Abstract
Background
Colistin resistance mediated by mcr-1-harbouring plasmids is an emerging threat in Enterobacteriaceae, like Salmonella. Based on its major contribution to the diarrhoea burden, the epidemic state and threat of mcr-1-harbouring Salmonella in community-acquired infections should be estimated.
Methods
This retrospective study analysed the mcr-1 gene incidence in Salmonella strains collected from a surveillance on diarrhoeal outpatients in Shanghai Municipality, China, 2006–2016. Molecular characteristics of the mcr-1-positive strains and their plasmids were determined by genome sequencing. The transfer abilities of these plasmids were measured with various conjugation strains, species, and serotypes.
Findings
Among the 12,053 Salmonella isolates, 37 mcr-1-harbouring strains, in which 35 were serovar Typhimurium, were detected first in 2012 and with increasing frequency after 2015. Most patients infected with mcr-1-harbouring strains were aged <5 years. All strains, including fluoroquinolone-resistant and/or extended-spectrum β-lactamase-producing strains, were multi-drug resistant. S. Typhimurium had higher mcr-1 plasmid acquisition ability compared with other common serovars. Phylogeny based on the genomes combined with complete plasmid sequences revealed some clusters, suggesting the presence of mcr-1-harbouring Salmonella outbreaks in the community. Most mcr-1-positive strains were clustered together with the pork strains, strongly suggesting pork consumption as a main infection source.
Interpretation
The mcr-1-harbouring Salmonella prevalence in community-acquired diarrhoea displays a rapid increase trend, and the ESBL-mcr-1-harbouring Salmonella poses a threat for children. These findings highlight the necessary and significance of prohibiting colistin use in animals and continuous monitoring of mcr-1-harbouring Salmonella.
Keywords: Salmonella, mcr-1, Multi-drug resistant, Children, Outbreak, Swine
Research in context.
Evidence before this study
We searched PubMed with the terms “prevalence and mcr-1 and salmonella” for reports published up to January 20, 2019, with no language restrictions. We found 18 results of relevance to this study, but none of these publications reported the prevalence and dissemination of mcr-1-positive Salmonella based on systematic clinical monitoring. The most relevant report detailed the molecular characteristics of mcr-1-positive Salmonella strains isolated from humans in China; however, it focused on a microbiological analysis of the isolates rather than on the prevalence and dissemination of these strains. Furthermore, that study lacked a systemic epidemiological surveillance and reported only two complete plasmid sequences. Thus, our search results indicated that no reports assessing the prevalence of mcr-1-positive Salmonella strains isolated from outpatients have been published previously.
Added value of this study
This study covering the population of a city for 11 consecutive years is the largest-scale study based on laboratory surveillance of community-acquired Salmonella infections. Our surveillance data allowed us to estimate the prevalence rate of mcr-1-harbouring Salmonella strains and revealed that the mcr-1-harbouring Salmonella in this study predominantly belonged to serovar Typhimurium and were mainly isolated from patients aged <5 years old. In addition to short-read sequencing, long-read sequencing with the MinION and PacBio RSII platforms was used to obtain the complete sequences of all 37 mcr-1-carrying plasmids, which enhanced the genome comparisons of the mega-plasmids. Data based on genome-scale comparisons indicate the presence of mcr-1-harbouring Salmonella outbreaks in the community and suggest that pork consumption is likely the contamination source for the epidemic mcr-1-positive S. Typhimurium strains.
Implications of all the available evidence
The present study revealed the low but rapidly increasing prevalence of mcr-1-harbouring Salmonella in community-acquired diarrhoea cases, highlighting mcr-1-harbouring Salmonella as an emerging threat in enteric infection and food safety. Additionally, this work provides a baseline for the prevalence of mcr-1-harbouring Salmonella before colistin withdrawal in China. To assess the effect of prohibiting the use of colistin as a feed additive in China, surveillance on diarrhoeal outpatients should be continued. Additionally, our results show that the spread of mcr-1-harbouring MDR Salmonella poses a threat for young children. Therefore, the risk factors of such infection patterns, as well as the transmission mode(s) and contamination source(s) should be determined through further epidemiological investigations.
Alt-text: Unlabelled Box
1. Introduction
Bacterial drug resistance is an ongoing severe public health problem. This issue concerns not only the bacteria causing nosocomial infections but also the pathogenic and non-pathogenic bacteria that spread in the community. Antibiotics use in hospitals and farms has caused the increasing emergence of multi-drug resistant (MDR, defined as resistant to more than three kinds of antibiotics) and pan-resistant strains, while global trade and travel contribute to their worldwide spread [1]. Additionally, there are fundamental driving forces that aggravate the spread of antibiotic resistance, including the competitive advantage for antibiotic-resistant bacteria under antibiotic pressure and the active horizontal transfer of antibiotic resistance gene-harbouring plasmids among bacteria [2]. In 2016, a plasmid encoding mcr-1-mediated polymyxin resistance in Enterobacteriaceae was newly recognized; its existence presents a severe threat in the global confrontation with antibiotic resistance [3]. MCR-1, coded by the mcr-1 gene, is predicted to be an integral membrane protein with the catalytic activity of phosphoethanolamine transferases [4] that modifies the chemical structure of lipid A via the addition of phosphoethanolamine, resulting in a reduction in the binding affinity of lipopolysaccharide (LPS) to colistin [[3], [4], [5]]. Polymyxins are used as the last resort antibiotics for clinical infections caused by MDR Gram-negative bacteria, especially carbapenem-resistant Enterobacteriaceae [6]; however, colistin use in the farming sector has promoted the selection and transmission of mcr-1 and mcr-like gene-mediated drug-resistant strains [7]. mcr-1-positive plasmids can also carry other antibiotic resistance genes, notably ESBL genes; this can generate resistance to multiple drugs and contribute to the spread of MDR bacteria in human populations [8].
Among the reported bacteria with mcr-1-mediated colistin resistance, most were isolated from animal sources, whereas only a few were isolated from hospital patients with nosocomial infections [9,10]; this uneven distribution is likely due to the asymmetric polymyxin use in these two sectors. Healthy carriers of these bacteria have also been reported [11]. Epidemiological investigations found that most mcr-1-harbouring plasmids appeared to be restricted to two Enterobacteriaceae species (Escherichia coli and Klebsiella pneumoniae). However, mcr-1-harbouring Salmonella have been detected from animals, food, and patients in Europe [12], as well as from swine, poultry, and carriers in China [11,13].
Salmonella are a common concern in food safety, as their frequent transmission from agricultural animals and food to humans causes numerous gastroenteritis cases, and these pathogens are responsible for >600,000 deaths annually [14]. Surveillance in the United States showed that non-typhoid Salmonella infections have been the leading cause of death among foodborne bacterial pathogens, with the highest incidence among children aged <5 years old (69.5 infections per 100,000 children) [15,16]. Furthermore, MDR Salmonella, which are important agents in the transmission of antibiotic resistance genes, have become a severe problem in the animal breeding sector as well as a medical threat to people [17,18].
To estimate the trend and threat of colistin resistance mediated by plasmids in community-acquired Salmonella infections, a population- or outpatient-based continuous epidemiological surveillance is necessary. In Shanghai Municipality in China, a laboratory surveillance project on Salmonella infection has been underway since 2006. Here, we conducted a retrospective study to determine the prevalence of mcr-1-positive Salmonella in community-acquired infections and to ascertain the molecular characteristics of the epidemic mcr-1-harbouring strains.
