Abstract
Background/Aim: Head and neck cancers are a heterogenous group of epithelial tumors represented mainly by squamous cell carcinomas (HNSCC), which are the sixth most common type of cancer worldwide. Surgery together with radiotherapy (RT) is among the basic treatment modalities for most HNSCC patients. Various biomarkers aiming to predict patients’ response to RT are currently investigated. The reason behind this effort is, on one hand, to distinguish radioresistant patients that show weak benefit from RT and, on the other hand, reduce the ionizing radiation dose in less aggressive radiosensitive HNSCC with possibly less acute or late toxicity. Materials and Methods: A total of 94 HNSCC patients treated by definitive intensity-modulated radiotherapy were included in our retrospective study. We used a global expression analysis of microRNAs (miRNAs) in 43 tumor samples and validated a series of selected miRNAs in an independent set of 51 tumors. Results: We identified miR-15b-5p to be differentially expressed between patients with short and long time of locoregional control (LRC). Kaplan–Meier analysis confirmed that HNSCC patients with higher expression of miR-15b-5p reach a significantly longer locoregional relapse-free survival compared to patients expressing low levels. Finally, multivariable Cox regression analysis revealed that miR-15b-5p is an independent predictive biomarker of LRC in HNSCC patients (HR=0.25; 95% CI=0.05-0.78; p<0.016). Conclusion: miR-15b-5p represents a potentially helpful biomarker for individualized treatment decisions concerning the management of HNSCC patients.
Keywords: Head and neck cancer, microRNA, miR-15b-5p, locoregional control, radiotherapy, IMRT
Head and neck cancer (HNC) is the sixth most common type of cancer worldwide and represents a heterogeneous group of diseases, including several classifications according to the anatomic location, the etiology, as well as molecular features (1,2). The vast majority of HNCs are squamous cell carcinomas (HNSCCs) developing in the oral cavity, the pharynx, or the larynx (2). Etiological factors of HNSCCs development include excessive use of tobacco and alcohol, as well as infection with human papilloma and Epstein-Barr viruses (2). The gold-standard therapy protocol consists of radical surgical resection followed by adjuvant radiotherapy (RT) as monotherapy, followed by chemotherapy or targeted therapy (3). Although ionizing radiation plays a key role in the oncological management of HNSCC patients, there is a significant portion of patients weakly responding to it. Importantly, RT is frequently associated with acute and late toxicity. For all these reasons, there is an effort to identify biomarkers that can predict the treatment outcomes of RT in HNSCC patients and help with the optimization of their dose in a case-specific manner. Several recent studies describe immunomarkers such as p16, EGFR and/or CD44 as predictive and prognostic for HNSCCs (4-6). However, these show a limited specificity and sensitivity and, thus, the results from the histopathology are not capable of precisely predicting the biopathological behavior of HNSCCs or their response to the administered therapy (4,6). This combined with the fact that cell sensitivity/resistance to ionizing radiation is affected by different cellular mechanisms, as well as the tumor microenvironment, a great focus is currently set on identifying novel useful microRNAs (miRNAs) for the management of HNSCCs (2). MiRNAs are endogenous, evolutionary-conserved, small non-coding RNA molecules 18-25 nucleotides long, that regulate the translation of mRNA molecules by binding to their 3’UTR, and can thus play key roles in cell signaling (2). These molecules are not only considered as very promising diagnostic biomarkers, but also as therapeutic targets (7). The latter is supported by the fact that they can retain high stability in the external environment (8). To date, many studies have focused on the role of miRNAs in the biopathology of HNSCCs, including their role in regulating cell behavior in response to ionizing radiation (2). There is currently no study analyzing original miRNA data dealing with the response of HNSCC patients to the adjuvant radiotherapy. The aim of this study was to analyze the global expression profiles of miRNAs in HNSCCs diagnostic tissue biopsies from patients consequently treated with definitive intensity-modulated radiotherapy (IMRT), and identify miRNAs that can predict the time of locoregional control (LRC).
Materials and Methods
The study was approved by the ethical committee of the Masaryk Memorial Cancer Institute (MMCI).
