Abstract
Pyrimidine bases are rapidly catabolized in growing plant tissues. The final enzyme of the catabolic pathway, β-ureidopropionase (β-UP; EC 3.5.1.6), was partially purified from the shoots of etiolated maize (Zea mays) seedlings. The enzyme had a Km for β-ureidopropionate (the substrate derived from uracil) of 11 μm. Only one enantiomer of racemic β-ureidoisobutyrate (derived from thymine) was processed with a Km of 6 μm. The enzyme was inactivated by dialysis against 1,10-phenanthroline and activity could be partially restored by addition of Zn2+. Maize β-UP was very sensitive to inactivation by iodoacetamide. This could be prevented by addition of substrate, indicating the presence of an active site Cys. The enzyme was strongly inhibited by short chain aliphatic acids and aryl propionates, the most potent inhibitor of which was 2-(2, 6-dinitrophenoxy)-propionate (I50 = 0.5 μm). A gene for Arabidopsis β-UP encodes a polypeptide of 405 amino acids and has about 55% homology with the enzymes from other eukaryotic organisms. Several highly conserved residues link the plant β-UP with a larger class of prokaryotic and eukaryotic amidohydrolases. An Arabidopsis cDNA truncated at the N terminus by 14 residues was cloned and overexpressed in Escherichia coli. The recombinant enzyme (43.7 kD) was soluble, functional, and purified to homogeneity with yields of 15 to 20 mg per 30 g fresh weight of E. coli cells. The recombinant enzyme from Arabidopsis and the native enzyme from maize had molecular masses of approximately 440 kD, indicating the enzyme is a decamer at pH 7.
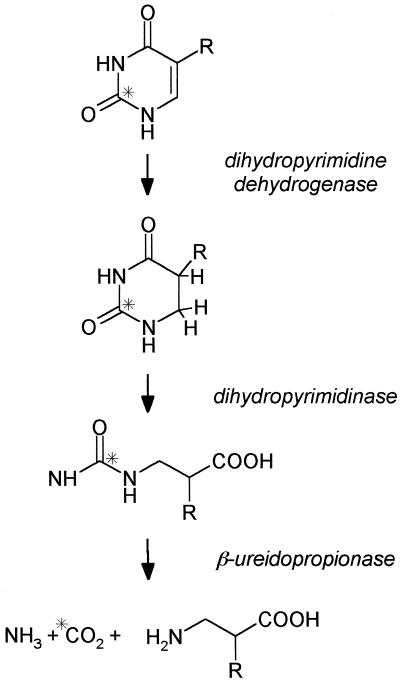
Plant cells can efficiently catabolize uracil, thymine, uridine, and thymidine to CO2, ammonia, and small amino acids (Barnes and Naylor, 1962; Tsai and Axelrod, 1965; Lesley et al., 1980; Slabas et al., 1980). Although there are pathways to recover pyrimidine bases and nucleosides, their predominant metabolic fate in plant cells appears to be catabolic. This is in contrast to the efficient and well-characterized salvage of the purine, adenine (Ashihara et al., 2000). The presence of this degradative activity for pyrimidines accounts for the relatively poor incorporation of exogenous radiolabeled thymidine into DNA in dividing plant cells (Lesley et al., 1980; Slabas et al., 1980). The pathway for the catabolism of pyrimidine bases is widespread and has been found in many eukaryotic and prokaryotic organisms (Wasternack, 1978). It consists of three steps, a reduction of the pyrimidine ring catalyzed by dihydropyrimidine dehydrogenase followed by a ring-opening hydrolysis catalyzed by dihydropyrimidinase, then hydrolysis of the resulting ureide group by β-ureidopropionase (β-UP, EC 3.5.1.6; also called β-Ala synthase or N-carbamoyl-β-Ala amidohydrolase in some studies) to produce CO2, ammonia, and a β-amino acid (Fig. 1). The end product of the pathway derived from uracil is β-Ala and from thymine is β-aminoisobutyrate. In the organisms so far tested each enzyme of the pathway can process the substrates derived from uracil and thymine.
Figure 1.
The reductive pathway for the catabolism of pyrimidine bases found in plants, mammals, and microorganisms. R, H for uracil and CH3 for thymine. The asterisk denotes the position of the carbon atom eventually released as CO2. The end product of the pathway from uracil is β-Ala and from thymine is β-isobutyrate
The pyrimidine catabolic pathway has been investigated in most detail in mammalian liver where it is responsible for the inactivation of the anticancer drug, 5-fluorouracil (Wasternack, 1980). However, almost no characterization of the pathway components has been reported in plants. Although the pathway is generally characterized as catabolic, it also acts as a significant biosynthetic source of β-Ala. In mammals β-Ala has a physiological role in neurotransmission and is a component of neural dipeptides such as carnosine and anserine. As a consequence, genetic disruptions in normal β-Ala metabolism can lead to severe neurological problems (Traut and Jones, 1996). In plants β-Ala is an essential component of the pantothenate moiety of coenzyme A. The betaine derivative of β-Ala is also a primary osmoprotectant in some plant families such as the Plumbaginacae (Rathinasabapathi et al., 2000). The pyrimidine catabolic pathway may therefore be a significant source of β-Ala for these important compounds in plants.
Our interest in the pyrimidine catabolic pathway was stimulated by noting the rapid and extensive release of 14CO2 from the addition of uracil or thymine labeled at C-2 to plant tissues during investigations of plant pyrimidine metabolism. This, combined with the potential importance of β-Ala as a biosynthetic precursor for pantothenate, prompted us to characterize the key enzyme in the pathway responsible for the release of CO2 and production of β-Ala from pyrimidines, β-UP. The enzyme has previously been studied from some microbes including Euglena gracilis (Wasternack et al., 1979; Ogawa and Shimizu, 1994) and from mammalian liver (Tamaki et al., 1987; Matthews et al., 1992). Genes encoding the rat and human β-UPs have been isolated and characterized (Kvalnes-Krick and Traut, 1993; Vreken et al., 1999). The rat enzyme has a subunit molecular mass of 42 kD and in the native state is a hexamer of 240 kD. However, the enzyme undergoes significant changes in oligomerization state in the presence of substrate or product that may also act as regulatory mechanisms. For example, it associates to a more active decamer or dodecamer (410–430 kD) in response to low concentrations of β-ureidopropionate (Matthews and Traut, 1987). In common with some other amidohydrolases, rat β-UP appears to be a Zn2+-dependent enzyme (Kvalnes-Krick and Traut, 1993).
In contrast to the mammalian enzyme, very few details regarding the enzyme from higher plants are available, although the pathway is clearly very active in plant tissues. In this study we have characterized the native β-UP from maize (Zea mays), cloned an Arabidopsis cDNA encoding β-UP, and functionally over-expressed the enzyme at high levels in Escherichia coli. This is the first detailed characterization of an enzyme synthesizing β-Ala in plants.
