SUMMARY
Itch is an unpleasant skin sensation that can be triggered by exposure to many chemicals, including those released by mast cells. The natriuretic polypep-tide b (Nppb)-expressing class of sensory neurons, when activated, elicits scratching responses in mice, but it is unclear which itch-inducing agents stimulate these cells and the receptors involved. Here, we identify receptors expressed by Nppb neurons and demonstrate the functional importance of these receptors as sensors of endogenous pruritogens released by mast cells. Our search for receptors in Nppb neurons reveals that they express leukotriene, serotonin, and sphingosine-1-phosphate receptors. Targeted cell ablation, calcium imaging of primary sensory neurons, and conditional receptor knockout studies demonstrate that these receptors induce itch by the direct stimulation of Nppb neurons and neurotransmission through the canonical gastrin-releasing peptide (GRP)-dependent spinal cord itch pathway. Together, our results define a molecular and cellular pathway for mast cell-induced itch.
Graphical Abstract
In Brief
Solinski et al. describe a cellular and molecular pathway that couples mast cell activation to itch behavior. They show that mast cell mediators, serotonin, leukotriene-C4, and sphingosine-1 phosphate directly stimulate Nppb sensory neurons via Htr1f, Cysltr2, and S1pr1 receptors, and this results in itch responses that are elicited through a canonical spinal cord-GRP pathway.
INTRODUCTION
Itch is an irritating cutaneous sensation that leads to the desire to scratch (Savin, 1998), a system thought to have developed as a means to rapidly remove potentially harmful objects or substances from the skin (Stante et al., 2005). To accomplish this protective response, sensory neurons detect cutaneous pruritogens, and these cells relay sensory information via a common and dedicated spinal pathway (Mishra and Hoon, 2013; Sun et al., 2009) to higher centers in the brain, producing an itch percept.
Both exogeneous compounds (e.g., the protease mucunain present in spicules from the cowhage plant; Reddy et al., 2008) and endogenous agents released from cells in the skin can trigger pruriception (Azimi et al., 2016; Oetjen et al., 2017). Skin-resident mast cells are thought to be key players in the latter indirect itch pathway, since they are ideally localized to detect and respond to intruder agents and organisms such as parasites (Metcalfe et al., 1997; Tay et al., 2013). In the past, work has focused on histamine released by mast cells, for instance in the context of type 1 hypersensitivity reactions (Modena et al., 2016). However, mast cells are also activated under nonallergic conditions (Krystel-Whittemore et al., 2016). Importantly, in most clinically relevant settings, including certain chronic diseases, antihistamines are ineffective at relieving itch (Kini et al., 2011; Yosipovitch et al., 2003), suggesting that molecules other than histamine serve as endogenous pruritogens. However, potential roles of non-histamine mast cell-derived mediators in pruriception are not well characterized, and the neurons and receptors involved in detection of these compounds are also not well understood.
In mice, pruritogens are detected by two nonoverlapping classes of sensory neurons that themselves are part of a larger population of neurons that express the transient receptor potential cation channel subfamily V member 1 (Trpv1) (Liu et al., 2009; Mishra and Hoon, 2013). One of these classes of neurons is marked by the transmitter natriuretic polypeptide b (Nppb), and the second is defined by the expression of the Mas-related G-protein-coupled receptor a3 (Mrgpra3) (Han et al., 2013; Mishra and Hoon, 2013). It was demonstrated that activation of Nppb and Mrgpra3 neurons is sufficient for the generation of itch behavior (Han et al., 2013; Huang et al., 2018). Further corroborating a role of Mrgpra3 neurons in pruriception, several itch receptor proteins, including Mrgpra3 itself, the receptor for the anti-malaria drug chloroquine, and Mrgprc11, the receptor for bovine adrenal medulla peptide 8–22 and several cysteine proteases, were shown to be expressed by Mrgpra3 neurons (Han et al., 2013; Liu et al., 2009), and elimination of these cells attenuated sensitivity to pruritogens (Han et al., 2013).
The transmitter Nppb is required for itch responses (Mishra and Hoon, 2013), but it is unclear which itch-inducing agents stimulate Nppb cells and the receptors involved. Interleukin-31 (IL-31), a cytokine associated with chronic itch (Sonkoly et al., 2006), might be sensed by Nppb neurons, as they express the IL-31 receptor IL-31Ra (Chiu et al., 2014). However, IL-31 alone does not provoke acute itch in healthy humans, questioning its role as an endogenous pruritogen (Hawro et al., 2014). In addition, the serotonin receptor Htr1f was reported to be expressed in Nppb cells, but ablation of these cells partially reduced itch responses to serotonin and other pruritogens, calling into question the contribution of both this receptor and Nppb cells to itch (Stantcheva et al., 2016). Lastly, transcriptomic studies suggest that Nppb neurons express select G-protein-coupled receptors (GPCRs) that may be involved in itch, but these receptors have not been validated (Li et al., 2016; Nguyen et al., 2017; Usoskin et al., 2015). Thus, to date, it remains unclear how Nppb neurons participate in the detection of pruritogens. To better understand this process, we sought to identify the itch receptors Nppb neurons express and thereby determine the origin of the pruritogens that stimulate these cells. We used an unbiased transcriptomic approach in conjunction with expression studies, calcium imaging, and mouse behavior to define the mechanism by which Nppb neurons couple activation of mast cells to itch.
RESULTS
Mast Cell Activation Induces Itch
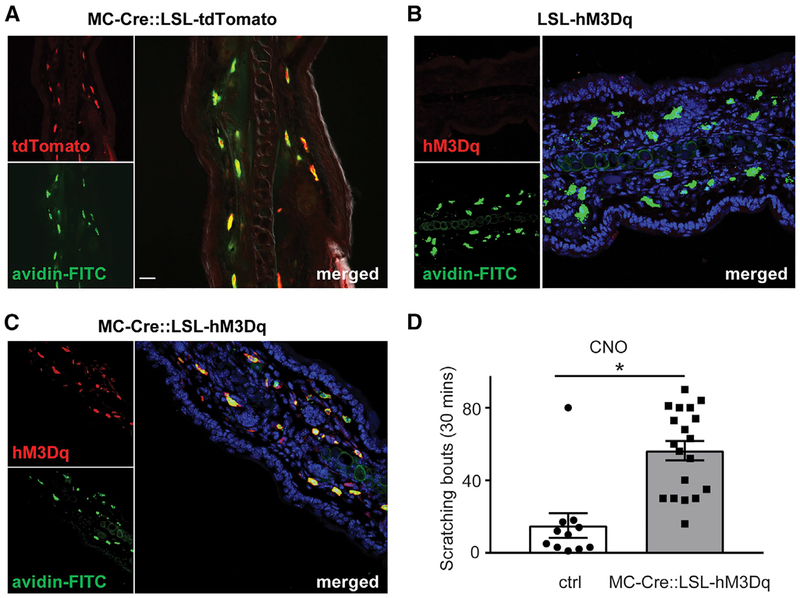
There are multiple chemicals, with varied sources, that can elicit the sensation of itch. Compounds from mast cells (Moon et al., 2014; Sjoerdsma et al., 1957; Steinhoff et al., 2018; Sumpter et al., 2019; Wernersson and Pejler, 2014) are regarded as a class of endogenous pruritogens; for instance, histamine is widely used in rodent itch behavioral tests (Bautista et al., 2014; Han and Dong, 2014). However, although mast cells are thought to cause itch, the release of chemicals from them has not been studied in detail because of challenges in solely activating these cells. This is because the high-affinity immunoglobulin E (IgE) receptor FcεRI, whose ligation causes degranulation, is expressed on several cell types in addition to mast cells (Gould and Sutton, 2008). Additionally, mediators that can cause IgE-independent mast cell degranulation have been shown to directly activate sensory neurons, raising doubts about the specificity of this group of agonists in vivo (Kido-Nakahara et al., 2014; Matsushima et al., 2004; McNeil et al., 2015; Schemann et al., 2012). Therefore, to characterize the contribution of mast cells to itch in more detail, we examined, using a targeted genetic approach, the behavior caused by stimulation of these cells. The strategy we employed utilized the mast cell-specific Cre-recombinase driver mouse line mast cell protease 5 (Mcpt5)-Cre (MCCre), together with the Cre-dependent R26-LSL-hM3Dq line (Scholten et al., 2008; Zhu et al., 2016). This permitted the expression of the GPCR, hM3Dq, only in mast cells. hM3Dq is an artificial receptor that has been widely employed to stimulate cells using the synthetic ligand clozapine-N-oxide (CNO), a compound that does not activate endogenous GPCRs (Armbruster et al., 2007). To confirm the specificity of this strategy, we crossed MC-Cre animals with the Cre-dependent tdTomato reporter line Ai9 (Madisen et al., 2010). As expected, we found very high co-localization between tdTomato reporter expression and mast cells in the skin (290 of 304 cells double labeled; Figure 1A). Further, we found almost complete co-expression between hM3Dq and mast cells that was strictly dependent on the expression of Cre (557 of 644 cells double labeled; Figures 1B and 1C). If activation of mast cells causes itch, then we would anticipate that intradermal administration of CNO should induce scratching. Exactly as expected, mice injected with CNO into the nape of the neck exhibited scratching responses toward the injected area, demonstrating that activation of mast cells is sufficient to provoke itch behavior. In contrast, Cre-negative control littermates failed to exhibit scratch responses when challenged with CNO (Figure 1D). These results suggest that the activation of mast cells likely leads to the release of agents, and this is sufficient to evoke itch behavior.
Figure 1. Chemogenetic Activation of Mast Cells Triggers Scratching.
(A) Native fluorescence (tdTomato; red) and avidin staining (avidin-FITC; green) of the ear of MCCre∷Ai9 mice shows almost perfect correspondence between expression of fluorescent trans-gene and mast cells.
(B and C) Immunostaining for hM3Dq in tissue from MC-Cre∷LSL-hM3Dq (C) and MC-Cre negative littermate animals (B) reveals that expression of hM3Dq is restricted to mast cells and strictly dependent on Cre expression. Similar results were obtained from at least 3 animals.
(D) Activation of hM3Dq with CNO (20 μg, intradermal) triggers significantly greater scratch responses in MC-Cre∷LSL-hM3Dq animals than in littermate controls. Significant differences between indicated treatment groups were assessed using unpaired, two-tailed Student’s t test (*p < 0.0001). Data represent means ± SEM (n = 11 control n = 19 MC-Cre∷LSL-hM3Dq mice). Scale bar, 20 μm.
