Abstract
Background:
Acinetobacter baumannii (A. baumannii) is a bothersome fatal pathogen, particularly in healthcare system. Persistence and successful invasion of A. baumannii in vertebrate host cells largely depends on iron acquisition methods. Siderophore molecules and Iron-Regulated Outer Membrane Proteins (IROMPs) are the two essential members of iron acquisition system. Siderophores are secreted by bacteria to bind peripheral ferric iron and the IROMPs are expressed at the bacterial outer membrane as the receptor of ferric-siderophore complex. BauA is the corresponding siderophore receptor of A. baumannii. In this study, an attempt was made to assess the immunogenicity of antigenic domains of BauA which could be effective in iron uptake restriction and protection against bacterial invasion of the host cells.
Methods:
The antigenic domains of bauA were amplified from A. baumannii ATCC-19606. The PCR products were ligated into pET32a and expressed in Escherichia coli (E. coli) BL21 (DE3). Purification of recombinant domains was done by Nickel-Nitri-lotriacetic Acid (Ni-NTA) affinity chromatography. The recombinant domains were injected into BALB/C mice separately and in combination. Sero-reactivities of the recombinant proteins and mouse challenge tests were carried out.
Results:
The antibodies raised in mice could successfully recognize and bind antigenic domains. Passive immunization studies accomplished by immune rabbit serum inhibited the establishment of infection in mice.
Conclusion:
The results adapted from the present study disclose the protective role of functional domains of BauA, especially the cork domain, suggesting a novel recombinant immunogen candidate.
Keywords: Acinetobackr baumannii, Immune sera, Iron, Siderophore
Introduction
Acinetobacter baumannii (A. baumannii) is a strictly aerobic, gram-negative, non-motile, oxidase negative coccobacillus 1. This opportunistic pathogen is widely notorious for its hospital-acquired outbreaks which constitute a growing public-health dilemma 2,3. The major clinical syndromes are pneumonia and bacteremia, together with infections in skin and soft tissue, urinary tract and surgical sites 4. A. baumannii can also be the major cause of meningitis, especially in patients with ventricular draining tubes 5. Several outbreaks due to A. baumanii have been reported 6–8.
Furthermore, MultiDrug-Resistant (MDR) A. baumannii has an extraordinary capacity to develop resistance against even the most potent antimicrobialcompounds, including carbapenems; hence, main infections have become incurable within recent years 9. The common carbapenem resistance mechanisms in A. baumannii include acquisition of carbapenemases, β-lactam-ases which are capable of hydrolyzing carbapenems, reduced affinity of penicillin binding proteins and low permeability of outer membrane proteins 1,10,11.
Iron is a fundamental nutrient required for the bacterial growth and virulence in the host cells 12. However, as a defense mechanism, most of the iron is combined with host iron storage proteins, such as transferrin, hemoglobin and hemosiderin and is therefore unattainable for bacteria 13. To be able to establish an infection, iron-starved pathogenic bacteria must synthesize, excrete and retrieve iron-scavenging low molecular weight proteins with high affinity to Fe (III), siderophores and hereby compete with and obtain iron from the host's iron binding proteins 14. In 1994, acinetobactin, the specific siderophore of A. baumannii, was isolated from the iron deficient cultures of A. baumannii ATCC19606 and its chemical structure was suggested by way of chemical degradation, FAB-MS spectrometry, and 1 H- and 13 CNMR spectroscopy methods 15,16. During a non-enzymatic isomerization in the extracellular space, pre-acinetobactin changes to acinetobactin. While the pH range <6 is optimum for the persistence of pre-acinetobactin, acinetobactin formation needs pH >7. Such an isomerization affords two siderophores for the price of one, allowing A. baumannii to absorb iron over a wide pH range that is probable during the infection period 17.
In order to set up infections, expression of the acinetobactin-mediated iron acquisition system is crucial for A. baumannii 18,19. Another group of this system in A. baumannii are Iron-Regulated Outer Membrane Proteins (IROMPs), which are expressed at the bacterial surface 20. The most important IROMP in A. baumannii is baumannii acinetobactin utilization A protein (Bau-A), which is pivotal for uptake of acinetobactin in complex with iron 12,21.
Disruption of BauA function has been shown to exert a bacteriostatic effect under iron-restricted milieu 19,22. BauA is a monomeric protein, belonging to TonB-dependent transporter protein family. The whole protein is composed of 2 domains; a cork domain at N terminal of the protein, and a trans-membrane barrel domain at the C terminal. The 22 transmembrane β-strands form barrel domain and 11 extracellular loops connect neighbor strands at the external side of the membrane. Also, 10 periplasmic turns exist in periplasmic part of the membrane 20. Cork domain comprises 168 residues from the N terminal of BauA and acts as a plug within the barrel, occluding the opening of β-barrel. Four to five anti parallel hydrophobic β-strands make up this domain 23. Ligand binding sites containing conserved residues are determined in the cork domain, suggesting a substantial role in iron entrance. Therefore, blocking of cork could have a bacteriostatic effect. Moreover, the surface-located loops which are highly exposed to the environment suggest their role in initial binding events with Fe-siderophore complex. As the extracellular part of IROMPs, loop regions are attractive targets for antigen-antibody interaction studies 24.