RESULTS
Native β-UP Characterization
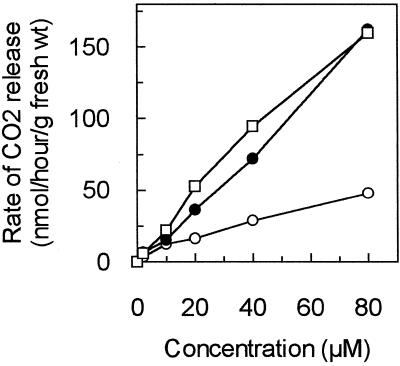
Figure 2 shows the release of 14CO2 when etiolated maize shoots are incubated with increasing concentrations of exogenous pyrimidines labeled in the C-2 position. Thymine was more efficiently processed by the pathway than uracil, as it produced a consistently higher rate of 14CO2 release than uracil, similar to previous studies in E. gracilis (Wasternack et al., 1977). Dihydrouracil also produced a higher rate of CO2 release than uracil, suggesting that the first reductive step in the pathway may be somewhat rate-limiting in these conditions. Similar experiments with Arabidopsis seedlings showed even higher rates of CO2 release from pyrimidines on a gram fresh weight basis (data not shown). However, etiolated maize shoots were chosen as the preferred source for enzyme isolation as large amounts of tissue could be grown that contained high levels of β-UP without significant interference from photosynthetic pigments. Partially purified preparations of β-UP that were isolated from etiolated maize shoots by ammonium sulfate precipitation and anion-exchange chromatography proved to be quite stable. There was only about 10% loss in activity after storage for 7 d at 4°C in the presence of Mg2+ and dithiothreitol (DTT). The enzyme preparations were stable for several months when frozen at −70°C in the presence of 10% (w/v) glycerol.
Figure 2.
Catabolism of exogenous pyrimidines by etiolated maize seedling tissue to release CO2. Etiolated maize shoots were incubated in vials containing varying amounts of uracil (○), thymine (●), or dihydrouracil (□) at 30°C for 2 h. Each vial contained 0.4 μCi [2-14C]pyrimidine. The assays were quenched by injection of 4 n perchloric acid into the vial and the 14CO2 released was trapped in a well containing 4 n KOH. The radioactivity in the wells was counted in a liquid scintillation counter to quantitate the amount of CO2 released. Each point is the average of three replicates.
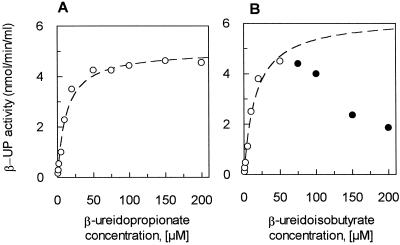
Maize β-UP had a Km value of 11 μm for β-ureidopropionate (Fig. 3A). β-Ureidoisobutyrate was also a substrate for the enzyme, and was a competitive inhibitor of the reaction with β-ureidopropionate (data not shown). The radiolabeled β-ureidoisobutyrate synthesized for the experiments was racemic, so we first determined if maize β-UP had a stereospecific preference by adding a large excess of the enzyme to a sample of racemic [6-14C]β-ureidoisobutyrate. Table I shows that a maximum of just under 50% of the racemic substrate could be hydrolyzed by the enzyme strongly, suggesting that only one enantiomer of β-ureidoisobutyrate is a competent substrate. In contrast, almost 90% of the non-chiral substrate β-ureidopropionate was hydrolyzed under the same conditions. The unhydrolyzed enantiomer of β-ureidoisobutyrate may account for the enzyme inhibition seen at high substrate concentrations of racemic β-ureidoisobutyrate seen in Figure 3B. Using the data points measured at low concentrations, a Km value of 6 μm for the active β-ureidoisobutyrate enantiomer was determined. To eliminate issues of chirality, β-ureidopropionate was used as the primary substrate for further analysis of β-UP activity.
Figure 3.
Km determinations for maize β-UP substrates. The β-UP activity was determined by measuring the amount of 14CO2 released from [5-14C]β-ureidopropionate (A) or racemic [6-14C]β-ureidoisobutyrate (B) at pH 7 over 30 min at varying substrate concentrations. ○, Data points used to fit the curves to simple Michaelis-Menten equations (Km for β-ureidopropionate = 11 μm and for β-ureidoisobutyrate = 6 μm; the latter value is calculated based on a single enantiomer being processed). Inhibition of the reaction seen at high concentrations of β-ureidoisobutyrate may be caused by the non-hydrolyzed enantiomer of the racemic substrate. These points (●) were not used to generate the fitted curve.
Table I.
Product formation by maize β-UP from racemic β-ureidoisobutyrate
| Reaction Time | Amount of
14CO2 Formed
|
|
|---|---|---|
| [6-14C]β-Ureidoisobutyrate (15,468 total dpm added) | [5-14C]β-Ureidopropionate (25,386 total dpm added) | |
| dpm (% total) | ||
| 2 h | 7,125 (46%) | 22,500 (89%) |
| 3 h | 7,423 (48%) | 22,350 (88%) |
A large excess of maize β-UP was used to drive product formation rapidly toward completion. The total volume of the reactions was 0.2 mL and the specific activity of the substrates was 5.7 mCi mmol−1. Assays were performed in duplicate.
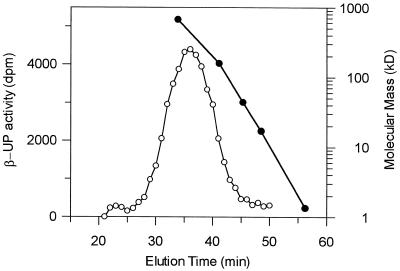
The pH optimum for the enzyme was broad at mildly acidic pH values as there was little change in the level of enzyme activity between pH values of 6.0 to 7.2. However, enzyme activity declined at more alkaline pH values. The native molecular mass of the enzyme was estimated to be 440 ± 40 kD by size exclusion chromatography (Fig. 4).
Figure 4.
Molecular mass of native maize β-UP determined by size-exclusion chromatography. A sample of partially purified β-UP was applied onto a Superose 12 column (Pharmacia) equilibrated in 0.1 m potassium phosphate, pH 7.0, 1 mm MgCl2, and 2 mm DTT at a flow-rate of 0.4 mL min−1. Fractions of 0.4 mL were collected and assayed for β-UP activity. The amount of radiolabel lost from the assay as CO2 relative to a control is shown on the left axis. The position of Bio-Rad native molecular mass standards (670, 158, 44, 17, and 1.35 kD) run under the same elution conditions is shown on the right axis.
Effect of Metal Ion Chelators
To assess whether maize β-UP is a Zn2+-requiring enzyme like the mammalian enzyme (Kvalnes-Krick and Traut, 1993), the enzyme was extensively dialyzed against buffer solutions containing several different metal ion chelators at 1 mm concentrations. The chelators were then removed by further dialysis against buffer alone and the residual enzyme activity was measured. Dialysis against EDTA, 2,6-dipicolinic acid, or 8-hydroxyquinoline lowered the enzyme activity by 56%, 33%, and 29%, respectively, relative to a control, whereas 1,10-phenanthroline lowered the activity by 96%. Enzyme that had been completely inactivated by treatment with 1,10-phenanthroline could be reactivated by addition of 20 μm Zn2+. After 5 h, 30% of the original enzyme activity was recovered and after 24 h, 46% was recovered. This suggests that maize β-UP is a Zn2+-dependent enzyme.
