Abstract
Background
Genome-scale metabolic models (GEMs) offer insights into cancer metabolism and have been used to identify potential biomarkers and drug targets. Drug repositioning is a time- and cost-effective method of drug discovery that can be applied together with GEMs for effective cancer treatment.
Methods
In this study, we reconstruct a prostate cancer (PRAD)-specific GEM for exploring prostate cancer metabolism and also repurposing new therapeutic agents that can be used in development of effective cancer treatment. We integrate global gene expression profiling of cell lines with >1000 different drugs through the use of prostate cancer GEM and predict possible drug-gene interactions.
Findings
We identify the key reactions with altered fluxes based on the gene expression changes and predict the potential drug effect in prostate cancer treatment. We find that sulfamethoxypyridazine, azlocillin, hydroflumethiazide, and ifenprodil can be repurposed for the treatment of prostate cancer based on an in silico cell viability assay. Finally, we validate the effect of ifenprodil using an in vitro cell assay and show its inhibitory effect on a prostate cancer cell line.
Interpretation
Our approach demonstate how GEMs can be used to predict therapeutic agents for cancer treatment based on drug repositioning. Besides, it paved a way and shed a light on the applicability of computational models to real-world biomedical or pharmaceutical problems.
Keywords: Genome-scale metabolic models, Drug repositioning, Drug repurposing, Prostate cancer, Approved drugs
Research in context.
Evidence before this study
Genome-scale metabolic models (GEMs) have been used to study metabolism in clinically relevant applications such as stratifying the patients, determining pivotal mediator genes or biological pathways in disease state and discovery of potential biomarkers and drug targets. GEMs also offer the opportunity for prediction of relevant drug effect or side effect for a target cell or tissue.
Added value of this study
In this study, we first reconstructed PRAD-specific GEM by integrating tissue-specific proteomics and transcriptomics data. After interpretation of drug-induced transcriptomics data through ConnectivityMap2, we predicted novel gene-drug interactions to reveal drug off-targets. To the best of our knowledge, this study is the first GEM application that used drug repositioning and in silico cell viability assay, simultaneously. We also performed in vitro cell viability assay to show inhibitory effect of ifenprodil as a novel drug candidate for prostate cancer treatment.
Implications of all the available evidence
We demonstrated that GEMs can be used as promising tools for real-world biomedical and pharmaceutical applications. Organ- or tissue-specific constraint-based models might be a crucial facet to determine drug effects in treatment of different diseases. Not only approved drugs, abandoned or withdrawn drugs might be used in this application for novel drug repositioning candidates for different diseases.
Alt-text: Unlabelled Box
1. Introduction
Prostate cancer (PRAD) is one of the most prevalent cancers without the distinction of gender and it is the second-most prevalent cancer among men, according to the latest worldwide cancer statistics [1]. Recently, PRAD has been reported in the United States as the first most common cancer type in estimated new cases and the second most common cancer type leading to deaths after lung and bronchus cancer in men [2].
Genome-scale metabolic models (GEMs) have been used to study cancer metabolism using either generic/personalized or tumor/cell-specific types, which might translate into clinically relevant applications [3,4]. These tools can also be used to identify drug targets leading to inhibition of cancer-related phenotypes or prediction of drug resistance in cancer therapy [5,6].
GEMs can be used as powerful tools to stratify the patients or determine pivotal mediator genes in cancer metabolism, intertwining biological pathways, and metabolites that supply energetic and biosynthetic demands in cancer cells [7,8]. Additionally, GEMs can be applied to identify novel targets and drugs and to create and/or test the hypotheses in the context of drug discovery or drug repositioning [9,10]. The prediction power of GEMs in determination of adverse drug effects and critical metabolic reactions using machine learning approaches was also investigated [11]. Emphasis on organ- or tissue-specific constraint-based models might be a crucial facet in obtaining more accurate and relevant drug effect or side effect predictions for the target tissue [12,13]. In describing the metabolic organization consisting of different components such as enzymes, metabolites, etc., GEMs offer the opportunity for dynamic assessment of metabolism to investigate drug action and even side effects using various computational modeling methods [14]. Since the metabolic alterations have the utmost importance in diagnosis or treatment of PRAD, regardless of disease stage [15], GEMs are also powerful tools for PRAD studies as well.
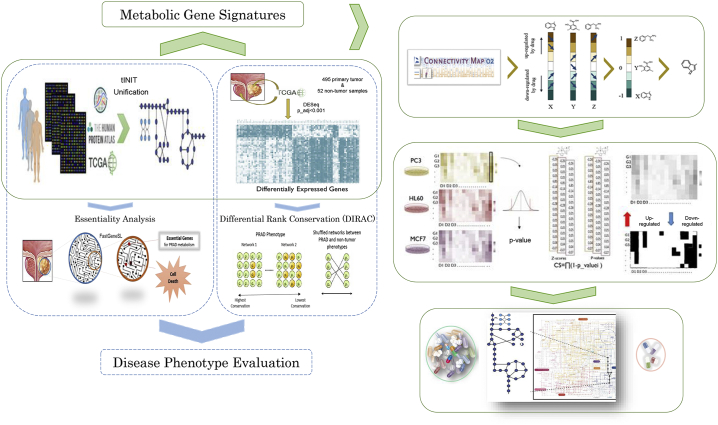
In this study, we reconstructed a disease-specific GEM for PRAD by combining personalized GEMs (n > 450) and prostate cancer-specific proteomics data. Subsequently, the representative potential of the metabolic model for the disease phenotype was evaluated through essentiality analysis applied to the metabolic model and evaluation of transcriptome data to explore tightly connected pathways via differential rank conservation (DIRAC) [16] analysis. After interpretation of the metabolic phenotype of the disease, we determined the metabolic gene signatures for drug repositioning. Gene expression data from drug-perturbed cancer cell lines was acquired from ConnectivityMap2 (CMap2) [17] and recapitulated to reveal candidate drugs that might reverse metabolic gene expressions and predict gene-drug interactions. In addition, drug action simulations were performed via reconstructed PRAD-specific GEM and resulted in four drugs repurposed for the treatment of prostate cancer based on in silico cell viability assays. We validated one of the candidate drugs in vitro and showed that ifenprodil is a promising candidate that has an inhibitory effect on the PC3 cancer cell line (Fig. 1).
Fig. 1.
Identification of potential therapeutic targets in PRAD using in silico modeling and drug signatures. We performed two independent analyses for disease phenotype evaluation. Reconstructed models were applied to explore essential genes for PRAD metabolism. Gene expressions from tumor and nontumor tissues were used to find tightly regulated pathways in prostate tumors through GEMs (blue dashed boxes). We subsequently selected metabolic gene signatures based on the differentially expressed genes in the reconstructed model for drug repositioning. Drugs that show reversal of gene expression for tumor vs. nontumor tissues were identified together with predicted gene-drug interactions from CMap2. Drugs with reversal effects and their predicted gene partners were implemented into GEMs as in silico viability tests and further selected for experimental validation due to their in silico growth-inhibiting effects (green boxes). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2. Results
2.1. Reconstructed PRAD-specific GEM comprises individual transcriptome and tissue-specific proteome data
Significant heterogeneity among prostate tumors might hinder characterization of PRAD and its biological pathways compared with the noncancerous prostate. Therefore, we reconstructed a consensus PRAD-specific GEM based on >450 personalized PRAD GEMs that contains all metabolic reactions and respective gene expressions in prostate cancer patients.
