Abstract
Trehalase is ubiquitous in higher plants. So far, indications concerning its function are scarce, although it has been implicated in the detoxification of exogenous trehalose. A putative trehalase gene, T19F6.15, has been identified in the genome sequencing effort in Arabidopsis. Here we show that this gene encodes a functional trehalase when its cDNA is expressed in yeast, and that it is expressed in various plant organs. Furthermore, we present results on the distribution and activity of trehalase in Arabidopsis and we describe how inhibition of trehalase by validamycin A affects the plants response to exogenous trehalose (α-d-glucopyranosyl-[1, 1]-α-d-glucopyranoside). Trehalase activity was highest in floral organs, particularly in the anthers (approximately 700 nkat g−1 protein) and maturing siliques (approximately 250 nkat g−1 protein) and much lower in leaves, stems, and roots (less than 50 nkat g−1 protein). Inhibition of trehalase in vivo by validamycin A led to the accumulation of an endogenous substance that had all the properties of trehalose, and to a strong reduction in sucrose and starch contents in flowers, leaves, and stems. Thus, trehalose appears to be an endogenous substance in Arabidopsis, and trehalose and trehalase may play a role in regulating the carbohydrate allocation in plants.
Trehalose (α-d-glucopyranosyl-[1, 1]-α-d-glucopyranoside; Tre) is a disaccharide widespread among microorganisms and invertebrates (Elbein, 1974), where it plays a role in stress protection, particularly with regard to desiccation, freezing, and heat stress (Crowe et al., 1984, 1998; Wiemken 1990; Ribeiro et al., 1997). Tre has, so far, not been conclusively identified as an endogenous compound in vascular plants, except for the two well-documented cases of “resurrection plants,” Selaginella lepidophylla and Myrothamnus flabellifolia (for review, see Müller et al., 1995a). The presence of functional genes encoding the enzymes of Tre synthesis (Tre-6-P synthase and Tre-6-P phosphatase) indicates, however, that higher plants potentially have the ability to synthesize Tre (Blázquez et al., 1998; Goddijn and Smeekens, 1998; Vogel et al., 1998; Goddijn and van Dun, 1999; Müller et al., 1999a). It is interesting that trehalase, the enzyme activity that hydrolyses Tre, is present in all organs of higher plants, with the highest activities in flowers (Müller et al., 1995a, 1999a). In Arabidopsis a putative trehalase isolog has been identified in the genome sequencing effort (Gene Bank accession no. BAC T19F06). This gene, T19F6.15, is closely homologous to various trehalases, including the recently identified trehalase GMTRE1 from soybean (Aeschbacher et al., 1999).
Here we present an analysis of the activity of trehalase in the model plant Arabidopsis in an effort to obtain a better insight into its function. We show that T19F6.15 encodes a functional trehalase when its cDNA is expressed in yeast, and that its expression pattern is similar to the activity pattern of trehalase in Arabidopsis. Furthermore, we corroborate results of previous studies in soybean and cowpea (Müller et al., 1995b) with validamycin A (Val), a strong trehalase inhibitor in vitro (see Müller et al., 1992), demonstrating that in Arabidopsis, too, Val inhibits trehalase activity in vivo and leads to alterations in carbohydrate allocation. In the presence of Val, a substance with all the properties of trehalose accumulates in Arabidopsis plants grown under sterile conditions, suggesting that endogenous trehalose could be involved in partitioning and carbohydrate allocation.
RESULTS
Spatial Pattern of Trehalase Activity in Arabidopsis
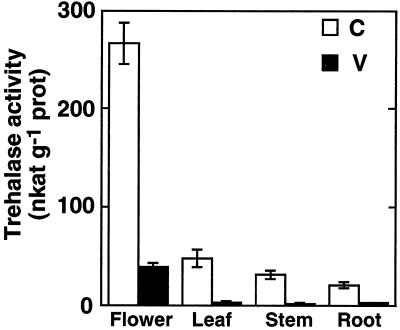
The distribution and activity of trehalase was measured in mature Arabidopsis grown under sterile conditions without a carbon source. In these plants a strong trehalase activity was found in mature flowers (280 nkat g−1 protein), whereas leaves, stems, and roots had significantly lower activities (ANOVA, P < 0.01; Fig. 1). In plants grown on the same media supplemented with 10 μm Val, a strong inhibitor of trehalases, activities were 10-fold reduced in the crude extracts prepared from flowers and reduced to background level in the extracts of leaves and roots, indicating that Val has been taken up by the plants. The morphology and the development of Val-treated plants was indistinguishable from untreated controls.
Figure 1.
Trehalase activity in various organs of flowering Arabidopsis. Arabidopsis plants were grown for 8 weeks on media in the absence (C; white bars) or presence (V; black bars) of the trehalase inhibitor Val (10 μm). Trehalase activity was measured (pH 6.3, 37°C) in crude extracts of various organs. Mean values ± se are given for four independent samples.
