Abstract
Aims
Alzheimer's disease (AD) is one of the leading causes of death in elderly people. Its pathogenesis is greatly associated with the abnormality of immune system. However, only a few immune‐relevant AD drug target genes have been discovered up to now, and it is speculated that there are still many potential drug target genes of AD (at least immune‐relevant genes) to be discovered. Thus, this study was designed to identify novel AD drug target genes and explore their biological properties.
Methods
A combinatorial approach was adopted for the first time to discover AD drug targets by collectively considering ontology inference and network analysis. Moreover, a novel strategy limiting the distance of reasoning and in turn reducing noise interference was further proposed to improve inference performance. Potential AD drug target genes were discovered by integrating information of multiple popular databases (TTD, DrugBank, PharmGKB, AlzGene, and BioGRID). Then, the enrichment analyses of the identified drug targets genes based on nine well‐known pathway‐related databases were conducted to explore the function of the identified potential drug target genes.
Results
Eighteen potential drug target genes were finally identified, and 13 of them had been reported to be closely associated with AD. Enrichment analyses of these identified drug target genes, based on nine pathway‐related databases, revealed that the enriched terms were primarily focus on immune‐relevant biological processes. Four of those identified drug target genes are involved in the classical complement pathway and process of antigen presenting.
Conclusion
The well‐reproducible results showed the good performance of the combinatorial approach, and the remaining five new targets could be a good starting point for our understanding of the pathogenesis and drug discovery of AD. Moreover, this study supported validity of the combinatorial approach integrating ontology inference with network analysis in the discovery of novel drug target for neurological diseases.
Keywords: Alzheimer's disease, drug target, immune, network analysis, ontology inference
1. INTRODUCTION
Alzheimer's disease (AD) is the most common type of dementia and one of the leading causes of death in elderly people, which is characterized by neurofibrillary tangles and amyloid β‐peptide (Aβ) deposits in brain.1, 2 Based on two independent studies, the number of people newly diagnosed with AD is expected to reach 135 million around the world and 16 million in USA by 2050, respectively.3, 4
So far, the deficiency of innate immune ability to clear Aβ deposits (rather than overproduction of them) has been widely accepted as one of the most important causes of AD pathogenesis, and current research was gradually moved from blocking the production of Aβ deposits to rebalancing the immune system of AD patients.5, 6, 7 For example, interleukin (IL)‐10, one of the best known inflammatory cytokines, was a major regulator of macrophages.8 Recent study showed that the inhibition of the IL‐10 pathway could rebalance the innate immunity and mitigate Alzheimer‐like pathology.9 Another member of interleukin family, IL‐33, was found to play a potential therapeutic role for AD. A previous study reported that IL‐33 could modulate the innate immune responses to reduce the accumulation of Aβ deposits and reverse the impairment of memory and synaptic plasticity in AD mouse.10 The genome‐wide association study found that a cluster of genes implicated in innate immune pathways was upregulated together with the downregulation of synaptic plasticity genes in AD patients.11, 12 Moreover, two lymphatic systems involved in the clearance of Aβ deposits in AD were reported. In particular, glymphatic (glial+lymphatic) system was a critical contributor to the clearance of interstitial solutes (including Aβ deposits) from the brain,13 and the function of glymphatic transport was suppressed in a mouse AD model.14 The malfunction of meningeal lymphatic vessel (the other cleaner of Aβ deposits closely related to lymphatic system) was found to be greatly associated with AD pathogenesis.15, 16, 17 In the meantime, some immunotherapies and drugs for Aβ clearance have attempted to control progress of AD.18, 19, 20 For example, a monoclonal antibody‐based immunotherapeutic drug was reported under development in recent year, and it selectively targeted and cleared the aggregated Aβ in brain and effectively relieved the symptom of AD.21, 22, 23
However, only a few of drug target genes of AD implicated in immunity had been discovered up to now. Specifically, among the known target genes of therapeutic drugs for AD, only four of them (about 10.5%) belonged to the family of innate immune genes (Table S1, based on the information of InnateDB with over 1500 innate immune genes collected by literature review).24 Moreover, the studies showed that the total number of the known AD drug targets had appeared to be also not complete enough, considering the wide range of pathologic features.25 In fact, only five drugs have got FDA approval for the treatment of AD, and they target primarily on two therapeutic targets: Acetylcholinesterase and NMDA receptor.26 Since 2003, no new target has been approved for treating AD. All the facts implied that there were still potential target genes of AD (especially the innate immune genes) remaining to be discovered.
Due to the time‐consuming and extremely high cost of modern drug discovery, computational methods have emerged as one of the most effective approaches for the discovery of new targets.27, 28, 29, 30, 31, 32 However, these computational methods focused mainly on single biologic perspective, such as pathway profile‐based,33 gene expression‐based,34, 35, 36 and similarity‐based37, 38, 39, 40 methods. Important information might be neglected by these methods, as many essential relationships (such as gene‐gene, gene‐disease, and disease‐drug) systematically contributed to the association between the disease and their corresponding drug targets.41 As reported, target druggability was found to be collectively defined by target's disease relevance and its roles in human protein‐protein interaction network,42 and a novel strategy integrating ontology inference and network analysis was thus proposed to predict the candidate targets of colorectal cancer (CRC).41 In this study, the inference performance of this published method41 was substantially enhanced by limiting the distance of reasoning and in turn reducing noise interference (prediction accuracy of ontology‐based inference was reported to be highly dependent on the distance from initial nodes).43 Then, relationships among drug, gene, SNP, disease, and haplotype were integrated by combining ontology‐based inference and biological network analysis to discover potential target of AD. Finally, the enrichment analysis to the in‐depth investigation of the biological functions for AD was conducted. In conclusion, this was the first discovery of drug targets for neurological disease by improving the inference performance of the newly proposed combinatorial method, and the novel candidate target genes identified in this study did provide significantly added values to the discovery of drugs for treating AD.