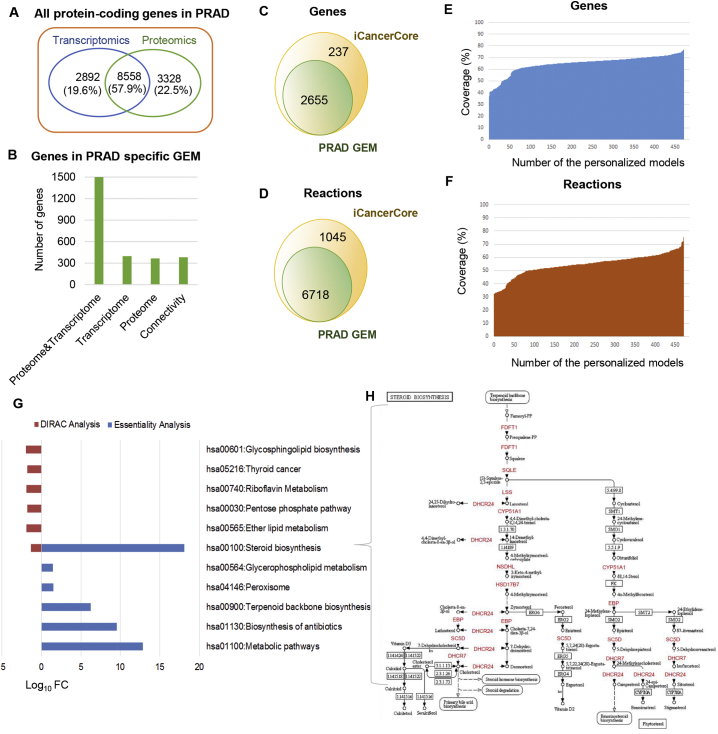
After following the existing RNA-seq data analysis protocol [8], PRAD tissue proteome from the Human Protein Atlas (HPA) [18] was used to make the model more representative and robust. The expression profiles for 14,778 protein-encoding genes were obtained either at the transcriptome and/or proteome levels. In addition, the expressions of 8558 (57.9% of all protein-coding genes) genes were detected in PRAD at both the transcriptome and proteome levels (Fig. 2A). A total of 3328 proteome-based and 2892 transcriptome-based signatures were also included in the model reconstruction practice as input (Fig. 2A). As a result, the PRAD-specific GEM consists of 2655 genes and 6718 reactions, which represent individual variations in PRAD metabolism. The majority of the genes in the reconstructed model was incorporated in both proteomic and transcriptomic data (Fig. 2B). iCancerCore [18] is designed as a generic cancer metabolism model, and a PRAD-specific GEM was reconstructed based on this generic model. The generic and PRAD-specific models share common signatures for 2655 genes and 6718 reactions (Fig. 2C–D). However, the personalized PRAD GEMs retrieved from the Human Pathology Atlas have different numbers of genes (978–2031) and reactions (2179–5053). The PRAD-specific GEM covers all genes and reactions represented in personalized models, but each personalized model might represent 37 to 77% of genes and 33 to 76% of reactions in the PRAD-specific GEM (Fig. 2E–F). These results recapitulated the cancer heterogeneity, as previously shown [18].
Fig. 2.
In silico modeling highlights steroid biosynthesis as a key pathway in PRAD. A. Number of protein-coding genes attained from transcriptomics data via TCGA (blue), and proteomics data from HPA (green), which were integrated during model reconstruction. Genes were excluded when not detected at the protein level or with FPKM < 1. B. Metabolic genes with evidence at the protein or transcriptomic levels and connectivity genes required to achieve metabolic feasibility (see Methods). C and D. Number of genes (C) and reactions (D) in PRAD-specific GEM and iCancerCore model. E and F. Coverage of PRAD-specific GEM in terms of gene (E) and reaction (F) numbers. G. Pathway-level fold changes (FC) after DIRAC analysis (tumors vs. nontumor models) and DAVID functional enrichment analysis for essential genes (p-value < 0.05). H. KEGG steroid biosynthesis pathway highlighting that 11 out of 23 essential genes (red) are found. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. Network-based independent analyses of PRAD metabolism consistently highlight altered lipid metabolism/steroid biosynthesis pathways
To reveal the global biological differences between PRAD tumors and nontumor prostate tissue, we first identified differentially expressed genes (DEGs) using differential expression sequencing (DESeq) and found a total of 3776 DEGs (adj. p-value < 0.001), of which 2266 genes were downregulated and 1510 genes were upregulated (Table S1).
To reveal the alterations in metabolism, we also identified reporter metabolites using the network topology supplied by the reconstructed PRAD-specific GEM. The reporter metabolites algorithm was developed to detect the hotspot of metabolism associated with up- or downregulated genes between two phenotypes [19]. Simulations resulted in 86 upregulated metabolites and 76 downregulated metabolites regardless of their cellular compartments (Table S2). As an example of upregulated reporter metabolites, folate moderates the invasiveness of PRAD cells [20], and elevated physiological levels of folic acid stimulate both growth and invasiveness of various prostate cancer cell lines [21] in parallel with our results. Another upregulated metabolite, prostaglandin E2, is a bioactive lipid produced from arachidonic acid by cyclooxygenase enzymes (e.g., COX2) and has a positive correlation with prostate tumor growth and angiogenesis [22]. Moreover, other reporter metabolites involved in fatty acid biosynthesis, primary bile acid biosynthesis, fatty acid elongation in mitochondria, one carbon pool affected by folate, and calcium signaling pathways were associated with upregulated genes in PRAD. Notably, reporter metabolites such as 4-alpha-methylzymosterol, 14-demethyllanosterol, 4-alpha-methylzymosterol-4-carboxylate and 4,4-dimethyl-5alpha-cholesta-8-en-3beta-ol participating in steroid biosynthesis were associated with the downregulated genes in PRAD as well as purine-pyrimidine metabolism, nicotinate and nicotinamide metabolism (Table S2).
In addition to the frequently used analyses (i.e., differential gene expression analysis and determination of reporter metabolites), we assessed the reconstructed metabolic model with two network-associated analysis frameworks to evaluate prostate cancer metabolism in detail.
First, we performed essentiality analysis (Edwards and Palsson, 2000) in which GEMs are used to determine essential genes or reactions whose knock-out or blocking have disabling effects on a specific biological function [23]. The objective function of the metabolic model was set to biomass maximization to find which genes are essential for tumor growth. In this study, we determined 23 essential genes (Table S3) by implementing logic transformation of the model and the FastGeneSL function [23] with the reconstructed PRAD-specific model. These essential genes were predicted as key genes with an inhibitory effect on tumor growth regardless of differential expression. Functional enrichment analysis (Table S3) performed for essential genes indicates that most were enriched in the steroid biosynthesis pathway (Fig. 2H), including genes associated with zymosterol biosynthesis (FDFT1, SQLE, LSS, CYP51A1, NSDHL, HSD17B7, DHCR24) and cholesterol production (DHCR24, DHCR7, SC5D, EBP). MVD, HMGCR, MVK and PMVK are genes enriched in both terpenoid backbone biosynthesis and in the mevalonate pathway.
Second, we used the DIRAC algorithm [16], which allows assessment of the combinatorial interactions for quantification of various biological pathways in a comparative sense at the population level to quantify conservation differences between networks for two phenotypes, including tumor and nontumor samples. The analyses revealed that the lipid metabolism (and more specifically glycosphingolipid biosynthesis, ether lipid metabolism and steroid biosynthesis) is variably expressed in PRAD compared with noncancerous prostate tissue. Additionally, riboflavin metabolism, pentose phosphate pathway, and thyroid cancer were emphasized in tumor samples (Fig. 2G).
These observations highlighted the steroid biosynthesis pathway as a mutual result related to lipid metabolism. This pathway has utmost importance not only for normal prostate development but also for prostate cancer progression [24]. Consistent results supplied from different analyses revealed the representative functionality of the reconstructed metabolic model. Previous studies also showed that a significant proportion of these genes leading to cholesterol and steroid biosynthesis were abundant in the peripheral zone of prostate tumors, where prostate tumors primarily arise [25,26]. In addition to consistency of the inter- and intra- studies of PRAD metabolism, these key genes and their associated pathways offer an opportunity for drug discovery or drug repositioning. For instance, lipid-lowering drugs such as statins showed a beneficial effect in reducing PRAD-related mortality as well as overall PRAD risk [14].