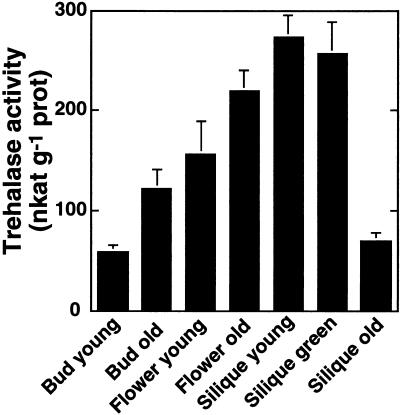
To get more detailed information about trehalase activity in flowers, different developmental stages were harvested between 4 and 6 weeks after germination from greenhouse-grown plants. The developmental stages were classified as “bud young” (sepals still closed, petals not visible), “bud old” (tips of petals visible), “flower young” (petals open), “flower old” (stigma beginning to elongate, but petals still present), “silique young” (siliques < 1 cm long, green), “silique green” (siliques < 1 cm long, green), and “silique old” (yellow-brown). Trehalase activity was highest in mature flowers and in green siliques (more than 250 nkat g−1 protein in mean). Young buds and old siliques had lower trehalase activities (Fig. 2). It is interesting that developing seeds prepared from green siliques had very high trehalase activities, namely 710 ± 20 nkat g−1 protein for three independent batches of seeds. This result was contradictory to former observations where trehalase was described as a pollen enzyme (Gussin et al., 1969). Therefore, flowers were collected and dissected to see whether only anthers or also other parts had higher trehalase activities than vegetative leaves. It is interesting that not only anthers, but also stigmas and to a lesser extent sepals and petals had higher trehalase activities than vegetative leaves (Table I). The highest activities per fresh weight were found in stigmas.
Figure 2.
Trehalase activity in various developmental stages of flowers. Arabidopsis plants were grown in a greenhouse. Flowers of different developmental stages were harvested between 4 and 6 weeks after germination. These stages are referred to as bud young (sepals still closed, no petals visible), bud old (tips of petals visible), flower young (petals open), flower old (stigma beginning to elongate, but petals still present), silique young (siliques <1 cm long, green), silique green (siliques >1 cm long, green), and silique old (yellow-brown). Trehalase activity was measured (pH 6.3, 37°C) in crude extracts. Mean values ± se are given for six independent samples.
Table I.
Trehalase activity in different parts of Arabidopsis flowers
| Tissue | Trehalase Activity | |
|---|---|---|
| nkat g−1 fresh wt | nkat g−1 protein | |
| Entire flower | 1.63 ± 0.19c | 140 ± 6d |
| Stigma | 2.19 ± 0.23c | 83 ± 13c |
| Anther | 1.96 ± 0.23c | 693 ± 72c |
| Petal | 0.31 ± 0.10b | 170 ± 20d |
| Sepal | 0.61 ± 0.19b | 30 ± 10b |
| Leaf | 0.13 ± 0.03a | 6 ± 3a |
Flowers were collected from flowering Arabidopsis plants (Wassiliewskaja) and dissected into sepals, petals, anthers, and stigmas. Leaves from the flower shoot were included. Trehalase activity was measured (pH 6.3, 37°C) in crude extracts. Mean values ± se are given for three independent samples. Values superscribed by the same letter (a–d) are not significantly different (ANOVA, followed by Student-Newman-Keuls test, P < 0.05).
Identification of T19F6.15 as an Arabidopsis Gene Encoding a Functional Trehalase
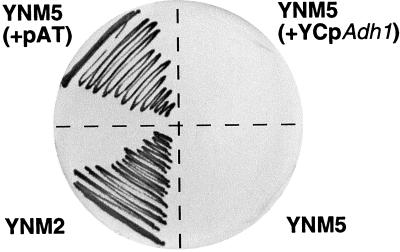
To test whether T19F6.15 encodes a functional Arabidopsis trehalase, primers were designed that map to both ends of the expected coding region and by reverse PCR, a cDNA named cAtTRE1 was amplified. This cDNA was cloned into the yeast YCpADH1 expression vector, and the corresponding construct, named pAT, was transformed into the Saccharomyces cerevisiae YNM5 mutant in which the acid trehalase gene ATH1 had been disrupted (Nwaka et al., 1996). This construct restored growth of the YNM5 mutant when Tre was the sole carbon source (Fig. 3). In protein extracts of this transformant, a strong trehalase activity at pH 4.5 and 6.3 was identified (Table II). In extracts from the untransformed YNM5 mutant, as well as YNM5 clones transformed with the empty vector, trehalase activities were below detection level (Table II). Thus, we conclude that cAtTRE1 encodes a functional trehalase. We, therefore, propose to rename T19F06.15 AtTRE1, for Arabidopsis trehalase 1 gene.
Figure 3.
Complementation of a trehalase-deficient yeast mutant by the Arabidopsis trehalase gene AtTRE1. A mutant of S. cerevisiae deficient in acid trehalase (strain YNM5) was transformed with an “empty vector” and with the corresponding vector expressing the Arabidopsis trehalase cDNA cAtTRE1. Wild-type yeast, the YNM5 mutant, and both transformed strains were then grown on SD minimal plates containing the appropriate auxotrophic additions and Glc. After 3 d of growth at 27°C about five individual colonies were mixed and streaked out on SD minimal plates containing the appropriate auxotrophic additions with Tre as the sole carbon source. After 4 d of further growth at 27°C a photograph was taken. YNM2, YNM2 (wild-type control); YNM5 (+pAT), YNM5 transformed with AtTRE1 expression plasmid pAT; YNM5 (acid trehalase mutant; background control); and YNM5 (+YcpAdh1), YNM5 transformed with YCpADH1 (empty vector control).
Table II.
