 and rs11568142) in ALOX15. Concordance between WES and genotyping data ranged from 92% to 95%. Given that similar results were obtained in association analyses for continuous Pmax z-scores and z-scores ≤ −2, only the latter are presented. The rs2001616 in the DUOX2 gene was associated with Pmax according to a dominant model of inheritance (P=2×10−4, Table 2). Carriers of a minor G allele were more frequent among participants with a z-score ≤ −2 as compared with those carrying the AA genotype (Odds ratio (OR)=3.4, 95% confidence interval (CI)=1.7–6.6). Likewise, the minor C allele of the rs41270041 in ADAMTS4 was associated with Fmax (z-score ≤ −2) according to a dominant model (OR=2.8, 95% CI, 1.2–6.8, P=0.02, Table 2). We next explored whether this association is influenced by clinical and treatment characteristics. Sex, prognostic risk groups, cranial radiation therapy (CRT), BMI, as well as cumulative GC and MTX dose were all significantly associated with Pmax Z score ≤ −2 (P≤0.02, Table S3). Pmax Z score reduction was seen more frequently in female survivors, in obese subjects and participants who were treated according to the high risk protocol. A significant association was noted as well with cumulative MTX and GC doses and with CRT. This reflected more frequent CRT, higher GC doses, but lower MTX doses administered to patients assigned to the high risk group (P<0.00001, Table S4). No significant association with demographics or treatment characteristics was found for Fmax. A logistic regression model was thus derived only for DUOX2 rs2001616, which remained significant in the multivariate model (P=0.003, Table 3) that included, besides genotype, sex, BMI at visit, and risk status (as a surrogate for CRT and higher GC doses). Subsequent stratification analyses were performed and showed that the association of both the DUOX2 rs2001616 and ADAMTS4 rs41270041 polymorphisms with phenotypes was modulated by demographics and treatment characteristics. The association signal for DUOX2 rs2001616 was limited to females (P=0.001), non-obese participants (P=0.0001) and patients assigned to the HR group (P=0.004) (Figure 1), whereas that of the rs41270041 in ADAMTS4 was more apparent among males (P=0.02) and participants that received the HR treatment (P=0.0001) (Figure 2). Given the strong correlation between HR and exposure to CRT (Table S4), as well as with higher GC cumulative doses, the similar modulation of the effect is seen when CRT or GC doses are used as stratification variables (data not shown).
Table 2.
Top-ranking common SNPs, genotyping data, and associated phenotypes
| PHENOTYPE | GENE_SNP | COMPLICATION | MODEL | COMPLICATION | OR (95% - CI) | P VALUE | ||
|---|---|---|---|---|---|---|---|---|
| Unaffected | Affected | Unaffected | Affected | |||||
| Pmax (Z≤-2) |
rs2001616DUOX2 A > G |
|||||||
| AA | 135 (84.9%) | 40 (62.5%) | AA | 135 (84.9%) | 40 (62.5%) | 1 | 0.0002 | |
| AG | 20 (12.6%) | 23 (35.9%) | AG + GG | 24 (15.1%) | 24 (37.5%) | 3.4 (1.7 - 6.6) | ||
| GG | 4 (2.5%) | 1 (1.6%) | ||||||
| Fmax (Z≤-2) |
rs41270041ADAMTS4 G > C |
|||||||
| GG | 146 (84.4%) | 19 (65.5%) | GG | 146 (84.4%) | 19 (65.5%) | 1 | 0.015 | |
| GC | 26 (15.0%) | 10 (34.5%) | GC + CC | 27 (15.6%) | 10 (34.5%) | 2.8 (1.2 - 6.8) | ||
| CC | 1 (0,6%) | 0 | ||||||
Abbreviations: OR; odds ratio, CI; confidence interval.
Table S3.
