Abstract
A cDNA coding for 3-ketoacyl-acyl-carrier protein (ACP) synthase III (KAS III) from spinach (Spinacia oleracea; So KAS III) was used to isolate two closely related KAS III clones (Ch KAS III-1 and Ch KAS III-2) from Cuphea hookeriana. Both Ch KAS IIIs are expressed constitutively in all tissues examined. An increase in the levels of 16:0 was observed in tobacco (Nicotiana tabacum, WT-SR) leaves overexpressing So KAS III when under the control of the cauliflower mosaic virus-35S promoter and in Arabidopsis and rapeseed (Brassica napus) seeds overexpressing either of the Ch KAS IIIs driven by napin. These data indicate that this enzyme has a universal role in fatty acid biosynthesis, irrespective of the plant species from which it is derived or the tissue in which it is expressed. The transgenic rapeseed seeds also contained lower levels of oil as compared with the wild-type levels. In addition, the rate of lipid synthesis in transgenic rapeseed seeds was notably slower than that of the wild-type seeds. The results of the measurements of the levels of the acyl-ACP intermediates as well as any changes in levels of other fatty acid synthase enzymes suggest that malonyl-ACP, the carbon donor utilized by all the 3- ketoacyl-ACP synthases, is limiting in the transgenic plants. This further suggests that malonyl-coenzyme A is a potential limiting factor impacting the final oil content as well as further extension of 16:0.
Fatty acid biosynthesis in higher plants is catalyzed by a set of enzymes located in plastids. Once malonyl-coenzyme A (CoA) is produced by acetyl-CoA carboxylase (ACCase), the fatty acid synthase (FAS) transfers the malonyl moiety to acyl-carrier protein (ACP) to use it as a carbon source for the synthesis of long chain fatty acids, mainly 16:0 and 18:0. Each cycle of C2 addition is initiated by a reaction catalyzed by a 3-ketoacyl-ACP synthase (KAS) and involves the condensation of a malonyl-ACP with an acyl acceptor. The discovery and subsequent studies of KAS III resulted in significant changes in the understanding of the initial reaction of the fatty acid biosynthesis in plants. The in vitro (Jaworski et al., 1989; Clough et al., 1992) and in vivo (Jaworski et al., 1993) studies established that KAS III initiates the fatty acid synthesis in plants by catalyzing the condensing reaction of acetyl-CoA and malonyl-ACP. Subsequent condensation reactions are catalyzed by other members of KAS family, namely KASI, II, and IV (Shimakata and Stumpf, 1982; Kauppinen et al., 1988; Dehesh et al., 1998). Contrary to KASIII, these other condensing enzymes utilize only acyl-ACPs for elongation by malonyl-ACP. Biochemical analyses of the plant condensing enzymes indicate that KASI catalyzes the majority of condensations using acyl-ACPs up to 14:0 as substrates, whereas KASII functions primarily in stearic acid synthesis and is most active with 14:0 through 16:0 substrates (Shimakata and Stumpf, 1982).
Despite recent progress in detailed characterization of many enzymes involved in the plant fatty acid synthesis, the regulation of the plant fatty acid synthesis is not well understood (Ohlrogge and Jaworski, 1997). One significant site of regulation appears to be ACCase, but the regulatory role of other enzymes has yet to be determined. One other candidate is KAS III, an enzyme first purified to homogeneity from spinach (Spinacia oleracea; Clough et al., 1992) and its respective cDNAs from several plant species have since been cloned (Tai and Jaworski, 1993; Tai et al., 1994; Slabaugh et al., 1995; Chen, 1996). Studies carried out by Jaworski et al. (1989) indicated that rate of KAS III enzyme activity matches well with the rate of fatty acid synthesis measured in spinach homogenates, suggesting that KAS III may have a rate-limiting role in fatty acid synthesis.
In some well-characterized metabolic pathways, the concept of a single rate-limiting step does not seem to apply. For example, in the glycolytic pathway of yeast, increasing the expression of the putative rate-limiting enzyme phosphofructokinase showed no effect on the rate of ethanol formation (Schaaff et al., 1989). Furthermore, simultaneous increase in the activities of pairs of the enzymes such as phosphofructokinase and pyruvate kinase or pyruvate decarboxylase and alcohol dehydrogenase did not increase the ethanol production (Schaaff et al., 1989). In a similar manner, in a study involving the Trp synthesis pathway of yeast (Niederberger et al., 1992), increasing Trp flux was achieved only by increasing all five enzymes of the pathway, whereas single or comanipulation of up to four enzymes did not affect the flux. These results suggest that the in vivo regulation of a pathway may depend more on the coordinate expression of the enzymes in a pathway rather than on kinetically defined “rate-limiting” steps.
There are limited examples of increasing the expression of FAS genes in plants. Two Escherichia coli FAS genes, fabD (encoding malonyl-CoA-ACP transacylase [MCAT]) and fabH (encoding KAS III), have been expressed in plants while under control of a napin promoter, a seed-specific promoter that, in these studies, increased the level of enzyme activity 3- to 4-fold (Verwoert et al., 1994; Verwoert et al., 1995). Although increased levels of MCAT had no effect on fatty acid composition, higher levels of E. coli KAS III resulted in a small shift in the fatty acid composition to higher levels of polyunsaturated fatty acids in the seed. ACCase, the proposed regulating enzyme in FAS, has also been overexpressed in plants. Overexpression of the Arabidopsis cytosolic ACCase gene resulted in a 10- to 20-fold increase of the enzyme activity in rapeseed (Brassica napus), yet only a very small increase in the total oil content was observed (Roesler et al., 1997). Although genetic manipulation of some enzymes such as thioesterases and desaturases altered fatty acid composition significantly (Knutzon et al., 1992; Voelker et al., 1992), the evidence to date suggests that the enzymes in FAS may be coordinately regulated to counteract changes brought about by genetic overexpression, and thus maintain the flux of fatty acid production.
In the present study, we attempted to determine how the introduction of different KAS IIIs into either tobacco (Nicotiana tabacum) or rapeseed and Arabidopsis, expressed under a constitutive promoter, cauliflower mosaic virus (CaMV) 35S or napin, a seed-specific promoter, would affect the pathway of fatty acid biosynthesis in plants. In all cases, regardless of the origin of the enzyme, organ, or plant species where KASIII is overexpressed, there is an increase in the levels of palmitic acid. It is interesting, however, that this increase in 16:0 levels did not translate into higher oil content in seeds. To the contrary, transgenic seeds contained lower levels of oil as well as reduced rates of the lipid biosynthesis as compared with the wild-type seeds. Analyses of these transgenic plants were extended by measuring the changes in the acyl-ACP intermediates as well as any changes in levels of other FAS enzymes. These data suggest that increasing fatty acid biosynthesis in plants is not controlled by one rate-limiting enzyme.
