Summary
Aims
This study was to determine whether curcumin had any effect on the proliferation of neural stem cell (NSC), analyze the expression of glucocorticoid receptor (GR), signal transducer and activator of transcription 3 (STAT3), and Notch1 at transcription and protein level, and discuss the related mechanisms.
Methods and results
NSCs were harvested from E15 SD rat brain and cultured. All experiments were performed at the second passage. Cell cytotoxicity, cell viability, and proliferation assays were used to figure out the optimal concentration of curcumin, which can be used for the protein and mRNA studies. The results showed that by downregulation of GR and STAT3 expression, 0.5 μmol L−1 curcumin exhibited the most pronounced effect in promoting the proliferation of NSCs, which were also induced by antagonists of GR and STAT3, but was inhibited by GR agonist.
Conclusion
This study shows that low‐dose curcumin stimulates the proliferation of NSCs, which is probably by inhibiting the mRNA and protein expressions of GR and directly or indirectly regulating the STAT3 via the synergistic effect of GR and STAT3 pathways and its related signal pathways.
Keywords: curcumin, glucocorticoid receptor, neural stem cell, proliferation, signal transducer and activator of transcription 3
1. INTRODUCTION
Curcumin is the major component of turmeric that is widely used in Indian curries and many other dishes and has also long been used for the treatment for inflammatory diseases.1 Numerous studies2 have demonstrated that curcumin possesses many pharmacological activities including anti‐inflammatory, antioxidative stress, and antitumor properties. Therefore, curcumin has been widely used in cancer chemoprevention and tumor growth suppression studies.3 Both in vivo and in vitro experiments have demonstrated the neuroprotective effects of curcumin. For instance, curcumin can protect against neurodegenerative conditions such as Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), and focal cerebral ischemia4 by attenuating neuronal apoptosis and inducing neuronal differentiation.5, 6, 7 Recently, some reports have revealed8, 9 that curcumin could stimulate proliferation of neural progenitor cells (NPCs) and adult hippocampal neurogenesis.
NSCs exist in both the developing nervous system and adult hippocampus and subventricular region of the cerebral cortex system.10 They have the ability to self‐renew and differentiate into neurons, astrocytes, and oligodendrocytes, playing an important role in neurogenesis of the normal mature brain and after injury.11 Factors are known to affect the division and differentiation of NSCs, including environmental stimuli, injuries, and medications.12 A deeper understanding of factors affecting neurogenesis would help to improve the treatment of central nervous system (CNS) injuries and neurodegenerative disorders. Curcumin has been reported to exert biphasic effects on the proliferation of some stem cells including spinal cord NPCs,13 mice embryonic NPCs,8 and 3T3‐L1 preadipocytes,14 in which low concentrations of curcumin could activate signaling pathways including mitogen‐activated protein kinase (MAPK) signaling pathway13 in particular involving with p‐ERK and p‐38 signals8 to stimulate cell proliferation. On the other hand, other studies argued that spinal cord NPCs apoptosis could be mediated by glucocorticoid receptor (GR),15 and also the regulation of stem cell self‐renewal involved many other signaling pathways such as Jak/STAT316 and Notch1.17 It has been reported curcumin inhibited GR‐mediated transcription in HeLa cells18 and downregulated the expression of STAT3 in adult brain.19 The MAPK, GR, and Notch signaling pathways all play important roles in neural development and work as a network, but exact mechanisms about how curcumin stimulates NSC proliferation are complicated and remain unclear. The aim of this study was to explore the involvements of GR, STAT3, and Notch1 to the effect of curcumin on rat embryonic NSC proliferation.
2. MATERIALS AND METHODS
2.1. Animals
Timed pregnant embryonic day 15 (E15) Sprague Dawley (SD) rats were purchased from Shanghai Animal Center (Shanghai, China), and all animal procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals according to the protocols approved by the Animal Care and Use Committee of Wenzhou Medical University (Wenzhou, China).
2.2. Reagents
Reagents used in this study were as follows: Hank's balanced salt solution (HBSS), serum‐free Dulbecco's modified Eagle's medium (DMEM)/F12 medium, and B27 supplement without vitamin A (Gibco, USA); basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF) (Cali‐Bio, USA); CytoTox 96® Non‐Radioactive Cytotoxicity Assay (Promega, USA); corticosterone, mifepristone, and cryptotanshinone (Target Molecule, China); curcumin, dimethyl sulfoxide (DMSO), MTT (3‐(4,5‐dimethyl‐2‐thiazolyl)‐2,5‐diphenyl‐2‐H‐tetrazolium bromide), and poly‐L‐ornithine (Sigma, USA); BrdU Cell Proliferation Assay Kit (Merck Millipore, USA); BCA Protein Assay Kit (Thermo, USA); anti‐NESTIN and anti‐SOX2 antibodies (Santa Cruz, USA); GR antibody (BuGR2) (Pierce, USA); anti‐STAT3 and anti‐p‐STAT3 (Abcam, UK); TRIzol (Invitrogen, USA); SsoFast EvaGreen Supermix (Bio‐Rad, USA); and Reverse Transcription System (Promega, USA).