Trehalase activity in yeast expressing AtTRE1 cDNA
| Yeast Strain | Trehalase Activity at pH 4.5 | Trehalase Activity at pH 6.3 |
|---|---|---|
| nkat mg−1 dry wt | ||
| YNM5 (+pAT) | 10.0 | 6.53 |
| YNM5 (+YcpAdh1) | <0.05 | <0.05 |
| YNM5 | <0.05 | <0.05 |
Trehalase activity in the following derivatives of the S. cerevisiae strain YNM5, lacking the ATH1 gene encoding acid trehalase: YNM5 (+pAT) (YNM5 transformed with the AtTRE1 expression plasmid pAT), YNM5 (untransformed mutant), and YNM5 (+YcpAdh1) (YNM5 transformed with YCpADH1; empty vector control). The cells had been grown to mid-log phase in liquid culture in synthetic dextrose minimal medium with the appropriate auxotrophic additions. Activity was measured at pH 4.5 and 6.3.
Expression of AtTRE1
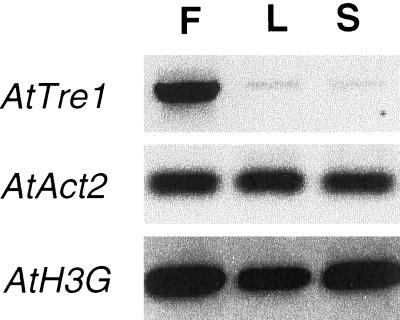
The availability of the AtTRE1 cDNA sequence provided a tool to test the expression of this trehalase gene to compare it with the trehalase enzyme activity. RNA hybridization experiments were performed, but signals could not be detected. Therefore, reverse transcriptase-PCR experiments were performed to analyze the expression of this gene. As positive controls, expression of an actin and a histone gene (AtACT2 and AtH3G) were also analyzed; as expected, these genes showed similar expression levels in all samples tested. AtTRE1 expression was high in flowers and much lower in leaves and stems thus corresponding to the activity pattern. (Fig. 4).
Figure 4.
Expression pattern of Arabidopsis trehalase AtTRE1. Expression of AtTRE1, AtAct2, and At-His3G was analyzed in flowers (F), leaves (L), and stems (S) of mature plants by reverse-transcriptase PCR. Tre, Arabidopsis trehalase isolog AtTRE1; Act, Arabidopsis Actin2 gene AtACT2. His, Arabidopsis Histone3G gene AtH3G.
Uptake of Tre
To examine whether trehalase is also inhibited by Val in vivo in Arabidopsis plants (compare with Müller et al., 1995b), seedlings were grown under sterile conditions on Murashige-Skoog agar medium, complemented with Tre alone (10 and 25 mm) or with Tre (10 mm) in combination with Val (10 μm).
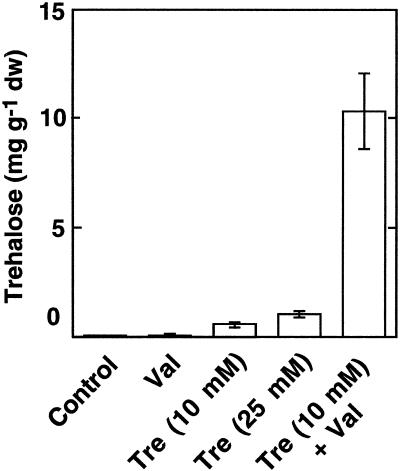
Tre accumulated in leaves of Arabidopsis plants grown on Tre containing media in a concentration-dependent manner. The presence of Val led to a 20-fold increase in Tre accumulation in Arabidopsis leaves grown on 10 mm Tre, indicating that trehalase could be effectively inhibited by Val in vivo (Fig. 5). When Val was added to the media in the absence of Tre a very weak signal with the retention time of Tre could be observed in two of four samples (Fig. 5).
Figure 5.
Tre accumulation in Arabidopsis leaves. Arabidopsis plants were grown under sterile conditions on 0.5× Murashige-Skoog medium without any carbohydrate (control) or supplemented with Tre (10 or 25 mm) and/or Val (10 μm). After 8 weeks, leaves were harvested and their Tre contents were analyzed using HPLC. Mean values ± se are given for four independent samples.
Alterations of Carbohydrate Partitioning by Val
In previous experiments it had been shown that the presence of Val in the culture medium of nodulated soybean and cowpea plants, grown under sterile conditions, led to alterations of carbohydrate pool sizes without impairing vegetative growth. Therefore, it was tempting to investigate whether such effects could be observed in Arabidopsis, where trehalase could be inhibited by Val in vivo.
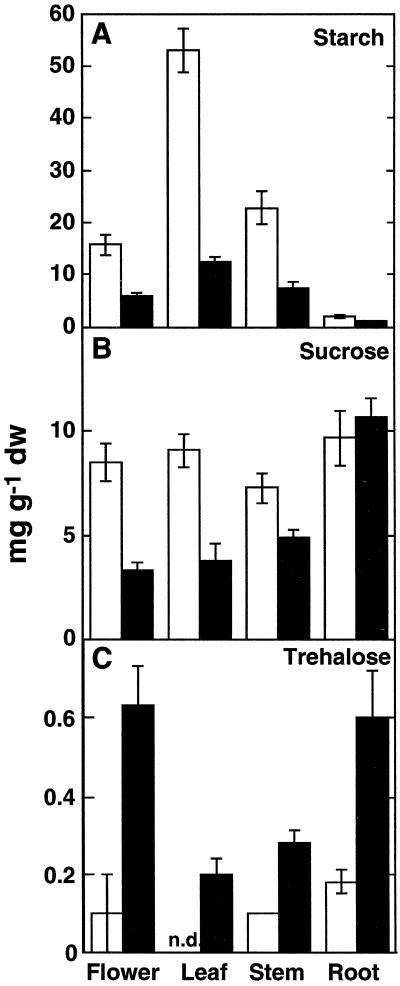
Val let to strong alterations in non-structural carbohydrate contents in different plant tissues as compared with untreated control plants (Fig. 6). Starch (Fig. 6A) and Suc (Fig. 6B) contents were significantly reduced in flowers, leaves, and stems. Starch, in addition, was also significantly reduced in Val treated roots. Glc, Fru, and raffinose pools were also analyzed, but no significant alterations between the treatments with or without Val were found. It is interesting that Val-treated plants showed alterations in fruiting having much less siliques and no seeds (data not shown). Therefore, this stage of development was not included in these analyses.