2. MATERIALS AND METHODS
2.1. Collection of AD drugs and their known targets
The drugs approved by US FDA or in clinical trial for the treatment of AD were first collected from the Therapeutic Target Database (TTD)44 which contained 31 614 drugs and 2589 targets covering over 125 diseases after its last update in 2018.26 Then, the drugs that did not belong to PharmGKB database were removed, and full list of AD drugs was obtained for further analyses. The PharmGKB was a preeminent worldwide resource and web interactive tool for the knowledge of pharmacogenomics. It was funded by the National Institute of General Medical Sciences (NIGMS) and the National Institutes of Health (NIH), and the relationship data of PharmGKB could be used to research how the genetic variations affected the response of drugs.45 For example, rs1800460 polymorphism was related to azathioprine, mercaptopurine, thioguanine based on the data of PharmGKB, and many studies reported this variant could cause adverse reactions to these drugs.45, 46, 47 In this study, PharmGKB relationship data between drugs and drugs, drugs and haplotypes, drugs and SNPs, drugs and genes, diseases and genes, diseases and haplotypes, diseases and SNPs, diseases and diseases, genes and haplotype were collected for subsequent study. Finally, the known targets of the selected AD drugs were extracted from both TTD and DrugBank.48, 49
2.2. Discovery of the candidate AD target genes using ontology inference
Ontology was a hierarchically organized structure about existences, types of them and their relationships, according to their innate logic. Based on ontology, semantic web technology was developed to integrate and reason the heterogeneous data. As a machine‐readable standard ontology language of semantic web technique, the web ontology language (OWL) effectively inherited triple model (subject‐predicate‐object) from resource description framework (RDF).50, 51, 52, 53, 54 Thus, through integrating and reasoning relationships among drugs, genes, SNPs, diseases, and haplotypes, the OWL could find the genes directly or indirectly connected with an AD drug, and these genes were defined as the candidate AD target genes. According to the newly proposed strategy,41 in OWL network, all of the drugs, genes, SNPs, diseases, haplotypes from PharmGKB were defined as nodes and the relationships of them were defined as links based on the Portégé (an editor and reasoner based on OWL). Then, the AD‐related drug (ADDrug), gene (ADGene), SNP (ADSNP), disease (ADDisease), and haplotype (ADHaplotype) were defined according to the OWL description logic rule. Specifically, a gene belonged to the class of ADGene if it was associated with one of these defined concepts (ADDrug, ADGene, ADSNP, ADDisease, and ADHaplotype) according to the relationships in PharmGKB, where other concepts were defined similar as the ADGene. The distance of reasoning for an AD drug (ie the number of nodes away from the AD drug node) was limited according to the location containing the most number of their known target genes.
2.3. Collecting the AD disease genes
To further identify the potential AD target genes from AD target genes, the levels of association between known AD disease genes and each of the candidate AD target genes were compared and ranked. The AD disease genes were searched in the AlzGene, a publicly available database collecting AD genetic variants from the publications of genetic association study about AD. To the best of our knowledge, the AlzGene is so far the only database specialized in the selection and analysis of AD risk genes, and its latest version (updated in 2011) contained 695 disease genes, 2973 polymorphisms, and 320 systematic meta‐analyses performed for genotype data (including at least three case‐control samples).55 To ensure the credibility of this study, the significantly associated genes with AD (95% confidence interval of OR values should not include 1)56 were selected for the subsequent analyses based on the results of the meta‐analyses.
2.4. Discovering potential AD target genes via association level test and aggregation rank
Based on the hypothesis that the drugs will more efficiently work on some disease genes if they show a tighter connection,41 the levels of association between each of the candidate AD target genes and the AD disease genes were assessed to identify potential AD target genes. The human protein‐protein interaction (PPI) network was thus collected for conducting such calculation. The BioGRID (Mount Sinai Hospital, Toronto, ON, Canada) was a monthly updated comprehensive PPI network database.57 It captured 836 212 nonredundant biological interactions from 57 058 published biomedical articles involving all major organisms and human beings by September 2016. Moreover, the different reliability of each interaction was provided in the BioGRID based on available evidences that how many independent studies consistently support the result. First, human PPI data were download from BioGRID (version 3.4.140), and the high reliability data supported by at least two independent studies were selected. Second, after removing self‐interaction data, the human PPI networks which centered on each of the candidate target genes were built. Within these networks, the nodes directly linked with the candidate target genes were defined as the first‐degree neighbor. Similarly, the nodes linked with the first‐degree neighbors (except the candidate AD target genes) were defined as the second‐degree neighbors, and the nodes linked with the second‐degree neighbors (except first‐degree neighbors) were defined as the third‐degree neighbors, etc. Thirdly, the previous studies showed that the disease genes were mainly enriched in the first three degree neighbors of the drug target genes.58, 59 Thus, the AD disease genes to the first‐, second‐, third‐degree neighbors of the PPI networks were mapped, and their percentages in each neighbor to assess the levels of association between each of the candidate AD target genes and AD disease genes were calculated. Specifically, for a given AD candidate gene, there were N genes in its first‐degree neighbor, and of which n genes belong to AD disease genes. Then, the closer that this ratio (n/N) was to 1, the more significant association was built between this given AD candidate gene and AD disease genes. Thus, this given candidate gene more likely becomes a potential drug target. Similarly, the ratio in other degree neighbors was calculated by this approach. Finally, based on the ratios in the three degree neighbors, the candidate AD target genes were ranked in three lists.
To identify the potential AD drug targets, robust rank aggregation (RRA) method was used to integrate the three lists of rank order. RRA was a computationally efficient and statistically stable order algorithm, and it assigned the P‐value to measure how well a candidate gene was positioned in the ranked lists than expected by chance.60 The R package of RRA (RobustRankAggreg) was available at the Comprehensive R Archive Network (https://www.icesi. edu.co/CRAN/web/packages/RobustRankAggreg/).
