Abstract
Background
Breast cancers show variations in the number and biological aggressiveness of cancer stem cells that correlate with their clinico-prognostic and molecular heterogeneity. Thus, prognostic stratification of breast cancers based on cancer stem cells might help guide patient management.
Methods
We derived a 20-gene stem cell signature from the transcriptional profile of normal mammary stem cells, capable of identifying breast cancers with a homogeneous profile and poor prognosis in in silico analyses. The clinical value of this signature was assessed in a prospective-retrospective cohort of 2, 453 breast cancer patients. Models for predicting individual risk of metastasis were developed from expression data of the 20 genes in patients randomly assigned to a training set, using the ridge-penalized Cox regression, and tested in an independent validation set.
Findings
Analyses revealed that the 20-gene stem cell signature provided prognostic information in Triple-Negative and Luminal breast cancer patients, independently of standard clinicopathological parameters. Through functional studies in individual tumours, we correlated the risk score assigned by the signature with the proliferative and self-renewal potential of the cancer stem cell population. By retraining the 20-gene signature in Luminal patients, we derived the risk model, StemPrintER, which predicted early and late recurrence independently of standard prognostic factors.
Interpretation
Our findings indicate that the 20-gene stem cell signature, by its unique ability to interrogate the biology of cancer stem cells of the primary tumour, provides a reliable estimate of metastatic risk in Triple-Negative and Luminal breast cancer patients independently of standard clinicopathological parameters.
Keywords: Breast cancer, Cancer stem cells, Prognostic signatures, Biomarkers, Metastasis
Research in context.
Evidence before this study
The increasingly recognized relevance of cancer stem cells to breast cancer heterogeneity argues that they might hold the key to inform the individualized management of breast cancer patients. We searched PubMed for systematic reviews and research articles reporting high-throughput screening, development of genomic predictors, validation of predictive and/or prognostic biomarkers in retrospective and prospective cohort studies in breast cancer published up to December 31, 2018, with the keywords “breast cancer”, “genomic predictors”, multigene assays”, “cancer stem cells”, “distant metastasis”. No publication date or language restrictions were applied. We found that, despite the number of putative breast cancer stem cell markers described in several studies, and the long purported correlation between the presence and biological characteristics of cancer stem cells in the primary tumour and clinical outcome, so far, no multigene assays able to interrogate the intrinsic degree of stemness of the primary tumour is available for the clinical management of breast cancer.
Added value of this study
Stratification of breast cancer patients for their intrinsic risk of recurrence and for selection of the optimal therapy, while avoiding overtreatment, demands biomarkers that rely on the underlying biology of each individual tumour. Most of existing genomic tools for breast cancer prognostication have been developed empirically by selecting multigene marker panels, or comparing the genomic profiles of breast cancer specimens from patients with or without disease recurrence. This implies that their predictive prognostic power derives from their capacity to measure the expression of genes at the level of the bulk tumour population. These genes are often associated with the same tumour characteristics interrogated by the standard clinicopathological parameters, namely, hormonal status or proliferation, thereby failing to capture the full complexity of intra-tumoural heterogeneity. Not surprisingly, these signatures have limited prognostic value for certain subtypes of breast cancers, such as Triple-Negative breast cancers, which are generally highly proliferative and hormone receptor-negative. We report here the identification and extensive clinical validation of a 20-gene signature that predicts risk of distant metastases in patients with different breast cancer subtypes, including Luminal and Triple-Negative breast cancers, by measuring the intrinsic content and degree of biological aggressiveness of the cancer stem cell population of the primary tumour. This study, to our knowledge, is the first translational assessment combining molecular profiling data with high-quality clinical data in the analysis of a large retrospective consecutive cohort for the development of a stem cell-based prognostic test for breast cancer. Our results demonstrate the discovery, assessment, and clinical validation of a new multigene assay based on the biology of cancer stem cells, which represents a novel concept in the landscape of genomic predictors in breast cancer.
Implications of all the available evidence
Considering the increasingly recognized role of cancer stem cells in driving tumour progression, therapy resistance and metastasis, a prognostic model based on the molecular information captured at the level of cancer stem cells in the primary tumour has the potential to be transformative for clinical decision-making in breast cancer, when used as a standalone test or in combination with other genomic predictors. Based on the analysis of a large prospective-retrospective cohort, we submit that our genomic predictor might prove clinically valuable for the stratification of patients with negligible metastatic risk, who might safely benefit from de-escalating regimens of chemotherapy and/or endocrine therapy, thus avoiding overtreatment. On the other hand, this genomic tool could help identify patients at high risk of recurrence who might benefit from more aggressive treatments. Additionally, our results highlight a set of genes with a likely mechanistic role in the metastatic process, which could represent novel molecular targets for the development of drugs counteracting metastatic progression of breast cancer.
Alt-text: Unlabelled Box
1. Introduction
Tumour heterogeneity may represent a major hurdle in the clinical management of breast cancer (BC). The identification of molecular subtypes of BC – Luminal-A, Luminal-B, Basal-like and HER2-positive (HER2+) – has provided molecular foundations for the clinical and pathological heterogeneity of this disease. The integration of this new taxonomy with traditional clinicopathological parameters has proved invaluable for informing clinical decision-making [1].
One source of phenotypic and functional heterogeneity in BC, and in other cancers, is thought to reside in a subpopulation of tumour cells with exclusive self-renewal and tumourigenic capacity, i.e., the cancer stem cells (CSCs) [2,3]. The relevance of CSCs to the natural history of tumours is manifold: they not only fuel the continuous growth of the cancer but represent also the prime suspect for its metastatic ability, and hence adverse clinical outcome, and – in certain cases – for refractoriness to therapies [[4], [5], [6]]. It is widely believed, therefore, that advancements in the understanding of the molecular and biological properties of CSCs might benefit all aspects of the management of cancer patients.
Previously, we demonstrated that a stem cell (SC)-specific transcriptional profile, obtained by comparing the transcriptome of normal mammary SCs (MaSCs) with that of their progeny, could predict the biological (grade) and the molecular subtype of BCs [7]. This finding argued that different BCs, profiled as a whole, display variable “degrees of stemness” (defined as extent of molecular resemblance to normal MaSCs), which, in turn, correlates with certain biological and molecular features. The “degree of stemness” most likely reflects the number of CSCs within a tumour, and hence their propensity to self-renew and proliferate. In support of this, we showed that poorly differentiated BCs contain more CSCs (measured as tumour-initiating cells in xenotransplantation assays) than well-differentiated BCs [7]. Nevertheless, direct evidence that the “degree of stemness” of a tumour is a measure of the intrinsic content of CSCs, or of their self-renewal/proliferative behaviour, is lacking. Furthermore, it is not known whether the “degree of stemness” of a BC is predictive of metastatic ability or clinical outcome. If this were the case, it should be possible to extract from the MaSC transcriptional profile, robust and clinically manageable signatures for prognostic stratification in BC. This would add a novel dimension to our ability to stratify BCs [8], by allowing direct and quantitative measurements of the impact of subversion of the SC compartment, on the natural history and clinical outcome of a tumour. The present studies were undertaken to investigate these possibilities.
2. Materials and methods
2.1. Study design and patients
Our study started with a series of in silico analyses of different public BC datasets, which were interrogated with the set of SC-specific genes previously identified as overexpressed in normal MaSCs vs. progenitors [7]. These analyses resulted in the identification and analytical validation of a 20-gene SC signature with independent predictive prognostic power in the four different BC datasets analysed. We next assessed the clinical relevance of the in silico findings, using a large prospective-retrospective cohort of 2453 female BC patients with early stage, operable BC and no history of a previous malignancy, operated at the European Institute of Oncology (IEO) in Milan between years 1997 and 2000 (the “IEO BC 97-00” cohort) (see Supplementary Methods for details on the selection, characterization and follow-up of this cohort). Finally, we used a prospective consecutive series of 90 BC patients, for whom it was possible to obtain sufficient amounts of fresh biopsy tissue amenable to functional in vitro studies, to assess the correlation between 20-gene SC risk score and the self-renewing proliferative behaviour of CSCs, through the execution of the serial tumoursphere propagation assay (see Supplementary Methods for details).
