Abstract
Despite previous reports on the genotypic variation of architectural and functional traits in fruit trees, phenotyping large populations in the field remains challenging. In this study, we used high-throughput phenotyping methods on an apple tree core-collection (1000 individuals) grown under contrasted watering regimes. First, architectural phenotyping was achieved using T-LiDAR scans for estimating convex and alpha hull volumes and the silhouette to total leaf area ratio (STAR). Second, a semi-empirical index (IPL) was computed from chlorophyll fluorescence measurements, as a proxy for leaf photosynthesis. Last, thermal infrared and multispectral airborne imaging was used for computing canopy temperature variations, water deficit, and vegetation indices. All traits estimated by these methods were compared to low-throughput in planta measurements. Vegetation indices and alpha hull volumes were significantly correlated with tree leaf area and trunk cross sectional area, while IPL values showed strong correlations with photosynthesis measurements collected on an independent leaf dataset. By contrast, correlations between stomatal conductance and canopy temperature estimated from airborne images were lower, emphasizing discrepancies across measurement scales. High heritability values were obtained for almost all the traits except leaf photosynthesis, likely due to large intra-tree variation. Genotypic means were used in a clustering procedure that defined six classes of architectural and functional combinations. Differences between groups showed several combinations between architectural and functional traits, suggesting independent genetic controls. This study demonstrates the feasibility and relevance of combining multi-scale high-throughput methods and paves the way to explore the genetic bases of architectural and functional variations in woody crops in field conditions.
Subject terms: High-throughput screening, Plant stress responses
High-throughput phenotyping for plants
A high-throughput method for determining plant characteristics offers a viable way to track competitive traits in the field. Increasingly difficult agricultural conditions are spurring plant breeding programs to prioritize plants with strong adaptations for healthy metabolism, but no current technologies can rapidly assess plant traits in the field. Now, Benoît Pallas, of the French National Institute for Agricultural Research, and his team have adapted tools such as infrared and laser-light scanning, and drone imagery, to take define and correlate metrics of 1000 apple trees, such as measurements of volume, leaf area, photosynthetic activity, and water usage. With these results and prior knowledge of the trees’ genotype, the team was able to group the trees into six classes with distinct physical and functional trait combinations. These techniques may open doors for discovering the genetic bases of plant traits.
Introduction
The biophysical approach proposed by Monteith (1977)1 states that plant production potential relies on traits associated with plant capacity to intercept solar radiation, to convert this energy into carbohydrates and allocate them to the harvested fruits or seeds. In fruit trees, breeding programs did not account for these elementary traits and rather focused on disease resistance, fruit organoleptic properties, and yield2. Moreover, in the current climatic context characterized by an increase in temperature and a risk of water scarcity, targeting new varieties with high performance even under constraining conditions becomes crucial for breeding programs. Potted experiments in phenotyping platforms have arisen as performing approaches to phenotype perennial species3. These platforms have led to encouraging results in young potted trees through the identification of large genotypic variability in architectural (plant height, total leaf area) and functional (transpiration, water-use efficiency) traits4. These studies were made possible by using new technologies provided in those platforms5, i.e., Red Green Blue (RGB) images for plant growth estimation, and automated plant transpiration measurements. Nevertheless, such technologies are not available in field conditions and new developments are necessary to enable the phenotyping of both architectural and functional traits of woody crops at different developmental stages, including adult and fruiting.
Plant architecture determines many traits associated with plant performance such as flowering intensity and intra-tree location, or light interception efficiency. Most of the studies that have dealt with genetic variations of architectural traits have focused on simple plant traits, such as height or trunk diameter6, whose measurements are feasible on large populations. More local traits, such as maximal internode length along trees, branching density or mean axis length were also considered on young trees7. These studies have shown that architectural traits are genetically controlled, and identified quantitative trait loci (QTL) associated with their variations although these traits display high polygenetic determinisms. Nevertheless, other properties of tree canopies such as the efficiency of architectures for intercepting solar radiation or for optimizing intra-canopy microclimate (temperature, humidity) could be relevant for describing between-trees variability. High-throughput (HT) estimations of traits such as the silhouette to total leaf area ratio (STAR8) or the leaf density within the tree canopy could help characterizing genotype efficiencies. Methods based on three-dimensional (3D) digitizing were successfully used for characterizing whole tree architecture8 but they remain time-consuming and not compatible with HT phenotyping. Terrestrial LiDAR (T-LiDAR) scanners have recently emerged as promising tools for measuring 3D vegetation structures. T-LiDAR scans have been used in many studies, mainly in forest but also in agronomic contexts9, notably for evaluating individual crown structure10 or leaf area density11. More recently, a study reported alpha hull volume estimation based on merging RGB images of tree orchards for assessing tree space occupation12. However, to the best of our knowledge, T-LiDAR has never been deployed in a context of HT phenotyping for genetic studies.
Measurements of carbon and water-related processes in plants are frequently carried out with InfraRed Gas-Analyzers (IRGAs). These devices allow precise assessment of leaf functions, but are time-consuming. Other variables, associated with chlorophyll fluorescence have been proposed to quickly estimate leaf photosynthesis through direct measurements on leaves or by chlorophyll fluorescence emission imaging13. The most used variables were the effective quantum efficiency of photosystem II (PSII), the derived electron transport rate and the photochemical and non-photochemical quenching coefficients13,14. These parameters have been used in genetic studies in which associated QTLs have been detected15. Nevertheless, variation in the relationship between these variables and photosynthetic rate has been observed, mainly under water stress due to complex regulation of stomatal closure and subsequent repercussions on leaf fluorescence16. Losciale et al.17. recently proposed a fast method combining measurements, without dark adaptation, of chlorophyll fluorescence, carboxylative activity of the RuBisCo and temperature-based variables. The authors have shown that a resulting variable, the IPL index, was strongly and linearly correlated to net photosynthesis in apple and pear under contrasted soil water supply. The IPL thus appears as a promising indicator, but has never been tested at HT.
Remotely sensed imagery in field crops have received large interest for rapid estimation of yield or responses to soil water deficit18. Recently, both multispectral (MS) and thermal infrared (TIR) imaging have been used for phenotyping large populations of individuals in annual species and performing genetic analyses on the estimated traits19,20. However, the related developments have mainly been tackled toward homogeneous plant cover and are not fully adapted to non-fully covering crops, like orchards. Analytical adaptations were developed in apple tree to estimate water stress indicators at the tree scale within biparental populations21, on which genetics analyses22 were performed. In these studies, the impact of water stress was computed based on the water deficit index (WDI) for partially soil covering vegetation. Although promising, these studies did not estimate other indicators provided by MS imaging. Among others, NDVI23 (Table 1) is considered as a proxy for leaf area index, and intercepted solar radiation; GNDVI24 and MCARI225 are quite similar to NDVI but display more (GNDVI) or less (MCARI2) sensitivity to chlorophyll concentration; while PRI26 accounts for the state of PSII and is considered as a proxy for radiation-use efficiency.
Table 1.
Description of the variables used in the study, with their respective methods of measurement and units
| Trait abbreviations | Definition | Unit | Method |
|---|---|---|---|
| TCSA | Trunk cross sectional area | cm² | Manual measurements |
| TLA | Tree leaf area | m² | |
| c_volume | Volume of the 3D convex hull | m3 | T-LiDAR |
| a_volume | Volume of the 3D alpha hull | m3 | |
| c i | Convexity index | ||
| STAR | Silhouette to total leaf area ratio | ||
| NDVI | Normalized difference vegetation index | Multispectral imaging | |
| GNDVI | Green NDVI | ||
| MCARI2 | Modified chlorophyll absorption ratio index improved | ||
| PRI | Photochemical reflectance index | ||
| WDI | Water deficit index | ||
| pix_num | Vegetation pixel number | ||
| Tsurf –Tair | Difference between canopy surface and air temperatures | °C | Thermal infrared imaging |
| A n | Net photo-assimilation | µmol m−2 s−1 | Infrared gas-analyzer (IRGA) |
| I PL | Photo-assimilation performance index | µmol m−2 s−1 | IRGA with fluorimeter |
| Tleaf -Tair | Difference between leaf surface and air temperature | °C | |
| P KO/KC | Computed leaf fluorescence parameter | µmol m−2 s−1 | |
| g s | Stomatal conductance | mmol m−2 s−1 | Porometer |
In this article, we describe for the first time in a woody plant in an orchard, the deployment of in field multi-scale and multi-objectives HT measurements on a core-collection of 241 genotypes representative of the European diversity of apple tree27. Fluorescence, airborne imagery and T-LiDAR techniques were deployed on the whole core-collection, and complemented by fine, in planta characterization (stomatal conductance, photosynthesis, leaf area) on a subset of contrasted genotypes. Effects of the genotype and watering scenario were dissected and a wide genotypic variability was identified for all traits. A clustering method was then used for determining genotype classes based on combinations of architectural and functional traits.