2.3. Elucidation of metabolic gene signatures to discover drugs with reversal expression effect
Metabolic gene signatures were common gene signatures if considering the intersection of genes in the reconstructed GEM and DEG list, and this group consists of 274 up- and 367 downregulated metabolic genes.
Investigation of these genes in the KEGG pathways shows that they tend to be enriched (p-value < 0.05) in lipid-specific pathways (e.g., sphingolipid, ether lipid, glycerophospholipid, and glycerolipid metabolism) and in biosynthesis of antibiotics and peroxisome, similar to the observations elicited by DIRAC and the essentially analysis above. In addition, these genes are also involved in other pathways such as glutathione, β-alanine, purine metabolism, arginine-proline metabolism, mucin type O-glycan biosynthesis and drug metabolism-cytochrome P450 (Table S4). These results indicate that despite representing a subset of all protein-coding genes, the metabolic genes found in the PRAD GEM are still able to capture the disease metabolism and key metabolic pathways for therapeutic targeting.
The metabolic gene signatures were used as input to query CMap2 for elucidating the differences and similarities between drug-induced expression profiles and disease expression. We queried the CMap database and identified drugs that showed correlated action mechanisms with the metabolic gene signatures in either direction. However, we considered the drugs with negative similarity scores because these drugs show reversal effects on gene expression in PRAD. As a result, we identified 81 significantly associated drugs (permutated p-value < 0.05), of which 43 drugs present as drug candidates with reversal effects of metabolic gene signatures (Table S5).
2.4. Identification of potential repositioning drugs for PRAD treatment based on perturbagen analysis
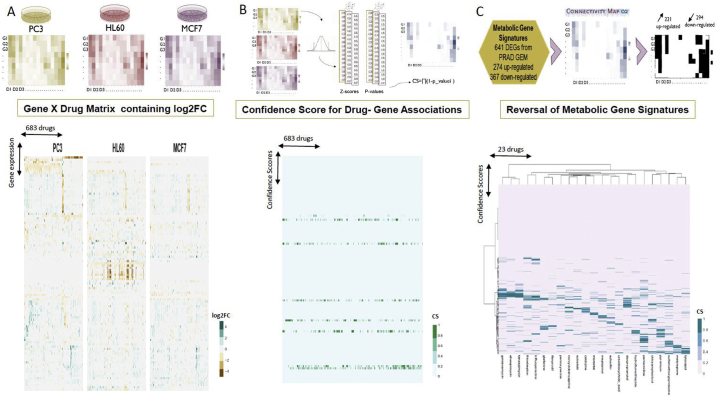
Seeking to further filter the predicted drugs and determine drug-gene interactions as well as the drug effects on gene expression, we used the drug-perturbed gene expression data of three cell lines (PC3, HL60, and MCF7). After data harmonization and processing (see Methods), we determined the Log2 fold changes (Log2FC) in response to drugs for approximately 12,300 genes in 683 drugs in three cell lines (Fig. 3A). We used the 3 cell lines since we were seeking to have a more robust response to the drugs, thus decreasing the potential false positives that would stem from using only one cell line - i.e. genes that would show a consistent response to the same drug, regardless of embryonic cell origin. Thus, genes whose Log2FC in response to the same drug differed in 2 cell lines were discarded. Because similar gene responses to drugs may nevertheless occur, we further combined the data from each cell line to calculate the confidence scores (CS) for each drug-gene interacting pair (Fig. 3B). As a proof of concept, a previous study showed that over 70% of drug-induced gene expressions were mutual for multiple cell lines [63]. Although the direction of the expressions was similar, the calculated CS values approximating 1 were not abundant. The number of genes with CS >0.5 for each drug was between 24 and 97 out of 12,228. Mapping of metabolic signatures on the drug-gene matrix containing the confidence score yielded an interaction matrix of 515 genes with 683 drugs (Fig. 3C; Table S6). After processing (see Methods), we selected gene-drug interactions with CS > 0.5 as those capable of altering the reaction fluxes in the metabolic model.
Fig. 3.
Integration of computationally predicted potential therapeutic targets and drug-gene association profiles that induce reversal of metabolic signatures predict 23 drugs for treatment of PRAD. A. Heatmaps showing gene expression responses to drugs (Log2FC) generated for 3 cell lines (PC3, HL60, MCF7) to identify common drug responses across cell lines. This analysis resulted in 683 drugs found in the 3 cell lines after quality control. Only genes with mean absolute log2FC > 0.2 are shown. B. Confidence scores were calculated for each drug-gene association in each cell line. Z-scores were computed, and the p-values were integrated by the naïve Bayesian formula. C. Confidence scores for interactions among 23 drugs with reversal metabolic effect and 515 of metabolic gene signatures.
Drug targets of the purposed interactions in this study share a similar protein family with known anti-cancer drug targets such as solute carrier (SLC) transporters, growth factors or enzymes such as carbonic anhydrases and glutathione S-transferases.
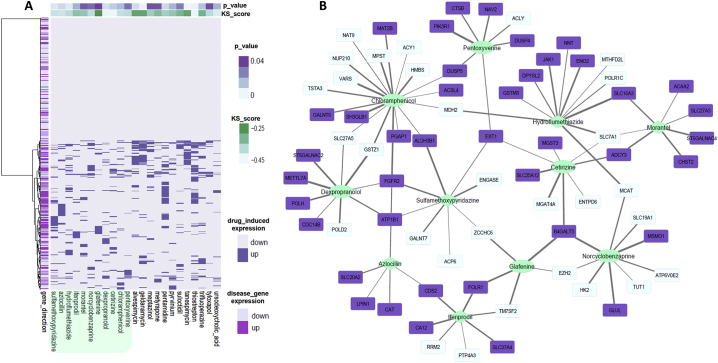
SLCs are transporter membrane proteins that are primarily involved in the uptake of the drug and thus might influence chemotherapy efficacy and/or toxicity. The members of the SLC protein family showed substantially different expression profiles in cancer cells compared with healthy cells, and in this manner, they are discussed as potential drug targets in cancer therapy [27]. Among all identified targets (CS > 0.5), 10% are from SLC protein family. For instance, SLC19A1 is one of the major facilitative folate and thiamine transporters [28] and is a predicted target of norcyclobenzaprine (Fig. 4). Carbonic anhydrases (CAs, EC 4.2.1.1), another predicted drug target group, were previously used to treat several diseases such as glaucoma, obesity, infections, and cancer and especially applied to overcome chemoresistance [29]. To the best of our knowledge, carbonic anhydrase CA1 has been mentioned as a potential plasma biomarker in prostate cancer diagnosis [30] but not as a drug target for treatment. This study proposed that CA12 interacts with ifenprodil. Fibroblast growth factor receptor (FGFR) was associated with azlocillin, sulfamethoxypyridazine, chloramphenicol, and dexpropranolol in the study (Fig. 4). FGFR is one of the studied drug targets for multiple cancer types [31]. The involvement of FGF/FGFR pathways in prostate cancer and their associations with angiogenesis and disease progression offer a rationale for FGFR as potential therapeutic target. FGF/FGFR pathways targeting dovitinib and nintedanib are two small-molecule tyrosine kinase inhibitors in clinical development for advanced prostate cancer [32]. Glutathione S-transferases (GSTs, EC 2.5.1.18) are represented by GSTM3, GSTM4, GSTZ1 and MGST3 in drug targets (Table S7). Different GSTs polymorphisms and overexpression have already been identified as potential biomarkers in different cancers, including fatal prostate cancer [33]. Recently, GSTM3 polymorphism has been observed in prostate cancer patients and associated with resistance to hormonal therapy through oxidative stress [34]. Genetic polymorphisms of GSTs and their functional reflections in cancer patients were correlated with chemotherapeutic efficacy. Therefore, GSTs have gained importance as candidate targets for design of specific drugs to solve the drug resistance problem [35,36]. Because the overexpression of GSTs and efflux pumps cause increased discharge of anti-neoplastic drugs, the use of GST inhibitors such as ethacraplatin, ethacrynic acid and analogs, auranofin or piperlongumine might address drug resistance and increase anti-cancer drug sensitivity in cancer cells. Another opportunity for cancer therapy is the use of prodrugs such as canfosfamide, nitric oxide prodrugs, metformin and doxorubicin analogs [37].