Co-variable association with Pmax (Z-score ≤ −2)
| Category | Group | Affected (%) | Unaffected (%) | P-value | OR (95%-CI) |
|---|---|---|---|---|---|
| Sex | Male | 22 (20.4) | 86 (79.6) | 0.008 | 2.2 (1.2–4.1) |
| Female | 43 (33.9) | 75 (63.6) | |||
| Risk level | Standard | 17 (16.2) | 88 (83.8) | 0.0001 | 3.4 (1.8–6.4) |
| High | 48 (39.7) | 73 (60.3) | |||
| MTX cumulative dose a (mg/m2) | ≤6723 | 40 (35.1) | 74 (64.9) | 0.03 | 0.5 (0.3–0.9) |
| >6723 | 24 (21.8) | 86 (78.2) | |||
| GC cumulative dose (pred. Eq. mg/m2) | ≤9025 | 23 (20.4) | 90 (79.6) | 0.006 | 2.3 (1.3–4.2) |
| >9025 | 41 (36.9) | 70 (63.1) | |||
| CRT | No | 15 (16.3) | 77 (83.7) | 0.001 | 3.1 (1.6–5.9) |
| Yes | 50 (37.3) | 84 (62.7) | |||
| BMI category at visit | Normal | 31 (20.7) | 119 (79.3) | 0.001 | |
| Overweight | 21 (47.7) | 23 (52.3) | |||
| Obese | 13 (40.6) | 19 (59.4) |
Note: aCumulative MTX and CS doses are recoded as below and above median.
Abbreviations: CRT, cranial radiation therapy; MTX, methotrexate; GC, glucocorticoids; BMI, body mass index.
Table S4.
Association of risk groups with CRT and cumulative MTX and GC dose
| Risk | P-value | |||
|---|---|---|---|---|
| Standard | High | |||
| CRT | No | 90 (81.1%) | 9 (6.9%) | 1.3×10−31 |
| Yes | 21 (18.9%) | 122 (93.1%) | ||
| MTX Cumulative1 (mg/m2) | ≤6723 | 34 (30.6%) | 86(66.4%) | 2.6×10−8 |
| >6723 | 77(69.4%) | 43 (33.6%) | ||
| GC Cumulative (mg/m2) | ≤9025 | 106 (95.5%) | 14 (10.9%) | 4.5×10−39 |
| >9025 | 5 (4.5%) | 115 (89.1%) | ||
Note: 1Cumulative MTX and GC doses are recoded as above and below median.
Abbreviations: CRT, cranial radiation therapy; MTX, methotrexate; GC, glucocorticoids.
Table 3.
Regression model
| Phenotype | Co-variable | B | P-value | OR (95%-CI) |
|---|---|---|---|---|
| Pmax (Z≤−2) | DUOX2 | 1.08 | 0.003 | 2.9 (1.4–6.1) |
| Sex | 0.87 | 0.009 | 2.4 (1.2–4.6) | |
| BMI at visit | 0.61 | 0.004 | 1.8 (1.2–2.8) | |
| Risk | 1.1 | 0.001 | 3.0 (1.5–5.9) |
Notes: DUOX2 gene: dominant model (AG and GG vs AA). Sex: Females are compared to males; BMI: participants given normal (≤24.9 kg/m2), overweight (≥25 kg/m2, <30 kg/m2), or obese status (≥30 kg/m2); risk: standard or high.
Figure 1.
Stratification result of association for DUOX2 rs2001616. Frequencies of AA and AG+GG genotypes (gray bars) and unaffected (black bars) individuals, as defined by Pmax Z≤−2 SD and Z>−2 SD, respectively. P-value with OR (odds radio) and 95% CI (confidence interval) is presented for each comparison. Number of patients in each genotype group is indicated in brackets with percentages. BMI; non obese: <30 kg/m2, obese: ≥30 kg/m2.
Figure 2.
Stratification result of association for ADAMTS rs4127041. Frequencies of GG and GC+CC genotypes for affected (grey bars) and unaffected (black bars) individuals, as defined by Fmax Z≤−2 SD and Z>−2SD, respectively. P-value with OR (odds radio) and 95% CI (confidence interval) is presented for each comparison. Number of patients in each genotype group is indicated on each bar.
An analysis of the four different combinations of the rs34210653, rs113604586 and rs11568142 rare variants in the ALOX15 gene revealed that the association signal resulted from the rs113604586 and rs11568142 combination, with a lowest P-value of 0.001 (Table 4). The low frequency of rare variant carriers precluded a stratified analysis.
Table 4.
Top-ranking rare SNPs, genotyping data, and associated phenotypes
| Phenotype | Pmaxa | OR (95%-CI) | P-value | ||
|---|---|---|---|---|---|
| Unaffected | Affected | ||||
| Genotype | ALOX15 (rs113604586_rs11568142) | ||||
| No minor allele ≥1 minor allele |
150 (100%) 0 |
57 (90.5%) 6 (9.5%) |
- | 0.001 | |
Note: aAffected individuals have the Z score for Pmax ≤ −2.