2.3. Cell culture
NSCs were harvested from E15 SD rat brain and cultured as described.20 Briefly, the embryo cortices were dissected in ice‐cold dissection buffer (HBSS), and single‐cell suspensions were obtained by mechanical dissociation. Primary cells were cultured at 1 × 106 cells/mL on 25 cm2 cell culture flasks in serum‐free DMEM/F12 medium supplemented with bFGF (20 ng/mL), EGF (20 ng/mL), penicillin (100 U/mL), and streptomycin (100 ug/mL) and 2% B‐27 supplement without vitamin A and then transferred to a humidified cell culture incubator at 37°C with 5% CO2. Half of the fresh medium was changed once every 2 days, and cells were passaged after 5‐7 days. All experiments were performed at the second passage. 5 × 104 cells in 100 μL medium were added to a well of poly‐l‐ornithine‐coated 96‐well microplates. The primary cells cultured in flasks were spherical, but after passage, they grew as individual undifferentiated cells on coverslips, and the NSCs were identified by NESTIN and SOX2 antibody staining. The result showed that more than 98% of them were NESTIN/SOX2 positive.
2.4. Curcumin administration
Curcumin was dissolved in DMSO (0.1%), which was proved no cytotoxicity.21 Cells were divided into seven groups at random: control group (C), 0.1% DMSO group (D), and 0.1, 0.5, 2.5, 12.5, and 62.5 μmol L−1 curcumin groups (0.1, 0.5, 2.5, 12.5, and 62.5 group). Cells were cultured in the plates for 24 hours and treated with curcumin for another 24 hours. Corticosterone (CORT, 10 μmol L−1) and mifepristone (Mife, 10 μmol L−1) as the agonist and antagonist of GR, respectively, and cryptotanshinone (CTS, 5 μmol L−1) as the antagonist of STAT3 were added 1 hour prior to curcumin exposure.
2.5. Lactate dehydrogenase assay
Lactate dehydrogenase (LDH) has been widely used to estimate cell cytotoxicity due to its ability of early detection of low‐level cell cytotoxicity. LDH was quantified using a commercially available CytoTox 96® Non‐Radioactive Cytotoxicity Assay according to the manual. Briefly, cells were seeded at a density of 5 × 104 cells in 96‐well plates and then administered with curcumin as previously described. Twenty four hours after curcumin treatment, 10 μL lysis 10× solutions per 100 μL of culture medium was added, followed by incubation at 37°C for 45‐60 min. Finally, the absorbance at 490 nm wavelength was measured with an enzyme‐linked immunosorbent assay (ELISA) microplate reader.
2.6. Cell viability and proliferation assay
Cell viability and proliferation were assessed by MTT assay and BrdU immunocytochemistry assay, respectively. Cells were seeded at a density of 2 × 104 cells in 96‐well plates and then administered with curcumin as previously described. 20 μL 5 mg mL−1 MTT was added to each well per 100 μL culture medium at the end of curcumin treatment. The plate was incubated at 37°C for 4 hours, and the absorbance at 560 nm wavelength was measured with an ELISA microplate reader. Cell proliferation was analyzed with BrdU Cell Proliferation Assay Kit. Twenty four hours after treatment, 20 μL BrdU was added to each well per 100 μL culture medium and incubated at 37°C for 24 hours. Finally, the absorbance at 490 nm was measured with an ELISA microplate reader.
2.7. Real‐time PCR
Cells were seeded at a density of 1 × 106 cells in 6‐well plates and then administered with curcumin as previously described. Total RNA was extracted with TRIzol. These RNA samples were used to generate cDNA using Reverse Transcription System according to the operating manual. Gene expression level was detected by quantitative real‐time PCR (qRT‐PCR) with relative standard curve method. The related primer sequences are as follows: Reference gene RS16, forward (F) AAGTCTTCGGACGCAAGAAA, reverse (R) TTGCCCAGAAGCAGAACAG. Target genes GR, (F) ATACAGCATCCCTTTCTCAGCA, (R) CTTGGCACCTATTCCAGTTTTC. STAT3, (F) TACCAGCAAAATCAGGTTGCT, (R) ACATCCCCAGAGTCCTTATCAA. Notch1, (F) ACATCCCCAGAGTCCTTATCAA, (R) GAAAAGCCACCGAGATAGTCAG.
