Summary
Although it is generally believed that genetic and developmental factors play critical roles in pathogenesis of schizophrenia, however, the precise etiological mechanism of schizophrenia remains largely unknown. Over past decades, miRNAs have emerged as an essential post‐transcriptional regulator in gene expression regulation. The importance of miRNA in brain development and neuroplasticity has been well‐established. Abnormal expression and dysfunction of miRNAs are known to involve in the pathophysiology of many neuropsychiatric diseases including schizophrenia. In this review, we summarized the recent findings in the schizophrenia‐associated dysregulation of miRNA and functional roles in the development and pathogenesis of schizophrenia. We also discussed the potential therapeutic implications of miRNA regulation in the illness.
Keywords: miRNAs, neuroplasticity, schizophrenia, single nucleotide polymorphism
1. INTRODUCTION
Schizophrenia is a disabling disorder characterized by complex symptoms of abnormal social behavior, mental disturbance, thinking disruption, and impaired cognition. It affects nearly 1% of the world population.1 It is generally believed that the interplay between developmental and environmental factors contributes to the pathogenesis of this disease; however, the precise etiology remains unknown. From the view of pathophysiology, abnormal neurotransmission, particularly disturbed dopamine‐glutamate transmission and altered prefrontal dysfunction may play an important role in the majority of the symptoms of schizophrenia.2, 3, 4 Recent emerging evidences suggest that small noncoding RNAs known as microRNAs (miRNAs) are involved in pathogenesis and pathological process of the illness.5, 6, 7, 8, 9 miRNAs are a class of ~22 nucleotides noncoding RNAs, which can functionally silence gene in a post‐transcriptional manner by complementary base‐pairing with target mRNA. Each miRNA can potentially regulate many downstream target genes through intracellular gene silencing machinery.10, 11 The importance of miRNA regulatory network in neuronal development and brain function has been widely studied. Its potential roles have become the focal point in understanding the pathogenesis and development of many neuropsychiatric diseases including schizophrenia. In this review, we summarize the recent development in miRNA regulation and potential functional implications in schizophrenia, with emphasizing on the recent findings of miRNA as novel biological markers and potential therapeutic approach for the disease.
2. miRNA BIOGENESIS AND FUNCTIONS IN THE CNS
The first miRNA was discovered in Caenorhabditis elegans in 1993.12 It took 7 years for the second miRNA let‐7 to be reported.13, 14 Since 2000, thousands of miRNAs have been identified in animals, plants, virus, as well as in mammalian central nervous system. The functional roles of miRNAs have been widely studied in cell proliferation, differentiation, and apoptosis.15, 16, 17, 18, 19 miRNAs are produced from either their own genes or from introns of coding or noncoding genes. Like the protein‐coding genes, miRNAs are transcribed into primary transcripts (pri‐miRNA) from genomic DNA by RNA polymerase II or RNA polymerase III. The pri‐miRNAs usually possess a 5′ CAP and a 3′ poly A tail and can be folded into a double‐stranded RNA hairpin with a stem‐loop structure in the nucleus.20, 21 The stem‐loop structure can be recognized by the microprocessor complex containing RNaseIII endonuclease Drosha and nuclear protein DGCR8 (DiGeorge syndrome critical region 8) in vertebrates. Followed the cleavage of 5′ CAP and a 3′ poly A tail by Drosha, the precursor‐miRNA (pre‐miRNA) with 70~100 nt hairpin shaped is released and exported into the cytoplasm, where the pre‐miRNA is cleaved by type III ribonuclease Dicer and resulted in a ~22nt double‐stranded RNA duplex: miRNA/miRNA*22, 23, 24, 25, 26 (as shown in Figure 1). During the unwinding process, one strand of the duplex, the mature miRNA, can be loaded into the RNA‐induced silencing complex (RISC), whereas the other strand is usually degraded.27 In addition to miRNA, RISC also contains Dicer, the RNA‐binding protein Argonaute (AGO), and the adaptor protein TRBP.28, 29 The miRNA‐containing RISC is able to recognize the complementary sequences in the 3′UTR of target mRNAs, consequently results in translational repression or degradation of the target mRNAs.30 The recognition of the miRNA with its target mRNAs is guided by the initial 2‐7 bases of miRNA.31 Because of the complementary silencing mechanism, one miRNA could target to hundreds or even thousands of mRNAs, whereas different miRNAs can also bind to the same mRNA, which forms a complex miRNAs‐genes regulation network.
Figure 1.
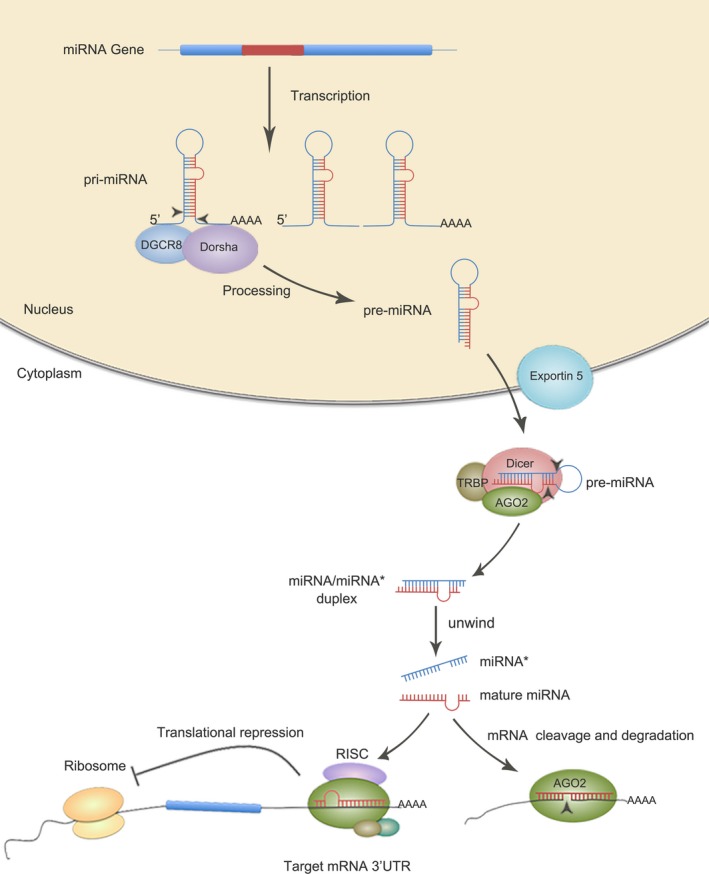
Schematic overview of miRNA biogenesis and function miRNAs are transcribed into primary transcripts (pri‐miRNA) from genomic DNA by RNA polymerase. PrimiRNAs are cleaved in the nucleus by Drosha to generate miRNA precursor (pre‐miRNAs), then exported in the cytoplasm by Exportin‐5and further cleaved by Dicer to produce a ~22nt double‐stranded RNA duplex: miRNA/miRNA*. One strand of the duplex, the mature miRNA can associate to the RNA‐induced silencing complex (RISC) complex and guide translational repression of target mRNAs, whereas the other strand is usually degraded
Approximately, nearly 70% of the known miRNAs are expressed in the mammalian brain.32, 33, 34 Bak et al studied the miRNA expression profiles from 13 distinct areas of the adult mouse brain. They detected 44 miRNAs displaying tissue‐specific enrichment, suggesting that CNS miRNAs may be associated with specific function within respective brain region. Although the mature miRNA sequences are conservative between mouse and zebrafish, more than 50% of the identified mouse CNS‐enriched miRNAs showed different expression patterns.18 Recently, Zaits et al presented a comprehensive assessment on spatio‐temporal miRNA expression in 18 human donor brains with age ranged from infancy to adolescence. Using RNA sequencing, they reported the expression patterns of miRNAs across both temporal (developmental stages) and spatial dimensions (prefrontal cortex, hippocampus, and cerebellum). The data showed a dramatic shift in miRNA expression shortly after birth. It is noted that most of their target genes for those miRNAs are related to transcriptional regulation, neurodevelopmental processes and common neurodevelopmental disorders, highlighting the central function of these miRNAs in brain transcription networks.35 The temporal or brain region‐specific manner of miRNA expression may imply the important roles in brain development and neuronal differentiation.36, 37, 38, 39, 40
The first evidence depicted miRNA role in the CNS was from the Dicer‐deficient study in zebrafish in which the animals were found unable to produce matured miRNAs. Dicer mutant zebrafish displayed severe brain developmental abnormality and other malformations including heart. Injection of miR‐430, a brain abundant miRNA, was found to rescue many of these defects.41 In mammalians, Dicer mutant mice died at embryonic day 7.5 before neurulation.42 Conditional knockout of Dicer in cortex and hippocampus results in the phenotypes of microcephaly, alterations in dendritic branch elaboration and dendritic spine length.43 Since then, there are numerous reports that revealed the roles of various miRNA in neuronal development and differentiation. The important functional roles of miRNA in neurogenesis, neuroprotection, survival, and pathogenesis of neuropsycho‐disorders have been also extensively studied and reviewed.7, 44, 45, 46, 47, 48, 49 In addition, alterations of miRNA are reported to be associated with many neuropsychiatric diseases. For instance, selective Dicer depletion in cerebellar Purkinje neurons results in neurodegenerative change, a pathological process similar to progressing neurodegenerative diseases such as Alzheimer's and Parkinson's diseases. More specifically, inactivation of Dicer in the midbrain dopaminergic neurons results in progressive neuron death. This phenotype is significantly rescued by transfection of miRNAs obtained from embryonic mouse midbrain, suggesting that miRNAs play essential roles in midbrain dopamine neuronal differentiation and survival.50, 51 There are other reports depicted various roles of miRNAs on dopaminergic neuron.52, 53, 54, 55 Our group also reported the critical roles of miR‐let‐7c‐5p and miR‐3473b in the protective effects of ischemic brain damage via regulating the microglia activation.56 Given the importance of miRNA in brain functional regulation, it is not surprising that dysregulation of miRNAs may contribute to a pathogenesis and pathological process of many neuropsychiatric disorders. Here, we will further discuss the role of miRNA in schizophrenia.