Nppb Neurons Express Potential Pruriceptive GPCRs
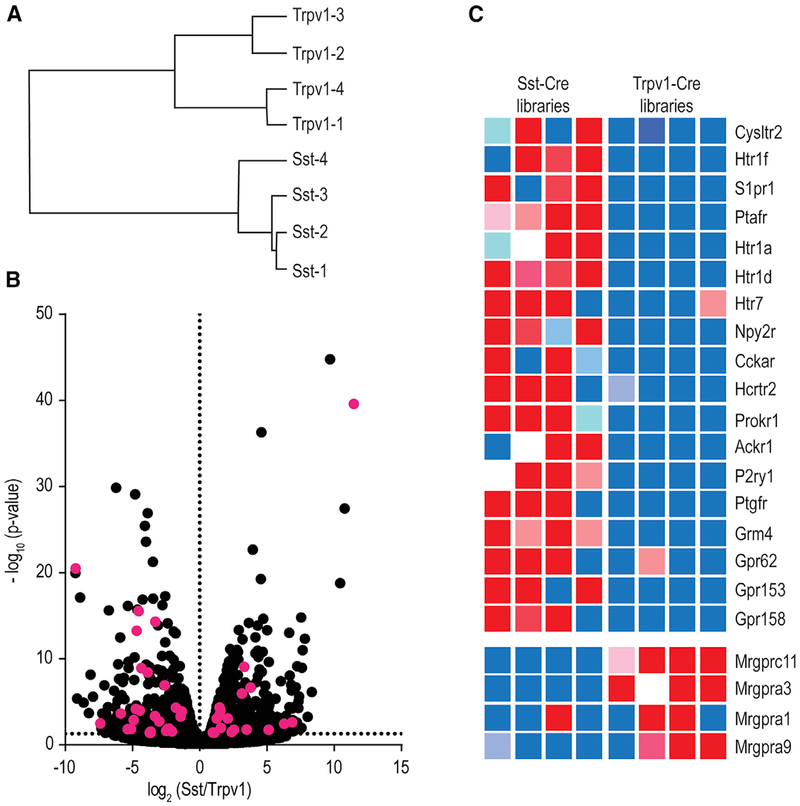
Although some of the receptors for compounds released by mast cells are known (Bautista et al., 2014; Reddy et al., 2010), it is unclear if there are additional itch receptors for less well-characterized chemicals. Therefore, we designed a screen to search for potential receptors. Because Nppb and Mrgpra3 cells have been shown previously to be responsible for itch in mice and when stimulated can elicit scratching responses (Han et al., 2013; Huang et al., 2018; Mishra and Hoon, 2013), we hypothesized that there might be unidentified itch receptors expressed in these populations of neurons. We further hypothesized that additional receptors might be differentially expressed between these two classes of cells because the Mrgpra3 and Mrgprc11 itch receptors are expressed preferentially by Mrgpra3 neurons (Usoskin et al., 2015). Since both Nppb and Mrgpra3 cells co-express the Trpv1 ion channel (Liu et al., 2009; Mishra and Hoon, 2013), we examined the genes expressed in Nppb cells, taking advantage of Sst-Cre mice, which permit the genetic labeling of Nppb neurons, (Huang et al., 2018), and compared them with genes expressed in Trpv1 neurons, thereby allowing us to search for receptors expressed in these two populations of cells.
In order to sample all (or close to all) genes expressed in these cells, we generated cDNA libraries from RNA collected from ~200 hand-picked labeled neurons (see STAR Methods for details). Four independent Sst+ and four Trpv1+ cDNA libraries were generated, sequenced, and subsequently compared. As expected, and confirming that the transcriptomes of Sst and Trpv1 cells differ significantly from each other, hierarchical clustering of transcriptome data showed that the largest determinant for difference between samples was cell type (Figure 2A). Comparison of data from Sst and Trpv1 samples revealed ~1,600 genes with significant differences in gene expression between cell types (Figure 2B; EdgeR, p < 0.05). Almost 1,000 genes were expressed ~2-fold higher in Sst compared to Trpv1 samples, and ~600 genes were enriched in Trpv1 versus Sst libraries (Table S1). From these lists of genes, we choose to focus our attention on GPCRs, since many pruritogens are detected by this class of receptors (Han et al., 2006; Imamachi et al., 2009; Ru et al., 2017) (Figure S1).
Figure 2. Identification of Potential Itch Receptors Expressed in Nppb Neurons.
(A) Dendrogram of the hierarchical clustering of gene expression between four Sst- and four Trpv1-cDNA libraries shows that the largest determinant for clustering is cell type.
(B) Volcano plot comparing gene expression between Sst- and Trpv1-cDNA libraries. The likelihood of a significant expression difference was assessed with EdgeR and accepted at p < 0.05 (a complete listing of differentially expressed genes is presented in Table S1). GPCRs in the group of significantly expressed genes are highlighted in magenta.
(C) Heatmap displays the relative gene expression in each cDNA library of some of these GPCRs.
Consistent with our postulate and with previous reports (Huang et al., 2018), the itch-related GPCRs Mrgprc11, Mrgpra3, Mrgpra1, and Mrgpra9 were enriched in Trpv1 libraries relative to Sst libraries (Figure 2C). While the specific expression of multiple receptor molecules in Mrgpra3 cells has been functionally linked to pruritogen-evoked scratching responses (Liu et al., 2009, 2011), the receptor repertoire for Nppb neurons is still incompletely understood. Therefore, we next concentrated our attention on the GPCRs enriched in Nppb cells and identified 18 candidate GPCRs (Figure 2C). Based on the GPCRs enriched in Nppb neurons, we sought to directly determine if mediators for some of these receptors are released by mast cells. We used peritoneal-derived mast cells (PDMCs), which have frequently been used to model skin mast cells (Metcalfe et al., 1997), to examine some of the substances they release upon stimulation. Figure S2 shows that after PDMC stimulation, leukotriene C4 (LTC4), serotonin, and sphingosine-1-phosphate (S1p), in agreement with previous reports (da Silva et al., 2014; Moon et al., 2014; Olivera and Rivera, 2005), are released, suggesting that these agents may be involved in mast cell-dependent itch via activation of their cognate receptors in Nppb neurons.
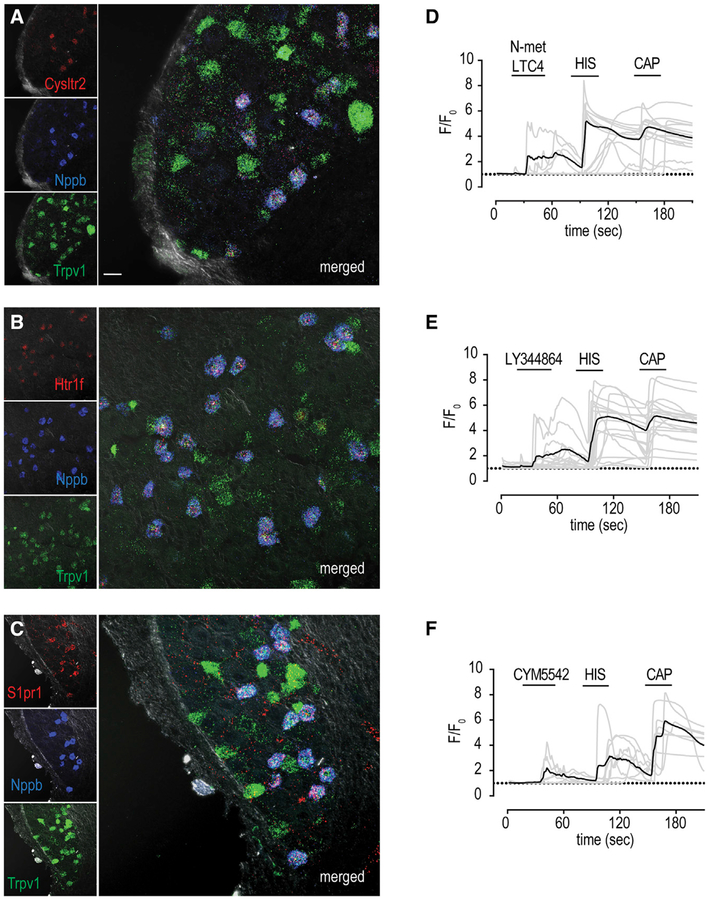
Cysltr2, Htr1f, and S1pr1 Elicit Itch via the Spinal Cord-GRP Pathway
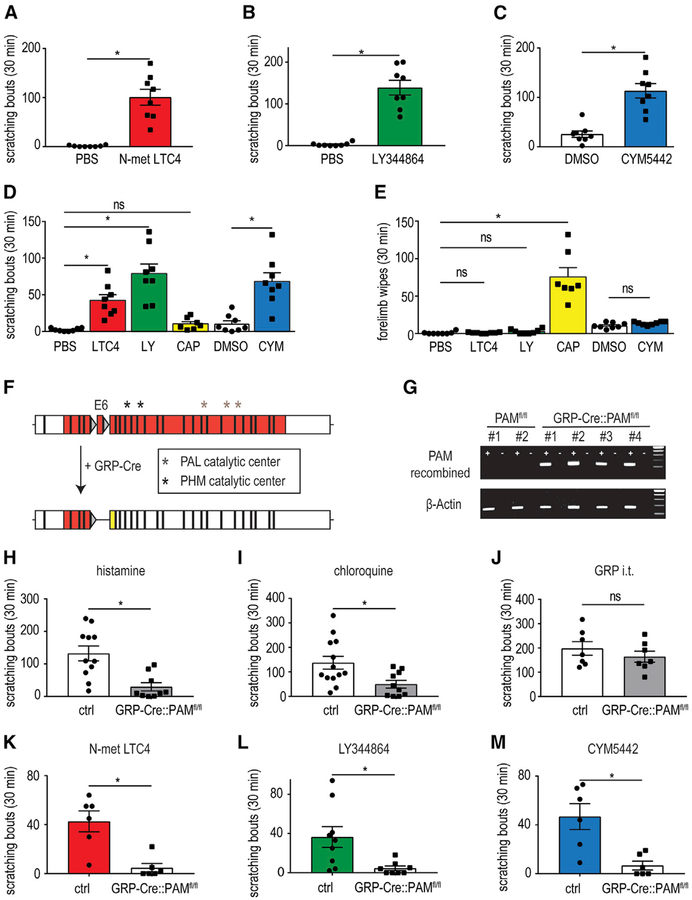
Since we found that the release of LTC4, serotonin, and S1p from mast cells is correlated with mast cell-induced itch behavior, we focused our attention on the receptors for these compounds (Cysltr2, Htr1f, and S1pr1). For these receptors to be involved in itch, the intradermal injection into the nape of the neck of specific ligands for them, would be expected to induce scratching. Indeed, exactly as anticipated, all of these compounds elicited vigorous scratch responses (Figures 3A–3C). The rodent cheek behavioral assay permits the distinction of itch from nociceptive responses, with pruritogens evoking scratching with the hindlimb while algogens trigger wiping with the forelimb (Shimada and LaMotte, 2008). In contrast to the wiping evoked by capsaicin, N-met LTC4, LY344864, and CYM5442 produced intense scratching and did not elicit wiping, suggesting that these agents are pruritogens (Figures 3D and 3E).
Figure 3. LTC4-,Serotonin-, and S1p-Evoked Itch Responses Are Mediated through the Spinal Cord-GRP-Neuron Pathway.
(A–C) Intradermal injection into the nape of the neck of N-met LTC4 (0.75 μg; A), LY344864 (40 μg; B), and CYM5442 (2.23 μg; C) induces scratching bouts that are significantly greater than following administration of vehicle. Notably, these concentrations are not the maximal potencies for these agents to induce scratching. Significant differences between indicated treatment groups were assessed using unpaired, two-tailed Student’s t test (*p = 0.0005 N-met LTC4, *p = 0.0001 LY344864, *p = 0.0003 CYM5442). Data represent means ± SEM (A; n = 8 PBS n = 8 N-met LTC4, B; n = 8 PBS n = 8 LY344864, C; n = 8 0.5% DMSO n = 8 CYM5442).