2.2. Meta-analysis of published BC datasets
For the analysis of the Ivshina, Pawitan, Loi KI, and METABRIC datasets [[9], [10], [11], [12]], original RAW data (CEL files) or processed data were downloaded from the GEO database (Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/) accession code GSE4922, GSE1456 and GSE6532 or from the cBioPortal for Cancer Genomics (http://www.cbioportal.org/). The datasets (see Supplementary Table S1 and S2) used for the unsupervised analyses were built by extracting, from the original datasets, information for those patients for whom a follow-up of at least 5 years was available (Ivshina: 227 of 249 patients; Pawitan: 153 of 159 patients; Loi KI: 119 of 149 patients; METABRIC: 1825 of 1989 patients). With the exception of the METABRIC dataset, Affymetrix GenGhip CEL files were reprocessed with the Affymetrix's proprietary MAS5 pre-processing algorithm, in order to make all samples comparable with those used in the present study. Processed files were then imported into GeneSpring GX software version 7.3.1 (Agilent Technologies, Santa Clara, CA). According to the GeneSpring normalization procedure, in each analysis the 50th percentile of all measurements was used as a positive control, within each hybridization array, and each measurement for each gene was divided by this control. The bottom 10th percentile was used for background subtraction. Among different hybridization arrays, each gene was divided by the median of its measurements in all samples. Data were then log transformed for subsequent analysis. All clustering analyses were performed with GeneSpring, using the Standard Correlation as a similarity measure and Average Linkage as a clustering algorithm for both genes and samples. All statistical analyses were performed using JMP 10.0 statistical software (SAS Institute, Inc).
2.3. Quantitative real-time PCR analysis
Total mRNA was extracted from formalin-fixed paraffin-embedded (FFPE) samples and RT-qPCR reactions were performed with an in-house custom designed TaqMan® Array. Each target was assayed in triplicate and average Cq (AVG Cq) values were calculated and normalized using four reference genes (HPRT1, GAPDH, GUSB and TBP) to compensate for possible variations in the expression of single reference genes and in RNA integrity due to tissue fixation. Normalized data were then processed for statistical analysis. Based on the distribution of the reference genes, we applied the Tukey's interquartile rule for outliers to identify poor quality RT-qPCR data [13]. After exclusion of patients with insufficient or poor quality RNA from the “IEO BC 97-00” study cohort of 2453 patients, a total of 2316 patients were finally included in the statistical analyses (see Supplementary Methods for details).
2.4. Development of the 20-gene SC signature and of StemPrintER risk scores
Using expression levels of the 20 SC genes obtained by RT-qPCR on paraffin samples, we generated two different prognostic models: i) the 20-gene SC signature, based on expression data of the 20 SC genes in a training set of patients from the entire “IEO BC 97-00” cohort; ii) StemPrintER, a Luminal BC-specific risk model, based on expression data of the 20 SC genes in a training set from the subgroup of Luminal BC patients. In the respective training sets, the prognostic models were derived using the ridge penalized Cox regression model, considering the normalized gene expression values of the 20 SC genes as continuous covariates with log-linear effect. Cross-Validated (10-fold) log-Likelihood (CVL) with optimization of the tuning penalty parameter was applied. Tuning of the penalty parameter was repeated 500 times using a different folding at each simulation and the model associated with the highest CVL was selected [[14], [15], [16]]. A continuous risk score was assigned to each patient based on the following formula: Risk score = ∑i (βi*Cqnormalized), where: i is the summation index for the 20 target genes; β is the ridge penalized Cox model coefficient for each target gene; Cqnormalized is the normalized average Cq for each target gene. Minimum and maximum risk scores from the training set were used to scale risk scores in a 0–100 range. For StemPrintER, the median of the continuous risk score of the training set was used to identify two classes of risk (Low and High).
2.5. Statistical analyses
In prognostic studies, primary endpoint was the cumulative incidence of distant metastasis (DM), defined as the time from surgery to the appearance of a metastasis or death from BC as a first event [17]. Local or regional recurrence, second primary cancer, death for unknown causes or other causes were considered as competing events. Considering first events, median follow-up for censored patients was 14·1 years (interquartile range [IQR] 12·1–15·7). One hundred and eighty-five (7·5%) patients were lost at 10 years of follow-up.
For the estimation of the primary endpoint, we used the Cumulative Incidence Function (CIF), according to the methods described by Kalbfleisch and Prentice [18], taking into account the competing causes of DM. Hazard ratios were estimated, both in the entire follow-up and in the early (0–5 years) or late (5–10 years) time intervals, using a Cox proportional hazards model. Multivariable models were adjusted for Grade (G1, G2 and G3), Ki-67 (Ki-67 < 14% and Ki-67 ≥ 14%), HER2 status (positive and negative), ER/PgR status [not expressed (Both 0) and expressed (ER > 0 or PgR > 0)], tumour size (pT1 and pT2-3-4), number of positive lymph nodes (pN0, pN1-2-3 and pNX) and age at surgery (<50 and ≥ 50) (as appropriate). Subgroup analysis was performed to investigate possible differences in the prognostic power of the risk models in the different sub-populations. Differences in the distribution of clinicopathological features between groups were evaluated by the Chi-square test. Differences in the distribution of continuous risk score between groups were evaluated using a linear regression model. A logistic regression model was used to establish association between CSC proliferative/self-renewal phenotype and continuous risk score in the consecutive cohort of 90 BC patients. All analyses were carried out with the SAS software (SAS Institute, Cary, NC). All reported p-values are two-sided.
3. Results
3.1. Identification and in silico validation of a prognostic 20-gene SC signature
To derive a prognostic SC-based predictor, we performed a stepwise series of in silico analyses in published BC datasets (schematically depicted in Fig. 1, a and b) employing the previously described panel of genes (1059 Affymetrix probestes) that were significantly overexpressed between human normal MaSC vs. progenitors [7]. In particular, we initially performed unsupervised hierarchical clustering of the BC dataset published by Ivshina et al. [9] (described in Supplementary Table S1). This allowed for the extraction, from the original list of 1059 probesets, of a discernible panel of 329 probesets that were highly and homogeneously expressed in a subgroup of BC patients (Supplementary Fig. S1a). When used alone to re-clusterize BC patients of the same dataset, this 329-probeset signature clearly distinguished between BCs displaying a “SC-like” profile (H, for High similarity to SCs) and a “non-SC-like profile (L, for Low similarity to SCs, Supplementary Fig. S1b). Interestingly, BCs displaying a high “SC-like” profile displayed worse prognosis both in univariate (HRH vs. L 2·30, 95% CI 1·50–3·59; p = 0·0001) and in multivariable analyses adjusted for all the standard clinicopathological parameters (HRH vs. L 1·83, 95% CI 1·15–2·95; p = 0·010) (Supplementary Fig. S1c, and Supplementary Table S1). Finally, from the 329-probeset signature, we identified a minimal cluster of 20 genes (henceforth, the “20-gene SC signature”) that displayed the highest differential expression between “SC-like” vs. “non-SC-like” BCs, and improved the independent predictive prognostic power of the parental 329-signature in the multivariate analysis of the Ivshina dataset (HRH vs. L 2·05, 95% CI 1·23-3·53; p = 0·0054) (Supplementary Fig. S1d, and Supplementary Table S1).