3. DYSREGULATION OF mIRNAS IN SCHIZOPHRENIA
3.1. Altered miRNA expression in schizophrenia
Many studies analyzed the miRNA expression profiles using the postmortem brain samples. Perkins et al examined the miRNA profiles in postmortem prefrontal cortex (PFC) from 13 schizophrenia patients by a customized microarray (miRBase version 7.0). In comparison with 21 psychiatrically unaffected individuals, they found that 15 miRNAs were differentially expressed, in which 14 miRNAs were downregulated and one miRNA (miR‐106b) was upregulated in schizophrenia patients.57 Significant upregulation of miR‐181b was also reported in cortical gray matter from the superior temporal gyrus (STG), a brain region involved in the generation of auditory hallucinations of schizophrenia.58 Two target genes of miR‐181b, the calcium sensor gene visinin‐like 1 (VSNL1) and the ionotropic glutamate receptor subunit (GRIA2) were identified.58 The same group further observed a significant schizophrenia‐associated increase in global miRNA expression in postmortem tissue in both the STG and the dorsolateral prefrontal cortex (DLPFC).59 This elevated expression of miRNA is suggested to attribute to an elevation of primary miRNA processing and upregulation of the microprocessor component DGCR8.59 However, data from Berveridge's study are not consistent with Perkin's report. Some miRNAs such as miR‐26b, miR‐29c and miR‐195 reported to be downregulated in Perkins’ study were, however, found to be upregulated in Berveridge's results. Reports on alterations in miRNA expression in schizophrenia are continuously to accumulate. However, the results of miRNA expression profiles are somewhat controversial.58, 59, 60, 61, 62, 63, 64, 65, 66, 67 The discrepancy of miRNA expression data may be explained by differences in sample size, therapeutic protocol, gender factors, and technical application, such as different miRNA extraction methods and miRBase test version. In addition, the temporal expression patterns of miRNAs in human brains could also contribute to the conflict results.68, 69
To explore the potential role of miRNA as biomarkers, several studies focused on the peripheral blood miRNA expression in schizophrenia patients. Gardiner et al examined miRNA expression in peripheral blood mononuclear cells (PBMCs) from 112 schizophrenia patients and 76 controls. They identified a significant reduction in 83 miRNAs in schizophrenia patients.70 Lai et al71 also carried an analysis of genome‐wide miRNA expression profile in the mononuclear leukocytes from schizophrenia patients and control groups. Seven miRNAs (upregulated: miR‐34a, miR‐449a, miR‐564, miR‐548d, miR‐572 miR‐652; downregulated: miR‐432) associated with negative symptoms and cognitive performance were identified as predictive biomarkers of schizophrenia. Interestingly, they found that the expressions of the seven miRNAs in PBMC were not affected by 2 months of hospitalization, even with a significant improvement of clinical symptoms.72 It is also noted that the altered expression of hsa‐miR‐34a and hsa‐miR‐548d in the blood was not presented in the brain samples.72 Wei et al73 also screened the plasma miRNA profiles in a larger sample and identified eight differentially expressed miRNAs (miR‐122, miR‐130a, miR‐130b, miR‐193a‐3p, miR‐193b, miR‐502‐3p, miR‐652, miR‐886‐5p) in patients with schizophrenia. They also found that the increased levels of miR‐130b and miR‐193a‐3p in patient plasma disappeared after 1 year of treatment with aripiprazole and risperidone and proposed the potential role as biomarkers for prognosis of schizophrenia. In addition, Gallego et al compared the miRNA expression profiles between cerebrospinal fluid (CSF) and the whole blood from schizophrenia patients and healthy controls. However, the miRNA expression levels in CSF and blood were poorly correlated.74 Although the potential biomarker of miRNAs in schizophrenia patients have been suggested, it is clear that more studies are needed. Indeed, serum miRNAs measurement provides a feasible way for clinical diagnosis and prognosis of schizophrenia including the therapeutic responses.
3.2. Dysregulation of miRNA biogenesis in schizophrenia
Aberrations in miRNA biogenesis and processing pathway are considered to be involved in the pathological process of schizophrenia. Generation of 22q11.2‐deletion mouse was one of the strongest evidence that associated schizophrenia with dysregulated miRNA biogenesis.75 Microdeletion of human 22q11.2 locus leads to behavioral and cognitive deficits, and a high risk of schizophrenia. Stark et al generated a mouse strain (Df(16)A+/− mice) carrying a hemizygous chromosomal deficiency corresponding to human 22q11.2 microdeletion, and observed schizophrenia‐like behaviors in mice. They reported that altered miRNA biogenesis in the brain and found that the dysregulated biogenesis was due to haploinsufficiency of the Dgcr8 gene, which contributes to the abnormal behavior and neuronal deficits in schizophrenia.75 In further study, the same group found a drastic reduction in miR‐185 in Df(16)A+/− mice, which resides within the 22q11.2 locus. The reduction (~70%‐80%) of miR‐185 in both hippocampus and PFC was more than expected by a hemizygous deletion (~50%) and contributed to the deficits of dendritic and spine development. miR‐185 was demonstrated to repress a previously uncharted inhibitor Mirta22 (miRNA target of the 22q11.2microdeletion) which resides in the Golgi apparatus with higher expression in prenatal brain. Reduction in miR‐185 expression in the brains of Df(16)A(+/−) mice leads to the sustained derepression of Mirta22 after birth and results in the structural alterations in the hippocampus and cognitive function.5 Schofield et al76 examined the Dgcr8+/− mice and found that the reduced expression of Dgcr8 and miRNA emerged over postnatal development during pyramidal neuron maturation rather than in neonatal mice. Altered electrophysiological properties, decreased complexity of basal dendrites, and reduced excitatory synaptic transmission were observed in layer V pyramidal neurons in Dgcr8+/− mice.76, 77 Earls et al reported an age‐dependent increase in long‐term potentiation in the hippocampus of Dgcr8+/− mice. This increase was attributed to the loss of two miRNAs (miR‐25 and miR‐185) which target the sarco (endo) plasmic reticulum Ca2+ ATPase (SERCA2). Elevated expression of SERCA2 was found in postmortem sample in PFC and hippocampus of schizophrenia patients.78 The decreased expressions in Dgcr8 and miR‐185 were also confirmed in peripheral leukocytes in individuals with 22q11 deletion syndrome.79 These findings suggest a pathogenic association between miRNAs and schizophrenia.78, 80