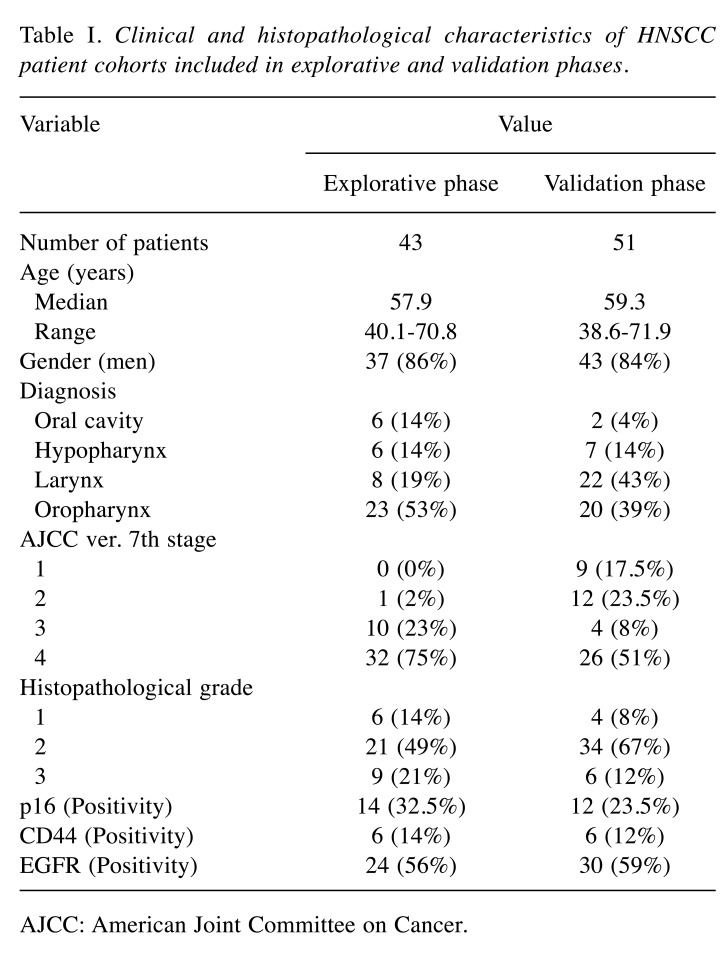
Patients. The retrospective mono-institutional cohort study included patients with histopathologically confirmed head and neck squamous cell carcinomas (HNSCC) of the oral cavity, the oropharynx, the hypopharynx, and the larynx. These patients were treated at the MMCI from January 1st, 2007 to December 31st, 2012. All HNSCC patients included in this study signed informed consent. Detailed clinical and histopathological characteristics of HNSCC patients included in both explorative and validation phases are summarized in Table I.
Table I. Clinical and histopathological characteristics of HNSCC patient cohorts included in explorative and validation phases.
AJCC: American Joint Committee on Cancer.
Therapeutic regime. All patients were treated irrespective of clinical trials, by intensity modulated radiotherapy (IMRT); some were also placed under systemic therapy (platinum-based chemotherapy or targeted therapy by EGFR inhibitor cetuximab). In the IMRT protocol the highest dose of 70 Gy or a biologically equivalent was administered on the site of the primary tumor and involved the nodes or the entire larynx in case of T1b laryngeal cancer. Alternatively, 66 Gy was administered to the entire larynx in the case of T1a laryngeal cancer. Intermediate or low risk areas were treated with doses of 60 Gy or 50 Gy, respectively. RT was performed 5 days a week either by standard normo-fractionated method, employing a shrinking field technique, or by a simultaneous integrated boost technique, with total doses of 69.96/62.7/56.1 Gy (daily fractions of 2.12/1.9/1.7 Gy); or alternatively, with 66/60/54 Gy (daily fractions of 2.2/2.0/1.8 Gy). Additional boost therapy with a second daily fraction of 1.2 Gy from the 17th fraction (total number of 42 fractions) was another option. All medically fit patients with tumor stage III or IV received either a neoadjuvant or a concurrent platinum-based chemotherapy or cetuximab.
Follow-up treatment. The follow-up treatment involved ENT examinations, held every 2 months during first two years and later every 3-4 months at minimum. Computed tomography (CT) or positron emission tomography (PET/CT) scan was performed 2-3 months after the end of the radiation therapy and it was subsequently adjusted according to the clinical image. The occurrence of LCR was checked twice and was approved by two experienced radiation oncologists (M.S. and R.C.) to ensure that the recurrences appeared within the irradiated areas.