RESULTS
Isolation and Sequence Analysis of Ch KAS III-1 and -2 cDNAs
Screening of the Cuphea hookeriana developing seed cDNA library with the So KAS III probe led to isolation of several clones. Based on sequencing data, these were grouped into two closely related classes designated as Ch KAS III-1 and Ch KAS III-2. The Ch KAS III-1 was found to encode a predicted polypeptide of 398 amino acids with a molecular mass of 42 kD and pI of 6.8, whereas Ch KAS III-2 encodes a predicted polypeptide of 406 amino acids with a molecular mass of 43 kD and pI of 5.9. The size difference between these two polypeptides is due to the length of the putative transit peptide, as identified by amino acid sequence comparison of these and prokaryotic enzymes (Fig. 1). The putative transit peptide of Ch KAS III-1 is 66 amino acids long versus 74 amino acids of Ch KAS III-2. The amino acid comparison (Fig. 1) between these and other KASs available in the database indicate that Ch KAS III-1 shares higher similarity with Cw KAS IIIA (accession no. U15935; 96% similarity) than with the Ch KAS III-2 (87% similarity), and Ch KAS III-2 shares 84% similarity with Cw KAS IIIB (accession no. U15934). The comparisons also indicate that Ch KAS III-1 and -2 share 70% similarity with So KAS III (accession no. Z22771) and At KAS III (accession no. L31891), and that they are equally distant (35% similarity) from H. influenza fabH (accession no. U32701), P. purpurea chloroplast fabH (accession no. U38804), and E. coli fabH (accession no. M77744). All the examined KAS IIIs from higher plants share 48% to 50% similarity with the Synechocystis sp. fabH (accession no. 1653102).
Figure 1.
The derived amino acid sequences of spinach (So KAS III), C. hookeriana (Ch KAS III 1 and 2), Cuphea wrightii (Cw KAS III a and b), Arabidopsis (At KAS III), E. coli (Ec KAS III), Hemophilus influenza (Hin KAS III), Porphyra purpurea (Por KAS III), and Synechocystis sp. (Syn KAS III) aligned using the Clustal algorithm (Higgins and Sharp, 1989) in the default setting of the computer software Megalign (DNASTAR, Inc., Madison WI). Identical residues are indicated as reverse contrast letters.
Ch KAS III-1 and CH KAS III-2 Are Constitutively Expressed
Ch KAS III-1 and Ch KAS III-2 are low abundant transcripts, not easily detectable on northern blots performed on total RNA. Thus, we performed quantitative reverse transcriptase (RT)-PCR using gene-specific probes to determine levels and patterns of expression of these two genes. The data from these studies (Fig. 2) show that both KAS III-1 and -2 are constitutively expressed in all tissues examined from C. hookeriana, namely seed, leaf, flower, and root. Furthermore, these data suggest that KAS III-1 is expressed at higher levels than KAS III-2 is. However, the difference in the expression levels of these two genes could be the result of preferential amplification of one gene versus the other, an intrinsic property of either RT and/or Taq polymerase.
Figure 2.
Quantitative RT-PCR analysis of Ch KAS III-1 and Ch KAS III-2 transcript levels. The amplified products were separated on agarose gel, blotted, and probed with the respective KAS III coding region.
Expression of KAS IIIs in Transgenic Plants Leads to Accumulation of 16:0
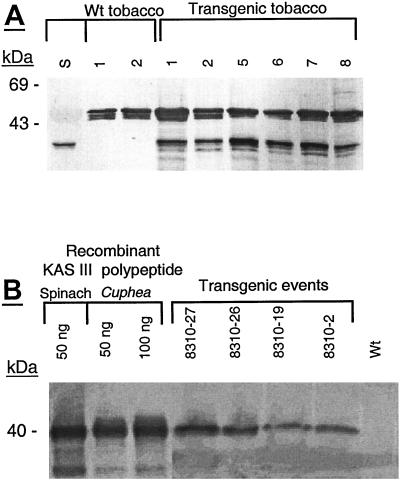
Agrobacterium-mediated transformation was employed to produce transgenic tobacco, rapeseed, and Arabidopsis plants. Transformation of tobacco was carried out using pKAS III, a transcriptional fusion of So KAS III with CaMV-35S promoter, and transformation of rapeseed as well as Arabidopsis was with pCGN 8310 and pCGN 8320, a transcriptional fusion of Ch KAS III-1 and Ch KAS III-2 with the seed-specific napin gene promoter derived from B. rapa (Kridl et al., 1991), respectively. Overall, a total of 55 independent transformation events composed of 10 tobacco, 30 rapeseed, and 15 Arabidopsis plants were examined. The insertion of the spinach KAS III sequence in the putative transgenic tobacco was initially confirmed by gene-specific amplification of spinach KAS III using wild-type and transgenic genomic DNA (data not shown). The levels of expression of So KAS III and Ch KAS III-1 were studied by immunoblot analysis carried out on transgenic tobacco leaves and rapeseed developing seeds respectively (Fig. 3, A and B). In each case, a cross-reacting band, with mobility similar to that predicted for the recombinant spinach and C. hookeriana KAS III polypeptides, was detected in transgenic but not the wild-type plants. In the tobacco leaves, an additional band with higher mobility than that of the introduced polypeptide was detected in both the transgenic and the wild-type plant (Fig. 3A). The presence of such an additional band is not unexpected because the So KAS III antiserum cross reacts equally well with Ch KAS III antigen (Fig. 3B) and thus one may predict that the cross-reacting band is the endogenous tobacco KAS III. The lack of any detectable additional bands on western blot performed with rapeseed developing seeds may be the result of very low levels of expression of endogenous KAS III in these tissues.
Figure 3.
Western-blot analysis of KAS III in transgenic and wild-type plants. A, So KAS III recombinant protein (S); protein extracts from the wild-type (Wt); and transgenic tobacco leaves. B, Fifty and 100 ng of affinity-purified recombinant spinach and C. hookeriana KAS III and protein extract from transgenic and wild-type rapeseed developing seeds. Proteins were blotted onto nitrocellulose paper and probed with polyclonal antibody directed to spinach KAS III.
Three developmentally synchronized tobacco transformants with the highest levels of KAS III enzymes, as measured by western-blot analysis, were employed in a detailed analysis of changes in the levels and composition of fatty acids in their leaves and seeds. These data show a modest but significant increase in the relative levels of 16:0 (18% to 24%) compensated by a decrease in the levels of 18:3 (55% to 50%) fatty acids in leaves (Table I). In contrast, tobacco seeds showed no changes in their fatty acid composition (data not shown). Altered levels of 16:0 and 18:3 did not result in any apparent changes in the in appearance of these plants. However, detection of any structural changes awaits detailed electron microscopy.
Table I.
Analysis of fatty acid methyl esters (FAME) of the total lipid extract from leaves of wild-type and transgenic tobacco plants transformed with So KAS III
| Fatty Acid | % Fatty Acid (sd)
|
||||
|---|---|---|---|---|---|
| WT-1a | WT-2a | Trans-5a | Trans-6a | Trans-7a | |
| 16:0b | 18.4 (1.3) | 18.2 (0.8) | 24.0 (1.3) | 24.5 (0.8) | 23.8 (1.1) |
| 16:1 | 2.0 (1.3) | 4.6 (3.2) | 3.2 (1.8) | 3.6 (0.3) | 3.6 (0.6) |
| 16:3 | 4.2 (1.4) | 6.3 (0.5) | 5.9 (0.3) | 4.4 (2.9) | 6.0 (1.4) |
| 18:0 | 3.5 (0.5) | 3.2 (0.1) | 4.2 (0.2) | 3.9 (0.5) | 3.9 (0.2) |
| 18:1 | 1.9 (0.4) | 1.0 (0.6) | 1.4 (0.1) | 1.5 (0.1) | 1.4 (0.1) |
| 18:2 | 13.3 (2.2) | 10.3 (0.5) | 8.8 (0.7) | 10.0 (0.8) | 10.0 (1.3) |
| 18:3 | 55.0 (2.2) | 54.6 (3.0) | 50.4 (2.2) | 50.5 (2.1) | 49.1 (2.1) |
| TFAc (mg g fresh wt−1) | 3.3 | 3.2 | 3.3 | 2.6 | 2.6 |
Four different samples were taken from each plant.