2.5. Enrichment analysis of the potential AD drug target genes
AD was reported to be closely related to immune system,5, 61 but very little of immune‐relevant AD drug target genes had been discovered comparing with other target genes. Therefore, it was possible that there were more potential immune‐relevant genes than others among the undiscovered AD drug target genes. However, it was improper to simply compare the numbers of them due to the different backgrounds (the various sizes of gene sets). So far, these backgrounds (such as human disease, organism system, cellular processes and signaling pathways) were provided in Kyoto Encyclopedia of Genes and Genomes (KEGG) for achieving this comparison.62 Apart from KEGG, there were several other data sources providing such signaling pathway information, including MetaCyc63 (database of the metabolic pathways and enzymes), NetPath64 (resource of curated signal transduction pathways), PathWhiz65 (database of biology pathways and web server for creating biologically accurate pathway diagram), Pathway Interaction Database PID66 (collection of curated and peer‐reviewed pathways composed of human molecular signaling, regulatory event and cellular process), and WikiPathways67 (multifaceted pathway database bridging metabolomics to other omics research). Moreover, there were three ontology‐based databases offering pathway‐related data, including Gene Ontology GO68 (gene ontology organized by biological process, molecular function and cellular component), PANTHER69 (gene products organized by biological function), and Reactome70 (molecular detail of signal transduction, transport, metabolism, and other cellular processes). Apart from PID, the comprehensive data of the remaining eight databases were fully downloadable, and the enrichment analyses of the potential AD drug target genes based on data from these eight databases were thus conducted using the R package clusterProfiler71 to validate the presumption of this study.
3. RESULTS AND DISCUSSION
3.1. Collecting AD drugs and limiting search criteria using their known targets
First, by selecting AD drugs from TTD database72 and removing the drugs not belonging to PharmGKB database,73 five approved and one phase III clinical trial drugs were obtained. Second, 23 known targets of these drugs were collected from TTD and DrugBank.48, 49 Third, to limit the distance of reasoning, the number of the known AD target genes was counted in each degree of OWL network. As demonstrated in Table 1, for the selected drugs, their known target genes were mainly concentrated in a specific degree. These results implied that for a given drug, its target genes were mainly distributed in a specific scope of OWL network. In this study, to reduce the effect of noise, the reasoning distance of each drug was thus limited to the degree where most of its known target genes can be found.
Table 1.
Summary of the final list of AD drugs and their known target genes
| Drug Name |
TTD PharmGKB |
Drug Status | Known drug target genes | Mode Value of Degrees | Average Value of Degrees | ||||
|---|---|---|---|---|---|---|---|---|---|
| Degree 1 | Degree 2 | Degree 3 | Degree 4 | Degree 5 | |||||
| Donepezil |
DAP000560 PA449394 |
Approved | ACHE | NaN | HTR2A | NaN | NaN | 1, 3 | 2 |
| Rivastigmine |
DAP000149 PA451262 |
Approved | ACHE, BCHE | NaN | NaN | NaN | NaN | 1 | 1 |
| Memantine |
DAP000493 PA10364 |
Approved | NaN | NaN | GRIN3A, GRIN2A, GRIN2B, HTR3A, CHRNA7, DRD2, GRIN1 | NaN | NaN | 3 | 3 |
| Galantamine |
DAP000559 PA449726 |
Approved | CHRNA4, CHRNA7, CHRNB2, ACHE, BCHE | NaN | CHRNA1, CHRNB1, CHRNA2, CHRNA3, CHRNA5, CHRNA6, CHRNB3, CHRNB4 | NaN | CHRND, CHRNG | 3 | 3 |
| Tacrine |
DAP000558 PA451576 |
Approved | NaN | NaN | ACHE, BCHE | NaN | NaN | 3 | 3 |
| Rosiglitazone |
DCL000633 PA451283 |
Phase III | PPARG | NaN | NaN | NaN | NaN | 1 | 1 |
The nodes directly linked with a certain drug according to the relationships of PharmGKB are defined as degree 1. The nodes linked with this drug through degree 1 are defined as degree 2. Degree 3, degree 4, and degree 5 are defined by similar way. NaN means that the known drug target gene is not in this degree. “Mode value of degrees” means that most of the known drug target genes are found in this degree.
3.2. Discovering the candidate AD target genes using OWL ontology method
An OWL ontology of AD drug, gene, SNP, disease, and haplotype was built based on their relationship in PharmGKB, and the reasoning was performed using Portégé in a specific scope limited by the previous step. The process of reasoning and the result is displayed in Figures 1 and S1. After eliminating duplicate data, 623 candidate AD target genes directly or indirectly connected with one of the AD drugs were discovered, and 109 of them had been reported to be closely related to AD.
Figure 1.
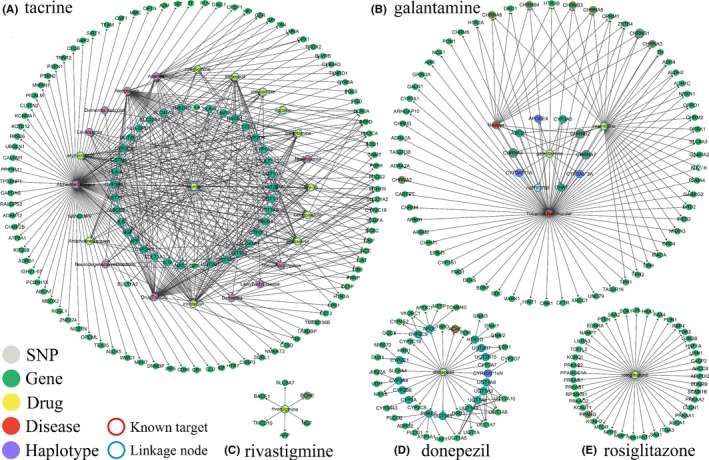
Process of discovering the candidate AD target genes from each final selected AD drug. Each part of this figure shows the process of discovery from one of the drugs, respectively. The process of discovery from memantine is shown in Figure S1 because it is relatively complex. Different colors of the nodes mean different data types. In particular, green represents gene, yellow represents drug, red represents disease, purple represents haplotype, and gray represents SNP. The nodes with red and blue circles represent known targets and linkage nodes, respectively. The annulus areas constituted of these nodes represent different discovery degrees. Only the linkage nodes and the discovered candidate target genes are retained in this figure. The duplicate data have been eliminated. This figure can be viewed more clearly by enlarging in the electronic version
3.3. Identifying the potential AD target genes by AD disease genes and RRA method
First, 41 AD disease genes confirmed by meta‐analysis (including at least three case‐control experiments) were collected from AlzGene database (Table S2). Second, 623 human PPI networks of candidate AD target genes were built by BioGRID data. Third, the 41 AD disease genes were mapped to the first‐, second‐, and third‐degree neighbors of the 623 PPI networks, and the percentages of AD disease genes in each degree neighbor were calculated to rank these candidate target genes at 3 different levels. Finally, these three levels of rank were integrated using RRA method. As shown in Table 2, 18 potential AD target genes with significant P‐values (<0.05) were identified, and of which 13 genes were reported to be closely associated with AD by previous studies.
