Abstract
Gestational age estimation at time of birth is critical for determining the degree of prematurity of the infant and for administering appropriate postnatal treatment. We present a fully automated algorithm for estimating gestational age of premature infants through smartphone lens imaging of the anterior lens capsule vasculature (ALCV). Our algorithm uses a fully convolutional network and blind image quality analyzers to segment usable anterior capsule regions. Then, it extracts ALCV features using a residual neural network architecture and trains on these features using a support vector machine-based classifier. The classification algorithm is validated using leave-one-out cross-validation on videos captured from 124 neonates. The algorithm is expected to be an influential tool for remote and point-of-care gestational age estimation of premature neonates in low-income countries. To this end, we have made the software open source.
1. Introduction
Premature birth is a significant cause of mortality in neonates and occurs in over 10% of all births annually [1]. Complications associated with premature birth are the second-leading cause of death for children under 5 years, causing over 1 million deaths in 2015 [1]. Accurate gestational age estimation is crucial for optimal neonatal outcomes [2], and a segment of infant deaths could be prevented with accurate estimation of prematurity and corresponding suitable treatment [1].
Currently, the accepted standard for estimating gestational age is prenatal ultrasound of fetal biometry, based on the high correlation between fetal size and gestational age during pregnancy [3]. However, in low-income countries, accessibility to ultrasound is limited. In these areas, gestational age is assessed via menstrual history or, postnatally, by neuromuscular and biophysical scoring systems, such as the Ballard or Dubowitz scores [3–5]. However, menstrual history is inaccurate in low literacy settings, and the complexity and subjectivity of scoring yields high error rates in estimating the degree of prematurity [6]. Thus, for a significant subset of developing regions in the world, there is a need for gestational age estimation methods that do not require expensive medical devices or expert clinical intervention.
Studies have demonstrated that the disappearance of vasculature in the anterior lens capsule is correlated with increasing gestational age by expert clinicians using the Dubowitz scoring system as gold standard [7]. In addition, inspection of the anterior lens capsule can be conducted using an ophthalmoscope connected to a smart phone camera, which is portable and easy to use. Therefore, assessing neonatal anterior lens capsule vasculature (ALCV) has the potential to serve as an alternative low-cost, portable method to estimate gestational age.
While there is a large body of work on image analysis of retinal images for diagnosis and prognosis of pediatric [8–10] and adult [11,12] ophthalmic diseases using fundus imaging, automated analysis of lens vasculature in premature infants has not been investigated. Furthermore, cellphone camera imaging of pediatric eyes yields low-quality images with a large diversity of imaging artifacts, complicating the development of reliable automated image segmentation and analysis techniques using traditional machine learning algorithms [13,14].
Fortunately, deep learning-based convolutional neural networks (CNNs) [15] have been demonstrated to be a robust tool in ophthalmic image analysis [11,16–23]. CNNs utilize hierarchal learning, in which relevant features are learned at different fields of view to optimize classification and segmentation decisions. Recent works have extended CNNs to successfully automate complex ophthalmic tasks such as identifying ellipsoid zone defects [21], cone localization [24], and retinal and fluid layer boundary segmentation [25,26]. Moreover, newer network principles, such as atrous (dilated) convolutions, batch normalization, and dropout, have helped decrease statistical variability and reduce parameter size while maintaining similar spatial fields of view. These developments have been demonstrated to reduce overfitting to small, homogenous training sets, which is often characteristic of medical data sets [27–29].
In this paper, we propose a fully automatic algorithm to segment anterior lens regions in smartphone images and classify neonatal gestational age using ALCV. This method captures videos of neonatal ALCV and uses a fully convolutional network (FCN) based on the DeepLabv3 + architecture [29] and blind image quality analyzers to extract the anterior lens region of the best frame. For classification, the method uses CNN and support vector machine (SVM) techniques to suggest a binary classification of gestational age. The performance of this algorithm was compared to ground truth, a manual method based on direct segmentation of the ALCV. Section 2 explains the segmentation and classification algorithm, Section 3 demonstrates the accuracy of the segmentation and classification models compared to ground truth and manual classification, and Section 4 describes our conclusions and future directions.
2. Methods
This section introduces our fully automatic method for classifying neonatal gestational age using ALCV images. The core steps are outlined in Fig. 1 and are described in detail in the following subsections. Finer implementation details of the algorithm are excluded from the text as they can be found in the open source software code.
Fig. 1.
Overview of algorithm for classification of pediatric eye videos by ALCV.
2.1 Data set
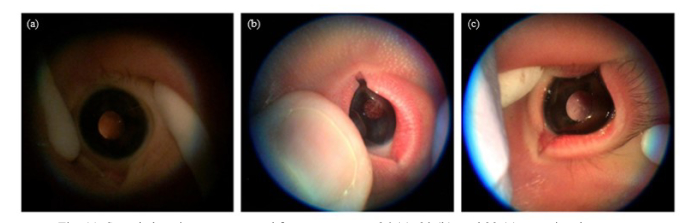
We collected a novel anterior lens video data set prospectively in a multi-institute clinical trial (ClinicalTrials.gov Identifier: NCT02346214) from September 2016 to September 2017. Neonatal patients were recruited from Loma Linda University, the University of North Carolina in Chapel Hill, and the University of Iowa. In total, the data set included 124 neonates between the ages of 26 and 42 gestational weeks. From each neonate, one to five ALCV videos between 10 and 80 seconds were acquired using a PanOptic ophthalmoscope with a Welch Allyn iExaminer attached to the lens of an iPhone 6 Plus as seen in Fig. 2(a). One video per subject was randomly chosen for analysis. Sample frames from a representative video of a neonate at 32.9 gestational weeks, which are shown in Fig. 2, demonstrate the challenges in analyzing ALCV. While frame (b) has a clear lens region, frames (c) and (d) do not show the anterior capsule region, and the anterior capsule region in frame (e) is out of focus. The median gestational age was 38.6 (IQR = 5.3) gestational weeks.