In the current study, the immunogenic effect of cork and loop domains were investigated separately and also in a cork-loop mixed form.
Materials and Methods
Bacterial strains, plasmids and media
The bacterial strains involved in this study were A. baumannii (ATCC19606), Pseudomonas aeruginosa (P. aeruginosa) (ATCC27853), Staphylococcus aureus (S. aureus) (ATCC25923), Escherichia coli (E. coli) (ATCC25922) and E. coli strain BL21 (DE3). The pET32a plasmid was made by Novagen (USA). Luria-Bertani (LB) broth and LB agar used to cultivate the bacterial strains were Merck (Germany) products.
Cloning of selected domains
The forward and reverse primers for the cork (5′-GT TATGAATTCATGGATAATTCAAAAAAACTCTAGA AC-3′)/(5′-GTGCAAGCTTAAACGGTTCATCAGC-3′) and loop (5′-ATCAGGAATTCATGTTGATGGCATAT GCGG-3′)/(5′-CTTGGTCGACAGTATCCCACTCTGC TCCAA-3′) domains were designed, respectively. A downstream His-tag sequence was used on the vector to deactivate the stop codon in both genes. Amplification of coding regions corresponding to chosen antigenic domains of BauA was done using A. baumannii genomic DNA. The PCR process for the two genes was carried out in a mixture composed of 50 ng/ml of DNA, 4 pM of forward and reverse primers, 0.4 mM of dNTP mix, 3 mM of MgCl2, 1XPCR buffer and 4 U Taq DNA polymerase (Cinnagen Tehran, Iran) in 20 μl on a thermal cycler (Techne Gradient Staffordshire, UK). The procedure proceeded in the following path: initial denaturation at 94°C for 5 min and 35 cycles of 1 min at 94°C, 1 min at 60°C, 1.5 min at 72°C, and a final 5 min at 72°C.
After electrophoresis in 1% w/v agarose gel, PCR purification kits (Bioneer Daejeon, Korea) were applied to purify PCR products. EcoRI (forward) and HindIII (reverse) endonucleases (Fermentas Vilnius, Lithuania) undertook digestion of the cork fragment and for the loop fragment EcoRI (forward) and SalI (reverse) were responsible. The pET32a vector was digested with the same enzymes of the corresponding genes. Then, the sticky ended vector and PCR products were purified using gel extraction kit. In either case, in total 40 ng of vector and 25 ng insert were mixed with 2.5 U of T4 DNA ligase (Fermentas Vilnius, Lithuania) and T4 ligation buffer in a 25 μl reaction mixture and incubated at 12°C for 16 hr. The recombinant DNA was transformed into the expression host, E. coli BL21 (DE3) component cell. Then, the recombinant clones were selected on LB plates containing ampicillin (50 μg/ml). The plasmid-insert constructs were isolated via plasmid extraction kits and the existence of the recombinant genes was validated by clone PCR and sequencing.
Expression and purification of selected domains
For both domains, the process continued as follows: bacteria were grown in LB broth containing 50 μm/ml of ampicillin and the cultures were aerated on a shaker at 37°C to mid-log phase (OD600 of 0.6).
To maximize protein expression, 1 mM IPTG was added and further cultures were incubated for 4 hr. The cells were then harvested by centrifugation at 5000×g for 10 min at 4°C. The cell pellets were re-suspended in lysis buffer (100 m MNaH2PO4, 10 mM Tris.HCl, 8 M urea), followed by the addition of lysozyme at a 1 mg/ml concentration, and were sonicated 5 times for 1 min at 30 s intervals. After centrifugation at 12000 rpm at 4°C for 20 min, the pellet was collected for further investigation on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
The proteins were purified in batch under denaturing conditions with Ni-NTA agarose (Qiagen) using buffers of different pH, all containing 8 M urea. Recombinant proteins were eluted using buffer E (pH=4.5). The denaturant (8 M urea) was removed by step wise dialysis in a Spectra/Por 2 dialysis bag (Spectrum). Finally, the proteins were dialyzed twice against phosphate-buffered saline (PBS).
The protein fractions were concentrated through freeze-drying. Densely purified proteins were estimated by the Bradford method using Bovine Serum Albumin (BSA) concentrations as standard 25.
