Abstract
The SELEX (Systematic Evolution of Ligands by EXponential enrichment) method has been used successfully since 1990, but work is still required to obtain highly specific aptamers. Here, we present a novel approach called ‘Competitive non-SELEX’ (and termed as ‘SELCOS’ (Systematic Evolution of Ligands by COmpetitive Selection)) for readily obtaining aptamers that can discriminate between highly similar targets. This approach is based on the theoretical background presented here, in which under the co-presence of two similar targets, a specific binding type can be enriched more than a nonspecifically binding one during repetitive steps of partitioning with no PCR amplification between them. This principle was experimentally confirmed by the selection experiment for influenza virus subtype-specific DNA aptamers. Namely, the selection products (pools of DNA aptamers) obtained by SELCOS were subjected to a DEPSOR-mode electrochemical sensor, enabling the method to select subtype-specific aptamer pools. From the clonal analysis of these pools, only a few rounds of in vitro selection were sufficient to achieve the surprisingly rapid enrichment of a small number of aptamers with high selectivity, which could be attributed to the SELCOS principle and the given selection pressure program. The subtype-specific aptamers obtained in this manner had a high affinity (e.g., KD = 82 pM for H1N1; 88 pM for H3N2) and negligible cross-reactivity. By making the H1N1-specific DNA aptamer a sensor unit of the DEPSOR electrochemical detector, an influenza virus subtype-specific and portable detector was readily constructed, indicating how close it is to the field application goal.
Subject terms: DNA, Screening
Introduction
The rapid, precise, and selective detection of viruses is absolutely required to prevent breakouts/pandemics. This is especially true of the highly infectious influenza virus. Thus, various approaches have been explored for this purpose, including antibody engineering. Among the available methods, an in vitro selection termed SELEX (Systematic Evolution of Ligands by EXponential enrichment) has allowed researchers to identify a diversity of DNA/RNA aptamer molecules with potential use in virus detection. SELEX is operated using an iterative cycle of three fundamental steps, namely binding, partitioning, and amplification, and it can gradually enrich target-binding DNA/RNA molecules over the selection cycle1–3. Although the SELEX protocol has long been performed with success4,5, the difficulty involved in selecting aptamers with high specificity remains6–8. The current approach to this problem uses “negative selection,” which is universally applied to select aptamers that bind to a molecule of interest from a pool of non-bound molecules to a particular target of no interest (thus, they are negatively selected). This approach is widely applied, and in the case of SELEX, for example, there are reports that negative selection had the greatest positive results in selecting for cell-specific aptamers9. Although this approach is useful, in principle, it requires multiple rounds of negative and positive selections. The SELEX process essentially requires many rounds of selection using PCR, leading to the amplification of undesired biases10–13. Unfortunately, the final success ratio of SELEX-based experiments has not been high8,14,15 although some cases were clearly successful16,17. Therefore, SELEX-based technology requires some effective improvements.
Here, we propose a novel approach for obtaining selective aptamers without PCR amplification procedures, namely ‘SELCOS’ (Systemic Evolution of Ligands by COmpetitive Selection), in which in vitro selection is performed using a solution system containing all the positive and negative targets. In this paper, we showed the plausibility of using SELCOS on close targets of influenza virus subtypes (H1N1 and H3N2). We also introduced a DEPSOR-mode electrochemical sensing method (or Apta-DEPSOR)18,19 for readily evaluating the specific binding of aptamers. On the whole, a powerful approach for rapidly detecting various influenza subtypes with high sensitivity is presented here, and it addresses several theoretical considerations.
Results
Integration of competitive selection and electrochemical evaluation
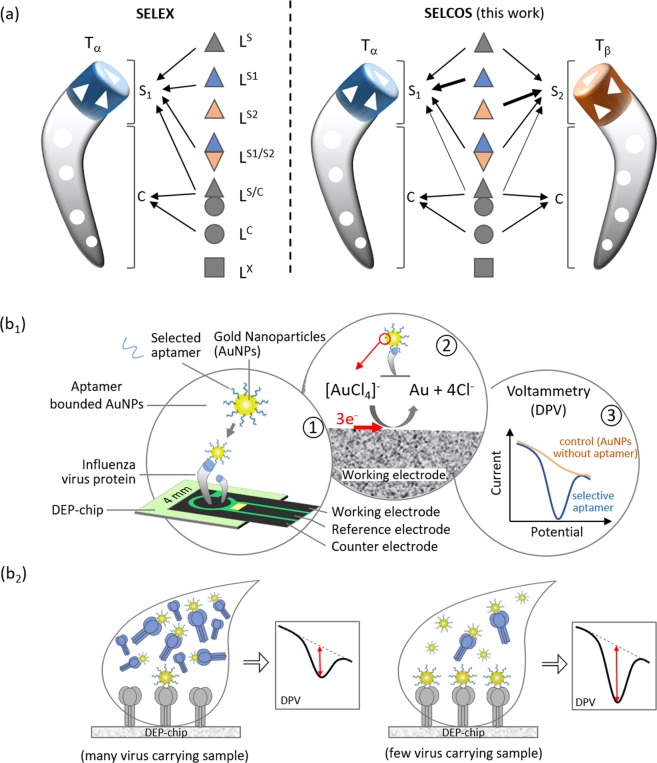
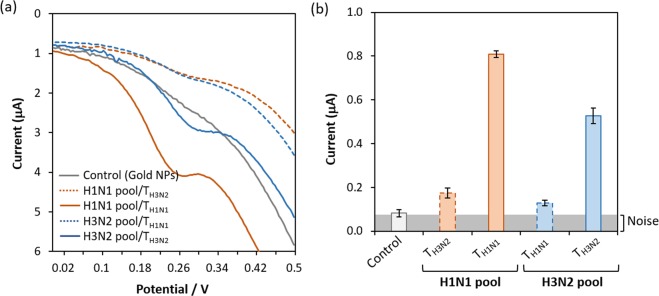