Fig. 2.
Sample imaging apparatus and video frames of 32-week, 6-day gestational aged neonate. (a) The imaging device consisted of a PanOptic ophthalmoscope using a Welch Allyn iExaminer attached to an iPhone 6 Plus. Sample raw video frames show (b) a frame, with a clear eye region, that was chosen by the automatic method, (c) frame with no eye, (d) frame with closed eye, and (e) frame with eye out of focus.
2.2 Data preprocessing – automatic coarse lens localizer (CALL)
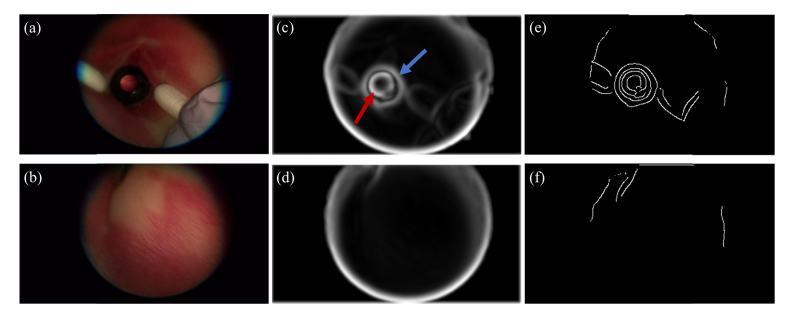
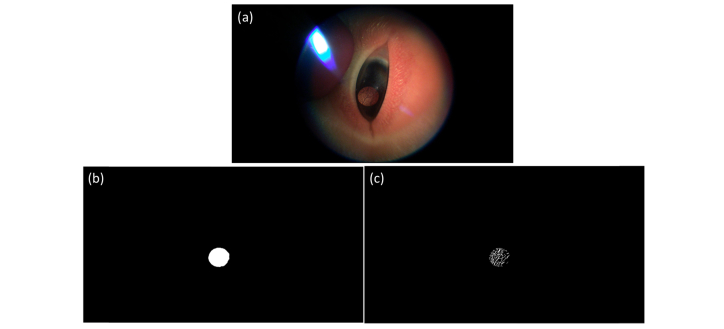
To identify frames of interest, we developed an automatic localization method termed coarse anterior lens localizer (CALL). Each subject video was partitioned into individual frames, which were then preprocessed independently. The input frame was first smoothed using a Gaussian low-pass filter with a standard deviation of 3. A 9 × 9 pixel sliding kernel was then used to locally normalize the image by dividing by the standard deviation of pixel intensity in the kernel field-of-view [30]. Finally, the Canny detection algorithm [31] was applied to extract the edges in the image frame. Figure 3 displays example outputs of image normalization and edge detection for input frames. Anatomically, the anterior lens region is located behind the pupil, which occupies a small area of the overall eye region. To ensure high quality images of the anterior lens regions were extracted, three primary criteria were used: the lens region should occupy over half of the eye region, should be a circular convex hull, and should have large intensity variations at the boundary between the anterior lens and the surrounding iris region. The frame in Fig. 3(a) has an identifiable lens region and was processed to produce the corresponding normalized frame (Fig. 3(c)) and the edge profile (Fig. 3(e)). In both the normalized and edge images, the circular lens region is seen to occupy over half of the overall eye region. In contrast, the normalized (Fig. 3(d)) and edge (Fig. 3(f)) images corresponding to the frame without a lens region (Fig. 3(b)) do not show these characteristics of the lens region.
Fig. 3.
Preprocessing of sample input frames (a) with visible lens and (b) without visible lens. (c) Normalized image of frame (a) in which the lens-iris boundary (red arrow) and iris-skin boundary (blue arrow) are visible. (d) Normalized image of frame (b) where anatomical eye boundaries are not visible. (e, f) Edge profiles of frames (a) and (b), respectively. Convex, circular regions corresponding to lens in (e) are not visible in (f).
To localize the circular regions of the frame, a circular Hough transform with a diameter greater than half of the width of the image was used. Circular edges corresponding to large global intensity variations were classified as edges of interest. The convex hull of the anterior lens was determined with MATLAB’s convhull function and was used in conjunction with the classified edges to extract the circular, convex region of the frame. Circular edges delineating a concave or flat region were ignored. If no region of the frame corresponded to both a circular and convex region, the frame was classified as not having the anterior lens region in its field of view and was not used for further analysis. For the remaining frames, the convex hull of the anterior lens defined the region of interest (ROI) and was extracted for further analysis.
2.3 Data preprocessing – automatic frame selection
If the lens region, which occupies a fraction of the frame, is out of focus from the CALL pre-processing, generic best frame selection algorithms may not be used directly for our application despite a high global quality of the video frame. Thus, the best frame in the video was automatically selected for analysis based on the quality of the lens region localized using CALL. We empirically determined that the best measure for quantifying the quality of the anterior lens region can be attained by combining two popular image quality metrics, i.e. the anisotropy-based metric of Gabarda, et al. [32] and the natural scene statistic model of Mittal, et al. [33]. We evaluated the image quality in the anterior lens capsule region by first normalizing each score to the same range and then averaging these two normalized scores. Up to the 50 highest quality lens ROIs and their corresponding frames were selected for further analysis. In the sample frames of the video shown in Fig. 2, frame (b) was automatically selected for further processing as it has a well-defined lens region, while deficient frames (c), (d), and (e) are rejected.
2.4 Automatic finer anterior lens localizer (FALL)
While CALL is a completely blind approach to lens localization and allows for frame pruning, the resulting ROIs may not be well localized, resulting in including extraneous information (false positives) or in missing the lens region (false negatives). Examples of both cases are shown in Fig. 4 and Fig. 5, respectively. Thus, a finer segmentation of the anterior lens region is needed to reduce extraneous data that could confound ALCV based estimation in a small low-quality image data set.
