Summary
The production of pentabromopseudilin and related brominated compounds by Pseudoalteromonas spp. has recently been linked to the bmp biosynthetic gene cluster. This study explored the distribution and evolutionary history of this gene cluster in the genus Pseudoalteromonas. A phylogeny of the genus revealed numerous clades that do not contain type strains, suggesting considerable species level diversity has yet to be described. Comparative genomics revealed four distinct versions of the gene cluster distributed among 19 of the 101 Pseudoalteromonas genomes examined. These were largely localized to the least inclusive clades containing the Pseudoalteromonas luteoviolacea and Pseudoalteromonas phenolica type strains and show clear evidence of gene and gene cluster loss in certain lineages. Bmp gene phylogeny is largely congruent with the Pseudoalteromonas species phylogeny, suggesting vertical inheritance within the genus. However, the gene cluster is found in three different genomic environments suggesting either chromosomal rearrangement or multiple acquisition events. Bmp conservation within certain lineages suggests the encoded products are highly relevant to the ecology of these bacteria.
Introduction
The structure of the bromine rich antibiotic pentabromopseudilin was the first marine microbial natural product described (Lovell, 1966). It was isolated from an obligate marine bacterium identified at the time as Pseudomonas sp. (Burkholder et al., 1966) but was likely a member of the genus now described as Pseudoalteromonas. Subsequently, pentabromopseudilin has been reported from multiple Pseudoalteromonas spp. (Isnansetyo and Kamei, 2003; Fehér et al., 2010; Vynne et al., 2011; Vynne et al., 2012; Whalen et al., 2015; El Gamal et al., 2016a, 2016b; Agarwal et al., 2017) and a single strain (MMB-1) of Marinomonas mediterranea (Agarwal et al., 2014). These two genera of Gammaproteobacteria belong to different Orders, raising the possibility that the associated biosynthetic gene cluster (BGC) was exchanged by horizontal gene transfer. Biosynthetically related brominated natural products (BNPs) have also been reported from diverse organisms ranging from sponge-associated cyanobacteria to seabirds (Agarwal et al., 2017). In addition to pentabromopseudilin, Pseudoalteromonas strains produce brominated phenol and pyrrole monomers and dimers as well as polybrominated diphenyl ethers (PBDEs). One brominated pyrrole monomer, tetrabromopyrrole, appears to play an important role in the settlement and development of coral larvae (Tebben et al., 2011; Sneed et al., 2014). PBDEs are also of particular interest due to their negative implications for human health via endocrine disruption and links to neurodevelopmental deficits and cancer (Hallgren et al., 2001; Zhou et al., 2002; Fernie et al., 2005; Richardson et al., 2008).
Starting in the late 1920s, PBDEs were produced industrially and used as flame-retardants in a wide variety of products such as electronics and textiles (Johnson-Restrepo and Kannan, 2009). While anthropogenic PBDEs can be introduced into the ocean via surface runoff, there is increasing evidence that these compounds are also produced naturally in the marine environment (Reddy et al., 2004; Teuten and Reddy, 2007). Due to their lipophilicity, these molecules bioaccumulate in the blubber and tissues of marine mammals, thus suggesting that a human diet rich in seafood could lead to increased PBDE exposure (Venkateswaran and Dohmoto, 2000; Malmvärn et al., 2005; Teuten et al., 2005).
The recent identification of the bmp gene cluster as the biosynthetic origin of pentabromopseudilin and related BNPs in M. mediterranea MMB-1 and select Pseudoalteromonas strains (Agarwal et al., 2014; El Gamal et al., 2016a) provides a unique opportunity to more broadly assess the biosynthesis of these compounds. The bmp cluster encodes the production of polybrominated phenol and pyrrole monomers (2–4), the phenolic homodimeric antibiotic bromophene (5), PBDEs (6), polybrominated biphenyls (7) and the brominated phenol/pyrrole heterodimer pentabromopseudilin (1) (Fig. 1). The gene cluster encodes an ACPTE di-domain protein (Bmp1) and two flavin-dependent brominases (Bmp2 and Bmp5) that catalyse the bromination of pyrrole and phenol monomers respectively (2–4). Bmp3, 4 and 8 play roles in bromopyrrole monomer synthesis and Bmp6 is involved in the biosynthesis of bromophene and PBDEs (6–7). Bmp7 catalyses the oxidative coupling of bromophenol and bromopyrrole monomers in the presence of Bmp9, Bmp10 and NADH.
Fig. 1.
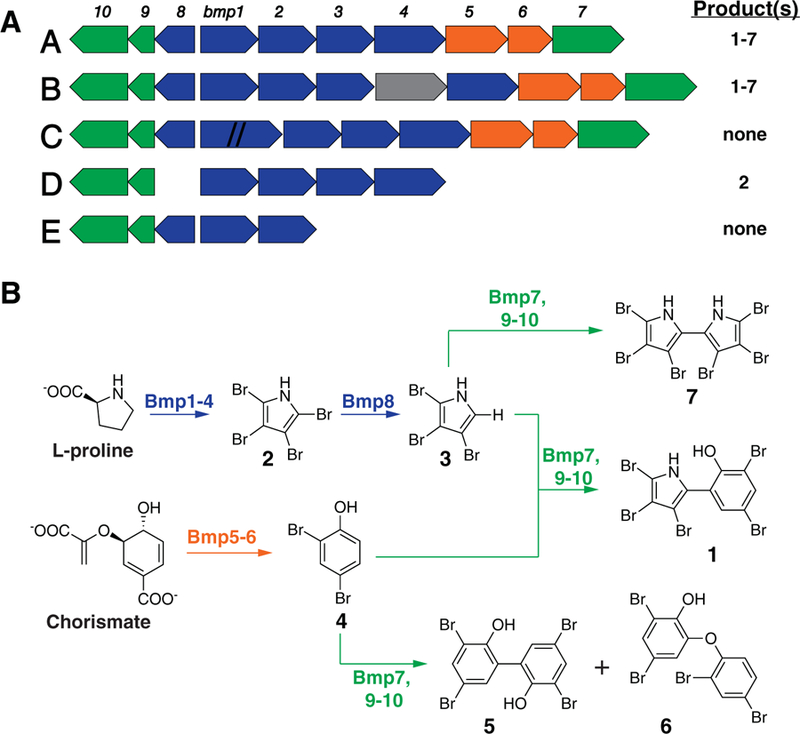
The bmp gene cluster and its products.
