Abstract
Background & Aims:
Transforming growth factor-β (TGFB) has key functions in fibrogenic cells, promoting fibrosis development in the liver and other organs. In contrast, the functions of TGFB in liver epithelial cells are not well understood, despite their high level of responsiveness to TGFB. We sought to determine the contribution of epithelial TGFB signaling to hepatic fibrogenesis and carcinogenesis.
Methods:
TGFB signaling in liver epithelial cells was inhibited by albumin-Cre-, K19-CreERT-, Prom1-CreERT2-, or AAV8-TBG-Cre-mediated deletion of the floxed TGFB receptor II gene (Tgfbr2). Liver fibrosis was induced by carbon tetrachloride, bile duct ligation, or disruption of the multidrug-resistance transporter 2 gene (Mdr2). Hepatocarcinogenesis was induced by diethylnitrosamine or hepatic deletion of PTEN.
Results:
Deletion of Tgfbr2 from liver epithelial cells did not alter liver injury, toxin-induced or biliary fibrosis, or diethylnitrosamine-induced hepatocarcinogenesis. In contrast, epithelial deletion of Tgfbr2 promoted tumorigenesis and reduced survival of mice with concomitant hepatic deletion of Pten, accompanied by an increase in tumor number and a shift from hepatocellular carcinoma to cholangiocarcinoma. Surprisingly, both hepatocyte-and cholangiocyte-specific deletion of Pten and Tgfbr2 promoted the development of cholangiocarcinoma, but with different latencies. The prolonged latency and the presence of hepatocyte-derived cholangiocytes after AAV8-TBG-Cre-mediated deletion of Tgfbr2 and Pten indicated that cholangiocarcinoma might arise from hepatocyte-derived cholangiocytes in this model. Pten deletion resulted in upregulation of Tgfbr2, and deletion of Tgfbr2 increased cholangiocyte but not hepatocyte proliferation, indicating that the main function of epithelial TGFBR2 is to restrict cholangiocyte proliferation.
Conclusion:
Epithelial TGFB signaling does not contribute to the development of liver fibrosis or formation of hepatocellular carcinomas in mice, but restricts cholangiocyte proliferation to prevent cholangiocarcinoma development, regardless of its cellular origin.
Keywords: TGF-beta, signal transduction, mouse model, liver cancer
INTRODUCTION
Transforming growth factor beta (TGFβ) is a key regulator of cell proliferation, differentiation, migration, survival, wound healing, angiogenesis, and immunosurveillance.1–3 Virtually every cell in the body, including epithelial and mesenchymal cells, produces TGFβ and/or has TGFβ receptors. In the liver, TGFβ levels increase in chronic disease states, and TGFβ signaling has been linked to the development of liver fibrosis in mice and patients.4–6 TGFβ is a well-characterized profibrogenic cytokine that promotes the activation of hepatic stellate cells (HSCs)5,7,8, the main profibrogenic cell type in the fibrotic liver.9
Although epithelial cells are highly responsive to TGFβ, the role of epithelial TGFβ signaling in chronic liver disease is not fully understood. In contrast to the well-known function of TGFβ signaling in HSCs, it is not clear how epithelial TGFβ signaling affects liver fibrosis development. In one study, hepatocyte-specific overexpression of Smad7 decreased liver fibrosis by preventing epithelial-mesenchymal transition (EMT)-like changes in hepatocytes.10 In another study, ablation of TGFβ receptor II (TBR2) or SMAD4 reduced the development of liver fibrosis induced by the hepatocyte-specific ablation of TAK1.11 This effect was mediated by a reduced hepatocyte cell death in TBR2-ablated hepatocytes. However, dual ablation of TAK1 and TBR2 is difficult to interpret as both are part of the TGFβ signaling cascade. Ablation of TBR2 in this model may modulate the phenotype of hepatic TAK1 deficiency rather than instructing about the role of TBR2 in epithelial responses to injury.
Besides the stimulation of wound healing, the suppression of proliferation through the mobilization of cyclin-dependent kinase inhibitors and suppression of c-Myc constitutes another key biological effect of TGFβ signaling.12 Accordingly, loss of epithelial TGFβ signaling has been linked to promotion of carcinogenesis in a wide range of cancers12,13, and occurs at the level of Smad4.14–17 Loss of Smad4 is common in cholangiocarcinoma18, but loss of TGFβ receptor II (TBR2) has also been described.19 TGFβ exerts a dichotomous role in carcinogenesis12, and epithelial TGFβ may also promote tumor development through induction of epithelial-mesenchymal transition.20 In addition, TGFβ in fibrogenic cells or the tumor microenvironment (TME) may promote remodeling and tissue fibrosis that contribute to carcinogenesis.13,21 Accordingly, the role of TGFβ in different cellular compartments is not easy to distinguish, particularly in human studies, as effects of increased TGFβ in the TME may mask the tumor-suppressive role of epithelial TGFβ signaling. Hence, cell-specific ablation strategies are required to specifically interrogate the role of TGFβ in the epithelial compartment.
Using multiple cell-specific Cre deleters to delete floxed TBR2 in hepatocytes, cholangiocytes or both populations, we demonstrate that epithelial TGFβ signaling does not affect liver fibrosis but that it suppresses the development of cholangiocarcinoma arising from hepatocytes and cholangiocytes by restricting cholangiocyte proliferation.
MATERIAL AND METHODS
Mice.
For liver-specific deletion of PTEN and TBR2, Albumin-Cre mice were crossed with floxed PTEN mice (both from Jackson, in C57Bl/6 background), floxed TBR2 mice (a gift from Hal Moses, Vanderbilt University, Nashville, TN; in C57Bl/6 background)22 or both. For hepatocyte-specific ablation of TBR2 and/or PTEN, mice were infected with an AAV8-TBG-Cre (1×1011 genome copies i.v.) as described23, and sacrificed at different time points after infection. For cholangiocyte-specific ablation of TBR2 and/or PTEN, floxed PTEN and/or floxed TBR2 mice were crossed with mice expressing K19-CreERT24 (a gift of Dr. Guoqiang Gu, Vanderbilt University) or Prom1-CreERT225 (a gift from Dr. Richard Gilbertson, St. Jude’s Children’s Research Hospital). K19-CreERT or Prom1-CreERT2 activity was induced by intraperitoneal injection of tamoxifen (80 mg/kg). Mdr2ko mice (a gift from Dr. Detlef Schuppan) were crossed with TBR2 floxed and Albumin-Cre mice. mTom/mGFP Cre reporter mice (from Jackson) were used for fate tracing. The following primers were used for genotyping: AAGTTTTTGAAGGCAAGATGC and CAAGCACTCTGCGAACTGAG (PTEN), ACTTCTGCAAGAGGTCCCCT and TAAACAAGGTCCGGAGCCCA (TBR2), ATGAAATGCGAGCTAAGTATGG and CGCCGCATAACCAGTGAAAC (Albumin-Cre), GCAGAATCGCCAGGAATTGACC and GTTCTTGCGAACCTCATCACTC (K19-CreERT), CAGGCTGTTAGCTTGGGTTC and AGGCAAATTTTGGTGTACGG (Prom1-CreERT2).