Tissue sample preparation and nucleic acid extraction. All FFPE HNSCC tissue samples were independently reviewed by two experienced pathologists and only samples with over 90% of tumor tissue were analyzed. Total RNA with enriched fraction of small RNAs was purified from FFPE samples by xylene deparaffinization using the mirVana miRNA Isolation Kit (ThermoFisher Scientific, Carlsbad, CA, USA). Nucleic acid concentrations and purities were assessed using both UV spectrophotometry (Nanodrop ND-1000, ThermoFisher Scientific) and fluorometry (Qubit 2.0, ThermoFisher Scientific).
MicroRNA microarray analysis. The samples were analyzed using Affymetrix GeneChip miRNA 4.0 arrays (ThermoFisher Scientific) containing 5607 probe sets for human small RNAs. Out of these, the 2578 probe sets of human mature miRNAs were filtered. All the steps of the procedure were performed according to the Affymetrix standardized protocol for miRNA 4.0 arrays. Intensity values for each probe cell (.cel file) were calculated using the Affymetrix GeneChip Command Console (AGCC). Quality control of the microarray was performed using the Affymetrix miRNA QC Tool, version 1.1.1.0. All data were pre-processed and were further analyzed using the software packages included in the R/Bioconductor. Pre-processing was performed by the RMA method using the default parameters as implemented in the Bioconductor package oligo. All data were log2-transformed. To identify differentially expressed miRNAs, the LIMMA approach for paired samples was applied with an additional Benjamini-Hochberg correction of p-values.
Real-time quantification of miRNAs by stem-loop RT-PCR. Complementary DNA (cDNA) was synthesized from 10 ng of small RNA enriched total RNA using gene-specific primers and the TaqMan MicroRNA Reverse Transcription kit, according to the TaqMan MicroRNA Assay protocol (ThermoFisher Scientific). Real-Time PCR was performed using the QuantStudio 12K Flex Real-Time PCR System in accordance with TaqMan MicroRNA Assay protocol. The threshold cycle data were determined using the default threshold settings. All real-time PCR reactions were run in triplicates and the average Ct and SD values were calculated. The relative miRNA expression levels were determined by the 2−ΔCT method, where ΔCTs were calculated as follows: ΔCT=CT(miRNA of interest) − CT(let-7a). Reference miRNA let-7a was determined by analyzing the Affymetrix data using GeneNorm and NormFinder algorithms, as this was the most stable one for all the samples analyzed. Statistical analyses were performed using the GraphPad Prism 7.0a software (San Diego, CA, USA) (Mann-Whitney non-parametric tests, ROC analyses, Kaplan-Meier analyses) and the JMP 9.0.1 software (Cary, NC, USA) (Cox regression analyses). p-Values≤0.05 were considered as significant in all cases.
p16, EGFR, and CD44 immunohistochemical analysis. All immunohistochemical (IHC) analyses were performed as described previously (9). Briefly, sections from the FFPE primary biopsy samples, obtained prior to the treatment, were stained with hematoxylin-eosin and were independently reviewed by two pathologists. IHC analyses were performed using the Automated IHC/ISH Slide Staining System BenchMark XT (Ventana, Tuscon, AZ, USA). The ultraView Universal DAB detection Kit (Ventana) was used according to the manufacturer’s instruction for staining with the BenchMark Series. Four μm thick tumoral tissue sections were applied to positively charged slides TOMO Matsunami. Slides were incubated with a primary ready-to-use rabbit monoclonal anti-CD44 antibody (clone SP37, Ventana), a rabbit monoclonal anti-EGFR antibody (clone 5B7, Ventana) and a mouse monoclonal CINtec p16 antibody (Ventana). A system of negative and positive controls was used during each run.