Abbreviations: OR, odds ratio; CI, confidence interval is not calculated due to zero observation in one of cells.
Discussion
Studies exploring the role of a genetic component in LAE development in childhood ALL survivors are limited, and this study is the first to focus on skeletal muscle function deficit through a genomic approach.40,41
Our analysis revealed an association of rs2001616 in the DUOX2 gene with skeletal muscle Pmax deficit. Survivors carrying a minor allele had a higher probability of Pmax reduction than other genotype groups. DUOX2 (Dual oxidase 2) is involved in muscle and bone metabolisms through its role in thyroid hormone synthesis. The encoded protein acts as a functional constituent of a peroxide generating system located at the apical membrane of thyroid follicular cells.42 Several DUOX2 polymorphisms have been associated with congenital hypothyroïdia, and inactivating mutations of this gene have recently been shown to play a causal role in very early onset inflammatory bowel diseases.43,44
Thyrotoxicosis is known to be a risk factor for hypercalcemia, osteoporosis and increased bone turnover.45 A study investigating correlations between thyroid hormone levels and skeletal muscle regeneration found that high serum T3 and T4 levels could interfere with the inflammatory process following muscle injuries through up-regulation of macrophage accumulation and cell infiltration.46 The detected effect of rs2001616 polymorphisms could therefore be due to up-regulation of thyroid hormone production in carriers of the variant allele.
We thus evaluated correlations between the DUOX2 rs2001616 variant and thyroid function in study participants. TSH and free thyroxine (T4) levels were available for 232 and 229 participants with genotype data. A significant association (P=0.02) was found between free T4 values and the rs2001616 polymorphism, carriers of a variant allele having a higher median T4 level (12.8 vs 12.2 μmol/L). Associations with TSH were statistically insignificant. The clinical significance of this result is limited, however, as none of the study participants was diagnosed with dysthyroïdia as based on TSH and T4 levels following their ALL treatment. Furthermore, no significant association was found between thyroid hormone levels and skeletal muscle function phenotypes.
An association with altered skeletal muscle function (Fmax) was also demonstrated with rs41270041, located in the ADAMTS4 gene. ADAMTS4 (ADAM metallopeptidase with thrombospondin type 1 motif 4) is a protease-coding gene responsible for the degradation of aggrecan, a cartilage-specific proteoglycan core protein.47 This gene has been shown to contribute to osteoarthritic disorders and tendon injuries.48,49 An increased susceptibility to tendinopathies or painful joints could lead to limb immobilization and thus alter muscle mass through lower muscle strain. The ADAMTS4 protein has also been shown to be a risk factor for atherosclerosis plaque vulnerability in mice models.50
ALOX15 codes for arachidonate 15-lipoxygenase, a pro-inflammatory protein involved in the arachidonic acid metabolism pathway. It has been reported to play a role in inflammatory response and immunity, and in ischemic heart disease.51,52 Studies carried on mice models suggest that ALOX15 could also affect bone mass through its negative regulation of maximum bone mineral density.53 Associations have been detected between ALOX15 variants and bone micro-architecture in young women, while other variants in this gene seem to predispose to fractures in older women.54 We detected a significant association for two rare SNPs included in the ALOX15 locus, justifying further study of this gene in related contexts.
A likely explanation why top-ranking SNPs did not all correlate with both Fmax and Pmax phenotypes lies in the distinct skeletal muscle functions reflected by these two measurements. Muscle force represents the ability to accelerate a given mass (Force = Mass × Acceleration) whereas power is the ability to produce force at a given velocity (Power = Force × Velocity). Previous studies have demonstrated that peak power, but not peak force, depends on muscle fiber type.55 On the other hand, evidence suggests a strong correlation between peak muscle force and muscle size as defined by cross-sectional area measurements.56,57 We can thus hypothesize that the ADAMTS4 gene could modulate molecular pathways involved with skeletal muscle size, while ALOX15 and DUOX2 would affect muscle fiber contractile properties.