2.8. Western blotting
Cell culture and curcumin administration were the same as qRT‐PCR assay. The protein was extracted and quantified with BCA Protein Assay Kit according to the manufacturer's instructions. The protein was homogenized and boiled for 5 min. Equal amounts of protein were separated by SDS‐PAGE using 8%‐10% gels and transferred onto a polyvinylidene difluoride membrane. The membrane was immediately placed in a blocking solution (5% nonfat dry milk in 0.05% TBS‐Tween (TBS‐T) buffer) for 1 hour at room temperature, then washed 3 times in TBS‐T buffer, and incubated with primary antibodies overnight at 4°C in blocking solution. The primary antibodies were used including mouse anti‐GR, rabbit anti‐STAT3, rabbit anti‐p‐STAT3 antibodies, and rabbit anti‐β‐actin as a loading control. The membrane was then washed in TBS‐T buffer and incubated with peroxidase‐conjugated goat anti‐rabbit IgG or peroxidase‐conjugated goat anti‐mouse IgG for 2 hours at room temperature. The protein band was detected by enhanced chemiluminescence (ECL) and captured by the imager (GE Healthcare Life Sciences, ImageQuant LAS 4000 mini). The Quantity One 4.62 software was used to analyze the integrated absorbance (IA) of related protein band.
2.9. Statistical analysis
Results are presented as mean ± standard error of the mean (SEM). The statistical significance of the differences between the groups was determined by analysis of variance (ANOVA) with Fisher protected least significant difference (PLSD). All statistical analyses were performed using SPSS 19.0. Values of P < 0.05 were considered to be statistical significance.
3. RESULTS
NSCs were isolated from E15 rat brain, and the cells formed neurosphere in the flask. After dissociation and plated on poly‐l‐ornithine‐coated coverslips, the cells grew as undifferentiated monolayer. Immunofluorescence showed more than 98% were stained positive for both NESTIN and SOX2, confirming that they were NSCs (Figure 1).
Figure 1.
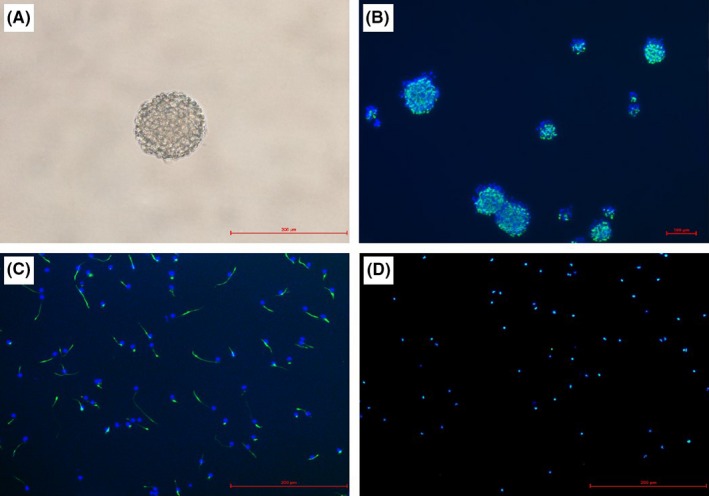
Photomicrographs of neural stem cells (NSCs) in culture. (A) The morphology of neurospheres. (B) Immunofluorescence of neurospheres merged image of NESTIN (green) and nuclear marker DAPI (blue). (C) Immunofluorescence of NSCs merged image of NESTIN (green) and nuclear marker DAPI (blue). (D) Immunofluorescence NSCs merged image of SOX2 (green) and nuclear marker DAPI (blue)
3.1. Effects of curcumin on NSC cytotoxicity
To determine the cytotoxicity of curcumin, colorimetric assay was used to measure the concentration of LDH in the culture medium (Figure 2A). There was no significant difference in the concentration of LDH between the control and DMSO groups (n = 9, P > 0.05), indicating that 0.1% DMSO was nontoxic to NSCs. However, the effects of curcumin on NSC cytotoxicity were significantly different between the curcumin‐treated groups, showing a concentration‐dependent manner. The concentrations of LDH in curcumin at low‐concentration (0.1, 0.5, and 2.5 μmol L−1) groups were similar to that in control group, and there was no significant difference between these groups (P > 0.05). In contrast, the concentration of LDH was significantly increased in 12.5 and 62.5 group as compared with that in control group (P < 0.05), but the difference between these two groups was not significant. These results showed that high‐dose curcumin may induced NSC cytotoxicity in a dose‐dependent manner.