Liver fibrosis and carcinogenesis models.
All animal procedures were in accordance with guidelines by the National Institutes of Health, and approved by the Institutional Animal Care and Use Committee at Columbia University. Toxic liver fibrosis was induced by intraperitoneal CCl4 injection (0.5 µ L/g, in corn oil at a ratio of 1:3) for 8 injections. For cholestatic liver fibrosis, mice underwent ligation of the common bile duct (BDL) for 21 days. Mdr2ko mice were used as second cholestatic liver fibrosis model. For genotoxic hepatocarcinogenesis, mice were injected with DEN (25 mg/kg i.p., day 15 postpartum). Liver-specific (via Albumin-Cre) or hepatocyte-specific (via AAV8-TBG-Cre) ablation of PTEN or PTEN and TBR2 were employed as genetic carcinogenesis models. For some induction of cholangiocarcinoma, mice with cholangiocyte-specific PTEN and TBR2 ablation were fed a 0.1% 3,5-diethoxycarbonyl-1,4-dihydro-collidin (DDC)-containing diet for 6–8 weeks. All mice for fibrosis and cancer studies were male with the exception of Prom1-CreERT2 mice for the induction of cholangiocarcinoma, which were all female.
Immunohistochemical staining and microscopy.
Immunohistochemistry was performed using primary antibodies against desmin (rabbit, Lab Vision Cat.No RB-9014-P, Thermo Fisher Scientific), αSMA (mouse, FITC-conjugated, Sigma-Aldrich F3777), keratin (rabbit, DAKO Z0622), keratin 19 (rat, Troma-IIIc, Developmental Studies Hybridoma Bank, University of Iowa), osteopontin (goat, R&D AF808), F4/80 (rat, AbD serotec MCA497A64), pSMAD2 (rabbit, Cell Signaling Technology, m3108, goat), pSMAD3 (rabbit, Abcam ab52903), GFP (rabbit, Abcam ab290) and HNF4α (goat, Santa Cruz Biotechnology SC-6556), and matching secondary anti-rabbit (donkey, A21207), anti-rat (chicken, A21472), anti-FITC (rabbit, A11090) and anti-goat (chicken, A21468) with various fluorescent conjugates (all Invitrogen), Confocal microscopy was performed on a Nikon A1 confocal laser microscope (Nikon Instruments) using a 20x lens or 40x and 60x oil immersion lenses.
Quantification of liver injury and fibrosis.
Hepatic injury was determined by quantification of serum ALT activity (Thermo Scientific). Hepatic fibrosis was determined by picrosirius red staining and αSMA immunohistochemistry as previously described.26 Pictures for quantification of picrosirius red staining were taken in >10 low-power fields/mouse using a polarized light filter and quantified by Adobe Photoshop software. αSMA immunohistochemistry was quantified by the same method but without polarized light. Hydroxyproline assays were performed as described.26
Quantification of liver tumors.
Immediately after sacrifice, tumor number and tumor size were determined as described.27 Livers were then digitally photographed and weighed to calculate the liver-body weight ratio. Liver lobes were fixed in 10% formalin for 24 hours and paraffin-embedded for further analysis.
Statistical evaluation
Statistical analysis was performed using Prism (GraphPad, San Diego, CA). Differences between two groups were calculated by Student’s t-test or Mann-Whitney U-test. Differences between multiple groups were determined by one-way ANOVA, followed by Dunnett’s post-hoc test. The significance of the overlap between gene sets was calculated by the χ® test. All data are expressed as means ± standard deviation.
Additional methods are described in the supplementary materials.
RESULTS
Epithelial TGFβ signaling is activated in murine and human fibrogenesis
To determine whether TGFβ signaling is activated within the epithelial compartment of the injured liver, we performed immunohistochemical staining of phosphorylated SMAD2 (pSMAD2) in different types of liver injury. In human cirrhosis as well as murine models of toxic and biliary liver fibrosis (CCl4 treatment, bile duct ligation and Mdr2ko), we detected abundant pSMAD2 expression in cells with characteristic hepatocyte and cholangiocyte morphology (Fig.1A–B). In CCl4-, BDL-and Mdr2ko-induced liver injury we observed a similar pattern of pSmad3 expression (Suppl.Fig.1A). Co-staining for pSMAD2 and HNF4α and keratin 19 as markers of hepatocytes and cholangiocytes, respectively, and subsequent confocal microscopy further confirmed TGFβ pathway activation in these two cell types (Fig.1C). As expected, pSmad2 expression was also seen in αSMA-positive hepatic stellate cells (Suppl.Fig.1B). Together, these data establish activation of TGFβ signaling in both the epithelial and mesenchymal cell compartments of the chronically injured liver.
Figure 1. Epithelial TGFβ signaling is activated in murine and human liver disease but does not contribute to toxic and biliary liver fibrosis.
A. Representative images of phospho-Smad2 staining show epithelial TGFβ pathway activation in cirrhotic human liver (n=5). B. Phospho-Smad2 staining shows activated TGFβ signaling in epithelial cells in CCl4-induced toxic liver injury as well as BDL-and Mdr2ko-induced biliary liver disease. C. Dual immunofluorescent staining and subsequent confocal microscopy demonstrate phospho-Smad2 in HNF4α-positive hepatocytes and keratin-positive cholangiocytes in CCl4-treated and BDL-treated livers, respectively. D-E. TBR2lko and TBR2f/f mice were subjected to 8 injections of CCl4 (0.5 µg/ml; TBR2lko n=10 and TBR2f/f n=11) or 15 days of BDL (TBR2lko n=16 abd TBR2f/f n=10). Hepatic fibrosis was determined by picrosirius red staining. Hepatic injury was determined by serum ALT measurement. F. Mdr2ko mice carrying floxed TBR2 in the presence (n=20) or absence (n=21) of Albumin-Cre were sacrificed at age 9 weeks. Liver fibrosis and injury were assessed as described above. Scale bars 100 µm (A,B,D-F), and 50 µm (C). ns, non-significant.
