SUMMARY
Chronic inflammation drives the progression of colorectal cancer (CRC). Increased expression of interleukin (IL)-17A is associated with poor prognosis, and IL-17A blockade curbs tumor progression in preclinical models of CRC. Here we examined the impact of IL-1 signaling, a key regulator of the IL-17 pathway, in different cells types within the CRC microenvironment. Genetic deletion of the IL-1 receptor (IL-1R1) in epithelial cells alleviated tumorigenesis in the APC model of CRC, demonstrating a cell-autonomous role for IL-1 signaling in early tumor seed outgrowth. T cell specific ablation of IL-1R decreased tumor-elicited inflammation dependent on IL-17 and IL-22, thereby reducing CRC progression. The pro-tumorigenic roles of IL-1 were counteracted by its effects on myeloid cells, particularly neutrophils, where IL-1R1 ablation resulted in bacterial invasion into tumors, heightened inflammation and aggressive CRC progression. Thus, IL-1 signaling elicits cell type-specific responses which, in aggregate, set the inflammatory tone of the tumor microenvironment and determine the propensity for disease progression.
Keywords: Cytokine, cell type specific signaling, tumor elicited inflammation, tumor microenvironment, Interleukin 1, colorectal cancer, microbiota
INTRODUCTION
A particularly important component of the pro-tumorigenic microenvironment is the presence of inflammatory cells and the subsequent chronic inflammatory response. Although chronic inflammation is a well-established driver in many cancers (Mantovani et al., 2008), inflammation also appears to play a deleterious role in cancers without overt underlying inflammatory etiologies. This phenomenon, characterized by a tumor’s ability to recruit inflammatory immune cells to the local microenvironment to drive both tumor growth and progression, is referred to as tumor-elicited inflammation (TEI) (Grivennikov et al., 2012). The relevance of TEI is illustrated by the efficacy of anti-inflammatory treatments in reducing mortality from cancers that are classically thought of as ‘non-inflammatory’ (Rothwell et al., 2012).
In sporadic colorectal cancer (CRC), the breakdown of the epithelial barrier subsequent to oncogenic transformation leads to the translocation of bacterial products and triggers the interleukin (IL)-23–IL-17A signaling pathway (Grivennikov et al., 2012). Increased amounts of these cytokines within tumors portends poor prognosis in CRC patients (Tosolini et al., 2011) and promotes CRC in various animal models (Blatner et al., 2012; Grivennikov et al., 2012; Huber et al., 2012; Kirchberger et al., 2013). In humans and mice, however, not all tumors expressing high amounts of the pro-tumorigenic cytokine IL-23 have commensurately high IL-17A expression – implying that other regulators of TEI exist. Overall, a complete mechanistic understanding of TEI induction, its maintenance, and its contribution to tumorigenesis is lacking.
IL-1 plays an essential role in inducing IL-17 production by T helper (Th17) cells in both humans and mice in the context of inflammation and autoimmunity (Coccia et al., 2012; Zhou and Littman, 2009). Approaches to block the IL-1 pathway are being used to treat inflammatory conditions such as rheumatoid arthritis, gout and familial Mediterranean fever (Dinarello et al., 2012). Despite its well-known role in maintaining immune-mediated inflammatory responses, the role of IL-1 signaling as a regulator of TEI and CRC in autochthonous models is obscure. IL-1 signaling is implicated in the generation of both pro- or anti-tumor immune responses, as well as in the development of intestinal inflammation linked to tumorigenesis (Bersudsky et al., 2014; Malik et al., 2016; Voronov et al., 2003). A recent CANTOS trial testing the ability of the anti-IL-1β antibody canakinumab to prevent adverse cardiovascular events revealed a protective effect of systemic IL-1β inhibition in lung cancer, but not colon cancer (Ridker et al., 2017).
Here, we used cell-type specific inactivation of IL-1R1 to examine the impact of IL-1 on distinct cells within the CRC tumor microenvironment. IL-1 signaling controlled CRC development and progression by three distinct mechanisms. In epithelial cells, IL-1 signaling directly drove tumorigenesis without affecting inflammatory responses. Deletion of Il1r1 in T cells blocked IL-17 and IL-22 production, reducing TEI and blunting CRC tumorigenesis. These pro-tumorigenic effects of IL-1 were countered by IL-1 signaling in myeloid cells, specifically neutrophils. Deletion of Il1r1 in myeloid cells resulted in increased infiltration of bacteria into the tumor tissue, and the triggering of a pro-tumorigenic inflammatory response resembling tumor elicited inflammation. The complex effects of IL-1 in distinct cell type within the tumor microenvironment may underlie the distinct results in bacteria-rich and other cancers upon IL-1 blockade.
RESULTS
Increased IL-1α and IL-1β expression in CRC tumors promotes IL-17A responses
We operated under the premise that potential regulators of TEI should be increased in tumor tissue and should be able to control processes associated with inflammatory responses in a variety of cell types. We used the CDX2Cre-Apcf/wt (CPC-APC) mouse model of conditional monoallelic APC loss in the colon to induce CRC. We found elevated Il1a, Il1b and Il17a gene expression in tumors compared to normal colon tissue (Figure 1A). Multiplex assay of tumor culture supernatants showed increased protein concentration of all three cytokines, compared to matched normal tissues (Figure 1B). Other members of the IL-1 cytokine family were detected in CRC (Figure S1A,B). FACS sorting of specific cell populations from normal and tumor tissue with subsequent Q-RT-PCR analysis revealed that monocytes are the main source of IL-1α and IL-1β (Figure S1C), although tumor epithelial and stromal cells also were producing IL-1 (Figure S1D).
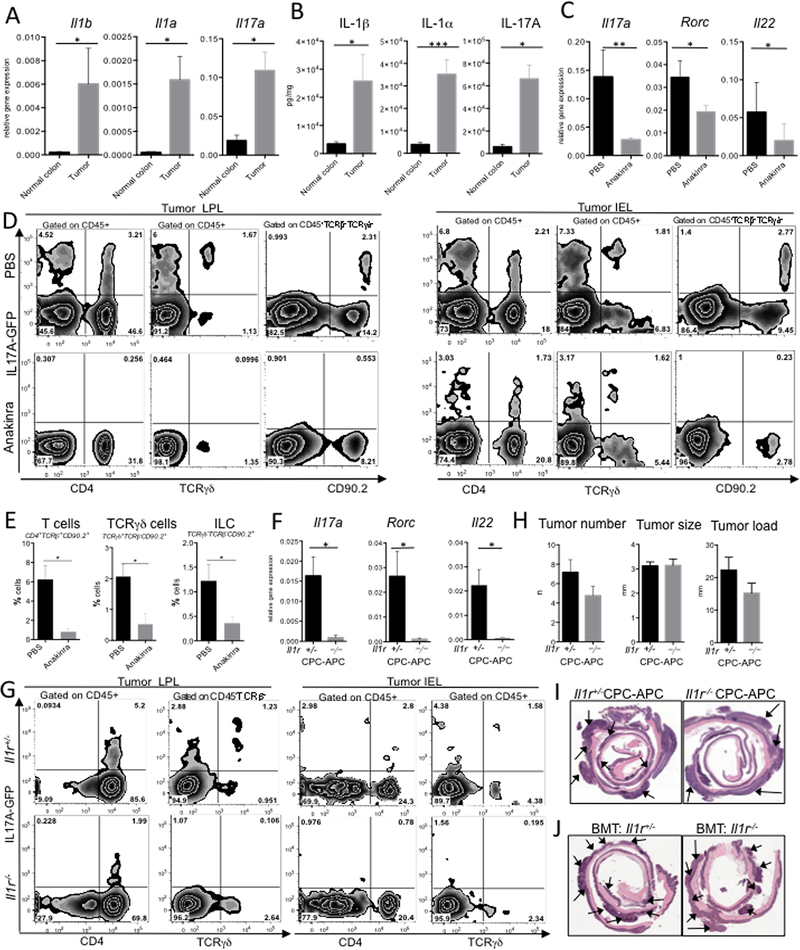
Figure 1. Global inactivation of IL-1R reduced IL-17A responses but only minimally affects CRC tumorigenesis.
(A) Il1α, Il1β and Il17a mRNA expression was normalized to housekeeping RpL32 gene in normal colon and CRC tumors from CDX2 (CPC)-APC mice. N≥10 (B) ELISA for IL-1α, IL-1β and IL-17A protein concentration in ex vivo normal colon and tumor culture supernatants (24h incubation), N≥5. (C-E) CRC tumors in CPC-APC-Il17aGFP mice treated with Anakinra or PBS control. (C) Q-RT-PCR analysis of Il17a, Rorc and Il22 mRNA expression in tumors, N≥7. (D) Representative FACS plot (N≥5) of GFP expression and (E) quantification in LPL and IEL fractions from CRC-bearing mice ± Anakinra. Gating strategy is indicated on the panels. (F-I) Analysis of tumor development in global Il1r1 (Il1r) −/− and Il1r1+/− six-month-old tumor bearing CPC-APC mice. (F) Q-RT-PCR analysis of TEI cytokines and markers in tumors, p*<0.05; N≥10. (G) LPL and IEL fractions of tumors from CPC-APC-Il17aGFP mice with Il1r−/− and Il1r+/− alleles were analyzed by FACS. Representative of three independent experiments, total N≥10. (H) Tumor multiplicity, load and size in Il1r+/− and Il1r−/− CPC-APC mice, N≥17 (I) H&E stained paraffin sections of colons from CPC-APC- Il1r−/− and Il1r+/− mice, tumors are pointed, representative of N≥17 (J) Analysis of CRC in 6–8 weeks old CPC-APC mice reconstituted with Il1r1−/− or Il1r1−/− bone marrow (BM) and allowed to develop CRC for 4 months. H&E stained paraffin sections of colons from Il1r1−/− or Il1r1−/− BM chimera CPC-APC mice, representative of N≥10. Data are mean ± SEM. Representative of 3 independent experiments. See also Figures S1, S2.
Data from in vitro models of mouse and human Th17 cell differentiation, and from in vivo models of chronic inflammation, positions IL-1 as an important regulator of Th17 differentiation and IL-17A production (Chung et al., 2009; Coccia et al., 2012). Therefore, we sought to examine whether IL-1 signaling is required for generating and maintaining the IL-17A TEI responses in CRC. To check whether pharmacological blockade of IL-1 signaling resulted in decrease of TEI cytokines in vivo, reporter CPC-APC-Il17agfp+/− mice were injected i.p. with the recombinant IL-1R antagonist Anakinra (IL-1Ra) for 3 consecutive days. Two hours after the last injection tumors and normal tissues were isolated and subjected to Q-RT-PCR and flow cytometry analyses. In tumor tissue, Q-RT-PCR analysis revealed a significant drop in mRNA expression for Il17a, its transcription factor Rorc and another TEI-related cytokine Il22 compared to normal colon (Figure 1C). IL-17A-GFP reporter expression was confined to various lymphoid (Figure 1D) but not epithelial, stromal or myeloid cells (Figure S1E). IL-1Ra treatment significantly reduced IL-17A-GFP reporter expression by the lymphoid cells (Figure 1D,E). Therefore, IL-1 signaling is required for IL-17A and TEI cytokine expression and even short-term inhibition of IL-1 resulted in reduction of intratumoral IL-17A and TEI in CRC.
Genetic inactivation of IL-1 signaling reduces IL-17A TEI but has only a limited effect on CRC tumorigenesis.
Next, we ablated IL-1R1 expression by crossing CPC-APC mice to Il1r1−/− background. Il1r1−/−CPC-APC mice and their co-housed littermate Il1r1+/−CPC-APC controls were allowed to spontaneously develop CRC. Genetic “whole body” ablation of IL-1R1 resulted in significant drop of Il17a, Rorc and Il22 mRNAs (Figure 1F). Flow cytometry analysis of intratumoral LPL and IEL fractions from Il1r1−/− CPC-APC-Il17aGFP+/− reporter mice revealed that IL-17A expression was reduced in the absence of IL-1 signaling (Figure 1G).
We and others previously reported that inactivation of IL-17A, RORγt or IL-22 results in a prominent reduction in CRC (Blatner et al., 2012; Grivennikov et al., 2012; Huber et al., 2012; Kirchberger et al., 2013; Wu et al., 2009). Thus, we expected that the significant decrease in IL-17A and IL-22 cytokine expression in Il1r1−/−CPC-APC mice would culminate in reduced CRC. However, tumor multiplicity was only modestly reduced and tumor size was not affected by IL-1R1 deficiency, leaving tumor load essentially unchanged (Figure 1H,I).
