Abstract
Here we report the complete mitochondrial sequences of 70 individual field collected mosquito specimens from throughout Sub-Saharan Africa. We generated this dataset to identify species specific markers for the following Anopheles species and chromosomal forms: An. arabiensis, An. coluzzii (The Forest and Mopti chromosomal forms) and An. gambiae (The Bamako and Savannah chromosomal forms). The raw Illumina sequencing reads were mapped to the NC_002084 reference mitogenome sequence. A total of 783 single nucleotide polymorphisms (SNPs) were detected on the mitochondrial genome, of which 460 are singletons (58.7%). None of these SNPs are suitable as molecular markers to distinguish among An. arabiensis, An. coluzzii and An. gambiae or any of the chromosomal forms. The lack of species or chromosomal form specific markers is also reflected in the constructed phylogenetic tree, which shows no clear division among the operational taxonomic units considered here.
Keywords: Mitogenome, species identification, Africa, malaria vector, mosquitoes, Anopheles, single nucleotide polymorphisms, phylogenomics
Introduction
Historically, mtDNA sequence has been used in taxonomy as a source of species diagnostic markers ( Cronin et al. (1991); De Barba et al. (2014); Pegg et al. (2006)) or in population genetics and evolutionary studies ( Fu et al. (2013); Harrison (1989); Llamas et al. (2016)). One advantage of using mitochondrial over nuclear DNA for such studies is that the mutation rate of mtDNA is about 10 times faster than nuclear DNA ( Brown et al. (1979); Haag-Liautard et al. (2008)), hence amplifying the evolutionary trajectory of populations and species. In addition, mtDNA is easy to amplify, because there are more copies of mitochondrial DNA relative to nuclear DNA. Also, universal primers can be applied to a wide range of species. Widely used universal primers target the cytochrome b and cytochrome oxidase 1 genes ( Tahir et al. (2016)), because both have conserved and highly variable regions. In addition to these, other genes as described in De Mandal et al. (2014), can also be used as markers. However, phylogenetic trees based on mtDNA can deviate from the ones that are derived from nuclear DNA ( Phillips et al. (2013); Shaw (2002); Sota & Vogler, 2001).
The Anopheles gambiae species complex consists of eight morphologically identical species that can only be distinguished with molecular markers ( Scott et al. (1993)) or, for some of the species, by cytological examination of polytene chromosomes ( Green, 1972; Pombi et al., 2008). The currently used molecular markers are located within genomic islands of divergence located proximal to the centromeres ( Lee et al. (2014); Turner et al. (2005)). Monitoring additional species-specific markers on mitochondrial DNA (mtDNA) could increase the ease of application and accuracy of species detection assays. In addition, mtDNA markers could enhance our understanding of divergence times among taxa within the complex.
In this study we wished to identify species-specific markers within the mtDNA for Anopheles arabiensis, An. coluzzii and An. gambiae, including among the chromosomal forms currently subsumed under the designations An. gambiae and An. coluzzii, with the goal of adding these to our existing Anopheles species detection assay ( Lee et al. (2014)). We sequenced the whole mitogenomes of 70 individual mosquito specimens collected throughout Sub-Saharan Africa. The raw Illumina sequencing reads were mapped to the AgamP4 reference sequence, which included both nuclear and mitochondrial sequences. We explore the relationship among An. arabiensis, An. coluzzii, An. gambiae and four of the sub-specific chromosomal form mitogenome sequences.
Methods
Sample collection
Anopheles arabiensis raw Illumina sequencing reads were obtained from our previous study ( Marsden et al. (2014)). These included 20 samples from three villages in Tanzania collected in 2012 (Lupiro ((-8.38000°N, 36.66912°W), Sagamaganga (-8.06781°N, 36.80207°W), and Minepa (-8.25700°N, 36.68163°W) in the Kilombero Valley) and 4 samples from Cameroon collected in 2005 (9.09957°N, 13.72292°W). The An. gambiae and An. coluzzii samples were collected as resting adults using mouth aspirators in Kela, Mali (11.88683°N, -8.44744°W) in 2012 and Mutengene, Cameroon (4.0994°N, 9.3081°W) in 2011. We subdivided the An. coluzzii specimen into the Forest and Mopti chromosomal forms. Similarly, we did this for the An. gambiae Savannah and Bamako chromosomal forms. We used the same definitions and methods to characterize the chromosomal forms as in Lanzaro & Lee, 2013.
Genome sequencing
Sequencing methods for An. arabiensis samples are as described in Marsden et al. (2014). In short, individually barcoded Illumina paired-end sequencing libraries, with insert sizes of 320-400 basepairs (bp) using NEXTflex Sequencing kits (NOVA-5144) and barcodes (NOVA-514102)(Bio Scientific, Austin, TX, USA), were sequenced on an Illumina HiSeq2000 (Illumina, San Diego, CA, USA) with 100-bp paired-end reads using twelve samples per lane. For the An. coluzzii and An. gambiae samples we used the same methods as described in Norris et al. (2015) and Main et al. (2015). For the latter species, libraries were created using the Nextera DNA Sample Preparation Kit (FC-121-1031) and TruSeq dual indexing barcodes (FC-121-103)(Illumina) and the samples were sequenced on an Illumina HiSeq2500 with 100-bp paired end reads.
Data analysis
De-multiplexed raw reads were trimmed using Trimmomatic ( Bolger et al. (2014)) version 0.36 and mapped to the mitogenome reference sequence of An. gambiae (Genbank accession number = NC_002084 ( Beard et al. (1993))). Freebayes (v1.0.1) ( Garrison & Marth, 2013) was used for mitochondrial variant calling assuming single ploidy and without population prior. Mapping statistics were calculated using qualimap version 2.2 ( Okonechnikov et al. (2016)) and the data is represented in Table 1. Following the recommendation of Crawford and Lazarro ( Crawford & Lazzaro, 2012), we used a minimum depth of 8 to call variants for each individual. Between positions 1-13,470bp of the mitogenome, we obtained consistently high quality reads for all samples, which were used for further analysis. An AT-rich region located between 13,471 and 15,388 suffers from low or zero coverage for sequences generated with the Nextera library preparation kit. Therefore, we excluded these regions from further analysis. The Vcf2fasta program ( Danecek et al. (2011)) was used to extract mitogenome sequences from vcf file to fasta format. Geneious version 10.1.3 was used for mitogenome alignments. The phylogenetic tree was generated using the Jukes-Cantor genetic distance model and Neighbor-Joining tree methods available in Geneious version 10.1.3. We used scikit-allel (v1.1.9), a software package for Python ( Miles & Harding (2017)), to identify species specific markers.
Table 1. List of samples that are used for the study.
Mapped reads indicates the reads that are mapped to the reference genome. Mean coverage indicates the average depth of reads on the mitochondrial DNA and standard deviation indicates the coverage deviation across the mitochondrial DNA.
