Abstract
Background
To identify a radiomics signature to predict local recurrence in patients with non-metastatic T4 nasopharyngeal carcinoma (NPC).
Methods
A total of 737 patients from Sun Yat-sen University Cancer Center (training cohort: n = 360; internal validation cohort: n = 120) and Wuzhou Red Cross Hospital (external validation cohort: n = 257) underwent feature extraction from the largest axial area of the tumor on pretreatment magnetic resonance imaging scans. Feature selection was based on the prognostic performance and feature stability in the training cohort. Radscores were generated using the Cox proportional hazards regression model with the selected features in the training cohort and then validated in the internal and external validation cohorts. We also constructed a nomogram for predicting local recurrence-free survival (LRFS).
Findings
Eleven features were selected to construct the Radscore, which was significantly associated with LRFS. For the training, internal validation, and external validation cohorts, the Radscore (C-index: 0.741 vs. 0.753 vs. 0.730) outperformed clinical prognostic variables (C-index for primary gross tumor volume: 0.665 vs. 0.672 vs. 0.577; C-index for age: 0.571 vs. 0.629 vs. 0.605) in predicting LRFS. The generated radiomics nomogram, which integrated the Radscore and clinical variables, exhibited a satisfactory prediction performance (C-index: 0.810 vs. 0.807 vs. 0.753). The nomogram-defined high-risk group had a shorter LRFS than did the low-risk group (5-year LRFS: 73.6% vs. 95.3%, P < .001; 79.6% vs 95.8%, P = .006; 85.7% vs 96.7%, P = .005).
Interpretation
The Radscore can reliably predict LRFS in patients with non-metastatic T4 NPC, which might guide individual treatment decisions.
Fund
This study was funded by the Health & Medical Collaborative Innovation Project of Guangzhou City, China.
Keywords: Nasopharyngeal carcinoma, T4 disease, Radiomics, Magnetic resonance imaging, Local recurrence
Research in context.
Evidence before this study
Approximately 20% of patients with non-metastatic stage T4 nasopharyngeal carcinoma (NPC) show local recurrence 10 years after intensity-modulated radiotherapy (IMRT) with or without chemotherapy. The accuracy of the current staging system and prognostic biomarkers is insufficient to identify patients at a high risk for recurrence. By extracting high throughput of quantitative imaging features from medical imaging, radiomics has shown potential for noninvasive characterization of intra-tumor heterogeneity and has been applied for prognostic evaluations in various types of cancer. However, no previous study has focused on predicting local recurrence in non-metastatic stage T4 NPC.
Added value of this study
In this multicenter large-scale study, we developed and validated a radiomics signature (named Radscore) built with 11 features profiled from pretreatment MRI as a noninvasive method to reliably predict local recurrence in patients with non-metastatic T4 NPC. The Radscore showed significantly better prognostic performance than that shown by other clinical variables in predicting local recurrence-free survival (LRFS). A radiomics nomogram that integrated Radscore and clinical variables showed a satisfactory prediction performance. The Radiomics nomogram-defined high-risk group had a shorter LRFS than did the low-risk group.
Implications of all the available evidence
The findings outline how radiomics-based approaches can be used to accurately determine the risk of local recurrence before treatment in non-metastatic T4 NPC. Our results may facilitate patient counselling and guide the treatment decision-making process in individual cases.
Alt-text: Unlabelled Box
1. Introduction
Nasopharyngeal carcinoma (NPC) is the most common head and neck malignancy in Southeast Asia and Southern China [1]. Radiotherapy is the mainstay treatment modality for non-disseminated NPC. With the application of precise imaging based on magnetic resonance imaging (MRI) technology and the advent of intensity-modulated radiotherapy (IMRT), the local control rate for NPC has improved significantly [2,3]. Patients with locally early-stage NPC can now achieve satisfactory 10-year local relapse-free survival (LRFS) rates (91.4%–94.2% for T1-T3 disease). However, the LRFS rate for locally advanced NPC remains poor (79.3% for T4 disease) [3]. Salvage treatments for recurrent NPC are challenging due to poor disease control and fatal late complications such as mucosa necrosis and massive hemorrhage [[4], [5], [6]]. Therefore, pretreatment prediction of a high risk of local recurrence is crucial for the development of individualized treatment strategies and reduction of the risk of local recurrence in T4 NPC.
Currently, risk assessment in local recurrence of NPC is primarily based on the clinical tumor-node-metastasis (TNM) staging. However, Chen et al. demonstrated that the TNM staging system had only 61% accuracy in predicting local recurrence [7]. This was mainly because the TNM staging system only describes the severity of the disease by evaluating the anatomical range of the existing tumors, but it is impossible to evaluate the intrinsic biological heterogeneity of tumors. In recent years, the promising field of “radiomics,” which aims to convert digitally encrypted medical images into mineable high-dimensional imaging features via automatic high-throughput quantitative image analyses [8,9], has emerged as a powerful new prognostic marker [[10], [11], [12], [13], [14], [15], [16], [17], [18]]. The central hypothesis of radiomics is that medical imaging features can capture crucial information regarding the intrinsic biological and physiological characteristics of tumors, making it possible to characterize intra-tumor heterogeneity [8,19,20]. Radiomics has been demonstrated to show profound significance in clinical practice such as cancer detection, staging, and prediction of prognosis in various types of cancer [[10], [11], [12], [13], [14], [15], [16],[21], [22], [23], [24]]. For NPCs, radiomics signatures can significantly predict responses to induction chemotherapy [22] and treatment failures in advanced NPC [[14], [15], [16]]. However, whether radiomics signatures could assist in identifying patients with a high risk of local NPC recurrence has not been reported.
Pretreatment MRI is routinely used to obtain morphological information with visual interpretation and tumor staging for NPCs, since MRI can provide superior soft-tissue resolution compared to that achieved with computed tomography (CT) [25]. The medical application potential of MRI has been recently explored using radiomics. However, it remains unclear whether the features selected from MRI of patients at one institution are still powerful prognostic markers for patients at other institutions because of the variability in MRI acquisition across MRI scanners from different institutions. The use of the ComBat method has been shown to be effective in correcting difference between scanners and may facilitate multicenter radiomic studies [[26], [27], [28]]. Thus, the current study sought to determine the association between pretreatment MRI-based radiomics signatures and local recurrence in non-metastatic T4 NPC, and then validate its effectiveness in patients from the same institution and an external institution. In addition, a nomogram was built by incorporating the radiomics signature and other clinical variables to accurately screen out patients at a high risk for local recurrence in non-metastatic T4 NPC.
2. Materials and methods
2.1. Patient selection
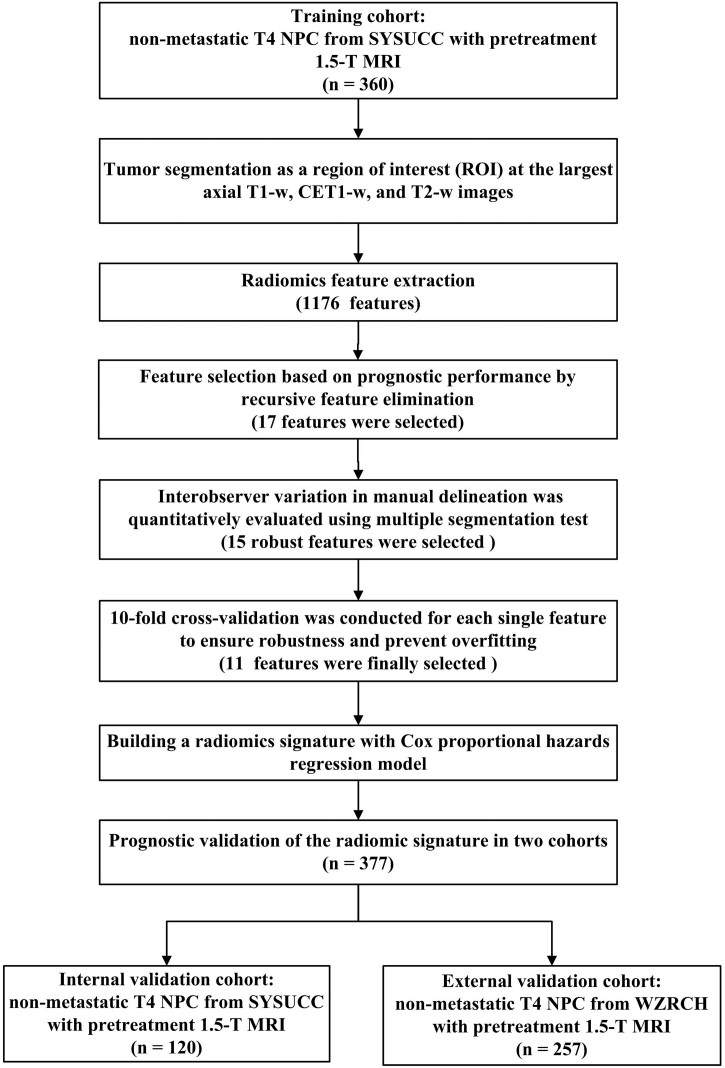
The current research was approved by the Research Ethics Committee of Sun Yat-sen University Cancer Center (SYSUCC) and Wuzhou Red Cross Hospital (WZRCH). Since this study was based on an analysis of routine MRI examination and clinical data, the requirement for individual informed consent was waived by the research ethics committees of SYSUCC (Approve number: YB2018-51) and WZRCH (Approve number: 2018-9). By using a prospective NPC-specific database from the well-established big-data intelligence platform at SYSUCC, a total of 2117 consecutive newly diagnosed patients with histologically proven T4N0-3M0 NPC (staged according to 8th American Joint Committee on Cancer /Union for International Cancer Control TNM staging system) treated with radical IMRT with/without chemotherapy between April 2009 and December 2015 were reviewed. In addition, a total of 1250 consecutive newly diagnosed patients with histologically proven T4N0-3M0 NPC treated with radical IMRT with/without chemotherapy between February 2012 and November 2014 were reviewed at WZRCH.