Western blot analysis
In order to evaluate the specificity of the recombinant domains, analysis of western blot was carried out with an Anti-His-Tag antibody. Protein samples were first electrophoresed on a 12% SDS-PAGE gel, then electro blotted into a nitro cellulose membrane at a constant current of 300 mA at 4oC for 1.5 hr. The membrane was blocked with 3% BSA with gentle shaking for 30 min at room temperature. The membrane was then washed 3 times with PBS-T (PBS + 0.05% Tween-20, pH=7.4) and mouse anti-His-tag mo-noclonal antibody (Abcam, USA) was added at 1:1,000 dilution for 1 hr at 37°C. Subsequently, the membranes were incubated with goat anti-mouse IgG Horseradish Peroxidase (HRP) (RAY, Biotech) at a dilution of 1:5000 for 1 hr. Finally, HRP staining solution (containing DAB and H2O2) led to detection of recombinant protein.
Animal immunization
Female BALB/c mice of 4–6 weeks old (20–25 g) and male New Zealand White rabbits were purchased from the Razi Institute (Tehran, Iran). Animals were kept under standard conditions in the animal care facility of Shahed University. The animal care rule was ethically verified by Shahed University. For each case (cork, loop and cork-loop cocktail), mice were divided into 4 groups (5 mice per group). As new vaccination strategy comprising the simultaneous co-administration by the nasal and parenteral routes of a multicomponent vaccine formulation in BALB/C and HBsAg-transgenic mice was reported to increase the immune response 26, the same strategy and the strategies recommended by others 27 were used to immunize our test animals by both intraperitoneal and subcutaneous routes of administration. Groups were immunized on days 0, 15 and 30 via intraperitoneal and subcutaneous routes at the same time to reach the highest level of immunity. In order to immunize, 20 μg of purified protein were mixed with the same volume of adjuvant and injected to the mice. While the first dose was accompanied with complete Freund’s adjuvant, subsequent doses contained incomplete adjuvant. The other 5 mice constituted the negative control group and were injected with PBS. The experimented rabbits were immunized in a similar process with 100 μg of recombinant proteins and the same amount of adjuvant. Blood samples were collected 7 days after the third injection.
Sero reactivity tests
Antibody titration was determined with indirect Enzyme-Linked Immunosorbent Assay (ELISA). Microtiter plates (Nunc) were coated with 5 μg of antigen (per well) together with 100 μl of coating buffer (bicarbonate/carbonate 100 mM, pH=9.6), overnight at 4°C. The wells were washed three times with PBS-T (PBS buffer 1X+0.05% Tween 20). Serially diluted sera (from 1:250 to 1:100000) were added to the wells and each plate was incubated for 1 hr at 37°C. Wells were rewashed with PBS-T. Primary antibodies were detected by adding 100 μl of HRP-conjugated anti-mouse or anti-rabbit antibodies to the wells for 30 min at 37°C. Finally, TMB, the chromogenic substrate, was added and the reaction was stopped with H2SO4.
Whole-cell ELISA
In order to confirm the specificity, anti-cork immune serum was selected and blotted against whole-cell extracts of A. baumannii (ATCC19606), P. aeruginosa) ATCC27853), E. coli (ATCC25922), and S. aureus (ATCC25923). The bacteria were grown under iron-limiting conditions, settled down at the bottom of sterile tubes and washed two times with sterile PBS. Bacteria were then diluted using 100 μl of coating buffer (bicarbonate/carbonate 100 mM, pH=9.6) and dried onto ELISA plates at 4°C overnight. The wells were supplemented with 100 μl of blocking buffer and incubated at 37°C for 1 hr and then rinsed with PBST. Serial dilutions of immune and un-immune mice-raised sera ranging from 1/100 to 1/1000 were prepared and added to the wells (100 μl per well) and the process was followed by 4 hr incubation at 37oC. Binding of mouse sera was detected by using 100 μl of anti-mouse immunoglobulin G (IgG)-HRP conjugate (RAY, Biotech) in each well, then the plates were incubated at 37°C for 1 hr. After washing, 100 μl of citrate buffer containing 0.06% (W/V) of O-phenylenediamine dihydrochloride (OPD) (SIGMA) and 0.06% (V/V) hydrogen peroxide were added to each well and incubated at room temperature for 15 min. The reaction was stopped with 100 μl of 2MH2SO4 and the OD492 was read on a microplate reader (Bio-Rad).
Determination of LD50 and challenge test
To calculate the 50% lethal Dose (LD50) in mice, A. baumannii dilutions in a range between l04 to l09 CFUs were prepared. Dilutions were intraperitoneally injected into six groups of BALB/c mice, 5 animals per group. For animal challenge test, immunized mice were dispensed into three random groups of five mice. Intraperitoneal injection of 10 8, 109 and 1010 CFU of A. baumannii to groups put them into challenge. LD50 was defined as the bacteria that are sufficient to kill 50 percent of a population of animals within 48 hr.