2. Materials and methods
2.1. Clinic-based salmonellosis surveillance on diarrhoeal outpatients in Shanghai and Salmonella strain identification
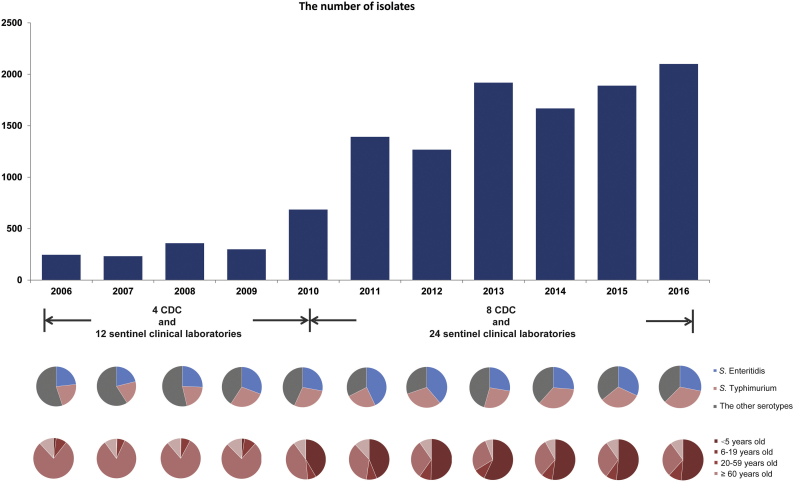
A clinic-based Salmonella infection surveillance of outpatients with diarrhoea was started in Shanghai city in 2006. Shanghai is a municipality in China that had a population of 19,640,000 in 2006 and of 24,200,000 in 2016 (National Bureau of Statistics of China, http://www.stats.gov.cn/tjsj/ndsj/2017/indexch.htm). From April 2006 to June 2010, four public health laboratories in four District Centers for the Disease Control and Prevention (CDC) and 12 sentinel clinical laboratories from 12 general hospitals participated in the project. In July 2010, the number of district public health laboratories was expanded to eight, and the number of sentinel clinical laboratories was increased to a total of 24 (22 general hospitals and two paediatric hospitals) (Supplementary Fig. 1). These hospitals and CDC laboratories covered 12 of the 16 Shanghai districts.
In this Salmonella surveillance, diarrhoea was defined as ≥3 abnormally loose stools in the previous 24 h. Faecal samples were collected from an average of one out of four diarrhoeal outpatients and cultured for Salmonella spp. isolation in accordance with the World Health Organization recommended protocol of Salmonella isolation from stool. From April 2006 to December 2016, a total of 538,852 diarrhoeal outpatients visited the sentinel hospitals; from these outpatients, 134,868 non-duplicate faeces samples were collected, and 12,053 Salmonella spp. strains were isolated. All these strains were serotyped by slide agglutination with commercial antiserum (S&A Reagents Laboratory, Thailand), and 108 serotypes were identified. All strains were preserved in lysogeny broth with 30% glycerol and stored at −80 °C. Basic clinical data were collected on the patients from whom the mcr-1-harbouring strains were isolated, including their age, sex, and date of Salmonella isolation.
2.2. mcr-1 gene screening
All Salmonella strains were recovered from the strain pool and isolated with CHROMagar Salmonella agar (CHROMagar Company, Paris, France). Suspected isolates were identified with the Vitek 2 system (BioMerieux, Lyon, France). The genomic DNA from each of the 12,053 strains was extracted by boiling and freeze-thawing processes, and the resulting supernatant was recovered for use as the PCR template. The mcr-1 gene was detected using real-time PCR assays.
2.3. Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed on the mcr-1-positive isolates for 23 antimicrobial agents (ampicillin (AMP), ampicillin/sulbactam 2:1 ratio (A/S2), tetracycline (TET), nalidixic acid (NAL), ciprofloxacin (CIP), azithromycin (AZI), chloramphenicol (CHL), cefazolin (FAZ), cefoxitin (FOX), ceftazidime (TAZ), cefotaxime (FOT), gentamicin (GEN), trimethoprim/sulfamethoxazole (SXT), cefazolin (CZO), cefuroxime (CXM), cefotaxime (CTX), cefotaxime/clavulanic acid (CTC), ceftazidime (CAZ), ceftazidime/clavulanic acid (CCV), aztreonam (AZT), cefepime (FEP), imipenem (IMI), and polymyxin B (PB)) using the reference broth microdilution method with custom plates (PRCDCN2, Thermo, USA) [19]. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints were used to assess the results. EUCAST defines colistin resistance as >2 μg/mL for Enterobacteriaceae [20].
2.4. Genome and plasmid sequencing
DNA was extracted from each of the mcr-1-positive strains using the Wizard Genomic DNA Extraction Kit (Promega, Madison, USA) and was sequenced using the HiSeq sequencer (Illumina). The plasmid sequences were determined using PacBio RSII (Pacific Biosciences) and MinION (Oxford Nanopore, Oxford, United Kingdom). Plasmid DNA was purified from 100 mL of liquid culture of the strain using the Qiafilter Plasmid max kit (Qiagen) as per the protocol for low-copy plasmids. For the PacBio RSII platform, a 10-kb DNA library was constructed and sequenced using single-molecule real-time sequencing technology. For the MinION platform, the library was prepared using the ONT 1D ligation sequencing kit (SQK-LSK108) with the native barcoding expansion kit (EXP-NBD103). Porechop (https://github.com/rrwick/Porechop) was used to trim off the adaptors from the raw nanopore sequencing reads as well as to split them into different samples. The short reads of <2 kb in length were filtered out for further analysis. We assembled each genome using a combination of short- and long-reads using the SPAdes and Unicycler hybrid assembler [21].
The complete sequences of all 37 detected plasmids have been deposited in GenBank (MH522409–MH522426 and MK477602–MK477620).
2.5. Molecular typing and drug-resistance gene analysis
Determination of the Multi-Locus Sequence Type (MLST), plasmid replicons, and drug-resistance gene content was performed in silico using online tools (http://www.genomicepidemiology.org/).
2.6. Phylogenetic analysis of the genomic sequences
SNP calling was performed using Snippy 3.1, and recombination variants were excluded using ClonalFrameML 1.0 [22]. Phylogenetic trees were constructed by the maximum likelihood method with RAxML (http://phylobench.vital-it.ch/raxml-bb/index.php) based on the recombination-free SNPs. The nucleotide sequences of the common gene regions from each of the selected plasmids were obtained by using cd-hit and blast+; the core genes were then aligned and merged.
To reveal the possible source for the epidemic of mcr-1-carrying S. Typhimurium strains, the 35 mcr-1-harbouring S. Typhimurium strains isolated from humans in this study, another 10 mcr-1-positive S. Typhimurium strains from swine obtained by our team, and 55 mcr-1-negative S. Typhimurium genomic sequences retrieved from GenBank (including 19 from poultry, 11 from swine, and 25 from humans) were included in the phylogenetic analysis.
2.7. Conjugation and transformation analysis
To test the host range and the transferability of each plasmid, conjugation experiments were performed using S. Typhi CT18, Escherichia coli J53 AziR (met pro; azide-resistant), and Klebsiella pneumoniae BJ1988 as recipients. Transfer of the colistin-resistance determinant by conjugation was assayed on LB agar plates (Oxoid) with an initial donor/recipient ratio of 1:1, using E. coli J53 as the recipient. After incubation at 37 °C for 4 h or 24 h, transconjugants were selected on LB agar supplemented with colistin (2 μg/mL) and sodium azide (100 μg/mL). When using S. Typhi CT18 or K. pneumoniae BJ1988 as recipients, the transformants were selected on LB agar with colistin (2 μg/mL) and streptomycin (5000 μg/mL). The transfer frequency is expressed as the number of transconjugants per total recipients. Positive transconjugants were confirmed by real-time PCR targeting the mcr-1 gene. The transfer frequency data were analysed by a multiple comparison with Kruskal-Wallis using the agricolae package in R language. The default alpha parameter is 0.05. Post hoc tests used the criterium for Fisher's least significant difference. The adjustment methods included the Bonferroni correction. The factors used in our analysis were: recipient type (E. coli J53, S. Typhi CT18, and K. pneumoniae BJ1988 strains were tested), replicon sequence type of the mcr-1-positive plasmids (IncI2, IncHI2, and IncX4 plasmids were tested), and transfer time (experiments were conducted for 4 h or 24 h).
To test the ability of the different Salmonella serotype strains to obtain a mcr-1-positive plasmid, conjugation experiments using four S. Derby, four S. Enteritidis, and five S. Typhimurium strains as recipients and using two E. coli J53 carrying the mcr-1-positive IncI2 plasmid, two E. coli J53 carrying the mcr-1-positive IncHI2 plasmid, and one E. coli J53 carrying the mcr-1-positive IncX4 plasmid as donors were performed. In total, 65 independent conjugative experiments were conducted. The resulting transformants were selected on LB agar with colistin (4 μg/mL) and streptomycin (1000 μg/mL). Positive transconjugants were confirmed by real-time PCR. The transfer frequency is expressed as the number of transconjugants per total recipients. The transfer frequency data were analysed as described above. The factors used in our analysis were: recipient strain (four S. Derby, four S. Enteritidis, and five S. Typhimurium strains were tested) and replicon sequence type of the donor mcr-1-positive plasmid (IncI2, IncHI2, and IncX4 mcr-1-positive plasmids were tested).
