Abstract
In recent years, our vision of lysosomes has drastically changed. From being considered as mere degradative compartments, they are now recognized as key players in many cellular processes. The ability of lysosomes to respond to different stimuli revealed a complex and coordinated regulation of lysosomal gene expression. This review discusses the participation of the transcription factors TFEB and TFE3 in the regulation of lysosomal function and biogenesis, and the role of the lysosomal pathway in cellular adaptation to a variety of stress conditions, including nutrient deprivation, mitochondrial dysfunction, protein missfolding, and pathogen infection. We also describe how cancer cells make use of TFEB and TFE3 to promote their own survival, and highlight the potential of these transcription factors as therapeutic targets for the treatment of neurological and lysosomal diseases.
Keywords: autophagy, lysosomes, mTOR, TFE3, TFEB, stress
INTRODUCTION
Lysosomes are the primary degradative compartment in all cells. Discovered in the early fifties by Christian De Duve, lysosomes are membrane-bound organelles containing over 50 acid hydrolases specialized in breaking-down different macromolecules, including proteins, lipids, carbohydrates, and nucleic acids (De Duve et al 1955). Lysosomes receive extracellular materials that enter the cell through endocytosis and phagocytosis, as well as intracellular materials that are delivered via autophagy (Luzio et al 2007).
Lysosomal function is critical for maintaining proper cellular homeostasis. Mutations in lysosomal proteins are the cause of a class of metabolic disorders known as Lysosomal Storage Diseases (LSDs); their combined prevalence is estimated at 1 in 5,000 births. LSDs are characterized by the progressive accumulation of undigested material in lysosomes that in turn disrupts cellular physiology. A common characteristic for most LSDs is the accumulation of autophagosomes, suggesting that healthy lysosomes are necessary for efficient autophagosome degradation (Lieberman et al 2012). Since basal autophagy is critical for elimination of damaged organelles and unfolded proteins, it is not surprising that accumulation of abnormal mitochondria and protein aggregates are also usually observed in LSDs (Vitner et al 2010).
Recent evidence indicates that the autophagic/lysosomal pathway is a highly dynamic system. Cells possess multiple mechanisms to rapidly and efficiently turn up or down this pathway, and this regulation is essential for cellular adaptation to different internal and external stresses. As a critical regulator of cellular survival, the autophagic/lysosomal pathway is often hijacked by pathogens and used by tumor cells for their own benefit. In addition, diminished or aberrant autophagic/lysosomal function is a common phenomenon during aging and has been linked to several neurodegenerative disorders. In this review we focus on the transcriptional regulation of lysosomal biogenesis and its implications for cellular clearance and response to stress, maintenance of energy homeostasis, and human disease.
TRANSCRIPTIONAL CONTROL OF AUTOPHAGY AND LYSOSOMAL BIOGENESIS
Cells must maintain a constitutive basal level of autophagy in order to preserve homeostasis, but equally important is their ability to effectively upregulate this process in response to different stress conditions, such as nutrient limitation, protein missfolding, oxidative stress, or organelle damage. Autophagy activation may be achieved by various mechanisms including post-translational modifications (e.g. phosphorylation, protein lipidation). However, it has been recently recognized that several transcription factors also play an important role in the transcriptional regulation of autophagy (reviewed in (Fullgrabe et al 2014)). These include transcription factors that promote autophagy activation (E2F1, GATA1, and members of the FOXO family), repression (GATA4), and those that have a dual inhibitory/activating function (TP53/p53 and NFKB).
Since efficient autophagosome degradation requires fusion with fully active lysosomes, it makes sense that cells possess mechanisms to increase the number and activity of lysosomes under stress conditions. Although a logical idea, the mechanism that governs lysosomal biogenesis remained uncharacterized until very recently. Lysosomal degradation was viewed as a housekeeping rather than a regulated process. In addition, the dynamic nature of lysosomes suggested that changes in the rate of endocytosis or the delivery of lysosomal enzymes through the secretory pathway might be sufficient to adjust lysosomal activity to cellular demands. Unexpectedly, analysis of the promoter region of many lysosomal genes revealed the presence of one or more 10 base-pair motif (GTCACGTGAC) typically localized within 200 base pairs of the transcription initiation site (Sardiello et al 2009). This motif, named Coordinated Lysosomal Expression and Regulation (CLEAR) element, constitutes a type of E-box (CANNTG), recognized by the MiTF/TFE family of basic helix-loop-helix (bHLH) transcription factors. It was, therefore, suggested that lysosomal biogenesis is transcriptionally regulated.
The MiTF/TFE family is composed of four different transcription factors, namely TFEB, TFE3, MITF, and TFEC. MITF and TFE3 had been implicated in promoting expression of several genes involved in melanosome biogenesis, which are considered lysosome-related organelles (Cheli et al 2010, Raposo & Marks 2007, Verastegui et al 2000). MITF also induces expression of particular lysosomal genes critical for osteoclast function, including CLCN7, OSTM1, ACP5, and cathepsin K (Hershey & Fisher 2004, Meadows et al 2007, Motyckova et al 2001). However, MITF and TFE3 were not initially considered to be broad regulators of lysosomal gene expression.
TFEB was the first member of the MiTF/TFE family identified as a master regulator of lysosomal biogenesis. TFEB over-expression in HeLa cells induces transcriptional activation of numerous lysosomal genes, including several subunits of the v-ATPase, lysosomal transmembrane proteins and lysosomal hydrolases, and results in a significant increase in the total number of lysosomes (Sardiello et al 2009). Genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) analysis showed a high enrichment in lysosomal genes and confirmed direct binding of TFEB to CLEAR elements (Palmieri et al 2011). TFEB also binds to the promoter of many other genes implicated in lysosomal-related processes, such as endocytosis, phagocytosis, and autophagy. The ability of TFEB to stimulate autophagy is of great importance, since it reveals a transcriptional co-regulation of two major cellular degradative pathways (Settembre et al 2011). More recently it was shown that TFE3 also binds CLEAR elements and induces lysosomal biogenesis and autophagy upon activation (Martina et al 2014). TFE3’s ability to increase number of lysosomes is TFEB-independent, since its effect is observed in TFEB-depleted cells (Martina et al 2014). Importantly, TFEB and TFE3 induce expression of a small subset of autophagy genes. However, these genes encode critical regulators of autophagosome formation and degradation (Martina et al 2014, Settembre & Ballabio 2011).
The participation of MITF in lysosomal biogenesis is less clear. Part of the difficulty is that MITF is subjected to alternative splicing and promoter usage, thus resulting in multiple isoforms. Over-expression of the ubiquitous MITF-A in ARPE-19 cells does not effectively activate expression of lysosomal genes but increases expression of several autophagy genes (Martina et al 2014). In contrast, analysis of microarray expression dataset of 51 different melanoma lines revealed a strong correlation between the expression of MITF-M, an isoform enriched in melanoma, and many, but not all, lysosomal genes containing CLEAR elements in their promoters (Ploper et al 2015). Furthermore, the ability of the members of the MiTF/TFE family hetero-dimerize with each other may influence the relative contribution of MITF to lysosomal gene expression depending on cell type or activation status. Therefore, this review will focus on the cellular functions of TFEB and TFE3, the two bona fide master regulators of lysosomal biogenesis.
NUTRIENT SENSING AND ENERGY HOMEOSTASIS
In order to survive nutrient deprivation, cells must inhibit protein synthesis and activate autophagy. Autophagy ensures delivery and degradation of cytosolic components within lysosomes, thus preventing accumulation of damaged proteins and organelles and promoting recycling of building blocks, such as fatty acids and amino acids, which are critical for maintaining ATP levels and synthesis of essential survival components. Lysosomes are also the site of activation of mTORC1, an evolutionary conserved serine/threonine kinase that regulates cell growth and division in response to energy levels, growth signals, and nutrients. Under conditions of nutrient and energy abundance, mTORC1 is recruited to the lysosomal surface and activated, thus promoting anabolic processes, such as protein synthesis and nutrient storage, and inhibiting autophagy. Conversely, when nutrients are scarce, mTORC1 is inactivated, leading to autophagy induction and utilization of energy stores. Therefore, lysosomes are intimately involved in nutrient response by facilitating autophagosome degradation and serving as signaling platforms for mTORC1 activation.
Mechanism of TFEB and TFE3 activation in response to nutrient levels
Elucidating the mechanism of TFEB/TFE3 activation was the key for understanding the conditions under which cells enhance lysosomal biogenesis (Figure 1). Perhaps not surprisingly, TFEB and TFE3 were shown to respond to the level of nutrients within cells (Martina et al 2014, Settembre & Ballabio 2011). More unexpected was the finding of a link between TFEB/TFE3 activation and their intracellular distribution. When cells are fully fed, TFEB and TFE3 remain excluded from the nucleus and accumulate in the cytosol. In contrast, following starvation they rapidly translocate to the nucleus and up-regulate multiple genes, thus promoting autophagy and lysosomal biogenesis as a way to help cells adapt and survive nutrient deprivation (Martina et al 2014, Settembre & Ballabio 2011). TFEB and TFE3 are partially redundant in terms of their ability to induce lysosomal biogenesis in response to starvation, but both must be present for a maximal response (Martina et al 2014).