Passive immunization
Four groups of healthy mice (5 mice per group) were selected. The first two groups were injected with 200 μl of immune rabbit serum at serial dilutions ranging from 1/250 to 1/10000 mixed with A. baumannii at 109 and 1010CFU concentrations, following incubation at 37°C for 30 min. A 105 CFU of S. aureus with LD50 of 104 was used to monitor the specificity of immune response against recombinant protein domains. S. aureus was mixed with 200 μl of pure immune rabbit serum and injected into mice in group 3. Group 4 was injected with only 200 μl of rabbit immune serum as control.
Investigation of A. baumannii establishment
At the same time with the death of control mice, injected mice were dissected and their liver and spleen were sterilely removed, homogenized and analyzed for the presence of A. baumannii. 1 gr of each homogenized organ was suspended in 10 ml of PBS. After that, 100 μl of each suspension was cultured on LB agar and incubated at 37oC. After 16 hr, bacterial colonies were counted and further tested for the identification of A. baumannii. Liver and spleen from mice injected only with rabbit immune serum were used as control.
Statistical analysis
Data were reported as mean±SD and statistical analysis was performed using One-Way ANOVA (SPSS 16.0). The significance (p<0.01) of differences was assessed by post hoc comparison of means using lowest significant differences (Dunkan).
Results
Gene cloning, protein expression and purification
The 948 bp and 501 bp DNA fragments encoding loop and cork domains (respectively) of BauA protein were amplified successfully (Figure 1) and cloned into pET32a expression vector. Over-expression in BL21 (DE3) led to the formation of corresponding 50 kDa and 35 kDa protein bands in SDS-PAGE gel (Figure 2). After successful purification (Figure 3) and dialysis steps, the identity of the proteins was verified via western blotting using anti-6X His tag antibodies (Figure 4).
Figure 1.
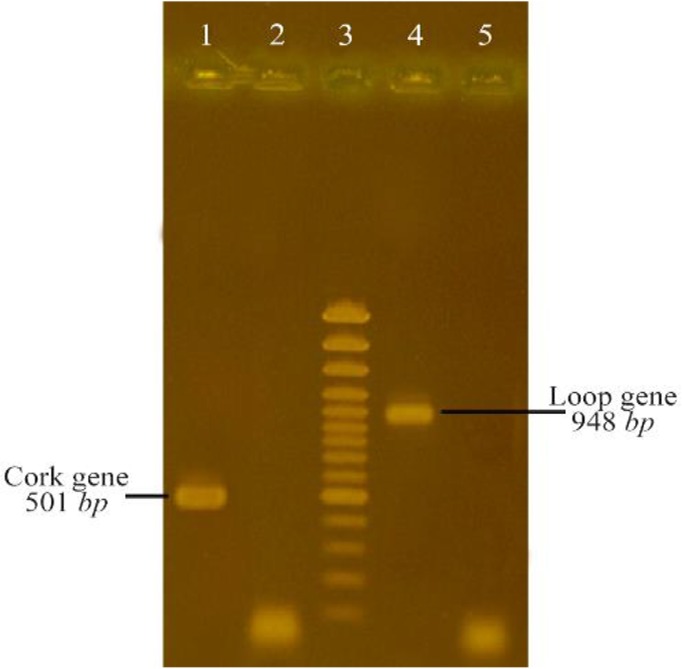
PCR amplifications of cork and loop DNA segment from BauA Lane 1: PCR product of cork, Lane 2: negative control (a PCR product without template), Lane 3:100 bp DNA plus ladder, Lane 4: PCR product of loop, Lane 5: negative control.
Figure 2.
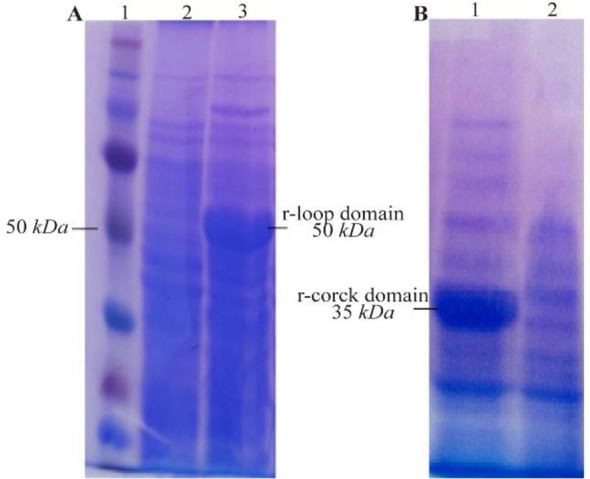
SD-PAGE analysis of the recombinant, A) loop and, B) cork domain expression. A) Lane 1: standard protein molecular weight marker, Lane 2: un-induced control, Lane 3: expression of recombinant loop domain induced with 1 mM IPTG, B) Lane 1: un-induced control, Lane 2: expression of recombinant cork domain.