Figure 2.
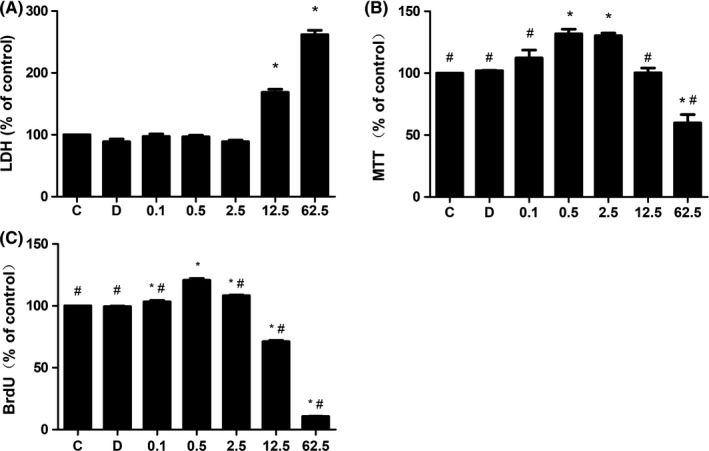
Effects of curcumin on LDH release, cell viability, and proliferation of NSCs. (A) The LDH release in NSCs after 24 h treatment of curcumin, which were increased in 12.5 and 62.5 group. (B) Cell viability was increased in 0.5 and 2.5 groups but decreased in 62.5 group. (C) Cell proliferation was especially increased in 0.5 group. *P < 0.05, compared with C group. # P < 0.05, compared with 0.5 group
3.2. Effects of curcumin on NSC viability and proliferation
We then analyzed the NSC viability and proliferation after curcumin treatment. NSC viability was evaluated by MTT proliferation assay (Figure 2B). Except in 0.1 group, cell viability was increased significantly in the other two curcumin low‐concentration (0.5 and 2.5 μmol L−1) groups (n = 9, P < 0.05), and there was no significant difference between them. However, cell viability was decreased significantly in 62.5 group (P < 0.05). BrdU is a thymidine analog that is incorporated into cells only during S‐phase of cell cycle, and it was used to monitor cell proliferation (Figure 2C). The result showed that curcumin at low concentrations (0.1, 0.5, and 2.5 μmol L−1) promoted cell proliferation, and the effect was most pronounced in 0.5 group compared with other groups. In contrast, curcumin at high concentrations (12.5 and 62.5 μmol L−1) suppressed cell proliferation significantly (n = 9, P < 0.05).
3.3. Effects of curcumin on NSCs proliferation‐related genes expression
To further explore the mechanism of curcumin‐induced proliferation in NSCs, 0.5 μmol L−1 curcumin was used to explore its effect on expression of related genes. As GR, STAT3, and Notch1 are all involved in neurodevelopment, their transcription after curcumin treatment was measured by qRT‐PCR. It showed that the mRNA expression of GR and STAT3 was decreased after 24 hours exposure to curcumin (Figure 3A,B, n = 8, P < 0.05). However, the mRNA expression of Notch1 in curcumin groups was similar to that in the control group, showing no significant difference (n = 8, P > 0.05) (Figure 4).
Figure 3.
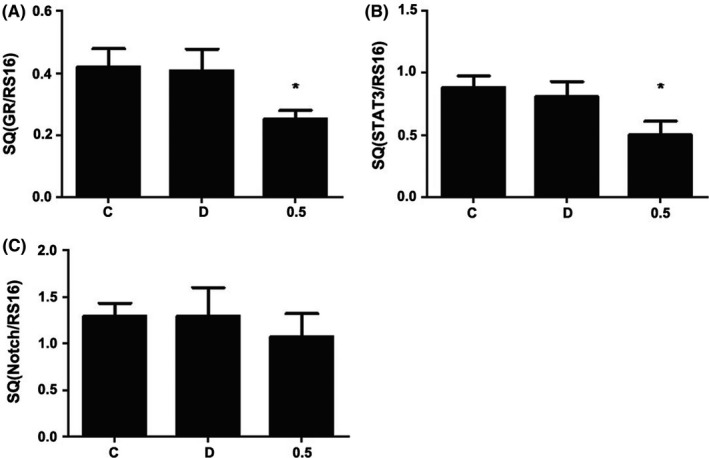
Effects of curcumin on mRNA expression of GR, STAT3, and Notch1 by RT‐PCR. NSCs were seeded into 6‐well culture plates and treated with curcumin for 24 h. The mRNA expression of GR (A) and STAT3 (B) in both C and D groups remained unchanged significantly, but it was significantly decreased in 0.5 group. However, the mRNA expression of Notch1 remained unchanged significantly in all groups (C). *P < 0.05, compared with C group
Figure 4.