Patients treated at SYSUCC and WZRCH were included in this study if they met the following criteria: (1) availability of pretreatment 1.5 telsa (1.5-T) MRI examination data within 3 weeks before treatment; (2) availability of clinical and treatment data; and (3) no history of malignant tumors or severe heart, lung, liver, and kidney diseases. Based on these criteria, a total of 480 patients at SYSUCC and 257 patients from WZRCH were included in the study. Patients from SYSUCC were randomly assigned to training (n = 360) and internal validation (n = 120) cohorts at a 3:1 ratio via computer software-generated random numbers. The generated random numbers were fixed for each run. Patients from WZRCH were defined as the external validation cohort (n = 257).
During the study period, the guidelines of SYSUCC and WZRCH recommended concurrent chemoradiotherapy (CCRT) with or without induction chemotherapy (IC) /adjuvant chemotherapy (AC) for stage IVA-B disease. All patients underwent radical IMRT, which was administered 5 days/week. The prescribed doses were 66–72 Gy/28–33 fractions to the planning target volume (PTV) of the primary gross tumor volume (GTVp), 64–70 Gy/28–33 fractions to the involved nodal gross tumor volume PTV (GTVnd), 60–63 Gy/28–33 fractions to the PTV of high-risk clinical target volume, and 54–56 Gy/28–33 fractions to the PTV of low-risk clinical target volumes. Overall, 95.5% of patients received platinum-based chemotherapy. Concurrent chemotherapy consisted of cisplatin administered weekly or on weeks 1, 4, and 7 of radiotherapy, beginning the first day of IMRT. Induction chemotherapy or adjuvant chemotherapy consisted of cisplatin with docetaxel, cisplatin with 5-fluorouracil, or cisplatin with 5-fluorouracil and docetaxel every 3 weeks for two to four cycles. Reasons for not receiving chemotherapy included age, organ dysfunction suggestive of intolerance to treatment, and an individual patient's refusal.
The duration of patient follow-up was measured from therapy initiation to the day of last examination or death. After treatment, the patients returned for follow-up examinations every 3–6 months during the first 3 years and every 6–12 months thereafter until death. Patients were followed-up by telephone when their recent attendance was not recorded in their medical records. Local recurrence for NPC was defined as complete response followed by recurrence at the primary site more than three months after treatment completion. Local recurrence was confirmed by biopsy and/or MRI of the nasopharynx; in addition, positron emission tomography/computed tomography imaging was used when necessary. The end-point in the current study was LRFS, which was calculated from the first day of therapy to the date of local recurrence or death.
2.2. MRI scan acquisition and tumor segmentation
Each subject was scanned with 1.5-T MRI (Signa EXCITE, General Electric Healthcare, Chalfont St. Giles, United Kingdom; Signa HDx, General Electric Healthcare, Chalfont St. Giles, United Kingdom; SIEMENS Espree, Siemens Healthcare, Erlangen, Germany) in SYSUCC and 1.5-T MRI in WZRCH (SIEMENS Novus15, Siemens Healthcare, Erlangen, Germany) to examine the area from the suprasellar cistern to the inferior margin of the sternal end of the clavicle by using a head-and-neck combined coil within 3 weeks before treatment. Before administration of the contrast material, T1-weighted (T1-w) images and T2-weighted (T2-w) images were obtained. Following injection of contrast material, contrast-enhanced T1-weighted (CET1-w) images.
MR imaging parameters at SYSUCC are as follows: section thickness, 5–6.0 mm; intersection gap, 0.5–1.6 mm; repetition time (TR) for axial T1-w images, 500.0, ranging from: 400.0 to 800.0 msec; echo time (TE) for T1-w images, 9.2, ranging from 7.1 to 15.2 msec; TR for T2-w images, 3140.0, ranging from 2259.0 to 8002.0 msec; TE for T2-w images, 84.5, ranging from 80.0 to 121.1 msec; TR for CET1-w images, 520.0, ranging from 300.0 to 700.0 msec; TE for CET1-w images, 9.8, ranging from 7.4 to 16.00 msec; matrix, 256 × 256 to 624 × 640; field of views (FOV), 200 × 200 to 280 × 280 mm; and in-plane resolution: 0.35 × 0.35 to 0.78 × 0.78 mm. MR imaging parameters at WZRCH are as follows: section thickness, 5–5.5 mm mm; intersection gap, 1.5–1.65 mm; TR for axial T1-w images, 500.0, ranging from: 400.0 to 500.0 msec; TE for T1-w images, 8.0, ranging from 8.2 to 11.0 msec; TR for T2-w images, 4000, ranging from 4000.0 to 5860.0 msec; TE for T2-w images: 99.0, ranging from 98.0 to 103.0 msec; TR for CET1-w images, 668.0, ranging from 500.0 to 1060.0 msec; TE for CET1-w images, 8.0, rangingfrom 8.0 to 11.00 msec; matrix, 256 × 192 to 512 × 512; FOV, 200 × 200 to 280 × 280 mm; and in-plane resolution, 0.37 × 0.37 to 0.97 × 0.97 mm.
For tumor segmentation, T1-w, CET1-w, and T2-w images in the Digital Imaging and Communications in Medicine (DICOM) format for each patient were retrieved from the picture archiving and communication system (PACS; GE healthcare centricity RIS CE Carestream, Ontario, Canada). A region of interest (ROI) was manually delineated around the tumor outline for the largest axial area on two-dimensional (2D) T1-w, CET1-w, and T2-w images. The process of segmentation was completed by an experienced radiologist with >15 years of work experience using the RadiAnt software (open-source software; available at http://www.radiantviewer.com). The accuracy of segmentation was then checked by a radiologist with 20 years of work experience.
2.3. Features extraction methodology
After tumor segmentation, the T1-w image, T2-w image, CET1-w and T1-w subtraction image, and the segmented tumor area were transferred to the imaging biomarker explorer software for feature extraction (http://bit.ly/IBEX_MDAnderson). Most feature extraction methods conform to the Imaging Biomarker Standardization Initiative (IBSI) standard [29]. A total of 1176 features were extracted from ROI. Among those, 392 features from T2-w images, 392 features from T1-w images and remaining 392 features from subtraction images (generated by CE-T1w image minus T1-w image and reflecting the level of tumor enhancement). All features were calculated using Matlab R2016a (MathWorks, Natick, MA, USA), detailed definition of features are described in Supplementary Methods. These 1176 features were divided into three groups as follows:
2.3.1. Shape features
Shape features describe the shapes of segmented ROI areas, including 11 features extracted from T2-w images, T1-w images and subtraction images, respectively (totaling 33 features) as follow: number of pixel, shape perimeter, surface density, sphericity, spherical disproportion, roundness, orientation, number of ROI, convex hull, shape convex and shape compactness.
2.3.2. Intensity features
Intensity features reflect directly to the distribution of gray-levels. We calculated the following typical features from segmented ROI areas: energy, entropy, maximum, mean, median, minimum, standard error, uniformity, quartile range and kurtosis, and we also exacted some features from intensity histogram and neighbor intensity difference matrix. Finally, 121 features were extracted from each MRI series (totaling 363 features). Intensity features were divided into four groups as follows:
-
(1)
Intensity Direct features: The first-order statistic of gray levels, including Energy, Global Entropy, Global Max, Global Mean, Global Median, Global Min, Global Standard Deviation, Global Uniformity, Inter Quartile Range, Kurtosis, Local Entropy Max, Local Entropy Mean, Local Entropy Median, Local Entropy Min, Local Entropy Standard Deviation, Local Range Max, Local Range Mean, Local Range Median, Local Range Min, Local Range Standard Deviation, Local Standard Deviation Max, Local Standard Deviation Mean, Local Standard Deviation Median, Local Standard Deviation Min, Local Standard Deviation Standard Deviation, Mean Absolute Deviation, Median Absolute Deviation, Percentile, Quantile, Range, Root Mean Aquare, Skewness and Variance. ‘Global’ means the whole interesting area, and ‘Local’ means the calculation is on each pixels' certain neighborhood, and then compute the statistic among all neighborhoods.
-
(2)
Intensity Histogram features: These features are calculated based on the gray level histogram from image inside the ROI, including Interquartile Range, Kurtosis, Mean Absolute Deviation, Median Absolute Deviation, Percentile, Percentile Area, Quantile, Range and Skewness of the histogram.
-
(3)
Intensity Histogram Gauss Fit features: These features are calculated based on the method that fitting the gray level histogram with Gaussian curves, including Amplitude, Area, Mean, Standard Deviation and the number of Gaussian curves to approximate the histogram.
-
(4)
Neighbor Intensity Difference features: These features compute the intensity difference matrix of each pixel in the interesting area.
2.3.3. Textural features
Textural features are used to quantify the spatial distribution of pixel intensities. There are mainly two types of textural features: gray level co-occurrence matrices (GLCM) and gray level run-length matrices (GLRLM).
