Abstract
The kidney vasculature facilitates the excretion of wastes, the dissemination of hormones, and the regulation of blood chemistry. To carry out these diverse functions, the vasculature is regionalized within the kidney and along the nephron. However, when and how endothelial regionalization occurs remains unknown. Here, we examine the developing kidney vasculature to assess its 3-dimensional (3D) structure and transcriptional heterogeneity. First, we observe that endothelial cells (ECs) grow coordinately with the kidney bud as early as E10.5, and begin to show signs of specification by E13.5 when the first arteries can be identified. We then focus on how ECs pattern and remodel with respect to the developing nephron and collecting duct epithelia. ECs circumscribe nephron progenitor populations at the distal tips of the ureteric bud tree and form stereotyped cruciform structures around each tip. Beginning at the renal vesicle stage, ECs form a continuous plexus around developing nephrons. The endothelial plexus envelops, and elaborates with, the maturing nephron, becoming preferentially enriched along the early distal tubule. Lastly, we perform transcriptional and immunofluorescent screens to characterize spatiotemporal heterogeneity in the kidney vasculature, and identify novel regionally enriched genes. A better understanding of development of the kidney vasculature will help instruct engineering of properly vascularized ex vivo kidneys and evaluate diseased kidneys.
Keywords: endothelium, epithelium, blood vessel, nephron, vascular patterning, endothelial cell heterogeneity
INTRODUCTION
During embryogenesis, the vasculature develops coordinately with its host tissues. Vascular development has been analyzed in different organisms, with a focus on how the first blood vessels take shape [1–4]. Mechanisms of blood vessel development during embryogenesis include vasculogenesis which is de novo formation from isolated endothelial progenitor cells (or angioblasts), and angiogenesis, which is the formation of new blood vessels from sprouting or remodeling of existing vessels. However, recent interest has turned to trying to understand the dynamic interface between blood vessels and surrounding tissues as they grow together. During pancreas formation, for instance, endothelial cells (ECs) are initially present in the mesoderm surrounding the budding and branching endodermal epithelium [5]. These vascular progenitor cells coalesce and significantly remodel in the growing organ as blood flow is initiated, leading to elaboration and differentiation of vessels. What remains unknown is whether similar mechanisms exist in other organs including the kidney. This question is relevant to tissue engineering, as vascularization of replacement tissues is critical for therapeutic success. Currently, significant efforts are directed at generating ex vivo nephrons for those affected with chronic kidney disease and end stage renal failure [6]. These efforts can be furthered by studying the process in murine mouse models, which unlike human tissue allow for developmental analyses using transgenic reporters and genetic modifications. A better understanding of murine kidney vasculature may also benefit therapeutic efforts in humans.
Murine kidney development begins at embryonic day 10.5 (E10.5) when the Wolffian duct buds into the metanephric mesenchyme, which is comprised of nephron progenitor cells (NPCs) expressing the transcription factor Six2 and stromal cells expressing the transcription factor FoxD1 [7]. Through reciprocal signaling, the Wolffian duct bud (termed the ureteric bud or UB) undergoes iterative branching events to form an epithelial tree, while NPCs aggregate at the UB tips to form “caps,” proliferate, and undergo mesenchymal-to-epithelial transition (MET) to begin nephrogenesis. NPCs that commit towards a nephron epithelial cell fate first condense to form pretubular aggregates (PTAs) at each distal ureteric tip. PTAs then undergo lumenogenesis to produce a renal vesicle (RV), which then elongates to form an S-shaped body, ultimately generating a mature nephron [8,9]. Nephrogenesis continues until NPCs are depleted at around post-natal day 3 (P3), at which point the kidney grows through proliferative expansion of the established renal architecture [7].
Despite the large body of knowledge documenting kidney epithelial patterning, less is known about the renal vasculature that develops in association with the epithelium. Flk1+ (also KDR or VEGFR2) ECs have been identified within the metanephric blastema at E10.5 and the forming metanephros at E12.5 [10]. Due to early technical limitations of interpreting 2D sections, previous studies considered the kidney prior to E14.5 as relatively avascular, or ‘prevascular’ [11]. Vascular structures appeared as isolated Flk1+ angioblasts rather than continuous vessels. Recently, studies have advanced our understanding of the early renal vasculature using 3D imaging [12]. In agreement with previous reports, they identify ECs around the E10.5 metanephric blastema but further show that they form a capillary plexus extending from vessels close to the nephric duct by E11.25. Therefore, ECs associate with the kidney from the onset of renal morphogenesis.
During the remainder of organogenesis, the vasculature develops in coordination with the maturing kidney by both organizing along maturing nephrons and undergoing arteriovenous (AV) specification; however, these processes remain poorly understood. Recent studies have shown that ECs circumscribe each Six2+ NPC cap, but do not penetrate them [12]. Indeed, during nephrogenesis, ECs are thought to first interact with the developing nephron at the S-shaped body stage in which presumptive podocytes express VEGF to induce EC migration into the cleft [11,13,14]. Whether ECs interact with earlier stages of nephron development and whether they are patterned along developing renal structures remain unclear. Additionally, as vessels remodel, AV specification and subsequent development of arteries was shown to occur by E13.5, as vessels begin to express Nrp1+ [15]. However, the structure and maturation of arteries, as well as the organization of veins, have not been assessed in the developing kidney. Many questions therefore remain regarding the developing architecture and fate of vascular cells within the developing kidney.
Here, we present an in-depth anatomical and molecular analysis of the developing renal endothelium. Our studies provide an independent validation of recent studies from Munro and colleagues [12], while extending their findings on several fronts. In addition, our transcriptomic data and spatiotemporal survey of vascular markers demonstrate molecular diversification of ECs in the developing kidney. We assess the organization of ECs around the ureteric tree, progenitor caps, and developing nephrons, as well as the establishment of EC heterogeneity and changes in EC identity over time. Using whole mount 3D imaging, we show that the renal vasculature grows coordinately with the kidney from as early as E10.5, forming a plexus within and surrounding the metanephric blastema. Analysis of ECs relative to other renal cell types reveals that vessels organize in highly stereotyped patterns around the ureteric tree, Six2+ NPCs and early nephron structures. Capillaries circumscribe NPCs, as well as differentiating RVs and emerging tubules, prior to the S-shaped body stage. Renal arterial development is first evident at E13.5, confirming previous studies [15,16], and exhibits a predictable branching pattern as it grows. We find that the renal veins largely follow arteries, despite few established markers available. Using transcriptional profiling, we identify novel renal EC-enriched genes expressed in regionally-defined and organ-specific patterns. We show that the renal endothelium displays marked heterogeneity as early as E15.5 and that many EC gene expression patterns are highly dynamic over time. This study will serve as an atlas to guide our understanding of the developing renal vasculature.
MATERIALS AND METHODS
Mice and embryo handling
Experiments were performed in accordance with protocols approved by the UT Southwestern Medical Center IACUC. E10.5 – E12.5 Flk1-eGFP embryos or E13.5-P5 kidneys were dissected and fixed in 4% PFA/PBS for overnight at 4°C except for β-galactosidase reaction.
β-galactosidase reaction
E12.5-E14.5 Kidneys from ephrinB2-LacZ or EphB4-LacZ mice were fixed using 4% PFA for 20 minutes, rinsed in PBS and stained for β-gal for 3 hours (hrs) to overnight as previously described [17]. Images were taken with a NeoLumar stereomicroscope (Zeiss) using a DP-70 camera (Olympus).
Immunofluorescence on sections and statistical analysis
Fixed embryos and kidneys were washed in PBS and cryoprotected in 30% sucrose overnight. Tissues were then embedded in Tissue-Tek O.C.T. Compound and sectioned at 10–20 μm on a cryostat. Frozen sections were washed in 1× PBS + 0.1% Triton X-100 and blocked for at least 1 hr at room temperature in 5% normal donkey serum. Certain slides were treated with heat-mediated antigen retrieval in 1 μM Tris pH 7.5, 5 μM EDTA pH 8.0 prior to blocking depending on the antibody. Primary antibody incubations were done at 4°C overnight (for antibody information, dilutions, and antigen retrieval conditions, see Table S1). Slides were then washed in PBS, incubated in secondary antibody for 1 hr at room temperature and subsequently incubated in DAPI. Slides were then washed in PBS and mounted using Prolong Gold Mounting Medium. Images were obtained using an A1R Nikon confocal microscope.
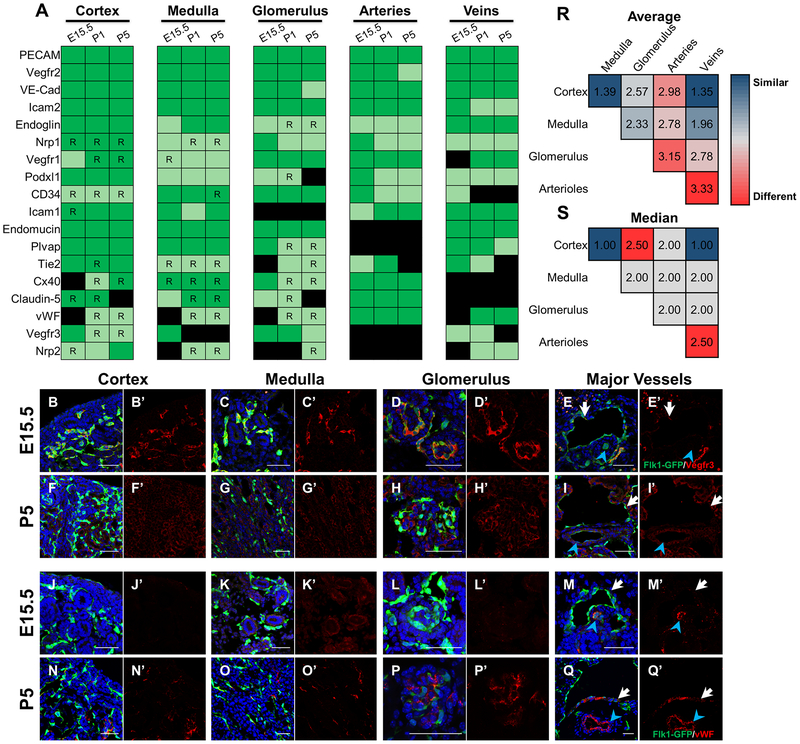
To obtain semi-quantitative differences in antigen intensity between kidney regions from the immunofluorescent screen in Fig. 5 and Supplemental Fig. 6, we developed a system to score fluorescence levels. First, each antibody staining at each time point for all 5 regions is assigned a score of 3 or 0 based on the presence or absence of fluorescence. Next, 1 point is taken away if fluorescence is low (light green), 1 point is taken away if fluorescence is restricted to a subset within a region (R), and 2 points are taken away if both are present (light green and R). Therefore, every cell will have an integer score between 0 and 3. The score is based on cumulative assessment from 3 separate and blinded analyses of multiple images of each region from 3 independent experiments. Each antibody was imaged at the same settings at all 3 time points. The scores for all 3 time points per gene in each region are then added together to give a “cumulative score” of the expression pattern ranging from 0 to 9. For example, the cumulative score for Vegfr1 in the cortex is 7 (2+2+3) and Vegfr1 in the medulla is 5 (1+2+2). Next, we calculated the differences in cumulative scores between two regions per gene (“difference score”). This number represents how disparate the expression patterns for a given gene are between the two regions, with a larger number indicating a more pronounced difference in expression patterns. The difference score can range from 0 (no difference) to 9 (completely opposite patterns). For example, the difference score for Vegfr1 between the cortex and medulla is 2 (|7–5|). Repeating this comparison for all 18 genes and taking the average and median of the difference scores in aggregate gives us an estimation for variations in immunostaining patterns between any two regions. This was then repeated for every combination of two regions (10 comparisons total) to give the final figure.