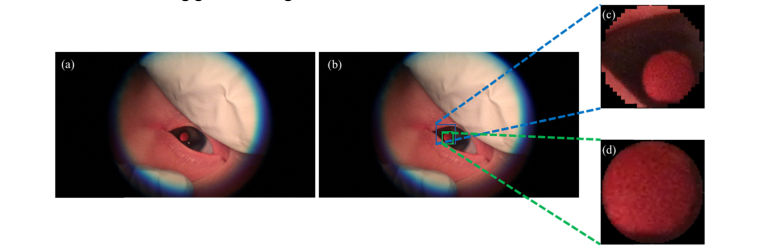
Fig. 4.
Example of CALL overestimating lens region (a) Sample frame extracted from video with (b) CALL bounding box (blue) and FALL bounding box (green). (c, d) Anterior capsule lens region segmented by CALL and FALL, respectively.
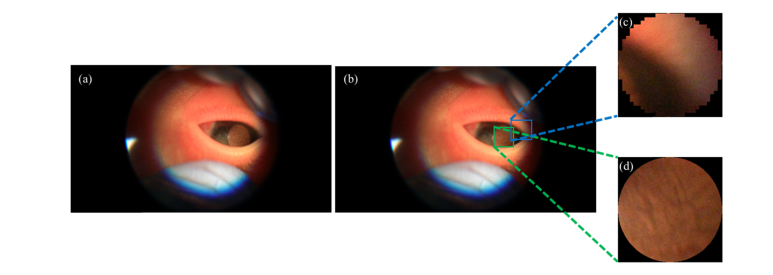
Fig. 5.
Example of CALL missing lens region (a) Sample frame extracted from video with (b) CALL bounding box (blue) and FALL bounding box (green) (c, d) Anterior capsule lens region segmented by CALL and FALL, respectively.
To better segment the anterior lens with the limitations of our data, we developed a finer anterior lens localizer (FALL) that utilizes the DeepLabv3 + CNN architecture [29]. The data set consisted of the video frames selected in 2.3, and the ground truth was generated by manually labelling the lens regions in these frames. The binary labels identified pixels that were the lens region (1) and the background (0). Due to the large dimensions of the original video frame, each frame and corresponding label mask were downsampled to dimensions of 216 × 384 pixels. The training and testing protocols are detailed in 2.5.
During inference, the frames in the test set were segmented to produce a pilot segmentation. During post processing, this pilot segmentation was refined based on the criteria that only one lens region exists in each frame, and that the lens is convex and solid. In the refining process, holes, or pixels classified as 0 surrounded by pixels classified as 1, were filled to produce fully connected components (FCCs). FCCs with a low percent solidity or insignificant area were discarded. The largest circular region inscribed in the raw segmentation was found and used as the refined mask.
Next, the refined mask was upsampled to the dimensions of the original input frame and multiplied with the frame to extract the lens region. The frame was then cropped around the lens region. These lens regions were again evaluated using the combination of blind image quality analyzers described in Section 2.3. The highest quality lens per subject was selected for analysis, resulting in 124 lens regions. Sampled images of the automatically extracted and selected anterior lens region can be seen in Fig. 6, in which an overall trend of decreasing ALCV with increasing gestational age is observed.
Fig. 6.
Sample segmented anterior lens capsule image data. Gestational age is indicated under each sample image in weeks (“w”) and days (“d”).
2.5 FALL segmentation protocol
To enforce the independence between the training, validation, and testing sets, we used 25-fold cross validation, in which the 124 subjects were split into 25 folds, each consisting of 4 to 5 subjects. Frames corresponding to subjects in 21 folds were used as the training set; those in 3 folds were used as the hold-out validation set; and those in 1 fold were used as the test set. Each fold consisted of the downsampled frame and mask pairs corresponding to the subjects in the fold. Kernel weights in the CNN were randomly initialized using Xavier initialization [34] and kernel biases were initialized to 0. Learning was optimized using Adam optimization [35] to minimize the dice loss of the lens region, , defined as
where pi is the prediction for pixel i ranging continuously from 0 to 1 and gi is the binary ground truth pixel value (0 or 1). M and N correspond to the pixel height and pixel width of the image, respectively. The loss will be minimized when pixel values in pi approach those of gi.
Training, validation, and testing mini-batch sizes were 20, 32, and 32, respectively. The initial learning rate was 0.001 and decayed by a factor of 0.2 when the loss on the validation set plateaued. The network was trained for 25 epochs and inferred on the validation set at the end of each epoch. The network state that produced the lowest loss on the validation set was used for inference on the test set. Training time was calculated per experiment (1 experiment per testing fold) and testing time was calculated per subject in the testing fold (124 subjects). The segmentation was implemented in Python using the TensorFlow [36] (version 1.4.0) and Keras [37] (version 2.1.5) frameworks.
2.6 Classification algorithm
After extracting and pre-processing the lens region of the frame, we incorporated a deep CNN-SVM-based classification system that used the extracted lens region for age-classification. We utilized the deep residual network model (Resnet-152) that was pre-trained on 1.28 million images with 1000 classes from the 2015 ImageNet Large Scale Visual Recognition Challenge [38]. In our experiments, end-to-end training of the model performed poorly due to the limited size of the data set (124 images). In addition, a transfer learning technique of fine-tuning parameters of the final fully connected layer also performed poorly. Given the limited amount of available data, we used the Resnet-152 residual CNN to extract representative features from images of the eye regions. The final softmax classification layer from the network was removed and the output of the residual network was used as a deep feature vector. The images were resized to 224 × 224 pixels centered around the center of the extracted ALCV eye region. The LIBSVM toolbox [39] was then used to classify the generated deep features using the leave-one-out-cross-validation strategy. The trained SVM used a radial basis kernel function with a cost of 1 to reduce overfitting of data to the training set. The SVM functioned as a binary classifier of gestational age across a threshold. The classes were split as follows: ≤ threshold and > threshold. The feature extraction and classification pipeline are shown in Fig. 7.