Table 2.
Summary of the 18 potential AD drug target genes identified in this study
| Gene symbol | Protein name | Gene ID | P‐value | Reported or NOT |
|---|---|---|---|---|
| RELN | Reelin | 5649 | 0.000546 | Reported |
| DNMBP | Dynamin binding protein | 23268 | 0.003415 | Reported |
| APOD | Apolipoprotein D | 347 | 0.003759 | Reported |
| LEP | Leptin | 3952 | 0.008264 | Reported |
| HLA‐B | Major histocompatibility complex, class I, B | 3106 | 0.011278 | Reported |
| C1QB | Complement C1q B chain | 713 | 0.015038 | Reported |
| HSPG2 | Heparan sulfate proteoglycan 2 | 3339 | 0.018797 | Reported |
| MTTP | Microsomal triglyceride transfer protein | 4547 | 0.022556 | NOT |
| IL−10 | Interleukin 10 | 3586 | 0.024793 | Reported |
| TF | Transferrin | 7018 | 0.026316 | Reported |
| NOS1AP | Nitric oxide synthase 1 adaptor protein | 9722 | 0.027164 | NOT |
| A2M | Alpha−2‐macroglobulin | 2 | 0.030075 | Reported |
| NEK4 | NIMA related kinase 4 | 6787 | 0.033058 | NOT |
| CD86 | CD86 molecule | 942 | 0.033835 | Reported |
| PIK3C2A | Phosphatidylinositol−4‐phosphate 3‐kinase catalytic subunit type 2 alpha | 5286 | 0.041353 | NOT |
| ADAM12 | ADAM metallopeptidase domain 12 | 8038 | 0.044259 | Reported |
| TARBP1 | TAR (HIV−1) RNA binding protein 1 | 6894 | 0.045113 | NOT |
| GABRG2 | Gamma‐aminobutyric acid type A receptor gamma2 subunit | 2566 | 0.048872 | Reported |
Taking RELN gene (coding reelin, an extracellular matrix glycoprotein) as an example, it participated in the regulation of neuronal migration, position, growth, and synaptic plasticity linking memory/learning formation,74 and it could prevent synaptic dysfunction induced by accumulation of Aβ deposits in AD.75 Some studies reported a significant association between RELN and AD among different populations.76, 77, 78 Another example could be DNMBP, which was a scaffold protein to bring dynamin and actin regulatory proteins together79 and participated in synaptic vesicle trafficking.80, 81, 82 This process could be disturbed by accumulation of Aβ deposits in AD.83 Previous studies have reported significant association between DNMBP and AD in Japanese population.84 The following studies also found a significant association between DNMBP and AD in Belgian and Chinese populations.85, 86
The distribution of 13 reported genes in the 18 identified potential AD target genes were calculated. The 18 potential target genes identified in this study were first arranged in the descending order of P‐values of robust rank aggregation (RRA) analysis. Second, the cumulative number N was defined as the first N potential target genes, and the percentage of reported genes in each cumulative number was calculated. As shown in Figure 2, the slope of the fitted curve is continually decreasing. This phenomenon reflected a good reliability of the applied prediction method because it was consistent between the significance of potential AD target genes and the possibility of it being an AD associated gene. Specifically, nearly half of the genes reported to be closely associated with AD were gathered in the first 30% identified potential target genes. Then, another phenomenon was observed by comparing candidate and potential AD target genes. Percentage of reported genes were significantly higher in potential AD target genes (about 72.2%) than it in candidate AD target genes (about 17.5%), which further reflect the reliability of the prediction.
Figure 2.
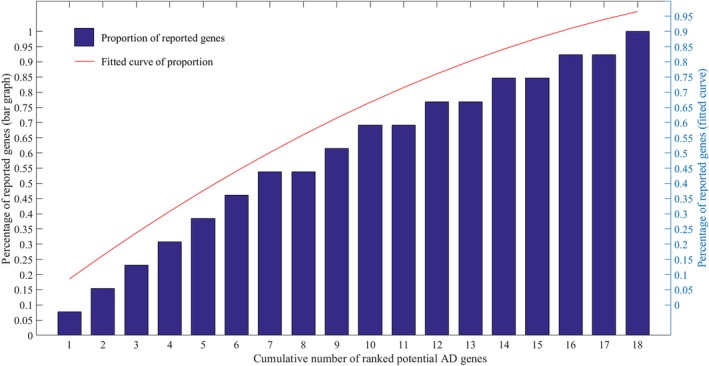
The distribution of reported genes among the ranked potential AD target genes. The bar graph shows percentage of reported genes in the first N identified potential AD genes which are ranked by RRA method. For example, there are 53.8% reported genes in first seven identified potential AD target genes. The red line is a fitted curve of the percentage values. The left and right ordinates show the percentage values of bar graph and fitted curve, respectively
3.4. Enrichment analysis of the potential target genes based on pathway‐related databases
To explore the function of those 18 identified potential AD target genes, the KEGG enrichment analysis was conducted. As shown in Table 3, two KEGG pathways were enriched in this analysis by setting the P‐value cutoff as 0.01, and both belonged to the human immune disease. If the P‐value cutoff was further set as 0.05, six KEGG pathways could be enriched, and they all belonged to immune disease or immune system. The remaining KEGG pathway (Type I diabetes mellitus) was also an autoimmune disease.87, 88 Altogether, these pathways contained four immune genes: C1QB, HLA‐B, IL‐10, and CD86. Among these genes, C1QB coded the B chain of complement C1q which was an activator in the classical complement pathway, and the remaining were involved in the process of antigen presenting. Major histocompatibility complex (MHC) is a cell surface protein to bind and present antigen peptide fragments to T cell. Studies demonstrated that the expression of both MHC class I and class II was markedly increased in AD.89, 90 HLA‐B is a member of the human leukocyte antigen gene family, and coded a part of heavy chain of the MHC class I.91 CD86 expresses on most antigen‐presenting cells and is a critical co‐stimulatory factors for antigen presenting from MHC to T cell.92 IL‐10 is an inhibitory factor of excessive immune response, and strongly downregulates the expression of both MHC class II and CD86.93, 94, 95 A recent study showed that inhibiting the IL‐10 pathway could relieve the symptom of AD.9
Table 3.