A. Five versions of the bmp gene cluster have been identified: four in Pseudoalteromonas spp. (A, C-E) and one in M. mediterranea MMB-1 (B). Hash marks indicate an ~150 bp insertion in bmp1 (version C). The small molecule products detected from each version are indicated.
B. Bmp functional roles in the biosynthesis of brominated natural products.
Previous studies of BNP production in Pseudoalteromonas spp. focused on a few specific strains (Isnansetyo and Kamei, 2003; Fehér et al., 2010). As a result, we lack a broader understanding of the diversity and distribution of the gene cluster within the genus and how this relates to compound production. Using metabolomics and a multilocus phylogeny generated from 144 Pseudoalteromonas strains, we mapped the distribution of bmp genes and pentabromopseudilin production. The results suggest a complex evolutionary history for the gene cluster and provide clues as to how this biosynthetic capacity evolved within the genus.
Results
Pseudoalteromonas species phylogeny
A Pseudoalteromonas species phylogeny that includes type strains for all 42 named species encompasses considerable diversity (Fig. 2). The primary bifurcation distinguishes the Pseudoalteromonas spongiae and Pseudoalteromonas piratica type strains from the remainder of the genus, which can be delineated into three major clades. Clade 3 contains the Pseudoalteromonas ulvae, Pseudoalteromonas tunicata and Pseudoalteromonas denitrificans type strains while the remainder of the type strains fall within clades 1 and 2. Numerous clades at varying depths in the phylogeny do not contain a type strain, suggesting some species level diversity has yet to be described. To further assess the potential for taxonomic novelty, average nucleotide identity (ANI) was calculated among all publicly available genome sequences (N = 91). At the suggested >95% ANI cut-off for species designations (Goris et al., 2007), 42 species level groups were resolved of which 14 contained a type strain (Fig. 3). Two of these (Pseudoalteromonas elyakovii/Pseudoalteromonas piscicida and Pseudoalteromonas flavipulchra/Pseudoalteromonas distincta) contain more than one type strain suggesting these lineages have been over-described. The 28 ANI groups that lacked a type strain were assigned Roman numerals (I-XXVIII, Fig. 3). The 26 type strains for which genome sequences were not available likely correspond to many of these ANI groups, however at least two can be considered unnamed based on this metric. To further address taxonomic novelty, we searched the species tree for clades that lacked a type strain and were coherent with a 95% ANI group. Using this approach, we identified 17 candidate new species, which are indicated in Fig. 2 by the Roman numeral assigned to their ANI group. Of the 17 strains isolated as part of this study (CN designations, Fig. 2), seven originated from the United States and 10 from Fiji. All of the Fiji strains were isolated from macroalgae, and a majority clade with Pseudoalteromonas luteoviolacea while four clade with one of the new ANI species (XXVI). Given that the processing and isolation efforts were not consistent among sample types and locations, no correlations can be drawn between these variables and the species phylogeny.
Fig. 2.
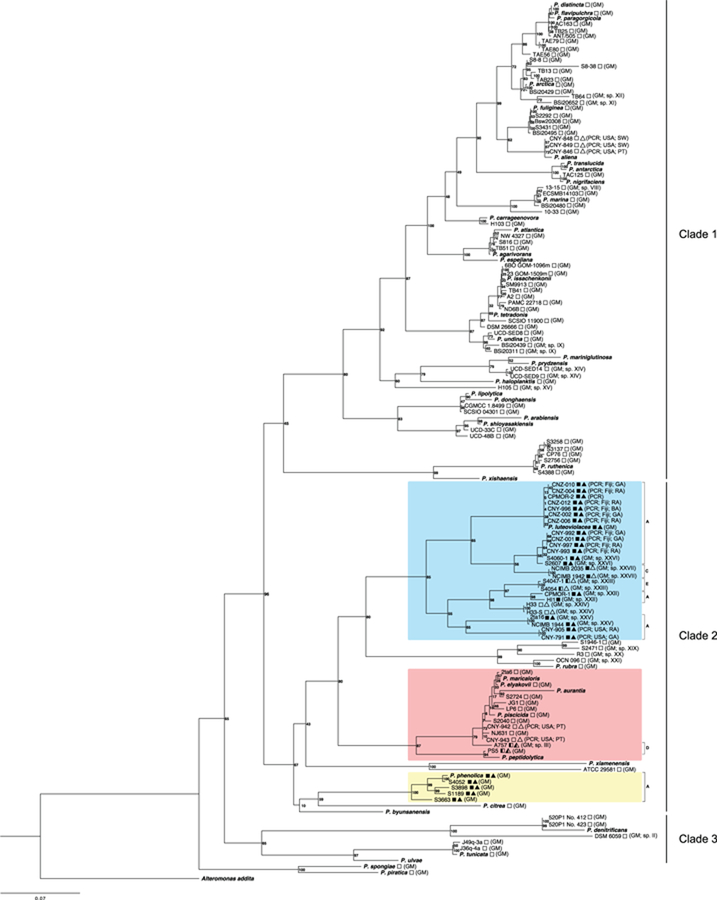
Pseudoalteromonas species phylogeny and bmp gene cluster distribution. A maximum likelihood phylogeny generated from concatenated 16S, gyrB, pyrH, recA and rpoD nucleotide sequences. The three least inclusive clades containing the bmp cluster are highlighted in color. Lettered brackets (A, C, D and E) indicate the version of the bmp cluster. The 42 type strains are indicated in bold italics and the 17 strains isolated as part of this study by CN prefixes followed by country of origin (USA or Fiji) and source (SW = seawater, PT = plankton tow, GA = green alga, RA = red alga, BA = brown alga). Squares indicate bmp distribution (filled = present, empty = absent) and bmp version (fully filled = version A, half-filled = partial BCG). Method of detection (GM = genome sequence or PCR) indicated. Filled triangles indicate the detection of pentabromopseudilin, half-filled triangles indicate the detection of tetrabromopyrrole but not pentabromopseudolin and empty triangles indicate that no compounds associated with the bmp gene cluster were detected. Roman numerals (as designated in Fig. 3) indicate 17 candidate new species identified based on coherence between 95% ANI groupings and the species phylogeny.