As shown in Fig. 1, the pool of ligands (aptamer candidates) consists of various molecules that can be named LS, LS1, LS2, LS1/S2, LC, LS/C, and LX depending on their binding nature in relation to the target molecules Tα and Tβ (see details in the legend to Fig. 1). Clearly, there is a difference in their behaviors under conventional SELEX and SELCOS, which holds two or more target molecules. Those targets compete with one another for common ligands (especially, LS, LS1/S2 and LS/C) that can bind both targets Tα and Tβ during SELCOS but exclusively Tα in conventional SELEX. This characteristic is the origin of the name “SELCOS”. For this reason, the ligands that bind to the S1 site (i.e., a Tα-specific site) are decreased to half except LS1 (which binds exclusively to S1 site), resulting in enriched LS1. Clearly, this effect cannot be expected from conventional SELEX. Therefore, in the equilibrium state of the interaction between the targets and the pool of ligands, we can expect a more LS1-enriched (in other words, Tα-specific ligand-enriched) result from SELCOS than SELEX. Under our experimental conditions (see the protocol in Methods and Supplementary Fig. S1), the near-saturation of binding sites with ligands is expected to be attained (an 8-fold excess of ligands against a target molecule at the final stage). The selection products (ligands) obtained in this way were processed for a negative selection (the selected ligands were treated with a mixture of all the possible targets except the genuine one and then the nonbinding ligands were collected), although this process is theoretically omittable (see Discussion). To monitor the quality of the products rapidly, we introduced a DEPSOR-mode electrochemical sensing component (Apta-DEPSOR: see Fig. 1-b1). Using two subtypes of influenza A virus as targets, we performed an entire SELCOS procedure and monitored the products with the Apta-DEPSOR. As in Fig. 2, the products thus obtained (and confirmed in Supplementary Fig. S2) provided the DPV response curves (Panel a) and the corresponding bar charts (Panel b) for the combination of targets (TH1N1 and TH3N2) and ligands (ligand pools against TH1N1 and against TH3N2), showing that this approach can measure the relative binding strength: the proper matching of a target and a ligand pool provided a far higher signal than those of improper matching, indicating that both SELCOS and Apta-DEPSOR are working sufficiently well. As described in Methods, the electrochemical sensing is very simple, and this integrated method is very promising for rapid and selective aptamer selection.
Figure 1.
Schematic drawing of SELCOS (competitive non-SELEX) and the electrochemical sensing application. (a) Comparison of (conventional) SELEX and SELCOS in the ligand binding mode to the target protein. A pool of ligands is classified into 7 types in their binding mode to two different targets (Tα and Tβ), which are composed of the common site (C) and the specific site (S1 or S2) as follows: LS, LS1, LS2, LS1/S2, LS/C, LC, and LX. As shown in the figure, each ligand binds to its own binding site(s). For example, LS is a ligand that can bind to the specific site of both targets (Tα and Tβ), while LS1 and LS2 bind to the S1 or S2 sites only, respectively. This result indicates that the same site can be recognized differently depending on a ligand. LS1/S2 binds to both S1 in Tα and S2 in Tβ. LS/C binds to both site S (i.e., S1 and S2) and site C. LC binds to the common site of Tα and Tβ. LX does not bind to either Tα or Tβ. (b1) A schematic drawing of the event on the Apta-DEPSOR electrode (Aptamer-based Disposable Electrochemical Printed Sensor) in which the anti-target (influenza virus protein)-DNA aptamer-coated gold nanoparticles (AuNP) bind to the target loaded onto the working electrode of the sensor chip, followed by the electron transfer between the AuNP and the sensor surface, resulting in the generation of the DPV (differential pulse voltammetry) pattern. (b2) A virus protein concentration-dependent measurement of the DPV. The AuNPs are carried away with free protein when the flowing sample solution contains a large amount of virus protein. Note that the dent in the DPV curve is the signal in proportion to the bound AuNP.
Figure 2.
SELCOS products. Aptamer pools obtained against TH1N1 (i.e., target H1N1, in red) and TH3N2 (blue) were subjected to the electrochemical measurement using Apta-DEPSOR. (a) For each sample, the DPV was measured against both TH1N1 and TH3N2. (b) The Ipc (current for the signal peak) data are presented in a bar chart (using the average taken from 3 independent experiments).
Note that SELCOS procedure does not depend on the PCR amplification, which is a prominent difference from conventional SELEX (see Supplementary Fig. S1) and as also discerned earlier by protocol of non-SELEX20. This property simplifies the whole procedure and saves experimental cost when selecting DNA aptamers. Incidentally, several studies have supported the idea that the presence of competitor molecules can enhance the specificity of the selected candidate21–23, though none has highlighted on the competitive effect pointed out in the work.