Figure 5. Standard endothelial markers display heterogeneous spatiotemporal expression in the developing kidney.
a Chart summarizing expression patterns for each endothelial gene in the kidney cortex, medulla, glomerulus, arteries, and veins at E15.5, P1, and P5. A gene was considered to be expressed in a region if the gene clearly colocalized with at least 1 Flk1-eGFP+ EC within a region. Strength of expression was determined qualitatively by comparing fluorescent intensity across different regions per gene. Expression in surrounding tissues was not considered. Dark green = strong expression, light green = weak expression, black = no expression, R = expression restricted to a subset of ECs within a region. *Vegfr3 and Nrp2 show strongest expression in lymphatics at all 3 timepoints. b-q Representative images for Vegfr3 (b-i) and vWF (j-q) at E15.5 (b-e, j-m) and P5 (f-i, n-q) costained with Flk1-eGFP (green) and DAPI (blue) to demonstrate changes in gene expression by region over time. ECs of arteries and veins are marked with a blue arrowhead and white arrows, respectively. r-s Average (r) and median (s) of the difference score between the regions (refer to Methods for how the score was determined). A score of 0 represents completely identical expression patterns while a score of 9 represents completely opposite expression patterns. Cells are shaded based off score. Red = higher score (more different) and blue = lower score (more similar). Scale bar = 50 μm.
Whole-mount immunofluorescence
Fixed E10.5-E14.5 embryos and kidneys were washed in PBS and dehydrated to 100% methanol and then rehydrated in PBS with a 1 hr wash in 50% methanol. Tissues were then incubated with 1% Triton X-100 in PBS for 2 hrs and blocked in CAS-Block (Invitrogen) for at least 1 hr at room temperature. The kidneys were incubated in primary antibodies overnight in CAS-Block (ThermoFisher) at 4°C. Tissues were washed in PBS and incubated in secondary antibodies overnight at 4°C and then washed and dehydrated in methanol. Tissues were cleared by incubating them in a 1:2 mixture of benzyl alcohol/benzyl benzoate (BABB) for at least 10 minutes. Kidneys were visualized mounted in BABB using an LSM710 Meta Zeiss confocal to take optical sections every 2.5–3 μm.
Whole mount imaging processing, analysis, and statistical analysis
Confocal tiled z-stacks were rendered in 3D and analyzed using Bitplane Imaris v8.4.1 software. Generation of randomly generated spots and distribution of Six2+ NPCs and random spots to the nearest blood vessel were determined as described previously [18]. Briefly, PECAM+/Emcn + vessels and Six2+ NPCs were represented as surfaces and spots, respectively, using automatic functions to generate an initial structure, then manually edited to only those surfaces and spots within the kidney proper. Random spots were generated as described previously [18] and restricted to the kidney proper through manual circumscribing. Spots were generated in sufficient numbers such that the total spots remaining after editing was within ~5% of the total number of Six2+ NPCs. To determine the distribution of the two spot populations to the nearest blood vessel, we first performed a distance transformation of the PECAM+/Emcn+ blood vessel surface, which generates a new channel with intensity directly proportional to distance from the surface. The distances between both spot populations and the area of max intensity of the generated channel were then calculated and either grouped into bins of 2 μm or averaged. The data grouped into bins were converted into a proportion of the total number of spots to better assess distribution.
Significance in Supplemental Fig. 4d (n=3) and 4e (n=5–7) was determined using unpaired student’s t test and Ordinary one-way ANOVA with Tukey’s multiple comparison test, respectively. Error bars indicate standard deviation.
Digoxigenin-labeled RNA probes and in situ hybridizations
cDNA templates for digoxigenin-labeled probes were acquired from plasmid purchased from Dharmacon (GE) (Rsad2) or generated from E18.5 whole kidney cDNA by PCR (Gimap4). Rsad2 plasmid was linearized using a one-cutter restriction enzyme. For PCR-based synthesis, we first isolated mRNA from E18.5 embryonic kidneys using the RNeasy Mini kit (Qiagen) and then generated cDNA using the Superscript III First-Strand Synthesis System kit (Invitrogen) following manufacturer’s instructions. To generate the cDNA template containing a T7 promoter site in the anti-sense direction, PCR was performed on the cDNA using the gene-specific primers listed below. PCR program: 1) 94°C for 5 minutes, 2) 35 cycles of 94°C or 30 seconds, 60°C for 45 seconds, 72°C for 3 minutes, and 3) 72°C for 5 minutes. The PCR product was purified by phenol:chloroform extraction. Probe synthesis for both genes was performed as described previously [5]. Briefly, probes were synthesized at 37°C for 2–4 hrs in digoxigenin-synthesis reaction mixture with T7 RNA polymerase (Roche). After synthesis, DNA was eliminated by adding RQ1 DNase I (Promega) and RNA probes were purified using Micro Bio-spin columns (Bio-RAD). 10× hybridization stock was prepared at 10 μg/mL by adding the appropriate volume of pre-hybridization buffer.
Accession numbers for clones: Rsad2 (BC057868). Primers for Gimap4: Forward: 5’-CTGGGATGGGAAAGAGCTTGT-3’. Reverse: 5’-TAATACGACTCACTATAGTCAAGGCAGCAGGCAGTAAT-3’.
In situ hybridizations were performed as described previously [5]. Briefly, fixed E15.5 or E18.5 kidneys were dehydrated to 100% ethanol and embedded in paraffin before sectioning with a microtome. Paraffin sections were de-paraffinized in xylene, then rehydrated to PBS before being treated with 15 μg/mL proteinase K for 15 minutes and fixed in 4% PFA. Slides were then washed and incubated with pre-hybridization buffer for 1 hr at room temperature before being hybridized with the specific probe at 1 μg/mL overnight at 65°C. Next day, slides were washed in 0.2× SSC then transferred to MBST before blocking with 2% blocking solution (Roche) for at least 1 hr at RT. Slides were then incubated with Anti-Dig alkaline phosphatase conjugated antibody (Roche, 1:4000) overnight at 4C. Next day, slides were washed in MBST 3× and NTMT 3× before incubating with BM purple (Roche) for color reaction. After color reaction, slides were fixed with 4% PFA and mounted using Permount. Images were taken using a Zeiss Axiovert 200M scope and a DP-70 camera (Olympus).
Fluorescent in situ hybridizations were performed following the above in situ hybridization protocol up to the 0.2× SSC washes. After washes, slides were transferred to TNT and treated with 0.3% H2O2 for 30 minutes before washing again in TNT and blocking in 1% blocking buffer (Perkin Elmer) for at least 1 hr at RT. Slides were then incubated with Anti-Dig peroxidase (Roche, 1:500), Rat anti-PECAM, and Rat anti-Endomucin overnight at 4C. Next day, slides were washed in TNT 3× before incubating with TSA Fluorescein Amplification Reagent (1:50 in Amplification Diluent, Perkin Elmer) for 15 minutes. Slides were washed in TNT following TSA incubation, incubated with Goat anti-rat Alexa Fluor 555 for 2 hrs at RT, and subsequently incubated in DAPI. Slides were then washed in TNT and mounted using Prolong Gold Mounting Medium. Images were obtained using an A1R Nikon confocal microscope.
RNA-seq of ECs from developing kidneys, lung, and pancreas
E12.5, E15.5, and E18.5 Flk1-eGFP kidneys, lungs, and pancreases were individually dissected in cold PBS, pooled, manually disaggregated with forceps, and digested in a mixture of 0.5% collagenase (Roche) and 20 U/mL RQI DNAse I (Promega) on a shaker for 40 minutes at 37°C. Digestion was stopped by adding ice cold FBS to a final concentration of 2%. Cell suspension was triturated with a progression of 18-, 23, and 25-gauge needles and filtered through a 70 μm filter yielding a single suspension. Cells were then washed twice with PBS + 0.5% BSA (Sigma Aldrich) + 5mM EDTA + 1mM CaCl2 before proceeding to flow cytometry. GFP+ cells were isolated by flow cytometry using a FACS AriaII and sorted into PBS + 0.5% BSA (Sigma Aldrich) + 5mM EDTA + 1mM CaCl2. Sorted cells were lysed and the RNA was cleaned using miRVANA kit (Invitrogen) following manufacturer’s instructions. RNA was isolated from cell lysates using RNAqueous Micro Total RNA Isolation kit (Invitrogen) following manufacturer’s instructions. RNA sample quality and quantity was analyzed using Bioanalyzer with RNA 6000 Pico Kit (Agilent). Mouse TotalScript mRNA-seq libraries were combined into 10 nM pooled stocks, denatured and diluted to 7 pM with pre-chilled hybridization buffer and loaded into TruSeq SE v3 flowcells on an Illumina cBot followed by indexed single-end sequencing (50+7 bp) on an Illumina HiSeq 1500 using TruSeq SBS Kit v3 chemistry (Illumina). Base call files were generated by Real Time Analysis (RTA) software (Illumina), which were then used to generate FASTQ files using bcl2fastq software (v. 1.8.4). Demultiplexed FASTQ files from RNA-seq libraries were trimmed for stretches of adapter sequences, and quality trimmed during the import to CLC Genomics Workbench 7.5 for alignment and quantification.