(D and E) Intradermal injection into the cheek of N-met LTC4 (0.75 μg), LY344864 (40 μg), and CYM5442 (4.46 μg) elicits scratching bouts (D), but not forelimb wipes (E), that are significantly greater than following administration of vehicle. Capsaicin (1 μg, CAP), a prototypic algogen, induces forelimb wipes, but not hindlimb scratching. Significant differences between indicated treatment groups were assessed using 1-way ANOVA with Sidak’s multiple comparisons test (D; *p = 0.0052 N-met LTC4, *p < 0.0001 LY344864, nonsignificant [ns] [p = 0.9377] capsaicin, *p < 0.0001 CYM5442) (E; ns [p > 0.9999] N-met LTC4, ns [p = 0.9972 LY344864], *p < 0.0001 capsaicin, ns [p = 0.9803 CYM5442]). Data represent means ± SEM (D; n = 8 PBS, n = 8 N-met LTC4, n = 8 LY344864, n = 8 capsaicin, n = 8 1% DMSO and n = 8 CYM5442) (E, n = 8 PBS, n = 8 N-met LTC4, n = 8 LY344864, n = 7 capsaicin, n = 8 1% DMSO and n = 8 CYM5442).
(F) Schematic of the genetic strategy for generation of conditional knockout of the PAM gene in GRPCre neurons. The location of the loxP sites (gray triangles) surrounding exon 6 is indicated as well as the positions of the coding sequence (red) and sites of catalytic activity (PAL, peptidyl-α-hydroxyglycine α-amidating lyase; PHM, peptidylglycine α-hydroxylating monooxygenase). Cre-mediated recombination of the floxed allele results in the production of an out-of-frame coding sequence after exon 5 (yellow) producing a missense coding sequence, early termination of transcript, and disruption of sequences encoding critical catalytic protein domains.
(G) Confirmation of gene disruption of PAM in spinal cord tissue. PCR was performed with primers designed to only amplify a fragment from cDNA produced from a PAM allele where recombination excised exon 6 (top; see STAR Methods for details). Amplification only occurred in reactions containing template cDNA (+) and not those lacking template (−). In contrast, a β-actin amplicon (bottom) was amplified from control and mutant cDNA samples.
(H–J) PAM mice displayed significantly attenuated itch responses to intradermal injection of histamine (200 μg; H) and chloroquine (100 μg; I) compared to control littermate animals. By contrast, scratching evoked by intrathecal (i.t.) administration of GRP (2.9 μg; J) was the same in PAM mice and control mice. Significant differences between indicated genotypes were assessed by unpaired, two-tailed Student’s t test (*p = 0.0012; H, *p = 0.0152; I, ns [p = 0.3728]; J). Data represent means ± SEM (H; n = 11 control n = 9 PAM mice, I; n = 13 control n = 10 PAM mice, J; n = 7 control n = 7 PAM mice).
(K–M) PAM mice exhibited significantly reduced itch responses to intradermal administration of N-met LTC4 (0.75 μg; K), LY344864 (60 μg; L), and CYM5442 (2.23 μg; M) compared to control littermate animals. Significant differences between indicated genotypes were assessed by unpaired, two-tailed Student’s t test (*p = 0.0022; K, *p = 0.017; L, *p = 0.0049; M). Data represent means ± SEM (K; n = 6 control n = 6 PAM mice, L; n = 9 control n = 8 PAM mice, M; n = 6 control n = 6 PAM mice).
Itch responses have been shown to be transmitted through a spinal cord circuit that requires the neuropeptide gastrin-releasing peptide (GRP) and GRP neurons (Mishra and Hoon, 2013; Sun et al., 2009). This circuit has been shown to be specific for itch, and therefore, to obtain evidence that the Cysltr2, Htr1f, and S1pr1 are pruriceptors, we tested if itch responses evoked by them are dependent on signaling through spinal cord-GRP neurons. To accomplish this, we used a targeted genetic strategy where we interfered with the biosynthesis of neuropeptides, including GRP, in spinal cord-GRP neurons. Specifically, we generated mice lacking the critical neuropeptide-modifying enzyme peptidylglycine alpha-amidating monooxygenase (PAM) in GRP-Cre neurons (Gong et al., 2007), GRP-Cre∷PAMfl/fl mice (PAM-mice) (Figure 3F). PAM is the sole enzyme responsible for C-terminal deglycination and amidation of peptides, a common modification of many neuropeptides including GRP (Merkler, 1994). In the absence of modification by PAM, GRP has two orders of magnitude less potency for its receptor than the amidated peptide (Patel et al., 2004, 2006; Perkins et al., 1990). We note that the resulting PAM mice were born at normal Mendelian ratios and displayed no overall gross phenotype. Confirming successful elimination of PAM from GRP neurons, we were only able to amplify the mutant PAM allele (with Cre-mediated deletion of exon 6) from cDNA derived from the spinal cord samples from PAM mice (Figure 3G). Further, as expected for interference with spinal GRP signaling, histamine-, chloroquine-, and compound-48/80-induced itch was profoundly reduced (Figures 3H, 3I, and S3A), while responses to other somatosensory stimuli were unaffected by this mutation (Figures S3B–S3E; Sun and Chen, 2007; Sun et al., 2009). Additionally, itch induced by intrathecal administration of GRP and injection of nor-Binaltorphimine were unaffected, consistent with the PAM genetic lesion only affecting signaling upstream of spinal cord GRPR neurons and not effecting other GRP neurons in the CNS (Figures 3J and S3F; Guo et al., 2015; Huang et al., 2018). Thus, PAM mice are an effective genetic method to test if compounds are involved in itch. To investigate if the GRP-dependent spinal circuit is required to evoke itch responses for ligands to Cysltr2, Htr1f, and S1pr1, we performed experiments with PAM mice. We hypothesized that if these receptors are required for itch, then spinal cord-GRP signaling would be required. Just as expected, itch behavioral responses mediated through Cysltr2, Htr1f, and S1pr1 were strongly attenuated in PAM mice compared to littermate controls (Figures 3K–3M).
Trpv1-Lineage Neurons Are Necessary for Cysltr2-, Htr1f-, and S1pr1-Evoked Itch
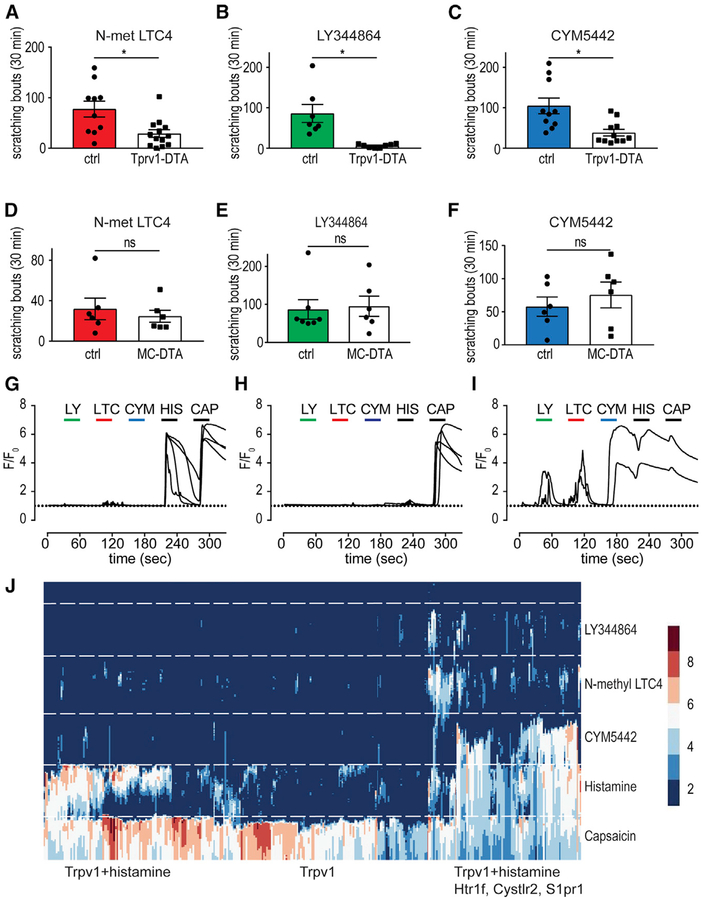
Our findings that Cysltr2-, Htr1f-, and S1pr1-induced itch-responses require spinal-GRP signaling suggest that they are itch receptors. A corollary of this result is that the elimination of the sensory neurons, which express Cysltr2, Htr1f, and S1pr1, should attenuate itch responses induced by the agonists for these receptors. To address this predicted consequence, we used animals that have lost the Trpv1 lineage of sensory neurons (Mishra et al., 2011) by crossing Trpv1-IRES-Cre animals (Cavanaugh et al., 2011) with a R26-floxed stop-diphtheria toxin A (DTA) line (Voehringer et al., 2008) to produce Trpv1-Cre∷LSLDTA mice (Trpv1-DTA). In agreement with previous reports (Mishra et al., 2011), we found that these mice have deficits in behavioral responses to the Trpv1 agonist capsaicin as well as in the detection of noxious heat and cold (Figures S4A–S4C). We verified histologically that Trpv1-DTA mice lack sensory neurons expressing Trpv1 and Nppb (Figures S4D–S4G) (Mishra and Hoon, 2013). Strikingly, Trpv1-DTA mice also exhibit severely reduced scratching responses to N-met LTC4, LY344864, and CYM5442 (Figures 4A–4C), indicating that these neurons are required to produce Cysltr2, Htr1f, and S1pr1 itch responses. However, even though Trpv1-lineage neurons are required for N-met LTC4-, LY344864-, and CYM5442-induced itch, this reaction may result via a deficit in an indirect itch pathway, e.g., via mast cells (Boyce, 2007). To test this possibility, we examined itch responses in mice lacking mast cells. Specifically, to ablate mast cells, we crossed MC-Cre∷LSL-tdTomato mice with the R26-floxed stop-DTA line (MC-DTA). Histochemical staining of skin from MC-DTA animals showed that they had no detectable mast cells (Figure S5; 0 mast cells in 7 sections from 3 mice). MC-DTA mice displayed itch responses indistinguishable from littermate control animals, showing that scratching induced by N-met LTC4, LY344864, and CYM5442 does not require mast cells (Figures 4D–4F).
Figure 4. Trpv1-Lineage Neurons Are Required in LTC4-, Serotonin-, and S1p-Evoked Itch Responses.
(A–C) Intradermal injection of N-met LTC4 (0.75 μg; A), LY344864 (50 μg; B), and CYM5442 (2.23 μg; C) elicits scratching bouts that are significantly reduced in Trpv1-DTA mice compared to control littermate animals, indicating that Trpv1-lineage neurons are required for itch responses to these agents. Significant differences between genotypes were assessed by unpaired, two-tailed Student’s t test (*p = 0.0071; A, *p = 0.0116; B, *p = 0.0043; C). Data represent means ± SEM (A; n = 10 control n = 13 Trpv1-DTA mice, B; n = 7 control n = 8 Trpv1-DTA mice, C; n = 10 control n = 12 Trpv1-DTA mice). (D–F) Intradermal injection of 3 μg N-met LTC4 (D), 60 μg LY344864 (E), and 2.23 μg CYM5442 (F) evoked scratching bouts that are not significantly different between MC-DTA mice and control animals, showing that mast cells are not required for itch responses to these agents. Significant differences between genotypes were assessed by unpaired, two-tailed Student’s t test (ns [p =0.5717]; D, ns [p = 0.8224]; E, ns [p = 0.4863]; F). Data represent means ± SEM (D; n = 6 control n = 6 MC-DTA mice, E; n = 7 control, n = 7 MC-DTA mice, F; n = 6 control n = 6 MC-DTA mice).
(G–I) Representative traces of calcium responses of Trpv1-lineage neurons (Trpv1-Cre∷Ai96) to pruritogens (LY, 1 μM LY344864; LTC, 100 nM N-met LTC4; CYM, 10 μM CYM5442; HIS, 10 μM histamine; CAP, 10 μM capsaicin). Note there are broadly three classes of neurons that respond to histamine and capsaicin (G), capsaicin alone (H), and N-met LTC4, LY344864, CYM5442, hista-mine, and capsaicin (I).