Figure 6.
Carbohydrate contents of mature, flowering Arabidopsis grown in the presence of Val. Plants were grown under sterile conditions with (black bars) or without (white bars) 10 μm Val. After 8 weeks, flowers, leaves, stems, and roots were harvested, and contents of non-structural carbohydrates were analyzed by capillary GC (Suc and Tre) or enzymatically (starch). A, Starch; B, Suc; C, Tre (tentatively identified, as described in “Results”). Mean values ± se are given for four independent samples. nd, Not detected.
During the analysis of non-structural carbohydrates, a substance was identified by capillary gas chromatography (GC) that was already present in untreated Arabidopsis tissues and that accumulated upon Val treatment in all tissues tested (Fig. 6C). This substance had the same retention time (16.8 min) as Tre and appeared as a single peak, as is typical for a non-reducing disaccharide; reducing disaccharides form double peaks under the conditions used for this study, due to differential retention times of the α and β anomers. To further test whether this substance could be Tre, carbohydrates were extracted from flowers of Arabidopsis plants, grown under sterile conditions, and were separated using thin-layer chromatography (TLC). A stained duplicate indicated a disaccharide with the same relative to front migration value as a Tre standard. This carbohydrate was eluted from the TLC plate and was analyzed by anion-exchange HPLC. Using this technique, a substance with the same relative to front migration value as Tre could be detected. Upon incubation of an aliquot of this sample by a commercially available purified porcine kidney trehalase (Sigma Chemie, Buchs, Switzerland), this substance disappeared (data not shown). Samples from the growth media were also analyzed, but did not contain this substance.
DISCUSSION
Arabidopsis Trehalase Occurs in Multiple Tissues and Is Highly Active in Flowers
We show that trehalase activity is present in multiple tissues of Arabidopsis and that it is particularly high in flowers. Previous studies have reported the presence of trehalase in pollen (see Müller et al., 1995a and refs. therein). Our results show that trehalase is particularly high in the anthers containing pollen, but that it is not confined to this tissue alone since siliques and even developing seeds have as high activities as the entire flowers, potentially indicating a role of trehalase in reproduction.
Inhibition of Trehalase Alters Carbohydrate Pool Sizes
Upon feeding Val together with Tre, Arabidopsis plants accumulate much more of this disaccharide than without this inhibitor. Thus, we can conclude that Arabidopsis trehalase is efficiently inhibited in vivo. All trehalases known from plants so far (Müller et al., 1992) and from other sources are very effectively inhibited by Val (e.g. Asano et al., 1987). Former results with cell cultures have shown that the major part of trehalase activity is extracellular (see Müller et al., 1995a). Thus, it is likely that a significant amount of Val reaches the site of trehalase in leaves via the transpiration stream. The residual trehalase activity in the flowers of plants treated with Val is probably due to a reduced uptake of Val by these organs. It is noteworthy to mention that Tre and Val normally have to cross the endodermis when taken up by the roots and therefore have to cross the plasma membrane. So far, how this may occur is unclear.
In previous experiments Val has not been found to have toxic effects on vegetative plant growth (Müller et al., 1995b; Wingler et al., 2000) nor on the expression of important metabolic enzymes (Wingler et al., 2000). Arabidopsis plants grown on media containing Val do not show an alteration in morphology (at least up to the flowering stage). Externally fed Tre, however, impairs growth of seedlings. This effect is enhanced in the presence of Val. At the same time, starch accumulates in the cotyledons (Wingler et al., 2000). In mature plants, application of Val alone strongly alters the pool sizes of soluble carbohydrates thus confirming former results obtained with cowpea and soybean (Müller et al., 1995b). Concerning starch, the effects observed here in flowering plants differ from data obtained with seedlings upon external feeding of Tre (Wingler et al., 2000).
Three possible explanations might be considered to explain these findings: (1) Val has effects apart from the inhibition of trehalase. Concerning enzyme activities, the only target of Val known so far are trehalases (Asano et al., 1987). Plant invertases are not inhibited by this substance (J. Miller, unpublished data). In fungi where Val alters developmental patterns (Robson et al., 1988) it has been shown to interfere with the expression of several enzymes (Robson et al., 1989). Therefore, it cannot be ruled out that Val also has some unknown deleterious effects on plants. (2) Trehalase is a sensor and regulates carbon allocation. Apoplastic trehalase may cleave endogenous Tre (if produced; see below) or an analogous substance or may have an unknown activity, e.g. bind to Suc or monosaccharides, undergo conformational changes, and transduce these changes to a cell surface protein. (3) Tre or Tre metabolites are signals in carbohydrate allocation. Given the presence of genes encoding enzymes for Tre synthesis (see Goodijn and van Dun, 1999; Müller et al., 1999a), it could well be that plants synthesize Tre in small amounts (see below).
