Abstract
Plants use a diverse array of cytochrome P450 monooxygenases in their biosynthetic and detoxification pathways. To determine the extent to which various maize P450s are induced in response to chemical inducers, such as naphthalic anhydride (NA), triasulfuron (T), phenobarbital, and bacterial pathogens (Erwinia stuartii, Acidovorax avenae), we have analyzed the response patterns of seven P450 transcripts after treatment of seedlings with these inducers. Each of these P450 transcripts has distinct developmental, tissue-specific, and chemical cues regulating their expression even when they encode P450s within the same biosynthetic pathway. Most notably, the CYP71C1 and CYP71C3 transcripts, encoding P450s in the DIMBOA biosynthetic pathway, are induced to the same level in response to wounding and NA treatment of younger seedlings and differentially in response to NA/T treatment of younger seedlings and NA and NA/T treatment of older seedlings. NA and T induce expression of both CYP92A1 and CYP72A5 transcripts in older seedling shoots, whereas phenobarbital induces CYP92A1 expression in older seedling shoots and highly induces CYP72A5 expression in young and older seedling roots. Expressed sequence tag (EST) 6c06b11 transcripts, encoding an undefined P450 activity, are highly induced in seedling shoots infected with bacterial pathogens.
Plants have evolved multiple strategies to protect themselves from animal, insect, and microbial invasions. The most general of these defense mechanisms exist in many plants and function against a range of organisms. Examples of these include the lignification of normal tissue to ward off passive microbial and fungal invaders, the lignification of damaged tissue to limit the extent of damage caused by active invaders, and the synthesis of toxic defense compounds capable of inhibiting common metabolic processes (e.g. psoralen derivatives that inhibit DNA replication [Berenbaum, 1991], monoterpene indole alkaloids that inhibit microtubule formation and cell division [Wink, 1993; Kutchan, 1995]). The more specific of these defense mechanisms exist in a limited number of plant species and/or function against select groups of pathogenic organisms (Seigler, 1998). Examples of these specific defenses include the synthesis of DIMBOA in maize (which inhibits proteases and oxidative enzymes in fungi, bacteria, and insects), the synthesis of cyanogenic glucosides in sorghum (which inhibit enzyme functions in insect and animal herbivores), the synthesis of nicotine alkaloids in tobacco (which inhibit neurotransmitter functions in insects), and the synthesis of monoterpenes in pines and mints (which act as feeding deterrents for insects and animals) (Niemeyer, 1988; Kutchan, 1995; Seigler, 1998).
A wide range of cytochrome P450 monooxygenases mediate the biosynthesis of lignins, terpenes, alkaloids, and the variety of other secondary compounds that act as these plant defense agents (Durst and O'Keefe, 1995; Schuler, 1996; Chou and Kutchan, 1998). Another subset of P450s, possibly including some P450s with biosynthetic functions, mediate the primary detoxification of natural and synthetic toxins encountered in the environment (Schuler, 1996; Werck-Reichhart et al., 2000). The relationships between these biosynthetic and detoxicative plant P450s are, in part, unknown because of the large number of enzymatic activities attributed to this expansive gene family and because the biological specificities of relatively few P450s have been defined. Studies examining P450-mediated metabolism of herbicides have indicated that in wheat (Triticum aestivum) and maize microsomes a range of P450s mediate N-demethylations and ring-methyl hydroxylations of phenylurea herbicides such as primisulfuron (Fonne-Pfister et al., 1990) and chlortoluron, the aryl hydroxylation of sulfonylurea herbicides such as triasulfuron (T) (Frear et al., 1991; Moreland et al., 1993; Thalacker et al., 1994; Persans and Schuler, 1995), and the aryl hydroxylation of other herbicides such as bentazon (McFadden et al., 1990). Among those which have been characterized in heterologous expression systems are the maize CYP71C1, CYP71C2, CYP71C3v1, and CYP71C4 sequences that mediate DIMBOA synthesis (Frey et al., 1997), the related maize CYP71C3v2 sequence that mediates T tolerance (M.W. Persans, J.M. Whitbred, and M.A. Schuler, unpublished data), the artichoke CYP73A1 that mediates t-cinnamic acid hydroxylation, and minimal chlortoluron ring-methyl hydroxylation (Pierrel et al., 1994; Urban et al., 1994), the artichoke CYP76B1 that effectively N-dealkylates a range of xenobiotics including chlortoluron (Robineau et al., 1998; Werck-Reichhart et al., 2000), and the soybean CYP71A10 that N-demethylates a range of phenylurea herbicides and ring-methyl hydroxylates chlortoluron (Siminszky et al., 1999).
Metabolic studies and mRNA analyses have suggested individual P450s or subsets of these P450s are induced in response to chemical inducers and varying environmental conditions. In artichoke tubers and pea (Pisum sativum) seedlings, CYP73A transcripts encoding t-CAH in the early phenylpropanoid pathway are expressed at a significant basal level and further induced in response to light (UV, high, blue, and red), wounding, pathogen attack, temperature stress, nutrient stress, and chemical (Mn, phenobarbital [PB], aminopyrine [AP]) application (Werck-Reichhart, 1995; Frank et al., 1996; Batard et al., 1997; Whitbred, 1998). In pea seedlings, transcripts encoding P450s downstream in the phenylpropanoid pathway (CYP82A1) are expressed at negligible basal levels and highly induced in response to light (UV-B), cold stress, wounding, and copper chloride treatment (simulating pathogen attack; Frank et al., 1996; Whitbred, 1998). In seedlings of maize and wheat, chemical inducers such as naphtalic anhydride (NA), PB, herbicides, ethanol, and Mn substantially induce some of the herbicide-detoxifying activities described above (Fonne-Pfister et al., 1990; Frear et al., 1991; Zimmerlin and Durst, 1992; Zimmerlin et al., 1992; Moreland et al., 1993; Barrett, 1995; Persans and Schuler, 1995) and an undefined number of other P450 transcripts (Potter et al., 1995; Persans, 1998). In the field-grown versions of these crop plants, the inducibilities of herbicide detoxification enzymes by NA are so significant that this compound is now designated as a plant safener or protectant against herbicide application (Hatzios and Wu, 1996; Davies and Caseley, 1999). Several of the compounds mentioned above (PB, AP, and Mn) have also been shown to induce 7-ethoxycoumarin O-deethylase and 7-ethoxyresorufin O-deethylase detoxifying activities and a range of P450 transcripts (CYP73A1, CYP76B1, and CYP81B1) in artichoke tuber tissues (Batard et al., 1997; Cabello-Hurtado et al., 1998; Robineau et al., 1998).
From several studies, it is clear that complex interactions exist between some of these inducers in that some chemical combinations (NA plus ethanol) additively enhance P450 metabolic activities and other chemical combinations (NA plus PB and NA plus T) synergistically enhance activities (Frear et al., 1991; Zimmerlin and Durst, 1992; Zimmerlin et al., 1992; Thalacker et al., 1994; Persans and Schuler, 1995). Studies in maize have made it apparent that developmental parameters substantially influence the responses to these types of chemical cues: The combination of NA plus T synergistically induces T hydroxylation in young (2.5-d-old) maize seedlings (88-fold over control levels), whereas only NA induces metabolism in older (6.5-d-old) seedlings (33-fold over control levels) (Persans and Schuler, 1995). RNA analyses evaluating the induction patterns of maize CYP71C3v2 transcripts in NA/T-treated 2.5-d-old and 6.5-d-old seedlings have correlate strongly with these developmentally-modulated T hydroxylase profiles (M.W. Persans, J.M. Whitbred, and M.A. Schuler, unpublished data), emphasizing the multiplicity of parameters controlling responses to chemical cues.
To further evaluate the importance of developmental parameters in the response to chemical cues, we have surveyed the expression patterns of a series of maize P450 transcripts in response to treatment with NA, T, PB, or infection with bacterial pathogens.
