Abstract
MicroRNAs are small non-coding RNAs that play important roles in many cellular processes including proliferation, metabolism and differentiation. They function by binding to specific regions within the 3’UTR of target mRNAs resulting in suppression of protein synthesis and modulation of potentially many cellular pathways. We previously showed that miRNA expression levels differed between cells from distinct regions of developing human embryonic long bones. Specifically, we found that miR-181a-1 was significantly more highly expressed in hypertrophic chondrocytes compared to proliferating differentiated or progenitor chondrocytes, suggesting a potential role in regulating chondrocyte hypertrophy and/or endochondral bone formation. The goal of this study was to determine how miR-181a-1 together with its clustered miRNA, miR-181b-1, regulates osteogenesis. We show that over-expression of the miR-181a/b-1 cluster enhanced osteogenesis and that cellular pathways associated with protein synthesis and mitochondrial metabolism were significantly up-regulated. Metabolic assays revealed that the oxygen consumption rate and ATP-linked respiration were increased by miR-181a/b-1. To further decipher a potential mechanism causing these metabolic changes, we showed that PTEN (phosphatase and tensin homolog) levels were suppressed following miR-181a/b-1 over-expression, and that PI3K/AKT signaling was subsequently increased. Over-expression of PTEN was found to attenuate the enhancing effects of miR-181a/b-1, providing further evidence that miR-181a/b-1 regulates the PTEN/PI3K/AKT axis to enhance osteogenic differentiation and mitochondrial metabolism. These findings have important implications for the design of miR-181a/b targeting strategies to treat bone conditions such as fractures or heterotopic ossification.
Keywords: microRNA, miR-181a/b, osteogenesis, PTEN, PI3K/AKT, mitochondrial metabolism
1. Introduction
MicroRNAs (miRNAs) are important regulators of many cellular processes including proliferation, apoptosis, metabolism and differentiation. They are initially transcribed by RNA polymerase II as large primary RNAs which are processed by Drosha to form shorter hairpin precursor RNAs. These pre-miRNAs are then further processed in the cytoplasm by Dicer to form a mature miRNA duplex consisting of a 5p and 3p strand. Generally, one of these single-stranded miRNAs (~20–24 nucleotides) will enter the RNA induced silencing complex (RISC) and bind to a specific region, via its seed sequence, within the 3’UTR of a target mRNA resulting in suppression of protein synthesis by either mRNA degradation or inhibition of translation (1). In general, protein suppression by miRNAs is not as robust compared to that mediated by siRNAs. However, one miRNA can potentially target many mRNAs within a given cell type, resulting in widespread alterations in cellular pathways and networks (2). There are many reports on altered expression of miRNAs in specific disease states, thus rendering these non-coding RNAs as potential biomarkers or therapeutic targets. As currently listed in the clinicaltrials.gov website (https://clinicaltrials.gov/), some miRNA mimics or antagomirs are currently under Phase I studies, including MRG-106 (a miR-155 antagomir) or MRG-201 (a miR-29 mimic) to determine effects in treating lymphoma or scar tissue formation, respectively.
Of relevance to this study, members of the miR-181 family have been reported to play regulatory roles in controlling bone or cartilage cell development/function as well as mature chondrocyte homeostasis (3–11). The miR-181 family consists of miR-181a-1, a-2, b-1, b-2, c and d. The mature functional 5p strands of miR-181a-1 and a-2 are identical as are the 5p strands of miR-181b-1 and b-2. miR-181a-1 is clustered with miR-181b-1 on chromosome 1 (miR-181a/b-1), miR-181a-2 and miR-181b-2 are clustered on chromosome 9 (miR-181a/b-2), and the miR-181c/d cluster is located on chromosome 19. Importantly, all family members contain the same seed sequence in the 5p strand suggesting a significant degree of functional redundancy (12). Mice lacking each of the miR-181 clusters have been generated (13,14) and display no obvious phenotype. However, closer examination reveals that mice devoid of miR-181a/b-1 have immune cell defects. Interestingly, mice lacking both miR-181a/b-1 and miR-181a/b-2 clusters have reduced survival rates while those that do survive are smaller in size than control littermates, suggesting a role for these paralogs in regulating skeletal development (13,15).
With respect to long bone development, we previously published a miRNA array dataset identifying miR-181a-1 as the most differentially-expressed miRNA in cells within specific regions of developing human long bones (16). Specifically, expression of this miRNA paralog was higher in hypertrophic chondrocytes compared to progenitor or proliferating chondrocytes of the growth plate. Similarly, expression of miR-181 family members were also found to increase during later phases of MSC chondrogenesis in vitro associated with increased markers of hypertrophy (17). These findings suggest a potential role for miR-181a-1 in regulating hypertrophic chondrocyte differentiation and/or processes of endochondral bone formation.
Analysis of immune cells from miR-181a/b-1 knock-out mice revealed PTEN (phosphatase and tensin homolog) to be a target gene (13). PTEN is the principal negative regulator of the PI3K/AKT pathway (18). Interestingly, there is enhanced bone formation in conditional PTEN knock-out mice (19,20) and a number of studies suggest that the PI3K/AKT pathway is a central nexus in the network of pathways that fine tunes osteoblast differentiation (21–25). In addition, many studies also reveal that PTEN as well as downstream mediators of the PI3K/AKT pathway can regulate cellular metabolism (26–30).
The goal of this study was to explore the function and mechanism of miR-181a-1 in regulating osteogenic differentiation of human primary skeletal cells. Given that miR-181a-1 is clustered closely with miR-181b-1 (~60 nucleotides apart), effects of over-expressing the miR-181a/b-1 cluster was investigated. This work presents new information on the role of miR-181a/b-1 in regulating osteogenesis, in part, by altering the PTEN/PI3K/AKT axis and mitochondrial metabolism. These findings have important implications toward developing new strategies to target the miR-181a/b-1 cluster as a potential therapeutic strategy to alter bone formation or repair processes in vivo.
2. Materials and Methods
2.1. Isolation and dedifferentiation of human primary chondrocytes
Utilization of human osteoarthritic (OA) articular cartilage tissue was approved by the Washington University Human Research Protection Office (IRB ID# 201104119). Knee joints were obtained following total knee arthroscopy. Regions of full thickness articular cartilage were removed from the joint surface, diced (approximately 2mm3) and digested in growth medium (DMEM/F12; 10% FBS) containing 0.025% collagenase and 0.025% pronase in cell spinner flasks overnight at 37°C. The resulting cell suspension was washed in HBSS and passed through a 70μm sterile filter. Primary chondrocytes (1 × 106 cells) were seeded in T75 cell culture flasks and cultured in growth medium until 90% confluent. Cells were passaged at least four times and dedifferentiation was confirmed by assessing the ratio of COL1A1: COL2A1 expression by qPCR.
2.2. Lentivirus production to over-express miR-181a/b-1
Human genomic pre-miR-181a-1 (NCBI Ref Seq: NR_029626.1), pre-miR-181b-1 (NCBI Ref Seq NR_029612.1) and intervening 61 nucleotide sequence was amplified by PCR (Table 1). The resulting miR-181a/b-1 amplicon was inserted into the pLemiR backbone (Addgene) using the Gibson Assembly Master Mix (New England Biolabs). Stocks of pLemir lentivirus expressing pre-miR-181a/b-1 or a non-silencing (NS) control RNA (31) were prepared as previously described (32) and titered using the Lenti-X p24 rapid titer ELISA (Clontech). DDCs were seeded at 2 × 105 cells/well in 12-well plates and transduced for 24 h with pLemiR lentiviruses expressing the NS control (LV-NS) or pre-miR-181a/b1 (LV-181a/b-1) at an MOI of 20 using growth medium containing 100 μg/mL protamine sulfate. Fresh growth medium was applied 24 h after transduction. Transduced DDCs were cultured in growth medium for an additional 48 h prior to addition of osteogenic induction medium.
Table 1:
Primer sequences and Life Technologies miRNA assay IDs used for vector cloning and quantitative PCR. Life Technologies = Life Technologies Inc, Grand Island, NY, USA; NCBI = National Center for Biotechnology Information
| Amplicon | Forward primer (5’ – 3’) | Reverse primer (5’ – 3’) | NCBI Reference |
|---|---|---|---|
|
miR-181-a/b-1 Genomic |
CTGGGGCACAGATAACCAATGTGATG TGGAGGTTTG |
AGGGGCGGAATTTGCTACAACAGTAG GAAGGTG |
NR_029612.1 |
| miR-181a-5p | Life technologies TaqMan miRNA assay ID 000480 |
NR_029626.1 | |
| miR-181b-5p | Life technologies TaqMan miRNA assay ID 001098 |
NR_029612.1 | |
| RNU44 | Life technologies TaqMan miRNA assay ID 001094 |
NR_002750 | |
| PPIA | TCCTGGCATCTTGTCCATG | CCATCCAACCACTCAGTCTTG | NM_021130.4 |
| RUNX2 | CATCACTGTCCTTTGGGAGTAG | ATGTCAAAGGCTGTCTGTAGG | NM_001024630.3 |
| OSX | CCACCTACCCATCTGACTTTG | CCTTCTAGCTGCCCACTATTT | AF477981.1 |
| OCN | AAATAGCCCTGGCAGATTCC | CAGCCTCCAGCACTGTTTAT | NM_199173.5 |
| PTEN | CCCACCACAGCTAGAACTTATC | TCGTCCCTTTCCAGCTTTAC | NM_000314.6 |
2.3. Osteogenic differentiation of DDCs in monolayer
Non-transduced or lentiviral transduced DDCs were seeded in 12-well plates (2 × 105 cells / well) in osteogenic differentiation medium (αMEM containing 10 % FBS, 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, 10 mM β-glycerol phosphate, 50 μM ascorbic acid, 10 nM dexamethasone) for up to 14 days with fresh medium changes every 3 days. To examine matrix mineralization, DDCs were fixed in 4% paraformaldehyde and stained for 30 min with 1% Alizarin Red ethanol solution. Alizarin Red staining was quantified using the Alizarin Red S Staining Quantification kit (ARed-Q #8678; ScienCell Research Laboratories) following the manufacturer’s instructions.
