Abstract
Background
Glioblastoma (GBM), a highly malignant brain tumor, invariably recurs after therapy. Quiescent GBM cells represent a potential source of tumor recurrence, but little is known about their molecular underpinnings.
Methods
Patient-derived GBM cells were engineered by CRISPR/Cas9-assisted knock-in of an inducible histone2B-GFP (iH2B-GFP) reporter to track cell division history. We utilized an in vitro 3D GBM organoid approach to isolate live quiescent GBM (qGBM) cells and their proliferative counterparts (pGBM) to compare stem cell properties and therapy resistance. Gene expression programs of qGBM and pGBM cells were analyzed by RNA-Seq and NanoString platforms.
Findings
H2B-GFP-retaining qGBM cells exhibited comparable self-renewal capacity but higher therapy resistance relative to pGBM. Quiescent GBM cells expressed distinct gene programs that affect cell cycle control, metabolic adaptation, and extracellular matrix (ECM) interactions. Transcriptome analysis also revealed a mesenchymal shift in qGBM cells of both proneural and mesenchymal GBM subtypes. Bioinformatic analyses and functional assays in GBM organoids established hypoxia and TGFβ signaling as potential niche factors that promote quiescence in GBM. Finally, network co-expression analysis of TCGA glioma patient data identified gene modules that are enriched for qGBM signatures and also associated with survival rate.
Interpretation
Our in vitro study in 3D GBM organoids supports the presence of a quiescent cell population that displays self-renewal capacity, high therapy resistance, and mesenchymal gene signatures. It also sheds light on how GBM cells may acquire and maintain quiescence through ECM organization and interaction with niche factors such as TGFβ and hypoxia. Our findings provide a starting point for developing strategies to tackle the quiescent population of GBM.
Fund
National Institutes of Health (NIH) and Deutsche Forschungsgemeinschaft (DFG).
Keywords: Glioblastoma, Tumor quiescence, H2B-GFP, GBM organoid, Proneural-mesenchymal transition, Stem cell niche
Research in context.
Evidence before this study
Glioblastoma (GBM) is a devastating primary brain cancer, and tumor recurrence after treatment is a main cause of its high lethality. Previous studies implicated a quiescent population in GBM with high therapy resistance as a potential source of tumor recurrence, but our knowledge about the cellular and molecular properties of quiescent GBM (qGBM) cells is limited.
Added value of this study
To gain new insights into the molecular underpinnings of qGBM cells, we took advantage of two recent experimental advances that allow tracking of quiescent cells during GBM expansion. First, we used CRISPR/Cas9-assisted gene targeting to insert a proliferation reporter (an inducible histone2B-GFP) into patient-derived GBM cells, and second, we utilized a 3D GBM organoid approach that mimics in vivo conditions of GBM in recapitulating intratumoral heterogeneity. We demonstrated that qGBM cells exhibit higher therapy resistance than their proliferative counterparts. Gene expression analysis revealed unique gene programs in qGBM cells that concern cell cycle control, metabolic adaptation, and interaction with extracellular matrix. Gene signatures also indicated a mesenchymal shift in qGBM cells. Bioinformatic analysis and functional assays established hypoxia and TGFβ signaling as potential niche factors that promote quiescence.
Implications of all the available evidence
Our approach of using a genetic proliferation reporter and a GBM organoid model for isolating live quiescent GBM cells opens new doors to identify molecular targets to tackle the quiescent population in GBM.