Dicer is another key gene in miRNA processing pathway. Deletion of Dicer in both zebrafish and mice displays severe defects in brain development.9 A genome‐wide scan for finding copy number variations (CNVs) in schizophrenia identified a de novo duplication in one individual that included DICER1 gene.81 In a case‐control analysis of Chinese population, the SNP (rs3742330) in DICER was reported to be highly associated with schizophrenia risk.82 The analysis for postmortem brain tissue of DLPFC in schizophrenia patients revealed an upregulation in Dicer expression as well as a global increase in miRNA expression.59, 64 Beveridge et al reported that global miRNA expression was at highest level in early year and declined significantly after adolescence. Dicer and Exportin‐5 were also age‐dependent but were not correlated with miRNA expression across the lifespan.83 They proposed that in schizophrenia, neurodevelopment‐associated miRNAs remain at a high level after adolescence instead of declining to a lower level in normal subject. The high levels of miRNA expression may cause inappropriate gene silencing that may consequently contribute to abnormal behaviors in schizophrenia.83 In support, Konopka et al84 found the enhanced learning and memory in adult mice with tamoxifen‐induced deletion of Dicer1 gene. However, when using knockout mouse models, some consideration should be taken into account as pointed out by Rajman.7 Indeed, Dicer1 has been proved to be essential for cell survival and embryonic development. Dicer1 gene deletion is often associated with massive cell apoptosis and developmental abnormalities,43, 50, 85, 86 which may complicate the phenotypes and relevance to schizophrenia. On the other hand, not all miRNAs are dependent on Dicer. Some miRNAs are generated through diverse Drosha‐independent and Dicer‐independent mechanisms.87, 88, 89 Therefore, it is possible that some phenotype observed in Dicer1‐deficient mice might be not directly related to the loss of miRNAs, and some functional miRNAs might be missed in these Dicer1 knockout models. It should be bear in mind that the miRNA network is a complicate regulatory system and caution should be taken to interpret functional role of a specific miRNAs underling a phenotype in Dicer knockout model. In this regards, miRNA families or clusters may help to provide more easily interpretable information.7
3.3. miRNA‐associated single nucleotide polymorphisms in schizophrenia
Single nucleotide polymorphisms (SNPs) or copy number variations (CNVs) are common DNA sequence variations within populations, which occur more frequently in noncoding region and contribute to human disease susceptibility.90, 91, 92 Using a case‐control study, several SNPs of miRNA genes associated with schizophrenia have been reported. SNP rs17578796 in miR‐206 showed a significant association with schizophrenia in Scandinavian (Danish and Norwegian) samples.93 Variant ss178077483 located in the pre‐mir‐30e was strongly associated with schizophrenia in Han Chinese population (allelic P = 0.00017; genotypic P = 0.00015).94 Watanabe et al95 replicated the association in Japanese population. The expression levels of mature miR‐30e in the peripheral leukocyte were significantly higher in schizophrenia patients,94 consistent with increased expression in the PFC of schizophrenia individuals.57 Also in Chinese samples, two SNP (hsa‐pre‐mir‐146a rs2910164 G>C and hsa‐mir‐499 rs3746444 T>C) were genotyped with susceptibility to schizophrenia from 268 patients and 232 controls. Patients carrying CC genotype of rs3746444 were more likely to develop hallucination and lack of motivation. However, there was no statistically significant association between these two SNPs and schizophrenia.96 Negative association was also observed in SNP rs7289941.97 Very recently, Yu et al performed a two‐stage GWAS of schizophrenia comprising 4384 cases and 5770 controls, followed by independent replications of 13 single nucleotide polymorphisms in an additional 4339 schizophrenia cases and 7043 controls of Han Chinese ancestry. They confirmed that three loci, at 2p16.1 (rs1051061, in an exon of VRK2), 6p22.1 (rs115070292 in an intron of GABBR1), and 10q24.32 (rs10883795 in an intron of AS3MT; rs10883765 at an intron of ARL3), are significantly associated with schizophrenia. These three loci are known to be involved in the regulation of GABAergic and dopaminergic signaling, cell adhesion molecules, and myelination pathways.98
The hairpin structure guides correct miRNA processing, and the 3′UTR of mRNA affects miRNA/mRNA interaction. SNPs in these regions may contribute to the risk of disease. Screening for the mutations in the susceptibility locus (22q11) of schizophrenia in Russian population identified a polymorphism in the 5′‐upstream of miR‐130b gene region, which contains DNA elements for putative transcription factors. However, genetic analysis did not show statistically significant association of miR‐130b variants with schizophrenia.99 Liu et al100 applied the MiRSNP database (http://cmbi.bjmu.edu.cn/mirsnp), a collection of human SNPs in predicted miRNA‐mRNA‐binding sites to GWAS for schizophrenia, and identified seven miRNA‐related SNPs. In another study, 803 SNPs from the 3′UTR of 425 schizophrenia‐associated genes were analyzed for Gibbs‐free energy of miRNA binding in silico. One uncharted SNP (rs3219151 of GABRA6) was reported to be significantly associated with the decreased risk of schizophrenia. Another SNP rs10759 (RGS4) interfered with the miR‐124 binding to RGS4, therefore could increase the risk of schizophrenia.101 Most of the SNP analysis was performed in Chinese Han population. John et al102 investigated the association of MiRSNPs in candidate genes with schizophrenia in a large population of genetically distinct north Indian cohorts(1017 cases and 1073 controls). They reported that five SNPs were associated with tardive dyskinesia and twelve SNPs were associated with strong schizophrenia genes. Recently, a genome‐wide investigation in Canadian patients revealed an enrichment of rare CNVs that overlap miRNAs by excluding the 22q11.2 CNVs, which is known to possess a high susceptibility for schizophrenia.103 The predicted target genes of the 25 CNV‐overlapped miRNA were tended be involved in neurodevelopmental processes.103 These studies suggest that miRNAs, by targeting schizophrenia risk genes, may contribute to this complex neuropsychiatric disorder.
Recently, a polymorphism (rs1625579) within an intron of primary transcript for miR‐137 has attracted lots of attention, which was found to be significantly associated in GWAS of schizophrenia (P = 1.6 × 10−11).104 Several studies replicated the correlation between miR‐137 polymorphisms and schizophrenia samples in Scottish,105 Canadian,106 Australian,107 and Chinese Han population.108, 109 However, negative association results were also reported in some case‐control studies.110, 111 In the study of schizophrenia Psychiatric GWAS Consortium, four putative targets gene (CSMD1, C10orf26, CACNA1C, and TCF4) were also reported to have genome‐wide significant association with schizophrenia. Luciferase report assay confirmed the interplay between miR‐137 and these four target genes,112 similar results were found for ZNF804A113 and CALN1.114 Using bioinformatics resources, several target genes were revealed, including ERBB4, GABRA1, GRIN2A, GRM5, GSK3B, NRG2, and HTR2C. These genes were identified to be involved in synaptic long‐term potentiation, a process that may underline mechanism of learning and memory which is impaired in schizophrenia patients.115 Using functional magnetic resonance imaging scans, van Erp et al116 reported that the rs1627759 TT (miR‐137 locus) is associated with DLPFC hyperactivity, which is a common measure of brain inefficiency. The relationship between rs1625579 genotypes and miR‐137 expression was reported by Guella et al117 They observed lower miR‐137 expression levels in the homozygous TT subjects compared to TG and GG subjects in the control group. The reduced miR‐137 levels in TT subjects corresponded to increased levels of the miR‐137 target gene TCF4.117 In SH‐SY5Y dopaminergic cell line, Strazisar et al118 also demonstrated that expressing the miR‐137 variants resulted in reduction in mature miR‐137 expression and lead to the deregulation of gene sets involved in synaptogenesis and neuronal transmission. However, Siegert et al observed gain of function of miR‐137 while carrying variants alleles. They found that the increased expression of miR‐137 caused the downregulation of the presynaptic target genes such as complexin‐1 (Cplx1), Nsf and synaptotagmin‐1 (Syt1), and lead to impaired vesicle release. In vivo, miR‐137 gain of function resulted in changes in synaptic vesicle pool distribution, impaired induction of mossy fiber long‐term potentiation and resulted in defects in hippocampus‐dependent learning and memory.119 All these observations implicate that altered miR‐137 may play a critical role in pathophysiology of schizophrenia.