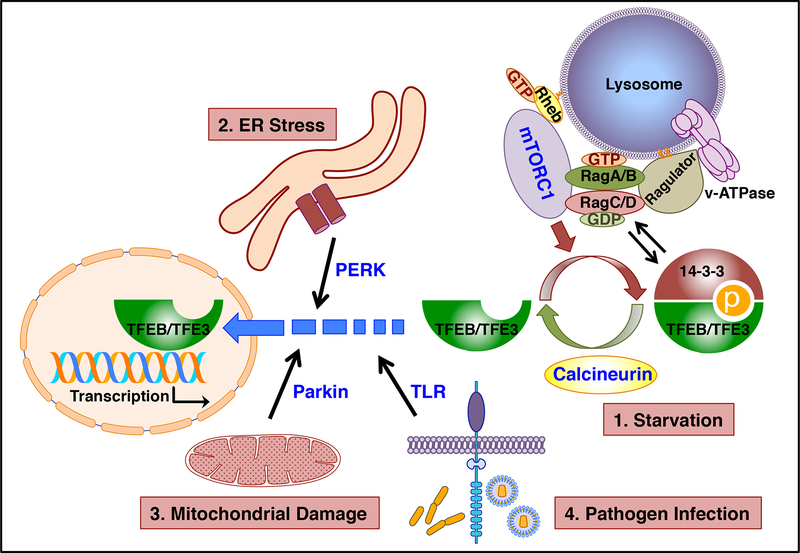
Figure 1. TFEB/TFE3 respond to different types of cellular stress.
Under normal conditions mTORC1 phosphorylates TFEB/TFE3, thus promoting their cytosolic retention. Following starvation (1), mTORC1 inactivation together with activation of specific phosphatases such as calcineurin, lead to TFEB/TFE3 nuclear translocation. TFEB/TFE3 activation is also observed in response to ER stress (2), mitochondrial damage (3), and pathogen infection (4). The proteins implicated in TFEB/TFE3 activation under different stress conditions are represented in blue.
Importantly, the intracellular localization of TFEB and TFE3 is regulated by mTORC1. In fully fed cells, TFEB and TFE3 are recruited to the lysosomal surface where they undergo mTORC1-dependent phosphorylation. mTORC1 phosphorylates TFEB and TFE3 at several resides but serine 211 (S211) in the case of TFEB and S321 in TFE3 are particularly relevant, since phosphorylation of these residues creates a binding site for the cytosolic chaperone 14–3–3. Interaction with 14–3–3 results in sequestration of these transcription factors in the cytosol (Martina et al 2012, Martina et al 2014, Roczniak-Ferguson et al 2012, Settembre et al 2012). Conversely, when nutrients are scarce, inactivation of mTORC1, together with de-phosphorylation of S211 and S321, prevent binding to 14–3–3, resulting in a rapid accumulation of TFEB and TFE3 in the nucleus.
Study of the mechanism that regulate recruitment of mTORC1, TFEB, and TFE3 to lysosomes further revealed that lysosomes do not function as mere signaling platform but play a more complex role in sensing and regulating energy homeostasis. It was shown that fluctuations in amino acids concentration inside the lysosomal lumen affect the conformation of v-ATPase. When the amino acid levels are high, v-ATPase interacts extensively with Ragulator, a pentameric protein complex associated with the outside of the lysosomal surface, thus activating its guanine nucleotide exchange factor (GEF) activity (Zoncu et al 2011). Ragulator then transmits the information regarding amino acid availability by modulating the nucleotide status of the small GTPases Rags (Bar-Peled et al 2012). Rag GTPases assemble as heterodimers consisting of either RagA or RagB bound to Rag C or RagD (Gao & Kaiser 2006, Sekiguchi et al 2001). Ragulator not only tethers Rag GTPases to the lysosome but also acts as a GEF for RagA/B, favoring the formation of RagA/BGTP-RagC/DGDP heterodimers (Bar-Peled et al 2012, Sancak et al 2010, Sancak et al 2008). It is in this particular conformation that Rags interact with TFEB/TFE3, promoting their recruitment to the lysosomal surface (Martina et al 2014, Martina & Puertollano 2013). The interaction between Rags and TFEB/TFE3 is mediated by a Rag-binding motif located within the 30 terminal residues of the transcription factors. Similar to TFEB/TFE3, mTORC1 is recruited to lysosomes in nutrient abundant conditions through the interaction of one of its subunits, Raptor, with RagA/BGTP-RagC/DGDP heterodimers (Sancak et al 2010). Once on the lysosome, mTORC1 is activated by the small GTPase Rheb, which requires the presence of growth factors for its own activation (Saucedo et al 2003, Stocker et al 2003). The synchronized, Rag-mediated recruitment of mTORC1 and its substrates to the same regions of the lysosomal surface in nutrient rich conditions provides a spatiotemporal regulation for the mTORC1-induced phosphorylation and inhibition of TFEB/TFE3. In fact, mutations in the TFEB/TFE3 Rag binding domain prevent their recruitment to lysosomes, mTORC1-mediated phosphorylation, and binding to 14–3–3, thus resulting in constitutive activation of these transcription factors even when nutrients are plentiful (Martina et al 2014, Martina & Puertollano 2013).
Under starvation conditions, the nucleotide state of Rags changes, resulting in the predominant formation of RagA/BGDP-RagC/DGTP heterodimers. Since Rags are unable to interact with mTORC1 or TFEB/TFE3 in this conformation, the mTORC1 inhibitory effect is relieved, allowing for TFEB/TFE3 nuclear translocation. Interestingly, RagA/BGDP-RagC/DGTP heterodimers recruit a different set of proteins to the lysosome in starvation conditions, including folliculin and tuberous sclerosis complex (TSC) (Demetriades et al 2014, Martina et al 2014, Petit et al 2013, Tsun et al 2013). While the TSC complex functions as a GTPase-activation protein (GAP) that inhibits Rheb activity further ensuring mTORC1 inactivation, folliculin shows GAP activity toward RagC/D and has been proposed to contribute to the rapid mTORC1 re-activation observed in refed conditions (Dibble et al 2012, Tsun et al 2013). Moreover, TFEB and TFE3 induce transcriptional up-regulation of folliculin and the folliculin interacting proteins FNIP1 and FNIP2, suggesting that these transcription factors not only help cells respond to starvation but prepare them for efficient transition to nutrient rich conditions (Martina et al 2014).
Apart from mTORC1, other kinases are likely to participate in TFEB/TFE3 regulation. Phosphorylation of TFEB-S142 by MAPK has been suggested to contribute to TFEB cytosolic retention (Settembre et al 2011). This is an interesting possibility considering that MAPK can also be recruited to lysosomes via interaction with specific Ragulator subunits (Nada et al 2009, Wunderlich et al 2001).
One important consideration is that efficient TFEB-S211 and TFE3-S321 dephosphorylation requires not only mTORC1 inhibition, but also activation of phosphatases that specifically target these residues. In this regard, it has been recently shown that calcineurin plays an important role; under starvation conditions, localized calcium release from lysosomes induces activation of calcineurin, which dephosphorylates TFEB critical serine residues, thus promoting its nuclear translocation (Medina et al 2015). These results open a possibility that calcium release form other compartments (e.g. endoplasmic reticulum, mitochondria) may also lead to calcineurin-mediated TFEB/TFE3 activation. Also, lysosomal calcium release may have a broader signaling impact by regulating additional calcineurin-dependent or independent pathways.
Finally, the mechanism of TFEB/TFE3 activation in response to starvation seems to be highly conserved among species. Mitf, the only member of the MiTF/TFE family in Drosophila melanogaster, is also retained in the cytosol through interactions with 14–3–3. Starvation-induced inactivation of TORC1 causes Mitf nuclear translocation and Mitf-dependent expression of all v-ATPase subunits, resulting in regulation of the v-ATPase activity. Of note, increased v-ATPase activity leads to enhanced TORC1 activity and promotes sequestration of Mitf back to the cytoplasm (Zhang et al 2015). This suggests the presence of a negative regulatory loop that maintains the activity of these critical regulators in balance and might have important implications in metabolic diseases and cancer.
TFEB and TFE3 as metabolic regulators
Metabolic regulation is essential to ensure survival during fasting. It has been suggested that autophagy plays an important role in lipid catabolism. Cells accumulate fat in the form of triglycerides (TGs) in specialized organelles named lipid droplets. Fasting triggers TGs breakdown into fatty acids that are eventually imported into mitochondria and incorporated into the TCA cycle to produce ATP. Autophagy participates in lipid catabolism by delivering fragments of lipid droplets to lysosomes, where the lysosomal acid lipase (LAP) mediates TGs degradation. TFEB contributes to the fasting response not only by up-regulating autophagy and lysosomal biogenesis, but also by increasing expression of key mediators of the lipid catabolism, including genes implicated in import of fatty acids, beta-oxidation of fatty acids in mitochondria and peroxisomes, LAP, and the master regulators of lipid catabolism PGC1-alpha and PPAR-alpha (Settembre et al 2013). Accordingly, liver-specific TFEB knockout results in accumulation of lipid droplets and defective lipid degradation during starvation. Conversely, TFEB over-expression enhances fatty acid catabolism and whole-body energy metabolism, while preventing obesity and metabolic syndrome in high-fat diet-fed mice (Settembre et al 2013).
The role of TFE3 in regulating expression of metabolic genes is less well characterized. TFE3 seems to play an important role in energy homeostasis in liver and muscle by regulating expression of several genes implicated in insulin signaling (Iwasaki et al 2012, Nakagawa et al 2006). Muscle-specific TFE3 transgenic mice display increased glycogen accumulation and high exercise endurance capacity. TFE3 also regulates expression of PGC1-alpha in muscle, further confirming the role of this transcription factor in energy metabolism (Salma et al 2015). Finally, it has been suggested that TFE3 regulates lipid metabolism in adipose tissues by modulating expression of key lipolysis regulators (Fujimoto et al 2013).