Alt-text: Unlabelled Box
1. Introduction
Glioblastoma (GBM), the most common malignant primary brain tumor in adults, has a median survival of <15 months despite maximal therapy [1,2]. A major determinant of the high lethality of GBM appears to be the presence of a therapy-resistant population with the capacity to spawn tumor recurrence, termed glioma stem cells [[3], [4], [5]]. Attempts to eradicate GBM stem cells have not yet been successful in clinical setting. It is now recognized that GBM comprises both a fast-dividing population and a relatively quiescent population with distinct characteristics. Quiescent GBM (qGBM) cells are thought to have higher therapy-resistance because conventional chemo and radiation therapies largely target the proliferative population [6]. Importantly, qGBM cells can be reawakened to initiate tumor re-expansion, hence targeting qGBM cells in combination with existing therapies against proliferative GBM (pGBM) cells may be critical to curb tumor recurrence [7]. However, our understanding of how tumor cells acquire and maintain quiescence, and how quiescent cells gain therapy resistance and malignant potency remains limited. This has led to the conceptual question of whether quiescence and stemness simply equate one another. However, quiescence and stemness may represent two distinct cellular characteristics in that the stem cell compartment in GBM may in fact contain both a quiescent and a proliferative subpopulation. Therefore, careful comparison of qGBM and pGBM cells for cellular characteristics and gene signatures is needed to distinguish quiescence and stemness, both of which may be critical features underlying GBM recurrence.
One challenge to study tumor cell quiescence is to establish a reporter system that can reliably track proliferative history during tumor expansion. In BrdU or EdU pulse/chase paradigms, label-retaining quiescent cells can only be isolated after fixation. Cell permeable fluorescent dyes have been used to identify live quiescent cell populations in GBM [6,8,9], however, this method is not suitable for long-term studies. Here, we utilized a knock-in approach for targeted insertion of a proliferation reporter, i.e. an inducible histone-2B fused with green fluorescent protein (iH2B-GFP). The H2B-GFP reporter was originally developed to label chromatin in live cells [10]. Taking advantage of the long-term stability of H2B-GFP protein, this reporter has since been adapted for label retention studies to track cell division and to identify live quiescent stem cells in a wide range of tissues and organs [11,12].
Here, we inserted the iH2B-GFP reporter into the “safe-harbor” AAVS1 locus in patient-derived GBM cell lines to track cell division during tumor expansion. We utilized a 3D organoid culture approach that recapitulates intratumoral heterogeneity [13] to isolate qGBM cells from their proliferative counterparts using doxycycline (Dox) pulse/chase paradigms. We found that qGBM cells exhibited self-renewal capacity comparable to pGBM cells, but higher resistance to temozolomide or radiation. Transcriptomic analysis revealed unique gene signatures in qGBM cells that that are linked to cell cycle control, metabolic adaptation, DNA repair/stress pathways, as well as extracellular matrix (ECM) interaction. Moreover, qGBM gene signatures also revealed a mesenchymal shift as a general feature of quiescent cell populations in both proneural and mesenchymal GBM subtypes. Bioinformatic analysis identified hypoxia and TGFβ signaling as potential niche factors, which were validated by functional assays. Finally, co-expression gene network analysis of TCGA patient data identified gene modules that are enriched for quiescence gene signatures but also associated with survival rate.
2. Materials and methods
2.1. GBM cells
GBM cell lines SD2 and SD3 have been established from resected tumor tissues in neural stem cell media in the laboratory Dr. Kesari at University of California, San Diego [14]. GBM cells were propagated as adherent cultures on laminin-coated dishes in Neurocult human NS-A proliferation media (Stemcell Technologies), containing 0·0002% Heparin, 10 ng/ml bFGF, and 20 ng/ml EGF. Cells were passaged by dissociation with Accutase (Gibco).
2.2. iH2B-GFP reporter knock-in
For knock-in of a doxycycline-inducible H2B-GFP reporter (iH2B-GFP) into the AAVS1 locus (gene symbol PPP1R12C), the targeting vector plasmid pAAVS1-Neo-M2rtTA-H2BGFP was assembled from plasmids pAAVS1-Neo-M2rtTA and pAAVS1-Puro-H2BGFP [15]. The CRISPR/Cas9 plasmid pX330-sgAAVS1 was generated by inserting oligonucleotides that target AAVS1 into the pX330 backbone [16]. Correct knock-in of the iH2B-GFP cassette at the AAVS1 locus destroys the AAVS1 sgRNA target site (Fig. S1a). Plasmids have been deposited at Addgene.org.