3.4. miRNA and synaptic plasticity in schizophrenia
Schizophrenia is considered as a complex neurodevelopmental disease. Substantial evidence suggests that dysfunction of several neurotransmitter systems (such as dopamine and glutamate) assemblies the pathophysiological processes of schizophrenia.54, 120 N‐methyl‐D‐aspartate‐glutamate (NMDA) receptor is an important regulator in synaptic plasticity. Hypo‐function of NMDA receptor signaling could alter the balance of excitation and inhibition in cortical circuits and produces behaviors resembling the symptom of schizophrenia.121, 122 Using a NMDA receptor antagonist dizocilpine, which could rapidly induce schizophrenia‐like behaviors, Kocerha et al examined the miRNA expression in different brain regions of mice. They found that mice treated with acute rather than chronic dizocilpine showed a significant decrease in a brain‐specific miRNA, miR‐219 in the PFC. Decreased miR‐219 expression was also observed in hypomorphic GRN1 (NR1) mutant mice. Pretreatment with antipsychotic drugs (haloperidol and clozapine) could prevent dizocilpine‐induced reduction of miR‐219. One of miR‐219 targets was identified as calcium/calmodulin‐dependent protein kinase II gamma subunit (CaMKIIγ), a component of the NMDA glutamate receptor signaling cascades. Inhibition of miR‐219 in mouse brain reduced the dizocilpine‐induced behavioral responses such as hyperlocomotion and stereotypies, suggesting the regulatory role of miR‐219 in NMDA receptor function.123 In support, miR‐219 was reported to be significantly upregulated in the DLPFC of postmortem brain tissue with schizophrenia.59 In addition, miR‐129 was found to participate in the regulation of oligodendrocyte differentiation and myelin maintenance,124 suggesting the importance of miR‐219 in synaptic structure and disease‐related function. Zhang et al performed an association analysis for 3 SNPs in hsa‐pri‐miR‐219/132/107 and 6 SNPs in 3′UTR of NMDAR signaling pathway genes (GRIN2A/2B/3A and CAMK2G) in a case‐control study of 1041 schizophrenia patients and 953 healthy controls, and confirmed that GRIN2B rs890 was significant associated with schizophrenia.73
Brain‐derived neurotrophic factor (BDNF) is the most prevalent growth factor in the CNS and plays an essential role in the brain development and neuronal plasticity.125 Accumulating evidence suggests that dysregulation of BDNF linked to multiple neuropsychiatric disorders.125, 126 Postmortem studies revealed altered BDNF expression level in certain brain regions of schizophrenia patients.127, 128, 129, 130 Mellios et al60 identified that two miRNAs, miR‐30a and miR‐195 directly targeted to BDNF 3′UTR and inhibited BDNF expression. They further reported that miR‐195 interaction with BDNF could subsequently regulate schizophrenia‐related gamma‐aminobutyric acid (GABA), that is, GABAergic gene expression, including neuropeptide Y (NPY) and somatostatin.131 miR‐30a‐5p was also demonstrated to control alcohol intake by regulating the BDNF signaling pathway.132
Cognitive impairment is one of the severe symptoms of schizophrenia.133 The analysis for the 3′UTR of 242 presynaptic and 304 postsynaptic proteins revealed that 91% of these proteins are predicted miRNA targets.134 The functional role of miR‐132 in learning and memory is suggested to be associated with its regulatory effects on synaptic plasticity. Some behavioral tasks associated with learning and memory were found to rapidly induce miR‐132 expression. Knockdown miR‐132 in vivo impaired the memory acquisition in trace fear‐conditioning paradigm,135, 136 while overexpression of miR‐132 in a transgenic mouse model showed increases in neuronal spine density and improvement in novel object recognition.137 Interestingly, Hansen group reported that mild upregulation of miR‐132 enhanced spatial learning of mice. However, a more than three‐fold increase in miR‐132 expression impaired learning.138 Double‐knockout miR‐132/212 impaired the long‐term potentiation as well as cognitive function in spatial memory, recognition memory, and in tests of novel object recognition, indicating an important role of miR‐132/212 in synaptic function.139 Compared the transcriptional profile of the hippocampus in respective miR‐132 and miR‐212 overexpression mouse and miR‐132/‐212 double‐knockout mice, RNA sequencing results revealed that 1138 genes expression were increased in miR‐132/‐212 deletion mice. Ninety‐six of those genes were downregulated in mice overexpressing miR‐132. Of the 58 genes that were decreased in overexpressing miR‐212 mice, only 4 of them were increased in the double‐knockout line.140 Although miR‐132 and miR‐212 share a seed sequence, these two miRNAs do not overlap greatly for mRNA targeting genes, suggesting a complex, nonredundant manner in transcriptional profile regulation.
Glial cells play a critical functional rile in brain function. Altered glial functions are known involved in the pathophysiological process in many neuropsychiatric diseases141, 142 including schizophrenia.143, 144, 145, 146 Previous work on miRNA in schizophrenia is focused on neuronal functions, how the glial miRNA contributes to the pathogenesis and development is largely unknown. We recently employed the GFAP‐GFP transgenic mice to profile the miRNA in astrocytes in response to acute phencyclidine (PCP) administration. Compared to the saline group, mice treated with PCP showed altered expression of miRNAs (miR‐143‐3p, miR‐212, miR‐127‐3p, miR‐183, miR‐298, miR‐381‐3p, miR‐338‐3p, miR‐let‐7d‐3p, miR‐132‐3p, etc.) in astrocytes of PFC (as shown in Table 1). In further study, we explore the functional role of miRNAs with schizophrenia and found the correlation between miR‐143‐3p dysregulation and PCP‐induced behavioral deficits in mice (T. Cao, P. Wang, C. Lu & X. Zhen, unpublished data).
Table 1.
MicroRNA sequencing analysis of the mouse prefrontal cortex (PFC) astrocytes in response to acute phencyclidine (PCP) treatment
| MATURE miRNA ID | PRE‐ACC | MATURE‐SEQ | PCP vs Control fold change |
|---|---|---|---|
| PCP vs Control 2.0‐fold change Upregulated miRNAs | |||
| mmu‐miR‐212‐5p | MI0000696 | ACCUUGGCUCUAGACUGCUUACU | 4.06 |
| mmu‐miR‐127‐3p | MI0000154 | UCGGAUCCGUCUGAGCUUGGCU | 2.90 |
| mmu‐miR‐183‐5p | MI0000225 | UAUGGCACUGGUAGAAUUCACU | 2.48 |
| mmu‐miR‐298‐5p | MI0000398 | GGCAGAGGAGGGCUGUUCUUCCC | 2.46 |
| mmu‐miR‐381‐3p | MI0000798 | UAUACAAGGGCAAGCUCUCUGU | 2.42 |
| mmu‐miR‐338‐3p | MI0000619 | UCCAGCAUCAGUGAUUUUGUUG | 2.40 |
| mmu‐let‐7d‐3p | MI0000405 | CUAUACGACCUGCUGCCUUUCU | 2.38 |
| mmu‐miR‐132‐3p | MI0000158 | UAACAUGCUACAGCCAUGGUCG | 2.22 |
| mmu‐miR‐130a‐3p | MI0000156 | CAGUGCAAUGUUAAAAGGGCAU | 2.19 |
| mmu‐miR‐744‐5p | MI0004124 | UGCGGGGCUAGGGCUAACAGCA | 2.17 |
| mmu‐miR‐330‐5p | MI0000607 | UCUCUGGGCCUGUGUCUUAGGC | 2.13 |
| mmu‐miR‐335‐3p | MI0000817 | UUUUUCAUUAUUGCUCCUGACC | 2.06 |
| mmu‐miR‐29c‐3p | MI0000577 | UAGCACCAUUUGAAAUCGGUUA | 2.04 |
| mmu‐miR‐181a‐1‐3p | MI0000697 | ACCAUCGACCGUUGAUUGUACC | 2.04 |
| mmu‐miR‐872‐5p | MI0005549 | AAGGUUACUUGUUAGUUCAGG | 2.00 |
| PCP vs Control 2.0‐fold change Downregulated miRNAs | |||
| mmu‐miR‐143‐3p | MI0000257 | UGAGAUGAAGCACUGUAGCUC | 0.26 |
| mmu‐miR‐15a‐5p | MI0000564 | UAGCAGCACAUAAUGGUUUGUG | 0.45 |
| mmu‐miR‐1968‐5p | MI0009965 | UGCAGCUGUUAAGGAUGGUGGACU | 0.50 |
| mmu‐miR‐582‐3p | MI0006127 | UAACCUGUUGAACAACUGAAC | 0.50 |
GFAP‐GFP transgenic mouse brains were rapidly removed 30 min after subcutaneous injection with PCP (4 mg/kg) or saline as control. GFP positive astrocytes were sorted by flow cytometry from prefrontal cortex. RNA was extracted from sorted astrocytes and used to prepare the miRNA sequencing libraries. The libraries were captured on Illumina flow cells, amplified in situ as clusters and finally sequenced for 36 cycles on Illumina HiSeq following the manufacturer's instructions. Trimmed reads were alignment to the miRBase pre‐miRNAs. miRNA read counts were normalized as tag counts per million miRNA alignments. Differentially expressed miRNAs (PCP vs Control >2.0 fold changes) were presented in the table.