2.4. Osteogenesis in 3D Bone Scaffolds
Human cancellous bone was harvested from OA knee joints following total knee replacement surgery, washed thoroughly with Dulbecco’s PBS (DPBS) and decalcified by using a mild Immunocal™ formic acid decalcifier solution (StatLab; 1414-X) for 3 days. Partially decalcified cancellous bone was diced into 5 mm3 pieces and further decalcified for 3 days. Resulting decalcified bone “scaffolds” were washed thoroughly in DPBS (3 × 6 h washes), immersed in an antibiotic/antifungal solution (10,000 unit/ml penicillin, 10,000 μg/ml streptomycin, 25ug/ml amphotericin B) for 6 h followed by additional washes in DPBS (3 × 30 min). Bone scaffolds were air dried and stored at room temperature before use. Scaffolds were imaged by micro-CT using a Scanco μCT40 scanner (Scanco Medical AG, Switzerland) to establish a baseline density prior to in vitro 3D osteogenesis. Scans were completed at 45kVp and 177μA with 300ms integration time at an effective voxel size of 6μm. 5 × 105 of non-transduced DDCs or those transduced with LV-NS or LV-181a/b-1 were seeded onto each scaffold and cultured in growth medium for 48 h prior to addition of osteogenic culture medium for 28 days. Micro-CT scans of day 28 scaffolds were collected and, using Scanco (V6.5) software, the morph tool was used to define a region of interest and contours were drawn every 20 slices. A threshold value of 180 was set for evaluation of all scanned images. VOX-BV (voxel of bone volume) and VOX-TV (voxel of total volume) values were used to calculate BV/TV. Bone mineral densities (BMDs) within the scaffolds were also calculated. 3D images and movies were created using DRAGONFLY software (Object Research Systems Inc. Canada).
2.5. RNA isolation and qPCR
Total RNA was extracted from cells using the Total RNA Purification Kit (Norgen). miRNAs were reverse-transcribed and quantified with the appropriate TaqMan primer/probe sets (Life Technologies; Table 1), using the TaqMan microRNA reverse transcription kit (Life Technologies) and TaqMan master mix with no UNG (Life Technologies). To determine actual copy numbers of miR-181a-5p and miR-181b-5p in DDCs following transduction with LV-NS or LV-181a/b-1, synthetic miRNAs corresponding to the 5p strand of mature miR-181a or miR-181b were generated (IDT) and known copy numbers (ranging from 100 million to 10 copies) were reverse-transcribed and amplified by qPCR. Standard curve plots showing miRNA copy number and corresponding Ct values were generated and these were used to calculate copy numbers of endogenous miR-181a or miR-181b in DDCs following transduction with LVNS or LV-181a/b-1. mRNAs were reverse-transcribed using Superscript RT II (Life Technologies), and qPCR was performed using PowerUp SYBR master mix (Life Technologies). PCR primer sequences are shown in Table 1. Fold changes were calculated using the 2-ΔΔCt method (33).
2.6. Western blot
Cell lysates were prepared with RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 % Triton-X-100, 0.1 % SDS, 0.5% sodium deoxycholate) supplemented with 1x cOmplete protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Pierce). Total protein concentration was measured using the Bio-Rad Protein Assay kit. Proteins were resolved by SDS-PAGE and transferred onto a PVDF membrane. Protein-containing membranes were blocked with Odyssey Blocking Buffer (Licor) containing 0.1 % Tween 20. Western blots were performed with the following rabbit polyclonal antibodies, all purchased from Cell Signaling Technology (anti-RUNX2 #8486; anti-PTEN #9552, anti-AKT #4691; anti-pAKT-T308 #2965; anti-pAKT-S473 #4060). For purposes of protein normalization, mouse anti-β-actin antibodies were used (Abcam ab6267). All primary antibodies were used at a 1:1000 dilution. Secondary antibodies (IRDye® 800CW-labeled anti-rabbit; IRDye® 680RD-labeled anti-mouse) (Licor) were used at a 1:10000 dilution following the manufacturer’s recommendation. Resulting band intensity was calculated and quantified using the Licor Odyssey software.
2.7. RNA-Seq and pathway analysis
Three biological replicates of DDCs (transduced with LV-NS or LV-181a/b-1) were induced in osteogenic media and RNA was extracted from day 7 cultures. RNA samples were prepared for sequencing with the Illumina oligo-dT priming system on an Illumina HiSeq 3000. RNA-Seq reads were demultiplexed with Illumina’s bcl2fastq2 and then aligned to the Ensembl release 76 top-level assembly with STAR version 2.0.4b (34). Gene counts were imported into the R/Bioconductor package EdgeR (35,36) and the trimmed mean of M-values normalization (TMM) method was used to adjust for differences in effective library size across all samples after removing low expressing genes and ribosomal genes. The TMM size factors and the matrix of counts were then imported into the R/Bioconductor package limma (37,38). Weighted likelihoods based on the observed mean-variance relationship of every gene and sample were then calculated for all samples with the voomWithQualityWeights function and additive generalized linear fitted models were then created to test for gene level differential expression. Results were filtered for genes with Benjamini-Hochberg FDR adjusted p-values ≤ 0.05. Global perturbations in known GO and KEGG pathways were detected using the R/Bioconductor package GAGE (39). The R/Bioconductor package Pathview (40) was used to generate annotated pathway maps on perturbed KEGG signaling and metabolism pathways. Significantly perturbed GO terms were illustrated with the R/Bioconductor package heatmap3 (41). Raw and processed data from this study has been uploaded to the GEO depository and assigned the accession code: GSE109108.
2.8. Cell metabolism analysis
DDCs were transduced with LV-NS or LV-181a/b-1 and cultured in osteogenic medium for 4 days. Cells were then harvested following trypsin digestion and re-seeded in Seahorse XF96 microplates at a density of 50,000 cells per well. Cells were further cultured in osteogenic medium overnight and then the oxygen consumption rate (OCR; mitochondrial respiration) or the extracellular acidification rate (ECAR; glycolysis) was measured using the Seahorse XF Cell Mito Stress Test kit (Agilent) or the Seahorse XF Glycolysis Stress Test kit (Agilent), respectively, following the manufacturer’s instructions. The Cell Mito Stress Test assay involves addition of oligomycin, FCCP and rotenone/antimycin A (all inhibitors of specific components of the electron transport chain) at set time points to permit measurement of ATP production and maximal mitochondrial respiration. The XF Glycolysis Stress Test assay involves sequential addition of glucose, oligomycin and 2DG (2-deoxy-glucose) to measure the extracellular acidification rate (ECAR) and provide information on the basal glycolysis rate and the maximal glycolytic capacity within cells.
2.9. Analysis of glucose uptake and lactate production
DDCs transduced with LV-NS or LV-181a/b-1 were seeded in 6-well plates (1 × 106 cells/well) containing 1ml phenol red-free osteogenic induction medium. After 24 h in culture, the medium was collected and the glucose concentration determined using the Glucose (HK) Assay Kit (Sigma, GAHK-20) following the manufacturer’s protocol. Lactate concentration in the culture medium was measured using the L-Lactate Assay Kit I (Eton Bioscience Inc; #1200011002) following the manufacturer’s recommendation.
2.10. miRNA Target Prediction
A search for predicted miR-181a/b-5p target mRNAs was performed using the databases TargetScan (http://www.targetscan.org/) (42), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) (43) and miRDB (http://www.mirdb.org/) (44).
2.11. Over-expression of PTEN in DDCs
Human PTEN cDNA (GeneCopoeia) was cloned into EX-LV105 lentiviral vector (Genecopoeia, EXI0450-Lv105) utilizing 5’-BamH I and 3’-Kpn I restriction enzyme sites. The EX-GFP-LV105 vector was also purchased and used as a control. Lentiviral preparations to over-express PTEN (LV-PTEN) or GFP control (LV-Ctl) were generated as described in Section 2.2. PTEN was over-expressed in DDCs in the presence or absence of LV-181a/b-1 and effects on PI3K/AKT signaling (via p-AKT-308 Western blotting), osteogenic differentiation, or mitochondrial metabolism (oxygen consumption rate) were examined.