Fig. 7.
Feature Extraction and Classification Pipeline – (a) resized images of anterior lens region, (b) feature extraction (Resnet-152) [38], (c) kernel-based SVM training, (d) leave-one-out validation.
2.7 Classification evaluation protocol
Six age-thresholds were chosen for SVM classification: 33, 34, 35, 36, 37, and 38 gestational weeks. Each training/testing experiment was run independently for each of these thresholds. Each of the 124 neonates were separated into two age-classes: less than or equal to the threshold and greater than the threshold. These results were compared with the output of the SVM.
In the experiment, 124 eye images (1 per subject) were selected for training/testing. The SVM was trained on the deep features extracted from the resized version of these images using Resnet-152 and was tested using a leave-one-out (124-fold) cross-validation protocol. There was no overlap between the training and test sets.
The sensitivity is defined as the fraction of subjects whose gestational age is less than or equal to the threshold that are correctly classified. The specificity is defined as the fraction of subjects whose gestational age is greater than the threshold that are correctly classified. Total accuracy (mean accuracy) is the fraction of total subjects that were correctly classified.
Average classification time was calculated for post-processing classification only (i.e. time after the eye regions have been extracted). This time includes the computational overhead of both extracting features using the deep CNN and training the SVM.
2.8 Alternative direct manual ALCV quantification
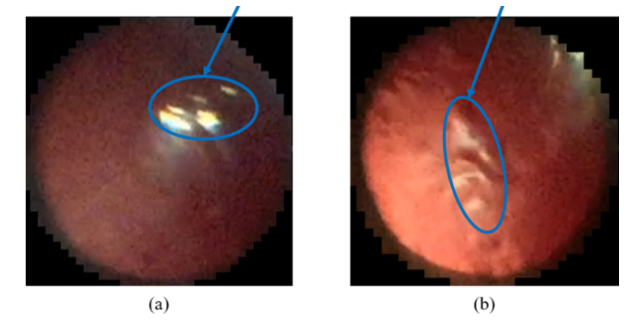
To further evaluate the effectiveness of our fully automatic method that embeds deep learning for feature selection, we devised alternative manual methods that directly quantify ALCV features. In this procedure, a user-friendly software was developed that utilizes the automatic procedure for extracting the “best” frame as discussed in Section 2.3. The software displayed the top 50 ranked frames in each video to the user. The user then selected which of these top 50 frames had the highest quality of the eye region. If none of these frames were of sufficient quality, the user could sift through the video to select the best frame. Following the selection of the best frame, the user marked the anterior lens region using the lasso tool in Adobe Photoshop. Note that some of the anterior lens region images are marred by imaging artifacts such as light reflection, as seen in Fig. 8. These artifact regions were detected using a brightness threshold calculated based on pixel intensity distribution in the image and were excluded from analysis. The user then used a stylus to annotate the anterior lens vessels using the paintbrush tool on a Microsoft Surface Tablet. These annotations were used by our ALCV analysis software to quantify key morphological features such as number of branches, branch tortuosity, branch length/width, and vessel density. The branch tortuosity was calculated using the arc length-to-chord ratio (L/C), where L is the length of the vessel branch and C is the distance between the two ends of the branch. Figure 9 shows an example of the manual annotations of the lens region mask and vasculature within the region.
Fig. 8.
Examples of light reflection artifacts in ALCV images.
Fig. 9.
Manual labelling of chosen eye frame (a) with eye region mask (b) and annotated vessels (c).
We used these features to build two comparative approaches to our fully automated algorithm. In the first method, we used the vasculature density for all subjects, fitted a linear regression of gestational age versus vasculature density, and used this regression to predict the gestational age of the neonates. In the second method, we used an SVM classification model to execute two different experiments. In the first experiment, only the vessel density was used as a feature to estimate gestational age. In the second experiment, the number of branches, median branch width, median branch length, median branch tortuosity, and vessel density were all used as features to estimate gestational age.
3. Results
3.1 ALCV region segmentation
We evaluated the segmentation pipeline after the refining protocol. As images were automatically discarded during the refining step, the number of evaluated images is smaller than the number of total segmented frames. However, to ensure that each subject was weighted evenly as well, the results were also evaluated on a per subject basis. Each subject’s score was the mean of the score of each of its refined frames. Table 1 details the segmentation performance in comparison to ground truth, where precision (P), recall (R), and dice score coefficient (DSC) [40] are defined as
In these equations TP, FN, and FP are true positive, false negative, and false positive, respectively.
Table 1. Segmentation performance metrics (mean ± standard deviation, median) per frame and per subject.
| Metric | Method | |||
|---|---|---|---|---|
|
| ||||
| Per Frame | Per Subject | |||
| Precision | 0.96 ± 0.05, 0.97 | 0.96 ± 0.04, 0.97 | ||
| Recall | 0.88 ± 0.07, 0.90 | 0.87 ± 0.07, 0.89 | ||
| DSC | 0.92 ± 0.05, 0.92 | 0.91 ± 0.04, 0.92 | ||
3.2 ALCV-based gestational age estimation
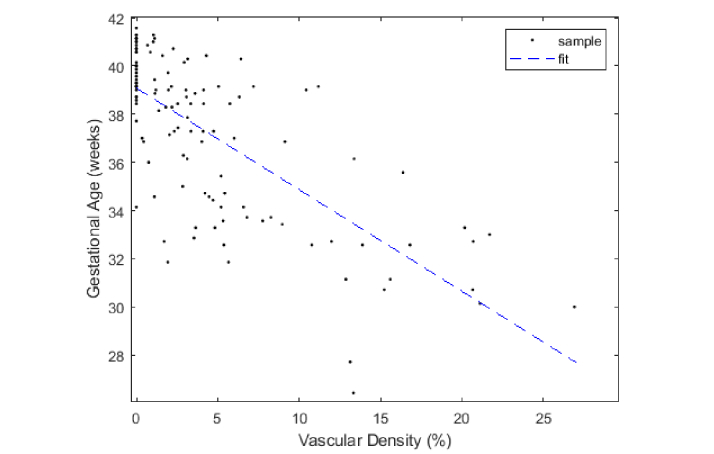
To determine the significance of ALCV as a gestational age predictor, we first quantified the correlation between vascular density and gestational age using the manual segmented ALCV data discussed in Section 2.8. The vascular density for each subject was plotted against gestational age and was fitted to a linear model, as seen in Fig. 10. A significant relationship between vascular density and gestational age was seen (p < 0.0001), with a correlation coefficient of −0.7199. To test the accuracy of the linear regression-based ALCV quantification method as a predictor of gestational age, we used leave-one-out cross validation on the 124 neonates. That is, when testing subject i, the information corresponding to subject i is removed from the training set to build the linear regression model, which is then used to predict the gestational age of subject i. The second manual model used the kernel-based SVM to classify neonatal gestational age in two experiments using different features (vasculature density alone versus all morphological features including vasculature density) as described in Section 2.6. The precision, recall, DSC, and weighted accuracy were measured for the three methods. Table 2 highlights the cross-validation results for the three different manual methods, of which the linear regression-based estimation method had the highest weighted accuracy.