Fig. 4.
Candidate drugs and their interacting genes that induce reversal effects in PRAD expression. A. Direction of 515 gene expressions induced by 23 drugs and drug-associated KS similarity score and p-value after random permutation testing. A total of 11 of candidate drugs, which have not been previously associated with PRAD cases, are highlighted (turquoise). B. Drug-gene association network composed of 11 candidate drugs and 71 genes with CS >0.5 and reversal expression patterns. Genes are highlighted if their expression is upregulated by the drug but downregulated in disease (purple) or vice versa (light blue). Edge represents confidence scores from 0.5 to 1. Several drugs (ifenprodil, azlocillin, sulfamethoxypyridazine, hydroflumethiazide, morantel) show inhibitory effect after in silico cell viability testing (diamonds). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.5. In silico cell viability test for each drug through GEMs
To test the drugs with reprogramming of PRAD metabolism back to nontumor phenotype, we sought to identify drugs that could reverse the expression effects in tumor vs. nontumor tissues as well as their drug-interacting gene partners. An examination of 683 drugs from the drug-gene interaction matrix and 43 drugs that show significant reversal of gene expression effects based on the modified KS-test and random permutations resulted in 23 drugs (Table S5; Table 1). We manually reviewed these candidate drugs on PubMed to find the association of drugs with prostate cancer. The literature review showed that 12 of 23 drugs have been already associated or tested for treatment of PRAD, further reinforcing our confidence in our observations. We considered these results as positive controls for the following analyses. The remaining 11 drugs have not been previously identified as anti-cancer agents. Among these 11, various drug groups were identified, including anti-bacterial, anti-helminthic and anti-protozoal agents, diuretics, opioids, β-adrenergic antagonist and NMDA receptor inhibitor types. For further investigation, we focused on 94 interactions among 11 PRAD non-associated drugs and 71 genes with reverse directionality of expression (Fig. 4).
Table 1.
In silico cell viability test results of 23 drugs which were predicted to reverse gene expression in prostate cancer regarding KS similarity score and permutation-based p-value from CMap2.
| Drugs | Permutation-based p-value | Similarity Score | References |
In silico Cell viability |
|---|---|---|---|---|
| Thiostrepton | 0.001 | −0.488 | [39] | ✖ |
| Geldanamycin | 0.001 | −0.283 | [64] | ✔ |
| Pentamidine | 0.008 | −0.383 | [65] | ✔ |
| Trifluoperazine | 0.014 | −0.317 | [66] | ✔ |
| Tanespimycin | 0.000 | −0.281 | [66] | ✔ |
| Meptazinol | 0.041 | −0.377 | [41] | ✖ |
| Alvespimycin | 0.008 | −0.280 | [67] | ✔ |
| Suloctidil | 0.031 | −0.324 | US 2006/0009506A1 | ✔ |
| Pyrvinium | 0.018 | −0.362 | [68] | ✔ |
| Metyrapone | 0.040 | −0.338 | [38] | ✖ |
| Tyloxapol | 0.041 | −0.409 | [69] | ✔ |
| Ursodeoxycholic acid | 0.018 | −0.463 | [40] | ✖ |
| Hydroflumethiazide | 0.003 | −0.408 | – | ✔ |
| Dexpropranolol | 0.007 | −0.489 | – | ✖ |
| Norcyclobenzaprine | 0.035 | −0.417 | – | ✖ |
| Glafenine | 0.046 | −0.239 | – | ✖ |
| Sulfamethoxypyridazine | 0.004 | −0.430 | – | ✔ |
| Cetirizine | 0.012 | −0.493 | – | ✖ |
| Azlocillin | 0.008 | −0.440 | – | ✔ |
| Morantel | 0.038 | −0.361 | – | ✔ |
| Chloramphenicol | 0.017 | −0.426 | – | ✖ |
| Pentoxyverine | 0.034 | −0.412 | – | ✖ |
| Ifenprodil | 0.019 | −0.353 | – | ✔ |
Drug targets (CS > 0.5) for these 11 drugs were enriched in KEGG pathways such as biosynthesis of antibiotics, biosynthesis of amino acids, glyoxylate and dicarboxylate metabolism, and central carbon metabolism in cancer, whereas O-glycan processing, protein glycosylation, long-chain fatty acid metabolism, sterol and cholesterol biosynthesis, lipid metabolism, glutathione-derivative biosynthesis, dephosphorylation, folic acid metabolism and transport were biological processes (Table S8).
The full list of 23 PRAD-associated drugs were analyzed through the PRAD-specific GEM, and eight of the 12 PRAD-associated drugs showed an inhibitory effect (a decrease in biomass production rate >1 × 10−8) for tumor growth through the metabolic model regardless of the threshold for drug effect between 15%–25% (Table 1). The missing PRAD-associated drugs, meptazinol, thiostrepton, metyrapone and ursodeoxycholic acid, were investigated in reference articles from the literature review. Metyrapone is applied for the temporary control of Cushing's syndrome as a paraneoplastic manifestation of prostate cancer in a patient and was found to be effective, with few side effects [38]. Thiostrepton decreases viability of various prostate cancer cell lines, but its apoptotic effectiveness increases in combination therapy with anticancer agents ABT-737 and bortezomib for treatment of PRAD [39]. Ursodeoxycholic acid showed no significant effects on growth of prostate cancer cells, whereas synthetic derivatives of the drug completely inhibited cell proliferation [40]. The opioid analgesic meptazinol has been mentioned as a significant computational prediction based on PRAD transcriptome dataset results queried on CMap2 [41]. To the best of our knowledge, no experimental validation exists for meptazinol against prostate cancer. Moreover, other PRAD-associated drugs showed in silico inhibitory effects on PRAD growth in parallel with previous studies (Table 1).
After the in silico cell viability test through PRAD-specific GEM in this study, 4 in 11 drugs were highlighted as novel candidates: sulfamethoxypyridazine, azlocillin, morantel, and ifenprodil. Later, we also checked the drug effects for 90% and 80% inhibition of tumor growth (Table S9). After sensitivity analysis composed of different growth-inhibition effects, another drug known as hydroflumethiazide also showed an effect on PRAD biomass. Furthermore, we simulated the effect of these on healthy prostate tissue to predict their lethal effects for each biological task (Table S10). Then, we evaluated results (Table S11) for our candidate drugs which were expected to inhibit tumor growth, while not being excessively lethal to healthy prostate cells.
