Abstract
Cytochrome P450 (CYP)3A is the most abundant CYP enzyme in the human liver, and a functional impairment of this enzyme leads to unanticipated adverse reactions and therapeutic failures; these reactions result in the early termination of drug development or the withdrawal of drugs from the market. The transcriptional regulation mechanism of the Cyp3a gene is not fully understood and requires a thorough investigation. We mapped the transcriptome of the Cyp3a gene in a mouse model. The Cyp3a gene was induced using the mPXR activator pregnenolone-16alpha-carbonitrile (PCN) and was subsequently downregulated using lipopolysaccharide (LPS). Our objective was to identify the transcription factors (TFs), epigenetic modulators and molecular pathways that are enriched or repressed by PCN and LPS based on a gene set enrichment analysis. Our analysis shows that 113 genes were significantly upregulated (by at least 1.5-fold) with PCN treatment, and that 834 genes were significantly downregulated (by at least 1.5-fold) with LPS treatment. Additionally, the targets of the 536 transcription factors were enriched by a combined treatment of PCN and LPS, and among these, 285 were found to have binding sites on Cyp3a11. Moreover, the repressed targets of the epigenetic markers HDAC1, HDAC3 and EZH2 were further suppressed by LPS treatment and were enhanced by PCN treatment. By identifying and contrasting the transcriptional regulators that are altered by PCN and LPS, our study provides novel insights into the transcriptional regulation of CYP3A in the liver.
Subject terms: Drug regulation, Toxicology, Toxicology, Toxicology, Drug regulation
Introduction
Cytochrome P450 3A (CYP3A) is the most abundant subfamily of the drug-metabolizing enzymes (DMEs) that are responsible for the disposition of more than 50% of the currently prescribed drugs1–4. A review of 121 new molecular entities (NMEs), approved by the FDA in 2003 and 2008, indicated that CYP3A was the main CYP enzyme involved in the disposition of these NMEs5. The clinical importance of CYP3A can be assessed from numerous reports showing that the downregulation of CYP3A expression and activity in infectious and inflammatory diseases and in liver cancer6,7 leads to the failure of therapy and/or potentially harmful adverse drug reactions8,9. On the other hand, the induction of CYP3A4 is associated with the reduced efficacy of clinically relevant medications10–12. Therefore, to reduce or prevent unwanted drug-drug interactions and adverse drug reactions, it is crucial to gain a comprehensive understanding of the molecular mechanisms that regulate the CYP3A enzyme.
CYP3A is both constitutively expressed and transcriptionally induced or inhibited by a variety of structurally diverse xenobiotics. Multiple signaling pathways contribute to the complex regulation of the CYP3A genes. The constitutive expression of CYP3A is regulated via basal transcription factors, such as HNF4, HNF1, AP1, C/EBPα, C/EBPβ, HNF3γ, and USF1, by binding to the constitutive liver enhancer module (CLEM4) and the distal enhancer module (XREM) of the CYP3A4 promoter13–18. The xenobiotic-mediated induction of CYP3A is indirect and involves the activation of nuclear receptors, such as pregnane X receptor (PXR), constitutive androstane receptor (CAR), glucocorticoid receptor (GR) and vitamin D receptor (VDR)19,20. However, PXR is considered the most important and critical determinant of hepatic CYP3A enzyme activity and expression21,22. PXR is expressed in the cytosol and is activated upon binding with structurally diverse drug ligands, including barbiturates, rifampicin, statins, pregnenolone 16α-carbonitrile (PCN) and many others. Upon activation, PXR is translocated to the nucleus, where it heterodimerizes with retinoid X receptor (RXR) and enhances CYP3A transcription by binding to AGGTCA-like direct repeat (DR-3) and everted repeat regions (ER-6) on the Cyp3a gene22–25. PXR activity can be modulated by phosphorylation through a number of cell signaling kinases, such as protein kinase A26,27, protein kinase C28, c-Jun-N-terminal kinase29, and this impacts its downstream transcriptional ability to induce CYP3A. Epigenetic changes, such as DNA methylation, histone protein modification and microRNAs (miRNAs) have also been implicated in the regulation of the CYP3A enzyme.
In contrast to induction of CYP3A being xenobiotic-mediated, the downregulation of hepatic CYP3A has mainly been reported in various pathophysiological conditions, especially infections and inflammation. Studies have shown that the gram-negative bacterial endotoxin lipopolysaccharide (LPS) induces an acute phase response30 in animals, and this response can lead to the decreased expression and activity of CYP3A1131,32; ultimately, this leads to a decrease in the hepatic drug metabolism33. Multiple mechanisms have been proposed to explain the effects of LPS on CYP3A downregulation. LPS treatment of mice suppresses the PXR mRNA levels and reduces the nuclear RXRα protein levels due to increased nuclear export34. The binding of PXR/RXRα to conserved sequences of Cyp3a11 was also reduced by LPS, thereby suppressing CYP3A11 mRNA34. LPS has also been shown to activate toll-like receptors (TLRs) on hepatocytes and Kupffer cells, leading to the induction of pro-inflammatory cytokines, such as IL-1β, IL-6 and TNF-α in immune cells35,36. In turn, these increased levels of cytokines downregulate Cyp3a gene expression by activating downstream mediators, such as JNK or NF-κB37–39. The translocation of NF-κB was shown to increase binding between NF-κB and RXRα, and this increase interfered with the formation of PXR-RXRα and suppressed CYP3A4 expression40.
Although numerous mechanisms, both in vitro and in vivo, have been proposed to explain the altered CYP3A expression levels, global transcriptome changes have not yet been investigated. We utilized the model of CYP3A upregulation by PCN (mouse-specific PXR activator) followed by CYP3A downregulation by LPS and performed a comprehensive transcriptome mapping and bioinformatics analysis to identify the novel mechanisms of CYP3A11 (mouse homolog of CYP3A4) regulation in vivo. Our study identified genes, gene pathways, transcription factors and epigenetic modulators that were significantly altered (induced or downregulated) by PCN and LPS. By comparing and contrasting the effects of PCN and LPS on the transcriptome as a whole, and by finding transcription factors and epigenetic modulators, which are either upregulated or downregulated by PCN or LPS, we identified potential regulators involved in the Cyp3a transcriptional machinery, which can be targeted for further investigation.
Results
CYP3A11 expression and activity
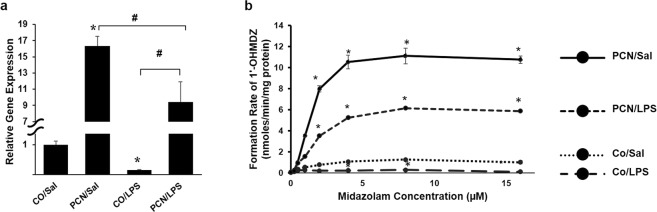
To validate our model, we analyzed the gene expression of Cyp3a11 in the mouse liver after treatment with PCN and LPS using RT-qPCR. We observed that treatment with PCN upregulated Cyp3a11 gene expression by 16-fold, whereas LPS treatment downregulated Cyp3a11 gene expression by 10-fold compared to the control gene expression (Fig. 1a). The combined treatment of PCN and LPS induced a significantly higher expression level of Cyp3a11 compared to that induced in the control; however, its expression was reduced by almost 1.7-fold by the combination of PCN and LPS compared to the expression by PCN treatment alone. A microarray analysis of the transcriptomic profile also showed that PCN significantly upregulated Cyp3a11 gene expression 2.41-fold, whereas LPS significantly downregulated Cyp3a11 gene expression 2.6-fold (Tables 1 and S1); these results are in concordance with the RT-qPCR data. The CYP3A11 activity was measured by the formation rate of the metabolite of midazolam (MDZ) as described in the Materials and Methods section. Consistent with the expression data, CYP3A11 activity was significantly induced by PCN and was downregulated by LPS (Fig. 1b). The combined treatment of PCN and LPS significantly induced Cyp3a11 activity compared to that in the control and attenuated Cyp3a11 activity compared to that in the individual PCN treatment.
Figure 1.