Evaluation of cloned DNA aptamers
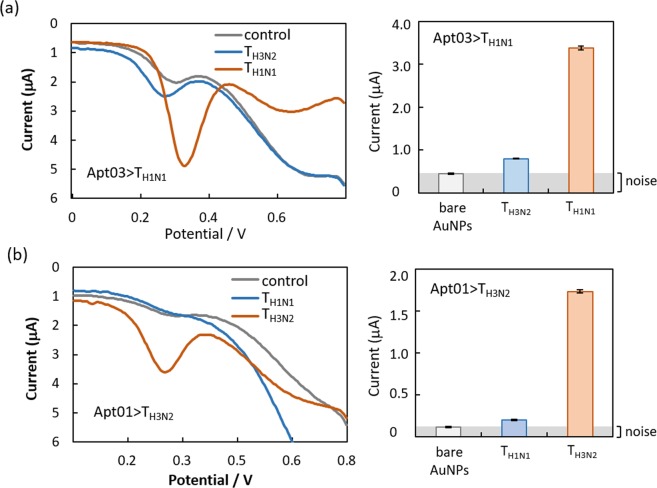
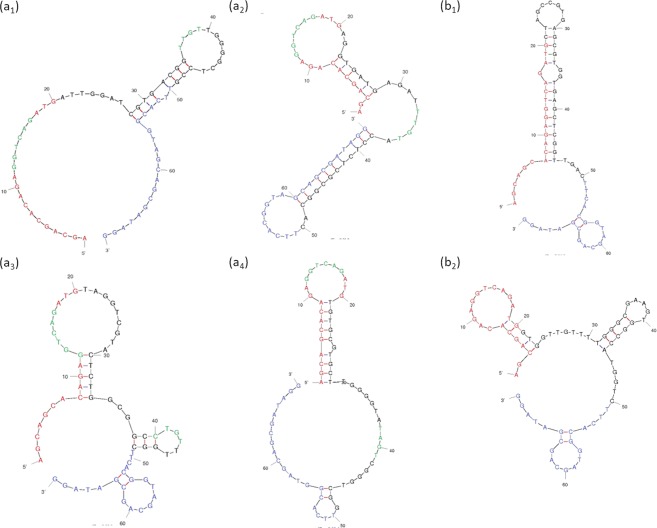
After SELCOS was performed with the targets TH1N1 and TH3N2, the selected pools were subjected to cloning and sequencing, providing multiple aptamers (Supplementary Fig. S3) with some of representative aptamers listed in Table 1. These aptamers were electrochemically analyzed separately as shown in Fig. 3 (with related data in Supplementary Fig. S4). The aptamer Apt01 > TH3N2 (denoting an aptamer named apt01 obtained in the selection targeting TH3N2) is shown to have an approximately 9-fold higher current signal (Ipc, cathodic peak current) against TH3N2 than against TH1N1. Similarly, the Apt03 > TH1N1 aptamer is more selective for its original TH1N1 target (near 5-fold higher current signal) than the nontarget TH3N2. The binding strength to its original target was tested by SPR for these aptamers (Fig. 4 and Table 1), for which the lpc values obtained from the Apta-DEPSOR method are also shown (giving a correlation score of −0.40 with the KD measured by SPR). From this result, the lpc value can be used to estimate the binding strength of aptamers though less exactly. The ∆G values for aptamer folding are shown in Table 1, and they present moderate stability values ranging from −6 to −12 kcal/mol with no significant correlation with the affinity KD (r = −0.11). Interestingly, the secondary structures of aptamers selected against the target H1N1 (i.e., Apt01~Apt04 > TH1N1) have a common motif of ‘Loop1-space-Loop2’ in which common sequences are involved (GGTCAG in Loop1 and T(or C)T(or A) GT in Loop2, although the GGTCAG sequence happens to come from the primer binding site), while the aptamers selected against TH3N2 have no similarly remarkable characteristics as far as the evidence shows (partly shown in Fig. 5). These conserved loop regions (Loop1 and Loop2) are highly expected to interact with the target molecules24,25. Although it is a much simpler and more rapid method than conventional SELEX, SELCOS can attain to, sometimes, find putatively functional motif as shown here. In Table 1, it is noteworthy that the frequency score that appeared for each selection has a relatively high correlation value (r = 0.55) with the binding affinity of KD, conforming to a rule that ‘the higher the affinity is, the higher the population’.
Table 1.
Properties of the aptamer DNAs selected against influenza virus proteins obtained by SELCOS.
| ID for cloned aptamerα | Aptamer sequencesβ (5′–3′) | Frequency of appearance (%) | ΔGχ (kcal/mol) | KDδ (M) | Ipcε (µA) |
|---|---|---|---|---|---|
| Apt03 > TH1N1 | 5′PBS-TAGGTCGTAC TCTGGCGGCC TGTTTGGC-3′PBS | 8.33 | −6.32 | 0.82×10−10 | 3.37 ± 0.035 |
| Apt04 > TH1N1 | 5′PBS-TGTGCGTGCT TGGGGTATAG TCGGGTCGG-3′PBS | 4.17 | −5.96 | 0.16×10−8 | 1.23 ± 0.011 |
| Apt02 > TH1N1 | 5′PBS-AGGTGATGAG ATTTGTACCT CTCGCGGCAC-3′PBS | 8.33 | −9.31 | 0.57×10−7 | 2.26 ± 0.011 |
| Apt01 > TH1N1 | 5′PBS-ATTGGATCGT GACGGTTGTT GGGGCTCCG-3′PBS | 12.5 | −5.33 | 0.35×10−4 | 0.85 ± 0.036 |
| Apt04 > TH3N2 | 5′PBS-TCTGCAGCGT GCAGGGCTGT GTGCTTACCC-3′PBS | 4.17 | −9.83 | 0.88×10−10 | 1.61 ± 0.48 |
| Apt01 > TH3N2 | 5′PBS-CTAGCCGTGA GCGTGGTGAG CTCGGTTGAC-3′PBS | 12.5 | −7.51 | 0.14×10−9 | 1.73 ± 0.032 |
| Apt03 > TH3N2 | 5′PBS-GCGCGGGCGG TGCGTCGGTG TCCCGCTGG-3′PBS | 4.17 | −12.50 | 0.60×10−9 | 1.11 ± 0.032 |
| Apt02 > TH3N2 | 5′PBS-GTGGTTGTTT TGGGCGAAGT GGCCATGGTC -3′PBS | 8.33 | −5.51 | 0.17×10−8 | 1.16 ± 0.026 |
αNomenclature for cloned aptamers were systematically assigned to be ‘serial#’ (e.g., Apt01) +‘>’ (a connector) +‘Target name’ (e.g., TH1N1), thus Apt01 > TH1N1.
β5′-PBS and 3′-PBS are primer binding sequences (AGCAGCACAG AGGTCAGATG and CCTATCGCTG CTACCGTGAA, respectively).
χdG (kcal/mol) is free energy value and calculated from Mfold online tool.
δKD (M) value is generated by BIACORE X100 using single cycle kinetics.
εIpc (µA) is average current value and obtained from DPV curves.
Aptamer DNAs were obtained from a single trial of SELCOS that offered two sets of aptamers, TH1N1-specific and TH3N2-specific ones. The ones listed here were chosen by the clustering analysis described in Supplementary Fig. S3. The sequence, frequency of appearance of the aptamer within a selected DNA pool, free energy for folding, dissociation constant (KD) for the binding of the target protein and aptamer, and peak current for the signal (Ipc) in the electrochemical analysis are listed.
Figure 3.
Validation of selected aptamer molecules by Apta-DEPSOR. (a) The aptamer, Apta03 > TH1N1 (namely, aptamer #03 selected against the target H1N1 protein (TH1N1)), was measured against TH1N1 and TH3N2. The DPV curves (left) and the corresponding bar graph (right) are shown. (b) The aptamer, Apt01 > TH3N2, was used here. “Control” (gray) indicates the signal from bare gold nanoparticles (AuNP). The concentrations of the target proteins, TH1N1 and TH3N2, were both 250 µg/mL.