2.6. Ifenprodil as a repositioned drug candidate inhibits prostate cancer metabolism
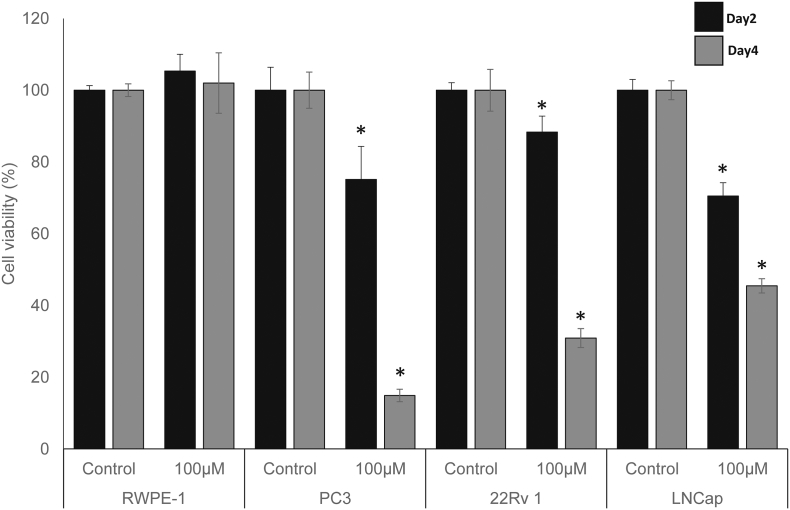
We used a CCK-8 assay to assess the effect of ifenprodil on the cell viability of the human prostate epithelial cell line (RWPE-1), and prostate cancer cell lines (LNCap, 22Rv1, PC3). All cell lines were treated with 100 μM concentration of ifenprodil. We determined cell viability after two days and four days and compared the inhibitory effect of ifenprodil between prostate epithelial cell line and cancer cell lines. The results showed that ifenprodil treatment with 100 μM concentrations significantly reduced three cancer cell lines viability time dependently (Fig. 5). Interestingly, 100 μM concentration of ifenprodil treatment did not reduced RWPE-1 cell viability but all prostate cancer cell lines (LNCap, 22 Rv1, and PC3). It showed statistical significantly reduced cell viability. In previous studies, ifenprodil showed an inhibitory effect on human esophageal cancer cell lines via NMDA receptor-mediated apoptosis [42]. There are also previous studies that NMDA receptor antagonists including ifenprodil have been tested in vitro and in vivo for ovarian cancer [43] and pancreatic cancer [44]. Ifenprodil has been reported more effective compared to other NMDA inhibitors within the well below doses known to have significant behavioral effects in rodents [44]. Treatment concentrations of ifenprodil were up to 800 μM for pancreatic cell lines (PanC-1, BXCPC3, HPAC) and ovarian cell lines (A2008, A2780, SKOV3) whereas the half-maximal inhibitory concentration of the drug was approximately 400 μM [43,44].
Fig. 5.
Experimental detection of the inhibitory effect of ifenprodil on the proliferation of the prostate fibroblast and prostate cancer cell lines. RWPE-1 human prostate epithelial fibroblast, and three prostate cancer cell lines were exposed to100μM of ifenprodil, and cell viability was tested after 2 and 4 days. Boxes show mean and standard deviation for cell viability as computed from the hexaduplicate experiments. * p-value < 10−5.
3. Discussion
GEMs have been used to study cancer metabolism using either generic/personalized or tumor/cell-specific types, which might translate into clinically relevant applications. These tools can also be used to identify drug targets leading to inhibition of cancer-related phenotypes or drug resistance in cancer therapy [5]. The prediction power of GEMs in determination of adverse drug effects and critical metabolic reactions using machine learning approaches was also investigated [11]. Emphasis on organ- or tissue-specific constraint-based models might be a crucial facet in obtaining more accurate and relevant drug effect or side effect predictions for the target tissue [12,13]. In this sense, we first reconstructed PRAD-specific GEM reinforced by tissue-specific proteome and individual transcriptome data and compared with previous PRAD GEM [45].
Our identification of altered lipid metabolism in PRAD GEM has recapitulated the fact that reprogramming of the lipid metabolism plays an important role in lipid signaling and meeting the energy demand for increased cell proliferation, macromolecule use in membrane synthesis and cell growth during tumorigenesis and tumor progression [26,46]. Increasing evidence exists for metabolic reprogramming in lipid pathways of different cancer types [47,48], including prostate cancer cells [49]. Key gene regulators of PRAD lipid metabolism such as fatty acid synthase (FASN) and alpha-methylacyl-CoA racemase (α- AMACR) were differentially expressed in this study, in parallel with the findings of previous studies [50,51].
To the best of our knowledge, no previous effort to repurpose a drug through PRAD-specific GEM has been reported. A recently published drug repositioning study for PRAD treatment via GEMs applied on two clonal subpopulations from a PC-3 cell line was used to uncover potential subpopulation-specific drug targets. Etomoxir, an inhibitor of long-chain fatty acid transport to the mitochondria, has been repurposed to treat PRAD subpopulations that have a similar phenotype of highly metastatic and low invasive type PC3 cells [52].
In parallel to increasing interest in drug off targets, prediction of gene-drug interactions becomes a challenging task that must be achieved using different approaches, including text mining, drug-induced gene expressions, drug binding sites, drug side effects, and therapeutic effects (Cheng et al., 2017). In this study, we used drug-induced gene expression profiles to test the drugs via GEM. However, other methods such as comparison of chemical structures and the moieties of repurposed drugs might also be emphasized. For instance, the moiety of sulfamethoxypyridazine is also present in other medications, including thiazide diuretics such as hydroflumethiazide repurposed in this study or sulfonylureas (e.g., glipizide) and certain COX-2 inhibitors (e.g., celecoxib) that are already repurposed for prostate cancer treatment [14]. Not only limited to structural similarities, drugs with similar functions such as anti-bacterial agents (e.g., clofoctol, nitroxoline) are also repurposed for prostate cancer treatment [14], including the azlocillin and sulfamethoxypyridazine drugs repurposed in this study.
Four candidate drugs elucidated after in silico cell viability test were discussed detailed (Text S1). Since ifenprodil has been tested in vitro in addition to in silico cell viability analysis, the potential use of NMDA receptor inhibitors in cancer treatment is investigated. Inappropriate expression of NMDA receptors on different cancer cell lines has been discussed and represented as a potential drug target to control dysregulated growth, division and invasiveness by regulation of mTOR signaling activity [53]. Moreover, blockade of NMDA receptor activity in cancer cells not only impairs phosphorylated activation of proteins in growth cascades but also prolongs survival in metastatic cancers [54]. To the best of our knowledge, Abdul and Hoosein [55] supplied evidence for NMDA receptor expression and activity in human prostate cancer and ten human cancer cell lines, including four prostate cancer cell lines that were inhibited by memantine as a NMDA-receptor antagonist at doses between 5 and 50 μg/ml (23 and 230 μM). We also repurposed the NMDA-receptor antagonist ifenprodil for treatment of PRAD but through predicted off-targets instead of targeting NMDA receptors directly.
4. Materials and methods
4.1. Genome-scale metabolic model reconstruction for PRAD
Transcriptome data from 495 individuals, composed of 495 primary prostate tumor and 52 matched nontumor tissues, were acquired from the NCBI Genome Data Commons [56] using the R/Bioconductor package “TCGAbiolinks” [57]. Expression data were converted to fragments per kilobase of transcript per million mapped reads (FPKM) and filtered using the criterion of median expression value ≥1 FPKM [8]. To integrate disease- and tissue-specific protein information into the GEM, proteome data were obtained from HPA version 18.0 [58]. Expression of each protein was integrated with transcriptome data via abundance levels such as low, medium and high for use as an input. The iCancerCore model [18] was selected as a template model for reconstruction via the tINIT (integrative network inference for tissues) algorithm [59] and the Mosek solver (version 8) in the RAVEN Toolbox [60]. A total of 471 personalized GEMs for PRAD were retrieved from the Human Pathology Atlas [18] and unified for covering individual metabolic variations in MATLAB (R2017b). Integration of proteome data into a unified model yielded a PRAD-specific GEM that captures individual metabolic variations. The biological tasks representing metabolic functions that occurred in all cell types (Table S10) were used to test the functionality of the metabolic model.
4.2. Differential expressed genes and defining metabolic signatures
Raw count data were used to detect differentially expressed genes (DEGs) between two phenotypes using the R/Bioconductor package “DESeq” manual [61]. Multiple hypothesis correction was conducted using the Benjamini and Hochberg false discovery rate, and those considered statistically significant were used as a multiple correlation test to calculate q-values. DEGs were filtered based on the threshold of the FDR adjusted p-value < 0.001. Fold-change cutoff of 1 was applied to determine up- and down- regulated genes (Table S1).