Fig. 10.
Scatter plot and linear fit of ALCV density vs gestational age (m = −0.4214, r = −0.7199).
Table 2. Fraction of subjects correctly classified using manual algorithms (bold indicates highest performing metric among algorithms).
| Threshold | 33 | 34 | 35 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Method a | LV | SV | SM | LV | SV | SM | LV | SV | SM |
| Precision | 0.80 | 0.71 | 0 | 0.87 | 0.68 | 0.50 | 0.79 | 0.61 | 0.50 |
| Recall | 0.42 | 0.53 | 0 | 0.48 | 0.48 | 0.07 | 0.42 | 0.56 | 0.11 |
| DSC | 0.55 | 0.61 | nan | 0.62 | 0.57 | 0.13 | 0.55 | 0.58 | 0.18 |
| Accuracy | 0.90 | 0.90 | 0.83 | 0.87 | 0.84 | 0.78 | 0.80 | 0.77 | 0.71 |
| Threshold | 36 | 37 | 38 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Method a | LV | SV | SM | LV | SV | SM | LV | SV | SM |
| Precision | 0.83 | 0.69 | 0.50 | 0.81 | 0.71 | 0.66 | 0.73 | 0.70 | 0.69 |
| Recall | 0.49 | 0.62 | 0.10 | 0.64 | 0.62 | 0.85 | 0.80 | 0.67 | 0.85 |
| DSC | 0.61 | 0.65 | 0.17 | 0.71 | 0.66 | 0.74 | 0.77 | 0.69 | 0.76 |
| Accuracy | 0.81 | 0.79 | 0.69 | 0.81 | 0.76 | 0.77 | 0.78 | 0.73 | 0.77 |
a LV = linear regression-based method utilizing only vascular density feature, SV = SVM utilizing only vascular density feature, SM = SVM utilizing all morphological features.
For assessing the ability of our fully automatic CNN/SVM classification algorithm to generalize to an independent data set, we again used leave-one-out-cross-validation to utilize all data sets and avoid overfitting our results. The SVM for each subject is trained on the features extracted from the remaining subset of 123 extracted eye images. Table 3 details the cross-validation results compared to the best performing manual method (linear regression).
Table 3. Fraction of subjects correctly classified during cross-validation – Automatic vs Manual (bold indicates highest performing metric among algorithms).
| Threshold | 33 |
34 |
35 |
36 |
37 |
38 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method a | A | LV | A | LV | A | LV | A | LV | A | LV | A | LV | |||||
| Precision | 0.70 | 0.80 | 0.76 | 0.87 | 0.81 | 0.79 | 0.82 | 0.83 | 0.85 | 0.81 | 0.83 | 0.73 | |||||
| Recall | 0.37 | 0.42 | 0.70 | 0.48 | 0.69 | 0.42 | 0.69 | 0.49 | 0.62 | 0.64 | 0.64 | 0.80 | |||||
| DSC | 0.48 | 0.55 | 0.73 | 0.62 | 0.75 | 0.55 | 0.75 | 0.61 | 0.72 | 0.71 | 0.72 | 0.77 | |||||
| Accuracy | 0.88 | 0.90 | 0.89 | 0.87 | 0.86 | 0.80 | 0.85 | 0.81 | 0.81 | 0.81 | 0.78 | 0.78 | |||||
a A = automatic, LV = manual linear regression-based method
3.3 Implementation and computational performance
The fully automated algorithm was implemented across two environments, in MATLAB (The MathWorks, Natick, MA) and in Python. Using a workstation with two Intel Xeon CPUs and four NVIDIA Titan V GPUs, the average testing time per video was 602.2 ± 233.6s. The average testing time per frame was 1.8 ± 0.3s. The GPU units were only used during the segmentation of the FALL stage. Table 4 details the computation time and environment details per stage in the algorithm. Both FALL segmentation and SVM classification training would be a one-time process and in practice would not factor into the overall runtime of the automatic algorithm.
Table 4. Computation details for different stages in automatic pipeline. Training times are absent for stages that did not require training.
| Stage | Environment | GPU used |
Runtime (mean ± std. dev., median) (seconds) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Training | Testing, per video | Testing, per frame | ||||||||
| CALL | MATLAB | No | —————— | 562.3 ± 233.4, 505.2 | 0.632 ± 0.300, 0.564 | |||||
| FALL, Segmentation | PYTHON | Yes | 4211.2 ± 58.6, 4213.7 | 0.463 ± 0.266, 0.388 | 0.011 ± 0.006, 0.008 | |||||
| FALL, Refining | MATLAB | No | —————— | 5.47 ± 2.82, 5.40 | 0.142 ± 0.063, 0.121 | |||||
| Top Frame Extraction | MATLAB | No | —————— | 33.7 ± 10.1, 39.0 | 0.693 ± 0.021, 0.688 | |||||
| Feature Extraction + Classification |
MATLAB | No | 0.606 ± 0.023, 0.602 | 0.294 ± 0.014, 0.292 | 0.294 ± 0.014, 0.292 | |||||
The automatic software introduced in this study is available at http://people.duke.edu/~sf59/Desai_BOE_2018.htm and on GitHub at https://github.com/ad12/ALCV-pipeline. Along with the open source code, we have included the segmented images of the anterior capsule region for the 124 neonates in this study.
