Abstract
The death toll of non-small cell lung cancer (NSCLC) patients is primarily due to metastases, which are poorly amenable to therapeutic intervention. In this review we focus on miRs associated with metastasis of NSCLC as potential new targets for anti-metastatic therapy. We discuss miRs validated as therapeutic targets by in vitro data, identification of target(s) and pathway(s) and in vivo efficacy data in at least one clinically-relevant metastasis-related model. A few of the discussed miRs correlate with the clinical status of NSCLC patients. Using miRs as therapeutic agents has the advantage that targeting a single miR can potentially interfere with several metastatic pathways. Depending on their mode of action, the corresponding miRs can be up- or down-regulated compared to normal matching tissues. Here, we describe therapeutic approaches for reconstitution therapy and miR inhibition, general principles of anti-metastatic therapy as well as current technical pitfalls.
Keywords: Epithelial mesenchymal transition (EMT), in vivo metastasis models (experimental, spontaneous, intracardiac, orthotopic), miR inhibition, miR reconstitution of function, target validation, review
The annual worldwide death toll due to lung cancer is 1.5 million patients (1). Lung cancer is classified into two groups: i) Non-Small Cell Lung Cancer (NSCLC) (85%) and ii) Small Cell Lung Cancer (SCLC) (15%). NSCLC cases are divided into: i) lung adeno-carcinomas (non-squamous) (40%), ii) squamous carcinoma (25-30%) and iii) large cell carcinoma (10-15%) subtypes (2). Actionable mutations or rearrangements have been detected in lung adenocarcinoma, whereas in squamous cell carcinoma, with the exception of fibroblast growth factor (FGFR) amplifications, these are rare (3). In lung adenocarcinoma, epidermal growth factor receptor (EGFR) mutations and rearrangements in ALK, RET and ROS act as oncogenic drivers while additional low-frequency mutations have also been identified (3). These driver mutations have paved the way for the discovery of approved drugs such as gefitinib, erlotinib and osimertinib (targeting EGFR), brigatinib (targeting EGFR and ALK) and crizotinib, ceritinib and alectinib (targeting ALK) (4). For NSCLC patients with BRAF V600E mutations, a combination of BRAF inhibitor Tafinlar (Dabrafenib) and MEK inhibitor Mekinist (Trametinib) has been approved (5). A subset of NSCLC patients is responsive to antibodies which target immune checkpoint inhibitors with ambiguous clinical results (6). Anti-PD1 antibodies Nivolumab and Pembrolizumab, as well as anti-PD-L1 antibodies Atezolizumab, Durvalumab and Avelumab have been approved for specific NSCLC-related clinical scenarios (6). Despite these remarkable achievements, the high death toll associated with lung cancer is due to metastases that poorly respond to these treatments in most patients (2).
NSCLC-related metastases. Ninety percent of NSCLC deaths are metastasis-related (1). A subgroup of cancer cells with an indefinite self-renewal potential referred to as cancer stem cells (CSC) has been strongly associated with metastasis (7). The metastatic process has been described in detail in several reviews and is briefly summarized herein (8-10). The process starts at the tumor rim by tumor cells that gain invasive properties as they undergo epithelial–mesenchymal transition (EMT), degrade the extracellular matrix (ECM) and form pseudopodia-like projections (lamellopodia). Following intravasation inside blood or lymphatic vessels, the survival of these tumor cells (TC) depends on mechanisms through which they evade anoikis and escape immuno-surveillance. Finally, these cells can extravasate and colonize distant organs. Two types of metastasizing TCs can be found in the circulation: i) circulating tumor cells (CTC) in the peripheral blood and ii) disseminated tumor cells (DTC), which aggregate in the bone marrow. DTC can re-enter the circulation and lead to metastasis in distant organs. CTC can mechanistically arrest in capillary beads or selectively arrest at specific organs in a non-random fashion. They can remain in a dormant state for varying periods of time as micrometastases before gaining the capability of colonizing distant organs. Following the investigation of 729 NSCLC patients, the preferred sites of metastases are: i) bone (34%), ii) lung (32%), iii) brain (28%), iv) adrenal gland (17%) and v) liver (13%) (11).
Micro RNAs and their role in cancer. Micro RNAs (miRs) are evolutionarily conserved 18-25 nucleotides (nts) comprising RNA molecules involved in repressing mRNA targets at the post-transcriptional level (12-14). Approximately 1,000 miR genes have been identified in humans. The genes encoding miRs can be found in exons, introns or in the non-coding regions of protein-encoding genes. A total of 30% of miRs are processed from introns of protein-encoding genes. miRs are transcribed by RNA polymerase II and the primary transcript, referred to as pri-mRNA, contains a 7-methylguanosine cap, a polyA-tail and it may additionally contain introns (12-14). Primary pre-miRs are cleaved by a complex, called Microprocessor, to 60-70 nts hairpin-looped precursors (pre-miRs), which are exported to the cytoplasm by the multi-protein complex DICER to produce mature miRs (15,16). These consist of a 20 bp RNA duplex with a 5’phosphate and two nts 3’ overhanging at each end. One strand of the mature miR (guide strand) is loaded onto the miR-induced silencing complex to target mRNA via sequence complementarity. miRs interact with a 6-8 nt seed sequence at the 5’end, by base-pairing with a complementary sequence located at the 3’-untranslated region (3’-UTR) of the mRNA targets. An alternative mechanism for miR processing has been identified, where mirtons arise from spliced out introns and circumvent the cleavage by a microprocessor complex. Gene suppression mediated by miRs can be obtained by degradation of the corresponding mRNA or by inhibiting its translation. A single miR can target hundreds or thousands of mRNAs and a single mRNA can be targeted by multiple miRs (17). Since miRs inhibit several targets, inhibition may result in the modulation of several regulatory pathways and disturb different regulatory networks (17).
The relevance of miRs in the pathogenesis of cancer has been demonstrated for miR-15a and miR-16-1 (18,19). Deletion of the miR-15a and miR-16-1 clusters in mice recapitulates the disease phenotype of B-cell chronic lymphatic leukemia (B-CLL) by circumventing the cleavage of the mRNA of the anti-apoptotic protein BCL2 (18,19). miRs are involved in many aspects of the pathogenesis and metastasis of cancer (20,21). We recently summarized their role in metastasis of breast, ovarian and prostate cancer (22-24). In this review we focus on the role of miRs on the metastasis of NSCLC with a documented efficacy in at least one metastasis-related in vivo model.
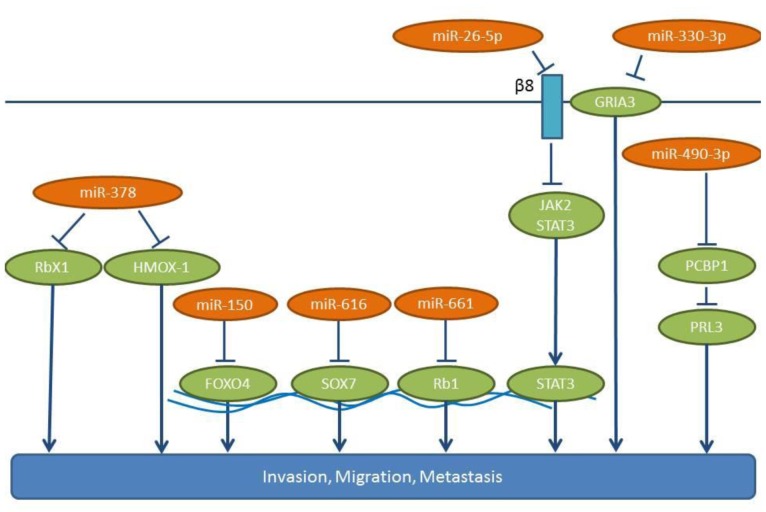
Up-regulated lung cancer-related miRs with an in vivo activity in experimental metastasis models. miR-150 promotes NSCLC migration and EMT of H460 cells in vitro (25). In an in vivo model, H460 cells overexpressing miR-150 grow faster than control cells when injected subcutaneously, while more metastatic nodules have been observed in the lungs of mice with miR-150 overexpressing cells (25). The same group has also identified Forkhead- box O 4 (FOXO4) as a direct target of miR-150 (Figure 1). FOXO4 is a member of the FOXO family of transcription factors which can positively and negatively affect gene expression. FOXO4 can induce cell cycle arrest and apoptosis as well as DNA damage repair and can function as a tumor suppressor in human cancer (26,27). Up-regulation of miR-150 and down-regulation of FOXO4 is frequently observed in metastatic lung cancer cells and NSCLC tissues (25).
Figure 1. Mode of action for miRs-26-5p, -150, -330-3p, -378, -490-3p, -616 and -661. These miRs are up-regulated in NSCLC tissues in comparison to matching normal tissues. The pro-metastatic pathways activated by these miRs are indicated. β8: Integrin β8, FOXO4: forkhead-box-protein 4, GRIA3: glutamate ionotropic receptor AMPA 3, HMOX1: hemoxygenase 1, JAK2: Janus kinase 2, PCBP1: poly(rc)-binding protein 1, PRL3: phosphatase of regenerating liver 3, Rb1: retinoblastoma protein 1, RbX: ring-box protein 1, SOX7: sry-box transcription factor 7, STAT3: signal transducer and activator of transcription 3.
miR-490-3p potentiates in vitro invasion and migration of the A545 NSCLC cell line (28), while an antisense-oligonucleotide (ASO) directed against miR-490-3p can reduce the in vivo lung colonization following tail vein injection of A549 lung cancer cells (28). Poly r(C) binding protein 1 (PCBP1), a negative regulator of lung cancer metastasis (29), has been identified as a direct target of miR-490-3p (28, Figure 1). PCBP1 acts in a transcript-selective translational pathway, in which a ribonucleoprotein complex containing PCBP1, silences the translation of mRNAs involved in EMT and metastatic progression. Prometastatic phosphatase of regenerating liver 3 (PRL3) is one of the proteins repressed by PCBP1 (30,31). miR-490-3p expression is inversely correlated with PCBP1 and correlates with disease progression in lung cancer patients (28).
miR-616 promotes proliferation, migration and invasion of NSCLC cell lines, such as H358 and A549 (32). Overexpression of miR-616 increases the subcutaneous growth of H358 cells in nude mice, and tail vein injection experiments have revealed that miR-616 overexpression significantly increases the frequency of lung metastases (32). SOX7 has been identified as a direct target of miR-616 in NSCLC cells (32). SOX7 belongs to the SRY-related HMG-box (SOX) family of transcription factors with tumor suppressing properties (33,34) (Figure 1). Transcript analysis, based on TCGA data, has revealed an up-regulation of miR-616 in lung cancer tissues compared to matching normal lung tissues (Figure 2).
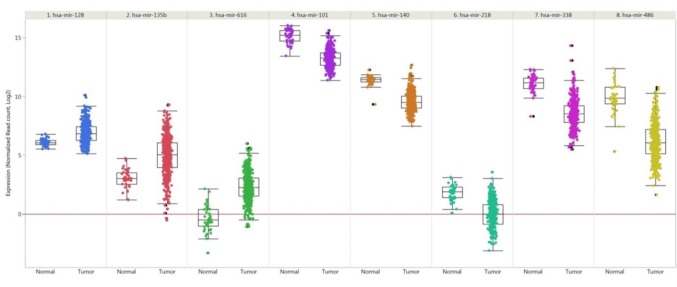
Figure 2. Expression of selected microRNAs in lung squamous carcinoma compared to matching normal tissues. Up-regulated miRs: -128, -135b, - 616. Down-regulated miRs: -101, -140, -218, -338 and -486. Data from 478 lung cancer samples and 45 matching normal samples derived from the LUSC cohort of The Cancer Genome Atlas (TCGA) are shown. miRNA expression was quantified by RNA sequencing and is shown as log2 of normalized read counts. The red lines indicate low versus higher expression. Expression data are shown as box plots. The line in the middle of the box represents the median values, the rectangles show the upper and lower 25% quartiles and 50% of all data points are included in the greater rectangle. All other data points, except for outliers are located within the upper and lower whiskers.
miR-661 is up-regulated in NSCLC and is an indicator of poorer prognosis (35). In A549 and SPC-A1 lung cancer cells, miR-661 enhances their migration (wound-healing assay, transwell assay), and promotes a significantly faster cell growth and colony formation (35). A549 cells overexpressing miR-661 give rise to more pulmonary metastases in nude mice following tail vein injection and form larger tumors compared to control cells following subcutaneous injection. Tumor suppressor Retinoblastoma 1 (Rb1) has been identified as a direct target of miR-661 (35) (Figure 1). Rb1 knock-down induces EMT by decreasing E-cadherin and EMT-related factors, such as Slug and Zeb1, in breast cancer cells (36). Furthermore, down-regulation of Rb1 is followed by up-regulation of the Rb-binding transcription factor E2F1, which promotes proliferation and invasion (37).
miRs promoting lung cancer metastasis identified in spontaneous metastasis models. miR-26a-5p enhances lung cancer cell invasion and migration following its transfection in A549, H1299 and H661 NSCLC cells (38). Integrin-β8 has been identified as a target for miR-26a-5p, a molecule which is overexpressed in several types of cancer together with integrin-αv (39,40). miR-26a-5p induces expression of prometastatic gene cluster of differentiation (CD44) and matrix metalloproteinase 9 (MMP-9) and activates Janus kinase 2/Signal transducer and activator 3 (JAK2/STAT3) signaling, which leads to an enhanced metastatic potential of NSCLC cell lines (38) (Figure 1). Knock-down of integrin-β8 has been shown to induce JAK2/STAT3 signaling (38). miR-26a-5p promotes tumor formation and metastases in the lungs of subcutaneously implanted xenografts (38). Data correlating the expression levels of miR-26a-5p with the clinical status of NSCLC patients are not yet available.
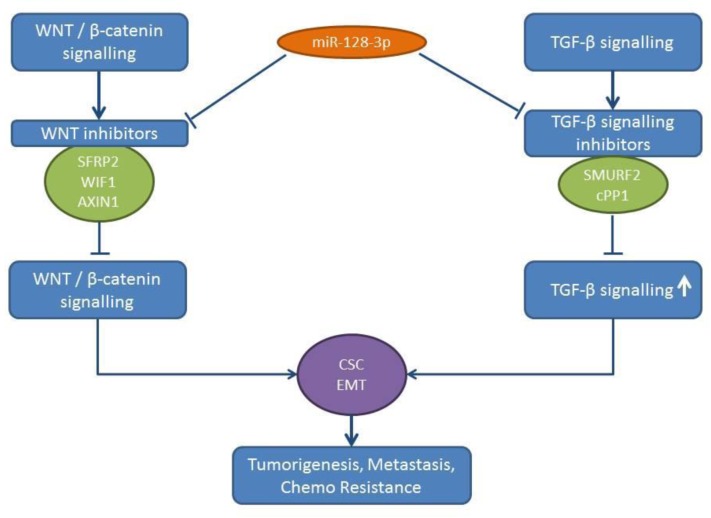
miR-128-3p induces EMT and CSC programming in lung cancer cells, such as A549 and Calu3D (41). A549 and H20 lung cancer cells transfected with miR-128-3p display an enhanced tumorigenicity and spontaneous metastatic colonization potential in multiple distal organs following their subcutaneous injection (41). miR-128 acts by targeting inhibitors of the Wnt/beta catenin pathway and activates the transforming growth factor-β (TGFβ) signaling by inhibiting negative regulators of the TGFβ signaling pathway (41). AXIN1, secreted frizzled related protein 2 (SFRP2) and Wnt inhibitory factor (WIF1) have all been identified as targets of the Wnt/beta catenin pathway (41,42). SMAD specific E3 ubiquitin protein ligase 2 (SMURF2) (43) and the catalytic subunit of protein phosphatase 1 (cPP1) (44) have emerged as targets of miR-128-3p in the TGFβ signaling pathway (45) (Figure 3). Expression of miR-128-3p correlates with chemoresistance, aggressiveness and poor prognosis in NSCLC patients (41). TCGA-related data reveal an up-regulation of miR-128-3p in lung cancer tissues compared to normal lung tissues (Figure 2).