The regulatory role of TFEB and TFE3 in metabolism is evolutionary conserved. HLH-30, the ortholog of TFEB and TFE3 in C elegans, translocates to the nucleus following starvation to up-regulate expression of the lysosomal lipases LIPL-1 and LIPL-3. Transcription of these lipases, as well as several autophagy genes, is impaired in hlh-30 mutants, resulting in defective degradation of lipid droplets (O’Rourke & Ruvkun 2013). In agreement with the recent suggestion that autophagy and nutrient-signaling pathways are linked to longevity in C. elegans (Lapierre & Hansen 2012), HLH30 over-expression was found to extend lifespan in this model (Lapierre et al 2013).
CELLULAR STRESS
Besides nutrient deprivation, cells must monitor and respond to various types of perturbations. The cellular response to stress involves numerous pathways including those that regulate protein folding, mitochondria homeostasis, cell fate and lineage decisions, growth control and cell cycle, and cellular survival/death programs. It is, therefore, not surprising that the signals that regulate these processes and those that control the autophagic/lysosomal pathway communicate with each other. Recent evidence indicates that TFEB and TFE3 are activated in response to mitochondrial and ER stress (Figure 1), suggesting a more general role in cellular adaptation to stress than previously anticipated.
Mitochondrial stress
Mitophagy is the process by which damaged mitochondria are eliminated via autophagy. Under conditions of loss of mitochondrial membrane potential, PINK1 kinase induces recruitment of the cytosolic E3 ligase Parkin to the outer mitochondrial membrane. Parkin-mediated ubiquitination of select outer mitochondrial membrane proteins, such as mitofusins and Miro1, initiates the recruitment of key regulators of autophagosome formation, leading to the elimination of impaired mitochondria (Narendra et al 2012). Interestingly, mitophagy induction by treatment with the ATP synthase inhibitor oligomycin and the complex III inhibitor antimycin A, results in translocation of TFEB and TFE3 to the nucleus in a process that requires PINK1, Parkin, Atg9A, and Atg5 but not mTORC1 inactivation. Conversely, Atg5 is not required for TFEB nuclear accumulation upon nutrient deprivation, suggesting that the mechanism of TFEB activation during starvation and mitophagy is different (Nezich et al 2015). Further pointing to a role for Parkin in TFEB regulation is the observation that Mutation Q311X in Parkin causes decreased degradation of PARIS, a transcriptional repressor of PGC1-alpha, leading to reduced levels of PGC1-alpha and TFEB (Siddiqui et al 2015). Depletion of TFEB alone does not result in mitophagy defects. However, depletion of all members of the MiTF/TFE family (TFEB/TFE3/MITF/TFEC) causes impaired degradation of damaged mitochondria (Nezich et al 2015), further confirming the redundancy among members of the MiTF/TFE family (Martina et al 2014, Steingrimsson et al 2002).
The positive transcriptional feedback loop between PGC1-alpha and TFEB is probably critical to modulate mitochondrial quality and function in different tissues. PGC1-alpha is a master regulation of mitochondrial biogenesis but it can also modulate mitophagy by regulating expression of TFEB (Tsunemi & La Spada 2012). Likewise, TFEB promotes mitochondria degradation but also biogenesis by inducing expression of PGC1-alpha (Settembre et al 2012). Accordingly, animals lacking PGC1-alpha exhibit myopathic characteristics reminiscent of those seen in autophagy-deficient muscle (Vainshtein et al 2015), whereas TFEB activation enhances removal of depolarized mitochondria, restores normally polarized mitochondria, and prevents ischemiareperfusion-induced cardiomyocyte death (Ma et al 2015). In addition, the cardioprotective effect of cobalt protoporphyrin IX (CoPPIX) has been linked to its ability to simultaneously activate TFEB and mitophagy (Unuma et al 2013). Finally, treatment with the TFEB/TFE3 activator rapamycin prevents losses in mitochondrial function and restores cell viability in mitochondrially compromised human iPSC-derived dopaminergic neurons (Siddiqui et al 2015).
ER stress
Accumulation of misfolded proteins in the ER is a potent stress signal that induces activation of stress responses, such as the unfolded protein response (UPR) and autophagy, with the goal of reestablishing cell homeostasis. Recent evidence indicates that TFEB and TFE3 are activated in response to ER stress (Martina et al 2016). TFE3 nuclear translocation under ER stress is mTORC1 independent but requires PERK, an ER integral membrane protein that senses protein missfolding in the ER lumen and activates UPR. ChIP-seq analysis of MEFs subjected to either starvation or tunicamycin treatment revealed a high degree of overlap between the genes regulated by TFE3 under each condition. TFE3 targets included not only autophagic/lysosomal genes, but also ATF4, an essential master regulator of the integrated stress response, and genes implicated in cell response to stress, signaling, and apoptosis (Martina et al 2016). Therefore, TFE3 may have an important role integrating cooperation between different cellular stress pathways. Of note, depletion of TFEB and TFE3 in MEFs results in increased resistance to apoptosis under conditions of prolonged ER stress. This suggests that TFEB and TFE3 might have a dual role in cell fate, promoting either survival or cell death depending on the duration and strength of the stress (Martina et al 2016).
Cell fate and lineage decisions
Cell lineage decisions are driven by the action of different transcription factors that promote stem cells’ commitment toward specific precursors. TFE3 has been implicated in differentiation of hematopoietic myeloid precursors. TFE3 activation in myoblasts is critical to promote their maturation toward the macrophage lineage (Zanocco-Marani et al 2006, Zanocco-Marani et al 2009). TFE3 also regulates osteoclast development by promoting transition of mono-nucleated to multi-nucleated osteoclasts. In this case, TFE3 activation occurs downstream of the growth factors M-CSF and RANK and requires MAPK-dependent TFE3 phosphorylation (Hershey & Fisher 2004). Conversely, a recent report suggested the involvement of TFE3 in the maintenance of pluripotency (Betschinger et al 2013). Translocation of TFE3 from the nucleus to the cytoplasm is required to allow embryonic stem cells to exit their naive pluripotent state and commit to cellular differentiation. Interestingly, cytoplasmic retention of TFE3 requires the folliculin/FNIP1/FNIP2 complex, which, as mention earlier, is also involved in bringing TFE3 back to the cytosol during re-feed conditions (Martina et al 2014).
Many cell types undergo a profound reorganization of their endo-lysosomal system during differentiation. This is the case of osteoclasts, a specialized cell type that secrete lysosomal hydrolases at the site of bone resorption and whose function is critical for skeletal formation and remodeling. It was recently shown that TFEB is required for osteoclasts function in vivo (Ferron et al 2013). Incubation of osteoclast with RANK promotes TFEB phosphorylation via activation of PKCβ kinase. Upon translocation to the nucleus, TFEB induces lysosomal biogenesis and facilitates secretion of lysosomal hydrolases. Accordingly, depletion of TFEB in osteoclasts results in decreased expression of lysosomal genes, reduced number of lysosomes, and defective resorption of the bone matrix (Ferron et al 2013).
Wnt signaling regulates cell differentiation, embryonic pattering and organogenesis during development. Wnt also participate in adult tissues homeostasis and its dysregulation often leads to cancer. Recent evidence suggests an important role of MITF in Wnt signaling (Ploper et al 2015). In the absence of Wnt, glycogen synthase kinase 3 (GSK3)-dependent phosphorylation of MITF promotes its degradation. In contrast, Wnt ligand binding induces GSK3 sequestration into multivesicular bodies (MVBs), resulting in MITF stabilization and nuclear transport. MITF activation expands the MVB compartment further enhancing Wnt signaling. Since most of the GSK3 phosphorylation sites identified in MITF are also present in TFEB and TFE3, it is possible that Wnt-dependent TFEB and TFE3 regulation also influence development and/or cancer progression.
IMMUNE RESPONSE
Numerous studies have demonstrated essential roles for the autophagy-lysosome system in macrophages and other cells of the immune system in response to pathogen exposure (Puleston & Simon 2014). These include degradation of phagocytosed pathogens, antigen processing and presentation, natural killer (NK) and T-cell cytotoxic granule secretion, and TLR signaling (Colbert et al 2009). In addition, autophagy is implicated in direct engulfment of intracellular pathogens and in modulation of inflammatory signaling through the inflammasome complex (Jo et al 2013, Shi et al 2012).
Considering the important role of TFEB/TFE3 as master regulators of autophagy induction and lysosomal biogenesis, their recently revealed participation in the transcriptional regulation of the immune response is not surprising. Activation of macrophages with different toll-like receptor (TLR) ligands or live bacteria has been recently shown to result in a robust and persistent accumulation of TFE3 in the nucleus. Similar to mitochondrial and ER stress conditions, TFE3 nuclear translocation is not accompanied by a noticeable reduction in mTORC1 activity in LPS-treated macrophages, further confirming that TFE3 can be activated in a mTORC1-independent manner (Pastore et al 2016). Generation of edited knockout cell lines and in vivo mouse models revealed that depletion of TFEB and TFE3 leads to a significant decrease in autophagosome/lysosomal biogenesis as well as inhibition of the late endosomal/lysosomal system remodeling normally associated with macrophage activation (Pastore et al 2016). Depletion of either TFEB or TFE3 results in a more subtle phenotype, indicating that the two transcription factors cooperate in the regulation of the innate immune response in activated macrophages. ChIP-seq analysis confirmed increased binding of TFE3 to the promoter of autophagic/lysosomal genes following LPS stimulation. Notably, TFE3 also binds to the promoter of numerous immune genes, suggesting a more direct role of TFE3 in inflammatory response. Accordingly, transcriptional up-regulation and secretion of several key cytokines and chemokines are severely reduced in TFEB/TFE3 knockout cells both in vitro and in vivo (Pastore et al 2016). These observations are in agreement with a report from the Irazoqui laboratory showing that HLH-30, the ortholog of TFEB and TFE3 in C elegans, regulates expression of numerous cryoprotective and antimicrobial genes in response to S aureus infection (Visvikis et al 2014), indicating that the role of TFEB/TFE3 in innate immune response is conserved through evolution.