Expression of p16 was analyzed in tumor cells (TC), and tumors with diffuse and intense nuclear and cytoplasmic immunostaining with a presence of TC at 70% minimum were evaluated as positive. EGFR staining was semi-quantitatively evaluated using the scoring system, analogous to the HER2 IHC evaluation. For the CD44 IHC, we assessed the positivity of the cell membrane and calculated the percentage of CD44 positive tumor cells. The intensity of immunostaining in immuno-positive cases was evaluated semiquantitatively and was scored as weak, moderate or strong. Tumors with moderate or strong intensity of immunostaining of ≥90 % CD44 positive TC were labeled as positive.
Results
Global expression analysis of microRNAs in tumor samples obtained from HNSCC patients with short and long locoregional control. Forty-six out of 2,578 analyzed miRNAs showed significant differential expression in HNSCC tumor tissues between patients with short (n=22; median of LRC=5.1 months) and long (n=21; 60.4) LRC (p<0.05; median of average expressions=2.038 [interquartile range (IQR)=3.373]; median of log fold change (logFC) absolute values=0.526 [IQR=0.294]) (Table II). Among these miRNAs, eleven showed lower expression and thirty-five showed higher expression in tumor tissues from HNSCC patients with short LRC, as compared to patients with long LRC. Subsequent univariate Cox regression analysis showed that forty-one out of these forty-six miRNAs were significantly associated with LRC (p<0.05); eleven miRNAs decreased (median of hazard ratio (HR)=0.367 [IQR=0.184]) and thirty miRNAs increased (median of HR=2.381 [IQR=1.073]) the risk of LRC in HNSCC patients (Table II).
Table II. List of the significantly deregulated tumor tissue miRNAs between HNSCC patients with short and long time of LRC and Cox proportional hazards of LRC according to these miRNAs.
LogFC: Decadic logarithm of fold change; AveExpr: average expression; HR (CI): hazard ratio (confidence interval); LIMMA: linear models for microarray data.
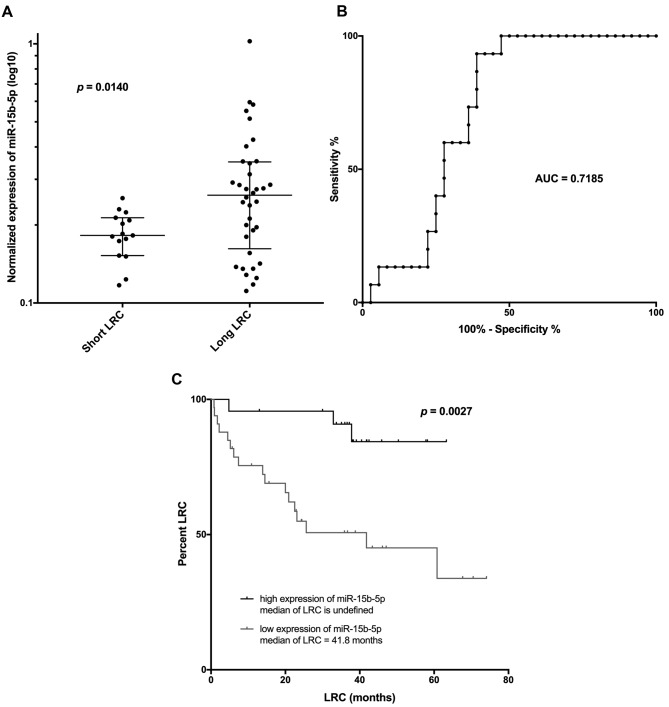
Validation of expression of selected microRNAs in the independent set of HNSCC positive tissue samples. Based on the global expression analysis results, five miRNAs (miR-15b-5p, miR-3613-3p, let-7c-5p, let-7b-5p, and miR-4668-5p) were selected for validation of their expression in tumor tissues and their ability to predict LRC in the independent set of HNSCC patients. The criteria for their selection were as follows: i) p-value should be <0.01 for the miRNA expression comparative analysis between short and long LRC HNSCC patient groups; and ii) average expression of miRNA across all analyzed samples (n=43) should be higher than four (based on the GeneChip miRNA 4.0 array analysis; Affymetrix), which, based on our experience, can enable consequent detection by qPCR during the validation phase. The miRNA analysis using the TaqMan stem-loop qPCR assays revealed that only miR-15b-5p was differentially expressed between short (n=15; median of LRC=6.1 months) and long (n=36; median of LRC=41.2 months) LRC HNSCC patients (p=0.014; Figure 1A). Subsequent ROC analysis showed that expression of miR-15b-5p in tumor samples is indicative of short LRC HNSCC patients’ classification with 93% sensitivity and 61% specificity (cut off value of miR-15b-5p normalized expression>0.2341; AUC=0.719; Figure 1B). These results were also confirmed using the Kaplan-Meier analysis on the extended set of HNSCC patients. Patients with lower expression of miR-15-5p (n=33; median of LRC=41.8 months) relapsed significantly sooner as compared to patients who exhibited higher expression of miR-15b-5p (n=23; median of LRC is not reached) (p=0.0027, Log rank test; Figure 1C). Finally, the univariable Cox regression analyses of miR-15b-5p, as well as other clinical and pathological markers, revealed that miR-15b-5p and p16 are both significantly associated with LRC. However, the multivariable analysis showed that only miR-15b-5p is significantly associated with LRC (Table III).