Tre has been shown to interfere with carbohydrate-mediated gene regulation in barley (Wagner et al., 1986; Müller et al., 2000), soybean (Müller et al., 1998), and Arabidopsis (Wingler et al., 2000). In Arabidopsis, Tre induces starch biosynthesis enzymes, but Val alone not (Wingler et al., 2000). This may explain apparent contradictions between both studies. It is interesting that transgenic plants expressing the Escherichia coli or S. cerevisiae genes encoding the enzymes involved in Tre synthesis exhibit drastic developmental alterations such as disturbed root systems and stunted growth. Tre itself or its precursor, Tre-6-P, is thought to be responsible for these effects (Goddjin et al., 1997; Romero et al., 1997; for review, see Goddjin and Smeekens, 1998; Goodjin and van Dun, 1999; Müller et al., 1999a). In S. cerevisiae, hexokinase II is inhibited by Tre-6-P (Blázquez et al., 1993).Whether Tre-6-P is produced in plants and whether it might affect the sugar sensing system by altering the hexokinase activity in Arabidopsis is, however, unknown (for review, see Müller et al., 1999a).
Trehalose Is an Endogenous Substance in Arabidopsis
By the combination of capillary GC, TLC, and HPLC analyses, as well as the disappearance of the potential Tre peak after trehalase treatment, we have shown that Arabidopsis contains very small amounts of a substance that behaves identically to Tre. It is unlikely that this product is a metabolite of Val. Val is metabolized into validoxylamine in plants and both substances are removed by the ion-exchange step used during the sample preparation. Since our plant material was grown under sterile conditions we can exclude a microbial origin of Tre, and therefore conclude that Tre is an endogenous substance in Arabidopsis. Earlier reports have already indicated that Tre is probably an endogenous compound in tobacco (Goddjin et al., 1997) although in this case, plants were not grown under sterile conditions, and a microbial origin of Tre was therefore not fully excluded.
CONCLUSION
Understanding the exact role of the endogenous Tre metabolism in carbohydrate allocation and plant development may be crucial when trying to exploit Tre as a stress protectant in transgenic crops that overproduce Tre (for review, see Goodijn and Van Dun, 1999; Müller et al., 1999a). A low, constitutive trehalase activity in Arabidopsis could be sufficient to protect the plant from the growth inhibition induced by exogenously supplied Tre (Wingler et al., 2000). Trehalase, in this respect, may provide a safeguard to the plant from being exposed to Tre from soil-borne microorganisms, for example. Furthermore, the function of trehalases in plants could be the degradation of endogenous Tre or related substances. In this context it is interesting that trehalase is present at very high levels in flowers and in developing seeds of Arabidopsis (Gussin et al., 1969; for review, see Müller et al., 1995a). Therefore, it will be interesting to analyze the role of trehalase in the context of flowering using local applications of Val.
MATERIALS AND METHODS
Plant Material
Seedlings of Arabidopsis (ecotype Columbia-0, obtained from the Nottingham Arabidopsis Stock Centre, Nottingham, UK) were surface-sterilized and germinated on vertically oriented Petri dishes in 0.5× Murashige-Skoog media solidified with purified agar (Unipath Ltd., Basingstoke, Hampshire, UK) as previously described (Benfey et al., 1993). Val (Asano et al., 1987) and Tre were added under sterile conditions after autoclaving and cooling down the media. Plants were grown under daily cycles of 18 h of light (40 μE m−2 s−1) at 22°C and 6 h of darkness at 18°C. Flowering plants were transferred into large plastic containers with lids that allowed ventilation (Sigma Chemie) to reduce condensation in the vessel. Greenhouse-grown plants had 14-h days and 10-h nights at 20°C. For the flower dissection, Arabidopsis (ecotype Wassiliewskaja) were grown under the same conditions in a phytotron (gift by Dr. O. Mittelsten Scheid, FMI, Basel). Plant organs were harvested as indicated, were immediately frozen in liquid nitrogen, and lyophilized.
Trehalase Assay
Thirty to 40 mg of lyophilized, powdered plant material was suspended in 400 μL of extraction buffer containing morpholinoethane sulfonic acid/K+ (50 mm, pH 6.3), EDTA (1 mm), phenylmethyl-sulfonylfluoride (1 mm), Triton X-100 (0.01%, w/v), and insoluble polyvinylpyrrolidone (1%, w/v; Polyclar AT). The suspension was incubated for at least 2 h at 0°C and centrifuged (13,000 rpm, 15 min). For trehalase assays from various parts of flowers, 0.3 to 1 mg of fresh material was homogenized with 15 μL of extraction buffer in a small microfuge tube. Trehalase activity was assayed in the supernatants at pH 6.3 (Müller et al., 1995b) and the Glc formed by the reaction was assayed using HPLC. Each reaction was corrected by subtracting enzyme and substrate blanks. Soluble proteins were determined as previously described (Bradford, 1976).