2.12. Statistics
All experiments were carried out in triplicate with DDCs derived from at least three independent biological replicates. Data are presented as means ± SD and statistical comparisons were made using the unpaired Student’s t test. For experiments involving PTEN over-expression, multiple comparisons were made using one-way ANOVA. Probability values were considered statistically significant at p ≤ 0.05.
3. Results
3.1. miR-181a/b-1 expression during osteogenesis
Given our unlimited access to human OA articular cartilage, we decided to utilize DDCs derived from these tissues as a tool to study osteogenesis. Supplemental Fig. 1 shows tri-lineage (chondrogenic, osteogenic, adipogenic) potential of DDCs and bone marrow-derived MSCs in our laboratory. We consistently achieve efficient chondrogenic and osteogenic differentiation of DDCs and MSCs, yet always find adipogenic differentiation to be more robust in MSC cultures. While DDCs from healthy articular cartilage have also been reported to have osteogenic potential (45), we recently published data showing successful osteogenic differentiation of DDCs harvested from human OA cartilage (46). In this work, increased expression of osteoblast-related proteins, RUNX2 and osteocalcin, was found in addition to production of a mineralized extracellular matrix (ECM) during osteogenic induction of DDCs. We also found that transduction of DDCs with a lentivirus expressing a non-silencing (NS) control RNA (LV-NS) did not affect the ability of these cells to differentiate and form a calcified ECM (46). Therefore, in these studies, LV-181a/b-1-transduced cultures were always compared to control LV-NS transduced cells.
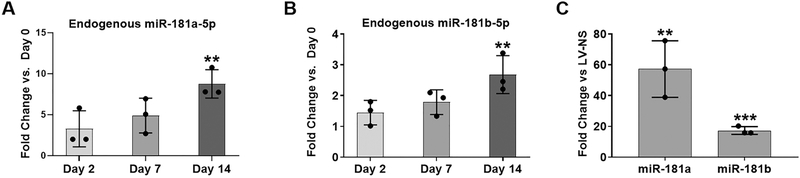
During DDC osteogenesis, endogenous expression levels of both miR-181a and miR-181b increased over time compared to day 0 cultures, and reached significance at day 14 (Figs. 1A, B). Note that we refer to miR-181a here since the 5p strands derived from pre-miR-181a-1 or its paralog, pre-miR-181a-2, are identical. Similarly, we refer to miR-181b since the mature 5p strands are identical whether derived from pre-miR-181b-1 or pre-miR-181b-2. Following transduction of DDCs with LV-181a/b-1, we found a significant increase in the expression of both miR-181a-5p and miR-181b-5p at 48 h as expected (Fig. 1C). Over-expression of miR-181a and miR-181b persisted until day 14 of osteogenesis, albeit over-expressed levels were lower than that found at 48 h (results not shown).
Fig. 1. Expression of miR-181a/b during osteogenesis in vitro.
Endogenous expression of miR-181a-5p or miR-181b-5p at day 2, 7 or 14 of DDC osteogenic differentiation (A, B). Fold change increase in miR-181a-5p and miR-181b-5p expression following transduction of LV-181a/b-1 after 48 h. We consistently find lower levels of over-expression of the clustered miR-181b-5p compared to miR-181a-5p. Data are expressed ± SD; n = 3. **p < 0.01; *** p < 0.001.
3.2. miR-181a/b-1 over-expression enhances osteogenesis
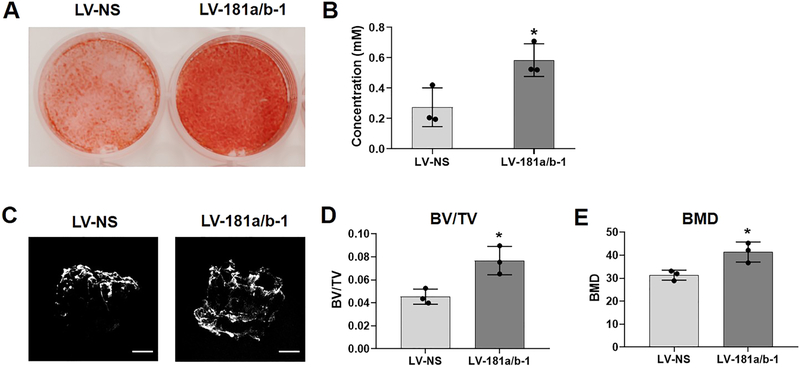
Following osteogenic induction (14 days) of cells transduced with LV-NS or LV-181a/b-1, Fig. 2A shows increased Alizarin Red staining in LV-181a/b-1 transduced cultures compared to LV-NS controls, indicating enhanced calcified ECM production. Levels of Alizarin Red stained ECM were quantified (Fig. 2B). DDCs transduced with LV-181a/b-1 or LV-NS were also seeded onto 3D demineralized human bone scaffolds and cultured in osteogenic induction medium for 28 days. Representative μ-CT images shows increased matrix mineralization in the scaffold containing LV-181a/b-1-transduced DDCs compared to LV-NS-transduced cultures (Fig. 2C). Quantification of bone volume / tissue volume (BV/TV) and total bone mineral density (BMD) within the scaffolds (Fig. 2D, E) confirms the enhancing effect of miR-181a/b-1 over-expression on osteogenesis in this 3D system.
Fig. 2. Enhanced matrix mineralization by miR-181a/b-1.
Representative images of Alizarin red-stained cultures of osteogenic-induced DDCs transduced with either LV-NS or LV-181a/b-1 at day 14 (A). Quantification of Alizarin red staining is shown (B). Representative μCT images of human demineralized, decellularized bone scaffolds seeded with LV-NS or LV-181a/b-1-transduced cells and cultured in osteogenic medium for 28 days (C). Mineralization within the scaffolds was quantified by measuring bone volume / tissue volume (BV/TV) and bone mineral density (BMD) levels (D, E). Data in B, D, E expressed ± SD; n = 3. *p < 0.05. Scale bars in (C) = 1mm.
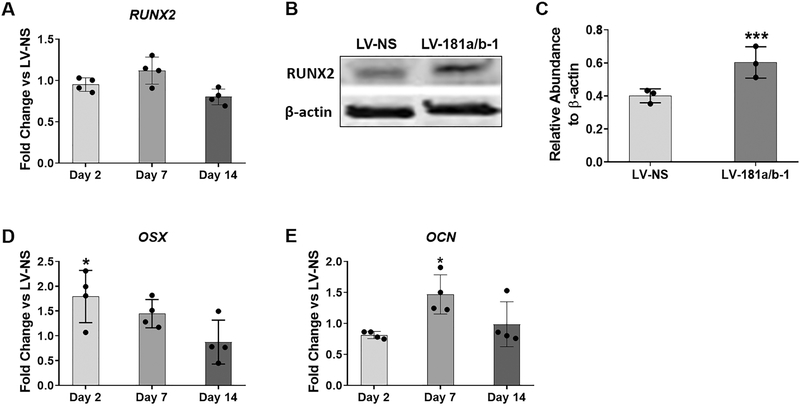
Analysis of osteoblast-related gene expression in monolayer osteogenesis assays did not show significant changes in RUNX2 levels due to miR-181a/b-1 over-expression (Fig. 3A), but we did detect an increase in RUNX2 protein expression at day 4 of osteogenesis in the cultures transduced with LV-181a/b-1 compared to control LV-NS-transduced cells (Figs. 3B, C). Modest yet significant increases in both osterix (OSX) and osteocalcin (OCN) gene expression were found at day 2 and day 7, respectively, due to miR-181a/b-1 over-expression (Fig. 3D–E).
Fig. 3. Effects of miR-181a/b-1 on osteogenic gene and protein expression.
All gene expression analyses were calculated using the unpaired t-test to compare fold change levels in expression between LV-181a/b-1-transduced DDCs and LV-NS-transduced cells at each indicated time point. Expression of RUNX2 at indicated time points during osteogenesis (A). Representative Western blot showing RUNX2 protein levels in LV-NS or LV-181a/b-1 transduced cultures at day 4 of osteogenesis (B). Quantification of RUNX2 expression from three independent Western blots is shown in panel (C). Fold change expression changes in osterix (OSX) and osteocalcin (OCN) expression in miR-181a/b-1-transduced cells compared to control cultures at indicated time points during osteogenesis are shown in (D) and (E) respectively. RUNX2: 56kDa; β-actin: 42kDa. Data are expressed ± SD; n = 4 (A, D, E); n = 3 (B, C). *p < 0.05.
3.3. RNA-Seq and pathway analysis suggests miR-181a/b-1 over-expression enhances mitochondrial metabolism
To begin to decipher how miR-181a/b-1 enhances osteogenesis, RNA-Seq was performed on RNA isolated from day 7 DDC osteogenesis cultures transduced with either LV-NS or LV-181a/b-1. Raw data files corresponding to this RNA-Seq study can be found in the online Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/) under the accession code: GSE109108. Overall modest changes in gene expression due to miR-181a/b-1 over-expression suggested that the observed effects of this miRNA cluster on osteogenesis were likely due to the combination of small, additive effects on biological pathways. This makes sense when considering that miRNAs function to modestly alter expression of numerous target genes within a given cell type, the cumulative effect of which can then lead to significant alterations in cellular pathways and networks. Therefore, gene expression data was further interrogated using the published GAGE method of gene set enrichment analysis (39). Using this approach, statistically significant changes in gene sets can be identified in scenarios where changes in individual genes are not significant. Tables 2 and 3 show the top 20 or top 10 significantly perturbed (enhanced) GO (Biological Function) pathways or KEGG (Signaling and Metabolism) pathways, respectively due to miR-181a/b-1 over-expression during osteogenesis. A clear pattern was identified whereby pathways involved in mitochondrial metabolism and protein synthesis were significantly upregulated.