TFEB has also been implicated in the expression of interferon-stimulated genes (ISGs). The exonuclease TREX1 degrades host cytosolic DNA as a way to prevent autoimmunity. Mutations in TREX1 result in ISGs expression though the activation of an interferon-independent, TFEB-dependent pathway (Hasan et al 2013). It was proposed that TFEB activation in TREX1-deficient cells causes an expansion of the lysosomal system, resulting in activation of STING, TBK1 and the transcription factors IRF3 and IRF7, and leading to ISGs expression (Hasan et al 2013). However, based on the aforementioned observations of Pastore et al. and Visvikis et al., it is tempting to speculate that TFEB may also directly contribute to the transcriptional up-regulation of ISGs. Importantly, mutations in TREX1 have been linked to lupus erythematosus and other inflammatory disorders. Therefore, TFEB is likely to play a role in the pathogenesis of autoimmune diseases.
TFEB and TFE3 also contribute to the adaptive immune response. TFEB is activated during dendritic cell maturation, leading to increased phagosomal acidification, increased protein degradation, and enhanced antigen presentation by the major histocompatibility complex (MHC) type II, a process that is critical for initiating the T-cell response to pathogen infection (Samie & Cresswell 2015). Moreover, B-lymphocytes isolated from TFE3 knockout mice show defects consistent with impaired B-cell activation, including reduced surface expression of CD23 and CD24 and decreased antibody production (Merrell et al 1997); a combined depletion of TFE3 and TFEB in activated CD4(+) T lymphocytes reduces expression of CD40 ligand, causing an aberrant antibody response (Huan et al 2006).
An additional indication that TFEB and TFE3 are key players in the host response to infection is the finding that some pathogens have developed systems to modulate their activation. For example, shortly after macrophage exposure to HIV, TFEB is activated in a process that is dependent on TLR8. This leads to a transient increase in autophagy that is critical for HIV replication (Campbell & Spector 2013). However, since sustained autophagy may increase HIV degradation, the virus has developed ways to down-regulate autophagy in chronic infection conditions. Campbell et al. recently showed that the HIV protein Nef directly binds Beclin, resulting in mTOR activation, TFEB phosphorylation and cytosolic retention, and consequent autophagy inhibition (Campbell et al 2015). Therefore, regulation of TFEB activation during HIV infection is critical for virus survival. In agreement with this, Apolipoprotein L1 (APOL1), a major component of the innate immune response, contributes to HIV suppression not only by increasing degradation of the viral proteins Vif and Gag and but also by inducing TFEB-dependent lysosomal biogenesis (Taylor et al 2014).
TFEB is also involved in other so far poorly understood inflammatory processes. For example, LPS stimulation results in extrusion of mitochondrial contents from hepatocytes and embryonic fibroblasts via a process that requires both autophagy and TFEB activation, and this mitochondrial content is sufficient to induce an inflammatory response (Unuma et al 2015). Another study reported that activation of protease-activated receptors (PARs) in urinary bladder leads to a concomitant increase in inflammation and TFEB expression (Saban et al 2007). Finally, TFEB over-expression reduced lysosomal cell death in macrophages activated under lipotoxic conditions, suggesting that TFEB regulates the cross talk between lipid metabolism, lysosomes, and immune response (Schilling et al 2013).
It is likely that TFEB and TFE3 make additional contributions to the immune response. Over-expression of TFEB or TFE3 is sufficient to induce fusion of lysosomes with the plasma membrane (Martina et al 2014, Medina et al 2011), and TFEB-mediated lysosomal exocytosis in osteoclasts and hepatocytes plays an important role in bone remodeling and copper homeostasis, respectively (Ferron et al 2013, Polishchuk et al 2014). Many immune cells use exocytosis of lysosomes and lysosome-related organelles as a mechanism for specialized secretion. For example, cytotoxic T-lymphocytes and natural killer (NK) cells store granzymes and perforin in secretory lysosomes. Controlled spatiotemporal release of these molecules is critical to eliminate infected host cells (Luzio et al 2014). It is, therefore, plausible that TFEB and TFE3 participate in the biogenesis, trafficking, and/or exocytosis of secretory lysosomes. In agreement with this idea is the finding of reduced degranulation of mast cells isolated from TFE3 knockout mice. Furthermore, histamine levels in plasma following an allergic trigger were reduced in TFE3 knockouts, suggesting that TFE3 is an important mediator of allergic response (Yagil et al 2012).
In summary, the unique ability of TFEB and TFE3 to simultaneously modulate autophagy induction, lysosomal biogenesis and exocytosis, cytokine expression, and antibody production, establishes them as key players in the transcriptional regulation of the immune response.
TFEB AND TFE3 IN CANCER
Dysregulation of MiTF/TFE factors can lead to different type of cancers (Haq & Fisher 2011). Cancer cells depend on effective lysosomal function, and multiple changes in the lysosomal composition and number happen during the oncogenic process. Although it is still unclear how the function of TFEB/TFE3 factors may help promote the oncogenic state, the emerging evidence suggests that cancer cells may exploit the TFEB/TFE3-mediated transcriptional activation of lysosome-dependent degradative pathway for their survival.
Renal cell carcinomas
Germline mutations in multiple genes confer susceptibility to renal cell carcinomas (RCC), a heterogeneous group of tumors arising from renal tubular epithelium (reviewed in (Linehan & Ricketts 2013)). Gene fusions involving members of MiTF/TFE family, TFE3, and less frequently TFEB, define a distinct subset of RCC that is referred to as translocation RCC (t-RCC). These rare tumors, seen more often in children and adults, account for less than 5% of all sporadic kidney cancers (reviewed in (Kauffman et al 2014, Linehan & Ricketts 2013, Magers et al 2015).
Since the first described fusion of TFE3 on the short arm of the X chromosome (Xp11.2) to Papillary renal cell carcinoma (PRCC) on chromosome 1q21.2 [PRCC-TFE3 t(X;1)(p11.2;q21)](Sidhar et al 1996), several other TFE3 translocation partners have been identified. These tumors, designated as ASPSCR1-TFE3 t(X;17)(p11.2;q25), SFPQTFE3 t(X;1)(p11.2;q34), CLTC-TFE3 t(X;17)(p11.2;q23), and NONO-TFE3 inv(X)(p11.2;q12), tend to be aggressive and are seen more often in women. The latter fusion is generated by inversion of the TFE3 and NONO loci. Two novel TFE3 fusion partners, RBM10 and DVL2, have been recently described (Linehan et al 2016); a fusion of TFE3 with unknown genes on chromosome 3 t(X;3)(p11.2;q23) and 10 t(X;3)(p11.2;q23) has also been reported, each in a single patient (Argani 2015). Xp11 translocation/TFE3 gene fusions are not restricted to RCC; for example, the ASPSCR1-TFE3 gene fusions were found in alveolar soft part sarcomas, a rare pediatric lung cancer without kidney involvement (Ladanyi et al 2001).
The breakpoint sites in TFE3 fusions differ even for the same partners, thus generating various isoforms. The chimeric transcripts contain the N-terminal part of the fusion partner connected to different C-terminal coding exons of TFE3; exons 6–10 of TFE3, which include the bHLH/LZ and transcriptional activation domains, are always preserved (Kauffman et al 2014). The resulting chimeric proteins are overexpressed and exhibit strong TFE3 nuclear staining.
Until recently, a single TFEB fusion partner – MALAT1 – was known (originally called Alpha; (Davis et al 2003)). MALAT1-TFEB t(6.11)(p21;q12)(Argani et al 2001) is an extremely rare condition with only two dozen cases described. Most of the reported TFEB breakpoints are in the cluster upstream of the ATG in exon 3 leading to the retention and overexpression of the full-length wild-type TFEB coding region (Kuiper et al 2003); a single case of TFEB breakpoint in exon 4 has also been reported (Inamura et al 2012). Two novel TFEB fusion partners, COL21A1 and CADM2 (the latter generates a truncated version of TFEB), have been identified in a large study using a comprehensive genomic approach (Linehan et al 2016).
The oncogenic potential of the fusion proteins is thought to arise from the augmentation of the intrinsic pro-oncogenic properties of TFE3/TFEB themselves; that is to say, the oncogene is highly expressed due to the introduction of a new, more active promoter (Kauffman et al 2014). MALAT1-TFEB is particularly revealing, since the MALAT1 provides a much stronger promoter without changing the TFEB’s protein coding sequence.
In addition to TFE3 overexpression brought about by Xp11.2 translocations, folliculin (FLCN)-induced phosphorylation-dependent nuclear translocation and activation of TFE3 is linked to renal tumor development in Birt-Hogg-Dube (BHD) syndrome (Hong et al 2010), a rare autosomal dominant disorder in which carriers of germline FLCN mutations are at risk for developing RCC, fibrofolliculomas, lung cysts, and spontaneous pneumothorax (Nickerson et al 2002, Schmidt & Linehan 2015, Schmidt et al 2005).
The relevance of MITF - a designated melanoma oncogene - to RCC has been recently demonstrated by the identification of a germline heterozygous MITF missense mutation (pE318K) that confers an increased risk for developing RCC and/or melanoma. The mutation does not affect MITF nuclear translocation, but rather weakens its interaction with small-ubiquitin-like modifier (SUMO) proteins leading to transcriptional activation of MITF and upregulation of its multiple downstream targets, including those involved in melanocyte and kidney tumorigenesis (Bertolotto et al 2011).