Analysis of Soluble Carbohydrates
For analysis of soluble carbohydrates, 1 to 5 mg of lyophilized, powdered plant material was mixed with 10 mg of insoluble polyvinylpyrrolidone (Polyclar AT) suspended in 80% (v/v) methanol and was incubated at 70°C for 10 min. After centrifugation (10 min at 13,000 rpm) the supernatants were removed and the pellets were re-extracted twice. The pellets were dried and conserved for the subsequent analysis of starch. The supernatants were combined, vacuum-dried, and resuspended in 0.6 mL of ultrapure water (MilliQ, Millipore, Molsheim, France). Fifty microliters of a wet mixed-bed ion-exchanger (Serdolit microblue:red, 3:1, v/v) was added to remove charged compounds. When setting up the procedure, this ion-exchanger-mix had been checked for hydrolysis of disaccharides. After centrifugation (10 min, 13,000 rpm) the supernatant was removed and the ion-exchanger was washed once with 0.4 mL of ultrapure water and centrifuged. The supernatants were combined and lyophilized. Pellets were dissolved in 0.1 mL of methanol (50%, v/v) and were further analyzed by capillary GC (Müller et al., 1995b).
Quantification of Starch
The insoluble pellets remaining from the carbohydrate extraction were resuspended in 0.2 mL of NaOH (0.5 m) and incubated at 60°C for 1 h. HCl (0.2 mL, 0.5 m) was subsequently added. After cooling down to room temperature, 0.6 mL of acetate/Na+ buffer (0.2 m, pH 4.5) containing 1 unit of amyloglucosidase (special quality for starch determination, Boehringer Mannheim, Germany) was added and the samples was incubated overnight at 37°C. The reaction was stopped by boiling for 2 min. The samples were centrifuged (10 min at 10,000g), the supernatants were 10 times diluted, and were analyzed for Glc formation using HPLC.
Capillary GC
For capillary GC, soluble carbohydrate extracts (50 μL) obtained as described above were completely dried, derivatized as described (Müller et al., 1995b), and were subsequently analyzed using capillary GC. The gas chromatograph (Carlo Erba Mega 3500, Brechbühler, Zurich) was equipped with a glass column (capillary DB-17 column, 30 m × 0.323 mm, J&W Scientific, Brechbühler, Zurich) and a flame-ionization detector (340°C). After injection (0.3 μL;injector at 320°C), the column was kept at 70°C for 2 min and then heated progressively with a rate of 25°C min−1 to 170°C, followed by a rate of 7°C min−1 to 300°C. The column was kept at 300°C for 5 min. Carbohydrates were quantified by comparison with an internal standard (mannoheptulose) and 11 external standards (arabitol, mannitol, Fru, Glc, myo-inositol, glycerol-Glc, Suc, arbutin, Tre, maltose, and raffinose). Chromatograms were integrated and subsequently analyzed using the Maxima software package (Brechbühler).
Analysis of Carbohydrates Using HPLC
The products formed in the enzyme reactions described above (trehalase and starch analysis) were separated using HPLC on a PA-10 column (Dionex, Olten, Switzerland) using a binary gradient of NaOH (0.1–0.3 m from 0–5 min, then 0.3 m until 15 min) and Na-acetate (0–0.3 m from 5–15 min; flow 1 mL min−1). Carbohydrates were quantified as previously described (Müller et al., 1999b) with a pulse-amperometric detector (Dionex, Olten, Switzerland) using a series of standards (Tre, Glc, Fru, and Suc).
Molecular Biology Techniques
If not otherwise mentioned, standard molecular biology techniques were performed according to Sambrook et al. (1989). Sequence analysis was done using the Genetics Computer Group software (GCG Wisconsin Package, Version 9.0, 1996). For reverse transcriptase-PCR, total RNA was extracted from sterilely grown Arabidopsis using the RNeasy kit (Qiagen, Basel). The RNA was reverse-transcribed using a reverse-transcription kit (Boehringer Mannheim) and a random as well as an oligo-dT primer in the reaction. PCR amplification was done using pairs of one forward and one reverse primer on this first strand cDNA. One microliter of the cDNA preparations was used per PCR reaction in a total volume of 30 μL. For detection of the Arabidopsis trehalase a total of 36 cycles were performed. Actin and histone cDNA fragments were amplified with 26 cycles. At these cycle numbers, amplifications did not reach saturated conditions. Primers used for the amplification were designed to have similar annealing temperatures and to span at least one intron to be able to distinguish the amplified cDNA from any potential genomic DNA contaminants. Genes tested, primers used, cDNA fragment sizes, and GenBank accession numbers are the following: actin AtACT2, 5′-GGAAGGATCTGTAC- GGTAAC-3′ and 5′-TGTGAACGATTCCTGGACCT-3′ (247 bp; accession no. U41998); histone AtH3G, 5′-AACCAC- TGGAGGAGTCAAGA-3′ and 5′-CAATTAAGCACGTTC- TCCTCTG-3′ (249 bp; accession no. X60429); trehalase T19F6.15, 5′-GAGGAAAGCCAGTAATCCAG-3′ and 5′-GTCTCTGACTCAGT-AAGAGAG-3′ (785 bp; accession no. O02343).
Amplifications were done under the following conditions (thermal cycler PTC-100, MJ Research, Inc., Watertown, MA). Initial denaturation at 94°C for 2 min followed by the indicated number of cycles with the following steps: 45-s denaturation at 94°C, 45-s annealing at 52°C, and extension for 1 min and 30 s at 72°C. A final extension for 8 min at 72°C was performed. Taq polymerase I from Pharmacia (Dübendorf, Switzerland) was used. Aliquots of each PCR reaction were analyzed by agarose gel electrophoresis using ethidium bromide to visualize amplified products under UV light.