-
(1)
GLCM: GLCMis defined over a grayscale image to calculate the co-occurrence between two gray levels from certain angle and length. In our study, we chose directions from (0°, 45°, 90°, 135°) and distance from (2 pixel, 4pixel, 8 pixel) to get different matrices, then each matrix's autocorrelation, cluster prominence, cluster shade, cluster tendency, contrast, correlation, difference entropy, dissimilarity, energy, entropy, homogeneity, information measures of correlation, inverse difference moment normalized, inverse variance, maximum probability, sum average, sum entropy, sum variance and variance were calculated. Finally we obtained 216 GLCM features for each MRI series (totaling 648 features).
-
(2)
GLRLM: As for GLRLM, it represents the number of runs with pixels of certain gray level and run length. Choosing directions from (0°, 45°, 90°, 135°) and calculating properties including gray level nonuniformity, high gray-level run emphasis, long run emphasis, long run high gray-level emphasis, long run low gray-level emphasis, low gray-level run emphasis, run length nonuniformity, run percentage, short run emphasis, short run high gray-level emphasis, short run low gray-level emphasis, we got 44 features of GLRLM (totaling 132 features).
2.4. MRI normalization
Considering that our MRI images were obtained using four different scanners, which might have caused inconsistency in acquisition and reconstruction parameters, a harmonization method named ComBat [[26], [27], [28],30,31] was used to correct the scanner effect based on the observed feature values. The harmonization method of ComBat was used to correct the scanner effect based on the observed feature values. We divided our data into four batches according to the different types of MRI scanners, one batch obtained using Signa EXCITE at SYSUCC (Signa CV/i; General Electric Healthcare, Chalfont St. Giles, United Kingdom), one batch obtained using Signa HDx at SYSUCC (Signa CV/i; General Electric Healthcare, Chalfont St. Giles, United Kingdom), one batch obtained using SIEMENS Espree at SYSUCC (Siemens Healthcare, Erlangen, Germany), and one batch obtained using SIEMENS Novus15 at WZRCH (Siemens Healthcare, Erlangen, Germany). The harmonization methods have been described in detail in Supplementary Methods.
2.5. Methodology of feature selection
Feature selection was conducted based on prognostic performance and stability of features in the training cohort (n = 360). The workflow for feature selection is detailed in Supplementary Fig. 1 a. The ratio of patients who did not experience local recurrence (n = 324) to those who experienced local recurrence (n = 36) was 9:1. Considering the imbalance of data, random undersampling was used to balance class distribution by randomly eliminating majority class examples [32]. Undersampling is a popular technique for unbalanced datasets to reduce the skew in class distributions. After random undersampling, 135 patients were randomly selected from 324 non-recurrent cases. These non-recurrent 135 cases and 36 recurrent cases constituted a new training set (n = 171).
The recursive feature elimination (RFE) [33] algorithm was employed to select the most powerful features to predict local recurrence using Python software (version 3.6.0; https://www.python.org). The RFE method is also a kind of wrapper. It uses a base model to perform multiple rounds of training. After each round of training, the features with the smallest weight coefficients are eliminated, and the next round of training is based on the new feature set. In our paper, the classifier used in RFE was the regularized logistic regression imposing L2-norm as the penalty term [34]. The use of the penalty term reduces overfitting and makes the solution much more stable and fast.
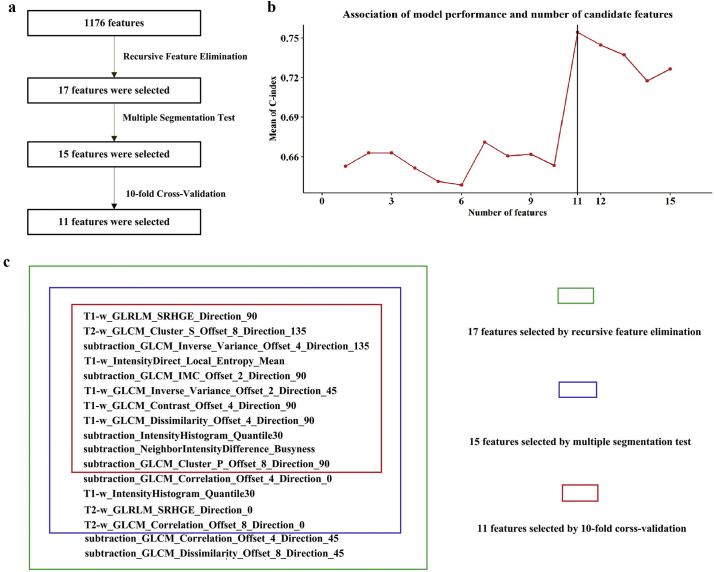
Among the 1176 extracted features, the remaining number of selected features was initially set to 17 [35], which is one-tenth the number of the new training set (n = 171). The specific steps implemented were as follows. First, the estimator of the logistic regression model was trained on 1176 features, and the importance of each feature was determined using the “coef_” attribute. Second, the feature with the smallest absolute value coefficient was pruned from the current set of features. Third, the new model was trained with the new 1175 features. The procedure was recursively repeated using the pruned set until the desired number of 17 features for selection was eventually reached. The detailed information of the 17 selected features is shown in our Supplementary Fig. 1 b.
After preliminary feature selection, interobserver variation in manual segmentation was quantitatively evaluated using the multiple segmentation test to measure the stability of the 17 selected features. Four radiation oncologists from our institute manually contoured the ROIs at the largest axial T1-w, CET1-w, and T2-w scans from 30 patients. For results about ROI delineation, the four radiation oncologists were blinded to each other. The Friedman test was applied in the multiple segmentation test to verify the stability of the 17 features by using R software (version 3.4.4; http://www.Rproject.org). The Friedman test (non-parametric repeated measurement test) is a non-parametric test for testing the difference between several related samples. The null hypothesis for the Friedman test is that there are no differences between the samples. If the calculated probability is low (P value less than the selected significance level, 0.05), the null-hypothesis is rejected and it can be concluded that at least 2 of the samples are significantly different from each other, namely the feature is unstable, otherwise the feature is stable. According to the results of Friedman test for four different segmentations, we selected the top 15 features with a P value >.5, which we believed were stable and not affected by variability in segmentation, and removed two unstable feature. The detail information of the remaining 15 features and their respective P values is shown in Supplementary Fig. S1b.
Supplementary Fig. S1.
Fig S1. Feature selection methodology. (a) Workflow for feature selection. (b) A total of 17 features were selected from 1176 features using recursive feature elimination methods. After preliminary feature selection, interobserver variation in manual segmentation was quantitatively evaluated using the multiple segmentation test to measure the stability of those 17 features. We selected the top 15 features with P values > 0.5 in the Friedman test that we believe were stable and not affected by variability in segmentation. To ensure robustness and prevent overfitting, we utilized a 10-fold cross-validation for features. On the basis of the P values of each variable after univariate Cox proportional hazards regression, 15 variables are sorted. (c) Association between model performance and the number of candidate features. The multivariate Cox model consisted of the top 11 features that achieved the highest performance; thus, the 11 features were finally selected.
Finally, to ensure robustness and prevent overfitting, we used a 10-fold cross-validation for features (R 3.4.4). According to the P value of each variable after the univariate Cox proportional hazards regression analysis, 15 variables were sorted. Then,the multivariate Cox model consisted of the top 11 features with the highest performance. As shown in Supplementary Fig. 1 c, when the number of variables increased to 11, the average value of the C index was the highest. Therefore, we choose the top 11 variables after sorting as the final model variables.
2.6. Building and validation of radiomics signature and radiomics nomogram
The radiomics score (Radscore, which was defined as the radiomics signature in the current research) was computed in the training cohort by determining a linear combination of the above finally selected features with their respective coefficients weighted by the Cox proportional hazards regression model. The correlations between the Radscore and LRFS were first assessed with the Cox proportional hazard regression model in the training cohort and then validated in the internal and external validation cohorts.
We further built a radiomics nomogram that integrated both the Radscore and other significant clinical risk factors for predicting local recurrence in the training cohort. By applying X-tile software (version 3.6.1; Yale University School of Medicine, New Haven, CT, USA), we identified the cut-off value in the training cohorts for the radiomics nomogram-defined score, after which this cutoff score was applied in the two validation cohorts. Based on the cutoff values, patients in the three cohorts were divided into high-risk and low-risk groups. The potential association of the radiomics nomogram-defined score and LRFS was first assessed in three training cohorts and then validated in the internal validation and external validation cohorts by using Kaplan-Meier survival analysis in the high-risk and low-risk groups.
2.7. Statistical analysis
Statistical analyses were conducted using R software (version 3.4.4) and SPSS version 23.0 (IBM Corp, Armonk, NY USA). LRFS was calculated using the Kaplan-Meier method and survival curves were compared using log-rank tests. Univariate and multivariate Cox proportional hazard regression was performed using SPSS to identify independent prognostic factors for LRFS. Radscore and 14 other candidate predictors as shown in Table 1 were included in univariate Cox proportional hazard regression analysis for constructing a nomogram. Age and GTVp were classified into two groups according to the cutoff value determined by X-tile. The other continuous clinical variables were translated into categorical variables on the basis of routine cutoff points in clinical application [36]. Clinical and characteristics and treatment methods between the groups were compared using the Chi-square test. Variables achieving significance at a level of P < .05 in univariate Cox proportional hazard regression were included in multivariate Cox proportional hazard regression for selection of independent prognostic factors. The relative hazard ratio (HR) and its 95% confidence interval (CI) were calculated from Cox regression analysis.
Table 1.