| Species | Banked_id | Year | Country | Village | Mapped bases | Mean
coverage |
Standard
deviation |
|---|---|---|---|---|---|---|---|
| An. coluzzii-Forest | 11MUTE470 | 2011 | Cameroon | Mutengene | 4265836 | 277.7 | 144.5 |
| An. coluzzii-Forest | 11MUTE472 | 2011 | Cameroon | Mutengene | 1862892 | 121.3 | 23 |
| An. coluzzii-Forest | 11MUTE476 | 2011 | Cameroon | Mutengene | 2130531 | 138.7 | 50.5 |
| An. coluzzii-Forest | 11MUTE477 | 2011 | Cameroon | Mutengene | 806611 | 52.5 | 16.7 |
| An. coluzzii-Forest | 11MUTE480 | 2011 | Cameroon | Mutengene | 804015 | 52.3 | 21 |
| An. coluzzii-Forest | 11MUTE483 | 2011 | Cameroon | Mutengene | 1702247 | 110.8 | 42.9 |
| An. coluzzii-Forest | 11MUTE487 | 2011 | Cameroon | Mutengene | 812839 | 52.9 | 21.2 |
| An. coluzzii-Forest | 11MUTE490 | 2011 | Cameroon | Mutengene | 1882088 | 122.5 | 52.4 |
| An. coluzzii-Forest | 11MUTE491 | 2011 | Cameroon | Mutengene | 1422997 | 92.6 | 46.6 |
| An. coluzzii-Forest | 11MUTE493 | 2011 | Cameroon | Mutengene | 627590 | 40.9 | 17.3 |
| An. coluzzii-Mopti | 12KELA022 | 2012 | Mali | Kela | 3695920 | 240.6 | 64.4 |
| An. coluzzii-Mopti | 12KELA024 | 2012 | Mali | Kela | 574282 | 37.4 | 30.8 |
| An. coluzzii-Mopti | 12KELA046 | 2012 | Mali | Kela | 4152520 | 270.3 | 87.2 |
| An. coluzzii-Mopti | 12KELA085 | 2012 | Mali | Kela | 10883282 | 708.4 | 345 |
| An. coluzzii-Mopti | 12KELA087 | 2012 | Mali | Kela | 3351158 | 218.1 | 79.8 |
| An. coluzzii-Mopti | 12KELA088 | 2012 | Mali | Kela | 1704283 | 110.9 | 91.3 |
| An. coluzzii-Mopti | 12KELA099 | 2012 | Mali | Kela | 349531 | 22.8 | 11 |
| An. coluzzii-Mopti | 12KELA112 | 2012 | Mali | Kela | 8550102 | 556.5 | 198.2 |
| An. coluzzii-Mopti | 12KELA161 | 2012 | Mali | Kela | 33794208 | 2199.7 | 629.3 |
| An. gambiae-Savannah | 12KELA210 | 2012 | Mali | Kela | 3007375 | 195.8 | 53.3 |
| An. gambiae-Bamako | 12KELA214 | 2012 | Mali | Kela | 26441050 | 1721.1 | 566.4 |
| An. gambiae-Bamako | 12KELA219 | 2012 | Mali | Kela | 3617355 | 235.5 | 130.2 |
| An. gambiae-Savannah | 12KELA228 | 2012 | Mali | Kela | 7783776 | 506.7 | 262.8 |
| An. gambiae-Savannah | 12KELA233 | 2012 | Mali | Kela | 7827363 | 509.5 | 138.6 |
| An. gambiae-Savannah | 12KELA234 | 2012 | Mali | Kela | 6721204 | 437.5 | 205.9 |
| An. gambiae-Bamako | 12KELA239 | 2012 | Mali | Kela | 6683521 | 435 | 126.4 |
| An. gambiae-Bamako | 12KELA240 | 2012 | Mali | Kela | 15131480 | 984.9 | 270.8 |
| An. gambiae-Bamako | 12KELA244 | 2012 | Mali | Kela | 12851754 | 836.5 | 306.5 |
| An. gambiae-Savannah | 12KELA285 | 2012 | Mali | Kela | 407888 | 26.6 | 119.8 |
| An. gambiae-Savannah | 12KELA321 | 2012 | Mali | Kela | 1034014 | 67.3 | 43.8 |
| An. gambiae-Savannah | 12KELA334 | 2012 | Mali | Kela | 20949015 | 1363.6 | 400.4 |
| An. gambiae-Savannah | 12KELA348 | 2012 | Mali | Kela | 12053890 | 784.6 | 280.9 |
| An. gambiae-Bamako | 12KELA367 | 2012 | Mali | Kela | 12109235 | 788.2 | 240.1 |
| An. coluzzii-Mopti | 12KELA400 | 2012 | Mali | Kela | 13707820 | 892.3 | 398.2 |
| An. gambiae-Bamako | 12KELA406 | 2012 | Mali | Kela | 17605437 | 1146 | 463.2 |
| An. gambiae-Savannah | 12KELA409 | 2012 | Mali | Kela | 10526480 | 685.2 | 259.1 |
| An. coluzzii-Mopti | 12KELA420 | 2012 | Mali | Kela | 31785953 | 2069 | 845.5 |
| An. gambiae-Bamako | 12KELA443 | 2012 | Mali | Kela | 25740781 | 1675.5 | 669.1 |
| An. gambiae-Bamako | 12KELA457 | 2012 | Mali | Kela | 1360654 | 88.6 | 36.6 |
| An. coluzzii-Mopti | 12KELA458 | 2012 | Mali | Kela | 153686 | 10 | 10.4 |
| An. gambiae-Bamako | 12KELA467 | 2012 | Mali | Kela | 10499093 | 683.4 | 249.1 |
| An. gambiae-Savannah | 12KELA468 | 2012 | Mali | Kela | 10315033 | 671.4 | 197.1 |
| An. gambiae-Bamako | 12KELA481 | 2012 | Mali | Kela | 20308589 | 1321.9 | 307.6 |
| An. coluzzii-Mopti | 12KELA496 | 2012 | Mali | Kela | 2975297 | 193.7 | 162.9 |
| An. gambiae-Bamako | 12KELA651 | 2012 | Mali | Kela | 376689 | 24.5 | 11.3 |
| An. gambiae-Savannah | 12KELA812 | 2012 | Mali | Kela | 799071 | 52 | 29.3 |
| An. arabiensis | 12LUPI001 | 2012 | Tanzania | Lupiro | 2843317 | 185.1 | 34.9 |
| An. arabiensis | 12LUPI007 | 2012 | Tanzania | Lupiro | 6288802 | 409.3 | 40 |
| An. arabiensis | 12LUPI024 | 2012 | Tanzania | Lupiro | 6328898 | 412 | 78.5 |
| An. arabiensis | 12LUPI056 | 2012 | Tanzania | Lupiro | 5440256 | 354.1 | 39.2 |
| An. arabiensis | 12LUPI059 | 2012 | Tanzania | Lupiro | 39721262 | 2585.5 | 801.8 |
| An. arabiensis | 12LUPI071 | 2012 | Tanzania | Lupiro | 3433158 | 223.5 | 59.2 |
| An. arabiensis | 12LUPI074 | 2012 | Tanzania | Lupiro | 10096062 | 657.2 | 100.5 |
| An. arabiensis | 12LUPI082 | 2012 | Tanzania | Lupiro | 5732773 | 373.2 | 69.6 |
| An. arabiensis | 12MINE001 | 2012 | Tanzania | Minepa | 7768923 | 505.7 | 66.9 |
| An. arabiensis | 12MINE040 | 2012 | Tanzania | Minepa | 2784428 | 181.2 | 54.9 |
| An. arabiensis | 12MINE100 | 2012 | Tanzania | Minepa | 10753877 | 700 | 93.9 |
| An. arabiensis | 12MINE101 | 2012 | Tanzania | Minepa | 5684230 | 370 | 41.9 |
| An. arabiensis | 12MINE105 | 2012 | Tanzania | Minepa | 1526829 | 99.4 | 32.8 |
| An. arabiensis | 12MINE111 | 2012 | Tanzania | Minepa | 5578562 | 363.1 | 76.3 |
| An. arabiensis | 12SAGA066 | 2012 | Tanzania | Sagamaganga | 12745079 | 829.6 | 142.3 |
| An. arabiensis | 12SAGA107 | 2012 | Tanzania | Sagamaganga | 14460217 | 941.2 | 259.2 |
| An. arabiensis | 12SAGA131 | 2012 | Tanzania | Sagamaganga | 15333239 | 998.1 | 282.9 |
| An. arabiensis | 12SAGA133 | 2012 | Tanzania | Sagamaganga | 3792945 | 246.9 | 62.5 |
| An. arabiensis | 12SAGA134 | 2012 | Tanzania | Sagamaganga | 2439101 | 158.8 | 34.5 |
| An. arabiensis | 12SAGA141 | 2012 | Tanzania | Sagamaganga | 3130504 | 203.8 | 33.3 |
| An. arabiensis | 05OKJ017 | 2005 | Cameroon | Ourodoukoudje | 9041052 | 588.5 | 78.8 |
| An. arabiensis | 05OKJ042 | 2005 | Cameroon | Ourodoukoudje | 148752684 | 9682.5 | 785.7 |
| An. arabiensis | 05OKJ045 | 2005 | Cameroon | Ourodoukoudje | 35514980 | 2311.7 | 262.8 |
| An. arabiensis | 05OKJ070 | 2005 | Cameroon | Ourodoukoudje | 22847478 | 1487.2 | 400.5 |
Copyright: © 2018 Hanemaaijer MJ et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Results and Discussion
We identified a total of 783 single nucleotide polymorphisms (SNPs) over the entire mitogenome. The majority of these (58.7%) were singletons (found on one of the 70 mitogenomes). We did not identify any SNPs unique to the species or chromosomal forms ( Table 2) and therefore conclude that mtDNA is not suitable for Anopheles gambiae complex species identification.