3. Results
mcr-1-harbouring Salmonella emerged after 2012 and were mainly isolated from the youngest age group of outpatients.
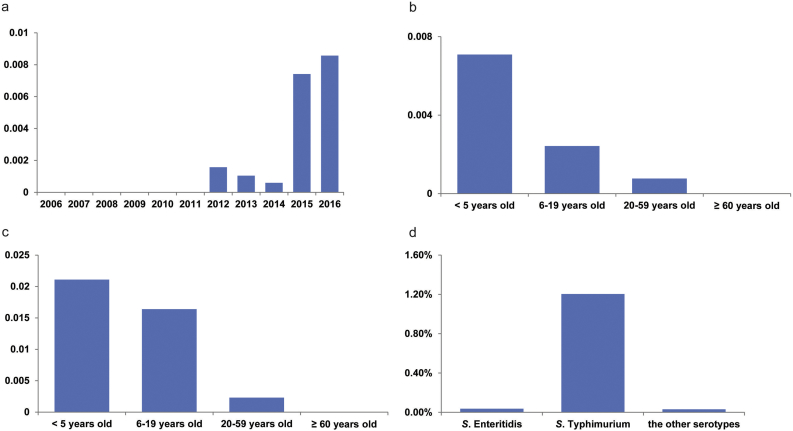
Salmonella infections were surveyed yearly from 2006 in diarrhoeal outpatients in Shanghai. Following the expansion of the surveillance laboratories in June 2010, the number of strains isolated in 2011 increased (Fig. 1). Over the course of the study, 12,053 Salmonella strains were collected, and 37 of these were found to harbour the mcr-1 gene. The minimal inhibitory concentration (MIC) for polymyxin B of each of these strains was 4–8 μg/mL. The first isolation of a mcr-1-positive strain occurred in 2012, and increasing isolation rates were observed in 2015 and 2016 (Table 1, Fig. 2a). After the initial appearance of mcr-1-positive Salmonella in 2012, its prevalence rate through 2016 in Shanghai was 4.2/1000 (37/8842) (Table 1, Fig. 2a).
Fig. 1.
Epidemiological description of the Salmonella isolates used in this study. From top to bottom: (1) the amount of isolates in each year from 2006 to 2016; (2) the serotype composition ratios of S. Typhimurium, S. Enteritidis, and the group of other serotypes in each year; (3) the composition ratios of each age group in each year.
Table 1.
Brief information about the 37 mcr-1-harbouring Salmonella strains identified in this study and their mcr-1 plasmids.
| Strains | Sampling date | Sex of patients | Age of patients | Serotypes | MIC of polymyxin B | Phenotype Resistance to third and fourth-generation cephalosporinsa | Phenotype Resistance to ciprofloxacinb | Levels of antibiotic resistancec | Strategies of sequencing | Replicon sequence type of mcr-1-positive plasmids |
|---|---|---|---|---|---|---|---|---|---|---|
| SH12G402 | 2012/6/7 | female | 1 year | Enterica | 8 | CAZ, CTX, FEP | R | MDR | PacBio RSII | IncI2 |
| SH12G950 | 2012/7/28 | male | 10 months | Typhimurium | 8 | CTX | R | MDR | MinION+HiSeq | IncI2 |
| SH13G1582 | 2013/7/31 | male | 2 years | Wandsworth | 8 | CTX | R | MDR | PacBio RSII | IncI2 |
| SH13G841 | 2013/10/20 | male | 9 months | Typhimurium | 8 | CAZ, CTX, FEP | S | MDR | HiSeq+Sanger | IncI2 |
| SH14G1670 | 2014/6/1 | female | 2 years | Typhimurium | 4 | CTX, FEP | R | XDR | PacBio RSII | IncHI2 |
| SH15G1169 | 2015/8/26 | male | 34 years | Typhimurium | 8 | CTX, FEP | S | MDR | MinION+HiSeq | IncHI2 |
| SH15G1219 | 2015/9/7 | male | 1 year | Typhimurium | 4 | CTX, FEP | R | MDR | MinION+HiSeq | IncHI2 |
| SH15G1397 | 2015/12/10 | male | 8 years | Typhimurium | 4 | CAZ, CTX, FEP | S | XDR | MinION+HiSeq | IncX4 |
| SH15G1428 | 2015/9/7 | female | 1 year | Typhimurium | 8 | CTX, FEP | R | XDR | MinION+HiSeq | IncHI2 |
| SH15G1450 | 2015/9/7 | female | 3 years | Typhimurium | 8 | CTX, FEP | R | MDR | MinION+HiSeq | IncHI2 |
| SH15G1452 | 2015/10/13 | male | 1 year | Typhimurium | 4 | CTX, FEP | R | MDR | MinION+HiSeq | IncHI2 |
| SH15G1493 | 2015/9/28 | male | 4 years | Typhimurium | 8 | CAZ, CTX | R | MDR | MinION+HiSeq | IncI2 |
| SH15G1527 | 2015/10/19 | male | 10 months | Typhimurium | 8 | S | S | MDR | PacBio RSII | IncI2 |
| SH15G1531 | 2015/10/27 | male | 8 months | Typhimurium | 8 | S | S | MDR | HiSeq+sanger | IncI2 |
| SH15G1571 | 2015/11/14 | female | 10 months | Typhimurium | 8 | CTX, FEP | R | MDR | MinION+HiSeq | IncHI2 |
| SH15G1598 | 2015/12/28 | female | 2 years | Typhimurium | 8 | S | R | MDR | MinION+HiSeq | IncHI2 |
| SH15G2167 | 2015/7/15 | female | 9 months | Typhimurium | 8 | S | R | MDR | MinION+HiSeq | IncX4 |
| SH15G2169 | 2015/7/15 | male | 2 years | Typhimurium | 8 | S | R | MDR | MinION+HiSeq | IncX4 |
| SH15G2194 | 2015/10/28 | female | 1 year | Typhimurium | 4 | CAZ, CTX | R | XDR | MinION+HiSeq | IncHI2 |
| SH16G0600 | 2016/5/10 | male | 3 months | Typhimurium | 4 | CTX, FEP | S | MDR | MinION+HiSeq | IncHI2 |
| SH16G0612 | 2016/6/12 | male | 9 months | Typhimurium | 8 | CTX, FEP | S | MDR | MinION+HiSeq | IncHI2 |
| SH16G0648 | 2016/6/17 | male | 1 year | Typhimurium | 4 | CTX | S | MDR | MinION+HiSeq | IncHI2 |
| SH16G1369 | 2016/6/26 | female | 32 years | Typhimurium | 4 | CTX, FEP | S | MDR | MinION+HiSeq | IncHI2 |
| SH16G1394 | 2016/8/1 | female | 2 years | Typhimurium | 4 | CTX, FEP | S | MDR | MinION+HiSeq | IncHI2 |
| SH16G1508 | 2016/7/18 | male | 8 months | Typhimurium | 8 | S | S | MDR | MinION+HiSeq | IncHI2 |
| SH16G1509 | 2016/8/16 | male | 8 years | Typhimurium | 8 | CTX, FEP | S | MDR | MinION+HiSeq | IncX4 |
| SH16G2456 | 2016/6/19 | male | 1 year | Typhimurium | 4 | CTX, FEP | R | XDR | MinION+HiSeq | IncHI2 |
| SH16G2457 | 2016/8/17 | female | 1 year | Typhimurium | 4 | CTX, FEP | R | MDR | MinION+HiSeq | IncHI2 |
| SH16G2543 | 2016/8/14 | male | 2 years | Typhimurium | 4 | S | S | MDR | MinION+HiSeq | IncX4 |
| SH16G4466 | 2016/6/21 | female | 9 months | Typhimurium | 4 | CTX, FEP | S | MDR | MinION+HiSeq | IncHI2 |
| SH16G4498 | 2016/8/23 | male | 10 months | Typhimurium | 4 | CTX, FEP | R | MDR | MinION+HiSeq | IncHI2 |
| SH16G4511 | 2016/7/4 | male | 1 year | Typhimurium | 8 | CTX, FEP | S | XDR | MinION+HiSeq | IncHI2 |
| SH16G4525 | 2016/8/25 | male | 1 year | Typhimurium | 8 | CTX, FEP | R | MDR | MinION+HiSeq | IncHI2 |
| SH16G4909 | 2016/8/10 | male | 3 years | Typhimurium | 8 | CTX, FEP | S | MDR | MinION+HiSeq | IncHI2 |
| SH16G4918 | 2016/8/16 | female | 9 months | Typhimurium | 4 | CTX, FEP | S | MDR | MinION+HiSeq | IncHI2 |
| SH16G4926 | 2016/8/27 | male | 7 months | Typhimurium | 8 | CTX, FEP | S | XDR | MinION+HiSeq | IncHI2 |
| SH16G4928 | 2016/8/31 | male | 1 year | Typhimurium | 4 | CTX | S | MDR | MinION+HiSeq | IncI2 |
Resistance to the third- and fourth-generation cephalosporins, including ceftazidime (CAZ), cefotaxime (CTX), or cefepime (FEP), in the antimicrobial susceptibility testing.