As hematopoietic cells are the main source of IL-17A and IL-22 (Figure 1D and S1E), hypothetically the ability of IL-1 to regulate TEI should be more evident within the hematopoietic compartment. Therefore, we lethally irradiated 6-week old CPC-APC mice and reconstituted them with bone marrow (BM) from Il1r1−/− and Il1r1+/+ control mice. Similarly, to what we observed in the whole-body Il1r1 deficiency, lack of IL-1R1 on hematopoietic cells led to a significant decrease in Il17a, Rorc and Il22 mRNA expression and protein release (Figure S2A, B and not shown) in CRC tumors. Intracellular cytokine staining confirmed the reduction in pro-tumorigenic IL-17A (Figure S2C, D), however hematopoietic IL-1R1 deficiency did not affect CRC tumor development (Figure 1J, S2E).
To confirm these unexpected results, we used an inducible CRC model, CDX2-ERCre (CDX2ERT)-Apcf/f, where bi-allelic deletion of the Apc gene and rapid development of CRC tumors are driven by injection of tamoxifen (Feng et al., 2013; Wang et al., 2014). Recipient CDX2ERT-Apcf/f mice were lethally irradiated, injected with Il1r1−/− or Il1r1+/− BM, allowed to recover for 2 months during which their hematopoietic system was replaced (over 90% chimerism), and then injected with tamoxifen. No reduction in CRC was found in mice with hematopoietic deficiency of IL-1 signaling (Figure S2F) whereas TEI factors Il17a, Rorc and Il22 expression were reduced (Figure S2G), as was the production of the IL-17A protein (Figure S2H).
Together, the results from two models of CRC revealed that IL-1 signaling in hematopoietic cells controlled key components of TEI, but global or hematopoietic cell specific “blanket” inactivation of IL-1R did not significantly alter CRC tumorigenesis.
IL-1R1 expressed by epithelial cells is required for CRC tumorigenesis, NF-κB activation and cell proliferation independent of TEI
We next hypothesized that IL-1R signaling in particular cells may account for the pro-tumorigenic inflammatory responses, but IL-1 activity in other, distinct cell types may have contrasting effects on CRC, ultimately neutralizing each other. We ablated IL-1R1 expression in colonic and tumor epithelial cells (Il1r1D/f-CDX2Cre-Apcf/WT – mice with epithelial deletion of Il1r1, Il1r1D/WT-CDX2Cre-Apcf/WT-controls, where ‘D” is deleted allele originated from loxP/Cre mediated excision of “floxed” allele). Q-RT-PCR analysis for Il1r1 exon 5 (excised by loxP/Cre recombination) demonstrated efficient deletion of Il1r1 in intestinal epithelial cells (Figure S3A). We found that epithelial specific IL-1R1 inactivation resulted in a pronounced reduction in CRC multiplicity and load (Figure 2A-C). These changes were not linked to alterations in TEI cytokine responses, as intratumoral mRNA expression of Il17a, Il17f, Rorc and Il22 were indistinguishable between epithelial Il1r1 deficient mice (D/f) and control (D/WT) mice (Figure 2D). Therefore, epithelial-specific Il1r1 deletion uncouples the role of IL-1 signaling in TEI induction from its role in direct tumor promotion in colonic epithelial cells.
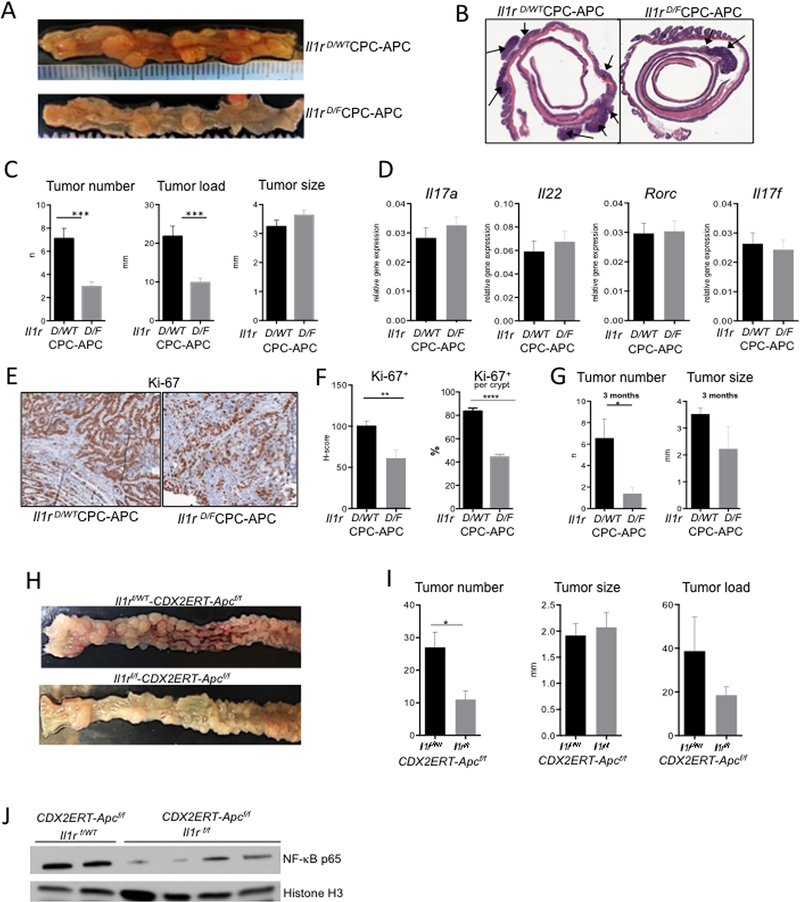
Figure 2. IL-1R in colonic epithelial cells plays a direct pro-tumorigenic role independent of inflammation.
CRC development in 6-month-old Il1r D/F-CPC-APC (epithelial deficiency) and Il1r D/WT (control) mice. (A) Representative macroscopic images of tumor-bearing colons and (B) microphotographs of H&E stained colons of indicated genotypes. (C) CRC tumors were assessed macroscopically upon necropsy of mice with epithelial IL-1R deficiency and controls, N≥21, ***p<0.0001. (D) Q-RT-PCR analysis of Il17a, Rorc, Il22 and Il17f mRNA expression in tumors, N=14, NS (E) Representative images of Ki-67 staining in tumors at 3 months of age. (F) Quantification (H-score) of Ki-67+ cell density per crypt in tumor area (N=4 mice) per genotype in CRC tumors of “young” Il1r D/F -CPC-APC and Il1r D/WT mice **p<0.01 ****p<0.0001. (G) CRC tumors of 3-month-old Il1r D/F-CPC-APC and Il1r D/WT control mice, N=8 *p<0.01. (H-J) CRC analysis in Il1rf/f-CDX2ERT-Apcf/f and Il1rf/WT control mice, injected with tamoxifen and allowed to develop tumors for 5–6 weeks. (H) Representative images of CRC bearing colons (I) Tumor analysis N≥5, *p<0.05 (J) Western Blot analysis of nuclear lysates of intestinal epithelial tumor cells (IECs) from indicated Il1r-CDX2ERT-Apcf/f mice for NF-κB (p65) expression. Loading was normalized to histone H3. Each lane shows tumors from individual mice. Data are mean ± SEM. Representative of 3 independent experiments. See also Figure S3.
The direct effect of IL-1 signaling in epithelial cells on CRC was linked to epithelial cell proliferation, as Ki-67 staining revealed that tumors from young CDX2Cre-Apcf/WT-Il1r1D/f (Il1r1D/f -CPC-APC) mice were less proliferative (Figure 2E, F), resulting in tumor size trending lower than in controls (Figure 2G). Therefore, epithelial IL-1 signaling regulates early proliferation and survival of CRC cells. This difference in proliferation and tumor size was no longer observed at 6 months (Figure 2C, S3B). The inducible model of CRC with Il1r1 epithelial deletion, Il1r1f/f-CDX2ERT-Apcf/f mice, also showed decreased tumor number and load compared to littermate Il1r1f/WT-CDX2ERT-Apcf/f- controls (Figure 2H, I).
Mechanistically, epithelial Il1r1 deficiency led to a decreased nuclear accumulation of the NF-κB p65 subunit in the tumor epithelial fraction in both the inducible (Figure 2J) and spontaneous models (Figure S3C). Thus, IL-1R in a cell autonomous manner is essential for NF-κB activation in the tumor epithelium. Proliferation of early tumor seeds was also reduced in vivo in the absence of epithelial IL-1R signaling (Figure S3D, E). As NF-κB (Schwitalla et al., 2013) and IL-1β in vitro (Kaler et al., 2009) enhance β-catenin signaling, we assessed whether the loss of epithelial IL-1R would reduce β-catenin activation. Immunofluorescent analysis had shown decreased expression and nuclear accumulation of β–catenin in early transformed epithelial tissues of tamoxifen-treated Il1r1f/f-CDX2ERT-Apcf/f mice (Figure S3F, G). Overall data suggested that IL-1 signaling in epithelial cells plays an essential role in tumor initiation.
IL-1 signaling in T cells drives IL-17A TEI and CRC
We next sought to resolve how hematopoietic IL-1 signaling could control CRC-promoting TEI, but not CRC itself. As CD4+ Th17 cells are essential for producing IL-17A, we assessed CRC tumorigenesis in CRC-prone mice devoid of IL-1R expression specifically in T cells (CD4Cre-Il1r1f/f). Because Cre is required both to induce colon-specific Apc deletion, and in CD4Cre-Il1r1f/f mice to delete Il1r1 in T cells, we could not intercross mice without also introducing an Il1r1 mutation in the epithelium and an Apc mutation in the T cells. To circumvent this issue, we transferred BM from in CD4Cre-Il1r1f/f (Cre+ (Il1r1 deficiency in T cells) and Cre- (control)) mice into lethally irradiated CPC-APC recipients. This allowed us to achieve specific deletion of IL-1R1 in T cells, particularly intratumoral T cells, as sorted T cells from CRC tumors of CD4Cre+-Il1r1f/f->CPC-APC mice displayed a significant deletion of Il1r1 gene exon 5 deletion (Figure S4A, B). We found that inactivation of IL-1R1 in T cells reduced CRC (Figure 3A-C). This coincided with the reduction of mRNA and protein expression of tumor-promoting IL-17A, RORγt and IL-22 (Figure 3D, E). Furthermore, use of RORγt-GFP reporter mouse strain revealed that blocking IL-1R signaling with Anakinra/IL-1Ra specifically reduced RORγt reporter expression in CD4+ intratumoral T cells (Figure 3F); and mRNA of Rorc and Il17a in T cells were correspondingly reduced (Figure S4C, D).
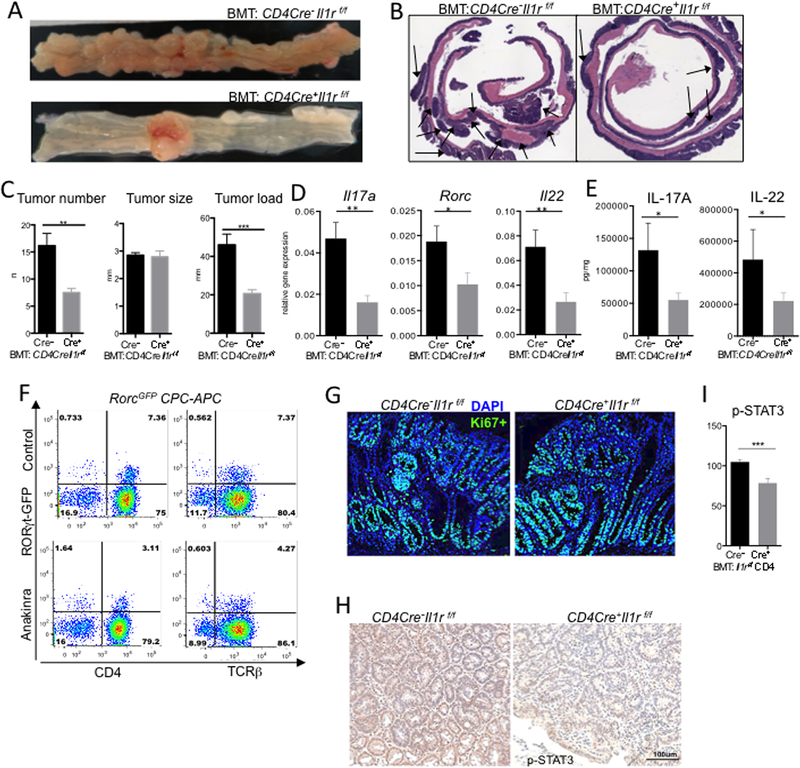
Figure 3. IL-1R signaling in T cells promotes IL-17A dependent TEI and CRC.
CPC-APC mice were transplanted with BM from CD4Cre+Il1rf/f or control mice and allowed to develop CRC for 4.5 months, representative of 3 experiments with 6–8 mice each (A) Macroscopic images of CPC-APC mice transplanted with CD4Cre+-Il1rf/f or control BM. (B) H&E stained CRC bearing colonic rolls (C) CRC tumor multiplicity, load and size, N≥6 per group, **p<0.01 ***p<0.001. (D) Q-RT-PCR analysis of Il17a, Rorc and Il22 mRNA expression in tumors N≥28 *p<0.01 **p<0.01. (E) Multiplex ELISA analysis of tumor culture supernatants (24h) for IL-17A and IL-22 proteins. (F) Representative FACS plot (N=5) of LPL tumor fraction from RorcGFP-CPC-APC reporter mice treated with Anakinra or PBS (control) for 3 days. (G) Representative IF images (N≥5) of CRC tumors stained for Ki-67. (H,I) IHC staining of paraffin-embedded CRC tumors for p-STAT3 (I) Quantification of p-STAT3+ cell density (H-score) in CRC tumors from BM-transplanted CPC-APC mice. N=4, ***p<0.001. Data are means ± SEM. See also Figure S4.