Epithelial TGFβ signaling does not contribute to toxic and biliary liver fibrosis.
To determine whether epithelial TGFβ signaling affects the development of liver fibrosis, we generated double transgenic mice co-expressing floxed TBR2 and Albumin-Cre (TBR2lko) leading to the deletion of TBR2 in hepatocytes and cholangiocytes. Ablation was highly efficient, as determined by qPCR, and the absence of pSmad2 in both hepatocytes and cholangiocytes in chronically injured livers (Suppl. Fig.1C–D). Following eight injections of CCl4, both floxed controls and TBR2lko mice developed significant accumulation of fibrillar collagen and αSMA-positive myofibroblasts as assessed by picrosirius red staining (Fig. 1D) and αSMA immunohistochemistry (Suppl. Fig.2A). However, there was no difference in fibrosis development between both groups. Moreover, we found no differences in liver injury (Fig.1D). To address the possibility that epithelial TBR2 might play a role in cholangiocytes rather than in hepatocytes, we additionally investigated biliary liver fibrosis induced by bile duct ligation (BDL) or by knockout of Mdr2, a model of progressive cholestatic liver disease that closely resembles human disease.28 Again, in both models we observed no differences in liver injury, as determined by serum ALT levels, or liver fibrosis, as determined by quantification of picrosirius red staining (Fig.1E–F) and αSMA immunohistochemistry (Suppl.Fig.2.B). However, we observed an increased expression of keratin, suggesting that TBR2-induced signals control the expansion of cholangiocytes (Suppl.Fig.2C). Together, our findings demonstrate that epithelial TGFβ signaling does not contribute to hepatic fibrogenesis but that it possibly regulates epithelial proliferation.
Epithelial TGFβ signaling does not affect DEN-induced hepatocarcinogenesis
Because of these findings, we next investigated the hypothesis that the function of epithelial TBR2 in the liver lies in the protection from cancer-promoting proliferation. To test this hypothesis, TBR2lko and floxed littermate controls were subjected to diethylnitrosamine (DEN) at day 15 postpartum, and sacrificed 10 months later. We observed no differences in the number of tumor nodules, tumor size or the liver body weight ratio, suggesting that epithelial TGFβ signaling does not alter genotoxic hepatocarcinogenesis (Fig.2A). Moreover, qPCR showed no differences in the expression of Afp, Cd133, and mKi67 (Fig.2A) demonstrating that TGFβ signaling does not affect tumor progenitors and proliferation.
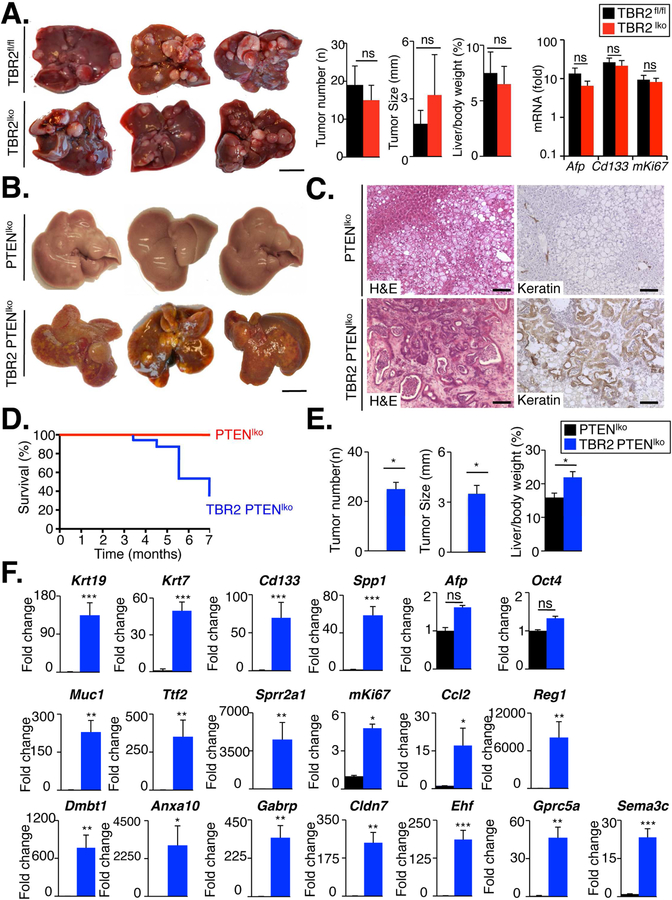
Figure 2. Epithelial TGFβ signaling does not contribute to genotoxic hepatocarcinogenesis but protects liver from cholangiocarcinoma development in the presence of concomitant PTEN loss.
A TBR2lko (n=14) and TBR2f/f mice (n=12) were injected with DEN (25 mg/kg i.p) at day 15 postpartum and sacrificed 10 months later. Tumor number, tumor size and liver body weight ratio were compared between TBR2lko and TBR2f/f mice (middle panel). mRNA expression of Afp, Cd133 and mKi67 was determined by qPCR (right panel). B-E. In contrast to PTENlko mice (n=9), TBR2 PTENldko mice (n=12) developed keratin-positive tumors with typical features of cholangiocarcinoma and increased mortality. F. mRNA expression of markers for cholangiocytes, progenitor cells, biliary injury and proliferation was determined by qPCR in tumors from PTENlko and TBR2 PTENlko mice. Scale bar 100 mm (A,D) and 100 µm (E). ns, non-significant. *p<0 .05, **p<0.01, ***p<0.001
Loss of epithelial TGFβ promotes cholangiocarcinoma development in the presence of concomitant PTEN loss.