Material and methods
Plant material and experimental set-up
This study was carried out on an apple tree core-collection27 previously characterized for architectural and functional traits at 1-year-old stage into a HT phenotyping platform in controlled conditions4. The 241 genotypes of the core-collection were re-multiplied at INRA Angers and grafted onto M9 Pajam® 2 rootstock before being planted in field in February 2014 at the INRA experimental unit ‘DiaScope’’ in Montpellier, France (43°36 N, 03°58E). Trees were planted at 5 × 2 m distances, irrigated using micro-sprayers located between-trees, and left unpruned. Trees were not thinned from planting until 2016. In 2017, trees were manually thinned in mid-June to maintain one fruit per inflorescence. The orchard (1.2 ha) comprised ten rows of 100 trees, embedding two replications of two trees per genotype randomly distributed within the field. Well-watered (WW) and water deficit (WD) tree rows alternated within the trial, one WW tree facing one WD for each genotype. During the two first years after planting, no limiting watering regime was implemented to ensure a proper development of all trees. Differential watering scenarios were set-up on WW and WD lines in 2016 and 2017 and consisted in withholding irrigation on WD lines for one month during summer. Soil water potential (Ψsoil) was monitored by Watermark® tensiometric probes (−30 and −60 cm depth) connected to Agribase® dataloggers on a subset of cultivars having shown contrasted vigor in a previous greenhouse experiment4. Results reported here are focused on the 2017 experimental campaign only. That year, the mean value of harvest fruit weight per tree over the population was 8.43 kg with a maximum of 39.48 kg. Although this 4th year after planting corresponded to the first year of significant fruit production for most trees, some of them (8%) did not bear any fruit. From 7th July to 2nd August 2017, WW trees received 2 h irrigation per day, while WD trees were watered 2 h per day twice a week only. The water deficit was gradually established after irrigation withholding and Ψsoil reached up to –120 kPa for WD trees at the end of this period (late July 2017, Supplementary Fig. S1) while Ψsoil on WW trees was maintained close to 0 kPa. All measurements presented hereafter were carried out at the end of July 2017, when contrast in Ψsoil between WW and WD trees nearly reached its maximum, except for the T-LiDAR acquisition, which was carried out in early October 2017.
Thermal infrared and multispectral imaging
Image acquisitions and experimental set-up
Airborne imaging acquisition was carried out on clear sunny days, on 27th July from 1 to 2 p.m. and on 28th July from 9:30 to 10:30 a.m. (UTC). The vector was a Mikrokopter® hexa-rotor drone (www.phenome-fppn.fr/phenome_eng/Facilities/Montpellier-Field) operating at 25 m height. Three successive elevations were performed each day to cover the entire field, hence around 45 min were needed between the first and last image acquisition in the field. Zenithal images were acquired at frequency allowing image along-track and across-track overlaps of 80% and 70%, respectively. The on-ground field set-up comprised (i) gray targets for radiometric correction, (ii) contrasted thermal targets (2 m²) used during the flights and measured by IR120 thermoradiometers (Campbell Sci.), (iii) ground control points GPS-RTK (Global Position System–Real Time Kinematic) geo-referenced (2.5 cm accuracy), and (iv) a meteorological station acquiring air temperature and relative humidity, wind speed and direction, and global radiation at 10 s time-step. TIR and MS images were acquired, respectively, by a FLIR® TAU2.7 uncooled camera (7.5–13 µm, resolution of 640 × 512 pixels) and an AirPhen v3 camera (www.hiphen-plant.com) measuring reflectance in blue (450 nm), green (530 and 570 nm), red (675 nm), red-edge (730 nm), and NIR (850 nm) with a bandwidth close to 10 nm.
Vegetation and water stress index computation
Data post-processing (radiometric correction, image ortho-rectification, geolocation and mosaicking) was performed with Erdas Imagine Pro, ExifTool and Agisoft Photoscan softwares. Linear regressions between contrasted targets and IR120 temperatures allowed transforming the numerical values of image pixels into temperatures28. GPS coordinates were used to accurately localize each single tree of the experiment and a 0.70 m radius zone (buffer zone, Supplementary Fig. S2) was delineated around tree centers. For the canopy temperature (Tsurf) calculation, the leaf MS signature was used to classify images and discard soil pixels. Differences between surface and air temperatures (Tsurf –Tair)29 were then computed by subtracting the mean air temperature during the acquisition. MS-based index values (NDVI, GNDVI, PRI, and MCARI2) were extracted for each pixel within the buffer zone (Supplementary Table S1). For each index, mean and standard deviation of pixels values were calculated inside the buffer zone, considering vegetation pixels for Tsurf – Tair or all the pixels for MS-based indices. The WDI (Supplementary Table S1) was computed based on a trapezoid shape built from the scatterplot between Tsurf – Tair and NDVI21,30. WDI is an indicator used for estimating the effect of water deficit on leaf transpiration, varying from 0 (maximal transpiration) to 1 (no transpiration).
High-throughput estimation of leaf photosynthesis
Estimation of a semi-empirical index (IPL)
Following Losciale et al.17, the semi-empirical index IPL was used as a proxy for net leaf photosynthetic rate (An). It was computed as a linear combination of the electron transport rate exiting from PSII (JPSII, derived from the effective PSII quantum efficiency, ΦPSII) aggregated with the Michaelis–Menten constants for carboxylation (KC) and photorespiration (KO) forming a new variable, PKO/KC = JPSII × KO/KC, and the leaf-to-air temperature difference (Tleaf – Tair). Calibration of the linear regression was achieved through measurements on a subset of six genotypes (24 trees). Leaf gas-exchanges were measured using an open circuit InfRared Gas-exchange Analyzer (IRGA) fitted with a leaf fluorimeter and a light-emitting diode light source (LI-6400, LI-COR Inc., Lincoln, NE, USA) on 30th July. Measurements were performed with two LI-COR devices on fully expanded, fully exposed leaves and repeated at three time-periods: morning, midday, afternoon with actinic light set to 1500 μmol m−2 s−1. During each measurement, we recorded An, stomatal conductance (gs), leaf (Tleaf) and air (Tair) temperatures, maximum fluorescence with light-adapted leaf (Fm’), and steady state fluorescence (Fs) in order to compute ΦPSII. We collected with each device, respectively, 98 (LI-COR 1) and 132 (LI-COR 2) measurements. In the calibration process, two thirds of the data were used for estimating model coefficients in the two datasets. For each device, linear models were established gathering all the data collected, and their validity was assessed across watering scenarios or time of the day. Model validation was performed on the remaining third of the datasets, using the coefficients determined in the calibration process.
High-throughput measurements of IPL
HT measurements were performed on 4 trees × 195 genotypes during 2 consecutive days (27th and 28th July) with the two LI-COR devices complementarily. Measurements consisted in rapid (ca. 30 s per leaf) acquisition of ΦPSII, Tleaf, and Tair, performed during a maximum period of 3 h around solar midday. Parameters calculated in the calibration process were then used for each LI-COR to calculate IPL values.
In planta measurement of leaf stomatal conductance
Stomatal conductance (gs) was measured on 27th July on a subset of eight genotypes (two WW and two WD trees per genotype) with a porometer (AP4; Delta-T Devices Ltd). Measurements were performed on three fully expanded, fully sun-exposed leaves per tree, during a period of 1 h 30 min centered around solar midday. Stomatal conductance values were then compared to Tsurf – Tair and WDI estimated from TIR imaging and to Tleaf – Tair estimated with IRGAs to assess the ability of the difference between air and vegetation temperatures to account for functional differences.
High-throughput phenotyping of whole-tree leaf area and light interception
T-LiDAR data acquisition
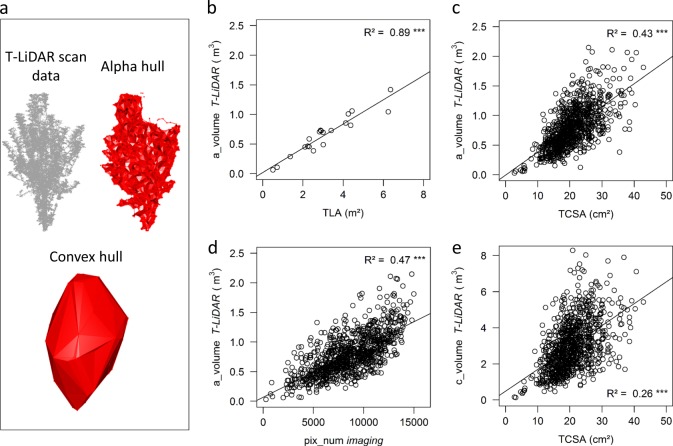
Data acquisition was performed on the whole population in October 2017 after harvest, with a RIEGL VZ400 terrestrial laser scanner (RIEGL Laser Measurement Systems GmbH, Horn, Austria). Scans were carried out with a 360° view each ten meters on every row with an angular resolution of 0.04°. Scans were then registered in a common coordinates system with the Riegl software (RiSCAN PRO v2.0.1) and point clouds were processed using the Computree platform (http://computree.onf.fr/) (Fig. 1a) to remove noise and outlier points.