Significantly different (P < 0.01) between wild-type and transgenic plants.
TFA, total fatty acids.
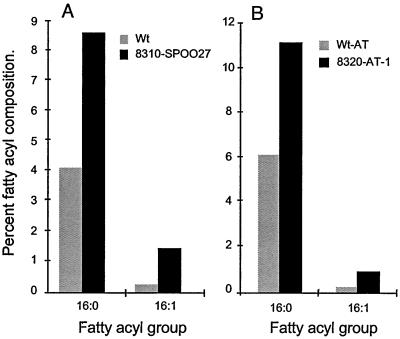
We have generated transgenic rapeseed and Arabidopsis plants overexpressing either Ch KASIII-1 or -2. In all instances, overexpression of these two enzymes in both plant species resulted in similar changes in the oil phenotype. For simplicity, here we are presenting one set of data for each case, namely the fatty acid composition of mature rapeseed seeds derived from 30 transformants, expressing Ch KAS III-1 and Arabidopsis seeds from 15 transformants expressing Ch KAS III-2. Seeds of most of the primary transformants (T2 seeds) from both species accumulated higher levels of 16:0 and 16:1 than the wild-type seeds. The highest expressing line of rapeseed (pCGN 8310-Spoo27) accumulated 8.7 and 1.39 mol% 16:0 and 16:1, respectively, compared with wild type where the levels of 16:0 and 16:1 were 4.2 and 0.22 mol%, respectively (Fig. 4A). Similar results were obtained from Arabidopsis, where 12 out of the 15 transformants accumulated substantially higher levels of 16:0 than the wild type. The highest expressing line of Arabidopsis (pCGN 8320-AT-1) accumulated 11 and 0.8 mol% 16:0 and 16:1, respectively, whereas the levels of 16:0 and 16:1 in the control plants were 6 and 0.2 mol%, respectively (Fig. 4B). In rapeseed, the increase in the 16:0 and 16:1 levels is at the expense of 18:1. In contrast, in Arabidopsis this increase is compensated solely by reduction in the levels of 18:2.
Figure 4.
Relative levels of 16:0 and 16:1 in wild-type and transgenic seeds. A, Proportions of 16:0 and 16:1 in wild-type (Wt) and transgenic rapeseed (line 8310-SPOO27) seeds overexpressing Ch KAS III-1. B, Proportions of 16:0 and 16:1 in wild-type (wt-At) and transgenic Arabidopsis (8320) seeds overexpressing Ch KAS III-2. The fatty acid composition of mature seeds was analyzed to determine the proportion of 16:0 and 16:1 (mol%).
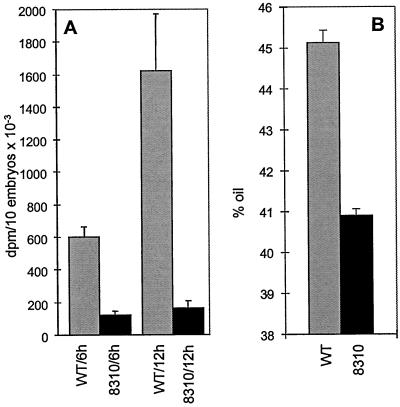
Measurements of the Rate of the Fatty Acid Biosynthesis and Oil Content in Transgenic and Wild-Type Rapeseed Seeds
Developing embryos dissected from wild type and the T3 generation of transgenic rapeseed seeds were employed in measurement of the rate of fatty acid biosynthesis. To reduce possible environmental queues impacting these measurements, wild-type and transgenic plants were grown simultaneously in the same greenhouse. Furthermore, a large number of embryos (10 replicates with 10 embryos per sample) were used to minimize variations due to developmental stages of the organ. The results of these experiments (Fig. 5A) clearly indicate that the rate of fatty acid biosynthesis in transgenic embryo is 5-fold lower than that of the wild type. Furthermore, this decrease in the rate of fatty acid biosynthesis was accompanied by a greater than 4% decrease in the percentage of the oil content in the transgenic seeds as compared with that of wild type (Fig. 5B).
Figure 5.
A, Analysis of [3H]-water into fatty acids of developing rapeseed seed embryos. Seeds from wild type and T3 generation of pCGN 8310 lines were dissected and embryos incubated for 6 and 12 h. Each data point is the average of 10 replicate samples containing 10 embryos per sample, and standard deviations are indicated by error bars. B, Percent oil content by weight of seeds from wild type and T3 generation of pCGN 8310 lines.
Activity of the Spinach KAS III in Transgenic Tobacco
Western-blot analysis data indicate that rapeseed seeds and tobacco leaves (Fig. 3, A and B) express comparable levels of KAS III enzyme, yet the increase in 16:0 levels in transgenic rapeseed seeds is 100% compared with the 30% increase in transgenic tobacco leaves. There may be many factors contributing to this difference, among them enzyme activity, the nature and function of the tissue where it is expressed, and appropriate integration of the introduced enzyme in the fatty acid machinery of the transgene.
KAS III enzyme activity in leaf homogenates of transgenic tobacco, measured under initial velocity conditions, was dramatically higher (100–300-fold) than KAS III activities of the wild-type plants (Table II). These data, in combination with immunoblot analysis, indicated that the spinach KAS III sequence was highly expressed as a fully active enzyme in tobacco.
Table II.
KAS III activity in leaves of wild-type and transgenic tobacco plants transformed with So KAS III
| Plant | KAS III Activity (pmol min−1 mg−1) |
|---|---|
| Wild-type 1 | 26 |
| 2 | 186 |
| Transgenic 5 | 7,880 |
| 6 | 3,950 |
| 7 | 6,630 |
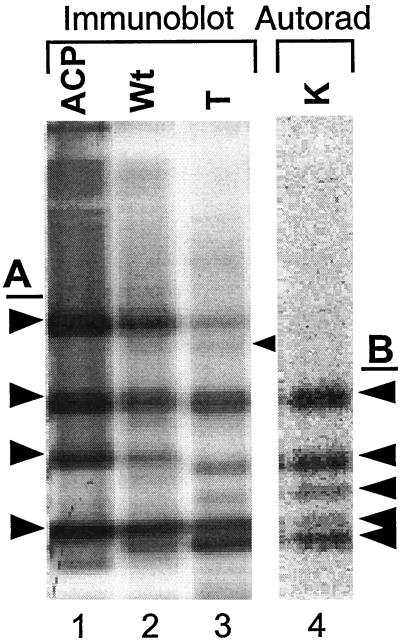
To determine whether the introduced KAS III enzyme has been integrated into the fatty acid biosynthesis pathway, changes in the acyl-ACP pools were examined using immunoblot analysis (Post-Beittenmiller et al., 1991). Interpretation of the immunoblot was complicated by the presence of four isoforms of ACP in tobacco and the additional difficulties associated with overlapping mobilities of their acyl-ACPs on native and denaturing gel electrophoresis. However, we were able to identify the positions of each of the unesterified ACPs (Fig. 6, lane 1), and their respective malonyl- and acetyl-ACPs using standards (data not shown). In addition, we performed an in vitro KASIII assay in the presence of tobacco ACP isoforms and generated a family of radiolabeled tobacco acyl-ACPs. We previously had characterized these immediate products of the KAS III reaction as primarily 4:0-ACPs (Jaworski et al., 1989; Clough et al., 1992). These data indicate that the ACP in the wild-type plants was mostly unesterified (Fig. 6, compare lanes 1 and 2), whereas in the transgenic plants the level of free ACP was greatly reduced and replaced by a family of bands that comigrate with the [14C]acyl-ACPs generated in vitro by a KAS III reaction (Fig. 6, compare lanes 3 and 4). In addition, an acetyl-ACP band detected among the transgenic tobacco acyl-ACPs was missing from the wild-type tobacco. Malonyl-ACP was not detectable in either transgenic or the wild-type plants, confirming the previous finding reported for tobacco suspension cells (Shintani and Ohlrogge, 1995).