Clinical characteristics and treatment parameters of patients with T4 disease nasopharyngeal carcinoma in the training and validation cohorts.
| Characteristic | Training cohort from SYSUCC (n = 360) No. (%) | Internal validation cohort from SYSUCC (n = 120) No. (%) | External validation cohort from WZRCH (n = 257) No. (%) | P value |
|---|---|---|---|---|
| Age (years) | 0.307 | |||
| ≥62 | 41 (11.4) | 13 (10.8) | 39 (15.2) | |
| <62 | 319 (88.6) | 107 (89.2) | 218 (84.8) | |
| Gender | 0.999 | |||
| Male | 270 (75.0) | 90 (75.0) | 193 (75.1) | |
| Female | 90 (25.0) | 30 (25.0) | 64 (24.9) | |
| WHO pathology | <0.001 | |||
| Type I | 3 (0.8) | 2 (1.7) | 16 (6.2) | |
| Type II-III | 357 (99.2) | 118 (98.3) | 241 (93.8) | |
| Family history of NPC | <0.001 | |||
| No | 265 (73.6) | 81 (67.5) | 249 (96.9) | |
| Yes | 95 (26.4) | 39 (32.5) | 8 (3.1) | |
| Smoking | <0.001 | |||
| No | 212 (58.9) | 72 (60.0) | 208 (80.9) | |
| Yes | 148 (41.1) | 48 (40.0) | 49 (19.1) | |
| Drinking | 0.524 | |||
| No | 299 (83.1) | 102 (85.0) | 222 (86.4) | |
| Yes | 61 (16.9) | 18 (15.0) | 35 (13.6) | |
| Pretreatment HGB (g/L) | <0.001 | |||
| ≥120 | 333 (92.5) | 107 (89.2) | 200 (77.8) | |
| <120 | 27 (7.5) | 13 (10.8) | 57 (22.2) | |
| Pretreatment CRP (mg/L) | 0.012 | |||
| ≥8.2 | 67 (18.6) | 10 (8.3) | / | |
| <8.2 | 293 (81.4) | 110 (91.7) | / | |
| EBV-DNA (copies/mL) | <0.001 | |||
| ≥4000 | 218 (60.6) | 74 (61.7) | 45 (17.5) | |
| <4000 | 142 (39.4) | 46 (38.3) | 212 (82.5) | |
| LDH, U/L | ||||
| ≥245 | 34 (9.4) | 9 (7.5) | / | |
| <245 | 326 (90.6) | 111 (92.5) | / | |
| ALB, g/L | 0.164 | |||
| ≥35 | 352 (97.8) | 118 (98.3) | 245 (95.3) | |
| <35 | 8 (2.2) | 2 (1.7) | 12 (4.7) | |
| Cumulative dose to GTVp, Gy | 0.838 | |||
| ≥68 | 333 (92.5) | 109 (90.8) | 236 (91.8) | |
| <68 | 27 (7.5) | 11 (9.2) | 21 (8.2) | |
| GTVp, cm3 | <0.001 | |||
| ≥93.7 | 138 (38.3) | 37 (30.8) | 136 (52.9) | |
| <93.7 | 222 (61.7) | 83 (69.2) | 121 (47.1) | |
| Chemotherapy | 0.035 | |||
| No | 13 (3.6) | 2 (1.7) | 18 (7.0) | |
| Yes | 347 (96.4) | 118 (98.3) | 239 (93.0) |
Statistical comparisons between the training and two validation cohorts were computed using the Chi-square test. A P-value of 0.05 indicates a significant difference. Abbreviations: SYSUCC, Sun Yat-sen University Cancer Center; WZRCH, Wuzhou Red Cross Hospital; WHO, World Health Organization; LDH, lactate dehydrogenase; HGB, hemoglobin; ALB, albumin; CRP, C-reaction protein; EBV-DNA, Epstein-Barr Virus DNA; GTVp, primary gross tumor volume; /: missing data.
We built a radiomics nomogram with the coefficients weighted by the multivariate Cox proportional hazard regression model in the training cohort using R software. Calibration curves, which indicated the calibration ability of the nomogram, were assessed graphically by plotting the actual observed survival rates and the nomogram-predicted survival rates. The discrimination performance of nomograms was measured quantitatively by Harrell's concordance indices (C-index), which was measured using R software (version 3.4.3.) and the Hmisc package. We also developed a clinical nomogram using significant clinical factors without Radscore. We further assessed the prognostic performance of Radscore, clinical features, and nomograms by calculating the area under the receiver operator characteristic (ROC) curve (AUC). A two-sided P value was always computed, with statistical significance set at 0.05.
3. Results
3.1. Clinical characteristics
The workflow of the current study is shown in Fig. 1. The detailed clinical characteristics and treatment parameters of patients with T4 NPC in the training cohort and the two validation cohorts are shown in Table 1 and Supplementary Table S1. The median follow-up duration was 46.2 months (6.9–96.4) for the training cohort, 45.6 months (10.8–95.6) for the internal validation cohort, and 45.3 months (7.2–69.3) for the external validation cohort. In the last follow-up, 36 (10.0%) patients in the training cohort, 12 (10.0%) in the internal validation cohort, and 17 (6.6%) in the external validation cohort experienced local recurrence.
Fig. 1.
Study workflow.
NPC = nasopharyngeal carcinoma; SYSUCC = Sun Yat-sen University Cancer Center; MRI = magnetic resonance imaging; T1-w = T1-weighted; CET1-w = contrast-enhanced T1-weighted; T2-w = T2-weighted; WZRCH = Wuzhou Red Cross Hospital. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Radscore building and validation
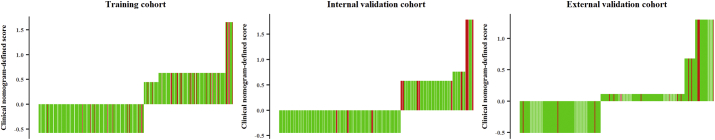
A total of 1176 features were extracted from the defined tumor, which was contoured on the MR tumor image in the training cohort: 392 features from T2-w images, 392 features from T1-w images, and the remaining 392 features from the CET1-w and T1-w subtraction images. We identified 17 features (3, 6, and 8 features selected from the T2-w, T1-w, and subtraction images, respectively) associated with LRFS using recursive feature elimination analysis. Among these 17 features, 15 features (3, 6, and 6 features selected from the T2-w, T1-w, and subtraction images, respectively) were shown to be robust in multiple segmentation tests. To ensure robustness and prevent overfitting, these 15 features were subjected to 10-fold cross-validation individually. Finally, 11 features (1, 5, and 5 features selected from T2-w, T1-w, and subtraction images, respectively) were selected (Supplementary Fig. S1). The detailed descriptions of these 11 features and the corresponding coefficients for the selected features in the formula for Radscore are shown in Supplementary Table S2. The calculation formulas for Radscore are listed in Supplementary Methods. The Radscores for each patient in the training cohort are presented in Fig. 2a–c. The Radscore yielded a C-index of 0.741 (95% CI: 0.635–0.848) for the training cohort. The good prognostic performance of the Radscore was further validated in the two independent validation cohorts: the Radscore yielded a C-index of 0.753 (95% CI: 0.618–0.887) for the internal validation cohort and 0.730 (95% CI: 0.587–0.873) for the external validation cohort (Table 2).
Fig. 2.
Radscore and radiomics nomogram-defined scores for each patient with non-metastatic T4 NPC.
Radscore in the training cohort, N = 360 (a); Radscore in the internal validation cohort, N = 120 (b); Radscore in the external validation cohort, N = 257(c); radiomics nomogram-defined score in the training cohort (d); radiomics nomogram-defined score in the internal validation cohort, N = 120 (e); radiomics nomogram-defined score in the external validation cohorts (f). Green bars represent the scores for patients who did not show local recurrence, while red bars represent the scores for those who showed local recurrence. NPC = nasopharyngeal carcinoma. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Performance of models.
| Models | C-index (95% CI) |
||
|---|---|---|---|
| Training cohort from SYSUCC (n = 360) | Internal validation cohort from SYSUCC (n = 120) | External validation cohort from WZRCH (n = 257) | |
| Radscore | 0.741 (0.635, 0.848) | 0.753 (0.618, 0.887) | 0.730 (0.587, 0.873) |
| Clinical nomogram | 0.696 (0.533 0.860) | 0.726 (0.528, 0.923) | 0.666 (0.429, 0.904) |
| Radiomics nomogram (Radiomics + clinical) | 0.810 (0.692,0.928) | 0.807 (0.700, 0.914) | 0.753 (0.654, 0.851) |
Abbreviations: SYSUCC, Sun Yat-sen University Cancer Center; WZRCH, Wuzhou Red Cross Hospital; C-index, Harrell's concordance indices; CI, confidence interval.