GBM cells were co-transfected with the targeting and CRISPR/Cas9 plasmids with Neon electroporation system (Invitrogen) using the following parameters: tip size: 100 μl; cell number: 2 × 106; DNA: 7 μg pAAVS1-Neo-CAG-M2rtTA-H2BGFP and 3 μg pX330-sgAAVS1; pulse settings: 3 pulses, 10 ms, and 1300 V. Cells were seeded after electroporation on laminin-coated 10 cm dishes, and selection with 150 μg/ml G418 (Gibco) was started 24 h after seeding. After 2 weeks, clones were picked for expansion and analysis by genomic PCR to confirm correct targeting (Fig. S1b).
2.3. 3D GBM organoids
3D GBM organoids were prepared as described [13]. Briefly, 1000 dissociated GBM cells were embedded in a Matrigel (BD Biosciences) droplet of 20 μl, and cultivated for 4 days in neural stem cell media (Neurocult NS-A proliferation media (human), Stemcell Technologies, with 0·0002% Heparin, 10 ng/ml bFGF, and 20 ng/ml EGF) in 6 cm dishes. Cultures were then transferred for further growth over several weeks onto an orbital shaker inside a tissue incubator. For +Dox pulsing, doxycycline (MP Biomedicals) was added to the culture media for a final concentration of 1 μg/ml.
2.4. Immunofluorescence and immunohistochemistry
The following primary antibodies were used for immunofluorescence (IF) or immunohistochemistry (IHC):
anti-CD31 (host species: rat), BD biosciences 553,370, 1:300 for IF, RRID:AB_394816;
anti-CD44 (mouse), Thermo MS-668, 1:100 for IF, RRID:AB_2335741;
anti-Collagen, Type IV (mouse), Sigma-Aldrich C1926, 1:100 for IF, RRID:AB_476828;
anti-FN1 (rabbit), EMD Millipore AB2033, 1:100 for IF/IHC, RRID:AB_2105702;
anti-HIF1A (rabbit), Cell Signaling Technologies 36,169, 1:100 for IF;
anti-Ki67 (rabbit), abcam ab15580, 1:500 for IF, RRID:AB_443209;
anti-Nanog (rabbit), abcam ab109250, 1:200 for IF, RRID:AB_10863442;
anti-Nestin (mouse), abcam ab6142, 1:200 for IF, RRID:AB_305313;
anti-OCT4 (mouse), abcam ab184665, 1:500 for IF;
anti-Olig2 (rabbit), EMD Millipore AB9610, 1:500 for IF, RRID:AB_570666;
anti-p57/Kip2 (rabbit), Sigma-Aldrich P0357, 1:200 for IF, RRID:AB_260850;
anti-PAX6 (mouse), abcam ab78545, 1:200 for IF, RRID:AB_1566562;
anti-SOX2 (rabbit), Stemcell Technologies 60,055, 1:200 for IF;
anti-SOX9 (mouse), abcam ab58191, 1:100 for IF, RRID:AB_945591;
anti-SPP1 (rabbit), proteintech 25,715, 1:100 for IF/IHC;
anti-TLX (rabbit), LifeSpan Biosciences LS-B4564, 1:100 for IF, RRID:AB_10796976;
anti-TNC (rabbit), EMD Millipore AB19011, 1:100 for IF/IHC, RRID:AB_2203804;
anti-Vimentin (phospho S55) (mouse), abcam ab22651, 1:100 for IF, RRID:AB_447222.
GBM organoids were fixed in 4% PFA/PBS at room temperature for 12 min, and then prepared for cryosectioning by successive overnight incubations in 12·5% and 25% sucrose/PBS at 4 °C. Cryosections were cut at a thickness of 12 μm and stored on glass slides at −20 °C. For IF staining, sections were blocked for 1 h (blocking buffer: PBS with 5% donkey serum and 0·3% Triton X-100), then incubated overnight with primary antibodies in antibody dilution buffer (PBS with 1% BSA and 0·3% Triton X-100), followed by staining with Alexa-labeled secondary antibodies (Jackson ImmunoResearch) for 2 h, and counterstaining with DAPI (Invitrogen). Sections were washed in PBS and mounted with Fluoromount G (Southern Biotech). For IHC of tissue microarrays (TMA), arrays from series BS17016c (US Biomax) containing cores from glioblastoma, astrocytoma, and normal brain were analyzed. TMAs were subjected to antigen retrieval with Basic Antigen Retrieval Reagent (R&D Systems), immunostained with primary antibodies and DAB kit (R&D Systems), and counterstained with Gill's #2 hematoxylin (Thermo Scientific).