Figure 3.
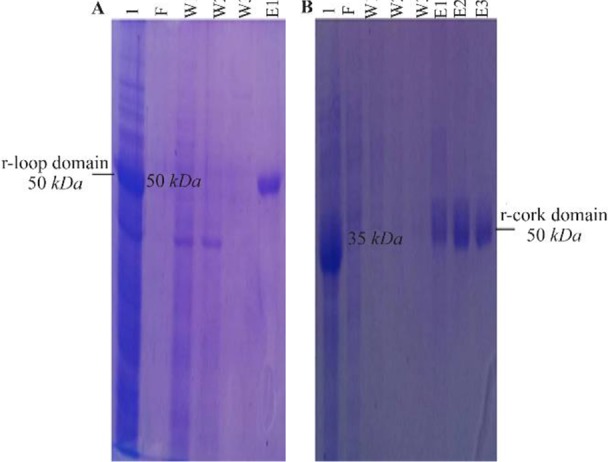
SDS-PAGE analysis of purification of recombinant (a) loop and (b) cork domain. (a) Lane 1: expression of recombinant loop domain, Lane F: unbound protein flow, Lanes W1–W3: column washed with buffer, Lane E1: column washed with elution buffer. (b) Lane 1: expression of recombinant cork domain, Lane F: unbound protein flow, Lanes W1–W3: column washed with buffer, Lanes E1–E3: column washed with elution buffer.
Figure 4.

Western blot analysis of the recombinant loop and cork domains from BauA. Lane 1: molecular weight marker, Lane 2: purified loop domain, Lane 3: purified cork domain
Immune response
ELISAs demonstrated that vaccination against selected cork and loop domains of BauA protein elicited significant levels of antibodies, whereas control mice had no detectable antigen-specific antibody (p<0.001; Student’s t test). The highest immune response in BALB/c mice was achieved after the third booster injection. The highest level of IgG between different injections was detected in case of cork domain. Mice and rabbit antibody titers against selected domains are presented in figure 5. Mice antibody titers against cork domain during three injections are shown in figure 6.
Figure 5.
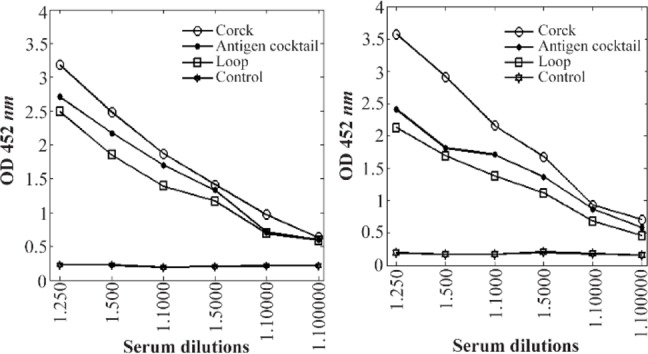
Mice (left) and rabbit (right) antibody titers against cork and loop domains of BauA.
Figure 6.
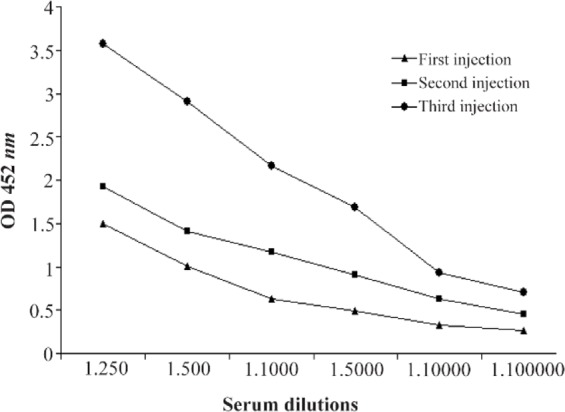
Mice antibody titers against cork domain during three injections
Whole-cell ELISA
To investigate the specificity of the proteins, five different strains were grown under iron limitation in batch cultures and tested in whole-cell ELISAs, using anti-cork antibodies raised against A. baumannii 196-06. The antibodies reacted strongly with strains 19606 and also an acceptable reaction was seen with the clinical isolate. Some very weak reaction was observed with strains E. coli, P. aeruginosa and S. aureus (Figure 7). A similar reaction was also seen with these bacteria grown with excess iron.
Figure 7.
