Abstract
The Rhabdochlamydiaceae family is one of the most widely distributed within the phylum Chlamydiae, but most of its members remain uncultivable. Rhabdochlamydia 16S rRNA was recently reported in more than 2% of 8,534 pools of ticks from Switzerland. Shotgun metagenomics was performed on a pool of five female Ixodes ricinus ticks presenting a high concentration of chlamydial DNA, allowing the assembly of a high-quality draft genome.
About 60% of sequence reads originated from a single bacterial population that was named “Candidatus Rhabdochlamydia helvetica” whereas only few thousand reads mapped to the genome of “Candidatus Midichloria mitochondrii,” a symbiont normally observed in all I. ricinus females. The 1.8 Mbp genome of R. helvetica is smaller than other Chlamydia-related bacteria. Comparative analyses with other chlamydial genomes identified transposases of the PD-(D/E)XK nuclease family that are unique to this new genome. These transposases show evidence of interphylum horizontal gene transfers between multiple arthropod endosymbionts, including Cardinium spp. (Bacteroidetes) and diverse proteobacteria such as Wolbachia, Rickettsia spp. (Rickettsiales), and Caedimonas varicaedens (Holosporales). Bacterial symbionts were previously suggested to provide B-vitamins to hematophagous hosts. However, incomplete metabolic capacities including for B-vitamin biosynthesis, high bacterial density and limited prevalence suggest that R. helvetica is parasitic rather than symbiotic to its host.
The identification of novel Rhabdochlamydia strains in different hosts and their sequencing will help understanding if members of this genus have become highly specialized parasites with reduced genomes, like the Chlamydiaceae, or if they could be pathogenic to humans using ticks as a transmission vector.
Keywords: chlamydia, HGT, comparative genomics, shotgun metagenomics, tick symbiont
Introduction
Ticks are the most common arthropod vector of human and animal diseases (Moutailler et al. 2016).They also frequently carry mutualist symbionts; all females and nearly 50% of the males of the European tick Ixodes ricinus carry the Rickettsiales symbiont “Candidatus Midichloria mitochondrii.” The high prevalence of this symbiont suggests an obligate association between the two species, but the role of the symbiont in the biology of I. ricinus remains unknown (Ahantarig et al. 2013; Moutailler et al. 2016). As confirmed by whole genome sequencing, ticks are incapable of de novo heme synthesis and acquire heme from exogenous source (Gulia-Nuss et al. 2016; Perner et al. 2016). Hence, tick symbionts such as Coxiella, Rickettsia, and Francisella-like may be involved in the provisioning of nutrients like B-vitamins and cofactors lacking from their exclusively hematophagous diet (Rio et al. 2016; Duron et al. 2018; Husnik 2018).
Chlamydiae DNA was identified in approximately 0.89% of I. ricinus ticks in Switzerland based on a large-scale screen of 62,889 I. ricinus ticks sampled from 172 different sites (Croxatto et al. 2014; Pilloux et al. 2015). Based on the 16S rRNA gene sequence of 359 positive samples, over 29% (105/359) belonged to the Rhabdochlamydiaceae family and exhibited high amounts of chlamydial DNA (up to 8 × 106 copies/µl, see Pilloux et al. 2015). Additional studies reported the presence of Rhabdochlamydia spp. in various tick populations around the world (Hokynar et al. 2016; Burnard et al. 2017). However, no Rhabdochlamydia isolate has been cultivated from ticks yet. The two other described species, were also identified in arthropods, suggesting that Rhabdochlamydia spp. may infect a wide range of arthropods. “Candidatus Rhabdochlamydia porcellionis” was isolated from hepatopancreatic cells of the terrestrial crustacean Porcellio scaber (Kostanjsek 2004) and “Candidatus Rhabdochlamydia crassificans” was identified in the cockroach Blatta orientalis (Corsaro et al. 2007). Up to now, only “Candidatus Rhabdochlamydia porcellionis” could be cultured in SF-9 cells (Sixt et al. 2013). Both R. porcellionis and R. crassificans species exhibited a particular cell morphology; elementary bodies presented a five layered cell wall and oblong translucent structures in the cytoplasm (Corsaro et al. 2007; Kostanjsek 2004). Older records describe very similar bacteria with pentalaminar cell walls infecting the spider Pisaura mirabilis (Morel 1978), the scorpion Buthus occitanus (Morel 1976), and the midge larvae Chironomus dorsalis (Morel 1980). No hybridization between the DNA from the scorpion and the midge larvae parasites could be observed and Guanine–Cytosine (GC)-content and genome size estimations were significantly different (1,550 kb [kilobases], 41% GC vs. 2,650 kb, 36.7% GC, respectively) for these two putative chlamydia-related bacteria of arthropods, suggesting that they likely belong to different genera (Frutos et al. 1989, 1994). The proposition to classify these bacteria in the genus Porochlamydia (Morel 1976) was not recognized and Porochlamydia were considered to be part of the Rickettsiella genus (Fournier and Raoult 2005). More recent metagenomics analyses of the spider Oedothorax gibbosus microbiome revealed a high prevalence of Rhabdochlamydia spp. in one spider population (Vanthournout and Hendrickx 2015), suggesting that these so-called “Porochlamydia” might in fact represent two members of the Rhabdochlamydiaceae family.
If R. porcellionis and R. crassificans were shown to be detrimental to their host (Radek 2000; Kostanjšek and Pirc Marolt 2015), the nature of the relationship between Rhabdochlamydia spp. and ticks, as well as their pathogenicity toward humans, remain to be investigated. In the present study, we bypassed the difficult step of cell-culture by sequencing the genome of a Rhabdochlamydia within its host I. ricinus, allowing us to identify distinctive features and to learn about the biology of this largely unknown chlamydial family.
Materials and methods
Genome Sequencing, Assembly, and Annotation
A sample with five female ticks from Lucerne area (Switzerland) in the work of Pilloux et al. (2015) was selected for high throughput sequencing. Genomic DNA was extracted and purified using Wizard Genomic DNA purification kit (Promega, Duebendorf, Switzerland). Genomic libraries were constructed using Nextera XT library kit (Illumina), normalized on beads and pooled into a single library for 150 base pairs (bp) paired-end sequencing using the MiSeq (Illumina, San Diego, CA).