KEGG enrichment results of potential AD drug target genes
| KEGG ID | KEGG pathway | Class | Nominal P‐value | Adjusted P‐value | Enriched genes |
|---|---|---|---|---|---|
| Cutoff P‐value = 0.01 | |||||
| hsa05330 | Allograft rejection | Immune diseaseD | 3.87 × 10− 5 | 2.13 × 10− 3 | HLA‐B, IL−10, CD86 |
| hsa05320 | Autoimmune thyroid disease | Immune diseaseD | 1.06 × 10− 4 | 2.91 × 10− 3 | HLA‐B, IL−10, CD86 |
| Cutoff P‐value = 0.05 | |||||
| hsa05322 | Systemic lupus erythematosus | Immune diseaseD | 1.66 × 10− 3 | 2.93 × 10− 2 | C1QB, IL−10, CD86 |
| hsa05332 | Graft‐versus‐host disease | Immune diseaseD | 2.42 × 10− 3 | 2.93 × 10− 2 | HLA‐B, CD86 |
| hsa04940 | Type I diabetes mellitus | Endocrine and metabolic diseaseD | 2.66 × 10− 3 | 2.93 × 10− 2 | HLA‐B, CD86 |
| hsa04672 | Intestinal immune network for IgA production | Immune systemO | 3.45 × 10− 3 | 3.16 × 10− 2 | HLA‐B, CD86 |
The capital letters beside the class name represent KEGG categories. Among them, D means human disease; O means organismal systems. The adjusted P‐values are obtained through multiple hypothesis testing to correct the nominal P‐values.
Extensive enrichment analyses on these potential drug target genes based on eight databased providing the pathway information (MetaCyc, NetPath, PathWhiz, PID, and WikiPathways) and the ontology‐based data (Gene Ontology, PANTHER and Reactome) were further conducted to reveal their functions. Apart from PID, the data of the remaining seven databases were fully downloadable, and the enrichment analyses of the potential AD target genes based on the data from the seven databases were thus conducted using R package clusterProfiler to validate the presumption of this study. As a result, no term was enriched based on the data of PANTHER, PathWhiz, MetaCyc, and NetPath, and the enriched terms based on GO, Reactome, and WikiPathways databases are provided in Tables 4, S3 and S4, respectively. As shown, the enriched terms based on GO and Reactome are focused on the biological process of lipoproteins (eg phosphatidylinositol) with P‐value <0.05, that is significantly associated with the immunity.96 Particularly, the activity of phosphatidylinositol 3‐kinase affects the expression of MHC class II gene.97 Moreover, one enriched terms based on WikiPathways is related to lipid metabolism, and the remaining enriched terms are immunity related pathways (Allograft Rejection & SIDS Susceptibility Pathways). SIDS neuroimmune disorder is reported as a neuroimmune disorder in brain, which is involved in the T‐cell deficiency in immune inflammatory response.98 In sum, these in‐depth analyses further supported the discovery of KEGG enrichment and further associated AD pathogenesis with the immunity.
Table 4.
GO enrichment results of the potential AD drug target genes identified in this study
| GO ID | GO pathway | Subontology | Nominal P‐value | Adjusted P‐value | Enriched Genes |
|---|---|---|---|---|---|
| GO:0042157 | Lipoprotein metabolic process | Biological Process | 1.26E−05 | 1.39E−02 | APOD, LEP, HSPG2, MTTP |
| GO:0048015 | Phosphatidylinositol mediated signaling | Biological Process | 5.29E−05 | 2.06E−02 | RELN, LEP, CD86, PIK3C2A |
| GO:0048017 | Inositol lipid‐mediated signaling | Biological Process | 5.60E−05 | 2.06E−02 | RELN, LEP, CD86, PIK3C2A |
| GO:0050746 | Regulation of lipoprotein metabolic process | Biological Process | 1.15E−04 | 2.28E−02 | APOD, LEP |
| GO:0032682 | Negative regulation of chemokine production | Biological Process | 1.48E−04 | 2.28E−02 | APOD, IL10 |
| GO:0030258 | Lipid modification | Biological Process | 1.53E−04 | 2.28E−02 | APOD, LEP, CD86, PIK3C2A |
| GO:0043235 | Receptor complex | Cellular Component | 2.49E−04 | 1.93E−02 | MTTP, NOS1AP, TF, GABRG2 |
| GO:0072562 | Blood microparticle | Cellular Component | 3.71E−04 | 1.93E−02 | C1QB, TF, A2M |
The adjusted P‐values are obtained through multiple hypothesis testing to correct the nominal P‐values.
4. CONCLUSIONS
A total of 18 potential AD target genes were identified by the combinatorial method of ontology‐based inference and biological network analysis. Further, the results of enrichment analysis showed that these 18 potential AD target genes were significantly enriched in the immune‐related pathways, and of which C1QB, HLA‐B, IL‐10, and CD86 were involved in the process of antigen presenting from MHC to T cell. These results implied that the pathogenic mechanism of AD may be relevant to the abnormal process of antigen presenting and may be an effective point of further drug development. In summary, our findings showed the importance of immune‐related drug target genes to the therapy of AD and would benefit to the AD research in the future.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
This work was funded by the research support of Precision Medicine Project of National Key Research and Development Plan of China (2016YFC0902200); the Innovation Project on Industrial Generic Key Technologies of Chongqing (cstc2015zdcy‐ztzx120003); and Fundamental Research Funds for Central Universities (10611CDJXZ238826, CDJZR14468801, CDJKXB14011).