(J) Raster plot of calcium responses (F/F0, see legend on the right) in 353 neurons to the indicated agents (same concentrations as in G–I). Neurons that responded to capsaicin and histamine are grouped to the left of the panel (80 neurons), capsaicin-only responsive cells are grouped in the middle of the panel (174 neurons), and those cells responding to agonists for Cysltr2, Htr1f, S1pr1, and capsaicin and histamine are grouped to the right of the panel (99 neurons).
A second prediction of Cysltr2, Htr1f, and S1pr1 being itch receptors is that application of their corresponding ligands should stimulate the neurons in which they are expressed. Since Trpv1-lineage neurons are required for itch responses mediated by these receptors, the cells that express these receptors must be within this population of neurons. Therefore, we performed calcium imaging on DRG neurons collected from mice where the Trpv1 lineage of cells expresses the calcium sensor GCaMP6s, Trpv1-Cre∷Ai96 mice (Madisen et al., 2015). Since this cell lineage contains multiple different classes of cells, not all of which are involved in itch (Mishra and Hoon, 2013; Mishra et al., 2011), sensory neurons were sequentially exposed to N-met LTC4, LY344864, CYM5442, histamine, and capsaicin to determine cellular specificity. With this testing regimen, we could classify neurons that express histamine and capsaicin (Trpv1) receptors and determine the neurons that were responsive to ligands for Cysltr2, Htr1f, and S1pr1. As expected, calcium imaging revealed a class of non-itch neurons that were stimulated by capsaicin but were unresponsive to all the pruritogens tested (Figure 4H and Figure 4J, middle). Imaging also revealed two different classes of neurons that reacted to pruritogens. The first of these reacted to histamine and capsaicin but was unresponsive to N-met LTC4, LY344864, and CYM5442 (Figure 4G and Figure 4J, left). The second class of neurons displayed largely overlapping responsiveness to N-met LTC4, LY344864, and CYM5442 as well as to histamine and capsaicin (Figure 4I and Figure 4J, right).
Nppb Neurons Co-express Cysltr2, Htr1f, and S1pr1
Our transcriptomic data show that Cysltr2, Htr1f, and S1pr1 are enriched in Nppb-cells (Figure 2), ablation experiments demonstrate that Trpv1-neurons are required for itch mediated by these receptors, and cellular imaging assays establish that a subpopulation of Trpv1-neurons responds to the ligands for these receptors (Figure 4), suggesting that Nppb-neurons co-express Cysltr2, Htr1f, and S1pr1. Recently, it was reported that Cysltr2 and Htr1f are found in Nppb-neurons (Li et al., 2016; Nguyen et al., 2017; Stantcheva et al., 2016; Usoskin et al., 2015). However, the extent of overlap in expression of these receptors with Nppb was not examined nor was the co-expression of receptors with each other and with other cell types determined. Therefore, to understand more about the neurons that express Cysltr2, Htr1f, and S1pr1, we performed triple-label in situ hybridization (ISH) using probes to individual receptors, Nppb, and Trpv1. Virtually all Cysltr2-positive neurons express Nppb-Trpv1 (87.2% ± 2.8%; 266 neurons from 17 sections/3 animals), and similarly, almost all Htr1f-positive and S1pr1-positive cells express Nppb-Trpv1 (86.3% ± 6.3%; 199 neurons from 12 sections/3 animals and 98.9% ± 1.3%; 89 neurons from 8 sections/3 animals), demonstrating a remarkably tight co-expression of these receptors in Nppb cells (Figures 5A–5C).
Figure 5. Cysltr2, Htr1f, and S1pr1 Are Co-expressed in Nppb Neurons.
(A–C) Triple label ISH reveals that Cysltr2 (A, red), Htr1f (B, red), and S1pr1 (C, red) are co-expressed with Nppb (blue), demonstrating that these three receptors are only expressed by Nppb-neurons. Note, all Nppb cells co-express Trpv1 (green). Similar results were obtained from at least 3 animals.
(D–F) Calcium imaging of neurons from Sst-Cre∷Ai96 mice showed that they respond to N-met LTC4 (100 nM), histamine (His, 10 μM), and capsaicin (CAP, 10 μM) (D); LY344864 (1 μM), histamine (His, 10 μM), and capsaicin (CAP, 10 μM) (E); or CYM5542 (10 μM), histamine (His, 10 μM), and capsaicin (CAP, 10 μM) (F). Traces of responses of individual neurons are indicated as gray traces, and the averaged response is indicated by the overlaid black trace. Scale bar, 20 μm.
The co-expression of Cysltr2, Htr1f, and S1pr1 in Nppb neurons predicts that these neurons should be activated by agonists for these receptors. To address this, we performed calcium imaging of primary DRG neurons derived from mice where the majority of Nppb neurons express the calcium reporter GCaMP6s, Sst-Cre∷Ai96 mice (Huang et al., 2018; Taniguchi et al., 2011). As predicted from the co-expression of Cysltr2, Htr1f, and S1pr1, almost all Sst-Cre∷Ai96 neurons responsive to capsaicin and histamine also responded to N-met LTC4 (7/11 cells), LY344864 (13/16 cells), and CYM5442 (5/7 cells) (Figures 5D–5F). These results suggest that Nppb neurons are a largely homogeneous population of cells that react to multiple pruritogens.
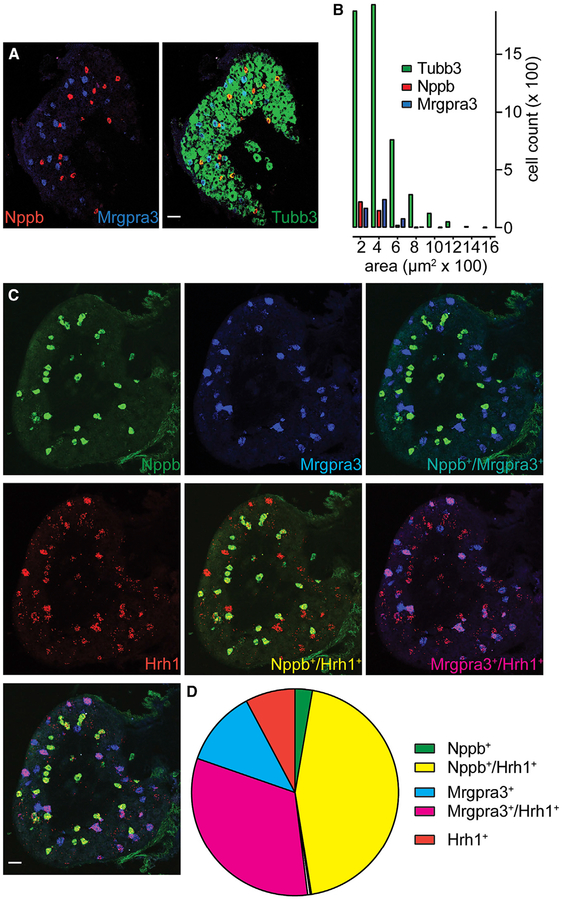
Our functional imaging experiments revealed that Nppb neurons are activated by histamine as well as N-met LTC4, LY344864, and CYM5442 (Figures 4I and 5D–5F). However, comparison of the transcriptomes of Nppb and Trpv1 neurons did not reveal differential expression of histamine receptors between these classes of cells. We wondered if this might be because histamine receptor is expressed in both Nppb and Mrgpra3 neurons. To better characterize these two classes of neurons, we determined that they are both predominantly small diameter and that Nppb neurons are 7.5% ± 0.7% and Mrgpra3-cells are 8.4% ± 2.8% of DRG cells (Figure 6A; 5,059 Tubb3+ neurons) (Figure 6B). Next, to determine if Nppb and Mrgpra3 neurons express histamine receptor, we performed ISH for the histamine receptor Hrh1 and compared its co-expression with Nppb and Mrgpra3. Interestingly, results from triple-label ISH revealed that Hrh1 is expressed by both Nppb and Mrgpra3 neurons (93.3% ± 0.8% of Nppb+ cells and 73.4% ± 2.3% of Mrgpra3+ cells; 1,644 and 1,185 neurons, respectively), as well as by an additional smaller population of cells (9.1% ± 1.4% of Hrh1+ cells negative for Nppb and Mrgpra3; 286 neurons) (Figures 6C and 6D). Together, these results demonstrate that while Nppb and Mrgpra3 cells have distinct repertoires of itch receptors, they both express histamine receptors.
Figure 6. The Histamine Receptor Hrh1 Is Expressed by Both Nppb and Mrgpra3 Neurons.
(A) Triple-label ISH reveals that Nppb (red) and Mrgpra3 (blue) are expressed in non-overlapping neuronal (Tubb3, green) populations.
(B) Quantification of soma size reveals that both itch-neuron populations belong to the group of small-diameter neurons.
(C) Triple-label ISH reveals that Hrh1 (red) is co-expressed by both Nppb (green) and Mrgpra3 (blue) cells. Similar results were obtained from 4 animals.
(D) Schematic of the overlap between markers (>4 sections from n = 4 mice). Scale bars, 50 μm.
Direct Activation of Nppb Neurons by S1p Induces Itch
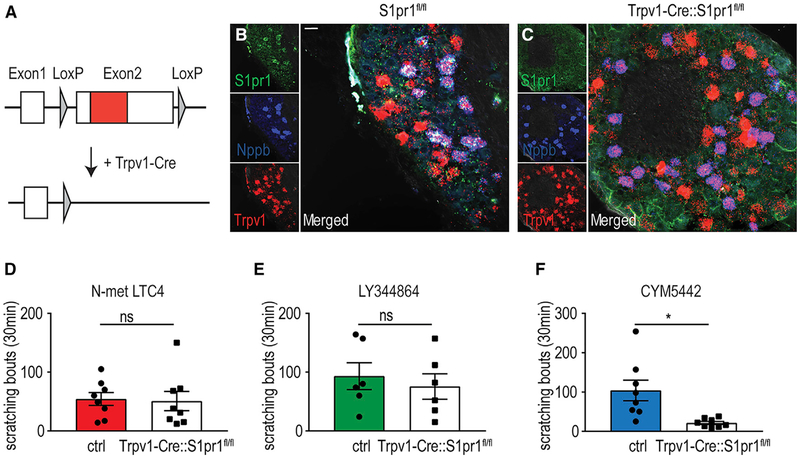
Results from analysis of gene expression, calcium imaging, ISH, and behavioral experiments strongly support Nppb neurons being sensors of mast cell-induced itch and substantiate Cysltr2, Htr1f, and S1pr1 as itch receptors. To obtain further in vivo validation that these receptors are itch sensors, we focused our attention on whether S1pr1 is essential for S1p-induced itch. In order to do this, we generated a mouse null for S1pr1 in only sensory neurons, Trpv1-Cre∷ S1pr1fl/fl mice (S1pr1 cKO) (Allende et al., 2003). As S1pr1 is expressed only in Nppb neurons (Figure 5), this intersectional approach enabled the specific deletion of S1pr1 in Nppb-expressing neurons. We reasoned that if S1pr1 functions in vivo as an itch receptor, then a sensory-neuron-specific knockout (KO) of this GPCR should produce mice lacking itch responses to S1p (CYM5442). Figure 7A illustrates the genetic strategy employed to eliminate expression of S1pr1 in sensory neurons. Notably, S1pr1 cKO mice were born at normal Mendelian ratios, and ISH experiments demonstrated a complete lack of specific S1pr1 staining in homozygous KO animals (Figures 7B and 7C; 0 S1pr1+ cells on 13 sections/3 mice). The loss of S1pr1 did not affect the expression of Nppb and Trpv1 (Figures 7B and 7C; control versus S1pr1 cKO mice 32.2 ± 9.8 versus 29.7 ± 12.4 Trpv1+ cells, p = 0.6927, and 16.2 ± 4.1 versus 12.7 ± 6.1 Nppb+ cells, p = 0.2577). We next investigated if S1pr1 cKO mice have sensory deficits, testing several acute somatosensory behaviors. No differences between mutants and controls were observed for behavioral responses to noxious heat, noxious cold, and innocuous mechanical stimuli or for motor coordination, itch responses to histamine and chloroquine, and ongoing nociceptive responses after intraplantar injection of capsaicin and allyl isothiocyanate, showing that the S1pr1 is not required for these behaviors (Figure S6). Responses to N-met LTC4 and LY344864 were also normal in mutant mice (Figures 7D and 7E). By contrast, scratching responses to intradermal injection of the S1pr1 agonist CYM5442 were virtually absent in S1pr1 cKO mice (Figure 7F). These results demonstrate that CYM5442-evoked itch requires the direct activation of S1pr1 in sensory neurons.