Figure 1. Mann-Whitney analysis of normalized miR-15b-5p expression in tumor tissues between HNSCC patients with short and long time of LRC (A); Receiver operating characteristic (ROC) curve of the normalized miR-15b-5p expression to detect HNSCC patients with short time on LRC (B); and Kaplan–Meier curves of locoregional relapse-free survival for HNSCC patients with low and high expression of tumor tissue miR-15b-5p (C). LRC: Locoregional control; AUC: area under the curve.
Table III. Cox hazard regression analyses of common clinico-histopathological characteristics and miR-15b-5p expression effects on locoregional relapse-free survival of HNSCC patients.
HR: Hazard ratio; CI: confidence interval; KI: Karnofsky index; Pos: positive; Negat: negative.
Discussion
Although radiotherapy is a key treatment modality in HNSCC management, an inconsiderably large percentage of patients do not benefit from this (10). Certain studies estimate that in over 25% of patients’ treatment with radiotherapy fails due to tumoral radio-resistance (11). Unfortunately, there is currently no clinically useful biomarker available for predicting the response of HNSCC patients to the ionizing radiation with sufficient analytical parameters. Such a biomarker would allow for a personalized therapeutic approach, including the determination of an optimal radiation dose. Therefore, novel methods that can reliably predict individual responses to radiotherapy are much needed.
Several recent studies suggest that promising biomarkers that can provide reliable information on the therapeutic responses of HNSCC patients could be certain miRNAs (12,13). These molecules have significant potential, not only as predictive, prognostic and diagnostic biomarkers, but also as therapeutic targets in many types of cancers and in other diseases, with some of them already being tested in clinical trials (14,15). In our study, we first examined the global expression profiles of miRNAs in FFPE tumor biopsies obtained from HNSCC patients with short and long time of LRC. For this purpose, we used an miRNA microarray hybridization analysis that has been previously reported to be suitable for FFPE cases (16,17). Compared to these studies, our data show quite a similar distribution of miRNA expression profiles in all of the samples examined.
Although statistical analysis revealed significance for forty-six differentially expressed miRNAs between both groups examined, only five miRNAs were selected for subsequent validation as potential biomarkers. The criteria for their selection were both the highly differential expression between the short and long LRC groups, and their high average expression in all our samples. This allowed us to use a wide range of quantification methods, and through these, to confirm the potential use of these miRNAs as biomarkers. Unfortunately, only miR-15b-5p showed a differential expression profile between patients with short and long LRC, including the ones from the independent sample set. The statistically insufficient size of the samples, as well as the variability of patients during the explorative and validation phases could be considered as the main limitations of our study. Quite possibly, this is the main reason why we could successfully validate only one miRNA molecule. In the explorative phase, the time to local relapse in the short LRC group ranged between 1.3 and 18.6 months, and only patients without a local relapse that were monitored for more than 46.8 months were included in the long LRC group. In the validation phase, only fifteen patients with time to local relapse ranging between 0.8 and 23.1 months were included in the short LRC group, and thirty-six patients with time to local relapse ranging between 24.3 and 74.1 months (out of whom only thirty-one were still alive without a local relapse at the time of the study) were included in the long LRC group. This suggests, that especially for the long LRC patients assessed during both phases, their differential distribution could be responsible for the low success rate of miRNA validation.