Illumina reads were filtered and trimmed with fastq-mcf 1.1.2 (Aronesty 2013), keeping 150 bp reads with an average Phred quality score higher than 30 (--max-ns 1 -l 150 -L 150 --qual-mean 30). Filtered reads were assembled with Edena 3 (Hernandez et al. 2008) and metaSPAdes 3.10.1 (Nurk et al. 2017) with k-mer or overlap size ranging from 51 to 127. In addition, reads that did not map on published genome assemblies (GCA_000973045.1 and GCF_000208615.1) from the ticks I. ricinus and Ixodes scapularis (Pagel Van Zee et al. 2007; Quillery et al. 2014) were assembled using MaSuRCA 2.1.9 (Zimin et al. 2013) and Velvet 1.2.09 (Zerbino and Birney 2008). Scaffolds were blasted against a database including the I. ricinus and I. scapularis genome assemblies with BlastN (e-value ≥ 1e−5, identity ≥ 80%) to remove contaminants. Scaffolds that did not show significant similarity to Ixodes sequences were further investigated as follows: (1) Coding sequences were predicted with prodigal 2.6 (parameters: -c -m -g 11 -p meta -f sco -q) (Hyatt et al. 2010). (2) Each predicted coding sequences (CDS) was assigned to a taxonomic rank with Kaiju 1.6.2 (Menzel et al. 2016) and the proGenomes database (Mende et al. 2017). (3) A bacterial phylum was assigned to each scaffold based on the most frequent phylum assigned to its predicted CDSs. The assembly graph produced by metaSPAdes was visualized with Bandage (Wick et al. 2015) in order to investigate the connectivity of scaffolds assigned to the Chlamydiae phylum. Finally, 8 assemblies from the 4 assemblers presenting between 107 and 66 scaffolds were compared and combined with Consed (Gordon et al. 1998). The combined assembly was split in case of disagreement between assembly methods. Only scaffolds larger than 1,000 bp exhibiting no significant similarity to Ixodes sequences, consistent GC-content and sequencing depth were retained for further analyses. The completeness of the final genome assembly was estimated based on the identification of 104 nearly universal single-copy genes with CheckM (Parks et al. 2015).
Raw reads were mapped to the assembled Rhabdochlamydia helvetica genome as well as to I. scapularis, I. ricinus, and “Candidatus Midichloria mitochondrii” (GCA_000973045.2, GCF_000208615.1, and GCF_000219355.1). The mapping was done with BWA mem version 0.7.17 (Li and Durbin 2009) with a minimum seed length of 23 and all 4 genomes combined into a single fasta file.
Scaffolds were reordered based on the reference genome of Simkania negevensis Z (NC_015713) using Mauve v. 2.3.1 (Darling et al. 2004). Genome annotation was performed using Prokka v. 1.10 (Seemann 2014). Domains were annotated using InterProScan 5.18-57 (Jones et al. 2014). Predicted coding sequences were compared with the COG database (Galperin et al. 2014) using BlastP 2.2.28+ (Camacho et al. 2009) with an e-value cutoff of 1e−5, and to the RefSeq database release 79 (Haft et al. 2018) using PLAST 2.3.1 (parameters: -a 8 -M BLOSUM62 -e 1e-3 -G 11 -E 1 -force-query-order 1000 -max-hit-per-query 100 -max-hsp-per-hit 1) (Van Nguyen and Lavenier 2009). The coding density was calculated based on predicted open reading frames (ORF), or published annotations. For draft assemblies, only contigs larger than 10 kb were used to estimate the coding density.
Confirmation of Tick Species from Genomic Data
To confirm that the host species was I. ricinus, raw reads were mapped on the 18S rRNA sequence of 82 different tick species (merged into a single fasta file, supplementary table S1, Supplementary Material online). Mapped reads were extracted and remapped individually on each 18S rRNA sequence and inspected with IGV (Robinson et al. 2011).
Metabolism and Respiratory Chains Annotation
Metabolic pathways and modules were investigated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Ogata et al. 1999). KEGG orthology assignments were done using the GhostKOALA web server (Kanehisa et al. 2016). Mapping between KEGG orthologs and pathways/modules was done using the KEGG API (http://www.kegg.jp/kegg/docs/keggapi.html; last accessed April 10, 2019). Specific focus was made on basic energy metabolism, amino acids, cofactors, and vitamins biosynthesis. Transporters were annotated using gBlast3 from the transporter classification database (Saier et al. 2016), with an e-value cutoff of 10−5 and 50% coverage of both query and hit. When proteins of interest could not be identified, pseudogenes remnants or unpredicted ORFs were searched in raw contigs of the Rhabdochlamydia assembly using TBlastN and in the raw reads using Diamond (Buchfink et al. 2015).
Cluster of Orthologs and Phylogenetic Reconstructions
Protein sequences of 61 PVC (Planctomycetes, Verrucomicrobia, and Chlamydiae) superphylum strains, including 31 members of the Chlamydiales order, were downloaded from RefSeq (supplementary table S2, Supplementary Material online) and grouped into orthologous groups using OrthoFinder 0.4.0 with default parameters (Emms and Kelly 2015). The complete orthology table is reported in supplementary table S3 (Supplementary Material online). The genome assemblies of the “Candidatus Limichlamydia” strain SM23_39 (marine sediments metagenome) and “Candidatus Hydrochlamydia” strain Ga0074140 (drinking water treatment plant metagenome), were not considered for genome content analyses. Unannotated genome assemblies were annotated with Prokka v.1.10. In addition, all predicted protein sequences were annotated using the same tools as for R. helvetica (see above). Protein identities were calculated based on multiple sequence alignments built using MAFFT 7.058b (Katoh and Standley 2013). Gaps were not considered in pairwise identity calculations. Genome maps were generated using Circos (Krzywinski et al. 2009). A phylogenetic tree was reconstructed based on 172 conserved single-copy proteins with RAxML 8.2.0 (Stamatakis 2014) using the LG substitution matrix (PROTGAMMALG) with distinct partitions for each of the 172 alignments and 100 rapid bootstrap replicates. This model was preferred based on previous experience with similar data sets (Pillonel et al. 2015). Figures with phylogenies and associated data were drawn with the Python package ete2 (Huerta-Cepas et al. 2010).
Proteins harboring a PD-(D/E)XK nuclease family transposase domain (Pfam accession PF12784) were searched in 6661 reference and representative genomes (downloaded from RefSeq, September 2017) with hmmsearch (HMMER v. 3.1b2 [Eddy 2011] with “–—cut_tc” parameter). The 1,911 identified proteins were further filtered based on similarity to at least 1 of the 6 R. helvetica homologs (filtered with BlastP v. 2.7.1 with an e-value of 1e−5 and 80% of hsp query coverage) and clustered at 90% identity with cd-hit v. 4.6 (Fu et al. 2012) to reduce redundancy. The phylogeny was reconstructed with FastTree v.2.1.9 (Price et al. 2010) double precision with default parameters.