RESULTS
To maximize opportunities for cloning P450 isozymes involved in herbicide metabolism, poly(A)+ mRNA prepared from 6.5-d-old NA-treated maize seedlings was used in an initial RT/PCR cloning described in detail by M.W. Persans, J.M. Whitbred, and M.A. Schuler (unpublished data). Biochemical analysis had previously (Persans and Schuler, 1995) demonstrated that T hydroxylation was induced 33-fold in NA-treated 6.5-d-old seedlings and 88-fold in NA/T-treated 2.5-d-old seedlings compared with the same age untreated seedlings. In this initial cloning, mRNA was reverse transcribed and PCR amplified using a 3′-oligo(dT) primer complementary to the poly(A) tract of mRNAs and a 1,024-fold degenerate primer encoding part of a conserved amino acid sequence (EEF-PERF) located approximately 30 amino acids upstream from the heme-binding Cys. The resulting RT-PCR products were cloned using BamHI and EcoRI sites included in the 5′- and 3′-RT-PCR primers and transformants with inserts in the appropriate size range were sequenced from their 5′ end to identify clones containing the conserved F-G-R-C-G P450 signature motif. Of the five distinct P450 clones identified in this process, one clone (NA PCR 1) shared extremely close amino acid identity (93%) with the C terminus of the CYP71C3v1 sequence encoding the fourth hydroxylase in the DIMBOA biosynthetic pathway (Frey et al., 1997), substantial identity (61%–62%) with the second and third hydroxylases in this pathway and lesser identity (47%) with the first hydroxylase in the pathway. NA PCR 3 shared 70% to 71% amino acid identity with the tobacco CYP92A2 and CYP92A3 sequences (Czernic et al., 1996), 57% amino acid identity with the artichoke CYP76B1 sequence (Batard et al., 1997), and 42% amino acid identity with the maize CYP71C3 sequences (Frey et al., 1995; M.W. Persans, J.M. Whitbred, and M.A. Schuler, unpublished data). NA PCR 4 shared 59% amino acid identity with the tobacco CYP72A2 sequence (LaRosa and Smigocki, 1995), 51% to 53% amino acid identity with Catharanthus roseus CYP72A1 sequences (Vetter et al., 1992) and 28% to 30% amino acid identity with other sequenced maize P450s.
Longer cDNAs corresponding to the truncated NA PCR 3 and NA PCR 4 clones were isolated from a cDNA library constructed with 2.5-d-old NA/T-treated seedling mRNA in the pYES yeast expression vector (Invitrogen, Carlsbad, CA). Further characterization of the full-length CYP71C3v2 cDNA corresponding to NA PCR 1 is described by M.W. Persans, J.M. Whitbred, and M.A. Schuler (unpublished data). One full-length (1.6 kb) clone, designated CYP92A1 (Fig. 1), was detected with the 32P-labeled NA PCR 3 probe at high stringency, and four partial (ranging in size to 1.3 kb) clones, designated CYP72A5 (Fig. 2) were detected with the 32P-labeled NA PCR 4 probe. Sequence comparisons indicated that the full-length CYP92A1 cDNA was absolutely identical at the nucleotide level to the NA PCR 3 clone. Alignment of the CYP92A1 amino acid sequence with other P450s (Fig. 1) indicates that CYP92A1 shares 57% and 54% identity with the tobacco CYP92A2 and CYP92A3 sequences (Czernic et al., 1996), respectively, and 42% identity with the avocado CYP71A1 sequence (Bozak et al., 1990). Sequence comparisons of the partial maize CYP72A5 cDNA with NA PCR 4 clone indicated that they differ in one coding nucleotide and have identical 3′-non-translated nucleotides. This single coding nucleotide difference corresponds to an Ile-to-Thr replacement in the 78 amino acid C-terminal region encompassed by these two clones. Comparisons of the CYP72A5 amino acid sequence with the tobacco CYP72A2 and C. roseus CYP72A1 sequences (Fig. 2) indicated that 205 N-terminal amino acids were not encoded in the CYP72A5 cDNA and that the C-terminal sequences shown in Figure 2 share 53% to 54% identity with CYP72A1 sequences and 53% identity with the CYP72A2 sequence.
Figure 1.
Alignment of CYP92A1 with related P450s. Alignments of the CYP92A1 amino acid sequence with the related tobacco CYP92A2 and CYP92A3 sequences (Czernic et al., 1996) generated using the Clustal W multiple sequence alignment program (Thompson et al., 1994) are shown.
Figure 2.
Alignment of CYP72A5 with related P450s. Alignments of the partial CYP72A5 amino acid sequence with the full-length C. roseus CYP72A1 and N. plumbaginifolia CYP72A2 sequences (Vetter et al., 1992; GenBank accession no. U35226) generated using the Clustal W multiple sequence alignment program (Thompson et al., 1994) are shown. In the NA PCR 4 clone, Ile242 encoded in the CYP72A5 cDNA is replaced by Thr.
In other clonings, control mRNAs from 6.5-d-old etiolated seedlings were RT-PCR amplified using a 5′-primer encoding amino acids 320 to 326 upstream of the conserved heme-binding region in pea t-CAH (Frank et al., 1996) and a 3′ primer complementary to nucleotides encoding amino acids 463 to 469 downstream of the conserved heme-binding region. Cloning of individual RT-PCR products identified several clones identical to the maize CYP73A7 sequence encoding t-CAH and one clone identical to the maize CYP73A6 sequence encoding a slightly different t-CAH (Potter et al., 1995).
Responses to Chemical Inducers
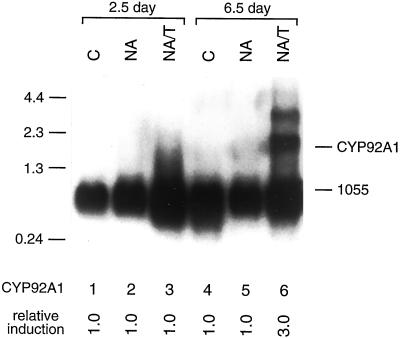
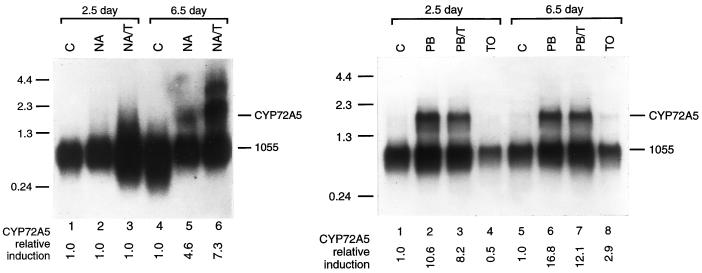
To determine the expression patterns of these various P450 transcripts in maize seedlings, poly(A)+ mRNAs from 2.5-d-old and 6.5-d-old control, NA-treated and NA/T-treated maize seedlings were hybridized with individual P450 cDNA probes using high stringency conditions that prevent cross-hybridization of super-ficially-related sequences. Subsequent hybridizations with a maize 1055 cDNA probe for a constitutive transcript (Sachs, 1991) normalized the level of induction for each transcript relative to the levels found in control seedlings of the same age. Phosphorimager quantification of these northerns indicate that CYP92A1 transcripts are virtually undetectable in 2.5-d-old NA- or NA/T-treated seedlings and in 6.5-d-old NA-treated but that they are induced at least 3.0-fold in NA/T-treated 6.5-d-old seedlings (Fig. 3; Table I). CYP92A1 transcripts were also undetectable in 2.5-d-old and 6.5-d-old seedlings treated with T alone and in 14-d-old seedling shoots. CYP72A5 transcripts are undetectable in 2.5-d-old NA- or NA/T-treated seedlings and in control seedlings (Fig. 4; Table I) and are induced 4.6-fold in 6.5-d-old NA-treated seedling shoots and 7.3-fold in 6.5-d-old NA/T-treated seedling shoots. CYP72A5 transcripts are undetectable in root tissues isolated from NA-, NA/T-, or T-treated 2.5-d-old and 6.5-d-old seedlings and in 14-d-old seedling shoots (data not shown).
Figure 3.