4. Discussion
This paper presented a fully automatic method for segmenting ALCV lens regions and estimating gestational age using smartphone imaging of the anterior lens capsule. It also presented three manual methods for gestational age estimation using ALCV density and morphology quantification. The combination of the CALL and FALL protocols automated the process for high quality frame selection and fine segmentation of the anterior lens. By reducing low-quality, extraneous information, the output of the segmentation passed relevant data to the classification stage. In addition, unlike the time-consuming manual methods, the fully automatic classification method did not rely on segmentation of ALCV, but rather used deep feature descriptors and SVM classifiers to estimate gestational age. This was particularly important in the case of low-quality images, where automatic vascular segmentation is difficult due to low contrast between vasculature and the background. The high accuracy in 2-bin classification of gestational age showed that our algorithm is robust to the low-quality smartphone images of the anterior lens capsule region.
In the linear regression-based method, the significance of the correlation coefficient suggests that there exists a strong relationship between ALCV density and gestational age. However, manual segmentation also revealed problems with annotating vasculature in low quality, occlusion, and artifact-prone images. In addition, due to the low-quality images and the inherent differences in gestational development of different subjects, not all captured images of the anterior capsule region follow the trend of high ALCV at a low gestational age and low ALCV at increasing gestational ages. In Fig. 11, the image captured for the neonate of 39 gestational weeks appears to have more vasculature than that of the 26-gestational-week neonate. As a result, while ALCV density does have a significant correlation with gestational age, even the manual ALCV estimation method could not perfectly predict gestational age. In low-income settings, the low image quality would limit the extent to which medical professionals can be accurate in manually estimating gestational age using smartphone imaging of ALCV. Differences in patient motion, lighting conditions, and scan times during acquisition can generate frames with different vessel-to-background contrast, which can impact clinician’s perception-based decisions for estimating prematurity. Moreover, the lack of availability of expert clinicians in low-income settings and the requirement of on-site training impedes the ability to scale a diagnostic pipeline for estimating prematurity. In contrast, an automated pipeline that looks at deep image features would provide a scalable, centralized process for estimating gestational age. Parallel to our clinical trial, a recent deep learning-based method has utilized images of newborns’ faces, feet, and ears to determine gestational age [41]. We expect that image features from the face, feet, and ears will be complementary to the deep ALCV features extracted in this algorithm and other potential imaging biomarkers. The potential orthogonality of the additional features can yield a more robust age estimation algorithm.
Fig. 11.
Sample best images extracted from neonates at 26 (a), 30 (b), and 39 (c) gestational weeks. Though (a) is expected to have highest vascularity in the anterior lens capsule, by inspection, the number of vessels seems larger in (b) than seen in (a).
The Resnet-152 CNN extracts patch-based features [38] that provide information about the anterior lens structure and vasculature beyond that which a simplistic ALCV density metric provides. Thus, despite fully automated execution, our fully automated gestational estimation results were overall on par with the time-consuming manual methods’ results. Indeed, as Resnet-152 was pre-trained on the ImageNet data set, the features optimized for natural images in the ImageNet data set likely did not correspond to the features of the anterior capsule region. Access to larger data sets are expected to improve the performance of this algorithm.
This fully automated method is potentially valuable for both remote and point-of-care gestational age classification. The computational efficiency of both feature extraction and classification makes this tool viable for rapid stand-alone processing in portable devices, such as smartphones. Image analysis time can be significantly reduced if shorter videos are captured (or only a small time portion of the captured video is passed to the algorithm). Open-source frameworks, such as OpenCV [42] and TensorFlow Mobile [43], have been demonstrated to be effective in optimizing image processing and machine learning inference tasks on mobile devices [43] and can be used to deploy the fully automated algorithm locally on the smartphone. However, as most smartphones do not have the computational power of workstations, the computational time may increase. To eliminate local processing, cloud-based computing platforms can be used to securely transmit video data and parallelize video preprocessing and analysis across multiple workstations.
We expect the core machine vision algorithms developed in this work to be generalizable to the task of analyzing images of neonates from different regions of the world. However, it is possible that minor modifications may be needed to adapt these algorithms for different populations. As part of our future work, we will conduct a large-scale clinical trial to collect videos from neonates in low-income countries and test the generalizability of our method to these subjects. If needed, we will also fine-tune our algorithm with data representative of populations with different geographical, racial, and socioeconomic backgrounds (e.g. by utilizing region specific training sets for SVMs). In another study, we will examine the effect of utilizing different cameras for imaging ALCV. Once again, while the core algorithms are expected to be the same, fine-tuning the machine-learned models on images captured by a specific camera may be required for optimal performance. Despite the encouraging results of our paper, there is a long road ahead in developing a robust method for newborn gestational age assessment through new combinations of existing and new non-invasive biomarkers or technology. As part of our image processing work, we are developing an image fusion technique to enhance the quality of the anterior lens region, which will improve the current CNN-based feature extraction method. We are also building a new multi-channel residual-network model that can be trained end-to-end with a small data set and that would reduce overfitting, to which deep-layered CNNs are prone. High image quality, utilization of multiple imaging biomarkers, and reducing overfitting during training are critical to developing robust CNN-based classification models. Finally, we will also assess the feasibility of emulating the techniques commonly used in eye trackers or pupillometers to eliminate the need for the time-consuming CALL stage.