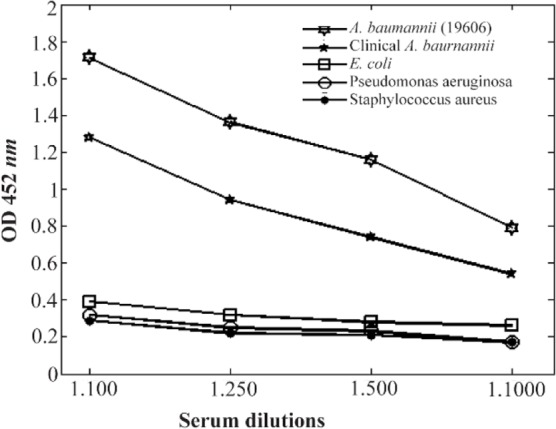
Antibody titers against whole-cell extracts of some gram −/+ strains. Mice anti-cork serum was used as antiserum against whole cells.
LD50 determination and animal challenge test
LD50 was determined as 108 CFU/ml at intraperitoneal injection and mice mortality was recorded for two consecutive post-challenge days. In challenge studies, the immunized mice survived in all groups while the non-immune control groups died within 24–48 hr.
Tracing bacterial infection
No bacterial colony was detected in immunized mice spleen and liver samples; however, samples from common control group revealed 16×104/g and 23.5× 104/g bacteria in spleen and liver cultures, respectively.
Passive immunization
Administration of 109 CFU from fresh live A. baumannii mixed with pure immune rabbit sera led to the survival of mice groups, while all higher sera dilutions did not impart protection.
Discussion
It has been shown that denatured forms of outer membrane proteins of gram-negative bacteria are able to stimulate mice immune system 28,29. IROMPs, which are expressed under iron-deficient conditions, have long been considered as desirable targets for vaccine development 30.
BauA is the most important A. baumannii IROMP, containing conserved regions in functional domains at the cell surface 31. BauA is involved in iron uptake of A. baumannii to survive in harsh environments and during host infection. Hence, functional blockade of the protein could have a cidal effect on the pathogen 18.
Here, the immunogenic capability of the recombinant cork and loop domains of BauA from A. baumannii 19606 was investigated in vivo. Although the selected domains were cloned, expressed and purified under denaturing conditions 32,33, after a dialysis step, renatured forms of proteins were injected to the animals. Immunogenicity of the recombinant domains was inspected by injecting BALB/c mice and rabbits. The antibodies produced against injectants could successfully recognize and bind them. Immunized animals challenged with bacterial doses higher than A. baumannii LD50 (108) survived. BauA is highly conserved in amino acid sequence and 3D structure of ligand binding sites of its functional outer membrane regions 20. Most of the functional conserved amino acids are located in plug domain and outer membrane loops. The functional sites on the surface of protein structure include cork domain and a part of barrel covering L5, L6, L7, L8 and L9. Therefore, it could be inferred that these regions are the most important functional sites of the protein. The important issue is accessibility of the antibodies to cork domain which in normal conditions is located within periplasm. The immunity generated thereby against this region could be explained by hypothesizing random or occasional exposure of this domain. This exposure facilitates their accessibility to antibodies produced against them.
In the present study, ELISAs showed the highest level of antibodies raised regarding cork domain. Our earlier bioinformatics study also introduces the cork domain at the beginning of protein sequence as the most conserved unit, with a high level of surface accessibility 23. Sequence-based and structure-based B-cell epitope prediction tools show that the cork domain holds the majority as well as the best B cell epitopes in BauA structure 23. Despite antigenic index plots which demonstrated that regions containing loops have a high antigenic propensity among BauA 23, in the current study, this domain held the lowest level of antigenic capacity in vivo. It could be due to the specific in vivo interactions between several loop structures in this domain. The cocktail of cork-loop domains showed an average level of protection among three injectants. It could be deduced that the presence of loop domains has diminished the antigenicity of cocktail during a negative interference. Moreover, acidic nature of BauA as well as its localization is an attractive factor for B-cell responses 34. B-cell epitope prediction survey has suggested that the best B cell epitopes in BauA are located in amino acids 26–191 of cork domain and amino acids 321–635 of part of the barrel domain including L4–L9. Therefore, these regions are selected as suitable vaccine candidates 23.
Passive immunization with rabbit anti-BauA antiserum also protected mice from the following infection. Post-injection bactericidal assay revealed 16×104/g and 23.5×104/g bacteria in spleen and liver cultures of un-immune group, respectively. The number of bacterial colonies was not significant in immune groups, showing the role of humoral immunity raised against A. baumannii infection. In accordance with our studies, Larrie et al showed that passive immunization with serum raised against an E. coli O157:H7 IROMP protects rabbits from subsequent infection 30.