Figure 4.
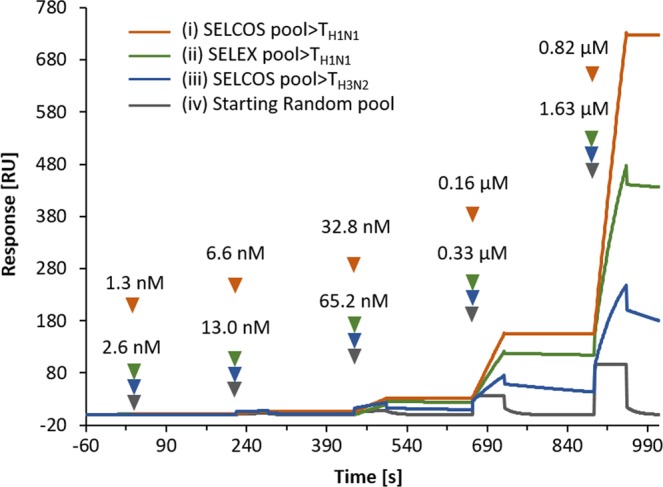
SPR analysis of the selection products with ligand TH1N1. Selected aptamer DNA pools were anlayzed by single-cycle kinetics SPR using a BiacoreX100. For DNA pools, a successive injections of five increasing concentrations (0.0299, 0.149, 0.746, 3.73, and 18.66 µg/mL for analyte sample (i) and 0.0592, 0.296, 1.48, 7.4, and 37 µg/mL for analyte samples (ii), (iii), and (iv) were used. The target protein binding capacity on the sensor chip surface was in levels of 2500–3000 RU (response unit). The X-axis and Y-axis represent the response (RU) and time (s) of the single-cycle kinetics sensogram, respectively. The sensograms were obtained by fitting the data using a 1:1 binding model (BioEvaluation software).
Figure 5.
Predicted secondary structures of cloned aptamers. Some of the aptamers obtained against TH1N1 and TH3N2 were analyzed with Mfold, a secondary structure-computing program. (a1–a4) For the aptamers selected against TH1N1 (namely, Apt01 > TH1N1, Apt02 > TH1N1, Apt03 > TH1N1, Apt04 > TH1N1). (b1,b2) For the aptamers Apt01 > TH3N2 and Apto2 > TH3N2. Commonly appearing sequences in loop regions are highlighted for the aptamers against TH1N1 (incidentally, no such sequences were found in the aptamers obtained against TH3N2). Note that the sequence regions of 1–20 and 51–70 over the entire sequence (70 nucleotides) are primer-binding sites, and they are constant.
As shown in Fig. 4, from the kinetics analysis with SPR (Biacore X100) when employing the single cycle mode analysis, data in Table 2 were obtained. Under our experimental conditions, the selected aptamer pool against TH1N1 in the SELCOS had a 300-fold stronger KD than the value selected by conventional SELEX, and this KD (1.01 × 10−10 M) is already close to that of the cloned aptamer Apt03 > TH1N1 (0.82 × 10−10 M). The Apt03 > TH1N1 aptamer is more than 100-fold stronger than that of the previously reported DNA aptamer RHA000626 that was selected against influenza virus subtype H1N1. For reference, the commercial monoclonal antibody was also measured by SPR, showing the strongest affinity (2.53 × 10−13 M), which was surprisingly sophisticated. Interestingly, the Apt03 > TH1N1 aptamer exhibited a similar pattern of fitted curve, showing a high resemblance of the calculated kinetic parameters (for details, see Supplementary Fig. S5). This important data addresses some key aspects for future approaches involving the replacement of antibodies with aptamers. In any case, SELCOS provided a sufficiently competent aptamer in terms of its binding affinity.
Table 2.
SPR analysis on the binding of selection products with the target protein used for the selection (TH1N1).
| Ligand/Target | kon (M−1s−1) | koff (M−1s−1) | KD (M) | Rmax (RU) |
|---|---|---|---|---|
| Random ligand pool/TH1N1 | not sufficiently bound | 16 | ||
| SELEX pool for TH1N1/TH1N1 | 6.30×103 | 1.89×10−4 | 2.99×10−8 | 809 |
| Compe-SELEX pool for TH1N1/TH1N1 | 9.34×103 | 9.41×10−7 | 1.01×10−10 | 2651 |
| Compe-SELEX pool for TH3N2/TH1N1 | 9.06×103 | 1.80×10−3 | 1.99×10−7 | 321 |
| TH1N1-Apta03/TH1N1 | 3.74×104 | 3.08×10−6 | 0.82×10−10 | 603 |
| Monoclonal antibody/TH1N1 | 2.33×105 | 5.90×10−8 | 0.25×10−12 | 694 |
| RHA0006/TH1 (Ref.*) | NA | NA | 1.53×10−8 | NA |
‘Pool’ indicates a set of DNA aptamers that were just selected. A single cycle kinetics analysis was adopted for the SPR (surface plasmon resonance).
Quantitation of an influenza virus subtype using Apta-DEPSOR
An influenza subtype H1N1-specific aptamer (Apt03 > TH1N1)-loaded Apta-DEPSOR was fabricated as shown in Fig. 1-b1 and b2. First, the working electrode surface of a DEP chip was covered with the free target H1N1 protein (TH1N1), and, then, a fixed amount of AuNP (gold nanoparticles) mixed with a virus sample is introduced to the electrode, where, when there is excess AuNP relative to the free TH1N1 of virus sample, the excess amount of free AuNP coated with the aptamer (Apt03 > TH1N1) can bind to TH1N1 on the surface of the electrode and then be trapped and detected by the sensor. In this experimental system, the electrochemical signal will increase depending on the amount of AuNP captured by the TH1N1 on the electrode surface, and it will decrease proportionally depending on the amount of target TH1N1 protein in the sample solution. Figure 6 shows the dependence of the DEPSOR signal (current) on the concentration of applied TH1N1. From the resulting calibration curve, the dynamic range of the measurements ranged from 0.4 to 100 µg/mL in 5% human serum and we successfully detected TH1N1 in as little as 1.23 ng/L.
Figure 6.