Associations of skeletal muscle deficits with common variants were more significant in groups known to suffer more from treatment-related complications (e.g. CRT, HR, and high GC doses groups). In fact, various studies demonstrated that muscle function is substantially more degraded in patients exposed to a highly aggressive treatment regimen, including higher glucocorticoid doses and CRT, which may explain the interaction with risk status noted for associated SNPs of DUOX2 and ADAMTS4 genes.58,59
A study carried in the St Jude Lifetime Cohort demonstrated severe anterior pituitary deficit related to CRT exposure in childhood cancer survivors.60 However, none of the patients in the PETALE cohort were diagnosed with secondary clinical hypopituitarism, which may be due to lower overall CRT doses received. It remains possible that a higher proportion of participants in the CRT-positive group suffered from CRT-induced skeletal muscle deficit due to alternative mechanisms involved with neuroendocrine signalling. Indeed, low dose ionising radiation therapy has been demonstrated to rapidly alter synaptic and mitochondrial signalling pathways in murine models, which could account for poorer outcomes, particularly apparent in patients with a given genetic background.61
The impact of obesity seems to overcome genetic predisposition, as a higher risk of skeletal muscle function deficit in the case of the DUOX2 SNP was seen only among non-obese patients. Studies investigating muscle pathologies in obese individuals have found that dysregulated adipokines production in hypertrophied adipose tissue decreased the expression of contractile proteins in myotubes, pointing towards an alternative mechanism for poorer skeletal muscle function in this group.39
A limitation of this study results from our representation of skeletal muscle function deficit through muscle power and force phenotypes. Additional measures of muscle quality, architecture, and function and myosteatosis such as muscle cross-sectional area, lean body mass, and metabolic indices could have been used to complement the study phenotypes and better grasp the issue of sarcopenia in childhood leukemia survivors.62
It is not excluded that variables not considered in this study, such as nutritional status, level of physical activity, as well as concomitant endocrine or chronic inflammatory conditions, could have been confounding factors in genetic associations. Some of these factors were not included due to the subjectivity of self-reported phenotypic data (e.g. level of physical activity), and others because none of the PETALE cohort participants presented evidence of such conditions (e.g. hypopituitarism). Likewise, although we controlled the analyses as much as possible for potential major confounders, a number of covariables could have still influenced the analyses, such as wide age range, duration of follow-up, or different complications that occurred during the treatment.63,64
It is worth noting that the limited sample size might have reduced the accuracy of the stratified analyses, and may have affected the detection of lower risk effects (the lowest OR that can be detected for common variants after correction for multiple testing in WES dataset is 2.6). On the other hand, cohort homogeneity with respect to treatment and ethnic background reduced the impact of confounding variables on the study results.10 While the candidate gene approach is generally biased towards rational gene selection, the investigation of many such genes allowed the identification of novel variants associated with genetic predictability of skeletal muscle deficit in the targeted population. The analyses of WES data were corrected for multiple testing using a 5% FDR, which in this dataset corresponded well with Bonferroni-adjusted P-values for the number of common variants tested or the number of genes in rare variants testing.
A replication analysis should follow this study to validate findings, whereas future studies conducted at the exome or genome level may detect in an unbiased fashion new genetic predictors underlying skeletal muscle deficit in childhood ALL survivors. Together, they may explain the observed heterogeneity of these late-adverse effects in ALL survivors, and could eventually lead to personalised prevention strategies.
Acknowledgments
The authors thank all childhood ALL survivors and their parents who consented to take part in the PETALE study, as well as all study collaborators for their precious involvement in the production of this manuscript. The PETALE study is funded by the Canadian Institutes of Health Research in collaboration with the Cancer Research Society Inc. (CRS), the Garron Family Cancer Center of the Hospital for Sick Children, the Paediatric Oncology Groups of Ontario (POGO), the Canadian Cancer Society Research Institute (CCSRI), the C17 Research Network (C17), the Sainte-Justine Hospital Foundation and the FRQS Applied Medical Genetics Network.
Abbreviation list
ALL, acute lymphoblastic leukemia; LAE, late adverse effects; GC, glucocorticoid; BMI, body mass index; FDR, false discovery rate; PETALE, Prévenir les effets tardifs des traitements de la leucémie aiguë lymphoblastique chez l’enfant; SNP, single nucleotide polymorphism; WES, whole exome sequencing; SJUHC, Sainte-Justine University Hospital Center; DFCI, Dana-Farber Cancer Institute; MSHC, Montreal Shriners Hospital for Children; Fmax, peak muscle force; Pmax, peak muscle power; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval; CRT, cranial radiation therapy.