2.5. Flow cytometry and FACS
GBM cells grown as 2D cultures on laminin-coated dishes were dissociated with Accutase and resuspended in FACS buffer (Hibernate-E low fluorescence (BrainBits) with 0·2% BSA and 20 μg/ml DNase (Worthington)). GBM cells of 3D organoids were first minced and incubated in Accutase with DNase (25 μg/ml) on a rotator for 30 min at RT, and then gently triturated with Pasteur pipettes of decreasing tip diameter. Cells were pelleted and resuspended in FACS buffer, and debris was removed by pelleting cells at 155 g for 6 min through a phase of 2% BSA in Hibernate-E low fluorescence buffer. Cells were resuspended in FACS buffer and passed through a 70 μm mesh filter. DAPI (Invitrogen) was added to cell suspensions at a concentration of 5 μg/ml to stain dead cells. Cell suspensions were analyzed by flow cytometry (BD LSRII) or sorted by FACS (BD FACSAria IIu), and data was evaluated with FACSDiva and FlowJo software.
2.6. Gliomasphere assay
Sphere forming potential of GBM cells was analyzed by extreme limiting dilution analysis (ELDA) [17]. Briefly 1, 5, 10, or 50 cells were seeded in wells of 96 well low-attachment plates, and after 10 days of culture the wells containing at least one sphere were scored as positive. Quantitative analysis of sphere formation frequency was performed at http://bioinf.wehi.edu.au/software/elda. Size of gliomaspheres was quantified by measuring diameter of spheres in wells with 10 seeded cells after 10 days of culture.
2.7. Irradiation and temozolomide treatment of GBM organoids
About 10–12 organoids were treated as a group in a 6-cm dish and used for XRT irradiation with 5 Gy in an X-RAD 320 device (Precision X-Ray). At 48 h after XRT, GBM organoids were dissociated and GFPhigh and GFPlow populations were quantified by flow cytometry. For temozolomide (TMZ) treatment, 10–12 organoids were treated with 250 μM TMZ (Sigma-Aldrich) or vehicle dimethyl sulfoxide (DMSO) in neural stem cell media for 5 days. GBM organoids were dissociated and GFPhigh and GFPlow populations were quantified by flow cytometry.
2.8. Functional quiescence assay with hypoxia and TGFβ inhibitor
About 10–12 organoids were treated as a group in a 6-cm dish and used for functional assays. For hypoxia studies, organoids were placed for one week either in a hypoxic chamber (Biospherix) with a setting of 3% oxygen or in a regular incubator with ~21% environmental oxygen level (control condition). For TGFβ inhibition, organoids were cultured for one week in presence of inhibitor SB-431542 (Selleckchem) at a concentration of 2 μM or with DMSO as vehicle control. Organoids were dissociated and GFPhigh and GFPlow populations were quantified by flow cytometry.
2.9. RNA-Seq
Total RNA from FACS sorted GFPhigh and GFPlow GBM cells was isolated with RNeasy (Qiagen), and cDNA libraries for Illumina next-generation sequencing were prepared with NEBNext Ultra Directional RNA Library Prep Kit (NEB E7420) for 2 week chase samples, or with NuGEN Ovation amplification followed by NEBNext Ultra DNA Library Prep Kit (NEB 7370) for 4 week chase samples. Sequencing was performed on Illumina HiSeq2500 devices in rapid run mode with paired 50 bp reads. The RNA-Seq data has been deposited at the NCBI Gene Expression Omnibus (GEO) under accession number GSE114574.
2.10. Nanostring platform
For NanoString gene expression analysis, the gene panel nCounter PanCancer Progression was used (catalog #XT-CSO-PROG1–12), which contains probes for 740 test genes and 30 housekeeping genes. RNA was purified with RNAeasy (Qiagen) from FACS sorted GFPhigh and GFPlow cells and analyzed with the nCounter platform. The nSolver software 2.0 was used for analysis of NanoString gene expression values and for principal component, fold change heatmap, and Pathway Score analysis.