Figure 3. miR-128-3p mode of action. miR-128b is up-regulated in NSCLC samples compared to matching normal tissues. miR-128-3p activates Wnt/β- catenin and TGFβ signaling, resulting in pro-metastatic activity. Axin 1: WNT pathway inhibitor, cPP1: dual specificity protein tyrosine phosphatase 1. CSC: cancer stem cell, EMT: epithelial mesenchymal transition, SFRP2: secreted frizzled-related protein 2, SMURF2: smad ubiquitin regulatory factor 2, TGFβ: transforming growth factor β, WIF-1: WNT inhibitory factor 1, WNT: wingless-type MMTV integration site family member.
miR-378 promotes the migration of A549 lung cancer cells as shown by wound healing assay (46). miR-378 mediates the expression of angiogenesis-promoting factors, such as vascular endothelial growth factor (VEGF), MMP2, MMP9, Interleukin 8 (IL8), Angiopoietin 1 and Stromal-derived factor -1 (SDF-1) (46). In A549 and NCI-292 lung cancer cells miR-378 induces formation of vascular tubes. In a zebrafish xenograft model, miR-378 inhibitors reduce the metastatic potential of A549 cells throughout the body and to the lungs (46). RBX1 has been identified as a direct target of miR-378 (46-48). Ring box protein X1 (RBX1), also referred to as regulator of cullins 1 (ROC1), is the ring component of the Cullin-based RING Ligase (CRL), the largest family of E3 ubiquitin ligases (47) (Figure 1). The details of the mechanism of miR-378/RBX1 signaling are not delineated yet. Swiss immunodeficient mice subcutaneously implanted with NCI-292 cells show that miR-378 enhances the vascularization of the xenografts as well as the distal metastasis to the brain and the lungs (49). In this system, the mode of action (MOA) might be based on the interaction of miR-378 with hemoxygenase 1 (HMOX1) (50), but this needs to be supported by additional studies. miR-378 is a potential biomarker in high-risk NSCLC patients for brain metastasis (46).
Identification of miRs promoting lung cancer in orthotopic metastasis models. miR-93 up-regulation promotes cell proliferation, migration and invasion in NSCLC cell lines, such as A549, H1975 and H1299 (51). miR-93 directly binds to the 3’-UTR of the tumor suppressor gene liver kinase B1 (LKB1) mRNA (51-53). It has been shown that miR-93 activates the PI3K/AKT pathway by inhibiting LKB1, Phosphatase and tensin homolog (PTEN) and protein 21 (p21) (51). Down-regulation of miR-93 in A549 cells orthotopically implanted in the lungs gave rise to a lower rate of pleural dissemination while no hepatic metastases could be identified with these cells compared to A549 cells (51).
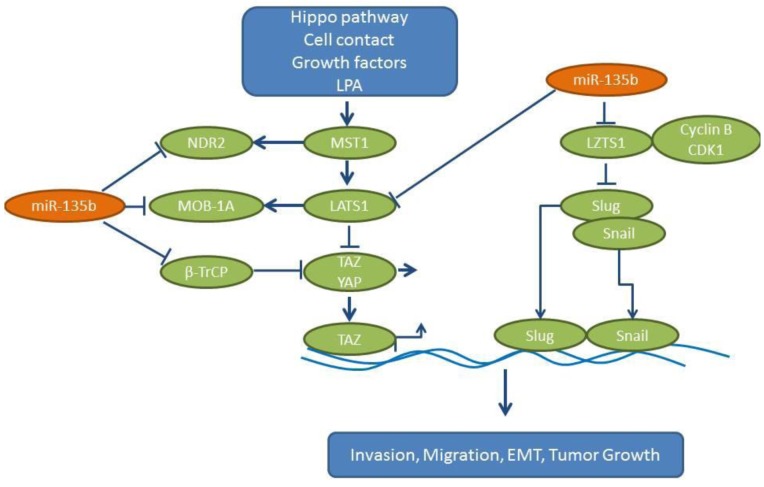
Expression of miR-135b correlates with invasivity in several lung cancer cell lines and promotes growth, invasion and EMT in vitro (54). CL1-0 cells transfected with miR-135b have shown increased tumor growth and metastases to the lungs and soft tissues of non-obese diabetic – severe combined immuno-deficient (NOD-SCID) mice following tail vein injection. A miR-related antagomir can inhibit orthotopic lung cancer growth of CL1-5 lung cancer cells and decrease the number of lung metastases (54). Leucine putative tumor suppressor (LZTS1), which may interact with the cdk1/cyclin B complex, has been identified as one of the targets of miR-135b (55). In addition, miR-135b targets several components of the Hippo pathway (Figure 4). This pathway has an impact on cancer stem cells, tumor growth, EMT, invasion and metastases of tumor cells (56). The following Hippo pathway components are direct targets of miR-135b: i) large tumor suppressor gene 1 (LATS1), which functions as a serine-threonine kinase, ii) MOB kinase activator 1A (MOB-1A), a non-catalytic protein, iii) nuclear Dbf2-related protein kinase 2 (NDR2), which binds to MOB-activator proteins (57) and finally iv) ubiquitin ligase β transducin containing protein (βTrCP) (58). Collectively, these modulations lead to increased levels of dephosphorylated oncoprotein transcriptional co-activator with PDZ-binding motif (TAZ), which translocates in the nucleus and acts as a transcriptional co-activator of genes involved in cell proliferation and survival (59,60). Interestingly, the expression levels of miR-135b, LZTS1 and nuclear TAZ predict the outcome in NSCLC patients. TCGA-related data reveal an up-regulation of miR-135b in lung cancer tissues compared to normal lung tissues (Figure 2) (54).
Figure 4. miR-135b mode of action. miR-135b is up-regulated in NSCLC samples compared to matching normal tissues. miR-135b exhibits a prometastatic activity through activation of the Hippo pathway and induction of EMT- mediating transcription factors Slug and Snail. β-TrCP: β- Transducin repeat containg protein, cdk1: cyclin-dependent kinase 1, LAP: lysophosphatic acid, LATS1: large tumor suppressor kinase 1, LZTS1: leucine zipper tumor suppressor 1, MOB-1A: MOB kinase activator 1A, MST1: mammalian sterile 20-like kinase 1, NDR2: nuclear Dbf2-realted 2, Slug, Snail: transcription factors, TAZ: transcriptional activator with a PDZ binding domain, YAP: Yes-associated protein.
The miR-330 promotes proliferation, migration and invasion of H460 and H1975 NSCLC cell lines in vitro (61). In addition, it promotes tube formation and angiogenesis of endothelial cells (61). The miR-330 also promotes the growth of H460 and H1975 cells in xenograft models, as following their orthotopic implantation in the brain, many metastatic foci containing H460 or H1975 NSCLC cells overexpressing miR-330 have been identified. It has been shown that miR-330 can directly target the 3’-UTR of the mRNA for the glutamate ionotropic receptor AMPA type subunit 3 (GRIA3), a ligand-gated ion channel of the central nervous system with a role in excitatory synaptic transmission (62,63) (Figure 1). Furthermore, miR-330 up-regulates total DNA methylation in NSCLC cells (61). Immuno-precipitation experiments have indicated that GRIA3 is associated with DNA methyltransferases DNMT1 and DNMT3 (61). miR-330 might be a biomarker for the identification of NSCLC patients with a brain metastasis potential and could play a role as a therapeutic target for the prevention and inhibition of brain metastasis of NSCLC (61).
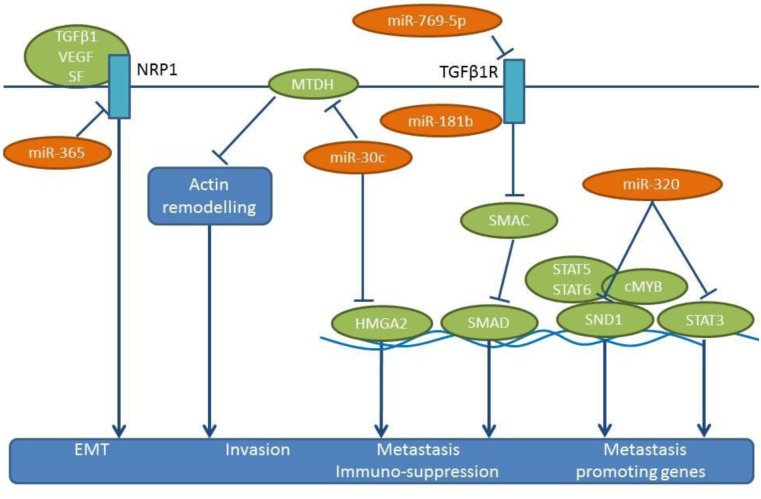
Identification of down-regulated miRs preventing lung cancer metastasis in models of experimental metastasis. miR-30c is activated by Fragile Histidine Triad (FHIT) and inhibits EMT by suppressing direct targets vimentin and fibronectin in A549 cells (64). FHIT reduces the mobility and invasiveness of lung cancer cells in vitro through miR-mediated suppression of EMT (64). In addition, miR-30c targets the metastases-related genes metadherin (MTDH) and high-mobility group AT-hook 2 (HMGA2) (65,66) (Figure 5). Overexpression of miR-30c in parental A549 cells suppresses the experimental metastases in lungs and brain by inhibiting the expression of MTDH and HMGA2 (64). MTDH regulates metastasis by acting on the actin cytoskeletal remodeling of NSCLC cells (65). HMGA2 has an impact on lung cancer cell proliferation and metastasis (66), by affecting cell apoptosis through caspases 3/9 and Bcl2, and it regulates EMT by targeting Twist. As indicated by immuno-histochemistry, comparing non-metastatic and metastatic lung tissues, FHIT is a negative regulator of metastasis in NSCLC (64).
Figure 5. miRs-30c, -181b, -320, -365 and -769-5p: mode of action. These miRs are down-regulated in NSCLC tissues compared to matching normal tissues. The corresponding pro-metastatic pathways activated by their down-regulation are shown. HMGA2: High mobility group A2, MTDH: metadherin, MYB: transcription factor MYB encoded by the MYB gene, NRP1: neuropilin 1, SF: semaphorin, SMAD: signal transducers of the TGFβ superfamily, SND1: staphylococcal nuclease and tudor domain containing 1, STAT 3,5,6: signal transducer and activator of the TGFβ superfamily, TGFβ: transforming growth factor β, VEGF: vascular endothelial growth factor.
miR-181b has been identified as a suppressor of metastasis and inhibits the proliferation of NSCLC cell lines, such as A549, H1650 and A549/DDP (67). In addition, miR-181b attenuates the migration, invasion and EMT of these cell lines (67). Transforming growth factor β receptor 1 (TGFβR1) has been identified as a direct target of miR-181b (67) (Figure 5). Suppression of TGFβR1 results in the inhibition of Smad-dependent and –independent signaling (67), while TGFβ signaling has been shown to exert metastatic and immuno-suppressive effects (68,69). In vivo, miR-181 suppresses the formation of lung metastases following tail vein injection of A549/DDP cells (70).
miR-320 suppresses the growth and invasion of NSCLC cell lines A549/34R cells and increases apoptosis of them (69,71). miR-320 targets STAT3 and staphylococal nuclease domain containing protein 1 (SND1) (Figure 5) (72,73). A549/34R NSCLC cells transfected with miR-320 exhibit considerably lower tumor volumes in mice compared to non-transfected cells (69). Following tail vein injection, miR-320-transfected A549/34R cells display reduced numbers of metastatic nodules in the lungs (69). miR-320 affects the nuclear translocation of pSTAT3, with an impact on downstream effectors, such as BCL2 (down-regulated) and BAX (up-regulated) (69). SND1 (p100), the other direct target of miR-320, is a multi-functional protein that regulates gene expression at both transcriptional and translational levels (73). SND1 can interact with c-MYB, STAT5 and STAT6 as a co-activator (73). In the context of metastasis, SND1 has been identified as a metadherin interacting protein (74). SND1 also acts as a component of the RNA-induced silencing complex, which is involved in the degradation of edited double-stranded RNA molecules (75).
miR-365 is down-regulated in lung cancer cell lines and lymph node metastases, while its expression inversely correlates with clinical stage, overall survival and relapse-free interval of NSCLC patients (76). miR-365 inhibits cell migration, invasion, growth and soft agar colony formation, and incuces apoptosis in A549 and BE1 NSCLC cell lines (76). In vivo, miR-365 inhibits the growth of subcutaneously implanted A549 cells and reduces lung metastases of tail-vein injected A549 cells transfected with an expression vector containing miR-365 (76). Neuropilin 1 (NRP1) has been identified as a direct target for miR-365 (76) (Figure 5). NRP1 is a transmembrane glycoprotein that acts as a co-receptor for several extracellular targets, such as diverse isoforms of VEGF, semaphorins and TGFβ (77-79). Functionally, NRP1 promotes EMT in the context of TGFβ, hedgehog and HGF/c-MET signaling (77-79).
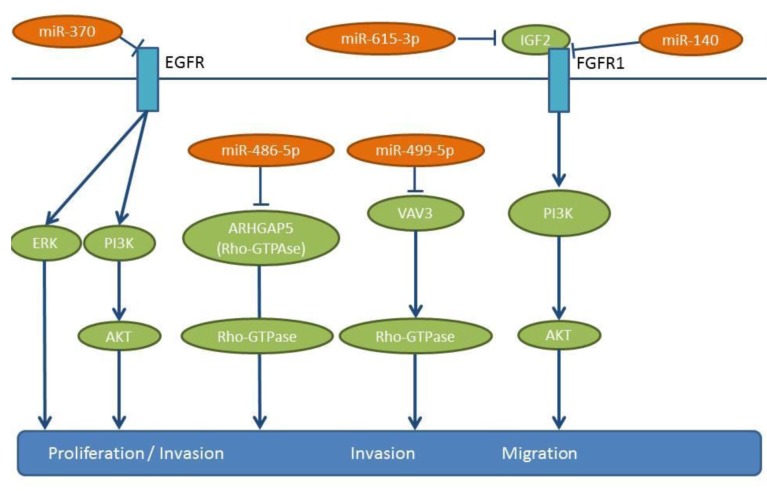
miR-370 overexpression inhibits the proliferation, clonogenicity, wound-healing and invasion of XWLC-05 and X157 NSCLC cells (80). miR-370 overexpression inhibits the growth and angiogenesis of XWLC-05 cells following their implantation in mice, as well as lung metastasis following tail vein injection. miR-370 targets the 3’-UTR of EGFR, a known promoter of proliferation and metastasis (81,82) (Figure 6). In XWLC-05 and X157 cells, miR-370 attenuates their expression of EGFR and down-regulates Hypoxia inducible factor 1α (HIF-1α), Extracellular regulated protein kinase (ERK) and AKT signaling. Patient related data in NSCLC patients are not yet available for miR-370 (80).