Figure 7. S1p-Induced Itch Requires Expression of S1pr1 by DRG Neurons.
(A) Schematic of the genetic strategy employed to generate conditional knockout of S1pr1 in Trpv1-Cre (sensory) neurons. The location of the loxP sites (gray triangles) surrounding exon 2 is indicated as well as the position of the coding sequence of the receptor (red). Cre-mediated recombination of the floxed allele results in the complete elimination of the coding sequence of S1pr1.
(B and C) Confirmation that gene disruption abolishes S1pr1 expression in the DRG. Triple-label ISH with probes to S1pr1 (green), Nppb (blue), and Trpv1 (red) demonstrates that S1pr1 is expressed in ganglia of littermate control mice (B) but is lost in the DRG of S1pr1 cKO animals (C).
(D–F) S1pr1 cKO mice display normal itch responses to intradermal injection of N-met LTC4 (0.75 μg; D) and LY344864 (60 μg; E) compared to control littermate animals. By contrast, scratching evoked by intradermal administration of CYM5442 (2.23 μg; F) was significantly attenuated in S1pr1 cKO mice compared to control mice. Significant differences between genotypes were assessed by unpaired, two-tailed Student’s t test (ns [p = 0.8616]; D, ns [p = 0.5849]; E, *p =0.0074; F). Data represent means ± SEM (D; n = 8 control n = 8 S1pr1 cKO mice, E; n = 6 control n = 6 S1pr1 cKO mice, F; n = 8 control n = 8 S1pr1 cKO mice). Scale bar, 20 μm.
DISCUSSION
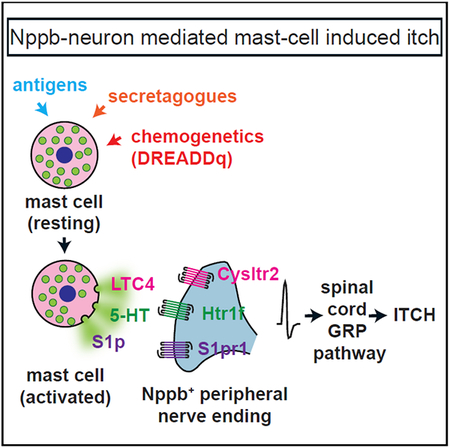
Here, using chemogenetics, transcriptome sequencing, calcium imaging, ISH, and selective genetic manipulations, we define a pathway by which itch is induced by mast cell activation. First, chemogenetic activation of mast cells produces itch behavior. Second, we show that mast cells, upon stimulation, can release serotonin, LTC4, and S1p. Third, we demonstrate that receptors for these three ligands are expressed selectively by Nppb neurons. Fourth, genetic disruption of spinal cord GRP-neuron neuropeptide-mediated itch establishes that Cysltr2-, Htr1f-, and S1pr1-induced scratch responses require the canonical GRP-spinal cord itch circuit. Fifth, calcium imaging results corroborate that Nppb neurons directly respond to agents released by mast cells. Lastly, we engineered and tested mice specifically lacking S1pr1 in sensory neurons, showing that these mice selectively lose itch responses to S1p. Together, our studies establish a cellular and molecular mechanism by which mast cell activation induces itch.
There has been much interest in identifying and characterizing receptors responsible for itch sensation in the last decade (Bautista et al., 2014; Han and Dong, 2014). Itch receptors include the GPCRs Mrgprc11, a receptor for Bam8–22 and activated by cysteine proteases (Lembo et al., 2002; Reddy et al., 2015); Mrgpra3, a receptor for chloroquine (Liu et al., 2009); Mrgprd, a receptor for b-alanine (Liu et al., 2012); Htr7 and Htr2b, receptors for serotonin (Lee et al., 2018; Morita et al., 2015); S1pr3, a receptor for S1p (Hill et al., 2018) and the protease-activated receptors (PARs) 2 and 4 (Reddy et al., 2008, 2010; Reddy and Lerner, 2010), as well as non-GPCR itch receptors, including IL-31Ra, thymic stromal lymphopoietin receptor (TSLPR), TLR7, and IL-4Ra (Cevikbas et al., 2014; Liu et al., 2010a; Oetjen et al., 2017; Wilson et al., 2013). Here, we identify and functionally characterize three additional itch receptors, Cysltr2, Htr1f, and S1pr1, that are specifically co-expressed in Nppb neurons. In addition to these three GPCRs, our transcriptomic analysis of Nppb neurons (Figure S1) revealed the existence of other receptors that might modulate Nppb-neuron activity. For instance, we found the serotonin receptors Htr7, Htr1a, and Htr1d to be enriched in Nppb neurons relative to Trpv1 neurons, some of which were previously reported to contribute to itch (Morita et al., 2015). Furthermore, our sequencing results suggest that the platelet-activating factor receptor (Ptafr) is also likely expressed in Nppb neurons. Interestingly, the platelet-activating factor lipid moiety was described to be produced by mast cells (Nakajima et al., 1997), hinting that it may be involved in mast cell-mediated itch as well. The large number of identified receptors suggests that there is redundancy in pathways for induction of itch, and almost all pruriceptors described so far have been reported to be coupled to Trpv1 and/or Trpa1 ion channels. It has also been described that the direct activation of Trpa1 can evoke itch in different animals (Esancy et al., 2018). Nevertheless, it is likely that itch and pruriceptors developed to respond to specific dermatological challenges. Additionally, even though agonists to the proposed itch receptors reported here evoke itch responses in experimental settings, they may not all have primary roles in itch detection.
The Cysltr2, Htr1f, and S1pr1 group of receptors we identified is co-expressed in a population of neurons that express the neuropeptide transmitter Nppb (Figure 5), while Mrgpra3 itch neurons express a different class of itch receptors but do not express Nppb, insinuating that there is specialization between these cell types. A caveat to this partition of itch receptors is the common expression of the histamine receptor Hrh1 (and other itch receptors; data not shown) by both Nppb and Mrgpra3 neurons (Figure 6). Additionally, we previously showed that deletion of Sst from sensory neurons can impact behavioral responses to the Mrgpra3 agonist chloroquine (Huang et al., 2018), indicating functional cross-talk between both groups of itch neurons. Future studies should resolve if there is itch-cell specialization between Nppb and Mrgpra3 neurons and an analysis of genes expressed by these cells (Table S1) may provide a means to better address this question.
We employed a mutation in a critical enzyme in the maturation pathway for neuropeptides to disrupt the GRP-itch circuit in the spinal cord (Merkler, 1994; Patel et al., 2004, 2006; Perkins et al., 1990). The elimination of the PAM enzyme in GRP-Cre-expressing cells produced a specific defect in itch for all of the itch-inducing agents we tested, including histamine and chloroquine, while leaving responses to all other somatosensory modalities tested normal (Figures 3 and S3). This highly selective effect on itch is similar to that seen when GRPR signaling was manipulated previously (Mishra and Hoon, 2013; Sun and Chen, 2007; Sun et al., 2009). Compared to many other neural circuits, the striking feature of the itch pathway is its reliance on neuropeptides (Liu et al., 2010b; Mishra and Hoon, 2013; Sun and Chen, 2007; Sun et al., 2009). This contrasts with the contribution of neuropeptides in other somatosensory synaptic signaling pathways where neuropeptides are thought to have modulatory effects and their loss produces only modest deficits in animal behavior (De Felipe et al., 1998; Ishida et al., 2014). The difference in timing and duration of itch sensation compared to other somatosensory modalities might be a result of this difference in neurotransmission.
The results we present in this study focus on acute itch responses evoked by injection of pruritogens or the chemogenetic activation of mast cells. The mechanisms underlying itch responses for both are likely similar to those which occur for normal itch responses, for instance after an insect bite. However, the agents we identified in this study are unlikely to be the only compounds that are released by mast cells and induce acute itch. For instance, mast cell-derived proteases are known to activate PARs, thereby inducing scratching responses (Ui et al., 2006). In addition to a role in mast cell-mediated acute itch, the endogenous ligands we characterized here may also be involved in persistent itch conditions. High levels of circulating S1p have been reported to be associated with psoriasis (Checa et al., 2015), and similarly, elevated LTC4 is associated with skin inflammation, including atopic dermatitis and psoriasis (Sadik et al., 2013). Furthermore, platelet-activating factor antagonist was reported to produce some relief in patients with itch caused by atopic dermatitis (Abeck et al., 1997). These lipid mediators, in addition to being released by mast cells, may be released by keratinocytes or other types of immune cells. Collectively, these studies suggest that S1pr1 and Cysltr2 in Nppb neurons, in addition to having roles in acute itch produced by mast cell activation, may also contribute to some forms of chronic itch.
Mast cells are thought to be immune sentinels that survey the skin for intruders. Classically, their function is believed to involve the detection of invading parasites and the recruitment of other immune cell types via paracrine signaling (Metcalfe et al., 1997; Tay et al., 2013). It was also known that the release of some agents from mast cells leads to the sensation of itch. Here, we demonstrate a mechanism by which mast cell activation produces sensory nerve stimulation by identifying ligands and several receptors responsible for this coupling. These results highlight a mechanism by which neural and immune systems cooperate to provide a unified defense of the skin and offer insights into the potential ways by which this cascade may become dysfunctional during chronic itch.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests should be directed to the Lead Contact, Mark Hoon (mark.hoon@nih.gov). Reagents will be distributed in accordance with the rules and procedures of the National Institute of Health.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All experiments using mice followed NIH guidelines and were approved by the National Institute of Dental and Craniofacial Research ACUC. Mice were housed in small social groups (4–5 animals) in individually ventilated cages under 12-hour light/dark cycles and fed ad libitum. 6–10-week old animals of both genders were used in all experiments. C57BL/6N wild-type mice were purchased from Envigo (Indianapolis, IN) and all other genetically modified mice were bred in house. Sst-IRES-Cre knock-in (Taniguchi et al., 2011), Trpv1-IRES-Cre knock-in (Cavanaugh et al., 2011), GRP-Cre BAC-transgenic (Gong et al., 2007) and Mcpt5-Cre BAC-transgenic (Scholten et al., 2008) mice that drive expression of Cre recombinase in genetically identifiable cell populations have been described previously. These Cre-driver lines were crossed to conditional alleles, as described throughout the text, to enable the Cre-dependent expression of tdTomato (Ai9) (Madisen et al., 2010), GCaMP6s (Ai96) (Madisen et al., 2015), hM3Dq-mCitrine (Zhu et al., 2016), or DTA (Voehringer et al., 2008) from the R26 locus. The LSL-hM3Dq-mCitrine allele was targeted for integration into the R26 locus but, as established by the Jackson Laboratory (Bar Harbor, ME), it integrated elsewhere in the genome without affecting functionality of the receptor. Trpv1-IRES-Cre animals were also bred to a floxed S1pr1 allele (Allende et al., 2003), allowing a conditional deletion of S1pr1 in sensory neurons. Pam mice (Pamtm1a(EUCOMM)Wtsi) were obtained from EuMMCR (Skarnes et al., 2011) and crossed with FLP deleter mice (B6N.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/J) to obtain PAMfl/fl mice. GRP-Cre BAC-transgenic mice were bred to PAMfl/fl mice to conditionally delete PAM in GRP-expressing spinal cord neurons. Genotyping of offspring from all breeding steps was performed with genomic DNA isolated from tail snips and allele-specific primer pairs (see Table S2).