2.11. Bioinformatics
For processing of RNA-Seq raw data, quality of sequencing reads was assessed using fastQC [18]. Reads were mapped to the human genome (hg19) and human rRNA sequences with ContextMap version 2.7.9 [19] (using BWA [20] as short read aligner and default parameters). Number of read fragments per gene was determined from the mapped RNA-Seq reads using featureCounts (strand-specific for stranded libraries, non-strand-specific otherwise) [21] and Ensembl (v75) annotations. Differential gene expression analysis was performed using edgeR [22]. P-values were adjusted for multiple testing using the method by Benjamini and Hochberg [23] and genes with an adjusted p-value <0·01 were considered significantly differentially expressed. The RNA-Seq analysis workflow was implemented and run using the workflow management system Watchdog [24]. PCA analysis of RNA-Seq data was performed on batch-corrected log2 (x + 1) transformed FPKM data using the R statistical software environment.
Gene ontology analysis of common DEG (significance cut off p < 0·01) was performed with the ENRICHR resource (http://amp.pharm.mssm.edu/Enrichr/; accessed 07/2017) [25], and results were ranked by combined score (adjusted p-value multiplied by z-score); adjusted p-values are indicated by asterisks in bar graphs.
Gene set enrichment analysis (GSEA) using all genes ranked by their differential expression as input was performed for Hallmark gene sets (tool GSEAPreranked; enrichment statistic: classic; http://software.broadinstitute.org/gsea/index.jsp; accessed 07/2017) [26]. GSEA with gene sets related to proneural-mesenchymal transition was performed with gene sets for GBM subtypes from ref. (27) (MSigDB M2121, M2122, M2116, M2115) and ref. (28) (table S3 therein), a gene set for PMT and multitherapy resistance from ref. (29) (Fig. 5 therein), and a gene set for mesenchymal transition and tumor invasion from ref. (30) (MSigDB M M2572). The assembled GSEA gene set matrix file that was used for this analysis is included as supplemental data File_S1_PMT.gmx.
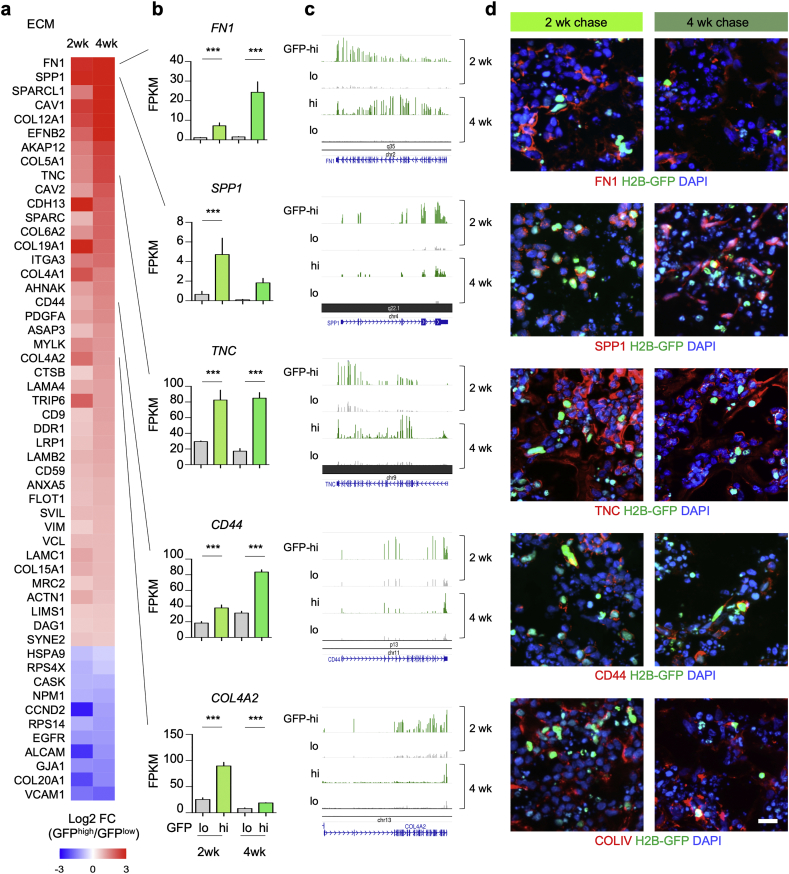
Fig. 5.