Recently, several studies have focused on the specific miRNA expression patterns in HNSCC and the application of these molecules in the evaluation of patients’ therapeutic responses (11,18). Unfortunately, there is only little evidence on the association between miR-15b-5p with the HNSCC biopathology and none on its role on radio-resistance in this type of tumors. Lu et al. determined a panel of specific expression miRNA profiles associated with oral cancer cell lines, in which miR-15b-5p was found upregulated in tumor cells compared to normal keratinocytes (19). These findings suggest a rather oncogenic function of miR-15b-5p that is, however, inconsistent with our observation. Yang et al. described the same phenomenon in hepatocellular carcinoma (HCC) (20), where miR-15b-5p levels were also reported high in HCC tissues, albeit this molecule negatively correlates with HCC recurrence (21,22). The authors suggested that miR-15b-5p directly targets Rab1A, a previously reported oncogene that induces endoplasmic reticulum stress and leads to HCC cell apoptosis, as well as inhibition of tumor growth in vivo (20). Association of miR-15b-5p with therapeutic responses was also described in colorectal carcinoma (CRC) as well as in breast cancer, where miR-15b-5p enhances the tumor sensitivity to the ionizing radiation (23). Specifically, miR-15b-5p expression was shown to positively correlate with the sensitivity to neoadjuvant radiotherapy (nRT) in rectal cancer patients (23). High miR-15b-5p expression in pre-treated tumor biopsies was associated with a higher tumor regression grading score after nRT. Patients with high tumor levels of miR-15b-5p were found to survive disease-free for longer. Moreover, patients who responded to the 5-FU-based therapy exhibited higher expression levels of miR-15b-5p compared to patients who did not respond to the treatment. The authors suggested that this response is mediated through the direct targeting of DCLK1 by miR-15b-5p (23). DCLK1 has been previously associated with self-renewal and tumorigenic activity in colorectal cancer tumor initiating cells (23). Interestingly, breast cancer stem cells show decreased levels of miR-15b/16 expression, while overexpression of miR-15a/15b/16 can induce breast cancer cell sensitivity to radiation by affecting their cell cycle (24,25). Finally, another study has showed that miR-15b-5p expression levels increase following the exposure of normal human fibroblasts to radiation as a response to oxidative stress (26).
In oral tongue squamous cell cancer, overexpression of miR-15b-5p promotes mesenchymal-epithelial transition (MET) and inhibits the formation of cancer stem cells, leading to their sensitization to cisplatin, despite the fact that they are otherwise resistant to it (27). The authors suggest that this effect is very likely due to miR-15b-5p targeting TRIM14, which induces epithelial-mesenchymal transition (EMT) in tongue squamous cell carcinoma cells and progression to development of cancer stem cell phenotypes (27). Similar results have also been published by Zhou et al., who describe a signaling pathway including miR-15b-5p/TRIM-29/PTEN/AKT/mTOR, involved in EMT, proliferation, and metastasis of nasopharyngeal carcinoma (28). Finally, Sun et al. has shown that reduced expression of miR-15b promotes the mechanism of EMT in tongue cancer cells by targeting BMI1, and leads to the chemotherapeutic resistance of tongue squamous cell carcinomas and poor patient survival (29).
Taken together, our data suggest that miR-15b-5p is associated with the time to LRC and could be a promising predictive biomarker for radiation responses in HNSCC patients. Based on the aforementioned studies, the involvement of miR-15b-5p in the regulation of cancer stem cell survival could serve as a possible explanation for miR-15b-5p’s protective role in the LRC of HNSCC.
Conclusion
One of the challenges of the current care for HNSCC patients is to individualize the radiotherapy protocol so as to both increase the benefit from it as well as reduce the acute and late toxicity. We performed global expression profiling of miRNA molecules from tumor samples from HNSCC patients and we independently validated high-throughput results so as to find diagnostic miRNAs capable of predicting the time of locoregional control and, thus, the response of the patients to RT. Our data show that miR-15b-5p is differentially regulated between patients with short and long time of LRC, while patients with higher levels of expression of this miRNA achieve a significantly longer locoregional relapse-free survival. The predictive value of miR-15b-5p seems to be independent of other clinicopathological characteristics, including the stage or the p16 status. Taken together, miR-15b-5p could be a promising predictive biomarker of the RT response and its full potential should be definitively validated in a large independent study.