3.3. Univariate and multivariate analyses of the risk factors for LRFS
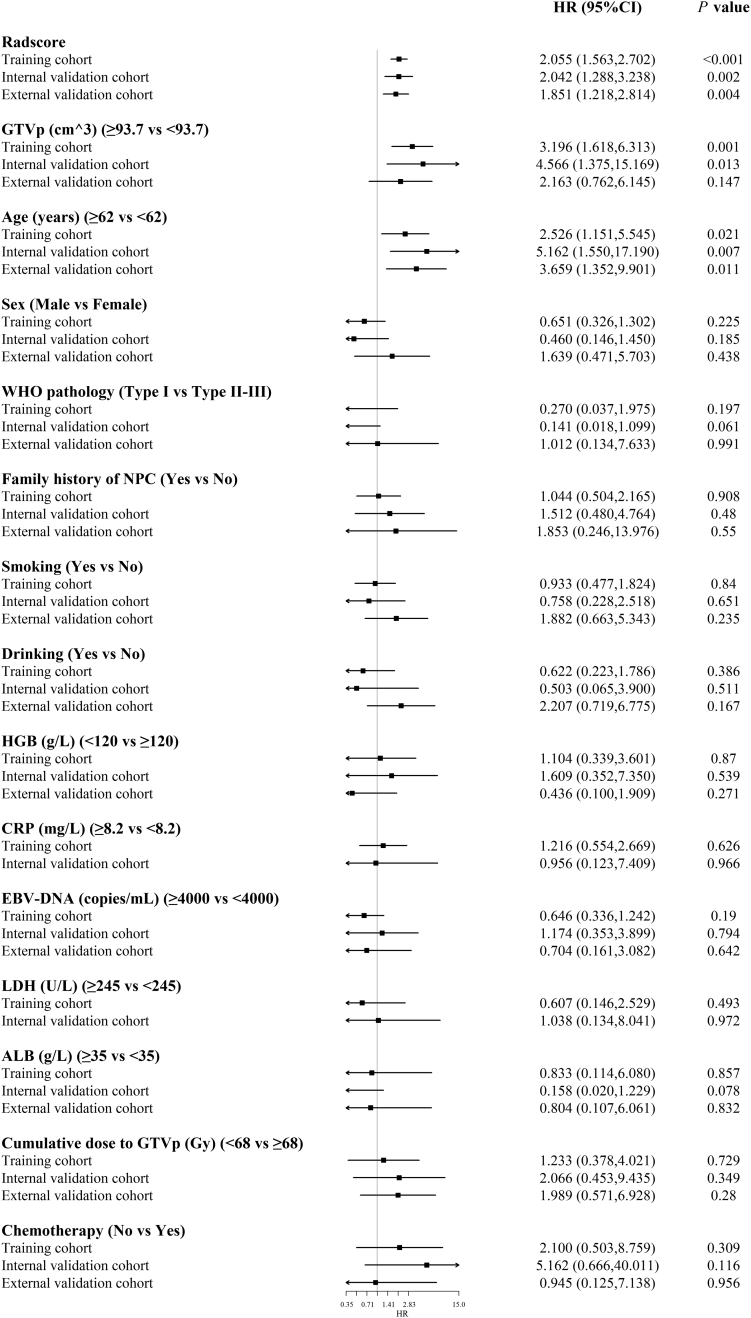
Univariate analysis results for LRFS in the three cohorts are shown in Supplementary Fig. S2, which showed a significant association between the Radscore and LRFS in all cohorts. The results of multivariate analysis indicated that for the training cohort, Radscore (HR = 2.050, 95% CI: 1.542–2.726, P < .001), GTVp (HR = 2.641, 95% CI: 1.330–5.244, P = .006), and age (HR =3.066, 95% CI: 1.385–5.244, P = .006) were independent risk factors; for the internal validation cohorts, Radscore (HR = 1.684, 95% CI: 1.068–2.653, P = .025) and GTVp (HR = 3.567, 95% CI: 1.062–11.976, P = .040) were independent risk factors; and for the external validation cohort, Radscore (HR = 1.736, 95% CI: 1.150–2.620, P = .009), and age (HR = 3.032, 95% CI: 1.104–8.321, P = .031) were independent risk factors (Supplementary Table S3).
Supplementary Fig. S2.
Fig S2. The forest plot showing univariate association of Radscore and clinicopathological characteristics in non-metastatic T4 NPC Radscore = Radiomics score; HGB = hemoglobin; CRP = C-reactive protein; EBV-DNA = Epstein-Barr virus DNA; LDH = lactate dehydrogenase; ALB = albumin; GTVp = primary gross tumor volume; HR = hazard ratio; CI = confidence interval; NPC = nasopharyngeal carcinoma; LRFS = local recurrence-free survival.
3.4. Radiomics nomogram building and validation
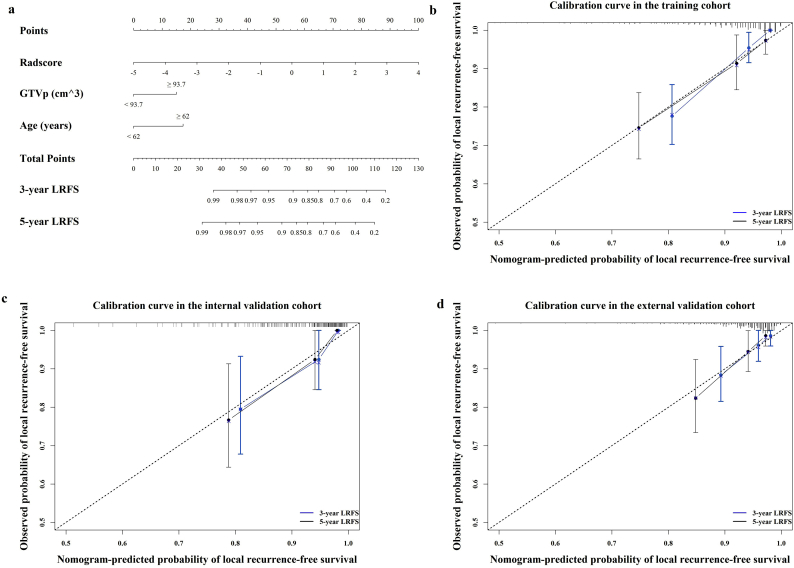
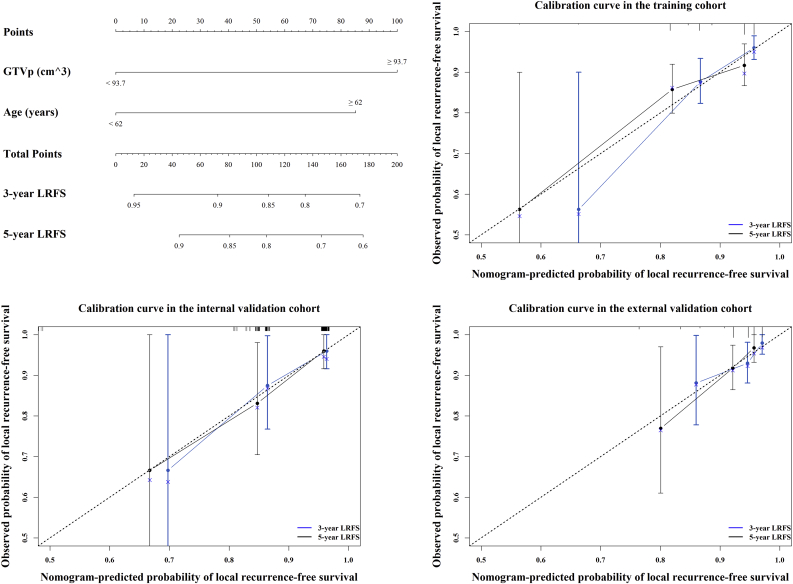
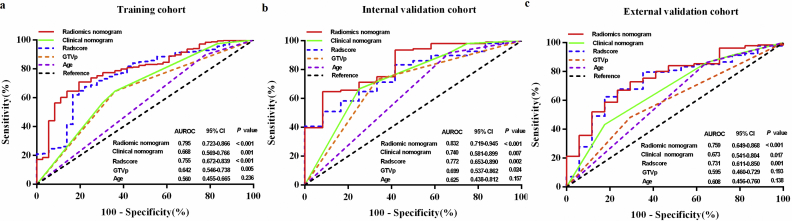
We further visualized the results of multivariate analysis of LRFS as a radiomics nomogram to predict the LRFS in the training cohort, as shown in Fig. 3a. The variables in the radiomics nomogram included Radscore, GTVp, and age. For the training, internal, and external validation cohorts, the C-index of the radiomics nomogram for LRFS prediction was 0.810 (95% CI, 0.692–0.928), 0.807 (95% CI, 0.700–0.914), and 0.753 (95% CI, 0.654–0.851), respectively (Table 2). Among single variables, the Radscore (C-index: 0.741, 95% CI: 0.635–0.848; 0.753, 95% CI: 0.618–0.887; 0.730, 95% CI: 0.587–0.873) outperformed GTVp (C-index: 0.665, 95% CI: 0.518–0.813; 0.672, 95% CI: 0.488–0.857; 0.577, 95% CI: 0.423–0.713) and age (C-index: 0.571, 95% CI: 0.427–0.714; 0.629, 95% CI: 0.488–0.857; 0.605, 95% CI: 0.455–0.755) in predicting LRFS (Supplementary Table S4). The calibration plot for the probability of LRFS presented excellent agreement between nomogram prediction and the actual observed LRFS in the training and two independent cohorts (Fig. 3b–d). The radiomics nomogram-defined scores for each patient in the three cohorts are presented in Fig. 2d–f.
Fig. 3.
Radiomics nomogram to predict the risk of local recurrence in patients with non-metastatic T4 NPC.
Radiomics nomogram to predict local recurrence-free survival (a); calibration curves of the radiomics nomogram to predict local recurrence-free survival probability at 3 and 5 years in the training cohort, N = 360 (b); the internal validation cohort, N = 120 (c); and the external validation cohort, N = 257 (d). Nomogram-predicted probability of local recurrence is plotted on the x-axis; actual observed probability is plotted on the y-axis. NPC = nasopharyngeal carcinoma.