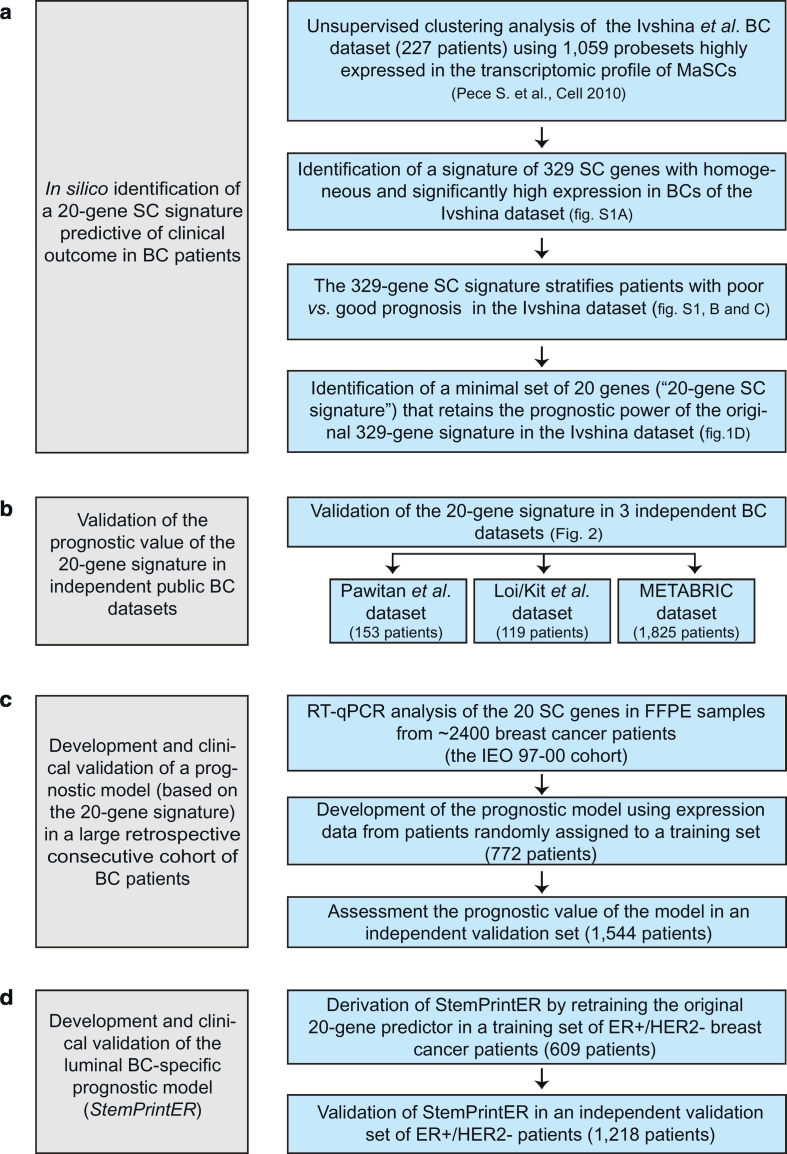
Fig. 1.
Schematic flowchart of the study design.
a. Stepwise in silico analyses in the public BC dataset published by Ivshina et al. [9] that led to the identification of a set of 20 genes (20-gene SC signature) derived from the transcriptomic profile of MaSCs with predictive prognostic power in BC. b. Analytical validation of the 20-gene SC signature in the indicated three independent public datasets. c. Generation and clinical validation of a prognostic model (20-gene SC risk model) based on transcript expression levels of the 20 SC genes assessed by RT-qPCR in FFPE samples from the “IEO 97–00” BC cohort. d. Development and clinical validation of the Luminal BC-specific prognostic model (StemPrintER) by retraining the original 20-gene SC risk model in Luminal ER+/HER2- BC patients.
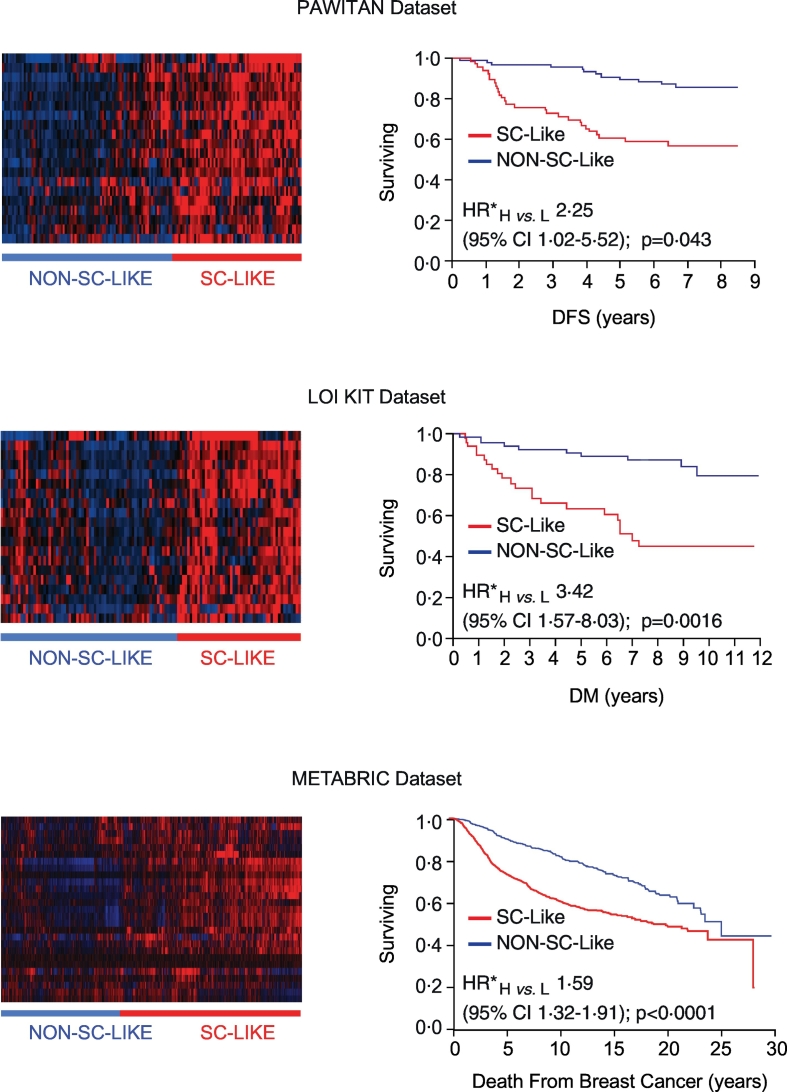
We validated the 20-gene SC signature in three independent BC expression datasets: the Pawitan et al. [10], the Loi et al. [11], and the METABRIC [12] datasets. In all cases, the signature was a predictor of poor prognosis, independently of standard clinicopathological parameters (Fig. 2; see also Supplementary Table S2 for detailed description of the datasets and statistical analyses).
Fig. 2.
Analytical validation of the 20-gene SC signature in independent BC datasets.
Left panels, Identification of “SC-like” and “Non-SC-like” subgroups of BCs by unsupervised hierarchical clustering analysis of the expression of the 20-gene SC signature in the BC datasets published by Pawitan et al. [10] (endpoint: disease-free survival, DFS), Loi et al. [11] (endpoint: time to distant metastasis, DM) and in the METABRIC dataset [12] (endpoint: death from breast cancer). Right panels, Prognostic significance of the “SC-like” (H, for High similarity to SCs) vs. “Non-SC-like” (L, for Low similarity to SCs) classification determined by Kaplan-Meier analysis in the same BC datasets using the endpoints specified above. Multivariable Cox proportional hazards model (see Supplementary Table S2 for details on the multivariable analyses in the different datasets). HR*, multivariable hazard ratio; CI, confidence interval; p, p-value.
Clinical validation of the 20-gene SC signature in a prospective-retrospective cohort study.