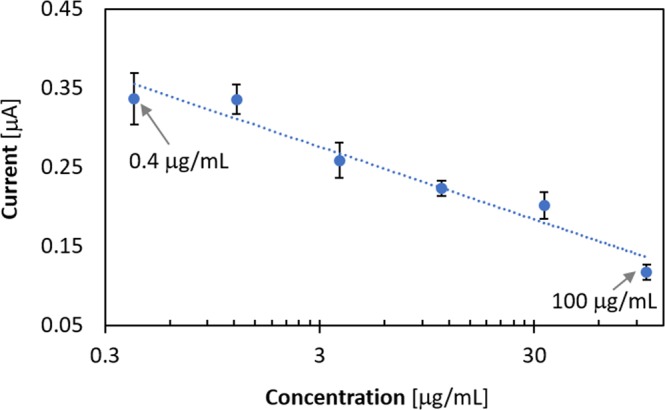
Calibration curve for measuring influenza virus A subtype H1N1 in human serum. A bound-and-free virus (protein) competition assay using Apta-DEPSOR was performed as explained in Fig. 1-b2. In this assay, there is competition for aptamer-coated gold nanoparticles (i.e., TH1N1-apt03-coated AuNP) by two phases of proteins (free and electrode surface-bound). From the regression analysis, the correlation coefficient r was −0.88. The lower detection limit was 0.51. The figure shows the averages taken from three trials.
Discussion
In this article, Competitive non-SELEX (SELCOS) was shown to be effective at enriching aptamers that were specific to a target protein, for which evaluations can be empowered by the introduction of the Apta-DEPSOR, a kind of electrochemical sensing device. Among the various detection devices, such as SPR, Apta-DEPSOR is useful for the current purpose due to its detectability of ligand-target interactions, readiness for operation and portability, and potential use in constructing a POC device for detecting infectious viruses with high selectivity.
The SELCOS principle is explained briefly in Fig. 1. Here, we further deepen this explanation in comparison to the conventional SELEX. The typical difference between these two methods relates to whether multiple targets can coexist or not. The current SELCOS application was operated under the experimental conditions described in Methods. There were 4 steps of imposed selection pressure (a step-wise reduction in the binding time during the increase in binding ligands with a gradual weakening from the washing effect), the successive addition of a ligand library (without PCR amplification) and one step of negative selection at the final stage (see Supplementary Fig. S1). Originally, SELEX (Systematic Evolution of Ligands by EXponential enrichment) had the following essential properties: it employs an RNA/DNA library; i) under selection pressures with increasing stringency, ii) it partitions materials into two groups that are ‘binding’ and ‘nonbinding’ (usually ‘solid phase of beads or resin’ and ‘bulk solution’), and iii) it involves the amplification of the library (usually by PCR). SELCOS has a different mode than conventional SELEX as the intermediate amplification step, i.e., PCR, of conventional SELEX is excluded in SELCOS and addition of stock supply from the original library was used between repetitive steps of partitioning. However, we must pay careful attention to the difference in the PCR amplification step of conventional SELEX and stock supply of SELCOS. First, we would like to point out that the naming of SELEX has been successful due to the appealing nature of its methodological details and the term´s nice compact ring. We have no doubt that it was a wonderful invention in the field of molecular evolution27. However, the words contained in “SELEX” might have been confusing since “exponential enrichment” is not correct if one believes that the molecules to be selected are enriched in an exponential mode; although the library components are amplified exponentially by PCR, no relative enrichment of a particular element occurs during this exponential amplification stage. The PCR amplification of a pool of ligands is expressed as shown below.
| 1 |
| 2 |
| 3 |
where Li(t), L1(t), and L2(t) represent the concentrations of ligands Li, L1, and L2 at time t, respectively, and Li(0), L1(0), and L2(0) are the ligand concentrations at time 0, and they are constant. The letter e(t) signifies the amplification efficiency at time t (the time is the integer). In PCR, ideally, e = 1. However, this value usually depends on the time due to the inactivation of polymerase, the decrease in primers, the increase in reactants, and so on. Since these factors are commonly influential for each template, the same e(t) value can be expected for each template. Thus, Eq. 3 holds true, indicating that the PCR does not change the ratio of constitutive elements before and after the PCR. In other words, no enrichment occurs during PCR, but the simple amplification of each component by the same factor (i.e., ·(1 + e(t))t) does lead to enrichment. Notably, PCR is well-known for generating alterations in the population of a library; the ratio change in the population and the mutation of DNA/RNA have no relation to the enrichment of the fitting aptamers. Parameter e also depends on the template DNA/RNA itself since these nucleic acids generate secondary and tertiary structures intrinsic to their sequence, and those structures are often unfavorable for the polymerization reaction (thus lessening the parameter e value). This effect can generate a bias in the population, but it does not denote an enrichment of the fitting aptamers; the axis of selection is quite different. Therefore, “exponential enrichment” is image-inducing wording but it is not realistic. However, since PCR, a typical process of SELEX, is not included in our method, we adopted the name “SELCOS” for our technology.