Differential gene expression from RNA-seq
Transcript abundance was estimated without aligning reads using Salmon [19] in R (v. 3.4.3) against an index of coding sequences from the Ensembl GRCm38 assembly. Transcript-level abundance was imported and count and offset matrices generated using the tximport R/Bioconductor package [20]. Differential expression analysis was performed using the DESeq2R/Bioconductor package [21]. To identify differentially expressed genes EC-specific, transcript abundance from FACS isolated ECs was compared to that from FACS isolated non-ECs from the same tissue and comparable developmental stage using publicly available datasets from the NCBI GEO database (listed below).
| Series | Run | Cell type | Developmental stage |
|---|---|---|---|
| GSE78772 | SRR3195205 | Nephron progenitor | E12.5 |
| SRR3195206 | Nephron progenitor | E12.5 | |
| GSE64959 | SRR1758421 | Collecting duct | E15.5 |
| SRR1758423 | Collecting duct | E15.5 | |
| SRR1758428 | Collecting duct | E15.5 | |
| GSE78772 | SRR3195211 | Nephron progenitor | P1 |
| SRR3195212 | Nephron progenitor | P1 |
Correlation patterns among genes across RNA-seq data sets were assessed using weighted correlation network analysis (WGCNA) [22]. A gene significance measure, GSi, of the correlation between expression change across samples and the organ/cell type of the samples was calculated for each gene i. A p-value measuring the statistical significance of this correlation was obtained from a univariate regression model between the gene’s expression across samples and the tissue/cell type of the samples. Overlap between gene sets whose expression were increased, at an FDR < 0.1, relative to non-endothelial comparator at each developmental stage was determined. Code for analysis provided in supplemental data.
RESULTS
Developing renal vasculature architecture is highly dynamic
To determine when, where, and how blood vessels arise in the kidney, we visualized embryonic vessels using a Flk1-eGFP reporter in both whole mount and sectioned tissue [23]. Flk1 is known to be expressed widely throughout the embryonic vasculature, in both progenitor and mature ECs [24].
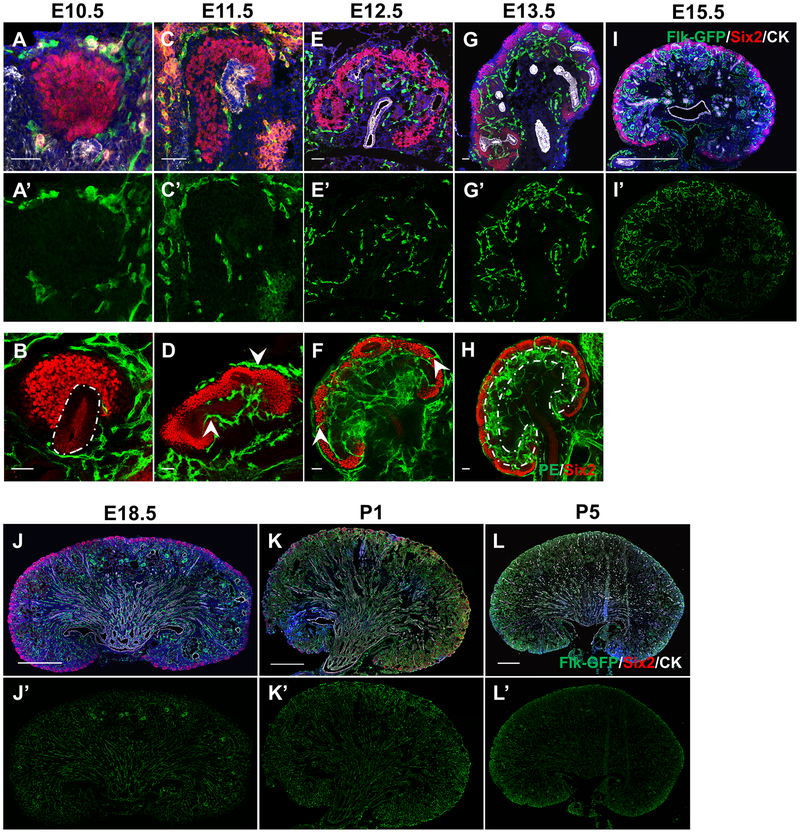
Kidney development begins at E10.5 when the UB evaginates from the Wolffian duct into the neighboring metanephric mesenchyme. Although ECs marked by Flk1-eGFP circumscribe the nephron progenitor population, they do not appear to invade it, in agreement with previous data [12] (Fig. 1a–a’, Movie 1). In addition, capillaries largely remain approximately a few cell diameters away from the Six2+ NPCs. Our observations show that ECs surround but do not penetrate caps of NPCs, although they are in proximity to developing kidney structures as early as E10.5.
Figure 1. Basic anatomy of the vasculature during development of the kidney.
a-h Sections (a, c, e, g, i-l) or whole mount stains (b, d, f, h) of kidneys from E10.5-P5 stained with Flk1-eGFP, Six2, and Cytokeratin to delineate the ECs, NPCs, and UB, respectively. White outline in b marks budding nephric duct (white outline). Arrowheads in d mark ECs surrounding UB or Six2 region, but not penetrating Six2 cells. Arrowheads in f highlight two examples of ECs that cross from outside to inside the kidney. Outline region in h marks area of enriched ECs deep to the NPCs. Scale bar = 50 μm (a-g) or 500 μm (i-l).
Kidney development proceeds through iterative rounds of UB branching, while NPCs within the mesenchyme proliferate, condense, epithelialize, and differentiate into RVs. At E11.5, we observed a continuous vascular plexus around progenitor cells and the branching UB (Fig. 1c–d, Movie 2). We observed that the plexus surrounds the NPC caps, but does not invade them confirming previous findings [12]. Whole mount and section staining shows that as the NPCs expand and split following branching of the UB, ECs can be identified between the two epithelial populations (Fig. 1d,f, arrowheads). By E12.5, we find ECs connecting across (peripheral to) the Six2+ cell aggregates and along the outer periphery of the kidney to the interior plexus (Fig. 1e–f, arrows, Movie 3). As previously shown, these connections bisect the Six2+ caps in a reiterative process [12]. By E13.5 through E15.5, the subcortical region (where nephron development occurs) is more highly vascularized than the outermost nephron progenitor and innermost medullary regions (Fig. 1g–i’, Movie 4).
Throughout late gestation, we find that the vasculature continues to pervade the entirety of the kidney. Blood vessels orient longitudinally from the medulla out towards the cortex (Fig 1j–j’). This organization is maintained after birth and after nephrogenesis is completed (Fig. 1k–l).
Arteries and veins of the kidney differentiate during midgestation
In the early embryo, ECs interconnect to form networks of cords prior to blood flow. Once cords open lumens and blood flow begins to exert hemodynamic force, vessels progressively remodel and differentiate into arteries and veins [25]. Studies have reported the onset of arterial specification and the presence a perfused arterial tree in the kidney as early as E13.5 [26,16,15]. However, it has been unclear if arterial differentiation occurs in a stereotyped manner during kidney development.
To assess AV development in the early embryonic kidney, we first examined expression of reporter lines previously shown to be arterial-specific, ephrinB2-LacZ, or venous-specific, EphB4-LacZ. Interestingly, neither line showed significant endothelial expression in the E12.5 kidney (Supplemental Fig. 1 a, c). By E14.5, renal arteries exhibited ephrinB2-LacZ expression; however, LacZ expression was also present in the UB tree, preventing clear assessment of the arterial tree (Supplemental Fig. 1b). Low EphB4-LacZ expression was detected in developing veins starting at E14.5, where it delineated three large branches (Supplemental Fig. 1d). Thus, we found neither reporter, ephrinB2-lacZ nor EphB4-lacZ, to be a useful tool to assess AV specification due to low specificity or expression in early arteries or veins.
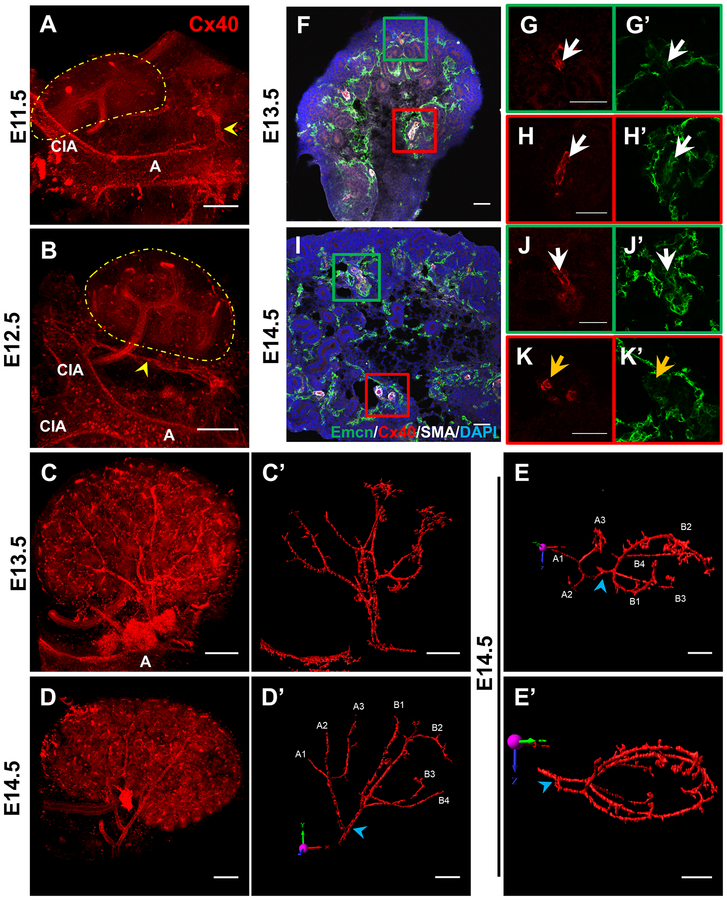
To better elucidate differentiation and architecture of forming arteries, we performed whole mount immunofluorescent staining with the known arterial-specific marker Connexin40 (Cx40) to examine developing arterial vessels in 3D with confocal microscopy [27]. Cx40 whole mount stains revealed arterial connections extending from the aorta towards both the mesonephros and metanephros including an ephrinB2-LacZ+ vascular cord that connects the aorta and common iliac artery; however, these did not visibly penetrate the metanephros at E11.5 or E12.5, suggesting that AV fate within the kidney has not been specified at this stage (Fig. 2a–b, Supplemental Fig. 1). The first evidence of arterial specification within the kidney was observed at E13.5, where a tree with multiple branches became distinguishable (Fig. 2c), in line with previous reports [26,15].