Table 2. List of SNP variants in the different Anopheles species and chromosomal forms.
| Position | Reference
nucleotide |
Alternative
nucleotides |
SNP variant counts per group (first value is count of reference allel) | ||||
|---|---|---|---|---|---|---|---|
|
An.
arabiensis |
An. coluzzii-
Forest |
An. coluzzii-
Mopti |
An. gambiae-
Bamako |
An. gambiae-
Savannah |
|||
| 55 | C | T/-/- | 21/3/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 71 | C | T/-/- | 0/24/0/0 | 0/11/0/0 | 0/13/0/0 | 1/11/0/0 | 0/11/0/0 |
| 117 | A | G/-/- | 23/1/0/0 | 9/2/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 126 | T | C/-/- | 11/13/0/0 | 1/10/0/0 | 0/13/0/0 | 0/12/0/0 | 1/10/0/0 |
| 205 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 12/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 223 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 12/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 248 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 12/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 250 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 298 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 340 | T | C/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 373 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 391 | T | C/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 396 | C | T/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 409 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 441 | T | C/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 469 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 488 | T | C/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 502 | A | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 503 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 514 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 520 | T | A/C/- | 22/0/2/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 526 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 562 | A | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 572 | G | A/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 583 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 10/3/0/0 | 12/0/0/0 | 11/0/0/0 |
| 611 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 665 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 685 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 688 | T | A/-/- | 24/0/0/0 | 10/1/0/0 | 12/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 700 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 706 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 712 | A | C/-/- | 24/0/0/0 | 10/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 757 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 763 | A | G/-/- | 11/13/0/0 | 3/8/0/0 | 0/13/0/0 | 2/10/0/0 | 1/10/0/0 |
| 787 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 788 | T | C/-/- | 24/0/0/0 | 9/1/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 796 | A | G/-/- | 22/2/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 811 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 821 | T | C/-/- | 11/13/0/0 | 1/9/0/0 | 1/12/0/0 | 0/12/0/0 | 1/10/0/0 |
| 832 | A | G/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 887 | T | C/-/- | 16/8/0/0 | 10/0/0/0 | 12/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 908 | A | G/-/- | 18/6/0/0 | 10/0/0/0 | 12/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 934 | G | A/-/- | 22/2/0/0 | 9/1/0/0 | 12/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 949 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 11/2/0/0 | 11/1/0/0 | 11/0/0/0 |
| 955 | T | C/-/- | 12/12/0/0 | 5/5/0/0 | 9/4/0/0 | 4/8/0/0 | 2/9/0/0 |
| 1012 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1029 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1042 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 11/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1045 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 11/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1114 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1150 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1153 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1155 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1163 | T | C/-/- | 0/24/0/0 | 0/11/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 1189 | G | A/-/- | 24/0/0/0 | 9/2/0/0 | 11/2/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1198 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1308 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1465 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1474 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1486 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1504 | A | G/-/- | 23/1/0/0 | 9/2/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 1507 | G | A/-/- | 23/1/0/0 | 9/2/0/0 | 11/2/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1523 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1538 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 1540 | A | T/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 1543 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1546 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1549 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 1558 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1561 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 1567 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1618 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 1624 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1636 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1639 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1642 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1645 | A | G/T/ | 24/0/0/0 | 9/1/0/0 | 13/0/0/0 | 12/0/0/0 | 10/0/1/0 |
| 1648 | G | A/-/- | 23/1/0/0 | 10/0/0/0 | 9/4/0/0 | 12/0/0/0 | 10/1/0/0 |
| 1681 | A | C/-/- | 24/0/0/0 | 9/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1693 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1729 | G | A/-/- | 23/1/0/0 | 7/4/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 1735 | T | C/-/- | 23/1/0/0 | 9/2/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 1738 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1751 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1765 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1780 | G | A/-/- | 23/1/0/0 | 10/1/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1792 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1807 | T | C/-/- | 23/1/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1837 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 11/1/0/0 | 10/1/0/0 |
| 1876 | A | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1879 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1903 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1906 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1918 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1930 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 1933 | G | A/-/- | 23/1/0/0 | 10/0/0/0 | 11/2/0/0 | 7/5/0/0 | 7/4/0/0 |
| 1964 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1972 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 1978 | G | A/-/- | 22/2/0/0 | 11/0/0/0 | 9/4/0/0 | 8/4/0/0 | 11/0/0/0 |
| 2026 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2068 | A | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 2071 | T | C/G/- | 23/1/0/0 | 11/0/0/0 | 12/0/1/0 | 12/0/0/0 | 10/0/1/0 |
| 2080 | T | C/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 2098 | T | C/-/- | 23/1/0/0 | 10/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2101 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2110 | T | C/-/- | 23/1/0/0 | 10/1/0/0 | 11/2/0/0 | 11/1/0/0 | 10/1/0/0 |
| 2116 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2149 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2164 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2179 | T | C/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 2182 | G | A/-/- | 22/2/0/0 | 6/5/0/0 | 11/2/0/0 | 4/8/0/0 | 8/3/0/0 |
| 2206 | A | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2209 | G | A/-/- | 19/5/0/0 | 10/1/0/0 | 12/1/0/0 | 11/1/0/0 | 9/2/0/0 |
| 2227 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2233 | A | G/T/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/0/1/0 | 11/0/0/0 |
| 2239 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 2260 | C | T/-/- | 2/22/0/0 | 0/11/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 2269 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2284 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2329 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2359 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 2374 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 10/3/0/0 | 11/1/0/0 | 10/1/0/0 |
| 2401 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2410 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2411 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2413 | G | A/-/- | 0/24/0/0 | 0/11/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 2426 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2432 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2434 | G | A/-/- | 22/2/0/0 | 9/2/0/0 | 11/2/0/0 | 11/1/0/0 | 10/1/0/0 |
| 2449 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2473 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2474 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 2482 | A | G/-/- | 20/4/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2521 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 12/1/0/0 | 11/1/0/0 | 10/1/0/0 |
| 2530 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2558 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2569 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2599 | A | G/T/- | 23/1/0/0 | 10/1/0/0 | 13/0/0/0 | 11/0/1/0 | 11/0/0/0 |
| 2608 | T | C/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2612 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2615 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2635 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2692 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2719 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2722 | A | G/-/- | 23/1/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2728 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2731 | A | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2737 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 2752 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2755 | C | A/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2758 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 2774 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 11/2/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2782 | T | A/C/- | 23/0/1/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 2791 | T | A/C/- | 23/0/1/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2797 | C | T/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 2806 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 2818 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2848 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2851 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2859 | C | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2872 | T | A/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2902 | C | T/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 2908 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 2938 | G | A/-/- | 23/1/0/0 | 10/1/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 2945 | C | T/-/- | 15/9/0/0 | 9/2/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 2975 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 3055 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 3124 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3136 | T | C/-/- | 24/0/0/0 | 9/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3199 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3202 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3205 | A | G/-/- | 24/0/0/0 | 9/2/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3238 | T | A/C/ | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 3277 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 11/1/0/0 | 11/1/0/0 | 8/3/0/0 |