Disclosure
The abstract for this manuscript was presented at the 2017 International Workshop on Musculoskeletal and Neuronal Interactions, Montreal, October 6–8, and was published in the Journal of Musculoskeletal and Neuronal Interactions (J Musculoskelet Neuronal Interact 2017; 17(4):347–357). Dr Nathalie Alos reports grants from CIHR, grants from Cancer Research Society Inc., grants from Pediatric Oncology Groups of Ontario (POGO), grants from Canadian Cancer Society Research Institute (CCSRI), grants from C17 Research Network (C17), grants from Sainte-Justine Hospital Foundation, nothing from FRQS Applied Medical Genetics Network, grants from Garron Family Cancer Center of the Hospital for Sick Children, during the conduct of the study; other from Novartis, other from Amgen, other from Alexion, other from novonrdisk, outside the submitted work. The other authors report no conflict of interests in this work.
Supplementary materials
References
- 1.Canadian Cancer Society’s Advisory Committee on Cancer Statistics. Canadian Cancer Statistics 2016 . Toronto (ON): Canadian Cancer Society; 2017. [Google Scholar]
- 2.American Cancer Society. Cancer facts and figures 2014. Available from: http://www.cancer.gov/types/leukemia/hp/child-all-treatment-pdq#cit/section_1.5. Accessed November17, 2017.
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 4.Ness KK, Baker KS, Dengel DR, et al. Body composition, muscle strength deficit and mobility limitations in adult survivors of childhood acute lymphoblastic leukemia. Pediatr Blood Cancer. 2007;49:975–981. doi: 10.1002/pbc.21091 [DOI] [PubMed] [Google Scholar]
- 5.Casillas J, Sakamoto KM. Topics in paediatric leukemia–acute lymphoblastic leukemia and late effects in long-term survivors. MedGenMed. 2005;7:22–37. [PMC free article] [PubMed] [Google Scholar]
- 6.Karakurt H, Sarper N, Kilic SC, Gelen SA, Zengin E. Screening survivors of childhood acute lymphoblastic leukemia for obesity, metabolic syndrome, and insulin resistance. Pediatr Hematol Oncol. 2012;29:551–561. doi: 10.3109/08880018.2012.708892 [DOI] [PubMed] [Google Scholar]
- 7.Krajinovic M, Elbared J, Drouin S, et al. Polymorphisms of ABCC5 and NOS3 genes influence doxorubicin cardiotoxicity in survivors of childhood acute lymphoblastic leukemia. Pharmacogenomics J. 2016;16:530–535. doi: 10.1038/tpj.2015.63 [DOI] [PubMed] [Google Scholar]
- 8.Jacola LM, Krull KR, Pui CH, et al. Longitudinal assessment of neurocognitive outcomes in survivors of childhood acute lymphoblastic leukemia treated on a contemporary chemotherapy protocol. J Clin Oncol. 2016;34:1239–1247. doi: 10.1200/JCO.2015.64.3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akyay A, Olcay L, Sezer N, Atay Sonmez C. Muscle strength, motor performance, cardiac and muscle biomarkers in detection of muscle side effects during and after acute lymphoblastic leukemia treatment in children. J Pediatr Hematol Oncol. 2014;36:594–598. doi: 10.1097/MPH.0000000000000067 [DOI] [PubMed] [Google Scholar]
- 10.Marcoux S, Drouin S, Laverdiere C, et al. The PETALE study: late adverse effects and biomarkers in childhood acute lymphoblastic leukemia survivors. Pediatr Blood Cancer. 2016;64:1–8. [DOI] [PubMed] [Google Scholar]
- 11.Vuotto SC, Krull KR, Li C, et al. Impact of chronic disease on emotional distress in adult survivors of childhood cancer: a report from the Childhood Cancer Survivor Study. Cancer. 2017;123:521–528. doi: 10.1002/cncr.30348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheede-Bergdahl C, Jagoe RT. After the chemotherapy: potential mechanisms for chemotherapy-induced delayed skeletal muscle dysfunction in survivors of acute lymphoblastic leukaemia in childhood. Front Pharmacol. 2013;4:49–71. doi: 10.3389/fphar.2013.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gouspillou G, Scheede-Bergdahl C, Spendiff S, et al. Anthracycline-containing chemotherapy causes long-term impairment of mitochondrial respiration and increased reactive oxygen species release in skeletal muscle. Sci Rep. 2015;5:8717–8740. doi: 10.1038/srep08717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jn R, Dl W, Ta G. Skeletal muscle Ca(2+) mishandling: another effect of bone-to-muscle signaling. Semin Cell Dev Biol. 2016;49:24–29. doi: 10.1016/j.semcdb.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bianchi L, Volpato S. Muscle dysfunction in type 2 diabetes: a major threat to patient’s mobility and independence. Acta Diabetol. 2016;53:879–889. doi: 10.1007/s00592-016-0880-y [DOI] [PubMed] [Google Scholar]