Figure 2. AV differentiation is first established at E13.5 and progresses towards the kidney periphery.
a-d Whole mount imaging of E11.5-E14.5 kidneys stained with Cx40 to mark the developing arterial tree. a Yellow arrowheads mark Cx40+ vessels extending from the aorta to the mesonephros but not penetrating the metanephros (marked by yellow dotted line). b Yellow arrowhead marks vascular cord extending from aorta to common iliac artery running alongside the E12.5 metanephros (marked by yellow dotted line). A = aorta, CIA = common iliac artery. c’, d’, e, e’ 3D reconstruction of arterial surface at E13.5 (c’) and E14.5 (d’, e, e’) using Imaris software with manual editing to highlight arterial vasculature. e 90° rotation of d’ in x-axis (red arrow) to better demonstrate arterial branching morphogenesis. e’ Caudal view of the E14.5 kidney (90° rotation of e in z-axis [blue]) to highlight that renal arteries do not penetrate the renal medulla. Blue arrowheads mark first bifurcation point. Individual lobar branches are labeled by a letter to signify which major artery from which it arises and a number. f-k Analysis of arterial differentiation on E13.5 (f-h) and E14.5 (i-k) kidney sections. Zoomed in images of boxed areas in the cortex (g, j) and medulla (h, k) show staining for arterial development in different parts of the kidney. White arrows demarcate Emcn+, Cx40+ vessels while orange arrows mark Emcn−, Cx40+ vessels. Scale bar = 150 μm (a-e) or 50 μm (f-k’).
By E14.5, the renal arterial tree was further defined. We expanded on previous work by characterizing branching morphogenesis of the arterial tree at E14.5. The renal artery branched into two major arteries, each undergoing two to three additional branching events, leading to an additional three or four lobar arteries (Fig. 2d). Of note, these branches extended away from the renal hilum to the periphery of the organ, avoiding the central region of the kidney containing the papilla/medulla, but wrapping around the entire organ (Fig. 2e–e’). We found that, although these basic patterns were grossly similar, branching patterns differed slightly between age-matched individual kidneys (Supplemental Fig. 3). This suggests that renal arterial patterning follows a predictable, but not stereotyped, ontogeny.
Next, we sought to determine if the arterial vessels exhibit properties of fully matured arteries. Mature arteries in the embryonic trunk express Cx40, but not Endomucin (Emcn) [27] (Supplemental Fig. 2). Characterization of smaller arteries at E13.5 revealed that they express both Cx40 and Emcn, suggesting they have not fully matured (Fig. 2f–h’). At E14.5, arteries close to the renal hilum cease expression of Emcn, while those further from the hilum remain double-positive for Cx40 and Emcn (Fig. 2i–k’). Thus, arterial fate is specified in a centripetal wave in the kidney, where differentiation progresses outward from the renal hilum towards the distal periphery of the organ.
Venous fate in the developing kidney proved to be difficult to assess, as we found no markers unique to venous cells. Nrp2, a known venous marker [28], did not exclusively label veins in the early embryonic kidney as it does in other tissues (data not shown) [12]. Although there are generally fewer useful vein markers available for the study of embryonic vessels [27], morphological differences between arteries and veins can distinguish them. Arteries display cuboidal ECs surrounded by a layer of smooth muscle cells (SMCs), while veins are larger caliber vessels with thinner ECs that lack a clear outer layer of SMCs [29]. By using pan-endothelial markers (PECAM or Flk1), we observed that renal arteries and veins often developed adjacent to one another, appearing as “artery-vein” doublets on sectioned tissue (Supplemental Fig. 2b–b’’). The co-alignment of arteries and veins, supported by observations of EphB4-LacZ whole mount stains and frequently observed in other tissues [30], suggest that branching morphogenesis of the venous tree largely mirrors that of the arterial tree in the developing kidney.
ECs closely associate with the UB, but not NPCs
ECs are known to regulate epithelial branching and progenitor populations during pancreas and lung development [31–33]. Evidence of EC-tissue crosstalk suggests that an understanding of the anatomy of the organ vasculature may shed new light on developmental signaling events. We therefore asked how ECs organize around the collecting ducts, nephron progenitor cells, and developing nephrons.
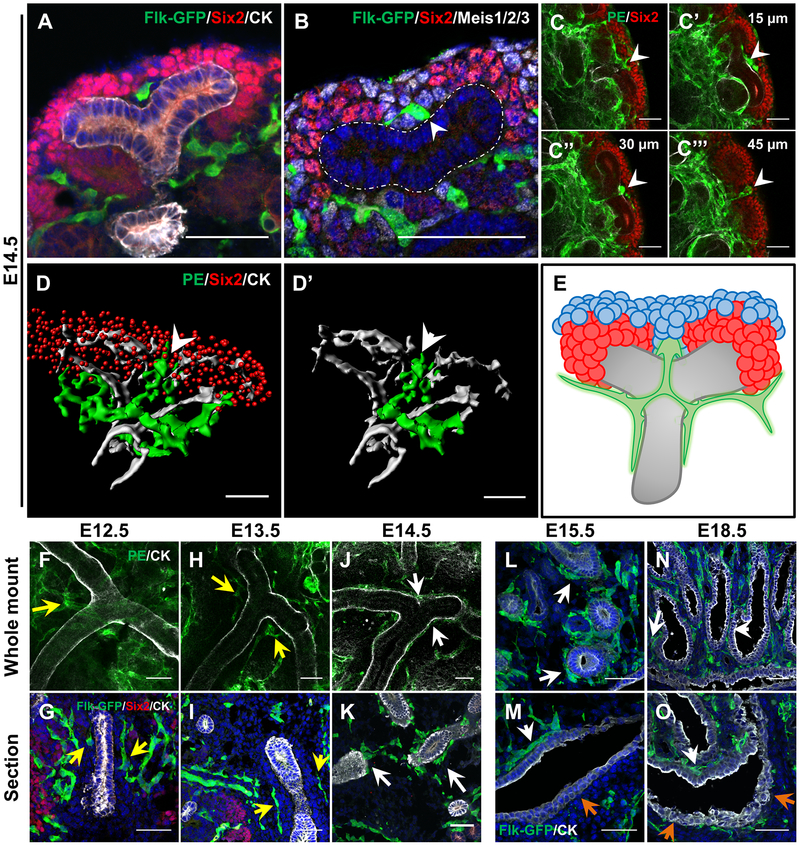
We first examined endothelial organization around NPC caps located at UB tips. Throughout development (E10.5 to P3), each UB tip forms via the bifurcation and extension of existing tips, giving rise to “flattened Y”-shaped structures [34]. At the tip of each UB, NPCs aggregate into distinct “caps.” Munro and colleagues previously characterized EC organization around the tips [12]. In line with their data, we found ECs present at the point of each bifurcation, between two UBs, separating NPC caps (Fig. 3a). Using whole mount 3D imaging and viewing the NPC caps en face, we observed ECs circumscribing individual caps, but never invading the clusters of Six2+ cells that form the caps (Supplemental Fig. 4a–b). To quantitatively demonstrate that NPCs cluster away from ECs, we assessed the distribution of NPCs to their nearest blood vessel compared to randomly distributed spots. This analysis in E13.5 whole mount kidneys indicated that NPCs preferentially localize away from ECs (Supplemental Fig. 4c–d). During the early stages of renal morphogenesis, this average distance from NPCs to the nearest blood vessel remains relatively stable at ~12–13 μm (Supplemental Fig. 4e).
Figure 3. ECs avoid proximal ureteric stalk until 14.5 and bisect distal UB tips.
a E14.5 Distal tips of UB with EC in between the two NPC caps, bisecting the UB tips. b Distal UB tip stained for the stromal marker Meis1/2/3 (white) demonstrating that the bisecting EC is surrounded mostly by stromal cells. White outline marks UB. White arrow marks connection between UB and bisecting EC. c Whole mount analysis of EC looping around the distal UB (white arrowheads). Numbers in c’-c’’’ represent distance from c in Z-axis. d 3D render of ECs around the distal UB tip with Six2+ NPCs. d’ highlights a single EC loop around the UB tip. White arrowhead marks bisecting EC. e Model of EC organization at the distal UB tips. ECs bisect the distal tips and circumscribe each branch of the UB tree, forming a cruciform structure around the bifurcation point. The EC that bisects the distal tip is almost entirely surrounded by stroma cells. Green = ECs, Red = NPCs, Blue = stromal cells, Grey = UB. Whole mount (f, h, j) and section (g, i, k) stains of the main trunk of the UB in E12.5 – E14.5 kidneys demonstrate ECs are separated from the UB trunk up to E13.5 (yellow arrows), but become tightly associated at E14.5 (white arrows). Immunofluorescent analysis on E15.5 (l, m) and E18.5 (n, o) sections show that ECs remain attached to the UB tree throughout development except on the side of the ureter facing the aorta (orange arrows). Scale bar = 50 μm.