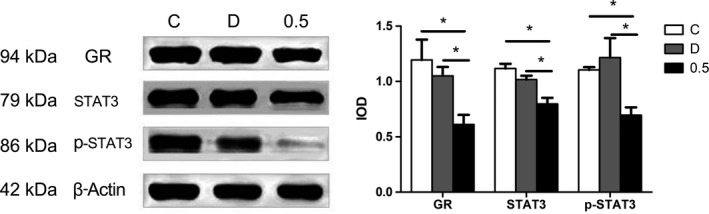
Effects of curcumin on protein expression of GR, STAT3, and p‐STAT3 by Western blotting. NSCs were seeded into 6‐well culture plates and treated with curcumin for 24 h, and then protein was extracted for immunoblot analysis using antibodies against GR, STAT3, and phospho‐STAT3. β‐actin level was determined as a control for possible protein loading variability. The protein expression of GR, STAT3, and p‐STAT3 remained unchanged in both C and D groups, but was significantly decreased in 0.5 group. *P < 0.05, compared with C group
3.4. Effects of curcumin on NSCs proliferation‐related protein expression
To further explore the effect of curcumin on NSC proliferation, we detected the protein expression of GR and STAT3 because their mRNA expression levels underwent change in RT‐PCR research. First of all, both GR and STAT3 proteins were expressed in our cultured NSCs (Figure 5A). Western blotting analysis showed that the expression of GR protein was suppressed in 0.5 group (Figure 5B), which was in accordance with the gene expression (n = 8, P < 0.05). In addition, the protein expression of both STAT3 (Figure 5C) and p‐STAT3 (Figure 5D) in 0.5 group was decreased when compared with C group (n = 8, P < 0.05). However, there was no difference between the three groups about the ratio of p‐STAT3/STAT3.
Figure 5.
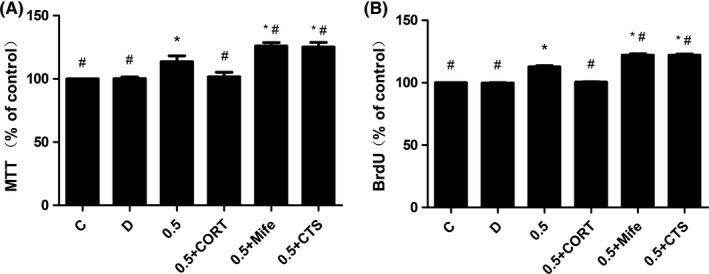
Effects of curcumin coincubate with corticosterone, mifepristone, and cryptotanshinone on cell viability and proliferation of NSCs. CORT, Mife, and CTS were added prior to curcumin exposure. Cell viability (A) and cell proliferation (B) were both increased in 0.5 + Mife and 0.5 + CTS group, but decreased in 0.5 + CORT group when compared with 0.5 group. *P < 0.05, compared with C group, # P < 0.05, compared with 0.5 group
3.5. Inhibition of GR and STAT3 signaling increased NSC viability and proliferation
We then set to investigate the effects of GR and STAT3 signaling on curcumin‐induced NSC viability and proliferation. Pretreatment with mifepristone and cryptotanshinone prior to curcumin exposure resulted in significant augment of cell viability (Figure 5A) and cell proliferation (Figure 5B) (0.5, 0.5 + Mife, and 0.5 + CTS group). Also, there was significant difference between 0.5 group and 0.5 + Mife and 0.5 + CTS group. However, pretreatment with CORT prior to curcumin exposure resulted in significant reduction in cell viability (Figure 5A) and cell proliferation (Figure 5B) (0.5 + CORT group), when compared with 0.5 group. (n = 9, P < 0.05).