Aneuploidy
Activation of TFEB has been recently reported in aneuploid cells as a cellular response to lysosomal stress following lysosomal accumulation of undegraded autophagic cargo (Santaguida & Amon 2015, Santaguida et al 2015). Altered protein composition, accumulation of misfolded/aggregated proteins, and proteotoxicity are critical components of the complex phenotype of aneuploid cells (Oromendia & Amon 2014, Stingele et al 2012, Tang et al 2011). Chromosome mis-segregation (induced in hTERT immortalized nontransformed RPE-1 cells) resulted in the accumulation of protein aggregate-containing autophagosomes within lysosomes, leading to lysosomal stress response - mTORC1-independent nuclear translocation and activation of TFEB and its downstream targets. This response became obvious only after 2–3 cell divisions subsequent to the persistent generation of misfolded and/or aggregated proteins. Of note, lysosomal function did not seem to be compromised in these cells, as indicated by normal pH and activity of cathepsins, suggesting that the capacity of lysosomes may be limited (dubbed “lysosomal saturation”) (Santaguida & Amon 2015). Remarkably, activation of TFEB does not appear to translate into lysosomal expansion, perhaps because the TFEB response in aneuploid cells is wired to stimulate autophagosomal rather than lysosomal biogenesis (Santaguida & Amon 2015) (Santaguida et al 2015).
Aneuploidy, a common feature of human cancers, is present in ~ 90% of all solid tumors and ~ 50% of hematopoietic cancers (Gordon et al 2012). However, many aneuploid cancer cell lines do not exhibit “lysosomal saturation”, suggesting that tumor cells develop additional mechanisms to enhance their degradative capacity (Santaguida et al 2015). Pancreatic ductal adenocarcinoma (PDA), the most common and highly lethal pancreatic cancer, may be a case in point.
Pancreatic ductal adenocarcinoma
PDA tumors exhibit high basal autophagy, which appears to be a prerequisite for the tumorigenic growth (Yang et al 2011b). In addition, an uncharacteristic predominant nuclear localization of TFEB has been reported in fully fed human pancreatic cancer cells (PANC1).
A recent study by Perera et al. (Perera et al 2015) greatly expanded these findings and documented the following: an increase in size and number of both autophagosomes and lysosomes; an augmentation of autophagic flux; high levels of expression of MiTF/TFE (although somewhat less than in melanoma and RCC) with each family member-MITF, TFE3 or TFEB - dominating in different samples; and upregulation of CLEAR gene network. Furthermore, all three transcription factors escaped mTORC1-dependent inactivation, and remained nuclear-localized and constitutively active in PDA irrespective of nutrient status due to their interaction with the nucleocytoplasmic transporters, importin 8 (IPO8) or 7 (IPO7). Accordingly, knockdown of these transporters decreased the levels of nuclear TFE3, TFEB, and MITF in PDA cell lines. Also, MiTF/TFE factors in PDA cells were shown to stimulate lysosomal breakdown of the cargos delivered through both autophagy and macropinocytosis, thus providing the tumor cells with intracellular and extracellular nutrients and allowing them to survive under nutrient deprivation (Perera et al 2015) (Alderton 2015)). This unusual transcriptional activity of TFEB in pancreatic cancer is also supported by the increased number of TFEB-regulated proteins in human PDA samples, and an increase in the N-glycosylation of TFEB-controlled glycoproteins (Pan et al 2014).
TFEB AND TFE3 AS THERAPEUTIC TARGETS
Recent studies have demonstrated the potential of modulating TFEB/TFE3 activity as a therapeutic strategy for several major neurodegenerative and lysosomal disorders, in which defective autophagy-lysosomal pathway is an important contributor to the disease pathogenesis (reviewed in (Menzies et al 2015) (Damme et al 2015).
Neurological Disorders
Alzheimer’s disease (AD) is the most prevalent age-related neurodegenerative disorder. It is characterized by abnormal deposition of β-amyloid (Aβ) in neuritic plaques and the formation of intraneuronal neurofibrillary tangles (NFTs) (reviewed in (Himmelstein et al 2012)) (Peric & Annaert 2015)). Defective lysosomal clearance of both, Aβ and phosphorylated Tau (p-Tau; the main component of NFTs), underlies the mechanism of their accumulation in AD (Yang et al 2011a) (Polito et al 2014). Disruption of lysosomal biogenesis in AD has been recently linked to the increased levels of lysosomal acid sphingomyelinase (ASM) leading to low levels of TFEB and LAMP1 in tissues from AD patients and APP/PS1 (amyloid precursor protein/precenelin-1) mice (Lee et al 2014). Normalization of ASM activity in mice by partial genetic deletion of the ASM gene or by amitriptyline inhibition of the enzyme restored the levels of TFEB and LAMP1, ameliorated autophagic defect, decreased Aβ load, and improved the phenotype (Lee et al 2014).
Activation of TFEB has been suggested as an effective strategy to attenuate the amyloid plaque deposition; the effect was cell-type specific depending on the role of autophagic-lysosomal pathway in APP processing and Aβ clearance. In astrocytes, TFEB reduced Aβ half-life in the interstitial fluid (ISF) by accelerating the uptake, trafficking, and lysosomal degradation of exogenous Aβ. In vivo, astrocyte-specific TFEB expression lowered the levels of ISF Aβ in young APP/PS1 mice (before the deposition of plaques) and alleviated amyloid plaque pathology in old ones (Xiao et al 2014). In neurons, where Aβ is generated, TFEB overexpression promoted APP proteolysis, thus limiting its availability for amyloidogenic processing into Aβ. TFEB-mediated increase in APP degradation in response to inhibition of GSK3, a well-established critical component of the AD pathogenesis ((Takashima 2006), reviewed in (Kremer et al 2011)) further supports the role of TFEB in AAP proteolysis (Parr et al 2012). AAV-TFEB neuronal transduction in young AD mice reduced steady-state ISF Aβ levels, and in older AD mice (at the stage of early plaque deposition) attenuated amyloid plaque load (Xiao et al 2015). Of note, a widespread and persistent AAV-mediated neuronal TFEB expression had no effect on amyloid plaque deposition in another model of AD (5xFAD; (Oakley et al 2006)) (Polito et al 2014). A more severe and early development of amyloid pathology in these mice and/or insufficient levels of TFEB may account for the negative result.
Beneficial effect of TFEB overexpression on Tau pathology has been shown both in vitro, in cells expressing mutant Tau (P301L), and in vivo, in transgenic Tau model (rTg4510; (Ramsden et al 2005) (Santacruz et al 2005)) (Polito et al 2014). TFEB promoted clearance of misfolded and p-Tau without affecting the levels of unphosphorylated Tau, suggesting that TFEB stimulates degradation rather than dephosphorylation of the abnormal Tau. TFEB-mediated clearance of p-Tau was shown to involve PTEN, which contains two putative CLEAR sequences and appears to be a direct target of TFEB (Polito et al 2014). TFEB-mediated reduction of p-Tau was also achieved in a neuroblastoma model system following treatment with flubendazole, an antiparasitic drug, which was identified through the screen of small molecule libraries that activate autophagy (Chauhan et al 2015).
Parkinson’s disease (PD), the second most common progressive neurodegenerative disorder, involves a selective loss of dopamine-producing neurons in the substantia nigra and the development of neuronal Lewy bodies composed of abnormal protein deposits including α-synuclein aggregates. The removal of excess α-synuclein depends on the proper function of the autophagic-lysosomal pathway (Dehay et al 2013, Ebrahimi-Fakhari et al 2011). The first indication of the protective role of TFEB activation in PD came from the study in the experimental neurotoxin-induced PD cell model (Dehay et al 2010).
Reduced TFEB activity leading to lysosomal depletion and defective autophagy was recently documented in an in vivo model of α-synuclein toxicity (Decressac et al 2013). The reduced nuclear TFEB levels were also seen in dopaminergic neurons from PD patients. Because of its structural homology with 14–3–3 proteins, α-synuclein (Ostrerova et al 1999, Perez et al 2002) can bind and sequester TFEB in the cytoplasm, thus inhibiting autophagy and its own lysosomal clearance. TFEB overexpression or activation through pharmacological inhibition of mTORC1 promoted clearance of excess α-synuclein and halted the progression of neurotoxicity (Decressac et al 2013).
The disease pathogenesis of spinal and bulbar muscular atrophy (SBMA) (Kennedy’s disease; an -X-linked neuromuscular disorder) caused by trinucleotide (CAG) repeat expansion in exon 1 of the androgen receptor (AR) gene, is defined by the loss of normal AR function and gain-of-function toxicity of the mutant protein (La Spada et al 1991). Both normal and mutant AR have been shown to directly interact with TFEB; however, only normal AR was able to promote TFEB activity, whereas mutant forms failed to act as TFEB co-activator leading to diminished TFEB function, reduction in the expression of TFEB target genes, and defective autophagic flux (Cortes et al 2014). Overexpression of TFEB in a SBMA stem cell model rescued the flux and reversed the mitochondrial depolarization, a new SBMA cytotoxicity phenotype uncovered in this model (Cortes et al 2014). Importantly, analysis of skeletal muscle from SBMA transgenic (YAC AR100; (Sopher et al 2004)) and AR113Q knock-in mice (Yu et al 2006) showed increased nuclear localization of TFEB and a dramatic up-regulation of TFEB target genes - the opposite of the findings in neurons (Chua et al 2014). Thus, it appears that mutant polyQAR repressed TFEB in neurons, whereas in muscle it acted as TFEB activator.
Since the first discovery of SBMA as a polyglutamine repeat disease more than twenty repeat expansion disorders have been described (La Spada et al 1991) (La Spada & Taylor 2010). In Huntington’s disease (HD), an autosomal dominant progressive form of dementia, the expansion of the polyQ tract in the N-terminal region of the huntingtin (Htt) protein gives rise to an aberrant misfolded protein that is prone to aggregation and neurotoxicity. Overexpression of TFEB in the rat striatal cell model of HD induced the degradation of the polyQ-expanded Htt, thus preventing its accumulation (Sardiello et al 2009).