Induction of CYP92A1 mRNA in response to NA/T treatment. One microgram of poly(A)+ mRNA isolated from control (C), NA-, or NA/T-treated 2.5- or 6.5-d-old seedlings were electrophoresed on 1.2% (v/v) agarose-formaldehyde gels, transferred to Hybond-N nylon membrane, and probed at high stringency with the 3′-terminal sequence (bp 1,200–1,600 relative to the first coding nucleotide) of the CYP92A1 cDNA and subsequently with the constitutive maize 1055 cDNA (Sachs, 1991). CYP92A1 mRNA levels were quantified by phosphorimager analysis and normalized relative to the 1055 mRNA levels. The relative induction levels for CYP92A1 mRNA after each treatment compared with the CYP92A1 mRNA in control seedlings of the same age are shown below each lane.
Table I.
Levels of P450 transcripts induced in shoot by NA and NA/T
| Shoots | 2.5-d-old
|
6.5-d-old
|
||||
|---|---|---|---|---|---|---|
| C | NA | NA/T | C | NA | NA/T | |
| 71C3 | 1.0 | 2.8 | 5.0 | 1.0 | 2.0 | 2.0 |
| 92A1 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 3.0 |
| 72A5 | 1.0 | 1.0 | 1.0 | 1.0 | 4.6 | 7.3 |
| 71C1 | 1.0 | 2.2 | 2.1 | 1.0 | 0.6 | 0.5 |
| 73A7 | 1.0 | 1.4 | 1.7 | 1.0 | 1.7 | 1.2 |
| 51 | 1.0 | 2.8 | 2.4 | 1.0 | 0.4 | 0.4 |
| 6C06b11 | 1.0 | 2.4 | 1.1 | 1.0 | 0.8 | 0.8 |
Figure 4.
Induction of CYP72A5 in response to chemical treatments. One microgram of poly(A)+ mRNA isolated from control (C), NA-, NA/T-, PB-, PB/T-, or T-treated 2.5- or 6.5-d-old seedling shoots (left) and roots (right) were electrophoresed on 1.2% (v/v) agarose-formaldehyde gels, transferred to Hybond-N nylon membrane, and probed at high stringency with the 3′-terminal sequence (approximately 400 bp) and subsequently with the constitutive maize 1055 cDNA (Sachs, 1991). CYP72A5 mRNA levels were quantified by phosphorimager analysis and normalized relative to the 1055 mRNA levels. The relative induction levels for CYP72A5 mRNA after each treatment compared with the CYP72A5 mRNA in control seedlings of the same age are shown below each lane.
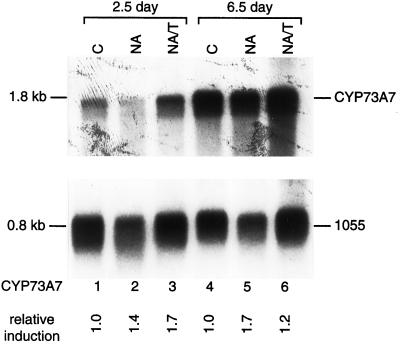
In contrast to these maize transcripts whose constitutive levels are not regulated at different stages in seedling growth, CYP73A7 transcripts encoding t-CAH are expressed in 6.5-d-old seedling shoots at a level 4.2-fold higher than in 2.5-d-old seedling shoots (Fig. 5). Also in contrast to the CYP92A1 and CYP72A5 transcripts, CYP73A7 transcripts are not significantly induced in either shoot or root tissue by NA or NA/T treatment (Table I and data not shown). Via northern analysis, CYP73A6 transcripts are undetectable in 2.5- and 6.5-d-old control, NA- and NA/T-treated seedling shoots (data not shown).
Figure 5.
Developmental induction of CYP73A7. One microgram of poly(A)+ mRNA isolated from control (C), NA-, or NA/T-treated 2.5- or 6.5-d-old seedlings were electrophoresed on 1.2% (v/v) agarose-formaldehyde gels, transferred to Hybond-N nylon membrane, and probed at high stringency with the 3′ terminal sequence (bp 960–1,410 relative to the first predicted coding nucleotide) of the CYP73A7 cDNA and subsequently with the constitutive maize 1055 cDNA (Sachs, 1991). CYP73A mRNA levels were quantified by phosphorimager analysis and normalized relative to the 1055 mRNA levels. The relative induction levels for CYP73A mRNA after each treatment compared with the CYP73A mRNA in control seedlings of the same age are shown below each lane.
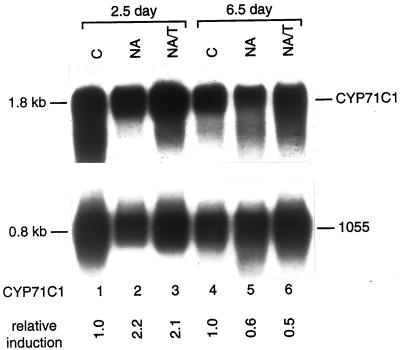
To further determine the range of other P450s induced by NA plus T, these northern blots were probed with maize EST clones agrc115 (encoding CYP71C1, the third P450 in the DIMBOA biosynthetic pathway; Frey et al., 1997), csu25 (encoding CYP51 potentially involved in sterol 14α-demethylation; Bak et al., 1997), and 6c06b11 and 7c02c12 (encoding P450s of unknown function). CYP71C1 and its related transcripts are expressed at a significant and equal level in 2.5- and 6.5-d-old seedling shoots (Fig. 6), induced 2.1-fold in NA-and NA/T-treated 2.5-d-old seedling shoots, and repressed in older NA- and NA/T-treated seedling shoots. In comparison, CYP71C3v2 transcripts encoding the fourth hydroxylase in the DIMBOA pathway are 2.8-fold induced in 2.5-d-old NA-treated seedling shoots, 2.0-fold induced in 6.5-d-old NA-treated seedling shoots, and 5.0-fold induced in 2.5-d-old NA/T-treated seedling shoots (Table I; M.W. Persans, J.M. Whitbred, and M.A. Schuler, unpublished data). In contrast to these inducibilities by NA in young shoot tissue, CYP71C1 and CYP71C3 transcripts are not induced in root tissues by either NA or NA/T treatment (Table I and data not shown). In response to these chemical inducers, CYP51 transcripts are induced to the same extent and in the same pattern in shoot tissue as CYP71C1 transcripts (Table I). Like t-CAH transcripts, P450 EST 6c06b11 transcripts are severalfold more abundant in 6.5-d-old seedling shoots than 2.5-d-old seedling shoots and not appreciably induced in response to NA or NA/T treatment of either young or old seedlings (Table I). P450 EST 7c02c12 transcripts are not detectable in 2.5- or 6.5-d-old seedling shoots and are not induced by NA or NA/T treatment (data not shown).
Figure 6.
Induction of CYP71C1 in response to NA and NA/T treatments. One microgram of poly(A)+ mRNA isolated from control (C), NA-, or NA/T-treated 2.5- or 6.5-d-old seedlings were electrophoresed on 1.2% (v/v) agarose-formaldehyde gels, transferred to Hybond-N nylon membrane, and probed at high stringency with the full-length EST sequence of the CYP71C1 cDNA and subsequently with the constitutive maize 1055 cDNA (Sachs, 1991). CYP71C1 mRNA levels were quantified by phosphorimager analysis and normalized relative to the 1055 mRNA levels. The relative induction levels for CYP71C1 mRNA after each treatment compared with the CYP71C1 mRNA in control seedlings of the same age are shown below each lane.
To determine the extent to which these P450 transcripts might be induced by non-safener treatments, several traditional P450 inducers including PB, PB plus T, wounding, and UV-C light were applied separately to root and shoot tissues. Of these, only PB treatment and wounding had any effect on the CYP92A1 transcript levels, and this effect was only observed in older shoots. Specifically, PB and wounding induced CYP92A1 transcripts 1.7- and 1.9-fold, respectively, in 6.5-d-old seedling shoots but not in 2.5-d-old shoots or any root tissues tested (Tables II and III). T and UV-C light treatments did not induce CYP92A1 transcripts in the shoots or roots at either stage of development. PB treatment exerted a unique and considerably stronger effect on the CYP72A5 transcript levels in root tissues (Fig. 4, right) with CYP72A5 transcripts induced 10.6-fold in 2.5-d-old roots and 16.8-fold in 6.5-d-old roots (Tables II and III). PB treatment in combination with T somewhat repressed this induction resulting in 8.2- and 12.1-fold inductions in 2.5- and 6.5-d-old seedling roots. No comparable induction occurred in shoot tissue in response to PB or PB/T treatment (Table II). Wounding had no significant influence on the levels of CYP72A5 transcripts in shoots or roots at any stage of development (Tables II and III).