PRAD-specific GEMs comprise 2655 genes, and an intersection of these genes and DEGs were assigned as “metabolic gene signatures” for PRAD in this study (Table S4). This set is composed of 641 genes, including 274 up- and 367 downregulated genes.
4.3. Reporter metabolites
To identify significantly enriched metabolites based on association with gene expression changes, we recruited the reporter metabolite algorithm embedded RAVEN Toolbox [60], which uses DEGs information through the network topology of the reconstructed model. These metabolites highlight the central components of the metabolic network that are affected by perturbations between different conditions [19].
5. Functional enrichment analyses
Functional enrichment analyses were performed in the DAVID functional annotation tool [62] using DEGs as input to identify the enriched KEGG pathways and GO biological processes.
5.1. DIRAC algorithm
DIRAC [16] was implemented in MATLAB to analyze gene order within the pathways of the reconstructed model together with other gene sets. The central concept of the algorithm is based on the relative expression ranks of the participating genes. The results show a quantitative measurement of how pathway rankings differ both within and between phenotypes.
5.2. Essentiality analysis
MATLAB incorporated with the RAVEN Toolbox [60] and Mosek solver (version 8) was applied for GEM operation and analysis of synthetic lethal genes. To determine essential genes, we used logical transformation of the model and FastGeneSL, as previously described [23].
5.3. Drug repositioning based on reversal of metabolic gene signatures
For robust representation of PRAD metabolism, we identified the genes that are differentially expressed inside the reconstructed prostate cancer metabolic model. DEGs from PRAD GEM were defined as metabolic gene signatures. A total of 641 DEGs, including 274 up- and 367 downregulated genes, were used as a query on Connectivity Map2 (CMap2).
CMap2 consists of 6100 gene expression profiles for 1309 drug perturbation experiments performed on different cancer cell lines [17]. The Kolmogorov-Smirnov statistic and random permutation tests are applied in CMap2 to calculate the similarity scores for a drug-perturbed expression profile with respect to gene expression profiles queried as input and their statistical significance (p-value), respectively. A positive similarity score denotes the similarity of the drug-perturbed expression profile to the query, whereas a negative score indicates a reverse pattern. Therefore, the p-value shows the probability of finding the same association when a random signature is supplied. Significant drugs (p-value < 0.05) with negative mean values are collected as drug candidates for reversal of metabolic signatures in PRAD.
5.4. Interpretation of CMap2 data to repurpose novel gene-drug interactions
To test the drug efficacy in silico, we aimed to quantify the drug-gene associations, therefore drug-associated gene expression profiles were included as follows: Connectivity Map (CMap Build 02, https://portals.broadinstitute.org/cmap/) data from production batches (HT-HG U133A) was downloaded for 3 cell lines: HL60 (Human promyelocytic leukemia), MCF7 (Human breast adenocarcinoma), and PC3 (Human prostate cancer). All analyses were performed individually for each cell line. Raw data were processed in R using the gcrma package (https://bioconductor.org/packages/release/bioc/html/gcrma.html), and single drug treatment and control were selected similarly to [63]. Briefly, after normalization, a single treatment with the highest concentration was selected based on those treatments whose gene expression displayed the highest correlations (results not shown). Outliers based on hierarchical clustering, and mean gene expression was computed across controls. This approach resulted in 683 treatment instances and a single control per cell line. For each cell line, gene Log2FC were computed by comparison between treatment instances and its respective controls.
Afterward, the confidence score was calculated per each drug-gene interaction using the Log2FC values from three cell lines. Drug-gene matrix comprising Log2FC of approximately 12,300 genes and 683 drugs were assessed for each drug and found normally distributed. Standard z-scores were calculated and converted p-value for each drug-gene pair in the matrix for three cell lines (n), separately. Then, the naive Bayesian formula was modified to calculate a confidence score for each drug-gene pair based on the integration of the information coming from each cell line by using following formula:
))
Since lower p-value ratios were favorable, an approximation of confidence score to 1 was assumed as the higher confidence level.
5.5. In silico drug efficacy test by using PRAD specific GEMs
Before testing drug efficacy in silico, metabolic gene signatures were mapped to the gene-drug interaction confidence score matrix, and 526 of 641 DEGs were matched. The final matrix of drug-gene interacting pairs with confidence scores was for 515 genes and 683 drugs (Table S6).
The intersection of significant drug candidates with KS similarity score below zero and 683 drugs interacting genes with calculated confidence scores yielded twenty-three drug candidates to test in silico drug effect through the metabolic model. Interacting gene partners of each drug were extracted regarding the criteria of confidence score > 0.5 and the reversal drug effect of metabolic gene signatures (Table S7).
First, biomass production was set as an objective function to test drug effect on prostate tumor metabolic model. Reactions which include drug interacting genes were assigned as up-reactions and down-reactions regarding drug perturbed gene expression profiles. We estimated the upper and lower bounds for these up- and down-reactions by random sampling 1000 times. The estimated lower and upper bounds of up-reactions were increased 20% whereas down-reactions were decreased by 20% for toxicity test. We also performed a sensitivity analysis to test the dependency of the results on the chosen threshold by simulations between 15%–25%. After linear programming algorithm was applied to solve the model, the drugs showed inhibitory effect (a decrease in biomass production >1 × 10−8) on the growth of the cancer metabolic model were collected as drug candidates for prostate cancer treatment.
We also evaluated the toxic effects of candidate drugs on healthy prostate tissue. We used the healthy prostate tissue GEM acquired from Human Metabolic Atlas (http://www.metabolicatlas.org) [58] to check drug effects for each biological task using same approach mentioned above. We predicted no toxic effect of the drugs in case central tasks classified as being involved in energy and redox balancing are still enabled in the model as previously done in another study [59].
5.6. In vitro cell viability assay
All cells were cultured followed by ATCC instruction. PC3 cells culture media formulation is F12K Nutrient mix supplemented with 10% FBS and 1% Penicillin/Streptomycin, RWPE-1 cells was cultured with Keratinocyte Serum Free Medium (K-SFM) supplemented with Bovine Pituitary Extract (BPE) and Human recombinant Epidermal Growth Factor (EGF) (Kit Catalog Number 17005–042). 22 Rv1, and LNCap cells were cultured with RPMI 1640 (R2405, Sigma-Aldrich) supplemented with 10% fetal bovine serum, 1% penicillin and streptomycin. For cell viability assay, cell lines were seeded to 96 well plate as hextuple, as in 100,000 cells per well. After 24 h of cell seeding, drugs were treated by media change for two and four days. Cell Counting Kit-8 (Sigma-Aldrich) was used to measure cell viability as manufacturers' instruction.
The following are the supplementary data related to this article.
Differential expressed genes in PRAD
Reporter metabolites in PRAD
Essential genes in PRAD specific GEM and their enriched pathways
Metabolic gene signatures and their enriched pathways
Drugs from CMap2 with similarity scores and permutated p-value
Drug-Gene matrix with confidence scores
Twenty-three repurposed drugs with reversal expression pattern of the disease and their interacting gene partners
Enrichment of interacting genes with confidence score higher than 0.5 for 11 novel drug candidates
Sensitivity analysis of in silico cell viability test via GEMs
Biological tasks used in model reconstruction
The effects of twenty-three repurposed drugs on biological tasks of healthy prostate GEM
Detailed information of five repurposed drugs after in silico cell viability test.
Acknowledgments
Acknowledgments
This work was supported by TUBITAK, 2211A and 2214A fellowship programs and project number 117S489, and funded by Knut and Alice Wallenberg Foundation.
Author contributions
B.T. and A.M. designed the project, B.T. wrote the manuscript. B.T., C.Z., and R.B. performed the computational analyses, and W.K. performed in vitro experiments. A.M., K.Y.A. and M.U. supervised the study. All authors were involved in editing the paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Contributor Information
Kazim Yalcin Arga, Email: kazim.arga@marmara.edu.tr.