To test the role of epithelial TGFβ signaling in a second model of liver carcinogenesis, we generated triple transgenic mice expressing Albumin-Cre, floxed PTEN and floxed TBR2 (TBR2 PTENldko). We chose this model as PTEN deletion led to a significant upregulation of Tgfbr2 mRNA and protein (Supp.Fig.3A–C), suggesting that TBR2 provides a protective signal that restricts the proliferation of PTEN-deleted cells. Hepatic deletion of PTEN and TBR2 was at the expected rate of ≈80%, representing the percentage of hepatocytes and cholangiocytes in the total liver (Suppl.Fig.3A–B). Single and double-knockout mice were born normally without any apparent phenotype, but all double knockout mice developed multiple tumors and died around age 5–7 months (Fig.2B,D,E), often exhibiting severe cholestasis shortly before death. In contrast, PTENlko mice displayed no tumors or mortality at this age (Fig.2B,D,E). Tumors in double knockout livers were macroscopically different from tumors developing in older PTENlko mice. Microscopically, tumors from TBR2 PTENldko displayed typical features of cholangiocarcinoma in the H&E sections (Fig.2C), positive keratin staining of virtually all tumor cells (Fig.2C), high expression of cholangiocyte markers including Krt7, Krt19, Cd133, Muc1, Tff2, and Sprr2a1 mRNA, expression of cholangiocarcinoma/tumor enriched genes/markers including Ehf, Reg1, Dmbt1, Sema3c, Gprc5a, Gabrp, Cldn7, and Anxa10 (Fig.2F) as well as accumulation of αSMA-positive cancer-associated fibroblasts (Suppl.Fig.3D). Together, these data suggest that the loss of TBR2 in the epithelial compartment promotes a shift from hepatocellular carcinoma to cholangiocarcinoma when there is loss of PTEN expression, a common event in human hepatocarcinogenesis.29 These relevance of these findings is emphasized by the high percentage of mutations in the TGFβ signaling pathway, in particular Smad4, in most studies of human cholangiocarcinoma18,30. Our findings therefore suggest that TGFβ-mediated activated of Smad4 represents a mechanism that protects from cholangiocarcinoma development, and that loss of this protective pathway promotes cancer development.
Loss of TGFβ signaling promotes cholangiocyte but not hepatocyte proliferation.
Of note, our deletion strategy using Albumin-Cre mice targeted both the hepatocellular and biliary compartments, as demonstrated by Albumin-Cre-mediated recombination of a fluorescent Cre reporter gene in both hepatocytes and cholangiocytes (Suppl.Fig.3E). To determine in which epithelial cell type TGFβ signaling acted, we next examined proliferation of hepatocytes and cholangiocytes by co-staining Ki-67 with HNF4α and K19, respectively, in livers of two month-old mice, i.e. at a time point when they did not yet display tumor formation. Whereas we observed no increased hepatocyte proliferation in TBR2 PTENldko in comparison to PTENlko mice, there was a strong expansion of the pool of keratin-positive cells as well as increased cholangiocyte proliferation and mRNA expression of cholangiocyte markers in the double knockout mice (Fig.3A–D). Together with our finding that epithelial TBR2 deletion results in expansion of keratin-positive cells in biliary fibrosis (Suppl.Fig.2C), these findings indicate that TBR2 restricts proliferation in cholangiocytes, suggesting that loss of this restriction after TBR2 deletion may promote the development of cholangiocarcinoma.
Figure 3. Loss of TGFβ signaling promotes cholangiocyte but not hepatocyte proliferation.
PTENlko mice and TBR2 PTENldko mice were sacrificed at the age of two months. A. Cholangiocyte expansion in PTENlko and TBR2 PTENldko mice was assessed by keratin immunohistochemistry. B. Hepatocyte proliferation was compared between PTENlko and TBR2 PTENldko mice by dual immunofluorescent staining for HNF4α and Ki-67 in combination with confocal microscopy. C. Hepatocyte proliferation was compared between PTENlko and TBR2 PTENldko mice by dual immunofluorescent staining for HNF4α and Ki-67 in combination with confocal microscopy. D. mRNA expression of progenitor and biliary injury markers was compared between PTENlko and TBR2 PTENldko mice by qPCR. Scale bar: 100 µm (A,C) and 200 µm (B). ns, non-significant.*p<0.05,**p<0.01,***p< 0.001
Loss of TGFβ signaling in cholangiocytes promotes cholangiocarcinoma through increased cholangiocyte proliferation.
To further address the cell type in which TBR2 signaling restricts cholangiocarcinoma development, we next employed cell-type specific ablation strategies that selectively target hepatocytes and cholangiocytes. Of note, previous studies demonstrated that intrahepatic cholangiocarcinoma may not only arise from cholangiocytes but also from hepatocytes.25,31,32 To determine whether loss of TGFβ signaling in cholangiocytes is responsible for the cholangiocarcinoma development observed in TBR2 PTENldko mice, we generated two different types of triple-transgenic mice, either co-expressing K19-CreERT with floxed PTEN and TBR2 (TBR2 PTEN∆chol(K19)) or Prom1-CreERT2 with floxed PTEN and TBR2 (TBR2 PTEN∆chol(Prom1)). These approaches permitted to delete PTEN and TBR2 in cholangiocytes without affecting hepatocytes as demonstrated in Prom1-CreERT2 mice with fluorescent Cre reporters (Suppl. Fig.4A) and previous published papers with K19-CreERT.24,33 As keratin 19 and prominin are not only expressed in cholangiocytes but also in many cell types of other organs, we needed to accelerate cholangiocyte proliferation and turnover by additional treatment with DDC diet, a well-established trigger of cholestatic liver injury, in order to avoid tumor development in other organs such as the pancreas, which preceded cholangiocarcinoma development in both with K19-CreERT (Suppl.Fig.4B) and Prom1-CreERT2 (data not shown) mice, and rapidly led to death. DDC-treated TBR2 PTEN∆chol(Prom1) mice indeed developed keratin-positive tumors with typical features of cholangiocarcinoma 10–12 weeks after the initial tamoxifen dose (Fig.4A–B). K19-positive tumors co-localized with Prom1CreERT2-induced GFP (Suppl.Fig.4C), thereby proving cholangiocyte-origin of these tumors. Similar results with rapid development of cholangiocarcinoma and presence of abundant αSMA-positive CAF were seen in DDC-treated TBR2 PTEN∆chol(K19) mice (Fig.4D–E, Suppl.Fig.4D). We observed an increase in the total cholangiocyte area as determined by GFP and keratin staining (Suppl.Fig.4E–F) and of Ki-67 positive cholangiocytes (Fig.4C and F) but not hepatocytes (data not shown) in both TBR2 PTEN∆chol(Prom1) mice and TBR2 PTEN∆chol(K19) mice. Together these data not only suggest that cholangiocytes are the cellular origin of cholangiocarcinomas in this model, but that the loss of TGFβ-mediated growth restriction in cholangiocytes promotes cancer development.