Figure 6. miRs-140, -370, -486-5p, -499-5p and -615-3p: mode of action. These miRs are down-regulated in NSCLC tissues compared to matching normal tissues. The pro-metastatic pathways activated by down-regulation of these miRs are shown. ARHGAP5: Rho GTPase-activating protein 5, AKT: serine-threonine kinase AKT, EGFR: epidermal growth factor receptor, ERK: extracellular regulated kinase, IGF2: insulin growth factor 2, IGFR1: insulin-like growth factor receptor 1, PI3K: phosphatidyl-inositol 3-kinase, VAV3: VAV3 guanine nucleotide exchange factor.
miR-486-5p expression inversely correlates with advanced stage and lymph node metastasis of NSCLC (83). Ectopic expression of miR-486-5p reduces proliferation and migration in cell lines A549 and H157 (83). Tail vein injection with bioluminscent H460 NSCLC cells transfected with a miR-486-5p mimic can give rise to reduced lung metastases (83). Protumorigenic RhoA GTPase (ARHGAP5), a negative regulator of GTPase RhoA has been identified as a target of miR-486-5p (84) (Figure 6). Rho GTPases are regulators of cellular adhesion, motility and polarity (85,86). Up-regulation of ARHGAP5 correlates with the down-regulation of miR-486-5p in clinical specimens of NSCLC (83). TCGA-related data reveal a down-regulation of miR-486 in lung cancer tissues compared to matching normal lung tissues (Figure 2).
miR-499-5p expression is significantly reduced in NSCLC tissues compared to matching normal lung tissues, while its expression inversely correlates with a poor clinical outcome (87). miR-499-5p overexpression inhibits cell proliferation and invasion in vitro and in vivo and induces apoptosis in NSCLC cell lines, such as A549, H23, H522 and HOP62 (87). A549 cells transfected with miR-499-5p display reduced lung metastasis in mice following tail vein injection (87). VAV3 has been identifed as a direct target of miR-499-5p (Figure 6). VAV3 is a guanine nucleotide exchange factor for RhoGTPases (88). VAV proteins are involved in a variety of cellular processes, including phagocytosis, cytoskeletal organization and transformation (89). VAV3 expression is decreased in breast cancer patients and it regulates the dissemination of NSCLC to the lungs (89). However, the mechansism behind this remains elusive.
miR-615-3p expression is down-regulated in NSCLC specimens compared to corresponding matching normal tissue (90). In A549 and H1975 NSCLC cells, miR-615-3p inhibits cell proliferation, migration and invasion (90). Following subcutaneous implantation of A549 cells overexpressing miR-615-3p tumor growth is inhibited (90). On the contrary, inhibition of mR-615-3p in A549 cells increases the lung metastatic burden following tail vein injection (90). Insulin-like growth factor 2 (IGF2) has been identified as a direct target of miR-615-3p (90,91) (Figure 6), and it can rescue the in vitro effects of miR-615-3p when it is overexpressed (90). A negative correlation between miR-615-3p levels and IGF2 has been identified in NSCLC specimens, with IGF2/IGFR1 interaction acting as a mediator of metastasis in several types of tumors (92).
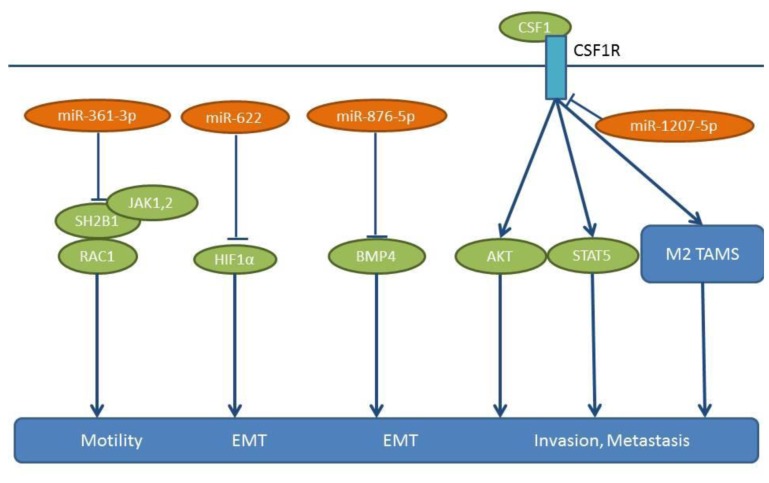
miR-622 transfection of NSCLC cell lines A549 and H1299 show ≥50% inhibition of migration and invasion compared to untransfected cells (93). In the tail vein injection model, miR-622 transfected A549 cells show significantly fewer metastases in the lungs compared to untransfected cells. miR-622 suppresses HIF-1α expression by directly targeting the 3’-UTR of HIF-1α mRNA (93) (Figure 7). miR-622 is up-regulated by Fork head box 3A (FOXO3A) and ERK activation down-regulates the FOXO3A/miR-622 axis to increase HIF-1α expression. HIF-1α acts as a promoter of EMT through the down-regulation of E-cadherin and up-regulation of β-catenin and vimentin expression (94,95).
Figure 7. miRs-361-3p, -622, -876-5p and 1207-5p: mode of action. These miRs are down-regulated in NSCLC specimens compared to matching normal tissues. The pro-metastatic pathways activated due to their down-regulation are shown. AKT: Serine-threonine kinase AKT, CSF1: colony stimulating factor 1, CSFR1: colony stimulating factor receptor 1, BMP4: bone morphogenetic protein 4, EMT: epithelial mesenchymal transition, HIF-1 alpha: hypoxia inducible factor 1 alpha, JAK1,2: Janus kinase 1,2, M2 TAMS: M2 tumor-associated macrophages, RAC1: Ras-related C3 botulinum toxin substrate 1, SH2B1: SH2B adaptor protein 1.
miR-876-5p has been found to be down-regulated in NSCLC cell lines and in tumor samples (96). miR-876-5p inhibits the migration and invasion of NSCLC cell lines A549 and HCC827, as assessed by wound healing invasion assay (96). Bone morphogenetic protein 4 (BMP4) has been identified as a direct target of miR-876-5p (96) (Figure 7) and is known as an inducer of EMT (97-99). Re-introduction of BMP4 into the cell lines as outlined above restores proliferation and invasion (96). NSCLC cell lines A549 and HCC827 expressing miR-876-5p give rise to fewer lung metastases compared to mice injected intravenously with control cell lines in the tail vein (96).
Another molecule, miR-1207-5p, is down-regulated in NSCLC cell lines and it inhibits the proliferation and invasion of NSCLC cell lines A549 and H358 (100). Colony stimulating factor-1 (CSF-1) has been identified as a direct target of miR-1207-5p (100) (Figure 7). miR-1207-5p inhibits CSF1/CSFR1-mediated AKT and STAT3 signaling and suppresses the tumorigenicity of A549 cells, while supernatants of A549 cells transfected with miR-1207-5p mimics inhibit tube formation of endothelial cells (100). miR-1207-5p agomirs inhibit lung metastasis of A549 cells following tail vein injection, while in NSCLC patients expressing high levels of miR-1207-5p the overall survival rates are significantly higher than in those expressing low miR-1207-5p levels (100). CSF-1 can be secreted by macrophages, epithelial cells, fibroblasts and tumor cells and it can stimulate the proliferation and invasion of tumor cells directly. In addition, CSF-1 can modify the Tumor microenvironment (TME), by promoting Tumor associated macrophages (TAMs) (M2 phenotype, high levels of IL10, VEGF and Arg-1), which promote tumor growth and metastasis (101-103).
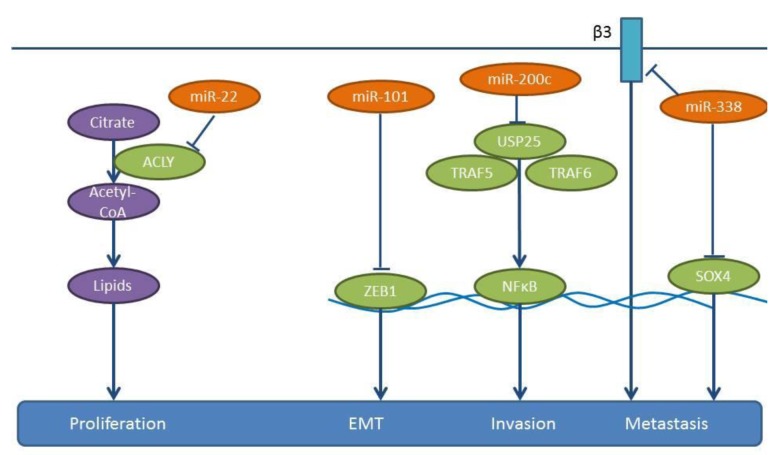
miRs inhibiting lung cancer metastasis identified in spontaneous models of metastasis. miR-22 reduces the proliferation, invasion and migration of A549 cells and induces their apoptosis (104). A549 cells, subcutaneously implanted into immune-deficient mice and subsequently treated intratumorally with a lentivirus-expressing miR-22, show an inhibition of tumor growth and a reduced probability of metastasis to distant organs, such as lung, liver, kidney, spleen and intestine compared to the control cell line A549 (104). ATP-citrate lyase (ACLY) has been identified as a direct target of miR-22 (104) (Figure 8). ACLY generates acetyl-CoA from citrate as a substrate. Acetyl-CoA is a substrate for fatty acid synthase and is involved in cholesterol synthesis via the mevalonate pathway (105,106), which can mediate tumor growth and invasion (105,106). Correlation between expression of miR-22 and ACLY in clinical NSCLC specimens needs further investigation.
Figure 8. miRs-22, -101, -200c and -338: mode action These miRs are down-regulated in NSCLC samples compared to matching normal tissues. Their down-regulation results in activation of pro-metastatic pathways shown here. ACLY: ATP citrate lyase, β3: integrin-β3, EMT: epithelial mesenchymal transition, NfkappaB: nuclear factor kappa B, SOX4: SRY-related HMG-box4, TRAF5,6: TNF receptor associated factor 5,6, USP25: ubiquitin specific lyase peptidase 25, ZEB1: Zinc finger E-box binding homeobox 1.
miR-101 suppresses the proliferation, migration and apoptosis resistance in A549 cells (107). Expression of miR-101 in A549 cells inhibits tumor growth following their subcutaneous implantation into severe combined immuno-defiecient (SCID) mice as well as lung metastases following their tail vein injection, as revealed by in vivo luciferase imaging. Zinc finger E-box binding homeobox 1 (ZEB1), a member of the ZEB family of transcription factors, has been identified as a target of miR-101 (107,108) (Figure 8). ZEB1 promotes EMT in cancer cells by suppressing E-cadherin (107,108). Expression levels of miR-101 inversely correlate with lymph-node metastasis and poor prognosis in NSCLC patients (107). TCGA-related data reveal that miR-101 is down-regulated in lung cancer tissues compared to normal lung tissues (Figure 2).
Expression of miR-140 is decreased in five NSCLC cell lines in comparison to the normal human bronchial cell line BEAS-2B (109), while down-regulation of miR-140 significantly correlates with late tumor stage and metastases (109). miR-140 inhibits the proliferation and migration of A549 and H157 cells (109) and down-regulates IGFR-1R by directly targeting its 3’-UTR (109) (Figure 6). miR-140 inhibits tumor growth and the metastatic potential of transfected A549 cells subcutaneously implanted to the lungs of nude mice (109). Interestingly, the effects of miR-140 expression can be recapitulated by the inhibition of Insulin-like growth factor receptor 1 (IGF-1R) (109). IGF-1R is an important regulator of cell proliferation, survival and metastasis in many types of malignancies (110,111). TCGA-related data confirm the lower expression levels of miR-140 in lung cancer tissues compared to normal lung tissues (Figure 2).
miR-200c suppresses the migration and invasion of NSCLC cells, such as A549, H1299 and SPC-A-1 (112) (Figure 8). In a model of experimental metastasis, SPC-A-1 cells transfected with a miR-200c inhibitor, the number of lung metastases increased compared to the control group (112). In a xenograft model, implanted SPC-A-1 cells overexpressing miR-200c through a lentiviral vector also impair the spontaneous metastasis to the lungs (112). Ubiquitin specific peptidase 25 (USP25) has been identified as a direct target of miR-200c (112). USP25 is involved in the negative regulation of IL17-mediated signaling and inflammation, by interacting with Tumor necrosis factor receptor associated factors 5 and 6 (TRAF5 and TRAF6) and by regulating Toll-like receptor 4 (TLR4)-dependent innate immune responses, via the de-ubiquitination of the adapter protein TRAF3 (112,113). The role of miR-200c in the metastasis of NSCLC requires further investigation. Knock-down of USP25 in A549 cells inhibits NSCLC metastasis in vivo (112). NSCLC specimens expressing high levels of miR-200c tend to express low levels of USP25 (112). Importantly, miR-200c negatively regulates tumor metastases in NSCLC patients by targeting USP25 (112).
miR-338 is down-regulated in highly metastatic lung cancer cells and lung cancer tissue specimens (114,115). The five-year survival rate of the low miR-338 expressing group is significantly lower than that of the high miR-338 expressing group (114). miR-338 inhibits proliferation, adhesion, tumor migration and invasion of NSCLC cell lines A549, NCI-H292 and 9981 in vitro (114,115). It was shown that 9,981 cells overexpressing miR-338-3p and injected into the posterior flank of mice give rise to less metastatic lung nodules compared to the untransfected control cell line (114). Similarly, fewer lung metastases can be identified with NCI-H292 cells overexpressing miR-338 compared to the untransfected cell line, following tail vein injection (115). TCGA-related data also support the lower expression levels of miR-338 in lung cancer tissues compared to normal lung tissues (Figure 2). SR4-related HMG-box (SOX4) and integrin-β3 have been identified as direct targets of miR-338 (114,115) (Figure 8). The transcription factor SOX4 is an inducer of EMT and has been found to be overexpressed in several types of cancer (116). Integrin-β3 is a receptor of the ECM of tumor cells and plays a role in tumor invasion and metastasis (117). In addition, increased expression of integrin-β3 correlates with a strong metastatic potential in Colorectal carcinoma (CRC) (118).