Han Z‐J, Xue W‐W, Tao L, Zhu F. Identification of novel immune‐relevant drug target genes for Alzheimer's Disease by combining ontology inference with network analysis. CNS Neurosci Ther. 2018;24:1253–1263. 10.1111/cns.13051
REFERENCES
- 1. Pate KM, Rogers M, Reed JW, et al. Anthoxanthin polyphenols attenuate abeta oligomer‐induced neuronal responses associated with Alzheimer's disease. CNS Neurosci Ther. 2017;23:135‐144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu S, Wang J, Zhang Y, et al. The role of neuroinflammation and amyloid in cognitive impairment in an APP/PS1 transgenic mouse model of Alzheimer's disease. CNS Neurosci Ther. 2017;23:310‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vaudano E, Vannieuwenhuyse B, Van Der Geyten S, et al. Boosting translational research on Alzheimer's disease in Europe: the innovative medicine initiative AD research platform. Alzheimers Dement. 2015;11:1121‐1122. [DOI] [PubMed] [Google Scholar]
- 4. Alzheimer's AA. 2017 Alzheimer's disease facts and figures. Alzheimer's Dementia. 2017;13:325‐373. [Google Scholar]
- 5. Guillot‐Sestier MV, Doty KR, Town T. Innate immunity fights Alzheimer's Disease. Trends Neurosci. 2015;38:674‐681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yao PL, Zhuo S, Mei H, et al. Androgen alleviates neurotoxicity of beta‐amyloid peptide (Abeta) by promoting microglial clearance of Abeta and inhibiting microglial inflammatory response to Abeta. CNS Neurosci Ther. 2017;23:855‐865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rong XF, Sun YN, Liu DM, et al. The pathological roles of NDRG2 in Alzheimer's disease, a study using animal models and APPwt‐overexpressed cells. CNS Neurosci Ther. 2017;23:667‐679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murray PJ. Understanding and exploiting the endogenous interleukin‐10/STAT3‐mediated anti‐inflammatory response. Curr Opin Pharmacol. 2006;6:379‐386. [DOI] [PubMed] [Google Scholar]
- 9. Guillot‐Sestier MV, Doty KR, Gate D, et al. Il10 deficiency rebalances innate immunity to mitigate Alzheimer‐like pathology. Neuron. 2015;85:534‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fu AK, Hung KW, Yuen MY, et al. IL‐33 ameliorates Alzheimer's disease‐like pathology and cognitive decline. Proc Natl Acad Sci U S A. 2016;113:E2705–E2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gjoneska E, Pfenning AR, Mathys H, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature. 2015;518:365‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao LX, Wang Y, Liu T, et al. alpha‐Mangostin decreases beta‐amyloid peptides production via modulation of amyloidogenic pathway. CNS Neurosci Ther. 2017;23:526‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iliff JJ, Wang M, Liao Y. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci Transl Med. 2012;4:147ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peng W, Achariyar TM, Li B, et al. Suppression of glymphatic fluid transport in a mouse model of Alzheimer's disease. Neurobiol Dis. 2016;93:215‐225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Louveau A, Smirnov I, Keyes TJ, et al. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523:337‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang FY, Fu TT, Zhang XY, et al. Comparison of computational model and X‐ray crystal structure of human serotonin transporter: potential application for the pharmacology of human monoamine transporters. Mol Simul. 2017;43:1089‐1098. [Google Scholar]
- 17. Zheng G, Xue W, Wang P, et al. Exploring the inhibitory mechanism of approved selective norepinephrine reuptake inhibitors and reboxetine enantiomers by molecular dynamics study. Sci Rep. 2016;6:26883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jabir NR, Khan FR, Tabrez S. Cholinesterase targeting by polyphenols: A therapeutic approach for the treatment of Alzheimer's disease. CNS Neurosci Ther. 2018. 10.1111/cns.12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Torika N, Asraf K, Apte RN, Fleisher‐Berkovich S. Candesartan ameliorates brain inflammation associated with Alzheimer's disease. CNS Neurosci Ther. 2018;24:231‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singh Y, Gupta G, Shrivastava B, et al. Calcitonin gene‐related peptide (CGRP): A novel target for Alzheimer's disease. CNS Neurosci Ther. 2017;23:457‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sevigny J, Chiao P, Bussiere T, et al. The antibody aducanumab reduces Abeta plaques in Alzheimer's disease. Nature. 2016;537:50‐56. [DOI] [PubMed] [Google Scholar]
- 22. Zhu F, Han L, Zheng C, et al. What are next generation innovative therapeutic targets? Clues from genetic, structural, physicochemical, and systems profiles of successful targets. J Pharmacol Exp Ther. 2009;330:304‐315. [DOI] [PubMed] [Google Scholar]
- 23. Yu CY, Li XX, Yang H, et al. Assessing the performances of protein function prediction algorithms from the perspectives of identification accuracy and false discovery rate. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Breuer K, Foroushani AK, Laird MR, et al. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228–D1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Silva T, Reis J, Teixeira J, Borges F. Alzheimer's disease, enzyme targets and drug discovery struggles: from natural products to drug prototypes. Ageing Res Rev. 2014;15:116‐145. [DOI] [PubMed] [Google Scholar]
- 26. Li YH, Yu CY, Li XX, et al. Therapeutic target database update 2018: enriched resource for facilitating bench‐to‐clinic research of targeted therapeutics. Nucleic Acids Res. 2018;46:D1121–D1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yao L, Evans JA, Rzhetsky A. Novel opportunities for computational biology and sociology in drug discovery. Trends Biotechnol. 2010;28:161‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fu J, Tang J, Wang Y, et al. Discovery of the consistently well‐performed analysis chain for SWATH‐MS based pharmacoproteomic quantification. Front Pharmacol. 2018;9:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang P, Fu T, Zhang X, et al. Differentiating physicochemical properties between NDRIs and sNRIs clinically important for the treatment of ADHD. Biochim Biophys Acta. 2017;1861:2766‐2777. [DOI] [PubMed] [Google Scholar]