A significantly reduced TFEB expression and its target genes were shown in both neuronal cells and HD mice, and these defects were rescued by PGC-1α (Tsunemi et al 2012), a well-established culprit in the HD pathogenesis (Weydt et al 2006) (Cui et al 2006). Inducible expression of PGC-1α in HD mice (Schilling et al 1999) not only restored mitochondrial function, but also reduced Htt protein aggregates in hippocampus and cortex and alleviated neurotoxicity. Furthermore, the experiments with TFEB silencing in combination with PGC-1α overexpression and vice versa demonstrated that PGC-1α acts upstream of TFEB, and that the PGC-1α-mediated activation of TFEB explains the clearance of mutant Htt aggregates (La Spada 2012, Tsunemi et al 2012).
Lysosomal Storage Diseases
The ability of TFEB to enhance lysosomal capacity and improve autophagic flux has been exploited in LSDs (Medina et al 2011, Sardiello et al 2009), many of which exhibit defective autophagic flux (Lieberman et al 2012). Perhaps even more relevant to LSDs is the capacity of TFEB to induce lysosomal exocytosis (Medina et al 2011), a process of attachment/fusion of lysosomes with plasma membrane followed by a discharge of lysosomal content outside the cell (Andrews 2000). TFEB regulates this process by increasing the number of lysosomes ready to dock to and fuse with the plasma membrane and by eliciting the release of intracellular Ca2+ through the activation of its target gene, MCOLN1 (Medina et al 2011).
Overexpression of TFEB stimulated lysosomal exocytosis and promoted cellular clearance in several LSDs cell models: neuronal stem cells from mouse models of Mucopolysaccharidosis-IIIA (Sanfilippo syndrome; a deficiency of heparan N-sulfatase) and Multiple Sulfatase Deficiency (MSD; deficiency of all sulfatases); cells from a murine model of juvenile form of Neuronal Ceroid lipofuscinosis (Batten disease; mutations in CLN3 gene), and in immortalized muscle cells from a mouse model of Pompe disease (deficiency of glycogen-degrading acid-alpha glucosidase) (Medina et al 2011) (Spampanato et al 2013). Consistent with the role of MCOLN1 in TFEB-mediated lysosomal exocytosis, TFEB failed to promote cellular clearance in human cells that harbor loss of function mutations of MCOLN1 (Medina et al 2011). Lysosomal exocytosis and glycogen clearance was also documented in Pompe muscle cells overexpressing TFE3 (Martina et al 2014).
The effect of TFEB was also documented in vivo, in models of MSD and Pompe disease. AAV-mediated TFEB delivery into MSD mice not only cleared the storage material in liver and muscle, but also reduced inflammation and cell death, the two secondary abnormalities in this disorder (Medina et al 2011). Similarly, TFEB activation in Pompe muscle cleared excess glycogen and alleviated autophagic buildup, a major secondary pathology in this tissue (Spampanato et al 2013). Interestingly, lysosomal exocytosis was much less efficient in the setting of muscle-specific suppression of autophagy in Pompe mice (Feeney et al 2013). A potential role of TFEB as a therapeutic target for LSDs is further supported by the data showing that the biological effects of two candidate compounds, 2-Hydroxypropyl-β-cyclodextrin (HPβCD) for treatment of Niemann-Pick type C and Genistein for Mucopolysaccharidoses, are in part TFEB-mediated (Song et al 2014) (Moskot et al 2014).
Thus, TFEB/TFE3-mediated stimulation of lysosomal exocytosis represents a conceptually new and attractive therapeutic avenue for LSDs. However, the benefit of TFEB/TFE3 activation in LSDs does not end here. Given the role of TFEB/TFE3 in the transcriptional regulation of many lysosomal genes (Palmieri et al 2011), activation of these transcription factors holds promise to enhance the levels of disease causing mutant proteins with residual enzyme activity and amplify the effect of the recombinant therapeutic enzymes (Awad et al 2015, Song et al 2013).
CONCLUDING REMARKS
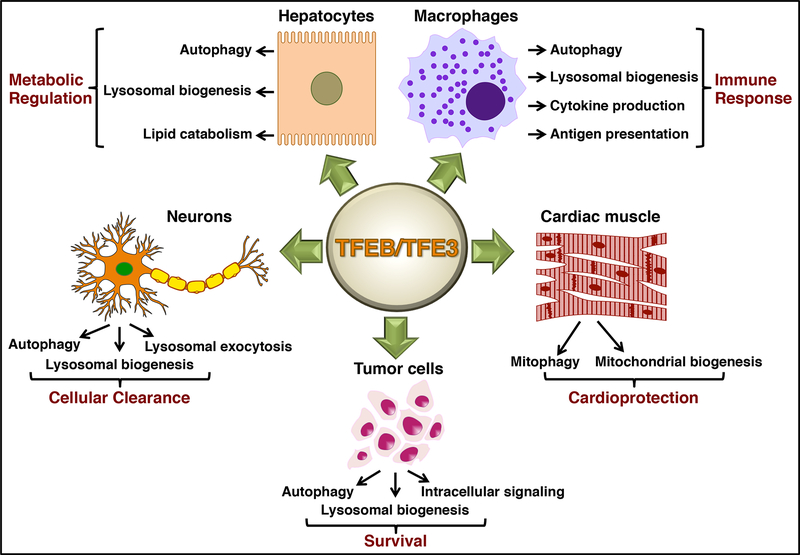
We have only recently begun to understand the contribution of TFEB and TFE3 to cellular response to stress. The role of these transcription factors in cellular adaptation to a wide variety of internal stresses and environmental fluctuations is inextricably linked to their unique ability to globally regulate the autophagic/lysosomal system. TFEB and TFE3 also control expression of key genes involved in the modulation of mitochondrial function, metabolism, unfolded protein response, apoptosis, and inflammatory response, revealing their broad functions. Moreover, the primary role of these transcription factors may well be cell type-specific (Figure 2): their activation appears critical for lipid catabolism in hepatocytes, for immune response in macrophages, and for efficient mitochondrial function in muscle. Further understanding of the TFEB/TFE3-mediated transcriptional network and the degree of redundancy of the two proteins in different cell types/conditions would be extremely useful.
Figure 2. TFEB/TFE3 regulate a complex transcriptional network critical for cellular adaptation to a variety of perturbations.
The primary role of TFEB/TFE3 may vary depending on cell type and includes metabolic regulation (hepatocytes), inflammatory response (macrophages), mitochondrial function (muscle), cellular clearance (neurons), and cell growth and survival (cancer cells).
The relevance of TFEB/TFE3 to human disease is no less critical. Activation of these transcription factors seems beneficial in many neurological and lysosomal disorders, but may confer an adaptive advantage to cancer cells. The development of small molecules that modulate TFEB/TFE3 activity in an accurate temporal- and tissue-specific manner is a rewarding area for future studies. These molecules have the potential to be used for a plethora of human diseases, including metabolic, immune, neurological, and oncogenic disorders, and would no doubt improve our understanding of the complex regulation of cellular adaptation to stress.