Funding
Bill and Melinda Gates Foundation (OPP1119360), Google Faculty Research Award, and NIH P30-EY005722. The funding bodies did not participate in the design, data collection, analysis, interpretation of the data, or in the writing of this manuscript.
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References
- 1.World Health Organization , “Preterm Birth” (2017), retrieved http://www.who.int/mediacentre/factsheets/fs363/en/.
- 2.Skupski D. W., Owen J., Kim S., Fuchs K. M., Albert P. S., Grantz K. L., “Estimating Gestational Age From Ultrasound Fetal Biometrics,” Obstet. Gynecol. 130(2), 433–441 (2017). 10.1097/AOG.0000000000002137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee A. C., Panchal P., Folger L., Whelan H., Whelan R., Rosner B., Blencowe H., Lawn J. E., “Diagnostic Accuracy of Neonatal Assessment for Gestational Age Determination: A Systematic Review,” Pediatrics 140(6), e20171423 (2017). 10.1542/peds.2017-1423 [DOI] [PubMed] [Google Scholar]
- 4.Ballard J. L., Novak K. K., Driver M., “A simplified score for assessment of fetal maturation of newly born infants,” J. Pediatr. 95(5), 769–774 (1979). 10.1016/S0022-3476(79)80734-9 [DOI] [PubMed] [Google Scholar]
- 5.Dubowitz L. M., Dubowitz V., Goldberg C., “Clinical assessment of gestational age in the newborn infant,” J. Pediatr. 77(1), 1–10 (1970). 10.1016/S0022-3476(70)80038-5 [DOI] [PubMed] [Google Scholar]
- 6.Blencowe H., Cousens S., Chou D., Oestergaard M., Say L., Moller A.-B., Kinney M., Lawn J., “Born too soon: the global epidemiology of 15 million preterm births,” Reprod. Health 10(Suppl 1), S2 (2013). 10.1186/1742-4755-10-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hittner H. M., Hirsch N. J., Rudolph A. J., “Assessment of gestational age by examination of the anterior vascular capsule of the lens,” J. Pediatr. 91(3), 455–458 (1977). 10.1016/S0022-3476(77)81324-3 [DOI] [PubMed] [Google Scholar]
- 8.Campbell J. P., Ataer-Cansizoglu E., Bolon-Canedo V., Bozkurt A., Erdogmus D., Kalpathy-Cramer J., Patel S. N., Reynolds J. D., Horowitz J., Hutcheson K., Shapiro M., Repka M. X., Ferrone P., Drenser K., Martinez-Castellanos M. A., Ostmo S., Jonas K., Chan R. V., Chiang M. F., “Expert diagnosis of plus disease in retinopathy of prematurity from computer-based image analysis,” JAMA Ophthalmol. 134(6), 651–657 (2016). 10.1001/jamaophthalmol.2016.0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrada R., Tomasi C., Cabrera M. T., Wallace D. K., Freedman S. F., Farsiu S., “Enhanced video indirect ophthalmoscopy (VIO) via robust mosaicing,” Biomed. Opt. Express 2(10), 2871–2887 (2011). 10.1364/BOE.2.002871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estrada R., Tomasi C., Cabrera M. T., Wallace D. K., Freedman S. F., Farsiu S., “Exploratory Dijkstra forest based automatic vessel segmentation: applications in video indirect ophthalmoscopy (VIO),” Biomed. Opt. Express 3(2), 327–339 (2012). 10.1364/BOE.3.000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulshan V., Peng L., Coram M., Stumpe M. C., Wu D., Narayanaswamy A., Venugopalan S., Widner K., Madams T., Cuadros J., Kim R., Raman R., Nelson P. C., Mega J. L., Webster D. R., “Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs,” JAMA 316(22), 2402–2410 (2016). 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 12.van Grinsven M. J., Theelen T., Witkamp L., van der Heijden J., van de Ven J. P., Hoyng C. B., van Ginneken B., Sánchez C. I., “Automatic differentiation of color fundus images containing drusen or exudates using a contextual spatial pyramid approach,” Biomed. Opt. Express 7(3), 709–725 (2016). 10.1364/BOE.7.000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.N. Anantrasirichai, A. Achim, J. E. Morgan, I. Erchova, and L. Nicholson, “SVM-based texture classification in optical coherence tomography,” in Biomedical Imaging (ISBI), 2013 IEEE 10th International Symposium on, (IEEE, 2013), 1332–1335. [Google Scholar]
- 14.Srinivasan P. P., Kim L. A., Mettu P. S., Cousins S. W., Comer G. M., Izatt J. A., Farsiu S., “Fully automated detection of diabetic macular edema and dry age-related macular degeneration from optical coherence tomography images,” Biomed. Opt. Express 5(10), 3568–3577 (2014). 10.1364/BOE.5.003568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeCun Y., Bengio Y., Hinton G., “Deep learning,” Nature 521(7553), 436–444 (2015). 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 16.Asaoka R., Murata H., Iwase A., Araie M., “Detecting preperimetric glaucoma with standard automated perimetry using a deep learning classifier,” Ophthalmology 123(9), 1974–1980 (2016). 10.1016/j.ophtha.2016.05.029 [DOI] [PubMed] [Google Scholar]
- 17.Cunefare D., Fang L., Cooper R. F., Dubra A., Carroll J., Farsiu S., “Open source software for automatic detection of cone photoreceptors in adaptive optics ophthalmoscopy using convolutional neural networks,” Sci. Rep. 7(1), 6620 (2017). 10.1038/s41598-017-07103-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang L., Cunefare D., Wang C., Guymer R. H., Li S., Farsiu S., “Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search,” Biomed. Opt. Express 8(5), 2732–2744 (2017). 10.1364/BOE.8.002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gargeya R., Leng T., “Automated identification of diabetic retinopathy using deep learning,” Ophthalmology 124(7), 962–969 (2017). 10.1016/j.ophtha.2017.02.008 [DOI] [PubMed] [Google Scholar]