- 30. Zhu F, Zheng CJ, Han LY, et al. Trends in the exploration of anticancer targets and strategies in enhancing the efficacy of drug targeting. Curr Mol Pharmacol. 2008;1:213‐232. [DOI] [PubMed] [Google Scholar]
- 31. Li YH, Wang PP, Li XX, et al. The human kinome targeted by FDA approved multi‐target drugs and combination products: a comparative study from the drug‐target interaction network perspective. PLoS One. 2016;11:e0165737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen HM, Sha ZQ, Ma HZ, He Y, Feng T. Effective network of deep brain stimulation of subthalamic nucleus with bimodal positron emission tomography/functional magnetic resonance imaging in Parkinson's disease. CNS Neurosci Ther. 2018;24:135‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ye H, Yang L, Cao Z, Tang K, Li Y. A pathway profile‐based method for drug repositioning. Chin Sci Bull. 2012;57:2106‐2112. [Google Scholar]
- 34. Hu G, Agarwal P. Human disease‐drug network based on genomic expression profiles. PLoS One. 2009;4:e6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iorio F, Bosotti R, Scacheri E, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci U S A. 2010;107:14621‐14626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xue W, Yang F, Wang P, et al. What contributes to serotonin‐norepinephrine reuptake inhibitors' dual‐targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem Neurosci. 2018;9:1128‐1140. [DOI] [PubMed] [Google Scholar]
- 37. Li J, Lu Z. A new method for computational drug repositioning using drug pairwise similarity. Proceedings IEEE Int Conf Bioinformatics Biomed. 2012;2012:1‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side‐effect similarity. Science. 2008;321:263‐266. [DOI] [PubMed] [Google Scholar]
- 39. Zhu F, Li XX, Yang SY, Chen YZ. Clinical success of drug targets prospectively predicted by in silico study. Trends Pharmacol Sci. 2018;39:229‐231. [DOI] [PubMed] [Google Scholar]
- 40. Xue W, Wang P, Tu G, et al. Computational identification of the binding mechanism of a triple reuptake inhibitor amitifadine for the treatment of major depressive disorder. Phys Chem Chem Phys. 2018;20:6606‐6616. [DOI] [PubMed] [Google Scholar]
- 41. Tao C, Sun J, Zheng WJ, Chen J, Xu H. Colorectal cancer drug target prediction using ontology‐based inference and network analysis. Database Oxford. 2015;2015:pii: bav015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dixon SJ, Stockwell BR. Identifying druggable disease‐modifying gene products. Curr Opin Chem Biol. 2009;13:549‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hobbs JK, Prentice EJ, Groussin M, Arcus VL. Reconstructed ancestral enzymes impose a fitness cost upon modern bacteria despite exhibiting favourable biochemical properties. J Mol Evol. 2015;81:110‐120. [DOI] [PubMed] [Google Scholar]
- 44. Yang H, Qin C, Li YH, et al. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016;44:D1069–D1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thorn CF, Klein TE, Altman RB. PharmGKB: the Pharmacogenomics Knowledge Base. Methods Mol Biol. 2013;1015:311‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu F, Qin C, Tao L, et al. Clustered patterns of species origins of nature‐derived drugs and clues for future bioprospecting. Proc Natl Acad Sci U S A. 2011;108:12943‐12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhu F, Han LY, Chen X, et al. Homology‐free prediction of functional class of proteins and peptides by support vector machines. Curr Protein Pept Sci. 2008;9:70‐95. [DOI] [PubMed] [Google Scholar]
- 48. Zhu F, Shi Z, Qin C, et al. Therapeutic target database update 2012: a resource for facilitating target‐oriented drug discovery. Nucleic Acids Res. 2012;40:D1128–D1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Knox C, Law V, Jewison T, et al. DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berners‐Lee T, Hendler J, Lassila O. The semantic web. Sci Am. 2001;284:28‐37. [Google Scholar]
- 51. Neumann EK, Miller E, Wilbanks J. What the semantic web could do for the life sciences. Drug Discovery Today BIOSILICO. 2004;2:228‐236. [Google Scholar]
- 52. Wang P, Zhang X, Fu T, et al. Differentiating physicochemical properties between addictive and nonaddictive ADHD drugs revealed by molecular dynamics simulation studies. ACS Chem Neurosci. 2017;8:1416‐1428. [DOI] [PubMed] [Google Scholar]
- 53. Xu J, Wang P, Yang H, et al. Comparison of FDA approved kinase targets to clinical trial ones: insights from their system profiles and drug‐target interaction networks. Biomed Res Int. 2016;2016:2509385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhu F, Ma XH, Qin C, et al. Drug discovery prospect from untapped species: indications from approved natural product drugs. PLoS One. 2012;7:e39782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta‐analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17‐23. [DOI] [PubMed] [Google Scholar]
- 56. Carlos J, Wolfe M, Zambon J, Kingman A. Periodontal disease in adolescents: some clinical and microbiologic correlates of attachment loss. J Dent Res. 1988;67:1510‐1514. [DOI] [PubMed] [Google Scholar]
- 57. Chatr‐Aryamontri A, Breitkreutz BJ, Oughtred R, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xiao Q, Liu ZJ, Tao S, et al. Risk prediction for sporadic Alzheimer's disease using genetic risk score in the Han Chinese population. Oncotarget. 2015;6:36955‐36964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li B, Tang J, Yang Q, et al. NOREVA: normalization and evaluation of MS‐based metabolomics data. Nucleic Acids Res. 2017;45:W162–W170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta‐analysis. Bioinformatics. 2012;28:573‐580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fu T, Zheng G, Tu G, et al. Exploring the binding mechanism of metabotropic glutamate receptor 5 negative allosteric modulators in clinical trials by molecular dynamics simulations. ACS Chem Neurosci. 2018;9:1492‐1502. [DOI] [PubMed] [Google Scholar]