ACKNOWLEDGEMENTS
N.R. was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin diseases (NIAMS) of the NIH and a CRADA between NIH and Genzyme Corporation. R.P. was supported by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute (NHLBI).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
REFERENCES
- Alderton GK. 2015. Autophagy: Surviving stress in pancreatic cancer. Nature Reviews. Cancer 15: 513. [DOI] [PubMed] [Google Scholar]
- Andrews NW. 2000. Regulated secretion of conventional lysosomes. Trends Cell Biol 10: 316–21 [DOI] [PubMed] [Google Scholar]
- Argani P 2015. MiT family translocation renal cell carcinoma. Seminars in diagnostic pathology 32: 103–13 [DOI] [PubMed] [Google Scholar]
- Argani P, Hawkins A, Griffin CA, Goldstein JD, Haas M, et al. 2001. A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1;q12) chromosome translocation. Am J Pathol 158: 2089–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad O, Sarkar C, Panicker LM, Miller D, Zeng X, et al. 2015. Altered TFEB-mediated lysosomal biogenesis in Gaucher disease iPSC-derived neuronal cells. Hum Mol Genet 24: 5775–88 [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150: 1196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, et al. 2011. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 480: 94–8 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. 2013. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153: 335–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, Rawat P, Bruckman RS, Spector SA. 2015. Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration. PLoS pathogens 11: e1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, Spector SA. 2013. Inhibition of human immunodeficiency virus type-1 through autophagy. Current Opinion in Microbiology 16: 349–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Ahmed Z, Bradfute SB, Arko-Mensah J, Mandell MA, et al. 2015. Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. Nature Communications 6: 8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli Y, Ohanna M, Ballotti R, Bertolotto C. 2010. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment cell & melanoma research 23: 27–40 [DOI] [PubMed] [Google Scholar]
- Chua JP, Reddy SL, Merry DE, Adachi H, Katsuno M, et al. 2014. Transcriptional activation of TFEB/ZKSCAN3 target genes underlies enhanced autophagy in spinobulbar muscular atrophy. Hum Mol Genet 23: 1376–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert JD, Matthews SP, Miller G, Watts C. 2009. Diverse regulatory roles for lysosomal proteases in the immune response. European Journal of Immunology 39: 2955–65 [DOI] [PubMed] [Google Scholar]
- Cortes CJ, Miranda HC, Frankowski H, Batlevi Y, Young JE, et al. 2014. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nature Neuroscience 17: 1180–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. 2006. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell 127: 59–69 [DOI] [PubMed] [Google Scholar]
- Damme M, Suntio T, Saftig P, Eskelinen EL. 2015. Autophagy in neuronal cells: general principles and physiological and pathological functions. Acta Neuropathol 129: 337–62 [DOI] [PubMed] [Google Scholar]
- Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, et al. 2003. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A 100: 6051–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F. 1955. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J 60: 604–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A. 2013. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A 110: E1817–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Bove J, Rodriguez-Muela N, Perier C, Recasens A, et al. 2010. Pathogenic lysosomal depletion in Parkinson’s disease. JNeurosci 30: 12535–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Martinez-Vicente M, Caldwell GA, Caldwell KA, Yue Z, et al. 2013. Lysosomal impairment in Parkinson’s disease. Movement disorders : official journal of the Movement Disorder Society 28: 725–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, Teleman AA. 2014. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156: 786–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, et al. 2012. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell 47: 535–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Cantuti-Castelvetri I, Fan Z, Rockenstein E, Masliah E, et al. 2011. Distinct roles in vivo for the ubiquitin-proteasome system and the autophagy-lysosomal pathway in the degradation of alpha-synuclein. J Neurosci 31: 14508–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney EJ, Spampanato C, Puertollano R, Ballabio A, Parenti G, Raben N. 2013. What else is in store for autophagy? Exocytosis of autolysosomes as a mechanism of TFEB-mediated cellular clearance in Pompe disease. Autophagy 9: 1117–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron M, Settembre C, Shimazu J, Lacombe J, Kato S, et al. 2013. A RANKL-PKCbeta-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev 27: 955–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y, Nakagawa Y, Satoh A, Okuda K, Shingyouchi A, et al. 2013. TFE3 controls lipid metabolism in adipose tissue of male mice by suppressing lipolysis and thermogenesis. Endocrinology 154: 3577–88 [DOI] [PubMed] [Google Scholar]
- Fullgrabe J, Klionsky DJ, Joseph B. 2014. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol 15: 65–74 [DOI] [PubMed] [Google Scholar]
- Gao M, Kaiser CA. 2006. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol 8: 657–67 [DOI] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D. 2012. Causes and consequences of aneuploidy in cancer. Nature Reviews. Genetics 13: 189–203 [DOI] [PubMed] [Google Scholar]
- Haq R, Fisher DE. 2011. Biology and clinical relevance of the micropthalmia family of transcription factors in human cancer. Journal of Clinical Oncology : official journal of the American Society of Clinical Oncology 29: 3474–82 [DOI] [PubMed] [Google Scholar]
- Hasan M, Koch J, Rakheja D, Pattnaik AK, Brugarolas J, et al. 2013. Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes. Nature Immunology 14: 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey CL, Fisher DE. 2004. Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function. Bone 34: 689–96 [DOI] [PubMed] [Google Scholar]
- Himmelstein DS, Ward SM, Lancia JK, Patterson KR, Binder LI. 2012. Tau as a therapeutic target in neurodegenerative disease. Pharmacology & therapeutics 136: 8–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM. 2010. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One 5: e15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan C, Kelly ML, Steele R, Shapira I, Gottesman SR, Roman CA. 2006. Transcription factors TFE3 and TFEB are critical for CD40 ligand expression and thymus-dependent humoral immunity. Nature Immunology 7: 1082–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura K, Fujiwara M, Togashi Y, Nomura K, Mukai H, et al. 2012. Diverse fusion patterns and heterogeneous clinicopathologic features of renal cell carcinoma with t(6;11) translocation. The American Journal of Surgical Pathology 36: 35–42 [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Naka A, Iida KT, Nakagawa Y, Matsuzaka T, et al. 2012. TFE3 regulates muscle metabolic gene expression, increases glycogen stores, and enhances insulin sensitivity in mice. American Journal of Physiology. Endocrinology and metabolism 302: E896–902 [DOI] [PubMed] [Google Scholar]
- Jo EK, Yuk JM, Shin DM, Sasakawa C. 2013. Roles of autophagy in elimination of intracellular bacterial pathogens. Front Immunol 4: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman EC, Ricketts CJ, Rais-Bahrami S, Yang Y, Merino MJ, et al. 2014. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nature Reviews. Urology 11: 465–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Louis JV, Jaworski T, Van Leuven F. 2011. GSK3 and Alzheimer’s Disease: Facts and Fiction. Frontiers in Molecular Neuroscience 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, van den Berg E, et al. 2003. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum Mol Genet 12: 1661–9 [DOI] [PubMed] [Google Scholar]
- La Spada AR. 2012. PPARGC1A/PGC-1alpha, TFEB and enhanced proteostasis in Huntington disease: defining regulatory linkages between energy production and protein-organelle quality control. Autophagy 8: 1845–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. 2010. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nature Reviews. Genetics 11: 247–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. 1991. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352: 77–9 [DOI] [PubMed] [Google Scholar]
- Ladanyi M, Lui MY, Antonescu CR, Krause-Boehm A, Meindl A, et al. 2001. The der(17)t(X; 17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene 20: 48–57 [DOI] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, et al. 2013. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nature Communications 4: 2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Hansen M. 2012. Lessons from C. elegans: signaling pathways for longevity. Trends EndocrinolMetab 23: 637–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Jin HK, Park MH, Kim BR, Lee PH, et al. 2014. Acid sphingomyelinase modulates the autophagic process by controlling lysosomal biogenesis in Alzheimer’s disease. The Journal of Experimental Medicine 211: 1551–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. 2012. Autophagy in lysosomal storage disorders. Autophagy 8: 719–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan WM, Ricketts CJ. 2013. The metabolic basis of kidney cancer. Seminars in cancer biology 23: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, et al. 2016. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med 374: 135–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Hackmann Y, Dieckmann NM, Griffiths GM. 2014. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harbor perspectives in biology 6: a016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA. 2007. Lysosomes: fusion and function. Nat Rev Mol Cell Biol 8: 622–32 [DOI] [PubMed] [Google Scholar]
- Ma X, Liu H, Murphy JT, Foyil SR, Godar RJ, et al. 2015. Regulation of the transcription factor EB-PGC1alpha axis by beclin-1 controls mitochondrial quality and cardiomyocyte death under stress. Mol Cell Biol 35: 956–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magers MJ, Udager AM, Mehra R. 2015. MiT Family Translocation-Associated Renal Cell Carcinoma: A Contemporary Update With Emphasis on Morphologic, Immunophenotypic, and Molecular Mimics. Archives of pathology & laboratory medicine 139: 1224–33 [DOI] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8: 903–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Brady OA, Puertollano R. 2016. TFEB and TFE3 are novel components of the integrated stress response Embo J In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Li L, Lim J-A, Patange S, et al. 2014. The Nutrient-Responsive Transcription Factor TFE3 Promotes Autophagy, Lysosomal Biogenesis, and Clearance of Cellular Debris. Sci Signal 7: ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Puertollano R. 2013. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol 200: 475–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows NA, Sharma SM, Faulkner GJ, Ostrowski MC, Hume DA, Cassady AI. 2007. The expression of Clcn7 and Ostm1 in osteoclasts is coregulated by microphthalmia transcription factor. J Biol Chem 282: 1891–904 [DOI] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, et al. 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17: 288–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, et al. 2011. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 21: 421–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies FM, Fleming A, Rubinsztein DC. 2015. Compromised autophagy and neurodegenerative diseases. Nature Reviews. Neuroscience 16: 345–57 [DOI] [PubMed] [Google Scholar]
- Merrell K, Wells S, Henderson A, Gorman J, Alt F, et al. 1997. The absence of the transcription activator TFE3 impairs activation of B cells in vivo. Mol Cell Biol 17: 3335–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskot M, Montefusco S, Jakobkiewicz-Banecka J, Mozolewski P, Wegrzyn A, et al. 2014. The phytoestrogen genistein modulates lysosomal metabolism and transcription factor EB (TFEB) activation. J Biol Chem 289: 17054–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motyckova G, Weilbaecher KN, Horstmann M, Rieman DJ, Fisher DZ, Fisher DE. 2001. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc Natl Acad Sci U S A 98: 5798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada S, Hondo A, Kasai A, Koike M, Saito K, et al. 2009. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. Embo J 28: 477–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, Shimano H, Yoshikawa T, Ide T, Tamura M, et al. 2006. TFE3 transcriptionally activates hepatic IRS-2, participates in insulin signaling and ameliorates diabetes. Nat Med 12: 107–13 [DOI] [PubMed] [Google Scholar]