Effect of Thiol Reagents
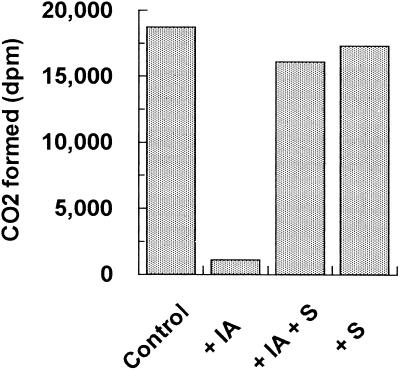
β-UP from maize was very sensitive to inhibition by thiol derivatizing reagents. Addition of 200 μm Cu2+ ion, 1 mm iodoacetamide, or p-chloromercuribenzene-sulfonic acid completely eliminated the activity of the enzyme. Further experiments showed that addition of 75 μm iodoacetamide for 30 min was sufficient to completely inactivate the enzyme. Inhibition by iodoacetamide was completely prevented by addition of substrate (Fig. 5), indicating that the sensitive thiol group is at the active site of the enzyme.
Figure 5.
Substrate protects maize β-UP from inactivation by iodoacetamide. Samples of partially purified maize β-UP were treated with 1 mm iodoacetamide (IA) in the presence or absence of 10 mm β-ureidopropionate (S) for 30 min. After quenching the iodoacetamide by adding DTT to 10 mm, the samples were extensively dialyzed against 0.1 m potassium phosphate buffer containing 2 mm DTT and were then assayed for β-UP activity. The control sample was treated identically but had no iodoacetamide or substrate added.
Inhibitor Studies
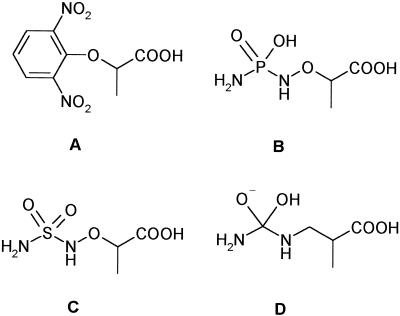
A variety of compounds were tested as probes of the mechanism or substrate recognition of β-UP. Hydroxamates and benzenesulfonamides can be effective inhibitors of metal ion-dependent hydrolases through interaction with the metal ion, but none of the examples tested (hydroxyurea, acetohydroxamic acid, benzene sulfonamide, 4-carboxybenzene-sulfonamide, or 4-nitrobenzenesulfonamide) inhibited maize β-UP at concentrations up to 1 mm. An aminophosphoramide and an aminosulfonamide analog of the substrate were tested as mimics of a possible transition state for hydrolysis (Fig. 6). Similar compounds can be extremely potent inhibitors of certain peptide hydrolases (Hanson et al., 1989), however these analogs were also poor inhibitors of plant β-UP. In contrast to the mechanism-based inhibitors that we tested, a number of short chain aryl carboxylic acids were surprisingly effective inhibitors of β-UP. I50 values for several of these compounds are shown in Table II. For example, 3-phenylpropionate had an I50 value of 5.6 μm. (S)-2-phenylpropionate was also an effective inhibitor of the enzyme (I50 = 5.7 μm), whereas the (R)-enantiomer was 20-fold less potent (I50 = 115 μm). The most potent inhibitor we discovered was 2-(2, 6-dinitrophenoxy)-propionate. This compound had an I50 of 0.5 μm and the inhibition was completely reversible by passing the enzyme-inhibitor complex over a gel filtration column. Several short chain aliphatic acids also inhibited β-UP, with progressively weaker inhibition observed with extended chain lengths (Table II). The aliphatic acid portion of the substrate appears to significantly contribute to substrate recognition, whereas mimics of the ureide portion of substrate (urea and N-methylurea) were ineffective as inhibitors of the enzyme. Several substrate analogs such as α-ureidopropionate, α-ureidoisobuty-rate, α-ureido-n-butyrate, N-carbamyl-Asp, and the reaction products β-Ala and β-ureidoisobutyrate had no effect on enzyme activity at concentrations up to 2 mm.
Figure 6.
Structures of compounds tested for inhibition of maize β-UP. A, 2-(2, 6-Dinitrophenoxy)-propionic acid; B, 2-aminophospho-ramidoxypropionic acid; C, 2-aminosulfonamidoxypropionic acid; D, possible oxyanion tetrahedral transition state of the β-UP reaction.
Table II.
Inhibition of maize β-UP by aryl propionic and aliphatic acids
| Compound | I50 Value |
|---|---|
| μm | |
| 3-Phenylpropionic acid | 5.6 |
| (S)-2-Phenylpropionic acid | 5.7 |
| (R)-2-Phenylpropionic acid | 115.0 |
| (RS)-2-Benzylpropionic acid | 6.0 |
| (RS)-2-(2,6-dinitrophenoxy)-propionic acid | 0.5 |
| Cyclopropanecarboxylic acid | 8.0 |
| Isobutyric acid | 18.0 |
| Propionic acid | 25.0 |
| N-Butyric acid | 55.0 |
I50 values were determined at a substrate concentration of 8 μM.
Arabidopsis β-UP cDNA Cloning and Overexpression
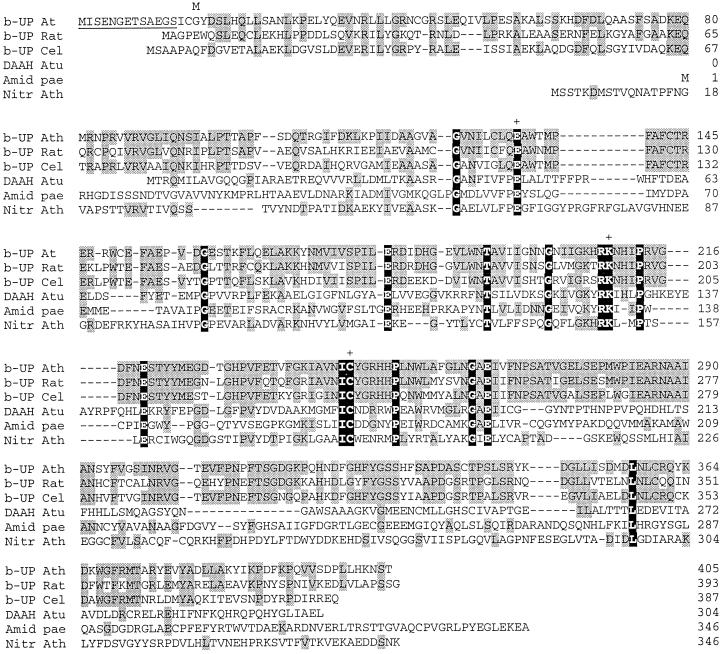
The published amino acid sequence of β-UP from rat liver (Kvalnes-Krick and Traut, 1993; GenBank accession no. Q03248) was used to identify a homologous Arabidopsis expressed sequence tag (EST; GenBank accession no. F20060). The EST was obtained from the Arabidopsis Biological Resource Center and was completely sequenced. The EST encoded a polypeptide of 392 amino acids, but no starting Met residue was apparent. However, alignment with the rat β-UP and a β-UP sequence from Caenorhabditis elegans (GenBank accession no. AAC46683) showed that the coding sequence completely overlapped these sequences (Fig. 7). An open reading frame (ORF) was therefore constructed by inserting an ATG codon at the beginning of the Arabidopsis EST. This eliminated the first three amino acid residues and replaced them with a Met residue to create an ORF encoding a polypeptide of 390 amino acids. An ORF with an identical sequence to the EST subsequently emerged from the Arabidopsis genomic sequencing project located on chromosome 5 (Nakamura et al., 1997; GenBank accession no. BAB09868). Inspection of this ORF indicated that the EST sequence was missing 13 N-terminal amino acids (shown in Fig. 7).