The reason (Supplementary Note) that SELCOS is superior at finding target-selective aptamers relative to conventional SELEX is discussed in the theoretical note. In brief, SELCOS can find more target-selective aptamers (LS1 and LS2 drawn in Fig. 1) due to the competitive effect of the ligand-binding between target molecules (Tα and Tβ). To be sure to obtain this effect, a thermodynamic equilibrium must be attained. In SELEX, successive subtraction is repeated by washing (partitioning) to enrich the aptamers of interest. During this process, due to the kinetic effect, a large population of fitting aptamers (such as LS1 and LS2) could be irreversibly lost by washing, which can be effectively circumvented by SELCOS due to the successive addition of the entire population of ligands. This approach can rationalize the experimental results in which SELCOS succeeded in yielding a higher affinity ligand pool than SELEX, as shown in Table 2 (although using a computer simulation that assumes a set of parameters might be more persuasive28). Surface plasmon resonance (SPR) has been extensively used to monitor binding events between analyte and ligand molecules. Thus, utilizing the similar approach for our study we compared the SPR analysis of selection pool products obtained by mode of SELCOS with the selection pool products obtained by mode of SELEX (Fig. 4). Fundamentally, we aim to compare the enrichment of the pool selected by SELCOS in presence of two targets with the pool selected by PCR-based SELEX for one target. As shown in Table 2, our observation suggested that the response value generated against ligand TH1N1 for initial random library was 16 RU which is negligible when compared to the SELEX pool showing 809 RU and 2.99×10−8 M KD which exhibits enrichment to certain extent compared with initial random library. However, to our astonishment the SELCOS pool for ligand TH1N1 showed a relative higher response value of 2651 RU with the KD 1.01×10−10 M. Specificity being an important aspect, we decided to check the analyte pool selected for ligand TH3N2 by SELCOS against ligand TH1N1 and observed a response of 321 RU with the KD 1.99×10−7 M relatively lower thus indicating specificity of the selected pool by SELCOS. Henceforth, the preliminary findings from the SPR data were supportive of our theoretical understanding of SELCOS mode of action. The above-mentioned observations were determining and motivating to proceed with further analysis of candidate aptamers selected via SELCOS. At the same time, the effect decreases the relative amount of ligands (LS/C and LC in Fig. 1) since they bind common sites, which is multiplied when there are multiple targets, which must also contribute to the relative enrichment of target-selective aptamers. These effects are usually obtainable when the negative selection (a selection that eliminates the ligands that bind non-authentic targets) is performed. Thus, SELCOS has the ability to provide a negative selection in parallel with a positive selection. This property confers SELCOS with cost savings relative to SELEX since it can provide M-multiple different aptamers at once when M-tiple targets are adopted (although in this article, M = 2).
Clearly, in such an M-tiple target system, the ultimately selected aptamers can be expected to be exclusively selective for the relevant target with nonbinding with the other targets. Future studies about these topics are very exciting for the development of the SELCOS field.
The electrochemical sensing device introduced here (Apta-DEPSOR) for the quantitative monitoring of aptamer-target binding can generally be used for these purposes. This tool is sufficiently powerful, as shown in this study, and it has the merits of being portable and having a high cost performance (due to the disposable sensor chip used here18). In particular, it is favorable that its sensing part is composed of an aptamer that can be selected and evaluated by this device. Therefore, the application of this device for POC purposes (such as detecting the influenza virus at the spot of contagion) is very promising for use in the near future. Finally, we must note that developing SELCOS solely for selecting DNA aptamers is, in principle, also applicable to other selection categories such as the in vitro selection of peptides/proteins29,30 and the DNA-encoded library (DEL) selection of small molecules31,32. For these selections, the PCR-free nature of SELCOS is very convenient because the troublesome retagging process (such as puromycin-linker ligation to mRNA) required for those technologies, can thus be discarded (SELEX is, conveniently, free from this tagging process).
Conclusions
The competition-driven selection of DNA aptamers using multiple targets (termed as SELCOS, Systemic Enrichment of Ligands by COmpetitive Selection) was first introduced in this study. The experimental results confirmed our success in obtaining influenza virus subtype-selective aptamers using SELCOS, which could be readily monitored with an electrochemical sensing tool (Apta-DEPSOR) as introduced here. By loading a selective aptamer as obtained (Apt03 > TH1N1) on its sensor unit, the feasibility of detecting the virus subtype was examined, and the detectability of subtype H1N1 ranged from 0.4–100 µg/mL. Although the situation in which the apta-DEPSOR can be useful is limited at present due to its sensitivity in the sub-µg/mL range, its portability (a merit of DEPSOR) enables us to collect important data at the POC (point of care). The theoretical consideration of SELCOS revealed its potential difference relative to conventional SELEX. In particular, its methodological advantages will be reinforced by multiple target selection, with the simultaneous acquisition of multiple aptamers of high selectivity. SELCOS, which is PCR-free, has appropriate properties for wider categories of selection such as the in vitro selection of peptides/proteins.
Methods
In vitro selection of DNA aptamers by competitive enrichment
Immobilization of target molecules on Ni-NTA beads: To perform SELCOS, we used the closely related subtypes of the influenza A virus H1N1 and H3N2. The targets H1N1 (abbreviated as TH1N1) and H3N2 (abbreviated as TH3N2) were immobilized onto Ni-NTA magnetic beads (20–70 µm) and Ni-NTA agarose resin beads (45–165 µm), respectively, according to the protocol for immobilizing the protein target stated by the manufacturer.
Library Design and Primers: The DNA library used for the selection was made up of a random 30-nucleotide region flanked by a 20-nucleotide primer region on both sides, specifically, 5′-AGCAGCACAGAGGTCAGATG(N30)CCTATGCGTGCTACCGTGAA-3′. For PCR amplification, the forward primer 5′-AGCAGCACAGAGGTCAGATG-3′ and the biotinylated reverse primer 5′-TTCACGGTAGCAGCGATAGG-3′ were used.
Selection Process: The plus strand ssDNA pool was heated to 90 °C for 5 min and immediately cooled to 4 °C and placed for 15 min, followed by incubation at 25 °C for 15 min. Following this step, the targets TH1N1 and TH3N2 that were immobilized on the Ni-NTA beads were incubated with 100 pmol of the ssDNA initial pool in the presence of the binding buffer (PBS buffer (pH 7.4), 100 mM NaCl, 5 mM KCl, 2 mM MgCl2, 1 mM CaCl2) for 60 min. The supernatant was then removed by washing three times with washing buffer (PBST buffer (pH 7.4 with 0.05% Tween20), 100 mM NaCl, 5 mM KCl, 2 mM MgCl2, 1 mM CaCl2). In each wash, the sample solution was briefly centrifuged at 1000 g for 10 s and the supernatant was removed carefully. The same procedure was repeated by 4 rounds with a successive addition of 200 pmol, 400 pmol, and 800 pmol of the ssDNA pool, changing the incubation time and washing frequency 30 min (3 times washing), 15 min (2 times washing), and 7.5 min (1 time washing), respectively. Finally, both the targets immobilized on the magnetic or nonmagnetic beads were separated by magnetic force using a magnet stand or centrifugation force (1000 g for 10 s), respectively, followed by the removal of the supernatant. The selected aptamer DNA pools, which are bounded on beads, were recovered by heat treatment (90 °C for 5 min followed by immediate removal of the supernatant). The selected aptamer DNA pools for TH1N1 and TH3N2 were then briefly incubated with the crude Ni-NTA beads for 15–20 min in order to remove any nonspecific candidates, if exists. The specific DNA pools selected against TH1N1 and TH3N2 were then briefly incubated with the different target solution, TH3N2 or TH1N1, to remove false positives. The specific pools for each target selected by SELCOS were amplified by PCR (initial incubation at 98 °C for 2 min, followed by 20 cycles of 98 °C for 10 s, 59 °C for 5 s, and 72 °C for 10 s, and finally, 72 °C for 4 min). Gel electrophoresis was used to monitor the successful amplifications using 8% polyacrylamide gel with 8 M urea at a temperature of 60 °C. The details of the cloning and candidate determination are mentioned in the Supporting Information section. In brief, all the selected pools were then cloned, and 20 clones were picked for sequencing. Sequence analyses were performed using the web-based tools ClustalW33 and Mfold34, for multiple sequence alignment (for details, see Supplementary Fig. S3) and secondary structure analysis, respectively.