Fig. 1. Representation of T-LiDAR variables, and comparisons with imaging data and direct plant measurements.
a Example of three-dimensional (3D) alpha and convex hulls computed on one 4-years-old tree scanned with T-LiDAR, in autumn 2017. b Correlation between alpha hull volume (a_volume) and total leaf area (TLA) computed on a subset of 20 trees in autumn 2017. c Correlation between a_volume and trunk cross sectional area (TCSA) measured on the whole core-collection in autumn 2017 (n = 930 trees). d Correlation between a_volume and vegetation pixel numbers (pix_num) in the airborne image buffer zone, on the whole core-collection. e Correlation between convex hull volume (c_volume) and TCSA. Coefficients of determination and their significance are indicated in each panel
Plant shape and light interception descriptors
Volumes of convex hulls (c_volume) of the tree point clouds were computed using the PlantGL library31 within OpenAlea platform32 and volumes of alpha hulls (a_volume) with R software33 using the alphashape3d library. The c_volume reflects the maximal space occupation of the tree, while a_volume is an extension of the convex hull allowing the creation of concave envelopes around the point cloud and is consequently more associated with the space really occupied by the trees. Creation of concavities in the alpha hull depends on a parameter (α) whose value was estimated on a subset of 20 trees on which the total tree leaf areas (TLAs) were estimated by randomly collecting one leaf over five within the canopy. Individual areas of collected leaves were then measured using a leaf area meter (LI-COR 3100, Area Meter; Lincoln, NE, USA). The chosen α value (0.15) was the one maximizing the correlation between a_volume and TLA on this subset of trees. A convexity index (ci) was computed as the ratio of a_volume to c_volume to evaluate the density of tree space occupation within its convex hull. A proxy of light interception efficiency, the STAR, was computed in PlantGL following previously published methods34.
The trunk cross sectional area (TCSA) was computed from the trunk circumference acquired with a tape measure on the whole core-collection in September 2017. As TCSA is known as a relevant indicator of plant vigor35, it was directly compared to the canopy volumes (T-LiDAR) and to the vegetation indexes (MS imaging).
Statistical analyses
Analyses were performed using R software33. For each trait, models were selected based on the lowest Bayesian Information Criterion, among several mixed-effect models (Supplementary Table S2). Models always included a random genotypic effect and a fixed-effect of the scenario, completed or not by other fixed effects (line, daytime period, gas-exchange device, climatic conditions). The best linear unbiased predictors (BLUPs) of genetic values were estimated for each trait with the selected model. Variance components were then used to estimate the broad-sense heritability (H2) as:
| 1 |
with the genetic variance, the residual variance, n the number of replicates per genotype.
Multivariate analyses were performed on the genetic values. Pearson coefficients of correlation between variables were evaluated, considering phenotypic and genotypic values. A PCA and a Hierarchical Ascendant Classification based on Ward method were performed, using variables related to architectural and functional traits and estimated from the three different HT methods. Some variables closely correlated to other ones were not considered in these multivariate analyses to limit redundancies.
Results
Characterization of tree architecture and effect on light interception efficiency
Correlation between a_volume and TLA (Table 1), assessed on 20 trees with TLA ranging from 0.3 to 6.2 m², was highly significant (R² = 0.89, Fig. 1b). a_volume was positively, highly significantly correlated to TCSA on the whole core-collection (Fig. 1c, R² = 0.43). a_volume was also highly correlated to pix_num estimated from the zenithal airborne images (Fig. 1d, R² = 0.47). By contrast, the correlation between c_volume and other variables related to tree vigor were lower (R² = 0.26, for both TCSA and pix_num, Fig. 1e and Supplementary Fig. S3).
All parameters estimated from T-LiDAR exhibited strong effect of the genotype and high heritability values (H² from 0.77 to 0.86; Table 2). A slight but significant effect of the watering scenario was found on all traits (except a_volume), associated with a moderate decrease in the size of the trees under WD as compared to WW conditions (e.g., –3.7% and –10.4% for a_volume and c_volume, respectively). Higher ci was observed under WD, suggesting a higher leaf area density. Consistently, higher leaf area density led to WD trees with lower STAR values (Table 2).
Table 2.
Range of variability for the traits (mean ± SD) measured on the core-collection of apple trees, with effects of the genotype, of the watering scenario, and broad-sense heritability
| Traits | Mean±SD | P G | P S | P D | H² | |
|---|---|---|---|---|---|---|
| WW | WD | |||||
| TCSA (cm2) | 20.7 ± 0.47 | 19.6 ± 0.45 | *** | *** | – | 0.73 |
| c_volume (m3) | 3.17 ± 1.44 | 2.84 ± 1.32 | *** | *** | – | 0.82 |
| a_volume (m3) | 0.81 ± 0.36 | 0.78 ± 0.33 | *** | ns | – | 0.77 |
| c i | 0.27 ± 0.09 | 0.30 ± 0.10 | *** | *** | – | 0.86 |
| STAR | 0.75 ± 0.15 | 0.71 ± 0.14 | *** | *** | – | 0.78 |
| NDVI | 0.34 ± 0.09 | 0.29 ± 0.09 | *** | *** | *** | 0.85 |
| GNDVI | 0.23 ± 0.09 | 0.24 ± 0.10 | *** | ns | *** | 0.79 |
| MCARI2 | 0.55 ± 0.13 | 0.43 ± 0.14 | *** | *** | ns | 0.87 |
| PRI | −0.048 ± 0.049 | −0.061 ± 0.042 | *** | *** | *** | 0.69 |
| WDI | 0.18 ± 0.18 | 0.30 ± 0.08 | *** | *** | *** | 0.70 |
| pix_num | 9582 ± 2512 | 8466 ± 2786 | *** | *** | *** | 0.84 |
| Tsurf – Tair (°C) | 0.82 ± 2.15 | 2.88 ± 2.21 | *** | *** | *** | 0.63 |
| I PL | 14.0 ± 5.93 | 9.72 ± 5.61 | *** | *** | – | 0.43 |
Acronyms as in Table 1. The significance of genotype (PG), watering scenario (PS), and date effects (PD) was tested considering a linear fixed-effect model without interaction. ****p ≤ 0.001; ns, non-significant. Date effect is considered for imaging data, only
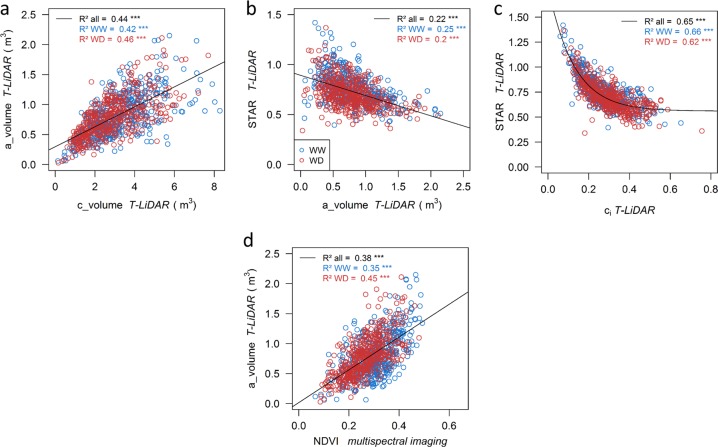
We explored the correlations between the variables derived from T-LiDAR measurements to evaluate the relationship between tree vegetative architecture and light interception efficiency as estimated by STAR values. a_volume was positively correlated to c_volume whatever the watering scenario with R² = 0.44 (Fig. 2a) showing however a range of contrasted combinations of a_volume and c_volume values within the core-collection. STAR was negatively correlated with ci (Fig. 2c; R² = 0.65, power function) and weakly, negatively correlated with a_volume (Fig. 2b; R² = 0.22). Finally, a_volume was significantly, but not tightly, correlated to NDVI estimated from MS images (Fig. 2d, R² = 0.38).