Figure 7.
Multiple sequence alignments of the amino acid sequences of Arabidopsis β-UP with β-UPs from rat and C. elegans, and with two amidohydrolases and a nitrilase. Arabidopsis β-UP (b-UP At) was aligned with the rat and C. elegans β-UPs (b-UP Rat and b-UP Cel; GenBank accession nos. Q03248 and AAC46683, respectively) using the MegAlign program of Lasergene (DNAstar, Madison, WI). The alignment of the N-carbamyl-d-amino acid amidohydrolase from Agrobacterium sp. (DAAH Atu; GenBank accession no. JW0082), Pseudomonas aeruginosa aliphatic amidase (Amid Pae; GenBank accession no. P11436), and Arabidopsis nitrilase I (Nitr Ath; GenBank accession no. CAA45041) sequences was based on that in Nakai et al. (2000). Residues conserved with Arabidopsis β-UP are shaded gray and residues that are completely conserved in all six sequences are shaded black. The Cys, Glu, and Lys residues that comprise the catalytic triad (Nakai et al., 2000) are marked by +. The underlined residues at the beginning of the Arabidopsis β-UP are derived from the genomic sequence (GenBank accession no. BAB09868). The M above residue 16 denotes the N terminus of the truncated polypeptide that was functionally overexpressed in E. coli.
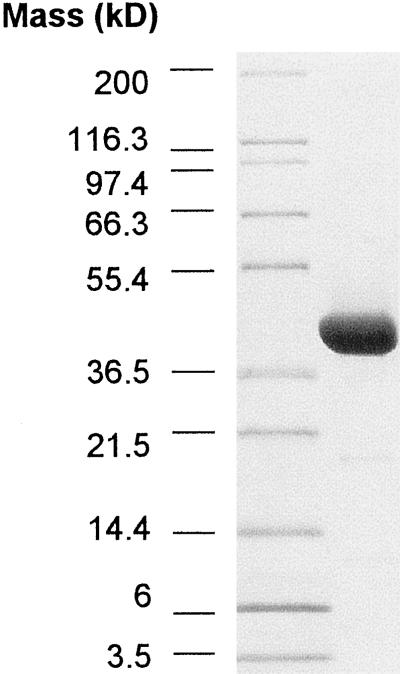
The truncated Arabidopsis gene was inserted into a pET24d expression vector and overexpressed in E. coli BL21 using T7 RNA polymerase directed expression after induction by isopropylthio-β-galactoside (Studier et al., 1990). Extracts from these E. coli contained high levels of β-UP activity, indicating that truncation of the amino acid sequence by 15 residues did not inactivate the enzyme activity. Purification of the recombinant β-UP routinely produced 15 to 20 mg of β-UP from 30 g wet weight of E. coli cells, with a final purity of >90% as judged by SDS-PAGE analysis (Fig. 8). The identity of the purified polypeptide was confirmed by accurate determination of the molecular mass of the purified protein by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. The experimentally measured molecular mass was 43,723.1 g/mol, which compared favorably with the theoretical value of 43,732.8 g/mol. The molecular masses of 13 tryptic peptides from the purified protein also closely agreed with the theoretical fragment masses calculated from the deduced amino acid sequence (data not shown).
Figure 8.
SDS-PAGE of purified recombinant Arabidopsis β-UP. Seventy-five micrograms of purified recombinant Arabidopsis β-UP was applied to a 10% to 20% (w/v) SDS-PAGE gel. The left lane contains molecular mass standards. The gel was stained with Coomassie Blue.
The addition of a thiol-reducing agent such as DTT was essential to maintain catalytic activity of recombinant β-UP and addition of glycerol to the buffers after the Mono Q chromatography step also increased the stability of the enzyme activity, similar to observations made for rat liver β-UP (Tamaki et al., 1987). The purified recombinant protein had a specific activity of 30 nmol min−1 mg−1 protein at a substrate concentration of 8 μm. This was significantly higher than the activity obtained from corn seedlings. There was some loss of overall activity during purification so this value does not represent the maximum specific activity. Attempts at purifying the protein by hydrophobic interaction chromatography column greatly reduced the recovery and specific activity of the enzyme. This lability may be associated with the hydrophobic properties noted for the rat enzyme (Matthews et al., 1992).
Isoelectric focusing of purified recombinant Arabidopsis β-UP gave a single band of pI 6.2 (compared with a value of 6.6 calculated from the deduced amino acid sequence). Size-exclusion chromatography indicated that the molecular mass of the recombinant enzyme at pH 7.3 (in the presence of 5% [v/v] glycerol) was similar to that of the native maize enzyme (>400 kD). Higher oligomeric species were also observed that eluted toward the void volume of the Superose 6 columns. Native PAGE analysis of these fractions showed three high molecular mass bands were present, but they contained a single 43-kD band by SDS-PAGE, suggesting the presence of higher aggregation states of β-UP (data not shown). Size-exclusion chromatography at pH 5.6 in the absence of glycerol gave a single peak corresponding to a dimer (approximately 90 kD).
DISCUSSION
Maize seedlings contained high levels of the pyrimidine catabolic pathway and were a convenient source of β-UP. It appears that most actively growing plant tissues have substantial pyrimidine catabolic capacity, as high levels of pathway activity were also found in Arabidopsis seedlings, and other workers have noted active pathways in a variety of other species (Wasternack, 1978; Ashihara et al., 2000). Despite the ubiquitous presence of the pathway in plants, its vital function is unclear. It may simply serve to recycle carbon and nitrogen from pyrimidine bases during conditions of high DNA and RNA turnover in actively dividing tissue. However, one end-product of the pathway, β-Ala, is an important constituent of pantothenate and coenzyme A and the betaine derivative of β-Ala is a primary osmoprotectant in some plant families such as the Plumbaginacae (Rathinasabapathi et al., 2000). β-Ala can potentially be synthesized via other pathways such as decarboxylation of Asp or transamination of malonate semialdehyde, but several studies have been unable to detect any α-decarboxylation of Asp in tissues from a variety of plant species (Naylor et al., 1958; Barnes and Naylor, 1962; Rathinasabapathi et al., 2000). The amino acid derived from thymine, β-ureidoisobutyrate, has no known significant metabolic function. In mammalian metabolism the level of excreted β-ureidoisobutyrate is used as an indicator of DNA turnover via thymine degradation (Nielsen et al., 1974), and it would be of interest to ascertain whether β-ureidoisobutyrate levels in plant tissues are similarly correlated.