Validation of Cyp3a11 modulation by PCN and LPS. (a) The real-time RT-qPCR analysis of Cyp3a11 gene expression is shown. (b) The CYP3A11 enzyme activity is shown from the livers of mice treated with corn oil or PCN (50 mg/kg/day) for 3 days followed by saline or LPS (2 mg/kg) for 16 h. The error bars represent the standard deviation from three independent experiments performed in quadruplicate. *p < 0.05 compared to the control treatment. #p < 0.05 compared to PCN or LPS treatment alone.
Table 1.
Top 15 differentially regulated genes (represented by their gene symbols) by PCN, LPS and combined PCN/LPS treatments compared to those in the control.
| PCN vs Control | LPS vs Control | PCN/LPS vs Control | PCN/LPS vs PCN/Sal | PCN/LPS vs CO/LPS | |||||
|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | Up | Down |
| GSTA1 | CYP4A14 | REG3B | CAR3 | S100A8 | HSD3B5 | CXCL9 | THRSP | CYP3A11 | CYP4A14 |
| GSTM3 | CML2 | S100A8 | HSD3B5 | S100A9 | HAMP2 | SAA3 | CAR3 | CES6 | FDPS |
| CYP2C55 | EGFR | S100A9 | CPS1 | SAA3 | CAR3 | S100A9 | AQP8 | GSTA1 | IDI1 |
| CES6 | EGR1 | SAA3 | THRSP | REG3B | AQP8 | S100A8 | HAMP2 | CYP3A25 | REG3A |
| GSTA2 | ARRDC3 | CXCL9 | HAMP2 | CRYBB3 | THRSP | REG3B | HSD3B5 | GSTM3 | SC4MOL |
| CYP2B10 | IDB2 | CRYBB3 | ELOVL3 | CXCL9 | VSIG4 | CRYBB3 | ELOVL6 | GSTA2 | SLC25A25 |
| AKR1B7 | CYP2C67 | CXCL1 | VSIG4 | CXCL1 | ELOVL3 | CD14 | GSTA1 | POR | CYP51 |
| CYP3A11 | GNAT1 | CD14 | INMT | CD14 | CPS1 | CXCL1 | CYP2C55 | HSD17B6 | PPP1R3C |
| GSTM6 | G0S2 | LCN2 | ACSS2 | LCN2 | ACSS2 | CCL5 | G6PC | CYP2C55 | LSS |
| GSTM2 | RNASE4 | ADH7 | AQP8 | MT2 | G6PC | LCN2 | ACSS2 | AKR1B7 | CRELD2 |
| HSD17B6 | ACOT1 | MT2 | CLEC4G | SAA2 | CLEC4G | SAA2 | GSTM3 | CYP2B10 | |
| CYP2B23 | AOX3 | CPNE8 | CYP2A5 | CHI3L3 | SLC2A2 | MT2 | SLC2A2 | GSTM2 | |
| DDIT4 | CYP2C70 | MT1 | UPP2 | CES6 | ACAA1B | CYP17A1 | CHRNA4 | SLCO1A4 | |
| CYP3A25 | HSD3B | SAA2 | G6PC | ADH7 | NUDT7 | GBP2 | CPS1 | GSTM6 | |
| CSAD | MUG2 | STEAP4 | GSTM6 | MT1 | AOX3 | CHI3L3 | GSTM6 | CES3 | |
Differential gene expression analysis
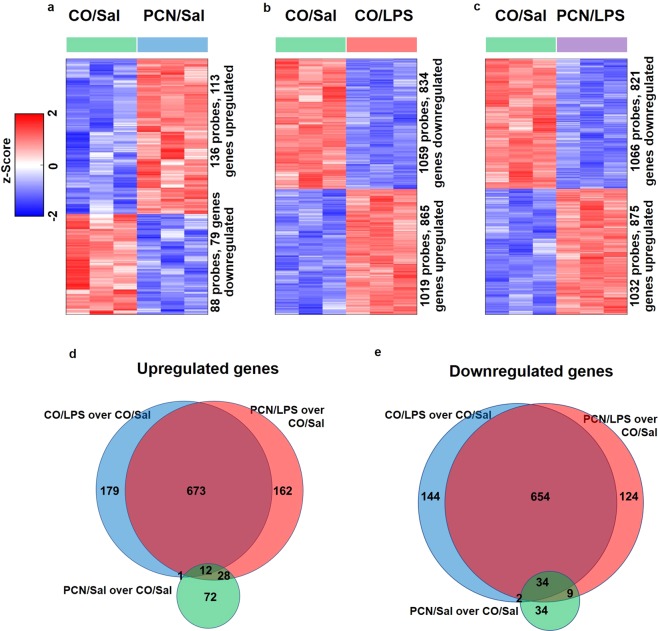
A gene expression analysis using a DNA microarray was carried out to identify genes and pathways that are upregulated by PCN and downregulated by LPS or downregulated by PCN and upregulated by LPS. After three days of PCN treatment, a total of 79 genes were downregulated (DR: 79), and 113 genes were upregulated (UR: 113) (Fig. 2a). However, after a 16 h LPS treatment, 834 genes were downregulated, and 865 genes were upregulated (Fig. 2b). With the combined PCN and LPS treatment, a total of 821 genes were downregulated, and 875 genes were upregulated compared to those in the control group (Fig. 2c). Figure 2d,e show the number of upregulated and downregulated genes in the three treatments compared to those in the control. Among these total changes, Table 1 represents the top 15 genes with the highest fold change; these genes were differentially expressed among all the global changes upon PCN and/or LPS treatment compared to those in the control. PCN treatment led to significant alterations in the gene expression of numerous drug metabolizing enzymes, such as glutathione S transferases, CYP3A11, CYP2B10, and carboxylesterases. On the other hand, LPS upregulated many inflammatory mediators, such as chemokines and CD14. Interestingly, with the combined treatment of PCN and LPS, we observed similarities in the top up- and downregulated genes as observed in genes treated with LPS alone.
Figure 2.
PCN and LPS treatments lead to robust yet distinct transcriptomic changes. Heatmaps of the differentially expressed genes (DEGs) with (a) exclusive PCN treatment, (b) exclusive LPS treatment and (c) combined PCN/LPS treatment are shown. (d–e) The number of upregulated and downregulated DEGs by PCN treatment, LPS treatment and combined PCN/LPS treatment compared to those in the control are shown. The genes were considered to be differentially expressed with a p-value < 0.05 and a fold change greater than or equal to 1.25-fold or less than or equal to 0.8-fold. Heatmaps were generated using mean-centered normalized expression values (z-scores), employing the Euclidean distance metric, the average clustering method and R statistical software.
Pathway analysis
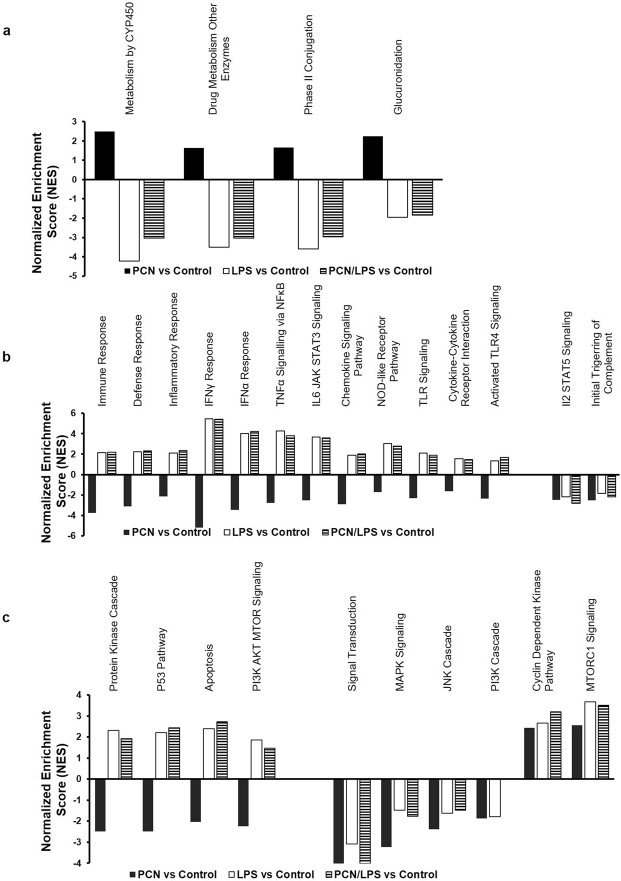
Biological processes that were significantly enriched but differentially modulated in the transcriptome footprint of the two treatments, PCN and LPS, were identified using gene set enrichment analysis (GSEA)41. Among the overall enriched pathways, we identified those that were regulated in opposite directions by each of the PCN and LPS treatments (Q < 0.25; normalized enrichment score/NES has opposite signs between the PCN and LPS treatments). In addition, we studied the effect of the cotreatment of PCN and LPS on these differentially regulated pathways, as shown in Fig. 3. We focused on three groups of major biological pathways involved in drug response biology: (a) drug metabolism pathways, including P450-dependent metabolism or glucuronidation (Fig. 3a); (b) inflammatory pathways, including interferon-γ, interferon-α, and TNF-α signaling (Fig. 3b); and (c) signal transduction pathways, including the protein kinase cascade and mitogen-activated protein kinase signaling (Fig. 3c). The drug metabolism pathways were positively enriched by PCN and were attenuated by LPS, whereas the LPS/PCN combination suppressed the effects of the single PCN treatment. On the other hand, both the inflammatory pathways and the signal transduction pathways were mainly negatively enriched by PCN and were positively enriched by LPS. Similar to the pattern observed in DEGs with the combined PCN and LPS treatment, most of the pathways were enriched in the same direction as the LPS treatment alone. Only a handful of inflammatory pathways were modulated in the same direction by both PCN and LPS, such as IL2 and STAT5 signaling. However, among the signal transduction processes, most of the pathways, including MAPK signaling, the JNK cascade, and the PI3K cascade, were similarly suppressed by both PCN and LPS treatments, whereas cyclin-dependent kinase and MTORC1 signaling were induced by both PCN and LPS treatments.