- 20.He Y., Carass A., Yun Y., Zhao C., Jedynak B. M., Solomon S. D., Saidha S., Calabresi P. A., Prince J. L., “Towards Topological Correct Segmentation of Macular OCT from Cascaded FCNs,” in Fetal, Infant and Ophthalmic Medical Image Analysis (Springer, 2017), pp. 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo J., Fang L., Cunefare D., Jaffe G. J., Farsiu S., “Deep longitudinal transfer learning-based automatic segmentation of photoreceptor ellipsoid zone defects on optical coherence tomography images of macular telangiectasia type 2,” Biomed. Opt. Express 9(6), 2681–2698 (2018). 10.1364/BOE.9.002681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venhuizen F. G., van Ginneken B., Liefers B., van Grinsven M. J. J. P., Fauser S., Hoyng C., Theelen T., Sánchez C. I., “Robust total retina thickness segmentation in optical coherence tomography images using convolutional neural networks,” Biomed. Opt. Express 8(7), 3292–3316 (2017). 10.1364/BOE.8.003292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao S., Bucher F., Wu Y., Rokem A., Lee C. S., Marra K. V., Fallon R., Diaz-Aguilar S., Aguilar E., Friedlander M., Lee A. Y., “Fully automated, deep learning segmentation of oxygen-induced retinopathy images,” JCI Insight 2(24), e97585 (2017). 10.1172/jci.insight.97585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunefare D., Langlo C. S., Patterson E. J., Blau S., Dubra A., Carroll J., Farsiu S., “Deep learning based detection of cone photoreceptors with multimodal adaptive optics scanning light ophthalmoscope images of achromatopsia,” Biomed. Opt. Express 9(8), 3740–3756 (2018). 10.1364/BOE.9.003740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy A. G., Conjeti S., Karri S. P. K., Sheet D., Katouzian A., Wachinger C., Navab N., “ReLayNet: retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks,” Biomed. Opt. Express 8(8), 3627–3642 (2017). 10.1364/BOE.8.003627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlegl T., Waldstein S. M., Bogunovic H., Endstraßer F., Sadeghipour A., Philip A.-M., Podkowinski D., Gerendas B. S., Langs G., Schmidt-Erfurth U., “Fully automated detection and quantification of macular fluid in OCT using deep learning,” Ophthalmology 125(4), 549–558 (2018). 10.1016/j.ophtha.2017.10.031 [DOI] [PubMed] [Google Scholar]
- 27.Ioffe S., Szegedy C., “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” arXiv preprint arXiv:1502.03167 (2015).
- 28.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R., “Dropout: a simple way to prevent neural networks from overfitting,” J. Mach. Learn. Res. 15, 1929–1958 (2014). [Google Scholar]
- 29.L.-C. Chen, Y. Zhu, G. Papandreou, F. Schroff, and H. Adam, “Encoder-decoder with atrous separable convolution for semantic image segmentation,” arXiv preprint arXiv:1802.02611 (2018).
- 30.Wang Z., Bovik A. C., “A universal image quality index,” IEEE Signal Process. Lett. 9(3), 81–84 (2002). 10.1109/97.995823 [DOI] [Google Scholar]
- 31.Canny J., “A computational approach to edge detection,” IEEE Trans. Pattern Anal. Mach. Intell. 8(6), 679–698 (1986). 10.1109/TPAMI.1986.4767851 [DOI] [PubMed] [Google Scholar]
- 32.Gabarda S., Cristóbal G., “Blind image quality assessment through anisotropy,” J. Opt. Soc. Am. A 24(12), B42–B51 (2007). 10.1364/JOSAA.24.000B42 [DOI] [PubMed] [Google Scholar]
- 33.Mittal A., Soundararajan R., Bovik A. C., “Making a “Completely Blind” Image Quality Analyzer,” IEEE Signal Process. Lett. 20(3), 209–212 (2013). 10.1109/LSP.2012.2227726 [DOI] [Google Scholar]
- 34.Glorot X., Bengio Y., “Understanding the difficulty of training deep feedforward neural networks,” in Proceedings of the thirteenth international conference on artificial intelligence and statistics, 2010), 249–256. [Google Scholar]
- 35.D. P. Kingma and J. Ba, “Adam: A method for stochastic optimization,” arXiv preprint arXiv:1412.6980 (2014).
- 36.Abadi M., Barham P., Chen J., Chen Z., Davis A., Dean J., Devin M., Ghemawat S., Irving G., Isard M., “Tensorflow: a system for large-scale machine learning,” in OSDI, 2016), 265–283. [Google Scholar]
- 37.Chollet F., “Keras: Deep learning library for theano and tensorflow,” URL: https://keras.io/k7 (2015).
- 38.He K., Zhang X., Ren S., Sun J., “Deep residual learning for image recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016), 770–778. [Google Scholar]
- 39.Chang C.-C., Lin C.-J., “LIBSVM: a library for support vector machines,” ACM transactions on intelligent systems and technology (TIST) 2, 27 (2011). [Google Scholar]
- 40.Dice L. R., “Measures of the amount of ecologic association between species,” Ecology 26(3), 297–302 (1945). 10.2307/1932409 [DOI] [Google Scholar]
- 41.Torres M. T., Valstar M. F., Henry C., Ward C., Sharkey D., “Small Sample Deep Learning for Newborn Gestational Age Estimation,” in 12th IEEE International Conference on Automatic Face & Gesture Recognition, 2017), 79–86. 10.1109/FG.2017.19 [DOI] [Google Scholar]
- 42.Bradski G., Kaehler A., “OpenCV,” Dr. Dobbs J. Softw. Tools Prof. Program. 3, 2000 (2000). [Google Scholar]
- 43.A. Ignatov, R. Timofte, P. Szczepaniak, W. Chou, K. Wang, M. Wu, T. Hartley, and L. Van Gool, “AI Benchmark: Running Deep Neural Networks on Android Smartphones,” arXiv preprint arXiv:1810.01109 (2018).