- 16.Hirsch JA, Winters M, Sims-Gould J, et al. Developing a comprehensive measure of mobility: mobility over varied environments scale (MOVES). BMC Public Health. 2017;17:513–537. doi: 10.1186/s12889-017-4450-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janiszewski PM, Oeffinger KC, Church TS, et al. Abdominal obesity, liver fat, and muscle composition in survivors of childhood acute lymphoblastic leukemia. J Clin Endocrinol Metab. 2007;92:3816–3821. doi: 10.1210/jc.2006-2178 [DOI] [PubMed] [Google Scholar]
- 18.Sato AY, Richardson D, Cregor M, et al. Glucocorticoids induce bone and muscle atrophy by tissue-specific mechanisms upstream of E3 ubiquitin ligases. Endocrinology. 2017;158:664–677. doi: 10.1210/en.2016-1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Jia Q, Xiao J, Jiao H, Lin H. Glucocorticoids retard skeletal muscle development and myoblast protein synthesis through a mechanistic target of rapamycin (mTOR)-signaling pathway in broilers (Gallus gallus domesticus). Stress. 2015;18:686–698. doi: 10.3109/10253890.2015.1083551 [DOI] [PubMed] [Google Scholar]
- 20.Dong Y, Silva KA, Dong Y, Zhang L. Glucocorticoids increase adipocytes in muscle by affecting IL-4 regulated FAP activity. FASEB J. 2014;28:4123–4132. doi: 10.1096/fj.14-254011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veilleux LN, Rauch F. Reproducibility of jumping mechanography in healthy children and adults. J Musculoskelet Neuronal Interact. 2010;10:256–266. [PubMed] [Google Scholar]
- 22.Sumnik Z, Matyskova J, Hlavka Z, Durdilova L, Soucek O, Zemkova D. Reference data for jumping mechanography in healthy children and adolescents aged 6–18 years. J Musculoskelet Neuronal Interact. 2013;13:297–311. [PubMed] [Google Scholar]
- 23.Kaji H. Interaction between muscle and bone. J Bone Metab. 2014;21:29–40. doi: 10.11005/jbm.2014.21.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerrero F, Herencia C, Almaden Y, et al. TGF-beta prevents phosphate-induced osteogenesis through inhibition of BMP and Wnt/beta-catenin pathways. PLoS One. 2014;9:e89179–98. doi: 10.1371/journal.pone.0089179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sims NA, Vrahnas C. Regulation of cortical and trabecular bone mass by communication between osteoblasts, osteocytes and osteoclasts. Arch Biochem Biophys. 2014;561:22–28. doi: 10.1016/j.abb.2014.05.015 [DOI] [PubMed] [Google Scholar]
- 26.Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501. doi: 10.1038/ng.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunton JE, Girgis CM, Baldock PA, Lips P. Bone muscle interactions and vitamin D. Bone. 2015;80:89–94. doi: 10.1016/j.bone.2015.02.029 [DOI] [PubMed] [Google Scholar]
- 28.Karol SE, Yang W, Van Driest SL, et al. Genetics of glucocorticoid-associated osteonecrosis in children with acute lymphoblastic leukemia. Blood. 2015;126:1770–1776. doi: 10.1182/blood-2015-05-643601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao W, Cheng Z, Busse C, Pham A, Nakamura MC, Lane NE. Glucocorticoid excess in mice results in early activation of osteoclastogenesis and adipogenesis and prolonged suppression of osteogenesis: a longitudinal study of gene expression in bone tissue from glucocorticoid-treated mice. Arthritis Rheum. 2008;58:1674–1686. doi: 10.1002/art.23454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riksen NP, Rongen GA, Smits P, van Riel P, Barrera P. Effect of the 34C>T variant in the AMPD1 gene on the clinical response to methotrexate in patients with rheumatoid arthritis: comment on the article by Wessels et al. Arthritis Rheum. 2007;56:694 author reply 694–5. doi: 10.1002/art.22862. [DOI] [PubMed] [Google Scholar]
- 31.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48–61. doi: 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spinella JF, Healy J, Saillour V, et al. Whole-exome sequencing of a rare case of familial childhood acute lymphoblastic leukemia reveals putative predisposing mutations in Fanconi anemia genes. BMC Cancer. 2015;15:539–552. doi: 10.1186/s12885-015-1549-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picard: set of command line tools for manipulating high-throughput sequencing (HTS) data. Available from: https://broadinstitute.github.io/picard/. Accessed November20, 2017.