In mice lacking IL-1R1 on T cells, well developed CRC tumors did not show obvious differences in Ki-67 expression compared to wild-type controls (Figure 3G, H). However, activation of the oncogenic factor STAT3 in tumor cells was reduced (Figure 3I), presumably via loss of STAT3 activating cytokine expression. A reduction in CRC was also observed in the inducible model of CRC (Figure S4E-G) accompanied by reduced Il17a, Rorc and Il22 expression (Figure S4H). Therefore, IL-1 signaling in T cells was required for the expression of RORγt in tumors, which induced pro-tumorigenic cytokines IL-17A and IL-22 and TEI-driven tumorigenesis.
Myeloid cell specific IL-1 signaling controls tumor microbial infiltration and prevents CRC development
Hematopoietic deletion of IL-1R1 did not alter CRC, however its specific inactivation in T cells did reduce tumors. We therefore hypothesized that IL-1 signaling in an as-yet-unidentified hematopoietic population possesses an anti-tumorigenic role. We inactivated IL-1R1 in myeloid cells (monocytes, macrophages and neutrophils) by creating CD11bCre-Il1r1f/f mice and confirmed specific Il1r1 exon 5 deletion (Figure S5A, B). We then used these mice to produce BM chimeras in CDX2ERT-Apcf/f mice, generating CRC-prone mice lacking IL-1R1 on myeloid cells (CD11bCre+Il1r1f/f⟶CDX2ERT-Apcf/f) and controls (CD11bCre+Il1r1f/WT (or WT) ⟶ CDX2ERT-Apcf/f). Surprisingly, mice lacking Il1r1 in myeloid cells had a higher tumor multiplicity and size (Figure 4A-C) accompanied by increased cell proliferation (Figure 4D). Moreover, these tumors displayed increased inflammation, as determined by Q-RT-PCR analysis for Il17a, Il17f, Rorc, Ptgs2 (encodes for COX2) and Il22 (Figure 4E). In agreement, increased protein levels of IL-17A and IL-22 were detected in ex vivo tumor culture supernatants (Figure 4F).
Figure 4. IL-1R signaling in myeloid cells is a negative regulator of bacterial invasion and TEI.
CDX2ERT-Apcf/f mice were transplanted with BM from CD11bCre+Il1rf/f (myeloid IL-1R deletion) or CD11bCre+Il1rf/w (heterozygous, control) mice, allowed to reconstitute BM for 2 months, injected with tamoxifen and analyzed in 6 weeks. (A) Representative images of CRC bearing colons from CDX2ERT-Apcf/f mice transplanted with indicated BM (B) Representative microphotographs of H&E stained colon sections. (C) Tumor analysis, N≥9, p<0.03. (D) IF Ki-67 staining of paraffin-embedded CRC sections from indicated genotypes, representative of N≥5 (E) Q-RT-PCR analysis of Il17a, Rorc, Il22, Il17f and Ptgs2 (encodes for COX2) gene expression in CRC, N=18 **p<0.0019. (F) Multiplex ELISA analysis of CRC tumor lysates for IL-17A and IL-22 proteins, N=13 *p<0.05. (G) Q-RT-PCR for bacterial infiltration with primers specific for 16S rRNA from indicated bacteria, normalized to mouse housekeeping gene (RpL32), N=18 *p<0.05. (H) Scheme of experiment for antibiotic-mediated microbiota depletion. CD11bCre+Il1rf/f or control BM-chimeric CDX2ERT-Apcf/f mice were injected with tamoxifen and put on broad-spectrum antibiotics or regular water for 4 weeks. (I) Representative images of CRC bearing colons from mice treated or untreated with antibiotics. (J) Tumors upon necroscopy after 4 weeks of antibiotics treatment, N≥6 (K) Q-RT-PCR analysis of total eubacteria 16S rRNA, SFB and Il17a mRNA levels in CRC tumors of mice with or without antibiotics; N≥6, **p<0.008. Data are mean ± SEM. Representative of 3 (A-G) or 2 (G-K) experiments. See also Figure S5.
Enhancement of Th17 inflammatory responses could not be explained by the lack of direct action of IL-1 on myeloid cells, as myeloid cells did not directly contribute to IL-17A production (Figure S1D). We therefore reasoned that IL-1R1 deficiency in myeloid cells triggered additional stimuli promoting IL-17A production. As various intestinal microbial species are essential for induction of IL-17A (Ivanov et al., 2009; Wu et al., 2009), we tested if alterations in CRC-associated microbes were responsible for the heightened CRC. Q-RT-PCR for generic bacterial 16S rRNA revealed an increased bacteria presence in tumors from mice lacking IL-1R1 on myeloid cells (Figure 4G) - with specific increase in bacteria included E. coli, Enterobacteroides and SFB (Figure 4G). We further tested whether short term IL-1 blockade with Anakinra (IL-1RA) would induce the same bacterial perturbations. Anakinra treatment for 12 days did increase bacterial infilitration of CRC tumors, particularly with E. coli (Figure S5C), but did not alter CRC tumorigenesis (Figure S5D, E).
Tumor associated bacteria are essential for driving enhanced CRC in the absence of IL-1 signaling in myeloid cells.
To establish a functional and causative relationship between myeloid IL-1R1 inactivation, increased bacterial presence, inflammation and CRC, we treated CD11bCre+Il1r1f/f ⟶CDX2ERT-Apcf/f and control mice with broad spectrum antibiotics for 4 weeks, starting after the epithelial transformation (Figure 4H). Ablation of intestinal and tumor associated microbiota resulted in a drastic reduction of CRC tumorigenesis in both control and myeloid IL-1R deficient mice (Figure 4 I-K). Bacterial 16S RNA, SFB 16S RNA and Il17a mRNA in tumors from myeloid IL-1R1 deficient mice treated with antibiotics were strongly reduced compared to the untreated group (Figure 4K). Importantly, difference in CRC was no longer observed between antibiotic-treated control and myeloid IL-1R1 deficient animals (Figure 4J-K).
To ascertain whether increased IL-17A/IL-22 response in tumors in mice lacking IL-1R1 in myeloid cells was driven in part by increased IL-1 action in T cells and increased IL-17A production, we created CD11bCre-CD4Cre-Il1r1f/f “double Cre” mice and then generated CRC-bearing BM chimeras. Additional ablation of IL-1R signaling in T cells reduced the heightened tumorigenesis observed in myeloid Il1r1−/− mice, and reduced the production of pro-tumorigenic IL-17A (Figure S5F, G). Moreover, in independent azoxymethane/dextrane sulfate sodium (AOM/DSS) induced model of colitis-associated cancer CD4Cre+Il1r1f/f mice displayed lower tumorigenicity, while CD11bCre+Il1r1f/f mice showed higher tumorigenicity in comparison with controls (data not shown).
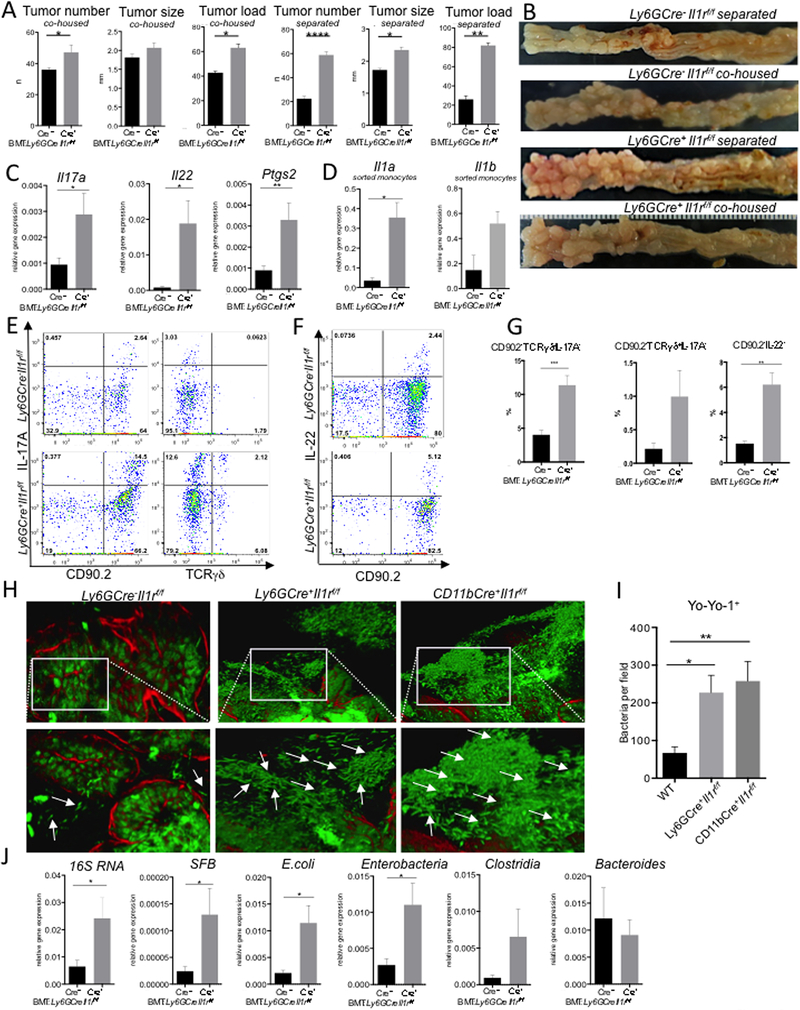
To independently confirm protective role of IL-1R1 in myeloid cells, we next generated LysMCreCre/WT (referred to as LysMCre+) and LysMCre− Il1r1f/f (control) mice and used their BM to create chimeric CDX2ERT-Apcf/f mice, to delete IL-1R1 using an overlapping Cre-deleter. LysMCre, similarly to CD11bCre, excised Il1r1 in monocytes, macrophages and neutrophills (Figure S5A, B). LysMCre+Il1r1f/f->CDX2ERT-Apcf/f mice also developed more CRC (Figure 5A, B). The difference was even more notable when myeloid IL-1R1 deficient mice and controls were housed separately as co-housing allowed microbiota equalization (Figure 5 A, B). LysMCre+Il1r1f/f->CDX2ERT-Apcf/f mice also demonstrated increased mRNA expression of Il17a, Il22 and Ptgs2 (Figure 5C) as well as bacterial 16S rRNA, E. coli and SFB in tumors (Figure 5D). Moreover, Yo-Yo 1 DNA staining of whole mount colons revealed an increase in bacteria located on the immediate surface of the tumor (Figure 5E).
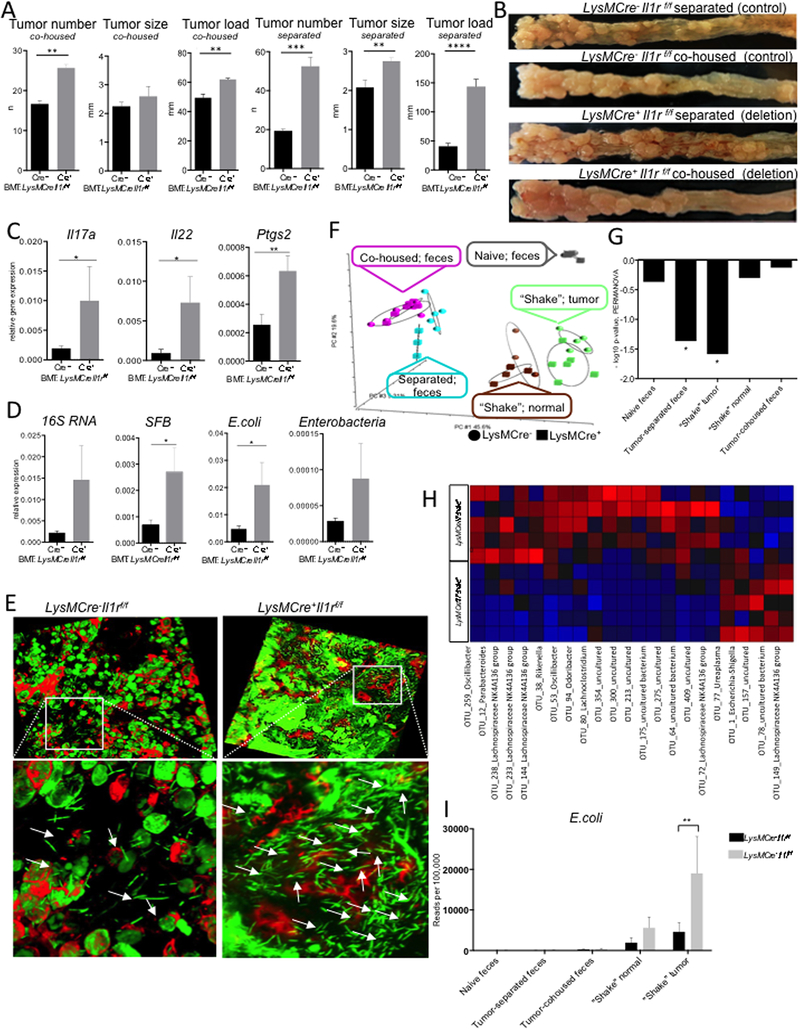
Figure 5. Myeloid IL-1R signaling prevents tumor associated dysbiosis.