Figure 3.
Gene set enrichment analysis (GSEA) reveals a distinct modulation of pathways between the PCN and LPS treatments. The mice were treated with corn oil or PCN (50 mg/kg/day) for 3 days followed by saline or LPS (2 mg/kg) for 16 h. The biological processes enriched in the transcriptome footprint of liver mRNA from the treated mice were identified using gene set enrichment analysis (GSEA). The normalized enrichment score (NES) is reported for select enriched pathways (FDR-adjusted Q-value < 0.25). The key differences were observed in (a) drug metabolism pathways, (b) inflammatory pathways and (c) signal transduction pathways.
Transcription factor analysis
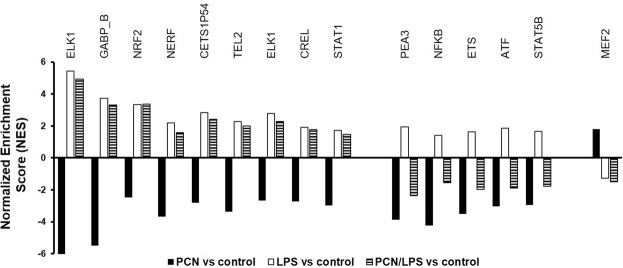
Next, we analyzed the transcription factors (TFs) that may have a role in mediating the changes in the expression of hepatic genes in mice treated with PCN +/− LPS. Using GSEA, we identified transcription factors whose targets were differentially enriched by the individual PCN or LPS treatment (Fig. 4). The top transcription factors that may be involved in the expression of upregulated and downregulated genes are shown in Table 2. After three days of PCN treatment alone, a total of 563 transcription factors were negatively enriched, and only 3 transcription factors were positively enriched, i.e., myocyte enhancer factor 2 (MEF2), nuclear factor erythroid 2 (NFE2) and peroxisome proliferator-activated receptors γ (PPARγ). Among these three, MEF2 was the only transcription factor that was also negatively enriched with LPS. However, after 16 h of LPS treatment, 472 transcription factors were negatively enriched, and 65 transcription factors were positively enriched. In the PCN/LPS group, 536 TFs were differentially expressed (upregulated: 35, downregulated: 501) compared to those in CO/Sal. Using a TRANSFAC-based motif analysis, we identified TFs that are altered by PCN or LPS (in the same or opposite direction) and might bind to Cyp3a11 (Table 3), and to the CYP3A4 promoter sequence (Supplementary Table S2). We found that among the 536 TFs, 285 had potential binding sites on Cyp3a11, including Stat1, Stat5b, Pax4, Mycmax and Pea3, along with some known mediators, such as HNF1, HNF3 and CREB.
Figure 4.
Gene set enrichment analysis (GSEA) reveals the distinct modulation of transcriptional regulators. The enrichment of transcriptional regulators in the transcriptomic response of mouse livers exposed to corn oil or PCN (50 mg/kg/day) for 3 days followed by saline or LPS (2 mg/kg) for 16 h was assessed using GSEA. An extensive search was carried out for transcriptional regulators that were enriched (FDR-adjusted Q-value < 0.25) but with targets changed in the opposite direction between the PCN and LPS treatments compared to those in the control. We report transcriptional regulators with a positive NES (acting primarily as transcriptional activators for Cyp3a11) and those with a negative NES (acting primarily as transcriptional repressors for Cyp3a11).
Table 2.
Top 15 differentially enriched transcription factors by PCN, LPS and combined PCN/LPS treatments compared those in the control.
| PCN vs Control | LPS vs Control | PCN/LPS vs Control | PCN/LPS vs PCN/Sal | PCN/LPS vs CO/LPS | |||||
|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | Up | Down |
| MEF2 | SP1 | ELK1 | FOXO4 | ELK1 | FOXO4 | ELK1 | FOXO4 | SP1 | |
| PPARG | ELK1 | GABP_B | SP1 | NRF2 | FREAC2 | GABP_B | SP1 | MAZ | |
| NFE2 | FOXO4 | NRF2 | FREAC2 | GABP_B | SP1 | NRF2 | FREAC2 | E12 | |
| MAZ | CETS1P54 | E12 | CETS1P54 | NFAT | ETS2_B | MAZ | LEF1 | ||
| ETS2_B | TEL2 | NFAT | TEL2 | MAZ | STAT5B | LEF1 | NFY | ||
| LEF1 | NFKB | MYC | NFKAPPAB65 | LEF1 | HNF4 | AP4 | FOXO4 | ||
| E12 | COUP | MAZ | NFE2 | E12 | CETS1P54 | NFAT | NFAT | ||
| GABP_B | AP1 | AP4 | CREL | AP4 | NERF | MEF2 | ELK1 | ||
| NFAT | PEA3 | LEF1 | IRF | MYOD | TEL2 | E12 | MYOD | ||
| FREAC2 | CREL | HNF3 | MAX | ERR1 | IRF | MYOD | LEF1 | ||
| AP4 | ATF | ERR1 | BACH1 | LEF1 | STAT5A | MYC | MEIS1 | ||
| HNF3 | CREB | HNF1 | USF | MYC | NFKB | ERR1 | AP4 | ||
| PU1 | BACH1 | CHX10 | ICSBP | CHX10 | PU1 | CHX10 | FREAC2 | ||
| PAX4 | MYCMAX | MYOD | AP1 | HNF3 | ICSBP | NFY | PU1 | ||
| MYC | ATF6 | NKX25 | NERF | MEF2 | SP1 | MEIS1 | E4F1 | ||
Table 3.