To determine the complementarity of the prognostic performance of Radscore to other clinical risk factors, we developed another clinical nomogram based on age and GTVp but without Radscore in the training cohort (Supplementary Fig. S3). The C-index of the clinical nomogram for LRFS prediction was 0.696 (95% CI, 0.533–0.860) in the training cohort, 0.726 (95% CI, 0.528–0.923) for the internal validation cohort, and 0.666 (95% CI, 0.429–0.904) for the external validation cohort (Table 2). The radiomics nomogram showed obviously better predictive performance in comparison with the clinical nomogram. The clinical nomogram-defined scores for each patient in the three cohorts are presented in Supplementary Fig. S4.
Supplementary Fig. S3.
Fig S3. Clinical nomogram to predict the risk of local recurrence in non-metastatic T4 NPC Clinical nomogram to predict local recurrence-free survival (a); calibration curves of the clinical nomogram to predict local recurrence-free survival probability at 3 and 5 years in the training cohort (b), the internal validation cohort (c), and the internal validation cohort (d). Nomogram-predicted probability of local recurrence is plotted on the x-axis; actual observed probability is plotted on the y-axis. NPC = nasopharyngeal carcinoma.
Supplementary Fig. S4.
Fig S4. Clinical nomogram-defined scores for each patient with non-metastatic T4 NPC Training cohort, N = 360; (b) internal validation cohort, N = 120; (c) external validation cohort, N = 257. Green bars represent scores for patients who did not show local recurrence, while red bars represent scores for those who showed local recurrence. NPC = nasopharyngeal carcinoma.
To substantiate the prognostic value of the radiomics nomogram, ROC analysis was conducted to evaluate the sensitivity and specificity of Radscore (areas under the curve [AUCs]: 0.756, 0.772, and 0.731; sensitivity: 0.778, 0.666, and 0.824; specificity: 0.676, 0.676, and 0.625), GTVp (AUCs: 0.642, 0.699, and 0.595; sensitivity: 0.639, 0.667, and 0.706; specificity: 0.945, 0.731, and 0.484), age (AUCs: 0.560, 0.625, and 0.608; sensitivity: 0.222, 0.333, and 0.353; specificity: 0.898, 0.917, and 0.863), clinical nomogram (AUC: 0.668, 0.740, and 0.673; sensitivity: 0.645, 0.750, and 0.824; specificity: 0.639, 0.667, and 0.433), and radiomics nomogram (AUC: 0.794, 0.832, and 0.759; sensitivity: 0.806, 0.648, and 0.765; specificity: 0.710, 0.917, and 0.671) in the training, internal validation, and external validation cohorts. Among these, the radiomics nomogram showed the highest accuracy (Fig. 4). Additionally, the balanced accuracy, sensitivity, specificity, and AUC for the training cohort after random undersampling (Supplementary methods) were 0.749, 0.778, 0.741, and 0.807, respectively.
Fig. 4.
Receiver operating characteristic (ROC) curves comparing the predictive power of the radiomics nomogram, clinical nomogram, and individual factors of Radscore, GTVp, and age in predicting local recurrence in non-metastatic T4 NPC.
(a) Training cohort, N = 360; (b) internal validation cohort, N = 120; (c) external validation cohort, N = 257; GTVp = primary gross tumor volume; NPC = nasopharyngeal carcinoma.
3.5. Subgroup survival analysis stratified by the radiomics nomogram
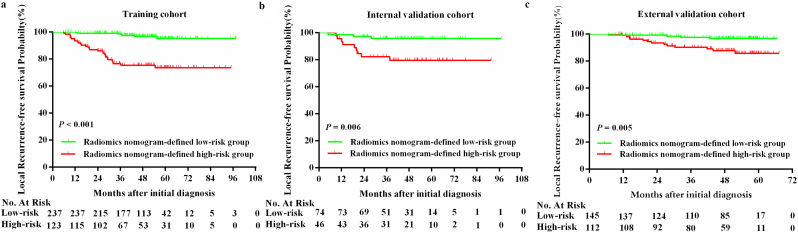
Using X-tile software, the optimal radiomics nomogram-defined score value of 3.0 (Supplementary Fig. S5) for LRFS prediction was determined as the cutoff value in the training cohort and the patients were assigned to high-risk and low-risk groups. Accordingly, 123 (34.2%), 46 (38.3%), and 112 (43.6%) patients were categorized into the high-risk group for the training, internal validation, and external validation cohorts, and 237 (65.8%), 74 (61.7%), and 145 (56.4%) patients were assigned to the low-risk group, respectively. The number of events is further listed in Supplementary Table S5. A subgroup survival analysis showed that patients in the radiomics nomogram-defined high-risk group showed a poor LRFS in the training cohort (3-year LRFS: 76.5% vs. 98.6%; 5-year LRFS: 73.6% vs. 95.3%; HR 8.921, 3.905–20.382, P < .001), and this result was verified in the internal validation (3-year LRFS: 82.2% vs. 95.8%; 5-year LRFS: 79.6% vs. 95.8%; HR 5.169, 1.399–19.100, P = .006) and external validation cohorts (3-year LRFS: 90.2% vs. 97.6%; 5-year LRFS: 85.7% vs. 96.7%; HR 4.385, 1.430–13.451, P = .005, Fig. 5, Supplementary Table S5).
Supplementary Fig. S5.
Fig S5. Optimum cutoff values for the radiomics nomogram-defined scores determined using the X-tile software The optimum cut-off point for the radiomics nomogram-defined score was calculated using the X-tile plot in the training cohort. The strength of the association between score and local recurrence-free survival was visualized by various colors in the plot. Red represents an inverse association between the Radscore and local recurrence-free survival, and green indicates a direct association.
Fig. 5.
Kaplan-Meier curves of local recurrence-free survival between high-risk and low-risk groups stratified by the radiomics nomogram in patients with non-metastatic T4 NPC.
Training cohort, N = 360 (a); internal validation cohort, N = 120 (b); external validation cohort, N = 257 (c). P values were calculated using the log-rank test, HRs were calculated using univariate Cox regression analysis. NPC, nasopharyngeal. The log-rank test was used to calculate P-values.
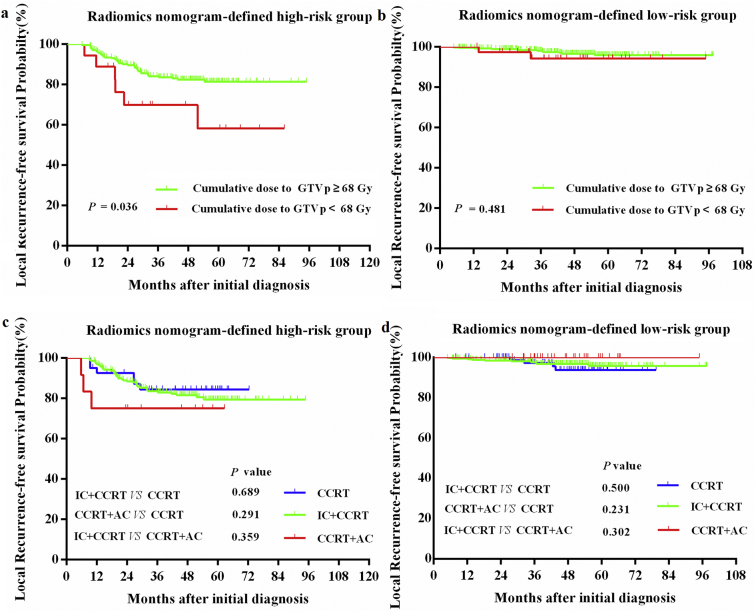
For the whole cohort, survival outcomes were comparable between the IC+ CCRT (n = 529), CCRT+AC (n = 47), and CCRT-alone groups (n = 128; 3-year LRFS: 91.9% vs 93.6% vs. 93.0%; 5-year LRFS: 89.1% vs. 93.6% vs. 90.6%; IC + CCRT vs. CCRT+AC: P = .598; IC + CCRT vs. CCRT: P = .684; CCRT vs. CCRT+AC: P = .828). We applied our radiomics nomogram to predict if patients could benefit from a specific therapy regimen. Within the high-risk group, patients receiving a cumulative GTVp dose of ≥68 Gy achieved significantly better LRFS than that achieved by those who received a cumulative GTVp dose of <68 Gy (3-year LRFS: 84.1% vs. 69.8%; 5-year LRFS: 81.5% vs. 58.2%, P = .036; Supplementary Fig. S6a). There was no difference in the LRFS between patients who received a cumulative GTVp dose of ≥68 Gy and those who received a cumulative GTVp dose of <68 Gy in the low-risk group (3-year LRFS: 98.1% vs. 94.2%; 5-year LRFS: 95.8% vs. 94.2%, P = .481; Supplementary Fig. S6b). Furthermore, both within the high-risk (3-year LRFS: 83.5% vs 75.0% vs. 84.4%; 5-year LRFS: 79.5% vs. 75.0% vs. 84.4%; IC + CCRT vs. CCRT+AC: P = .359; IC + CCRT vs. CCRT: P = .689; CCRT vs. CCRT+ AC: P = .291; Supplementary Fig. S6c) and low-risk groups (3-year LRFS: 97.6% vs 100.0% vs. 97.2%; 5-year LRFS: 95.8% vs. 100.0% vs. 93.7%; IC + CCRT vs. CCRT+AC: P = .302; IC + CCRT vs. CCRT: P = .500; CCRT vs. CCRT+AC: P = .231; Supplementary Fig. S6d), there was no difference in the LRFS between patients who received IC + CCRT, CCRT+AC, and CCRT alone.