(A-I) CDX2ERT-Apcf/f mice were reconstituted with BM from LysMCre+Il1rf/f or Cre−(control) mice and CRC was induced with 2 injections of tamoxifen. (A) Macroscopic tumor multiplicity, size, and load analysis upon necropsy 6 weeks after last tamoxifen injection, for mice which are either co-housed or housed separately according to BM genotype. N≥5, p<0.05. (B) Representative images of CRC bearing colons of − BM transplanted mice (C) Q-RT-PCR analysis of Il17a, Il22 and Ptgs2 mRNA expression in CRC tumors, N=20, *p<0.05, **p<0.007. (D) Q-RT-PCR analysis of CRC tumor lysates for specific bacterial content indicated; results are normalized to RpL32 expression N=20 *p<0.05. (E) IF images of adhesive bacteria (white arrows) associated with tumors from LysMCre+Il1rf/f or control mice. YoYo-1 and phalloidin staining of whole colon tissue from indicated mice. Representative of 3 independent experiments, each of at least 5 tumors. (F-I) 16S RNA sequencing of fecal, normal tissue and tumor adhesive bacteria isolated from naive and tumor bearing LysMCre+Il1rf/f and control mice, which were either co-housed or separately housed (F) Principal component analysis of different groups of bacteria (G) PERMANOVA analysis between the groups, * p<0.05 (H) Heatmap of altered (p<0.05) bacteria in tumor-adhesive fraction of LysMCre+Il1rf/f mice “Red”-overrepresented; “Blue- “underrepresented”. (I) Specific representation of E. coli bacteria in different groups, N=5. See also Figure S5.
As CRC load in genetically identical animals were affected by their housing status and bacteria attached to the tumors of myeloid Il1r1−/− mice, we next characterized the microbial alteration induced by the absence of IL-1R in myeloid cells. We performed Illumina MiSeq 16S rRNA sequencing on DNA samples from the colonic contents of naive mice, co-housed and separated tumor bearing mice, as well as from the epithelial „shakes” (mechanical isolation of epithelium-adherent bacteria) of normal and tumor tissue from control and LysMCre+Il1r1f/f mice. Principal component analysis (PCA) demonstrated that myeloid specific IL-1R ablation did not lead to a global changes in fecal microbiota in naive mice (Figure 5F). This was largely confirmed by 16S Q-RT-PCR of naive colonic tissue (Figure S5H). CRC formation did induce differences in fecal microbiota between wild type and myeloid Il1r1−/− mice, but that difference was not evident if mice were co-housed (Figure 5F). On the other hand, significant differences were detected in the microbiota composition of bacterial adherent to the tumor tissue, but not to the normal tissue in LysMCre+ mice (Figure 5F,G). PERMANOVA analysis established that the only differences between LysMCre+Il1r1f/f mice and controls mice was in tumor-adherent bacteria and in fecal microbiota, when mice were housed separately (Figure 5G). Among increased bacteria we found Escherichia-Shigella genera, represented by E. coli in the tumor-adherent fraction of myeloid cell specific Il1r1−/− mice (Fig 5H, I), also detected by Q-RT-PCR analysis of tumor tissue (Fig 4G, 5D). It was likely that differences in fecal but not in tumor-associated microbiota could be mitigated by co-housing. Together this suggested that myeloid IL-1R signaling repressed bacterial invasion and tumor associated infammation.
Neutrophil specific IL-1R signaling controlled bacterial invasion and tumor elicited inflammation.
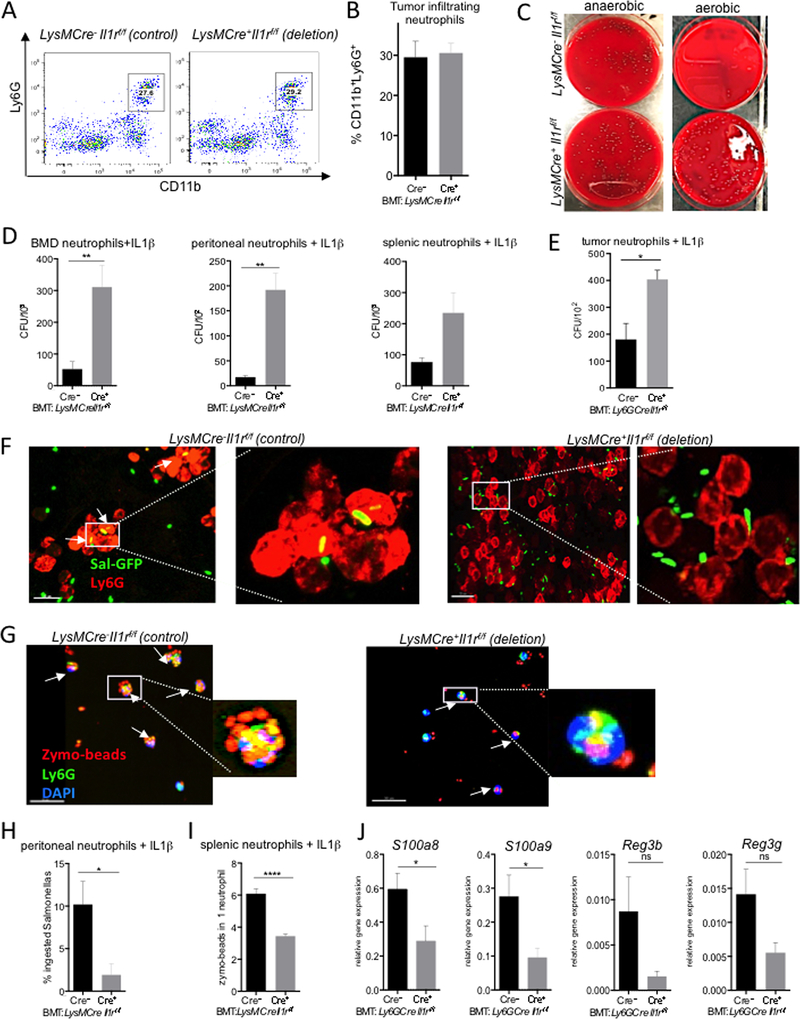
To address whether IL-1R expression by monocytes/macrophages or by neutrophils control CRC, we first targeted IL-1R1 in monocytes and macrophages by injecting BM from CX3CR1CreCre/WT(Cre+)Il1rf/f or CX3CR1CreWT/WT(Cre−)Il1rf/f control mice into CDX2ERT-Apc recipients. We did not detect any differences in tumor number or size (Figure S6A,B). Next, using Ly6Gcre strain, we inactivated IL-1R1 specifically in neutrophils. Similar to total myeloid Il1r−/−, mice lacking IL-1R1 only in neutrophils demonstrated an increase in CRC when IL-1R deficient and control mice were co-housed. Even stronger acceleration of CRC was detected when mice were housed separate (Figure 6A,B). Il17a, Il22 and Ptgs2 expression in tumors, as well as Il1α and Il1β expression in monocytes was increased in the absence of neutrophil-specific IL-1R signaling (Figure 6C,D) as was protein expression of both IL-17A and IL-22 (Figure 6E-G). We also detected an association of bacteria with CRC tumors (Figure 6H,I) and an increased presence of distinct bacteria, including E. coli, inside CRC tumors (Figure 6J). No significant differences in bacteria adherent to the colonic epithelium in naive mice were detected (Figure S7A) Thus, increased infiltration of CRC tumors by bacteria in the absence of IL-1R signaling in neutrophils results in enhanced bacteria-driven inflammation and CRC tumorigenesis.
Figure 6. IL-1R signaling in neutrophils is essential for control of bacterial invasion and excessive inflammation in CRC.
CRC analysis in CDX2ERT-Apcf/f mice transplanted with BM from Ly6GCre+Il1rf/f or Ly6GCre- controls. (A) Macroscopic tumor multiplicity, size, and load analysis upon necropsy 5–6 weeks after last tamoxifen injection. N=7, *p<0.04 ***p<0.001; data for co-housed or separated by genotype mice is shown separately. (B) Representative images of CRC bearing colons (C) Q-RT-PCR analysis of Il17a, Il22 and Ptgs2 mRNA expression in tumors, N=20, *p<0.05, **p<0.002. (D) Q-RT-PCR analysis of mRNA expression for Il1a and Il1b in FACS sorted monocytes (CD45+CD11b+Ly6G−Ly6C+) from CRC tumors, N=5, *p<0.04. (E-G) Intracellular cytokine staining of cells isolated from tumors in mice with indicated genotypes, representative of N=6, quantified on (E) N=6, **p<0.002, ***p<0.0004 (H) Increased bacteria (arrows) associated with tumors. Yo-Yo-1 staining of whole tumor-bearing colon tissue, quantified in (I), N≥5. (J) Q-RT-PCR analysis of CRC tumor lysates for specific bacterial content with 16S rRNA specific primers; normalized to RpL32 expression, N=17 *p<0.05. Data are means ± SEM. Representative of 3 independent experiments. See also Figures S6, S7.
IL-1R signaling controls anti-bacterial functions of neutrophils.
Neutrophil recruitment into the tumors was not affected by myeloid specific or neutrophil specific IL-1R ablation, while donor BM neutrophils exhibited profound deletion of exon 5 of the Il1r1 gene indicating efficient cell recruitment and cell specific gene deletion (Figure S7B-D). (Figure 7A,B and S7B-D). We also did not find any significant changes in the presence of neutrophil extracellular traps (NET’s) in CRC tumors of LysMCre+Il1r1f/f and controls (Figure S7E). Also, ex vivo experiments did not detect significant differences in intracellular levels of bactericidal ROS in neutrophils (Figure S7F), although for these experiments intratumoral neutrophils undergo mechanical isolation possibly resulting in tehir activation. CD11b+Ly6G+ neutrophils, enriched or sorted from spleens, bone marrow, thioglycollate elicited peritoneal exudate or from CRC tumors, were able to kill various bacteria, including lab strain of E. coli and bulk fecal bacteria, upon IL-1β treatment. That ability was impaired when neutrophils were rendered IL-1R deficient (Figure 7C-E). We further found that the absence of functional IL-1R signaling led to a decreased ability of neutrophils to ingest zymosan particles or GFP-labeled Salmonella typhimurium bacteria (Figure 7F-I). Expression of antimicrobial peptides was also reduced in the absence of IL-1R in neutrophils (Figure 7J).
Figure 7. IL-1R signaling in neutrophils controlled anti-bacterial response.
(A-B) FACS analysis of tumor-infiltrating neutrophils from LysMCre+Il1rf/f or control BM transplanted CDX2ERT-Apcf/f mice. (A) Representative FACS plots and (B) quantification, N=5. (C) Splenic neutrophils isolated from LysMCre+Il1rf/f or control BM transplanted tumor bearing mice were treated with IL-1β and incubated with fecal bacteria from tumor-bearing mice. Representative images of anaerobic (left) and aerobic (right) colonies on blood agar plates (D,E) Neutrophils derived from BM and peritoneal cavities of LysMCre+Il1rf/f or control mice and neutrophils from spleen of tumor bearing mice (D), intratumoral neutrophils from CRC bearing mice with Ly6GCre+Il1rf/f or control BM (E) were stimulated with IL-1β and incubated with E. coli for 1 hour, followed by bacteria plating. Quantification of CFU on agar plates after serial dilutions. N≥3. (F) Representative confocal images of peritoneal neutrophils (“red” Ly6G) ingesting Salmonella-GFP (“green”). (G) Representative confocal images of spleen-derived neutrophils from tumor bearing mice (“green” Ly6G) ingesting zymosan beads (“red”) counterstained with DAPI. (H) Quantification of neutrophils with ingested Salmonella, N≥10 (I) Quantification of zymosan beads number ingested per neutrophil isolated from indicated mice, N≥10. (J) Q-RT-PCR analysis of the expression of indicated antibacterial peptides in tumors lysates of Ly6GCre+Il1rf/f or control BM transplanted mice, N≥8. Data are means ± SEM. Representative of at least 2 independent experiments. See also Figure S7.
Overall our data suggested that neutrophils lacking IL-1R signaling have defective anti-bacterial functions. Together, these results underscore the importance of direct IL-1R signaling in neutrophils preventing tumor associated dysbiosis, over inflammation and CRC development.