Previously, the immunogenicity of whole BauA protein 33 was investigated and also a recombinant type was constructed by the coding regions from cork domain to loop 9 of bauA gene which revealed significant protection levels after the third injection dose 35.
BauA is highly prevalent in pathogenic strains of A. baumannii 18; hence, BauA based developed vaccines could be effective against all pathogenic strains of the bacterium. Immune serums were also used in whole-cell ELISAs using the five strains described earlier. This antiserum did not react significantly with any strains except strain 19606 and its clinical isolate, showing a specific bind of anti-BauA antiserum.
Conclusion
In conclusion, the results adapted from the present study disclose the protective role of functional domains of BauA, especially the cork domain, suggesting a novel recombinant vaccine candidate.
Acknowledgement
The authors wish to thank Shahed University for supporting this work.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1. Vergidis P, Falagas ME. Control of Multi-Drug Resistant Acinetobacter Infections. In: Antibiotic Policies, Edition Springer, 2012, p. 117–125. [Google Scholar]
- 2. Camp C, Tatum OL. A review of Acinetobacter baumanniias a highly successful pathogen in times of war. Lab Med 2015;41(11):649–657. [Google Scholar]
- 3. García-Quintanilla M, Pulido MR, López-Rojas R, Pachón R, McConnell MJ. Emerging therapies for multi-drug resistant Acinetobacter baumannii. Trend Microbiol 2013;21(3):157–163. [DOI] [PubMed] [Google Scholar]
- 4. Dexter C, Murray GL, Paulsen IT, Peleg AY. Community-acquired Acinetobacter baumannii: clinical characteristics, epidemiology and pathogenesis. Expert Rev Anti Infect Ther 2015;13(5):567–573. [DOI] [PubMed] [Google Scholar]
- 5. De Bonis P, Lofrese G, Scoppettuolo G, Spanu T, Cultrera R, Labonia M, et al. Intraventricular versus intravenous colistin for the treatment of extensively drug resistant Acinetobacter baumannii meningitis. Eur J Neurol 2016; 23(1):68–75. [DOI] [PubMed] [Google Scholar]
- 6. Hsu LY, Apisarnthanarak A, Khan E, Suwantarat N, Ghafur A, Tambyah PA. Carbapenem-resistant Acineto-bacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin Microbiol Rev 2017;30(1):1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mosqueda N, Espinal P, Cosgaya C, Viota S, Plasensia V, Alvarez-Lerma F, et al. Globally expanding carbapenemase finally appears in Spain: nosocomial outbreak of Acinetobacter baumannii producing plasmid-encoded OXA-23 in Barcelona, Spain. Antimicrob Agents Chemother 2013;57(10):5155–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Romanelli RM, Jesus LA, Clemente WT, Lima SS, Rezende EM, Coutinho RL, et al. Outbreak of resistant Acinetobacter baumannii: measures and proposal for prevention and control. Braz J Infect Dis 2009;13(5):341–347. [DOI] [PubMed] [Google Scholar]
- 9. Farshadzadeh Z, Hashemi FB, Rahimi S, Pourakbari B, Esmaeili D, Haghighi MA, et al. Wide distribution of carbapenem resistant Acinetobacter baumannii in burns patients in Iran. Front Microbiol 2015;6:1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fernández-Cuenca F, Martínez-Martínez L, Conejo MC, Ayala JA, Perea EJ, Pascual A. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of car-bapenems against clinical isolates Acinetobacter baumannii. J Antimicrob Chemother 2003;51(3):565–574. [DOI] [PubMed] [Google Scholar]
- 11. Zarrilli R, Pournaras S, Giannouli, Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 2013;41(1):11–19. [DOI] [PubMed] [Google Scholar]
- 12. Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA. Role of acinetobactn-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun 2012;80(3):1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abu Kwaik Y, Bumann D. Microbial quest for food in vivo:‘nutritional virulence’as an emerging paradigm. Cell Microbiol 2013;15(6):882–890. [DOI] [PubMed] [Google Scholar]
- 14. Olive AJ, Sassetti CM. Metabolic crosstalk between host and pathogen: sensing, adapting and competing. Nature Rev Microbiol 2016;14(4):221–234. [DOI] [PubMed] [Google Scholar]