Table II.
Level of P450 transcripts induced in shoots by PB, PB/T, and T
| Shoots | 2.5-d-Old
|
6.5-d-Old
|
2.5-d-Old Wounding | 6.5-d-Old Wounding | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C | PB | PB/T | T | C | PB | PB/T | T | |||
| 71C3 | No response | No response | No response | No response | No response | No response | No response | No response | 1.4 | 1.7 |
| 92A1 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.7 | 1.7 | 1.0 | 1.0 | 1.9 |
| 72A5 | No response | No response | No response | No response | No response | No response | No response | No response | No response | No response |
| 73A9 | No response | No response | No response | No response | No response | No response | No response | No response | 1.6 | 2.1 |
| 71C1 | No response | No response | No response | No response | No response | No response | No response | No response | 1.3 | 1.3 |
| 51 | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked |
| 6C06b11 | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked |
Table III.
Levels of P450 transcripts induced in roots by PB, PB/T, and T
| Roots | 2.5-d-Old
|
6.5-d-Old
|
2.5-d-Old Wounding | 6.5-d-Old Wounding | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C | PB | PB/T | T | C | PB | PB/T | T | |||
| 71C3 | No response | No response | No response | No response | No response | No response | No response | No response | No response | No response |
| 92A1 | No response | No response | No response | No response | No response | No response | No response | No response | No response | No response |
| 72A5 | 1.0 | 10.6 | 8.2 | 0.5 | 1.0 | 16.8 | 12.1 | 2.9 | No effect | No effect |
| 73A9 | No response | No response | No response | No response | No response | No response | No response | No response | No response | No response |
| 71C1 | No response | No response | No response | No response | No response | No response | No response | No response | No response | No response |
| 51 | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked |
| 6C06b11 | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked | Not checked |
CYP73A7 and CYP71C1 transcript levels in shoots and roots were unaffected by PB treatment with or without T. Wounding induced CYP73A7 transcript levels 1.6-fold in 2.5-d-old seedling shoots and 2.1-fold in 6.5-d-old seedling shoots but did not affect CYP73A7 transcript accumulation in root tissues (Tables II and III). Wounding induced CYP71C1 transcript levels to a much lesser degree (1.3-fold) in both 2.5- and 6.5-d-old seedling shoots and did not affect CYP71C1 transcript levels in root tissues (Tables II and III).
Responses to Bacterial Pathogens
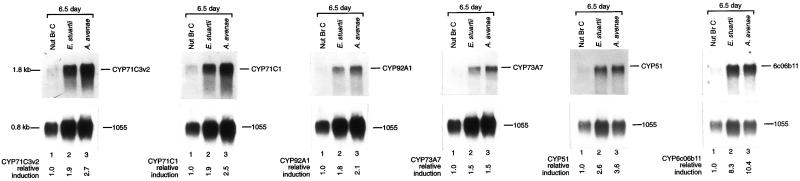
To determine the responses of these genes to challenge with pathogens, the mRNA expression patterns in 6.5-d-old seedlings were analyzed after 12- to 16-h infection with maize bacterial pathogens Erwinia stuartii and Acidovorax avenae (Fig. 7). Relative to control seedlings treated with nutrient broth, the pathogen E. stuartii induced CYP71C3v2, CYP71C1, CYP92A1, CYP73A7, CYP51, and CYP6c06b11 transcripts 1.9-fold, 1.9-, 1.8-, 1.5-, 2.6-, and 8.3-fold, respectively (Fig. 7). The pathogen A. avenae induced CYP71C3v2, CYP71C1, CYP92A1, CYP73A7, CYP51, and CYP6c06b11 transcripts 2.7-, 2.5-, 2.1-, 1.5-, 3.6-, and 10.4-fold, respectively. Neither of these pathogens induce CYP72A5 transcripts to any extent in 6.5-d-old maize shoots (data not shown).
Figure 7.
Pathogen induction. One microgram of poly(A)+ mRNA isolated from nutrient broth control, E. stuartii-treated, or A. avenae-treated 6.5-d-old seedling shoots were electrophoresed on 1.2% (v/v) agarose-formaldehyde gels, transferred to Hybond-N nylon membrane, and probed at high stringency with CYP92A1, CYP72A5, CYP71C3, CYP71C1, CYP73A7, CYPS1, or CYP6c06b11 probes and subsequently normalized with the 1055 cDNA. The relative induction levels for each transcript compared with control seedlings of the same age are shown below each lane.
Genomic Analysis
The genomic copy numbers of the CYP92A1, CYP72A5, CYP71C3v2, and CYP73A7 genes were assessed using restriction enzymes that do not cleave within individual cDNAs and gene-specific probes representing the 3′-coding sequences. By this analysis, CYP92A1 sequences are contained within a single large HindIII (10–12 kb) and two smaller EcoRI (6.5 kb, 9.0 kb) fragments, CYP72A5 sequence are contained within single small EcoRI (3.6 kb) and XhoI (4.2 kb) fragments, CYP71C3v2 sequences are contained within single EcoRI (7.5 kb) and XbaI (7.0 kb) fragments, and CYP73A7/CYP73A6 sequences are contained within two EcoRI (4.0 kb, 6.5 kb) fragments (Persans, 1998). This level of analysis suggests that CYP92A1 is encoded by a small group of closely spaced genes, that CYP72A5 and CYP71C3v2 are encoded by unique genes, and that the CYP73A7/CYP73A6 proteins are encoded by a small group of unlinked genes.
Sequence and database comparisons suggest that the small cluster of CYP92A1 genes actually encompass a unique CYP92A1 gene, which exhibits no sequence variation between our cDNA and RT-PCR clones, and at least one closely-related CYP92A4 gene (95% identical; M. Barrett, unpublished data). Additional comparisons are not possible since neither CYP92A sequence is represented in existing EST databases. Sequence and database comparisons support the singularity of the CYP71C3v2 gene in the maize inbred B73 genome: No sequence variations occur between the CYP71C3v2 cDNA and RT-PCR clones and the only available EST clone in the maize B73 EST database is identical to the CYP71C3v2 cDNA sequence. More expansive database and genomic DNA comparisons indicate that a single related CYP71C3v1 gene (99% identical) exists in the maize inbred CI31A genome (Frey et al., 1997). Sequence comparisons also support the singularity of the CYP72A5 gene in the maize inbred B73 genome: A single nucleotide difference occurs between the CYP72A5 cDNA and RT-PCR clones, and the single available EST clone in the maize B73 EST database is identical to the CYP72A5 cDNA sequence.
To define the internal organization of these genes in the B73 line, maize genomic DNA was PCR-amplified with primers spanning different regions of the gene. PCR amplification of genomic DNA with the INT PR1 and INT PR3 primers positioned internally within the CYP92A1 coding sequence generates products that are the same size as products generated from the CYP92A1 cDNA (Persans, 1998). This result indicates that there is no intron positioned between these two primers. PCR amplification of genomic DNA with INT PR1 and INT PR2 primers positioned within the CYP72A5 coding sequence generate a single product that is approximately 370 nucleotides larger than that generated from the CYP72A5 cDNA (data not shown). The subsequent cloning and sequencing of the largest amplified product indicated that one intron of 371 nucleotides occurs within the CYP72A5 coding sequence between amino acids 172 and 173.