| 3286 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 3304 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 3307 | A | C/G/T | 24/0/0/0 | 11/0/0/0 | 9/0/1/3 | 12/0/0/0 | 11/0/0/0 |
| 3310 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 3325 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 10/2/0/0 | 11/0/0/0 |
| 3331 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 3334 | T | C/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3337 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 3352 | T | C/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 3400 | A | G/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3401 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 3403 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3412 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 3413 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3424 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3451 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3457 | G | A/T/ | 12/12/0/0 | 6/4/1/0 | 12/1/0/0 | 3/9/0/0 | 5/6/0/0 |
| 3458 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3466 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3508 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 11/1/0/0 | 10/1/0/0 |
| 3511 | C | A/T/- | 22/1/1/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3514 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3523 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 3544 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 3595 | A | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3613 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 3619 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 3652 | G | A/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 3773 | A | G/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3838 | G | A/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 3870 | C | T/-/- | 12/12/0/0 | 3/8/0/0 | 6/7/0/0 | 1/11/0/0 | 3/8/0/0 |
| 3882 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3924 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 3960 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 10/3/0/0 | 11/1/0/0 | 11/0/0/0 |
| 3976 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4002 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4011 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4034 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 4037 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4052 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 4100 | C | T/-/- | 24/0/0/0 | 9/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4104 | G | T/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 4112 | C | T/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 4115 | A | G/-/- | 11/13/0/0 | 1/9/0/0 | 0/13/0/0 | 0/12/0/0 | 2/9/0/0 |
| 4121 | A | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4145 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4157 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4181 | T | A/-/- | 3/21/0/0 | 0/11/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 4190 | A | G/-/- | 0/24/0/0 | 1/10/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 4196 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4206 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4208 | A | G/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 4214 | T | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4220 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4256 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 4259 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4292 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 4310 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 4332 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 4352 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 4367 | A | G/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4368 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4373 | T | C/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 4430 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4442 | G | A/-/- | 14/10/0/0 | 10/1/0/0 | 11/2/0/0 | 12/0/0/0 | 9/2/0/0 |
| 4451 | G | A/-/- | 24/0/0/0 | 10/1/0/0 | 12/1/0/0 | 11/1/0/0 | 11/0/0/0 |
| 4484 | A | T/-/- | 20/4/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4487 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4490 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 4494 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 4526 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4529 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 4538 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4559 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4562 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 4565 | T | A/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 4566 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 4579 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4603 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 4620 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4637 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4649 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4652 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4715 | G | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 4747 | C | A/T/- | 24/0/0/0 | 10/0/1/0 | 13/0/0/0 | 10/1/1/0 | 11/0/0/0 |
| 4750 | A | G/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 4756 | C | T/-/- | 21/3/0/0 | 9/2/0/0 | 13/0/0/0 | 9/3/0/0 | 10/1/0/0 |
| 4774 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 4777 | A | C/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4795 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4846 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 4847 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4867 | G | A/-/- | 24/0/0/0 | 9/1/0/0 | 12/1/0/0 | 11/1/0/0 | 11/0/0/0 |
| 4885 | T | C/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 4969 | A | C/-/- | 16/8/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 5011 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 5020 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 5029 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5044 | A | G/-/- | 24/0/0/0 | 9/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5056 | T | C/-/- | 23/1/0/0 | 10/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5059 | A | T/-/- | 11/13/0/0 | 1/9/0/0 | 0/13/0/0 | 0/12/0/0 | 1/10/0/0 |
| 5062 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 5065 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 5072 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5074 | A | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5077 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 5080 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 5128 | A | G/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5131 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5137 | C | T/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5165 | G | A/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5167 | T | A/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5173 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 5182 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5215 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5216 | G | A/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5221 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 11/1/0/0 | 10/1/0/0 |
| 5254 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 5257 | T | C/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 5272 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 5278 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5279 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 5287 | A | G/-/- | 19/5/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5308 | A | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5314 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 5320 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 5323 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5359 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 11/1/0/0 | 11/0/0/0 |
| 5398 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5428 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 5431 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5513 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5540 | C | T/-/- | 23/1/0/0 | 7/4/0/0 | 7/5/0/0 | 7/5/0/0 | 9/2/0/0 |
| 5564 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 12/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5572 | A | G/-/- | 21/3/0/0 | 10/1/0/0 | 11/1/0/0 | 11/1/0/0 | 11/0/0/0 |
| 5622 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5652 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5661 | A | G/-/- | 20/4/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5664 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 5670 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5679 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 10/3/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5682 | G | A/-/- | 23/1/0/0 | 7/4/0/0 | 5/8/0/0 | 6/6/0/0 | 11/0/0/0 |
| 5688 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5691 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5700 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5712 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 5850 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 5865 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 5871 | C | T/-/- | 16/8/0/0 | 10/1/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 5883 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 5898 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 5941 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6021 | A,T | A/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 6054 | G | A/-/- | 21/3/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6222 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6252 | A | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6277 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 6289 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 6315 | A | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6388 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6421 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6425 | A | G/-/- | 9/15/0/0 | 1/10/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 6430 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6439 | C | T/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 11/1/0/0 | 11/0/0/0 |