4. PERSPECTIVE OF miRNA THERAPIES
miRNAs were predicted to regulate 20%‐30% of human genes.147, 148 The involvement of miRNAs in many aspects of psychiatric disorder makes them potential candidates as biomarkers or targets for clinical diagnosis and treatment.6, 7, 9, 149 Although the importance of miRNAs in brain functions has been well‐documented, more efforts are required to understand the full extent of miRNA regulatory mechanism and their pathological effects in neuropsychiatric disorders. The first challenge remained is to identify functional miRNAs located in human genome associated with schizophrenia. Project such as the Encyclopedia of DNA Elements project (ENCODE), which aims to map all functional elements in the human genome, has made great progress.150 Methods of RNA microarray and novel high‐throughput technologies such as next‐generation sequencing (NGS) provide powerful tools for the analysis of miRNA transcription profiles and their target genes in diseases.151, 152, 153, 154 miRNAs regulate target genes through the base pair interaction of seed region (nucleotide 2‐7 of the miRNA) with target mRNA 3′UTR. A sequence of this length will occur with much high frequency in the whole genome; therefore, the prediction of functional miRNA target sites is challenging but will be critical task. Currently, the most comprehensive and wildly applied prediction programs are TargetScan and PicTar. However, two‐thirds of their predicted targets appeared nonresponsive to the miRNA.155 Thus, developing or continuing improvement in experimental tools to understand the miRNA effect on target genes is definitely needed.
Given the abnormal expression of miRNAs in many psychiatric disorders, inhibition or overexpression of miRNAs may be a potential approach in clinical treatment.156, 157, 158, 159, 160 Much effort has been taken to develop high efficiency and nontoxic oligonucleotide mimetics or antisense oligonucleotides to regulate miRNA expression levels. In associated with these efforts, many chemical modifications such as 2′‐O‐methoxyethyl and locked nucleic acid (LNA) were developed in to enhance the stability of the RNAs.161, 162, 163, 164, 165 Systemic delivery of LNA‐antimiR‐212 in the liver of African green monkeys leads to a long‐lasting and reversible decrease in total plasma cholesterol without any evidence for LNA‐associated toxicities.162 Treatment of hepatitis C virus (HCV)‐infected chimpanzees with LNA‐antimiR‐212 (SPC3649) also leads to long‐lasting suppression of HCV viremia, with no evidence of viral resistance or side effects in the animals.166 The first‐in‐human study tested a miR‐16‐based miRNA mimic packaged in TargomiRs‐EDVs targeted to EGFR in patients with malignant pleural mesothelioma. The results showed that TargomiRs at a dose of 5 × 109 per week with full dexamethasone prophylaxis were well tolerated and was accompanied by early signs of antitumor activity. This is an open‐label study, and a randomized phase 2 study with larger population is needed to confirm the observation.160
When exploring such therapy for neurotherapeutics, the major barrier is the blood‐brain barrier (BBB) in the CNS. Several approaches for delivering drugs to the CNS have been developed to enhance the capacity of therapeutic molecules to cross the BBB by modifying the drug itself, or by coupling it to a vector.167 The mediator of exosomes used as a nano‐delivery system has several advantages as delivery vehicles.168, 169 Exosomes are the smallest membranous vesicles with homogenous shape and are secreted by diverse mammalian cells. Due to low immunogenicity, remarkable delivering properties and the ability to cross the BBB, exosomes have been considered as efficient delivery mediators for RNA therapy.168, 170 Whether the exosomes can be applied to deliver miRNAs into CNS still need to be tested. Hwang et al have developed a brain‐specific nanocarrier, RVG‐SSPEI (rabies virus glycoprotein‐disulfide linked polyethyleneimine) to successfully deliver miR‐124a in mouse brain.171 Other approaches such as viral delivery systems, chemical modification and conjugation strategies, aptamers, and nanotechnologies have been studied. Although the technological innovations are promising, selective delivery into the CNS remains challenging. Meanwhile, the functional activities of the delivered miRNAs in those systems are difficult to be evaluated. There will be a long way to go for establishing a genuine and practical miRNA treatment in clinical practice.
5. CONCLUDING REMARKS
Exploring miRNAs biogenesis and function in the CNS demonstrate that miRNAs play essential roles in the pathophysiology of schizophrenia. The precise profiles of changes in miRNA in schizophrenia patients and its association with the prognosis and therapeutic response remain largely unknown. The future work should focus on identifying specific‐disease‐related miRNA and understand its precise mechanism in regulating the biological pathway and contribution in pathological process. With the technological progress, targeted delivery of miRNA to CNS may provide a potential novel therapeutic approach for the treatment of psychiatric diseases such as schizophrenia.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
This study was supported by grants from the National Science Foundation of China [No. 81773702, 81372688, and 81373382] Supports from the Priority Academic Program Development of the Jiangsu Higher Education Institutes (PAPD) are also appreciated.
Cao T, Zhen X‐C. Dysregulation of miRNA and its potential therapeutic application in schizophrenia. CNS Neurosci Ther. 2018;24:586–597. 10.1111/cns.12840
REFERENCES
- 1. Tandon R, Keshavan MS, Nasrallah HA. Schizophrenia, “just the facts” what we know in 2008. 2. Epidemiology and etiology. Schizophr Res. 2008;102:1‐18. [DOI] [PubMed] [Google Scholar]
- 2. Goto Y, Yang CR, Otani S. Functional and dysfunctional synaptic plasticity in prefrontal cortex: roles in psychiatric disorders. Biol Psychiatry. 2010;67:199‐207. [DOI] [PubMed] [Google Scholar]
- 3. Lisman J. Excitation, inhibition, local oscillations, or large‐scale loops: what causes the symptoms of schizophrenia? Curr Opin Neurobiol. 2012;22:537‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Howes O, McCutcheon R, Stone J. Glutamate and dopamine in schizophrenia: an update for the 21st century. J Psychopharmacol. 2015;29:97‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu B, Hsu PK, Stark KL, et al. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia‐related microdeletion. Cell. 2013;152:262‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nadim WD, Simion V, Benedetti H, et al. MicroRNAs in neurocognitive dysfunctions: new molecular targets for pharmacological treatments? Curr Neuropharmacol. 2017;15:260‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rajman M, Schratt G. MicroRNAs in neural development: from master regulators to fine‐tuners. Development. 2017;144:2310‐2322. [DOI] [PubMed] [Google Scholar]
- 8. Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. 2012;46:263‐271. [DOI] [PubMed] [Google Scholar]
- 9. Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228‐234. [DOI] [PubMed] [Google Scholar]
- 11. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281‐297. [DOI] [PubMed] [Google Scholar]
- 12. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell. 1993;75:843‐854. [DOI] [PubMed] [Google Scholar]
- 13. Slack FJ, Basson M, Liu Z, et al. The lin‐41 RBCC gene acts in the C. elegans heterochronic pathway between the let‐7 regulatory RNA and the LIN‐29 transcription factor. Mol Cell. 2000;5:659‐669. [DOI] [PubMed] [Google Scholar]
- 14. Reinhart BJ, Slack FJ, Basson M, et al. The 21‐nucleotide let‐7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901‐906. [DOI] [PubMed] [Google Scholar]
- 15. Griffiths‐Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154‐D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lagos‐Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853‐858. [DOI] [PubMed] [Google Scholar]
- 17. Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans . Science. 2001;294:862‐864. [DOI] [PubMed] [Google Scholar]
- 18. Bak M, Silahtaroglu A, Møller M, et al. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dogini D, Ribeiro P, Rocha C, et al. MicroRNA expression profile in murine central nervous system development. J Mol Neurosci. 2008;35:331‐337. [DOI] [PubMed] [Google Scholar]
- 20. Kim V. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376‐385. [DOI] [PubMed] [Google Scholar]
- 21. Cai X, Hagedorn C, Cullen B. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957‐1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kwak P, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol. 2012;19:145‐151. [DOI] [PubMed] [Google Scholar]