Our characterization of β-UP from maize indicated that the native enzyme was relatively large, having an estimated molecular mass of 440 kD. The recombinant enzyme also had a similar high molecular mass, and size-exclusion chromatography and native PAGE analysis indicated the presence of even higher oligomerization states. Based on a subunit molecular mass of 44 kD, this suggests that the native enzyme is at least a decamer. This is larger than native rat β-UP (240-kD hexamer), but is similar in size to the rat enzyme in the presence of substrate (Matthews et al., 1992). Thus, the native state of the plant enzyme appears to be at a high oligomerization state and does not require substrate or product to induce this level of subunit oligomerization, as has been found with the rat enzyme (Matthews and Traut, 1987).
The enzyme exhibited a strict stereospecific preference for one enantiomer of the thymine-derived substrate β-ureidoisobutyrate. Thus, only one enantiomer of the product β-isobutyrate is likely to be formed from the pyrimidine catabolic pathway in vivo. In the mammalian liver, both preceding enzymes of the pathway have been shown to have a stereospecific preference to eventually yield the R isomer of β-ureidoisobutyrate (Gani and Young, 1983; Kikugawa et al., 1994). The stereospecificity of plant β-UP was also manifested in the effect of chiral inhibitors, for example R-2-phenylpropionate was 20-fold more inhibitory than the S-enantiomer.
We were interested in finding inhibitors of β-UP that would be useful to probe the in vivo metabolic function of the enzyme and give clues as to its biochemical mechanism. The inactivation of β-UP by 1,10-phenanthroline and its subsequent reactivation by added Zn2+ suggest that maize β-UP requires this metal ion as a catalytic cofactor. Hydroxamates and benzene sulfonamides are often inhibitors of hydrolases that cleave amide or urea bonds using a non-redox-active divalent metal ion such as Zn2+ as a cofactor. Examples are urease, a Ni2+-containing hydrolase (Dixon et al., 1980), Zn2+-containing metalloproteases (Browner et al., 1995), and dihydropyrimidinase, the preceding enzyme in the pyrimidine catabolic pathway, which is a Zn2+-containing hydrolase (Brooks et al., 1983). However, the compounds that we tested did not inhibit maize β-UP significantly, suggesting that β-UP is mechanistically different from these amidohydrolases or that the Zn2+ is more inaccessible to the inhibitors. We also reasoned that the enzyme may hydrolyze the ureide group via a hydroxylated tetrahedral intermediate equivalent to metal ion-assisted mechanisms of peptide hydrolysis (Hanson et al., 1989), but mimics of this transition state were also not inhibitory. This could be because the active site Cys may be used as a nucleophile to generate a thioadduct tetrahedral intermediate rather than the ureide being directly attacked by hydroxide. This mechanism has been proposed for two enzymes catalyzing similar ureide hydrolysis reactions, N-carbamoylsarcosine amidohydrolase (Romao et al., 1992) and N-carbamoyl-d-amino acid amidohydrolase (Nakai et al., 2000). It is interesting that neither of these enzymes has been reported to contain a metal ion cofactor. The best inhibitors of maize β-UP were simple aliphatic acids and arylpropionic acids, suggesting that β-UP primarily recognizes the acidic portion of the substrate and the aryl groups of these inhibitors may occupy the ureide binding region.
The published sequence of rat β-UP was used to identify an Arabidopsis EST encoding a putative β-UP that encoded a truncated polypeptide of 392 residues. Subsequent to our cloning and overexpression of this protein, an ORF identical to the EST was identified from the Arabidopsis genome sequencing project encoding a polypeptide of 405 residues. The β-UP we overexpressed was truncated by 14 amino acid residues, but the protein was functional, soluble, and as stable as the native protein indicating that these residues are not required for activity. The N-terminal residues do not appear to have the characteristics of a transit peptide, which is consistent with the reported cytosolic subcellular location of the enzyme in tomato cell cultures (Tintemann et al., 1987).
Alignment of the Arabidopsis β-UP amino acid sequence with sequences from rat β-UP and a putative β-UP from the nematode genome show that 55% to 61% of residues are conserved, respectively, particularly in the central portion of the sequences (Fig. 7). Several ESTs with high degrees of homology to the Arabidopsis β-UP sequence can also be identified from cotton, soybean, tomato, oat, and rice using BLAST searches of EST databases in GenBank, indicating that the enzyme is widespread and well conserved in plants. An additional cDNA sequence encoding a 300-amino acid residue polypeptide from tomato (GenBank accession no. CAB45873) has been annotated as a putative β-UP, but this sequence had significantly lower homology (25% identity) to all of the other β-UP sequences, including the ones from plants. This suggests that this tomato protein may not be a true β-UP, but perhaps is a closely related ureidohydrolase with a different substrate specificity.
The sequences of β-UPs from many organisms are quite highly conserved. However, there is weaker, but significant, homology to a larger class of enzymes that include nitrilases, cyanide hydratases, some aliphatic amidases, and some ureidohydrolases (Bork and Koonin, 1994; Novo et al., 1995). These enzymes all catalyze the hydrolysis of carbon-nitrogen bonds to release ammonia and the corresponding acid. Residues that are completely conserved within this class of enzymes are highlighted in Figure 7. This homology is centered around a conserved Cys that has been shown by site directed mutagenesis studies to be essential for catalytic activity (Novo et al., 1995). Based on the recently published crystal structure of N-carbamoyl-d-amino acid amidohydrolase from Agrobacterium sp. (Nakai et al., 2000), this conserved Cys residue forms a catalytic triad with an adjacent Glu and Lys residue in the active site of the enzyme. The homologous residues in the Arabidopsis β-UP sequence are Cys-246, Glu-134, and Lys 209, and these residues are also conserved in other β-UP sequences (Fig. 7). As a consequence, it is likely that Cys-246 is the active site thiol that is susceptible to iodoacetamide modification in our studies.
The presence of the conserved residues of the amidohydrolase class of enzymes accounts for the weak, but significant, homology of β-UP to many plant nitrilases such as those hydrolyzing indoleacetonitrile to indoleacetic acid. The mechanism of hydrolysis employed by auxin-producing nitrilases is therefore likely to be similar to that of β-UP and other members of this class of amidohydrolases, although the substrates are superficially quite different. These overlapping mechanisms may allow for the design of specific inhibitors or mutated enzymes with altered potencies or specificities that could affect many aspects of plant metabolism.
MATERIALS AND METHODS
Plants
Maize (Zea mays) was grown at 25°C for 5 to 8 d in potting soil under a black cloth sheet or in a darkroom to induce etiolation.