Expression of ECM associated genes in GBM organoids.
a) Heatmap of expression changes of DEGs associated with ECM interaction in SD3 qGBM relative to pGBM cells from 2 and 4 week -Dox chase organoids.
b-d) Selected qGBM DEGs associated with ECM interaction denoted in (a) are shown by absolute transcription levels (b), RNA-Seq read coverage tracks (c), and immunofluorescence images in GBM organoids at 2 or 4 week -Dox chase (d). Data were combined from three independent experiments per paradigm. Statistical analysis was performed using edgeR analysis of read counts, with Benjamini-Hochberg correction. SPP1 gene expression was not detectable in GFPlow cells at 4 week -Dox chase, hence no p-value could be calculated.
Scale bar: 20 μm (d).
Ingenuity pathway analysis for upstream regulators was performed with DEG lists of 2 week and 4 week -Dox chase paradigms (IPA; https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/; accessed 01/2018).
For network analysis of TCGA glioma data, level 3 GBM/LGG TCGA RNA-seqV2 normalized count data was downloaded from the Broad Institute GDAC Firehose (https://gdac.broadinstitute.org). Recurrent tumors were removed, and data was log2 (x + 1) transformed, quantile-normalized, and corrected for age, gender, and batch. Co-expression network analysis was performed using the MEGENA R package [31] with Spearman correlations. Enrichment of MEGENA modules for qGBM signature was assessed using Fisher's Exact Test. Functional annotation of modules was carried out in the R statistical computing environment using a one-sided Fisher's Exact Test for overrepresentation of the MSigDB v6·1 gene sets (Hallmark, Canonical Pathways, Gene Ontology) [32] and WikiPathways (release 03/10/2018) [33], with a significance threshold of Benjamini-Hochberg p ≤ 0·05. Survival analysis was performed using the Survival R package using the log-rank test and Cox regression. All p-values were adjusted using Benjamini-Hochberg correction. Modules were subsequently ranked by the sum of the following -log10 (adjusted p-values): enrichment for qGBM DEG 2 week up, qGBM DEG 2 week down, qGBM DEG 4 week up, qGBM DEG 4 week down, and Cox and log-rank tests for association of module first principal component (PC1) with survival. For module ranking and visualization, module survival p-values below 1e-15 were set to 1e-15 to match the scale of qGBM DEG enrichment p-values. PubMed queries of MEGENA hub genes from modules of interest were performed using the RISmed and bayesbio R packages.
Evaluation of TCGA glioma patient gene expression data and survival analysis was performed with the TCGA GBM-LGG dataset at the web platform GlioVis (http://gliovis.bioinfo.cnio.es/; accessed 01/2019) [34]. For Kaplan-Meier survival analysis, median gene expression was selected as cutoff to split high and low expresser populations.
Cell lines were classified for transcriptional subtypes with the SubtypeME function of GlioVis. As input for SubtypeME, we prepared normalized read counts of RNA-Seq data of GBM cell lines SD2 and SD3 (obtained from three independent replicates for each line, cultured on laminin as proliferating cultures). Raw read counts were first filtered to keep genes containing non-zero counts in at least one sample. Counts were then normalized by library size using the edgeR R package and log2 transformed with an added offset of 0·5. The log2 transformed expression values were then quantile normalized using the limma R package. We applied the 3-Way analysis function of SubtypeME, which uses three algorithms for subtype calculation (support-vector machine, k-nearest neighbors, and single sample gene set enrichment) and calculates a majority call for subtype prediction.
2.12. Statistical analysis
For gliomasphere size analysis, boxplots were created to check data distribution. The boxplots show the 25th, 50th, and 75th percentiles of the sphere diameters and whiskers represent the min and max values. The dots are values of sphere diameters. The distribution was found to be skewed so Mann-Whitney tests were used to compare sphere diameters. The Mann-Whitney tests were conducted using GraphPad Prism 7. Because this was an exploratory experiment, the p-values from the Mann-Whitney tests were not adjusted for multiple comparisons [35].