Figure 4. Concomitant loss of TBR2 and PTEN in cholangiocytes promotes cholangiocarcinoma development.
A. Quadruple transgenic mice expressing PromCreERT2, PTENf/f and mTom/mGFP or PromCreERT2, TBR2f/f, PTENf/f and mTom/mGFP were treated with DDC diet and tamoxifen as indicated. Two months after treatment initiation, TBR2 PTEN∆Chol/Prom (n=4) but not PTEN∆Chol/Prom (n=4) mice displayed multiple GFP-positive hepatic tumors B. Tumors from TBR2 PTEN∆Chol/Prom mice displayed typical features of cholangiocarcinoma as demonstrated by H&E staining and immunohistochemical staining for keratin. C. Cholangiocyte proliferation was ompared between PTEN∆Chol/Prom and TBR2 PTEN∆Chol/Prom mice using dual immunofluorescent staining for Ki67 and GFP, and confocal microscopy. D. Mice expressing K19CreERT and PTENf/f (PTEN∆Chol/K19) (n=8), or K19Cre-ERT, TBR2f/f and PTENf/f (PTEN TBR2∆Chol/K19) (n=9) were treated with DDC diet and tamoxifen as above. Nine weeks after treatment initiation, PTEN TBR2∆Chol/K19 but not PTEN∆Chol/K19 mice displayed hepatic tumors. E. Tumors from TBR2 PTEN∆Chol/K19 mice displayed typical features of cholangiocarcinoma seen by H&E staining and immunohistochemistry for keratin. F. Cholangiocyte proliferation was compared between PTEN∆Chol/K19 and TBR2 PTEN∆Chol/K19 mice using dual immunofluorescent staining for Ki67 and keratin 19 and confocal microscopy. Scale bar 100 mm (A) and 100 µm (B, C).*p<0.05
Loss of TGFβ signaling in hepatocytes promotes cholangiocarcinoma but only at later stages.
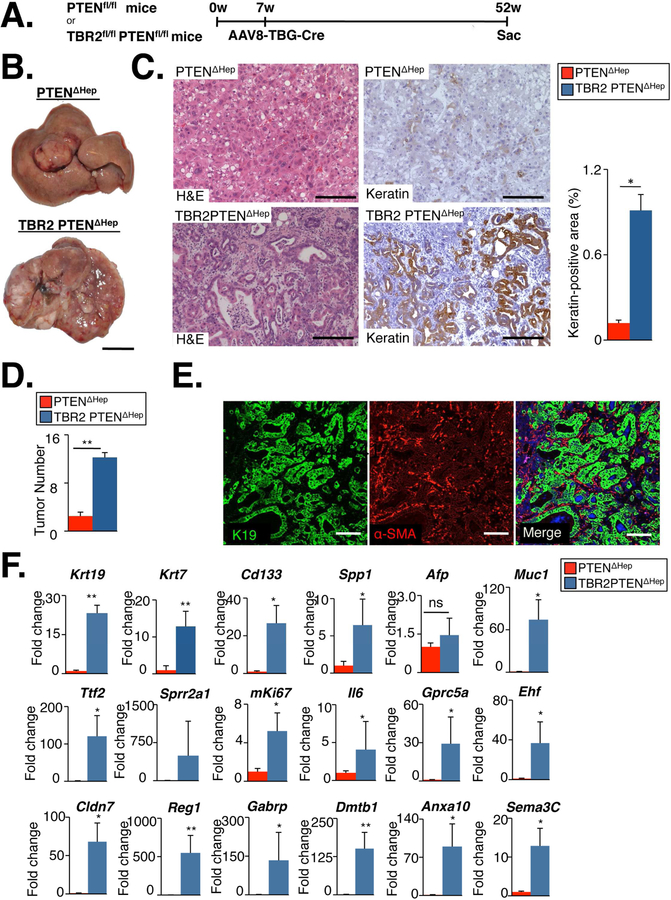
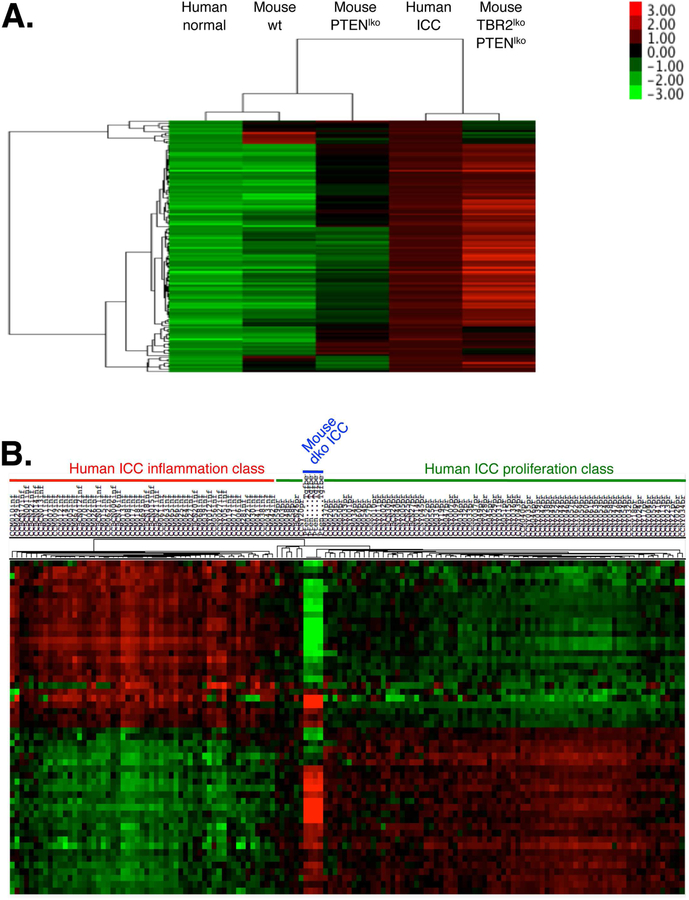
To determine whether the combined deletion of TBR2 and PTEN in hepatocytes was responsible for cholangiocarcinoma development, we employed AAV8-TBG-Cre23, allowing for selective ablation of TBR2 and PTEN in hepatocytes (Fig.5A). As previously reported, AAV8-TBG-Cre-mediated deletion was highly efficient with a 94.4% and 71.3% reduction of Pten and Tgfbr2 mRNA expression, respectively, in whole liver (Suppl.Fig.5A). The lower reduction of Tgfbr2 mRNA is likely due to its high expression in other liver cell types such as HSCs. In constrat to mice with liver-specific TBR2 and PTEN deletion via Albumin-Cre, all mice with hepatocyte-specific AAV8-TBG-Cre-mediated deletion of TBR2 and PTEN survived for one year. However, when mice were sacrificed at this age, they displayed tumors with clear features of cholangiocarcinoma (Fig.5B–D), as evidenced by pronounced keratin staining (Fig. 5C), significantly increased expression of progenitor markers Krt19, Krt7, Spp1 and Cd133, biliary injury markers of Muc1 and Tff2, cholangiocarcinoma/tumor markers Reg1, Gprc5a, Ehf, Cldn7, Gabrp, Dmbt1, Anxa10 and Sema3c (Figure 5F) and accumulation of αSMA-positive CAFs (Figure 5E). Occasional development of tumors with features of HCC or mixed tumors were also noted (data not shown). Of note, keratin-positive cholangiocarcinoma cells in mice with hepatocyte-specific AAV8-TBG-Cre-mediated deletion of TBR2 and PTEN co-labeled with Cre reporter GFP, demonstrating hepatocyte origin (Suppl.Fig.5B–C). To further confirm that these tumors were true cholangiocarcinomas, we performed microarray studies and compared their gene expression profiles to human cholangiocarcinoma. Of 131 genes that were at least 4-fold and significantly (FDR<0.05) upregulated in human cholangiocarcinoma vs normal human liver, 69 were also upregulated in murine cholangiocarcinoma (p<2.2×10−16). This similarity was also revealed by clustering of tumor from double knockout mice with human cholangiocarcinoma (Fig.6A). Further comparison to the two recently described classes of human cholangiocarcinoma revealed that this murine cholangiocarcinoma model tightly clustered with the “proliferation” class (Fig.6B). To fit these results with the seemingly contrasting results from cholangiocyte-specific deletion of PTEN and TBR2, we determined whether cholangiocytes or progenitors in the AAV8-TBG-Cre-deleted mice were derived from hepatocytes as suggested by recent studies.23 Indeed, we detected intermediate hepatocyte-derived progenitors co-expressing GFP and keratin, or GFP and osteopontin in non-tumor areas of both PTEN∆Hep and TBR2 PTEN∆Hep mice albeit at low numbers (Suppl.Fig.5, and Suppl.Fig.6). Moreover, we also detected intermediate hepatocyte-derived progenitors in both PTEN∆Hep and TBR2 PTEN∆Hep mice 8 weeks after AAV8-TBG-Cre-mediated deletion (Suppl.Fig.7). Together, these data suggest that loss of TBR2 in hepatocyte-derived cholangiocytes results in cholangiocarcinoma development through an increase in hepatocyte-derived cholangiocyte proliferation (rather than an increase of transdifferentiation). The relatively low number of hepatocyte-derived cholangiocytes is consistent with the long latency of cholangiocarcinoma development in this model. This observation fits well with the fact that mice with liver-specific deletion of PTEN develop cholangiocarcinomas, but that these are typically overshadowed by the strong and more rapid HCC development.34,35.
Figure 5. Concomitant loss of TBR2 and PTEN in hepatocytes promotes cholangiocarcinoma development.
A. PTENf/f (n=6) and double transgenic mice expressing TBR2f/f and PTENf/f (n=12) were infected with AAV8-TBG-Cre (1×1011 genome copies/mouse, i.v.) for hepatocyte-specific ablation of PTEN (PTEN∆Hep) or PTEN and TBR2 (TBR2 PTEN∆Hep), and sacrificed 10 months later. B. Tumors developed from PTEN∆Hep and TBR2PTEN∆Hep mice, albeit with different macroscopic appearance. C. H&E and immunohistochemistry for keratin demonstrated cholangiocarcinoma development in TBR2 PTEN∆Hep but not in PTEN∆Hep mice. D. Tumor number was quantified in PTEN∆Hep and TBR2 PTEN∆Hep mice. E. The presence of αSMA-positive CAF in cholangiocarcinoma from TBR2 PTEN∆Hep mice was determined by dual immunohistochemistry for K19 (marking tumor cells) and αSMA (marking CAF). F. Gene expression of progenitor cell/cholangiocyte (Krt19, Krt7, Cd133, Spp1 and Afp), biliary injury (Muc1, Tff2, Sprr2a) and cholangiocarcinoma markers (Gprc5a, Ehf, Cldn7, Reg1, Gabrp, Anxa10, Dmbt1, Sema3c,) was compared between tumors from PTEN∆Hep and TBR2PTEN∆Hep mice, using qPCR. Scale bar 100 mm (B), 100 µm (C,E). ns, non-significant. *p<0.05,**p<0.01,***p<0.001
Figure 6. Comparison of cholangiocarcinoma developing in mice with hepatocyte-specific deletion of PTEN and TBR2 to human cholangiocarcinoma.
A. Microarray from paired human non-tumor liver tissues (n=23), human intrahepatic cholangiocarcinoma (ICC, n=23), normal mouse liver (n=3), liver tumors arising in PTEN∆Hep mice (n=4) and liver tumors arising in TBR2PTEN∆Hep mice (n=4). Displayed is a heatmap of all genes with a least 4-fold and significant (FDR<0.05) upregulation in human cholangiocarcinoma vs. human normal liver, demonstrating clustering of human ICC and cholangicarcinoma from TBR2PTEN∆Hep mice. B. Comparison of cholangiocarcinoma from TBR2PTEN∆Hep mice (n=4) to human ICC (n=136) demonstrated clustering with the proliferation subclass.
DISCUSSION
TGFβ is a key cytokine in the regulation of wound healing processes and in the activation of fibrogenic cells, and has been implicated in both liver fibrosis as well as HCC development. Our study provides strong evidence that TGFβ signaling also occurs in both cholangioyctes and hepatocytes, and that these signals play important roles in the injured liver, but only in distinct settings. Importantly, we did not find a role for epithelial TGFβ signaling in the development of liver fibrosis, as determined in three different fibrosis models, suggesting that TGFβ signals in HSCs represent the key mechanism through which TGFβ promotes liver fibrosis. Our findings contrast data from a recent study in which the contribution of epithelial TGBβ signaling to liver fibrosis and hepatocarciongenesis was studied in mice with liver-specific knockout of TGFβ-activated kinase 1 (TAK1). As TAK1 is part of the TGFβ pathway and exquisitely sensitizes cells to cell death, it is possible that the cell-death promoting effects of TGFβ may have been revealed in this specific setting. Moreover, the overactivation of TAK1-independent TGFβ signaling events in TAK1-knockout mice are likely linked to the phenotype of this mouse, and blocking TGFβ may have inhibited this pathway. Nonetheless, as TAK1 deletion is not a component of normal liver fibrosis, it can be concluded that epithelial TGFβ signaling does not contribute to the development of toxic or biliary liver fibrosis.