DISCUSSION
Inflammatory cytokines are emerging as regulators of the tumor microenvironment through multiple mechanisms of action. Cytokines are also increasingly attractive targets in cancer as they are amenable to antibody-mediated neutralization without the toxicities of common chemotherapies. As more cytokines are found to be essential for tumor development, at least in pre-clinical models, it is expected that the number of cytokine blockers tested in cancer clinical trials will increase. However, whether the efficacy of anti-cytokine therapy might be suboptimal because of cytokine signals operating within a particular spatio-temporal context, remains unexplored.
Previous studies have demonstrated that genetic or pharmacologic blockade of a single pro-tumorigenic cytokine can cause an unambiguous reduction in CRC, because these TEI cytokines act on epithelial and cancer cells and drive tumor growth (Chae et al., 2010; Grivennikov et al., 2009; Wang et al., 2014). Based on a vast literature implicating IL-1 signaling in the regulation of autoimmunity, inflammation and specific induction of IL-17 production and differentiation of Th17 cells (Carmi et al., 2011; Nakae et al., 2003; Shaw et al., 2012), we hypothesized that IL-1 driven signals would be essential for controlling TEI and thereby facilitating CRC growth. We found that whole body and hematopoietic cell specific IL-1R1 ablation did decrease expression of TEI cytokines, but unexpectedly caused very limited, if any, decrease in CRC. This presented us with a conundrum of why inactivation of IL-1R signaling in the hematopoietic system did not tame CRC, despite a reduction in pro-tumorigenic cytokines whose inactivation reproducibly reduce CRC in a variety of experimental conditions (Blatner et al., 2012; Grivennikov et al., 2012; Kirchberger et al., 2013). Given the multifaceted roles of IL-1R1 and the fact that IL-1R1 is expressed by virtually all cell types in the body, it was conceivable that IL-1 signaling may have opposing roles in different cell types. In this work we showed that in the CRC tumor microenvironment, IL-1 signaling plays distinct roles, depending on cell type and established the cellular and molecular mechanisms governing how this particular cytokine regulated multipronged processes during intestinal tumorigenesis.
Analysis of epithelial-specific IL-1R1 deletion revealed a decrease in CRC tumor multiplicity, slower proliferation of early tumor seeds and decreased activation of NF-κB. NF-κB is an essential regulator of cell survival and proliferation, and its inactivation in intestinal epithelial cells by deletion of IKKβ kinase during the development of colitis-associated cancer decreases tumor multiplicity (Greten et al., 2004). These observations are in line with reports that IL-1α and IL-1β stimulate growth in colon cancer cell lines in vitro and in xenograft transplants in vivo (Voronov et al., 2003; Holen et al., 2016). While NF-κB can be activated by a plethora of stimuli, IL-1R signaling may be essential for activating NF-κB in CRC because IL-1α and IL-1β are rapidly released by transformed and stressed cells, as well as by cells stimulated with microbes infiltrating early tumors. IL-1 signaling in epithelium regulated early CRC, while being dispensable for the expression of inflammatory cytokines, indicating that IL-1 signaling controlled CRC in an inflammation-independent manner.
In T cells, IL-1 signaling regulated CRC in an inflammation-dependent manner. Lymphoid cells, including T cells and ILC3, are important producers of IL-17A and other TEI cytokines during host defense and chronic inflammation. These lymphoid cells infiltrate CRC tumors and their increased presence correlates with poor prognosis in CRC patients, according to CRC “Immunoscore” (Bindea et al., 2013; Tosolini et al., 2011). Here we found that IL-1 signaling in T cells was essential for the production of IL-17A and IL-22 cytokines in CRC tumors, consistent with what has been observed in other inflammatory conditions in mice and humans (Shaw et al., 2012; Zhou and Littman, 2009). Ablation of IL-1R in T cells did not result in a complete absence of IL-17A and IL-22 in tumors. Likely, this residual IL-17 and IL-22 was produced by ILC3 and γδT cells.
The novel observation of this work was that IL-1 signaling in myeloid cells was anti-tumorigenic, at least in the context of tumors growing at “microbial-rich” surfaces. IL-1R signaling in myeloid cells kept specific species/genera of tumor infiltrating microbes at bay, and prevented local, tumor-specific dysbiosis and excessive amounts of pro-tumorigenic inflammatory cytokines. We propose that increased infiltration of “bad” (pro-tumorigenic) and a decrease in “good” (anti-tumorigenic) gut bacteria led to an increased bacterial recognition by myeloid cells and production of IL-17A, which induces IL-1β and IL-23, along with other tumor-promoting inflammatory entities, which drive CRC. 16S rRNA sequencing of the microbiome had shown that the observed dysbiosis did not occur in naive mice and was induced in a tumor-specific manner.
Neutrophils and not monocytes or macrophages were essential to deter tumor-infiltrating bacteria and dampen CRC promoting inflammation. Neutrophils represent an important component of the tumor microenvironment, where they can be immunosuppressive as a form of myeloid derived suppressor cells or directly tumor promoting or anti-tumorigenic (Fridlender et al., 2009; Gabrilovich and Nagaraj, 2009). While neutrophils are a key component of anti-microbial defense, here we found a tumor specific, anti-microbial role for neutrophils in cancer. Previous work on Il1r1−/− mice shows defective neutrophil recruitment and higher susceptibility to infection, with no change in bacterial killing assessed primarily in macrophages (Miller et al., 2006). Here, we found that while lack of IL-1R on neutrophils did not alter neutrophil recruitment and its ability to form NET’s, however, it led to defective bacterial killing. Thus, neutrophil-specific IL-1 signaling is critical for preventing tumor-associated dysbiosis, TEI and CRC, where bacteria are invasive due to the increased epithelial permeability and decreased mucus coverage. Significant alterations of commensal microbiota are present in mouse mutants devoid of IL-18 signaling and in various mutants affecting the inflammasome pathway (Elinav et al., 2011; Franchi et al., 2010) essential for IL-1β and IL-18 cytokine processing and activation; but altered microbiota have not been found in mice with global IL-1R1 deficiency.
The IL-1 pathway has been extensively studied in the context of cancer, but often using cell line based non-autochthonous models, immunodeficient mice or cancer models where epithelial surfaces are not rich in microbes. Levels of IL-1β are elevated in multiple cancers in human and mouse models, including colon, gastric, breast, non-small-cell lung carcinoma and melanomas (Elaraj et al., 2006). Genetic inactivation of IL-1R1 or caspase 1, a key enzyme in IL-1β processing, reduced tumorigenesis in a two-step skin cancer model (Drexler et al., 2012). Inactivation of IL-1α and MyD88 reduces liver carcinogenesis in DEN model (Sakurai et al., 2008). Blocking IL-1 signaling in human breast cancer xenografts or a mouse model of breast cancer results in variable effects on tumor growth and metastatic spread and decreased infiltration by myeloid CD11b+Gr1+ cells (Dagenais et al., 2017; Holen et al., 2016). Neutrophils and IL-1 signaling are critical for breast cancer metastasis but not primary tumor growth (Coffelt et al., 2015), and environmental and dietary factors can drive inflammasome/IL-1 dependent breast cancer (Kolb et al., 2016). Furthermore, in transplantable B16 melanoma and prostate cancer models, IL-1β was found to be essential for invasiveness and metastatic growth while the same function has been assigned to IL-1α in breast cancer (Voronov et al., 2003). IL-1α and MyD88 signaling drives skin tumorigenesis (Cataisson et al., 2012), however in the model of MCA-induced fibrosarcoma, IL-1α is essential for activation of antitumor responses (Elkabets et al., 2009). Moreover IL-1 can play a role of a “danger signal”, when produced from necrotizing cells (Ghiringhelli et al., 2009). It still remains to be determined whether IL-1 exerts its pro-tumorigenic signaling by induction of TEI or whether IL-1 signaling is largely autonomous to cancer cells. Recently, an IL-1α neutralizing antibody was tested in clinical trials for advanced CRC (Hickish et al., 2017); meanwhile a complete blockade of IL-1 signaling has been proposed but not thoroughly examined.
Propelled by numerous studies implicating inflammatory IL-1 signaling in cancer, several clinical studies with IL-1 antagonists as monotherapies and combination therapies for human cancers are underway. Our results, however, suggest that a more cautious approach may be required when considering the use of IL-1 blockade in CRC and urges caution against a “one size fits all” global blockade of IL-1R signaling. We propose that in some microbe-rich cancers a complete inhibition of IL-1R signaling would not be as beneficial as specific blockade of this axis in T cells or epithelial cells, for example via antibody-based approaches that deliver IL-1 blockers specifically to T cells. Such next-generation approaches will allow blocking IL-1R signaling only at “pro-tumorigenic” cell types, while preserving “anti-tumorigenic”, protective signaling. Alternatively, approaches aimed at enhancing intra-tumoral anti-bacterial functions or depleting potentially pathogenic microbiota should be considered along with IL-1R signaling blockade. Finally, the role of individual IL-1α and IL-1β ligands and the potential for their neutralization over complete IL-1R blockade needs to be further assessed.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, S.I. Grivennikov, Sergey.Grivennikov@fccc.edu.
Experimental models
Mice
Using Il1r1f/f conditional allele (Bruttger et al., 2015), we generated a panel of mice in which IL-1R1 expression was ablated in colonic epithelial and cancer celIs. Il1r1f/f mice were crossed to different Cre-recombinases to get cell-specific deletion of Il1r1 gene, including CD4Cre, CX3CR1Cre, CD11bCre, LysMCre and Ly6GCre. Cre-mediated recombination of these sites resulted in a frame shift and early termination of protein translation in exon 5. CX3CR1Cre, CD4Cre, Il17aGFP, RorcGFP and LysMCre mice were obtained from Jackson Laboratory. Apcf/f, CDX2Cre (CPC) and CDX2ERT mice have been provided by E. Fearon. Ly6GCre (‘catchup”) mice were from Matthias Gunzer (Hasenberg et al., 2015). For further information and MGI numbers identifying afore-mentioned mouse strains please see Key Resources Table.
For conditional ablation of IL-1R1 in colonic epithelial and tumor cells of CPC-APC mice, Il1rf/fApcf/f mice were crossed to CDX2Cre-Il1D/WT mice, where the null (D, deleted) Il1r allele is derived from the whole-body knockout strain. All mice were maintained in filter-topped cages on autoclaved food and water at the Fox Chase Cancer Center Animal Facility (FCCC), and all experiments were approved by FCCC IACUC. All experiments used co-housed littermates, unless indicated otherwise, so that consistency of common microflora and genetic background/alterations would be ensured. For tamoxifen-inducible tumorigenesis, CDX2ERT-Apcf/f mice were given 2 injections of tamoxifen 2 mg and 1.5 mg per mouse (Sigma; dissolved in 5% ethanol, 95% corn oil (Sigma)) intraperitoneally (i.p.) every other day. Mice were sacrificed 4–6 weeks after the last dose of tamoxifen for histological analysis and tumor statistics.
Bone Marrow Transplantation
Six- to eight-week-old recipient mice were irradiated twice (600 rad and 500 rad) using cesium irradiator at FCCC during one day to achieve a lethal dose and intravenously injected with a single-cell suspension of donor bone-marrow cells. Recipients were co-housed littermates, which were transplanted with both gene-deficient and wild type bone marrow. After transplantation, the recipients were placed on neomycin and polymixin B in drinking water for two weeks, followed by transfer to dirty cages that were from the same room and rack to reconstitute potentially depleted microbiota. Mice were sacrificed and analyzed for tumor development 4 months after transplantation; for inducible CDX2ERT-Apcf/f model mice were allowed to reconstitute BM, injected with tamoxifen (as above) and then allowed to develop CRC for 4–6 weeks.
Antibiotic treatment
To deplete gut microbiota mice were given mix of antibiotics in drinking water for 4 weeks. Water was changed every 3–4 days. Antibiotics cocktail included: Ciprofloxacin (0.2g/L), Neomycin (1g/L), Vancomycin (0.5g/L), Ampicilin (1g/L), Metronidazol (0.5g/L), Primaxin (Imipenem) (0.5g/L). Antibiotics treatments started after last injection of Tamoxifen and after 4 weeks of treatment mice were sacrificed for tumor analysis.
Immune cells isolation, ex vivo cell stimulation, Flow Cytometry and Cell Sorting
Lamina propria lymphocytes (LPL) and Intraepithelial lymphocytes (IEL) cells from colonic tumors and normal tissues were isolated. Briefly, colonic tumors and normal tissues were cut into small pieces and briefly washed in 1mM DTT-HBSS, followed by mechanical shaking with 10mM EDTA-HBSS in a shaker (200 rpm) at 37°C for 20 min for 2 rounds total. Then IEL/IEC fraction was passed through a metal mesh and cells collected, while the remaining tissue was digested in 20 ml of Collagenese Type VIII (Sigma) solution in 1xHBSS in the shaker (180rpm) at 37°C for 30 min. Digested tissue was passed through a 70 μm Cell Strainer and cell suspension was collected (unpurified LPL). IEL/IEC and LPL fractions were further enriched by Percoll gradient centrifugation (20min, 2200rpm, no brake; Percoll gradient 40%/ 80% in HBSS). Lymphoid cell ring on a border of fraction was collected, washed and re-suspended in FACS buffer (1xPBS, 1mM EDTA, 2%FBS). Single cells suspension was subjected then to flow cytometry analysis, cytokine stimulation or cell sorting.