Adil Mardinoglu, Email: adilm@scilifelab.se.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Mardinoglu A., Nielsen J. New paradigms for metabolic modeling of human cells. Curr Opin Biotechnol. 2015;34:91–97. doi: 10.1016/j.copbio.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 4.Mardinoglu A., Boren J., Smith U., Uhlen M., Nielsen J. Systems biology in hepatology: approaches and applications. Nat Rev Gastroenterol Hepatol. 2018;15:365–377. doi: 10.1038/s41575-018-0007-8. [DOI] [PubMed] [Google Scholar]
- 5.Yizhak K., Chaneton B., Gottlieb E., Ruppin E. Modeling cancer metabolism on a genome scale. Mol Syst Biol. 2015;11:817. doi: 10.15252/msb.20145307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nam H., Campodonico M., Bordbar A., Hyduke D.R., Kim S., Zielinski D.C. A systems approach to predict oncometabolites via context-specific genome-scale metabolic networks. PLoS Comput Biol. 2014 doi: 10.1371/journal.pcbi.1003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benfeitas R., Bidkhori G., Mukhopadhyay B., Klevstig M., Arif M., Zhang C. Characterization of heterogeneous redox responses in hepatocellular carcinoma patients using network analysis. EBioMedicine. 2019;40:471–487. doi: 10.1016/j.ebiom.2018.12.057. (doi:S2352396418306352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bidkhori G., Benfeitas R., Elmas E., Kararoudi M.N., Arif M., Uhlen M. Metabolic network-based identification and prioritization of anticancer targets based on expression data in hepatocellular carcinoma. Front Physiol. 2018;9 doi: 10.3389/fphys.2018.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mardinoglu A., Nielsen J. Systems medicine and metabolic modelling. J Intern Med. 2012;271:142–154. doi: 10.1111/j.1365-2796.2011.02493.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C., Hua Q. Applications of genome-scale metabolic models in biotechnology and systems medicine. Front Physiol. 2016;6:1–8. doi: 10.3389/fphys.2015.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaked I., Oberhardt M.A., Atias N., Sharan R., Ruppin E. Metabolic network prediction of drug side effects. Cell Syst. 2016;2:209–213. doi: 10.1016/j.cels.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Jerby L., Shlomi T., Ruppin E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol. 2010;6:1–9. doi: 10.1038/msb.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folger O., Jerby L., Frezza C., Gottlieb E., Ruppin E., Shlomi T. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:1–10. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turanli B, Grøtli M, Boren J, Nielsen J, Uhlen M, Arga KY, et al. Drug Repositioning for Effective Prostate Cancer Treatment 2018;9:1–20. 10.3389/fphys.2018.00500. [DOI] [PMC free article] [PubMed]
- 15.Eidelman E., Twum-Ampofo J., Ansari J., Siddiqui M.M. The metabolic phenotype of prostate cancer. Front Oncol. 2017;7:1–6. doi: 10.3389/fonc.2017.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eddy J.A., Hood L., Price N.D., Geman D. Identifying tightly regulated and variably expressed networks by Differential Rank Conservation (DIRAC) PLoS Comput Biol. 2010;6:1–17. doi: 10.1371/journal.pcbi.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J. The connectivity map : using. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. (80-) [DOI] [PubMed] [Google Scholar]
- 18.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G. A pathology atlas of the human cancer transcriptome. Science (80-) 2017:357. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 19.Patil K.R., Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci U S A. 2005;102:2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green T., Chen X., Ryan S., Asch A.S., Ruiz-Echevarría M.J. TMEFF2 and SARDH cooperate to modulate one-carbon metabolism and invasion of prostate cancer cells. Prostate. 2013;73:1561–1575. doi: 10.1002/pros.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen L.F., Brockton N.T., Bakkar A., Liu S., Wen J., Weljie A.M. Elevated physiological levels of folic acid can increase in vitro growth and invasiveness of prostate cancer cells. BJU Int. 2012;109:788–795. doi: 10.1111/j.1464-410X.2011.10437.x. [DOI] [PubMed] [Google Scholar]
- 22.Jain S., Chakraborty G., Raja R., Kale S., Kundu G.C. Prostaglandin E2 regulates tumor angiogenesis in prostate cancer. Cancer Res. 2008;68:7750–7759. doi: 10.1158/0008-5472.CAN-07-6689. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C., Ji B., Mardinoglu A., Nielsen J., Hua Q. Logical transformation of genome-scale metabolic models for gene level applications and analysis. Bioinformatics. 2015;31:2324–2331. doi: 10.1093/bioinformatics/btv134. [DOI] [PubMed] [Google Scholar]
- 24.Sharifi N., Auchus R.J. Steroid biosynthesis and prostate cancer. Steroids. 2012;77:719–725. doi: 10.1016/j.steroids.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 25.Sinnott J.A., Rider J.R., Carlsson J., Gerke T., Tyekucheva S., Penney K.L. Molecular differences in transition zone and peripheral zone prostate tumors. Carcinogenesis. 2015;36:632–638. doi: 10.1093/carcin/bgv051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al Kadhi O., Traka M.H., Melchini A., Troncoso-Rey P., Jurkowski W., Defernez M. Increased transcriptional and metabolic capacity for lipid metabolism in the peripheral zone of the prostate may underpin its increased susceptibility to cancer. Oncotarget. 2017;8:84902–84916. doi: 10.18632/oncotarget.17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q., Shu Y. Role of solute carriers in response to anticancer drugs. Mol Cell Ther. 2014;2:15. doi: 10.1186/2052-8426-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin L., Yee S.W., Kim R.B., Giacomini K.M. SLC transporters as therapeutic targets: emerging opportunities. Nat Rev Drug Discov. 2015;14:543–560. doi: 10.1038/nrd4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopecka J., Campia I., Jacobs A., Frei A.P., Ghigo D., Wollscheid B. Carbonic anhydrase XII is a new therapeutic target to overcome chemoresistance in cancer cells. Oncotarget. 2015;6:6776–6793. doi: 10.18632/oncotarget.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takakura M., Yokomizo A., Tanaka Y., Kobayashi M., Jung G., Banno M. Carbonic anhydrase I as a new plasma biomarker for prostate cancer. ISRN Oncol. 2012;2012:1–10. doi: 10.5402/2012/768190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wesche J., Haglund K., Haugsten E.M. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437:199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 32.Corn P., Wang F., McKeehan W., Navone N. Targeting fibroblast growth factor pathways in prostate cancer. Clin Cancer Res. 2013;6:247–253. doi: 10.1158/1078-0432.CCR-13-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agalliu I., Lin D.W., Salinas C.A., Feng Z., Stanford J.L. Polymorphisms in the glutathione S-transferase M1, T1, and P1 genes and prostate cancer prognosis. Prostate. 2006;66:1535–1541. doi: 10.1002/pros.20491. [DOI] [PubMed] [Google Scholar]
- 34.Shiota M., Fujimoto N., Itsumi M., Takeuchi A., Inokuchi J., Tatsugami K. Gene polymorphisms in antioxidant enzymes correlate with the efficacy of androgen-deprivation therapy for prostate cancer with implications of oxidative stress. Ann Oncol Off J Eur Soc Med Oncol. 2017;28:569–575. doi: 10.1093/annonc/mdw646. [DOI] [PubMed] [Google Scholar]
- 35.Di Pietro G., Magno L.A.V., Rios-Santos F. Glutathione S-transferases: an overview in cancer research. Expert Opin Drug Metab Toxicol. 2010;6:153–170. doi: 10.1517/17425250903427980. [DOI] [PubMed] [Google Scholar]