PRIMARY CELL CULTURES
Primary cultures of DRG neurons were generated from Trpv1-Cre∷Ai96 and Sst-Cre∷Ai96 mice of both genders. DRG were dissected and dissociated as described previously (Li et al., 2017). Briefly, DRG were harvested and incubated in 5 mg/mL Collagenase/Dispase (10269638001, Millipore-Sigma) for 30 minutes. Cells were mechanically dissociated and seeded on poly-D lysine-coated coverslips and cultured for 48 hours before the experiment (DMEM/F12, 10% FBS, penicillin/streptomycin, 100 ng/mL NGF, 50 ng/mL GDNF).
Primary cultures of peritoneum-derived mast cells (PDMC) were prepared as described previously (Tsvilovskyy et al., 2018). Briefly, mast cells were isolated by peritoneal lavage from C57BL/6N mice of both genders and cultured for two weeks in medium (DMEM, 2 mM L-glutamine, penicillin/streptomycin, 5% FBS, 10 mM HEPES, 50 mg/ml gentamicin) containing IL3 (20 ng/ml) and SCF (30 ng/ml).
METHOD DETAILS
RNA-sequencing
Sst-IRES-Cre and Trpv1-IRES-Cre pups at P2 were intraperitoneally injected with AAV9-CAG-Flex-eGFP or AAV9-CAG-Flex-tdTomato (10 μL, 1×1013 vg/mL). From these mice, DRG neurons were acutely dissociated as described previously (Li et al., 2017). Between 150–250 eGFP- or tdTomato-positive cells per mouse were hand-picked. Each picking session included one sample per genotype to potentially identify experimentally induced biases. A total of 4 independent samples per genotype were generated. Total RNA was extracted with a PicoPure RNA isolation kit (ThermoFisher Scientific, Waltham, MA) using the manufacturer’s instructions. cDNA was produced and amplified using the Ovation® RNA-seq system V2 (NuGEN, San Carlos, CA) according the manufacturer’s protocol. Sequencing libraries were generated using the Nextera XT method (Illumina, San Diego, CA), pooled and sequenced in 126 bp paired-end mode on a HiSeq 1000 instrument (Illumina). Data was demultiplexed, trimmed and mapped to the mouse genome GRCm38 using star (Dobin et al., 2013). Read counting was performed with featureCounts (Liao et al., 2014) and only genes with at least 1 read in 3 out of 4 replicates were further analyzed. Differential gene expression was assessed with EdgeR (Robinson et al., 2010) and genes with p < 0.05 were included in downstream analyses. Hierarchical clustering of samples was performed with Morpheus (Broad institute, Cambridge, MA) using the settings “One minus Pearson correlation” and “linkage method complete”. GPCRs in the set of differentially expressed genes were identified using gene ontology. Expression differences in this GPCR gene set were visualized as a heatmap using Morpheus. Prior to plotting, row median reads were subtracted from raw reads and subsequently divided by the row median absolute deviation, to enhance the visibility.
ISH
DRGs were dissected from mice with various genotypes. Multi-labeling ISH was performed using the RNAscope® technology (ACD, Newark, CA) according to the manufacturer’s instructions. Probes against Trpv1, Nppb, Mrgpra3, Tubb3, Hrh1, Htr1f, Cysltr2, and S1pr1 in conjunction with the RNAscope® multiplex fluorescent development kit were used. Images were collected on an Eclipse Ti (Nikon, Melville, NY) confocal laser-scanning microscope. Soma size was determined with ImageJ (NIH, Bethesda, MD).
Mast cell staining in the skin
Ear skin from MC-Cre∷LSL-tdTomato, LSL-hM3Dq, MC-Cre∷LSL-hM3Dq, and MC-DTA mice was fixed in 4% (w/v) paraformalde-hyde (PFA) overnight at 4°C, cryoprotected in 30% sucrose for 2 days, embedded in Tissue-Tek® O.C.T. (Sakura Finetek, Torrance, CA) and cut on a cryostat (Leica Biosystems, Buffalo Grove, IL) at 20 mm. Slides were warmed at 37°C for 30 minutes before fixing sections for 10 minutes in 4% PFA. Sections were blocked (10% normal goat serum in PBS/0.2% Triton X-100) for 2 hours at room temperature. Mast cells were visualized by incubation with avidin-FITC (1:500 in blocking solution, Millipore-Sigma, Burlington, MA) for 60 minutes and three subsequent washing steps in PBS. In some cases, mast cell visualization was combined with immunohistochemical staining for hM3Dq-mCitrine. Here, sections were incubated overnight at 4°C with a polyclonal anti-GFP antiserum (A6455, ThermoFisher Scientific) at a 1:500 dilution in PBS/0.2% Triton X-100. Mast cell visualization was then performed during incubation with the donkey-anti-rabbit-Cy3 secondary antibody (711-165-152, Jackson ImmunoResearch, West Grove, PA) at a 1:200 dilution in PBS/0.2% Triton X-100. All sections were coverslipped in Fluoromount G with DAPI (SouthernBiotech, Birmingham, AL) and imaged on an Eclipse Ti (Nikon) confocal laser-scanning microscope with appropriate filter sets. Co-localization of FITC- and tdTomato/Cy3-dependent fluorescence was analyzed with ImageJ.
Calcium Imaging
After 48 hours of culture, coverslips with primary DRG neurons were mounted on a DMi8 microscope (Leica, Allendale, NJ) in HBSS buffer (in mM: 140 NaCl, 2 CaCl2, 10 HEPES, 4 KCl, 1 MgCl2, pH 7.4) and constantly super fused from a gravity-fed six-channel system (VC-6, Warner Instruments, Hamden, CT). Imaging was done with an ORCA-Flash 4.0 C1440 digital CMOS camera (Hamamatsu, Bridgewater, NJ) at 1 Hz. Fluorescence intensity in hand-drawn regions of interest was extracted using HCImage (Hamamatsu) and plotted against time. Raster plots to visualize population responses were produced in R (R Core Team, 2013).
Reverse Transcription-PCR
Spinal cords from PAMfl/fl and GRP-Cre∷PAMfl/fl mice were dissected and total RNA extracted using a RNeasy lipid tissue mini kit (QIAGEN, Germantown, MD) following the manufacturer’s instructions. During RNA extraction, on column DNase treatment was performed. Total RNA was reverse transcribed using SMARTScribe reverse transcriptase (Takara Bio, Mountain View, CA) and oligo(dT) 18 primer according to the manufacturer’s protocol. As a control, mock reactions lacking reverse transcriptase were performed and used as no template control during PCR. Cre-recombined PAM transcripts (for primer: CACTGGGAGTTACTGGTGTTGGAT, rev primer: TAAGGACACACCGGAACAGTCTT) or b-actin transcripts as a control (for primer: CTGGCTCCTAGCACCATGAAGATC, rev primer: CTAGAAGCACTTGCGGTGCACG) were amplified by PCR using a Phusion® hot start flex polymerase (New England Biolabs, Ipswich, MA) according to the manufacturer’s instructions and detected on 2% agarose gels.
ELISA of PDMC supernatants
After two weeks in culture and before stimulation with compound 48/80 (50 μg/ml), cells were washed twice in growth factor-free medium and 1.5 × 104 (for quantification of LTC4 and serotonin release) or 1.5 × 105 (for quantification of S1p release) cells were seeded in round-bottom 96-well plates. Release of LTC4 (501070, Cayman Chemical, Ann Arbor, MI), serotonin (ADI-900–175, Enzo Life Sciences, East Farmingdale, NY) and S1p (CEG031Ge, Cloud-Clone Corp., Katy, TX) were measured in supernatants using competition ELISAs according to the manufacturer’s instructions.
Mouse behavioral measurements
For all behavioral paradigms, the experimenter was blind to the genotype of mice under study. Ear-tag numbers were read after the experiment and results were unblinded at the end of testing sessions. All behavioral experiments were conducted during the light cycle at ambient temperature (~23°C). Behavioral assessment of scratching behavior was conducted as described previously (Mishra et al., 2011). Briefly, mice were injected subcutaneously into the nape of the neck with histamine, chloroquine, compound 48/80, LY344864 (all Millipore-Sigma), nor-Binaltorphimine, CYM5442 (both Tocris, Minneapolis, MN), CNO (Abcam, Cambridge, MA), or N-met LTC4 (Cayman Chemical). Compounds were diluted in PBS except CYM5442 which was diluted in dH2O. Higher doses of CNO led to anaphylaxis-like reactions, most likely because of the systemic dispersion of highly lipophilic CNO. Therefore, we limited the CNO dose we employed in our studies to prevent anaphylaxis and consequently the conditions we used may not have produced fully activated mast cells. In a separate experiment, GRP was injected intrathecally. Scratching behavior was recorded for 30 minutes and is presented in bouts per 30 minutes. One bout was defined as scratching behavior toward the injection site between lifting the hind leg from the ground and either putting it back on the ground or guarding the paw with the mouth. Injections of LY344864, CYM5442, N-met LTC4 and capsaicin in the mouse cheek itch/pain model were performed as described previously (Shimada and LaMotte, 2008). Injection volume was always 10 μl.
Ablation of Trpv1 lineage neurons was functionally verified by testing the sensitivity of mice toward application of capsaicin onto the cornea as described previously (Mishra et al., 2011). Briefly, 50 μl of a 50 μM capsaicin solution were applied onto the cornea and wiping behavior was counted for 30 s. Responses to heat were tested as described previously with minor modifications (Hargreaves et al., 1988). Hind paws of mice habituated in small plastic enclosures on a plantar test instrument (Ugo Basile, Gemonio, Italy) were stimulated with a radiant heat beam and time to withdrawal was measured. Withdrawal was tested 5 times for each hind paw, consecutive tests of the same paw were separated by at least 3 minutes. Cold responses were tested as described previously (Brenner et al., 2012). Briefly, a dry ice pellet was applied below the hind paw of a mouse sitting on a glass surface and time to withdrawal was measured. Withdrawal was tested 5 times for each hind paw, consecutive tests of the same paw were separated by at least 3 minutes. Mechanical sensitivity thresholds were assessed using calibrated von Frey filaments employing the simplified up-down method (Bonin et al., 2014). Motor coordination was tested by measuring the performance on an accelerating rota-rod, as previously described (Jones and Roberts, 1968). Each mouse performed the task 3 times and the rod speed at failure was averaged. Ongoing spontaneous pain responses were quantified by counting paw flinches for 15 or 30 minutes after intraplantar injection of capsaicin or allyl isothiocyanate, respectively.