- 35.McKenna A, Hanna M, Banks E, et al. the genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genomes PC, Gr A, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–81. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pellegrinelli V, Rouault C, Rodriguez-Cuenca S, et al. Human adipocytes induce inflammation and atrophy in muscle cells during obesity. Diabetes. 2015;64:3121–3134. doi: 10.2337/db14-0796 [DOI] [PubMed] [Google Scholar]
- 40.Krull KR, Bhojwani D, Conklin HM, et al. Genetic mediators of neurocognitive outcomes in survivors of childhood acute lymphoblastic leukemia. J Clin Oncol. 2013;31:2182–2188. doi: 10.1200/JCO.2012.46.7944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross JA, Oeffinger KC, Davies SM, et al. Genetic variation in the leptin receptor gene and obesity in survivors of childhood acute lymphoblastic leukemia: a report from the Childhood Cancer Survivor Study. J Clin Oncol. 2004;22:3558–3562. doi: 10.1200/JCO.2004.11.152 [DOI] [PubMed] [Google Scholar]
- 42.Bethesda NCBI. U.S. National Library of Medicine. DUOX2. Available from:https://www.ncbi.nlm.nih.gov/gene/50506. Accessed November 20, 2017.
- 43.Kizys MML, Louzada RA, Mitne-Neto M, et al. DUOX2 mutations are associated with congenital hypothyroidism with ectopic thyroid gland. J Clin Endocrinol Metab. 2017;102:4060–4071. doi: 10.1210/jc.2017-00832 [DOI] [PubMed] [Google Scholar]
- 44.Parlato M, Charbit-Henrion F, Hayes P, et al. First identification of biallelic inherited DUOX2 inactivating mutations as a cause of very early onset inflammatory bowel disease. Gastroenterology. 2017;153(609–11):e603. doi: 10.1053/j.gastro.2016.12.053 [DOI] [PubMed] [Google Scholar]
- 45.Mosekilde L, Eriksen EF, Charles P. Effects of thyroid hormones on bone and mineral metabolism. Endocrinol Metab Clin North Am. 1990;19:35–63. [PubMed] [Google Scholar]
- 46.Leal AL, Albuquerque JP, Matos MS, et al. Thyroid hormones regulate skeletal muscle regeneration after acute injury. Endocrine. 2015;48:233–240. doi: 10.1007/s12020-014-0271-5 [DOI] [PubMed] [Google Scholar]
- 47.Bethesda NCBI. U.S. National Library of Medicine. ADAMTS4. Available from: http://www.ncbi.nlm.nih.gov/gene/9507. Accessed November 20, 2017.