To analyze therapy resistance of quiescent GBM cells, mixed effect models with beta distribution and logit link [36,37] were fit for each chase time and treatment (TMZ and XRT). The dependent variables in the models were the proportion of GFPhigh cells. The models included treatment group (treatment and control) as the fixed effects and sample ID as the random effect. The mixed effect beta regression models were fit using PROC GLIMMIX procedure in SAS 9.4. Because this was an exploratory experiment, p-values calculated from the models without multiple comparisons were provided [35].
The significance levels for the statistical analysis mentioned above were all set as 0·05. All bar graphs represent mean values and error bars the standard error of the mean. * p-value <0·05, ** < 0·01, *** < 0·001.
3. Results
3.1. Engineering GBM cells with iH2B-GFP reporter
To track cell division during GBM expansion, we carried out CRISPR/Cas9-assisted knock-in of a Dox-inducible H2B-GFP (iH2B-GFP) reporter into the human AAVS1 locus, which serves as a safe harbor for transgene expression [15] (Fig. 1a; Fig. S1a). The iH2B-GFP reporter enables labeling of cells with stable, chromatin-bound H2B-GFP during +Dox pulsing, and then dilution of the H2B-GFP label by cell division during -Dox chase periods (Fig. 1b, c). We engineered the iH2B-GFP reporter into patient-derived GBM cell lines that had been established in neural stem cell culture media, which are conditions that preserve critical features of GBM pathophysiology [[38], [39], [40]]. After co-electroporation of GBM cells with a CRISPR/Cas9 plasmid targeting AAVS1 and a donor plasmid with the iH2B-GFP cassette, targeted GBM cell lines were isolated by G418 drug selection and screened by genomic PCR for correct donor integration (Fig. S1b). We selected two GBM cell lines for further in-depth analysis, i.e. SD3-iH2B-GFP (parental line SD3 classified as IDH1/2 wild-type GBM cell line, proneural molecular subtype, with elevated PDGFRα expression), and SD2-iH2B-GFP (parental line SD2 characterized as IDH1/2 wild-type, mesenchymal subtype, with elevated EGFR and MET expression) (Fig. S2).
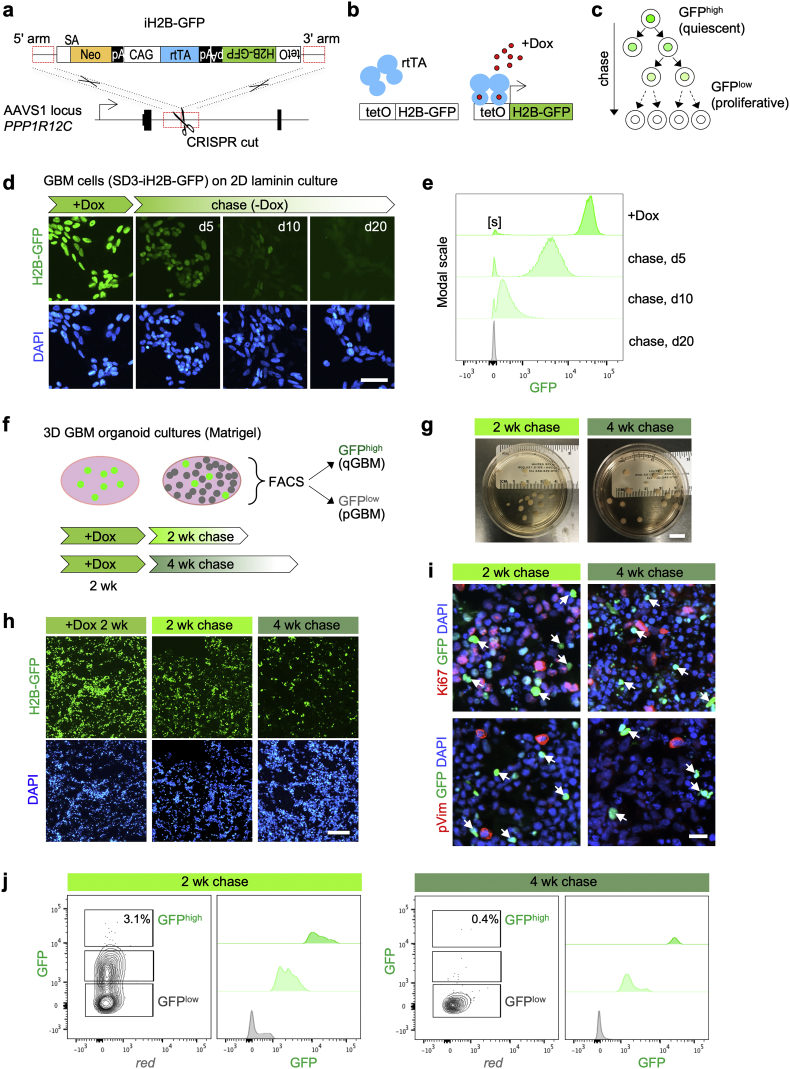
Fig. 1.