Conflicts of Interest
No conflict of interests.
Acknowledgements
This study was supported by Ministry of Health of the Czech Republic, by the Czech Health Research Council project, No. 15-31627A.
References
- 1.Klussmann JP. Head and neck cancer-new insights into a heterogeneous disease. Oncol Res Treat. 2017;40(6):318–319. doi: 10.1159/000477255. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad P, Sana J, Slavik M, Slampa P, Smilek P, Slaby O. Micrornas involvement in radioresistance of head and neck cancer. Dis Markers. 2017;2017:8245345. doi: 10.1155/2017/8245345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo Nigro C, Denaro N, Merlotti A, Merlano M. Head and neck cancer: Improving outcomes with a multidisciplinary approach. Cancer Manag Res. 2017;9:363–371. doi: 10.2147/CMAR.S115761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen ER, Reis IM, Gomez C, Pereira L, Freiser ME, Hoosien G, Franzmann EJ. Immunohistochemistry analysis of cd44, egfr, and p16 in oral cavity and oropharyngeal squamous cell carcinoma. Otolaryngol Head Neck Surg. 2017;157(2):239–251. doi: 10.1177/0194599817700371. [DOI] [PubMed] [Google Scholar]
- 5.Bossi P, Resteghini C, Paielli N, Licitra L, Pilotti S, Perrone F. Prognostic and predictive value of egfr in head and neck squamous cell carcinoma. Oncotarget. 2016;7(45):74362–74379. doi: 10.18632/oncotarget.11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baschnagel AM, Tonlaar N, Eskandari M, Kumar T, Williams L, Hanna A, Pruetz BL, Wilson GD. Combined cd44, c-met, and egfr expression in p16-positive and p16-negative head and neck squamous cell carcinomas. J Oral Pathol Med. 2017;46(3):208–213. doi: 10.1111/jop.12478. [DOI] [PubMed] [Google Scholar]
- 7.Sethi N, Wright A, Wood H, Rabbitts P. Micrornas and head and neck cancer: Reviewing the first decade of research. Eur J Cancer. 2014;50(15):2619–2635. doi: 10.1016/j.ejca.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, Jung K. Robust microrna stability in degraded rna preparations from human tissue and cell samples. Clin Chem. 2010;56(6):998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- 9.Gurin D, Slavik M, Hermanova M, Shatokhina T, Sana J, Kazda T, Selingerova I, Ahmad P, Smilek P, Horakova Z, Hendrych M, Slampa P, Slaby O. Prognostic impact of combined immunoprofiles in oropharyngeal squamous cell carcinoma patients with respect to ajcc 8th edition. J Oral Pathol Med. 2018;47(9):864–872. doi: 10.1111/jop.12772. [DOI] [PubMed] [Google Scholar]
- 10.Graboyes EM, Garrett-Mayer E, Ellis MA, Sharma AK, Wahlquist AE, Lentsch EJ, Nussenbaum B, Day TA. Effect of time to initiation of postoperative radiation therapy on survival in surgically managed head and neck cancer. Cancer. 2017;123(24):4841–4850. doi: 10.1002/cncr.30939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Wen Y, Zhang J, Sun W, Lui VWY, Wei Y, Chen F, Wen W. Prediction of radiotherapy response with a 5-microrna signature-based nomogram in head and neck squamous cell carcinoma. Cancer Med. 2018;7(3):726–735. doi: 10.1002/cam4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courthod G, Franco P, Palermo L, Pisconti S, Numico G. The role of microrna in head and neck cancer: Current knowledge and perspectives. Molecules. 2014;19(5):5704–5716. doi: 10.3390/molecules19055704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John K, Wu J, Lee BW, Farah CS. Micrornas in head and neck cancer. Int J Dent. 2013;2013:650218. doi: 10.1155/2013/650218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding rnas-an update. Nat Rev Clin Oncol. 2018;15(9):541–563. doi: 10.1038/s41571-018-0035-x. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Chen J, Sen S. Microrna as biomarkers and diagnostics. J Cell Physiol. 