Reagents
[2-14C]Uracil, [2-14C]thymine, [2-14C]dihydrothymine, and potassium [14C]cyanate were purchased from Moravek Biochemicals (Brea, CA) or New England Nuclear (Boston) with initial specific activities of about 56 mCi mmol−1. 2-Aminosulfonamidoxypropionic acid and 2-aminophos-phoramidoxypropionic acid were provided by Dr. M. Hopkins and 2-(2, 6-dinitrophenoxy)-propionic acid was provided by R. Johnston of Dow AgroSciences Agricultural Chemical Discovery Research (Indianapolis). All other chemicals were purchased from Sigma Chemical (St. Louis).
Partial Purification of Maize β-UP
Shoot tissue from 5- to 8-d-old etiolated maize seedlings (approximately 250 g) was homogenized in 3 volumes of 0.1 m potassium phosphate, pH 7.0, 1 mm MgCl2, and 2 mm DTT (buffer A). The homogenate was filtered through four layers of cheesecloth and was made 30% (w/v) in ammonium sulfate. After standing for 20 min, the homogenate was centrifuged. The pellet was discarded and the supernatant made 55% (w/v) in ammonium sulfate. After standing for 20 min, the suspension was centrifuged. The supernatant was discarded and the pellet was dissolved in 20 mm Tris, pH 7.2, 1 mm MgCl2, and 2 mm DTT (buffer B). The solution was dialyzed overnight in a 50,000-kD cut-off dialysis membrane against three changes of 2 L of buffer B. The dialysate was clarified by centrifugation and the supernatant was applied to a 2.5 × 15 cm column of diethylaminoethyl-Sepharose equilibrated in buffer B. The column was washed with 45 mL of buffer B at 1.5 mL/min, and was then eluted with 0% to 100% gradient of 0.5 m KCl in buffer B over 4 h at the same flow rate. Fractions of 5 mL were collected and assayed for β-UP activity. Active fractions (eluting at buffer volumes of 200–240 mL after gradient initiation) were pooled and concentrated in a CentriPrep-100 (Amicon, Beverly, MA) to approximately 3 mL. The pooled, concentrated enzyme was made 10% (v/v) in glycerol, frozen at −70°C, and was used for β-UP assays and characterization. All steps were performed at 4°C.
The molecular mass of maize β-UP was estimated by applying a 0.5-mL sample of partially purified β-UP onto a Superose 12 column (Pharmacia, Piscataway, NJ) equilibrated in buffer A at a flow-rate of 0.4 mL/min. Fractions of 0.4 mL were collected and assayed for β-UP activity. The column was calibrated under the same elution conditions with Bio-Rad (Hercules, CA) native Mr standards.
Synthesis of 14C-Labeled Substrates
A convenient and accurate method to assay β-UP activity is to monitor the release of radiolabeled CO2 from substrate labeled in the ureido group. Preparation of [5-14C]β-ureidopropionate was based on the method described in Traut and Loechel (1984). In a screw-top Eppendorf vial, 0.02 mL of 0.5 m β-Ala was mixed with 0.02 mL of 0.5 m [14C]potassium cyanate (5.6 mCi mmol−1) and was incubated at 50°C for 3 h. The solution was then spotted on to silica thin-layer chromatography plates (2.5 μL/lane) and the plates were developed in 70% (v/v) isopropanol/water. [5-14C]β-Ureidopropionate was located by phosphorimaging of the plate using a phosphorimager (Molecular Dynamics, Sunnyvale, CA) and the spots were scraped off and eluted with four washes of 70% (v/v) isopropanol/water. The solution was concentrated under vacuum to approximately 1 mL and stored at −20°C. A similar protocol using β-aminoisobutyrate as starting material was used to synthesize racemic [6-14C]β-ureidoisobutyrate. Both radiolabeled substrates were greater than 98% pure by thin-layer chromatography analysis.
Radiometric Assay of β-UP
β-UP assays were performed at 25°C by mixing 0.94 mL of buffer A, 0.02 mL of β-ureidopropionase extract, and 0.02 mL of a test solution in a 20-mL scintillation vial. The mixture was pre-equilibrated for 30 min and the reaction was initiated by addition of 0.02 mL of [5-14C]β-ureidopropionate (approximately 100,000 dpm, 5.7 mCi mmol−1, corresponding to a final substrate concentration of 8 μm in the assay). The vial was immediately capped with a rubber septum that had a small plastic well (Kontes part no. 882320) containing 0.05 mL of 4 n KOH attached to it. The reaction vials were incubated at 30°C for 30 min, then 0.25 mL of 4 n perchloric acid was injected into the vial using a repeating pipettor with a needle attached. The vials were incubated for a further 20 min, the septum was removed, and the well was placed into another scintillation vial containing 15 mL of Hionic-Fluor (Packard, Meriden, CT). After thorough shaking, the vials were counted in a liquid scintillation counter to quantitate the amount of 14CO2 released. (Background reactions containing no enzyme additions were run in all experiments and subtracted from the enzyme reaction values. Nonenzymatic decomposition was routinely less than 0.5% of the added radioactivity.) Concentrations that inhibited the reaction by 50% (I50 values) were determined using a dilution series of test compounds in this assay. Standard kinetic analyses and curve-fitting to derive kinetic constants was done using Grafit 4 (Erithacus Software Ltd., Staines, UK).
Although the radiometric 14CO2 capture assay was accurate and sensitive, it was not convenient for the rapid determination of enzyme activity in large numbers of samples (e.g. column fractions) due to the manipulations involved in assembling the vials with septa and CO2 trapping wells. A simpler, qualitative assay format was devised that monitored the depletion of substrate as follows: 65 μL of the solution to be assayed was placed in a 7-mL scintillation vial and 65 μL of buffer A was added. The reaction was started by addition of 20 μL of [5-14C]β-ureidopropionate (approximately 10,000 dpm; 5.7 mCi/mmol). The vials were incubated at 30°C for 30 min, then 0.05 mL of 4 n perchloric acid was added. After leaving the vials uncapped in a fume hood for 30 min to allow 14CO2 to be released, 5 mL of scintillation cocktail was added to the vials, mixed thoroughly, and the residual radioactivity was counted in a liquid scintillation counter for 1 min. The presence of enzyme activity could be readily determined by the reduction in the amount of residual substrate remaining in the vial.
Protection of β-UP from Iodoacetamide Modification
A sample of partially purified maize β-UP was exchanged into 0.1 m potassium phosphate, pH 7, by passage over a PD10 column (Pharmacia) to remove reductants. Then iodoacetamide was added to 1 mm final concentration in the presence or absence of 10 mm β-ureidopropionate. After 30 min, DTT was added to 10 mm final concentration and the samples were dialyzed against two changes of 2 L of buffer A. The samples were then assayed for β-UP activity. Control samples untreated with iodoacetamide were run in parallel.