To detect intracellular cytokine expression in tumor infiltrating immune cells, isolated LPL and IEL cells were stimulated in RPMI 1640 High glucose activation media (10% FBS, 1× Glutamax, HEPES 1mM, 1× penicillin/streptavidin, β-mercaptoethanol) for 5.5 hours with PMA 500 ng/ml and ionomycin 100 ng/ml at 37 C. To inhibit protein transport, Monensin and Brefeldin A at concentration 5 μg/ml were added in activation media after first 40 min of stimulation. Cells were collected, stained for surface markers, washed, fixed and permeabilized (BD Cytofix/Cytoperm Kit), stained for intracellular cytokines and analyzed by flow cytometry. All flow samples were acquired on a LSRII (BD Biosciences) and analyzed using FlowJo v. 10 (TreeStar). For cell sorting, samples were stained and sorted on BD FACSAria II sorter (BD Biosciences).
Intestinal epithelial cell isolation for protein analysis
Colonic tumors and normal tissues were cut into small pieces followed by mechanical shaking (200 rpm) with 10mM EDTA in Ca2+ Mg2+ free 1× HBSS at 4°C for 15 min; 3 rounds total. After each round of shaking cells were vortexed for 20 seconds and put through metal strainer; cell suspension was collected and centrifuged at 1600 rpm for 10 min at 4C. Pellets were resuspended in ice cold PBS containing inhibitors of proteases and phosphatases and then subjected to protein nuclear extraction.
Nuclear protein extraction and Western blot
Intestinal epithelial cells (IEC) were re-suspended in buffer A (10mM HEPES (pH=7.9), 10mM KCl, 10mM MgCl2, 1mM DTT and 1× protease inhibitor cocktail (Roche)) and incubated on ice for 15 min followed by adding 10% NP40 to total concentration of 1%. Content was vortexed for 20 seconds and spun down at 12000 rpm for 30 seconds. Supernatants were collected as cytoplasmic protein extracts. Pellets were re-suspended again in buffer A and centrifuged. Washed pellets were re-suspended in buffer C (20mM HEPES (pH=7.9), 25% Glycerol, 0.4M NaCl, 1.5mM MgCl2 and 1× protease inhibitor cocktail) and incubated on ice for 15 min, then centrifuged at 12000 rpm for 5 min. Supernatants were collected as nuclear protein extract, mixed with RIPA buffer with protease inhibitors and subjected to western blot analysis with anti-NF-κB p65 (Cell Signaling) antibody and anti-Histone H3 (Cell Signaling) antibody for loading control.
In Vivo Treatment with Anakinra
Mice were injected intraperitoneally for three consecutive days with 2.5 mg recombinant human IL-1Ra (Anakinra/Kineret, Rx, FCCC Pharmacy), or PBS for control group.
RNA extraction and quantitative Real Time-PCR Analysis
Total RNA was extracted with the RNAeasy Plus kit (QIAGEN) and reversely transcribed using IScript kit (Biorad). Quantitative RT-PCR (qRT-PCR) was performed with iTaq Universal SYBR Green Supermix (Biorad) on a Biorad CFX96 machine. Expression data were normalized to RpL32 mRNA levels. The data are presented in arbitrary units and were calculated as 2(Ct (RpL32 – gene of interest)). Primer sequences generally were obtained from the NIH Mouse qPrimerDepot website repository and are listed in Key Resources Table.
Analysis of Cytokine Production by Multiplex or ELISA
Small pieces of tumors were incubated for 24h in complete RPMI 1640 media. Supernatants were collected and cytokine secretion was measured by mouse Procarta 17-Plex cytokine array (eBioscience) on MagPix instrument (Luminex) according to manufacturer’s instructions. For ELISA analysis of IL-17A, colonic tumors were cultured in DMEM containing 10% fetal bovine serum (FBS) and antibiotics overnight, and supernatant was analyzed with IL-17A ELISA kit (eBioscience). Cytokine concentration was normalized to the weight of tumors in each well.
Immunohistology and immunofluorescence
For immunohistology, immediately after euthanasia, colon rolls were fixed with 10% buffered formalin and kept overnight at RT. After fixation samples were transferred in 70% ethanol prior to paraffin embedding. 5 μm thick sections were cut in the sagital plane for staining with hematoxylin and eosin. Images were acquired and quantified with slide scanning Vectra microscope (Vectra).
Sections were de-paraffinased in 4 changes of xylene, then dehydrated and re-hydrated in 100%->70% ethanol changes. Antigen retrieval of samples was performed in 1× Citrate buffer (10mM Citrate, 0.05% Tween 20, pH 6.0) at 95C for 1h. Then slides were rinsed in dH2O, dehydrated in 100% ethanol, then blocked with 3% H2O2 in PBS, rinsed with water, blocked in PBS-5% BSA-5% normal goat serum for 20 min, then incubated with primary antibody anti-Ki-67 (1:250) or anti-p-STAT3 (1:100) overnight at 4C, then washed 3× in PBS – 1% BSA. Next, secondary biotinylated anti-rabbit antibodies were applied at 1:200 dilution and incubated at RT for 1h, rinsed in PBS. Next, Streptavidin-HRP was applied at dilution 1:250 in PBS-1%BSA for 30 min at room temperature, slides were washed 3× in PBS, then developed with chromogen DAB solution for 7 min at room temperature. Next, slides were rinsed and counterstained with hematoxylin, rinsed with water and immersed into 0.25% ammonium hydroxide, dehydrated with 4 changes of ethanol 70%->100%, followed by 4 changes of xylene for 5 minutes each, and mounted.
Images were acquired with slide scanning Vectra microscope (Vectra). Immunofluorescencent staining was performed in the same way with IHC protocol with few exceptions- blocking buffer was containing 5% goat serum, 10% BSA, 0.1% Triton-X-100 in 1xPBS. Goat-anti-rabbit AF488 was used for secondary antibody, and then samples were washed follow by staining with DAPI for 10 min at RT. Slides were mounted with VectorShield and covered with cover slip. After drying for 1–2h at RT slides were analyzed on Leica SP8 Confocal Microscope.
Whole tissue IF staining for bacteria with Yo-Yo 1 dye
For whole-mount staining, colons were fixed in buffered 4% paraformaldehyde for 8h at 4C. Tissues were permeabilized by incubation with 0.5% (wt/vol) saponin, 2% (vol/vol) FBS, and 0.09% (wt/vol) sodium azide in PBS for at least 18 hours. The same buffer was used for subsequent incubations with antibodies. Colon fragments were incubated with AlexaFluor 647 phalloidin (1:200) and Yo-Yo1 (1:500) dye for 12–16 hours at 4C followed by 1–2 hour incubation at 37C. Samples were washed with PBS, mounted flat in PBS and imaged with water dipping lens by Leica SP8 Confocal microscope.
Neutrophils or macrophage isolation from bone marrow, spleen, peritoneal cavities and tumors
To isolate neutrophils from peritoneal cavities, we injected i.p. naïve LysMCre+Il1rf/f mice and their wild type Cre negative controls with 1 ml of 3% thioglycollate broth solution (BD Biosciences) and sacrificed mice in 4–5 hours.
To isolate neutrophils from spleens and bone marrow we used tumor bearing CDX2ERT-Apcf/f mice with bone marrow from LysMCre+Il1rf/f or control mice. Single cell suspensions were prepared from spleens, flushed tibia and femurs. Cells were filtered through 70 μm Cell Strainer (BD Biosciences). For neutrophil purification, Easy Mouse Neutrophil Enrichment kit (StemCell Technologies #19762A) was used for enrichment, followed by FACS sorting for cells with phenotype CD11b+Ly6G+CD4−TCRβ−F4/80−.
Bacteria killing assay
Sorted neutrophil populations from peritoneal cavities, spleen or bone marrow were pretreated with 100ng/ml of recombinant IL-1β for 1h in complete antibiotic free RPMI media at 37C. Meanwhile, bacteria (suspension of E. coli) were grown to the approximately 1.6 × 108 cells/ml (OD 600 nm=0.2) or stool bacteria were prepared to similar concentration by disrupting feces in ice cold PBS and filtering through 40 mcm strainer. Next, E. coli or “bulk” stool bacteria was incubated for 30 minutes with 20% of normal mouse serum for opsonization and after that added to neutrophils in the approximate of 50:1 bacteria/neutrophil ratio, incubated for 1 h at 96-well plate, then spin down for 5 min at 1500 rpm and lysed in DI water with 0.1% Triton-100. Lysates were diluted serially (1:10, 1:100, 1:1000) for plating on LB or blood agar. Plates were incubated in anaerobic (BD BBL GasPak Anaerobic Systems) or regular aerobic conditions overnight at 37C. Colonies were counted manually.
Phagocytosis assay
Sorted neutrophil populations from peritoneal cavities or spleens were pretreated with 100ng/ml of recombinant IL-1β for 1 h in complete RPMI media at 37C, then suspension of zymosan beads conjugated with Alexa Fluor 594 dye (Invitrogen) or Salmonella-GFP was added and cultured with neutrophils for 2h. Cell suspensions were transferred on Poly-L-lysine coated glass slides, fixed with 4% PFA for 5 min and stained with anti-Ly6G (A18)-FITC or -APC antibodies in 1:75 ratio for 20 mins and counterstained with DAPI for confocal imaging.
Microbiome analyses
Bacterial DNA was isolated from a fecal content or epithelial “shakes” of various naïve or CRC bearing mouse strains. Samples were homogenized with RNAse/DNase free 2.8mm Ceramic Beads using Omni Bead Ruptor 24 in ASL buffer (Stool lysis buffer; Qiagen) followed by DNA isolation using QIAamp DNA stool Mini Kit (Qiagen) according to manufacturer’s protocol. Library preparation, sequencing and data analysis were carried out essentialy as previously described (Fatkhullina, 2018). Briefly, QPCR was carried out using universal or bacterial strain-specific primers for 16S rRNA genes. The V4 region of the 16S rDNA gene using two round amplification PCR using Phusion High-Fidelity DNA-polymerase (NEB) with 100ng of input DNA. First round PCR (15 cycles):
515F TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG
806R GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC
Second round PCR (10 cycles) with i5 and i7 Illumina barcode sequences:
AATGATACGGCGACCACCGAGATCTACAC-i5-TCGTCGGCAGCGTC
CAAGCAGAAGACGGCATACGAGAT-i7-GTCTCGTGGGCTCGG
Amplicones were purified between and after the PCR reactions with Agencourt AMPure XP (Beckman Coulter, Inc.) and libraries were quantified using KAPA Library Quantification Kit for Illumina Platforms (Kapa Biosystems) and size was estimated using Agilent Tapestation 4200 (Agilent). Libraries were pooled at equimolar concentration using Epmotion 5075 tmx automatic liquid handling machine (Eppendorf) and sequenced with 5% PhiX library added to the pool on Illumina MiSeq machine according to manufacture instructions using v3 kit (600 cycles). Fastq files were generated on Basespace (Illumina). On average 100,000 reads were obtained per sample, and after quality control filtering; on average 90,000 reads per sample were processed further. Chimeric sequences were filtered out of the FASTQ files containing the 16S rRNA gene sequences using the USEARCH (version 8.1) utility’s UCHIME implementation and the ‘gold’ database (version microbiomeutil-r20110519). The reads, thus filtered, were then binned into operational taxonomic units (OTUs) at 97% similarity using USEARCH’s cluster_otus command. QIIME (1.9.1) scripts were used to classify and align the obtained OTUs. The assign_taxonomy.py script was used to assign taxonomy using the default RDP method and GreenGenes database.
Quantification and Statistical Analysis
Data are presented as mean ± SEM, as indicated in the legend of each figure. The significance of the differences between groups was evaluated using unpaired t test, p value < 0.05 was considered significant. Statistical parameters including the precision measures (mean ± SEM) and statistical significance are reported in the Figures and the Figure Legends when necessary. In figures, asterisks denote statistical significance (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001), unless indicated otherwise. Experiments were repeated at least twice. Statistical analysis was performed in GraphPad PRISM 6.
For microbiome analyses the statistical differences between the groups were estimated with non-parametric Mann-Whitney test, PERMANOVA test and p-values were corrected for multiple tests using q-value 0.05 as a cutoff and data was visualized using heatmaps and PCA plots (Partek 6.6).