- 23. Morlando M, Ballarino M, Gromak N, et al. Primary microRNA transcripts are processed co‐transcriptionally. Nat Struct Mol Biol. 2008;15:902‐909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carthew R, Sontheimer E. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Denli A, Tops B, Plasterk R, et al. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231‐235. [DOI] [PubMed] [Google Scholar]
- 26. Gregory R, Yan K, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235‐240. [DOI] [PubMed] [Google Scholar]
- 27. Yates L, Norbury C, Gilbert R. The long and short of microRNA. Cell. 2013;153:516‐519. [DOI] [PubMed] [Google Scholar]
- 28. Chendrimada T, Gregory R, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740‐744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheloufi S, Dos Santos C, Chong M, et al. A dicer‐independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597‐610. [DOI] [PubMed] [Google Scholar]
- 31. Doench J, Sharp P. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504‐511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cao X, Yeo G, Muotri A, et al. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77‐103. [DOI] [PubMed] [Google Scholar]
- 33. Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401‐1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim J, Krichevsky A, Grad Y, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360‐365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zaits MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry. 2014;19:848‐852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Babak T, Zhang W, Morris Q, et al. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813‐1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barad O, Meiri E, Avniel A, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486‐2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miska E, Alvarez‐Saavedra E, Townsend M, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sempere L, Freemantle S, Pitha‐Rowe I, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain‐expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hohjoh H, Fukushima T. Expression profile analysis of microRNA (miRNA) in mouse central nervous system using a new miRNA detection system that examines hybridization signals at every step of washing. Gene. 2007;391:39‐44. [DOI] [PubMed] [Google Scholar]
- 41. Giraldez A, Cinalli R, Glasner M, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833‐838. [DOI] [PubMed] [Google Scholar]
- 42. Murchison E, Partridge J, Tam O, et al. Characterization of Dicer‐deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135‐12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davis T, Cuellar T, Koch S, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322‐4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Motti D, Bixby J, Lemmon V. MicroRNAs and neuronal development. Semin Fetal Neonatal Med. 2012;17:347‐352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bian S, Xu T, Sun T. Tuning the cell fate of neurons and glia by microRNAs. Curr Opin Neurobiol. 2013;23:928‐934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu Z, Li Z. miRNAs in synapse development and synaptic plasticity. Curr Opin Neurobiol. 2017;45:24‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alural B, Genc S, Haggarty SJ. Diagnostic and therapeutic potential of microRNAs in neuropsychiatric disorders: past, present, and future. Prog Neuropsychopharmacol Biol Psychiatry. 2017;73:87‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang F, Xu Y, Shugart YY, et al. Converging evidence implicates the abnormal microRNA system in schizophrenia. Schizophr Bull. 2015;41:728‐735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun E, Shi Y. MicroRNAs: small molecules with big roles in neurodevelopment and diseases. Exp Neurol. 2015;268:46‐53. [DOI] [PubMed] [Google Scholar]
- 50. Schaefer A, O'Carroll D, Tan C, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553‐1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim J, Inoue K, Ishii J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220‐1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saba R, Störchel P, Aksoy‐Aksel A, et al. Dopamine‐regulated microRNA MiR‐181a controls GluA2 surface expression in hippocampal neurons. Mol Cell Biol. 2012;32:619‐632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiao J, Li Y, Prandovszky E, et al. MicroRNA‐132 dysregulation in Toxoplasma gondii infection has implications for dopamine signaling pathway. Neuroscience. 2014;268:128‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Bartolomeis A, Iasevoli F, Tomasetti C, et al. MicroRNAs in schizophrenia: implications for synaptic plasticity and dopamine‐glutamate interaction at the postsynaptic density. New avenues for antipsychotic treatment under a theranostic perspective. Mol Neurobiol. 2015;52:1771‐1790. [DOI] [PubMed] [Google Scholar]
- 55. Hauberg ME, Roussos P, Grove J, et al. Analyzing the role of microRNAs in schizophrenia in the context of common genetic risk variants. JAMA Psychiatry. 2016;73:369‐377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ni J, Wang X, Chen S, et al. MicroRNA let‐7c‐5p protects against cerebral ischemia injury via mechanisms involving the inhibition of microglia activation. Brain Behav Immun. 2015;49:75‐85. [DOI] [PubMed] [Google Scholar]
- 57. Perkins DO, Jeffries CD, Jarskog LF, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beveridge NJ, Tooney PA, Carroll AP, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156‐1168. [DOI] [PubMed] [Google Scholar]
- 59. Beveridge NJ, Gardiner E, Carroll AP, et al. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15:1176‐1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mellios N, Huang HS, Grigorenko A, et al. A set of differentially expressed miRNAs, including miR‐30a‐5p, act as post‐transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030‐3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim AH, Reimers M, Mahera B, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124:183‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Han J, Kim HJ, Schafer ST, et al. Functional implications of miR‐19 in the migration of newborn neurons in the adult brain. Neuron. 2016;91:79‐89. [DOI] [PubMed] [Google Scholar]
- 63. Kim Y, Zhang Y, Pang K, et al. Bipolar disorder associated microRNA, miR1908‐5p, regulates the expression of genes functioning in neuronal glutamatergic synapses. Exp Neurobiol. 2016;25:296‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Santarelli DM, Beveridge NJ, Tooney PA, et al. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex brodmann area 46 in schizophrenia. Biol Psychiat. 2011;69:180‐187. [DOI] [PubMed] [Google Scholar]
- 65. Smalheiser NR, Lugli G, Zhang H, et al. Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PLoS ONE. 2014;9:e86469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scarr E, Craig JM, Cairns MJ, et al. Decreased cortical muscarinic M1 receptors in schizophrenia are associated with changes in gene promoter methylation, mRNA and gene targeting microRNA. Transl Psychiat. 2013;3:e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Moreau MP, Bruse SE, David‐Rus R, et al. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiat. 2011;69:188‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mellios N, Sur M. The emerging role of microRNAs in schizophrenia and autism spectrum disorders. Front Psychiatry. 2012;3:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moreau MP, Bruse SE, Jornsten R, et al. Chronological changes in microRNA expression in the developing human brain. PLoS ONE. 2013;8:e60480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gardiner E, Beveridge NJ, Wu JQ, et al. Imprinted DLK1‐DIO3 region of 14q32 defines a schizophrenia‐associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry. 2012;17:827‐840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lai CY, Yu SL, Hsieh MH, et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS ONE. 2011;6:e21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lai CY, Lee SY, Scarr E, et al. Aberrant expression of microRNAs as biomarker for schizophrenia: from acute state to partial remission, and from peripheral blood to cortical tissue. Transl Psychiat. 2016;6:e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wei H, Yuan Y, Liu S, et al. Detection of circulating miRNA levels in schizophrenia. Am J Psychiatry. 2015;172:1141‐1147. [DOI] [PubMed] [Google Scholar]
- 74. Gallego J, Gordon M, Claycomb K, et al. In vivo microRNA detection and quantitation in cerebrospinal fluid. J Mol Neurosci. 2012;47:243‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stark KL, Xu B, Bagchi A, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11‐deletion mouse model. Nat Genet. 2008;40:751‐760. [DOI] [PubMed] [Google Scholar]