Figure 6.
Analysis of short-chain acyl-ACPs of wild-type and transgenic tobacco. Immunoblot is of Lane 1, tobacco ACP-SH isoforms (ACP); 2, plant extracts of wild type (Wt); and 3, transgenic line 5(T) tobacco. A, Left arrows point to the four isoforms of tobacco ACP. Lane 4 (K) is an autoradiogram of [14C]-acyl-ACPs produced in vitro by a KAS III reaction using tobacco ACPs. B, Right arrows next to lane 4 point to major tobacco acyl-ACPs produced by a KAS III reaction. The right arrow next to lane 3 points to an acetyl-ACP accumulating in the transgenic tobacco.
Increased Levels of ACP in Plants Overexpressing KAS III
Visual inspection of numerous immunoblots during the acyl-ACP analysis suggested that the level of ACP was higher in the transgenic plants than in the wild type. Coomassie-stained protein gels, run as controls, confirmed equal loading of the proteins in each lane. We also measured independently the level of total ACP in plant extracts using an acyl-ACP synthetase assay. The levels were then calculated as relative values to either the total protein or tissue weight of the wild-type and the transgenic plants (Table III). Based on either total protein or tissue mass, the amount of ACP in the transgenic plants was 2- to 3-fold higher than in the wild-type plants.
Table III.
Amount of ACP measured in wild type and transgenic tobacco plants by E. coli acyl-ACP synthetase reaction
| Plant | ACP (ng)/Protein (μg) | ACP (ng)/Tissue (mg) |
|---|---|---|
| Wild-type 1 | 1.81 | 4.20 |
| Wild-type 2 | 2.29 | 4.72 |
| Transgenic 5 | 5.09 | 12.52 |
| Transgenic 8 | 3.43 | 8.43 |
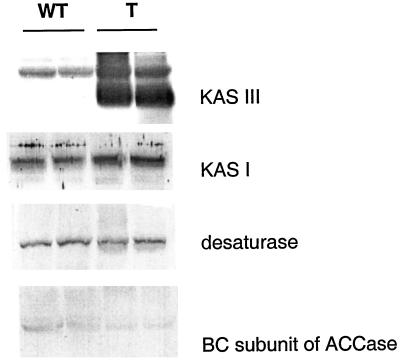
To address the question of whether the elevated levels of ACP is a specific response to KAS III overexpression, or is the result of a general increase in other enzymes associated with fatty acid biosynthesis, the levels of several enzymes in fatty acid biosynthesis pathway were measured using immunoblots analysis (Fig. 7). These data suggest that overexpression of KAS III did not result in any drastic changes in the levels of KAS I, stearoyl-ACP desaturase, or the biotin carboxylase (BC) of the plastid ACCase.
Figure 7.
Western-blot analysis of the FAS component enzymes from Wild-type and transgenic tobacco plants. Wt, Wild type; T, transgenic plants.
DISCUSSION
To examine a potential role of KAS III in the overall regulation of the fatty acid biosynthetic pathway, So KAS III under control of the CaMV-35S promoter was overexpressed in tobacco. In addition, two new KAS III cDNAs were cloned from C. hookeriana, and their respective proteins, Ch KAS III-1 and Ch KAS III-2, were expressed in Arabidopsis and rapeseed under control of napin, a strong seed-specific promoter.
In all instances, overexpression of KAS III enzymes resulted in an increase in levels of 16:0, albeit to different extents, reflecting the general role of this enzyme in fatty acid biosynthesis, irrespective of the plant species or tissue where it is expressed. The extent of increase in 16:0 levels in tobacco leaves (30%) was much less than that of rapeseed seeds (100%), despite evidence from western-blot analyses that KAS III was expressed strongly and equally in both tissues. This discrepancy does not appear to be the result of lack of enzymatic activity in the transgenic tobacco, as leaves extract exhibited a 300-fold increase in activity above the wild type. Several factors could contribute to the accumulation of higher levels of 16:0 in transgenic rapeseed seeds than in transgenic tobacco leaves. In seeds, the 16:0 fatty acid is predominantly exported from the plastid and incorporated into triacylglycerol, whereas in leaves, a large portion of de novo synthesized fatty acids are directly transferred to plastidial lipids by acyl transferases. Therefore, one contributing factor could be the differences between overall activities of downstream enzymes, such as thioesterses and acyl transferases in rapeseed seed versus tobacco leaves. Enzymes of different specificities could be another factor responsible for the pronounced increase in levels of 16:0 in transgenic seeds versus leaves; for example, acyl-transferases that channel 16:0 into triacylglycerol versus enzymes that are involved in polar lipid biosynthesis. Another possible reason for the discrepancy in the levels of 16:0 between the two transgenic tissues could be the difference in the overall capacity of the fatty acid biosynthetic machinery between the two systems. In an earlier study, overexpression of an E. coli KAS III in rapeseed did not lead to an increase in the levels of 16:0 (Verwoert et al., 1995). However, in that study, the maximum level of overexpression was relatively low (3.7-fold), and the heterologous expression of the bacterial KAS III may have mitigated against changes in the 16:0 level.
The increase in 16:0 fatty acid, accompanied by a reduction in the rate of lipid synthesis and relative levels of oil content in the seeds transgenic plants, suggested that overexpressing the KAS III resulted in changes in the activity of a cascade of enzymes of FAS. The amount of 16:0 fatty acid is largely determined by the relative activities of KAS II versus acyl-ACP thioesterase and/or acyl-ACP acyl transferase. The KAS II extends the 16:0-ACP to an 18:0 fatty acid or the 16:0-ACP is hydrolyzed by acyl-ACP thioesterase or transferred into plastidial lipids by an acyl transferase, and in each case thereby preventing further extension. Thus, the percentage of 16:0 would increase if either KAS II activity decreases or the thioesterase and acyl transferase activities increase in a plant. In this study, the overexpression of KAS III resulted in increased amounts of 16:0, indicating either decreased 16:0-ACP extension by KAS II, or increased 16:0-ACP utilization by a thioesterase or acyl transferase. However, the decreased rate of fatty acid synthesis in the transgenic rapeseed seeds clearly indicates a decrease in the KAS II activity in these plants. The source of carbon in fatty acid chain elongation is malonyl-ACP, which is used in the condensation reactions catalyzed by KAS III as well as by KAS I and KAS II. In the acyl-ACP blot, malonyl-ACPs were undetectable in both wild-type and transgenic plants, suggesting that the level of malonyl-ACP was generally quite low and that newly synthesized malonyl-ACP was quickly utilized in the fatty acid biosynthesis. The high levels of KAS III in transgenic plants most probably reduced the amount of malonyl-ACP available to KAS II, reducing its activity. This would have resulted in the thioesterase out-competing KAS II for additional 16:0-ACP, increasing the amount of 16:0 integrated into total lipids of these transgenic plants. In addition, the reduced level of malonyl-ACP would also decrease the KAS I activity and account for the overall reduced rate of fatty acid synthesis.