Fig. 2. Correlations between T-LiDAR and multispectral (MS) imaging indices.
a–c Correlations between T-LiDAR variables on the whole core-collection (n = 930 trees): alpha hull (a_volume) and convex hull volumes (c_volume), silhouette to total leaf area ratio (STAR) and convexity index (ci). d Correlation between a_volume and NDVI estimated from MS imaging. Blue and red points represent trees subjected to well-watered (WW) or water deficit (WD) conditions, respectively. For each correlation, the coefficient of determination and its significance was computed considering either the whole dataset (all), or each watering scenario independently (WW or WD)
Variability of the multispectral indices
Vegetation indices resulting both from functional and architectural plant features were computed from MS imaging. All indices (NDVI, GNDVI, MCARI2, and PRI, Tables 1 and 2) exhibited a wide range of variability within the core-collection. They displayed a strong effect of the genotype and high heritabilities (0.70 < H² < 0.87). High-positive phenotypic correlations were found between NDVI and GNDVI on one hand (R² = 0.52), and between NDVI and MCARI2 on the other hand (R² = 0.76, Table 3). By contrast, the correlation between GNDVI and MCARI2 was low, although significant (R² = 0.09). Importantly, these correlations were conserved when considering genotypic values, evidencing the consistent behaviors across trees of the same genotype. PRI was positively correlated with GNDVI (R² = 0.40), slightly with MCARI2 (negative correlation) and not correlated with NDVI if genotypic correlations were considered (Table 3). Water deficit generally had a low but significant effect on all the MS imaging indices, except GNDVI, with a relative variation between WW and WD of about 10% (Table 2).
Table 3.
Determination coefficients for the correlations between the indices estimated from multispectral and thermal imaging and the IPL
| Genotypic values | |||||||
|---|---|---|---|---|---|---|---|
| Phenotypic values | NDVI | ( + )0.49*** | ( + )0.85*** | ( + )0.0022 ns | (−)0.15*** | ( + )0.19*** | ( + )0.044** |
| ( + )0.52*** | GNDVI | ( + )0.15*** | ( + )0.28*** | (−)0.063*** | ( + )0.12*** | ( + )0.017 ns | |
| ( + )0.76*** | ( + )0.090*** | MCARI2 | (−)0.04** | (−)0.13*** | ( + )0.16*** | ( + )0.036** | |
| ( + )0.044*** | ( + )0.40*** | (−)0.023*** | PRI | ( + ) 0.032** | ( + )0.053*** | ( + )0.006 ns | |
| (−)0.12*** | (−)0.0044* | (−)0.20*** | ( + )0.048*** | Tsurf – Tair | ( + )0.41*** | ( + )0.0001 ns | |
| ( + )0.012** | ( + )0.029*** | ( + )0.001 ns | ( + )0.023*** | ( + ) 0.64*** | WDI | ( + )0.026* | |
| (−)0.0061* | (−)0.029*** | ( + )0.0009 ns | (−)0.084*** | (−) 0.11*** | (−)0.11*** | I PL | |
Acronyms as in Table 1. Above the diagonal, correlations calculated on the genotypic values (BLUPs, n = 241 genotypes). Below the diagonal, correlations calculated on the individual phenotypic values (n = 930 trees). For each correlation, the determination coefficient is indicated together with the sign ( + , positive, −, negative) of the correlation. The correlation coefficients are displayed with their significance level: *p ≤ 0.08; **p ≤ 0.05; ***p ≤ 0.01; ****p ≤ 0.001; ns, non-significant.
Using fluorescence measurements for estimating genotypic variation of photosynthesis
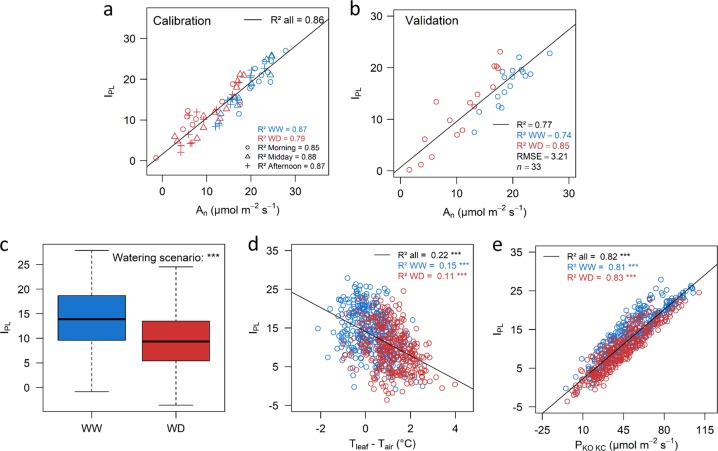
In this study, leaf fluorescence measurements were used as a proxy for photosynthesis activity in order to extend the functional phenotyping of the core-collection. The calibration dataset for the IPL model covered a wide range of variation for An (from 2 to 30 µmol m−2 s−1, Fig. 3a and Supplementary Fig. S4a). The device (two IRGAs were used) impacted the range of values recorded for Tair, Tleaf, and ΦPSII (Supplementary Table S3), likely due to internal calibration proper to each. Thus, a specific set of parameters was chosen for each device. Both PKO/KC and Tleaf – Tair (Table 1) had highly significant effects on IPL (p < 0.0001), confirming the relevance of using both parameters to estimate photosynthesis accurately. Calibrations were robust (R² = 0.86 and 0.84, Fig. 3a and Supplementary Fig. S4a) and a unique set of parameter values was relevant whatever the scenario or period (R² > 0.79 when splitting the calibration dataset per scenario or daytime). Accuracy was confirmed by the high values of R² and root mean square errors (RMSE) obtained in the validation datasets (Fig. 3b and Supplementary Fig. S4b, R² = 0.77 and 0.74, respectively, and RMSE = 3.1 and 3.2, respectively). IPL values were then estimated at HT on the core-collection, from the calibrated model, and ranged from 0 to 27 among the WW trees and from –5 to 25 for WD ones, with a significant effect of the genotype (Table 2). WD decreased IPL values by 30% on average as compared to WW conditions (Table 2 and Fig. 3c). Its variation within the core-collection was mostly driven by variability in PKO/KC (Fig. 3e, R² = 0.82), which combines electron transport rate exiting from PSII (JPSII), in turn dependent on the absorbed photosynthetically active radiation and effective PSII efficiency, and, to a lesser extent, by variability in Tleaf – Tair (Fig. 3d, R² = 0.22), itself related to the degree of stomatal closure. Variations in air VPD along the measurement duration also significantly affected IPL values (Supplementary Fig. S5). We thus integrated these undesired effects (device, VPD) into the mixed-models used for BLUPs and heritability calculation (Supplementary Table S2). This yielded medium heritability for IPL (H² = 0.43, Table 2). Low correlation coefficients, although sometimes significant due to the large number of individuals, were found between the IPL and the vegetation indices derived from MS imaging (Table 3).
Fig. 3. Calibration and validation of the photo-assimilation performance index (IPL) model, and variability of the IPL within the core-collection.
a, b The calibration was obtained by measuring net photosynthesis and fluorescence on 48 trees in a one-day measurement during summer 2017. Calibration model was built using 2/3 of the data (n = 93, a), and model was then validated on the other 1/3 of the data (n = 33, b). Data collected at three measurement period (morning, midday, afternoon) are identified with different symbols. In a and b, calibration and validation are presented for one of the two IRGA devices used (see Supplementary Fig. S4 for the second one). c Boxplot representation of the IPL measured at high-throughput on the whole core-collection (n = 800 trees). The significance of the watering scenario was assessed with a one-way ANOVA. *** significant at p < 0.001. d, e Correlations between the IPL and its two components, Tleaf – Tair (d) and PKO/KC (e) on the whole core-collection. Blue and red points represent trees subjected to well-watered (WW) or water deficit (WD) conditions, respectively. Coefficients of determination and their significances were computed considering either the whole dataset (all), or within each watering scenario independently (WW or WD). Root mean square errors were estimated to assess the consistency of correlations between An and IPL
High-throughput characterization of whole-plant response to water deficit
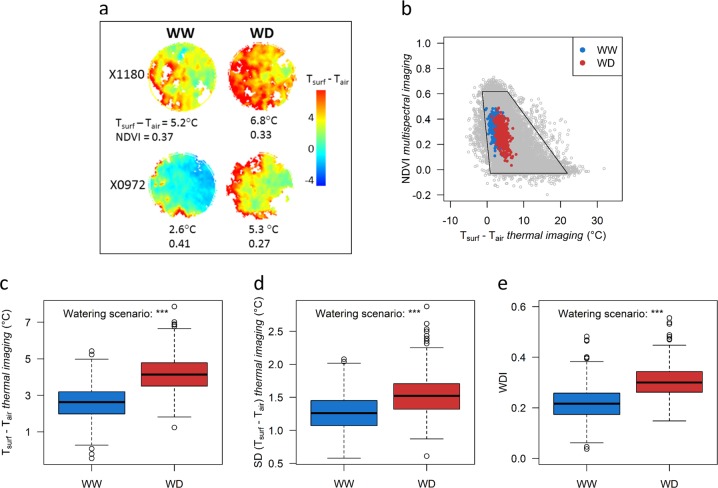
Our experimental design allowed to further investigate the response of genotypes to soil water deficit as two plants per genotype were subjected to soil water deficit conditions. Whole-plant sensitivity to water deficit was analyzed with two main variables computed from airborne images: Tsurf – Tair and WDI (Fig. 4a), over two consecutive dates (27th July at midday and 28th July morning). The acquisition date had a highly significant effect on both variables, with lower values on 28th July (mean Tsurf – Tair = 3.07 and 0.64 °C, and WDI = 0.27 and 0.22 for all the trees in 27th July and 28th July, respectively). The lower values on 28th July were likely due to the daytime period differing between both dates, with lower temperature and VPD (Supplementary Fig. S6) in the morning conditions that led to higher stomatal conductance and transpiration rate. Moreover, as image acquisition extended over >45 min with three different elevations for each flight date, some variations also existed within each day of measurement (Supplementary Fig. S6). Despite changes in absolute values, correlations between data collected on the two dates and corrected based on the fixed effects of day and flight within the day remained significant although with medium correlation coefficients (Supplementary Fig. S7).