Table 2:
Top 20 significantly perturbed (enhanced) “GO Biological Function” pathways following miR-181a/b-1 over-expression at day 7 of osteogenic induction.
| Pathway | Mean Linear FC |
P-Value |
|---|---|---|
| Translational termination (GO:0006415) | 25.79 | 2.27e-06 |
| Translational elongation (GO:0006414) | 23.92 | 3.48e-06 |
| Translational initiation (GO:0006413) | 20.93 | 7.52e-06 |
| Mitochondrial translational initiation (GO:0070124) | 17.77 | 3.25e-05 |
| Cellular respiration (GO:0045047) | 17.63 | 2.38e-05 |
| Mitochondrial translation (GO:0032543) | 16.86 | 3.65e-05 |
| Mitochondrial translational elongation (GO:0070124) | 16.86 | 4.31e-05 |
| Respiratory electron transport chain (GO:0022904 | 16.77 | 3.78e-05 |
| Mitochondrial translational termination (GO:0070126) | 14.95 | 8.00e-05 |
| Cellular protein complex disassembly (GO:0043624) | 14.37 | 7.02e-05 |
| Electron transport chain (GO:0022900) | 13.05 | 1.41e-04 |
| Protein complex disassembly (GO:0043241) | 12.57 | 1.47e-04 |
| Macromolecular complex disassembly (GO:0032984) | 12.27 | 1.66e-04 |
| Cotranslational protein targeting to membrane (GO:0006613) | 11.38 | 2.97e-04 |
| Generation of precursor metabolites and energy (GO:0006091) | 11.02 | 2.87e-04 |
| Protein folding (GO:0006457) | 10.77 | 3.49e-04 |
| SRP-dependent cotranslational protein targeting to membrane (GO:0006614) | 10.65 | 4.14e-04 |
| Glycosyl compound metabolic process (GO: 1901657) | 10.58 | 3.59e-04 |
| Energy derivation by oxidation of organic compounds (GO:0015980) | 10.46 | 3.81e-04 |
| Protein localization to endoplasmic reticulum (GO:0070972) | 10.31 | 4.63e-04 |
Table 3:
Top 10 significantly perturbed (enhanced) KEGG Signaling and Metabolism pathways following miR-181a/b-1 over-expression at day 7 of osteogenic induction.
| Pathway | Mean Linear FC |
P-Value |
|---|---|---|
| Oxidative phosphorylation (hsa00190) | 24.69 | 4.17e-06 |
| Ribosome (hsa0310) | 19.84 | 1.35e-05 |
| Spliceosome (hsa03040) | 6.46 | 3.89e-03 |
| Purine metabolism (hsa00230) | 5.09 | 9.91e-03 |
| Proteasome (hsa03050) | 4.81 | 1.39e-02 |
| Pyrimidine metabolism (hsa00240) | 4.32 | 1.82e-02 |
| Protein processing in endoplasmic reticulum (hsa04141) | 3.77 | 2.84e-02 |
| Protein export (hsa03060) | 3.74 | 4.66e-02 |
| Glutathione metabolism (hsa00480) | 3.23 | 4.84e-02 |
| Mineral absorption (hsa04978) | 2.83 | 6.89e-02 |
3.4. miR-181a/b-1 increases mitochondrial respiration during osteogenic induction
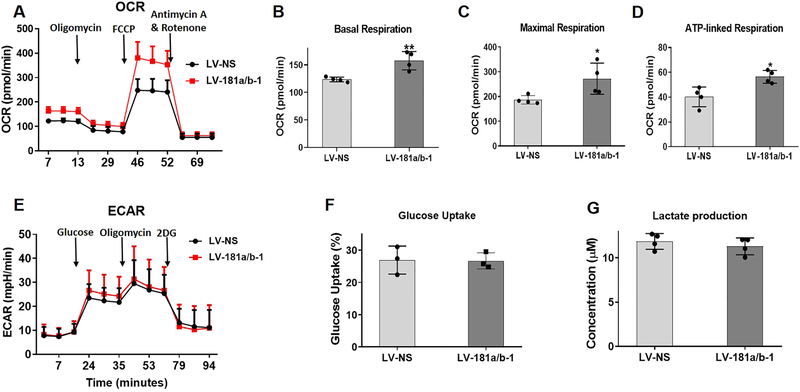
To follow on from the pathway analyses suggesting enhanced mitochondrial metabolism, we explored potential cellular metabolic changes using the Seahorse XF Cell Mito Stress Test Kit (Agilent). This kit uses modulators of mitochondrial respiration that target components of the electron transport chain to permit measurement of basal and maximal respiration as well as ATP production. We found that miR-181a/b-1 over-expression significantly increased the basal and maximal oxygen consumption rates (OCR) as well as ATP production in DDCs following 4 days of osteogenic induction (Fig. 4A–D). Using the Seahorse XF Glycolysis Stress Test Kit, no differences in the extracellular acidification rate (ECAR; an indication of glycolysis) were found between LV-NS and LV-181a/b-1 transduced cultures (Fig. 4E). In addition, no significant differences were found in either glucose uptake or lactate production by cells transduced with LV-181a/b-1 compared to LV-NS control cultures at 24 h (Fig. 4F, G). Overall, these findings, together with the RNA-Seq and pathway analyses data, strongly suggest that one of the mechanisms by which miR-181a/b-1 enhances osteogenesis is by increasing mitochondrial respiration.
Fig. 4. Effects of miR-181a/b-1 on cellular metabolism during osteogenic induction.
Mitochondrial oxygen consumption rate in DDCs transduced with LV-NS or LV-181a/b-1 at day 4 of osteogenesis following addition of specific modulators of respiration targeting the electron transport chain (A). From the data in (A), changes in basal respiration (B), maximal respiration (C) and ATP-linked respiration (D) between cells transduced with LV-NS and LV-181a/b-1 are shown. The extracellular acidification rate (ECAR) was measured following addition of modulators known to affect this process as shown in panel (E). No significant changes in ECAR was noted at any point during the assay between LV-NS and LV-181a/b-1-transduced cells following 4 days of osteogenic induction. Measurement of glucose uptake (F) or lactate production (G) in transduced DDCs cultured in osteogenic medium for 24 h. Data expressed ± SD; n = 4 (A-D, G); n = 3 (E, F). *p < 0.05; **p < 0.01.
3.5. miR-181a/b-1 targets PTEN and increases PI3K/AKT signaling in DDCs
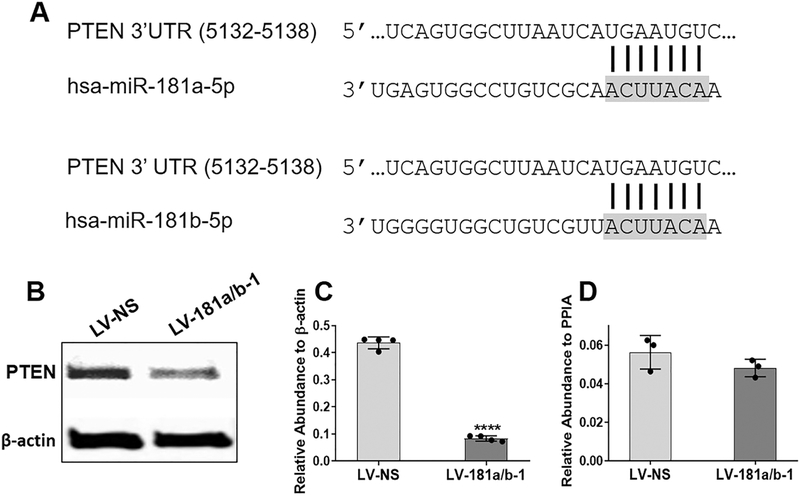
We utilized three online databases (TargetScan, miRTarBase and miRDB) to identify potential miR-181a/b-5p target genes. Of all the target genes listed, we explored PTEN further given its reported role in altering bone formation as well as metabolism in other studies. An interaction between miR-181a/b and the complimentary site within the 3’UTR of PTEN (Fig. 5A) has been confirmed in other cell types by luciferase assays (5,47). Figs. 5B and C show that miR-181a/b-1 targets PTEN in DDCs given the significant reduction in PTEN protein levels following miR-181a/b-1 over-expression. No difference in PTEN mRNA expression was found (Fig. 5D) suggesting that miR-181a/b-1 inhibits PTEN at the level of translation in DDCs.
Fig. 5. PTEN is a target of miR-181a/b in DDCs.
A conserved binding site in the 3’UTR of PTEN is shown to interact with the seed sequence (grey shaded area) of the mature 5p strand of human (hsa) miR-181a or miR-181b (A). Representative Western blot image showing decreased protein levels of PTEN in DDCs following 24 h transduction with LV-181a/b-1 compared to LV-NS (B). Quantification of PTEN protein expression relative to β-actin (C) or PTEN mRNA expression relative to PPIA (D). Data in (C) and (D) are expressed ± SD; n = 4 (C); n = 3 (D). ****p < 0.0001.