Altered transcription factors predicted to have putative binding sites on the Cyp3a11 promoter using TRANSFAC analysis.
| PCN↓ LPS↓ | PCN↓ LPS↑ |
|---|---|
| FREAC2 | NFKB |
| NFAT | PEA3 |
| LEF1 | STAT1 |
| HNF1 | STAT5B |
| CHX10 | AP2 |
| ERR1 | CETS1P54 |
| HNF3 | CREB |
| MYOD | PAX4 |
| GATA1 | COUP |
| FOXO4 | HSF |
| USF2 | MYCMAX |
| PAX4 | PTF1BETA |
| NFY | |
| NF1 | |
| SREBP1 |
Analysis of epigenetic changes
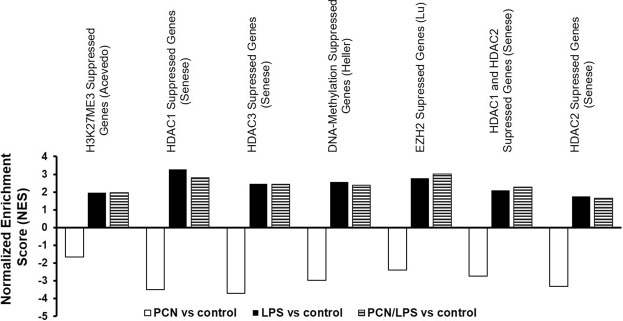
To understand the role of epigenetic modulators in regulating Cyp3a11 at the transcriptional level, a GSEA was carried out against the Molecular Signature Database (MSigDB) compendium of annotated gene sets42. Figure 5 shows the targets of epigenetic modulators that are significantly enriched but in opposite directions by a single treatment with either PCN or LPS. The targets of methylation by modulators, such as HDAC1, HDAC3, EZH2, H3K27ME3, were suppressed by PCN, but the suppression was reversed by the combined LPS and PCN treatment. Changes by these epigenetic modulators have been reported in numerous in vitro and in vivo models, and we believe that the same epigenetic modulators could also be involved in the regulation of Cyp3a11 expression and activity in models treated with PCN and LPS.
Figure 5.
Gene set enrichment analysis (GSEA) reveals distinct patterns of change for epigenetic modulators. The enrichment of epigenetic mechanisms in the mouse livers exposed to corn oil or PCN (50 mg/kg/day) for 3 days followed by saline or LPS (2 mg/kg) for 16 h was assessed using GSEA. An extensive search was carried out for epigenetic regulators that were enriched (FDR-adjusted Q-value < 0.25) but with targets changed in the opposite direction between the PCN and LPS treatments compared to those in the control.
Real-time qPCR
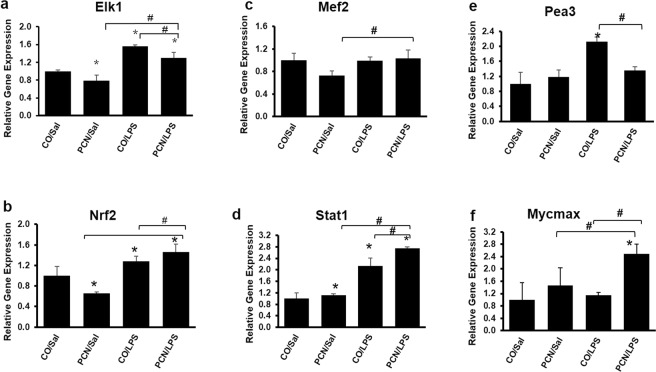
The microarray results were validated by RT-qPCR. Instead of choosing targets based solely on fold change for validation, we selected candidate genes from the list of transcription factors and epigenetic modulators that were differentially regulated between the PCN and LPS treatment groups. Two transcription factors, Elk1 and Nrf2, were selected for validation because they were maximally enriched in opposite directions by PCN and LPS. As shown in Fig. 6a,b, there was good concordance between the microarray and the RT-qPCR data for both Elk1 and Nrf2, as both are significantly downregulated by PCN and are upregulated by LPS. We also validated the expression levels of Mef2, Stat1, Pea3 and Mycmax because these transcription factors were differentially enriched by PCN and LPS in opposite directions and also might bind to the Cyp3a11 promoter according to TRANSFAC analysis. The expression of both Stat1 and Pea3 was induced by LPS treatment, and there was good concordance between the microarray analysis and RT-qPCR data, suggesting that Stat1 and Pea3 might be negative regulators of Cyp3a.
Figure 6.
Real-time qPCR analysis for the validation of gene expression of the following transcription factors: (a) Elk1, (b) Nrf2, (c) Mef2 (d) Stat1, (e) Pea3, and (f) Mycmax. The mice were treated with corn oil or PCN (50 mg/kg/day) for 3 days followed by saline or LPS (2 mg/kg) for 16 h. A few select candidate genes from the list of transcription factors that were differentially regulated between the PCN and LPS treatments were chosen for the validation of gene expression. *p < 0.05 compared to the control treatment. #p < 0.05 compared to PCN or LPS treatment alone.
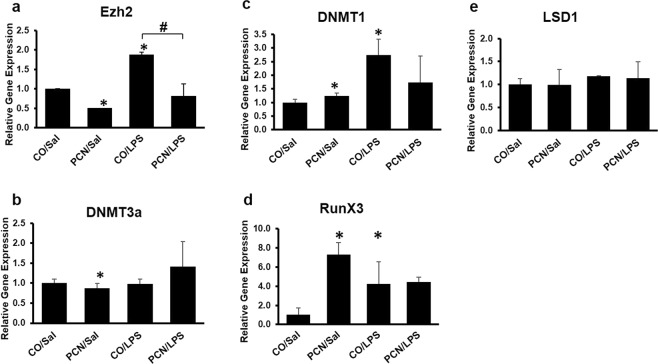
We also carried out an RT-qPCR analysis to investigate whether the actual gene expression of epigenetic markers is altered by PCN and/or LPS. We found that both EZH2 and DNMT3a were significantly downregulated with PCN treatment compared to those in the control; this is in accordance with our GSEA data (Fig. 7a,b). The combined treatment of PCN and LPS significantly reduced the gene expression of EZH2, thereby showing that PCN can attenuate the effect of LPS by regulating targets of EZH2. However, the gene expression of DNMT1 and RunX3 was significantly induced by PCN and LPS (Fig. 7c,d).
Figure 7.
Real-time qPCR analysis for the validation of gene expression of the following epigenetic modulators: (a) Ezh2, (b) DNMT3a, (c) DNMT1, (d) RunX3 and (e) LSD1. The mice were treated with corn oil or PCN (50 mg/kg/day) for 3 days followed by saline or LPS (2 mg/kg) for 16 h. A few select candidate genes from the list of epigenetic factors that were differentially regulated between the PCN and LPS treatments were chosen for the validation of gene expression. *p < 0.05 compared to the control treatment. #p < 0.05 compared to PCN or LPS treatment alone.
Discussion
In this study, we identified novel signaling pathways, transcription factors and epigenetic mechanisms that are potentially involved in the regulation of Cyp3a11 (mouse homolog of CYP3A4). Numerous mechanisms that alter the drug metabolizing enzymes (DMEs), especially CYP3A11, have been reported, but a comprehensive study of all the transcriptomic changes associated with the upregulation and downregulation of CYP3A11 has not been carried out. We found robust changes in the mouse genomic profile upon treatment with PCN, the Cyp3a11 inducer, and LPS, the endotoxin responsible for the downregulation of Cyp3a11. PCN treatment leads to the increased binding of PXR to the Cyp3a promoter/enhance region leading to Cyp3a induction, while LPS attenuates this induction; this is probably due to the reduced binding of PXR and/or additional transcription factors and co-regulators. Therefore, our analysis has led to the identification of genes encoding signaling pathways, transcription factors, coregulators, and epigenetic factors, which regulate changes in Cyp3a gene expression. Common genes that are changed by PCN and LPS in opposite directions will likely be involved in modulating Cyp3a upregulation and downregulation. These results will provide the foundation for further studies to identify changes in the binding of these factors to the Cyp3a gene and to understand how signaling pathways modulate these factors to change their expression and binding in relation to Cyp3a expression.
Genes that had the largest upregulation due to PCN treatment mainly included DMEs, such as Cyp2c55, carboxylesterases, Cyp2b10, glutathione S-transferases, and aldo-keto reductases. These results are consistent with previous studies that used chromatin immunoprecipitation sequencing (ChIP-Seq) and reported PXR binding sites on glutathione S-transferases43, carboxylesterases and most other DMEs. Most studies that showed the effects of PCN on the liver have traditionally focused on the function and inducibility of enzymes involved in drug metabolism. Interestingly, in our transcriptome analysis, PXR simultaneously induced and repressed hundreds of genes apart from DMEs, including epidermal growth factor receptor (EGFR), early growth response protein 1 (EGR1), arrestin domain containing protein 3 (ARRDC3), and cysteine sulfinic acid decarboxylase (CSAD). We therefore believe that further analysis is warranted to confirm our gene expression analysis of the regulators listed in Table 1, as the drugs that bind directly or indirectly (by the activation of PXR) to these novel regulators could further alter the expression of the Cyp3a11 gene. Since multiple EGR1 binding sites have previously been identified within the 5′-regulatory promoter region of the CYP2B6 gene44,45, its altered gene expression may imply potential involvement in the regulation of Cyp3a11. With LPS treatment alone, the gene expression pattern of numerous inflammatory mediators was differentially regulated, including serum amyloid A3 (SAA3), chemokine ligand 9 (CXCL9), cluster of differentiation 14 (CD14), and metallothionein 2 (MT2). The accurate and comprehensive knowledge of regulators that are differentially regulated by LPS can help identify factors that may potentially affect Cyp3a11.