Unlike most metabolic pathways, KAS III shares one of its substrates, malonyl-ACP, with the other two condensing enzymes. The level of KAS III's other substrate, acetyl-CoA, far exceeds the level of the acyl-ACPs used by KAS I and KAS II (Post-Beittenmiller et al., 1992). Under those circumstances, it might have been expected that if malonyl-ACP was equally available to all plastidic KASs, then the KAS III could have starved KAS I and KAS II for malonyl-ACP much more effectively than the rates of overall FAS suggest. Again, in leaf, the KAS III has increased 300-fold, but in that tissue, overall fatty acid levels were unchanged and there was only a small increase in the level of 16:0. The in vitro assays and the analysis of the in vivo pools of short-chain acyl-ACPs confirm that the activity of the KAS III was high in the transgenic leaves. Therefore, it appears that a significant percentage of the malonyl-ACP was available to KAS I and KAS II despite the very high levels of KAS III. One of the possible explanations for this is that there may be a “channeling” of substrates for the FAS. It has been demonstrated that osmotically disrupted spinach chloroplasts, where enzymes and cofactors were diluted at least 100-fold, were capable of high rates of fatty acid synthesis (Roughan and Ohlrogge, 1996). This and the results of our study support the idea that FAS enzymes in the chloroplast may be organized into a multienzyme complex that can channel substrates to produce long chain fatty acids. Another possible explanation is that the accumulated 4:0-ACP might inhibit the surplus of the KAS III activity. Although excess amounts of spinach 4:0-ACP or 10:0-ACP failed to inhibit spinach-purified KAS III activity in vitro (B. Hinneburg-Wolf and J. G. Jaworski, unpublished data), E. coli 10:0-ACP has been reported to inhibit the Cuphea lancedata KAS III in crude seed homogenates (Bruck et al., 1996). If the kinetic constants for the KASs were dramatically different, where the Km of KAS III for malonyl-ACP was significantly higher than that of KAS I and KAS II, then the latter condensing enzymes might effectively compete. The Km of KAS III from E. coli for malonyl-ACP is 5 μm, but the Km is not available for the other KASs. However, it seems very unlikely that the KAS III would have a much higher Km than KAS I or II because it would likely then function very poorly in the wild-type plants. The presence of acyl-ACPs of all chain lengths at nearly equal levels in spinach indicates that the KASs of that plant at least are functioning in vivo at comparable activities (Post-Beittenmiller et al., 1991).
In our study, the analysis of FAS intermediates by acyl-ACP immunoblot provided clear evidence that the spinach KAS III was integrated and functioning in the tobacco FAS system. The most striking difference in the acyl-ACP profiles was that the majority of the acyl-ACP detected in the transgenic plant was the KAS III reaction product, i.e. 4:0-ACP, whereas most of the ACP observed in the wild type was free ACP-SH (Fig. 6). The other notable acyl-ACP observed in the transgenic leaves was acetyl-ACP. One of the secondary activities of KAS III is acetyl-CoA-ACP transacylase activity (Clough et al., 1992). Under normal conditions, the level of acetyl transacylase activity and accumulation of acetyl-ACP are quite low, as KAS III primarily acts as a condensing enzyme. Acetyl-ACP normally accumulates under conditions where malonyl-CoA or malonyl-ACP levels are limiting (Post-Beittenmiller et al., 1991), and thus its appearance in the transgenic leaves further supports the suggestion that KAS III activities in these plants were sufficiently high to significantly affect the level of malonyl-ACP available for fatty acid synthesis.
In addition to providing evidence that the spinach KAS III was functional in tobacco FAS, this finding was significant because it demonstrated that in wild-type plants, KAS III reaction was not operating near equilibrium. If the KAS III reaction were near equilibrium, large increase in the KAS III activity would result in only small changes in the ratio of its substrate and product concentrations. The significant changes in the relative amount of acyl-ACPs observed in the transgenic plant clearly indicated that the KAS III was far from equilibrium. This suggests that a large increase in FAS activity may not be possible without a corresponding increase in KAS III. Accumulation of 4:0-ACP in the transgenic plant also indicated that KAS I, the enzyme that carries out the subsequent condensation reaction using 4:0-ACP, had become limiting in transgenic tobacco leaves. The immunoblot using KAS I antibody demonstrated that the level of KAS I was not changed in the transgenic plants (Fig. 7).
The results obtained in our study, as well as other studies in which plant FAS genes were overexpressed, imply that increasing the expression of a single gene in a complex metabolic pathway such as fatty acid synthesis is not effective in changing the final product, unless concomitant changes in other enzymes involved is achieved. Overexpression of spinach ACP in the tobacco plant (Post-Beittenmiller et al., 1989) or E. coli fabD gene encoding MCAT expressed in tobacco and rapeseed (Verwoert et al., 1994) did not bring about significant changes in fatty acid profile. ACCase, which has been suggested as a rate-limiting enzyme in plant fatty acid biosynthesis, also did not alter fatty acid composition when overexpressed in a photosynthetic alga, Cyclotella cryptica.(Roessler et al., 1995). In a more recent study, Roesler et al. (1997) successfully overexpressed Arabidopsis homomeric ACCase in rapeseed to increase the enzyme activity by 10- to 20-fold, but only 5% increase in total oil content was achieved in the transgenic seeds. These results are consistent with what has been found in other pathways.
The results of our study suggest that the regulation of plant fatty acid biosynthetic pathway is spread out and coordinated among the many enzymes involved in the pathway. In E. coli FAS, although it is a dissociated system (type II) as in plant, several FAS genes were found to be clustered and each gene in the cluster is cotranscribed with at least one other gene (Zhang and Cronan, 1996), suggesting that they might be controlled by common factors. Not much is known about structural and/or genomic organization of plant FAS. In that context, the elevated level of ACP observed in the transgenic plants is intriguing. However, no such elevation was observed for other fatty acid biosynthetic enzymes and the significance of the ACP level is unknown. Future studies including determination of genomic loci and regulatory elements of FAS genes will help understand the coordination of plant fatty acid biosynthesis regulation.
MATERIALS AND METHODS
Plant Material
Cuphea hookeriana plants were propagated from seeds obtained from the U.S. Department of Agriculture (Ames, IA). Plants were grown under similar conditions as described previously (Dehesh et al., 1996b). Tissue for RNA isolation were frozen in liquid nitrogen and kept at −70°C.
Tobacco (Nicotiana tabacum, WT-SR [Maliga et al., 1973]) seeds were kind gifts from Dr. Douglas Furtek (Pennsylvania State University, State College).
RNA Isolation, cDNA Library Construction, and Screening
Total cellular RNA was isolated from C. hookeriana developing seeds as described previously (Dehesh et al., 1996a) and used in a commercial kit to prepare cDNA libraries (Uni-ZAP, Stratagene, La Jolla, CA). Plating the library and hybridization of filters were carried out as previously described (Dehesh et al., 1996a). Using the So KAS III coding region (Tai and Jaworski, 1993) as a probe led to isolation of a series of cDNA clones. The cDNA clones were recovered as excised plasmids and the DNA sequence was obtained from the 3′ end and 5′ end of each insert. Comparison of the DNA sequences grouped all the clones into two closely related classes designated as Ch KAS III-1 and Ch KAS III-2.
DNA Sequencing and Sequence Analyses
The cDNAs were sequenced in both directions using an automated ABI 373A sequencer (Applied Biosystems, Foster City, CA).
Quantitative RT-PCR Analysis
Total cellular RNA was isolated from C. hookeriana developing seeds, leaf, flower, and root tissue, as described according to (Dehesh et al., 1996a) and used in a commercial kit to prepare first strand cDNA libraries (Smart PCR cDNA synthesis, CLONTECH Laboratories, Palo Alto, CA). This template was used for amplification of Ch KAS III-1 and Ch KAS III-2 transcripts using the following gene-specific oligonucleotides: Ch KAS III-1 5′, CAAATCCTGCGGTTGCAA; Ch KAS III-1 3′, CGCCGAGGCTTGTGAGTA; Ch KAS III-2 5′, CTTACTGTGTTGAGACAC; and Ch KAS III-2 3′, CTCTGTTGCTTGCTGCTTTG.