Fig. 3.
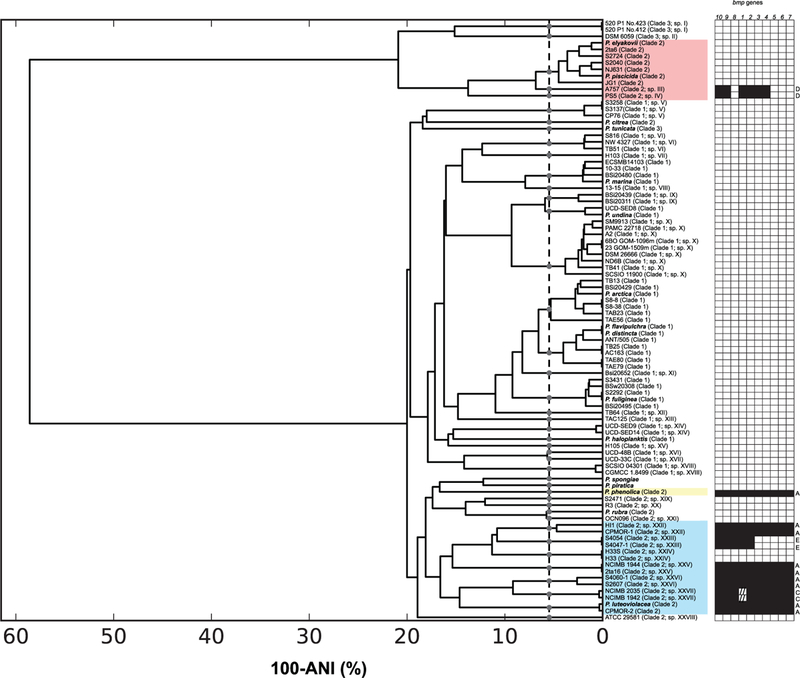
ANI analysis. Average nucleotide identity of 91 Pseudoalteromonas genome sequences. The vertical dashed line specifies 95% ANI and the grey circles designate ANI species groupings. Strain names are followed by clade designations (from Fig. 2) and the 28 ANI groups (95%) that lack type strains indicated by Roman numerals (I–XXVIII). Coloured blocks correspond to sub-clade designations (from Fig. 2). The distribution of the 10 genes in the bmp gene cluster are displayed on the right with the version of the gene cluster (from Fig. 1) indicated on the right. Hash marks indicate an ~150 bp insertion in bmp1.
Distribution of the bmp gene cluster among Pseudoalteromonas spp.
We next mapped the distribution of the bmp gene cluster onto the Pseudoalteromonas species phylogeny. Of the 101 Pseudoalteromonas genomes mined (91 published, 10 unpublished), 19 contain some version of this gene cluster and all of these fall within clade 2 (Fig. 2). Three least inclusive sub-clades containing the bmp BGC can be identified: one containing P. luteoviolacea, one containing P. phenolica, and one containing five named species. In cases where genome sequences were not available, PCR was used to test for the presence of the bmp BGC. The 12 strains that were PCR positive and sequence verified for the bmp cluster all fall within the least inclusive clade containing P. luteoviolacea. Pseudoalteromonas luteoviolacea and P. phenolica are the only named species whose type strains contain the BGC. Six of the candidate ANI species also contain the BGC (sp. III, XXII, XXIII, XXV, XXVI and XXVII).
A comparative analysis of the bmp gene cluster revealed four versions (A, C–E) among the Pseudoalteromonas strains and a fifth version (B) in M. mediterranea strain MMB-1 (Fig. 1). Version A contains 10 genes and has been experimentally linked to the production of pentabromopseudilin and related BNPs (Agarwal et al., 2014). It was observed in 25 of the clade 2 strains including the P. phenolica and P. luteoviolacea type strains and five of the candidate ANI species (Fig. 2). Version B was only observed in a single M. mediterranea strain (MMB-1) and has one additional gene annotated as a putative permease. Version C has an ~150 bp insertion in bmp1 suggesting it may be non-functional. This insertion was sequence verified by PCR in the two strains (NCIMB 1942 and NCIMB 2035) in which it was observed. The genome sequences for PS5 (ANI sp. IV) and A757 (ANI sp. III) contain a partial bmp cluster that lacks bmp5–8 (version D) (Fig. 3). These are the only two strains with any form of the BGC in the least inclusive clade containing 2ta6 and P. peptidolytica (Fig. 2). Strains S4047 and S4054 (both ANI sp. XXIII) possess version E of the gene cluster, which lacks bmp3–7 (Fig. 3). This truncated version, along with a pair of strains that lack the entire gene cluster (H33 and H33-S), are part of a well-supported sub-clade that is dominated by version A (Fig. 2), suggesting they result from gene loss events. Strains isolated as part of this study and for which genomes were not available were assigned version A if sequence verified PCR products from both bmp2 and bmp5 were detected. They were not assigned version B, since this has only been detected in a single strain of M. mediterranea, or versions C–E based on the results of the metabolomics analyses (below).