Supplementary Material
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD45 - PerCP | Biolegend | Cat#103130 |
| Anti-mouse CD45.2 (104)- PacBlue | eBioscience | Cat#48-0454-82 |
| Anti-mouse CD45.1- APC | Invitrogen | Cat#17-0453-82 |
| Anti-mouse CD4 (GK1.5) - APC | eBioscience | Cat#17-0041-83 |
| Anti-mouse CD4 – FITC | Biolegend | Cat#100406 |
| Anti-mouse CD4 (GK1.5) - PE | Biolegend | Cat#100408 |
| Anti-mouse CD90.2(53-2.1)-APC-Cy7 | eBioscience | Cat#47-0902-82 |
| Anti-mouse TCRβ (H57-597) – PE-Cy7 | eBioscience | Cat#25-5691-82 |
| Anti-mouse TCRβ (H57-597) – APC | eBioscience | Cat#17-5961-83 |
| Anti-mouse TCRβ (H57-597) – PacBlue | eBioscience | Cat#48-5961-82 |
| Anti-mouse TCRγδ – FITC | Biolegend | Cat#107504 |
| Anti-mouse TCRγδ (eBioGL3) – APC | eBioscience | Cat#17-5711-82 |
| Anti-mouse TCRγδ (eBioGL3)- PE | eBioscience | Cat#12-5711-82 |
| Anti-mouse IL17A (eBio17B7) – PE | eBioscience | Cat#12-7177-81 |
| Anti-mouse IL17A (eBio17B7) – PE-Cy7 | eBioscience | Cat#25-7177-82 |
| Anti-mouse IL22 (IL22JOP) – APC | eBioscience | Cat#17-7222-82 |
| Anti-mouse CD11b (M1/70)-PacBlue | Invitrogen, eBioscience | Cat#48-0112-82 |
| Anti-mouse CD11b (M1/70) – PE | eBioscience | Cat#12-0112-82 |
| Anti-mouse CD11b (M1/70) – FITC | Biolegend | Cat#101206 |
| Anti-mouse Ly6C (HK1.4) –APC-Cy5.5 | Biolegend | Cat#128024 |
| Anti-mouse Ly6G (1A8) –APC-Cy7 | Biolegend | Cat#127624 |
| Anti-mouse Ly6G (1A8) – PE-Cy7 | Biolegend | Cat#127617 |
| Anti-mouse Ly6C (HK1.4)- AF488 | Biolegend | Cat#128022 |
| Anti-mouse Ly6C (HK1.4)-APC-Cy7 | Biolegend | Cat#128026 |
| Anti-mouse Ly6C (HK1.4) – PE-Cy7 | Biolegend | Cat#128018 |
| Anti-mouse CD11c (N418) – APC | Biolegend | Cat#117310 |
| Anti-mouse CD11c (N418)-PE-Cy7 | eBioscience | Cat#25-0114-82 |
| Anti-mouse MHC II – APC-Cy5.5 | eBioscience | Cat#56-5321-82 |
| Anti-mouse F4/80 (BM8) – APC | eBioscience | Cat#17-4801-82 |
| Anti-mouse F4/80 (BM8) – PE-Cy7 | eBioscience | Cat#25-4801-82 |
| Anti-mouse F4/80 (BM8) – PE | Biolegend | Cat#123110 |
| Anti-mouse CD8β (eBioH35-17.2) – PacBlue | eBioscience | Cat#48-0083-82 |
| Anti-mouse CD8α (53-6.7) – APC-Cy5.5 | eBioscience | Cat#56-0081-82 |
| Rabbit monoclonal Ki67 antibody (SP6) | Genetex | Cat#16667 |
| Rabbit monoclonal NF-kB p65 (D14E12) | Cell signaling | Cat#8242S |
| Rabbit monoclonal phospho-STAT3 (Y705) | Cell signaling | Cat#9131S |
| Rabbit monoclonal Histone H3 | Cell signaling | Cat#9715S |
| Rabbit monoclonal Non-P (active) beta-catenin (D2U8Y) | Cell signaling | Cat#19807T |
| YoYo-1 dye | ThermoFisher Scientific | Cat#Y3601 |
| Goat polyclonal Anti-h/mMPO antibody | R&D System | Cat#AF3667 |
| Rabbit polyclonal CitH3 antibody | Abcam | Cat#Ab5103 |
| Alexa Fluor 647 Phalloidin | Invitrogen by ThermoFisher Scientific | Cat#A22287 |
| Goat anti-Rabbit Secondary antibody AF488 | Invitrogen by ThermoFisher Scientific | Cat#A-11008 |
| Donkey anti-Goat Secondary antibody AF594 | Invitrogen by ThermoFisher Scientific | Cat#A-11058 |
| Biotin Goat Anti-Rabbit Ig | BD Pharmingen | Cat#550338 |
| Bacterial and Virus Strains | ||
| Salmonella-GFP | S. Balachandran (FCCC) from M. O’Riordan lab (University of Michigan) | (Radtke et al., 2007) |
| E.coli | Zymo Research | Cat#T3003 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| IL-1β recombinant protein | Peprotech | Cat #211-11B |
| CM-H2DCFDA dye | Invitrogen by ThermoFisher Scientific | Cat #C6827 |
| Zymosan A BioParticles, Alexa Fluor 594 conjugate | Invitrogen by ThermoFisher Scientific | Cat #Z23374 |
| Yellow “Live/Dead” fluorescent reactive dye | Invitrogen by ThermoFisher Scientific | Cat #L34968 |
| PMA (phorbol 12-myristate 13-acetate) | Sigma | Cat #P8139 |
| Ionomycin | Invitrogen by ThermoFisher Scientific | Cat #I24222 |
| Monensin 1000x Solution | Biolegend | Cat #420701 |
| Brefeldin A 1000x Solution | eBioscience | Cat #00-4506-51 |
| Fixation/Permeabilization kit (RUO) | BD Biosciences | Cat #554714 |
| Percoll | GE Healthcare | Cat #17-0891-01 |
| BD BBL Stacker Plate (CDC Anaerobe blood agar) | BD Biosciences | Cat #221733 |
| Opti-MEM I+ Glutamax-I Medium | Gibco | Cat. #51985-034 |
| Collagenese from Clostridium histolyticum (Type VIII) | Sigma | Cat. #C2139-5G |
| Anakinra (Kineret) | Amgen/ FCCC Pharmacy | Rx |
| Vancomycin Hydrochloride for Injection, USP | FCCC Pharmacy | NDC 0409-6531-02 |
| Primaxin (Imipinem) | NOVAPLUS | NDC 63323-322-93 |
| Neomycin trisulfate salt hydrate | Sigma | Cat #N1876 |
| Ciprofloxacin | Hospira | Cat #17850-25G-F |
| Ampicillin sodium salt | Sigma | Cat #A9518 |
| Metronidazole | Sigma | Cat #M3761 |
| Polymixin B Sulfate | Milipore | Cat #M5291 |
| Tamoxifen | Sigma | Cat #T5648 |
| Critical Commercial Assays | ||
| QIAamp DNA Stool Mini kit | Qiagen | Cat # 51504 |
| RNeasy Plus Mini kit | Qiagen | Cat# 74136 |
| EasySep Mouse Neutrophil Enrichment Kit | StemCell technologies | Cat#19762A |
| Trans-Blot Turbo 5x Transfer Buffer | Bio-Rad | Cat #10026938 |
| Trans-Blot Turbo Transfer Pack | Bio-Rad | Cat #1704156 |
| Mini-PROTEAN TGX Gels | Bio-Rad | Cat #456-1094 |
| Mouse IL1b ELISA Ready-SET-Go! | eBioscience | Cat #88-7013-22 |
| DuoSet Mouse IL22 | R&D Systems | Cat #DY582-05 |
| Mouse IL17A ELISA Ready-SET-Go! | eBioscience | Cat #88-7371-88 |
| Streptavidin HRP | BD Pharmingen | Cat #554066 |
| iScript Reverse Transcription Supermix for RT-qPCR | Bio-Rad | Cat #170-8841 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat #172-5121 |
| Deposited Data | ||
| 16S RNA microbiome sequencing | NCBI SRA Database | BioProject:PRJNA480459; SRA Accession Number: SRP152945 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Apcf/f | Eric Fearon (University of Michigan) | (Hinoi et al., 2007) |
| Mouse: Il17aGFP | Jackson Laboratories | Stock 018472 - C57BL/6-Il17atm1Bcgen/J |
| Mouse: CDX2ERT | Eric Fearon (University of Michigan) | (Feng et al., 2013) |
| now available from Jackson Laboratories | B6.Cg-Tg(CDX2-Cre/ERT2)752Erf/J Stock No: 022390; CDX2P-CreER | |
| Mouse: CDX2Cre | Jackson Laboratories | B6.Cg-Tg(CDX2-cre)101Erf/J Stock No: 009350; CDX2P-NLS Cre |
| Mouse: RorcGFP | Jackson Laboratories | B6.129P2(Cg)-Rorctm2Litt/J Stock 007572 |
| Mouse: Il1r1f/f | Ari Waisman (University of Mainz) | (Bruttger et al., 2015) MGI: 5792908 |
| Mouse: Ly6GCre-Il1r1f/f | Ari Waisman and Matthias Gunzer (University Mainz and University Duisburg-Essen) | (Hasenberg et al., 2015); (Bruttger et al., 2015) |
| Mouse: CD4Cre | Jackson Laboratories | Tg(Cd4-cre)1Cwi/BfluJ Stock 017336 MGI:2386448 |
| Mouse: CD11bCre | (Ferron and Vacher, 2005) MGI:3577104 | |
| Mouse: LysMCre | Jackson Laboratories | B6.129P2-Lyz2tm1(cre)Ifo/J Stock 004781 MGI:1934631 |
| Mouse: CX3CR1Cre | Jackson Laboratories | B6J.B6N(Cg)-Cx3cr1tm 1.1(cre)Jung/J Stock No: 025524 |
| Mouse: CD45.1 | Jackson Laboratories | B6.SJL-Ptprca Pepcb/BoyJ Stock No: 002014; B6 Cd45.1 |
| Oligonucleotides | ||
| L32 FW: TTCCTGGTCCACAATGTCAA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| L32 REV: GGCTTTTCGGTTCTTAGAGGA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL1b FW: GGTCAAAGGTTTGGAAGCAG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL1b REV: TGTGAAATGCCACCTTTTGA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL1a FW: CCAGAAGAAAATGAGGTCGG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL1a REV: AGCGCTCAAGGAGAAGACC | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL17A FW: TGAGAGCTGCCCCTTCACTT | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL17A REV: ACGCAGGTGCAGCCCA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL22 FW: CATGCAGGAGGTGGTACCTT | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL22 REV: CAGACGCAAGCATTTCTCAG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| RORc FW: CCGCTGAGAGGGCTTCAC | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| RORc REV: TGCAGGAGTAGGCCACATTACA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL17F FW: CCTCCCCTGGAGGATAACAC | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL17F REV: CATGGGGAACTGGAGCGGTT | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| Ptgs2 (COX2) FW: TGAGCAACTATTCCAAACCAGC | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| Ptgs2 (COX2) REV: GCACGTAGTCTTCGATCACTATC | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| 16S bacterial RNA FW: AGAGTTTGATCMTGGCTCAG | Ordered from IDT | |
| 16S bacterial RNA REV: TACGGYTACCTTGTTACGACTT | Ordered from IDT | |
| SFB FW: GACGCTGAGGCATGAGAGCAT | Ordered from IDT | |
| SFB REV: GACGGCACGGATTGTTATTCA | Ordered from IDT | |
| E.Coli FW: CATGCCGCGTGTATGAAGAA | Ordered from IDT | |
| E.Coli REV: CGGGTAACGTCAATGAGCAAA | Ordered from IDT | |
| Enterobacteria FW: GTGCCAGCMGCCGCGGTAA | Ordered from IDT | |
| Enterobacteria REV: GCCTCAAGGGCACAACCTCCAAG | Ordered from IDT | |
| Clostridia FW: CGCATAACGTTGAAAGATGG | Ordered from IDT | |
| Clostridia REV: CCTTGGTAGGCCGTTACCC | Ordered from IDT | |
| Bacteroides FW: GGTTCTGAGAGGAGGTCCC | Ordered from IDT | |
| Bacteroides REV: GCTGGCTCCCGTAGGAGT | Ordered from IDT | |
| S100A8 FW CCAATTCTCTGAACAAGTTTTCG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| S100A8 REV TCACCATGCCCTCTACAAGA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| S100A9 FW: GTCCAGGTCCTCCATGATGT | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| S100A9 REV: GAAGGAAGGACACCCTGACA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| Reg3b FW: ACTCCCTGAAGAATATACCCTCC | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| Reg3b REV: CGCTATTGAGCACAGATACGAG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| Reg3g FW: TGCTGCTCTCCTGCCTGATG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| Reg3g REV: ATAGGAGCCATAGGCACGGG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL33 FW: TCCAACTCCAAGATTTCCCCG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL33 REV: CATGCAGTAGACATGGCAGAA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL18 FW: TCCTTGAAGTTGACGCAAGA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL18 REV: TCCAGCATCAGGACAAAGAA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL1F10 FW: TGTCTGAATCAGGGTCTCCC | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL1F10 REV: TGAAGACCAGACACTCCCAA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL36a FW: GGCTTTTACAGGTTCCTTTTTG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL36a REV: GATGAGCTGCCTGTTCTGC | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL36g FW: GTCCGGGTGTGGTAAAACAG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL36g REV: GGACACCCTACTTTGCTGCT | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL36b FW: AGAGTATTCAAATGTGGGAACCG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL36b REV: GACCCATACCATCTGTTGTGAG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL36Ra FW: GCAGCTGGTTATTGTGCAGAT | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL36Ra REV: TGGAGCTCATGATGGTTCTG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL36R FW: TCCCCGCTTTAATCTCTTCA | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL36R REV: ATCGGGCAGTCTGAATTGTC | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL18bp FW: TCTCCAGCAGTCCCAACTAAGC | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL18bp REV: AGGCAGTACAGGACAAGGTCAG | Ordered from IDT | mouseprimerdepot.nci.nih.gov |
| IL1R exon5 FW: TGGAAGTCTTGTGTGCCCTT | Ordered from IDT | This paper |
| IL1R exon5 REV: ACTCCGAAGAAGCTCACGTT | Ordered from IDT | This paper |
| Recombinant DNA | ||
| Software and Algorithms | ||
| Flow Jo 9.7.6 | FlowJo LLC | https://www.flowjo.com/ |
| Vectra 3.0 | Perkin Elmer | www.perkinelmer.com/product/vectra-3-0-6-slide-cls142568 |
| Imaris | Bitplane | http://www.bitplane.com/ |
| UCHIME | https://www.drive5.com/usearch/manual/uchime_algo.html | |
| QIIME (1.9.1) | http://qiime.org/ | |
| Graph Prism 6.0 | GraphPad | https://www.graphpad.com/ |