- 62. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Caspi R, Billington R, Fulcher CA, et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46:D633–D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kandasamy K, Mohan SS, Raju R, et al. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pon A, Jewison T, Su Y, et al. Pathways with PathWhiz. Nucleic Acids Res. 2015;43:W552–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schaefer CF, Anthony K, Krupa S, et al. PID: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Slenter DN, Kutmon M, Hanspers K, et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46:D661–D667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. The Gene Ontology Consortium . Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mi H, Huang X, Muruganujan A, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fabregat A, Jupe S, Matthews L, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhu F, Han B, Kumar P, et al. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 2010;38:D787–D791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hewett M, Oliver DE, Rubin DL, et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30:163‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lakatosova S, Ostatnikova D. Reelin and its complex involvement in brain development and function. Int J Biochem Cell Biol. 2012;44:1501‐1504. [DOI] [PubMed] [Google Scholar]
- 75. Durakoglugil MS, Chen Y, White CL, Kavalali ET, Herz J. Reelin signaling antagonizes beta‐amyloid at the synapse. Proc Natl Acad Sci U S A. 2009;106:15938‐15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bufill E, Roura‐Poch P, Sala‐Matavera I, et al. Reelin signaling pathway genotypes and Alzheimer disease in a Spanish population. Alzheimer Dis Assoc Disord. 2015;29:169‐172. [DOI] [PubMed] [Google Scholar]
- 77. Feher A, Juhasz A, Pakaski M, Kalman J, Janka Z. Genetic analysis of the RELN gene: Gender specific association with Alzheimer's disease. Psychiatry Res. 2015;230:716‐718. [DOI] [PubMed] [Google Scholar]
- 78. Yang F, Zheng G, Fu T, et al. Prediction of the binding mode and resistance profile for a dual‐target pyrrolyl diketo acid scaffold against HIV‐1 integrase and reverse‐transcriptase‐associated ribonuclease H. Phys Chem Chem Phys. 2018. 10.1039/c8cp01843j. [DOI] [PubMed] [Google Scholar]
- 79. Salazar MA, Kwiatkowski AV, Pellegrini L, et al. Tuba, a novel protein containing bin/amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J Biol Chem. 2003;278:49031‐49043. [DOI] [PubMed] [Google Scholar]
- 80. Casoli T, Di Stefano G, Fattoretti P, et al. Dynamin binding protein gene expression and memory performance in aged rats. Neurobiol Aging. 2012;33(618):e615–e619. [DOI] [PubMed] [Google Scholar]
- 81. Li B, Tang J, Yang Q, et al. Performance evaluation and online realization of data‐driven normalization methods used in LC/MS based untargeted metabolomics analysis. Sci Rep. 2016;6:38881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li YH, Xu JY, Tao L, et al. SVM‐Prot 2016: a web‐server for machine learning prediction of protein functional families from sequence irrespective of similarity. PLoS One. 2016;11:e0155290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kelly BL, Vassar R, Ferreira A. Beta‐amyloid‐induced dynamin 1 depletion in hippocampal neurons. A potential mechanism for early cognitive decline in Alzheimer disease. J Biol Chem. 2005;280:31746‐31753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kuwano R, Miyashita A, Arai H, et al. Dynamin‐binding protein gene on chromosome 10q is associated with late‐onset Alzheimer's disease. Hum Mol Genet. 2006;15:2170‐2182. [DOI] [PubMed] [Google Scholar]
- 85. Yang P, Xu M, Liu Z, et al. Genetic association of CUGBP2 and DNMBP with Alzheimer' s disease in the Chinese Han population. Curr Alzheimer Res. 2015;12:228‐232. [DOI] [PubMed] [Google Scholar]
- 86. Wang P, Yang F, Yang H, et al. Identification of dual active agents targeting 5‐HT1A and SERT by combinatorial virtual screening methods. Biomed Mater Eng. 2015;26(Suppl 1):S2233‐S2239. [DOI] [PubMed] [Google Scholar]
- 87. Addobbati CJ, de Azevedo SJ, Tavares NA, et al. Short Communication FYB polymorphisms in Brazilian patients with type I diabetes mellitus and autoimmune polyglandular syndrome type III. Genet Mol Res. 2015;14:29‐33. [DOI] [PubMed] [Google Scholar]
- 88. Zheng G, Xue W, Yang F, et al. Revealing vilazodone's binding mechanism underlying its partial agonism to the 5‐HT1A receptor in the treatment of major depressive disorder. Phys Chem Chem Phys. 2017;19:28885‐28896. [DOI] [PubMed] [Google Scholar]
- 89. Tooyama I, Kimura H, Akiyama H, McGeer PL. Reactive microglia express class I and class II major histocompatibility complex antigens in Alzheimer's disease. Brain Res. 1990;523:273‐280. [DOI] [PubMed] [Google Scholar]
- 90. Yang YM, Shang DS, Zhao WD, Fang WG, Chen YH. Microglial TNF‐alpha‐dependent elevation of MHC class I expression on brain endothelium induced by amyloid‐beta promotes T cell transendothelial migration. Neurochem Res. 2013;38:2295‐2304. [DOI] [PubMed] [Google Scholar]
- 91. Raghavan M, Geng J. HLA‐B polymorphisms and intracellular assembly modes. Mol Immunol. 2015;68:89‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Brzostek J, Gascoigne NR, Rybakin V. Cell type‐specific regulation of immunological synapse dynamics by B7 ligand recognition. Front Immunol. 2016;7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Redpath S, Angulo A, Gascoigne NR, Ghazal P. Murine cytomegalovirus infection down‐regulates MHC class II expression on macrophages by induction of IL‐10. J Immunol. 1999;162:6701‐6707. [PubMed] [Google Scholar]
- 94. Xue W, Wang P, Li B, et al. Identification of the inhibitory mechanism of FDA approved selective serotonin reuptake inhibitors: an insight from molecular dynamics simulation study. Phys Chem Chem Phys. 2016;18:3260‐3271. [DOI] [PubMed] [Google Scholar]
- 95. Tao L, Zhu F, Xu F, et al. Co‐targeting cancer drug escape pathways confers clinical advantage for multi‐target anticancer drugs. Pharmacol Res. 2015;102:123‐131. [DOI] [PubMed] [Google Scholar]
- 96. Tall AR, Yvan‐Charvet L. Cholesterol, inflammation and innate immunity. Nat Rev Immunol. 2015;15:104‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hardy PO, Diallo TO, Matte C, Descoteaux A. Roles of phosphatidylinositol 3‐kinase and p38 mitogen‐activated protein kinase in the regulation of protein kinase C‐alpha activation in interferon‐gamma‐stimulated macrophages. Immunology. 2009;128:e652–e660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Reid GM. Sudden infant death syndrome (SIDS): T‐cell immunodeficiency–Part 1. Med Hypotheses. 2001;56:256‐258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