| 6442 | T | C/G/- | 14/9/1/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 8/3/0/0 |
| 6450 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6466 | A | G/-/- | 16/8/0/0 | 7/4/0/0 | 13/0/0/0 | 7/5/0/0 | 6/5/0/0 |
| 6469 | G | A/-/- | 21/3/0/0 | 9/2/0/0 | 11/2/0/0 | 12/0/0/0 | 10/1/0/0 |
| 6482 | G | A/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6488 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 6501 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6517 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6529 | G | A/-/- | 12/12/0/0 | 1/10/0/0 | 0/13/0/0 | 0/12/0/0 | 1/10/0/0 |
| 6551 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 6556 | G | A/-/- | 9/15/0/0 | 1/10/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 6563 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 6571 | G | A/-/- | 23/1/0/0 | 9/2/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 6578 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 6580 | G | A/-/- | 21/3/0/0 | 7/4/0/0 | 11/2/0/0 | 10/2/0/0 | 6/5/0/0 |
| 6616 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 6619 | G | A/-/- | 21/3/0/0 | 9/2/0/0 | 13/0/0/0 | 11/1/0/0 | 7/4/0/0 |
| 6628 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 11/1/0/0 | 10/1/0/0 |
| 6631 | T | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6637 | G | A/-/- | 23/1/0/0 | 9/2/0/0 | 11/2/0/0 | 10/2/0/0 | 11/0/0/0 |
| 6649 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6661 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6662 | G | A/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6670 | C | T/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 6684 | G | A/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 6687 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 6699 | C | T/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6718 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 6726 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6730 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 6731 | C | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6733 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6753 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6764 | C | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 6791 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6793 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 10/3/0/0 | 11/1/0/0 | 11/0/0/0 |
| 6805 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 6808 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6829 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6841 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 6847 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6928 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 6933 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 6982 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 6985 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 7027 | C | T/-/- | 24/0/0/0 | 9/2/0/0 | 13/0/0/0 | 10/2/0/0 | 10/1/0/0 |
| 7045 | G | A/-/- | 15/9/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 7075 | T | C/-/- | 24/0/0/0 | 8/2/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7090 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 7099 | T | C/G/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 9/2/1/0 | 11/0/0/0 |
| 7105 | T | C/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 7108 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 10/2/0/0 | 10/1/0/0 |
| 7116 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 7135 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 7168 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7195 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 7207 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 7231 | C | T/-/- | 24/0/0/0 | 9/2/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 7240 | G | A/-/- | 9/15/0/0 | 1/10/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 7255 | G | A/-/- | 3/21/0/0 | 1/10/0/0 | 0/13/0/0 | 1/11/0/0 | 1/10/0/0 |
| 7261 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7282 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7290 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7351 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7360 | T | C/-/- | 11/13/0/0 | 2/9/0/0 | 4/9/0/0 | 1/11/0/0 | 1/10/0/0 |
| 7381 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 7396 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7402 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7423 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 7432 | A | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 7450 | G | A/-/- | 23/1/0/0 | 10/1/0/0 | 12/1/0/0 | 12/0/0/0 | 9/2/0/0 |
| 7478 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7479 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 7483 | C | G/T/ | 24/0/0/0 | 10/1/0/0 | 11/0/2/0 | 11/1/0/0 | 11/0/0/0 |
| 7486 | C | T/-/- | 11/13/0/0 | 5/6/0/0 | 4/9/0/0 | 0/12/0/0 | 2/9/0/0 |
| 7525 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7555 | A | G/-/- | 24/0/0/0 | 9/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7578 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7588 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7603 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7606 | G | A/-/- | 24/0/0/0 | 10/1/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7627 | A | G/-/- | 13/11/0/0 | 7/4/0/0 | 12/1/0/0 | 5/7/0/0 | 5/6/0/0 |
| 7672 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7687 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7693 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 7735 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 11/1/0/0 | 11/0/0/0 |
| 7777 | C | T/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 7797 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7798 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7822 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7861 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 7923 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 10/3/0/0 | 11/1/0/0 | 11/0/0/0 |
| 7952 | A | G/-/- | 0/24/0/0 | 0/11/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 8001 | T | C/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8034 | C | T/-/- | 13/11/0/0 | 6/4/0/0 | 11/1/0/0 | 3/9/0/0 | 5/6/0/0 |
| 8091 | A | AT/-/- | 22/2/0/0 | 9/1/0/0 | 12/1/0/0 | 12/0/0/0 | 9/2/0/0 |
| 8122 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8123 | C | T/-/- | 22/2/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8149 | G | A/-/- | 2/22/0/0 | 0/10/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 8196 | G | A/-/- | 23/1/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 8201 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8261 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8297 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8302 | G | A/-/- | 23/1/0/0 | 10/0/0/0 | 10/3/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8303 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8309 | C | T/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8339 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8342 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 10/3/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8372 | C | T/-/- | 24/0/0/0 | 9/2/0/0 | 11/2/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8375 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8382 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 8384 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8393 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 8399 | G | A/-/- | 2/22/0/0 | 0/11/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 8404 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8414 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8425 | A | G/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8450 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8465 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8471 | G | A/T/- | 24/0/0/0 | 6/5/0/0 | 7/6/0/0 | 10/2/0/0 | 10/1/0/0 |
| 8513 | A | G/-/- | 22/2/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8528 | A | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8546 | A | T/-/- | 24/0/0/0 | 10/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8564 | C | A/T/- | 23/0/1/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8582 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 8588 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 9/4/0/0 | 11/1/0/0 | 10/1/0/0 |
| 8612 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8645 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8657 | T | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8663 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8678 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8684 | T | A/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 8714 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8720 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 8722 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8725 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8726 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8740 | G | A/-/- | 24/0/0/0 | 10/1/0/0 | 9/4/0/0 | 9/3/0/0 | 10/1/0/0 |
| 8744 | G | A/-/- | 23/1/0/0 | 8/3/0/0 | 11/2/0/0 | 10/2/0/0 | 10/1/0/0 |
| 8765 | G | A/-/- | 22/2/0/0 | 10/1/0/0 | 12/1/0/0 | 11/1/0/0 | 8/3/0/0 |
| 8807 | T | C/-/- | 22/2/0/0 | 9/2/0/0 | 12/1/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8819 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 8/3/0/0 |
| 8840 | C | T/-/- | 11/13/0/0 | 1/10/0/0 | 0/13/0/0 | 0/12/0/0 | 1/10/0/0 |
| 8842 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 10/3/0/0 | 12/0/0/0 | 9/2/0/0 |
| 8855 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 8882 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 9/2/0/0 |
| 8893 | G | A/-/- | 24/0/0/0 | 7/4/0/0 | 8/6/0/0 | 9/3/0/0 | 11/0/0/0 |
| 8903 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 14/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8913 | A | C/-/- | 24/0/0/0 | 11/0/0/0 | 14/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8915 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 14/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 8927 | A | G/-/- | 11/13/0/0 | 2/9/0/0 | 0/14/0/0 | 1/11/0/0 | 2/9/0/0 |