In Vivo Assay of the Pyrimidine Catabolic Pathway
Five-day-old etiolated maize shoots were sliced into 3- to 5-mm segments and briefly vacuum infiltrated with water. Two hundred milligrams fresh weight of tissue was then placed in 20-mL scintillation vials containing 4 mL of water and 0.4 μCi [2-14C]uracil, thymine, or dihydrothymine was added. The vial was immediately capped with a rubber septum that had a well containing 0.04 mL of 4 m KOH attached to it. The reaction vials were incubated at 30°C for 2 h, then 1 mL 4 m perchloric acid was injected into the vial using a repeating pipettor with a needle attached. The vials were incubated for a further 30 min, then the septum was removed, and the well was placed into a scintillation vial containing 15 mL Hionic-Fluor. After thorough shaking, the radioactivity in the vials was counted in a liquid scintillation counter to quantitate the amount of CO2 released.
Gene Cloning and Overexpression of Arabidopsis β-UP
An Arabidopsis EST with significant homology to rat β-UP was obtained from the Arabidopsis Biological Resource Center (Columbus, OH; stock no. ATTS6099) and was completely sequenced. DNA from this clone was purified and the insert was PCR amplified using the forward primer TCTTCTCATATGTATGATTCGCTGCATCAA and the reverse primer AGAAGAGTCGACTTATGTAGAATTCTTGTG. This added a Met codon and an NdeI restriction site to the 5′ end of the gene and a SalI restriction site to the 3′ end. It also deleted the nucleotides ATTTGCGGC from the 5′ end of the coding sequence of the EST. The amplified fragment was inserted into the intermediate vector pCR 2.1 (Invitrogen, Carlsbad, CA) and was then transferred as an NdeI/SalI fragment into the expression vector pET24a (Novagen, Madison, WI). The insert was sequenced to ensure no mutations were introduced during the cloning process. Overexpression of the recombinant protein from this vector was performed in E. coli BL2 with isopropylthio-β-galactoside induction using standard protocols (Studier et al., 1990). E. coli cell pellets were routinely frozen in liquid nitrogen and stored at −80°C for up to a month without any apparent loss in yield.
Purification of Recombinant Arabidopsis β-UP
E. coli BL21 cells containing recombinant β-UP were pelleted by centrifugation and resuspended in one volume of extraction buffer containing 100 mm Tris-HCl, pH 7.3, 20 mm DTT, 1 mm EDTA, 10 mm MgCl2, 1 mm KCl, and 10 μm leupeptin. The cells were then ruptured in a French press under a pressure of 20,000 psi. (Initial attempts at extracting β-UP using lysozyme to lyse the E. coli cells produced relatively low yields of β-UP.) DNase was added to the cell extract to a final concentration of 0.02 mg/mL and was incubated at room temperature for 5 min. The extract was centrifuged to remove cell debris and protamine sulfate (88 μL of a 5% [w/v] solution) was added to each milliliter of the supernatant while stirring at 4°C. The precipitate was removed by centrifugation and the supernatant was brought to 60% saturation with saturated ammonium sulfate (pH 7.0), and then stirred for 20 min at 4°C. The precipitated material was collected by centrifugation and the pellet was dissolved in 20 mm Tris-HCl, pH 7.3, containing 20 mm DTT, and was then dialyzed for 16 h against the same buffer using 100-kD cut-off dialysis tubing. The dialyzed sample was applied to a Mono Q 16/10 column (Pharmacia) equilibrated with 20 mm Tris-HCl, pH 7.3, 20 mm DTT, and 5% (v/v) glycerol (buffer C). The protein was eluted at 3 mL/min with a 0% to 30% gradient of 1 m KCl in buffer C over 50 min. Fractions containing β-UP activity were pooled and concentrated using Centriprep-50 centrifugal concentrators (Amicon). The concentrated sample was exchanged into 20 mm malonate buffer, pH 5.6, containing 20 mm DTT and 5% (v/v) glycerol (buffer D) using a Fast Desalt column (Pharmacia) at 1 mL/min and applied to a Mono S 5/5 column (Pharmacia) equilibrated with buffer D. The protein was eluted at 0.5 mL/min with a 0% to 30% gradient of 1 m KCl in buffer D over 30 min. The fractions containing β-UP activity were pooled, concentrated, and applied to two Superose 6 columns (Pharmacia) connected in series that were equilibrated with buffer C at a flow rate of 0.3 mL/min. Active fractions were pooled, concentrated, and stored at −70°C.
Protein Analysis
SDS-PAGE was performed using 10% to 20% (w/v) Tris-Gly gradient gels and Mark 12 molecular mass standards from Novex (San Diego). Native gel electrophoresis was performed using 4% to 20% (w/v) SDS-Tris-Gly gradient gels and Pharmacia HMW electrophoresis calibration standards. Isoelectric focusing was performed using Novex pH 3–7 gels. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis was performed by L. Alward and tryptic digestion was performed by V. Patterson of the Dow AgroSciences Biotechnology Group.
ACKNOWLEDGMENT
We thank Dr. David McCaskill of Dow AgroSciences for helpful comments.
LITERATURE CITED
- Ashihara H, Stasolla C, Loukanina N, Thorpe TA. Purine and pyrimidine metabolism in cultured white spruce (Picea glauca) cells: metabolic fate of 14C-labeled precursors and activity of key enzymes. Physiol Plant. 2000;108:25–33. [Google Scholar]
- Barnes RL, Naylor NW. Formation of beta-alanine by pine tissues supplied with intermediates in uracil and orotic acid metabolism. Plant Physiol. 1962;37:171–175. doi: 10.1104/pp.37.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Koonin EV. A new family of carbon-nitrogen hydrolases. Protein Sci. 1994;3:1344–1346. doi: 10.1002/pro.5560030821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks KP, Jones EA, Kim BD, Sander EG. Bovine liver dihydropyrimidine amidohydrolase: purification, properties, and characterization as a zinc metalloenzyme. Arch Biochem Biophys. 1983;226:469–483. doi: 10.1016/0003-9861(83)90316-8. [DOI] [PubMed] [Google Scholar]
- Browner MF, Smith WW, Castelhano AL. Crystal structures of matrilysin-inhibitor complexes. Biochemistry. 1995;34:6602–6610. doi: 10.1021/bi00020a004. [DOI] [PubMed] [Google Scholar]
- Dixon NE, Hinds JA, Fihelly AK, Gazzola C, Winzor DJ, Blakeley RL, Zerner B. Jack bean urease (EC 3.5.1.5): IV. The molecular size and the mechanism of inhibition by hydroxamic acids: spectrophotometric titration of enzymes with reversible inhibitors. Can J Biochem. 1980;58:1323–1334. doi: 10.1139/o80-180. [DOI] [PubMed] [Google Scholar]