Expression of another molecule, miR-361, is inversely associated with clinico-pathological characteristics and its expression correlates with a good prognosis of NSCLC, as shown by comparing frozen NSCLC tissues to the corresponding normal lung tissues (119). As shown in NSCLC cell lines A549, HTB-182 and SPC-A-1, miR-361-3p inhibits their proliferation, migration and invasion in vitro (119), by binding to the 3’-UTR of its target SH2B1 adaptor protein 1 (SH2B1) (Figure 7) (119). NSCLC cell lines A549 and HTB-182 stably expressing miR-361-3p exhibit reduced tumor growth and lung metastasis following subcutaneous and orthotopic implantation (119). Gain and loss-of function SH2B1 mutations can abrogate or mimick the impact of miR-361 on cell proliferation, migration and invasion in cell lines A549, HT-182 and SPC-A-1 (119). SH2B1 has a src homology 2 and pleckstrin homology (PH) domain and functions as an adaptor protein that can bind to janus kinases 1 and 2 (JAK1 and JAK2), fibroblast growth factor receptor, insulin receptor and insulin receptor substrate 1 (120-122). As an adaptor/scaffolding protein, it can recruit Rac GTPases to activated membrane receptor JAK complexes or to receptor tyrosine kinases to regulate the cytoskeleton and promote membrane ruffling and cell motility (120-122).
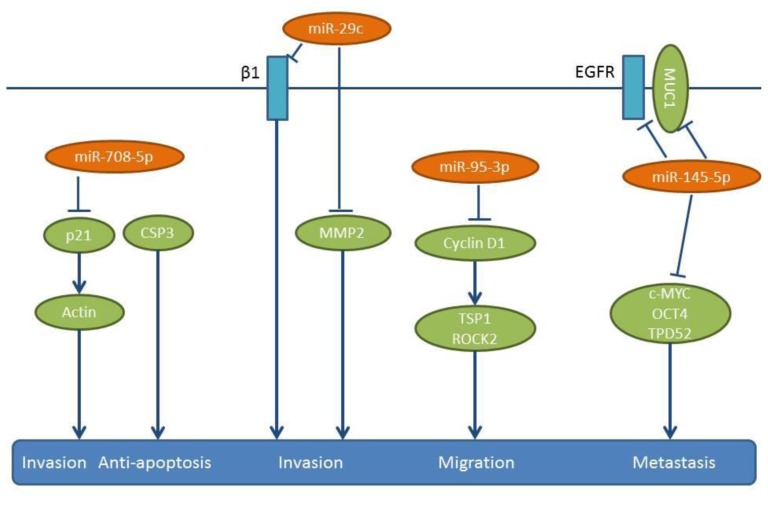
miR-708-5p expression inversely correlates with the metastatic potential of NSCLC cell lines and tissue samples (123). Transfection of NSCLC cell lines PG, A549 and H1299 with miR-708-5b induces apoptosis and inhibits their migration and invasion (123). The anti-metastatic activity of miR-708-5p has been evaluated in two in vivo models (123). In a tail vein injection model, a “sponge”, which contains multiple binding elements for miR-708-5p increased metastasis to the lungs and the heart, while in a spontaneous metastasis model of subcutaneously implanted A549 cells, miR-708-5p delivered intravenously twice weekly for three weeks attenuated lung metastasis significantly (123). The same group has identified cyclin-dependent kinase inhibitor p21 as as a direct target of miR-708-5p (Figure 9), which suppresses not only cytoplasmic p21, but also its translocation to the nucleus (123). p21–caspase 3 complexes inhibit apoptosis, while down-regulation of p21 in the cytoplasm weakens actin rearrangement and leads to decreased cell motility (124,125).
Figure 9. miRs-29c, 95-3p, -145-5p and -708-5p: mode of action. These miRs are down-regulated in NSCLC compared to matching normal tissues. Their down-regulation results in the activation of pro-metastatic pathways as shown here. β1: Integrin-β1, CSP3: caspase 3, EGFR: epidermal growth factor receptor, MMP2: matrix metalloproteinase 2, MYC: transcription factor MYC, Oct 4: octamer-binding transcription factor 4, p21: protein 21, ROCK2: Rho-associated coiled-coil containing protein kinase 2, TPD52: tumor protein D52, TSP1: Thrombospondin 1.
miR-769-5p is down-regulated in NSCLC compared to matching normal tissues and is a predictor of poor prognosis (126). The use of miR-769-5p mimics and inhibitors has shown that miR-769-5p inhibits proliferation, migration and invasion of several NSCLC cell lines, such as A549, H157, A973 and GLC82 (126). The tumor growth and metastasis-related properties of miR-769-5p have been assessed by subcutaneous implantation of A973 cells in nude mice and by intravenous injection with a miR-769-5p antagomir. Tumor volume, as well as prevalence of metastasis in the lungs were significantly enhanced in the presence of the antagomir (126). TGFβR1 has been identified as a direct target of miR-769-5p (126) (Figure 5). The role of the TGFβ1/TGFβR1 signaling in metastasis has been described for several tumor entities (127-129).
Lung cancer metastasis inhibitory miRs identified following intracardiac injection of lung cancer cells. mir29c is down-regulated in the metastatic NSCLC cell line 95D, and it suppresses proliferation, adhesion to ECM as well as invasion and migration of these cells in vitro (130). In an in vivo mouse model of metastasis, nude mice injected intracardially with 95D cells transfected with miR-29c mimics, have shown reduced metastasis to the bones and the liver (130). miR-29c directly targets the 3’-UTR of integrin-β1, as well as of the MMP2 gene (130) (Figure 9), whose enzymatic activity is inhibited by miR-29c (130) MMP2, also referred to as gelatinase A, is an inducer of EMT, but also a mediator of metastasis through the degradation of ECM components of the basement membrane (131). Integrin-β1 has been shown to facilitate cancer cell proliferation, adhesion, migration as well as metastasis (132,133).
Lung cancer inhibitory miRs identified in intracardiac injection/orthotopic models or in orthotopic models. miR-95-3p is another molecule that is down-regulated in brain metastatic lesions of NSCLC and correlates with poor prognosis (134). miR-95-3p inhibits metastasis of orthotopically implanted PC14PE6/LvBr4 cells to whole body and brain (134). Expression of miR-95-3p suppresses the invasiveness, proliferation and clonogenicity of brain metastatic PC14PE6/LvBr4 cells, through the down-regulation of cyclin D1 (134) (Figure 9). Cyclin D1 is a direct target of miR-95-3p (134) and is involved in cancer progression, including metastasis (135). Cyclin D1 increases cellular migration by down-regulating thrombospondin 1 and Rho-activated Kinase 2 (ROCK2) and by interacting with the cell-cycle inhibitor p27 (136). Cyclin D1 is induced by TGFβ and is reponsible for TGFβ-mediated migration in cooperation with p21 (137).
In highly metastatic lung cancer cells, miR-145-5p expression is maintained at low levels by epigenetic modifications, such as hypermethylation and deacetylation of its promoter (138). Expression of miR-145-5p impairs cell migration of H1299 NSCLC cells (138). In similar experiments, intracranial orthotopic injection of A549 and A549 transfected with miR-145-5p restrains their invasive capacity both in vitro and in vivo (138). Several direct targets of miR-145-5p have been identified, such as c-MYC, EGFR, Mucin-1 (MUC-1), Octamer binding transcription factor 4 (OCT-4) and Tumor Protein D52 (TPD52) (138-141) (Figure 9). Out of these targets, the down-regulation of MUC-1 protein is overexpressed in many types of carcinomas and known to prevent EGFR nuclear localization and thus has an inhibitory effect on EGFR signaling (138). Taken together, the down-regulation of miR-145-5p leads to activation of a network of oncogenic proteins involved in metastasis.
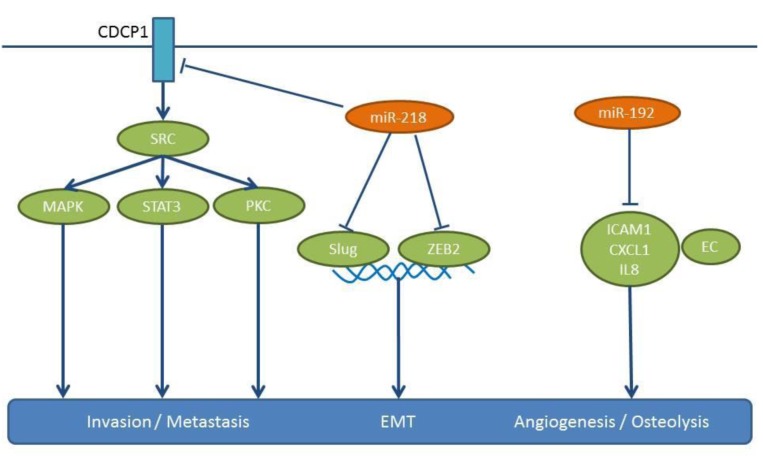
In the highly metastatic subpopulations of the NSCLC cell line A549, an inverse correlation between expression of miR-192 and in vitro invasion has been found (142). Intracardiac injection of cells overexpressing miR-192 has revealed a significant reduction of tumor burden and bone osteolytic lesions (142). A549 cells overexpressing miR-192 can release exosome-like vesicles (ELV) containing miR-192, which can deliver their cargo into the cytosol of neighbouring cells, such as endothelial cells (142). miR-192-enriched cells can modulate angiogenesis and impact tumor-induced osteolysis, as they are transferred into the bone marrow via the systemic circulation. Intratibial injection of highly metastatic M1 cells and intravenous injection of ELVs from A549 cells can lead to a six-fold decrease in osteolytic lesions of mice (142). In this context it has been shown that miR-192 down-regulates mediators of angiogenesis, such as Intercellular cell adhesion molecule1 (ICAM1), Chemokine (CXC) motif ligand 1 (CXCL1) and IL8 in endothelial cells (Figure 10). In conclusion, miR-192 mediates inhibition of angiogenesis as well as bone resorption, however, the direct targets of miR-192 in this context remain to be identified.
Figure 10. miRs-192 and -218: mode of action. These miRs are down-regulated in NSCLC in comparison to matching normal tissues. Their downregulation results in the activation of pro-metastatic pathways, as shown here. CDCP1: CUB domain containing protein 1, CXCL1: chemokine (C-XC motif) ligand 1, EC: endothelial cell, EMT: epithelial mesenchymal transition, ICAM1: intercellular adhesion molecule 1, IL8: interleukin 8, MAPK: mitogen-activated protein kinase, PKC: protein kinase C, Slug: transcription factor slug, SRC: tyrosine kinase SRC, STAT3: signal transducer and activator of transcription 3, ZEB2: zinc finger E-box binding homeobox 2.
miR-218 has been validated as a target for treatment of NSCLC with CUB domain containing protein 1 (CDCP1) and independently with Slug and Zinc finger box 2 (ZEB2) as its targets (143,144) (Figure 10). miR-218 is down-regulated in advanced clinical stage and metastases of NSCLC specimens compared to matching normal tissues (143,144). In B7brmx2 cells, doxocycline-dependent expression of miR-218 inhibits tumor cell motility, anchorage-dependent-free survival and tumor sphere formation (143). Furthermore miR-218 negatively regulates the migration, invasion and EMT of NSCLC cells, such as A549, H1299, PL9 and SPA-1 (143). Intracardially injected B7brmx2 cells into SCID mice with an inducible expression of mir-218 exhibit reduced lung metastasis compared to the control group. In another model, mice treated with a systemic delivery of liposomal complex plasmids expressing miR-218 twice weekly for three weeks have shown a significantly prolonged survival time compared to the control group (143). In addition, miR-218 inhibited the growth of H1299 and A549 cells injected into the posterior flanks of nude mice (143). Finally, in an orthotopic model in which trimmed tumor pieces were transplanted into the lungs of mice, fewer metastatic foci were found with miR-218 transfected cells compared to the control cell line (144). miR-218 targets CDCP1, a protein with an Extracellular domain (ECD) containing two CUB domains, which mediates loss of cell adhesion (145,146). CUB1 has a documented role in metastases (145,147), while miR-218 targets Slug and ZEB2 are validated mediators of EMT (147). TCGA-related data support the down-regulation of miR-218 in lung cancer tissues compared to normal lung tissues (Figure 2).
Therapeutic aspects. As outlined in the previous chapters, several up- or down-regulated miRs have been identified as mediators of metastasis in several NSCLC cell lines in nude mouse models. Some of these are associated with clinical parameters, such as metastasis to distant organs, prognosis and patient survival. Their identification relies on the use of different NSCLC cell lines and in vivo metastasis assays, such as tail vein injection and metastasis to the lungs, metastasis to distant organs following intracardiac or subcutaneous injection, as well as orthotopic bone or brain models followed by intraosseous dissemination or intracranial dissemination and metastasis to distant organs. Despite the identification of one or several targets for each of the miRs described here, their mode of action in defined steps of the metastatic cascade remains unknown and is dependent on the corresponding cell lines and on the type of the in vivo system. Experimental metastasis (tail vein injection and metastasis to the lungs) is the most frequently used in vivo system. This system assesses the process of metastasis from the blood to the lungs with the objective to identify genes and pathways that mediate metastasis of NSCLC from the lungs to distant organs. This may sound counterintuitive, however, the process of cancer dissemination through blood vessels is shared by both metastatic stages and many targets whose expression correlates with clinical parameters, such as prognosis and survival, as outlined in previous chapters, have been identified by this approach.
Anti-metastatic compounds to be used in clinical settings should have an impact on the migration, proliferation and survival of disseminated tumor cells rather than merely attempting to block the escape of these cells from the primary tumors. Compounds that only affect migration are confined to long-lasting preventive clinical protocols with permanent toxicities, which should not be acceptable. For example, the src inhibitor Dasatinib antagonizes bone metastasis of breast cancer cells in xenograft models by impairing the survival of already extravasated tumor cells, before they colonize the bone marrow (148). In the best case, anti-metastatic agents directly affect the growth of already established metastases and lead to their eradication (148). There are many hurdles to tackle this problem, however, there are examples of adoptive immunotherapy or immune checkpoint inhibitors combined with other immunotherapy-based strategies that can achieve this objective in some patients (149,150). Unfortunately, none of the described in vivo metastasis systems is reflecting this scenario in clinical practise.