| 8942 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 14/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8995 | A | G/-/- | 21/3/0/0 | 11/0/0/0 | 14/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 8996 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9014 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 14/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 9035 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 14/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 9041 | C | T/-/- | 3/21/0/0 | 0/11/0/0 | 0/14/0/0 | 0/12/0/0 | 0/11/0/0 |
| 9062 | C | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9122 | T | C/-/- | 24/0/0/0 | 9/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9152 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 9158 | C | T/-/- | 23/1/0/0 | 7/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 9210 | G | A/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9236 | C | T/-/- | 24/0/0/0 | 9/2/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9244 | G | A/-/- | 22/2/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9245 | C | A/T/- | 23/0/1/0 | 10/0/1/0 | 13/0/0/0 | 12/0/0/0 | 9/1/1/0 |
| 9278 | G | A/-/- | 24/0/0/0 | 10/1/0/0 | 12/1/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9287 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9311 | G | A/-/- | 9/15/0/0 | 1/10/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 9353 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 11/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9359 | G | A/-/- | 9/15/0/0 | 1/10/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 9365 | T | C/-/- | 24/0/0/0 | 9/2/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9421 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9422 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9459 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 10/3/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9493 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 9529 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9532 | C | A/-/- | 24/0/0/0 | 9/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9535 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9550 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9575 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9583 | A | G/-/- | 20/4/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9611 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9631 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 9660 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9688 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 9690 | T | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9711 | T | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9724 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 9795 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 9808 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9911 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 9930 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 9976 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 14/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10011 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 10047 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10063 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10068 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10104 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 14/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10156 | G | A/C/- | 24/0/0/0 | 8/1/2/0 | 13/1/0/0 | 10/1/1/0 | 8/3/0/0 |
| 10168 | C | A/T/- | 24/0/0/0 | 7/4/0/0 | 9/5/0/0 | 9/2/1/0 | 10/0/1/0 |
| 10176 | A | G/-/- | 11/13/0/0 | 2/9/0/0 | 3/11/0/0 | 0/12/0/0 | 2/9/0/0 |
| 10177 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 10185 | C | A/T/- | 16/0/8/0 | 11/0/0/0 | 13/1/0/0 | 12/0/0/0 | 10/0/1/0 |
| 10187 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 14/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10252 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 14/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 10253 | C | T/-/- | 23/1/0/0 | 10/0/0/0 | 14/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10269 | C | T/-/- | 24/0/0/0 | 7/3/0/0 | 9/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 10284 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 14/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10311 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 14/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 10324 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 14/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 10332 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10341 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 10/3/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10371 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10383 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10396 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10401 | G | A/-/- | 24/0/0/0 | 9/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10424 | T | A/-/- | 16/8/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 10427 | T | C/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10454 | C | T/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 10/2/0/0 | 10/1/0/0 |
| 10463 | T | C/-/- | 24/0/0/0 | 7/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 10466 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 10481 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10482 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10499 | C | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10502 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10535 | C | T/-/- | 23/1/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10536 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10542 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 9/2/0/0 |
| 10559 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10566 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10568 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10583 | C | T/-/- | 23/1/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10598 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10607 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10628 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10640 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10664 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10670 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 10691 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10700 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10715 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10718 | A | G/-/- | 24/0/0/0 | 9/2/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10719 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10724 | T | C/-/- | 22/2/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10727 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10737 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10742 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10763 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10784 | A | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 10796 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 10805 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 10808 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10820 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10841 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10863 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10872 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10877 | T | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10880 | C | T/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 10881 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10886 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 10889 | G | A/-/- | 22/2/0/0 | 9/2/0/0 | 11/2/0/0 | 9/3/0/0 | 11/0/0/0 |
| 10892 | T | C/-/- | 23/1/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10898 | T | A/C/- | 11/0/13/0 | 2/0/9/0 | 0/0/13/0 | 1/0/11/0 | 1/0/10/0 |
| 10910 | A | G/-/- | 20/4/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10913 | G | A/-/- | 22/2/0/0 | 9/2/0/0 | 10/3/0/0 | 10/2/0/0 | 8/3/0/0 |
| 10916 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 10/2/0/0 | 11/0/0/0 |
| 10931 | C | T/-/- | 24/0/0/0 | 8/3/0/0 | 8/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 10934 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 10955 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10988 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 10997 | G | A/-/- | 23/1/0/0 | 9/1/0/0 | 11/2/0/0 | 10/2/0/0 | 10/1/0/0 |
| 10998 | A | G/-/- | 24/0/0/0 | 9/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11042 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11060 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11093 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11117 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 11/2/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11150 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11159 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11160 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11171 | T | C/-/- | 23/1/0/0 | 9/2/0/0 | 11/2/0/0 | 11/1/0/0 | 8/3/0/0 |
| 11174 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11204 | T | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11213 | A | G/T/- | 24/0/0/0 | 10/1/0/0 | 12/1/1/0 | 12/0/0/0 | 10/0/1/0 |
| 11216 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/1/0/0 | 10/2/0/0 | 10/1/0/0 |
| 11222 | A | G/-/- | 24/0/0/0 | 8/3/0/0 | 9/5/0/0 | 10/2/0/0 | 11/0/0/0 |
| 11234 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/2/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11249 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/1/0/0 | 12/0/0/0 | 9/2/0/0 |
| 11276 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 14/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11282 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 14/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11301 | C | T/-/- | 9/15/0/0 | 2/9/0/0 | 0/14/0/0 | 0/12/0/0 | 0/11/0/0 |
| 11318 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11339 | C | T/-/- | 24/0/0/0 | 8/3/0/0 | 7/6/0/0 | 10/2/0/0 | 11/0/0/0 |
| 11348 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11351 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 10/2/0/0 | 10/1/0/0 |
| 11354 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11363 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11390 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 14/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 11414 | G | A/-/- | 23/1/0/0 | 10/1/0/0 | 10/4/0/0 | 10/2/0/0 | 11/0/0/0 |
| 11415 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 14/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11432 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 14/0/0/0 | 10/2/0/0 | 10/1/0/0 |
| 11450 | C | T/-/- | 13/11/0/0 | 1/10/0/0 | 1/13/0/0 | 0/12/0/0 | 2/9/0/0 |
| 11475 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 14/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11492 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11537 | G | A/-/- | 24/0/0/0 | 9/2/0/0 | 8/5/0/0 | 10/2/0/0 | 8/3/0/0 |