R: resistance to ciprofloxacin; S: sensitivity to ciprofloxacin.
MDR: multidrug-resistant strains, defined as resistant to more than three kinds of antibiotics; XDR: extensively drug-resistant strains, defined as sensitive to less than three kinds of antibiotics.
Fig. 2.
The rate of mcr-1-harbouring isolates grouped by different factors. (a–b) The rates of mcr-1-harbouring isolates among the total Salmonella sp. strains in different years (a) and in different age groups (b). (c) The rates of mcr-1-harbouring S. Typhimurium strains among just the S. Typhimurium isolates in different age groups. (d) The rates of mcr-1-harbouring isolates among the total Salmonella sp. strains in different serotypes.
Among the 37 patients infected with mcr-1-positive Salmonella, 33 (89%) were aged <5 years old (Table 1, Supplementary Fig. 2). The detection rates of mcr-1 strains, both across all Salmonella serotypes and in only serotype Typhimurium, were much lower in the older age groups (Fig. 2b and c).
3.1. Serotypes of the mcr-1-harbouring Salmonella strains had a highly skewed distribution
Among the 37 mcr-1-positive Salmonella strains, three different serotypes were identified, but they showed a highly skewed distribution: S. Typhimurium (35 strains), S. Enteritidis (one strain), and S. Wandsworth (one strain) (Table 1). The mcr-1-positive rate of S. Typhimurium (1.24%) was significantly higher than that of S. Enteritidis (0.04%) (p < .001, chi-squared test) (Fig. 2d), despite similar percentages of the Salmonella strains isolated post-2012 belonging to each of these two serotypes (31.9% for S. Typhimurium and 30.0% for S. Enteritidis).
3.2. All mcr-1-harbouring Salmonella strains were multi-antibiotic resistant and most carried MDR plasmids
Antimicrobial resistance testing was performed on the 37 mcr-1-positive strains. Among them, 30 strains were MDR, and seven were extensively drug-resistant (XDR, defined as sensitive to less than three kinds of antibiotics) (Table 1). Additionally, 29 of these strains were ESBL-producing strains, 32 of them were resistant to fluoroquinolone, and 24 of them were resistant to both cephalosporins and fluoroquinolone; thus, these strains showed severe resistance to the antibiotics commonly used in the treatment of salmonellosis patients.
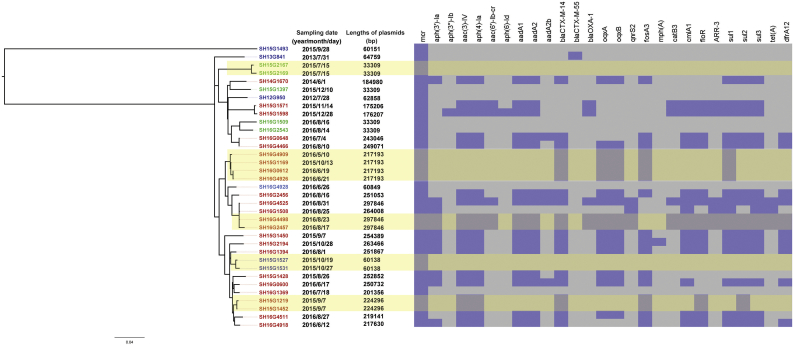
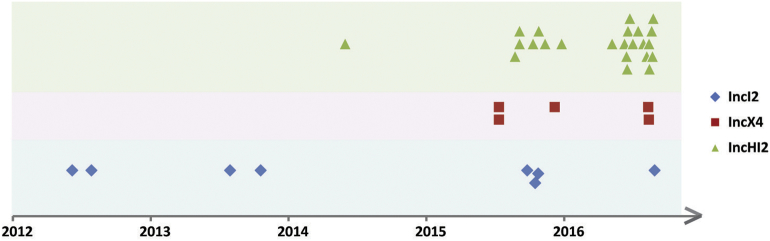
We sequenced the genomes of the mcr-1-positive strains and their mcr-1-harbouring plasmids, which yielded 37 complete mcr-1 plasmid sequences (Table 1). Based on these complete plasmid sequences, the co-existence of various resistance genes in the plasmids and the similarity among the different plasmids could be accurately determined. Among the 37 plasmids, the replicon types IncI2, IncHI2, and IncX4 were detected in 8, 24, and 5 plasmids, respectively (Table 1, Fig. 3). The earliest isolation of a strain carrying the IncHI2 type mcr-1 plasmid occurred in 2014; the detection of strains carrying this plasmid increased rapidly after the summer of 2015, and these strains were commonly isolated in 2015 and 2016 (Table 1, Fig. 3, 4).
Fig. 3.
Phylogenetic analysis of the mcr-1-harbouring S. Typhimurium isolates obtained in this study. From left to right: (1) Maximum likelihood (ML) tree of mcr-1-harbouring S. Typhimurium isolates. The font colour of the labels in the tree represents the replicon sequence type of the mcr-1-harbouring plasmids (blue: IncI2, red: IncHI2, green: IncX4); (2) The sampling dates of the outpatients; (3) The length of the mcr-1-harbouring plasmids; (4) A heatmap of the antimicrobial resistance genes on the mcr-1-harbouring plasmids. The yellowish shade indicates close genomic clonal clusters. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Distribution of the replicon sequence types of mcr-1-harbouring plasmids, based on the sampling dates.
The antibiotic resistance genes in the sequences of these mcr-1-harbouring plasmids were further identified. Twenty-four out of the 25 IncHI2 plasmids carried 4–21 resistance genes other than mcr-1. One of the eight IncI2 plasmids carried the blaCTX-M-55 gene, whereas none of the IncX4 plasmids were found to harbour other antibiotic resistance genes (Supplementary Table 1, Fig. 3). Searching the assembled genome contigs of these 37 strains revealed that 35 carried ESBL genes, including blaCTX-M-3/14/24/55, blaTEM-1B, blaCMY-2, blaDHA-1, and blaOXA-1. Therefore, among these strains, 23 were confirmed to harbour mcr-1 and ESBL genes (blaCTX-M-14/55 and blaOXA-1) on the same plasmid; this group of plasmids harbouring genes for both mcr-1 and ESBL was comprised of one IncI2 and 22 IncHI2 plasmids (Fig. 3, Supplementary Table 1). Additionally, four plasmids simultaneously harboured mcr-1 and qnrS2.