Our data and previous work establish that ECs circumscribe individual NPC caps, but some questions remain on how ECs organize with respect to the UB. We observed that bifurcating vascular cords rarely made contact with the UB tip epithelial cells (Fig 3a). Instead, these ECs were mostly surrounded by stromal cells, which separated ECs from UB epithelium (Fig 3b). Whole mount imaging of UB tips showed that ECs form loops not only through the bifurcation and around the epithelial stalk, but also around each tip separately (Fig. 3c–e). The same cruciform structure can be identified around each tip, indicating that the patterning of these vascular structures is predictable.
Next, we focused on endothelial organization around the UB trunk. We used a pan-cytokeratin antibody to delineate the UB and Flk1-eGFP or a combination of antibodies recognizing PECAM and Emcn (PE) to mark the ECs. We found that until E13.5, vessels surrounded the main trunk of each UB, but did not contact the epithelium (Fig. 3f–i). ECs surrounding the UBs remained at least 1–2 cell diameters away from UB stalks. At E14.5, this intervening distance disappeared, as the endothelium became intimately associated with the UB trunk and its early branches, coating most of the UB epithelium (Fig. 3j–k). This relationship was maintained throughout the rest of embryonic kidney development except at the renal pelvis, where ECs closely associate with epithelial cells in the renal papilla, but not those that comprise the ureter (Fig. 3l–o).
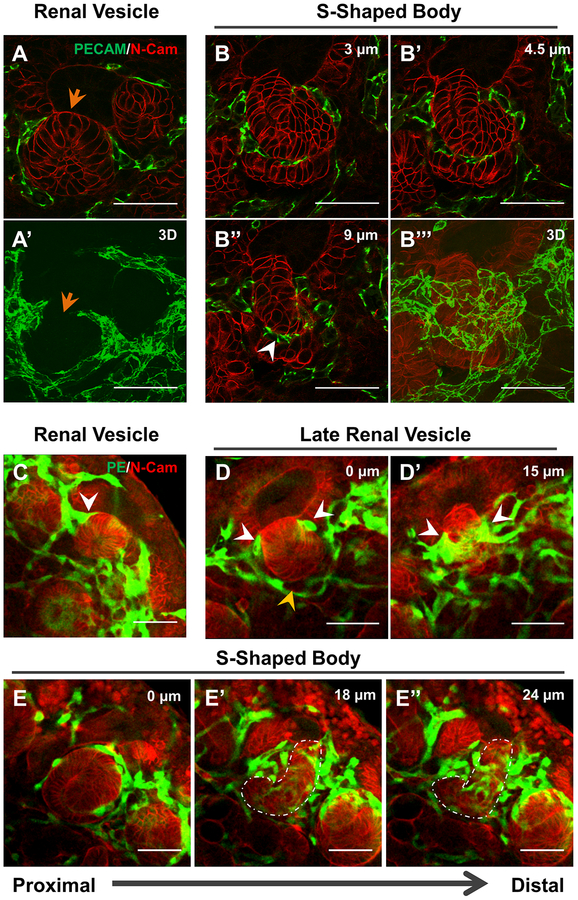
ECs form a plexus around the RV and S-shaped body
Nephron development begins when a subset of NPCs undergoes mesenchymal-to-epithelial transition and condenses into a PTA. This PTA undergoes tubulogenesis to form an RV, which then progresses to an S-shaped body as the nephron matures [8]. ECs have long been believed to associate with the developing nephron at the S-shaped body stage [14,35]. This cleft contains podocyte progenitors expressing VEGF which attracts ECs [36,11,35]. However, relatively little is known about how blood vessels arise in and around the S-shape body and how they take shape during different stages of nephron development.
To assess the fine capillary structures around developing nephrons at different stages, we performed 3D imaging of thick sections (20 μm). This analysis revealed that a net-like endothelial plexus forms around the developing nephron as early as the RV stage (Fig. 4a–a’). Notably, ECs surround the RV except in the region of direct cell-cell contact between the UB and RV epithelium, the point where these structures fuse (arrow, Fig 4a–a’). At the S-shaped body stage, we observed ECs within the cleft, as previously shown [35]. However, 3D imaging revealed that these ECs connect around the S-shaped body to form a plexus surrounding the developing nephron (Fig. 4b–b’’’, Supplemental Fig. 5).
Figure 4. ECs form a plexus around the developing nephron.
a-b 20 μm sections of a developing RV (a) and S-Shaped Body (b) with 3D reconstruction of the structure (a’, b’’’) demonstrating the developing endothelial plexus from the RV stage forward. Orange arrow marks region where RV and UB are fused together and no ECs can be seen. White arrowhead marks ECs in cleft of developing S-shaped body. c-e Whole mount stains for the RV (c), late RV – when the RV elongates and connects to the UB tip (d), and S-shaped body stages (e). White arrowheads mark ECs in between the RV and UB (in c) or looping around the connection between the RV and the UB (in d). Orange arrowhead in d indicates areas devoid of direct epithelial contact as part of the vascular basket. White outline in e’-e’’ marks distal portion of S-shaped body. Scale bar = 50 μm.
Using whole mount analysis, we were able to further characterize the changing anatomy of the renal vasculature at multiple stages of nephrogenesis. At the RV stage, when aggregated NPCs transform into an epithelial ball, ECs surround the RV on all sides, including between the lateral aspect of the vesicle and the UB (Fig. 4c, Supplemental Fig. 5a, Movie 5, arrow). As the RV elongates and fuses to the UB epithelium, an EC collar forms around the most distal part of the RV (Fig 4d–d’, white arrowheads, Movie 6, white arrows), while a vascular ‘basket’ remains surrounding the rest of the developing nephron that only occasionally made direct contact with the epithelium (Fig. 4d–d’, orange arrowheads Supplemental Fig. 5b, Movie 6, orange arrow). By the S-shaped body stage, the EC collar has elaborated into a plexus which envelopes the part of the S-shaped body destined to become the distal tubules (white outlines), while only sparsely covering the part that will form the glomerulus and proximal tubules (Fig. 4e–e’’, Supplemental Fig. 5c, Movie 7). Notably, this plexus is continuous with the surrounding endothelium, rather than consisting of a single invading sprout.
ECs of the developing kidney are heterogeneous
Recently, Lindstrom and colleagues utilized an immunofluorescent screen to demonstrate and map the heterogeneity of the epithelial cells in the RV and S-shaped body [37]. Given that ECs display a high degree of heterogeneity between different tissues and even within individual organs, we screened a variety of standard vascular markers in the developing kidney to assess potential regional differences as vessels take shape. We carried out a detailed qualitative characterization of expression patterns in E15.5, P1, and P5 kidneys, using Flk1-eGFP to confirm cells as endothelial. Marker fluorescence was scored for presence, intensity, and breadth of expression in capillaries of the renal cortex, medulla, and glomerulus, and ECs of the major vessels. Major vessels are defined as arteries and veins that appear as ‘doublets’, as shown above (Supplemental Fig. 2). Importantly, this analysis should only be used to compare staining intensities for the same protein in different regions at different time points, but should not be used to directly compare staining intensities between protein levels. For the purposes of this analysis, only expression in ECs was scored, although a few factors (E.g. Vegfr1, Claudin-5, Nrp1, Podxl1) also exhibited non-endothelial expression [38].
Strikingly, endothelial markers tested displayed highly dynamic expression patterns during murine kidney development (Fig. 5a). Only 1 gene – PECAM–was strongly expressed in all renal ECs, at all time points. An additional 3 genes – Vegfr2, VE-Cad and Icam2 – were expressed in all ECs at all time points, but displayed differences in expression intensity in specific regions as the kidney matures. Therefore, only these 4 genes are pan-endothelial up through P5 of kidney development.
The remaining 14 factors displayed restricted endothelial expression patterns. Changes in expression over time were gene-dependent. The expression pattern of some genes became regionalized over time (e.g., Vegfr3, Fig. 5b–i’), while that of other genes broadened as the kidney matured (e.g., vWF, Fig. 5j–q’). Other genes demonstrated region-specific differences, whereby expression expanded in one region but became more restricted in another over time (e.g. Cx40, Claudin-5).
To better assess differences in overall expression patterns between two given regions, we developed a qualitative scoring system to compare the overall expression profile of a specific gene, comparing levels in one region to a different region (refer to Methods). Our comparisons were arbitrary, but all assessments were carried out using identical experimental and imaging settings (as per [39]). Briefly, two regions that greatly differ in expression patterns will have a higher score (maximum of 9), while two regions with very similar expression patterns will have a lower score (minimum of 0). From this analysis, the two most similar regions are cortex and veins, while the two most divergent regions are arteries and veins (Fig. 5r–s, Supplemental Fig. 6). The comparisons can be organized into two groups based on the difference score: 1. arterial and glomerular ECs (score ≥ 2), and 2. cortical, medullary, and venous ECs (score < 2). Thus, arterial and glomerular ECs have the most distinct molecular signatures within the kidney endothelium.