| 11538 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11592 | G | A/-/- | 0/24/0/0 | 0/11/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 11599 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11610 | A | AT/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 11655 | G | A/-/- | 8/16/0/0 | 1/9/0/0 | 0/13/0/0 | 0/12/0/0 | 0/11/0/0 |
| 11673 | G | A/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 11729 | G | A/-/- | 24/0/0/0 | 9/2/0/0 | 11/2/0/0 | 12/0/0/0 | 8/3/0/0 |
| 11739 | A | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11745 | T | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11748 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11757 | C | A/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11760 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11763 | A | G/-/- | 18/6/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 11787 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11793 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11829 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11862 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11868 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 11899 | G | A/-/- | 24/0/0/0 | 9/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11907 | C | T/-/- | 20/4/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 11910 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11916 | G | A/-/- | 20/4/0/0 | 5/5/0/0 | 2/11/0/0 | 5/7/0/0 | 8/3/0/0 |
| 11925 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11931 | T | C/-/- | 23/1/0/0 | 9/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 11982 | A | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 11988 | A | C/-/- | 12/12/0/0 | 6/4/0/0 | 11/2/0/0 | 3/9/0/0 | 3/8/0/0 |
| 12005 | G | A/-/- | 23/1/0/0 | 9/1/0/0 | 12/1/0/0 | 9/3/0/0 | 8/3/0/0 |
| 12006 | C | T/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 12009 | G | A/T/- | 24/0/0/0 | 6/4/0/0 | 8/2/3/0 | 8/4/0/0 | 11/0/0/0 |
| 12015 | A | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 12020 | A | G/-/- | 24/0/0/0 | 9/1/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 12036 | A | G/-/- | 13/11/0/0 | 2/8/0/0 | 1/12/0/0 | 2/10/0/0 | 4/7/0/0 |
| 12039 | A | G/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12061 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12078 | A | G/-/- | 24/0/0/0 | 9/2/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 12084 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 10/3/0/0 | 11/1/0/0 | 11/0/0/0 |
| 12092 | C | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12159 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 12165 | C | A/T/- | 19/4/1/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12168 | C | T/-/- | 22/2/0/0 | 10/1/0/0 | 12/1/0/0 | 12/0/0/0 | 9/2/0/0 |
| 12171 | C | T/-/- | 24/0/0/0 | 9/2/0/0 | 13/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 12204 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12210 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 12225 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 11/2/0/0 | 12/0/0/0 | 10/1/0/0 |
| 12228 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12258 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 14/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 12273 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 14/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12278 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 14/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 12333 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 9/2/0/0 |
| 12336 | A | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 12339 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12348 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 12420 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12471 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 14/0/0/0 | 10/2/0/0 | 10/1/0/0 |
| 12480 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 14/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12499 | G | A/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12501 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12536 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 12558 | T | C/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12601 | T | TTA/-/- | 24/0/0/0 | 10/0/0/0 | 14/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 12607 | A | T/-/- | 24/0/0/0 | 10/0/0/0 | 11/3/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12633 | G | A/-/- | 23/1/0/0 | 9/1/0/0 | 12/2/0/0 | 11/1/0/0 | 10/1/0/0 |
| 12784 | C | T/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12890 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12964 | T | G/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 12967 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 13109 | A | G/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 13123 | G | A/-/- | 22/2/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 13310 | T | A/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 13311 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 13489 | G | A/-/- | 23/1/0/0 | 10/0/0/0 | 12/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 13579 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 11/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 13641 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 12/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 13683 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 12/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 13696 | G | A/-/- | 22/2/0/0 | 11/0/0/0 | 12/0/0/0 | 11/1/0/0 | 10/1/0/0 |
| 13741 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 12/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 13776 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 12/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 13876 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 13882 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 13941 | A | AT/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/0/0/0 |
| 13943 | T | A/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/0/0/0 |
| 13955 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 13989 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 14105 | G | A/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 14115 | G | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14194 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/1/0/0 |
| 14233 | T | C/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14291 | C | T/-/- | 21/3/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14359 | G | A/-/- | 23/1/0/0 | 10/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14373 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14384 | C | A/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14439 | A | G/-/- | 21/3/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14454 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14536 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 14540 | C | T/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 14543 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 10/1/0/0 |
| 14549 | A | T/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14582 | T | TTA/-/- | 24/0/0/0 | 11/0/0/0 | 12/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14602 | T | C/-/- | 24/0/0/0 | 9/2/0/0 | 12/1/0/0 | 11/1/0/0 | 11/0/0/0 |
| 14642 | C | T/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14686 | T | C/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14691 | T | A/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14741 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
| 14847 | T | C/-/- | 11/13/0/0 | 1/9/0/0 | 2/10/0/0 | 1/11/0/0 | 1/9/0/0 |
| 14875 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 10/0/0/0 |
| 14881 | T | TA/-/- | 24/0/0/0 | 11/0/0/0 | 11/1/0/0 | 12/0/0/0 | 9/0/0/0 |
| 14912 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 10/1/0/0 | 10/0/0/0 | 9/0/0/0 |
| 14916 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 11/0/0/0 | 10/0/0/0 | 8/1/0/0 |
| 14930 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 9/0/0/0 | 11/0/0/0 | 10/0/0/0 |
| 14936 | A | G/-/- | 24/0/0/0 | 11/0/0/0 | 9/1/0/0 | 11/0/0/0 | 11/0/0/0 |
| 14976 | T,A | T/-/- | 23/1/0/0 | 10/1/0/0 | 8/2/0/0 | 9/1/0/0 | 7/3/0/0 |
| 14978 | A | C/T/ | 20/4/0/0 | 3/8/0/0 | 4/6/0/0 | 3/7/0/0 | 5/4/1/0 |
| 14984 | A,T,T | TTT/AT/A | 14/2/7/1 | 2/2/0/3 | 3/2/0/0 | 5/2/0/1 | 4/0/0/2 |
| 15002 | A | T/-/- | 22/2/0/0 | 5/2/0/0 | 4/1/0/0 | 3/5/0/0 | 4/2/0/0 |
| 15012 | T | C/-/- | 24/0/0/0 | 9/0/0/0 | 11/0/0/0 | 11/1/0/0 | 10/0/0/0 |
| 15035 | T | TA/-/- | 23/1/0/0 | 10/0/0/0 | 12/0/0/0 | 9/0/0/0 | 11/0/0/0 |
| 15036 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 12/0/0/0 | 9/0/0/0 | 9/2/0/0 |
| 15038 | A | AATT/-/- | 22/2/0/0 | 7/3/0/0 | 10/0/0/0 | 11/0/0/0 | 7/2/0/0 |
| 15039 | A | G/-/- | 24/0/0/0 | 10/0/0/0 | 10/0/0/0 | 7/4/0/0 | 8/1/0/0 |
| 15049 | T | C/-/- | 24/0/0/0 | 10/0/0/0 | 10/0/0/0 | 11/0/0/0 | 7/2/0/0 |
| 15050 | T | TTA/-/- | 23/1/0/0 | 10/0/0/0 | 10/0/0/0 | 11/0/0/0 | 9/0/0/0 |
| 15052 | A | G/-/- | 23/1/0/0 | 10/0/0/0 | 7/1/0/0 | 11/0/0/0 | 9/0/0/0 |
| 15057 | C | T/-/- | 23/1/0/0 | 10/0/0/0 | 8/0/0/0 | 8/0/0/0 | 9/0/0/0 |
| 15058 | A | G/-/- | 22/2/0/0 | 5/5/0/0 | 3/5/0/0 | 4/4/0/0 | 7/2/0/0 |
| 15066 | T | C/-/- | 23/1/0/0 | 10/0/0/0 | 10/0/0/0 | 11/0/0/0 | 8/0/0/0 |
| 15069 | T | C/-/- | 23/1/0/0 | 10/0/0/0 | 10/0/0/0 | 11/0/0/0 | 8/1/0/0 |
| 15082 | A | G/-/- | 22/2/0/0 | 10/0/0/0 | 10/0/0/0 | 11/0/0/0 | 8/2/0/0 |
| 15098 | A | G/-/- | 23/1/0/0 | 9/1/0/0 | 10/0/0/0 | 8/1/0/0 | 8/0/0/0 |
| 15115 | T | C/-/- | 21/3/0/0 | 8/1/0/0 | 8/2/0/0 | 8/1/0/0 | 8/0/0/0 |
| 15116 | G | A/-/- | 14/10/0/0 | 9/1/0/0 | 10/0/0/0 | 8/1/0/0 | 3/5/0/0 |
| 15128 | A | G/-/- | 23/1/0/0 | 8/1/0/0 | 6/1/0/0 | 8/0/0/0 | 7/0/0/0 |
| 15129 | G | A/-/- | 1/23/0/0 | 0/9/0/0 | 0/7/0/0 | 1/7/0/0 | 0/7/0/0 |
| 15144 | T | A/-/- | 24/0/0/0 | 10/0/0/0 | 13/0/0/0 | 11/1/0/0 | 11/0/0/0 |
| 15189 | A | G/-/- | 18/6/0/0 | 10/1/0/0 | 11/0/0/0 | 10/0/0/0 | 7/0/0/0 |
| 15243 | T | C/-/- | 15/9/0/0 | 10/1/0/0 | 7/1/0/0 | 7/0/0/0 | 6/0/0/0 |
| 15282 | T | C/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 12/0/0/0 | 11/0/0/0 |
| 15301 | C | A/-/- | 24/0/0/0 | 11/0/0/0 | 12/1/0/0 | 11/0/0/0 | 11/0/0/0 |
| 15304 | A | G/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 11/0/0/0 | 11/0/0/0 |
| 15305 | T | TA/-/- | 24/0/0/0 | 10/1/0/0 | 13/0/0/0 | 11/0/0/0 | 11/0/0/0 |
| 15321 | C | T/-/- | 23/1/0/0 | 11/0/0/0 | 11/2/0/0 | 11/0/0/0 | 11/0/0/0 |
| 15327 | A | G/-/- | 23/1/0/0 | 11/0/0/0 | 13/0/0/0 | 12/0/0/0 | 11/0/0/0 |
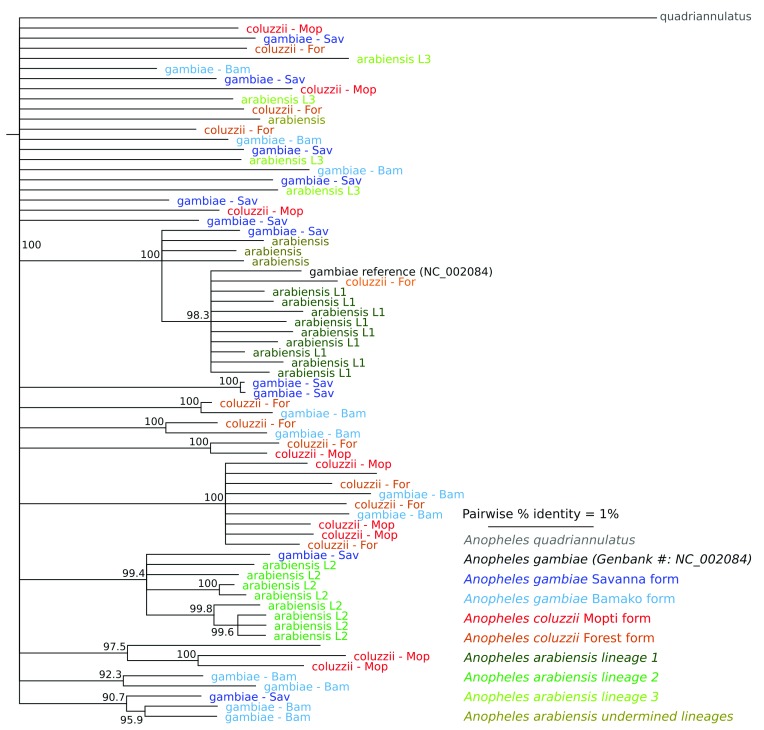
The lack of species-specific markers is also reflected in the phylogenetic tree ( Figure 1). An. arabiensis, An. coluzzii and An. gambiae did not cluster separately, which is consistent with previous reports that compared mitochondrial genome sequence data from specimens originating from Kenya, Senegal and South Africa ( Besansky et al. (1997)) and Burkina Faso, Cameroon, Kenya, Mali, South Africa, Tanzania and Zimbabwe ( Fontaine et al. (2015), supplemental material).