QUANTIFICATION AND STATISTICAL ANALYSIS
Prism 7.0 (GraphPad Software, La Jolla, CA) was used for statistical analyses. Differences between mean values were analyzed using unpaired two-tailed Student’s t test when two groups were compared or 1-way analysis of variances with Sidak’s multiple comparisons post hoc test when more than two data groups were compared. Differences were considered significant for *p < 0.05. Exact p values, definition and number of replicates as well as definitions of center and dispersion are given in the respective figure legend. No statistical method was employed to predetermine sample sizes. The sample sizes used in our experiments were similar to those generally used in the field.
DATA AND SOFTWARE AVAILABILITY
The RNaseq datasets generated in this paper have been deposited with GEO under ID code GSE125626.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | ThermoFisher | Cat#A-6455:RRID: AB_221570 |
| Bacterial and Virus Strains | ||
| AAV9-CAG-FLEX-eGFP-WPRE | Oh et al., 2014 | “Addgene” Cat#51502-AAV9 |
| AAV9-CAG-FLEX-tdTomato-WPRE | Oh et al., 2014 | “Addgene” Cat#51503-AAV9 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CYM5442 | Tocris | Cat#3601 |
| CNO | Abcam | Cat#ab141704 |
| N-methyl LTC4 | Cayman Chemical | Cat#13390 |
| LY344864 | Millipore-Sigma | Cat#SML0556 |
| Critical Commercial Assays | ||
| LTC4 ELISA | Cayman Chemical | Cat#501070 |
| Serotonin ELISA | Enzo Life Sciences | Cat#ADI-900–175 |
| S1p ELISA | Cloud-Clone Corp. | Cat#ECG031Ge |
| RNAscope® multiplex fluorescent development kit | ACD | Cat#320850 |
| PicoPure RNA isolation kit | ThermoFishe | Cat#KIT0204 |
| Ovation® RNA-seq system V2 | NuGEN | Cat#7102–08 |
| Deposited Data | ||
| Mouse reference genome GRCm38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/mouse |
| Raw and analyzed data | ||
| RNaseq datasets | This paper | GEO:GSE125626 |
| Experimental Models: Organisms/Strains | ||
| Mice: C57BL/6N | Envigo | Order code 044 |
| Mouse: Ssttm21(cre)Zjh/J | The Jackson Laboratory | JAX: 013044 |
| Mouse: Trpv1tm1(cre)Bbm/J | The Jackson Laboratory | JAX: 017769 |
| Mouse: Tg(Grp-Cre)KH107Gsat/Mmucd | MMRRC | 031183-UCD |
| Mouse: TG(Cma1-Cre)Aroer/D | Scholten et al., 2008 | Dr. Axel Roers |
| Mouse: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX: 007909 |
| Mouse: B6;129S6-Gt(ROSA)26Sortm96(CAG-GCaMP6s)Hze/J | The Jackson Laboratory | JAX: 024106 |
| Mouse: B6N;129-Gt(ROSA)26Sortm2(CAG-CHRM3*−mCitrine)1Ute/J | The Jackson Laboratory | JAX: 026220 |
| Mouse: Gt(ROSA)26Sortm1(DTA)Jpmb/J | The Jackson Laboratory | JAX: 006331 |
| Mouse: B6.129S6(FVB)-S1pr1tm21Rlp/J | The Jackson Laboratory | JAX: 019141 |
| Mouse: B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/RainJ | The Jackson Laboratory | JAX: 009086 |
| Mouse: PAMfl/fl | This paper | N/A |
| Oligonucleotides | ||
| Recombined PAM allele FOR primer for RT-PCR: CACTGGGAGTTACTGGTGTTGGAT | This paper | N/A |
| Recombined PAM allele REV primer for RT-PCR: TAAGGACACACCGGAACAGTCTT | This paper | N/A |
| β-actin FOR primer for RT-PCR: CTGGCTCCTAGCACC ATGAAGATC | This paper | N/A |
| b-actin REV primer for RT-PCR: CTAGAAGCACTTGCG GTGCACG | This paper | N/A |
| Primers for genotyping, see Table S2 | This paper | N/A |
| Software and Algorithms | ||
| star | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| featureCounts | Liao et al., 2014 | http://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| EdgeR | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Morpheus | Broad Institute | https://software.broadinstitute.org/morpheus/ |
Highlights.
Chemogenetic activation of mast cells induces itch responses
Receptors for mast cell mediators are specifically expressed by Nppb neurons
Serotonin, leukotriene, and sphingosine-1-phosphate stimulate Nppb neurons
Mast cell activation via GRP spinal cord signaling elicits itch behavior
ACKNOWLEDGMENTS
We wish to thank Dr. Axel Roers for generously sharing Mcpt5-Cre mice. PAMfl/fl mice (Pamtm1a(EUCOMM)Wtsi) were obtained from the European Mouse Mutant Cell Repository (EuMMCR) and engineered by the Welcome Trust Sanger Institute. We thank the NIMH Transgenic Core for IVF sperm recovery of PAMfl/fl mice. This work was supported by the intramural research program of the National Institute of Dental and Craniofacial Research, NIH (M.A.H.), the Lundbeck Foundation, Denmark (M.C.K.), and the National Center for Complementary and Integrative Health, NIH (A.T.C.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.celrep.2019.02.089.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abeck D, Andersson T, Grosshans E, Jablonska S, Kragballe K, Vahlquist A, Schmidt T, Dupuy P, and Ring J (1997). Topical application of a platelet-activating factor (PAF) antagonist in atopic dermatitis. Acta Derm. Venereol 77, 449–451. [DOI] [PubMed] [Google Scholar]
- Allende ML, Yamashita T, and Proia RL (2003). G-protein-coupled receptor S1P1 acts within endothelial cells to regulate vascular maturation. Blood 102, 3665–3667. [DOI] [PubMed] [Google Scholar]
- Armbruster BN, Li X, Pausch MH, Herlitze S, and Roth BL (2007). Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. USA 104, 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi E, Xia J, and Lerner EA (2016). Peripheral mechanisms of itch. Curr. Probl. Dermatol 50, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista DM, Wilson SR, and Hoon MA (2014). Why we scratch an itch: the molecules, cells and circuits of itch. Nat. Neurosci 17, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin RP, Bories C, and De Koninck Y (2014). A simplified up-down method (SUDO) for measuring mechanical nociception in rodents using von Frey filaments. Mol. Pain 10, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce JA (2007). Mast cells and eicosanoid mediators: a system of reciprocal paracrine and autocrine regulation. Immunol. Rev 217, 168–185. [DOI] [PubMed] [Google Scholar]
- Brenner DS, Golden JP, and Gereau RW 4th. (2012). A novel behavioral assay for measuring cold sensation in mice. PLoS ONE 7, e39765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh DJ, Chesler AT, Jackson AC, Sigal YM, Yamanaka H, Grant R, O’Donnell D, Nicoll RA, Shah NM, Julius D, and Basbaum AI (2011). Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. J. Neurosci 31, 5067–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevikbas F, Wang X, Akiyama T, Kempkes C, Savinko T, Antal A, Kukova G, Buhl T, Ikoma A, Buddenkotte J, et al. (2014). A sensory neuron-expressed IL-31 receptor mediates T helper cell-dependent itch: Involvement of TRPV1 and TRPA1. J. Allergy Clin. Immunol 133, 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checa A, Xu N, Sar DG, Haeggström JZ, Ståhle M, and Wheelock CE (2015). Circulating levels of sphingosine-1-phosphate are elevated in severe, but not mild psoriasis and are unresponsive to anti-TNF-a treatment. Sci. Rep 5, 12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu IM, Barrett LB, Williams EK, Strochlic DE, Lee S, Weyer AD, Lou S, Bryman GS, Roberson DP, Ghasemlou N, et al. (2014). Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. eLife 3, e04660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva EZ, Jamur MC, and Oliver C (2014). Mast cell function: a new vision of an old cell. J. Histochem. Cytochem 62, 698–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felipe C, Herrero JF, O’Brien JA, Palmer JA, Doyle CA, Smith AJ, Laird JM, Belmonte C, Cervero F, and Hunt SP (1998). Altered nociception, analgesia and aggression in mice lacking the receptor for substance P. Nature 392, 394–397. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esancy K, Condon L, Feng J, Kimball C, Curtright A, and Dhaka A (2018). A zebrafish and mouse model for selective pruritus via direct activation of TRPA1. eLife 7, e32036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, and Gerfen CR (2007). Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J. Neurosci 27, 9817–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould HJ, and Sutton BJ (2008). IgE in allergy and asthma today. Nat. Rev. Immunol 8, 205–217. [DOI] [PubMed] [Google Scholar]
- Guo M, Qu X, and Qin XQ (2015). Bombesin-like peptides and their receptors: recent findings in pharmacology and physiology. Curr. Opin. Endocrinol. Diabetes Obes 22, 3–8. [DOI] [PubMed] [Google Scholar]
- Han L, and Dong X (2014). Itch mechanisms and circuits. Annu. Rev. Biophys 43, 331–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Mancino V, and Simon MI (2006). Phospholipase Cbeta 3 mediates the scratching response activated by the histamine H1 receptor on C-fiber nociceptive neurons. Neuron 52, 691–703. [DOI] [PubMed] [Google Scholar]
- Han L, Ma C, Liu Q, Weng HJ, Cui Y, Tang Z, Kim Y, Nie H, Qu L, Patel KN, et al. (2013). A subpopulation of nociceptors specifically linked to itch. Nat. Neurosci 16, 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves K, Dubner R, Brown F, Flores C, and Joris J (1988). A new and sensitive method for measuring thermal nociception in cutaneous hyper-algesia. Pain 32, 77–88. [DOI] [PubMed] [Google Scholar]
- Hawro T, Saluja R, Weller K, Altrichter S, Metz M, and Maurer M (2014). Interleukin-31 does not induce immediate itch in atopic dermatitis patients and healthy controls after skin challenge. Allergy 69, 113–117. [DOI] [PubMed] [Google Scholar]
- Hill RZ, Morita T, Brem RB, and Bautista DM (2018). S1PR3 mediates itch and pain via distinct TRP channel-dependent pathways. J. Neurosci 38, 7833–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Polgar E, Solinski HJ, Mishra SK, Tseng PY, Iwagaki N, Boyle KA, Dickie AC, Kriegbaum MC, Wildner H, et al. (2018). Circuit dissection of the role of somatostatin in itch and pain. Nat. Neurosci 21, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamachi N, Park GH, Lee H, Anderson DJ, Simon MI, Basbaum AI, and Han SK (2009). TRPV1-expressing primary afferents generate behavioral responses to pruritogens via multiple mechanisms. Proc. Natl. Acad. Sci. USA 106, 11330–11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Kawamata T, Tanaka S, Shindo T, and Kawamata M (2014). Calcitonin gene-related peptide is involved in inflammatory pain but not in postoperative pain. Anesthesiology 121, 1068–1079. [DOI] [PubMed] [Google Scholar]
- Jones BJ, and Roberts DJ (1968). The quantiative measurement of motor inco-ordination in naive mice using an acelerating rotarod. J. Pharm. Pharmacol 20, 302–304. [DOI] [PubMed] [Google Scholar]
- Kido-Nakahara M, Buddenkotte J, Kempkes C, Ikoma A, Cevikbas F, Akiyama T, Nunes F, Seeliger S, Hasdemir B, Mess C, et al. (2014). Neural peptidase endothelin-converting enzyme 1 regulates endothelin 1-induced pruritus. J. Clin. Invest 124, 2683–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini SP, DeLong LK, Veledar E, McKenzie-Brown AM, Schaufele M, and Chen SC (2011). The impact of pruritus on quality of life: the skin equivalent of pain. Arch. Dermatol 147, 1153–1156. [DOI] [PubMed] [Google Scholar]