Transcriptome analysis of embryonic kidney ECs identifies regional molecular differences
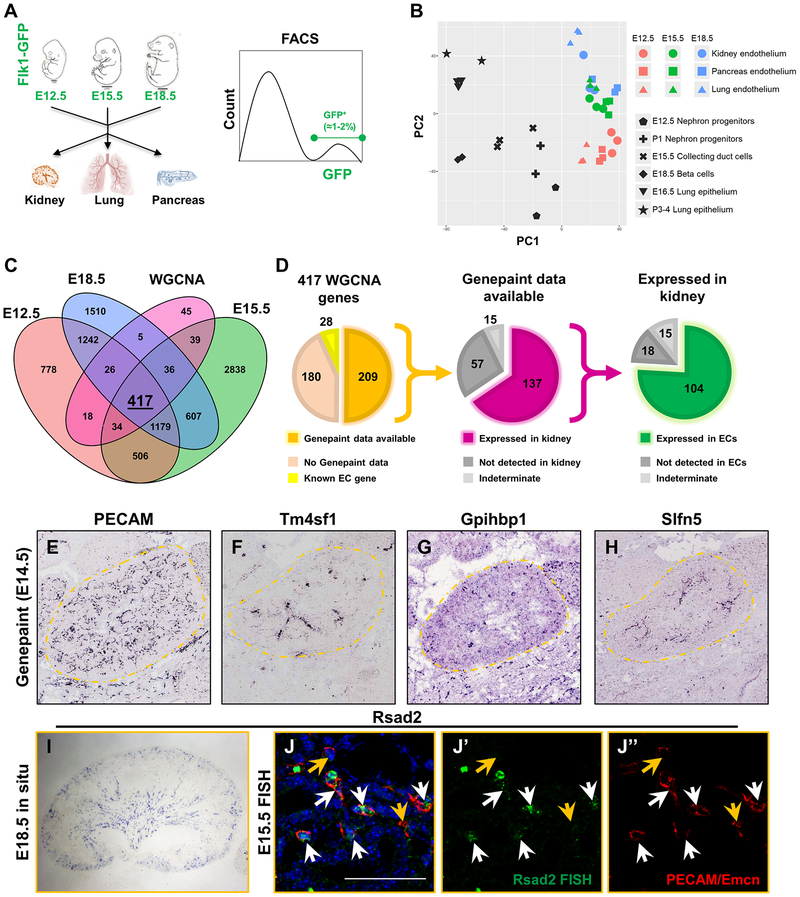
In order to better characterize the expression profile of renal vasculature and to identify potential kidney-specific EC genes, we carried out transcriptional analysis on ECs isolated from the embryonic kidney and other embryonic organs. Utilizing the Flk1-eGFP transgenic line, we first isolated eGFP+ ECs at three embryonic time points, E12.5, E15.5, and E18.5, from three different developing organs – kidney, lung, and pancreas – by FACS. We then performed RNA-seq (Fig. 6a). Bioinformatic analysis was used to compare these samples to each other and to publicly available datasets of non-EC types in the kidney (Gene Expression Omnibus). This analysis identified genes enriched in ECs that are specific to one organ or time point (Fig. 6b–c, Supplemental Fig. 7). Principal component analysis (PCA) verified the clustering of endothelial transcriptomes together, and that they were linearly separable from the comparator cell types along the first principal component (Fig. 6b). To further enrich this set of genes for those most likely to be expressed in the kidney endothelium, we performed Weighted Correlation Network Analysis (WGCNA) on the genes with increased expression in the kidney endothelium at all 3 time points. This analysis yielded 417 genes likely to be significantly expressed in the renal endothelium (Fig. 6c, see Supplemental Data for full list). Of these, 35 genes demonstrated a significant monotonic increase in expression from E12.5 to E18.5 while only 2 genes showed a significant monotonic decrease in expression within the kidney (Supplemental Fig. 7).
Figure 6. RNA-seq reveals endothelial spatiotemporal heterogeneity.
a Schematic for RNA-seq analysis. 3 organs – kidneys, lungs, and pancreas – were dissected out of E12.5, E15.5 and E18.5 Flk1-eGFP+ embryos (for a total of 9 samples) and digested to single cells. After digestion, GFP+ cells were isolated by FACS and subjected to RNA-seq analysis in bulk. b Principal component analysis of 9 endothelial populations (colored) compared to six different outgroups (black). Endothelial populations cluster away from outgroups and show clear stratification over time. c Venn diagram of genes enriched in the kidney endothelium between E12.5, E15.5, E18.5, and those identified through WGCNA (p-value < 0.05). d Graphs representing validation approach of the 417 genes identified through WGCNA of the gene enriched in the kidney endothelium at all 3 time points. Of the 417 genes identified, 28 of them are known EC genes (yellow) and 209 of them had data from genepaint.org available (orange, left graph). Of the 209 genes in which data was available, 137 of them were expressed in the kidney (middle graph, purple). Lastly, 104 of the 137 genes in kidney exhibited endothelial staining pattern in the kidney (green, right graph). e-h in situ hybridizations from genepaint.org. Kidneys are outlined in orange dotted lines to better visualize the kidney proper. i In situ hybridization of Rsad2 on E18.5 kidneys. j-j’’) Fluorescent in situ hybridizations on E15.5 kidneys demonstrating Rsad2 is restricted to ECs in the kidney (white arrows) but is not expressed in all ECs (orange arrows). Note that two large green dots on the FISH are autofluorescent blood cells. Scale bar = 50 μm.
We found that 28 of the 417 genes were established EC markers, demonstrating that our analysis properly identified endothelial-specific genes (Fig. 6d). We first validated expression of the remaining genes using the publicly available www.genepaint.org and Genitourinary Development Molecular Anatomy Project (GUDMAP) databases, which contain in situ hybridizations of E14.5 embryos. We found that 180 genes did not have reported Genepaint data, leaving 209 potential novel EC genes (Fig. 6d). Each of these genes was assessed for both expression in the kidney in general and expression in kidney ECs. PECAM in situ hybridization data from www.genepaint.org was used as a positive control to confirm endothelial expression (Fig. 6e).
Our analysis revealed that 66% (137/209) of the genes showed clear expression in the kidney with 76% (104/137) of those being expressed in renal ECs (Fig. 6d). The vast majority of these genes were endothelial-specific (97/104), while the rest exhibited both stromal and endothelial patterns. Through this analysis, we also identified 12 genes that exhibited restricted expression patterns within the kidney. These patterns include arterial (Tm4sf1, Fig. 6f), outer cortical (Gpihbp1, Fig. 6g), and corticomedullary (Slfn5, Fig. 6h). We performed additional in situ hybridizations on E15.5 - E18.5 kidneys to verify that the genes identified are expressed in the renal endothelium (Fig. 6i–j, Supplemental Fig. 8a–b) and validate genes not available on Genepaint (Supplemental Fig. 8c–d). Of the genes we screened, Rsad2 appeared to be the most restricted to the kidney endothelium in the embryo with moderate punctate expression in the liver (Supplemental Fig. 8a). FISH analysis revealed that Rsad2 is expressed in a subset of ECs in the E15.5 kidney (Fig. 6j–j’’). Together these data provide a toolkit of new vascular markers, both region- and organ-specific, that will be useful to those studying the kidney and its blood vessels. In addition, it maps out emergence of EC heterogeneity in the kidney, demonstrating transcriptional and possibly functional regionalization of the vasculature occurs early in development.
DISCUSSION
Recent interest has focused on development of blood vessels in developing organs, especially to identify ways to apply these results to generating ex vivo transplantable organs for tissue replacement [5,33]. Although there are important differences between murine and human kidney development [40,39,37], the studies performed in the mouse can instruct the approaches carried out in human samples [41]; therefore, annotation of mouse kidney development, understanding EC heterogeneity, and recognition of regionalized gene expression, will aid efforts to engineer ex vivo kidneys. To date, ontogeny of the renal vasculature has remained poorly understood. In part, this has been due to a lack of useful markers for the kidney vasculature. Here, we analyze renal blood vessel anatomy from the onset of kidney morphogenesis and create a comprehensive atlas of the developing vasculature. Using whole mount imaging, we visualize the kidney in 3D to identify novel paradigms underlying formation of renal blood vessels. Specifically, we identify several patterns of capillary organization around epithelial sub-structures during metanephric development, including the progenitor pools, the developing nephron, and the collecting ducts, expanding upon previous studies such as [12]. In addition, using transcriptional and immunofluorescent profiling, we identify both known and novel vascular markers and map their regionalized expression within the kidney to better illustrate renal EC heterogeneity.
Emergence of the kidney vasculature
Our detailed spatiotemporal analysis of the renal vasculature shows that the vasculature grows coordinately with each newly forming nephron. Fine renal capillaries form a meshwork that expands and remodels throughout metanephric development. Our findings are in line with recent studies [12], but extend our understanding of plexus remodeling in and around the early nephron. Fine capillaries emerge earlier than previously thought, and do not envelop all renal structures indiscriminately or uniformly. Vascular cords first surround the metanephros at E10.5, then expand around UBs and caps of NPCs from E11.5. We found no evidence for isolated angioblasts within or around the E11.5 metanephros in whole mount stained tissues. Instead, we observe in 3D elaboration of a capillary plexus around kidney progenitors. We note, however, that our results do not rule out the possibility of a more immature angioblast population that is Flk1, PECAM, and Emcn negative and can give rise to mature ECs. Thus the origin of ECs in the developing kidney remains uncertain.
Renal AV differentiation
In the present study, we also characterize AV differentiation during kidney development. We validate previous findings that the renal artery differentiation is first evident at E13.5, using an alternate arterial-specific marker [15]. At this time point, an arterial tree with at least three branching events can be distinguished. This organization is known to rely on cues from the surrounding stromal cells, as deletion of FoxD1-expressing stroma [42], FoxD1 [43], or Pbx1 [16] all drastically impair arterial organization. Because these arteries are perfused by E13.5 [26,16], we suggest that renal arterial maturation is – at least in part – dependent on blood flow, similar to many other vascular beds.
AV differentiation has been previously described in other systems. In the trunk of the early embryo and the pancreas, formation of arteries precedes that of veins, and developing vessels express for a time overlap of AV markers [5,27]. Venous development in the kidney occurs alongside arterial development. Although faint, EphB4-LacZ staining suggests that the venous tree mirrors the pattern of the arterial tree. This agrees with previous work in adult rat kidney that identified artery and vein co-alignment [44]. Our work demonstrates that this paradigm is established early during renal development. We further show that larger veins and arteries can be readily identified in cross sections of tissue as tubular “doublets”, providing an anatomical tool to demarcate renal veins given the lack of proper venous markers.
Endothelial coordination with UB and RV formation
In agreement with work done by Munro and colleagues, we observed ECs reiteratively bisect distal UB tips to circumscribe individual NPC caps [12]. We expanded on this work by characterizing endothelial organization in and around all parts of the developing renal epithelium and identifying several new paradigms of renal blood vessel formation. First, ECs form a highly stereotyped cruciform pattern around the distal UB tip. Notably, a layer of cells immediately surrounding the bisecting ECs are not UB cells nor NPCs, but rather stromal cells. It is unclear if the stroma directly communicates with ECs, UB, or NPCs to regulate endothelial organization, or if there are signals from the endothelium that pattern the UB, NPCs, or stroma. A number of studies have suggested kidney-EC crosstalk [45,46], but the focus has primarily been on epithelial VEGFA [35]. Additionally, the endothelium closely associates with the UB trunk beginning at E14.5, but not earlier, suggesting that the UB trunk and tips pattern the endothelium differently.