Results and Discussion
Sequencing the Endosymbiont Genome within Its Tick Host
Shotgun metagenomics of the tick pool DNA produced 25.7 million paired-ends reads in two sequencing runs. Among 80 tick species, I. ricinus and I. scapularis recruited the highest number of reads by mapping on the 18S rRNA (respectively 985 and 835). Because some regions of the 18S rRNA are strictly identical between ticks species, all recruited reads were mapped again individually on I. scapularis and I. ricinus sequences. The single base of 18S rRNA sequence enabling to distinguish both species was identical to I. ricinus variant, confirming that all five ticks in the pool indeed belonged to I. ricinus species. No other polymorphism could be identified except for one position exhibiting two populations of reads.
Sixteen million high-quality reads were used to assemble the genome de novo. Most raw reads mapped either to the R. helvetica assembly (59.96%) or to one of the two Ixodes genomes (40.03%). Only 8,087 reads mapped to “Ca. M. mitochondrii” genome. Scaffolds assembled de novo were carefully investigated to remove contaminants such as Ixodes sequences (low sequencing depth, GC-content >40%) (supplementary fig. S1, Supplementary Material online). All scaffolds larger than 12.5 kb were classified as belonging to the Chlamydiae phylum (Most of them have a median depth >1600x and a GC content <40%) (supplementary fig. S1, Supplementary Material online). Chlamydial scaffolds formed a highly interconnected network of about 1.88 Mbp in the assembly graph (supplementary fig. S2, Supplementary Material online). The final manually curated assembly of 38 scaffolds, comprising 1,830,543 bp, was estimated to be complete (104/104 CheckM markers identified) and exhibited no detectable contamination (no duplicated marker gene). The cumulative G + C skew of the reordered assembly exhibited the typical inverted “V” shape of bacterial genomes (supplementary fig. S3, Supplementary Material online). Altogether, these results indicate that Rhabdochlamydia DNA was largely dominant in the pool of ticks sequenced, and only few sequences from other bacterial species were obtained, allowing to assemble a high-quality draft genome. A putative 23,934 bp circular plasmid was identified thanks to its uniformly lower sequencing depth as compared with the rest of the assembly (supplementary fig. S4, Supplementary Material online). It encodes a homolog of the plasmid integrase pGP8-D commonly found in chlamydial plasmids.
A New Species of the Rhabdochlamydia Genus
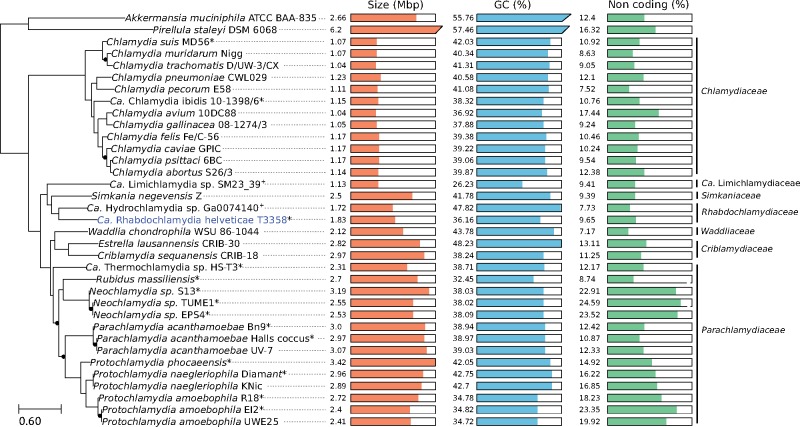
The 16S ribosomal sequence exhibited 97.88% and 97.98% identity with R. porcellionis (AY223862) and R. crassificans (AY928092), respectively. The conservation of this single gene is not sufficient to accurately classify Chlamydiae at the genus and species level (Pillonel et al. 2015). However, given the level of sequence divergence (>2%) and the lack of other Rhabdochlamydia genomes available, we considered this strain as a new Candidatus species named “Candidatus Rhabdochlamydia helvetica”. It exhibited 91.78% and 88.93% sequence identity respectively with the 16S and 23S rRNA of S. negevensis strain Z (supplementary table S4, Supplementary Material online). The reconstruction of the Chlamydiae phylogeny based on a concatenated set of 172 conserved protein sequences (fig. 1, supplementary table S5, Supplementary Material online) as well as the high number of orthologous protein families (supplementary figs. S4 and S5, Supplementary Material online) confirmed that S. negevensis is the closest sequenced relative of R. helvetica.
Fig. 1.
—Phylogeny and comparison of genome characteristics in the order Chlamydiales. The phylogeny was reconstructed using the Maximum Likelihood method implemented in RAxML. It was inferred based on the concatenated alignment of 172 single-copy protein sequences conserved in all genomes included in the phylogeny. Bootstrap supports lower than 100 are indicated with black dots. For draft genomes (indicated with asterisks), the proportion of noncoding sequences was estimated based on contigs larger than 10 kb. Plus symbols (+) indicate metagenome derived genome assemblies that were excluded from genome content analyses.