- 76. Schofield C, Hsu R, Barker A, et al. Monoallelic deletion of the microRNA biogenesis gene Dgcr8 produces deficits in the development of excitatory synaptic transmission in the prefrontal cortex. Neural Dev. 2011;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fénelon K, Mukai J, Xu B, et al. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short‐term plasticity in the prefrontal cortex. Proc Natl Acad Sci USA 2011;108:4447‐4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Earls LR, Fricke RG, Yu J, et al. Age‐dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J Neurosci. 2012;32:14132‐14144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sellier C, Hwang VJ, Dandekar R, et al. Decreased DGCR8 expression and miRNA Dysregulation in individuals with 22q11.2 deletion syndrome. PLoS ONE. 2014;9:e103884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Forstner A, Degenhardt F, Schratt G, et al. MicroRNAs as the cause of schizophrenia in 22q11.2 deletion carriers, and possible implications for idiopathic disease: a mini‐review. Front Mol Neurosci. 2013;6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xu B, Roos J, Levy S, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880‐885. [DOI] [PubMed] [Google Scholar]
- 82. Zhou Y, Wang J, Lu X, et al. Evaluation of six SNPs of microRNA machinery genes and risk of schizophrenia. J Mol Neurosci. 2013;49:594‐599. [DOI] [PubMed] [Google Scholar]
- 83. Beveridge NJ, Santarelli DM, Wang X, et al. Maturation of the human dorsolateral prefrontal cortex coincides with a dynamic shift in microRNA expression. Schizophr Bull. 2014;40:399‐409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Konopka W, Kiryk A, Novak M, et al. MicroRNA loss enhances learning and memory in mice. J Neurosci. 2010;30:14835‐14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. McLoughlin H, Fineberg S, Ghosh L, et al. Dicer is required for proliferation, viability, migration and differentiation in corticoneurogenesis. Neuroscience. 2012;223:285‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Huang T, Liu Y, Huang M, et al. Wnt1‐cre‐mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2010;2:152‐163. [DOI] [PubMed] [Google Scholar]
- 87. Yang J, Lai E. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892‐903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Marinaro F, Marzi M, Hoffmann N, et al. MicroRNA‐independent functions of DGCR8 are essential for neocortical development and TBR1 expression. EMBO Rep. 2017;18:603‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Burger K, Gullerova M. Swiss army knives: non‐canonical functions of nuclear Drosha and Dicer. Nat Rev Mol Cell Biol. 2015;16:417‐430. [DOI] [PubMed] [Google Scholar]
- 90. Hindorff L, Sethupathy P, Junkins H, et al. Potential etiologic and functional implications of genome‐wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362‐9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cooper G, Shendure J. Needles in stacks of needles: finding disease‐causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628‐640. [DOI] [PubMed] [Google Scholar]
- 92. Persengiev S, Kondova I, Bontrop R. Insights on the functional interactions between miRNAs and copy number variations in the aging brain. Front Mol Neurosci. 2013;6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hansen T, Olsen L, Lindow M, et al. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS ONE. 2007;2:e873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Xu Y, Li F, Zhang B, et al. MicroRNAs and target site screening reveals a pre‐microRNA‐30e variant associated with schizophrenia. Schizophr Res. 2010;119:219‐227. [DOI] [PubMed] [Google Scholar]
- 95. Watanabe Y, Iijima Y, Egawa J, et al. Replication in a Japanese population that a MIR30E gene variation is associated with schizophrenia. Schizophr Res. 2013;150:596‐597. [DOI] [PubMed] [Google Scholar]
- 96. Zou M, Li D, Lv R, et al. Association between two single nucleotide polymorphisms at corresponding microRNA and schizophrenia in a Chinese population. Mol Biol Rep. 2012;39:3385‐3391. [DOI] [PubMed] [Google Scholar]
- 97. Zhang F, Chen Y, Liu C, et al. Systematic association analysis of microRNA machinery genes with schizophrenia informs further study. Neurosci Lett. 2012;520:47‐50. [DOI] [PubMed] [Google Scholar]
- 98. Yu H, Yan H, Li J, et al. Common variants on 2p16.1, 6p22.1 and 10q24.32 are associated with schizophrenia in Han Chinese population. Mol Psychiatry. 2017;22:954‐960. [DOI] [PubMed] [Google Scholar]
- 99. Burmistrova OA, Goltsov AY, Abramova LI, et al. MicroRNA in schizophrenia: genetic and expression analysis of miR‐130b (22q 11). Biochemistry (Mosc). 2007;72:578‐582. [DOI] [PubMed] [Google Scholar]
- 100. Liu C, Zhang F, Li T, et al. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA‐related SNPs in GWAS SNPs and eQTLs. BMC Genom. 2012;13:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Gong Y, Wu CN, Xu J, et al. Polymorphisms in microRNA target sites influence susceptibility to schizophrenia by altering the binding of miRNAs to their targets. Eur Neuropsychopharmacol. 2013;23:1182‐1189. [DOI] [PubMed] [Google Scholar]
- 102. John J, Bhatia T, Kukshal P, et al. Association study of MiRSNPs with schizophrenia, tardive dyskinesia and cognition. Schizophr Res. 2016;174:29‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Warnica W, Merico D, Costain G, et al. Copy number variable microRNAs in schizophrenia and their neurodevelopmental gene targets. Biol Psychiat. 2015;77:158‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Schizophrenia Psychiatric Genome‐Wide Association Study (GWAS) Consortium . Genome‐wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Whalley HC, Papmeyer M, Romaniuk L, et al. Impact of a microRNA MIR137 susceptibility variant on brain function in people at high genetic risk of schizophrenia or bipolar disorder. Neuropsychopharmacology. 2012;37:2720‐2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lett TA, Chakravarty MM, Felsky D, et al. The genome‐wide supported microRNA‐137 variant predicts phenotypic heterogeneity within schizophrenia. Mol Psychiatry. 2013;18:443‐450. [DOI] [PubMed] [Google Scholar]
- 107. Green MJ, Cairns MJ, Wu J, et al. Genome‐wide supported variant MIR137 and severe negative symptoms predict membership of an impaired cognitive subtype of schizophrenia. Mol Psychiatry. 2013;18:774‐780. [DOI] [PubMed] [Google Scholar]
- 108. Guan F, Zhang B, Yan T, et al. MIR137 gene and target gene CACNA1C of miR‐137 contribute to schizophrenia susceptibility in Han Chinese. Schizophr Res. 2014;152:97‐104. [DOI] [PubMed] [Google Scholar]
- 109. Ma G, Yin J, Fu J, et al. Association of a miRNA‐137 polymorphism with schizophrenia in a Southern Chinese Han population. Biomed Res Int. 2014;2014:751267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Egawa J, Nunokawa A, Shibuya M, et al. Resequencing and association analysis of MIR137 with schizophrenia in a Japanese population. Psychiatry Clin Neurosci. 2013;67:277‐279. [DOI] [PubMed] [Google Scholar]
- 111. Yuan J, Cheng Z, Zhang F, et al. Lack of association between microRNA‐137 SNP rs1625579 and schizophrenia in a replication study of Han Chinese. Mol Genet Genomics. 2015;290:297‐301. [DOI] [PubMed] [Google Scholar]
- 112. Kwon E, Wang W, Tsai LH. Validation of schizophrenia‐associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR‐137 targets. Mol Psychiatry. 2013;18:11‐12. [DOI] [PubMed] [Google Scholar]
- 113. Kim AH, Parker EK, Williamson V, et al. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa‐miR‐137. Schizophr Res. 2012;141:60‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Xia S, Zhou X, Wang T, et al. Experimental validation of candidate schizophrenia gene CALNI as a target for microRNA‐137. Neurosci Lett. 2015;602:110‐114. [DOI] [PubMed] [Google Scholar]
- 115. Wright C, Turner JA, Calhoun VD, et al. Potential impact of miR‐137 and its targets in schizophrenia. Front Genet. 2013;4:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. van Erp TG, Guella I, Vawter MP, et al. Schizophrenia miR‐137 locus risk genotype is associated with dorsolateral prefrontal cortex hyperactivation. Biol Psychiatry. 2014;75:398‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Guella I, Sequeira A, Rollins B, et al. Analysis of miR‐137 expression and rs1625579 in dorsolateral prefrontal cortex. J Psychiatr Res. 2013;47:1215‐1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Strazisar M, Cammaerts S, van der Ven K, et al. MIR137 variants identified in psychiatric patients affect synaptogenesis and neuronal transmission gene sets. Mol Psychiatry. 2015;20:472‐481. [DOI] [PubMed] [Google Scholar]
- 119. Siegert S, Seo J, Kwon EJ, et al. The schizophrenia risk gene product miR‐137 alters presynaptic plasticity. Nat Neurosci. 2015;18:1008‐1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Moghaddam B, Javitt D. From revolution to evolution: the glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology. 2012;37:4‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhou Q, Sheng M. NMDA receptors in nervous system diseases. Neuropharmacology. 2013;74:69‐75. [DOI] [PubMed] [Google Scholar]