Amplification of the fragments was carried out for 30 cycles at 94°C for 30 s followed by 30 s at 55°C and 1 min at 72°C.
Southern-blot analysis was performed on the PCR products. Blots were probed with either Ch KAS III -1 or Ch KAS III-2 labeled full-length cDNA fragments. Hybridization and wash conditions were identical to those for screening the cDNA library.
Construction of Binary Vectors and Plant Transformation
DNA used for production of transgenic tobacco was cloned into the expression cassette (p1079), driven by tandem repeat of the CaMV-35S promoter. To clone So KAS III into the p1079 expression cassette, a PCR fragment using the following oligonucleotides was generated: So KAS III, 5′-GTCGACTGATCAATGGCGACTTCATA; and So KAS III, 3′-GTCGACTTTCTTTGTTTATCCCCATC-3′. This fragment was then digested with EcoRV and subcloned into the respective site of pBluescript SK+. The insert was then excised with SallI and cloned into the respective site of p1079 (p1079-KAS). The cleavage of p1079-KAS by XbaI resulted in a fragment that was isolated and inserted into the respective site in the plant binary vector, pCGN 1557 (Mcbride and Summerfelt, 1990), producing pKAS III.
Cloning of Ch KAS III-1 and -2 into the seed-specific expression cassette (pCGN 3223), driven by a napin gene promoter (Kridl et al., 1991), was achieved by producing a PCR fragment using the following oligonucleotides: Ch KAS III-1 and Ch KAS III-2 5′, GCGGCCGCAGATCTATGGCGAATGCATCTGGG; Ch KAS III-1 3′, GCGGCCGCCTCGAGGGCTCGGCTTCAGTCTTA; and Ch KAS III-2 3′, GCGGCCGCCTCGAGGCAGCTTCATTCTTATCA. These fragments generated from Ch KAS III-1 and Ch KAS III-2 were digested with BglII and XhoI and cloned into the respective sites of the 3223 cassette producing pCGN 8306 and 8305, respectively. The KpnI fragment of pCGN 8306 and 8305 were isolated and inserted into the binary vector pCGN 1557, resulting in construction of pCGN 8310 and 8320, respectively.
To transform tobacco plants, the pKAS III construct was introduced into Agrobacterium tumefaciens strain EHA 105 by direct transformation (An et al., 1988), which was subsequently used to infect tobacco leaf discs grown under sterile conditions (Rogers et al., 1986). The resulting putative transgenic plants were grown in a growth chamber under 16-h light and 8-h dark at 20°C.
Binary constructs pCGN 8310 and 8320 were electroporated into A. tumefaciens strain EHA 105 and rapeseed (Brassica napus cv Quantum; Radke et al., 1988); Arabidopsis plants were also transformed. Transformation of Arabidopsis was by vacuum infiltration (Bechtold et al., 1993).
Antibody Production and Western-Blot Analysis
The pGEX-KG vector (Guan and Dixon, 1991) expression system, driven by the tac promoter, was utilized for in-frame cloning of the So KAS III coding region and subsequent production of the recombinant protein. The cDNA region of So KAS III encoding the mature portion of the polypeptide was amplified and the generated fragment was cloned into the NcoI site of linearized pGEX-KG vector. The plasmid was used to transform Escherichia coli BL21 (LysS). Subsequent induction led to the high levels of expression of the soluble KAS III-glutathione S-transferase (GST) fusion protein (Guan and Dixon, 1991). The fusion protein was purified using a glutathione sepharose 4B column and cleavage of KAS III from the GST was carried out with thrombin (8 μg L−1 culture) as directed by the manufacturer (Pharmacia, Piscataway, NJ). The soluble KAS III fraction subsequently was collected by centrifugation and separated from the agarose-bound GST. The residual contaminating thrombin was removed by salt gradient elution from a monoQ column (Clough et al., 1992) and the resulting affinity-purified recombinant KAS III was used in raising rabbit polyclonal antiserum.
Immunoblot analysis was performed using the previously described procedures (Kyhse-Andersen, 1984; Post-Beittenmiller et al., 1991). Crude protein extracts were prepared from several different transgenic sources: tobacco leaves, rapeseed developing seeds, and E. coli overexpressing either So KAS III or Ch KAS III-1. The tobacco leaf extract was prepared by grinding two to three leaf discs (6-mm diameter, 7–9 mg fresh weight/disc) in approximately 5× volume of extraction buffer (50 mm Tris-HCl [pH 8.0], 5 mm EDTA, 2 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 mm ε-amino-n-caproic acid, and 1% [w/v] polyvinyl polypyrrolidone). Protein concentration was determined by a bicinchoninic acid protein assay procedure using the manufacturer's protocol (Pierce, Rockford, IL). The remaining material (namely rapeseed developing seeds and E. coli cultures) was harvested and frozen; plant tissues were pulverized in liquid nitrogen. Materials were then mixed with the suspension buffer (357 mm Tris [pH 6.8], 14.25% glycerol, and 4.2% [w/v] SDS) and immediately heated in boiling water for 10 min and centrifuged at 14, 000 rpm for 10 min. The pellet was discarded and an aliquot of supernatant was used for protein determination using a commercially available assay system (DC Protein Assay, Bio-Rad Laboratories, Hercules, CA). The remaining portion of the supernatant was mixed with an equal volume of Laemmli gel sample buffer (Laemmli, 1970), heated for 3 min, and centrifuged at 14,000 rpm for 10 min in a microcentrifuge. Immunoblot analyses were performed on equal amounts of protein extracts separated electrophoretically on a 12% (w/v) polyacrylamide SDS gel. All blots were probed with immune as well as pre-immune serum. Levels as low as 30 ng of KAS III recombinant protein were detected, using 1:3,000 dilution of the antiserum, whereas pre-immune serum did not result in any detectable signal from any of the samples. For screening of the transgenic plants, typically 1,000× dilution of the antibody was used. The immunoblot analyses of other enzymes were carried out with antibodies for barley KAS I, castor seed BC subunit of ACCase, and soybean stearoyl-ACP Δ9 desaturase, which were kindly provided by Drs. Penny von Wettstein-Knowles (Carlsberg Laboratory, Copenhagen), John Ohlrogge (Michigan State University, East Lansing), and Narendra Yadav (E.I. DuPont de Nemours and Co., Wilmington, DE), respectively.
Analysis of Transgenic Tobacco Plants
DNA samples were prepared from approximately 100 mg of the leaves of putative transgenic plants (Dellaporta et al., 1983) and subjected to PCR using spinach (Spinacia oleracea) KAS III-specific primers.
The KAS III enzyme activity measurements was performed on crude protein extracts (above section) according to the previously described method (Clough et al., 1992).
Analysis of Fatty Acids
FAME were prepared from fresh tobacco leaves as well as tobacco, Brasicca, and Arabidopsis mature seeds using the method described by Browse et al. (1986). Tri-17:0 triacylglycerol was included as an internal standard. Analyses of the FAME were performed on a gas chromatograph (series 5890, Hewlett-Packard, Palo Alto, CA) equipped with a mass spectrophotometer (series 5971).