4. DISCUSSION
Previous studies22 demonstrated that curcumin enhanced neurogenesis in neurodegenerative diseases. In addition, treatment of curcumin stimulated proliferation of stem cells both in vivo and in vitro.8, 9 However, responses to curcumin were reported to be different under various conditions with respect to the dose, time, and cell type.23 In the present research, we demonstrated that curcumin had a biphasic effect on the proliferation of rat NSCs. The low concentrations (0.1, 0.5, and 2.5 μmol L−1) of curcumin stimulated NSC proliferation, whereas high concentrations (12.5 and 62.5 μmol L−1) inhibited NSC proliferation. In view of the high concentrations of curcumin produced a cytotoxic effect on NSC, we chose the low concentrations for further investigation. According to the results of cell toxicity, vitality, and proliferation, 0.5 μmol L−1 curcumin was found to be atoxic on NSCs; and it strongly promoted NSC vitality and proliferation. Therefore, 0.5 μmol L−1 curcumin was selected as the optimal dose to explore the possible mechanism underlying the effect of curcumin on NSC proliferation. Division and differentiation of NSCs are known to be controlled by a network of transcriptional, growth, and hormonal factors, which is vital for the normal brain development including the embryo brain and adult CNS.10 GR and STAT3 are expressed in embryonic NSCs and are all involved in the development of the brain. Our qRT‐PCR and Western blotting data suggested that the concentration of 0.5 μmol L−1 curcumin significantly decreased the level of mRNA and protein both in GR and STAT3. To investigate curcumin function in depth, we performed proliferation assay with agonist/antagonist of GR and antagonist of STAT3. The agonist of GR effectively inhibited the cell viability and proliferation effect of curcumin on NSCs, whereas the inhibitors of GR and STAT3 augmented its effects, which suggested that GR and STAT3 were probably both involved in the effects of curcumin on NSC proliferation. Our results showed that GR and STAT3 signal pathways were involved in the biological effect of curcumin on NSCs.
GR is generally accepted as a cognate intracellular receptor that interacts with glucocorticoid hormones to mediate a wide range of biological processes, such as cell proliferation, inflammation, and immunity.24 Previous studies25 demonstrated that glucocorticoid hormones could reduce both neuronal proliferation and differentiation in human hippocampal progenitor cells by activating GR and induce NPC apoptosis in the mouse developing cerebellum.15 Exposure of NSCs to dexamethasone decreased their proliferation in a GR‐dependent manner.26 According to the previous research reports, the action of GR involved classical GR signaling and nonclassical steroid hormone signaling.27 The traditional view of GR transcriptional regulation is through the direct binding of glucocorticoid hormone (GC) to glucocorticoid response elements (GREs) or indirectly associated with other DNA‐bound transcription factors. However, an increasing number of studies suggested that GCs could mediate rapid cellular responses via transcription‐independent (nonclassical) mechanisms. The nonclassical existed a cross‐talk between GR and other signaling pathways, which include MAPK family, such as ERK, p38, and JNK/SAPK signaling pathways.28 A study in murine embryonic NPCs found that GR signaling pathway affected NPC proliferation through inhibition of gap junction intercellular communication (GJIC), which triggered to reduce the S‐phase progression, and the rapid activation of GR could alter the ERK‐1/2 in NPCs.29 Kim et al8 reported that curcumin stimulated NSC proliferation by activating the kinases of ERK and p38. However, ERK activation could block the serine phosphorylation of STAT3 in human neutrophils as it is the prevailing upstream kinase for phosphorylating STAT3 on Ser‐727.30 STAT3 pathway plays an important role in the maintenance of self in ‐renewal mouse embryonic stem cells,31, 32 and suppression of STAT3 could directly induce neurogenesis and inhibit astrogliogenesis in neural stem cells.33 Besides, Langlais et al34 reported that GR can behave as transcriptional repressor when tethering to DNA‐bound STAT3 and resulted in transcriptional repression. Therefore, the nonreciprocal interaction between GR and STAT3 could result in different outcome of transcription. Notch signaling is reported to be involved in cell maintenance and determination of NSCs.35 Notch1 is a stimulus for rat NSC development.36 But in this study, the expression of Notch1 gene did not undergo significant changes.
5. CONCLUSIONS
In summary, the present study showed that low‐dose curcumin was able to stimulate the proliferation of rat NSCs and both GR and STAT3 signaling were involved. The specific mechanism may be associated with the nonclassical signaling pathway of GR, which control the proliferation of NSCs by activating the ERK pathway to regulate STAT3 protein expression or through the transcriptional interaction between GR and STAT3. But there are many shortcomings of this experiment. In view of the complicated interactions between signal pathways, more accurate researches are required, such as the gene silencing technology. Meanwhile, in vivo experiments are needed to confirm our in vitro findings. Nevertheless, this experiment provides a theoretical basis for further research in this field.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
This study was partially supported by National Natural Science Foundation of China (81471448), Natural Science Foundation of Zhejiang Province (Y14H090071), and Science Technology Department of Zhejiang Province (2012R10073), Wenzhou Science & Technology Bureau (Y20150061).