Pentabromopseudilin production
Pseudoalteromonas strains possessing different versions of the bmp gene cluster were analysed for pentabromopseudilin (1) production (Supporting Information Fig. S1). Pentabromopseudilin is a heterodimer comprised of brominated pyrrole and phenol monomers. All 10 genes in version A of the gene cluster are required for the production of this compound in Pseudoalteromonas spp. (Agarwal et al., 2014). Therefore, any Pseudoalteromonas strains that produce this compound can be expected to contain bmp version A, since version B has not been observed in the genus. Of the 25 Pseudoalteromonas strains containing bmp version A, 24 were confirmed for pentabromopseudilin production (Fig. 2) thus matching metabolic predictions. The 25th strain (HI1) was not available for testing. H33 and H33-S are the only strains in clade 2 that are missing the entire bmp gene cluster and, as expected, no BNPs were detected (Vynne et al., 2012). We also failed to detect brominated compounds from the strains that possess bmp version C (NCIMB 1942 and NCIMB 2035). The 150 bp insertion in the ACP-TE encoding bmp1 gene distinguishes version C from A and may explain the lack of BNP production in these strains. Tetrabromopyrrole (2) was the only BNP detected from strains with bmp version D, which is consistent with the expected phenotype given that genes encoding production of the polybrominated phenol monomer (bmp5–6), the bromopyrrole homodimer (bmp7) and tribromopyrrole via the debromination of tetrabromopyrrole (bmp8) are missing (Fig. 1B). Additionally, BNPs were not detected from the two strains containing version E of the BGC (S4047 and S4054), which lacks bmp3–7. These results are consistent with the functional characterization of the gene cluster, as the products of the missing genes are required to synthesize brominated pyrrole and phenol monomers.
Evolutionary history of the bmp gene cluster
A concatenated phylogeny of the bmp genes derived from all 19 Pseudoalteromonas genome sequences that contained some version of the BGC was compared with a species tree generated for the same strains (Fig. 4). While no outgroup was used in the bmp phylogeny, the sequences from M. mediterranea strain MMB-1 were included and the tree was rooted consistently with the species tree (Fig. 4A). With the exception of the position of strains MMB-1 and S3663, the two trees are largely congruent. The bmp phylogeny shows that the sister clades containing P. luteoviolacea and A757 and the more distantly related P. phenolica clade are consistent with the species tree, supporting a single acquisition of the bmp cluster in the genus. The relationship between the bmp BGCs observed in M. mediterranea strain MMB-1 and Pseudoalteromonas spp. supports the hypothesis that it was acquired by horizontal gene transfer.
Fig. 4.
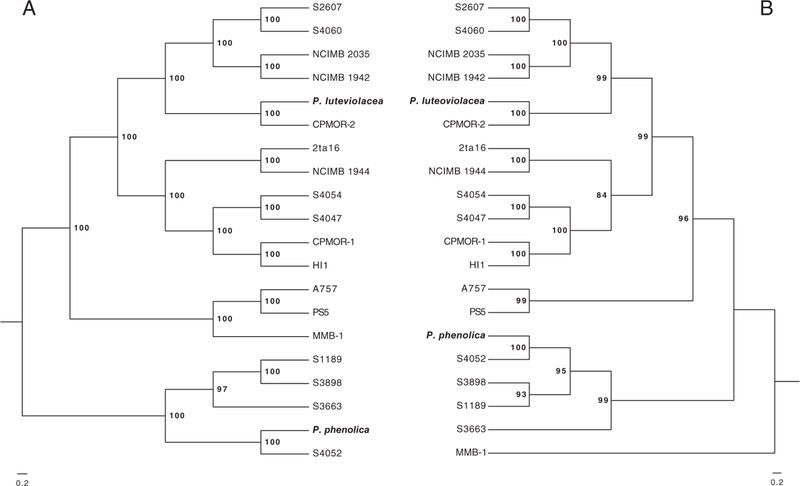
Bmp gene phylogeny.
A. Concatenated maximum likelihood phylogeny of bmp gene sequences.
B. Concatenated maximum likelihood phylogenies of 16S, gyrB, pyrH, recA and rpoD sequences derived from the 19 Pseudoalteromonas and one M. mediterranea strain (MMB-1) that contain the bmp gene cluster. The bmp gene tree is rooted based on the species phylogeny while the species tree is rooted with MMB-1. Type strains are indicated by species names.
To further assess if a single acquisition event could account for the distribution of the bmp gene cluster in the genus Pseudoalteromonas, we explored its genomic environment, expecting it to be similar among strains. Pairwise comparisons revealed high levels of sequence identity in and around the bmp BGC for strains within the large clade containing CPMOR-1 and −2 indicating positional conservation in this lineage (Fig. 5A). These results further support the hypothesis that strain S4054 experienced partial BGC loss while strain NCIMB 1942 experienced BGC modification resulting in bmp versions C and E respectively. The detection of a remnant of bmp7 in strain S4054 provides additional support for the gene loss hypothesis (Supporting Information Fig. S2), while it can be inferred that the complete BGC was lost in strain H33. Outside of this clade, the lack of sequence identity between strains A757 and H33 suggests that the BGC is in a different genomic environment in strain A757 and the related strain PS5, the only two strains that possess version D of the BGC. When more distantly related strains are compared (HI1 and P. phenolica, Fig. 5B), the lack of sequence identity outside the BGC is more difficult to interpret, but could also indicate different genomic environments. A comparison of strain HI1, which contains bmp version A, with strain A757, which contains version D, reveals little sequence identity outside of the BGC (Fig. 5C), again suggesting different genomic environments with version D representing a partial loss of the cluster. While congruence between the bmp gene phylogeny and the Pseudoalteromonas species phylogeny suggests a single acquisition event for the BGC, the genomic environments in which it occurs could be explained by either multiple acquisition events or intragenomic rearrangements. Regardless of which explanation is correct, there is clear evidence within sub-clades of the Pseudoalteromonas phylogeny that the gene cluster has been maintained, modified or lost following a model of vertical inheritance (Fig. 6).
Fig. 5.