Fig. 4. Impact of water deficit on the water deficit index (WDI) and the surface-to-air temperature difference (Tsurf – Tair).
a Thermal pixel array within the buffer zone estimated from thermal infrared (TIR) imaging on four trees belonging to two genotypes (X1180 and X0972) under well-watered (WW) and water deficit (WD) conditions on 27th July. Corresponding mean Tsurf – Tair and NDVI values are indicated below each tree. b Relationship between NDVI and Tsurf – Tair. Gray points represent all the pixels, including soil, weed, and trees, whereas red and blue points are the mean values of Tsurf – Tair and NDVI for each individual tree (blue: WW trees, red: WD trees). Solid lines represent the trapezoid shape used for computing WDI. Extremities of the trapezoid represent “well-watered vegetation” (top left), “water-stressed vegetation” (top right), “satured bare soil” (bottom left) and “dry bare soil” (bottom right) conditions. c–e Boxplot representations on the whole core-collection of the mean Tsurf – Tair values (c), the standard deviation of Tsurf – Tair within the canopy (d), and WDI (e) depending on watering scenarios. In c–e data are phenotypic values (n = 930), corrected for fixed effects of date and daytime period. The significance of the watering scenario was assessed with a one-way ANOVA. ***Significant at p < 0.001
On 27th July, the four temperatures used for estimating the trapezoid relationship between NDVI and Tsurf – Tair were, respectively, equal to 0.87, −1.34, 21.86, and 5.6 °C for saturated bare soil, well-watered vegetation, dry bare soil, and water-stressed vegetation, respectively (Fig. 4b). Consistently with the observed Tsurf –Tair, these temperatures were lower on 28th July (Supplementary Fig. S8). As expected, whatever the date, Tsurf –Tair and WDI (Fig. 4c, e and Supplementary Fig. S8b, c) increased under water deficit conditions. Moreover, large variations were observed for WD trees with a genotypic coefficient of variation equal to 0.41 and 0.32 under WD conditions for Tsurf –Tair and WDI, respectively. Tsurf –Tair was more heterogeneous within tree canopy under WD as compared to WW conditions, as shown by the higher standard deviation computed from all vegetation pixels of each tree (Fig. 4d and Supplementary Fig. S8d). Mixed-effect models accounting for scenario, date and elevation effects within each day were used for WDI and Tsurf –Tair on the dataset combining the 2 days of measurements (Supplementary Table S2), yielding high heritabilities for both traits (Table 2).
Assessments of high-throughput phenotyping methods for functional traits
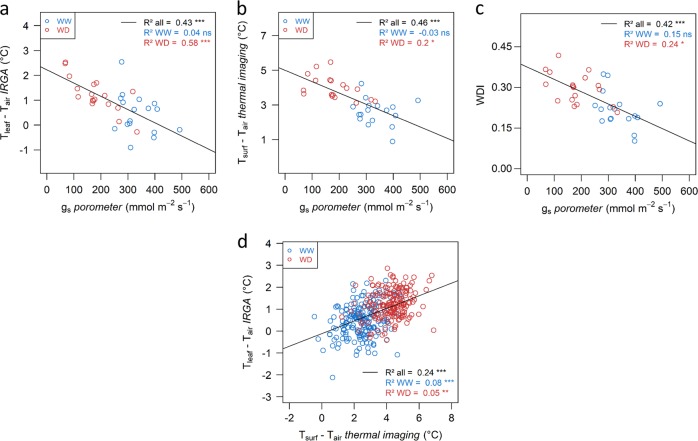
The relevance of HT measurements of functional traits was assessed by comparing Tsurf –Tair derived from imaging to low-throughput measurements with the IRGA of these variables and to stomatal conductance values. We thus explored the relation between gs and the leaf-to-air temperature difference considering the latter at different scales: (i) at the canopy level (mean value of Tsurf –Tair derived from imaging), and (ii) at the leaf level (Tleaf –Tair measured with the IRGA during the IPL measurements, on one of every three leaves per tree measured with the porometer). Significant correlations between Tsurf –Tair (respectively, Tleaf –Tair) or WDI and gs were observed when considering the whole dataset (WW and WD) or the WD plants only (Fig. 5a, c). Nevertheless, these correlations were no longer significant for the WW trees. Tleaf –Tair and Tsurf –Tair were significantly correlated (Fig. 5d) but this correlation was mostly driven by the contrast between WW and WD trees, whereas it was no longer significant to discriminate the individuals within each watering regime separately. Vegetation temperature was significantly lower when measured with the IRGA as compared to canopy imaging even though these temperatures were measured at the same time of the day (Fig. 5d). For most of the genotypes, the temperature measured with the IRGA ranked into the first quartile of the pixel canopy temperature estimated from airborne imagery (Supplementary Figs. S9 and 10).
Fig. 5. Assessment of high-throughput methods for computing leaf or surface temperatures.
a Correlation between leaf-to-air temperature difference (Tleaf – Tair) measured with the gas-exchange analyzer during the IPL measurement, and stomatal conductance (gs) measured with the porometer. b Correlation between the mean value of surface-to-air temperature difference (Tsurf – Tair) obtained by thermal infrared imaging (TIR) and gs measured with the porometer. c Correlation between water deficit index (WDI) and gs measured with the porometer. d Correlation between Tleaf – Tair measured with the gas-exchange analyzer during the IPL measurement, and the mean value of Tsurf – Tair obtained by TIR imaging. In a–c, n = 40 trees; in d, n = 400 trees. All measurements were performed on the same day. gs and Tleaf – Tair were measured, respectively, on three and one single leaf per tree. Mean Tsurf – Tair was calculated overall the pixels of each tree
Identification of genotype classes based on architectural and functional traits
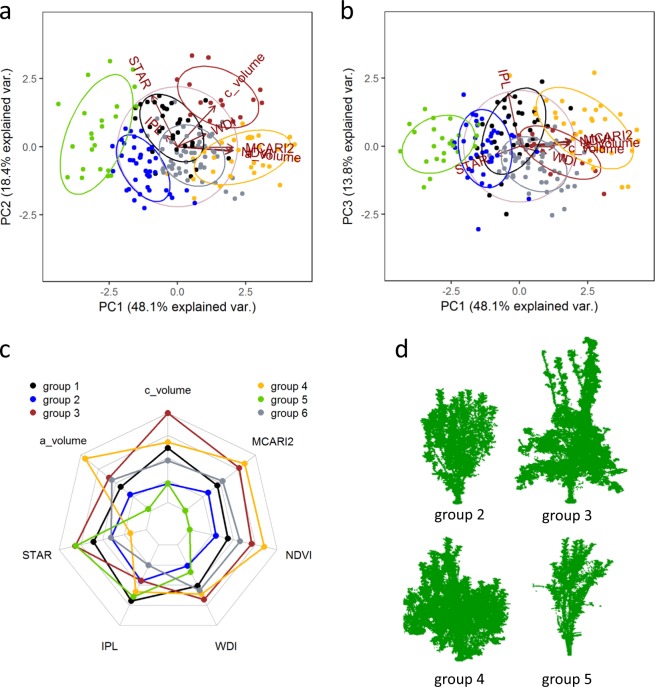
We performed a PCA analysis on the BLUPs estimated on the whole core-collection with mixed-effect models including, when significant, the watering scenario as fixed-effect to explore the quantitative relationships between variables and similarities between genotypes. Seven variables were chosen to encompass traits (i) related to plant architecture (c_volume, a_volume), (ii) combining plant architecture and functioning (NDVI, MCARI2, STAR), (iii) accounting for the limitation in leaf functioning due to soil water content (WDI) and (iv) representing leaf photosynthesis (IPL). The first two axes explained >66% of the variance and the first four axes >94% (46.1%, 18.4%, 15.2%, 14.6% for axes 1, 2, 3, and 4, respectively, Fig. 6a, b and Supplementary Fig. S11). The variability in the population was first explained by a combination of variables related to the size of the tree (a_volume, NDVI, MCARI2, PCA’s axis 1) and second by STAR, and to a lower extent c_volume (PCA’s axis 2). The third and fourth dimensions were almost fully explained by one variable, only (IPL or WDI, respectively). A hierarchical clustering (Fig. 6c, d) performed on the same variables as those used for PCA allowed the identification of six groups of genotypes. Consistently with the results of the PCA, the population was first separated into two parts depending on variables related to tree size with groups 2 and 5 displaying lowest values. Groups of large and small plants then differed depending on STAR values (strongly associated with the ratio of a_volume to c_volume) with groups 4 and 2 displaying low values, for small and large plants, respectively (Table 4). Among groups of large plants with high STAR values, group 6 had the lowest IPL values and group 2 was the least sensitive to water stress (low WDI value). This clustering analysis showed independence between architectural and functional traits. Indeed, many combinations of plant size and IPL values could be observed (e.g., group 5: small plants, high IPL; group 3: large plants, low IPL; group 2: small plants, low IPL; group 4: large plants, medium IPL).