Although transcriptome profiling using a DNA microarray has become a mainstay of genomics research46,47, the challenge no longer lies in obtaining differential gene expression patterns, but rather, is in interpreting the results to gain insights into the biological mechanisms. A powerful analytical tool is gene set enrichment analysis (GSEA), in which all expressed genes are ranked according to their differential expression, then the enrichment scores for each pathway or gene set of interest are computed. The normalized enrichment scores (NES) and the statistical measures of significance (P-value and FDR-adjusted Q-value) for specific pathways or gene sets are determined by performing 1000 permutations of the rank file, re-computing the enrichment score for each gene set and permutation, and finally integrating the results of all 1000 permutations. GSEA then determines the degree of representation of the members in a gene set by ranking them in a list from the top (positive enrichment) to the bottom (negative enrichment)41. The transcriptome profiling of mouse livers after PCN and LPS treatments robustly identified numerous genes, including some genes with functions related to drug metabolism, cell cycle kinetics and inflammation mediation. We broadly selected pathways that belong to the following three major mechanisms for enrichment analysis: drug metabolism (DM), inflammatory regulation (IR) or signal transduction (ST). We observed that most of the drug metabolism pathways were positively enriched by PCN and were downregulated by LPS; this is consistent with the changes in Cyp3a11 gene expression. Taking a closer look into the subsets of genes belonging to these pathways, we found that although Cyp3a11 is positively enriched, multiple drug metabolizing enzymes, such as glutathione S transferase A3, aldo-keto reductase 1C6 or alcohol dehydrogenase 1 are negatively enriched; this negatively shifts the total enrichment score of the pathway by treatment with either PCN or LPS. In contrast to the DM pathways, most of the IR and ST pathways were found to be negatively enriched by PXR activation and positively enriched by LPS treatment and by the combined treatment. Understanding which pathways are enriched by PCN and LPS is crucial because this may affect the components involved in the transcriptional regulation of not only Cyp3a11 but also other DMEs.
The gene transcription of Cyp3a11 is largely regulated by transcription factor (TF) proteins that bind to genomic cis-regulatory elements that are characterized by precise DNA motifs. Changes in the gene expression of TFs are usually not detected in microarray experiments, because their activity is primarily regulated by ligand binding or by posttranscriptional modifications48. We employed GSEA to find the top transcription factors whose targets were enriched in opposite directions by PCN and LPS. One of the top transcription factors was Elk1, an ETS family transcription factor that is responsible for target gene transcription upon mitogen-activated protein kinase-signaling pathway stimulation49. Elk1 was downregulated by PCN and was upregulated by LPS, and the combined treatment followed the LPS response. In fact, most of the transcription factors, such as Tel2, Pea3, Stat1, and Stat5b that were changed in opposite directions were suppressed by PCN and were positively enriched by LPS treatment; this suggests that these transcription factors might negatively regulate basal Cyp3a11 expression. This fact was strengthened by previous reports showing that the loss of Stat5b increased the gene expression of Cyp3a in mice50. LPS-mediated activation of NF-κB has also been shown to play a significant role in the downregulation of the Cyp3a enzyme40,51. On the other hand, Mef2 was the only transcription factor that was positively enriched by PCN and was attenuated by LPS in our study. Mef2 regulates cell differentiation, proliferation, morphogenesis, survival and apoptosis in a wide range of cell types52, and a previous microarray analysis has revealed that a few DMEs, such as CYP1B1, and nuclear receptors, such as Ahr, are downregulated in the absence of Mef253. Although the actual gene expression of Mef2 was not induced by PCN in our data, it might still be involved in altering the expression of its downstream genes. Further studies to understand the role of Mef2 in the regulation of the CYP3A enzyme need to be carried out.
Furthermore, a TRANSFAC analysis was carried out to investigate whether these enriched transcription factors have any binding sites on the Cyp3a promoter and enhancer regions. TRANSFAC (TRANScription FACtor database) is a manually curated database of eukaryotic transcription factors, their genomic binding sites and their DNA binding profiles. The contents of this database can be used to predict potential transcription factor binding sites. Some transcription factors that are already known to bind to Cyp3a, e.g., HNF1, HNF3, CREB and COUP, were also identified by TRANSFAC, validating our analysis. Other transcription factors that may have potential binding sites on Cyp3a are listed in Table 3, and RT-qPCR was performed to investigate whether PCN and LPS alter the gene expression of these transcription factors. The data from this analysis provide novel insights into the mechanisms involved in the regulation of human CYP3A4 and suggest new therapeutic targets to treat disorders that are caused by an altered drug metabolism.
Lastly, recent studies have demonstrated that many other factors, such as epigenetics54 and micro RNAs (miRNAs)55, may modulate DME gene expression and may cause variations in drug metabolism and toxicity. The effect of epigenetic processes on pharmacologically relevant genes and, ultimately, drug response is a relatively new area of research56. Among all the epigenetic changes, changes in the DNA methylation profiles determine whether there is a permissive chromatin state for the transcription machinery to access the gene promoter regions and to initiate transcription57,58. DNA methylation is a key epigenetic mechanism and a covalent modification that results in stable gene silencing59. In our data, we found that genes that are suppressed by DNA methylation by modulators, such as enhancer of zeste homolog 2 (Ezh2) in previous studies60, were further suppressed by PCN, and this effect was relieved by LPS. We further carried out an RT-qPCR analysis to measure the gene expression of the methylation modulator Ezh2 in our model. We found that Ezh2 gene expression was suppressed by PCN and was induced by LPS; these results indicate that Ezh2 could also have a significant role in the regulation of the CYP3A enzyme. Future studies to determine whether Ezh2 methylates Cyp3a11 need to be conducted. Enrichment of histone-3-lysine-27 trimethylation (H3K27me3) in promoters and gene bodies has also been associated with the inactivation of gene transcription61,62. Li et al. found that increased H3K27me3 within the margins of the Cyp3a16 gene may be responsible for switching off Cyp3a16 gene expression in the livers of adult mice63. In addition to being homologous to the human CYP3A isoforms in DNA and protein sequences, the mouse Cyp3a11 and Cyp3a16 homologues also mimic a developmental switch, such as human CYP3A4 and CYP3A7 64. In our GSEA, we found that the genes that were downregulated in liver tumors by H3K27me365 were further suppressed by PCN and relieved by LPS. This could imply that although H3K27me3 might be responsible for the switch of Cyp3a16 to Cyp3a11, high levels of H3K27me3 could be responsible for the decreased expression of Cyp3a11 in the adult liver. However, further methylation-specific studies need to be carried out to confirm the involvement of H3K27me3 in the regulation of Cyp3a11 in adult mice. Apart from epigenetic modulation, microRNA-27b (miR-27b) and mouse microRNA-298 (mmu-miR-298) have previously been shown to downregulate CYP3A4 expression66. Hence, GSEA was performed to understand the involvement of miRNAs in the regulation of Cyp3a11; however, PCN and LPS did not significantly enrich any miRNAs in opposite directions in our model (data not shown). It is possible that LPS can change the miRNA expression at different time-points, and LPS is known to have a temporal effect on the expression of miRNAs and genes.
In conclusion, we carried out a whole-transcriptome analysis to understand the novel molecular mechanisms that are associated with the downregulation of the Cyp3a11 enzyme. Using high-throughput microarray technology, we screened a large number of genes to detect changes stimulated by individual PCN or LPS treatments, as well as by their combined treatment. Potential transcription factors that are altered by PCN and LPS in opposite directions and might be involved in the regulation of the Cyp3a gene were identified, such as Pea3 and Stat1. Their differential expression was validated, and future studies will entail chromatin immunoprecipitation assays to investigate their binding to the Cyp3a promoter. The results from this study further enhance our understanding of the intricate network of different cell signaling pathways and epigenetic mechanisms with nuclear receptors, such as PXR. In addition, Cyp3a might be a potential target of DNA methylation by epigenetic modulators, such as Ezh2; hence, its exact role needs to be further investigated. Since PXR is involved in the regulation of several DMEs other than CYP3A, these novel pathways, transcription factors and epigenetic modulators could be involved in the regulation of numerous other genes controlled by PXR.