Figure 1. Phylogenetic tree inferred from mtDNA genome sequence data.
With the exception of the outgroup, An. quadriannulatis, this analysis fails to reveal a clear division of the operational taxonomic units included in this analysis. Colors indicate the species or chromosomal form and numbers at the branches indicate the accuracy of the inferred branches on a scale of 0–100, where 100 represents the highest confidence.
Of note, 36 of the samples that we used in our study originated from Kela (Mali). Kela is located near the village of Selinkenyi, where previous studies have shown a history of hybridization and introgression between An. gambiae and An. coluzzii ( Lee et al. (2013); Main et al. (2015); Norris et al. (2015)) , which may have resulted in shared polymorphisms in their mitochondrial genomes. Shared polymorphisms in their mitochondrial genomes, where history has not been reported, also appeared to have occurred in Mutengene (Cameroon), where both An. gambiae and An. coluzzii occur sympatrically. Hybridization between either An. coluzzii or An. gambiae with An. arabiensis yields sterile males ( Slotman et al. (2004)), but phylogenomic analysis of these species show patterns of introgression between all of them ( Fontaine et al. (2015)). Our mitochondrial genome study does not provide conclusive evidence for hybridization and introgression among the taxa under study. However, our data suggest that this is a possibility.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2018 Hanemaaijer MJ et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
Aligned sequences were submitted to the National Center for Biotechnology Information (NCBI) Accession number: MG930826 - MG930896
Dataset 1. Aligned FASTA file of mitogenome samples 10.5256/f1000research.13807.d192892 ( Hanemaaijer et al., 2018)
Funding Statement
We thank University of California - Irvine, Malaria Initiatives (UCIMI) for their support.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 1 approved
Acknowledgments
We thank Michelle Sanford for her assistance in the field collection in Cameroon in 2011. We thank Clare Marsden for providing the raw data of An. arabiensis samples.
References
- Beard CB, Hamm DM, Collins FH: The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 1993;2(2):103–124. 10.1111/j.1365-2583.1993.tb00131.x [DOI] [PubMed] [Google Scholar]
- Besansky NJ, Lehmann T, Fahey GT, et al. : Patterns of mitochondrial variation within and between African malaria vectors, Anopheles gambiae and An. arabiensis, suggest extensive gene flow. Genetics. 1997;147(4):1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Loshe M, Usadel B: Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WM, George M, Jr, Wilson AC: Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979;76(4):1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JE, Lazzaro BP: Assessing the accuracy and power of population genetic inference from low-pass next-generation sequencing data. Front Genet. 2012;3:66. 10.3389/fgene.2012.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin MA, Palmisciano DA, Vyse ER, et al. : Mitochondrial DNA in wildlife forensic science: species identification of tissues. Wildlife Soc B (1973-2006). 1991;19(1):94–105. Reference Source [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. : The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Barba M, Adams JR, Goldberg CS, et al. : Molecular species identification for multiple carnivores. Conserv Genet Resour. 2014;6(4):821–824. 10.1007/s12686-014-0257-x [DOI] [Google Scholar]
- De Mandal S, Chhakchhuak L, Gurusubramanian G, et al. : Mitochondrial markers for identification and phylogenetic studies in insects–A Review. DNA Barcodes. 2014;2(1). 10.2478/dna-2014-0001 [DOI] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, et al. : Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347(6217):1258524. 10.1126/science.1258524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Mittnik A, Johnson PLF, et al. : A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 2013;23(7):553–559. 10.1016/j.cub.2013.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G: Haplotype-based variant detection from short-read sequencing. arXiv: 1207.3907.Cornell University Library;2013. Reference Source [Google Scholar]
- Green CA: Cytological maps for the practical identification of females of the three freshwater species of the Anopheles gambiae complex. Ann Trop Med Parasitol. 1972;66(1):143–147. 10.1080/00034983.1972.11686806 [DOI] [PubMed] [Google Scholar]
- Haag-Liautard C, Coffey N, Houle D, et al. : Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 2008;6(8):e204. 10.1371/journal.pbio.0060204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanemaaijer MJ, Houston P, Collier TC, et al. : Dataset 1: Mitochondrial genomes of Anopheles arabiensis, An. gambiae and An. coluzzii show no clear species division. F1000Research. 2018. 10.5256/f1000research.13807.d192892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG: Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends Ecol Evol. 1989;4(1):6–11. 10.1016/0169-5347(89)90006-2 [DOI] [PubMed] [Google Scholar]
- Lanzaro GC, Lee Y: Speciation in Anopheles gambiae—The distribution of genetic polymorphism and patterns of reproductive isolation among natural populations. Anopheles mosquitoes-New insights into malaria vectors.InTech,2013. 10.5772/56232 [DOI] [Google Scholar]
- Lee Y, Marsden CD, Norris LC, et al. : Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci U S A. 2013;110(49):19854–19859. 10.1073/pnas.1316851110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Marsden CD, Nieman C, et al. : A new multiplex SNP genotyping assay for detecting hybridization and introgression between the M and S molecular forms of Anopheles gambiae. Mol Ecol Resour. 2014;14(2):297–305. 10.1111/1755-0998.12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas B, Fehren-Schmitz L, Valverde G, et al. : Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas. Sci Adv. 2016;2(4):e1501385. 10.1126/sciadv.1501385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main BJ, Lee Y, Collier TC, et al. : Complex genome evolution in Anopheles coluzzii associated with increased insecticide usage in Mali. Mol Ecol. 2015;24(20):5145–5157. 10.1111/mec.13382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden CD, Lee Y, Kreppel K, et al. : Diversity, differentiation, and linkage disequilibrium: prospects for association mapping in the malaria vector Anopheles arabiensis. G3 (Bethesda). 2014;4(1):121–131. 10.1534/g3.113.008326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles A, Harding N: cggh/scikit-allel: v1.1.8 (Version v1.1.8). Zenodo. 2017. 10.5281/zenodo.822784 [DOI] [Google Scholar]
- Norris LC, Main BJ, Lee Y, et al. : Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc Natl Acad Sci U S A. 2015;112(3):815–820. 10.1073/pnas.1418892112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Conesa A, García-Alcalde F: Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32(2):292–294. 10.1093/bioinformatics/btv566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg GG, Sinclair B, Briskey L, et al. : MtDNA barcode identification of fish larvae in the southern Great Barrier Reef–Australia. Sci Mar. 2006;70(S2):7–12. Reference Source [Google Scholar]
- Phillips MJ, Haouchar D, Pratt RC, et al. : Inferring kangaroo phylogeny from incongruent nuclear and mitochondrial genes. PLoS One. 2013;8(2):e57745. 10.1371/journal.pone.0057745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombi M, Caputo B, Simard F, et al. : Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions. BMC Evol Biol. 2008;8(1):309. 10.1186/1471-2148-8-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Brogdon WG, Collins FH: Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49(4):520–529. 10.4269/ajtmh.1993.49.520 [DOI] [PubMed] [Google Scholar]
- Shaw KL: Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: what mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proc Natl Acad Sci U S A. 2002;99(25):16122–16127. 10.1073/pnas.242585899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman M, Della Torre A, Powell JR: The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. arabiensis. Genetics. 2004;167(1):275–287. 10.1534/genetics.167.1.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sota T, Vogler AP: Incongruence of mitochondrial and nuclear gene trees in the Carabid beetles Ohomopterus. Syst Biol. 2001;50(1):39–59. 10.1093/sysbio/50.1.39 [DOI] [PubMed] [Google Scholar]
- Tahir HM, Mehwish, Kanwal N, et al. : Genetic diversity in cytochrome c oxidase I gene of Anopheles mosquitoes. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(6):4298–4301. 10.3109/19401736.2015.1082104 [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV: Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3(9):e285. 10.1371/journal.pbio.0030285 [DOI] [PMC free article] [PubMed] [Google Scholar]