Another issue is targeting the dormant tumor cells in metastatic niches and the bone marrow. These cells have already exited the cell-cycle and this renders them more resistant to almost all available therapies (151,152). Recently it was shown that dormant tumor cells can be re-activated by epigenetic therapy which can render them sensitive to cytotoxic therapy (151,152). The elimination process of dormant tumor cells can be monitored by measuring their number in bone marrow aspirates and at the same time determining their decrease in the peripheral blood (153,154). As an alternative to targeting the seed, interference with the soil, i.e. the tumor microenvironment, can result in the therapeutic benefit of patients with bone metastatic lesions. Targeting non-neoplastic stromal cells in the TME of metastases by agents, such as bisphosphonates, anti-RANK mAb Denosumab and TGFβ inhibitors, all of which prevent the osteoclast-mediated degradation of bone, have resulted in a therapeutic benefit of cancer patients (155,156).
miR-related therapy of metastatic lung cancer. As outlined in the previous chapters, NSCLC metastasis-modulating miRs can be up- or down-regulated in cancer tissues compared to matching normal tissues. If they have a validated role in in vitro and in vivo pre-clinical systems, either the inhibition or the reconstitution of their function can be used as a therapeutic option. In cases of non-resectable metastatic NSCLC or in metastasis to distant organs, local and intravenous delivery, respectively, are potential options for therapeutic miRs.
Inhibition of miRs can be achieved by ASOs, Locked nucleic acids (LNAs), decoy miRs or small-molecule drugs (157-159). Anti-miRs are single-stranded nucleic acids based on ASO or modified locked nucleic acids that block the function of the corresponding miRs by binding to them strongly via a complementary sequence (157-159). Decoy agents contain many artificial miR-binding sites functioning as sponges for the miRs to be inhibited (157-159). Considerable progress has been achieved by improving their binding affinity, stability and target modulation, as well as their impact on the pharmaco-kinetic and pharmaco-dynamic properties of nucleic acid-based agents through chemical modifications of the backbone and of the bases. Low molecular weight drugs can affect the transcription of miRs or interfere with their secondary structure, but specificity issues associated with such compounds remain to be solved (157-159).
Replenishing metastasis-suppressive miRs can be performed via their expression in tumor cells following their incorporation into plasmid- or virus-based expression systems or by using synthetic oligo duplexes, which mimic the function of the miRs to be substituted (157-159).
Considerable progress has been achieved in the field of delivery agents, such as viral vectors, neutral lipid emulsions, liposomes, Poly (ethylene glycol) and chitosan, dendrimers and N-acetyl-D-galactosamin, which can be modified and formulated to serve the optimal target strategy (160). Issues which must be addressed include: i) the hybridization-dependent and -independent off-target effects, ii) the hepatotoxicity and other associated toxicities, iii) immunomodulatory effects, iv) the design of efficient delivery systems, v) the rapid clearance by the reticulo-endothelial system, vi) the enhancement of circulation time when injected into the bloodstream, vii) the delivery to metastases and vii) the endosomal escape (161-163). Fast renal clearance can be prevented by coupling miRNA with polyethylene glycol (PEG), albumins or cholesterol, which mediate shielding from renal clearance (164-166). Shielding of nucleic acids ameliorates their immuno-genicity and their potential to activate a cytokine release syndrome (167). Co-administraion or fusion of nucleic acids with cell penetrating viral proteins can lower the endosomal barrier and promote endosomal escape (168-170). Combination of formulation of nucleic acids with lipophilic polymers and targeting entities can create cell-type specific delivery packages (171-174). The therapeutic efficacy in vivo could be triggered by a combination of efficient delivery approaches antd anti-tumoral immune responses directed against the tumor-related antigens hat are released by the apoptotic tumor cells.
Ten synthetic miRNAs of a pronounced therapeutic interest are summarized in a review by Titze-de-Almeida et al. (175). The therapeutic evaluation of miR-122 antagomir miravirsen (Santaris Pharma) has shown a meaningful activity by inhibiting HCV replication (176,177) RG 101 (Regulus Therapeutics), a more potent drug containing a GalNAC targeting moiety, displays an efficient targeting in the liver, however, it has been put on clinical hold due to jaundice in two patients (177).
In patients with solid and haematological cancers, a miR-34 mimetic, which restores a range of cell death and survival genes, has been abandoned after Phase I, due to cytokine release syndrome-related side-effects (177,178). In patients with malignant pleural mesothelioma, a combination of miR-15,16 mimetic drugs encapsulated in nonliving bacterial minicells (nanoparticles) cells encoated with an EGFR antibody (targomirs) that exert a tumor-suppressive activity, are presently evaluated in a Phase I study (179). Many other oncology-related studies will follow to get a clearer picture on the clinical performance of miR-related approaches.
Conflicts of Interest
Authors are or have been (UHW) employees of Roche.
Authors’ Contributions
AN and UHW wrote the manuscript, FB prepared the figures and performed the bioinformatics analysis.
References
- 1.Perilkos F, Harrington KJ, Syrigos KN. Key mechansisms in lung cancer invasion and metastasis: a comprehensive review. Crit Rev Oncol Hematol. 2013;87:1–11. doi: 10.1016/j.critrevonc.2012.12.007. PMID: 23332547. DOI: 10.1016/j.critrevonc.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Politi K, Herbst RS. Lung cancer in the era of precision medicine. Clin Cancer Res. 2015;21:2213–2220. doi: 10.1158/1078-0432.CCR-14-2748. PMID: 25979927. DOI: 10.1158/1078-0432.CCR-14-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herbst R, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. PMID: 29364287. DOI: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 4.Dholaria B, Hammond W, Shreders A, Lou Y. Emerging therapeutic agents for lung cancer. J Hematology and Oncology. 2016;9:138. doi: 10.1186/s13045-016-0365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotow J, Bivona TG. Understanding and targeting resistance mechansisms in NSCLC. Nat Rev Cancer. 2017;17:637–658. doi: 10.1038/nrc.2017.84. PMID: 29068003. DOI: 10.1038/ nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- 6.Chae YK, Arya A, Iams W, Cruz M, Mohindra N, Villaflor V, Giles FJ. Immune checkpoint pathways in non-small cell lung cancer. Ann Transl Med. 2018;6:88. doi: 10.21037/atm.2017.09.30. PMID: 29666811. DOI: 10.21037/atm.2017.09.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Zu ZY, Wang LF, Su W. CD133+ cancer stem cells in lung cancer. Front Biosci (Landmark Ed) 2013;18:447–453. doi: 10.2741/4113. PMID: 23276935. DOI: http://dx.doi.org/10.2741/4113. [DOI] [PubMed] [Google Scholar]
- 8.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. PMID: 28187288. DOI: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massague J, Obenauf AC. Metastatic colonization by circulating tumor cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. PMID: 28187288. DOI: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fidler IJ, Kripke ML. The challenge of targeting metastasis. Cancer Metastasis Rev. 2015;34:635–641. doi: 10.1007/s10555-015-9586-9. PMID: 2632852. DOI: 10.1007/s10555-015-9586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura T, Kurishima K, Kagohashi K, Ishikawa H, Satoh H, Hizawa K. Specific organ metastases and survival in metastatic non-small cell lung cancer. Mol and Clin Oncol. 2015;3:217–221. doi: 10.3892/mco.2014.410. PMID: 25469298. DOI: 10.3892/mco.2014.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicrRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. PMID: 14744438. DOI: org/10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. PMID: 28209991. DOI: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. Metazoan microRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. PMID: 25998712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. DOI: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creugny A, Fender A, Pfeffer S. Regulation of primary microRNA processing. FEBS Lett. 2018;592:1980–1996. doi: 10.1002/1873-3468.13067. PMID: 29683487. DOI: org/10.1002/1873-3468.13067. [DOI] [PubMed] [Google Scholar]
- 17.Pradhan AK, Emdad L, Das SK, Sarkar D, Fisher PB. The enigma of miRNA regulation in cancer. Adv Cancer Res. 2017;135:25–52. doi: 10.1016/bs.acr.2017.06.001. PMID: 28882224. DOI: 10.1016/bs.acr. 2017.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Calin CA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Ageilan RI, Alder H, Volinia S, Rassenti L, Liu X, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. PMID: 18362358. DOI: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pekarsky Y, Balatti V, Croce CM. Bcl2 and miR-15/16: from gene discovery to treatment. Cell Death Differ. 2018;25:21–26. doi: 10.1038/cdd.2017.159. PMID: 28984869. DOI: 10.1038/cdd.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. PMID: 23728460. DOI: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou L, Liu F, Wang X, Ouyang G. The roles of microRNAs in the regulation of tumor metastasis. Cell Biosci. 2015;5:32. doi: 10.1186/s13578-015-0028-8. PMID: 26146543. DOI: 10.1186/s13578-015-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weidle UH, Dickopf S, Hintermair C, Kollmorgen G, Birzele F, Brinkmann U. The role of microRNAs in breast cancer metastasis: preclinical validation and therapeutic targets. Cancer Genomics Proteomics. 2018;15:17–39. doi: 10.21873/cgp.20062. PMID: 29275360. DOI: 10.21873/cgp.20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weidle UH, Birzele F, Kollmorgen G, Nopora A. Potential microRNA-related targets for therapeutic intervention with ovarian cancer metastasis. Cancer Genomics Proteomics. 2018;15:1–15. doi: 10.21873/cgp.20061. PMID: 29275359. DOI: 10.21873/cgp.20061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weidle UH, Epp A, Birzele F, Brinkmann U. The functional role of prostate cancer-related micro-RNAs. Cancer Genomics Proteomics. 2019;16:1–19. doi: 10.21873/cgp.20108. PMID: 30587496. DOI: 10.21873/cgp.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Ouyang R, Wang Z, Zhou W, Chen H, Jiang Y, Zhang Y, Li H, Liao M, Wang W, Ye M, Ding Z, Feng X, Liu J, Zhang B. miR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci Rep. 2016;6:39001. doi: 10.1038/srep39001. PMID: 27976702. DOI: 10.1038/srep39001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coomans de Brachene A, Demoulin JB. FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci. 2016;73:1159–1172. doi: 10.1007/s00018-015-2112-y. PMID: 26686861. DOI: 10.1007/s00018-015-2112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su B, Gao L, Baranowski C, Gillard B, Wang J, Ransom R, Ho HK, Gelman IH. A genome-wide RNAi screen identifies FOXO4 as a metastasis suppressor through counteracting PI3K/AKT signal pathway in prostate cancer. PLoS One. 2014;9:e101411. doi: 10.1371/journal.pone.0101411. PMID: 24983969. DOI: 10.1371/ journal.pone.0101411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Feng Q, Wie X, Yu Y. Micro-RNA-490 regulates lung cancer metastasis by targeting poly r(C)-binding protein 1. Tumor Biol. 2016;37:15221–15228. doi: 10.1007/s13277-016-5347-9. PMID: 27683057. DOI: 10.1007/ s13277-016-5347-9. [DOI] [PubMed] [Google Scholar]
- 29.Song Q, Sheng W, Zhang X, Jiao S, Li F. ILEI drives epithelial to mesenchymal transition and metastatic progression in the lung cancer cell line A549. Tumor Biol. 2014;35:1377–1382. doi: 10.1007/s13277-013-1188-y. PMID: 24072492. DOI: 10.1007/s13277-013-1188-y. [DOI] [PubMed] [Google Scholar]
- 30.Liang F, Jiang J, Wang WQ, Sun JP, UDHO E, Zhang ZY. PRL3 promotes cell invasion and proliferation by downregulation of Csk leading to Src activation. J Biol Chem. 2007;282:5413–5419. doi: 10.1074/jbc.M608940200. PMID: 17192274. DOI: 10.1074/jbc.M608940200. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, Vardy LA, Tan CP, Loo JM, Guo K, Li J, Lim SG, Zhou J, Chng WJ, Ng SB, Li HX, Zeng Q. PCBP1 suppresses the translation of metastasis-associated PRL-2 phosphatase. Cancer Cell. 2010;18:52–62. doi: 10.1016/j.ccr.2010.04.028. PMID: 20609352. DOI: 10.1016/ j.ccr.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Cao Q, Qu M, Ziao Z, Zhang M, Di S. microRNA-616 promotes the growth and metastasis of non-small cell lung cancer by targeting SOX-7. Oncol Rep. 2017;38:2078–2086. doi: 10.3892/or.2017.5854. PMID: 28765960. DOI: 10.3892/or.2017.5854. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Li B, Ge Z, Song S, Zhang S, Yan H, Huang B, Zhang Y. Decreased expression of SOX7 is correlated with poor prognosis in lung adenocarcinoma patients. Pathol Oncol Res. 2012;18:1039–1045. doi: 10.1007/s12253-012-9542-8. PMID: 22777918. DOI: 10.1007/s12253-012-9542-8. [DOI] [PubMed] [Google Scholar]