AtTRE1 Cloning and Trehalase Assay in Transgenic Yeast
A cDNA from T19F6.15 (accession no. 002343) that encompasses the complete coding sequence, was amplified using the primers 5′-TCCACTAGTCCCGGGCTAGGCTTCAATG-CTAAGATGAG-3′ and 5′-TCCACTAGTCCCGGGATGTT-GGACTCGGACACAGACAC-3′ from cDNA prepared from RNA from Arabidopsis seedlings grown under sterile conditions. Both oligos have an anchor sequence at their 5′ end for subcloning. Forty cycles were used with the following profile: initial denaturation at 94°C for 2 min, 45-s denaturation at 94°C, 45-s annealing at 55°C, and extension for 2 min at 72°C. A final extension for 8 min at 72°C was performed. The amplified fragment was cloned into the pGEMT vector. The cDNA was then cut out by the SmaI restriction enzyme and cloned into the SmaI site of the YCpADH1 expression vector (Reinders et al., 1998) in the sense orientation to yield the plasmid pAT. Yeast strains were transformed and selected using standard methods of yeast genetics (Sambrook et al., 1989).
The untransformed yeast mutant YNM5, lacking the gene encoding acid trehalase (Nwaka et al., 1996), and YNM5 transgenic for pAT or the empty YCpADH1 expression vector were grown in 50 mL of SD minimal medium (Difco, Detroit) with the appropriate auxotrophic additions and with Glc as carbon source in 300-mL Erlenmeyer flasks at 27°C on a rotary shaker (170 rpm) until mid-log-phase was reached. Cells were harvested by centrifugation (4,000 rpm, 2 min), washed, and suspended in 0.5 mL of MES [2-(N-morpholino)-ethanesulfonic acid]/K+ (0.02 m, pH 6.3) supplemented with EDTA (1 mm), phenylmethylsulfonyl fluoride (1 mm), and Triton X-100 (0.01%, w/v), which has been shown to induce cell permeabilization during the subsequent freeze-thaw cycle (Miozzari et al., 1978). Suspensions were immediately frozen in liquid nitrogen and stored at −70°C until further treatment. To assay trehalase, suspensions were quickly thawed using a heat-block preheated at 30°C, and 10 μL of these suspensions was added to 0.2 mL of an incubation mix containing a ternary buffer (citrate-P-Gly, 50 mm each) adjusted with KOH or HCl to the desired pH (4.5 or 6.3) and 10 mm Tre. By using chelators during extraction and incubation, neutral trehalase, which depends on bivalent cations and which is the only remaining endogenous trehalase activity in this genetic context, was completely inhibited (compare with App and Holzer, 1989). The assay was incubated for 30 min at 37°C and was stopped by boiling. Glc released from Tre was determined using the Glc oxidase-peroxidase method as described by the supplier (Boehringer Mannheim). The values were corrected for substrate and enzyme blanks (Müller et al., 1992).
Chemicals
If not indicated otherwise, all chemicals were purchased from Fluka (Buchs, Switzerland). Val was a gift from Novartis (Basel).
Statistics
Analyses of variance and Student-Newman-Keuls tests were performed using the software SigmaStat (Jandel Scientific, San Rafael, CA).
ACKNOWLEDGMENTS
Arabidopsis seeds were provided by the Nottingham Arabidopsis Stock Centre. The yeast strain YNM 5 was a gift of Salomon Nwaka (University of Freiburg, Germany). We thank Dr. Otrun Mittelsten Scheid (FMI) for giving us a large number of Arabidopsis plants for flower dissection.
Footnotes
This work was supported by the Swiss National Science Foundation (grant nos. 3100–042535.94 to A.W. and 3100–040837.94 to T.B.) and by a fellowship from the Roche foundation (to J.M.).
LITERATURE CITED
- Aeschbacher R, Müller J, Boller T, Wiemken A. Purification of the trehalase GmTRE1 from soybean nodules and cloning of its cDNA: GmTRE1 is expressed at a low level in multiple tissues. Plant Physiol. 1999;119:489–496. doi: 10.1104/pp.119.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- App H, Holzer H. Purification and characterization of neutral trehalase from the yeast ABYS 1 mutant. J Biol Chem. 1989;264:17583–17588. [PubMed] [Google Scholar]
- Asano N, Yamaguchi T, Kameda Y, Matsui K. Effect of validamycins on glycohydrolases of Rhizoctonia solani. J Antibiot. 1987;40:526–532. doi: 10.7164/antibiotics.40.526. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Lagunas R, Gancedo C, Gancedo JM. Tre-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett. 1993;329:51–54. doi: 10.1016/0014-5793(93)80191-v. [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Santos E, Flores CL, Martínez-Zapater JM, Salinas J, Gancedo C. Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding Tre-6-phosphate synthase. Plant J. 1998;13:685–690. doi: 10.1046/j.1365-313x.1998.00063.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Carpenter JF, Crowe LM. The role of vitrification in anhydrobiosis. Annu Rev Physiol. 1998;60:73–103. doi: 10.1146/annurev.physiol.60.1.73. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Chapman D. Preservation of membranes in anhydrobiotic organisms: the role of trehalose. Science. 1984;223:701–703. doi: 10.1126/science.223.4637.701. [DOI] [PubMed] [Google Scholar]