Fig. 6. Results of the multivariate analyses performed on the genetic values of the architectural and functional traits on the whole population.
a, b Projections of individuals and variables on the three first axes of the principal component (PC) analysis. Groups represent the six groups identified using a Hierarchical Ascendant Classification. c Radar plot of the mean trait values computed using scale variables for the six groups. d T-LiDAR point clouds of 4 trees representative of groups 2, 3, 4 and 5
Table 4.
Mean values per group for the variables used for clustering analysis
| Groups | Number of genotypes | c_volume | a_volume | STAR | I PL | WDI | NDVI | MCARI2 |
|---|---|---|---|---|---|---|---|---|
| 1 | 32 | 3.34b | 0.76c | 0.77a | 12.7a | 0.24a,b | 0.31c | 0.48b |
| 2 | 39 | 2.21d | 0.68d | 0.71b | 11.7b | 0.22c | 0.29d | 0.44c |
| 3 | 15 | 4.45a | 0.87b | 0.83a | 11.8b,c | 0.26a | 0.35a,b | 0.56a |
| 4 | 32 | 3.52b | 1.08a | 0.64c | 12.3a,b | 0.25a | 0.37a | 0.58a |
| 5 | 24 | 2.21d | 0.52e | 0.83a | 12.5a,b | 0.23b,c | 0.25c | 0.36d |
| 6 | 52 | 2.94c | 0.84b | 0.71b | 10.9c | 0.25a | 0.33b | 0.50b |
| Group effect | *** | *** | *** | *** | *** | *** | *** |
The significance of the group effect was tested with a one-way ANOVA followed by a Tukey’s HSD test for pairwise comparison. ***significant at p < 0.001. Values followed by different letters are significantly different at p < 0.05
Discussion
An original combination of high-throughput tools to phenotype thousands of trees under contrasted watering regimes
HT phenotyping methods were combined to study the genotypic variability of variables related to plant architecture (a_volume, c_volume, ci, STAR), functioning (IPL), sensitivity to soil water deficit (WDI, Tsurf – Tair) and “mixed” traits accounting both for architectural and functional characteristics (NDVI, GNDVI, MCARI2). This study thus extends the domain of application of new HT methods, up to now mostly deployed on annual species18.
To assess the accuracy of these methods, the traits measured at HT were compared to in planta measurements, or to other traits acquired at HT with an alternative method. For architectural variables, the significant correlations between a_volume, TLA, TCSA, pix_num, and NDVI confirm the relevance of using T-LiDAR or airborne imagery for characterizing whole-tree development. Nevertheless, correlations between T-LiDAR variables and NDVI were quite low, likely due to the nature of NDVI, which also accounts for features not directly associated with plant architecture (i.e., chlorophyll content). Architectural variables were complemented with STAR estimations. STAR was negatively correlated with a_volume and ci. This first suggests that plants with large leaf area have lower radiation absorption per unit leaf area, probably due to leaf over-lapping, as previously observed36, and second, that ci is a good indicator of foliage 3D distribution within the canopy. Interestingly, large genotypic variability was also found among genotypes for GNDVI and MCARI2, which both depend on leaf area index and chlorophyll concentration24,25. PRI, which is less dependent on tree architecture but closely associated with radiation-use efficiency, also displayed large variability. Overall, our results underline the relevance of using a 6-channel multispectral acquisition in combination with thermal infrared imagery, which allowed a high spatial resolution and proved to be cost-effective37. A technical alternative could rely on hyperspectral acquisition, whose non a-priori approach and high spectral resolution is tempting but would imply lower spatial resolution and more challenging data analysis. Subsequently, in order to estimate photosynthesis at the leaf level, we computed a semi-empirical parameter (IPL) estimated using a calibration-validation procedure, which revealed the accuracy of this indicator independently of climate or watering scenario. While it was proposed that the model parameters were species specific17, we show that they also vary with the device used, hence the importance of running calibration measurements for any new experiment.
Relevant design and analytical methods allow detecting strong genotypic variability in response to water deficit
Experimental design and timing of measurements are critical to ensure successful HT characterization. In our case, due to local regulation, the drone flight had to be carried out at reduced altitude, thus requiring several successive elevations to cover the entire field. This results in variations of environmental conditions with Tsurf following the variations of Tair and VPD. By repeating measurements over 2 consecutive days at two distinct periods, we were able to compare the effect of external conditions. Measurements centered on solar midday offered more stabilized conditions. Similarly, for the leaf fluorescence measurements, we made sure to perform measurements under saturating solar radiation conditions to avoid any pitfalls frequently reported as fluorescence measurements are dependent on the ambient light levels13. Importantly, the use of mixed-effect models was crucial to get rid of the environmental, uncontrolled variability (e.g., fixed effects of elevation for the airborne imagery-derived indices; of the VPD for the IPL).
With this approach, we were thus able to evidence strong genotypic effects on all the studied variables, most of them displaying high heritabilities (H² > 0.6). Whereas strong genotypic effects on indices derived from airborne imagery were similarly found on annual crops38, such high values were not frequently reported on woody crops (e.g., no effect of the genotype from TIR imagery on a poplar biparental population39). For architectural traits, heritabilities were similar to or even higher than those obtained in previous studies in field on biparental population7 or on the same core-collection in controlled conditions4, thus confirming the strong genetic determinism of architectural traits.
Marked genotypic differences were observed between the effects of the watering regimes on the architectural and functional variables. Functional traits revealed average reductions by >30% in IPL and to a lesser extent in PRI. Leaf temperature strongly increased under water deficit (around + 2 °C) probably due to stomatal closure under these conditions. Such responses are consistent with rapid changes in leaf homeostasis upon soil drying, likely due to hormonal or hydraulic regulations40. On the other hand, vegetative and architectural traits (e.g., a_volume, NDVI) only showed a slight, not always significant reduction in WD as compared to WW trees. Indeed, these integrative traits were built over the whole 4-years tree life, while the WD condition was only established during one month in 2016 and 2017. Besides, in these 2 years, the WD occurred in July, when most of the vegetative development was already achieved.