Horizontal Gene Transfers Likely Occurred with Other Symbionts and Eukaryotic Hosts
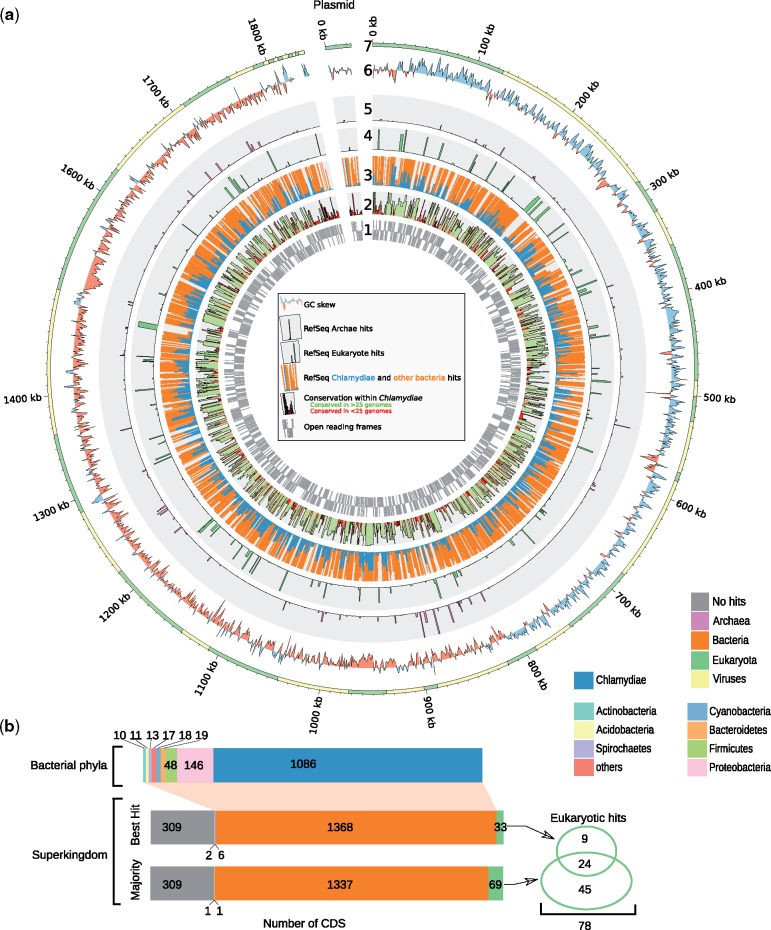
Over 62.8% of the 1,717 predicted proteins of R. helvetica exhibited a best RefSeq hit within the Chlamydiae phylum (Fig. 2). Comparative analyses of 31 chlamydial genomes from 6 families (supplementary table S1, Supplementary Material online, excluding the 2 metagenome-assembled genomes) revealed that 322 (18%) R. helvetica CDS exhibited no ortholog in other chlamydial genomes (supplementary fig. S4, Supplementary Material online, purple CDS). Among these, 73 had significant hits in RefSeq database (supplementary table S6, Supplementary Material online). Six are PD-(D/E)XK nuclease family transposases exhibiting more than 60% sequence identity with proteins of Occidentia massiliensis, a soft tick symbiont from the Rickettsiaceae family (Mediannikov et al. 2014). The transposase phylogeny including homologs identified in 6,661 RefSeq reference and representative genomes (supplementary fig. S6, Supplementary Material online) showed that the 6 R. helvetica homologs are monophyletic and cluster with phylogenetically diverse intracellular bacteria belonging to Bacteroidetes (Cardinium spp.) and Proteobacteria in the orders Rickettsiales (Wolbachia, Rickettsia, Orientia, and Occidentia spp.), Holosporales (Caedimonas varicaedens), and Myxococcales (Pajaroellobacter abortibovis). Highly similar sequences were also identified in two arthropod genomes (Cimex lectularius and Vollenhovia emeryi). This could represent an example of interkingdom horizontal gene transfer (HGT), but might also result from the contamination of the two arthropod genomes with DNA sequences of colonizing bacteria. Indeed, C. lectularius was shown to carry Wolbachia symbionts (Rasgon and Scott 2004). Interphylum transfer of transposases is not uncommon, particularly in case of shared habitats (Hooper et al. 2009), and was already observed in arthropod symbionts (Duron 2013). Several other proteins specific to R. helvetica showed significant sequence similarity to other obligate endosymbionts (supplementary table S6, Supplementary Material online). These results suggest the occurrence of HGTs with other bacteria sharing a similar niche, in this case ticks and other arthropods, as previously described for Parachlamydia and other ameba-infecting microorganisms (Gimenez et al. 2011).
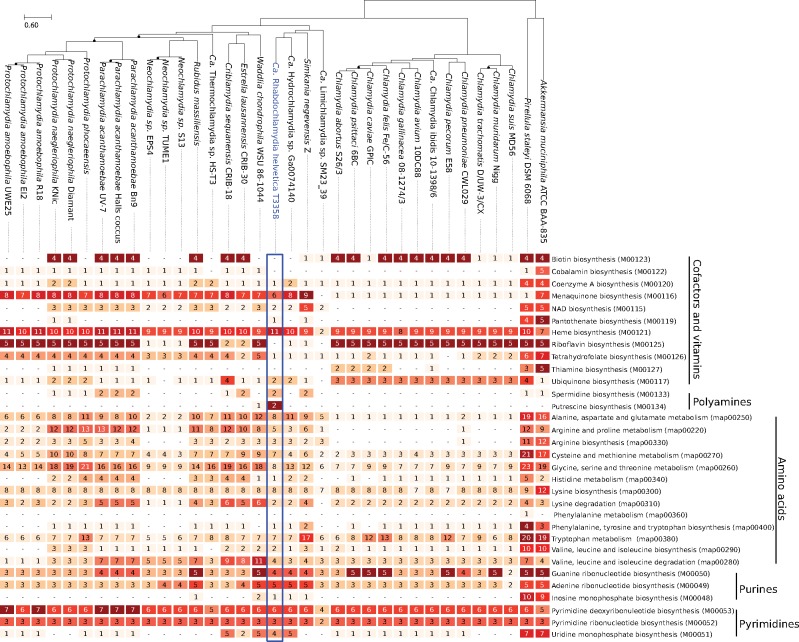
Fig. 3.
—Comparative analysis of cofactors, vitamins, amino acids, polyamines, and nucleotides metabolism. Each row reports the number of unique KEGG ortholog(s) for the corresponding module/pathway. NAD: nicotinamide adenine dinucleotide.
Seventy-eight R. helvetica proteins exhibited significant similarity to eukaryotic sequences (fig. 2, supplementary table S7, Supplementary Material online). These include known cases of chlamydial HGT such as multiple enzymes involved in the metabolism of C5 isoprenoid (ispD, ispG, ispE) (Lange et al. 2000), menaquinone (menA, menD), uridine monophosphate (pyrE, pyrF, discussed below), heme (hemY), and Pimeloyl-ACP (fabI, fabF) (Collingro et al. 2011). As previously shown (Schmitz-Esser et al. 2010), proteins with bacteria-host interaction domains such as ankyrin repeats, ubiquitin protease or BTB/POZ domains also exhibit high proportions of BLAST hits in eukaryotic sequences.
Fig. 2.