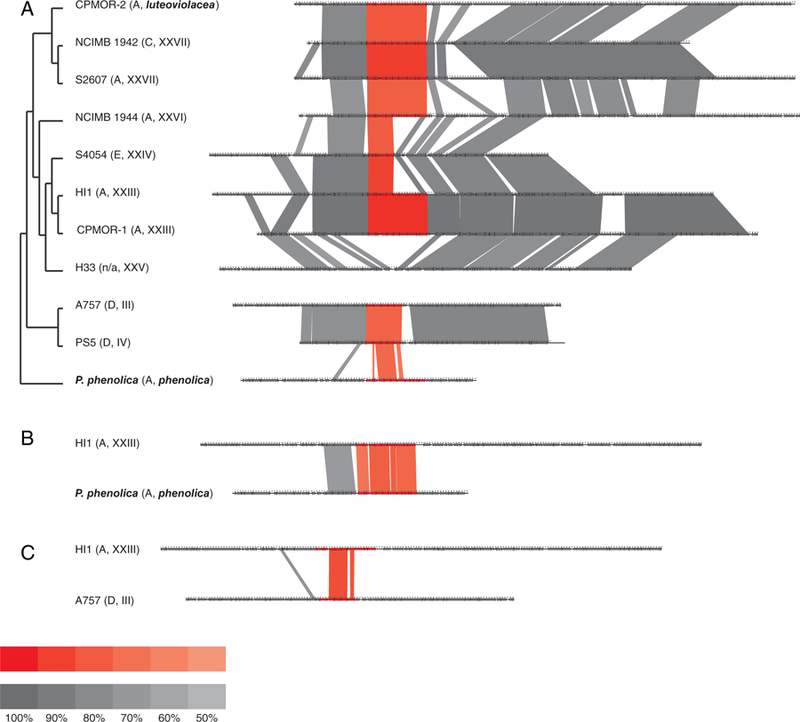
Genomic environment of the bmp gene cluster.
A. The species phylogeny is contrasted with conservation of the bmp gene cluster (red) and surrounding genomic environments (grey). Strain names are followed by the version of the gene cluster (A, C, D or E) and candidate ANI species identifiers. Opacity indicates percent sequence identity with anything below 50% not shown.
B. A high level of sequence identity is observed in the bmp gene cluster between two distantly related strains (HI1 and P. phenolica) but not in the surrounding genomic environment.
C. Strain A757 (bmp version D) reveals a partial loss of the bmp gene cluster.
Fig. 6.
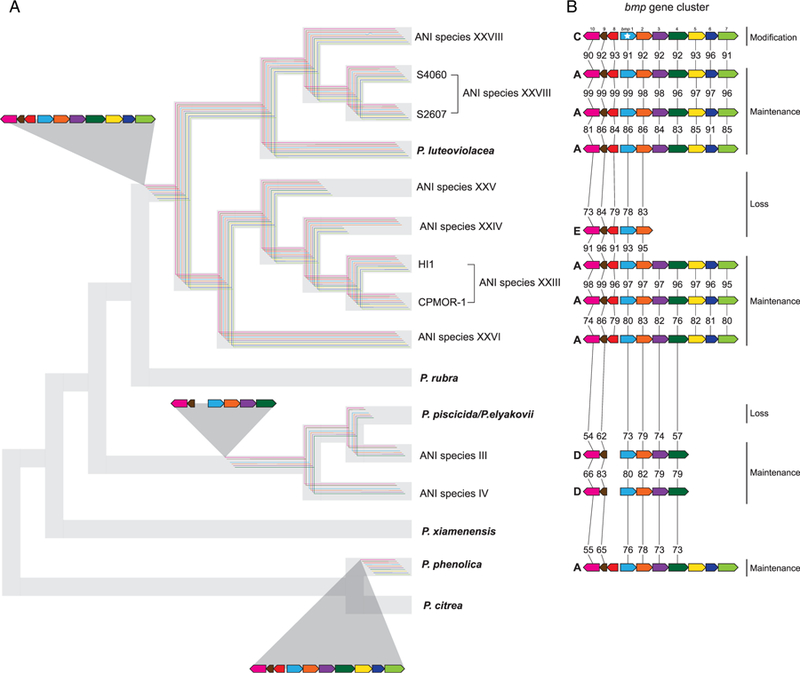
Evolutionary history of the bmp gene cluster in Pseudoalteromonas.
A. Inferred ancestral gene clusters are mapped onto a condensed species phylogeny (animated from Fig. 2). The distribution of each gene (coloured lines) is indicated based on presence or absence in that lineage (truncated or missing lines = absence).
B. Versions of the gene cluster (A, C, D or E) observed in relation to the tree with evolutionary events indicated as maintenance, loss or modification. Losses include complete or partial deletion of the gene cluster. An insertion in bmp1 is the only modification observed (indicated with a star). Percent sequence identities between homologues indicated.
Discussion
Characterization of the bmp BGC revealed it accounts for a diverse set of compounds including the potent antibiotic pentabromopseudilin (Burkholder et al., 1966), the coral larval settlement cue tetrabromopyrrole (El Gamal et al., 2016a) and PBDEs whose industrial production and use as flame-retardants have been banned in some countries due to human health implications (Johnson-Restrepo and Kannan, 2009). These compounds have primarily been reported from members of the genus Pseudoalteromonas, yet despite their ecological and environmental significance, compound production and the distribution of the associated biosynthetic genes have not been investigated.
The results presented here indicate a narrow distribution and complex evolutionary history for the bmp gene cluster that includes gene modifications and deletions that affect compound production. While congruence between the species and bmp phylogenies supports a single acquisition event (Fig. 4), this model requires eight complete or partial gene cluster losses to explain the observed distributions. Given that the gene cluster was observed in different genomic environments in some strains, the possibility of multiple acquisition events cannot be ruled out. However, different environments could also be accounted for by intragenomic rearrangements, as has previously been shown for the genus Salinispora (Letzel et al., 2017). While the evolutionary history of the bmp gene cluster remains unresolved at the genus level, there are three points from which vertical inheritance can be inferred with confidence (Fig. 6). Even within these sub-lineages, there is clear evidence of gene cluster loss or modification, attesting to its complex evolutionary history.
There is ample evidence that the gene clusters associated with natural product biosynthesis are exchanged by horizontal gene transfer. Despite this, there was a strong correlation between taxonomy, bmp composition and compound production within the genus Pseudoalteromonas. For example, all P. phenolica strains analysed maintained the same version of the BGC and produced the same natural products. The least inclusive clade that includes P. luteoviolacea is also highly conserved for these features, with the exception of a few loss events. The conservation of bmp within certain clades suggests that the products encoded provide an important selective advantage and add to growing evidence that natural products can be useful taxonomic markers for bacteria (Hoffmann et al., 2018).