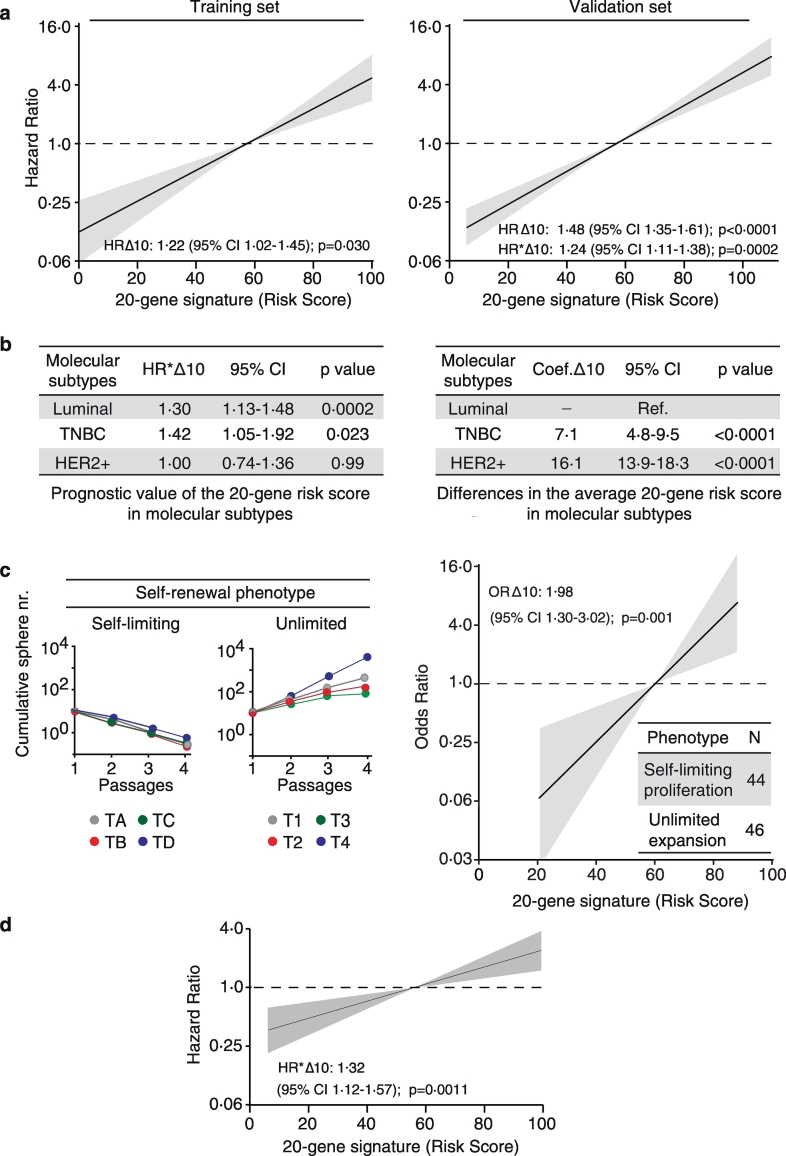
The clinical validity of the 20-gene SC signature was assessed using the “IEO BC 97-00” cohort of 2453 BC patients (described in Supplementary Table S3). Total mRNA was extracted from FFPE samples and used to perform RT-qPCR reactions (see Supplementary Table S4 for the detailed list of assays). RT-qPCR expression data for the 20 SC genes, obtained from a training set of 772 cases (one-third of the cohort), were used to develop a 20 SC gene-based risk model using a ridge-penalized Cox regression model (Fig. 1c; see also Supplementary Table S5 for description of the algorithm). The performance of the risk model was tested in a validation set composed of the remaining 1544 patients. The training and validation sets were balanced for clinicopathological features and showed no difference in the average risk score (Supplementary Table S6). In both the training and validation sets, the 20-gene SC risk model, used as a continuous variable over the entire follow-up period, behaved as an independent predictor of DM in a multivariable Cox regression analysis adjusted for tumour size (pT), number of positive lymph nodes (pN), tumour grade, Ki-67, ER/PgR or HER2 status, and age at surgery (Fig. 3a, and Supplementary Table S7).
Fig. 3.
Clinical validity and biological characterization of the 20-gene SC signature.
a. The 20-gene SC risk score, used as a continuous variable with a 10-unit increase (HRΔ10), predicts distant metastasis over the entire follow-up interval in the “IEO BC 00-97” cohort. Training set, N = 772 (left); validation set, N = 1544 (right). Univariate (HR) and multivariable (HR*) hazard ratios are in log scale. Cox proportional hazards model. Multivariable models were adjusted for standard clinicopathological parameters (see Materials and Methods). Grey shaded areas, 95% CI. p, p-value; CI, confidence interval. b. Left, Prognostic performance of the 20-gene SC continuous risk score in Luminal (N = 1213), Triple-Negative (TNBC) (N = 143), HER2-positive (HER2+) (N = 169) BC. Right, Differences in the average 20-gene SC continuous risk score (Coef. Δ10) estimated in TNBC and HER2+ BC, relative to Luminal BC (Ref.) in the validation set. Linear regression model. c. Left, Representative examples of tumoursphere assays displaying a self-limiting (TA-TD) or unlimited (T1-T4) CSC self-renewal phenotype performed on primary BC biopsies. Right, Logistic regression model showing the direct correlation between the 20-gene SC risk score, used as a continuous variable with a 10-unit increase (ORΔ10), and probability of CSCs displaying an unlimited self-renewal phenotype in the 90-BC patient cohort (Supplementary Table S9 for patients' characteristics). Inset, distribution of tumours showing the phenotype. Odds ratio (OR) values plotted in logarithmic scale (vertical axis). ∆10, ten unit-increase; N, number of patients; p, p-value; CI, confidence interval. d. The 20-gene SC risk score, used as a continuous function as in (a) over the entire follow-up interval is an independent predictor of distant metastasis in patients with LVI (N = 500). Multivariable Cox proportional hazards model. HR*, multivariable hazard ratio.
A time-varying analysis of the validation set revealed that the signature is also an independent predictor of early (0–5 years) and late (5–10 years) recurrence (Supplementary Fig. S2a). Furthermore, in a stratified analysis of the validation set by BC molecular subtype, the 20-gene SC continuous risk score was an independent predictor of the individual likelihood of developing DM in Luminal and TNBC, but not in HER2+, subtypes (Fig. 3b, and Supplementary Table S8 for complete analyses). Notably, compared to Luminal BCs, TNBCs showed a significantly higher average risk score (p < 0·0001), which was further significantly increased in HER2+ BCs compared to TNBCs (p < 0·0001) (Fig. 3b, and Supplementary Fig. S2b). We submit that the lack of predictive power of the 20-gene SC risk model in HER2+ BCs might reflect a homogeneously distributed high “degree of stemness” in these tumours compared to the more heterogeneous subgroups of TNBCs and Luminal BCs.
3.2. Assessment of the biological basis of the 20-gene SC signature
We exploited the tumoursphere serial propagation assay to investigate the biological bases of the 20-gene SC signature. This in vitro assay allows for the accurate estimation of the number and degree of biological aggressiveness of the CSCs of individual BCs [6,7], as it reflects the intrinsic propensity of CSCs to continually self-renew and proliferate (referred to as an “unlimited” phenotype) or to progressively extinguish (“self-limiting” phenotype) over several tumoursphere generations (Fig. 3c) (see also Supplementary Methods).
On the basis of this background, we subjected a consecutive series of 90 BC patients (described in Supplementary Table S9) to the tumoursphere propagation assay, to investigate the correlation between the 20-gene SC risk score and the “unlimited” vs. “self-limiting” self-renewal behaviour of CSCs. We found that, for every 10-unit increase in the risk score of the primary tumour, there was a ~2-fold increase in the probability of CSCs to display an unlimited self-renewal and proliferative phenotype, and therefore a propensity to expand in number (Fig. 3c). These findings argue that the 20-gene SC risk model provides a quantitative estimate of the metastatic risk of BCs by its ability to interrogate the number and biological characteristics of their CSCs. This also corroborates the notion that, in the context of the bulk tumour population, the metastatic potential likely resides in the subfraction of tumour cells that display CSC characteristics (see Discussion).
In support of the clinical relevance of this idea, we found that even in patients with lymphovascular invasion (LVI) – which is an initial critical step in metastasis – the risk of DM augments as a function of increasing levels of the 20-gene SC risk score (Fig. 3d). From a biological viewpoint, this finding argues that: i) the presence of tumour emboli in the lymphatic and/or blood vessels of the peritumoural area is not sufficient per se to predict the occurrence of clinically-relevant metastases in BC patients; ii) an increased probability that LVI areas contain cells with true metastatic potential correlates with a higher CSC burden of the primary tumour, reflected in a higher 20-gene SC risk score.