Loss of epithelial TGFβ signaling occurs in many types of cancers and is considered a key contributor to carcinogenesis in prostate, pancreatic, breast, colon cancer.14–17 However, we did not observe a role for epithelial TGFβ signaling in hepatocarcinogenesis as evidenced by unaltered HCC development by TBR2 deletion in two different models. Again, these data are in contrast to the study of Yang et al, in which HCC development in TAK1-deleted livers was reduced by TBR2 deletion. As discussed above, this result is most likely explained by the involvement of TAK1 in the TGFβ signaling cascade and the key role cell death in TAK1-deleted hepatocytes in this model.11 In contrast to our data on HCC, we observed a key role of the TGFβ pathway in the development of cholangiocarcinoma in mice with liver-specific ablation of TBR2 and PTEN. Of note, recent exome sequencing studies have revealed a high prevalence of mutations in Smad4, a key downstream mediator of TGFβ signals, in human cholangiocarcinoma.18,30 In contrast, human HCC is dominated by mutations in the β-catenin, p53 and chromatin remodeling pathways.36 Thus, it appears that TGFβ signaling has a more important role in cholangiocytes than in hepatocytes. This is likely explained by intrinsic differences between these two cell types, but requires further investigation of the underlying mechanisms. Moreover, it is also possible that the bioavailability of TGFβ, which is often stored with the ECM and could be more abundant in the cholangiocyte than in the hepatocyte niche due to more abundant ECM surrounding the portal tracts, could additionally contribute to this phenotype. It should be noted that TGFβ ablation also affected cholangiocyte expansion in Mdr2ko model of cholestatic fibrosis. Accordingly, cholangiocyte-specific combined ablation of PTEN and TBR2 by two different approaches resulted in cholangiocarcinoma formation as well as increased cholangiocyte proliferation. Together, these data imply a key restrictive function of TGFβ signaling on cholangiocyte proliferation in multiple settings. The more profound effect of TBR2 deletion in the setting of concomitant PTEN deletion is likely explained by the reactive TBR2 upregulation in cells with PTEN deletion. The deleterious effect of combined PTEN and TBR2 deletion has been found in a wide range of cancers including prostate, pancreas and stomach.14–16,37 Xu et al reported that combined deletion of PTEN and Smad4 by Albumin-Cre results in increased cholangiocarcinoma formation.34 While our study confirms the relevance of this pathway, it also links cholangiocarcinoma specifically to the TGFβ pathway, as Smad4 can be activated by a wide range of other receptors. Importantly, there is an abundance of activated TGFβ in the injured liver, suggesting that it not only functions to promote wound healing through HSC activation but also serves to restrict cholangiocyte proliferation – which often goes hand in hand with fibrosis in the “ductular reaction”.38 A recent study found a similar phenotype in mice with dual deletion of TBR2 and PTEN in the liver, but did not further investigate the involved cell types.39 Our data suggest that the upregulation of the TGFβ signaling pathway is a protective mechanism that is particularly important in chronic injury in order to prevent expansion of cells with genetic alterations.
Another novel aspect of our study lies in pinpointing the cell type through which the TGFβ pathway restricts cholangiocarcinoma development. We clearly show the key role of this pathway in cholangiocytes, as demonstrated by cholangiocarcinoma induction in mice with cholangiocyte-specific ablation of PTEN and TBR2 but not PTEN alone. In contrast, the deletion strategy employed by Xu et al. resulted in PTEN and Smad4 ablation in both hepatocytes and cholangiocytes.34 Our data also show that concomitant and selective PTEN and TBR2 ablation in hepatocytes (mediated by AAV8-TBG-Cre) can result in cholangiocarcioma formation. While this could be direct transdifferentiation of hepatocytes into cholangiocarcinoma, the presence of hepatocyte-derived cholangiocytes together with the slow development of cholangiocarcinoma in this model are consistent with the hypothesis that cholangiocarcinoma develops from hepatocyte-derived cholangiocytes in TBR2 PTEN-deleted mice – i.e. similar to the development of cholangiocarcinoma by cholangiocyte-specific deletion of TBR2 and PTEN, albeit much slower.
In addition to better understanding the multiple and cell-specific roles of TGFβ in the injured liver, results from our study have also one relevant clinical implication. TGFβ is a key pathway in the promotion of liver fibrosis and targeting TGFβ may inhibit the development of this significant clinical problem. However, our results suggest that non-targeted inhibition of TGFβ signaling may adversely affect cholangiocytes and increase the risk for cholangiocarcinoma development. Further studies are required to understand why loss of TGFβ affects cholangiocytes and cholangiocarcinoma formation so profoundly whereas it has little effect on hepatocytes and HCC development. Similar differences exist probably in other organs as TBR2 deletion results in increased tumor formation only in some cell types.
Supplementary Material
Acknowledgements:
We would like thank Drs. Daniela Sia and Josep Llovet (both Mount Sinai School of Medicine, New York, NY) for providing information about subclasses of human ICC.
Grant support: This study was supported by grants from NIH (5U54CA163111 and 1R01CA190844, to RFS; and 5F31DK0911980 to DHD). Xueru Mu was supported by a grant from the China Scholarship Council. Silvia Affò was supported by a grant from the American liver foundation.
Abbreviations:
- TGFβ
transforming growth factor beta
- TBR2
TGFβ receptor II
- DEN
diethylnitrosamine
- CCl4
carbon tetrachloride
- DDC
3,5-diethoxycarbonyl-1,4-dihydro-collidin
- BDL
bile duct ligation
- ALT
alanine transaminase
- HCC
hepatocellular carcinoma
- ICC
intrahepatic cholangiocarcinoma
- FDR
false discovery rate
- qPCR
quantitative real-time PCR
- CAF
cancer -associated fibroblasts
- TBR2 PTENldko
Albumin-Cre, floxed PTEN and floxed TBR2
- PTENldko
Albumin-Cre, floxed PTEN
- TBR2 PTEN∆chol(Prom1)
Prom1-CreERT2, floxed PTEN and floxed TBR2
- PTEN∆chol(Prom1)
Prom1-CreERT2, floxed PTEN
- TBR2 PTEN∆chol(K19)
K19-CreERT, floxed PTEN and floxed TBR2
- PTEN∆chol(K19)
K19-CreERT, floxed PTEN
- TBR2 PTEN∆Hep
AAV8-TBG-Cre-mediated deletion of floxed PTEN and floxed TBR2
- PTEN∆Hep
AAV8-TBG-Cre-mediated deletion of floxed PTEN
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: All authors have nothing to disclose.