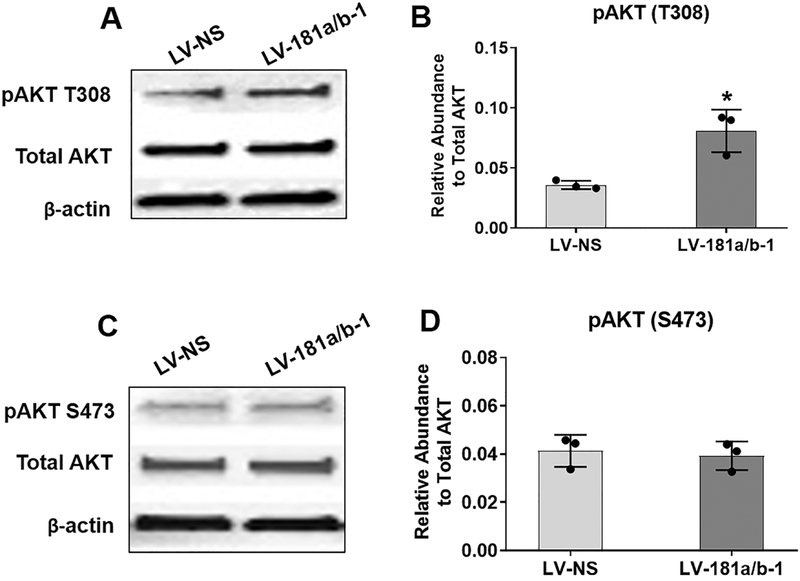
PTEN is the principal negative regulator of the PI3K/AKT signaling pathway. Therefore, we expected that suppression of PTEN by miR-181a/b-1 would enhance PI3K/AKT signaling. Indeed, we found an increase in levels of AKT T308 phosphorylation levels following miR-181a/b-1 over-expression in DDCs at 16 h (Fig. 6A, B). However, no changes in AKT S473 phosphorylation were detected (Fig. 6C, D).
Fig. 6. Modulation of AKT phosphorylation by miR-181a/b-1.
Representative Western blots show expression of phosphorylated AKT at Threonine-308 (T308) (A) or Serine-473 (S473) (C) following 16 h transduction of DDCs with either LV-NS or LV-181a/b-1. Quantification of protein expression of pAKT T308 or pAKT S473 from three biological replicates is shown in (B) and (D), respectively. Total AKT levels were first normalized to β-actin and then abundance of pAKT was calculated relative to normalized total AKT. (p)AKT: 60kDa; β-actin: 42kDa. Data in (B) and (D) are expressed ± SD; n = 3. *p < 0.05
3.6. Regulation of the PTEN/PI3K/AKT axis by miR-181a/b-1 enhances mitochondrial metabolism to increase osteogenesis
Given the central role for PI3K/AKT signaling in controlling a number of cellular processes including metabolism, it is likely that the effects of miR-181a/b-1 on regulating the PTEN/PI3K/AKT axis results in the observed changes in mitochondrial respiration and, subsequently, osteogenesis. In support of this statement, treatment with the PI3K inhibitor, LY294002, was found to inhibit both DDC osteogenesis as well as mitochondrial respiration during osteogenic induction of DDCs (Supplemental Fig. 2).
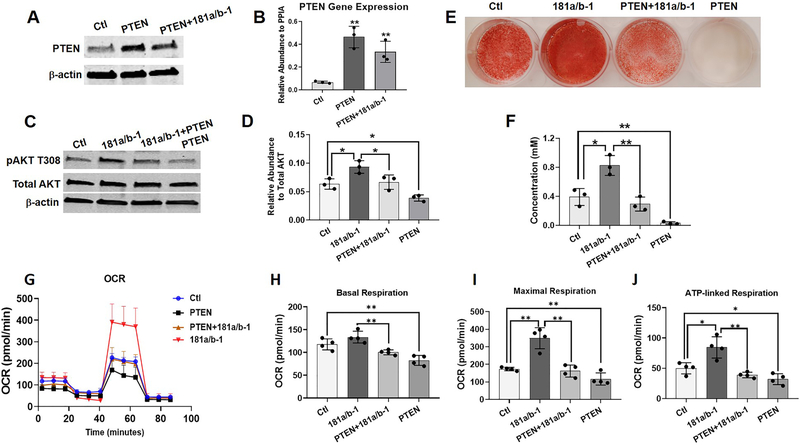
In addition, PTEN over-expression in DDCs (Fig. 7A) was found to suppress PI3K/AKT signaling (Fig. 7C, D) and osteogenesis (Fig. 7E, F) as expected, but we also found that it decreased mitochondrial respiration during 4 days of osteogenic induction (Fig. 7G–J). To further prove the link between miR-181a/b-1 and the regulation of PTEN/PI3K/AKT signaling to alter mitochondrial metabolism and osteogenesis, PTEN over-expression was carried out in DDCs transduced with LV-181a/b-1. We found that PTEN over-expression attenuated the enhancing effects of miR-181a/b-1 on PI3K/AKT signaling (Fig. 7C, D), osteogenesis (Fig. 7E, F) and mitochondrial metabolism (basal and maximal respiration as well as ATP production) in DDCs during osteogenic induction (Fig. 7G–J).
Fig. 7. PTEN over-expression attenuates the enhancing effects of miR-181a/b-1 on PI3K/AKT signaling, osteogenesis and mitochondrial metabolism.
Representative Western blot showing increased PTEN protein (A) or gene (B) expression following transduction of DDCs with lentivirus expressing PTEN ± lentivirus over-expressing miR-181a/b-1 compared to control cells transduced with lentivirus expressing GFP (Ctl). Representative Western blot showing expression of phosphorylated AKT T308 (pAKT T308), total AKT and β-actin following transduction of DDCs with lentivirus expressing GFP (Ctl), miR-181a/b-1, miR-181a/b-1 + PTEN or PTEN alone (C). Quantification of pAKT-T308 expression shown in D. Alizarin red-stained cultures following lentiviral transduction and osteogenic differentiation for 14 days (E). Quantification of Alizarin red is shown in F. Mitochondrial respiration (oxygen consumption rate; OCR) of transduced cells following 4 days of osteogenic induction (G). From the data in (G), changes in basal respiration (H), maximal respiration (I) and ATP-linked respiration (J) between treatment groups are shown. Data expressed ± SD; n = 3 (A-F), n = 4 (G-J) *p < 0.05; **p < 0.01.
4. Discussion
To address the role of miR-181a/b-1 in regulating osteogenesis, we utilized DDCs from human OA cartilage as a cell source with osteogenic potential. DDCs from non-diseased articular cartilage have been reported to undergo osteogenesis (45). Also, stem-like cells with tri-lineage potential (osteogenic, adipogenic, chondrogenic) have been shown to exist in both human healthy and OA articular cartilage (48–50). We recently published data on the inhibitory effects of miR-138 on osteogenic differentiation of DDCs (46). The same miR-138-mediated osteo-suppressive effect has also been reported using MSCs (51). Therefore DDCs can be regarded as an acceptable primary cell source to study processes regulating osteogenesis in vitro.
Given our previous findings that miR-181a-1 was more highly expressed in the hypertrophic zone of developing human long bones (16), we postulated this this miRNA paralog may regulate processes of hypertrophic chondrocyte differentiation and/or endochondral ossification processes that occur following production of hypertrophic cartilage. Also, the fact that viable mice devoid of both miR-181a/b-1 and a/b-2 clusters are phenotypically smaller than littermate controls (13,15) suggests potentially important roles for these paralogs in regulating cartilage and bone development. We therefore designed studies to examine the role of miR-181a-1 and its clustered miRNA, miR-181b-1 (miR-181a/b-1) in regulating osteogenesis. We chose to study the cluster since miR-181a-1 and b-1 are located very close together on chr 1 (~60nt apart) and thus would always be co-transcribed in vivo. While this study is focused on osteogenic processes, current work in our laboratory is also focused on investigating the potential role of miR-181a/b-1 in regulating hypertrophic chondrocyte differentiation.
We chose to over-express the precursor forms of the clustered miR-181a/b-1 using lentiviral technology to ensure stable over-expression during longer-term osteogenic differentiation assays in vitro. By over-expressing miR-181a/b-1, we are essentially also over-expressing miR-181a/b-2 since the mature, processed functional 5p strands of a1 and a2, or b1 and b2, are identical. However, the processed 3p strands between these paralogs are different which is why we specifically state that we are modulating expression of pre-miR-181a/b-1. In this study we focus only on the effects of miR-181a/b over-expression rather than knock-down. If antagomirs are designed to bind and inhibit activity of endogenous miR-181a/b, then there could be issues with redundancy due to the presence of miR-181c/d which contain the same functional seed sequence. Also, design of inhibitor sponge reagents (52,53) may lead to inhibition of the entire miR-181 family which could result in deleterious effects. Therefore, we anticipate that findings from such inhibition studies may be complex to decipher and not simply reveal opposite results to the effects of over-expression. However, determining how suppression of miR-181 paralogs affects osteogenic and chondrogenic differentiation is important to understand but is beyond the scope of this work. Current efforts are underway in our laboratory to investigate this in independent studies.