- Krystel-Whittemore M, Dileepan KN, and Wood JG (2016). Mast cell: a multi-functional master cell. Front. Immunol 6, 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Cho PS, Tonello R, Lee HK, Jang JH, Park GY, Hwang SW, Park CK, Jung SJ, and Berta T (2018). Peripheral serotonin receptor 2B and transient receptor potential channel 4 mediate pruritus to serotonergic antidepressants in mice. J. Allergy Clin. Immunol 142, 1349–1352.e16. [DOI] [PubMed] [Google Scholar]
- Lembo PM, Grazzini E, Groblewski T, O’Donnell D, Roy MO, Zhang J, Hoffert C, Cao J, Schmidt R, Pelletier M, et al. (2002). Proenkephalin A gene products activate a new family of sensory neuron–specific GPCRs. Nat. Neurosci 5, 201–209. [DOI] [PubMed] [Google Scholar]
- Li CL, Li KC, Wu D, Chen Y, Luo H, Zhao JR, Wang SS, Sun MM, Lu YJ, Zhong YQ, et al. (2016). Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res 26, 83–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tseng PY, Tiwari V, Xu Q, He SQ, Wang Y, Zheng Q, Han L, Wu Z, Blobaum AL, et al. (2017). Targeting human Mas-related G protein-coupled receptor X1 to inhibit persistent pain. Proc. Natl. Acad. Sci. USA 114, E1996–E2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, Kim A, Ru F, Guan Y, Weng HJ, Geng Y, et al. (2009). Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell 139, 1353–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Xu ZZ, Park CK, Berta T, and Ji RR (2010a). Toll-like receptor 7 mediates pruritus. Nat. Neurosci 13, 1460–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Abdel Samad O., Zhang L, Duan B, Tong Q, Lopes C, Ji RR, Lowell BB, and Ma Q (2010b). VGLUT2-dependent glutamate release from nociceptors is required to sense pain and suppress itch. Neuron 68, 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Weng HJ, Patel KN, Tang Z, Bai H, Steinhoff M, and Dong X (2011). The distinct roles of two GPCRs, MrgprC11 and PAR2, in itch and hyperalgesia. Sci. Signal 4, ra45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Sikand P, Ma C, Tang Z, Han L, Li Z, Sun S, LaMotte RH, and Dong X (2012). Mechanisms of itch evoked by b-alanine. J. Neurosci 32, 14532–14537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, et al. (2015). Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima H, Yamada N, Matsue H, and Shimada S (2004). The effects of endothelin-1 on degranulation, cytokine, and growth factor production by skin-derived mast cells. Eur. J. Immunol 34, 1910–1919. [DOI] [PubMed] [Google Scholar]
- McNeil BD, Pundir P, Meeker S, Han L, Undem BJ, Kulka M, and Dong X (2015). Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature 519, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkler DJ (1994). C-terminal amidated peptides: production by the in vitro enzymatic amidation of glycine-extended peptides and the importance of the amide to bioactivity. Enzyme Microb. Technol 16, 450–456. [DOI] [PubMed] [Google Scholar]
- Metcalfe DD, Baram D, and Mekori YA (1997). Mast cells. Physiol. Rev 77, 1033–1079. [DOI] [PubMed] [Google Scholar]
- Mishra SK, and Hoon MA (2013). The cells and circuitry for itch responses in mice. Science 340, 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Tisel SM, Orestes P, Bhangoo SK, and Hoon MA (2011). TRPV1-lineage neurons are required for thermal sensation. EMBO J 30, 582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modena BD, Dazy K, and White AA (2016). Emerging concepts: mast cell involvement in allergic diseases. Transl. Res 174, 98–121. [DOI] [PubMed] [Google Scholar]
- Moon TC, Befus AD, and Kulka M (2014). Mast cell mediators: their differential release and the secretory pathways involved. Front. Immunol 5, 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, McClain SP, Batia LM, Pellegrino M, Wilson SR, Kienzler MA, Lyman K, Olsen AS, Wong JF, Stucky CL, et al. (2015). HTR7 mediates serotonergic acute and chronic itch. Neuron 87, 124–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Murakami M, Yanoshita R, Samejima Y, Karasawa K, Setaka M, Nojima S, and Kudo I (1997). Activated mast cells release extracellular type platelet-activating factor acetylhydrolase that contributes to autocrine inactivation of platelet-activating factor. J. Biol. Chem 272, 19708–19713. [DOI] [PubMed] [Google Scholar]
- Nguyen MQ, Wu Y, Bonilla LS, von Buchholtz LJ, and Ryba NJP (2017). Diversity amongst trigeminal neurons revealed by high throughput single cell sequencing. PLoS ONE 12, e0185543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetjen LK, Mack MR, Feng J, Whelan TM, Niu H, Guo CJ, Chen S, Trier AM, Xu AZ, Tripathi SV, et al. (2017). Sensory neurons co-opt classical immune signaling pathways to mediate chronic itch. Cell 171, 217–228.e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. (2014). A mesoscale connectome of the mouse brain. Nature 508, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera A, and Rivera J (2005). Sphingolipids and the balancing of immune cell function: lessons from the mast cell. J. Immunol 174, 1153–1158. [DOI] [PubMed] [Google Scholar]
- Patel O, Dumesny C, Giraud AS, Baldwin GS, and Shulkes A (2004). Stimulation of proliferation and migration of a colorectal cancer cell line by amidated and glycine-extended gastrin-releasing peptide via the same receptor. Biochem. Pharmacol 68, 2129–2142. [DOI] [PubMed] [Google Scholar]
- Patel O, Shulkes A, and Baldwin GS (2006). Gastrin-releasing peptide and cancer. Biochim. Biophys. Acta 1766, 23–41. [DOI] [PubMed] [Google Scholar]
- Perkins SN, Husten EJ, and Eipper BA (1990). The 108-kDA peptidylglycine alpha-amidating monooxygenase precursor contains two separable enzymatic activities involved in peptide amidation. Biochem. Biophys. Res. Commun 171, 926–932. [DOI] [PubMed] [Google Scholar]
- R Core Team (2013). R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing)
- Reddy VB, and Lerner EA (2010). Plant cysteine proteases that evoke itch activate protease-activated receptors. Br. J. Dermatol 163, 532–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VB, Iuga AO, Shimada SG, LaMotte RH, and Lerner EA (2008). Cowhage-evoked itch is mediated by a novel cysteine protease: a ligand of protease-activated receptors. J. Neurosci 28, 4331–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VB, Shimada SG, Sikand P, Lamotte RH, and Lerner EA (2010). Cathepsin S elicits itch and signals via protease-activated receptors. J. Invest. Dermatol 130, 1468–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VB, Sun S, Azimi E, Elmariah SB, Dong X, and Lerner EA (2015). Redefining the concept of protease-activated receptors: cathepsin S evokes itch via activation of Mrgprs. Nat. Commun 6, 7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru F, Sun H, Jurcakova D, Herbstsomer RA, Meixong J, Dong X, and Undem BJ (2017). Mechanisms of pruritogen-induced activation of itch nerves in isolated mouse skin. J. Physiol 595, 3651–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadik CD, Sezin T, and Kim ND (2013). Leukotrienes orchestrating allergic skin inflammation. Exp. Dermatol 22, 705–709. [DOI] [PubMed] [Google Scholar]
- Savin JA (1998). How should we define itching? J. Am. Acad. Dermatol 39, 268–269. [DOI] [PubMed] [Google Scholar]
- Schemann M, Kugler EM, Buhner S, Eastwood C, Donovan J, Jiang W, and Grundy D (2012). The mast cell degranulator compound 48/80 directly activates neurons. PLoS ONE 7, e52104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten J, Hartmann K, Gerbaulet A, Krieg T, Müller W, Testa G, and Roers A (2008). Mast cell-specific Cre/loxP-mediated recombination in vivo. Transgenic Res 17, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada SG, and LaMotte RH (2008). Behavioral differentiation between itch and pain in mouse. Pain 139, 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoerdsma A, Waalkes TP, and Weissbach H (1957). Serotonin and hista-mine in mast cells. Science 125, 1202–1203. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. (2011). A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E, Muller A, Lauerma AI, Pivarcsi A, Soto H, Kemeny L, Alenius H, Dieu-Nosjean MC, Meller S, Rieker J, et al. (2006). IL-31: a new link between T cells and pruritus in atopic skin inflammation. J. Allergy Clin. Immunol 117, 411–417. [DOI] [PubMed] [Google Scholar]
- Stantcheva KK, Iovino L, Dhandapani R, Martinez C, Castaldi L, Nocchi L, Perlas E, Portulano C, Pesaresi M, Shirlekar KS, et al. (2016). A sub-population of itch-sensing neurons marked by Ret and somatostatin expression. EMBO Rep 17, 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stante M, Hanna D, and Lotti T (2005). Itch, pain, and metaesthetic sensation. Dermatol. Ther 18, 308–313. [DOI] [PubMed] [Google Scholar]
- Steinhoff M, Buddenkotte J, and Lerner EA (2018). Role of mast cells and basophils in pruritus. Immunol. Rev 282, 248–264. [DOI] [PubMed] [Google Scholar]
- Sumpter TL, Balmert SC, and Kaplan DH (2019). Cutaneous immune responses mediated by dendritic cells and mast cells. JCI Insight 4, 123947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YG, and Chen ZF (2007). A gastrin-releasing peptide receptor mediates the itch sensation in the spinal cord. Nature 448, 700–703. [DOI] [PubMed] [Google Scholar]
- Sun YG, Zhao ZQ, Meng XL, Yin J, Liu XY, and Chen ZF (2009). Cellular basis of itch sensation. Science 325, 1531–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay SS, Roediger B, Tong PL, Tikoo S, and Weninger W (2013). The skin-resident immune network. Curr. Dermatol. Rep 3, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvilovskyy V, Solis-Lopez A, Ohlenschlager K, and Freichel M (2018). Isolation of peritoneum-derived mast cells and their functional characterization with Ca2+-imaging and degranulation assays. J. Vis. Exp 4, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui H, Andoh T, Lee JB, Nojima H, and Kuraishi Y (2006). Potent pruritogenic action of tryptase mediated by PAR-2 receptor and its involvement in anti-pruritic effect of nafamostat mesilate in mice. Eur. J. Pharmacol 530, 172–178. [DOI] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, et al. (2015). Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci 18, 145–153. [DOI] [PubMed] [Google Scholar]
- Voehringer D, Liang HE, and Locksley RM (2008). Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J. Immunol 180, 4742–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson S, and Pejler G (2014). Mast cell secretory granules: armed for battle. Nat. Rev. Immunol 14, 478–494. [DOI] [PubMed] [Google Scholar]
- Wilson SR, Thé L, Batia LM, Beattie K, Katibah GE, McClain SP, Pellegrino M, Estandian DM, and Bautista DM (2013). The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell 155, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosipovitch G, Greaves MW, and Schmelz M (2003). Itch. Lancet 361, 690–694. [DOI] [PubMed] [Google Scholar]
- Zhu H, Aryal DK, Olsen RH, Urban DJ, Swearingen A, Forbes S, Roth BL, and Hochgeschwender U (2016). Cre-dependent DREADD (designer receptors exclusively activated by designer drugs) mice. Genesis 54, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNaseq datasets generated in this paper have been deposited with GEO under ID code GSE125626.