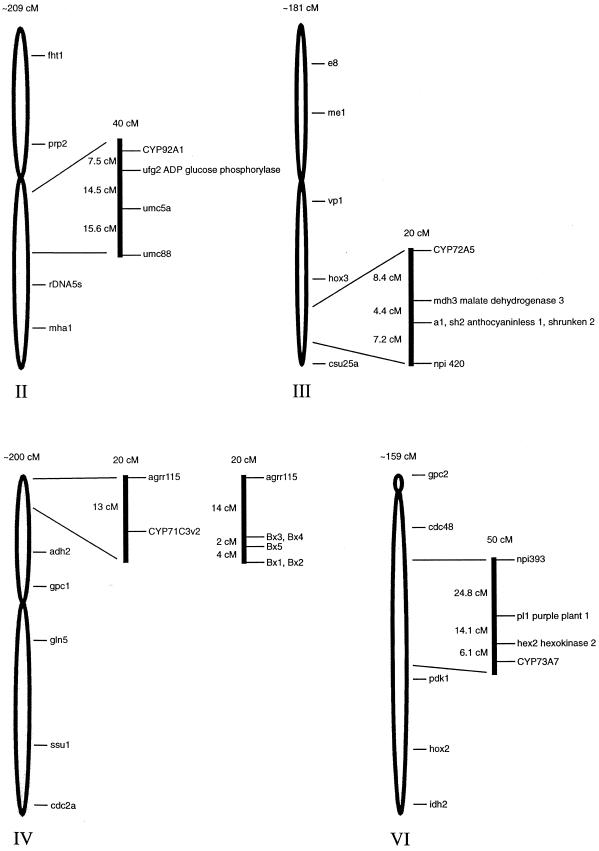
The map locations of the CYP92A1, CYP72A5, and CYP73A7 genes in recombinant maize inbred populations were determined as outlined in “Materials and Methods.” This analysis localizes the CYP92A1 gene to a single locus on the long arm of chromosome 2 (Fig. 8), the CYP72A5 gene to the long arm of chromosome 3, the CYP71C3v2 gene to the short arm of chromosome 4, and the CYP73A7 gene to the long arm of chromosome 6.
Figure 8.
P450 map locations.
To determine the extent to which the CYP92A1 and CYP72A5 genes are conserved between plant species, several genomes including pea, wheat, Arabidopsis, three different maize inbred lines (B73, CI31A, and Mo17), and teosinte (Zea mays var parviglumis) were subjected to high stringency PCR amplification with primers directed against coding sequences. Amplification with the CYP92A1 INT PR1 and INT PR3 primers, which do not span an intron, and hybridization with a 32P-labeled fragment of the CYP92A1 cDNA resulted in no discernible amplification products from pea, wheat, and Arabidopsis genomic DNA and a single cross-hybridizing product of approximately 550 bp from all of the maize varieties and teosinte (Persans, 1998). This product, which corresponds to the expected size of a PCR product spanning this intronless portion of the CYP92A1 gene, indicates that all of these varieties contain close CYP92A1 homologs. Amplification with the CYP72A5 INT PR1 and INT PR2 primers, which span the fourth intron in this gene (J. Wang, unpublished data) and hybridization with a 32P-labeled fragment of the CYP72A5 cDNA resulted in no discernible bands from pea, wheat, or Arabidopsis genomic DNA and a single 740- to 800-bp product from all of the maize varieties as well as teosinte (Persans, 1998). This product, which is the expected size of a PCR product spanning a variable length intron in this region of the CYP72A5 gene, again, indicates that each of these lines contain a single CYP72A5 gene. Similar amplification of the CYP71C3-related genes in these species with primers, which span a region of the CYP71C3v2 coding sequence containing two introns, results in a single 1,100-bp product from all of the maize varieties including teosinte (Persans, 1998).
DISCUSSION
Using a degenerate RT-PCR strategy, we have cloned four highly divergent P450 transcripts that are expressed in NA-induced maize coleoptiles. The derived amino acid sequences for these P450s, which share little amino acid identity (13%–23%) with one another, indicate that these transcripts represent distinct P450 subfamilies. Of these, three represent novel maize P450s not previously reported in the literature and one (CYP71C3v2) shares extensive homology with a maize P450 in the DIMBOA biosynthetic pathway (Frey et al., 1995, 1997).
Northern analyses of the transcripts complementary to CYP92A1, CYP72A5, and five other maize P450 ESTs indicates that most of these P450 transcripts are expressed with their own distinct pattern of RNA accumulation in response to chemical treatment and wounding. Of the P450s whose function has not yet been defined, CYP92A1 mRNAs show enhanced accumulation in response to NA/T treatment in older seedling shoots and CYP72A5 mRNAs show significant accumulation in response to NA treatment in older shoots and substantially more accumulation in response to NA/T treatment. Induction of these two P450 transcripts by this plant safener and herbicide is developmentally- and tissue-specifically regulated. Neither of these transcripts is detectable or inducible in 2.5-d-old seedling shoots or in any age root after exposure to these two chemicals. For both CYP92A1 and CYP72A5, the enhanced transcript accumulation at 6.5 d in response to NA and T is dependent on pretreatment with the plant safener; T treatment alone does not induce CYP92A1 transcripts at either 2.5 or 6.5 d. In contrast to their similar ability to be induced in older seedling shoots by the combination of NA and T, CYP92A1 transcripts are induced to a small extent in older seedling shoots in response to PB and PB in combination with T and CYP72A5 transcripts are very significantly induced in young and older seedling roots in response to PB and to a lesser extent in response to PB in combination with T. These variations indicate that NA and PB activate expression of these genes through different tissue-specific pathways and that T only influences expression in the presence of NA. Differential induction of the CYP92A1 and CYP72A5 transcripts is also apparent in their responses to wounding and bacterial pathogens: both of these biotic stimuli induce CYP92A1 mRNA in 6.5-d-old seedlings and neither stimulus induces CYP72A5 mRNA. Maize EST 6c06b11 transcripts, which show little identity (<30%) with other known P450 sequences, are not induced by either NA- or NA/T- treatment of older seedlings and, with respect to these chemical inducers, their expression contrasts with CYP92A1 and CYP72A5.
Of the P450s with tentative functions, CYP73A7 transcripts encoding t-CAH, are expressed at a high constitutive level in 2.5-d-old shoots and at a 4.2-fold higher level in control 6.5-d-old shoots. As expected for an enzyme needed for the production of lignin and other phenylpropanoid end products, wounding and bacterial infections induce CYP73A7 transcripts severalfold above these high constitutive levels, whereas NA and NA/T treatments exert more marginal effects on the CYP73A7 transcript levels (1.2- to 1.7-fold relative to control seedlings at each of these ages). These molecular analyses support previous conclusions, derived from metabolic studies (Zimmerlin and Durst, 1992; Moreland et al., 1993; Persans and Schuler, 1995), that t-CAH transcripts are not induced by NA and NA/T treatments. CYP73A6 transcripts were not detected at either 2.5 or 6.5 d in the shoots, suggesting that CYP73A7 transcripts encode the major t-CAH species present in these developmental stages. Maize EST csu25 transcripts, which are highly homologous to sorghum and wheat CYP51 transcripts and presumed to have a function in obtusifoliol 14-demethylation (Bak et al., 1997; Cabello-Hurtado et al., 1997), are induced in response to NA treatment of young seedlings and repressed in older seedlings.
CYP71C1 transcripts encoding the third P450 in the DIMBOA biosynthetic pathway are induced in NA-treated 2.5-d-old shoots in wounded and bacterially infected 6.5-d-old shoots to approximately the same extent as CYP71C3v2 transcripts encoding the fourth P450 in DIMBOA synthesis. But, with other inducers and stages in seedling development, CYP71C1 and CYP71C3v2 transcripts are expressed differently. With respect to the inducers that we have analyzed, the most apparent difference in these transcripts occurs in the response to NA/T treatment of younger shoots and in NA and in NA/T treatment of older seedlings: CYP71C1 transcripts are not induced in 2.5-d-old seedlings by T in combination with NA and are repressed in older seedlings by both NA and NA/T treatment; CYP71C3v2 transcripts are additively induced in young shoots by T in combination with NA and induced in older shoots by NA. These similarities in induction patterns with respect to wounding and bacterial pathogens are consistent with coordinate induction of consecutive enzymes in a pathway needed to block bacterial infections. These differences with respect to safener and herbicide induction are consistent with the recruitment of select P450s within a pathway for detoxification of one or more herbicides.