- 34.Stovall DB, Cao P, Sui G. SOX7: From a developmental regulator to an emerging tumor suppressor. Histol Histopathol. 2014;29:439–445. doi: 10.14670/hh-29.10.439. PMID: 24288056. DOI: 10.14670/HH-29.10.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu F, Cai Y, Rong X, Chen J, Zeng D, Chen L, Zhang J, Luo R, Zhao P, Ruan J. miR-661 promotes tumor invasion and metastasis by directly inhibiting RB1 in non-small cell lung cancer. Mol Cancer. 2017;16:122. doi: 10.1186/s12943-017-0698-4. PMID: 28716024. DOI: 10.1186/s12943-017-0698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arima Y, Inoue Y, Shibata T, Hayashi H, Nagano O, Saya H, Taya Y. Rb depletion results in deregulation of E-cadherin and induction of cellular phenotypic changes that are characteristic of the epithelial-to-mesenchymal transition. Cancer Res. 2008;68:5104–5112. doi: 10.1158/0008-5472.CAN-07-5680. PMID: 18593909. DOI: 10.1158/0008-5472.CAN-07-5680. [DOI] [PubMed] [Google Scholar]
- 37.Chong JL, Wenzel PL, Saenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, Wang SH, Trikha P, Culp B, Mezache L, Winton DJ, Sansom OJ, Chen D, Bremner R, Cantalupo PG, Robinson ML, Pipas JM, Leone G. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462:930–934. doi: 10.1038/nature08677. PMID: 20016602. DOI: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Q, Liu B, Li X, Zhang Q, Cao L, Xu M, Meng Z, Wu X, Xu K. miR-26a-5p potentiates metastasis of human lung cancer cells by regulating ITGβ8-JAK2/STAT3 axis. Biochem Biohys res Commun. 2018;501:494–500. doi: 10.1016/j.bbrc.2018.05.020. PMID: 29746867. DOI: 10.1016/j.bbrc.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 39.Zu Z, Wu R. Alteration in metastasis potential and gene expression in human lung cancer cells by ITGβ8 silencing. Anat Rec (Hoboken) 2012;295:1446–15454. doi: 10.1002/ar.22521. PMID: 22753015. DOI: 10.1002/ar.22521. [DOI] [PubMed] [Google Scholar]
- 40.Hayashido Y, Kitano H, Sakaue T, Fujjj T, Suematsu M, Sakurai S, Okamoto T. Overexpression of integrin αv facilitates proliferation and invasion of oral squamous cell carcinoma cells via MEK/ERK signaling pathway that is activated by interaction of αvβ8 with type I collagen. Int J Oncol. 2014;45:1875–1882. doi: 10.3892/ijo.2014.2642. PMID: 25190218. DOI: 10.3892/ijo.2014.2642. [DOI] [PubMed] [Google Scholar]
- 41.Cai J, Fang L, Huang Y, Li R, Xu X, Hu Z, Zhang L, Yang Y, Zhu X, Zhang H, Wu J, Huang Y, Li J, Zeng M, Song E, He Y, Zhang L, Li M. Simultaneous overactivation of Wnt/β-catenin and TGFβ signalling by miR-128-3p confers chemoresistanceassociated metastasis in NSCLC. Nat Commun. 2017;8:15870. doi: 10.1038/ncomms15870. PMID: 28627514. DOI: 10.1038/ncomms15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta catenin pathway in cancer. Cancer Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. PMID: 29169144. DOI: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blank M, Tang Y, Yamashita M, Burkett SS, Cheng SY, Zheng YE. A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and stability through RNF20. Nat Med. 2012;18:227–234. doi: 10.1038/nm.2596. PMID: 22231558. DOI: 10.1038/nm.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartscht T, Lehnert H, Gieseler E, Ungefroren H. The Src family kinase inhibitors PP2 and PP1 effectively block TGF-beta1-induced cell migration and invasion in both established and primary carcinoma cells. Cancer Chemother Pharmacol. 2012;70:221–230. doi: 10.1007/s00280-012-1904-0. PMID: 22699812. DOI: 10.1007/s00280-012-1904-0. [DOI] [PubMed] [Google Scholar]
- 45.Levy L, Hills CS. Alterations in components of the TGF-beta superfamily pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. PMID: 16310402. DOI: 10.1016/ j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Ho CS, Noor SM, Nagoor NH. miR-378 and miR-1827 regulate tumor invasion, migration and angiogenesis in human lung adenocarcinoma by targeting RBX1 and CRKL, respectively. J Cancer. 2018;9:331–345. doi: 10.7150/jca.18188. PMID: 29344280. DOI: 10.7150/jca.18188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei D, Sun Y. Small ring finger proteins RBX1 and RBX2 and SCF U3 ubiquitin ligases. The role in cancer and as cancer targets. Genes Cancer. 2010;1:700–707. doi: 10.1177/1947601910382776. PMID: 21103004. DOI: 10.1177/1947601910382776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Wang Y, Zang W, Du Y, Li M, Zhao G. mir-194 targets RBX1 to modulate proliferation and migration of gastric cancer cells. Tumor Biol. 2015;36:2393–2401. doi: 10.1007/s13277-014-2849-1. PMID: 25412959. DOI: 10.1007/s13277-014-2849-1. [DOI] [PubMed] [Google Scholar]
- 49.Skrzpek T, Tertil M, Golda S, Ciesla M, Weglarczyk K, Collet G, Guichard A, Kozakowska M, Boczkowski J, Was H, Gil T, Kuzdzal J, Muchova L, Vitek L, Loboda A, Jozkowicz A, Kieda C, Dulak J. Interplay between heme oxygenase-1 and miR-378 affects non-small cell lung carcinoma growth, vascularization and metastasis. Antioxid Redox Signal. 2013;19:644–660. doi: 10.1089/ars.2013.5184. PMID: 23617628. DOI: 10.1089/ars.2013.5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jozkowicz A, Was H, Dulak J. Heme oxygenase-1 in tumors: is it a false friend. Antioxid Redox Signal. 2007;9:2099–2117. doi: 10.1089/ars.2007.1659. PMID: 17822372. DOI: 10.1089/ars.2007.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li C, Lyu J, Meng QH. miR-93 promotes tumorigenesis and metastasis of non-small cell cancer cells by activating the PI3K/AKT pathway via inhibition of LKB1/PTEN/CDKN1A. J Cancer. 2017;8:870–879. doi: 10.7150/jca.17958. PMID: 28382150. DOI: 10.7150/ jca.17958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR siganling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. PMID: 15261145. DOI: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 53.Li N, Huang D, Lu N, Luo L. Role of the LKB1/AMPK pathway in tumor invasion and metastasis of cancer cells. Oncol Rep. 2015;34:2821–2826. doi: 10.3892/or.2015.4288. PMID: 26398719. DOI: 10.3892/ or.2015.4288. [DOI] [PubMed] [Google Scholar]
- 54.Li CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, Wu CT, Chen HY, Hong TM, Yang PC. microRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun. 2013;4:1877. doi: 10.1038/ncomms2876. PMID: 23695671. DOI: 10.1038/ncomms2876. [DOI] [PubMed] [Google Scholar]
- 55.Vecchione S, Baldasarre G, Ishhii H, Nicoloso MS, Belletti B, Petrocca F, Zanesi N, Fong LY, Battista S, Guarnieri D, Baffa R, Alder H, Farber JL, Donovan PJ, Croce CM. Fez1/Lzts1 absence impairs Cdk1/Cdc25C interaction during mitosis and predisposes mice to cancer development. Cancer Cell. 2007;11:275–289. doi: 10.1016/j.ccr.2007.01.014. PMID: 17349584. DOI: 10.1016/ j.ccr.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Avruch J, Zhou D, Fitamant J, Bardeesy N, Mou F, Barrufet LR. Protein kinases of the Hippo pathway: regulation and metastasis. Sem Cell Devel Biol. 2012;23:770–784. doi: 10.1016/j.semcdb.2012.07.002. PMID: 22898666. DOI: 10.1016/j.semcdb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avruch J, Zhou D, Fitamant J, Bardeesy N. Mst1/2 signaling to Yap: gatekeeper for liver size and tumor development. Br J Cancer. 2011;104:24–32. doi: 10.1038/sj.bjc.6606011. PMID: 21102585. DOI: 10.1038/ sj.bjc.6606011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ly X, Liu C, Zhang H, Jiang Y, Xiong Y, Lei QJ, Guan KL. The N-terminal phosphodegron targets TAZ/WWTR1 protein for SCFβ-TrC dependent degradation in response to phosphatidyl-3-kinase inhibition. J Biol Chem. 2012;287:26245–26253. doi: 10.1074/jbc.M112.382036. PMID: 22692215. DOI: 10.1074/jbc.M112.382036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu H, Du S, Lei T, Wang H, He X, Tong R, Wang Y. Multifaceted regulation and functions of YAP/TAZ in tumors. Oncol Rep. 2018;40:16–28. doi: 10.3892/or.2018.6423. PMID: 29749524. DOI: 10.3892/or.2018.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lo Sardo F, Strano S, Blandino G. YAP and TAZ in lung cancer: oncogenic role and clinical targeting. Cancers. 2018;10:E137. doi: 10.3390/cancers10050137. PMID: 29734788. DOI: 10.3390/cancers10050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wie CH, Wu G, Cai Q, Gao XC, Tong F, Zhou R, Zhang RG, Dong JH, Hu Y, Dong XR. microRNA-330-3p promotes cell invasion and metastasis in non-small cell lung cancer through GRIA3 by activating MAPK/ERK signaling pathway. J Hematol Oncol. 2017;10:125. doi: 10.1186/s13045-017-0493-0. PMID: 28629431. DOI: 10.1186/s13045-017-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Ripka S, Riedel J, Neese A, Griesmann H, Buchholz M, Ellenrieder V, Moeller F, Barth P, Gress TM, Michl P. Glutamate receptor GRIA3-target of CUX1 and mediator of tumor progression in pancreatic cancer. Neoplasia. 2010;12:659–667. doi: 10.1593/neo.10486. PMID: 20689760. DOI: PMC2915410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lipsky RH, Goldman D. Genomics and variation of ionotropic glutamate receptors. Ann NY. 2003;1003:22–35. doi: 10.1196/annals.1300.003. PMID: 14684433. DOI: https://doi.org/10.1196/annals.1300.003. [DOI] [PubMed] [Google Scholar]
- 64.Suh SS, Yoo JY, Cui R, Kaur B, Huebner K, Lee TK, Ageilan RI, Croce CM. HIT suppresses epithelial-mesenchymal transition (EMT) and metastasis in lung cancer through modulation of microRNAs. PLoS Genet. 2014;10:e1004652. doi: 10.1371/journal.pgen.1004652. PMID: 25340791. DOI: 10.1371/journal.pgen.1004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao Y, Gu X, Liu H, Wu G, Yuan D, Yang X, Song Y. Metadherin regulates proliferation and metastasis via actin cytoskeletal remodelling in non-small cell lung cancer. Br J Cancer. 2014;111:355–364. doi: 10.1038/bjc.2014.267. PMID: 24918821. DOI: 10.1038/ bjc.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao X, dai M, Li Q, Wang Z, Lu Y, Song Z. HMGA2 regulates lung cancer proliferation and metastasis. Thorac Cancer. 2017;8:501–510. doi: 10.1111/1759-7714.12476. PMID: 28752530. DOI: 10.1111/1759-7714.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vazquez PP, Carlini MJ, Daroqui MC, Colombo L, Dalurzo ML, Smith DE, Grasselli J, Pallotta MG, Ehrlich M, Bal de Kier Joffe ED, Puricelli L. TGF-beta specifically enhances the metastatic attributes of murine lung adenocarcinoma: imolications for human non-small lung cancer. Clin Exp Metastasis. 2013;30:993–1007. doi: 10.1007/s10585-013-9598-1. PMID: 23832740. DOI: 10.1007/ s10585-013-9598-1. [DOI] [PubMed] [Google Scholar]
- 68.Neuzillet C, Tijeras-Raballand A, Cohen R, Cros J, Faivre S, Raymond E, de Gramont A. Targeting the TGFbeta pathway for cancer therapy. Pharmacol Ther. 2015;147:22–31. doi: 10.1016/j.pharmthera.2014.11.001. PMID: 25444759. DOI: 10.1016/j.pharmthera.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 69.Lv Q, Hu JX, Li YJ, Xie N, Song DD, Zhao W, Yan YF, Li BS, Wang PY, Xie SY. miR-320a effectively suppresses lung adenocarcinoma cell proliferation and metastasis by regulating STAT3 signals. Cancer Biol Ther. 2017;18:142–151. doi: 10.1080/15384047.2017.1281497. PMID: 28106481. DOI: 10.1080/15384047.2017.1281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, Chen X, Meng Q, Jing H, Lu H, Yang Y, Cai L, Zhao Y. miR-181b regulates cisplatin chemosensitivity and metastasis by targeting TGFbetaR1/mad signaling in NSCLC. Sci Rep. 2015;5:17618. doi: 10.1038/srep17618. PMID: 26620926. DOI: 10.1038/srep17618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xing A, Pan L, Gao J. p100 functions as a metastasis activator and is targeted by tumor suppressing microRNA-320a in lung cancer. Thorac Cancer. 2018;9:152–158. doi: 10.1111/1759-7714.12564. PMID: 29159900. DOI: 10.1111/1759-7714.12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li CJ, Li YC, Zhang DR, Pan JH. Signal transducers and activators of transcription 3 function in lung cancer. J Cancer Res Ther. 2013;9 (Suppl 2):S67–73. doi: 10.4103/0973-1482.119100. [DOI] [PubMed] [Google Scholar]
- 73.Jariwala N, Rajasekaran D, Srivastava J, Gredler R, Akiel MA, Robertson CL, Emdad L, Fisher PB, Sarkar D. Role oft he staphylococcal nuclease and tudor domain containing 1 in oncogenesis. Int J Oncol. 2015;46:465–473. doi: 10.3892/ijo.2014.2766. PMID: 25405367. DOI: 10.3892/ijo.2014.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blanco MA, Aleckovic M, Hua Y, Li T, Wie Y, Xu Z, Cristea IM, Kang Y. Identification of staphylococcal nuclease-domain-containing 1 (SND1) as a metadherin-interacting protein with metastasis-promoting functions. J Biol Chem. 2011;286:19982–19992. doi: 10.1074/jbc.M111.240077. PMID: 21478147. DOI: 10.1074/jbc.M111.240077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsuchiya N, Ochiai M, Nakashima K, Ubagai T, Sugimura T, Nakagama H. SND1, a component of RNA-induced silencing complex, is up-regulated in human colon cancers and implicated in early stage colon carcinogenesis. Cancer Res. 2007;67:9568–9576. doi: 10.1158/0008-5472.CAN-06-2707. PMID: 17909068. DOI: 10.1158/0008-5472.CAN-06-2707. [DOI] [PubMed] [Google Scholar]
- 76.Cao G, Li F, Du T. microRNA-365 inhibits growth, invasion and metastasis of lung cancer by targeting NRP1 expression. Int J Clin Exp Pathol. 2016;9:1081–1092. [PMC free article] [PubMed] [Google Scholar]
- 77.Lampropoulou A, Ruhrberg C. Neuropilin regulation of angiogenesis. Biochem Soc Trans. 2011;42:1623–1638. doi: 10.1042/BST20140244. [DOI] [PubMed] [Google Scholar]
- 78.Glinka Y, Stoilova S, Mohammed N, Prud’homme GJ. Neuropilin-1 exerts co-receptor function for TGF-beta-1 on the membrane of cancer cells and enhances responses to both latent and active TGF-beta. Carcinogenesis. 2011;32:613–621. doi: 10.1093/carcin/bgq281. PMID: 21186301. DOI: 10.1093/carcin/bgq281. [DOI] [PubMed] [Google Scholar]