—(a) Protein homologs to the R. helvetica proteome in the National Center for Biotechnology Information (NCBI) RefSeq database. The inner gray circle (1) reports the localization of genes encoded on leading and lagging strands. The second circle (2) reports the conservation of the protein families in the 61 genomes included in the comparison (supplementary table S2, Supplementary Material online). Protein families conserved in <25 genomes are highlighted in red. The following circles report the taxonomic classification of the top 100 NCBI RefSeq hits of each R. helvetica protein: circle (3): number of chlamydial (blue) and other bacteria (orange) hits; circle (4): number of eukaryotic hits (green); circle (5): number of archaeal hits (pink). The “two outer circles” report (6) the GC-skew along the genome and (7) the localization of gaps in the genome assembly. (b) Summary of RefSeq hits taxonomy. Left part: consensus taxonomy of the top 100 best RefSeq hits (majority rule) and taxonomy of the best RefSeq hit of each R. helvetica protein. Right part: number of proteins with significant similarity to eukaryotic sequences.
Among the 78 proteins sharing similarity with eukaryotic sequences (fig. 2), 17 proteins are unique to R. helvetica. A “methyl-accepting chemotaxis” domain-containing protein presented more than 50% amino acid identity with arthropod sequences. Two lysine/ornithine decarboxylases (including a partial 145 amino-acids sequence) participating to the polyamine biosynthesis pathway (supplementary fig. S7, Supplementary Material online) exhibited 60.25% identity with a sequence from the common house spider Stegodyphus mimosarum (see phylogeny in supplementary fig. S8, Supplementary Material online). Polyamines are essential for cellular growth of both prokaryotes and eukaryotes, and may be involved in bacterial pathogenesis (Shah and Swiatlo 2008), but their role in Rhabdochlamydia biology remains unknown. The R. helvetica genome encodes all enzymes necessary for the biosynthesis of polyamines from l-arginine and l-methionine (fig. 3, supplementary fig. S7, Supplementary Material online). All enzymes show evidence of HGT with eukaryotes (rocF, speC, supplementary figs. S8 and S9, Supplementary Material online) and proteobacteria (metK, speD, and speE, supplementary figs. S10–S12, Supplementary Material online), which suggests again a strong impact of HGT in endosymbiont evolution (Bertelli and Greub 2012; Alsmark et al. 2013; Clarke et al. 2013).
Intriguingly, two enzymes of the tricarboxylic acid cycle (TCA) cycle and the subunit A of the V-type ATPase exhibited a best RefSeq hits with coding sequences of the ameba Acytostelium subglobosum. A comparison of the entire A. subglobosum genome assembly showed that about 70 kb of scaffold 25 exhibits strong similarity to Chlamydiae proteins, indicating that the assembly is probably contaminated with a fragment of an unknown chlamydial symbiont genome (supplementary fig. S13, Supplementary Material online).
Reduced Biosynthesis Capabilities Limit the Possible Mutualism with the Tick Host
Tick symbionts were previously suggested to provide essential nutrients to their host, however the metabolic repertoire of Chlamydia-related bacteria and Chlamydia sp. is known to be limited (fig. 3) (Omsland et al. 2014). Major differences comprise the presence of a glucokinase (RhT_00001) and a complete citric acid cycle. R. helvetica is able to synthesize aspartate (RhT_00062), asparagine, and glutamate and can further convert serine into glycine. Alanine production might be possible through the conversion of cysteine, but no alanine dehydrogenase could be identified. Noteworthy, no tryptophan operon—an important regulator of the chlamydial development cycle—could be identified although it is present in S. negevensis (Collingro et al. 2011; Omsland et al. 2014) and partially present in some genito-urinary strains of Chlamydia trachomatis (Omsland et al. 2014). An operon encoding the full pathway for the synthesis of pyrimidines could be identified, similar to those found in Waddlia chondrophila and Criblamydia sequanensis (supplementary fig. S14, Supplementary Material online) (Bertelli et al. 2010, 2015). The closest RefSeq homologs of proteins encoded in the operon belong to Eukaryotes such as Streptophyta and Arthropoda (pyrE, pyrF, supplementary figs. S15 and S16, Supplementary Material online), amebae (pyrD, supplementary fig. S17, Supplementary Material online), and gamma-proteobacteria such as Rickettsiella spp., obligate intracellular bacteria infecting a wide range of arthropods (pyrC, pyrB, carB, carA, supplementary figs. S18–S21, Supplementary Material online), suggesting that the operon may have been acquired through HGTs. Biosynthesis of vitamins and cofactors is restricted to heme and menaquinone (Figure 3), suggesting that R. helvetica could participate to the supply—albeit limited—of some essential factors to its host like other symbionts of blood-feeding parasite (Leclerque 2008; Duron et al. 2018; Husnik 2018). However, R. helvetica is not able to synthetize quite a number of vitamins and cofactors, limiting its possible mutualistic interactions with the host, and rather suggesting a pathogenic behavior.
Hallmarks of Chlamydial Parasitism
As mentioned in a previous publication and like all Chlamydiae sequenced to date, the genome of R. helvetica encodes homologs of the type three secretion system (T3SS) apparatus and chaperones (Pillonel et al. 2018). Homologs of several known T3SS effectors such as NUE, pkn5, and Lda2 were also identified (see supplementary table S8, Supplementary Material online, for detailed list of identified loci). Like its closest sequenced relative S. negevensis, R. helvetica does not encode a homolog of the protease-like activity factor (CPAF), a well-known chlamydial virulence factor. R. helvetica encodes about half as many transporters as S. negevensis (supplementary table S9, Supplementary Material online). It does not have a homolog of the sulfate permease (family 2.A.53) present in most chlamydial genomes nor the sodium-driven transporters found in Chlamydiaceae such as the anion/Na+ symporter SodiTl (2.A.47, CT_204), the amino acid symporters BrnQ (2.A.26, CT_554), and TnaT (CT_231). The loss of such transporters might be linked to the loss of the complex 1 of the respiratory chain involved in Na+ gradient generation. No homolog of the Na+/H+ antiporter NhaE (2.A.111: CT_805) was identified, but R. helvetica has a CPA2 family Na+/H+ antiporter homolog absent from the Chlamydiaceae. These transporters involved Na+ tolerance, pH homeostasis, tolerance to alkali, and fluctuations in osmolarity are infrequent in intracellular symbionts and pathogens (Krulwich et al. 2009). Some oligopeptide and amino acid transporters were identified in R. helvetica; A d-alanine/glycine sodium symporter (GlyP), an alanine sodium symporter (AgcS), two putative proton/glutamate-aspartate symporters (family 2.A.23), and an active oligopeptide transporter OppABCDF operon. The 13 periplasmic solute-binding proteins (OppA) constitute the largest group of paralogs in the R. helvetica genome.