We and others note that ECs circumscribe progenitor caps but avoid invading them. This organization raises the question of whether peripheral cap NPCs, which are located more closely to the endothelium, behave differently than those farther away. Hypoxia has been shown to be an important modulator of embryonic [47], neural [48], hematopoietic [49], and cardiac [50] stem cells. Some studies suggest vessels closest to NPCs are not perfused and hypoxia inhibits NPC differentiation in ex vivo kidney explants, suggesting that hypoxia may modulate NPCs [26]. Munro et al. did however identify red blood cells (RBCs) in these vessels. It remains an open question whether presence of RBCs definitively means that peripheral capillaries are perfused, as blood has been observed to arise in situ [51]. Given that there may be non-nutritional signals from ECs to peripheral cap NPCs, as observed in other tissues [31] or that oxygen levels may impact progenitors, future studies will be required to determine any functional relationship between the vasculature and NPCs.
Previous models of endothelial activity during nephrogenesis have posited that ECs first interact with the developing nephron during the S-shaped body stage, where they migrate into the cleft of the developing glomerulus [35,13]. These conclusions were based on 2D sections that can only image part of the nephron. However, imaging the entire nephron in 3D reveals new and unexpected microanatomy of endothelial and epithelial tissues. We find that a capillary plexus intimately surrounds the RV, prior to S-shaped body stage, and develops with the maturing nephron. Later, following formation of the S-shaped body, capillaries surround most of the developing nephron while maintaining connections to the surrounding plexus. These observations define clear patterns of capillary organization with developing nephrons. Due to the limitations of imaging fixed tissue, questions remain about how exactly ECs and developing nephrons dynamically remodel with respect to each other as they mature through these stages.
The close association between the RV and the endothelial plexus may point to active communication between these cell types. Indeed, ECs demonstrate clear differences in patterning along the developing nephron, showing greater density around the region fated to become distal tubule. Proximodistal polarity in the developing nephron is established early in nephrogenesis during the RV stage when we observe the first signs of endothelial patterning [34,37]. Whether endothelial patterning around the new nephron is regulated by the same mechanisms that establish nephron polarity remains to be determined. These findings also bring up the question of whether early fate commitment in the RV is regulated by ECs. Further studies are needed to uncover such potential functions of ECs in nephron development.
Transcriptional heterogeneity in ECs precedes functional heterogeneity
Solute movement and serum chemistries are carried out by the vasculature and regionalized along the nephron, implying that the associated endothelium is also specialized to specific renal compartments [52]. However, we have yet to understand the mechanisms that establish such differences between ECs. Previous work looking at endothelial heterogeneity have focused on the organism as a whole, either characterizing differences between arterial or venous ECs [53] or ECs across adult organs [54,55], but not across embryonic organs. One study aimed to characterize endothelial heterogeneity in different regions of the adult kidney by identifying genes expressed in specific regions using microarray analysis [56]. Although this study also screened E15.5 kidneys, the authors did not focus on distinct regions within the embryonic kidney.
Our transcriptional and immunofluorescent screens reveal that heterogeneity evident in adulthood is established early during development, providing an initial foundation for EC specification at early stages of metanephric development. Indeed, only a handful of known EC markers exhibited pan-endothelial expression throughout kidney development. Most were expressed in a regionally defined manner. Based on our data, it may be more appropriate to define each population of ECs by a panel of markers, rather than commonly used ones like PECAM or Flk1. Our data further show that the endothelial transcriptome is spatiotemporally dynamic, as most genes display highly distinct expression patterns across the embryonic and postnatal stages tested. Thus, the immunofluorescent screen provides a powerful toolbox of markers that can be used to assess endothelial identity in the developing kidney in the absence of region-specific markers. However, the conclusions that can be drawn from the immunofluorescent screen are inherently limited because 1. it is a qualitative assessment of gene expression by immunofluorescence, 2. we cannot strictly compare intensities between markers, but only for the same gene across regions and time points, and 3. the absence of antigen expression does not rule out gene expression at a level below the threshold detected by immunofluorescence. However, we suggest the data will be useful for those investigating kidney vasculature, as relative measures of marker levels.
Based on our RNA-seq analysis followed by Genepaint and GUDMAP screen, only a handful of validated genes exhibited some degree of regionality within the kidney. One potential explanation for this is that the resolution provided by Genepaint is not high enough to identify specific regional patterns unless the staining pattern is strong and specific. Additionally, vessels in E14.5 kidneys may not be as clearly differentiated and heterogeneous as their more mature counterparts. Consistent with this, in situ hybridization for Rsad2 at E15.5 appeared to be more broadly expressed in the kidney endothelium compared to that in the E18.5 kidney. Lastly, our analysis identified genes that were highly expressed at all 3 time points during development. Overall, this screen can provide a valuable tool for identification of novel functional genes that regulate kidney vascular development or genes important for endothelial-epithelial crosstalk to the nearby nephron or the UB as they develop.
SUMMARY
Our results indicate that endothelial development in the kidney is a highly dynamic process that coordinates with epithelial and stromal development and undergoes extensive remodeling as the kidney grows. These predictable patterns of endothelial development strongly suggest that endothelial patterning is tightly regulated. The data presented here will serve as a foundation to assess alterations in the renal vasculature in diseased or engineered nephrons, provide a useful toolkit for vascular assessment in the developing kidney, and identify genes important for renal vascular patterning and function.
Supplementary Material
HIGHLIGHTS.
Initial vessels in the kidney arise from the onset of ureteric budding from an existing vascular plexus within the mesoderm.
Arteriovenous (AV) differentiation initiates at E13.5, after which arteries and veins develop in predictable patterns.
Endothelial cells (ECs) organize around nephron progenitors, ureteric bud tree, and developing nephrons in defined manners.
Renal vascular patterning is highly dynamic throughout kidney development.
ACKNOWLEDGEMENTS
We thank Janet Rossant for the Flk1-eGFP mouse line, as well as members of the Cleaver lab, including Caitlin Braitsch, Xiaowu Gu, and David Barry, for discussions and critical reading of the manuscript. We thank the Genepaint.org database for in situ hybridization data (where noted).
FUNDING
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases DK099478 to D.K.M., DK080004, DK095057, DK106743 to T.J.C. and DK106743, DK079862 to O.C.; CPRIT RP110405; and National Institute of Heart, Lung and Blood HL113498 to O.C. Deposited in PMC for release after 12 months.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing or financial interests.
REFERENCES
- 1.Cleaver O, Tonissen KF, Saha MS, Krieg PA (1997) Neovascularization of the Xenopus embryo. Dev Dyn 210 (1):66–77 [DOI] [PubMed] [Google Scholar]
- 2.Herbert SP, Huisken J, Kim TN, Feldman ME, Houseman BT, Wang RA, Shokat KM, Stainier DY (2009) Arterial-venous segregation by selective cell sprouting: an alternative mode of blood vessel formation. Science 326 (5950):294–298. doi: 10.1126/science.1178577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coultas L, Nieuwenhuis E, Anderson GA, Cabezas J, Nagy A, Henkelman RM, Hui CC, Rossant J (2010) Hedgehog regulates distinct vascular patterning events through VEGF-dependent and -independent mechanisms. Blood 116 (4):653–660. doi: 10.1182/blood-2009-12-256644 [DOI] [PubMed] [Google Scholar]
- 4.Drake CJ, Fleming PA (2000) Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood 95 (5):1671–1679. [PubMed] [Google Scholar]
- 5.Azizoglu DB, Chong DC, Villasenor A, Magenheim J, Barry DM, Lee S, Marty-Santos L, Fu S, Dor Y, Cleaver O (2016) Vascular development in the vertebrate pancreas. Dev Biol 420 (1):67–78. doi: 10.1016/j.ydbio.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oxburgh L, Carroll TJ, Cleaver O, Gossett DR, Hoshizaki DK, Hubbell JA, Humphreys BD, Jain S, Jensen J, Kaplan DL, Kesselman C, Ketchum CJ, Little MH, McMahon AP, Shankland SJ, Spence JR, Valerius MT, Wertheim JA, Wessely O, Zheng Y, Drummond IA (2017) (Re)Building a Kidney. J Am Soc Nephrol 28 (5):1370–1378. doi: 10.1681/ASN.2016101077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien LL, McMahon AP (2014) Induction and patterning of the metanephric nephron. Semin Cell Dev Biol 36:31–38. doi: 10.1016/j.semcdb.2014.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Zimmerman S, Brakeman PR, Beaudoin GM 3rd, Reichardt LF, Marciano DK (2013) De novo lumen formation and elongation in the developing nephron: a central role for afadin in apical polarity. Development 140 (8):1774–1784. doi: 10.1242/dev.087957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao L, Yang Z, Hiremath C, Zimmerman SE, Long B, Brakeman PR, Mostov KE, Bryant DM, Luby-Phelps K, Marciano DK (2017) Afadin orients cell division to position the tubule lumen in developing renal tubules. Development 144 (19):3511–3520. doi: 10.1242/dev.148908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert B, St John PL, Abrahamson DR (1998) Direct visualization of renal vascular morphogenesis in Flk1 heterozygous mutant mice. Am J Physiol 275 (1 Pt 2):F164–172 [DOI] [PubMed] [Google Scholar]