During the course of this study, gene clusters related to bmp were detected in sponge cyanobacterial symbionts (Agarwal et al., 2017). While these gene clusters have yet to be fully characterized, they include bmp5–7 homologues, which encode the production of PBDEs, but not the genes required to make brominated pyrrole molecules or pentabromopseudilin. These results suggest that an even more complex evolutionary history remains to be resolved for the bmp gene cluster.
Remarkable new insight is being gained into the evolutionary processes that drive natural product biosynthesis (Medema et al., 2014; Ruzzini and Clardy, 2016; Lind et al., 2017). These studies have helped reveal the dynamic processes of gene gain, loss and degradation in BGCs (Letzel et al., 2017) and how subtle changes in regulatory genes can have substantial effects on both gene expression and compound production (Amos et al., 2017). This study adds to the growing number of examples in which the evolutionary history of a BGC provides insight into the processes that establish lineage specific chemical diversity (Freel et al., 2011). While multiple versions of the bmp cluster were observed, selection appears to favour the maintenance of version A, which encodes the biosynthesis of the potent antibiotic pentabromopseudilin (1). In cases where the BGC was lost, it may be complemented by the products of another BGC, as was shown with siderophore biosynthesis in marine actinomycetes (Bruns et al., 2017). Alternatively, relaxed selective pressures associated with some habitats may lead to BGC loss. Regardless of the factors driving loss events, their occurrence within well-supported lineages demonstrates the unpredictable nature of microbial natural product discovery and presents a caveat to their applications in chemotaxonomy. The bioinformatic analyses performed here provided a powerful predictor of chemical phenotypes, as there was a perfect correlation between the version of the BGC detected and BNP production among the strains analysed. Continued access to more genome sequences we will undoubtedly expand our understanding of the evolutionary history of the bmp gene cluster in Pseudoalteromonas and other taxa in which it may be observed.
Experimental procedures
Bacterial isolation
Seawater samples were collected at Scripps Institution of Oceanography (SIO), plankton tows (20 µm mesh size) were performed from the SIO pier, and algal samples were collected by SCUBA from Catalina Island, CA and the Coral Coast, Fiji (Supporting Information Table S1). The algae, which were identified as red, green or brown based on visual appearance, were gently shaken to remove excess water before a small piece of tissue was added to 10 ml of 0.2 µm-filtered seawater. The algal samples were mixed and diluted 1:10, 1:50 and 1:100 in sterile seawater, and 100 µl of each dilution spread onto Difco 2216 media with 15 g/l agar added. Seawater and plankton tow samples were diluted 1:2, 1:10 and 1:50 and similarly plated. Bacterial colonies were isolated based on morphology, specifically selecting for dark purple and other pigmentation that is characteristic of Pseudoalteromonas spp. Strains were identified at the genus level based on 16S rRNA sequence analysis (see below) and glycerol stocks frozen (−80 °C) in Difco 2216 broth with 25% glycerol.
Genome mining, primer design and PCR
Published genome sequences are available for 16 of 42 Pseudoalteromonas type strains while gyrB and 16S sequences are available for an additional 12 (Supporting Information Table S2). Our analysis also included an additional 75 published Pseudoalteromonas genomes (Supporting Information Table S3), 10 unpublished genomes and 17 strains isolated as part of this study. Genome sequences were acquired from the NCBI and JGI public databases or sequenced at SIO. They were uploaded and annotated in RAST (Aziz et al., 2008) and mined for 16S, gyrB, pyrH, recA and rpoD based on annotations. Bmp1–10 gene sequences were identified using the internal BLAST search tool in RAST with gene sequences from P. luteoviolacea strain 2ta16 as queries. Ten unpublished Pseudoalteromonas genomes sequenced at the Technical University of Denmark were similarly mined. PCR was used to generate sequences for strains isolated as part of this study but for which genome sequences were not available. Degenerate PCR primers were designed based on the translated and MUSCLE aligned gyrB, pyrH, recA, rpoD and bmp sequences (Edgar et al., 2004). The primers were validated across different Pseudoalteromonas spp. using Primaclad software and manual analyses (Supporting Information Table S4) (Gadberry et al., 2005). All of the mined and amplified sequences have been deposited in NCBI (MK421604-MK421630; MK469519-MK468590)
For genomic DNA extractions, strains were cultured overnight at room temperature in 10 ml of marine broth (Difco 2216) with shaking at 230 rpm. Cells were pelleted (16,000g, 2 min) from 1 ml of culture, re-suspended in 600 µl of Nuclei Lysis Solution (Promega), and incubated at 80 °C for 5 min after which 200 µl of Protein Precipitate Solution (Promega) was added. The samples were vortexed at high speed for 20s, incubated on ice for 5 min, centrifuged (16,000g, 10 min), and the supernatant transferred into 600 µl of isopropanol. The samples were mixed by gentle inversion, centrifuged (13,000 rpm, 10 min), and the DNA pellet washed once with 70% ethanol, dried and rehydrated in water. PCR thermocycling conditions were as follows: 5 min of initial denaturation at 95°C, 30 cycles of denaturation at 95°C for 1 min, annealing for 1 min at 52–55°C, extension at 72°C for 1 min, and a final extension at 72°C for 5 min. PCR products were sequenced using Sanger methods by Eton Biosciences (https://www.etonbio.com/) and trimmed for quality before analysis.
Phylogenetic analyses
Nucleotide sequences used to establish the species phylogeny were aligned using MAFFT (Katoh et al., 2002) then concatenated. Maximum likelihood analysis of the partitioned data was run using RAxML with 100 rapid bootstrap replicates and the GTR + G model (Stamatakis, 2014). To assess the phylogeny of the bmp cluster, the MUSCLE alignments of the bmp1, 2, 9 and 10 gene nucleotide sequences were concatenated and a maximum likelihood analysis performed using RAxML with 100 rapid bootstrap replicates and GTR + G model (Edgar et al., 2004; Stamatakis, 2014). All trees were visualized using FigTree v1.4.2.