Phosphorylated glucose can be imported through the UhpC transporter, a highly conserved transporter encoded in all sequenced chlamydia genomes (2.A.1.4.6). Like in other Chlamydiae, no phosphoenolpyruvate transport system was found in R. helvetica. Multiple cofactor and vitamin transporters were identified, including putative biotin transporters with low similarity to Vibrio cholerae, and three transporters with low similarity to riboflavin transporters from Ochrobactrum anthropi and V. cholerae (22%–28% identity). An S-adenosylmethionine transporter and a two-partner secretion operon with low similarity to a heme/hemopexin transport system (huxA/huxB) from Haemophilus influenzae were also identified (supplementary table S8, Supplementary Material online).
Seven ATP/ADP antiporter homologs were identified. One homolog (RhT_00329) is most closely related to S. negevensis SnNTT1 and C. trachomatis Npt1 which were shown to transport adenosine triphosphate (Adenosine diphosphate) and ATP/NAD, respectively (Knab et al. 2011; Fisher et al. 2013). Another homolog (RhT_01503) is more closely related to SnNTT2, a guanine nucleotide/ATP/H+ transporter (Knab et al. 2011). Three subsequent paralogs (RhT_00750, RhT_00751, RhT_00752) are phylogenetically most closely related to SnNTT4, whose substrate remains undetermined. The last putative ATP/ADP antiporters (RhT_00462, RhT_00945) are distant from all transporters characterized so far.
In summary, despite the presence of a larger metabolic repertoire than Chlamydiaceae, Rhabdochlamydia is probably highly dependent on its host, thus questioning the mutualistic or parasitic nature of the relationship with its host.
Rhabdochlamydia helvetica Membrane Proteins
The membrane of Chlamydia is thought to be stabilized by extensive disulfide bonds between the major outer membrane protein (MOMP) and two cysteine-rich proteins OmcB and OmcA (Hatch 1996). These three abundantly expressed proteins are the major constituents of the chlamydial outer membrane complex that also contains porins and polymorphic membrane proteins (Pmps) (Birkelund et al. 2009; Liu et al. 2010). The outer membrane protein composition of Chlamydia-related bacteria is very divergent, probably reflecting differences in their ecological niches, but all of them, with the notable exception of S. negevensis, contain OmcB, OmcA, and other cysteine-rich proteins (Collingro et al. 2011; Rusconi et al. 2013; Aistleitner et al. 2015). OmcB and OmcA could not be retrieved in the genome of R. helvetica that also encode few cysteine-rich membrane proteins of the OmpA family compared with W. chondrophila (n = 11) or Estrella lausannensis (n = 10) (Bertelli et al. 2010, 2015). However, the R. helvetica genome encodes one homolog of MOMP (RhT_00042) and five MOMP-like proteins (RhT_00150, RhT_00151, RhT_00153, RhTp_00022, RhT_01103). These latter have a 1.4%–2.7% higher cysteine content than the 37 S. negevensis MOMP and MOMP-like proteins (<1.1% cysteine) (Aistleitner et al. 2015). Like S. negevensis (Vouga et al. 2016), the membrane of R. helvetica might not be stabilized by a network of highly cross-linked proteins hence conserving more flexibility.
Chlamydiaceae bacteria express between 9 and 21 Pmps autotransporters that share little sequence similarity but play important roles in adhesion via conserved multiple repeats of the tetrapeptide motifs GGA(ILV) and FXXN (Mölleken et al. 2010; Becker and Hegemann 2014). The R. helvetica genome contains three Simkania PmpB homologs predicted to form a beta barrel structure in the outer membrane (RhT_00712, RhT_00713, and RhT_01040), but only RhT_01040 contains the GGAI sequence and none displays a FXXN motif. However, R. helvetica possesses one additional protein (RhT_00552) with an autotransporter beta-domain displaying similar structural features as the Pmp-like protein of W. chondrophila (wcw_0271); a signal peptide, a passenger domain, and a beta-barrel C-terminal domain. This protein containing five FXXN repeats and one repeat of the GGA(ILV) motif in the passenger domain could, by similarity with wcw_0271, be implicated in adhesion to the host cells (Kebbi-Beghdadi et al. 2015).
Conclusion
The 1.83 Mb genome of R. helvetica sequenced directly from a pool of ticks provides the first molecular data to investigate the biology of this widespread genus (Lagkouvardos et al. 2014) infecting a wide range of arthropods (Kostanjsek 2004; Corsaro et al. 2007; Pilloux et al. 2015; Vanthournout and Hendrickx 2015; Cooling et al. 2017). Evidence of multiple HGTs between Rhabdochlamydia, arthropods and several lineages of arthropod symbionts suggests that Rhabdochlamydiaceae are widely distributed arthropod parasites and highlights the importance of nonvertical inheritance in the evolution of symbionts and parasites. Although arthropods are frequently engaged in mutualistic associations with bacteria (Moran et al. 2008), the R. helvetica genome only shows reduced metabolic capacities limiting any potential mutualism. In contrast, it encodes several ATP/ADP transporters and multiple transporters of vitamins, cofactors, amino acids, and oligopeptides. Considering that the 2.65 Gb genome of I. scapularis (Cramaro et al. 2017) is over 1,000-fold larger than the R. helvetica genome and that 60% percent of the raw reads obtained by shotgun metagenomics belonged to R. helvetica, the bacterial density was likely extremely high and not without consequences for the host. Altogether, the limited metabolic capacities, high bacterial density and low prevalence suggest that R. helvetica is rather parasitic to its host.
Data Availability
The genome assembly of R. helvetica as well as raw sequencing reads are available under the European Nucleotide Archive project accession PRJEB24578.
Acknowledgments
This project was funded by the budget of the Center for Research on Intracellular Bacteria. The salary of some of the authors (Carole Kebbi-Beghdadi, Marie de Barsy and Manon Vouga) are from diverse Swiss National Science Foundation grants.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Footnotes
Data deposition: Data were submitted to the European Nucleotide Archive under the accession PRJEB24578. SRA Experiments ERX2858304 and ERX2858303. Assembly GCA_901000775.