3.3. Retraining the 20-signature to derive a specific genomic predictor for luminal BC patients
In ER+/HER2- Luminal BC patients, accurate prognostication based on the individual risk of early or late recurrence is key to tailor the use of chemotherapy and hormonal therapy, thus avoiding under−/over-treatment (see Discussion) [8,19]. To develop a genomic tool specifically designed for metastatic risk prediction in this group of patients, we randomly split the Luminal BC patients of the “IEO BC 97-00” cohort (N = 1827) into a training set (one-third, N = 609), that was used to derive a Luminal-specific risk model using the ridge penalized Cox regression model, and a validation set (two-thirds, N = 1218) (Fig. 1d). The two sets were balanced for clinicopathological features (Supplementary Table S10). This approach generated a Luminal-specific risk model that we named StemPrintER, based on its proposed use as a SC-based genomic predictor in ER+/HER2- Luminal BCs (Fig. 1d; see also Supplementary Table S11 and S12 for description of the algorithm and for patient stratification). The StemPrintER risk score correlated with clinicopathological parameters of biological aggressiveness and poor prognosis (Table 1, and Supplementary Tables S13). Used as continuous function, StemPrintER behaved as an independent predictor of DM over the entire follow-up interval (Supplementary Fig. S3a, and Supplementary Table S14). Moreover, in a time-varying analysis, the StemPrintER continuous risk score predicted both early (0–5 years) and late (5–10 years) risk of DM in a multivariable analysis of the validation set, adjusted for pT, pN, tumour grade, Ki-67, and age at surgery (Fig. 4a, and Supplementary Table S15).
Table 1.
Correlation between the StemPrintER 2-class risk categories (Low, High) and clinicopathological parameters in the Luminal validation set.
| Variable | ALL N (% col) |
2-class risk category |
p value | |
|---|---|---|---|---|
| Low N (% row) |
High N (% row) |
|||
| All | 1218 (100) | 644 (52·9) | 574 (47·1) | |
| Age at surgery | 0·51 | |||
| <50 | 453 (37·2) | 234 (51·7) | 219 (48·3) | |
| ≥50 | 765 (62·8) | 410 (53·6) | 355 (46·4) | |
| Histology | <0·0001 | |||
| Ductal | 937 (76·9) | 443 (47·3) | 494 (52·7) | |
| No Ductal | 281 (23·1) | 201 (71·5) | 80 (28·5) | |
| pT | <0·0001 | |||
| pT1a/b | 169 (13·9) | 117 (69·2) | 52 (30·8) | |
| pT1c | 677 (55·6) | 412 (60·9) | 265 (39·1) | |
| pT2 | 335 (27·5) | 101 (30·1) | 234 (69·9) | |
| pT3/pT4 | 37 (3·0) | 14 (37·8) | 23 (62·2) | |
| pN | <0·0001 | |||
| pN0 | 607 (49·8) | 360 (59·3) | 247 (40·7) | |
| pN+ | 579 (47·5) | 267 (46·1) | 312 (53·9) | |
| pNX | 32 (2·6) | 17 (53·1) | 15 (46·9) | |
| Grade | <0·0001 | |||
| 1 | 278 (22·8) | 219 (78·8) | 59 (21·2) | |
| 2 | 619 (50·8) | 350 (56·5) | 269 (43·5) | |
| 3 | 292 (24·0) | 60 (20·5) | 232 (79·5) | |
| n/a | 29 (2·4) | 15 (51·7) | 14 (48·3) | |
| LVI | <0·0001 | |||
| Absent | 852 (70·0) | 495 (58·1) | 357 (41·9) | |
| Present | 366 (30·0) | 149 (40·7) | 217 (59·3) | |
| Ki-67 | <0·0001 | |||
| <14% | 414 (34·0) | 336 (81·2) | 78 (18·8) | |
| ≥14% | 803 (65·9) | 307 (38·2) | 496 (61·8) | |
| n/a | 1 (0·1) | 1 (100) | 0 (0·0) | |
| CT/HT | <0·0001 | |||
| Nil | 55 (4·5) | 36 (65·5) | 19 (34·5) | |
| HT | 514 (42·2) | 322 (62·6) | 192 (37·4) | |
| CT | 40 (3·3) | 14 (35·0) | 26 (65·0) | |
| HT-CT | 609 (50·0) | 272 (44·7) | 337 (55·3) | |
| Surgery | <0·0001 | |||
| Quadrantectomy | 1024 (84·1) | 570 (55·7) | 454 (44·3) | |
| Mastectomy | 194 (15·9) | 74 (38·1) | 120 (61·9) | |
| Radiotherapy | 0·002 | |||
| No | 201 (16·5) | 86 (42·8) | 115 (57·2) | |
| Yes | 1017 (83·5) | 558 (54·9) | 459 (45·1) | |
The association between the StemPrintER 2-class risk categories (Low, High) and the demographic, clinical and pathological variables was evaluated with the chi-square test. The number (N) of patients and percentage (%) in each group is indicated. pT, primary tumour size; pN, nodal status; LVI, lymphovascular invasion; Ki-67, proliferation index; CT, adjuvant chemotherapy; HT, adjuvant hormone therapy; Nil, no adjuvant therapy; n/a, not available.
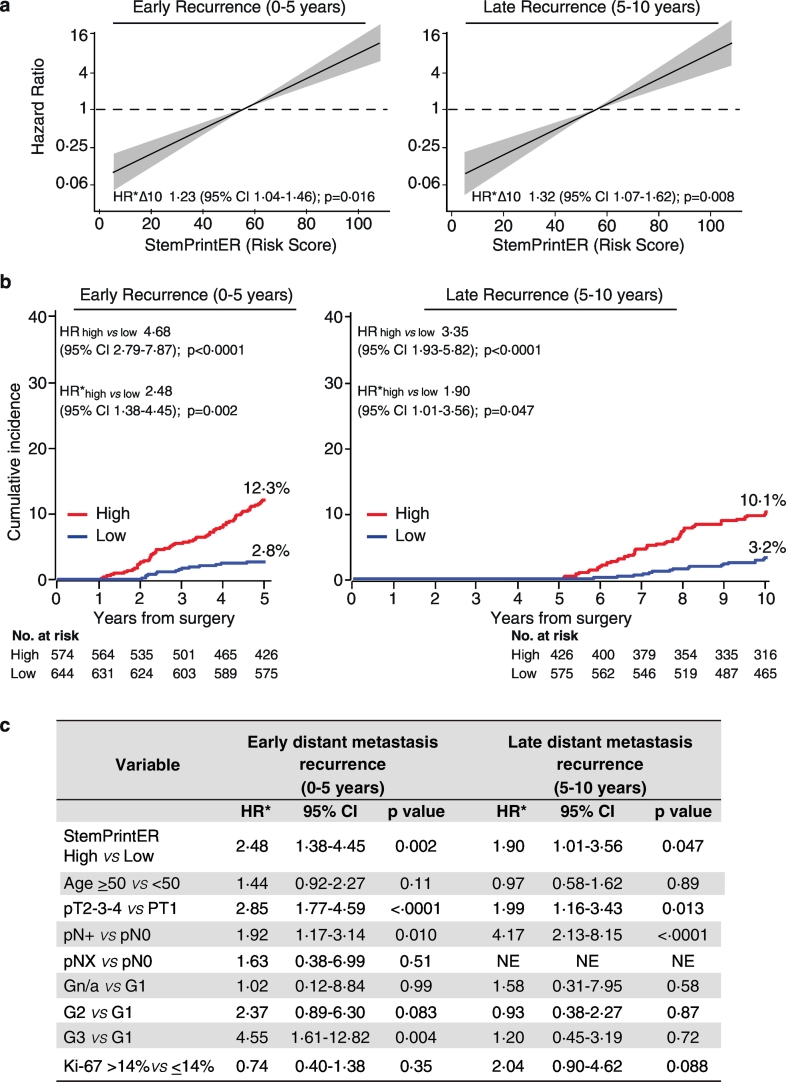
Fig. 4.