2016;231(1):25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leichter AL, Purcell RV, Sullivan MJ, Eccles MR, Chatterjee A. Multi-platform microrna profiling of hepatoblastoma patients using formalin fixed paraffin embedded archival samples. Gigascience. 2015;4:54. doi: 10.1186/s13742-015-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edward DP, Alkatan H, Rafiq Q, Eberhart C, Al Mesfer S, Ghazi N, Al Safieh L, Kondkar AA, Abu Amero KK. Microrna profiling in intraocular medulloepitheliomas. PLoS One. 2015;10(3):e0121706. doi: 10.1371/journal.pone.0121706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lubov J, Maschietto M, Ibrahim I, Mlynarek A, Hier M, Kowalski LP, Alaoui-Jamali MA, da Silva SD. Meta-analysis of micrornas expression in head and neck cancer: Uncovering association with outcome and mechanisms. Oncotarget. 2017;8(33):55511–55524. doi: 10.18632/oncotarget.19224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu YC, Chen YJ, Wang HM, Tsai CY, Chen WH, Huang YC, Fan KH, Tsai CN, Huang SF, Kang CJ, Chang JT, Cheng AJ. Oncogenic function and early detection potential of mirna-10b in oral cancer as identified by microrna profiling. Cancer Prev Res (Phila) 2012;5(4):665–674. doi: 10.1158/1940-6207.CAPR-11-0358. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Hou N, Wang X, Wang L, Chang S, He K, Zhao Z, Zhao X, Song T, Huang C. Mir-15b-5p induces endoplasmic reticulum stress and apoptosis in human hepatocellular carcinoma, both in vitro and in vivo, by suppressing rab1a. Oncotarget. 2015;6(18):16227–16238. doi: 10.18632/oncotarget.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rissland OS, Hong SJ, Bartel DP. Microrna destabilization enables dynamic regulation of the mir-16 family in response to cell-cycle changes. Mol Cell. 2011;43(6):993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung GE, Yoon JH, Myung SJ, Lee JH, Lee SH, Lee SM, Kim SJ, Hwang SY, Lee HS, Kim CY. High expression of microrna-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep. 2010;23(1):113–119. [PubMed] [Google Scholar]
- 23.Ji D, Zhan T, Li M, Yao Y, Jia J, Yi H, Qiao M, Xia J, Zhang Z, Ding H, Song C, Han Y, Gu J. Enhancement of sensitivity to chemo/radiation therapy by using mir-15b against dclk1 in colorectal cancer. Stem Cell Reports. 2018;11(6):1506–1522. doi: 10.1016/j.stemcr.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mei Z, Su T, Ye J, Yang C, Zhang S, Xie C. The mir-15 family enhances the radiosensitivity of breast cancer cells by targeting g2 checkpoints. Radiat Res. 2015;183(2):196–207. doi: 10.1667/RR13784.1. [DOI] [PubMed] [Google Scholar]
- 25.Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci USA. 2012;109(36):14470–14475. doi: 10.1073/pnas.1212811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, Degraff W, Cook J, Harris CC, Gius D, Mitchell JB. Ionizing radiation-induced oxidative stress alters mirna expression. PLoS One. 2009;4(7):e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Guo H, Yao B, Helms J. Mir-15b inhibits cancer-initiating cell phenotypes and chemoresistance of cisplatin by targeting trim14 in oral tongue squamous cell cancer. Oncol Rep. 2017;37(5):2720–2726. doi: 10.3892/or.2017.5532. [DOI] [PubMed] [Google Scholar]
- 28.Zhou XM, Sun R, Luo DH, Sun J, Zhang MY, Wang MH, Yang Y, Wang HY, Mai SJ. Upregulated trim29 promotes proliferation and metastasis of nasopharyngeal carcinoma via pten/akt/mtor signal pathway. Oncotarget. 2016;7(12):13634–13650. doi: 10.18632/oncotarget.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L, Yao Y, Liu B, Lin Z, Lin L, Yang M, Zhang W, Chen W, Pan C, Liu Q, Song E, Li J. Mir-200b and mir-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting bmi1. Oncogene. 2012;31(4):432–445. doi: 10.1038/onc.2011.263. [DOI] [PubMed] [Google Scholar]