Materials and Methods
Reagents and materials
5-Pregnen-3β-ol-20-one-16α-carbonitrile (#P0543) was purchased from Sigma-Aldrich (St. Louis, MO). Lipopolysaccharide (E. coli, #tlrl-pslta) was purchased from InvivoGen (San Diego, CA). The RNeasy Mini Kit (#74104) was obtained from Qiagen (Valencia, CA). A 96-well PCR plate, Roche PCR Master Mix (Roche Diagnostics), TaqMan® primer and probes for Cyp3a11 (FP: GGATGAGATCGATGAGGCTCTG, RP: CAGGTATTCCATCTCCATCACAGT) and cyclophilin (FP: GGCCGATGACGAGCCC, RP: TGTCTTTGGAACTTTGTCTGCA) were purchased from Sigma-Genosys (Houston, TX). A Mouse WG-6 v2.0 expression BeadChip Kit was obtained from Illumina (San Diego, CA). TaqMan® Gene Expression assays with primers and probes for mouse Elk1 (#Mm00468233_g1), Mef2 (#Mm01340842_m1), Nrf2 (#Mm00477784_m1), Pea3 (#Mm00476696_m1), Stat1 (#Mm01257286_m1), and Mycmax (#Mm00487804_m1) were obtained from Thermo Fisher Scientific (#4331182, Waltham, MA). The primers for the epigenetic markers Ezh2, DNMT1, DNMT3a, LSD1 and RunX3 were a kind gift from Dr. Moorthy from the Baylor College of Medicine, Houston, TX.
Animals and treatments
Adult C57BL/6 mice (~6 weeks, male, Jackson Labs, Stock no. 000664) were allowed to acclimate to the animal care facility for 7 days. The mice were maintained in a temperature- and humidity-controlled environment, and all animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Houston, Houston, TX. All experiments were performed in accordance with the relevant guidelines and regulations of the committee. The mice were fed a standard diet. Both the food and water could be accessed ad libitum, and the mice were maintained in a 12 h day/night cycle. The mice were treated with PCN (50 mg/kg/day) or corn oil I.P. for 3 days followed by LPS (2 mg/kg/day) or saline I.P. for 16 h. After treatment, the animals were anesthetized with isoflurane and euthanized by cervical dislocation under deep anesthesia. The liver tissues were harvested for further analysis.
RNA Isolation
We used a total of 4 animals per treatment group. The total RNA from the liver samples of mice treated with PCN/LPS was isolated using the RNeasy kit according to the manufacturer’s standard protocols (Qiagen, Valencia, CA). Following the total RNA isolation, the sample concentration was assayed using a NanoDrop-8000 (Thermo Scientific, Wilmington, DE, USA), and quality checks were performed using the NanoDrop spectrophotometer and the Agilent Bioanalyzer. RNA quality parameters were as follows: the 260/280 and 260/230 ratios needed to be greater than 1.8. Further, the RNA Integrity Number (RIN) was analyzed using the Agilent Bioanalyzer. The samples needed to have RIN values of 7–10 and with a range of 1–1.5.
Real-time qPCR analysis
The cDNA was synthesized from the isolated total mRNA using the High Capacity Reverse Transcription Kit from Applied Biosystems. Real-time PCR was performed using an ABI PRISM 7300 Sequence Detection System instrument and software (Applied Biosystems; Foster City, CA) as described previously34,39,67. In short, each reaction mixture (total volume of 25 ml) contained 50–100 ng cDNA, 300 nM forward primer, 300 nM reverse primer, 200 nM fluorogenic probe, and 15 ml TaqMan Universal PCR Master Mix. We extrapolated the quantitative expression values from the standard curves, and these values were normalized to the value of cyclophilin.
Immunoblotting
Whole liver extracts were prepared as described previously68, and the protein concentration was determined using the bicinchoninic acid assay according to the manufacturer’s protocol (Pierce, Rockford, IL). Equal amounts of protein (10 mg) were analyzed by SDS-polyacrylamide gel electrophoresis and were transferred onto a nitrocellulose membrane. The membranes were then probed with a rabbit anti-CYP3A11 antibody, followed by probing with a goat anti-rabbit IgG-alkaline phosphatase secondary antibody. The membranes were then washed and incubated with Tropix CDP star nitroblock II ECL reagent as per the manufacturers’ instructions (Applied Biosystems). The membranes were analyzed using a FlourChem FC imaging system (Alpha Innotech). The images were quantified by densitometry using AlphaEase software.
CYP3A11 enzyme activity assay
The mouse liver microsomes were prepared as described previously68, and the protein concentration of the microsomal fractions was determined using a BCA protein assay kit (Pierce, Rockford, IL, USA) using bovine serum albumin (BSA) as the standard. The CYP3A11 enzyme activity was determined in the mouse liver microsomes using the CYP3A substrate midazolam (MDZ) as described previously with minor modifications69. The formation of 1′-OHMDZ from MDZ was used as a specific indicator for mouse CYP3A11 activity. In brief, 0.1 mg of total microsomal protein was incubated with MDZ (0–16 μM), 1.3 mM NADPH and reaction cofactors in 50 mM potassium phosphate buffer (pH 7.4). The reaction was initiated by the addition of glucose-6-phosphate dehydrogenase (1-unit mL−1). After 5 min, the reactions were stopped by the addition of an equal volume of acetonitrile containing phenacetin as the internal standard (IS). The incubation mixture was centrifuged at 13,000 rpm at 4 °C for 10 min, and the supernatant was transferred to a 96-well autosampling plate for LC-MS/MS analysis. The identity of 1′OHMDZ and IS was verified by comparing with authenticated standards. The data were fit to the standard Michaelis-Menten rate equation.
Microarray analysis
A total of 250 ng of total RNA was reverse transcribed, and microarray hybridization was performed using the Illumina Gene Expression Mouse WG-6 v2.0 Expression BeadChip Kit at the Laboratory for Translational Genomics at Baylor College of Medicine. The transcriptome profile data were quartile-normalized by the Bioconductor Lumi package. The Lumi package implemented in the R statistical software, version 2.14.1, was used to perform quality control of the signal intensity data on the transcript probes, background adjustment, variance stabilization transformation, and rank invariant normalization. A detection p-value cutoff of 0.01 was required for the normalized intensities to consider a transcript as detected. The differentially expressed genes (DEGs) were selected following the t-test comparisons among the groups of interest using the R statistical system. The genes were considered to be differentially expressed for a p-value < 0.05 and a fold change greater than or equal to 1.25-fold or less than or equal to 0.8-fold. A graphical representation of the DEGs was generated in the form of heatmaps of mean-centered normalized expression values (z-scores) with the Euclidean distance metric and the average clustering method using R statistical software.
Pathway enrichment and transcription factor analysis
A rank file for each comparison was created based on the log2-fold change of each gene between the respective comparison groups. We next employed the gene set enrichment analysis (GSEA) methodology and software41 against the Molecular Signature database (MSigDB) compendium42 of gene sets. Gene set enrichment analysis first finds an aggregate gene set score (termed the enrichment score (ES)) and then runs 1000 permutations to establish a background distribution for the ES. The ratio between the ES and the average ES is termed the normalized enrichment score (NES). GSEA determines whether a key component of a pathway or biological process gene set is significantly and preferentially enriched in the upregulated genes (NES > 0, FDR-adjusted Q-value < 0.25) or in the downregulated genes (NES < 0, FDR-adjusted Q-value < 0.25). An established paradigm to generate a hypothesis is that if the NES values for a pathway that compares two different treatments are significant but have opposite signs, then the treatments might direct the pathways in opposite directions. The following pathways were used to determine the enriched pathways: KEGG, Reactome, Hallmark, and GOBP (Gene Ontology Biological Processes). We also used a compendium of putative transcription factors to identify enriched transcription factor targets in the transcriptome footprints analyzed. A TransFac analysis was employed to identify the list of transcription factors that might bind to Cyp3a11 or the CYP3A4 promoter regions.