Pentabromopseudilin analysis
Bacterial cultures were grown at room temperature with agitation (200 rpm) for 48 h in 50 ml of Difco 2216 media with 1 g/l of KBr added. The cultures were extracted twice with 50 ml ethyl acetate and the organic fractions combined and dried in vacuo. The dried extracts were dissolved in 500 µl of MeOH and analysed by LC/MS using previously described methods (Agarwal et al., 2014). Pentabromopseudilin production was verified in comparison to an authentic standard or based on previously reported results.
Comparative genomics
ANI was determined for 91 Pseudoalteromonas genomes using custom scripts available at https://github.com/juanu/ANI_analysis. A distance matrix was generated from all pairwise comparisons and ANI divergence (100 – ANI) was calculated. Pairwise comparisons between contigs to assess the genomic environment of the bmp BGC were performed using the Artemis Comparison Tool v13.0.0 (Carver et al., 2005).
Supplementary Material
Figure S1 Detection of pentabromopseudilin by LCMS. Strains isolated as part of this study were screened for pentabromopseudilin production. (A) Extracted ion chromatograms for the mass of pentabromopseudilin (553.67 g/mol) from a strain with version A of the BGC (CNY-791, top) and one that lacks the BGC (CNY-943, middle). Pentabromopseudilin standard (bottom). All strains containing bmp version A produced pentabromopseudilin with an absolute intensity of at least 1.4 × 106 (B) Isotopic pattern of a pentabromopseudilin standard and the compound detected from strain CNY-791 (C).
Fig. S2. Evidence for partial loss of the bmp cluster. A region immediately downstream of bmp2 in strain S4054 (version E) has high sequence identity to a portion of bmp7 in strain HI1 (version A), supporting gene loss in S4054.
Table S1. Source of Pseudoalteromonas strains isolated as part of this study.
Table S2. Type strains without published genome sequences.
Table S3. Pseudoalteromonas genomes used in this study. Type strains indicated in bold. Roman numerals (I-XXVIII) correspond to the 28 ANI groups (95%) that lack type strains (from Fig. 3). Asterisks (*) indicate 95% ANI groups where type strain genome sequences were unavailable.
Table S4. PCR primer sequences.
Acknowledgements
This work was jointly supported by the US National Science Foundation (OCE-1313747) and the US National Institute of Environmental Health Sciences (P01-ES021921) through the Oceans and Human Health program. We acknowledge N. Patin for providing strains and the National Institutes of Health award U19-TW007401–01 for access to samples from Fiji.
Footnotes
The authors declare no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
References
- Agarwal V, El Gamal A, Yamanaka K, Poth D, Kersten RD, Schorn M, et al. (2014) Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat Chem Biol 10: 640–647. 10.1038/nchembio.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Blanton JM, Podell S, Taton A, Schorn MA, Busch J, et al. (2017) Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat Chem Biol 13: 537–543. 10.1038/nature15540.Genetic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos GCA, Awakawa T, Tuttle RN, Letzel A-C, Kim MC, Kudo Y, et al. (2017) Comparative transcriptomics as a guide to natural product discovery and biosynthetic gene cluster functionality. Proc Natl Acad Sci 114: E11121–E11130. 10.1073/pnas.1714381115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards RA, et al. (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9: 1–15. 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns H, Crüsemann M, Letzel A-C, Alanjary M, McInerney JO, Jensen PR, et al. (2017) Function-related replacement of bacterial siderophore pathways. ISME J 12: 320–329. 10.1038/ismej.2017.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder PR, Pfister RM, and Leitz FH (1966) Production of a pyrrole antibiotic by a marine bacterium. Appl Microbiol 14: 649–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, and Parkhill J (2005) ACT: the Artemis comparison tool. Bioinformatics 21: 3422–3423. 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Drive RM, and Valley M (2004) MUSCLE : Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gamal A, Agarwal V, Diethelm S, Rahman I, Schorn MA, Sneed JM, et al. (2016a) Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase–thioesterase enzyme pair. Proc Natl Acad Sci 113: 3797–3802. 10.1073/pnas.1519695113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gamal A, Agarwal V, Rahman I, and Moore BS (2016b) Enzymatic reductive dehalogenation controls the biosynthesis of marine bacterial pyrroles. J Am Chem Soc 138: 13167–13170. 10.1021/jacs.6b08512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér D, Barlow R, McAtee J, and Hemscheidt TK (2010) Highly brominated antimicrobial metabolites from a marine Pseudoalteromonas sp. J Nat Prod 73: 1963–1966. 10.1021/np100506z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie KJ, Shutt JL, Mayne G, Hoffman D, Letcher RJ, Drouillard KG, and Ritchie IJ (2005) Exposure to polybrominated diphenyl ethers (PBDEs): changes in thyroid, vitamin A, glutathione homeostasis, and oxidative stress in American kestrels (Falco sparverius). Toxicol Sci 88: 375–383. 10.1093/toxsci/kfi295. [DOI] [PubMed] [Google Scholar]
- Freel KC, Nam SJ, Fenical W, and Jensen PR (2011) Evolution of secondary metabolite genes in three closely related marine Actinomycete species. Appl Environ Microbiol 77: 7261–7270. 10.1128/AEM.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadberry MD, Malcomber ST, Doust AN, and Kellogg EA (2005) Primaclade - a flexible tool to find conserved PCR primers across multiple species. Bioinformatics 21: 1263–1264. 10.1093/bioinformatics/bti134. [DOI] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, and Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57: 81–91. 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Hallgren S, Sinjari T, Håkansson H, and Darnerud P (2001) Effects of polybrominated diphenyl ethers (PBDEs) and polychlorinated biphenyls (PCBs) on thyroid hormone and vitamin A levels in rats and mice. Arch Toxicol 75: 200–208. 10.1007/s002040000208. [DOI] [PubMed] [Google Scholar]