Literature Cited
- Ahantarig A, Trinachartvanit W, Baimai V, Grubhoffer L.. 2013. Hard ticks and their bacterial endosymbionts (or would be pathogens). Folia Microbiol. 585:419–428. [DOI] [PubMed] [Google Scholar]
- Aistleitner K, et al. 2015. Conserved features and major differences in the outer membrane protein composition of Chlamydiae. Environ Microbiol. 17(4):1397–1413. [DOI] [PubMed] [Google Scholar]
- Alsmark C, et al. 2013. Patterns of prokaryotic lateral gene transfers affecting parasitic microbial eukaryotes. Genome Biol. 142:R19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty, E. 2013. Comparison of sequencing utility programs. Open Bioinforma J 7:1–8. [Google Scholar]
- Becker E, Hegemann JH.. 2014. All subtypes of the Pmp adhesin family are implicated in chlamydial virulence and show species-specific function. MicrobiologyOpen 34:544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, et al. 2015. Sequencing and characterizing the genome of Estrella lausannensis as an undergraduate project: training students and biological insights. Front Microbiol. 6:101.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, et al. 2010. The Waddlia genome: a window into chlamydial biology. PloS One 55:e10890.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, Greub G.. 2012. Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms. Front Cell Infect Microbiol. 2, doi: 10.3389/fcimb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkelund S, et al. 2009. Analysis of proteins in Chlamydia trachomatis L2 outer membrane complex, COMC. FEMS Immunol Med Microbiol. 552:187–195. [DOI] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 121:59–60. [DOI] [PubMed] [Google Scholar]
- Burnard D, et al. 2017. Novel Chlamydiales genotypes identified in ticks from Australian wildlife. Parasit Vectors 101:46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 101:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, et al. 2013. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 142:R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A, et al. 2011. Unity in variety—the pan-genome of the Chlamydiae. Mol Biol Evol. 2812:3253–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooling M, Gruber MAM, Hoffmann BD, Sébastien A, Lester PJ.. 2017. A metatranscriptomic survey of the invasive yellow crazy ant, Anoplolepis gracilipes, identifies several potential viral and bacterial pathogens and mutualists. Insectes Soc. 642:197–207. [Google Scholar]
- Corsaro D, et al. 2007. “Candidatus Rhabdochlamydia crassificans”, an intracellular bacterial pathogen of the cockroach Blatta orientalis (Insecta: Blattodea). Syst Appl Microbiol. 303:221–228. [DOI] [PubMed] [Google Scholar]
- Cramaro WJ, Hunewald OE, Bell-Sakyi L, Muller CP.. 2017. Genome scaffolding and annotation for the pathogen vector Ixodes ricinus by ultra-long single molecule sequencing. Parasit Vectors 101:71.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxatto A, et al. 2014. Presence of Chlamydiales DNA in ticks and fleas suggests that ticks are carriers of Chlamydiae. Ticks Tick-Borne Dis 54:359–365. [DOI] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT.. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 147:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron O. 2013. Lateral transfers of insertion sequences between Wolbachia, Cardinium and Rickettsia bacterial endosymbionts. Heredity 1114:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron O, et al. 2018. Tick-bacteria mutualism depends on B vitamin synthesis pathways. Curr Biol. 2812:1896–1902.e5. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 710:e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DJ, Fernández RE, Maurelli AT.. 2013. Chlamydia trachomatis transports NAD via the Npt1 ATP/ADP translocase. J Bacteriol. 19515:3381–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier PE, Raoult D.. 2005. Genus II. Rickettsiella Philip 1956, 267AL. In:Garrity G, Brenner DJ, Krieg NR, Staley JR. editors. Bergey’s Manual of Systematic Bacteriology. Springer US. Vol. 2, p. 241–247. [Google Scholar]
- Frutos R, Federici BA, Revet B, Bergoin M.. 1994. Taxonomic studies of Rickettsiella, Rickettsia, and Chlamydia using genomic DNA. J Invertebr Pathol. 633:294–300. [DOI] [PubMed] [Google Scholar]
- Frutos R, Pages M, Bellis M, Roizes G, Bergoin M.. 1989. Pulsed-field gel electrophoresis determination of the genome size of obligate intracellular bacteria belonging to the genera Chlamydia, Rickettsiella, and Porochlamydia. J Bacteriol. 1718:4511–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 2823:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI, Koonin EV.. 2014. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43(D1):D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez G, et al. 2011. Insight into cross-talk between intra-amoebal pathogens. BMC Genomics 12:542.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P.. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 83:195–202. [DOI] [PubMed] [Google Scholar]
- Gulia-Nuss M, et al. 2016. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 7:10507.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, et al. 2018. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 46(D1):D851–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch TP. 1996. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in Chlamydiae? J Bacteriol. 1781:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez D, François P, Farinelli L, Østerås M, Schrenzel J.. 2008. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 185:802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokynar K, et al. 2016. Chlamydia-like organisms (CLOs) in Finnish Ixodes ricinus ticks and human skin. Microorganisms 43:28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SD, Mavromatis K, Kyrpides NC.. 2009. Microbial co-habitation and lateral gene transfer: what transposases can tell us. Genome Biol. 104:R45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Gabaldón T.. 2010. ETE: a python environment for tree exploration. BMC Bioinformatics 11:24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F. 2018. Host-symbiont-pathogen interactions in blood-feeding parasites: nutrition, immune cross-talk and gene exchange. Parasitology 14510:1294–1303. [DOI] [PubMed] [Google Scholar]
- Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, et al. 2014. Sequence analysis InterProScan 5: genome-scale protein function classification. Bioinformatics 309:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K.. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 4284:726–731. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 304:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebbi-Beghdadi C, et al. 2015. OmpA family proteins and Pmp-like autotransporter: new adhesins of Waddlia chondrophila. Pathog Dis. doi: 10.1093/femspd/ftv035. [DOI] [PubMed] [Google Scholar]
- Knab S, Mushak TM, Schmitz-Esser S, Horn M, Haferkamp I.. 2011. Nucleotide parasitism by Simkania negevensis (Chlamydiae). J Bacteriol. 1931:225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostanjsek R. 2004. “Candidatus Rhabdochlamydia porcellionis”, an intracellular bacterium from the hepatopancreas of the terrestrial isopod Porcellio scaber (Crustacea: Isopoda). Int J Syst Evol Microbiol. 542:543–549. [DOI] [PubMed] [Google Scholar]
- Kostanjšek R, Pirc Marolt T.. 2015. Pathogenesis, tissue distribution and host response to Rhabdochlamydia porcellionis infection in rough woodlouse Porcellio scaber. J Invertebr Pathol. 125:56–67. [DOI] [PubMed] [Google Scholar]