- Narendra D, Walker JE, Youle R. 2012. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harbor perspectives in biology 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezich CL, Wang C, Fogel AI, Youle RJ. 2015. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J Cell Biol 210: 435–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, et al. 2002. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer cell 2: 157–64 [DOI] [PubMed] [Google Scholar]
- O’Rourke EJ, Ruvkun G. 2013. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 15: 668–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley H, Cole SL, Logan S, Maus E, Shao P, et al. 2006. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci 26: 10129–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia AB, Amon A. 2014. Aneuploidy: implications for protein homeostasis and disease. Disease models & mechanisms 7: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrerova N, Petrucelli L, Farrer M, Mehta N, Choi P, et al. 1999. alpha-Synuclein shares physical and functional homology with 14–3–3 proteins. J Neurosci 19: 5782–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, et al. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20: 3852–66 [DOI] [PubMed] [Google Scholar]
- Pan S, Chen R, Tamura Y, Crispin DA, Lai LA, et al. 2014. Quantitative glycoproteomics analysis reveals changes in N-glycosylation level associated with pancreatic ductal adenocarcinoma. Journal of Proteome Research 13: 1293–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr C, Carzaniga R, Gentleman SM, Van Leuven F, Walter J, Sastre M. 2012. Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-beta precursor protein. Mol Cell Biol 32: 4410–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore N, Brady OA, Diab HI, Martina JA, Sun L, et al. 2016. TFEB and TFE3 Cooperate in the Regulation of the Innate Immune Response in Activated Macrophages Autophagy In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, et al. 2015. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 524: 361–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez RG, Waymire JC, Lin E, Liu JJ, Guo F, Zigmond MJ. 2002. A role for alpha-synuclein in the regulation of dopamine biosynthesis. J Neurosci 22: 3090–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric A, Annaert W. 2015. Early etiology of Alzheimer’s disease: tipping the balance toward autophagy or endosomal dysfunction? Acta Neuropathol 129: 363–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit CS, Roczniak-Ferguson A, Ferguson SM. 2013. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 202: 1107–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploper D, Taelman VF, Robert L, Perez BS, Titz B, et al. 2015. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci U S A 112: E420–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polishchuk EV, Concilli M, Iacobacci S, Chesi G, Pastore N, et al. 2014. Wilson disease protein ATP7B utilizes lysosomal exocytosis to maintain copper homeostasis. Dev Cell 29: 686–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito VA, Li H, Martini-Stoica H, Wang B, Yang L, et al. 2014. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med 6: 1142–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston DJ, Simon AK. 2014. Autophagy in the immune system. Immunology 141: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden M, Kotilinek L, Forster C, Paulson J, McGowan E, et al. 2005. Age-dependent neurofibrillary tangle formation, neuron loss, and memory impairment in a mouse model of human tauopathy (P301L). J Neurosci 25: 10637–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Marks MS. 2007. Melanosomes--dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol 8: 786–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, et al. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5: ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saban R, Simpson C, Davis CA, Dozmorov I, Maier J, et al. 2007. Transcription factor network downstream of protease activated receptors (PARs) modulating mouse bladder inflammation. BMC Immunol 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salma N, Song JS, Arany Z, Fisher DE. 2015. Transcription Factor Tfe3 Directly Regulates Pgc-1alpha in Muscle. Journal of Cellular Physiology 230: 2330–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie M, Cresswell P. 2015. The transcription factor TFEB acts as a molecular switch that regulates exogenous antigen-presentation pathways. Nature Immunology 16: 729–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, et al. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, et al. 2005. Tau suppression in a neurodegenerative mouse model improves memory function. Science 309: 476–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Amon A. 2015. Aneuploidy triggers a TFEB-mediated lysosomal stress response. Autophagy 12: 2383–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Vasile E, White E, Amon A. 2015. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev 29: 2010–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, et al. 2009. A gene network regulating lysosomal biogenesis and function. Science 325: 473–7 [DOI] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 5: 566–71 [DOI] [PubMed] [Google Scholar]
- Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, et al. 1999. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet 8: 397–407 [DOI] [PubMed] [Google Scholar]
- Schilling JD, Machkovech HM, He L, Diwan A, Schaffer JE. 2013. TLR4 activation under lipotoxic conditions leads to synergistic macrophage cell death through a TRIF-dependent pathway. J Immunol 190: 1285–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LS, Linehan WM. 2015. Molecular genetics and clinical features of Birt-Hogg-Dube syndrome. Nature reviews. Urology 12: 558–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LS, Nickerson ML, Warren MB, Glenn GM, Toro JR, et al. 2005. Germline BHD-mutation spectrum and phenotype analysis of a large cohort of families with Birt-Hogg-Dube syndrome. Am J Hum Genet 76: 1023–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. 2001. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem 276: 7246–57 [DOI] [PubMed] [Google Scholar]
- Settembre C, Ballabio A. 2011. TFEB regulates autophagy: an integrated coordination of cellular degradation and recycling processes. Autophagy 7: 1379–81 [DOI] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, et al. 2013. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 15: 647–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. The EMBO J 31: 1095–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, et al. 2012. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nature Immunology 13: 255–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A, Bhaumik D, Chinta SJ, Rane A, Rajagopalan S, et al. 2015. Mitochondrial Quality Control via the PGC1alpha-TFEB Signaling Pathway Is Compromised by Parkin Q311X Mutation But Independently Restored by Rapamycin. J Neurosci 35: 12833–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhar SK, Clark J, Gill S, Hamoudi R, Crew AJ, et al. 1996. The t(X;1)(p11.2;q21.2) translocation in papillary renal cell carcinoma fuses a novel gene PRCC to the TFE3 transcription factor gene. Hum Mol Genet 5: 1333–8 [DOI] [PubMed] [Google Scholar]
- Song W, Wang F, Lotfi P, Sardiello M, Segatori L. 2014. 2-Hydroxypropyl-beta-cyclodextrin promotes transcription factor EB-mediated activation of autophagy: implications for therapy. J Biol Chem 289: 10211–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Wang F, Savini M, Ake A, di Ronza A, et al. 2013. TFEB regulates lysosomal proteostasis. Hum Mol Genet 22: 1994–2009 [DOI] [PubMed] [Google Scholar]
- Sopher BL, Thomas PS Jr., LaFevre-Bernt MA, Holm IE, Wilke SA, et al. 2004. Androgen receptor YAC transgenic mice recapitulate SBMA motor neuronopathy and implicate VEGF164 in the motor neuron degeneration. Neuron 41: 687–99 [DOI] [PubMed] [Google Scholar]
- Spampanato C, Feeney E, Li L, Cardone M, Lim JA, et al. 2013. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med 5: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, et al. 2002. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl AcadSci USA 99: 4477–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. 2012. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Molecular Systems Biology 8: 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, et al. 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol 5: 559–65 [DOI] [PubMed] [Google Scholar]
- Takashima A 2006. GSK-3 is essential in the pathogenesis of Alzheimer’s disease. Journal of Alzheimer’s disease : JAD 9: 309–17 [DOI] [PubMed] [Google Scholar]
- Tang YC, Williams BR, Siegel JJ, Amon A. 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HE, Khatua AK, Popik W. 2014. The innate immune factor apolipoprotein L1 restricts HIV-1 infection. J Virol 88: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, et al. 2013. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 52: 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, et al. 2012. PGC-1alpha rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Science Translational Medicine 4: 142ra97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, La Spada AR. 2012. PGC-1alpha at the intersection of bioenergetics regulation and neuron function: from Huntington’s disease to Parkinson’s disease and beyond. Progress in neurobiology 97: 142–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unuma K, Aki T, Funakoshi T, Hashimoto K, Uemura K. 2015. Extrusion of mitochondrial contents from lipopolysaccharide-stimulated cells: Involvement of autophagy. Autophagy 11: 1520–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unuma K, Aki T, Funakoshi T, Yoshida K, Uemura K. 2013. Cobalt protoporphyrin accelerates TFEB activation and lysosome reformation during LPS-induced septic insults in the rat heart. PLoS One 8: e56526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainshtein A, Desjardins EM, Armani A, Sandri M, Hood DA. 2015. PGC-1alpha modulates denervation-induced mitophagy in skeletal muscle. Skeletal muscle 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verastegui C, Bertolotto C, Bille K, Abbe P, Ortonne JP, Ballotti R. 2000. TFE3, a transcription factor homologous to microphthalmia, is a potential transcriptional activator of tyrosinase and TyrpI genes. Mol Endocrinol 14: 449–56 [DOI] [PubMed] [Google Scholar]
- Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AM, et al. 2014. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40: 896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitner EB, Platt FM, Futerman AH. 2010. Common and uncommon pathogenic cascades in lysosomal storage diseases. J Biol Chem 285: 20423–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, et al. 2006. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab 4: 349–62 [DOI] [PubMed] [Google Scholar]
- Wunderlich W, Fialka I, Teis D, Alpi A, Pfeifer A, et al. 2001. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J Cell Biol 152: 765–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Yan P, Ma X, Liu H, Perez R, et al. 2014. Enhancing astrocytic lysosome biogenesis facilitates Abeta clearance and attenuates amyloid plaque pathogenesis. J Neurosci 34: 9607–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Yan P, Ma X, Liu H, Perez R, et al. 2015. Neuronal-Targeted TFEB Accelerates Lysosomal Degradation of APP, Reducing Abeta Generation and Amyloid Plaque Pathogenesis. J Neurosci 35: 12137–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagil Z, Hadad Erlich T, Ofir-Birin Y, Tshori S, Kay G, et al. 2012. Transcription factor E3, a major regulator of mast cell-mediated allergic response. J Allergy Clin Immunol 129: 1357–66 e5 [DOI] [PubMed] [Google Scholar]
- Yang DS, Stavrides P, Mohan PS, Kaushik S, Kumar A, et al. 2011a. Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer’s disease ameliorates amyloid pathologies and memory deficits. Brain 134: 258–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, et al. 2011b. Pancreatic cancers require autophagy for tumor growth. Genes Dev 25: 717–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Dadgar N, Albertelli M, Gruis K, Jordan C, et al. 2006. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. The Journal of clinical investigation 116: 2663–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanocco-Marani T, Vignudelli T, Gemelli C, Pirondi S, Testa A, et al. 2006. Tfe3 expression is closely associated to macrophage terminal differentiation of human hematopoietic myeloid precursors. Exp Cell Res 312: 4079–89 [DOI] [PubMed] [Google Scholar]
- Zanocco-Marani T, Vignudelli T, Parenti S, Gemelli C, Condorelli F, et al. 2009. TFE3 transcription factor regulates the expression of MAFB during macrophage differentiation. Exp Cell Res 315: 1798–808 [DOI] [PubMed] [Google Scholar]
- Zhang T, Zhou Q, Ogmundsdottir MH, Moller K, Siddaway R, et al. 2015. Mitf is a master regulator of the v-ATPase, forming a control module for cellular homeostasis with v-ATPase and TORC1. J Cell Sci 128: 2938–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334: 678–83 [DOI] [PMC free article] [PubMed] [Google Scholar]