On the consistency and discrepancies between scales of analysis
The scale of analysis (from the leaf to the whole-plant) is a recurrent matter of debate for the study of functional traits. Complementarity of our approaches allows tackling this question within the apple core-collection. “Ground-truthing” measurements (e.g., stomatal conductance by porometry) are often conducted in parallel to airborne image acquisitions and confronted to TIR or MS indices. This results in a profusion of studies reporting a wide range of correlations between canopy indices and in planta measurements (e.g., R² between gs and Tsurf – Tair ranging from 0.27 to 0.92 in several woody crops41,42). In our study, the simultaneous measurement of Tleaf – Tair (IRGA) and of Tsurf – Tair (TIR imaging) reveals that a well exposed, well developed leaf typically chosen for gas-exchange measurements generally, but not systematically, falls within the first quartile of values reported from zenithal images on the same tree (Supplementary Figs. S9 and 10 and Fig. 5d). Consistently, the correlation found between Tsurf – Tair and gs was significant with a medium R² (0.43) mostly driven by the contrast between watering scenarios rather than genotypes (Fig. 5a), consistent with other reports39. Another example is the low correlation found between the IPL measured on single leaves and the PRI associated with leaf fluorescence but also known to be highly affected by factors such as canopy structure, viewing and illumination geometry effects, and background43. As leaves within a tree strongly differ in terms of size, age, position or nitrogen content, care should thus be taken when confronting scales and thinking in terms of comparison should be preferred rather than in terms of validation. Upscaling to the canopy level and assessing the sample heterogeneity, as authorized by imagery techniques, might be crucial to extract relevant information on tree functioning.
Toward genetic analyses of tree performances in a genotype × environment interaction context
Based on the BLUPs extracted for each trait, we were able to identify six distinct genotypic classes. These results demonstrate the absence of unique relationships between tree functioning, size and architecture, suggesting independent genetic controls. This result also suggests sink activity regulation or other developmental controls (transcriptome, hormones) as drivers of plant growth and phenotype construction44. Moreover, the large variations in tree crop loads in the core-collection could have hidden a relationship between architectural and functional traits due to feedback inhibition of photosynthesis under low crop load conditions45.
This work will benefit from recent advances in genotyping and the availability of a high-density map for the apple core-collection46 to explore the genetic bases of trait variations, using genome-wide association studies. Whereas classical QTL analyses led on fruit tree species often tackle a unique type of traits (e.g., architecture7; phenology47), our work will offer the opportunity to simultaneously decipher the genetic control of architectural and functional traits. The wide genetic background offered by the core-collection promises a gain for the power of detection and the genericity of detected QTLs as compared to classical approaches led on biparental populations48. The portability of our approach will also allow its deployment over other sites across Europe on the same core-collection47 and a unique opportunity to decipher the genotype-by-environment interactions ruling the traits of interest. Our protocol could be deployed throughout the gradual establishment of water deficit in order to compute response curve parameters depending on the genotype. Such an approach will complement the results of this study in which we considered the water stress treatment as a fixed-effect at one date. The robustness of our HT indices also suggest their relevance for studies oriented toward agronomical questions, e.g., orchard management on a few varieties. Nevertheless, their further deployment would require the creation of standardized protocols and common data analysis pipelines accessible to non-expert users. This could be achieved within open-access platforms dedicated to phenotyping data analyses (e.g., OpenAlea32). Finally, this study was carried out on fruiting adult trees but did not directly account for the HT evaluation of fruit production variation within the population. A number of imagery methods at HT were proposed to detect fruits within the canopy49. Nevertheless, this detection is not straightforward due to the large variability in fruit characteristics (size, color) within such a collection. To overcome this problem, new popular methods for remote sensing data based on deep learning could be used to improve the classification of fruits and leaves within the canopy50.
Supplementary information
Acknowledgements
We thank AFEF team for technical support, Stephan Feral (INRA Mauguio) for the phytosanitary monitoring of the core-collection, Benoit Rolland and Florence Meunier (INRA Mauguio) for UAS operating, and Myriam Dauzat (INRA Montpellier) for help in management of the IRGAs. We thank Cyrille Violle and Clement Piel (CNRS Montpellier) for the loan of their Li-COR 6400 devices. We are also thankful to Luca Corelli-Grappadelli, Luigi Manfrini, and Brunella Morandi (University of Bologna) for fruitful discussions about the IPL. Olivier Simler and Bilan Yonis-Omar are acknowledged for their help for developing the field protocol for T-LiDAR acquisition, and Laurent Albrech (IRSTEA), Cécile Madelaine-Antin (CIRAD), and Alexandre Piboule (ONF) for their technical assistance in post-processing analyses. A.C.-L. received a fellowship from the Agropolis Fondation (post-doctoral fellowship Outbound mobility 1502-306-APStress), and the support of the AgreenSkills + fellowship program (co-funded by the EU’s Seventh Framework Program under grant agreement No. FP7-609398 (AgreenSkills + contract)). This study was also funded by the ApCoreStress project (INRA-BAP department) and supported by GIS Fruit (Bilan Yonis-Omar’s internship).
Authors' contributions
B.P., A.C.-L., M.D, F.B., J.-L.R., and E.Co. designed research. A.C.-L., B.P., M.D., E.Ca., S.M., and J.-L.R. performed the experiments. M.D. performed the MS and TIR images post-processing. B.P., E.Ca., S.M., and F.B. performed the T-LiDAR data post-processing. A.C.-L. and B.P. analyzed data. A.C.-L. and B.P. wrote the paper with the contribution of J.-L.R., E.Co., M.D., and F.B.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-019-0137-3).
References
- 1.Monteith JL. Climate and the efficiency of crop production in Britain. Philos. Trans. R. Soc. Lond. B. 1977;281:277–294. doi: 10.1098/rstb.1977.0140. [DOI] [Google Scholar]
- 2.Laurens, F. et al. An integrated approach for increasing breeding efficiency in apple and peach in Europe. Hortic. Res.1, 5–11 (2018). [DOI] [PMC free article] [PubMed]
- 3.Coupel-Ledru A, et al. Reduced nighttime transpiration is a relevant breeding target for high water-use efficiency in grapevine. Proc. Natl Acad. Sci. 2016;113:8963–8968. doi: 10.1073/pnas.1600826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez G, et al. Genetic variation of morphological traits and transpiration in an apple core collection under well-watered conditions: Towards the identification of morphotypes with high water use efficiency. PLoS ONE. 2015;10:e0145540. doi: 10.1371/journal.pone.0145540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabrera‐Bosquet L, et al. High‐throughput estimation of incident light, light interception and radiation‐use efficiency of thousands of plants in a phenotyping platform. New Phytol. 2016;212:269–281. doi: 10.1111/nph.14027. [DOI] [PubMed] [Google Scholar]
- 6.Liebhard R, Kellerhals M, Pfammatter W, Jertmini M, Gessler C. Mapping quantitative physiological traits in apple (Malus × domestica Borkh.) Plant Mol. Biol. 2003;52:511–526. doi: 10.1023/A:1024886500979. [DOI] [PubMed] [Google Scholar]
- 7.Segura V, Cilas C, Costes E. Dissecting apple tree architecture into genetic, ontogenetic and environmental effects: mixed linear modelling of repeated spatial and temporal measures. New Phytol. 2008;178:302–314. doi: 10.1111/j.1469-8137.2007.02374.x. [DOI] [PubMed] [Google Scholar]
- 8.Massonnet C, Regnard JL, Lauri PEacute, Costes E, Sinoquet H. Contributions of foliage distribution and leaf functions to light interception, transpiration and photosynthetic capacities in two apple cultivars at branch and tree scales. Tree Physiol. 2008;28:665–678. doi: 10.1093/treephys/28.5.665. [DOI] [PubMed] [Google Scholar]
- 9.Dassot M, Constant T, Fournier M. The use of terrestrial LiDAR technology in forest science: application fields, benefits and challenges. Ann. For. Sci. 2011;68:959–974. doi: 10.1007/s13595-011-0102-2. [DOI] [Google Scholar]
- 10.Moorthy I, et al. Field characterization of olive (Olea europaea L.) tree crown architecture using terrestrial laser scanning data. Agric. For. Meteorol. 2011;151:204–214. doi: 10.1016/j.agrformet.2010.10.005. [DOI] [Google Scholar]
- 11.Hosoi F, Omasa K. Factors contributing to accuracy in the estimation of the woody canopy leaf area density profile using 3D portable lidar imaging. J. Exp. Bot. 2007;58:3463–3473. doi: 10.1093/jxb/erm203. [DOI] [PubMed] [Google Scholar]
- 12.Dong, W., Roy, P. & Isler, V. Semantic mapping for orchard environments by merging two-sides reconstructions of tree rows. arXiv:1809.00075 [cs] (2018).