| Other | ||
| Poly-L-Lysine Cellware 12mm round Coverslips | Corning | Cat. #354085 |
| Hard-Shell PCR Plates, 96-well, thin-wall for Q-RT-PCR | Bio-Rad | Cat. #HSP9601 |
| U bottom 96-well tissue culture plate | Olympus | Cat # 25-221 |
| RNAse/DNAse free 2.8mm Ceramic Beads | OMNI | Cat # 19-646-3 |
Acknowledgements
We thank S. Balachandran and G. Rall (FCCC), D. Mucida (Rockefeller University) and R. Locke for critical comments; Dr E. Koltsova (FCCC) for the help with confocal imaging and reading the manuscript; Aliia Fatkhullina (FCCC) for the help with graphical abstract and E. Fearon (U Michigan) for CDX2Cre, CDX2ERT and Apcf/f mice. We thank Histology laboratory of Department of Pathology at FCCC for tissue processing and slide preparation; and LAF, Flow Cytometry, Cell Culture and Microscopy Facilities at FCCC. Supported by NIH P30 CA-006927 to FCCC, RSF Grant N14-15-00894 to M.R.K.; NIH R00 DK088589 and R01CA227629, Pew Scholar in Biomedical Sciences and AACR-Landon Innovator Awards, ACS-IRG #15-175-22, PA DOH CURE and US-Israel BSF grants to S.G. This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Footnotes
Declaration of interest:
The Authors declare no competing interests
References
- Bersudsky M, Luski L, Fishman D, White RM, Ziv-Sokolovskaya N, Dotan S, Rider P, Kaplanov I, Aychek T, Dinarello CA, et al. (2014). Non-redundant properties of IL-1alpha and IL-1beta during acute colon inflammation in mice. Gut 63, 598–609. [DOI] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. (2013). Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795. [DOI] [PubMed] [Google Scholar]
- Blatner NR, Mulcahy MF, Dennis KL, Scholtens D, Bentrem DJ, Phillips JD, Ham S, Sandall BP, Khan MW, Mahvi DM, et al. (2012). Expression of RORgammat marks a pathogenic regulatory T cell subset in human colon cancer. Sci Transl Med 4, 164ra159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruttger J, Karram K, Wortge S, Regen T, Marini F, Hoppmann N, Klein M, Blank T, Yona S, Wolf Y, et al. (2015). Genetic Cell Ablation Reveals Clusters of Local Self-Renewing Microglia in the Mammalian Central Nervous System. Immunity 43, 92–106. [DOI] [PubMed] [Google Scholar]
- Carmi Y, Rinott G, Dotan S, Elkabets M, Rider P, Voronov E, and Apte RN (2011). Microenvironment-derived IL-1 and IL-17 interact in the control of lung metastasis. J Immunol 186, 3462–3471. [DOI] [PubMed] [Google Scholar]
- Cataisson C, Salcedo R, Hakim S, Moffitt BA, Wright L, Yi M, Stephens R, Dai RM, Lyakh L, Schenten D, et al. (2012). IL-1R-MyD88 signaling in keratinocyte transformation and carcinogenesis. J Exp Med 209, 1689–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae WJ, Gibson TF, Zelterman D, Hao L, Henegariu O, and Bothwell AL (2010). Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America 107, 5540–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, Kang HS, Ma L, Watowich SS, Jetten AM, Tian Q, and Dong C (2009). Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 30, 576–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia M, Harrison OJ, Schiering C, Asquith MJ, Becher B, Powrie F, and Maloy KJ (2012). IL-1beta mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4+ Th17 cells. The Journal of experimental medicine 209, 1595–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffelt SB, Kersten K, Doornebal CW, Weiden J, Vrijland K, Hau CS, Verstegen NJM, Ciampricotti M, Hawinkels L, Jonkers J, and de Visser KE (2015). IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenais M, Dupaul-Chicoine J, Douglas T, Champagne C, Morizot A, and Saleh M (2017). The Interleukin (IL)-1R1 pathway is a critical negative regulator of PyMT-mediated mammary tumorigenesis and pulmonary metastasis. Oncoimmunology 6, e1287247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello CA, Simon A, and van der Meer JW (2012). Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nature reviews. Drug discovery 11, 633–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler SK, Bonsignore L, Masin M, Tardivel A, Jackstadt R, Hermeking H, Schneider P, Gross O, Tschopp J, and Yazdi AS (2012). Tissue-specific opposing functions of the inflammasome adaptor ASC in the regulation of epithelial skin carcinogenesis. Proc Natl Acad Sci U S A 109, 18384–18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elaraj DM, Weinreich DM, Varghese S, Puhlmann M, Hewitt SM, Carroll NM, Feldman ED, Turner EM, and Alexander HR (2006). The role of interleukin 1 in growth and metastasis of human cancer xenografts. Clin Cancer Res 12, 1088–1096. [DOI] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, and Flavell RA (2011). NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabets M, Krelin Y, Dotan S, Cerwenka A, Porgador A, Lichtenstein RG, White MR, Zoller M, Iwakura Y, Dinarello CA, et al. (2009). Host-derived interleukin-1alpha is important in determining the immunogenicity of 3-methylcholantrene tumor cells. J Immunol 182, 4874–4881. [DOI] [PubMed] [Google Scholar]
- Fatkhullina AR, Peshkova IO, Dzutsev A, Aghayev T, McCulloch JA, Thovarai V, Badger J, Vats R, Sundd P,Tang HY, Kossenkov AV, Hazen SL, Trinchieri G, Grivennikov SI, Koltsova EK (2018). An interleukin-23- interleukin-22 axis regulates intestinal microbial homeostasis to protect from diet-induced atherosclerosis. Immunity 49, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Sentani K, Wiese A, Sands E, Green M, Bommer GT, Cho KR, and Fearon ER (2013). Sox9 induction, ectopic Paneth cells, and mitotic spindle axis defects in mouse colon adenomatous epithelium arising from conditional biallelic Apc inactivation. The American journal of pathology 183, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron M, and Vacher J (2005). Targeted expression of Cre recombinase in macrophages and osteoclasts in transgenic mice. Genesis 41, 138–145. [DOI] [PubMed] [Google Scholar]
- Franchi L, Munoz-Planillo R, Reimer T, Eigenbrod T, and Nunez G (2010). Inflammasomes as microbial sensors. European journal of immunology 40, 611–615. [DOI] [PubMed] [Google Scholar]
- Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, Worthen GS, and Albelda SM (2009). Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell 16, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI, and Nagaraj S (2009). Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 9, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, Vermaelen K, Panaretakis T, Mignot G, Ullrich E, et al. (2009). Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nature medicine 15, 1170–1178. [DOI] [PubMed] [Google Scholar]
- Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, and Karin M (2004). IKKb links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 118, 285–296. [DOI] [PubMed] [Google Scholar]
- Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, and Karin M (2009). IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, et al. (2012). Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenberg A, Hasenberg M, Mann L, Neumann F, Borkenstein L, Stecher M, Kraus A, Engel DR, Klingberg A, Seddigh P, et al. (2015). Catchup: a mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nat Methods 12, 445–452. [DOI] [PubMed] [Google Scholar]
- Hickish T, Andre T, Wyrwicz L, Saunders M, Sarosiek T, Kocsis J, Nemecek R, Rogowski W, Lesniewski-Kmak K, Petruzelka L, et al. (2017). MABp1 as a novel antibody treatment for advanced colorectal cancer: a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol 18, 192–201. [DOI] [PubMed] [Google Scholar]
- Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, Cho KR, and Fearon ER (2007). Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res 67, 9721–9730. [DOI] [PubMed] [Google Scholar]
- Holen I, Lefley DV, Francis SE, Rennicks S, Bradbury S, Coleman RE, and Ottewell P (2016). IL-1 drives breast cancer growth and bone metastasis in vivo. Oncotarget 7, 75571–75584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, Hedl M, Zhang W, O’Connor W Jr., Murphy AJ, et al. (2012). IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 491, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaler P, Augenlicht L, and Klampfer L (2009). Macrophage-derived IL-1beta stimulates Wnt signaling and growth of colon cancer cells: a crosstalk interrupted by vitamin D3. Oncogene 28, 3892–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, Harrison O, and Powrie F (2013). Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. The Journal of experimental medicine 210, 917–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb R, Phan L, Borcherding N, Liu Y, Yuan F, Janowski AM, Xie Q, Markan KR, Li W, Potthoff MJ, et al. (2016). Obesity-associated NLRC4 inflammasome activation drives breast cancer progression. Nat Commun 7, 13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A, Sharma D, Zhu Q, Karki R, Guy CS, Vogel P, and Kanneganti TD (2016). IL-33 regulates the IgA-microbiota axis to restrain IL-1alpha-dependent colitis and tumorigenesis. J Clin Invest 126, 4469–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Allavena P, Sica A, and Balkwill F (2008). Cancer-related inflammation. Nature 454, 436–444. [DOI] [PubMed] [Google Scholar]
- Miller LS, O’Connell RM, Gutierrez MA, Pietras EM, Shahangian A, Gross CE, Thirumala A, Cheung AL, Cheng G, and Modlin RL (2006). MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity 24, 79–91. [DOI] [PubMed] [Google Scholar]
- Nakae S, Saijo S, Horai R, Sudo K, Mori S, and Iwakura Y (2003). IL-17 production from activated T cells is required for the spontaneous development of destructive arthritis in mice deficient in IL-1 receptor antagonist. Proc Natl Acad Sci U S A 100, 5986–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke AL, Delbridge LM, Balachandran S, Barber GN, and O’Riordan MX (2007). TBK1 protects vacuolar integrity during intracellular bacterial infection. PLoS Pathog 3, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker PM, MacFadyen JG, Thuren T, Everett BM, Libby P, Glynn RJ, and Group CT (2017). Effect of interleukin-1beta inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet 390, 1833–1842. [DOI] [PubMed] [Google Scholar]
- Rothwell PM, Wilson M, Price JF, Belch JF, Meade TW, and Mehta Z (2012). Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet 379, 1591–1601. [DOI] [PubMed] [Google Scholar]
- Sakurai T, He G, Matsuzawa A, Yu GY, Maeda S, Hardiman G, and Karin M (2008). Hepatocyte necrosis induced by oxidative stress and IL-1 alpha release mediate carcinogen-induced compensatory proliferation and liver tumorigenesis. Cancer Cell 14, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. (2013). Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38. [DOI] [PubMed] [Google Scholar]
- Shaw MH, Kamada N, Kim YG, and Nunez G (2012). Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. The Journal of experimental medicine 209, 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pages F, and Galon J (2011). Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer research 71, 1263–1271. [DOI] [PubMed] [Google Scholar]
- Voronov E, Shouval DS, Krelin Y, Cagnano E, Benharroch D, Iwakura Y, Dinarello CA, and Apte RN (2003). IL-1 is required for tumor invasiveness and angiogenesis. Proc Natl Acad Sci U S A 100, 2645–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Kim MK, Di Caro G, Wong J, Shalapour S, Wan J, Zhang W, Zhong Z, Sanchez-Lopez E, Wu LW, et al. (2014). Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity 41, 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 15, 1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, and Littman DR (2009). Transcriptional regulatory networks in Th17 cell differentiation. Curr Opin Immunol 21, 146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.