3.3. All the mcr-1 plasmids were transferable, and S. Typhimurium acquired the mcr-1 plasmids more easily compared with other common serotypes
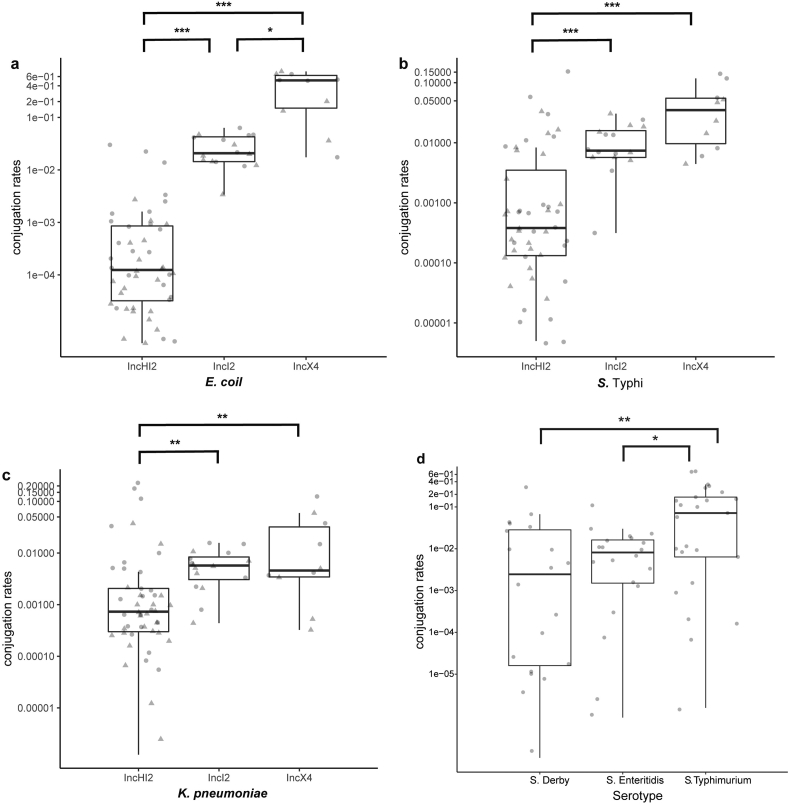
To evaluate the transferability of the mcr-1-harbouring plasmids obtained in this study, conjugation experiments were performed. Transconjugants were obtained from each of the mixtures of the recipient E. coli J53 with one of the 37 mcr-1-positive strains. All the tested transconjugants showed 32- to 64-fold higher polymyxin B MICs compared with that of E. coli J53. The transfer frequency of the mcr-1-positive IncX4 plasmid, which is the main mcr-1 gene carrier in E. coli, was higher than those of the mcr-1-positive IncI2 and IncHI2 plasmids (p < .05, Kruskal-Wallis test; Fig. 5a). All 37 mcr-1-harbouring plasmids were also capable of transferring from the S. Typhimurium strains to S. Typhi CT18 and K. pneumoniae BJ1988, and the transfer frequencies of both the IncX4 and IncI2 plasmids were higher than those of the IncHI2 plasmids (p < .01, Kruskal-Wallis test; Fig. 5b and c). Furthermore, when mcr-1 plasmids were transferred from E. coli to Salmonella spp., the conjugation rates for the S. Typhimurium strains were up to seven or ten times higher than those of S. Derby and S. Enteritidis, respectively (p < .05, Kruskal-Wallis test; Fig. 5d), suggesting that S. Typhimurium is more efficient at acquiring mcr-1-harbouring plasmids.
Fig. 5.
The conjugation rates of the mcr-1-harbouring plasmids isolated in this study.
(a–c) The 37 mcr-1-harbouring Salmonella strains were used as the donors, and E. coli J53 strain (a), S. Typhi CT18 strain (b), or K. pneumoniae BJ1988 strain (c) was used as the recipient. (d) Five E. coli J53 strains (two carrying the mcr-1 IncHI2 plasmid, two carrying the mcr-1 IncI2 plasmid, and one carrying the mcr-1 IncX4 plasmid) were used as the donors, and four S. Derby strains, four S. Enteritidis strains, and five S. Typhimurium strains were used as the recipients. The grey triangles and circles are the log10-transformation of the transfer frequency data after incubation at 37 °C for 4 h and 24 h, respectively.
3.4. Several of the mcr-1-carrying S. Typhimurium strains formed close genomic clonal clusters
To evaluate the genomic clonality and similarity of the 35 mcr-1-harbouring S. Typhimurium strains, their phylogenetic relationships were analysed based on 1231 recombination-free SNPs; these are presented in Fig. 3 as a maximum likelihood tree. Our analysis shows that the ST19 strain SH15G1493 formed a single branch 546 SNPs away from the ST34 strains. The other 34 ST34 strains comprised several different but closely related phylogenetic branches. At least five clonal clusters of strains were identified: SH15G2167/SH15G2169, SH16G4909/SH15G1169/SH16G0612/SH16G4926, SH16G4498/SH16G2457, SH15G1527/SH15G1531, and SH15G1219/SH15G1452 (Fig. 3). In each of these clusters, a limited number of core SNPs (1 to 10) existed in the chromosome genomes, which were each separated from the nearest neighbour isolate by >29 core SNPs. Additionally, the mcr-1-carrying plasmids within each cluster have the same lengths, the same antimicrobial resistance gene composition patterns, a similar presence of coding sequences, and a limited number of variations (0 to 4 SNPs) in the common genes of the plasmids (Fig. 3, 6), strongly supporting a close genetic relationship among the plasmids in each cluster. The intervals between the clinical visits of the corresponding patients were within 40 days for the mcr-1-positive isolates in each cluster, except for SH15G1169 in its cluster. This result could suggest that the strains within each cluster may have had a common source, and the corresponding cases likely had epidemiological associations.
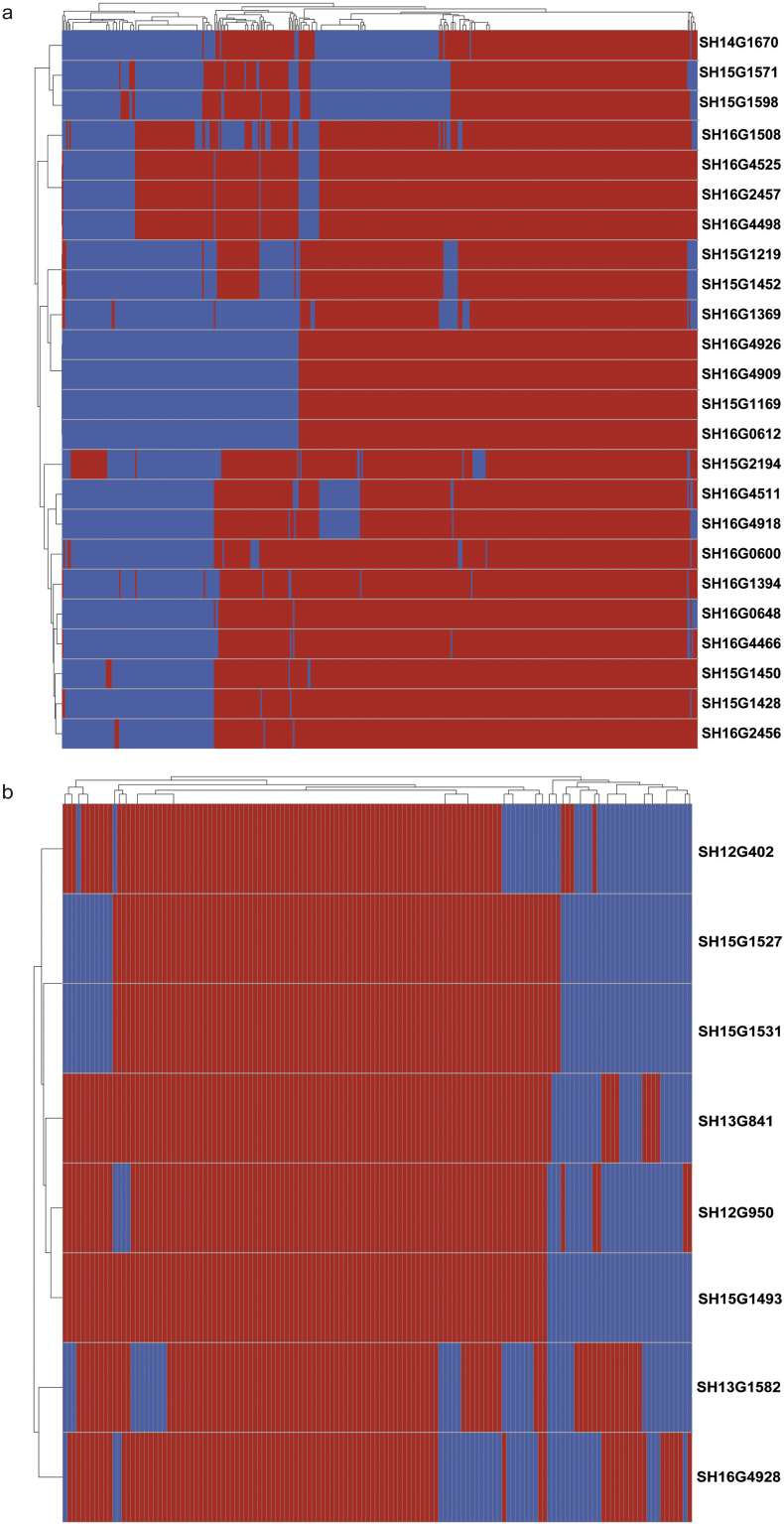
Fig. 6.