- Gani D, Young DW (1983) Stereochemistry of the dihydrouracil dehydrogenase reaction in metabolism of uracil to β-alanine. J Chem Soc Chem Commun 576–578
- Hanson JE, Kaplan AP, Bartlett PA. Phosphonate analogs of carboxypeptidase A substrates are potent transition-state analog inhibitors. Biochemistry. 1989;28:6294–6305. doi: 10.1021/bi00441a022. [DOI] [PubMed] [Google Scholar]
- Kikugawa M, Kaneko M, Fujimoto-Sakata S, Maeda M, Kawasaki K, Takagi T, Tamaki N. Purification, characterization and inhibition of dihydropyrimidinase from rat liver. Eur J Biochem. 1994;219:393–399. doi: 10.1111/j.1432-1033.1994.tb19951.x. [DOI] [PubMed] [Google Scholar]
- Kvalnes-Krick KL, Traut TW. Cloning, sequencing, and expression of a cDNA encoding β-alanine synthase from rat liver. J Biol Chem. 1993;268:5686–5693. [PubMed] [Google Scholar]
- Lesley SM, Maretzki A, Nickell LG. Incorporation and degradation of carbon-14 and 3H-labeled thymidine by sugarcane cells in suspension culture. Plant Physiol. 1980;65:1224–1228. doi: 10.1104/pp.65.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews MM, Liao W, Kvalnes-Krick KL, Traut TW. β-Alanine synthase: purification and allosteric properties. Arch Biochem Biophys. 1992;293:254–263. doi: 10.1016/0003-9861(92)90393-b. [DOI] [PubMed] [Google Scholar]
- Matthews MM, Traut TW. Regulation of N-carbamoyl-β-alanine amidohydrolase, the terminal enzyme in pyrimidine catabolism, by ligand-induced change in polymerization. J Biol Chem. 1987;262:7232–7237. [PubMed] [Google Scholar]
- Nakai T, Hasegawa T, Yamashita E, Yamamoto M, Kumasaka T, Ueki T, Nanba H, Ikenaka Y, Takahashi S, Sato M, Tsukihara T. Crystal structure of N-carbamyl-d-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure (London) 2000;8:729–738. doi: 10.1016/s0969-2126(00)00160-x. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Sato S, Kaneko T, Kotani H, Asamizu E, Miyajima N, Tabata S. Structural analysis of Arabidopsis thalianachromosome 5: III. Sequence features of the regions of 1,191,918 bp covered by seventeen physically assigned P1 clones. DNA Res. 1997;4:401–414. doi: 10.1093/dnares/4.6.401. [DOI] [PubMed] [Google Scholar]
- Naylor NW, Rabson R, Tolbert NE. Aspartic-14-C acid metabolism in leaves, roots and stems. Physiol Plant. 1958;11:537–547. [Google Scholar]
- Nielsen HR, Sjolin KE, Nyholm K, Baliga BS, Wong R, Borek E. Beta-aminoisobutyric acid, a new probe for the metabolism of DNA and RNA in normal and tumorous tissue. Cancer Res. 1974;34:1381–1384. [PubMed] [Google Scholar]
- Novo C, Tata R, Clemente A, Brown PR. Pseudomonas aeruginosa aliphatic amidase is related to the nitrilase/cyanide hydratase enzyme family and Cys166 is predicted to be the active site nucleophile of the catalytic mechanism. FEBS Lett. 1995;367:275–279. doi: 10.1016/0014-5793(95)00585-w. [DOI] [PubMed] [Google Scholar]
- Ogawa J, Shimizu S. β-Ureidopropionase with N-carbamoyl-α-L-amino acid amidohydrolase activity from an aerobic bacterium, Pseudomonas putidaIFO 12996. Eur J Biochem. 1994;223:625–630. doi: 10.1111/j.1432-1033.1994.tb19034.x. [DOI] [PubMed] [Google Scholar]
- Rathinasabapathi B, Sigua C, Ho J, Gage DA. Osmoprotectant β-alanine betaine synthesis in the Plumbaginaceae: S-adenosyl-l-methionine dependent N-methylation of β-alanine to its betaine is via N-methyl and N,N-dimethyl β-alanines. Physiol Plant. 2000;109:225–231. [Google Scholar]
- Romao MJ, Turk D, Gomis-Rueth FX, Huber R, Schumacher G, Moellering H, Ruessmann L. Crystal structure analysis, refinement and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobactersp. at 2.0 Å resolution. J Mol Biol. 1992;226:1111–1130. doi: 10.1016/0022-2836(92)91056-u. [DOI] [PubMed] [Google Scholar]
- Slabas AR, MacDonald G, Lloyd CW. Thymidine metabolism and the measurement of the rate of DNA synthesis in carrot suspension cultures: evidence for a degradation pathway for thymidine. Plant Physiol. 1980;65:1194–1198. doi: 10.1104/pp.65.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tamaki N, Mizutani N, Kikugawa M, Fujimoto S, Mizota C. Purification and properties of β-ureidopropionase from the rat liver. Eur J Biochem. 1987;169:21–26. doi: 10.1111/j.1432-1033.1987.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Tintemann H, Wasternack C, Helbing D, Glund K, Hause B. Pyrimidine degradation in tomato cell suspension cultures and in Euglena gracilis: localization of enzymes. Comp Biochem Physiol B Comp Biochem. 1987;88B:943–948. [Google Scholar]
- Traut TW, Jones ME. Uracil metabolism-UMP synthesis from orotic acid or uridine and conversion of uracil to β-alanine: enzymes and cDNAs. Prog Nucleic Acid Res Mol Biol. 1996;53:1–78. doi: 10.1016/s0079-6603(08)60142-7. [DOI] [PubMed] [Google Scholar]
- Traut TW, Loechel S. Pyrimidine catabolism: individual characterization of the three sequential enzymes with a new assay. Biochemistry. 1984;23:2533–2539. doi: 10.1021/bi00306a033. [DOI] [PubMed] [Google Scholar]
- Tsai CS, Axelrod B. Catabolism of pyrimidines inrape seedlings. Plant Physiol. 1965;40:39–44. doi: 10.1104/pp.40.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreken P, van Kuilenburg ABP, Hamajima N, Meinsma R, van Lenthe H, Gohlich-Ratmann G, Assmann BE, Wevers RA, van Gennip AH. cDNA cloning, genomic structure and chromosomal localization of the human BUP-1 gene encoding β-ureidopropionase. Biochim Biophys Acta. 1999;1447:251–257. doi: 10.1016/s0167-4781(99)00182-7. [DOI] [PubMed] [Google Scholar]
- Wasternack C. Degradation of pyrimidines: enzymes, localization and role in metabolism. Biochem Physiol Pflanz. 1978;173:467–499. [Google Scholar]
- Wasternack C. Degradation of pyrimidines and pyrimidine analogs: pathways and mutual influences. Pharmacol Ther. 1980;8:629–651. doi: 10.1016/0163-7258(80)90079-0. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Krauss G-J, Reinbothe H. Degradation of pyrimidines in Euglena gracilis: III. Ratio of uracil to thymine degradation. Plant Sci Lett. 1977;10:121–128. [Google Scholar]
- Wasternack C, Lippmann G, Reinbothe H. Pyrimidine-degrading enzymes: purification and properties of β-ureidopropionase of Euglena gracilis. Biochim Biophys Acta. 1979;570:341–351. doi: 10.1016/0005-2744(79)90154-2. [DOI] [PubMed] [Google Scholar]