- 79.Kwiatkowsky SC, Guerrero PA, Hirota S, Chen Z, Morales JE, Aghi M, McCarthy JH. Neuropilin-1 modulates TGFbeta signaling to drive glioblastoma growth and recurrence after anti-angiogenic therapy. Plos One. 2017;12:e0185065. doi: 10.1371/journal.pone.0185065. PMID: 28938007. DOI: 10.1371/journal.pone.0185065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu X, Huang YG, Jin CG, Zhou YC, Chen XQ, Li J, Chen Y, Li M, Yao Q, Li K, Lan M, Ye JG, Wang XC. microRNA-370 inhibits the growth and metastasis of lung cancer by down-regulating epidermal growth factor receptor expression. Oncotarget. 2017;8:88139–88151. doi: 10.18632/oncotarget.21537. PMID: 29152147. DOI: 10.18632/oncotarget.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kitano H, Chung JY, Ylaya K, Conway C, Takikita M. Fukuoka J, Doki Y, Hanaoka J, Hewitt SM. Profiling of phosho-AKT, phospho-mTOR, phospho-MAPK and EGFR in non-small cell lung cancer. J Histochem Cytochem. 2014;62:335–346. doi: 10.1369/0022155414523022. PMID: 24487999. DOI: 10.1369/0022155414523022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Diaz-Serrano A, Gella P, Jiminez E, Zugazagoitia J, Paz-Ares Rodriguez L. Targeting EGFR in lung cancer: current standards and developments. Drugs. 2018 doi: 10.1007/s40265-018-0916-4. PMID: 29915896. DOI: 10.1007/s40265-018-0916-4. [DOI] [PubMed] [Google Scholar]
- 83.Wang J, Tian X, Han R, Zhang X, Wang X, Shen H, Xue L, Liu Y, Yan X, Shen J, Manoor K, Deepak J, Donahue JM, Stass SA, Xing L, Jiang F. Downregulation of miR-486-5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene. 2014;33:1181–1189. doi: 10.1038/onc.2013.42. PMID: 23474761. DOI: 10.1038/onc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stiegler AL, Boggon TJ. p190RhoGAP proteins contain pseudo GTPase domains. Nat Commun. 2017;8:506. doi: 10.1038/s41467-017-00483-x. PMID: 28894085. DOI: 10.1038/s41467-017-00483-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bustelo XR. Rho GTPases in cancer: known facts, open questions and therapeutic challenges. Biochem Soc Trans. 2018;19:741–760. doi: 10.1042/BST20170531. PMID: 29871878. DOI: 10.1042/BST20170531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jansen S, Gosens R, Wieland T, Schmidt M. Paving the Rho in cancer metastasis: Rho GTPases and byond. Pharmacol Ther. 2018;183:1–21. doi: 10.1016/j.pharmthera.2017.09.002. PMID: 28911825. DOI: 10.1016/j.pharmthera. 2017.09.002. [DOI] [PubMed] [Google Scholar]
- 87.Li M, Zhang S, Wu N, Wu L, Wang C, Lin Y. Overexpression of miR-499-5p inhibits non-small cell lung cancer proliferation and metastasis by targeting VAV3. Sci Rep. 2016;6:23100. doi: 10.1038/srep23100. PMID: 26972445. DOI: 10.1038/srep23100. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Movilla N, Bustelo XR. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol Cell Biol. 1999;19:7870–7885. doi: 10.1128/mcb.19.11.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bustelo XR. Vav family exchange factors: an integrated regulatory and functional view. Small GTPases. 2014;5:9. doi: 10.4161/21541248.2014.973757. PMID: 25483299. DOI: 10.4161/21541248.2014.973757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu J, Jia L, Li T, Yang L, Zhang G. microRNA-615-3p inhibits the tumor growth and metastasis of NSCLC via inhibiting IGF2. Oncol Res. 2019;2:269–279. doi: 10.3727/096504018X15215019227688. PMID: 29562959. DOI: 10.3727/096504018X15215019227688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kessler SM, Haybaeck J, Kiemer AK. Insulin-like growth factor 2 – the oncogene and its accomplices. Curr Pharm Des. 2016;22:5948–5961. doi: 10.2174/1381612822666160713100235. PMID: 27412171. DOI: 10.2174/13816128226 66160713100235. [DOI] [PubMed] [Google Scholar]
- 92.Iams WT, Lovly CM. Molecular pathways: clinical applications and future direction of insulin-like growth factor-1 receptor blockade. Clin Cancer. 2015;21:4270–4377. doi: 10.1158/1078-0432.CCR-14-2518. PMID: 26429980. DOI: 10.1158/1078-0432.CCR-14-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng CW, Chen PM, Hsieh YH, Wenig CC, Chang CW, Yao CC, Hu LY, Wu PE, Shen CY. Foxo3a-mediated overexpression of microRNA-622 suppresses tumor metastasis by repressing hypoxia-inducible factor-1alpha in ERK-responsive lung cancer. Oncotarget. 2015;6:44222–44238. doi: 10.18632/oncotarget.5826. PMID: 26528854. DOI: 10.18632/oncotarget.5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang K, Huang G, Li X, Zhang Y, Jiang Y, Shen J, Liu J, Wang Q, Zhu J, Feng X, Dong J, Qian C. Hypoxia induces epithelial-mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor-1alpha in hepatocellular carcinoma. BMC Cancer. 2013;13:108. doi: 10.1186/1471-2407-13-108. PMID: 23496980. DOI: 10.1186/1471-2407-13-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsai YP, Wu KJ. Hypoxia-regulated target genes implicated in tumor metastases. J Biomed Sci. 2012;19:102. doi: 10.1186/1423-0127-19-102. PMID: 23241400. DOI: 10.1186/1423-0127-19-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bao L, Lv L, Feng J, Chen Y, Wang X, Han S, Zhao H. miR-876-5p suppresses epithelial-mesenchymal transition of lung cancer by directly down-regulating bone morphogenetic protein 4. J Biosc. 2017;42:671–681. doi: 10.1007/s12038-017-9722-5. PMID: 29229885. DOI: 10.1007/s12038-017-9722-5. [DOI] [PubMed] [Google Scholar]
- 97.Molloy EL, Adams A, Moore JB, Masterson JC, Madrigal-Estebas L, Mahon BP, O’Dea S. BMP4 induces epithelial-mesenchymal transition-like response in adult airway epithelial cells. Growth Factors. 2008;26:12–22. doi: 10.1080/08977190801987166. PMID: 18365875. DOI: 10.1080/08977190801987166. [DOI] [PubMed] [Google Scholar]
- 98.Hamada S, Satoh K, Hirota M, Kimura K, Kanno A, Masamune A, Shimosegawa T. Bone morphogenetic protein 4 induces epithelial-mesenchymal transition through MSX2 induction on pancreatic cancer cell line. J Cell Physiol. 2007;213:768–774. doi: 10.1002/jcp.21148. PMID: 17516553. DOI: 10.1002/jcp.21148. [DOI] [PubMed] [Google Scholar]
- 99.Kestens C, Siersema PD, Offerhaus GJ, van Baal JW. BMP4 signaling is able to induce an epithelial-mesenchymal transition-like phenotype in Barretts esophagus and esophageal adenocarcinoma through induction of SNAIL2. PLoS One. 2016;11:e0155754. doi: 10.1371/journal.pone.0155754. PMID: 27191723. DOI: 10.1371/journal. pone.0155754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dang W, Qin Z, Fan S, Wen Q, Lu Y, Wang J, Zhang X, Wie L, He W, Ye Q, Li G, Ma J. miR-1207-5p suppresses lung cancer growth and metastasis by targeting CSF1. Oncotarget. 2016;7:32421–3243. doi: 10.18632/oncotarget.8718. PMID: 27107415. DOI: 10.18632/ oncotarget.8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. PMID: 19279573. DOI: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Montovani A, Allavena P, Sica A, Balkwil F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. PMID: 18650914. DOI: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 103.Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. PMID: 15958574. DOI: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 104.Xin M, Qiao Z, Li J, Song S, Zhao X, Milao P, Tang T, Wang L, Liu W, Xang X, Dai K, Huang G. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget. 2016;7:44252–44265. doi: 10.18632/oncotarget.10020. PMID: 27317765. DOI: 10.18632/oncotarget.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bauer DE, Hatzivassillou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. PMID: 16007201. DOI: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 106.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–312. doi: 10.1016/j.ccr.2005.09.008. PMID: 16226706. DOI: 10.1016/ j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 107.Lv P, Zhang P, Li X, Chen Y. Micro ribonucleic acid (RNA)-101 inhibits cell proliferation and invasion of lung cancer by regulating cyclooxygenase-2. Thorac Cancer. 2015;6:778–784. doi: 10.1111/1759-7714.12283. PMID: 26557918. DOI: 10.1111/1759-7714.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guo F, Cogdell D, Hu L, Yang D, Sood AK, Xue F, Zhang W. miR-101 suppresses the epithlial-to-mesenchymal transition by targeting ZEB1 and ZEB2 in ovarian carcinoma. Oncol Rep. 2014;31:2021–2028. doi: 10.3892/or.2014.3106. PMID: 24677166. DOI: 10.3892/ or.2014.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yuan Y, Shen Y, Xue L, Fan H. miR-140 suppresses tumor growth and metastases of non-small cell lung cancer by targeting insulin-like growth factor 1 receptor. PLoS One. 2013;8:e73604. doi: 10.1371/journal.pone.0073604. PMID: 24039995. DOI: 10.1371/journal.pone.0073604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chitnis MM, Yuen JS, Protheroe AS, Pollak M, Macaulay VM. The type 1 insulin-like growth factor receptor pathway. Clin Cancer Res. 2008;14:6364–6370. doi: 10.1158/1078-0432.CCR-07-4879. PMID: 18927274. DOI: 10.1158/1078-0432.CCR-07-4879. [DOI] [PubMed] [Google Scholar]
- 111.Liefers-Visser JAL, Mejering RAM, Reyners AKL, van der Zee AGJ, de Jong S. IGF system targeted therapy: Therapeutic opportunities for ovarian cancer. Cancer Treat Rev. 2017;60:90–99. doi: 10.1016/j.ctrv.2017.08.012. PMID: 28934637. DOI: 10.1016/j.ctrv.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 112.Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F, Bao G, Kong H, Ge C, Zhang F, Yu T, Li J, He X, Yao M. miR-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol Cancer. 2014;13:155. doi: 10.1186/1476-4598-13-166. PMID: 24997798. DOI: 10.1186/1476-4598-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhong B, Liu X, Wang X, Liu X, Li H, Darnay BG, Lin X, Sun SC, Dong C. Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitinylation of the adaptor protein TRAF3. Sci Signal. 2013;6:ra35. doi: 10.1126/scisignal.2003708. PMID: 23674823. DOI: 10.1126/scisignal.2003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li Y, Chen P, Zu L, Weng M, Zhou Q. MicroRNA-338-3p suppresses metastasis of lung cancer cells by targeting the EMT regulator Sox4. Am J Cancer Res. 2016;6:127–140. [PMC free article] [PubMed] [Google Scholar]
- 115.Chen X, Wei L, Zhao S. miR-338 inhibits the metastasis of lung cancer by targeting integrin beta 3. Oncol Rep. 2016;36:1467–1474. doi: 10.3892/or.2016.4928. PMID: 27431198. DOI: 10.3892/or.2016.4928. [DOI] [PubMed] [Google Scholar]
- 116.Liu Y, Zeng S, Jiang X, Lai D, Su Z. SOX4 induces tumor invasion by targeting EMT-related pathway in prostate cancer. Tumor Biol. 2017;39:1010428317694539. doi: 10.1177/1010428317694539. PMID: 28466783. DOI: 10.1177/1010428317694539. [DOI] [PubMed] [Google Scholar]
- 117.Pan B, Guo J, Liao Q, Zhao Y. Beta 1 and Beta 3 integrins in breast, prostate and pancreatic cancer: a novel implication. Oncol Letters. 2018;15:5412–5416. doi: 10.3892/ol.2018.8076. PMID: 29556293. DOI: 10.3892/ol.2018.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sun C, Zargham R, Shao Q, Gui X, Marcus V, Lazaris A, Salman A, Metrakos P, Qu X, Gao Z. Association of CD98, integrin beta 1, integrin beta 3 and Fak with the progression and liver metastases of colorectal cancer. Pathol Res Pract. 2014;210:668–674. doi: 10.1016/j.prp.2014.06.016. PMID: 25041835. DOI: 10.1016/j.prp.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 119.Chen W, Wang J, Liu S, Wng S, Cheng Y, Zhiu W, Duan C, Zhang C. microRNA-361-3p suppresses tumor cell proliferation and metastasis by directly targeting SH2B1 in NSCLC. J Exp Clin Cancer Res. 2016;35:76. doi: 10.1186/s13046-016-0357-4. PMID: 27164951. DOI: 10.1186/s13046-016-0357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chua S. SH2B1 – the adaptor protein that could. Endocrinology. 2010;151:4100–4102. doi: 10.1210/en.2010-0572. PMID: 20736406. DOI: 10.1210/en.2010-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rider L, Tao J, Snyder S, Brinley B, Lu J, Diakonova M. Adapter protein SH2B1 cross-links actin filaments and regulates actin cytoskeleton. Mol Endocrinol. 2009;23:1065–1076. doi: 10.1210/me.2008-0428. PMID: 19342444. DOI: 10.1210/me.2008-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Diakonova M, Gunter DR, Herrington J, Carter-Su C. SH2-Bbeta is a Rac-binding protein that regulates cell motility. J Biol Chem. 2002;277:10669–10677. doi: 10.1074/jbc.M111138200. PMID: 11786545. DOI: 10.1074/ jbc.M111138200. [DOI] [PubMed] [Google Scholar]
- 123.Wu X, Liu T, Fang O, Dong W, Zhang F, Leach L, Hu X, Luo Z. MicroRNA-708-5p acts as a therapeutic agent against metastatic lung cancer. Oncotarget. 2016;7:2417–2432. doi: 10.18632/oncotarget.6594. PMID: 26678031. DOI: 10.18632/oncotarget.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu S, Bishop WR, Liu M. Differential effects of cell cycle regulatory protein p21 (WAF/Cip1) on apoptosis and sensitivity to cancer chemotherapy. Drug Resist Updat. 2003;6:183–195. doi: 10.1016/s1368-7646(03)00044-x. PMID: 12962684. DOI: 10.1016/S1368-7646(03)00044-X. [DOI] [PubMed] [Google Scholar]
- 125.Fan Y, Borowsky AD, Weiss RH. An antisense oligodeoxynucleotide to p21 (Waf1/Cip1) causes apoptosis in human breast cancer cells. Mol Cancer Ther. 2003;2:773–782. PMID: 12939467. [PubMed] [Google Scholar]
- 126.Wu X, Liu T, Fang O, Dong W, Zhang F, Leach L, Hu X, Luo Z. microRNA-708-5p acts as a therapeutic against against metastatic lung cancer. Oncotarget. 2016;7:2417–2432. doi: 10.18632/oncotarget.6594. PMID: 26678031. DOI: 10.18632/oncotarget.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. PMID: 12809600. DOI: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 128.Fang Y, Chen Y, Yu L, Zheng C, Qi Y, Li Z, Yang Z, Zhang Y, Shi T, Luo J, Liu M. Inhibition of breast cancer metastases by a novel inhibitor of TGFbeta receptor 1. J Natl Cancer Inst. 2013;105:47–58. doi: 10.1093/jnci/djs485. PMID: 23178439. DOI: 10.1093/jnci/djs485. [DOI] [PubMed] [Google Scholar]
- 129.Lei Z, Xu G, Wang L, Yang H, Liu X, Zhao J, Zhang HT. miR-142-3p represses TGF-beta induced growth inhibition through repression of TGFbetaR1 in non-small cell lung cancer. FASEB J. 2014;28:2696–2704. doi: 10.1096/fj.13-247288. PMID: 24558198. DOI: 10.1096/fj.13-247288. [DOI] [PubMed] [Google Scholar]