- Hoffmann T, Krug D, Bozkurt N, Duddela S, Jansen R, Garcia R, et al. (2018) Correlating chemical diversity with taxonomic distance for discovery of natural products in Myxobacteria. Nat Commun 9: 1–10. 10.1038/s41467-018-03184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isnansetyo A, and Kamei Y (2003) MC21-A, a bactericidal antibiotic produced by a new marine bacterium, Pseudoalteromonas Phenolica sp. Nov. O-BC30T, against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 47: 480–488. 10.1128/AAC.47.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Restrepo B, and Kannan K (2009) An assessment of sources and pathways of human exposure to polybrominated diphenyl ethers in the United States. Chemosphere 76: 542–548. 10.1016/j.chemosphere.2009.02.068. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K.-i., and Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzel AC, Li J, Amos GCA, Millán-Aguiñaga N, Ginigini J, Abdelmohsen UR, et al. (2017) Genomic insights into specialized metabolism in the marine actinomycete Salinispora. Environ Microbiol 19: 3660–3673. 10.1111/1462-2920.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind AL, Wisecaver JH, Lameiras C, Wiemann P, Palmer JM, Keller NP, et al. (2017) Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol 15: 1–26. 10.1371/journal.pbio.2003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell FM (1966) The structure of a bromine-rich marine antibiotic. Journal of The American Chemical Society 88: 4510–4511. [Google Scholar]
- Malmvärn A, Marsh G, Kautsky L, Athanasiadou M, Bergman A, and Asplund L (2005) Hydroxylated and methoxylated brominated diphenyl ethers in the red algae Ceramium tenuicorne and blue mussels from the Baltic Sea. Environ Sci Technol 39: 2990–2997. [DOI] [PubMed] [Google Scholar]
- Medema MH, Cimermancic P, Sali A, Takano E, and Fischbach MA (2014) A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput Biol 10: e1004016 10.1371/journal.pcbi.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy CM, Xu L, O’Neil GW, Nelson RK, Eglinton TI, Faulkner DJ, et al. (2004) Radiocarbon evidence for a naturally produced, bioaccumulating halogenated organic compound. Environ Sci Technol 38: 1992–1997. [DOI] [PubMed] [Google Scholar]
- Richardson VM, Staskal DF, Ross DG, Diliberto JJ, DeVito MJ, and Birnbaum LS (2008) Possible mechanisms of thyroid hormone disruption in mice by BDE 47, a major polybrominated diphenyl ether congener. Toxicol Appl Pharmacol 226: 244–250. 10.1016/j.taap.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Ruzzini AC, and Clardy J (2016) Gene flow and molecular innovation in bacteria. Curr Biol 26: R859–R864. 10.1016/j.cub.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneed JM, Sharp KH, Ritchie KB, and Paul VJ (2014) The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Proc R Soc B Biol Sci 281: 1–9. 10.1098/rspb.2013.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebben J, Tapiolas DM, Motti CA, Abrego D, Negri AP, Blackall LL, et al. (2011) Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS One 6: 1–8. 10.1371/journal.pone.0019082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuten EL, and Reddy CM (2007) Halogenated organic compounds in archived whale oil: a pre-industrial record. Environ Pollut 145: 668–671. 10.1016/j.envpol.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Teuten EL, Xu L, and Reddy CM (2005) Two abundant bioaccumulated halogenated compounds are natural products. Science 307: 917–920. 10.1126/science.1106882. [DOI] [PubMed] [Google Scholar]
- Venkateswaran K, and Dohmoto N (2000) Pseudoalteromonas peptidolytica sp. Nov., a novel marine musselthread-degrading bacterium isolated from the sea of Japan. Int J Syst Evol Microbiol 50: 565–574. [DOI] [PubMed] [Google Scholar]
- Vynne NG, Månsson M, Nielsen KF, and Gram L (2011) Bioactivity, chemical profiling, and 16S RRNA-based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Marine Biotechnol 13: 1062–1073. 10.1007/s10126-011-9369-4. [DOI] [PubMed] [Google Scholar]
- Vynne NG, Mansson M, and Gram L (2012) Gene sequence based clustering assists in dereplication of Pseudoalteromonas luteoviolacea strains with identical inhibitory activity and antibiotic production. Mar Drugs 10: 1729–1740. 10.3390/md10081729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen KE, Poulson-Ellestad KL, Deering RW, Rowley DC, and Mincer TJ (2015) Enhancement of antibiotic activity against multidrug-resistant bacteria by the efflux pump inhibitor 3,4-dibromopyrrole-2,5-dione isolated from a Pseudoalteromonas sp. J Nat Prod 78: 402–412. 10.1021/np500775e. [DOI] [PubMed] [Google Scholar]
- Zhou T, Taylor MM, DevVito MJ, and Crofton KM (2002) Developmental exposure to brominated diphenyl ethers results in thyroid hormone disruption. Toxicol Sci 116: 105–116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Detection of pentabromopseudilin by LCMS. Strains isolated as part of this study were screened for pentabromopseudilin production. (A) Extracted ion chromatograms for the mass of pentabromopseudilin (553.67 g/mol) from a strain with version A of the BGC (CNY-791, top) and one that lacks the BGC (CNY-943, middle). Pentabromopseudilin standard (bottom). All strains containing bmp version A produced pentabromopseudilin with an absolute intensity of at least 1.4 × 106 (B) Isotopic pattern of a pentabromopseudilin standard and the compound detected from strain CNY-791 (C).
Fig. S2. Evidence for partial loss of the bmp cluster. A region immediately downstream of bmp2 in strain S4054 (version E) has high sequence identity to a portion of bmp7 in strain HI1 (version A), supporting gene loss in S4054.
Table S1. Source of Pseudoalteromonas strains isolated as part of this study.
Table S2. Type strains without published genome sequences.
Table S3. Pseudoalteromonas genomes used in this study. Type strains indicated in bold. Roman numerals (I-XXVIII) correspond to the 28 ANI groups (95%) that lack type strains (from Fig. 3). Asterisks (*) indicate 95% ANI groups where type strain genome sequences were unavailable.
Table S4. PCR primer sequences.