Statistical analysis
The real-time PCR data are shown as the mean and are analyzed with Student’s t-test or one-way analysis of variance for all groups, followed by Tukey’s post-hoc test for pairwise comparisons. The significant values are represented as P < 0.05.
Supplementary information
Acknowledgements
This work was supported in part by PHS grants 1R21DA035751 to R.G. and B.M. and grants 1R01ES029382, 1R01HL129794, and 5R01ES009132 to B.M., C.C. and S.M. were partially supported by the Cancer Prevention and Research Institute of Texas (CPRIT) grant RP170005. C.C. was partially supported by the CPRIT grant RP170295 and NIH/NIDDK R01DK111522.
Author Contributions
Conceived and designed the experiments: G.T., B.M., C.C., R.G. Performed the experiments: G.T., W.J. Analyzed the data: G.T., S.M., C.C. Contributed reagents/materials/analysis tools: C.C., B.M., R.G. Contributed towards the writing of the manuscript: G.T., B.M., C.C., R.G.
Data Availability
The microarray dataset generated during the current study will be made available in the Gene Expression Omnibus (GEO).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bhagavatula Moorthy, Email: bmoorthy@bcm.edu.
Cristian Coarfa, Email: coarfa@bcm.edu.
Romi Ghose, Email: rghose@central.uh.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-43248-w.
References
- 1.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Veith H, et al. Comprehensive characterization of cytochrome P450 isozyme selectivity across chemical libraries. Nat. Biotechnol. 2009;27:1050–1055. doi: 10.1038/nbt.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson GR. Drug metabolism and variability among patients in drug response. N. Engl. J. Med. 2005;352:2211–2221. doi: 10.1056/NEJMra032424. [DOI] [PubMed] [Google Scholar]
- 4.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, Zhang YD, Zhao P, Huang S-M. Predicting drug-drug interactions: an FDA perspective. AAPS J. 2009;11:300–306. doi: 10.1208/s12248-009-9106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallick P, Shah P, Gandhi A, Ghose R. Impact of obesity on accumulation of the toxic irinotecan metabolite, SN-38, in mice. Life Sci. 2015;139:132–138. doi: 10.1016/j.lfs.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Mallick P, Taneja G, Moorthy B, Ghose R. Regulation of drug-metabolizing enzymes in infectious and inflammatory disease: implications for biologics-small molecule drug interactions. Expert Opin. Drug Metab. Toxicol. 2017;13:605–616. doi: 10.1080/17425255.2017.1292251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch T, Price A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Physician. 2007;76:391–396. [PubMed] [Google Scholar]
- 9.Palleria C, et al. Pharmacokinetic drug-drug interaction and their implication in clinical management. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2013;18:601–610. [PMC free article] [PubMed] [Google Scholar]
- 10.Sabo JP, et al. Clinical assessment of potential drug interactions of faldaprevir, a hepatitis C virus protease inhibitor, with darunavir/ritonavir, efavirenz, and tenofovir. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2014;59:1420–1428. doi: 10.1093/cid/ciu616. [DOI] [PubMed] [Google Scholar]
- 11.Benoist GE, et al. Drug–drug interaction potential in men treated with enzalutamide: Mind the gap. Br. J. Clin. Pharmacol. 2018;84:122–129. doi: 10.1111/bcp.13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, X. et al. Pharmacokinetic Interactions of Rolapitant With Cytochrome P450 3A Substrates in Healthy Subjects. J. Clin. Pharmacol, 10.1002/jcph.1339 (2018). [DOI] [PubMed]
- 13.Jover R, Bort R, Gómez-Lechón MJ, Castell JV. Down-regulation of human CYP3A4 by the inflammatory signal interleukin-6: molecular mechanism and transcription factors involved. FASEB. J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2002;16:1799–1801. doi: 10.1096/fj.02-0195fje. [DOI] [PubMed] [Google Scholar]
- 14.Rodríguez-Antona C, et al. Transcriptional regulation of human CYP3A4 basal expression by CCAAT enhancer-binding protein alpha and hepatocyte nuclear factor-3 gamma. Mol. Pharmacol. 2003;63:1180–1189. doi: 10.1124/mol.63.5.1180. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-Jiménez CP, Gómez-Lechón MJ, Castell JV, Jover R. Transcriptional regulation of the human hepatic CYP3A4: identification of a new distal enhancer region responsive to CCAAT/enhancer-binding protein beta isoforms (liver activating protein and liver inhibitory protein) Mol. Pharmacol. 2005;67:2088–2101. doi: 10.1124/mol.104.008169. [DOI] [PubMed] [Google Scholar]
- 16.Matsumura K, et al. Identification of a novel polymorphic enhancer of the human CYP3A4 gene. Mol. Pharmacol. 2004;65:326–334. doi: 10.1124/mol.65.2.326. [DOI] [PubMed] [Google Scholar]
- 17.Tirona RG, et al. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat. Med. 2003;9:220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 18.Biggs JS, et al. Transcription factor binding to a putative double E-box motif represses CYP3A4 expression in human lung cells. Mol. Pharmacol. 2007;72:514–525. doi: 10.1124/mol.106.033795. [DOI] [PubMed] [Google Scholar]
- 19.Luo G, Guenthner T, Gan L-S, Humphreys WG. CYP3A4 induction by xenobiotics: biochemistry, experimental methods and impact on drug discovery and development. Curr. Drug Metab. 2004;5:483–505. doi: 10.2174/1389200043335397. [DOI] [PubMed] [Google Scholar]
- 20.Pascussi JM, Gerbal-Chaloin S, Drocourt L, Maurel P, Vilarem MJ. The expression of CYP2B6, CYP2C9 and CYP3A4 genes: a tangle of networks of nuclear and steroid receptors. Biochim. Biophys. Acta. 2003;1619:243–253. doi: 10.1016/S0304-4165(02)00483-X. [DOI] [PubMed] [Google Scholar]
- 21.Kojima K, Nagata K, Matsubara T, Yamazoe Y. Broad but distinct role of pregnane x receptor on the expression of individual cytochrome p450s in human hepatocytes. Drug Metab. Pharmacokinet. 2007;22:276–286. doi: 10.2133/dmpk.22.276. [DOI] [PubMed] [Google Scholar]
- 22.Liu F-J, Song X, Yang D, Deng R, Yan B. The far and distal enhancers in the CYP3A4 gene co-ordinate the proximal promoter in responding similarly to the pregnane X receptor but differentially to hepatocyte nuclear factor-4alpha. Biochem. J. 2008;409:243–250. doi: 10.1042/BJ20070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodwin B, Hodgson E, Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol. Pharmacol. 1999;56:1329–1339. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- 24.Timsit YE, Negishi M. CAR and PXR: the xenobiotic-sensing receptors. Steroids. 2007;72:231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pascussi J-M, et al. The tangle of nuclear receptors that controls xenobiotic metabolism and transport: crosstalk and consequences. Annu. Rev. Pharmacol. Toxicol. 2008;48:1–32. doi: 10.1146/annurev.pharmtox.47.120505.105349. [DOI] [PubMed] [Google Scholar]
- 26.Ding X, Staudinger JL. Induction of drug metabolism by forskolin: the role of the pregnane X receptor and the protein kinase a signal transduction pathway. J. Pharmacol. Exp. Ther. 2005;312:849–856. doi: 10.1124/jpet.104.076331. [DOI] [PubMed] [Google Scholar]
- 27.Lichti-Kaiser K, Xu C, Staudinger JL. Cyclic AMP-dependent protein kinase signaling modulates pregnane x receptor activity in a species-specific manner. J. Biol. Chem. 2009;284:6639–6649. doi: 10.1074/jbc.M807426200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding X, Staudinger JL. Repression of PXR-mediated induction of hepatic CYP3A gene expression by protein kinase C. Biochem. Pharmacol. 2005;69:867–873. doi: 10.1016/j.bcp.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 29.Taneja G, Chu C, Maturu P, Moorthy B, Ghose R. Role of c-Jun-N-Terminal Kinase in Pregnane X Receptor-Mediated Induction of Human Cytochrome P4503A4 In Vitro. Drug Metab. Dispos. Biol. Fate Chem. 2018;46:397–404. doi: 10.1124/dmd.117.079160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crawford JM, Boyer JL. Clinicopathology conferences: inflammation-induced cholestasis. Hepatol. Baltim. Md. 1998;28:253–260. doi: 10.1002/hep.510280133. [DOI] [PubMed] [Google Scholar]