StemPrintER is an independent predictor of early and late metastasis in Luminal BC patients.
a. StemPrintER continuous risk score (10-unit increase, Δ10) in the prediction of early (0–5 years) and late (5–10 years) distant metastasis in the Luminal BC validation set (N = 1218). Multivariable Cox proportional hazards model (see Materials and Methods). Grey shaded areas, 95% CI. Multivariable hazard ratio (HR*) values were plotted in logarithmic scale (vertical axis); CI, confidence interval; p, p-value. b. Cumulative incidence of early (0–5 years) and late (5–10 years) distant metastasis for the StemPrintER 2-class risk model (High, Low). Univariate (HR) and multivariable (HR*) hazard ratios (High vs. Low) and the distant metastasis rate in the High- and Low-risk group for each time interval are indicated. Multivariable Cox proportional hazards model was adjusted for standard clinicopathological parameters (see Materials and Methods), as appropriate. CI, confidence interval; p, p-value; No. at risk, number of patients at risk. c. Prognostic performance of the StemPrintER 2-class risk model (High, Low), and of the indicated clinicopathological parameters in the prediction of the early (0–5 years) and late (5–10 years) distant metastasis risk in the ER+/HER- BC validation set (N = 1218) in a multivariate analysis as in (b). HR*, multivariable hazard ratio; CI, confidence interval; p, p-value; NE, not estimable; n/a, not available.
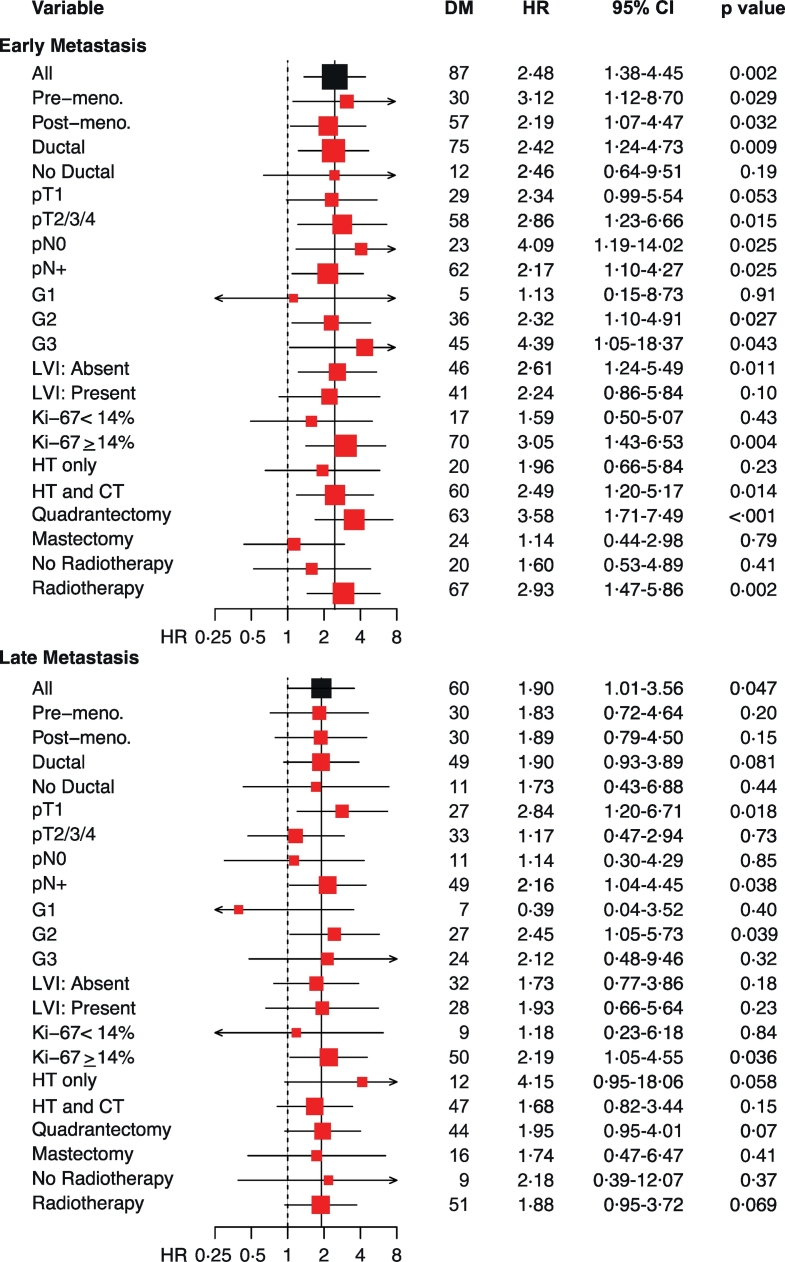
With the idea in mind to translate this tool into the clinical practice, we developed a 2-class risk model, based on the median of the StemPrintER continuous risk score in the training set (see Materials and Methods for details), which could be used to stratify Luminal BC patients into a high vs. low risk group. The 2-class categorization further confirmed the clinical value of StemPrintER as an independent predictor of DM in the entire follow-up (Supplementary Fig. S3b), and in the early or late time-interval (Fig. 4, b and c). In the low risk group, the cumulative incidence of distant metastasis was 2·8% before 5 years and 3·2% between 5 and 10 years after surgery; the cumulative incidence for the high-risk group was, respectively, 12·3% and 10·1% (Fig. 4b; see also Supplementary Table S14 for details on univariate and multivariate analyses). Finally, analysis of the Luminal BC validation cohort, stratified by clinicopathological characteristics, showed no evidence of substantial heterogeneity in the predictive power of StemPrintER among the different subgroups, regardless of whether StemPrintER was used as a continuous function (Supplementary Table S16) or as a 2-class risk model (Fig. 5, and Supplementary Table S17 and S18 for complete analyses). However, considering the importance of the patient's lymph node status for prognostic prediction and therapy decision-making, we note that StemPrintER is an independent predictor of early recurrence in lymph node-negative, and of both early and late recurrence in lymph node-positive Luminal BC patients.
Fig. 5.
Behaviour of the StemPrintER 2-class risk model in prespecified ER+/HER2- BC patient groups.
The forest plot shows multivariable hazard ratios (HR*) of early (0–5 years) and late (5–10 years) distant metastasis using the StemPrintER 2-class risk model (High, Low) in ER+/HER2- BC patients stratified according to their clinicopathological characteristics. Multivariable Cox proportional hazards model was adjusted for Grade (G1, G2 and G3), Ki-67 (Ki-67 < 14% and Ki-67 ≥ 14%), tumour size (pT1 and pT2–3-4), number of positive lymph nodes (pN0, pN1–2-3 and pNX), and age at surgery (<50 and ≥ 50) as appropriate in each group. HR > 1, unfavourable prognosis for high StemPrintER status. Horizontal axis shows hazard ratio values plotted in logarithmic scale. DM, number of distant metastasis; CI, confidence interval.
4. Discussion
The identification and development of multigene assays for accurate prognostication of individual BC patients has represented an expanding area of research for more than a decade. In this context, it has become progressively clear that biomarkers for the prediction of clinical outcome should be able to interrogate the underlying biology of the tumours of individual BC patients [20]. The increasingly recognized relevance of CSCs to BC heterogeneity and disease course [5] argues that the knowledge of the “degree of stemness” of a BC might substantially advance individualized patient management. Herein, we describe a novel genomic predictor based on a cluster of 20 SC genes whose high expression levels were capable of discerning a homogeneous group of patients with adverse clinical outcome in the meta-analysis of four distinct public breast cancer datasets. Through validation studies in a large prospective-retrospective cohort of BC patients with high-quality follow-up, and functional prospective studies based on the use of fresh tumour samples from an additional consecutive series of BC patients, we established that our 20-gene SC-based assay:
i) predicts the individual likelihood to develop distant metastasis in BC, in particular in TNBC and Luminal ER+/HER2- BC cancers; ii) does so, most likely, by interrogating the number and biological characteristics of their CSCs. Of note, our genomic predictor comprises a set of genes that do not belong (with one exception) to any other genomic tool or molecular classifier described for TNBC and Luminal BCs. Thus, we submit that we have developed a unique tool capable of probing into the “degree of stemness”, and hence into the clinical outcome, of BCs.
In our efforts, we started from genes discriminating MaSCs from progenitors in the normal gland [7]. Furthermore, we selected only those genes that were expressed at higher levels in MaSCs vs. progenitors. We did so by reasoning that: i) CSCs might display traits reminiscent of those present in normal MaSCs [3]; ii) since CSCs are rare, the selection of overexpressed genes (MaSCs vs. progenitors) afforded a higher likelihood of scoring differences, with respect to underexpressed genes. We believe that our findings have important implications from both the biological and clinical perspective.