Transcript profiling: Microarray data has been deposited in GEO (GSE66717).
REFERENCES
- 1.Mishra L, Derynck R, Mishra B. Transforming growth factor-beta signaling in stem cells and cancer. Science 2005;310:68–71. [DOI] [PubMed] [Google Scholar]
- 2.Rahimi RA, Leof EB. TGF-beta signaling: a tale of two responses. J Cell Biochem 2007;102:593–608. [DOI] [PubMed] [Google Scholar]
- 3.Massague J TGFbeta signalling in context. Nat Rev Mol Cell Biol 2013;13:616–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Border WA, Noble NA. Transforming growth factor beta in tissue fibrosis. N Engl J Med 1994;331:1286–92. [DOI] [PubMed] [Google Scholar]
- 5.Gressner AM, Weiskirchen R, Breitkopf K, et al. Roles of TGF-beta in hepatic fibrosis. Front Biosci 2002;7:d793–807. [DOI] [PubMed] [Google Scholar]
- 6.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest 2005;115:209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuoka M, Pham NT, Tsukamoto H. Differential effects of interleukin-1 alpha, tumor necrosis factor alpha, and transforming growth factor beta 1 on cell proliferation and collagen formation by cultured fat-storing cells. Liver 1989;9:71–8. [DOI] [PubMed] [Google Scholar]
- 8.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 2008;88:125–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mederacke I, Hsu CC, Troeger JS, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun 2013;4:2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dooley S, Hamzavi J, Ciuclan L, et al. Hepatocyte-specific Smad7 expression attenuates TGF-beta-mediated fibrogenesis and protects against liver damage. Gastroenterology 2008;135:642–59. [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Inokuchi S, Roh YS, et al. Transforming growth factor-beta signaling in hepatocytes promotes hepatic fibrosis and carcinogenesis in mice with hepatocyte-specific deletion of TAK1. Gastroenterology 2013;144:1042–1054 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massague J TGFbeta in Cancer. Cell 2008;134:215–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickup M, Novitskiy S, Moses HL. The roles of TGFbeta in the tumour microenvironment. Nat Rev Cancer 2013;13:788–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Ehdaie B, Ohara N, et al. Synergistic action of Smad4 and Pten in suppressing pancreatic ductal adenocarcinoma formation in mice. Oncogene 2010;29:674–86. [DOI] [PubMed] [Google Scholar]
- 15.Xue J, Lin X, Chiu WT, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. J Clin Invest 2014;124:564–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Z, Wu CJ, Chu GC, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 2011;470:269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voorneveld PW, Kodach LL, Jacobs RJ, et al. Loss of SMAD4 alters BMP signaling to promote colorectal cancer cell metastasis via activation of Rho and ROCK. Gastroenterology 2014;147:196–208 e13. [DOI] [PubMed] [Google Scholar]
- 18.Ong CK, Subimerb C, Pairojkul C, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet 2012;44:690–3. [DOI] [PubMed] [Google Scholar]
- 19.Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 2013;45:1470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumdar A, Curley SA, Wu X, et al. Hepatic stem cells and transforming growth factor beta in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2012;9:530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Achyut BR, Yang L. Transforming growth factor-beta in the gastrointestinal and hepatic tumor microenvironment. Gastroenterology 2011;141:1167–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chytil A, Magnuson MA, Wright CV, et al. Conditional inactivation of the TGF-beta type II receptor using Cre:Lox. Genesis 2002;32:73–5. [DOI] [PubMed] [Google Scholar]
- 23.Yanger K, Zong Y, Maggs LR, et al. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev 2013;27:719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Means AL, Xu Y, Zhao A, et al. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis 2008;46:318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu L, Gibson P, Currle DS, et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 2009;457:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradere JP, Kluwe J, De Minicis S, et al. Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology 2013;58:1461–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dapito DH, Mencin A, Gwak GY, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012;21:504–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trauner M, Fickert P, Wagner M. MDR3 (ABCB4) defects: a paradigm for the genetics of adult cholestatic syndromes. Semin Liver Dis 2007;27:77–98. [DOI] [PubMed] [Google Scholar]
- 29.Nault JC, Zucman-Rossi J. Genetics of hepatobiliary carcinogenesis. Semin Liver Dis 2011;31:173–87. [DOI] [PubMed] [Google Scholar]
- 30.Chan-On W, Nairismagi ML, Ong CK, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet 2013;45:1474–8. [DOI] [PubMed] [Google Scholar]
- 31.Fan B, Malato Y, Calvisi DF, et al. Cholangiocarcinomas can originate from hepatocytes in mice. J Clin Invest 2012;122:2911–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guest RV, Boulter L, Kendall TJ, et al. Cell lineage tracing reveals a biliary origin of intrahepatic cholangiocarcinoma. Cancer Res 2014;74:1005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanger K, Knigin D, Zong Y, et al. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 2014;15:340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, Kobayashi S, Qiao W, et al. Induction of intrahepatic cholangiocellular carcinoma by liver-specific disruption of Smad4 and Pten in mice. J Clin Invest 2006;116:1843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galicia VA, He L, Dang H, et al. Expansion of hepatic tumor progenitor cells in Pten-null mice requires liver injury and is reversed by loss of AKT2. Gastroenterology 2010;139:2170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng Y, Sun AN, Pan XC, et al. Synergistic function of Smad4 and PTEN in suppressing forestomach squamous cell carcinoma in the mouse. Cancer Res 2006;66:6972–81. [DOI] [PubMed] [Google Scholar]
- 38.Williams MJ, Clouston AD, Forbes SJ. Links between hepatic fibrosis, ductular reaction, and progenitor cell expansion. Gastroenterology 2014;146:349–56. [DOI] [PubMed] [Google Scholar]
- 39.Morris SM, Carter KT, Baek JY, et al. TGF-beta signaling alters the pattern of liver tumorigenesis induced by Pten inactivation. Oncogene 2014;0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.