Analyses of Acyl-ACPs
To isolate ACPs, transgenic and wild-type tobacco leaves were frozen and pulverized in liquid nitrogen, and homogenized in 5% (v/v) trichloroacetic acid (TCA). After centrifugation at 13,000g the resulting pellet was washed in 1% (v/v) TCA and resuspended in 50 mm MOPS [3-(N-morpholino)propanesulfonic acid] (pH 7.6) and 10 mm N-ethylmaleimide before loading onto a gel. For the preparation of free ACP (ACP-SH), the 1% (v/v) TCA pellet was dissolved in a deacylation buffer [100 mm Tris-HCl (pH 9.0) and 100 mm dithiothreitol] and incubated for 1 h at 37°C. The de-acylated tobacco ACP was further concentrated on a DEAE column and subsequently used for preparation of 14C-labeled acetyl-ACP, malonyl-ACP, and KAS III reaction products. The reaction products of KAS III as well as acetyl-ACP standards were prepared enzymatically as described previously (Clough et al., 1992), employing [1-14C]-acetyl-CoA and an enzyme extract from transgenic tobacco leaf containing high levels of KAS III activity. Reducing agents, malonyl-CoA, and MCAT were omitted from the reaction for the production of acetyl-ACP. Preparation of malonyl-ACP was carried out enzymatically, using [2-14C]-labeled malonyl-CoA and MCAT (Stapleton and Jaworski, 1984). The acyl-ACPs were separated on a 15% (w/v) native PAGE and analyzed as described previously (Post-Beittenmiller et al., 1991). The radioactive acyl-ACPs were analyzed using ImageQuant software on a phosphorimager (Molecular Dynamics, Inc., Sunnyvale, CA).
Analysis of Other FAS Components
The total amounts of ACPs in the highest expressing transgenic lines and wild-type tobacco leaves were measured by acyl-ACP synthetase assay (Rock et al., 1981). The same plants were also examined for the levels of other FAS related enzymes. KAS I, BC subunit of plastid ACCase, and stearoyl-ACP Δ9-desaturase were determined by immunoblot assays.
Measurement of the Rate of Fatty Acid Synthesis and Determination of the Percentage of the Lipid Content
The wild type and T3 generation of pCGN 8310 lines with highest levels of 16:0 in their seeds were grown simultaneously in the greenhouse. To measure the rate of fatty acid synthesis, the embryos of developing seeds (30–35 DPA) were dissected. During the dissection, the embryos were incubated on ice in a buffer containing 25 mm MES [2-(N-morpholino)ethanesulfonic acid]-NaOH (pH 6), 25 mm Suc, and 100 mm sorbitol. Upon completion of the task, the buffer was replaced with a fresh batch with the addition of 25 mCi mL−1 of [3H]-water (18.50 GBq). For each seed type and time point, 10 replicate samples (10 embryos per sample) were then incubated at 30°C shaking at 200 rpm in the light (130 μE m−2 s−1). Assays were terminated after 6 or 12h by the addition of HCl to the final concentration of 0.1 m. The samples were then extracted in a sealed tube with 2 mL of chloroform-methanol, (1:1 [v/v]) for 1 h at 75°C. The lipid extracts were separated from the embryos and 2 mL of 1.0 m NaCl was added to the extracts to affect solvent phase separation. The upper aqueous phase from each was removed by aspiration, and the lower chloroform phase containing the lipid was recovered. These lipid extracts were converted to FAME with 2% (w/v) sulfuric acid in methanol at 60°C for 1 h, and methyl esters extracted into hexane for tritium analysis by liquid scintillation counting.
The lipid content from 18 transgenic events and six wild-type plants were measured by near-infrared reflectance spectroscopy (NIR) using Foss Nir model 6500 (FOSS NIRSystems, Inc., Silver Spring, MD) This method is used for simultaneous nondestructive determination of the total percentage of oil, total glucosinolates, and total percentage of protein content. The infrared reflectance of the unknown sample is measured against a known calibration set and the values are calculated using the equation generated from the calibration samples. For each measurement 100 seeds were packed in the sample chamber and loaded on the NIR instrument. Oil sample measurement using oil calibration and equation were carried out according to the manufacturer's instructions.
Other Chemicals
All enzymes were purchased from Promega (Madison, WI) or New England Biolabs (Beverly, MA). [1-14C]-acetyl-CoA (58 Ci mol−1) and [2-14C]-malonyl-CoA (56 Ci mol−1) were prepared from sodium-[14C]-acetate and [14C]-malonic acid, respectively, as described previously (Rutkoski and Jaworski, 1978; Clough et al., 1989).
ACKNOWLEDGMENTS
We would like to thank Drs. Penny von Wettstein-Knowles (Carlsberg Laboratory), John Ohlrogge (Michigan State University), and Narendra Yadav (E.I. DuPont de Nemours and Co.) for providing antibodies. We would like to extend our thanks to Paul Bertain (Calgene) for his help in dissecting the embryos and Sanjay Panda (Calgene) for performing the NIR measurements.
Footnotes
This work was supported in part by the National Science Foundation (grant no. MCB–9728786).
LITERATURE CITED
- An G, Ebert PR, Mitra A, Ha SB. Binary Vectors. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Acadmic Publishers; 1988. pp. A3:1–19. [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- Browse J, McCourt PJ, Somerville CR. Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem. 1986;152:141–145. doi: 10.1016/0003-2697(86)90132-6. [DOI] [PubMed] [Google Scholar]
- Bruck FM, Brummel M, Schuch R, Spener F. In vitro evidence for feed-back regulation of β-ketoacyl-acyl carrier protein synthase III in medium-chain fatty acid biosynthesis. Planta. 1996;198:271–278. [Google Scholar]
- Chen JPBD. Molecular cloning of a cDNA encoding 3-ketoacyl-acyl carrier protein synthase III from leek. Gene. 1996;182:45–52. doi: 10.1016/s0378-1119(96)00472-6. [DOI] [PubMed] [Google Scholar]
- Clough RC, Barnum SR, Jaworski JG. Synthesis of radiolabeled acetyl-coenzyme A from sodium acetate. Anal Biochem. 1989;176:82–84. doi: 10.1016/0003-2697(89)90276-5. [DOI] [PubMed] [Google Scholar]
- Clough RC, Matthis AL, Barnum SR, Jaworski JG. Purification and characterization of 3-ketoacyl-acyl carrier protein synthase-III from spinach: a condensing enzyme utilizing acetyl-coenzyme-A to initiate fatty acid synthesis. J Biol Chem. 1992;267:20992–20998. [PubMed] [Google Scholar]
- Dehesh K, Edwards P, Fillatti J, Slabaugh M, Byrne J. KAS IV: a 3-ketoacyl-ACP synthase from Cuphea sp. is a medium chain specific condensing enzyme. Plant J. 1998;15:383–390. doi: 10.1046/j.1365-313x.1998.00218.x. [DOI] [PubMed] [Google Scholar]