From a biological viewpoint, our findings raise two connected questions: i) the relevance of the 20 SC genes to CSC phenotypes, in particular to their metastatic potential; ii) the relationship between their expression in the normal vs. the CSC compartment.
Based on extant literature, several of the 20 genes display evident connection to metastatic dissemination through their role in matrix degradation, migration, invasion and engraftment (e.g. MMP1, SNF, MIEN1, PHLDA2, EPB41L5) [[21], [22], [23], [24]]. For other genes (RACGAP1, H2AFZ, H2AFJ, APOBEC3B, CENPW, TOP2A CDK1), their implication in the establishment of CSC phenotypes might be linked to their involvement in genomic instability, which can be reasonably hypothesized based on their role in processes, such as DNA replication and repair, chromatin remodeling and mitotic control of chromosome segregation [[25], [26], [27], [28], [29]]. A final set of genes, whose putative role in metastasis is less obvious, might be linked – directly or indirectly - to the development of adaptive plastic responses required for CSCs to withstand and survive in hostile environmental conditions, such as hypoxia and nutrient deprivation, both at the primary tumour and metastatic site level, and/or to resist hormonal or chemotherapy treatments, often in the broader context of the activation of an epithelial-to-mesenchymal transition (EMT) program. These genes include those involved in: i) metabolism reprogramming and mitochondrial physiology (MRPS23, NDUFB10, PHB) [[30], [31], [32]]; ii) mRNA ribonucleoparticle biogenesis, mRNA transcription, splicing and export, and RNA processing and degradation events (ALYREF, EXOSC4) [[33], [34], [35]]; iii) survival/escape from apoptosis, which is connected to resistance to hormonal and/or chemotherapy through hijacking of signaling pathways, such as TGF-beta and PI3K-AKT-mTOR (NOL3, LY6E, EIF4EBP1) [[36], [37], [38]]. Additional evidence for a mechanistic link between the 20 genes and the CSC phenotype comes from the observation that these genes are frequently overexpressed in BC, sometimes as a consequence of gene amplification [31].
While further studies are needed to establish whether the genes of our signature are causal in the determination and/or maintenance of CSCs in BCs, and possibly of their metastatic potential, our observations support the idea that CSCs are not simply reminiscent of normal MaSCs; rather the emergence of CSC phenotypes is, directly or indirectly, connected to the aberrant function of one or more of the 20 SC genes. Furthermore, the ability of the 20-gene SC signature to predict DM in TNBC and Luminal BC patients points to the existence of common molecular workings underlying the metastatic potential of CSCs in different BC subtypes, regardless of the molecular and phenotypic differences that typically distinguish the different subtypes at the bulk tumour level. In this framework, it is not surprising that, with the sole exception of RACGAP1 (present in the Breast Cancer Index [39]) the genes of the 20-gene SC signature are not comprised in any of the already existing genomic predictors developed for prognostication of Luminal BC or in molecular classifiers that distinguish different subtypes in TNBC, considering that these genomic tools are all invariably based on the molecular profile of the bulk tumour mass [40,41].
Together, our findings also support the emerging notion that the metastatic potential of individual BCs can be traced back to the molecular characteristics of a rare subpopulation of tumour cells that display CSC traits [42]. In this context, it is worth noting that, even in patients with LVI, i.e., with the presence of emboli of frank tumour cells that have already invaded lymphatic and/or blood vessels of the peritumoural area, the likelihood of developing clinically evident DM correlates with the CSC content of the primary tumour.
From a clinical standpoint, although future studies in independent retrospective and/or prospective BC patient cohorts are warranted to increase the level of clinical evidence of the reliability and transportability of our 20-gene SC-based genomic tool, our results might have immediate relevance to the clinical management of BC patients, in particular for the subgroup of ER+/HER2- Luminal BC patients. These patients represent the majority (~75%) of the cases [43] and display high molecular heterogeneity and variability in their clinical behaviour. Therefore, Luminal BC patients can greatly benefit from accurate stratification of their risk of recurrence, for the administration of the optimal therapy, while avoiding under- or over-treatment [19,44]. In this direction, we developed - based on the 20-gene SC signature - StemPrintER, a specific risk model for Luminal BC. StemPrintER is an independent predictor of both early and late metastasis. This places StemPrintER among the more recently developed second generation multi-gene assays, such as Prosigna [45], BCI [39], and EndoPredict [46], which have been shown to outperform first generation BC prognostic tests - e.g., Oncotype DX [47] and Mammaprint [48] - in the prediction of the risk of late recurrence (5–10 years post-surgery). In particular, StemPrintER predicts early metastasis in lymph node-negative BC patients, and both early and late metastasis in lymph node-positive BC patients. StemPrintER could therefore find clinical application as a tool to tailor the administration of adjuvant chemotherapy, in addition to the standard endocrine therapy, in those Luminal BC patients at high risk of early recurrence, while sparing unnecessary chemotherapy to low risk patients [19].
On the other hand, StemPrintER could also represent a valuable tool to identify Luminal BC patients at high risk of late recurrence, who might benefit from prolongation of endocrine therapy beyond the standard 5 years of treatment. This is an important question in the clinical management of ER+/HER2- Luminal BC patients who remain at persistent risk of recurrence for at least 15–20 years [49], with >50% of relapses and more than two-thirds of deaths occurring >5 years after the original diagnosis. However, while continuation of endocrine therapy reduces the proclivity to develop late recurrences [50], its benefits must be weighed against side effects and quality of life, avoiding overtreatment through accurate patient stratification.
We therefore submit that, by its unique ability to interrogate the “stemness” of individual BCs, StemPrintER might prove clinically valuable, either as a standalone test or in combination with other genomic predictors or clinicopathological parameters, to guide individualized clinical decision-making in Luminal BC patients.
Acknowledgments
Acknowledgments
We thank the anonymous patients who donated their samples for research. We also thank G. Corso, M. Tillhon, S. Pirroni, C. Luise, G. Jodice, the Primary/Stem Cell, the Clinical Biomarker, the Imaging and the Molecular Pathology Infrastructures of the IEO Novel Diagnostics Program. G. Peruzzotti and the IEO Clinical Trial Office. R. Gunby for critically editing the manuscript. This study was approved by the IEO Institutional Ethical Board.
Funding sources
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC; IG 11904 to S.P., IG 18988 to P.P.D.F., and MCO 10.000), MIUR (the Italian Ministry of University and Scientific Research), the Italian Ministry of Health to S.P., P.P.D.F. and D.T. This work was also supported in part by a research grant from Tiziana Life Sciences PLC. The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Declaration of interest
The research grant from Tiziana Life Sciences PLC was part of a license agreement in which the rights for StemPrintER were licensed to Tiziana Life Sciences PLC. Authors declare no other competing financial interests related to this study.
Author contributions
D.D., D.T., M.V. and S.C. performed experimental work and analysed data. G.B. and G.V. collected and processed clinical samples and supervised the histopathological analyses. M.C., P.V. and V.G. sorted out clinical data. S.P. and P.P.D.F. designed the study and supervised the project, performed data analysis and wrote the manuscript. All authors were involved in the discussion of results and critical reading of the manuscript.
Funding
AIRC and others
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.02.036.
Contributor Information
Salvatore Pece, Email: salvatore.pece@ieo.it.