- 130.Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang L, Zhang H, Chen X, Yang Y, Liu G. miRNA-29c suppresses lung cancer cell adhesion to extracellular matrix and metastasis by targeting integrin beta1 and matrix metalloproteinase 2 (MMP2) PLoS One. 2013;8:e70192. doi: 10.1371/journal.pone.0070192. PMID: 23936390. DOI: 10.1371/ journal.pone.0070192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hua H, Li M, Luo T, Yin Y, Yiang Y. Matrix metallproteinases in tumorigenesis: an evolving paradigm. Cell Mol Life Sci. 2011;68:3853–3868. doi: 10.1007/s00018-011-0763-x. PMID: 21744247. DOI: 10.1007/s00018-011-0763-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kryczka J, Stasiak M, Dziki L, Mik M, Dziki A, Cierniewski C. Matrix metalloproteinase-2 cleavage of the beta1 integrin ectodomain facilitates colon cancer cell motility. J Biol Chem. 2012;287:36556–36566. doi: 10.1074/jbc.M112.384909. PMID: 22898815. DOI: 10.1074/ jbc.M112.384909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Grzesiak JJ, Tran Cao HS, Burton DW, Kaushal S, Vargas F, Clopton P, Snyder CS, Deftos LJ, Hoffman RM, Bouvet M. Knockdown of the beta1 integrin subunit reduces primary tumor growth and inhibits pancreatic cancer metastasis. Int J Cancer. 2011;129:2905–2915. doi: 10.1002/ijc.25942. PMID: 21491421. DOI: 10.1002/ ijc.25942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hwang SJ, Lee HW, Kim HR, Song HJ, Lee DH, Lee H, Shin CH, Joung JG, Kim DH, Joo KM, Kim HH. Overexpression of microRNA-95-3p suppresses brain metastasis of lung adenocarcinoma through downregulation of cyclin D1. Oncotarget. 2015;6:20434–20448. doi: 10.18632/oncotarget.3886. PMID: 25971210. DOI: 10.18632/oncotarget.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gautschi O, Ratschiller D, Gugger M, Betticher DC, Heighway J. Cyclin D1 in non-small cell lung cancer: a key driver of malignant transformation. Lung Cancer. 2007;55:1–14. doi: 10.1016/j.lungcan.2006.09.024. PMID: 17070615. DOI: 10.1016/j.lungcan.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 136.Li Z, Wang C, Jiao X, Lu Y, Fu M, Quong AA, Dye C, Yang J, Dai M, Ju X, Zhang X, Li A, Burbeto P, Stanley ER, Pestell RG. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol Cell Biol. 2006;26:4340–4356. doi: 10.1128/MCB.02124-05. PMID: 16705174. DOI: 10.1128/MCB.02124-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li Z, Jiao X, Wang C, Ju X, Lu Y, Yuan L, Lisanti MP, Katiyar S, Pestell RG. Cyclin D1 induction of cellular migration requires p27 /KIP1. Cancer Res. 2006;66:9986–9994. doi: 10.1158/0008-5472.CAN-06-1596. PMID: 17047061. DOI: 10.1158/0008-5472.CAN-06-1596. [DOI] [PubMed] [Google Scholar]
- 138.Donzelli S, Mori F, Bellissimo T, Sacconi A, Casini B, Frixa T, Roscilli G, Aurisiccho L, Facciolo F, Pompili A, Carosi MA, Pescarmona E, Segatto O, Pond G, Muti P, Telera S, Strano S, Yarden Y, Blandino G. Epigenetic silencing of miR-145-5p contributes to brain metastasis. Oncotarget. 2015;6:35183–35201. doi: 10.18632/oncotarget.5930. PMID: 26440147. DOI: 10.18632/ oncotarget.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pillai K, Pourgholami MH, Chua TC, Dorris DL. MUC1 as a potential target in anticancer therapies. Am J Clin Oncol. 2015;38:108–118. doi: 10.1097/COC.0b013e31828f5a07. PMID: 23608828. DOI: 10.1097/COC.0b013e 31828f5a07. [DOI] [PubMed] [Google Scholar]
- 140.Wang YH, Herlyn M. The emerging roles of Oct4 in tumor-initiating cells. Am J Physiol Cell Physiol. 2015;309:C709–C718. doi: 10.1152/ajpcell.00212.2015. PMID: 26447206. DOI: 10.1152/ajpcell.00212.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Byme JA, Frost S, Chen Y, Bright RK. Tumor protein D52 (TPD52) and cancer-oncogene understudy or understudied oncogene. Tumr Biol. 2014;35:7369–7382. doi: 10.1007/s13277-014-2006-x. PMID: 24798974. DOI: 10.1007/s13277-014-2006-x. [DOI] [PubMed] [Google Scholar]
- 142.Valencia K, Luis-Ravelo D, Boxy N, Anton I, Martin-Canarias S, Zandueta C, Ormazabal C, Struman I, Tabruyn S, Rebmann V, de las Rivas J, Guruceaga E, Bandres E, Lecanda F. miRNA cargo within exosome-like vesicle transfer influences metastatic bone colonization. Mol Oncol. 2014;8:689–703. doi: 10.1016/j.molonc.2014.01.012. PMID: 24593875. DOI: 10.1016/j.molonc.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chiu KL, Kuo TT, Kuok QY, Lin YS, Hua CH, Lin CY, Su PY, Lai LC, Sher YP. ADAM9 enhances CDCP1 protein expression by suppressing miR-218 for lung tumor metastasis. Sci Rep. 2015;5:16426. doi: 10.1038/srep16426. PMID: 26553452. DOI: 10.1038/ srep16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shi ZM, Wang L, Shen H, Jiang CF, Ge X, Li DM, Wen YY, Sun HR, Pan MH, Li W, Shu YQ, Liu LZ, Eiper SC, He J, Jiang BH. Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signailng. Oncogene. 2017;36:2577–2588. doi: 10.1038/onc.2016.414. PMID: 28192397. DOI: 10.1038/onc.2016.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Uekita T, Sakai R. Roles of CUB domain-containing protein 1 signaling in cancer invasion and metastasis. Cancer Sci. 2011;102:1943–1948. doi: 10.1111/j.1349-7006.2011.02052.x. PMID: 21812858. DOI: 10.1111/j.1349-7006.2011.02052.x. [DOI] [PubMed] [Google Scholar]
- 146.Deryugina EI, Conn EM, Wortmann A, Partridge JJ, Kupriyanova TA, Ardi VC, Hooper JD, Quigley JP. Functional role of cell surface CUB domain-containing protein 1 in tumor cell dissemination. Mol Cancer Res. 2009;7:1197–1211. doi: 10.1158/1541-7786.MCR-09-0100. PMID: 19671673. DOI: 10.1158/1541-7786.MCR-09-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Davis FM, Stewart TA, Thompson EW, Monteith GR. Targeting EMT in cancer: opportunities for pharmacological intervention. Trends in Pharmacol Sci. 2014;35:479–488. doi: 10.1016/j.tips.2014.06.006. PMID: 25042456. DOI: 10.1016/j.tips.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 148.Montero JC, Seoane S, Ocana A, Pandiella A. Inhibition of src family kinases and tyrosine receptor kinases by dasatinib: possible combinations in solid tumors. Clin Cancer Res. 2011;17:5546–5552. doi: 10.1158/1078-0432.CCR-10-2616. PMID: 21670084. DOI: 10.1158/1078-0432.CCR-10-2616. [DOI] [PubMed] [Google Scholar]
- 149.Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z, Pasetto A, Langhan M, Shelton T, Prickett T, Gartner J, Jia L, Trebska-Mc Gowan K, Sümerville RP, Robbins PF, Rosenberg SA, Goff SL, Feldman SA. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med. 2018;24:724–730. doi: 10.1038/s41591-018-0040-8. PMID: 29867227. DOI: 10.1038/s41591-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Sciene. 2015;348:62–68. doi: 10.1126/science.aaa4967. PMID: 25838374. DOI: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gomis RR, Gawrzak S. Tumor cell dormancy. Mol Oncol. 2017;11:62–78. doi: 10.1016/j.molonc.2016.09.009. PMID: 28017284. DOI: 10.1016/ j.molonc.2016. 09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li J, Wang X, Shangguan AJ, Zhang L, Yu Z. Dormant cells: The original cause of tumor recurrence and metastasis. Cell Biochem and Biophysics. 2015;72:317–320. doi: 10.1007/s12013-014-0477-4. PMID: 25556072. DOI: 10.1007/s12013-014-0477-4. [DOI] [PubMed] [Google Scholar]
- 153.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for tumor dormancy. Nat Rev Cancer. 2007;7:834–846. doi: 10.1038/nrc2256. PMID: 17957189. DOI: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Alix-Panabieres C, Riethdorf S, Pantel K. Circulating tumor cells and bone marrow micrometastasis. Clin Cancer Res. 2008;14:5013–5021. doi: 10.1158/1078-0432.CCR-07-5125. PMID: 18698019. DOI: 10.1158/1078-0432.CCR-07-5125. [DOI] [PubMed] [Google Scholar]
- 155.Erdogan B, Cicin I. Medical treatment of breast cancer bone metastasis: from bisphosphonates to targeted drugs. Asian Pac J Cancer Prev. 2014;15:1503–1510. doi: 10.7314/apjcp.2014.15.4.1503. PMID: 24641358. DOI: 10.7314/APJCP.2014.15.4.1503. [DOI] [PubMed] [Google Scholar]
- 156.Katz LH, Li Y, Chen JS, Munoz NM, Majumdar A, Chen J, Mishra L. Targeting TGF-β signalling in cancer. Exp Opin Ther Targets. 2013;17:743–760. doi: 10.1517/14728222.2013.782287. PMID: 23651053. DOI: 10.1517/14728222.2013.782287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. PMID: 24172333. DOI: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gambari R, Bognari E, Spandidos DA, Fabbri E. Targeting oncomirs and mimicking tumor suppressor miRNAs: New trends in the development of miRNA therapeutic strategies in oncology. Int J Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. PMID: 27175518. DOI: 10.3892/ ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Broderick JA, Zamore PD. microRNA Therapeutics. Gene Ther. 2011;18:1104–1110. doi: 10.1038/gt.2011.50. PMID: 21525952. DOI: 10.1038/ gt.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Goldberg MS. siRNA delivery for the treatment of ovarian cancer. Methods. 2013;63:95–100. doi: 10.1016/j.ymeth.2013.01.007. PMID: 23403216. DOI: 10.1016/j.ymeth.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 161.MacLeod AR, Crook ST. RNA Therapeutics in Oncology: Advances, challenges and future directions. J Clin Pharmacol. 2017;57(Suppl 10):S43–S59. doi: 10.1002/jcph.957. PMID: 28921648. DOI: 10.1002/jcph.957. [DOI] [PubMed] [Google Scholar]
- 162.Li Z, Rana TM. Therapeutic targeting of microRNAs: Current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. PMID: 25011539. DOI: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 163.Garafalo M, Leva GD, Croce CM. MicroRNAs as anti-cancer therapy. Curr Pharm Des. 2014;20:5328–5335. doi: 10.2174/1381612820666140128211346. PMID: 24479801. DOI: 10.2174/1381612820666140128211346. [DOI] [PubMed] [Google Scholar]
- 164.Malek A, Merkel O, Fink L, Czubayko F, Kissel T, Aigner A. In vivo pharmakinetics, tissue distribution and underlying mechansisms of various PE(-PEG) siRNA complexes. Toxicol Appl Pharmacol. 2009;236:97–108. doi: 10.1016/j.taap.2009.01.014. PMID: 19371615. DOI: 10.1016/j.taap.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 165.Jung S, Lee SH, Mok H, Chung HJ, Park TG. Gene silencing efficiency of siRNA-PEG conjugates: Effect of PEGylation site and PEG molecular weight. J Control Release. 2010;144:296–331. doi: 10.1016/j.jconrel.2010.03.002. PMID: 20230864. DOI: 10.1016/j.jconrel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 166.Mui BL, Tam YK, Jajaraman M, Ansell SM, Du X, Tam YY, Lin PJ, Chen S, Narayanannair JK, Rajeev KG, Manoharan M, Akinc A, Maier MA, Cullis P, Madden TD, Hope MJ. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Ther Nucleic Acids. 2013;2:e139. doi: 10.1038/mtna.2013.66. PMID: 24345865. DOI: 10.1038/ mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang AZ, Langer R, Farokhazad OC. Nanoparticle delivery of cancer drugs. Annu Rev Med. 2012;63:185–198. doi: 10.1146/annurev-med-040210-162544. PMID: 21888516. DOI: 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- 168.Guo X, MacKay JA, Szoka FC Jr. Mechanism of pH-triggered collapse of phosphatidylethanolamine liposomes stabilized by an ortho ester polyethylenglycol lipid. Biophys J. 2003;84:1784–1795. doi: 10.1016/s0006-3495(03)74986-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shin J, Shum P, Thompson DH. Acid-triggered release via dePEGylation of DOPE liposomes containing acid-labile vinyl ether PEG-lipids. J Control Release. 2003;91:187–200. doi: 10.1016/s0168-3659(03)00232-3. PMID: 12932651. DOI: 10.1016/S0168-3659(03)00232-3. [DOI] [PubMed] [Google Scholar]
- 170.Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. PMID: 8144628. [PubMed] [Google Scholar]
- 171.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. PMID: 15520166. DOI: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 172.Song E, Zhu P, Lee SK, Chowdury D, Kussmann S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. PMID: 15908939. DOI: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 173.Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. PMID: 16740739. DOI: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schneider B, Grote M, John M, Haas A, Bramlage B, Ickenstein LM, Jahn-Hofmann K, Bauss F, Cheng W, Croasdale R, Daub K, Dill S, Hoffmann E, Lau W, Burtscher H, Ludtke JL, Metz S, Mundigl O, Neal Z, Scheuer W, Stracke J, Herweijer H, Brinkmann U. Targeted siRNA delivery and mRNA knockdown mediated by bispecific digoxigenin antibodies. Mol Ther Nucleic Acids. 2012;18:e46. doi: 10.1038/mtna.2012.39. PMID: 23344238. DOI: 10.1038/ mtna.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Titze-de-Almeida R, David C, Titze-de-Almeida SS. The race of 10 synthetic RNAI- based drugs to the pharmaceutical market. Pharmaceutical Res. 2017;34:1339–1363. doi: 10.1007/s11095-017-2134-2. PMID: 28389707. DOI: 10.1007/s11095-017-2134-2. [DOI] [PubMed] [Google Scholar]
- 176.Jones D. Setbacks shadow microRNA therapies in the clinic. Nat Biotechnol. 2018;36:909–910. doi: 10.1038/nbt1018-909. PMID: 30307922. DOI: 10.1038/nbt1018-909. [DOI] [PubMed] [Google Scholar]
- 177.Borgia G, Maraolo AE, Buonomo AR, Scotto R, Gentile I. The therapeutic potential of new investigational hepatitis C virus translation inhibitors. Exp Op Invest Drugs. 2016;25:1209–1214. doi: 10.1080/13543784.2016.1225036. PMID: 27537604. DOI: 10.1080/13543784.2016.1225036. [DOI] [PubMed] [Google Scholar]
- 178.Hosseinahli N, Aghapur M, Duif PHG, Badaran B. Treating cancer with microRNA replacement therapy. A literature review. J Cell Physiol. 2018;233:5574–5588. doi: 10.1002/jcp.26514. PMID: 29521426. DOI: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 179.Viteri S, Rosell R. An innovative mesothelioma treatment based on miR-16 mimic loaded EGFR targeted minicells (TargomiRs) Transl Lung cancer Res. 2018;7:S1–S4. doi: 10.21037/tlcr.2017.12.01. PMID: 29531894. DOI: 10.21037/tlcr.2017.12.01. [DOI] [PMC free article] [PubMed] [Google Scholar]