- Krulwich TA, Hicks DB, Ito M.. 2009. Cation/proton antiporter complements of bacteria: why so large and diverse? Mol Microbiol. 742:257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 199:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, et al. 2014. Integrating metagenomic and amplicon databases to resolve the phylogenetic and ecological diversity of the Chlamydiae. ISME J. 81:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Rujan T, Martin W, Croteau R.. 2000. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci U S A. 9724:13172–13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerque A. 2008. Whole genome-based assessment of the taxonomic position of the arthropod pathogenic bacterium Rickettsiella grylli. FEMS Microbiol Lett. 2831:117–127. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Afrane M, Clemmer DE, Zhong G, Nelson DE.. 2010. Identification of Chlamydia trachomatis outer membrane complex proteins by differential proteomics. J Bacteriol. 19211:2852–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marreiros BC, et al. 2016. Exploring membrane respiratory chains. Biochim Biophys Acta 18578:1039–1067. [DOI] [PubMed] [Google Scholar]
- Mediannikov O, et al. 2014. High quality draft genome sequence and description of Occidentia massiliensis gen. nov., sp. nov., a new member of the family Rickettsiaceae. Stand Genomic Sci. 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende DR, et al. 2017. proGenomes: a resource for consistent functional and taxonomic annotations of prokaryotic genomes. Nucleic Acids Res. 45(D1):D529–D534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel P, Ng KL, Krogh A.. 2016. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 7:11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mölleken K, Schmidt E, Hegemann JH.. 2010. Members of the Pmp protein family of Chlamydia pneumoniae mediate adhesion to human cells via short repetitive peptide motifs. Mol Microbiol. 784:1004–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A.. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 421:165–190. [DOI] [PubMed] [Google Scholar]
- Morel G. 1978. Isolement de deux chlamydiales (rickettsies) chez un arachnide: l’araignée Pisaura mirabilis Cl [Two Chlamydiales (Rickettsias) in an Arachnida: the spider Pisaura mirabilis Cl]. Experientia 343:344–346. [Google Scholar]
- Morel G. 1976. Studies on Porochlamydia buthi g. n., sp. n., an intracellular pathogen of the scorpion Buthus occitanus. J Invertebr Pathol. 282:167–175. [Google Scholar]
- Morel G. 1980. Surface projections of a chlamydia-like parasite of midge larvae. J Bacteriol. 1443:1174–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutailler S, et al. 2016. Co-infection of ticks: the rule rather than the exception. PLoS Negl Trop Dis. 103:e0004539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA.. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 275:824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, et al. 1999. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 271:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, Sixt BS, Horn M, Hackstadt T.. 2014. Chlamydial metabolism revisited: interspecies metabolic variability and developmental stage-specific physiologic activities. FEMS Microbiol Rev. 384:779–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel Van Zee J, et al. 2007. Tick genomics: the Ixodes genome project and beyond. Int J Parasitol. 3712:1297–1305. [DOI] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW.. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 257:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner J, et al. 2016. Acquisition of exogenous haem is essential for tick reproduction. eLife. 5:e12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillonel T, Bertelli C, Greub G.. 2018. Environmental metagenomic assemblies reveal seven new highly divergent chlamydial lineages and hallmarks of a conserved intracellular lifestyle. Front Microbiol. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillonel T, Bertelli C, Salamin N, Greub G.. 2015. Taxogenomics of the order Chlamydiales. Int J Syst Evol Microbiol. 654:1381–1393. [DOI] [PubMed] [Google Scholar]
- Pilloux L, et al. 2015. The high prevalence and diversity of Chlamydiales DNA within Ixodes ricinus ticks suggest a role for ticks as reservoirs and vectors of Chlamydia-related bacteria. Appl Environ Microbiol. 8123:8177–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PloS One 53:e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillery E, Quenez O, Peterlongo P, Plantard O.. 2014. Development of genomic resources for the tick Ixodes ricinus: isolation and characterization of single nucleotide polymorphisms. Mol Ecol Resour. 142:393–400. [DOI] [PubMed] [Google Scholar]
- Radek R. 2000. Light and electron microscopic study of a Rickettsiella species from the cockroach Blatta orientalis. J Invertebr Pathol. 764:249–256. [DOI] [PubMed] [Google Scholar]
- Rasgon JL, Scott TW.. 2004. Phylogenetic characterization of Wolbachia symbionts infecting Cimex lectularius L. and Oeciacus vicarius Horvath (Hemiptera: Cimicidae). J Med Entomol. 416:1175–1178. [DOI] [PubMed] [Google Scholar]
- Rio RVM, Attardo GM, Weiss BL.. 2016. Grandeur alliances: symbiont metabolic integration and obligate arthropod hematophagy. Trends Parasitol. 329:739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. 2011. Integrative genomics viewer. Nat Biotechnol. 291:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi B, et al. 2013. Crescent and star shapes of members of the Chlamydiales order: impact of fixative methods. Antonie Van Leeuwenhoek 104:521–532. [DOI] [PubMed] [Google Scholar]
- Saier MH, et al. 2016. The transporter classification database (TCDB): recent advances. Nucleic Acids Res. 44(D1):D372–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Esser S, et al. 2010. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J Bacteriol. 1924:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069.24642063 [Google Scholar]
- Shah P, Swiatlo E.. 2008. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol. 681:4–16. [DOI] [PubMed] [Google Scholar]
- Sixt BS, Kostanjšek R, Mustedanagic A, Toenshoff ER, Horn M.. 2013. Developmental cycle and host interaction of Rhabdochlamydia porcellionis, an intracellular parasite of terrestrial isopods. Environ Microbiol. 1511:2980–2993. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 309:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nguyen H, Lavenier D.. 2009. PLAST: parallel local alignment search tool for database comparison. BMC Bioinformatics 10:329.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanthournout B, Hendrickx F.. 2015. Endosymbiont dominated bacterial communities in a dwarf spider. PloS One 102:e0117297.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouga M, Baud D, Greub G.. 2016. Simkania negevensis, an insight into the biology and clinical importance of a novel member of the Chlamydiales order. Crit Rev Microbiol. doi: 10.3109/1040841X.2016.1165650. [DOI] [PubMed] [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE.. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 185:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, et al. 2013. The MaSuRCA genome assembler. Bioinformatics 29:2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome assembly of R. helvetica as well as raw sequencing reads are available under the European Nucleotide Archive project accession PRJEB24578.