- 36.Ramsay E.E., Dilda P.J. Glutathione S-conjugates as prodrugs to target drug-resistant tumors. Front Pharmacol. 2014;5(JUL):1–16. doi: 10.3389/fphar.2014.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allocati N., Masulli M., Di Ilio C., Federici L. Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis. 2018;7 doi: 10.1038/s41389-017-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alshaikh O., Al-Mahfouz A., Al-Hindi H., Mahfouz A., Alzahrani A. Unusual cause of ectopic secretion of adrenocorticotropic hormone: Cushing syndrome attributable to small cell prostate Cancer. Endocr Pract. 2010;16:249–254. doi: 10.4158/EP09243.CR. [DOI] [PubMed] [Google Scholar]
- 39.Pandit B., Gartel A.L. New potential anti-cancer agents synergize with bortezomib and ABT-737 against prostate cancer. Anal Chem. 2015;25:368–379. doi: 10.1002/pros.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi Y.H., Im E.O., Suh H., Jin Y., Yoo Y.H., Kim N.D. Apoptosis and modulation of cell cycle control by synthetic derivatives of ursodeoxycholic acid and chenodeoxycholic acid in human prostate cancer cells. Cancer Lett. 2003;199:157–167. doi: 10.1016/s0304-3835(03)00351-3. [DOI] [PubMed] [Google Scholar]
- 41.Wen D.Y., Geng J., Li W., Guo C.C., Zheng J.H. A computational bioinformatics analysis of gene expression identifies candidate agents for prostate cancer. Andrologia. 2014;46:625–632. doi: 10.1111/and.12127. [DOI] [PubMed] [Google Scholar]
- 42.Kim M.S., Yamashita K., Baek J.H., Park H.L., Carvalho A.L., Osada M. N-methyl-D-aspartate receptor type 2B is epigenetically inactivated and exhibits tumor-suppressive activity in human esophageal cancer. Cancer Res. 2006;66:3409–3418. doi: 10.1158/0008-5472.CAN-05-1608. [DOI] [PubMed] [Google Scholar]
- 43.North W.G., Liu F., Tian R., Abbasi H., Akerman B. NMDA receptors are expressed in human ovarian cancer tissues and human ovarian cancer cell lines. Clin Pharmacol Adv Appl. 2015;7:111–117. doi: 10.2147/CPAA.S90367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.North W.G., Liu F., Lin L.Z., Tian R. NMDA receptors are important regulators of pancreatic cancer and are potential targets for treatment. Clin Pharmacol Adv Appl. 2017;9:79–86. doi: 10.2147/CPAA.S140057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agren R., Bordel S., Mardinoglu A., Pornputtapong N., Nookaew I., Nielsen J. Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Q., Luo Q., Halim A., Song G. Targeting lipid metabolism of cancer cells: a promising therapeutic strategy for cancer. Cancer Lett. 2017;401:39–45. doi: 10.1016/j.canlet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Özcan E., Çakir T. Reconstructed metabolic network models predict flux-level metabolic reprogramming in glioblastoma. Front Neurosci. 2016 doi: 10.3389/fnins.2016.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jerby L., Wolf L., Denkert C., Stein G.Y., Hilvo M., Oresic M. Metabolic associations of reduced proliferation and oxidative stress in advanced breast cancer. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-12-2215. [DOI] [PubMed] [Google Scholar]
- 49.Wu X., Daniels G., Lee P., Monaco M.E. Lipid metabolism in prostate cancer. Am J Clin Exp Urol. 2014;2:111–120. [PMC free article] [PubMed] [Google Scholar]
- 50.Swinnen J.V., Heemers H., Deboel L., Foufelle F., Heyns W., Verhoeven G. Stimulation of tumor-associated fatty acid synthase expression by growth factor activation of the sterol regulatory element-binding protein pathway. Oncogene. 2000;19:5173–5181. doi: 10.1038/sj.onc.1203889. [DOI] [PubMed] [Google Scholar]
- 51.Luo J., Zha S., Gage W.R., T a Dunn, Hicks J.L., Bennett C.J. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- 52.Mas D., Aguilar E., Zodda E., Balcells C., Marin S., Dallmann G. Model-driven discovery of long-chain fatty acid metabolic reprogramming in heterogeneous prostate cancer cells. PLoS Comput Biol. 2018:1–20. doi: 10.1371/journal.pcbi.1005914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deutsch S.I., Tang A.H., Burket J.A., Benson A.D. NMDA receptors on the surface of cancer cells: target for chemotherapy? Biomed Pharmacother. 2014;68:493–496. doi: 10.1016/j.biopha.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stepulak A., Sifringer M., Rzeski W., Endesfelder S., Gratopp A., Pohl E.E. NMDA antagonist inhibits the extracellular signal-regulated kinase pathway and suppresses cancer growth. Proc Natl Acad Sci U S A. 2005;102:15605–15610. doi: 10.1073/pnas.0507679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdul M., Hoosein N. N-methyl-D-aspartate receptor in human prostate cancer. J Membr Biol. 2005;205:125–128. doi: 10.1007/s00232-005-0777-0. [DOI] [PubMed] [Google Scholar]
- 56.Grossman R.L., Heath A., Murphy M., Patterson M., Wells W. A case for data commons: toward data science as a service. Comput Sci Eng. 2016;18:10–20. doi: 10.1109/MCSE.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A. Tissue-based map of the human proteome. Proteomics. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 59.Agren R., Mardinoglu A., Asplund A., Kampf C., Uhlen M., Nielsen J. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol Syst Biol. 2014;10:1–13. doi: 10.1002/msb.145122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agren R., Liu L., Shoaie S., Vongsangnak W., Nookaew I., Nielsen J. The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using\nDAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 63.Iskar M., Campillos M., Kuhn M., Jensen L.J., van Noort V., Bork P. Drug-induced regulation of target expression. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J., Li Z., Lin Z., Zhao B., Wang Y., Peng R. 17-DMCHAG, a new geldanamycin derivative, inhibits prostate cancer cells through Hsp90 inhibition and survivin downregulation. Cancer Lett. 2015;362:83–96. doi: 10.1016/j.canlet.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 65.Pathak M.K., Dhawan D., Lindner D.J., Borden E.C., Farver C., Yi T. Pentamidine is an inhibitor of PRL phosphatases with anticancer activity. Mol Cancer Ther. 2002;1:1255–1264. [PubMed] [Google Scholar]
- 66.Shigemizu D., Hu Z., Hung J.H., Huang C.L., Wang Y., DeLisi C. Using functional signatures to identify repositioned drugs for breast, myelogenous leukemia and prostate cancer. PLoS Comput Biol. 2012;8:1–9. doi: 10.1371/journal.pcbi.1002347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pacey S., Wilson R.H., Walton M., Eatock M.M., Hardcastle A., Zetterlund A. A phase I study of the heat shock protein 90 inhibitor alvespimycin (17-DMAG) given intravenously to patients with advanced solid tumors. Clin Cancer Res. 2011;17:1561–1570. doi: 10.1158/1078-0432.CCR-10-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Momtazi-borojeni A.A., Abdollahi E., Ghasemi F., Caraglia M., Sahebkar A. The novel role of pyrvinium in cancer therapy. J Cell Physiol. 2017 doi: 10.1002/jcp.26006. [DOI] [PubMed] [Google Scholar]
- 69.Chen H.-R., Sherr D.H., Hu Z., DeLisi C. A network based approach to drug repositioning identifies plausible candidates for breast cancer and prostate cancer. BMC Med Genomics. 2016;9 doi: 10.1186/s12920-016-0212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential expressed genes in PRAD
Reporter metabolites in PRAD
Essential genes in PRAD specific GEM and their enriched pathways
Metabolic gene signatures and their enriched pathways
Drugs from CMap2 with similarity scores and permutated p-value
Drug-Gene matrix with confidence scores
Twenty-three repurposed drugs with reversal expression pattern of the disease and their interacting gene partners
Enrichment of interacting genes with confidence score higher than 0.5 for 11 novel drug candidates
Sensitivity analysis of in silico cell viability test via GEMs
Biological tasks used in model reconstruction
The effects of twenty-three repurposed drugs on biological tasks of healthy prostate GEM
Detailed information of five repurposed drugs after in silico cell viability test.