SPR Measurements
The SPR measurement was performed using a BIACORE X100 instrument. A sensor Chip-NTA and NTA reagent kit (GE Healthcare, Uppsala, Sweden) were used for the immobilization of the His-tag protein target for the interaction studies according to the manufacturer’s instructions. The running buffer HBS-P was used for all the experiments and 0.35 M EDTA was used for regeneration. The single-cycle mode was performed to compare the pool for ligand TH1N1 selected by conventional methods and SELCO. For this, 4 independent experiments were performed for the immobilization of ligand TH1N1 (0.01 mg/mL) in the running buffer onto the sensor surface at a level of 2500–3000 RU, with a contact time of 60 s and stabilization period of 60 s. The different analytes used for comparison were the random library, pool for TH1N1 selected by conventional method, pool for TH1N1 selected by SELCO, and pool for TH3N2 selected by SELCO. The selected ssDNA pool solutions of the following concentrations: 37, 7.4, 1.48, 0.296, and 0.0592 µg/mL were prepared in the running buffer and sequentially injected, starting with lowest concentration, at a flow rate of 30 µL/min for 60 s, followed by 60 s of dissociation. The kinetics of the association and dissociation were studied and compared.
To study the interaction analysis (association/dissociation) of candidate aptamers selected via SELCOS, single-cycle mode was performed for the immobilization of the ligand-target protein H1N1 with the his-tag (0.01 mg/mL) in the running buffer onto the sensor surface at a level of 2000 RU, with a contact time of 120 s and a stabilization period of 60 s. Aptamer solutions of the following concentrations: 26.66, 5.33, 1.07, 0.213, and 0.0427 µg/mL were prepared in the running buffer and sequentially injected, starting with lowest concentration, at a flow rate of 30 µL/min for 60 s, followed by 60 s of dissociation. All measurements of the binding analysis were performed in triplicate and the fitting was done for the 1:1 binding model by BiacoreX100 Evaluation software. The single-cycle mode was performed for the immobilization of the ligand protein TH3N2 (0.01 mg/mL) in the running buffer onto the sensor surface at a level of 1000 RU, with a contact time of 120 s and stabilization period of 60 s. Aptamer solutions of the following concentrations: 44.44, 8.89, 1.78, 0.356 and 0.0711 µg/mL were prepared in the running buffer; similar conditions for injection were used and the resulting binding curves were studied.
Integration of SELCOS and an electrochemical sensing device (Apta-DEPSOR)
To evaluate and show the point-of-care applicability of SELCOS, we designed an electrochemical assay using our originally developed portable and disposable electrochemical printed (DEP) chip-based three-electrode sensing system DEPSOR, which functions on the principle of differential pulse voltammetry (DPV). A plot of the current relative to the voltage is generated by the electrochemical analyzer on the basis of the redox reaction18. As shown in Fig. 1-b1 and b2, the aptamers selected using SELCOS were integrated with DEPSOR. By performing a voltammetry assay with aptamer-conjugated AuNPs as a recognition element, a clear signal peak can be detected sensitively when a candidate aptamer binds with the target onto the working electrode of the DEP chip.
Electrochemical Measurements
A disposable three-electrode screen-printed (DEP) chip, which was obtained from Biodevice Technology, Co. (Ishikawa, Japan), was used for this experiment. The DEP chip works on the principle of the three-electrode system for electrochemical analysis, with a carbon-based working electrode (3 mm in diameter), a counter electrode, and an Ag/AgCl reference electrode. Two µL of the recombinant proteins H1N1 and H3N2 at a concentration of 0.25 µg/µL were dropped onto the working electrode of the DEP chip, which was then incubated for one hour at 4 °C. This incubation allowed for the passive adsorption of the target protein onto the working electrode surface. After the incubation, excess target protein was rinsed three times with 100 mM PBS, and the chip was dried by gentle-blowing air. To suppress nonspecific adsorption, 3.5 µL of blocking buffer (100 mM PBS containing 1% BSA) was added to the chip; it was then incubated overnight at 4 °C. For the electrochemical analysis, the chip was further rinsed three times with 100 mM PBS buffer and dried before it could be used for the assay. A 2-µL sample made up of Au nanoparticles conjugated to the selected DNA aptamer candidates was dropped onto the target-modified DEP chip surface; the chip was then incubated for 15 min at room temperature. It was then rinsed three times with 100 mM PBS buffer and connected to an electrochemical analyzer system (Model 650 A, CH Instruments, Inc., Austin, USA). Thirty µL of 0.1 M HCl was dispensed onto the DEP chip to electrooxidate the AuNPs at a constant potential of +1.4 V for 40 s, immediately followed by DPV detection from +0.6 V to 0 V, with a step potential of 4 mV, a pulse amplitude of 50 mV, and a pulse period of 0.2 s. The selected aptamer candidates were tested for their specific subtype, and the best aptamer was selected for further analysis. For the specificity validation, the selected aptamer for TH1N1 was reacted with the TH3N2-modified DEP chip and vice-versa. Additionally, a specificity check was performed for the TH1N1 and TH3N2 pool selected by SELCOS using the same protocol. All the experiments were repeated three times to confirm the consistency of the analysis. The details of the competitive detection in human serum samples are mentioned in the Supporting Information section.
Supplementary information
Acknowledgements
This work was supported by the MEXT Regional Innovation Strategy Support Program Hokuriku Life Science Cluster project.