- 48.Li W, Du C, Wang H, Zhang C. Increased serum ADAMTS-4 in knee osteoarthritis: a potential indicator for the diagnosis of osteoarthritis in early stages. Genet Mol Res. 2014;13:9642–9649. doi: 10.4238/2014.November.14.9 [DOI] [PubMed] [Google Scholar]
- 49.El Khoury LY, Rickaby R, Samiric T, Raleigh SM. Promoter methylation status of the TIMP2 and ADAMTS4 genes and patellar tendinopathy. J Sci Med Sport. 2018;21:378–382. doi: 10.1016/j.jsams.2017.08.016 [DOI] [PubMed] [Google Scholar]
- 50.Dong H, Du T, Premaratne S, et al. Relationship between ADAMTS4 and carotid atherosclerotic plaque vulnerability in humans. J Vasc Surg. 2018;67(1120–6). doi: 10.1016/j.jvs.2017.08.075. [DOI] [PubMed] [Google Scholar]
- 51.Namgaladze D, Snodgrass RG, Angioni C, et al. AMP-activated protein kinase suppresses arachidonate 15-lipoxygenase expression in interleukin 4-polarized human macrophages. J Biol Chem. 2015;290:24484–24494. doi: 10.1074/jbc.M115.678243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magnusson LU, Lundqvist A, Asp J, et al. High expression of arachidonate 15-lipoxygenase and proinflammatory markers in human ischemic heart tissue. Biochem Biophys Res Commun. 2012;424:327–330. doi: 10.1016/j.bbrc.2012.06.117 [DOI] [PubMed] [Google Scholar]
- 53.Klein RF, Allard J, Avnur Z, et al. Regulation of bone mass in mice by the lipoxygenase gene Alox15. Science. 2004;303:229–232. doi: 10.1126/science.1090985 [DOI] [PubMed] [Google Scholar]
- 54.Herlin M, McGuigan FE, Luthman H, Åkesson K. Polymorphisms in inflammation associated genes ALOX15 and IL-6 are associated with bone properties in young women and fracture in elderly. Bone. 2015;79:105–109. doi: 10.1016/j.bone.2015.05.035 [DOI] [PubMed] [Google Scholar]
- 55.Fitts RH, McDonald KS, Schluter JM. The determinants of skeletal muscle force and power: their adaptability with changes in activity pattern. J Biomech. 1991;24(Suppl 1):111–122. [DOI] [PubMed] [Google Scholar]
- 56.Bamman MM, Newcomer BR, Larson-Meyer DE, Weinsier RL, Hunter GR. Evaluation of the strength-size relationship in vivo using various muscle size indices. Med Sci Sports Exerc. 2000;32:1307–1313. [DOI] [PubMed] [Google Scholar]
- 57.Tonson A, Ratel S, Le Fur Y, Cozzone P, Bendahan D. Effect of maturation on the relationship between muscle size and force production. Med Sci Sports Exerc. 2008;4:918–925. doi: 10.1249/MSS.0b013e3181641bed [DOI] [PubMed] [Google Scholar]
- 58.Hanada M, Sakamoto N, Ishimatsu Y, et al. Effect of long-term treatment with corticosteroids on skeletal muscle strength, functional exercise capacity and health status in patients with interstitial lung disease. Respirology. 2016;21:1088–1093. doi: 10.1111/resp.12807 [DOI] [PubMed] [Google Scholar]
- 59.Braun TP, Szumowski M, Levasseur PR, et al. Muscle atrophy in response to cytotoxic chemotherapy is dependent on intact glucocorticoid signaling in skeletal muscle. PLoS One. 2014;9(e106489):1–21. doi: 10.1371/journal.pone.0106489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chemaitilly W, Li Z, Huang S, et al. Anterior hypopituitarism in adult survivors of childhood cancers treated with cranial radiotherapy: a report from the St Jude Lifetime Cohort study. J Clin Oncol. 2015;33:492–500. doi: 10.1200/JCO.2014.56.7933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kempf SJ, Moertl S, Sepe S, et al. Low-dose ionizing radiation rapidly affects mitochondrial and synaptic signaling pathways in murine hippocampus and cortex. J Proteome Res. 2015;14:2055–2064. doi: 10.1021/acs.jproteome.5b00114 [DOI] [PubMed] [Google Scholar]
- 62.Correa-de-Araujo R, Harris-Love MO, Miljkovic I, Fragala MS, Anthony BW, Manini TM. The need for standardized assessment of muscle quality in skeletal muscle function deficit and other aging-related muscle dysfunctions: a symposium report. Front Physiol. 2017;8:87–138. doi: 10.3389/fphys.2017.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frost RA, Lang CH. Multifaceted role of insulin-like growth factors and mammalian target of rapamycin in skeletal muscle. Endocrinol Metab Clin North Am. 2012;41(297–322). doi: 10.1016/j.ecl.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ten BRW, Grefte S, Jw VDH. Regulatory factors and cell populations involved in skeletal muscle regeneration. J Cell Physiol. 2010;224:7–16. doi: 10.1002/jcp.22127 [DOI] [PubMed] [Google Scholar]