Heatmap of mcr-1-harbouring plasmids based on the presence of coding sequences (cds). (a–b) Heatmap of mcr-1-harbouring IncHI2 plasmids (a) and mcr-1-harbouring IncI2 plasmids (b). Red represents the presence of similar cds. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Genomic comparisons reveal a probable swine source for the epidemic of mcr-1carrying S. Typhimurium strains
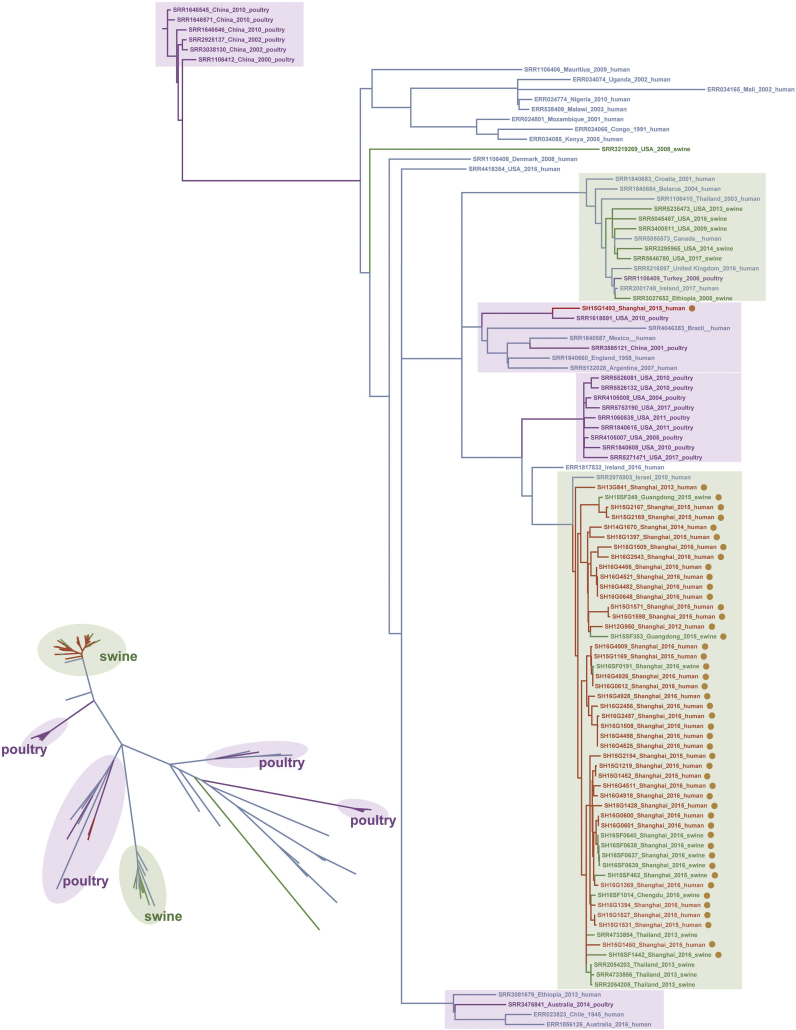
S. Typhimurium is a very common pathogen that can infect humans via the meat products of poultry and livestock. We selected S. Typhimurium strains from human, swine, and poultry sources (including 45 mcr-1-harbouring strains and 55 mcr-1-negative strains) (Supplementary Table 2) and performed a phylogenetic comparison based on the genomic sequences. In total, 7091 recombination-free SNPs were obtained. Among the 35 mcr-1-harbouring S. Typhimurium strains isolated from patients in our study, only one strain, SH15G1493, was clustered with the poultry-source strains, whereas the other 34 strains were clustered in the branch containing swine-source S. Typhimurium strains, particularly those from Thailand and China (Fig. 7). These data suggest the consumption of pork as the likely contamination source.
Fig. 7.
Dendrograms of mcr-1-positive S. Typhimurium isolates from different hosts. In addition to the 35 mcr-1-harbouring S. Typhimurium strains from humans found in this study, another 10 mcr-1-positive S. Typhimurium strains from swine obtained by our team and 55 mcr-1-negative S. Typhimurium genomic sequences retrieved from GenBank (comprised of 19 from poultry, 11 from swine, and 25 from humans) were also included in this analysis. Red: mcr-1-harbouring S. Typhimurium strains isolated from humans; Green: S. Typhimurium strains isolated from swine; Purple: S. Typhimurium strains isolated from poultry; Blue: mcr-1-negative S. Typhimurium strains isolated from humans. Right phylogenetic tree: the strains carrying mcr-1 plasmids are marked with light-brown spots, the branches containing swine-source strains are marked with light green boxes as the background, and the branches containing poultry-source strains are marked with light purple boxes. Bottom-left phylogenetic tree: the branches containing strains from different sources are marked with the same colour format as the phylogenetic tree on the right, to more clearly present the separation of the branches and locations of the mcr-1-harbouring strains. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Diarrhoea caused by Salmonella is a severe foodborne disease in humans. After the first report of mcr-1-mediated colistin resistance in Enterobacteriaceae, Salmonella were also surveyed for the presence of mcr-1-harbouring plasmids. A study in Portugal found a prevalence rate of 1.5% for mcr-1-harbouring Salmonella 1,4,[5],12:i:- strains in 2011–2015 [23], and mcr-1-harbouring Salmonella isolates have been reported in sporadic cases [24], healthy carriers [11], and non-human sources, such as food and farm animals [13]; however, the epidemiology-based prevalence of mcr-1-mediated colistin resistance in Salmonella spp. infecting outpatients remained unclear. Here, our 11-year retrospective analysis of Salmonella from a successive diarrhoeal outpatient surveillance revealed the appearance and increasing prevalence of mcr-1-mediated colistin resistance in community-acquired Salmonella infections. The strains screened in this study were collected via a designed, continuous laboratory-based surveillance; this approach can provide comparable data groups to assess the emergence, positive rates, age-group distribution, and strain-serotype distribution of mcr-1-harbouring Salmonella strains. Our study highlights the emerging problem of mcr-1-carrying Salmonella infection and spread in the community.
We observed that the mcr-1-positive Salmonella strains were recovered from outpatients in Shanghai only after 2012, with the most explosive increase in detection occurring after 2015. This pattern warns of the threat posed by an expansion in the community of Salmonella with mcr-1-mediated colistin resistance. Although the total positive rate of mcr-1-harbouring Salmonella strains in this study was low, our conjugation experiments show that all the plasmids harboured by these Salmonella strains can transfer from/to Salmonella, demonstrating the ability of Salmonella to spread these plasmids as either the recipient or vector. A similar dramatic increase in mcr-1 detection was also found in clinical E. coli isolates from China [25]. Together with our data, these findings may support a common expansion of mcr-1-harbouring strains in humans after 2012 and expose the potential threat presented by Salmonella strains carrying mcr-1 plasmids via their role in transmitting the mcr-1 resistance gene.
Our Salmonella surveillance covered all age groups of outpatients; however, 90% (33/37) of outpatients suffering from infection with mcr-1-harbouring Salmonella were children aged <5 years, revealing a much higher positive rate of mcr-1-harbouring Salmonella in the child group than in the adult group. Diarrhoea is the second leading cause of death in children under five years old and is responsible for killing 2195 children every day worldwide [26]. Furthermore, Salmonella is a prominent cause of bacterial diarrhoea in children aged <5 years. The higher positive rates of mcr-1-harbouring Salmonella in children highlights the threat posed by these pathogens. Notably, 26 of the 33 mcr-1-positive strains isolated in the present study simultaneously carried ESBL genes, which confer resistance to the common antibiotics used for the treatment of diarrhoea in children, such as cephalosporins [27]. Children are particularly vulnerable in MDR Enterobacteriaceae epidemics due to the lack of broad-spectrum antibiotics approved for use in this age group. Here, because of the co-resistance to third-generation cephalosporins and colistin in Salmonella strains isolated from children, the use of cephalosporin may result in treatment failure and enrich the mcr-1-positive strains in gut flora. These data indicate that the infection of children by bacteria with mcr-1-mediated colistin resistance has reached the forefront of the emerging antibiotic resistance problem.