Genomic DNA analysis that shows a single, large and multiple, small DNA fragments hybridizing with the CYP92A1 cDNA indicates that CYP92A1 is encoded within a small group of closely-spaced genes in the inbred B73 line. Genomic mapping places this subset of genes within a single locus on chromosome 2 near the maize ADP Glc phosphorylase gene. In justaposition to this copy number assessment is the fact that multiple primer sets directed against internal segments of the CYP92A1 gene generate single band PCR amplified products from maize genomic DNA. This latter data, which is indicative of a single copy CYP92A1 gene, are reconciled with the genomic DNA analysis by suggesting that the closely spaced genes detected by high stringency Southern analysis represent related, but nonidentical CYP92A sequences. The recent cloning of a closely related CYP92A4 cDNA supports this conclusion (M. Barrett, unpublished data). The fact that the full-length CYP92A1 cDNA and its corresponding RT-PCR clone (NA PCR 3) isolated, respectively, from NA/T- and NA-treated maize seedlings are identical and the fact that CYP92A1-related sequences are absent from available EST databases suggest that CYP92A1-related sequences (e.g. CYP92A4) are not expressed in response to these particular chemical inducers.
By similar analysis, CYP72A5 appears to be encoded by a single gene or a very small cluster of genes in the inbred B73 genome. The CYP72A5 cDNA, originally isolated from a cDNA library prepared with NA/T-treated 2.5-d-old seedling RNA, is only one nucleotide different from the NA PCR 4 clone amplified from NA-treated maize seedling mRNA. Based on its perfect identity in this region with an EST clone isolated from the maize inbred B73 line, we presume that this nucleotide difference in the original RT-PCR clone represents a PCR-generated mistake rather than an allelic variant of the CYP72A5 cDNA clone. In agreement with this, the CYP72A5 cDNA hybridizes with a single relatively small fragment of maize genomic DNA (var B73) that exhibits no apparent RFLP differences with the CYP72A5 cDNA. Also, PCR amplification across the intron positioned within the last one-half of the CYP72A5 coding sequence provides no evidence for additional CYP72A5 genes.
Splice site comparisons across the available sequence of this CYP72A5 intron indicate that when compared with other plant introns (Simpson and Filipowicz, 1996; Schuler, 1998), it is unusual in its lack of U1 snRNA complementarity upstream from the 5′-splice site and uridines upstream from the 3′-splice site. To compensate for this, the sequences immediately downstream from the 5′-splice site (ACT/GUAAGU; double underlines designate complementarity) share extended complementarity with the U1 snRNA used in recognition of this site, and the sequence immediately upstream from the 3′-splice site maintains the five nucleotides optimal for recognition of this site (UGCAG/; Baynton et al., 1996). In addition, the AU transitions between exon and intron sequences in this transcript are 11% and 13% across the 5′- and 3′-splice sites and in a range consistent with many other plant introns (Simpson and Filipowicz, 1996; Schuler, 1998).
The high stringency genomic PCR amplification of genomic DNA from pea, wheat, Arabidopsis, three different maize varieties (B73, CI31A, and Mo17), and teosinte and subsequent high stringency probing with CYP92A1 and CYP72A5 indicate that teosinte and the various maize varieties (B73, CI31A, and Mo17) possess closely related CYP92A1 and CYP72A5 genes. The size of the PCR amplification product and the absence of introns in this region of the CYP92A1 sequence suggest that the organization of the CYP92A1 gene is evolutionarily conserved in maize and teosinte. The failure to generate corresponding PCR amplification products using wheat, pea, and Arabidopsis genomic DNAs indicates that these species do not contain closely related CYP92A1 homologs. Likewise, the PCR product patterns indicate that the organizations of the maize and teosinte CYP72A5 genes are highly conserved and that closely related homologs are not present in wheat, pea, and Arabidopsis.
MATERIALS AND METHODS
Seed Growth and Herbicide Application
Approximately 100 g of captan-treated corn seeds (Zea mays inbred B73) was soaked overnight in a 1-L flask with flowing tap water. Seeds (not treated with captan) were sterilized using a 30% (v/v) bleach solution containing 0.05% (v/v) Tween 20 for 45 min and washed four times with 400 mL of sterile distilled water. For induction with NA, 100 g of seeds was coated with the inducer by shaking the seeds vigorously in a 100-mL bottle with 1 g of dry powdered NA. Seeds were then arranged in rows on sterile white teri towels and moistened with sterile distilled water. The teri towels were subsequently sandwiched between cafeteria trays and placed upright in a tub of distilled water. The seeds were grown at 25°C to 30°C for 2.5 or 6.5 d. Approximately 16 h before harvesting, the seedlings were T treated by soaking the teri towel with 25 mL of a solution containing (0.052%, w/v) of a 75% (w/w) powder of the commercial form of T (Amber, provided by Novartis, Research Triangle Park, NC) corresponding to a final concentration of 1 mm T. For RNA extraction, 5 to 10 g of maize coleoptiles was collected, frozen in liquid nitrogen, and stored at −80°C.
For induction with PB, the maize plants were grown as above but watered with an 8- to 10-mm PB solution (pH 7.0). For induction by wounding, maize shoots were sliced in 0.25-cm intervals but not so deep as to cut off the coleoptiles. UV-C light (254 nm) treatment was administered by directly placing the maize seedlings on a UV transilluminator (6,000 W/cm2) for 5 min.
RNA Extractions
Total RNA was isolated according to the method of Puissant and Houdebine (1990) from approximately 5 to 10 g of liquid nitrogen frozen maize coleoptiles ground to a fine powder in a mortar and pestle. Following lithium chloride extraction and isopropanol precipitation, each nucleic acid sample was resuspended in 300 μL of sterile water and frozen at −80°C. Typically, 1 mg of total RNA was recovered from 10 g of coleoptile tissue.
Poly(A)+ mRNA was isolated from approximately 1 mg of total RNA using the rapid mRNA purification kit (Amresco, Solon, OH) as outlined in the manufacturer's directions. The mRNA was resuspended in sterile water and stored at −80°C. Typically, 10 μg of mRNA was recovered per milligram of total RNA.
RT-PCR Cloning
One-hundred nanograms of mRNA isolated from NA-treated 6.5-d-old seedlings was reverse transcribed at 50°C for 30 min in a 50-μL reaction containing 4 units of avian myeloblasto-virus reverse transcriptase (Promega, Madison, WI) and 100 pmol oligo(dT) primer (Persans, 1998) in 1× PCR buffer (50 mm KCl, 10 mm Tris-HCl, pH 8.4, 200 μm dNTPs, and 50 μg/mL gelatin). The first-strand cDNA products were PCR amplified in a 50 μL reaction containing 2.5 units of Taq polymerase (Gibco BRL, Gaithersburg, MD), 100 pmol of the degenerate PN-3, and nondegenerate oligo(dT) primers (Persans, 1998). Twenty-five cycles of PCR amplification were performed with each consisting of 95°C denaturation for 1 min, 42°C or 60°C annealing for 2 min, and 72°C extension for 2 min. A final 5-min 72°C extension step was done to complete synthesis of all DNA strands.
For cloning, one-half of the RT-PCR products derived from a single amplification reaction were phenol:chloroform (1:1) extracted, ethanol precipitated, resuspended in sterile water, and digested with EcoRI and BamHI, and ligated into EcoRI-BamHI-cut pBluescript SK+ vector (Stratagene, La Jolla, CA). The EcoRI-BamHI inserts of 800 ampicillin-resistant transformants were sized on 2.2% (v/v) agarose gels and 90 clones in the 300- to 500-bp range were sequenced using T3 and T7 primers complementary to the Bluescript SK+ vector and a Sequenase 2.0 kit (U.S. Biochemicals, Cleveland).
A maize CYP73A7 RT-PCR clone was obtained by RT-PCR amplification using the conditions outlined above and degenerate tCAH 5′ and tCAH 3′ primers complementary/identical to conserved amino acids 320 to 326 and 463 to 469 in the pea (Pisum sativum) t-CAH (CYP73A9; Frank et al., 1996) sequence; these sequences are conserved in the maize t-CAH (CYP73A7; Potter et al., 1995) sequence.