- 15. Takahashi A, Yomoda S, Kobayashi I, Okubo T, Tsunoda M, Iyobe S. Detection of carbapenemase-producing acinetobacter baumannii in a hospital. J Clin Microbiol 2000;38(2):526–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wuest WM, Sattely ES, Walsh CT. Three siderophores from one bacterial enzymatic assembly line. J Am Chem Soc 2009;131(14):5056–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shapiro JA, Wencewicz TA. Acinetobactin isomerization enables adaptive iron acquisition in Acinetobacter baumannii through pH-triggered siderophore swapping. ACS Infect Dis 2015;2(2):157–168. [DOI] [PubMed] [Google Scholar]
- 18. Zimbler DL, Penwell WF, Gaddy JA, Menke SM, Tomaras AP, Connerly PL. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals 2009;22(1):23–32. [DOI] [PubMed] [Google Scholar]
- 19. Song WY, Jeong D, Kim J, Lee MW, Oh MH, Kim HJ. Key structural elements for cellular uptake of Acineto-bactin, a major siderophore of Acinetobacter baumannii. Org Lett 2017;19(3):500–503. [DOI] [PubMed] [Google Scholar]
- 20. Sefid F, Rasooli I, Jahangiri A. In silico determination and validation of baumannii acinetobactin utilization a structure and ligand binding site. Biomed Res Int 2013; 2013:17284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nwugo CC, Gaddy JA, Zimbler DL, Actis LA. Deciphering the iron response in Acinetobacter baumannii: a proteomics approach. J Proteomics 2011;74(1):44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goel VK, Kapil A. Monoclonal antibodies against the iron regulated outer membrane proteins of Acinetobacter baumannii are bactericidal. BMC Microbiol 2001;1:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sefid F, Rasooli I, Jahangiri A, Bazmara H. Functional exposed amino acids of BauA as potential immunogen against Acinetobacter baumannii. Acta Biotheor 2015;63 (2):129–149. [DOI] [PubMed] [Google Scholar]
- 24. Krewulak KD, Vogel HJ. Structural biology of bacterial iron uptake. Biochim Biophys Acta 2008;1778(9):1781–1804. [DOI] [PubMed] [Google Scholar]
- 25. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–254. [DOI] [PubMed] [Google Scholar]
- 26. Lobaina Y, Trujillo H, García D, Gambe A, Chacon Y, Blanco A, et al. The effect of the parenteral route of administration on the immune response to simultaneous nasal and parenteral immunizations using a new HBV therapeutic vaccine candidate. Viral Immunol 2010;23 (5):521–529. [DOI] [PubMed] [Google Scholar]
- 27. William LA, Larry KP, Benjamin S, Bruce GW, John KI, John CW. General recommendations on immunization. Morbidity and Mortality Weekly Report (MMWR) 2002; 51(RR02):1–36. [Google Scholar]
- 28. Kumar VS, Gautam V, Balakrishna K, Kumar S. Overexpression, purification, and immunogenicity of recombinant porin proteins of Salmonella enterica Serovar Typhi (S. Typhi). J Microbiol Biotechnol 2009;19(9): 1034–1040. [DOI] [PubMed] [Google Scholar]
- 29. Malickbasha M, Arunachalam R, Senthilkumar B, Raja-sekarapandian M, Annadurai G. Effect of ompR gene mutation in expression of ompC and ompF of Salmonella typhi. Interdiscip Sci 2010;2(2):157–162. [DOI] [PubMed] [Google Scholar]
- 30. Baghal SM, Gargari SL, Rasooli I. Production and immunogenicity of recombinant ferric enterobactin protein (FepA). Int J Infect Dis 2010;14 Suppl 3: e166–170. [DOI] [PubMed] [Google Scholar]
- 31. Esmaeilkhani H, Rasooli I, Nazarian S, Sefid F. In vivo validation of the immunogenicity of recombinant baumannii acinetobactin utilization a protein (rBauA). Microb Pathog 2016;98:77–81. [DOI] [PubMed] [Google Scholar]
- 32. Fattahian Y, Rasooli I, Mousavi Gargari SL, Rahbar MR, Darvish Alipour Astaneh S, Amani J. Protection against Acinetobacter baumannii infection via its functional deprivation of biofilm associated protein (Bap). Microb Pathog 2011;51(6):402–406. [DOI] [PubMed] [Google Scholar]
- 33. Toobak H, Rasooli I, Talei D, Jahangiri A, Owlia P, Darvish Alipour Astaneh S. Immune response variations to Salmonella enterica serovar Typhi recombinant porin proteins in mice. Biologicals 2013;41(4):224–230. [DOI] [PubMed] [Google Scholar]
- 34. Rahbar MR, Rasooli I, Mousavi Gargari SL, Amani J, Fattahian Y. In silico analysis of antibody triggering bio-film associated protein in Acinetobacter baumannii. J Theor Biol 2010;266(2):275–290. [DOI] [PubMed] [Google Scholar]
- 35. Sangroodi YH, Rasooli I, Nazarian S, Ebrahimizadeh W, Sefid F. Immunogenicity of conserved cork and ß-barrel domains of baumannii acinetobactin utilization protein in an animal model. Turk J Med Sci 2015;45(6):1396–1402. [DOI] [PubMed] [Google Scholar]