- 11.Tufro A, Norwood VF, Carey RM, Gomez RA (1999) Vascular endothelial growth factor induces nephrogenesis and vasculogenesis. J Am Soc Nephrol 10 (10):2125–2134 [DOI] [PubMed] [Google Scholar]
- 12.Munro DAD, Hohenstein P, Davies JA (2017) Cycles of vascular plexus formation within the nephrogenic zone of the developing mouse kidney. Sci Rep 7 (1):3273. doi: 10.1038/s41598-017-03808-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughan MR, Quaggin SE (2008) How do mesangial and endothelial cells form the glomerular tuft? J Am Soc Nephrol 19 (1):24–33. doi: 10.1681/ASN.2007040471 [DOI] [PubMed] [Google Scholar]
- 14.Gao X, Chen X, Taglienti M, Rumballe B, Little MH, Kreidberg JA (2005) Angioblast-mesenchyme induction of early kidney development is mediated by Wt1 and Vegfa. Development 132 (24):5437–5449. doi: 10.1242/dev.02095 [DOI] [PubMed] [Google Scholar]
- 15.Munro DAD, Hohenstein P, Coate TM, Davies JA (2017) Refuting the hypothesis that semaphorin-3f/neuropilin-2 exclude blood vessels from the cap mesenchyme in the developing kidney. Dev Dyn 246 (12):1047–1056. doi: 10.1002/dvdy.24592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurtado R, Zewdu R, Mtui J, Liang C, Aho R, Kurylo C, Selleri L, Herzlinger D (2015) Pbx1-dependent control of VMC differentiation kinetics underlies gross renal vascular patterning. Development 142 (15):2653–2664. doi: 10.1242/dev.124776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villasenor A, Chong DC, Cleaver O (2008) Biphasic Ngn3 expression in the developing pancreas. Dev Dyn 237 (11):3270–3279. doi: 10.1002/dvdy.21740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acar M, Kocherlakota KS, Murphy MM, Peyer JG, Oguro H, Inra CN, Jaiyeola C, Zhao Z, Luby-Phelps K, Morrison SJ (2015) Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526 (7571):126–130. doi: 10.1038/nature15250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14 (4):417–419. doi: 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soneson C, Love MI, Robinson MD (2015) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4:1521. doi: 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15 (12):550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ema M, Takahashi S, Rossant J (2006) Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood 107 (1):111–117. doi: 10.1182/blood-2005-05-1970 [DOI] [PubMed] [Google Scholar]
- 24.Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC (1995) Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376 (6535):62–66 [DOI] [PubMed] [Google Scholar]
- 25.Moyon D, Pardanaud L, Yuan L, Breant C, Eichmann A (2001) Plasticity of endothelial cells during arterial-venous differentiation in the avian embryo. Development 128 (17):3359–3370 [DOI] [PubMed] [Google Scholar]
- 26.Rymer C, Paredes J, Halt K, Schaefer C, Wiersch J, Zhang G, Potoka D, Vainio S, Gittes GK, Bates CM, Sims-Lucas S (2014) Renal blood flow and oxygenation drive nephron progenitor differentiation. Am J Physiol Renal Physiol 307 (3):F337–345. doi: 10.1152/ajprenal.00208.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong DC, Koo Y, Xu K, Fu S, Cleaver O (2011) Stepwise arteriovenous fate acquisition during mammalian vasculogenesis. Dev Dyn 240 (9):2153–2165. doi: 10.1002/dvdy.22706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klagsbrun M, Takashima S, Mamluk R (2002) The role of neuropilin in vascular and tumor biology. Adv Exp Med Biol 515:33–48 [DOI] [PubMed] [Google Scholar]
- 29.dela Paz NG, D’Amore PA (2009) Arterial versus venous endothelial cells. Cell Tissue Res 335 (1):5–16. doi: 10.1007/s00441-008-0706-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villasenor A, Wang ZV, Rivera LB, Ocal O, Asterholm IW, Scherer PE, Brekken RA, Cleaver O, Wilkie TM (2010) Rgs16 and Rgs8 in embryonic endocrine pancreas and mouse models of diabetes. Dis Model Mech 3 (9–10):567–580. 10.1242/dmm.003210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magenheim J, Ilovich O, Lazarus A, Klochendler A, Ziv O, Werman R, Hija A, Cleaver O, Mishani E, Keshet E, Dor Y (2011) Blood vessels restrain pancreas branching, differentiation and growth. Development 138 (21):4743–4752. doi: 10.1242/dev.066548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng X, Wert SE, Federici R, Peters KG, Whitsett JA (1998) VEGF enhances pulmonary vasculogenesis and disrupts lung morphogenesis in vivo. Dev Dyn 211 (3):215–227. [DOI] [PubMed] [Google Scholar]
- 33.Lazarus A, Del-Moral PM, Ilovich O, Mishani E, Warburton D, Keshet E (2011) A perfusion-independent role of blood vessels in determining branching stereotypy of lung airways. Development 138 (11):2359–2368. doi: 10.1242/dev.060723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costantini F, Kopan R (2010) Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell 18 (5):698–712. doi: 10.1016/j.devcel.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eremina V, Sood M, Haigh J, Nagy A, Lajoie G, Ferrara N, Gerber HP, Kikkawa Y, Miner JH, Quaggin SE (2003) Glomerular-specific alterations of VEGF-A expression lead to distinct congenital and acquired renal diseases. J Clin Invest 111 (5):707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitamoto Y, Tokunaga H, Tomita K (1997) Vascular endothelial growth factor is an essential molecule for mouse kidney development: glomerulogenesis and nephrogenesis. J Clin Invest 99 (10):2351–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindstrom NO, Tran T, Guo J, Rutledge E, Parvez RK, Thornton ME, Grubbs B, McMahon JA, McMahon AP (2018) Conserved and Divergent Molecular and Anatomic Features of Human and Mouse Nephron Patterning. J Am Soc Nephrol. doi: 10.1681/ASN.2017091036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z, Zimmerman SE, Tsunezumi J, Braitsch C, Trent C, Bryant DM, Cleaver O, Gonzalez-Manchon C, Marciano DK (2016) Role of CD34 family members in lumen formation in the developing kidney. Dev Biol 418 (1):66–74. doi: 10.1016/j.ydbio.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindstrom NO, McMahon JA, Guo J, Tran T, Guo Q, Rutledge E, Parvez RK, Saribekyan G, Schuler RE, Liao C, Kim AD, Abdelhalim A, Ruffins SW, Thornton ME, Basking L, Grubbs B, Kesselman C, McMahon AP (2018) Conserved and Divergent Features of Human and Mouse Kidney Organogenesis. J Am Soc Nephrol. doi: 10.1681/ASN.2017080887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindstrom NO, Guo J, Kim AD, Tran T, Guo Q, De Sena Brandine G, Ransick A, Parvez RK, Thornton ME, Basking L, Grubbs B, McMahon JA, Smith AD, McMahon AP (2018) Conserved and Divergent Features of Mesenchymal Progenitor Cell Types within the Cortical Nephrogenic Niche of the Human and Mouse Kidney. J Am Soc Nephrol. doi: 10.1681/ASN.2017080890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Brien LL, Guo Q, Lee Y, Tran T, Benazet JD, Whitney PH, Valouev A, McMahon AP (2016) Differential regulation of mouse and human nephron progenitors by the Six family of transcriptional regulators. Development 143 (4):595–608. doi: 10.1242/dev.127175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hum S, Rymer C, Schaefer C, Bushnell D, Sims-Lucas S (2014) Ablation of the renal stroma defines its critical role in nephron progenitor and vasculature patterning. PLoS One 9 (2):e88400. doi: 10.1371/journal.pone.0088400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sequeira-Lopez ML, Lin EE, Li M, Hu Y, Sigmund CD, Gomez RA (2015) The earliest metanephric arteriolar progenitors and their role in kidney vascular development. Am J Physiol Regul Integr Comp Physiol 308 (2):R138–149. doi: 10.1152/ajpregu.00428.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nordsletten DA, Blackett S, Bentley MD, Ritman EL, Smith NP (2006) Structural morphology of renal vasculature. Am J Physiol Heart Circ Physiol 291 (1):H296–309. doi: 10.1152/ajpheart.00814.2005 [DOI] [PubMed] [Google Scholar]
- 45.Tufro A (2000) VEGF spatially directs angiogenesis during metanephric development in vitro. Dev Biol 227 (2):558–566 [DOI] [PubMed] [Google Scholar]
- 46.Abrahamson DR (2009) Development of kidney glomerular endothelial cells and their role in basement membrane assembly. Organogenesis 5 (1):275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazumdar J, O’Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS, Simon MC (2010) O2 regulates stem cells through Wnt/beta-catenin signalling. Nat Cell Biol 12 (10):1007–1013. doi: 10.1038/ncb2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Culver JC, Vadakkan TJ, Dickinson ME (2013) A specialized microvascular domain in the mouse neural stem cell niche. PLoS One 8 (1):e53546. doi: 10.1371/journal.pone.0053546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, Suda T (2010) Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 7 (3):391–402. doi: 10.1016/j.stem.2010.06.020 [DOI] [PubMed] [Google Scholar]
- 50.Kimura W, Xiao F, Canseco DC, Muralidhar S, Thet S, Zhang HM, Abderrahman Y, Chen R, Garcia JA, Shelton JM, Richardson JA, Ashour AM, Asaithamby A, Liang H, Xing C, Lu Z, Zhang CC, Sadek HA (2015) Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature 523 (7559):226–230. doi: 10.1038/nature14582 [DOI] [PubMed] [Google Scholar]
- 51.Saxén L (1987) Vascularization of the nephron In: Barlow PW, Green PB, Wylie CC (eds) Organogenesis of the kidney. Cambridge University Press, Cambridge, pp 129–142 [Google Scholar]
- 52.Molema G, Aird WC (2012) Vascular heterogeneity in the kidney. Semin Nephrol 32 (2):145–155. doi: 10.1016/j.semnephrol.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 53.Aird WC (2007) Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res 100 (2):158–173. doi: 10.1161/01.RES.0000255691.76142.4a [DOI] [PubMed] [Google Scholar]
- 54.Aird WC (2012) Endothelial cell heterogeneity. Cold Spring Harb Perspect Med 2 (1):a006429. doi: 10.1101/cshperspect.a006429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nolan DJ, Ginsberg M, Israely E, Palikuqi B, Poulos MG, James D, Ding BS, Schachterle W, Liu Y, Rosenwaks Z, Butler JM, Xiang J, Rafii A, Shido K, Rabbany SY, Elemento O, Rafii S (2013) Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell 26 (2):204–219. doi: 10.1016/j.devcel.2013.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunskill EW, Potter SS (2010) Gene expression programs of mouse endothelial cells in kidney development and disease. PLoS One 5 (8):e12034. doi: 10.1371/journal.pone.0012034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.