Pier Paolo Di Fiore, Email: pierpaolo.difiore@ieo.it.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
References
- 1.Perou C.M., Sorlie T., Eisen M.B. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Beck B., Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13(10):727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 4.Diehn M., Cho R.W., Lobo N.A. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu S., Wicha M.S. Targeting breast cancer stem cells. J Clin Oncol. 2010;28(25):4006–4012. doi: 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tosoni D., Pambianco S., Ekalle Soppo B. Pre-clinical validation of a selective anti-cancer stem cell therapy for numb-deficient human breast cancers. EMBO Mol Med. 2017;9(5):655–671. doi: 10.15252/emmm.201606940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pece S., Tosoni D., Confalonieri S. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140(1):62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Coates A.S., Winer E.P., Goldhirsch A. Tailoring therapies--improving the management of early breast cancer: St Gallen international expert consensus on the primary therapy of early breast Cancer 2015. Ann Oncol. 2015;26(8):1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivshina A.V., George J., Senko O. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66(21):10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 10.Pawitan Y., Bjohle J., Amler L. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loi S., Haibe-Kains B., Desmedt C. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtis C., Shah S.P., Chin S.F. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tukey J.W. Addison-Wesley; 1977. Exploratory Data Analysis. [Google Scholar]
- 14.Hoerl A.E., Kennar R.W. Ridge regression: biased estimation for nonorthogonal problems. Technometrics. 1970;12:55–67. [Google Scholar]
- 15.van Wieringen W.N., Kun D., Hampel R., Boulesteix A.L. Survival prediction using gene expression data: a review and comparison. Comput Stat Data An. 2009;53:1590–1603. [Google Scholar]
- 16.Waldron L., Pintilie M., Tsao M.S., Shepherd F.A., Huttenhower C., Jurisica I. Optimized application of penalized regression methods to diverse genomic data. Bioinformatics. 2011;27(24):3399–3406. doi: 10.1093/bioinformatics/btr591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudis C.A., Barlow W.E., Costantino J.P. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol. 2007;25(15):2127–2132. doi: 10.1200/JCO.2006.10.3523. [DOI] [PubMed] [Google Scholar]
- 18.Kalbfleisch J.D., Prentice R.L. John Wiley & Sons Inc; New York: 1980. The statistical analysis of failure time data. [Google Scholar]
- 19.Goldhirsch A., Winer E.P., Coates A.S. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen international expert consensus on the primary therapy of early breast Cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gyorffy B., Hatzis C., Sanft T., Hofstatter E., Aktas B., Pusztai L. Multigene prognostic tests in breast cancer: past, present, future. Breast Cancer Res. 2015;17:11. doi: 10.1186/s13058-015-0514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Ye C., Lu D. Matrix metalloproteinase-1 expression in breast carcinoma: a marker for unfavorable prognosis. Oncotarget. 2017;8(53):91379–91390. doi: 10.18632/oncotarget.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao H.B., Zhang X.F., Wang H.B., Zhang M.Z. Migration and invasion enhancer 1 (MIEN1) is overexpressed in breast cancer and is a potential new therapeutic molecular target. Genet Mol Res. 2017;16(1) doi: 10.4238/gmr16019380. [DOI] [PubMed] [Google Scholar]
- 23.Moon H.G., Oh K., Lee J. Prognostic and functional importance of the engraftment-associated genes in the patient-derived xenograft models of triple-negative breast cancers. Breast Cancer Res Treat. 2015;154(1):13–22. doi: 10.1007/s10549-015-3585-y. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto A., Hashimoto S., Sugino H. ZEB1 induces EPB41L5 in the cancer mesenchymal program that drives ARF6-based invasion, metastasis and drug resistance. Oncogenesis. 2016;5(9):e259. doi: 10.1038/oncsis.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawson C.D., Der C.J. Filling GAPs in our knowledge: ARHGAP11A and RACGAP1 act as oncogenes in basal-like breast cancers. Small GTPases. 2016:1–7. doi: 10.1080/21541248.2016.1220350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vardabasso C., Hasson D., Ratnakumar K., Chung C.Y., Duarte L.F., Bernstein E. Histone variants: emerging players in cancer biology. Cell Mol Life Sci. 2014;71(3):379–404. doi: 10.1007/s00018-013-1343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns M.B., Lackey L., Carpenter M.A. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494(7437):366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prendergast L., van Vuuren C., Kaczmarczyk A. Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol. 2011;9(6) doi: 10.1371/journal.pbio.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handa H, Hashimoto A, Hashimoto S, Sabe H. Arf6 and its ZEB1-EPB41L5 mesenchymal axis are required for both mesenchymal- and amoeboid-type invasion of cancer cells. Small GTPases 2016: 1–7. [DOI] [PMC free article] [PubMed]
- 30.Celia-Terrassa T., Kang Y. Distinctive properties of metastasis-initiating cells. Genes Dev. 2016;30(8):892–908. doi: 10.1101/gad.277681.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gatza M.L., Silva G.O., Parker J.S., Fan C., Perou C.M. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat Genet. 2014;46(10):1051–1059. doi: 10.1038/ng.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hebert-Chatelain E., Jose C., Gutierrez Cortes N. Preservation of NADH ubiquinone-oxidoreductase activity by Src kinase-mediated phosphorylation of NDUFB10. Biochim Biophys Acta. 2012;1817(5):718–725. doi: 10.1016/j.bbabio.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 33.Dominguez-Sanchez M.S., Saez C., Japon M.A., Aguilera A., Luna R. Differential expression of THOC1 and ALY mRNP biogenesis/export factors in human cancers. BMC Cancer. 2011;11:77. doi: 10.1186/1471-2407-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee A., Ray S. Mutations and interactions in human ERalpha and bZIP proteins: an in silico approach for cell signaling in breast oncology. Gene. 2017;610:90–102. doi: 10.1016/j.gene.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Stefanska B., Cheishvili D., Suderman M. Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clin Cancer Res. 2014;20(12):3118–3132. doi: 10.1158/1078-0432.CCR-13-0283. [DOI] [PubMed] [Google Scholar]
- 36.Medina-Ramirez C.M., Goswami S., Smirnova T. Apoptosis inhibitor ARC promotes breast tumorigenesis, metastasis, and chemoresistance. Cancer Res. 2011;71(24):7705–7715. doi: 10.1158/0008-5472.CAN-11-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.AlHossiny M., Luo L., Frazier W.R. Ly6E/K Signaling to TGFbeta promotes breast Cancer progression, immune escape, and drug resistance. Cancer Res. 2016;76(11):3376–3386. doi: 10.1158/0008-5472.CAN-15-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armengol G., Rojo F., Castellvi J. 4E-binding protein 1: a key molecular "funnel factor" in human cancer with clinical implications. Cancer Res. 2007;67(16):7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 39.Sgroi D.C., Sestak I., Cuzick J. Prediction of late distant recurrence in patients with oestrogen-receptor-positive breast cancer: a prospective comparison of the breast-cancer index (BCI) assay, 21-gene recurrence score, and IHC4 in the TransATAC study population. Lancet Oncol. 2013;14(11):1067–1076. doi: 10.1016/S1470-2045(13)70387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burstein M.D., Tsimelzon A., Poage G.M. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21(7):1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann B.D., Bauer J.A., Chen X. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawson D.A., Bhakta N.R., Kessenbrock K. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526(7571):131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nadji M., Gomez-Fernandez C., Ganjei-Azar P., Morales A.R. Immunohistochemistry of estrogen and progesterone receptors reconsidered: experience with 5,993 breast cancers. Am J Clin Pathol. 2005;123(1):21–27. doi: 10.1309/4wv79n2ghj3x1841. [DOI] [PubMed] [Google Scholar]
- 44.Cardoso F., Harbeck N., Barrios C.H. Research needs in breast cancer. Ann Oncol. 2017;28(2):208–217. doi: 10.1093/annonc/mdw571. [DOI] [PubMed] [Google Scholar]
- 45.Sestak I., Dowsett M., Zabaglo L. Factors predicting late recurrence for estrogen receptor-positive breast cancer. J Natl Cancer Inst. 2013;105(19):1504–1511. doi: 10.1093/jnci/djt244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filipits M., Rudas M., Jakesz R. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin Cancer Res. 2011;17(18):6012–6020. doi: 10.1158/1078-0432.CCR-11-0926. [DOI] [PubMed] [Google Scholar]
- 47.Paik S., Shak S., Tang G. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 48.van 't Veer L.J., Dai H., van de Vijver M.J. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 49.Pan H., Gray R., Braybrooke J. 20-year risks of breast-Cancer recurrence after stopping endocrine therapy at 5 years. N Engl J Med. 2017;377(19):1836–1846. doi: 10.1056/NEJMoa1701830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goss P.E., Ingle J.N., Pritchard K.I. Extending aromatase-inhibitor adjuvant therapy to 10 years. N Engl J Med. 2016;375(3):209–219. doi: 10.1056/NEJMoa1604700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3