- 13.Murchie EH, Lawson T. Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications. J. Exp. Bot. 2013;64:3983–3998. doi: 10.1093/jxb/ert208. [DOI] [PubMed] [Google Scholar]
- 14.Baker NR. Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 2008;59:89–113. doi: 10.1146/annurev.arplant.59.032607.092759. [DOI] [PubMed] [Google Scholar]
- 15.Poormohammad Kiani S, Maury P, Sarrafi A, Grieu P. QTL analysis of chlorophyll fluorescence parameters in sunflower (Helianthus annuus L.) under well-watered and water-stressed conditions. Plant Sci. 2008;175:565–573. doi: 10.1016/j.plantsci.2008.06.002. [DOI] [Google Scholar]
- 16.Flexas J, Bota J, Escalona JM, Sampol B, Medrano H. Effects of drought on photosynthesis in grapevines under field conditions: an evaluation of stomatal and mesophyll limitations. Funct. Plant Biol. 2002;29:461–471. doi: 10.1071/PP01119. [DOI] [PubMed] [Google Scholar]
- 17.Losciale P, et al. A multivariate approach for assessing leaf photo-assimilation performance using the IPL index. Physiol. Plant. 2015;154:609–620. doi: 10.1111/ppl.12328. [DOI] [PubMed] [Google Scholar]
- 18.Araus JL, Kefauver SC, Zaman-Allah M, Olsen MS, Cairns JE. Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 2018;23:451–466. doi: 10.1016/j.tplants.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebetzke GJ, et al. Genomic regions for canopy temperature and their genetic association with stomatal conductance and grain yield in wheat. Funct. Plant Biol. 2013;40:14–33. doi: 10.1071/FP12184. [DOI] [PubMed] [Google Scholar]
- 20.Condorelli, G. E. et al. Comparative aerial and ground based high throughput phenotyping for the genetic dissection of NDVI as a proxy for drought adaptive traits in durum wheat. Front. Plant Sci. 9, 893 (2018). [DOI] [PMC free article] [PubMed]
- 21.Virlet N, et al. Stress indicators based on airborne thermal imagery for field phenotyping a heterogeneous tree population for response to water constraints. J. Exp. Bot. 2014;65:5429–5442. doi: 10.1093/jxb/eru309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Virlet N, Costes E, Martinez S, Kelner JJ, Regnard JL. Multispectral airborne imagery in the field reveals genetic determinisms of morphological and transpiration traits of an apple tree hybrid population in response to water deficit. J. Exp. Bot. 2015;66:5453–5465. doi: 10.1093/jxb/erv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouse JW. Monitoring the vernal advancement and retrogradation (green wave effect) of natural vegetation. Third ERTS Symp. 1974;1:309–317. [Google Scholar]
- 24.Gitelson AA, Kaufman YJ, Merzlyak MN. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996;58:289–298. doi: 10.1016/S0034-4257(96)00072-7. [DOI] [Google Scholar]
- 25.Haboudane D, Miller JR, Pattey E, Zarco-Tejada PJ, Strachan IB. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004;90:337–352. doi: 10.1016/j.rse.2003.12.013. [DOI] [Google Scholar]
- 26.Gamon JA, Peñuelas J, Field CB. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992;41:35–44. doi: 10.1016/0034-4257(92)90059-S. [DOI] [Google Scholar]
- 27.Lassois L, et al. Genetic diversity, population structure, parentage analysis, and construction of core collections in the french apple germplasm based on SSR Markers. Plant Mol. Biol. Report. 2016;34:827–844. doi: 10.1007/s11105-015-0966-7. [DOI] [Google Scholar]
- 28.Gómez-Candón D, Virlet N, Labbé S, Jolivot A, Regnard JL. Field phenotyping of water stress at tree scale by UAV-sensed imagery: new insights for thermal acquisition and calibration. Precis. Agric. 2016;17:786–800. doi: 10.1007/s11119-016-9449-6. [DOI] [Google Scholar]
- 29.Jones HG. Use of thermography for quantitative studies of spatial and temporal variation of stomatal conductance over leaf surfaces. Plant Cell Environ. 1999;22:1043–1055. doi: 10.1046/j.1365-3040.1999.00468.x. [DOI] [Google Scholar]
- 30.Moran MS, Clarke TR, Inoue Y, Vidal A. Estimating crop water deficit using the relation between surface-air temperature and spectral vegetation index. Remote Sens. Environ. 1994;49:246–263. doi: 10.1016/0034-4257(94)90020-5. [DOI] [Google Scholar]
- 31.Pradal C, Boudon F, Nouguier C, Chopard J, Godin C. PlantGL: A Python-based geometric library for 3D plant modelling at different scales. Graph. Models. 2009;71:1–21. doi: 10.1016/j.gmod.2008.10.001. [DOI] [Google Scholar]
- 32.Pradal C, Dufour-Kowalski S, Boudon F, Fournier C, Godin C. OpenAlea: a visual programming and component-based software platform for plant modelling. Funct. Plant Biol. 2008;35:751–760. doi: 10.1071/FP08084. [DOI] [PubMed] [Google Scholar]
- 33.R Development Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2013).
- 34.Sinoquet H, Roux XL, Adam B, Ameglio T, Daudet FA. RATP: a model for simulating the spatial distribution of radiation absorption, transpiration and photosynthesis within canopies: application to an isolated tree crown. Plant, Cell &. Environment. 2001;24:395–406. [Google Scholar]
- 35.Strong D, Azarenko AN. Relationship between trunk cross-sectional area, harvest index, total tree dry weight and yield components of ‘Starkspur Supreme Delicious’ apple trees. J. Am. Pomol. Soc. 2000;54:22–27. [Google Scholar]
- 36.Da Silva D, Han L, Costes E. Light interception efficiency of apple trees: A multiscale computational study based on MAppleT. Ecol. Model. 2014;290:45–53. doi: 10.1016/j.ecolmodel.2013.12.001. [DOI] [Google Scholar]
- 37.Reynolds, D. et al. What is cost-efficient phenotyping? Optimizing costs for different scenarios. Plant Sci. (2018). 10.1016/j.plantsci.2018.06.015 [DOI] [PubMed]
- 38.Inostroza L, et al. Using aerial images and canopy spectral reflectance for high-throughput phenotyping of white clover. Crop Sci. 2016;56:2629–2637. doi: 10.2135/cropsci2016.03.0156. [DOI] [Google Scholar]
- 39.Ludovisi, R. et al. UAV-based thermal imaging for high-throughput field phenotyping of black poplar response to drought. Front. Plant Sci. 8, 1681 (2017). [DOI] [PMC free article] [PubMed]
- 40.Zufferey V, Cochard H, Ameglio T, Spring JL, Viret O. Diurnal cycles of embolism formation and repair in petioles of grapevine (Vitis vinifera cv. Chasselas) J. Exp. Bot. 2011;62:3885–3894. doi: 10.1093/jxb/err081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez-Dugo V, et al. Almond tree canopy temperature reveals intra-crown variability that is water stress-dependent. Agric. For. Meteorol. 2012;154–155:156–165. doi: 10.1016/j.agrformet.2011.11.004. [DOI] [Google Scholar]
- 42.Gonzalez-Dugo V, et al. Using high resolution UAV thermal imagery to assess the variability in the water status of five fruit tree species within a commercial orchard. Precis. Agric. 2013;14:660–678. doi: 10.1007/s11119-013-9322-9. [DOI] [Google Scholar]
- 43.Ballester C, et al. Evaluating the performance of xanthophyll, chlorophyll and structure-sensitive spectral indices to detect water stress in five fruit tree species. Precis. Agric. 2018;19:178–193. doi: 10.1007/s11119-017-9512-y. [DOI] [Google Scholar]
- 44.Körner C. Paradigm shift in plant growth control. Curr. Opin. Plant Biol. 2015;25:107–114. doi: 10.1016/j.pbi.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Poirier-Pocovi M, Lothier J, Buck-Sorlin G. Modelling temporal variation of parameters used in two photosynthesis models: influence of fruit load and girdling on leaf photosynthesis in fruit-bearing branches of apple. Ann. Bot. 2018;121:821–832. doi: 10.1093/aob/mcx139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bianco L, et al. Development and validation of the Axiom®Apple480K SNP genotyping array. Plant J. 2016;86:62–74. doi: 10.1111/tpj.13145. [DOI] [PubMed] [Google Scholar]
- 47.Urrestarazu, J. et al. Genome-wide association mapping of flowering and ripening periods in apple. Front. Plant Sci. 8, 1923 (2017). [DOI] [PMC free article] [PubMed]
- 48.Xu Y, Li P, Yang Z, Xu C. Genetic mapping of quantitative trait loci in crops. The Crop J. 2017;5:175–184. doi: 10.1016/j.cj.2016.06.003. [DOI] [Google Scholar]
- 49.Cubero S, Aleixos N, Moltó E, Gómez-Sanchis J, Blasco J. Advances in machine vision applications for automatic inspection and quality evaluation of fruits and vegetables. Food Bioprocess Technol. 2011;4:487–504. doi: 10.1007/s11947-010-0411-8. [DOI] [Google Scholar]
- 50.Sa I, et al. DeepFruits: A fruit detection system using deep neural networks. Sensors. 2016;16:1222. doi: 10.3390/s16081222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.