Northern Analysis
One microgram of mRNA was electrophoresed on 1.2% (w/v) agarose-formaldehyde gels and capillary-blotted to Hybond-N nylon membranes (Amersham, Arlington Heights, IL) overnight using 10× SSC. Membranes were UV-crosslinked using a Stratalinker (Stratagene) and prehybridized in 200 mm Na2PO4, pH 7.2, 5% (v/v) SDS, 1 mm EDTA, 10 mg/mL bovine serum albumin, 0.1 mg/mL sheared salmon sperm DNA for at least 2 h at 65°C. Blots were probed with denatured 32P-labeled probes added directly to the prehybridization solution at 60°C to 65°C for 12 to 16 h. Blots were washed twice for 15 min at 60°C to 65°C with 40 mm Na2PO4, pH 7.2, 5% (v/v) SDS, 1 mm EDTA, 5 mg/mL bovine serum albumin, washed once for 5 to 30 min at 60°C to 65°C in 40 mm Na2PO4, pH 7.2, 1% (v/v) SDS, 1 mm EDTA and autoradiographed at −80°C. For quantification, hybridization signals were quantified by Phosphorimagery (Molecular Dynamics, Sunnyvale, CA) and compared after normalization with the level of constitutive maize 1055 mRNA (Sachs, 1991).
cDNA Library Construction and Screening
Three micrograms of mRNA from NA/T-treated 2.5-d-old maize seedlings was reverse transcribed at 42°C for 1.5 h using 33 units of avian myeloblasto-virus reverse transcriptase (Promega) in a 50-μL reaction containing 1× RT buffer (50 mm Tris-HCl, pH 8.3, 50 mm KCl, 10 mm MgCl2, 10 mm dithiothreitol, and 0.5 mm spermidine), 500 μm dNTPs, 20 units of RNAsin (Promega), and 5.7 μg NotI oligo(dT) primer (Persans, 1998). After reverse transcription, 72 μL of 5× second strand buffer (94 mm Tris-HCl, pH 8.0, 453 mm KCl, 23 mm MgCl2, 18.75 mm dithiothreitol, and 200 μm dNTPs), 3 units of RNase H (Boehringer Mannheim, Indianapolis), and 90 units of DNA polymerase I (Boehringer Mannheim) were added, and the second- strand reaction was incubated at 15°C for 2.5 h. The cDNA was phenol:chloroform (1:1) extracted, ethanol precipitated, and resuspended in 38 μL of sterile water, 10 μL of 5× end-polishing buffer [90 mm (NH4)2SO4, 330 mm Tris-HCl, pH 8.3, 33 mm MgCl2, 50 mm β-mercaptoethanol, and 200 μm dNTPs]. The repair reaction was carried out with 10 units of T4 DNA polymerase (Gibco BRL) for 1 h at 37°C, phenol:chloroform (1:1) extracted, and ethanol precipitated. A 30-μL adaptor ligation reaction was set up containing 2 μg of BstXI adaptors (Invitrogen, Carlsbad, CA) and 2.5 units of T4 DNA ligase. After an overnight 15°C ligation, excess adaptors were removed by centrifugation through a 0.5-mL Sephacryl S-400 (Promega) column. Test ligations containing 1, 2, or 3 μL of the cDNA insert and 100 ng of BstXI-cut pYES vector (Invitrogen) in 20 μL of ligation reactions were incubated overnight at 4°C, and 5 to 10 μL of each ligation reaction were electroporated into electrocompetent TOPP10F′ cells (Invitrogen). Approximately 40,000 primary transformants were screened by colony hybridization using inserts from the NA PCR 3, 4, and 5 clones and the conditions previously described for northern analysis. For the primary and secondary screens, equivalent counts of the NA PCR 1, 3, 4, and 5 probes were combined. For the tertiary screen, 32P-labeled probes representing individual NA PCR clones were used to screen restriction fragments derived from clones identified in the secondary screen.
Genomic Mapping Analysis
Genomic DNA isolated from 210 progeny of the F3 generation of the maize inbreds Tex6 and Mo17 was used for mapping. Forty micrograms of DNA was electrophoresed on a 0.8% (v/v) agarose gel, blotted to Hybond-N nylon membranes overnight using 20× SSC, and probed with 3′-terminal gene fragments (last 500 bp) of each P450 cDNA. The blots were prehybridized at 65°C for at least 2 h in 50 mm Tris-HCl, pH 8.0, 10 mm EDTA, pH 8.0, 5× SSC, 5× Denhardt's solution, 0.2% (v/v) SDS, 7.5% (v/v) dextran sulfate, and 100 μg/mL sheared salmon sperm DNA, then a 32P-labeled probe was added, and the blots were hybridized overnight at 65°C. After hybridization, the hybridization solution was drained off and the blots were washed with 2× SSC, 0.1% (v/v) SDS for 5 min at 25°C, and washed twice for 15 min at 65°C in 0.1× SSC, 0.1% (v/v) SDS. The blots were air-dried and autoradiographed at −80°C. The blots were scored, and the computer program Mapmaker/EXP3.0 (Lander et al., 1987) was used to generate the map locations of the P450 genes.
ACKNOWLEDGMENTS
We gratefully acknowledge Dr. Donald Moreland for scientific input, Dr. Martin Sachs for providing the maize 1055 cDNA probe, and Dr. Torbert Rocheford for mapping these loci.
Footnotes
This work was supported by U.S. Department of Agriculture Competitive Research Grants (nos. 92–37301–7748 and 98–35304–6683 to M.A.S.), by a National Institutes of Health Predoctoral Traineeship, and a University of Illinois Graduate College Fellowship (to M.W.P.).
LITERATURE CITED
- Bak S, Kahn RA, Olsen CE, Halkier BA. Cloning and expression in Escherichia coli of the obtusifoliol 14-α demethylase of Sorghum bicolor (L.) Moench, a cytochrome P450 orthologous to the sterol 14-α demethylases (CYP51) from fungi and mammals. Plant J. 1997;11:191–201. doi: 10.1046/j.1365-313x.1997.11020191.x. [DOI] [PubMed] [Google Scholar]
- Barrett M. Metabolism of herbicides by cytochrome P450 in corn. Drug Metab Drug Interact. 1995;12:299–315. doi: 10.1515/dmdi.1995.12.3-4.299. [DOI] [PubMed] [Google Scholar]
- Batard Y, Schalk M, Pierrel M-A, Zimmerlin A, Durst F, Werck-Reichhart D. Regulation of the cinnamate 4-hydroxylase (CYP73A1) in Jerusalem artichoke tubers in response to wounding and chemical treatments. Plant Physiol. 1997;113:951–959. doi: 10.1104/pp.113.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynton CE, Potthoff SJ, McCullough AJ, Schuler MA. U-rich tracts enhance 3′-splice site recognition in plant nuclei. Plant J. 1996;10:703–711. doi: 10.1046/j.1365-313x.1996.10040703.x. [DOI] [PubMed] [Google Scholar]
- Berenbaum MR. Coumarins. In: Rosenthal G, Berenbaum M, editors. Herbivores: Their Interactions with Secondary Plant Metabolites. Vol. 1. San Diego: Academic Press; 1991. pp. 221–249. [Google Scholar]
- Bozak KR, Yu H, Sirevag R, Christoffersen RE. Sequence analysis of ripening-related cytochrome P-450 cDNAs from avocado fruit. Proc Natl Acad Sci USA. 1990;87:3904–3908. doi: 10.1073/pnas.87.10.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Hurtado F, Batard Y, Salaun J, Durst F, Pinot F, Werck-Reichart D. Cloning, expression in yeast, and functional characterization of CYP81B1, a plant cytochrome P450 that catalyzes in-chain hydroxylation of fatty acids. J Biol Chem. 1998;273:7260–7267. doi: 10.1074/jbc.273.13.7260. [DOI] [PubMed] [Google Scholar]
- Cabello-Hurtado F, Zimmerlin A, Rahier A, Taton M, DeRose R, Nedelkina S, Batard Y, Durst F, Pallett KE, Werck-Reichhart D. Cloning and functional expression in yeast of a cDNA coding for an obtusifoliol 14-α demethylase (CYP51) in wheat. Biochem Biophys Res Commun. 1997;230:381–385. doi: 10.1006/bbrc.1996.5873. [DOI] [PubMed] [Google Scholar]
- Chou W-M, Kutchan T. Enzymatic oxidations in the biosynthesis of complex alkaloids. Plant J. 1998;15:289–300. doi: 10.1046/j.1365-313x.1998.00220.x. [DOI] [PubMed] [Google Scholar]