Supplementary Fig. S6.
Fig S6. Kaplan-Meier curves of local recurrence-free survival for the radiomics nomogram-defined high-risk and low-risk groups stratified by treatment regimen in patients with non-metastatic T4 NPC (a) Kaplan-Meier curves of local recurrence-free survival in patients receiving a cumulative GTVp dose of Gy and a cumulative dose of <68 Gy between the high-risk group and (b) low-risk group; (c) Kaplan-Meier curves of local recurrence-free survival in patients who received IC+CCRT, CCRT+AC, and CCRT alone between the high-risk group and (d) low-risk group; P values were calculated using the log-rank test, HRs were calculated using univariate Cox regression analysis. CCRT = concurrent chemoradiotherapy; IC = induction chemotherapy; AC = adjuvant chemotherapy; NPC = nasopharyngeal carcinoma.
4. Discussion
In this multicenter study, we developed and validated the feasibility of applying an MR imaging-based radiomics signature named Radscore for non-invasive prediction of local recurrence in T4 disease NPC before making treatment-related decisions. The Radscore developed in our study showed good prediction performance, and its prognostic performance was significantly better than those of the other clinical variables in predicting LRFS. We further presented and validated a radiomics nomogram that integrated Radscore and clinical variables for prediction of local recurrence; the nomogram achieved a satisfactory discrimination performance (C-index in the training, internal, and external validation cohorts: 0.810 vs. 0.807 vs. 0.753.
The emerging use of IMRT for the treatment of NPC has narrowed the prognostic differences between T1, T2, and T3 NPC (10-year LRFS: 94.2%, 92.5%, and 91.4% for T1, T2, and T3 disease, respectively) [3]. However, achievement of a favorable prognosis remains a challenge for T4 NPC (10-year LRFS: 79.3% for T4 disease). Although previous studies have indicated that a variety of clinical covariates such as T stage, age, WHO histologic type, primary gross tumor GTVp, lactate dehydrogenase (LDH) levels, and molecular biomarkers such as microRNA and leukemia inhibitory factor (LIF) are associated with local recurrence in NPC [[36], [37], [38], [39], [40], [41], [42]], none of them is sufficiently accurate in routine practice to identify patients at a high risk of local recurrence. In the current study, we determined the Radscore, which was powerful in predicting the risk of local recurrence in T4 NPC (C-index values for the training, internal validation, and external validation cohorts: 0.741, 0.753, 0.730), and was significantly better than the other clinical risk factors. The strong predictive performance of Radscore may be attributable to the fact that unlike previously reported clinical covariates and molecular biomarkers, radiomics may be an effective approach to visualize and quantify intra-tumor heterogeneity [8,19,20]. Considering the presence of intra-tumor heterogeneity, which was acknowledged as a feature of malignancy, the surviving tumor cell sub-lines that are resistant to treatment may cause local recurrence or distant metastasis [[43], [44], [45]]. Radiomics has emerged as a non-invasive and cost-effective method to interpret such intra-tumor heterogeneity by high-throughput and objective mining of quantitative image features from medical images, thereby providing valuable information for prognostication in clinical settings.
Several previous studies have reported the association between radiomics and clinical outcomes in a range of cancer types [[10], [11], [12], [13], [14], [15], [16], [17], [18]]. Although radiomics signatures have been identified to evaluate progression-free survival in patients with stage III-IV NPC as well as to predict early responses to induction chemotherapy in patients with stage II-IV NPC [[14], [15], [16],24], the association between radiomics and local recurrence for NPC has not yet been investigated. In addition, in comparison with previous studies on radiomics and PFS in NPC, which used small sample sizes for training (70 to 88 cases) and validation (30 to 33 cases), our study used large-scale training (360 cases) and two independent validation cohorts (120 and 257 cases, respectively).
One issue associated with the use of radiomics in MR imaging is the variability between imaging scanners and imaging acquisition parameters across hospitals. We applied a ComBat harmonization method, previously described for radiomic studies, to correct the so-called batch effect [[26], [27], [28]]. The ComBat method was first and has since been widely used to adjust for batch effects in genomics, where a batch refer to the differences of a technical nature between datasets but does not reflect biological variation [30,31]. This is similar to the scanner effect in radiomics. Several previous studies reported that Combat methods makes it possible to remove the multicenter effect for radiomics features selected from CT/ positron emission tomography (PET) /PET and MR images [[26], [27], [28]]. Consistent with previous studies, the MR imaging-based radiomics signature developed in our study was successfully validated in an independent external cohort after multicentric harmonization of the features.
To establish the radiomics signature, data dimensionality reduction was performed on 1176 extracted features by recursive feature elimination, multiple segmentation test, and 10-fold cross-validation. Tumor segmentation is critical for radiomics since features are extracted from segmented areas. ROIs are segmented manually, and the resultant variability in segmentation may introduce a bias in the extraction of features. However, most previous radiomics studies did not take this issue into account. In the current study, multiple segmentation tests were performed by four radiation oncologists who were blinded to each other's assessments to select robust features that are not affected by variability in segmentation. Through this process, we identified the Radscore consisting of 11 features that achieved good performance in predicting local recurrence in the training cohort and validated the results in the internal and external validation cohorts. To provide a clinically applicable method for individual prediction of local recurrence, we further generated a radiomics nomogram that integrated both the Radscore and other significant clinical risk factors in the training cohort. The C-index of the radiomics nomogram was higher than that of the clinical nomogram (which only consisted of clinical risk factors) and the Radscore, indicating that the prognostic performance of the radiomics nomogram was the highest. The performance of the nomogram was further verified in two validation cohorts. This conclusion was also supported by the results of the ROC analysis. In the current study, the radiomics nomogram-defined high-risk patients benefited from a cumulative GTVp dose of ≥68 Gy while the low-risk patients did not. These findings provided a new insight into the future delivery of IMRT; high-risk patients should receive more intense treatment. Future studies are needed to validate our findings.
There were two limitations of the current research that still need to be addressed. First, a prospective study is still warranted to confirm the conclusions of the current study. Second, in this research, features were extracted from the largest axial area of tumor on a 2D single slice, whereas previous radiomics studies were mainly based on three-dimensional (3D) image analysis. Theoretically, a 3D whole-tumor analysis can better represent tumor heterogeneity. However, a study conducted by Lubner et al. [46] revealed that a 2D single slice texture analysis affords fairly comparable results to those afforded by 3D whole-tumor analyses. Moreover, another study by Huang et al. [21] demonstrated that satisfactory results were achieved based on a 2D single slice analysis for predicting lymph node metastasis in patients with colorectal cancer. In addition, a whole-tumor analysis is more time-consuming. Although a 3D (semi)automatic segmentation tool could be considered to address this issue, the truth is that, semi-automatic segmentation is only applicable to tumors with large differences in signal intensity between lesions and surrounding tissues [47]. This is observed in lung tumors [[48], [49]]. However, semi-automatic augmentation may not be applicable to NPC due to the unclear borders between lesions and surrounding tissues, especially for the scope of skull base invasion. Therefore, we selected a 2D radiomics analysis in the current study. Similar to prior 2D radiomics analyses in various type of cancers, our results confirm the good predictive performance of 2D studies in T4 NPC. Considering the potential limitation of 2D radiomics analyses, a future radiomics analysis in full 3D for comparison is required.
In conclusion, we have identified and validated a 11-feature Radscore as a powerful prognostic tool for predicting local recurrence in patients with non-metastatic T4 NPC. This study also presented and validated a radiomics nomogram that integrated Radscore and clinical risk factors and can be conveniently used for accurate pretreatment individualized prediction of local recurrence. Our results may facilitate the treatment decision-making process in individual cases.
The following are the supplementary data related to this article.
Supplementary material
Declaration of interests
The authors declare no potential conflicts of interest.
Specific author contributions
Lu-Lu Zhang, Chuan-Miao Xie, Yao Lu and Ying Sun were involved in the conception and design of the study. Lu-Lu Zhang, Jin-Hui Liang, Tian-Sheng Gao, Bin Deng, Ji-Jin Yao, Li Lin, Fo-Ping Chen, Xiao-Dan Huang, Jia Kou and Chao-Feng Li were involved in the data collection. Chuan-Miao Xie, Yao Lu and Ying Sun supervised the study. Lu-Lu Zhang, Meng-Yao Huang and Yan Li were in charge of the statistical analysis of the study. Lu-Lu Zhang, Meng-Yao Huang and Yan Li was involved in manuscript writing, which was corrected and approved by all authors.
Funding and acknowledgments
This work was supported by Health & Medical Collaborative Innovation Project of Guangzhou City, China [grant number 201604020003], Special Support Program of Sun Yat-sen University Cancer Center [grant number 16zxtzlc06], Natural Science Foundation of Guangdong Province [grant number 2017A030312003], Program for Changjiang Scholars and Innovative Research Team in University [PCSIRT, grant number IRT_17R110], and Overseas Expertise Introduction Project for Discipline Innovation [grant number B14035].
None of these funding sources had any role in writing the manuscript or the decision to submit for publication. We sincerely thank Dr. Wei Liang and Dr. Lei Shi (Yidu Cloud Technology Ltd., Beijing, China) for providing technical support in extracting study data from their big-data intelligence database platform. This manuscript has been proofread by an English-speaking professional with a science background at Elixigen Corporation (Huntington Beach, California).