Author Contributions
M.B. conceived and designed the research; A.K. performed the experiments; M.B. contributed to the preliminary work; A.K., Y.T., K.N. and M.B. analyzed the data and wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-43187-6.
References
- 1.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment:RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 4.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song S, Wang L, Li J, Zhao J, Fan C. Aptamer-based biosensors. Trends Anal. Chem. 2008;27:108–117. doi: 10.1016/j.trac.2007.12.004. [DOI] [Google Scholar]
- 6.Zhou W, Huang PJ, Ding J, Liu J. Aptamer-based biosensors for biomedical diagnostics. Analyst. 2014;139:2627–2640. doi: 10.1039/c4an00132j. [DOI] [PubMed] [Google Scholar]
- 7.Bowser MT. SELEX: Just another separation? Analyst. 2005;130:128–130. doi: 10.1039/b412492h. [DOI] [PubMed] [Google Scholar]
- 8.Baird GS. Where are all the aptamers? Am. J. Clin. Pathol. 2010;134:529–531. doi: 10.1309/AJCPFU4CG2WGJJKS. [DOI] [PubMed] [Google Scholar]
- 9.Thiel WH, et al. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS ONE. 2012;7:e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiel HW, et al. Nucleotide Bias Observed with a short SELEX RNA Aptamer Librar. y. Nucleic Acid Ther. 2011;21:253–263. doi: 10.1089/nat.2011.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyce FG. Amplification, mutation and selection of catalytic RNA. Gene. 1989;82:83–87. doi: 10.1016/0378-1119(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 12.Tolle F, Wilke J, Wengel J, Mayer G. By-Product Formation in Repetitive PCR amplification of DNA Libraries during SELEX. Plos One. 2014;9:e114693. doi: 10.1371/journal.pone.0114693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djordjevic M. SELEX experiments: new prospects, aplications and data analysis in inferring regulatory pathways. Biomol Eng. 2007;24:1179–1189. doi: 10.1016/j.bioeng.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhuo Z, et al. Recent advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017;18:e2142. doi: 10.3390/ijms18102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn MR, Jimenez RM, Chaput JC. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017;1:76. doi: 10.1038/s41570-017-0076. [DOI] [Google Scholar]
- 16.Blind M, Blank M. Aptamer Selection Technology and Recent Advances. Mol Ther Nucleic Acids. 2015;4:e223. doi: 10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozer A, Pagano MJ, Lis TJ. New technologies provide quantum changes in the scale, speed and success of SELEX methods and aptamer characterization. Mol. Therapy. Nuc. Acids. 2014;3:e183. doi: 10.1038/mtna.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biyani M, et al. PEP-on-DEP: A competitive peptide-based disposable electrochemical aptasensor for renin diagnostics. Biosens. Bioelectron. 2016;84:120–125. doi: 10.1016/j.bios.2015.12.078. [DOI] [PubMed] [Google Scholar]
- 19.Biyani M, et al. Instant enumeration of total viable bacterial counts for food quality assurance using DEP-On-Go sensor. Anal. Methods. 2018;10:1585–1592. doi: 10.1039/C7AY02927F. [DOI] [Google Scholar]
- 20.Berezovski MV, Musheev MU, Drabovich AP, Jitkova JV, Krylov SN. Non-SELEX: selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat. Protoc. 2006;1:1359–1369. doi: 10.1038/nprot.2006.200. [DOI] [PubMed] [Google Scholar]
- 21.Boel E, et al. T. Phage antibodies obtained by competitive selection on complement-resistant Moraxella (Branhamella) catarrhalis recognize the high-molecular weight outer membrane protein. Infect. Immun. 1998;66:83–88. doi: 10.1128/iai.66.1.83-88.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogasawara D, Hasegawa H, Kaneko K, Sode K, Ikebukuro K. Screening of DNA aptamer against Mouse Prion Protein by Competitive Selection. Prion. 2007;4:248–254. doi: 10.4161/pri.1.4.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paramanathan T, Reeves D, Friedman LJ, Kondev J, Gelles J. A general mechanism for competitor-induced dissociation of molecular complexes. Nat. Commun. 2014;5:5207. doi: 10.1038/ncomms6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenleaf, W. J.; Frieda, K.L.; Foster, D. A. N.; Woodside, M. T. & Block, S. M. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 319, 630–633. [DOI] [PMC free article] [PubMed]
- 25.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genetics. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiratori I, Akitomi J, Boltz DA, Horii K, Furuichi M, Waga I. Selection of DNA aptamers that bind to influenza A viruses with high affinity and broad subtype specificity. Biochem. Biophys. Res. Commun. 2014;443:37–41. doi: 10.1016/j.bbrc.2013.11.041. [DOI] [PubMed] [Google Scholar]
- 27.Gibney E, Noorden R, Ledford H, Castelvecchi D, Warren M. Nobel for test-tube evolution. Nature. 2018;562:176. doi: 10.1038/d41586-018-06753-y. [DOI] [PubMed] [Google Scholar]
- 28.Aita T, Nishigaki K, Husimi Y. Theoretical consideration of selective enrichment in in vitro selection: optimal concentration of target molecules. Math Biosci. 2012;240:201–211. doi: 10.1016/j.mbs.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Crawford M, Woodman R, Ko Ferrigno P. Peptide aptamers: tools for biology and drug discovery. Brief. Funct. Genomic. Proteomic. 2003;2:72–79. doi: 10.1093/bfgp/2.1.72. [DOI] [PubMed] [Google Scholar]
- 30.Kitamura, et al. Development of systemic in vitro evolution and its application to generation of peptide-aptamer-based inhibitors of cathepsin E. J. Mol. Biol. 2009;387:1186–1198. doi: 10.1016/j.jmb.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 31.Litovchick, et al. Encoded Library Synthesis Using Chemical Ligation and the Discovery of sEH Inhibitors from a 334-Million Member Library. Sci. Rep. 2015;5:10916. doi: 10.1038/srep10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleiner RE, Dumelin CE, Liu DR. Small-molecule discovery from DNA-encoded chemical libraries. Chem. Soc. Rev. 2011;40:5707–5717. doi: 10.1039/c1cs15076f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.