- Czernic P, Huang HC, Marco Y. Characterization of hsr201 and hsr515, two tobacco genes preferentially expressed during the hypersensitive reaction provoked by phytopathogenic bacteria. Plant Mol Biol. 1996;31:255–265. doi: 10.1007/BF00021788. [DOI] [PubMed] [Google Scholar]
- Davies J, Caseley JC. Herbicide safeners: a review. Pestic Sci. 1999;55:1043–1058. [Google Scholar]
- Durst F, O'Keefe DP. Plant cytochrome P450: an overview. Drug Metab Drug Interact. 1995;12:171–187. doi: 10.1515/dmdi.1995.12.3-4.171. [DOI] [PubMed] [Google Scholar]
- Fonne-Pfister R, Gaudin J, Kreuz K, Ramsteiner K, Ebert E. Hydroxylation of primisulfuron by an inducible cytochrome P450-dependent monooxygenase system from maize. Pestic Biochem Physiol. 1990;37:165–173. [Google Scholar]
- Frank M, Deyneka JM, Schuler MA. Cloning of wound-induced cytochrome P450 monooxygenases expressed in pea. Plant Physiol. 1996;110:1035–1046. doi: 10.1104/pp.110.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frear DS, Swanson HR, Thalacker FW. Induced microsomal oxidation of diclofop, triasulfuron, chlorsulfuron, and linuron in wheat. Pestic Biochem Physiol. 1991;41:274–287. [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner G, Grun S, Winklmair A, Eisenreich W, Bacher A, Meeley R, Briggs S, Simcox K, Gierl A. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- Frey M, Kliem R, Saedler H, Gierl A. Expression of a cytochrome P450 gene family in maize. Mol Gen Genet. 1995;246:100–109. doi: 10.1007/BF00290138. [DOI] [PubMed] [Google Scholar]
- Hatzios KK, Wu J. Herbicide safeners: tools for improving the efficacy and selectivity of herbicides. J Environ Sci Health. 1996;B31:545–553. [Google Scholar]
- Kutchan TM. Alkaloid biosynthesis: the basis for metabolic engineering of medicinal plants. Plant Cell. 1995;7:1059–1070. doi: 10.1105/tpc.7.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- McFadden JJ, Gronwald JW, Eberlein CV. In vitro hydroxylation of bentazon by microsomes from naphthalic anhydride-treated corn shoots. Biochem Biophys Res Commun. 1990;168:206–213. doi: 10.1016/0006-291x(90)91695-o. [DOI] [PubMed] [Google Scholar]
- Moreland DE, Corbin FT, McFarland JE. Oxidation of multiple substrates by corn shoot microsomes. Pestic Biochem Physiol. 1993;47:206–214. [Google Scholar]
- Niemeyer HM. Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones), defense chemicals in the gramineae. Phytochemistry. 1988;27:3349–3358. [Google Scholar]
- Persans MW. Characterization of maize P450s. PhD dissertation. Urbana-Champaign, IL: University of Illinois; 1998. [Google Scholar]
- Persans MW, Schuler MA. Differential induction of cytochrome P450-mediated triasulfuron metabolism by naphthalic anhydride and triasulfuron. Plant Physiol. 1995;109:1483–1490. doi: 10.1104/pp.109.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel MA, Batard Y, Kazmaier M, Mignotte-Vieux C, Durst F, Werck-Reichhart D. Catalytic properties of the plant cytochrome P450 CYP73 expressed in yeast: substrate specificity of a cinnamate hydroxylase. Eur J Biochem. 1994;224:835–844. doi: 10.1111/j.1432-1033.1994.00835.x. [DOI] [PubMed] [Google Scholar]
- Potter S, Moreland DE, Kreuz K, Ward E. Induction of cytochrome P450 genes by ethanol in maize. Drug Metab Drug Interact. 1995;12:317–327. doi: 10.1515/dmdi.1995.12.3-4.317. [DOI] [PubMed] [Google Scholar]
- Puissant C, Houdebine L-M. An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Biotechniques. 1990;8:148–149. [PubMed] [Google Scholar]
- Robineau T, Batard Y, Nedelkina S, Cabello-Hurtado F, LeRet M, Sorokine O, Didierjean L, Werck-Reichhart D. The chemically inducible plant cytochrome P450 CYP76B1 actively metabolizes phenylureas and other xenobiotics. Plant Physiol. 1998;118:1049–1056. doi: 10.1104/pp.118.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MM. Molecular response to anoxic stress in maize. In: Jackson MB, Davies DD, Lambers H, editors. Plant Life under Oxygen Deprivation. The Netherlands: Academic Publishing; 1991. pp. 129–139. [Google Scholar]
- Schuler MA. Plant cytochrome P450 monooxygenases. Crit Rev Plant Sci. 1996;15:235–284. [Google Scholar]
- Schuler MA. Plant pre-mRNA splicing. In: Bailey-Serres JN, Gallie DR, editors. A Look Beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. Rockville, MD: American Society of Plant Physiologists Publications; 1998. pp. 1–19. [Google Scholar]
- Seigler DS. Plant Secondary Metabolism. Boston: Kluwer Academic Publishers; 1998. [Google Scholar]
- Siminszky B, Corbin FT, Ward EJ, Fleischmann TJ, Dewey RE. Expression of a soybean P450 monooxygenase cDNA in yeast and tobacco enhances the metabolism of phenylurea herbicides. Proc Natl Acad Sci USA. 1999;96:1750–1755. doi: 10.1073/pnas.96.4.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Filipowicz W. Splicing of precursors to messenger RNA in higher plants: mechanism, regulation and sub-nuclear organization of the spliceosomal machinery. Plant Mol Biol. 1996;32:1–41. doi: 10.1007/BF00039375. [DOI] [PubMed] [Google Scholar]
- Thalacker FW, Swanson HR, Frear DS. Characterization, purification, and reconstitution of an inducible cytochrome P450-dependent triasulfuron hydroxylase from wheat. Pestic Biochem Physiol. 1994;49:209–223. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P, Werck-Reichhart D, Teutsch HG, Durst F, Regnier S, Kazmaier M, Pompon D. Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast. Eur J Biochem. 1994;222:843–850. doi: 10.1111/j.1432-1033.1994.tb18931.x. [DOI] [PubMed] [Google Scholar]
- Vetter H-P, Mangold U, Schroder G, Marne F-J, Werck-Reichhart D, Schroder J. Molecular analysis and heterologous expression of an inducible cytochrome P-450 protein from periwinkle (Catharanthus roseus L.) Plant Physiol. 1992;100:998–1007. doi: 10.1104/pp.100.2.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D. Cytochromes P450 in phenylpropanoid metabolism. Drug Metab Drug Interact. 1995;12:221–243. doi: 10.1515/dmdi.1995.12.3-4.221. [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart D, Hehn A, Didierjean L. Cytochromes P450 for engineering herbicide tolerance. Trends Pharmacol Sci. 2000;5:116–123. doi: 10.1016/s1360-1385(00)01567-3. [DOI] [PubMed] [Google Scholar]
- Whitbred JM. Differential regulation of the CYP73A9 and CYP82A1 P450 genes involved in plant defense. MS dissertation. Urbana-Champaign: University of Illinois; 1998. [Google Scholar]
- Wink M. Allelochemical properties or the raison d'etre of alkaloids. In: Cordell GA, editor. The Alkaloids. Vol. 43. San Diego: Academic Press; 1993. pp. 1–118. [Google Scholar]
- Zimmerlin A, Durst F. Aryl hydroxylation of the herbicide diclofop by a wheat cytochrome P-450 monooxygenase. Plant Physiol. 1992;100:874–881. doi: 10.1104/pp.100.2.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerlin A, Salaun J-P, Durst F, Mioskowski C. Cytochrome P-450-dependent hydroxylation of lauric acid at the subterminal position and oxidation of unsaturated analogs in wheat microsomes. Plant Physiol. 1992;100:868–873. doi: 10.1104/pp.100.2.868. [DOI] [PMC free article] [PubMed] [Google Scholar]