Ma X‐X, Liu J, Wang C‐M, Zhou J‐P, He Z‐Z, Lin H. Low‐dose curcumin stimulates proliferation of rat embryonic neural stem cells through glucocorticoid receptor and STAT3. CNS Neurosci Ther. 2018;24:940–946. 10.1111/cns.12843
The first two authors contributed equally to this work.
Contributor Information
Zhen‐Zhou He, Email: sandyhezz@126.com.
Han Lin, Email: nanlinhannansh@gmail.com.
REFERENCES
- 1. Ammon HP, Wahl MA. Pharmacology of Curcuma longa. Planta Med. 1991;57:1‐7. [DOI] [PubMed] [Google Scholar]
- 2. Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Curcumin: from ancient medicine to current clinical trials. Cell Mol Life Sci. 2008;65:1631‐1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scarpa ES, Ninfali P. Phytochemicals as innovative therapeutic tools against cancer stem cells. Int J Mol Sci. 2015;16:15727‐15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ataie A, Shadifar M, Ataee R. Polyphenolic antioxidants and neuronal regeneration. Basic Clin Neurosci. 2016;7:81‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jurenka JS. Anti‐inflammatory properties of curcumin, a major constituent of Curcuma longa: a review of preclinical and clinical research. Altern Med Rev. 2009;14:141‐153. [PubMed] [Google Scholar]
- 6. Xu Y, Li S, Vernon MM, et al. Curcumin prevents corticosterone‐induced neurotoxicity and abnormalities of neuroplasticity via 5‐HT receptor pathway. J Neurochem. 2011;118:784‐795. [DOI] [PubMed] [Google Scholar]
- 7. Zhu YG, Chen XC, Chen ZZ, et al. Curcumin protects mitochondria from oxidative damage and attenuates apoptosis in cortical neurons. Acta Pharmacol Sin. 2004;25:1606‐1612. [PubMed] [Google Scholar]
- 8. Kim SJ, Son TG, Park HR, et al. Curcumin stimulates proliferation of embryonic neural progenitor cells and neurogenesis in the adult hippocampus. J Biol Chem. 2008;283:14497‐14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tiwari SK, Agarwal S, Seth B, et al. Curcumin‐loaded nanoparticles potently induce adult neurogenesis and reverse cognitive deficits in Alzheimer's disease model via canonical Wnt/beta‐catenin pathway. ACS Nano. 2014;8:76‐103. [DOI] [PubMed] [Google Scholar]
- 10. Kennea NL, Mehmet H. Neural stem cells. J Pathol. 2002;197:536‐550. [DOI] [PubMed] [Google Scholar]
- 11. Song HJ, Stevens CF, Gage FH. Neural stem cells from adult hippocampus develop essential properties of functional CNS neurons. Nat Neurosci. 2002;5:438‐445. [DOI] [PubMed] [Google Scholar]
- 12. Parent JM. Adult neurogenesis in the intact and epileptic dentate gyrus. Prog Brain Res. 2007;163:529‐540. [DOI] [PubMed] [Google Scholar]
- 13. Son S, Kim KT, Cho DC, Kim HJ, Sung JK, Bae JS. Curcumin stimulates proliferation of spinal cord neural progenitor cells via a mitogen‐activated protein kinase signaling pathway. J Korean Neurosurg Soc. 2014;56:1‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim JH, Park SH, Nam SW, et al. Curcumin stimulates proliferation, stemness acting signals and migration of 3T3‐L1 preadipocytes. Int J Mol Med. 2011;28:429‐435. [DOI] [PubMed] [Google Scholar]
- 15. Noguchi KK, Lau K, Smith DJ, Swiney BS, Farber NB. Glucocorticoid receptor stimulation and the regulation of neonatal cerebellar neural progenitor cell apoptosis. Neurobiol Dis. 2011;43:356‐363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hirai H, Karian P, Kikyo N. Regulation of embryonic stem cell self‐renewal and pluripotency by leukaemia inhibitory factor. Biochem J. 2011;438:11‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gangemi RM, Perera M, Corte G. Regulatory genes controlling cell fate choice in embryonic and adult neural stem cells. J Neurochem. 2004;89:286‐306. [DOI] [PubMed] [Google Scholar]
- 18. Aoyagi S, Archer TK. Differential glucocorticoid receptor‐mediated transcription mechanisms. J Biol Chem. 2011;286:4610‐4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tiwari SK, Agarwal S. B Seth. Curcumin‐loaded nanoparticles potently induce adult neurogenesis and reverse cognitive deficits in Alzheimer's disease model via canonical Wnt/β‐Catenin pathway. ACS Nano. 2013;8:76‐103. [DOI] [PubMed] [Google Scholar]