- 31.Morgan ET. Suppression of constitutive cytochrome P-450 gene expression in livers of rats undergoing an acute phase response to endotoxin. Mol. Pharmacol. 1989;36:699–707. [PubMed] [Google Scholar]
- 32.Renton KW, Nicholson TE. Hepatic and central nervous system cytochrome P450 are down-regulated during lipopolysaccharide-evoked localized inflammation in brain. J. Pharmacol. Exp. Ther. 2000;294:524–530. [PubMed] [Google Scholar]
- 33.Monshouwer M, et al. A lipopolysaccharide-induced acute phase response in the pig is associated with a decrease in hepatic cytochrome P450-mediated drug metabolism. J. Vet. Pharmacol. Ther. 1996;19:382–388. doi: 10.1111/j.1365-2885.1996.tb00068.x. [DOI] [PubMed] [Google Scholar]
- 34.Ghose R, Zimmerman TL, Thevananther S, Karpen SJ. Endotoxin leads to rapid subcellular re-localization of hepatic RXRalpha: A novel mechanism for reduced hepatic gene expression in inflammation. Nucl. Recept. 2004;2:4. doi: 10.1186/1478-1336-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renton KW. Cytochrome P450 regulation and drug biotransformation during inflammation and infection. Curr. Drug Metab. 2004;5:235–243. doi: 10.2174/1389200043335559. [DOI] [PubMed] [Google Scholar]
- 36.Aitken AE, Richardson TA, Morgan ET. Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu. Rev. Pharmacol. Toxicol. 2006;46:123–149. doi: 10.1146/annurev.pharmtox.46.120604.141059. [DOI] [PubMed] [Google Scholar]
- 37.Yu R, et al. Role of a mitogen-activated protein kinase pathway in the induction of phase II detoxifying enzymes by chemicals. J. Biol. Chem. 1999;274:27545–27552. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- 38.Tan Z, Huang M, Puga A, Xia Y. A critical role for MAP kinases in the control of Ah receptor complex activity. Toxicol. Sci. Off. J. Soc. Toxicol. 2004;82:80–87. doi: 10.1093/toxsci/kfh228. [DOI] [PubMed] [Google Scholar]
- 39.Ghose R, Guo T, Haque N. Regulation of gene expression of hepatic drug metabolizing enzymes and transporters by the Toll-like receptor 2 ligand, lipoteichoic acid. Arch. Biochem. Biophys. 2009;481:123–130. doi: 10.1016/j.abb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu X, et al. Role of NF-kappaB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. J. Biol. Chem. 2006;281:17882–17889. doi: 10.1074/jbc.M601302200. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinforma. Oxf. Engl. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui JY, Choudhuri S, Knight TR, Klaassen CD. Genetic and epigenetic regulation and expression signatures of glutathione S-transferases in developing mouse liver. Toxicol. Sci. Off. J. Soc. Toxicol. 2010;116:32–43. doi: 10.1093/toxsci/kfq115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoue K, Negishi M. Nuclear receptor CAR requires early growth response 1 to activate the human cytochrome P450 2B6 gene. J. Biol. Chem. 2008;283:10425–10432. doi: 10.1074/jbc.M800729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inoue K, Negishi M. Early growth response 1 loops the CYP2B6 promoter for synergistic activation by the distal and proximal nuclear receptors CAR and HNF4alpha. FEBS Lett. 2009;583:2126–2130. doi: 10.1016/j.febslet.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 47.Lockhart DJ, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 48.Everett L, Hansen M, Hannenhalli S. Regulating the regulators: modulators of transcription factor activity. Methods Mol. Biol. Clifton NJ. 2010;674:297–312. doi: 10.1007/978-1-60761-854-6_19. [DOI] [PubMed] [Google Scholar]
- 49.Besnard A, Galan-Rodriguez B, Vanhoutte P, Caboche J. Elk-1 a transcription factor with multiple facets in the brain. Front. Neurosci. 2011;5:35. doi: 10.3389/fnins.2011.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park SH, Liu X, Hennighausen L, Davey HW, Waxman DJ. Distinctive roles of STAT5a and STAT5b in sexual dimorphism of hepatic P450 gene expression. Impact of STAT5a gene disruption. J. Biol. Chem. 1999;274:7421–7430. doi: 10.1074/jbc.274.11.7421. [DOI] [PubMed] [Google Scholar]
- 51.Abdulla D, Goralski KB, Del Busto Cano EG, Renton KW. The signal transduction pathways involved in hepatic cytochrome P450 regulation in the rat during a lipopolysaccharide-induced model of central nervous system inflammation. Drug Metab. Dispos. Biol. Fate Chem. 2005;33:1521–1531. doi: 10.1124/dmd.105.004564. [DOI] [PubMed] [Google Scholar]
- 52.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Dev. Camb. Engl. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 53.Di-Luoffo M, Daems C, Bergeron F, Tremblay JJ. Novel Targets for the Transcription Factors MEF2 in MA-10 Leydig Cells. Biol. Reprod. 2015;93:9. doi: 10.1095/biolreprod.114.127761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ingelman-Sundberg M, et al. Potential role of epigenetic mechanisms in the regulation of drug metabolism and transport. Drug Metab. Dispos. Biol. Fate Chem. 2013;41:1725–1731. doi: 10.1124/dmd.113.053157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu A-M, Tian Y, Tu M-J, Ho PY, Jilek JL. MicroRNA Pharmacoepigenetics: Posttranscriptional Regulation Mechanisms behind Variable Drug Disposition and Strategy to Develop More Effective Therapy. Drug Metab. Dispos. Biol. Fate Chem. 2016;44:308–319. doi: 10.1124/dmd.115.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ingelman-Sundberg M, Gomez A. The past, present and future of pharmacoepigenomics. Pharmacogenomics. 2010;11:625–627. doi: 10.2217/pgs.10.59. [DOI] [PubMed] [Google Scholar]
- 57.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 58.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 60.Lu C, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 62.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Cui Y, Hart SN, Klaassen CD, Zhong X. Dynamic patterns of histone methylation are associated with ontogenic expression of the Cyp3a genes during mouse liver maturation. Mol. Pharmacol. 2009;75:1171–1179. doi: 10.1124/mol.108.052993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hart SN, Cui Y, Klaassen CD, Zhong X. Three patterns of cytochrome P450 gene expression during liver maturation in mice. Drug Metab. Dispos. Biol. Fate Chem. 2009;37:116–121. doi: 10.1124/dmd.108.023812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Acevedo LG, Bieda M, Green R, Farnham PJ. Analysis of the mechanisms mediating tumor-specific changes in gene expression in human liver tumors. Cancer Res. 2008;68:2641–2651. doi: 10.1158/0008-5472.CAN-07-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pan Y-Z, Gao W, Yu A-M. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab. Dispos. Biol. Fate Chem. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shah P, Guo T, Moore DD, Ghose R. Role of constitutive androstane receptor in Toll-like receptor-mediated regulation of gene expression of hepatic drug-metabolizing enzymes and transporters. Drug Metab. Dispos. Biol. Fate Chem. 2014;42:172–181. doi: 10.1124/dmd.113.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ghose R, Guo T, Vallejo JG, Gandhi A. Differential role of Toll-interleukin 1 receptor domain-containing adaptor protein in Toll-like receptor 2-mediated regulation of gene expression of hepatic cytokines and drug-metabolizing enzymes. Drug Metab. Dispos. Biol. Fate Chem. 2011;39:874–881. doi: 10.1124/dmd.110.037382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He P, Court MH, Greenblatt DJ, von Moltke LL. Factors influencing midazolam hydroxylation activity in human liver microsomes. Drug Metab. Dispos. Biol. Fate Chem. 2006;34:1198–1207. doi: 10.1124/dmd.105.008904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray dataset generated during the current study will be made available in the Gene Expression Omnibus (GEO).