- Elbein AD. The metabolism of α,α-trehalose. Adv Carbohyd Chem. 1974;30:227–256. doi: 10.1016/s0065-2318(08)60266-8. [DOI] [PubMed] [Google Scholar]
- GCG Wisconsin Package, Version 9.0 Genetics. Madison, WI: Computer Group; 1996. [Google Scholar]
- Goddijn OJ, Verwoerd TC, Voogd E, Krutwagen RW, de Graaf PT, van Dun K, Poels J, Ponstein AS, Damm B, Pen J. Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol. 1997;113:181–190. doi: 10.1104/pp.113.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddijn OJM, Smeekens S. Sensing trehalose biosynthesis in plants. Plant J. 1998;14:143–146. doi: 10.1046/j.1365-313x.1998.00140.x. [DOI] [PubMed] [Google Scholar]
- Goddijn OJM, van Dun K. Trehalose metabolism in plants. Trends Plant Sci. 1999;4:315–319. doi: 10.1016/s1360-1385(99)01446-6. [DOI] [PubMed] [Google Scholar]
- Gussin A, McCormack J, Yih-Lo Waung L, Gluckin D. Trehalase: a new pollen enzyme. Plant Physiol. 1969;44:1163–1168. doi: 10.1104/pp.44.8.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miozzari GF, Niederberger P, Hütter R. Permeabilization of microorganisms by Triton X-100. Anal Biochem. 1978;90:220–223. doi: 10.1016/0003-2697(78)90026-x. [DOI] [PubMed] [Google Scholar]
- Müller J, Aeschbacher R, Sprenger N, Boller T, Wiemken A. Disaccharide-mediated regulation of sucrose:fructan-6-fructosyltransferase (6-SFT), a key enzyme of fructan synthesis in barley leaves. Plant Physiol. 2000;123:265–273. doi: 10.1104/pp.123.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Boller T, Wiemken A. Trehalose and trehalase in plants: recent developments. Plant Sci. 1995a;112:1–9. [Google Scholar]
- Müller J, Boller T, Wiemken A. Effects of validamycin A, a potent trehalase inhibitor, and phytohormones on trehalose metabolism in roots and nodules of soybean and cowpea. Planta. 1995b;197:362–368. [Google Scholar]
- Müller J, Boller T, Wiemken A. Trehalose affects sucrose synthase and invertase activities in soybean (Glycine max L. Merr.) roots. J Plant Physiol. 1998;153:255–257. [Google Scholar]
- Müller J, Mohr U, Sprenger N, Bortlik K, Boller T, Wiemken A. Pool sizes of fructans in roots and leaves of mycorrhizal and non-mycorrhizal barley. New Phytol. 1999b;142:551–559. [Google Scholar]
- Müller J, Staehelin C, Mellor RB, Boller T, Wiemken A. Partial purification and characterization of trehalase from soybean nodules. J Plant Physiol. 1992;140:8–13. [Google Scholar]
- Müller J, Wiemken A, Aeschbacher RA. Trehalose metabolism in sugar sensing and plant development. Plant Sci. 1999a;147:37–47. [Google Scholar]
- Nwaka S, Mechler B, Holzer H. Deletion of the ATH1 gene in Saccharomyces cerevisiae prevents growth on trehalose. FEBS Lett. 1996;386:235–238. doi: 10.1016/0014-5793(96)00450-4. [DOI] [PubMed] [Google Scholar]
- Reinders A, Bürckert N, Boller T, Wiemken A, De Virgilio C. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 1998;12:2943–2955. doi: 10.1101/gad.12.18.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro MJS, Reinders A, Boller T, Wiemken A, De Virgilio C. Trehalose synthesis is important in the acquisition of thermotolerance in Schizosaccharomyces pombe. Mol Microbiol. 1997;25:571–581. doi: 10.1046/j.1365-2958.1997.4961856.x. [DOI] [PubMed] [Google Scholar]
- Robson G, Kuhn P, Trinci A. Effects of validamycin A on the morphology, growth and sporulation of Rhizoctonia cereali, Fusarium culmorum and other fungi. J Gen Microbiol. 1988;134:3184–3194. doi: 10.1099/00221287-134-12-3187. [DOI] [PubMed] [Google Scholar]
- Robson G, Kuhn P, Trinci A. Effect of validamycin A on the production of cellulase, xylanase and polygalacturonase by Rhizoctonia solani. J Gen Microbiol. 1989;135:739–750. [Google Scholar]
- Romero C, Bellés JM, Vayá JL, Serrano R, Culiáñez-Macià FA. Expression of the yeast Tre-6-phosphate synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance. Planta. 1997;201:293–297. doi: 10.1007/s004250050069. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Vogel G, Aeschbacher RA, Müller J, Boller T, Wiemken A. Tre-6-phosphate phosphatases from Arabidopsis thaliana: identification by functional complementation of the yeast tps2 mutant. Plant J. 1998;13:673–683. doi: 10.1046/j.1365-313x.1998.00064.x. [DOI] [PubMed] [Google Scholar]
- Wagner W, Wiemken A, Matile P. Regulation of fructan metabolism in leaves of barley (Hordeum vulgare L. cv Gerbel) Plant Physiol. 1986;81:444–447. doi: 10.1104/pp.81.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemken A. Trehalose in yeast: stress protectant rather than reserve carbohydrate. J Gen Mol Microbiol. 1990;58:209–217. doi: 10.1007/BF00548935. [DOI] [PubMed] [Google Scholar]
- Wingler A, Fritzius T, Wiemken A, Boller T, Aeschbacher R. Trehalose induces the ADP-glucose pyrophosphorylase gene, APL3, and starch synthesis in Arabidopsis. Plant Physiol. 2000;124:105–114. doi: 10.1104/pp.124.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]