The higher rate of mcr-1-harbouring Salmonella in the low-age group may be related to the diets, intra-familial transmissions, behaviours, and compromised immune systems of children. Previous work has reported some infection risk factors, such as immunosuppression associated with mcr-1-harbouring Enterobacteriaceae infection [25] and mother–child transmission as a potential means of new-born colonization by ESBL-producing strains [28]. Bed and toy sharing in the day-care setting can also be considered as a risk for community transmission of mcr-1-harbouring strains. Additionally, a survey conducted in a paediatric hospital in Hangzhou, a city near Shanghai, found that a high rate (9.8%) of non-diarrhoeal paediatric patients carried mcr-1-harbouring E. coli in their gut flora, and more than half of these strains also carried ESBL genes [29]. This indicates a relatively higher carriage of mcr-1-positive strains in children and may support the possibility of plasmid transfer between bacterial species in the intestine. To monitor the low-age-group infection preference of mcr-1-harbouring Salmonella and identify possible transmission factors, continuous surveillance and detailed epidemiological investigations are needed.
Adult gastroenteritis is usually treated with Norfloxacin, Ciprofloxacin, and Levofloxacin. In this study, most mcr-1-harbouring strains were resistant to fluoroquinolone, and four mcr-1-harbouring plasmids carried qnrS2 genes, with three of these four also simultaneously carrying ESBL genes. Fluoroquinolone-resistant Salmonella were regarded by the WHO in 2017 as priority pathogens for which new antibiotics were urgently needed [30]; therefore, the spread of mcr-1-ESBL-qnrS2 MDR Salmonella may lead to clinical treatment failure. Importantly, although colistin is rarely used in the treatment of salmonellosis, the administration of antibiotics that are used commonly in clinics may unintentionally promote the co-selection and preservation of colistin-resistant Salmonella strains in humans due to the presence of mcr-1-harbouring plasmids in Salmonella strains that are also resistant to other antibiotics.
Here, based on the results of short- and long-read sequencing, we assembled the complete sequences of all 37 mcr-1-harbouring plasmids. The use of complete plasmid sequences ensured accurate analyses of the co-existence of different resistance genes and accurate similarity comparisons between the MDR plasmids. Whole genome sequencing and comparisons among the mcr-1-harbouring plasmids and their hosts verified that a variety of mcr-1-carrying Salmonella clones have spread in the community in recent years. The same or highly similar plasmids were found in host strains belonging to different genomic subclades, showing the broad and active transfer abilities of these plasmids into different host strains. Bacterial genome sequencing can be applied to identifying genetic clusters as well as to detecting outbreaks and tracking sources [31]. Here, some clusters of cases were identified that had extremely similar chromosomal and plasmid sequences, the same antimicrobial resistance patterns, and even similar onset dates; together, these findings strongly suggest that mcr-1-harbouring S. Typhimurium triggered outbreaks and that the rapid expansion of colistin-resistant strains in the community presents a potential public health threat. The early identification of mcr-1-harbouring S. Typhimurium through active surveillance may help prevent their expansion.
Our genomic phylogeny analysis provided evidence of a clonal relationship among the mcr-1-harbouring strains from the outpatients in this study and those from swine. Agricultural animals, particularly pigs, have been singled out as the most likely reservoir for the amplification and spread of Enterobacteriaceae that are resistant to colistin and other antibiotics [[32], [33], [34]]. The same major mcr-1-carrying plasmid types as those in the present study were also detected in isolates from a pig farm located in Shanghai [35]. Therefore, our study provides strong genome epidemiology-based evidence that the consumption of pork is the likely contamination source of mcr-1-harbouring S. Typhimurium.
Since the first description of mcr-1, seven other mcr-like genes (mcr-2 to mcr-8) have been identified [[36], [37], [38], [39], [40], [41], [42]]. Despite the phylogenetic diversity of these mcr-like genes, it seems likely that functional unification occurs among them [7,[43], [44], [45], [46]]. Over the course of our study, several mcr-like genes have been reported in Salmonella, such as mcr-346, mcr-438, and mcr-539. Only the mcr-1 gene was screened in the Salmonella isolates in this study, but adding screening for other mcr-like genes in future work will provide more data for a better overview of the spread of mcr-mediated resistance.
In summary, our study revealed the increasing prevalence of mcr-1-harbouring Salmonella in outpatients with community-acquired diarrhoea and indicated that these strains were also found in potential outbreaks. The skewed distributions of the mcr-1-harbouring strains across outpatient age groups (more prevalent in patients aged <5 years) and Salmonella serotypes (more prevalent in S. Typhimurium) may provide insight into the distinct manner of their transmission; hence, further studies on the epidemiology and microbiology of these strains are needed. Importantly, MDR Salmonella strains that carry a mcr-1-ESBL-qnrS2 plasmid pose threats to both food safety and health management via two factors: 1) the active horizontal transfer of MRD genes in Enterobacteriaceae, and 2) their preservation through co-selection with other antibiotics, even if colistin is not used in treatment. China has taken a first step towards limiting the development of antimicrobial resistance by banning the use of colistin as a feed additive [47], but sustained surveillance needs be conducted to monitor the epidemic trends of Salmonella with mcr-1-mediated colistin resistance in animal products, food, the community, and hospitals and to estimate the effectiveness of control measures in the face of antibiotic selective pressure.
Author contributions
X Lu contributed to all the experiments, data analysis and interpretation, and manuscript writing; M Zeng contributed to the data collection, analysis, and interpretation, study design, and article writing; J Xu contributed to the gene detection, genome sequencing, and plasmid conjugation experiments, data analysis and interpretation, and article writing; H Zhou contributed to the data analysis and interpretation; B Gu, H Jin, and W Xiao contributed to the data collection, analysis, and interpretation; Z Li, W Zhang, and Y Hu contributed the genome sequence analysis and interpretation; X Wang contributed to the gene detection experiments and resistance analysis; B Zhu contributed to the data interpretation and manuscript revision; X Xu contributed to the data collection, analysis, and interpretation and the study design and supervision; B Kan contributed to the study conception, design, and supervision and manuscript revision.
Declaration of interests
We declare no competing interests.
Acknowledgments
Acknowledgments
We thank Bo Li from Beijing Applied Biological Technologies for support on the Nanopore sequence assembly and Katie Oakley from Liwen Bianji, Edanz Editing China for editing the English text of a draft of this manuscript.
Funding
Ministry of Science and Technology of the People's Republic of China and Chinese Center for Disease Control and Prevention.
Funding sources
This work was supported by the National Key Basic Research Program (2015CB554201) from Ministry of Science and Technology of the People's Republic of China; The National Key Research and Development Program (2016YFC1200103) from Ministry of Science and Technology of the People's Republic of China; The Foundation for Young Scholars (2018A103) from Chinese Center for Disease Control and Prevention.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.03.006.
Contributor Information
Xuebin Xu, Email: xuxuebin@scdc.sh.cn.
Biao Kan, Email: kanbiao@icdc.cn.