Our findings that the miR-181a/b-1 cluster enhances osteogenesis in vitro is in agreement with that reported by Bhushan et al (3). This study used mouse cell lines or primary murine calvarial osteoblasts to examine osteoblast differentiation following transient transfection of miR-181a mimics (i.e. the 5p strand equivalent to miR-181a-1 or miR-181a-2) while we used human primary cells and a stable lentiviral transduction system to over-express both mature 5p and 3p strands derived from precursor miR-181a-1 and miR-181b-1. The sequence of the mature 5p strands of miR-181a and miR-181b are conserved between human and mouse which can explain the similarities between our data and that of Bhushan et al with respect to osteo-induction effects. However, while Bhushan et al showed that miR-181a enhanced osteogenesis by repressing TGF-β signaling, we report a different mechanism involving modulation of PTEN/PI3K/AKT signaling. We hypothesize that alteration of this signaling pathway is partly responsible for the metabolic effects observed in our studies whereby mitochondrial metabolism is enhanced during early osteogenic induction, while glycolysis was not affected.
PTEN is a well-established tumor suppressor gene commonly found to be mutated in cancers (54). Subsequent decreases in PTEN expression/function have been associated with increased glycolysis in cancer cells (i.e. the Warburg effect) (55). However, it has also been reported that PTEN inhibition can enhance mitochondrial respiration (26,27). These latter findings agree with our data whereby miR-181a/b-1 over-expression increases mitochondrial metabolism, in part, by targeting and suppressing PTEN. The primary function of PTEN is to inhibit the PI3K/AKT pathway which is a critical regulator of many processes including cell growth, differentiation and metabolism (56,57). Our studies showed that miR-181a/b-1 over-expression increased PI3K/AKT signaling as would be expected following PTEN suppression. An indication of PI3K/AKT activation is the phosphorylation of two key residues on AKT: T308 and S473 (56). Phosphorylation of both residues is required for maximal activation of the kinase. We found that miR-181a/b-1 induced phosphorylation of T308, but not S473, indicating enhanced activity of PI3K as well as phosphoinositide-dependent protein kinase 1 (PDPK1) which is responsible for AKT T308 phosphorylation. Our data also suggest that the mechanistic target of rapamycin (mTOR) complex 2 (mTORC2), primarily responsible for AKT S473 phosphorylation, is not activated by miR-181a/b-1. It is not unusual, under specific stimuli, that one of the AKT residues is phosphorylated and not the other. For example, in response to insulin-like growth factor 2 (Igf2), AKT S473 phosphorylation is increased but not AKT T308 (58,59). In cancer cells, miR-181a has been shown to enhance glycolysis by targeting PTEN and increasing AKT S473 phosphorylation (47). Studies on miR-181a/b-1 knock-out cells also report an association between AKT S473 phosphorylation and altered glycolysis (13,60). Taken together, these findings agree with an earlier report that mTORC2 regulates cell glycolysis by phosphorylating AKT S473 (61).
Furthermore, in the context of osteoblast differentiation, Esen et al showed that Wnt signaling induced glycolysis via mTORC2 activation and AKT S473 phosphorylation (62). The discrepancy between these findings and our data showing increased AKT T308 phosphorylation and mitochondrial metabolism during osteogenesis, may be explained by the differences in stimuli tested (i.e. altered Wnt signaling versus miR-181a/b-1 over-expression), differences in time points analyzed (Esen et al measured OCR and ECAR following 6 h of Wnt3a treatment while we analyzed these metabolic parameters following 4 days in osteogenic induction medium), and perhaps also due to the fact that a mouse cell line was used by Esen et al while our studies involve human primary dedifferentiated chondrocytes. It should also be highlighted that the mature 5p strands of miR-181a/b over-expressed in our assays results in the targeting of many genes in addition to PTEN. While we believe that modulation of PTEN/PI3K/AKT signaling in our assays has significant effects on metabolism and osteogenesis, other pathways will most likely be affected which may also contribute to the observed phenotype. For example, the study by Bhushan et al showed decreased TGFβ signaling in their osteogenesis assays as a result of miR-181a over-expression. If this is also occurring in our system, it could further explain the lack of effects on glycolysis since other studies have reported TGFβ-enhancing effects on glycolysis (63,64).
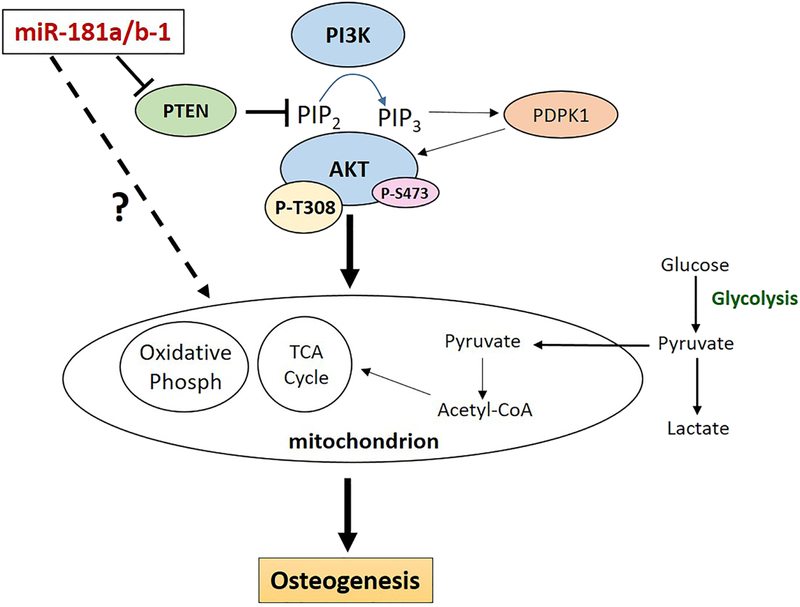
The bone enhancing effects of miR-181a/b over-expression via PTEN suppression and increased PI3K/AKT signaling makes sense from what has already been reported in the bone field. For example, Pten deletion in murine osteoprogenitors (20) or mature osteoblasts (19) resulted in increased bone production. A number of studies also show that PI3K/AKT signaling is critical for osteogenesis (21–25,65). In our hands, we show that PTEN over-expression not only inhibited osteogenesis as expected (Fig. 7E, F), but also suppressed mitochondrial metabolism during osteogenic induction (Fig. 7G–J). Similarly, while the PI3K inhibitor LY294002 suppressed DDC osteogenesis as expected, we also found that this treatment decreased mitochondrial metabolism during osteogenic induction of these cells (Supplemental Fig. 2). These findings provide further evidence linking modulation of PTEN activity and PI3K/AKT signaling to altered mitochondrial metabolism within the context of osteogenesis. Also, our data showing that PTEN over-expression can attenuate the enhancing effects of miR-181a/b-1 (Fig. 7) is further proof that miR-181a/b-1 acts upstream to suppress PTEN and subsequently enhance PI3K/AKT signaling, mitochondrial respiration and osteogenesis (Fig. 8).
Figure. 8. miR-181a/b-1 over-expression regulates the PTEN/PI3K/AKT signaling axis to enhance mitochondrial metabolism and osteogenesis.
Our studies show that over-expression of miR-181a/b-1 in DDCs results in targeting and suppression of PTEN which subsequently activates PI3K/AKT signaling by increasing phosphorylation of AKT-T308. Modulation of PTEN/PI3K/AKT signaling results in increased mitochondrial metabolism which has an overall enhancing effect on osteogenesis. In addition, miR-181a/b-1 may also function in the cytoplasm to modify other cellular pathways or even function directly within in the mitochondria to alter cellular respiration and osteogenic differentiation
From the published literature, it is apparent that cells rely on both mitochondrial respiration (oxidative phosphorylation) and glycolysis during osteoblast differentiation (66–75). The majority of studies analyzing bioenergetics during osteogenesis utilized murine progenitor cells, calvarial osteoblasts or human MSCs. To our knowledge this is the first report studying metabolic changes during osteogenic induction of human DDCs. We report a new epigenetic mechanism whereby over-expression of miR-181a/b-1 in DDCs increases osteogenesis by modulating the PTEN/PI3K/AKT axis to specifically enhance mitochondrial respiration. As already mentioned, we analyzed metabolic processes in cells during an early time point of osteogenic induction (day 4). The effects of miR-181a/b-1 on metabolism at later time points of osteogenesis (i.e. day 10 onwards) were not investigated because analysis by Seahorse technology may prove challenging due to the presence of a mineralized ECM. Therefore, whether or not miR-181a/b-1 over-expression continues to affect mitochondrial respiration or alter glycolysis in more differentiated cells in our system has yet to be determined.