- Dehesh K, Edwards P, Hayes T, Cranmer AM, Fillatti J. Two novel thioesterases are key determinants of the bimodal distribution of acyl chain length of Cuphea palustris seed oil. Plant Physiol. 1996a;110:203–210. doi: 10.1104/pp.110.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh K, Jones A, Knutzon DS, Voelker TA. Production of high levels of 8:0 and 10:0 fatty acids in transgenic canola by overexpression of Ch FatB2, a thioesterase cDNA from Cuphea hookeriana. Plant J. 1996b;9:167–172. doi: 10.1046/j.1365-313x.1996.09020167.x. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Guan K, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione s-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. Fast and sensitive multiple sequence alignments on a microcomputer. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Jaworski JG, Clough RC, Barnum SR. A cerulenin-insensitive short chain 3-ketoacyl-acyl carrier protein synthase in Spinacia oleracea leaves. Plant Physiol. 1989;90:41–44. doi: 10.1104/pp.90.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski JG, Post-Beittenmiller D, Ohlrogge JB. Acetyl-acyl carrier protein is not a major intermediate in fatty acid biosynthesis in spinach. Eur J Biochem. 1993;213:981–987. doi: 10.1111/j.1432-1033.1993.tb17843.x. [DOI] [PubMed] [Google Scholar]
- Kauppinen S, Siggaard-Anderson M, von Wettstein-Knowles P. β-Ketoacyl-ACP synthase I of Escherichia coli nucleotide sequence of the fabB gene and identification of the cerulenin binding residue. Carlsberg Res Commun. 1988;53:357–370. doi: 10.1007/BF02983311. [DOI] [PubMed] [Google Scholar]
- Knutzon DS, Thompson GA, Radke SE, Johnson WB, Knauf VC, Kridl JC. Modification of Brassica seed oil by antisense expression of a stearoyl-acyl carrier protein desaturase gene. Proc Natl Acad Sci USA. 1992;89:2624–2628. doi: 10.1073/pnas.89.7.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridl JC, McCarter DW, Rose RE, Scherer DE, Knutzon DS, Radke SE, Knauf VC. Isolation and characterization of an expressed napin gene from Brassica rapa. Seed Sci Res. 1991;1:209–219. [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maliga P, Sz-Breznovits A, Marton L. Streptomycin-resistant plants from callus culture of haploid tobacco. Nat New Biol. 1973;244:29–30. doi: 10.1038/newbio244029a0. [DOI] [PubMed] [Google Scholar]
- Mcbride K, Summerfelt K. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol Biol. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- Niederberger P, Prasad R, Miozzari G, Kacser H. A strategy for increasing an in vivo flux by genetic manipulations: the tryptophan system of yeast. Biochem J. 1992;287:473–479. doi: 10.1042/bj2870473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG. Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller D, Jaworski JG, Ohlrogge JB. In vivo pools of free and acylated acyl carrier proteins in spinach: evidence for sites of regulation of fatty acid biosynthesis. J Biol Chem. 1991;266:1858–1865. [PubMed] [Google Scholar]
- Post-Beittenmiller D, Roughan G, Ohlrogge J. Regulation of plant fatty acid biosynthesis: analysis of acyl-CoA and acyl-acyl carrier protein substrate pools in spinach and pea chloroplasts. Plant Physiol. 1992;100:923–930. doi: 10.1104/pp.100.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller MA, Schmid KM, Ohlrogge JB. Expression of holo and apo forms of spinach acyl carrier protein-I in leaves of transgenic tobacco plants. Plant Cell. 1989;1:889–899. doi: 10.1105/tpc.1.9.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke SE, Andrews BM, Moloney MM, Crouch ML, Kridl JC, Knauf VC. Transformation of Brassica napus L. using Agrobacterium tumefaciens: developmentally regulated expression of reintroduced napin gene. Theor Appl Genet. 1988;75:685–694. [Google Scholar]
- Rock CO, Garwin JL, Cronan J, John E. Preparative enzymatic synthesis of acyl-acyl carrier protein. Methods Enzymol. 1981;72:397–403. doi: 10.1016/s0076-6879(81)72029-9. [DOI] [PubMed] [Google Scholar]
- Roesler K, Shintani D, Savage L, Boddupalli S, Ohlrogge J. Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol. 1997;113:75–81. doi: 10.1104/pp.113.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler P, Schneider C, Dunahay T. Overexpression of acetyl-CoA carboxylase in the alga Cyclotella cryptica (O-14). CA: Biochemistry and Molecular Biology of Plant Fatty Acid and Glycerolipids Symposium, South Lake Tahoe; 1995. [Google Scholar]
- Rogers S, Horsch R, Fraley R. Gene transfer in plants: production of transformed plants using Ti plasmid vectors. Methods Enzymol. 1986;118:627–641. [Google Scholar]
- Roughan P, Ohlrogge J. Evidence that isolated chloroplasts contain an integrated lipid-synthesizing assembly that channels acetate into long-chain fatty acids. Plant Physiol. 1996;110:1239–1247. doi: 10.1104/pp.110.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkoski A, Jaworski JG. An improved synthesis of malonyl-coenzyme A. Anal Biochem. 1978;91:370–373. doi: 10.1016/0003-2697(78)90854-0. [DOI] [PubMed] [Google Scholar]
- Schaaff I, Heinisch J, Zimmermann FK. Overproduction of glycolytic enzymes in yeast. Yeast. 1989;5:285–290. doi: 10.1002/yea.320050408. [DOI] [PubMed] [Google Scholar]
- Shimakata T, Stumpf PK. Isolation and function of spinach leaf β-ketoacyl-[acyl-carrier-protein] synthases. Proc Nat Acad Sci USA. 1982;79:5808–5812. doi: 10.1073/pnas.79.19.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani DK, Ohlrogge JB. Feedback inhibition of fatty acid synthesis in tobacco suspension cells. Plant J. 1995;7:577–587. [Google Scholar]
- Slabaugh MB, Tai H, Jaworski J, Knapp SJ. cDNA clones encoding β-ketoacyl-acyl carrier protein synthase III from Cuphea wrightii. Plant Physiol. 1995;108:443–444. doi: 10.1104/pp.108.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton SR, Jaworski JG. Characterization and purfication of malonyl-coenzyme A: [acyl-carrier-protein] transacylases from spinach and Anabaena variabilis. Biochim Biophys Acta. 1984;794:240–248. [Google Scholar]
- Tai H, Post-Beittenmiller D, Jaworski JG. Cloning of a cDNA encoding 3-ketoacyl-acyl carrier protein synthase III from Arabidopsis. Plant Phys. 1994;106:801–802. doi: 10.1104/pp.106.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai HY, Jaworski JG. 3-Ketoacyl-acyl carrier protein synthase-III from spinach (Spinacia oleracea) is not similar to other condensing enzymes of fatty acid synthase. Plant Physiol. 1993;103:1361–1367. doi: 10.1104/pp.103.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoert IIGS, Linden KHvd, Walsh MC, Nijkamp HJJ, Stuitje AR. Modification of Brassica napus seed oil by expression of the Esherichia coli fabH gene, encoding 3-ketoacyl-acyl carrier protein synthase III. Plant Mol Biol. 1995;27:875–886. doi: 10.1007/BF00037016. [DOI] [PubMed] [Google Scholar]
- Verwoert IIGS, Vanderlinden KH, Nijkamp HJJ, Stuitje AR. Developmental specific expression and organelle targeting of the Escherichia coli fabD gene, encoding malonyl coenzyme A-acyl carrier protein transacylase in transgenic rape and tobacco seeds. Plant Mol Biol. 1994;26:189–202. doi: 10.1007/BF00039531. [DOI] [PubMed] [Google Scholar]
- Voelker TA, Worrell AC, Anderson L, Bleibaum J, Fan C, Hawkins DJ, Radke SE, Davies HM. Fatty acid biosynthesis redirected to medium chains in transgenic oilseed plants. Science. 1992;257:72–74. doi: 10.1126/science.1621095. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cronan JE. Polar allel duplication for transcriptional analysis of censecutive essential genes: Application to a cluster of Escherichia coli fatty acid biosynthetic genes. J Bacteriol. 1996;178:3614–3620. doi: 10.1128/jb.178.12.3614-3620.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]