Appendix A. Supplementary data
Supplementary Tables
Supplementary Figures
References
- 1.Laxminarayan R., Duse A., Wattal C. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013;13(12):1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 2.Laxminarayan R., Heymann D.L. Challenges of drug resistance in the developing world. BMJ. 2012;344:e1567. doi: 10.1136/bmj.e1567. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y.Y., Wang Y., Walsh T.R. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Ye H., Li Y., Li Z. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio. 2016;7(2) doi: 10.1128/mBio.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao R., Hu Y., Li Z. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog. 2016;12(11) doi: 10.1371/journal.ppat.1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaye K.S., Pogue J.M., Tran T.B., Nation R.L., Li J. Agents of last resort: polymyxin resistance. Infect. Dis. Clin. N. Am. 2016;30(2):391–414. doi: 10.1016/j.idc.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Sun J., Zhang H., Liu Y.H., Feng Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018;26(9):794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y.Q., Zhang A.Y., Ma S.Z. Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J. Antimicrob. Chemother. 2016;71(8):2336–2338. doi: 10.1093/jac/dkw243. [DOI] [PubMed] [Google Scholar]
- 9.Huang X., Yu L., Chen X. High prevalence of colistin resistance and mcr-1 gene in escherichia coli isolated from food animals in China. Front. Microbiol. 2017;8:562. doi: 10.3389/fmicb.2017.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian G.B., Doi Y., Shen J. MCR-1-producing Klebsiella pneumoniae outbreak in China. Lancet Infect. Dis. 2017;17(6):577. doi: 10.1016/S1473-3099(17)30266-9. [DOI] [PubMed] [Google Scholar]
- 11.Lu X., Hu Y., Luo M. MCR-1.6, a new MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica Serovar typhimurium isolate from a healthy individual. Antimicrob. Agents Chemother. 2017;61(5) doi: 10.1128/AAC.02632-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doumith M., Godbole G., Ashton P. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother. 2016;71(8):2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 13.Yi L., Wang J., Gao Y. mcr-1-harboring Salmonella enterica serovar typhimurium sequence type 34 in pigs, China. Emerg. Infect. Dis. 2017;23(2):291–295. doi: 10.3201/eid2302.161543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lokken K.L., Walker G.T., Tsolis R.M. Disseminated infections with antibiotic-resistant non-typhoidal Salmonella strains: contributions of host and pathogen factors. Pathog. Dis. 2016;74(8) doi: 10.1093/femspd/ftw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez-Vargas F.M., Abu-El-Haija M.A., Gomez-Duarte O.G. Salmonella infections: an update on epidemiology, management, and prevention. Travel. Med. Infect. Dis. 2011;9(6):263–277. doi: 10.1016/j.tmaid.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Barton Behravesh C., Jones T.F., Vugia D.J. Deaths associated with bacterial pathogens transmitted commonly through food: foodborne diseases active surveillance network (FoodNet), 1996–2005. J. Infect. Dis. 2011;204(2):263–267. doi: 10.1093/infdis/jir263. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez J., Guerra B., Rodicio M.R. Resistance to carbapenems in non-typhoidal Salmonella enterica Serovars from humans, animals and food. Vet. Sci. 2018;5(2) doi: 10.3390/vetsci5020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrijver R., Stijntjes M., Rodriguez-Bano J., Tacconelli E., Babu Rajendran N., Voss A. Clinical microbiology and infection : The official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2017. Review of antimicrobial resistance surveillance programmes in livestock and their meat in Europe, with a focus on antimicrobial resistance patterns in humans. [DOI] [PubMed] [Google Scholar]
- 19.(CLSI) CALSI. M07-A10 . 2015. Methods for Dilution Antimicrobial Susceptibility Test for Bacteria That Grow Aerobically; Approved Standard-Tenth Edition. [Google Scholar]
- 20.Testing. TECoAS . 2016. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 6.0. The Committee. [Google Scholar]
- 21.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13(6) doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Didelot X., Wilson D.J. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 2015;11(2) doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campos J., Cristino L., Peixe L., Antunes P. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill. Bull. Europeen sur les maladies transmissibles = Eur. Commun. Dis. Bull. 2016;21(26) doi: 10.2807/1560-7917.ES.2016.21.26.30270. [DOI] [PubMed] [Google Scholar]
- 24.Torpdahl M., Hasman H., Litrup E., Skov R.L., Nielsen E.M., Hammerum A.M. Detection of mcr-1-encoding plasmid-mediated colistin-resistant Salmonella isolates from human infection in Denmark. Int. J. Antimicrob. Agents. 2017;49(2):261–262. doi: 10.1016/j.ijantimicag.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Tian G.B., Zhang R. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 2017;17(4):390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 26.Liu L., Johnson H.L., Cousens S. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379(9832):2151–2161. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- 27.Hasman H., Mevius D., Veldman K., Olesen I., Aarestrup F.M. Beta-lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in the Netherlands. J. Antimicrob. Chemother. 2005;56(1):115–121. doi: 10.1093/jac/dki190. [DOI] [PubMed] [Google Scholar]
- 28.Lukac P.J., Bonomo R.A., Logan L.K. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in children: old foe, emerging threat. Clin. Infect. Dis. 2015;60(9):1389–1397. doi: 10.1093/cid/civ020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y.Y., Wang Y.L., Sun Q.L. Colistin resistance gene mcr-1 in gut flora of children. Int. J. Antimicrob. Agents. 2017;50(4):593–597. doi: 10.1016/j.ijantimicag.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Cuypers W.L., Jacobs J., Wong V., Klemm E.J., Deborggraeve S., Van Puyvelde S. Fluoroquinolone resistance in Salmonella: insights by whole-genome sequencing. Microb. Genom. 2018;4(7) doi: 10.1099/mgen.0.000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popovich K.J., Snitkin E.S. Whole genome sequencing-implications for infection prevention and outbreak investigations. Curr. Infect. Dis. Rep. 2017;19(4):15. doi: 10.1007/s11908-017-0570-0. [DOI] [PubMed] [Google Scholar]
- 32.Nordmann P., Poirel L. Plasmid-mediated colistin resistance: an additional antibiotic resistance menace. Clin. Microbiol. Infect. Off. Pub. Eur. Soc. Clin. Microbiol. Infect. Dis. 2016;22(5):398–400. doi: 10.1016/j.cmi.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Olaitan A.O., Thongmalayvong B., Akkhavong K. Clonal transmission of a colistin-resistant Escherichia coli from a domesticated pig to a human in Laos. J. Antimicrob. Chemother. 2015;70(12):3402–3404. doi: 10.1093/jac/dkv252. [DOI] [PubMed] [Google Scholar]
- 34.Hald T., Lo Fo Wong D.M., Aarestrup F.M. The attribution of human infections with antimicrobial resistant Salmonella bacteria in Denmark to sources of animal origin. Foodborne Pathog. Dis. 2007;4(3):313–326. doi: 10.1089/fpd.2007.0002. [DOI] [PubMed] [Google Scholar]
- 35.Wu R., Yi L.X., Yu L.F. Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro. Front. Microbiol. 2018;9:331. doi: 10.3389/fmicb.2018.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xavier B.B., Lammens C., Ruhal R. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro surveillance: bulletin Europeen sur les maladies transmissibles = Eur. Commun. Dis. Bull. 2016;21(27) doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 37.Yin W., Li H., Shen Y. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio. 2017;8(3) doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carattoli A., Villa L., Feudi C. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro surveillance: bulletin Europeen sur les maladies transmissibles = Eur. Commun. Dis. Bull. 2017;22(31) doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borowiak M., Fischer J., Hammerl J.A., Hendriksen R.S., Szabo I., Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017;72(12):3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 40.AbuOun M., Stubberfield E.J., Duggett N.A. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 2018;73(10):2904. doi: 10.1093/jac/dky272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y.Q., Li Y.X., Lei C.W., Zhang A.Y., Wang H.N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018;73(7):1791–1795. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Wang Y., Zhou Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microb. Infect. 2018;7(1):122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng Y. Transferability of MCR-1/2 polymyxin resistance: complex dissemination and genetic mechanism. ACS Infect. Dis. 2018;4(3):291–300. doi: 10.1021/acsinfecdis.7b00201. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H., Hou M., Xu Y. Action and mechanism of the colistin resistance enzyme MCR-4. Commun. Biol. 2019;2:36. doi: 10.1038/s42003-018-0278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y., Wei W., Lei S., Lin J., Srinivas S., Feng Y. An evolutionarily conserved mechanism for intrinsic and transferable polymyxin resistance. mBio. 2018;9(2) doi: 10.1128/mBio.02317-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y., Zhong L.L., Srinivas S. Spread of MCR-3 colistin resistance in China: an epidemiological, genomic and mechanistic study. EBioMedicine. 2018;34:139–157. doi: 10.1016/j.ebiom.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lingxian Y., Yiyun L., Renjie W., Zisen L., Jian-Hua L. Research progress on the plasmid-mediated colistin resistance gene mcr-1. Yi chuan = Hereditas. 2017;39(2):110–126. doi: 10.16288/j.yczz.16-331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables
Supplementary Figures