Future studies in our laboratory will focus on deciphering which downstream mediators of the PI3K/AKT pathway are regulated by miR-181a/b-1 over-expression, including mTORC1 and FoxO1/3 transcription factors. We predict that activation of mTORC1 is occurring in our system given the reports that this enzyme complex promotes general protein synthesis as well as mitochondrial respiration (28,29,76,77), the two processes that were predicted to be enhanced from our RNA-Seq and pathway analyses (Tables 2 and 3). Studies also show that modulating mTORC1 activity affects osteoblast differentiation (78,79). Also, while our work has focused on the cytoplasmic role of miR-181a/b-1, it is also intriguing that miR-181 family members have been identified within the mitochondria of various cell types (60,80–83). Preliminary studies in our laboratory have also identified miR-181a/b in the mitochondria of DDCs and MSCs (results not shown). These mitochondrial miR-181 family members could play a more direct role in controlling mitochondria function/respiration during osteogenesis and efforts are underway to investigate this further in our laboratory.
Of clinical relevance, and given the fact that miRNAs are attractive from a drug development standpoint, findings from these studies provide a strong rationale to design strategies to target miR-181a/b in vivo as a means to modulate bone formation to improve fracture healing or inhibit heterotopic ossification, for example.
Supplementary Material
HIGHLIGHTS.
Over-expression of miR-181a/b-1 induces osteogenic differentiation of human primary de-differentiated chondrocytes (DDCs).
Induction of osteogenesis by miR-181a/b-1 is due, in part, to suppression of PTEN, subsequent activation of PI3K/AKT signaling, and enhanced mitochondrial respiration.
The bone enhancing effects of miR-181a/b-1 reported here justifies future studies to target this miRNA cluster in vivo as a means to enhance bone formation in scenarios such as endochondral fracture repair.
Acknowledgements:
We acknowledge and thank Dr. Britta Anderson for cloning and preparation of the miR-181a/b-1 expression plasmid.
Funding sources: This work was supported by the National Institutes of Health [R01 AR064191; to AM and P30 AR057235; Washington University Musculoskeletal Research Center].
Abbreviations used are:
- PTEN
phosphatase and tensin homolog
- PI3K
phosphoinositide 3-kinase
- DDC
dedifferentiated chondrocyte
- PDPK1
3-phosphoinositide-dependent kinase-1
- mTOR
mammalian target of rapamycin (mTOR) complex 1 (mTORC1) or complex 2 (mTORC2)
- OSX
osterix
- OCN
osteocalcin
- OCR
oxygen consumption rate
- ECAR
extracellular acidification rate
- ECM
extracellular matrix
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhushan R, Grunhagen J, Becker J, Robinson PN, Ott CE, and Knaus P (2013) miR-181a promotes osteoblastic differentiation through repression of TGF-beta signaling molecules. Int J Biochem Cell Biol 45, 696–705 [DOI] [PubMed] [Google Scholar]
- 4.Sumiyoshi K, Kubota S, Ohgawara T, Kawata K, Abd El Kader T, Nishida T, Ikeda N, Shimo T, Yamashiro T, and Takigawa M (2013) Novel role of miR-181a in cartilage metabolism. J Cell Biochem 114, 2094–2100 [DOI] [PubMed] [Google Scholar]
- 5.Wu XF, Zhou ZH, and Zou J (2017) MicroRNA-181 inhibits proliferation and promotes apoptosis of chondrocytes in osteoarthritis by targeting PTEN. Biochem Cell Biol 95, 437–444 [DOI] [PubMed] [Google Scholar]
- 6.Song J, Lee M, Kim D, Han J, Chun CH, and Jin EJ (2013) MicroRNA-181b regulates articular chondrocytes differentiation and cartilage integrity. Biochem Biophys Res Commun 431, 210–214 [DOI] [PubMed] [Google Scholar]
- 7.Nakamura A, Rampersaud YR, Sharma A, Lewis SJ, Wu B, Datta P, Sundararajan K, Endisha H, Rossomacha E, Rockel JS, Jurisica I, and Kapoor M (2016) Identification of microRNA-181a-5p and microRNA-4454 as mediators of facet cartilage degeneration. JCI Insight 1, e86820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Tang C, Zhang Q, and Chen W (2014) Reduced miR-9 and miR-181a expression down-regulates Bim concentration and promote osteoclasts survival. Int J Clin Exp Pathol 7, 2209–2218 [PMC free article] [PubMed] [Google Scholar]
- 9.Shao B, Liao L, Yu Y, Shuai Y, Su X, Jing H, Yang D, and Jin Y (2015) Estrogen preserves Fas ligand levels by inhibiting microRNA-181a in bone marrow-derived mesenchymal stem cells to maintain bone remodeling balance. FASEB J 29, 3935–3944 [DOI] [PubMed] [Google Scholar]
- 10.McAlinden A, and Im GI (2018) MicroRNAs in orthopaedic research: Disease associations, potential therapeutic applications, and perspectives. J Orthop Res 36, 33–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura A, Rampersaud YR, Nakamura S, Sharma A, Zeng F, Rossomacha E, Ali SA, Krawetz R, Haroon N, Perruccio AV, Mahomed NN, Gandhi R, Rockel JS, and Kapoor M (2019) microRNA-181a-5p antisense oligonucleotides attenuate osteoarthritis in facet and knee joints. Ann Rheum Dis 78, 111–121 [DOI] [PubMed] [Google Scholar]
- 12.Sun X, Sit A, and Feinberg MW (2014) Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc Med 24, 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henao-Mejia J, Williams A, Goff LA, Staron M, Licona-Limon P, Kaech SM, Nakayama M, Rinn JL, and Flavell RA (2013) The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity 38, 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fragoso R, Mao T, Wang S, Schaffert S, Gong X, Yue S, Luong R, Min H, Yashiro-Ohtani Y, Davis M, Pear W, and Chen CZ (2012) Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLoS Genet 8, e1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams A, Henao-Mejia J, Harman CC, and Flavell RA (2013) miR-181 and metabolic regulation in the immune system. Cold Spring Harb Symp Quant Biol 78, 223–230 [DOI] [PubMed] [Google Scholar]
- 16.McAlinden A, Varghese N, Wirthlin L, and Chang LW (2013) Differentially expressed microRNAs in chondrocytes from distinct regions of developing human cartilage. PLoS One 8, e75012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabler J, Ruetze M, Kynast KL, Grossner T, Diederichs S, and Richter W (2015) Stage-Specific miRs in Chondrocyte Maturation: Differentiation-Dependent and Hypertrophy-Related miR Clusters and the miR-181 Family. Tissue Eng Part A 21, 2840–2851 [DOI] [PubMed] [Google Scholar]
- 18.Chow LM, and Baker SJ (2006) PTEN function in normal and neoplastic growth. Cancer Lett 241, 184–196 [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Bruxvoort KJ, Zylstra CR, Liu J, Cichowski R, Faugere MC, Bouxsein ML, Wan C, Williams BO, and Clemens TL (2007) Lifelong accumulation of bone in mice lacking Pten in osteoblasts. Proc Natl Acad Sci U S A 104, 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guntur AR, Reinhold MI, Cuellar J Jr., and Naski MC (2011) Conditional ablation of Pten in osteoprogenitors stimulates FGF signaling. Development 138, 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGonnell IM, Grigoriadis AE, Lam EW, Price JS, and Sunters A (2012) A specific role for phosphoinositide 3-kinase and AKT in osteoblasts? Front Endocrinol (Lausanne) 3, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh-Choudhury N, Abboud SL, Nishimura R, Celeste A, Mahimainathan L, and Choudhury GG (2002) Requirement of BMP-2-induced phosphatidylinositol 3-kinase and Akt serine/threonine kinase in osteoblast differentiation and Smad-dependent BMP-2 gene transcription. J Biol Chem 277, 33361–33368 [DOI] [PubMed] [Google Scholar]
- 23.Guntur AR, and Rosen CJ (2011) The skeleton: a multi-functional complex organ: new insights into osteoblasts and their role in bone formation: the central role of PI3Kinase. J Endocrinol 211, 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, and Hay N (2003) Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev 17, 1352–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulici V, Hoenselaar KD, Agoston H, McErlain DD, Umoh J, Chakrabarti S, Holdsworth DW, and Beier F (2009) The role of Akt1 in terminal stages of endochondral bone formation: angiogenesis and ossification. Bone 45, 1133–1145 [DOI] [PubMed] [Google Scholar]
- 26.Li Y, He L, Zeng N, Sahu D, Cadenas E, Shearn C, Li W, and Stiles BL (2013) Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) signaling regulates mitochondrial biogenesis and respiration via estrogen-related receptor alpha (ERRalpha). J Biol Chem 288, 25007–25024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goo CK, Lim HY, Ho QS, Too HP, Clement MV, and Wong KP (2012) PTEN/Akt signaling controls mitochondrial respiratory capacity through 4E-BP1. PLoS One 7, e45806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schieke SM, Phillips D, McCoy JP Jr., Aponte AM, Shen RF, Balaban RS, and Finkel T (2006) The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem 281, 27643–27652 [DOI] [PubMed] [Google Scholar]
- 29.Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, McLaughlan S, Nouet Y, Pause A, Pollak M, Gottlieb E, Larsson O, St-Pierre J, Topisirovic I, and Sonenberg N (2013) mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 18, 698–711 [DOI] [PubMed] [Google Scholar]
- 30.Morita M, Prudent J, Basu K, Goyon V, Katsumura S, Hulea L, Pearl D, Siddiqui N, Strack S, McGuirk S, St-Pierre J, Larsson O, Topisirovic I, Vali H, McBride HM, Bergeron JJ, and Sonenberg N (2017) mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol Cell 67, 922–935 e925 [DOI] [PubMed] [Google Scholar]