References
- 1.Torre L.A., Bray F., Siegel R.L. Global cancer statistics 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Lee A.W., Ma B.B., Ng W.T. Management of nasopharyngeal carcinoma: current practice and future perspective. J Clin Oncol. 2015;33(29):3356–3364. doi: 10.1200/JCO.2015.60.9347. [DOI] [PubMed] [Google Scholar]
- 3.Wu L.R., Liu Y.T., Jiang N. Ten-year survival outcomes for patients with nasopharyngeal carcinoma receiving intensity-modulated radiotherapy: an analysis of 614 patients from a single center. Oral Oncol. 2017;69:26–32. doi: 10.1016/j.oraloncology.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Hu J., Bao C., Gao J. Salvage treatment using carbon ion radiation in patients with locoregionally recurrent nasopharyngeal carcinoma: initial results. Cancer. 2018;124(11):2427–2437. doi: 10.1002/cncr.31318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hua Y.J., Han F., Lu L.X. Long-term treatment outcome of recurrent nasopharyngeal carcinoma treated with salvage intensity modulated radiotherapy. Eur J Cancer. 2012;48:3422–3428. doi: 10.1016/j.ejca.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 6.Han F., Zhao C., Huang S.M. Long-term outcomes and prognostic factors of re-irradiation for locally recurrent nasopharyngeal carcinoma using intensity-modulated radiotherapy. Clin Oncol (R Coll Radiol) 2012;24:569–576. doi: 10.1016/j.clon.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Chen F.P., Lin L., Qi Z.Y. Pretreatment nomograms for local and regional recurrence after radical radiation therapy for primary nasopharyngeal carcinoma. J Cancer. 2017;8(13):2595–2603. doi: 10.7150/jca.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aerts H.J., Velazquez E.R., Leijenaar R.T. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y., Liu Z., He L. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016;281(3):947–957. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]
- 11.Kickingereder P., Burth S., Wick A. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280(3):880–889. doi: 10.1148/radiol.2016160845. [DOI] [PubMed] [Google Scholar]
- 12.Li H., Zhu Y., Burnside E.S., Drukker K. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of Mammaprint, Oncotype DX, and PAM50 gene assays. Radiology. 2016;281(2):382–391. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe J., Lee S.M., Do K.H. Prognostic value of radiomic analysis of iodine overlay maps from dual-energy computed tomography in patients with resectable lung cancer. Eur Radiol. 2019;29(2):915–923. doi: 10.1007/s00330-018-5639-0. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B., Tian J., Dong D. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res. 2017;23(15):4259–4269. doi: 10.1158/1078-0432.CCR-16-2910. [DOI] [PubMed] [Google Scholar]
- 15.Ouyang F.S., Guo B.L., Zhang B. Exploration and validation of radiomics signature as an independent prognostic biomarker in stage III-IVb nasopharyngeal carcinoma. Oncotarget. 2017;8(43):74869–74879. doi: 10.18632/oncotarget.20423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B., Ouyang F., Gu D. Advanced nasopharyngeal carcinoma: pre-treatment prediction of progression based on multi-parametric MRI radiomics. Oncotarget. 2017;8(42):72457–72465. doi: 10.18632/oncotarget.19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y., Chen C., Xie J. Radiomics signature of computed tomography imaging for prediction of survival and chemotherapeutic benefits in gastric cancer. EBioMedicine. 2018;36:171–182. doi: 10.1016/j.ebiom.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S., Zheng J., Li Y. Development and validation of an MRI-based radiomics signature for the preoperative prediction of lymph node metastasis in bladder cancer. EBioMedicine. 2018;34:76–84. doi: 10.1016/j.ebiom.2018.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi E.R., Lee H.Y., Jeong J.Y. Quantitative image variables reflect the intratumoral pathologic heterogeneity of lung adenocarcinoma. Oncotarget. 2016;7(41):67302–67313. doi: 10.18632/oncotarget.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambin P., Rios-Velazquez E., Leijenaar R. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y.Q., Liang C.H., He L. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 22.Kim J.Y., Park J.E., Jo Y. Incorporating diffusion- and perfusion-weighted MRI into a radiomics model improves diagnostic performance for pseudoprogression in glioblastoma patients. Neuro Oncol. 2018 doi: 10.1093/neuonc/noy133. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Wu C.J., Bao M.L. Machine learning-machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PIRADS v2 in clinically relevant prostate cancer. Eur Radiol. 2017;27(10):4082–4090. doi: 10.1007/s00330-017-4800-5. [DOI] [PubMed] [Google Scholar]
- 24.Wang G., He L., Yuan C. Pretreatment MR imaging radiomics signatures for response prediction to induction chemotherapy in patients with nasopharyngeal carcinoma. Eur J Radiol. 2018;98:100–106. doi: 10.1016/j.ejrad.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Ng S.H., Chang T.C., Ko S.F. Nasopharyngeal carcinoma: MRI and CT assessment. Neuroradiology. 1997;39(10):741–746. doi: 10.1007/s002340050499. [DOI] [PubMed] [Google Scholar]
- 26.Orlhac F., Boughdad S., Philippe C. A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med. 2018;59(8):1321–1328. doi: 10.2967/jnumed.117.199935. [DOI] [PubMed] [Google Scholar]
- 27.Orlhac F., Frouin F., Nioche C. Validation of a method to compensate multicenter effects affecting CT Radiomics. Radiology. 2019;29 doi: 10.1148/radiol.2019182023. 182023. [DOI] [PubMed] [Google Scholar]
- 28.Lucia F., Visvikis D., Vallières M. External validation of a combined PET and MRI radiomics model for prediction of recurrence in cervical cancer patients treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging. 2018 doi: 10.1007/s00259-018-4231-9. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Zwanenburg A., Leger S., Vallières M. 2018. Image Biomarker Standardization Initiative. [Epub ahead of print] [Google Scholar]
- 30.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt F., List M., Cukuroglu E. An ontology-based method for assessing batch effect adjustment approachesin heterogeneous datasets. Bioinformatics. 2018;34(17):908–916. doi: 10.1093/bioinformatics/bty553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee P.H. Resampling methods improve the predictive power of modeling in class-imbalanced datasets. Int J Environ Res Public Health. 2014;11(9):9776–9789. doi: 10.3390/ijerph110909776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weston J., Barnhill S., Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46(1–3):389–422. [Google Scholar]
- 34.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc. 1996;24(3):267–288. [Google Scholar]
- 35.Chalkidou A., O'Doherty M.J., Marsden P.K. False discovery rates in PET and CT studies with texture features: a systematic review. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0124165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang L.Q., Li C.F., Li J. Establishment and validation of prognostic nomograms for endemic nasopharyngeal carcinoma. J Natl Cancer Inst. 2015;108(1) doi: 10.1093/jnci/djv291. [DOI] [PubMed] [Google Scholar]
- 37.Li J.X., Huang S.M., Jiang X.H. Local failure patterns for patients with nasopharyngeal carcinoma after intensity-modulated radiotherapy. Radiat Oncol. 2014;9:87. doi: 10.1186/1748-717X-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan X.B., Wei L., Li H. High pretreatment serum lactate dehydrogenase level correlates with disease relapse and predicts an inferior outcome in locally advanced nasopharyngeal carcinoma. Eur J Cancer. 2013;49(10):2356–2364. doi: 10.1016/j.ejca.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Cheng S.H., Tsai S.Y., Horng C.F. A prognostic scoring system for locoregional control in nasopharyngeal carcinoma following conformal radiotherapy. Int J Radiat Oncol Biol Phys. 2006;66(4):992–1003. doi: 10.1016/j.ijrobp.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Xu X., Lu J., Wang F., Liu X. Dynamic changes in plasma microRNAs have potential predictive values in monitoring recurrence and metastasis of nasopharyngeal carcinoma. Biomed Res Int. 2018;2018 doi: 10.1155/2018/7329195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S.C., Tsang N.M., Chiang W.C. Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. J Clin Invest. 2013;123(12):5269–5283. doi: 10.1172/JCI63428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng G., Cao R., Xue J. Increased expression of SHP-1 is associated with local recurrence after radiotherapy in patients with nasopharyngeal carcinoma. Radiol Oncol. 2014;48(1):40–49. doi: 10.2478/raon-2014-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mroz E.A., Tward A.D., Hammon R.J. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the cancer genome atlas. PLoS Med. 2015;12(2) doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Connor J.P.B., Rose C.J., Waterton J.C. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21(2):249–257. doi: 10.1158/1078-0432.CCR-14-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eskey C.J., Koretsky A.P., Domach M.M. 2H-nuclear magnetic resonance imaging of tumor blood flow: spatial and temporal heterogeneity in a tissue-isolated mammary adenocarcinoma. Cancer Res. 1992;52(21):6010–6019. [PubMed] [Google Scholar]
- 46.Lubner M.G., Stabo N., Lubner S.J. CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging. 2015;40(7):2331–2337. doi: 10.1007/s00261-015-0438-4. [DOI] [PubMed] [Google Scholar]
- 47.Limkin E.J., Sun R., Dercle L. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28(6):1191–1206. doi: 10.1093/annonc/mdx034. [DOI] [PubMed] [Google Scholar]
- 48.Tan Y., Schwartz L.H., Zhao B. Segmentation of lung lesions on CT scans using watershed, active contours, and Markov random field. Med Phys. 2013;40(4) doi: 10.1118/1.4793409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rios Velazquez E., Aerts H.J.W.L. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists' delineations and with the surgical specimen. Radiother Oncol. 2012;105(2):167–173. doi: 10.1016/j.radonc.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material