- 20. Culley DJ, Boyd JD, Palanisamy A, et al. Isoflurane decreases self‐renewal capacity of rat cultured neural stem cells. Anesthesiology. 2011;115:754‐763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu B, Chen X, Mao Z, et al. Perfluorooctane sulfonate disturbs Nanog expression through miR‐490‐3p in mouse embryonic stem cells. PLoS ONE. 2013;8:e74968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cole GM, Teter B, Frautschy SA. Neuroprotective effects of curcumin. Adv Exp Med Biol. 2007;595:197‐212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Attari F, Zahmatkesh M, Aligholi H, et al. Curcumin as a double‐edged sword for stem cells: dose, time and cell type‐specific responses to curcumin. Daru. 2015;23:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Periyasamy S, Sanchez ER. Antagonism of glucocorticoid receptor transactivity and cell growth inhibition by transforming growth factor‐beta through AP‐1‐mediated transcriptional repression. Int J Biochem Cell Biol. 2002;34:1571‐1585. [DOI] [PubMed] [Google Scholar]
- 25. Anacker C, Cattaneo A, Luoni A, et al. Glucocorticoid‐related molecular signaling pathways regulating hippocampal neurogenesis. Neuropsychopharmacology. 2013;38:872‐883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bose R, Moors M, Tofighi R, Cascante A, Hermanson O, Ceccatelli S. Glucocorticoids induce long‐lasting effects in neural stem cells resulting in senescence‐related alterations. Cell Death Dis. 2010;1:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samarasinghe RA, Witchell SF, DeFranco DB. Cooperativity and complementarity: synergies in non‐classical and classical glucocorticoid signaling. Cell Cycle. 2012;11:2819‐2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ayroldi E, Cannarile L, Migliorati G, Nocentini G, Delfino DV, Riccardi C. Mechanisms of the anti‐inflammatory effects of glucocorticoids: genomic and nongenomic interference with MAPK signaling pathways. FASEB J. 2012;26:4805‐4820. [DOI] [PubMed] [Google Scholar]
- 29. Samarasinghe RA, Di Maio R, Volonte D, et al. Nongenomic glucocorticoid receptor action regulates gap junction intercellular communication and neural progenitor cell proliferation. Proc Natl Acad Sci U S A. 2011;108:16657‐16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuroki M, O'Flaherty JT. Extracellular signal‐regulated protein kinase (ERK)‐dependent and ERK‐independent pathways target STAT3 on serine‐727 in human neutrophils stimulated by chemotactic factors and cytokines. Biochem J. 1999;341(Pt 3):691‐696. [PMC free article] [PubMed] [Google Scholar]
- 31. Sumi T, Fujimoto Y, Nakatsuji N, Suemori H. STAT3 is dispensable for maintenance of self‐renewal in nonhuman primate embryonic stem cells. Stem Cells. 2004;22:861‐872. [DOI] [PubMed] [Google Scholar]
- 32. Cao F, Hata R, Zhu P, Nakashiro K, Sakanaka M. Conditional deletion of Stat3 promotes neurogenesis and inhibits astrogliogenesis in neural stem cells. Biochem Biophys Res Commun. 2010;394:843‐847. [DOI] [PubMed] [Google Scholar]
- 33. Gu F, Hata R, Ma YJ, et al. Suppression of Stat3 promotes neurogenesis in cultured neural stem cells. J Neurosci Res. 2005;81:163‐171. [DOI] [PubMed] [Google Scholar]
- 34. Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell. 2012;47:38‐49. [DOI] [PubMed] [Google Scholar]
- 35. Ungerer P, Eriksson BJ, Stollewerk A. Unravelling the evolution of neural stem cells in arthropods: notch signalling in neural stem cell development in the crustacean Daphnia magna. Dev Biol. 2012;371:302‐311. [DOI] [PubMed] [Google Scholar]
- 36. Rodriguez‐Rivera NS, Molina‐Hernandez A, Sanchez‐Cruz E, Escalante‐Alcalde D, Velasco I. Activated Notch1 is a stronger astrocytic stimulus than leukemia inhibitory factor for rat neural stem cells. Int J Dev Biol. 2009;53:947‐953. [DOI] [PubMed] [Google Scholar]