- 31.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, and Crabtree GR (2011) MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richner M, Victor MB, Liu Y, Abernathy D, and Yoo AS (2015) MicroRNA-based conversion of human fibroblasts into striatal medium spiny neurons. Nat Protoc 10, 1543–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 34.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, and Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy DJ, Chen Y, and Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu R, Holik AZ, Su S, Jansz N, Chen K, Leong HS, Blewitt ME, Asselin-Labat ML, Smyth GK, and Ritchie ME (2015) Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res 43, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo W, Friedman MS, Shedden K, Hankenson KD, and Woolf PJ (2009) GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo W, and Brouwer C (2013) Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao S, Guo Y, Sheng Q, and Shyr Y (2014) Advanced heat map and clustering analysis using heatmap3. Biomed Res Int 2014, 986048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal V, Bell GW, Nam JW, and Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, Tsai TR, Ho SY, Jian TY, Wu HY, Chen PR, Lin NC, Huang HT, Yang TL, Pai CY, Tai CS, Chen WL, Huang CY, Liu CC, Weng SL, Liao KW, Hsu WL, and Huang HD (2016) miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 44, D239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong N, and Wang X (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43, D146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbero A, Ploegert S, Heberer M, and Martin I (2003) Plasticity of clonal populations of dedifferentiated adult human articular chondrocytes. Arthritis Rheum 48, 1315–1325 [DOI] [PubMed] [Google Scholar]
- 46.Zheng H, Ramnaraign D, Anderson BA, Tycksen E, Nunley R, and McAlinden A (2018) MicroRNA-138 inhibits osteogenic differentiation and mineralization of human dedifferentiated chondrocytes by regulating RhoC and the actin cytoskeleton. JBMR Plus 10.1002/jbm4.10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei Z, Cui L, Mei Z, Liu M, and Zhang D (2014) miR-181a mediates metabolic shift in colon cancer cells via the PTEN/AKT pathway. FEBS Lett 588, 1773–1779 [DOI] [PubMed] [Google Scholar]
- 48.Williams R, Khan IM, Richardson K, Nelson L, McCarthy HE, Analbelsi T, Singhrao SK, Dowthwaite GP, Jones RE, Baird DM, Lewis H, Roberts S, Shaw HM, Dudhia J, Fairclough J, Briggs T, and Archer CW (2010) Identification and clonal characterisation of a progenitor cell sub-population in normal human articular cartilage. PLoS One 5, e13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson L, McCarthy HE, Fairclough J, Williams R, and Archer CW (2014) Evidence of a Viable Pool of Stem Cells within Human Osteoarthritic Cartilage. Cartilage 5, 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fellows CR, Williams R, Davies IR, Gohil K, Baird DM, Fairclough J, Rooney P, Archer CW, and Khan IM (2017) Characterisation of a divergent progenitor cell sub-populations in human osteoarthritic cartilage: the role of telomere erosion and replicative senescence. Sci Rep 7, 41421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, and Kassem M (2011) MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A 108, 6139–6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barta T, Peskova L, and Hampl A (2016) miRNAsong: a web-based tool for generation and testing of miRNA sponge constructs in silico. Sci Rep 6, 36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebert MS, and Sharp PA (2010) MicroRNA sponges: progress and possibilities. RNA 16, 2043–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen CY, Chen J, He L, and Stiles BL (2018) PTEN: Tumor Suppressor and Metabolic Regulator. Front Endocrinol (Lausanne) 9, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ortega-Molina A, and Serrano M (2013) PTEN in cancer, metabolism, and aging. Trends Endocrinol Metab 24, 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manning BD, and Toker A (2017) AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franke TF (2008) PI3K/Akt: getting it right matters. Oncogene 27, 6473–6488 [DOI] [PubMed] [Google Scholar]
- 58.Shi Y, Chen J, Karner CM, and Long F (2015) Hedgehog signaling activates a positive feedback mechanism involving insulin-like growth factors to induce osteoblast differentiation. Proc Natl Acad Sci U S A 112, 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wan X, and Helman LJ (2003) Levels of PTEN protein modulate Akt phosphorylation on serine 473, but not on threonine 308, in IGF-II-overexpressing rhabdomyosarcomas cells. Oncogene 22, 8205–8211 [DOI] [PubMed] [Google Scholar]
- 60.Das S, Kohr M, Dunkerly-Eyring B, Lee DI, Bedja D, Kent OA, Leung AK, Henao-Mejia J, Flavell RA, and Steenbergen C (2017) Divergent Effects of miR-181 Family Members on Myocardial Function Through Protective Cytosolic and Detrimental Mitochondrial microRNA Targets. J Am Heart Assoc 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, and Hall MN (2012) Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab 15, 725–738 [DOI] [PubMed] [Google Scholar]
- 62.Esen E, Chen J, Karner CM, Okunade AL, Patterson BW, and Long F (2013) WNTLRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab 17, 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barter MJ, Tselepi M, Gomez R, Woods S, Hui W, Smith GR, Shanley DP, Clark IM, and Young DA (2015) Genome-Wide MicroRNA and Gene Analysis of Mesenchymal Stem Cell Chondrogenesis Identifies an Essential Role and Multiple Targets for miR-140–5p. Stem Cells 33, 3266–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guido C, Whitaker-Menezes D, Capparelli C, Balliet R, Lin Z, Pestell RG, Howell A, Aquila S, Ando S, Martinez-Outschoorn U, Sotgia F, and Lisanti MP (2012) Metabolic reprogramming of cancer-associated fibroblasts by TGF-beta drives tumor growth: connecting TGF-beta signaling with “Warburg-like” cancer metabolism and L-lactate production. Cell Cycle 11, 3019–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baker N, Sohn J, and Tuan RS (2015) Promotion of human mesenchymal stem cell osteogenesis by PI3-kinase/Akt signaling, and the influence of caveolin-1/cholesterol homeostasis. Stem Cell Res Ther 6, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Esen E, and Long F (2014) Aerobic glycolysis in osteoblasts. Curr Osteoporos Rep 12, 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Esen E, Lee SY, Wice BM, and Long F (2015) PTH Promotes Bone Anabolism by Stimulating Aerobic Glycolysis via IGF Signaling. J Bone Miner Res 30, 2137. [DOI] [PubMed] [Google Scholar]
- 68.Lee WC, Guntur AR, Long F, and Rosen CJ (2017) Energy Metabolism of the Osteoblast: Implications for Osteoporosis. Endocr Rev 38, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karner CM, and Long F (2017) Wnt signaling and cellular metabolism in osteoblasts. Cell Mol Life Sci 74, 1649–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shum LC, White NS, Mills BN, Bentley KL, and Eliseev RA (2016) Energy Metabolism in Mesenchymal Stem Cells During Osteogenic Differentiation. Stem Cells Dev 25, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shares BH, Busch M, White N, Shum L, and Eliseev RA (2018) Active mitochondria support osteogenic differentiation by stimulating beta-catenin acetylation. J Biol Chem 293, 16019–16027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Q, Gao Z, Chen Y, and Guan MX (2017) The role of mitochondria in osteogenic, adipogenic and chondrogenic differentiation of mesenchymal stem cells. Protein Cell 8, 439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guntur AR, Le PT, Farber CR, and Rosen CJ (2014) Bioenergetics during calvarial osteoblast differentiation reflect strain differences in bone mass. Endocrinology 155, 1589–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Komarova SV, Ataullakhanov FI, and Globus RK (2000) Bioenergetics and mitochondrial transmembrane potential during differentiation of cultured osteoblasts. Am J Physiol Cell Physiol 279, C1220–1229 [DOI] [PubMed] [Google Scholar]
- 75.Pietila M, Lehtonen S, Narhi M, Hassinen IE, Leskela HV, Aranko K, Nordstrom K, Vepsalainen A, and Lehenkari P (2010) Mitochondrial function determines the viability and osteogenic potency of human mesenchymal stem cells. Tissue Eng Part C Methods 16, 435–445 [DOI] [PubMed] [Google Scholar]
- 76.Saxton RA, and Sabatini DM (2017) mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371 [DOI] [PubMed] [Google Scholar]
- 77.Chen J, and Long F (2018) mTOR signaling in skeletal development and disease. Bone Res 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen J, and Long F (2015) mTORC1 Signaling Promotes Osteoblast Differentiation from Preosteoblasts. PLoS One 10, e0130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fitter S, Matthews MP, Martin SK, Xie J, Ooi SS, Walkley CR, Codrington JD, Ruegg MA, Hall MN, Proud CG, Gronthos S, and Zannettino AC (2017) mTORC1 Plays an Important Role in Skeletal Development by Controlling Preosteoblast Differentiation. Mol Cell Biol 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, and Gidrol X (2011) PremicroRNA and mature microRNA in human mitochondria. PLoS One 6, e20220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, and Mattick JS (2011) The human mitochondrial transcriptome. Cell 146, 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sripada L, Tomar D, Prajapati P, Singh R, Singh AK, and Singh R (2012) Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One 7, e44873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sripada L, Tomar D, and Singh R (2012) Mitochondria: one of the destinations of miRNAs. Mitochondrion 12, 593–599 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.