- 122. Laruelle M. Schizophrenia: from dopaminergic to glutamatergic interventions. Curr Opin Pharmacol. 2014;14:97‐102. [DOI] [PubMed] [Google Scholar]
- 123. Kocerha J, Faghihi MA, Lopez‐Toledano MA, et al. MicroRNA‐219 modulates NMDA receptor‐mediated neurobehavioral dysfunction. Proc Natl Acad Sci USA. 2009;106:3507‐3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Li JS, Yao ZX. MicroRNAs: novel regulators of oligodendrocyte differentiation and potential therapeutic targets in demyelination‐related diseases. Mol Neurobiol. 2012;45:200‐212. [DOI] [PubMed] [Google Scholar]
- 125. Autry AE, Monteggia LM. Brain‐derived neurotrophic factor and neuropsychiatric disorders. Pharmacol Rev. 2012;64:238‐258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Mitchelmore C, Gede L. Brain derived neurotrophic factor: epigenetic regulation in psychiatric disorders. Brain Res. 2014;1586:162‐172. [DOI] [PubMed] [Google Scholar]
- 127. Takahashi M, Shirakawa O, Toyooka K, et al. Abnormal expression of brain‐derived neurotrophic factor and its receptor in the corticolimbic system of schizophrenic patients. Mol Psychiatry. 2000;5:293‐300. [DOI] [PubMed] [Google Scholar]
- 128. Weickert CS, Hyde TM, Lipska BK, et al. Reduced brain‐derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8:592‐610. [DOI] [PubMed] [Google Scholar]
- 129. Hashimoto T, Bergen SE, Nguyen QL, et al. Relationship of brain‐derived neurotrophic factor and its receptor TrkB to altered inhibitory prefrontal circuitry in schizophrenia. J Neurosci. 2005;25:372‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Angelucci F, Brene S, Mathe AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10:345‐352. [DOI] [PubMed] [Google Scholar]
- 131. Mellios N, Huang HS, Baker SP, et al. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiat. 2009;65:1006‐1014. [DOI] [PubMed] [Google Scholar]
- 132. Darcq E, Warnault V, Phamluong K, et al. MicroRNA‐30a‐5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol Psychiatry. 2015;20:1219‐1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Ceaser A, Csernansky JG, Barch DM. COMT influences on prefrontal and striatal blood oxygenation level‐dependent responses during working memory among individuals with schizophrenia, their siblings, and healthy controls. Cogn Neuropsychiatry. 2013;18:257‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Paschou M, Paraskevopoulou MD, Vlachos IS, et al. miRNA regulons associated with synaptic function. PLoS ONE. 2012;7:e46189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Nudelman AS, DiRocco DP, Lambert TJ, et al. Neuronal activity rapidly induces transcription of the CREB‐regulated microRNA‐132, in vivo. Hippocampus. 2010;20:492‐498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Wang RY, Phang RZ, Hsu PH, et al. In vivo knockdown of hippocampal miR‐132 expression impairs memory acquisition of trace fear conditioning. Hippocampus. 2013;23:625‐633. [DOI] [PubMed] [Google Scholar]
- 137. Hansen KF, Sakamoto K, Wayman GA, et al. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE. 2010;5:e15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hansen KF, Karelina K, Sakamoto K, et al. miRNA‐132: a dynamic regulator of cognitive capacity. Brain Struct Funct. 2013;218:817‐831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Remenyi J, van den Bosch MW, Palygin O, et al. miR‐132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PLoS ONE. 2013;8:e62509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hansen KF, Sakamoto K, Aten S, et al. Targeted deletion of miR‐132/‐212 impairs memory and alters the hippocampal transcriptome. Learn Mem. 2016;23:61‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Abdul‐Muneer PM. MicroRNA in the pathophysiology of CNS injury: implication in neuroregenerative medicine. CNS Neurosci Ther. 2016;22:543‐545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Dong XH, Zhen XC. Glial pathology in bipolar disorder: potential therapeutic implications. CNS Neurosci Ther. 2015;21:393‐397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Goudriaan A, de Leeuw C, Ripke S, et al. Specific glial functions contribute to schizophrenia susceptibility. Schizophr Bull. 2014;40:925‐935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kim R, Healey K, Sepulveda‐Orengo M, Reissner KJ. Astroglial correlates of neuropsychiatric disease: from astrocytopathy to astrogliosis. Prog Neuropsychopharmacol Biol Psychiatry. 2017. [Epub ahead of print]. 10.1016/j.pnpbp.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Laskaris L, Di Biase M, Everall I, et al. Microglial activation and progressive brain changes in schizophrenia. Br J Pharmacol. 2016;173:666‐680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bernstein H, Steiner J, Guest P, et al. Glial cells as key players in schizophrenia pathology: recent insights and concepts of therapy. Schizophr Res. 2015;161:4‐18. [DOI] [PubMed] [Google Scholar]
- 147. Xie X, Lu J, Kulbokas EJ, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15‐20. [DOI] [PubMed] [Google Scholar]
- 149. Mahmoudi E, Cairns M. MiR‐137: an important player in neural development and neoplastic transformation. Mol Psychiatry. 2017;22:44‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Pantazatos S, Huang Y, Rosoklija G, et al. Whole‐transcriptome brain expression and exon‐usage profiling in major depression and suicide: evidence for altered glial, endothelial and ATPase activity. Mol Psychiatry. 2017;22:760‐773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Backes C, Meder B, Hart M, et al. Prioritizing and selecting likely novel miRNAs from NGS data. Nucleic Acids Res. 2016;44:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Roberts B, Hardigan A, Kirby M, et al. Blocking of targeted microRNAs from next‐generation sequencing libraries. Nucleic Acids Res. 2015;43:e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sand M, Bechara F, Gambichler T, et al. Next‐generation sequencing of the basal cell carcinoma miRNome and a description of novel microRNA candidates under neoadjuvant vismodegib therapy: an integrative molecular and surgical case study. Ann Oncol. 2016;27:332‐338. [DOI] [PubMed] [Google Scholar]
- 155. Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Janssen H, Reesink H, Lawitz E, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685‐1694. [DOI] [PubMed] [Google Scholar]
- 157. Nana‐Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther. 2013;93:98‐104. [DOI] [PubMed] [Google Scholar]
- 158. Kim M, Kasinski AL, Slack FJ. MicroRNA therapeutics in preclinical cancer models. Lancet Oncol. 2011;12:319‐321. [DOI] [PubMed] [Google Scholar]
- 159. Takeshita F, Patrawala L, Osaki M, et al. Systemic delivery of synthetic microRNA‐16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell‐cycle genes. Mol Ther. 2010;18:181‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. van Zandwijk N, Pavlakis N, Kao S, et al. Safety and activity of microRNA‐loaded minicells in patients with recurrent malignant pleural mesothelioma: a first‐in‐man, phase 1, open‐label, dose‐escalation study. Lancet Oncol. 2017;18:1386‐1396. [DOI] [PubMed] [Google Scholar]
- 161. Esau C, Davis S, Murray SF, et al. miR‐122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87‐98. [DOI] [PubMed] [Google Scholar]
- 162. Elmen J, Lindow M, Schutz S, et al. LNA‐mediated microRNA silencing in non‐human primates. Nature. 2008;452:896‐899. [DOI] [PubMed] [Google Scholar]
- 163. Davis S, Lollo B, Freier S, et al. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294‐2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Orom UA, Kauppinen S, Lund AH. LNA‐modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137‐141. [DOI] [PubMed] [Google Scholar]
- 165. Stenvang J, Silahtaroglu AN, Lindow M, et al. The utility of LNA in microRNA‐based cancer diagnostics and therapeutics. Semin Cancer Biol. 2008;18:89‐102. [DOI] [PubMed] [Google Scholar]
- 166. Lanford RE, Hildebrandt‐Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA‐122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Hossain S, Akaike T, Chowdhury EH. Current approaches for drug delivery to central nervous system. Curr Drug Deliv. 2010;7:389‐397. [DOI] [PubMed] [Google Scholar]
- 168. Alvarez‐Erviti L, Seow Y, Yin H, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341‐345. [DOI] [PubMed] [Google Scholar]
- 169. Thery C, Boussac M, Veron P, et al. Proteomic analysis of dendritic cell‐derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309‐7318. [DOI] [PubMed] [Google Scholar]
- 170. Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Hwang DW, Son S, Jang J, et al. A brain‐targeted rabies virus glycoprotein‐disulfide linked PEI nanocarrier for delivery of neurogenic microRNA. Biomaterials. 2011;32:4968‐4975. [DOI] [PubMed] [Google Scholar]