Tracking cell division with iH2B-GFP reporter identifies quiescent population in GBM organoids.
a) Targeting strategy for iH2B-GFP reporter knock-in by CRISPR-assisted homologous recombination into AAVS1 locus (gene symbol PPP1R12C). SA: splice acceptor; Neo: Neomycin resistance gene; pA: poly-adenylation signal; CAG: CAG promoter; rtTA: reverse tetracycline-controlled transactivator; H2B-GFP: histone2B-green fluorescent protein; tetO: tet operator.
b) Principle of doxycyline (Dox)-inducible expression of H2B-GFP.
c) Schematic depiction of divisional dilution of H2B-GFP label during -Dox chase period. Quiescent cells retain H2B-GFP label (GFPhigh), while proliferative cells dilute the label (GFPlow).
d) GBM cell line SD3-iH2B-GFP grown as proliferative culture on 2D laminin-coated dishes. In the presence of doxycycline (+Dox), nuclei are uniformly labeled with H2B-GFP. Cells dilute H2B-GFP label during -Dox chase periods (5, 10, and 20 days shown) by cell division. DAPI is used for nuclear counter staining.
e) Flow cytometry analysis of SD3-iH2B-GFP cells grown on 2D laminin for the indicated -Dox chase periods. A small fraction of SD3-iH2B-GFP cells remained GFP-negative even in +Dox conditions (denoted as “[s]”), possibly due to sporadic silencing of transgene. Histograms are normalized on y-axis to modal scale (FlowJo).
f) Experimental design for isolation of quiescent GBM cells from 3D GBM organoids. GBM organoids are generated by seeding cells in Matrigel droplets and expanding them as floating cultures. After growth for 2 weeks with +Dox pulse, organoids are chased for 2 or 4 weeks in -Dox conditions. Dissociated cells are separated into GFPhigh and GFPlow populations by FACS.
g) Images of GBM organoids in culture dishes, after 2 or 4 week -Dox chase periods.
h) Fluorescence images of sections of 3D GBM organoids show a declining number of label-retaining GFPhigh cells during organoid expansion.
i) Immunofluorescence images show absence of proliferation markers Ki67 and phospho-Vimentin (pVim) in GFPhigh cells (arrows), confirming slow dividing nature of GFPhigh cells. Notice debris from dead cells accumulates during organoid culture, which is more prominent after 4 week chase.
j) Representative FACS results of GBM organoids analyzed after 2 or 4 week -Dox chase. After 2 week -Dox chase, 3·1% of cells remained GFPhigh; after 4 week chase, only 0·4% of cells remained GFPhigh. Three independent experiments (10–12 pooled organoids per experiment) yielded similar results. X-axis in left histograms shows red auto-fluorescence of cells. Histograms are normalized on y-axis to modal scale (FlowJo).
Scale bars: 50 μm (d), 10 mm (g), 200 μm (h), 20 μm (i).(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
Uniform labeling and subsequent divisional dilution of H2B-GFP during +Dox pulse and -Dox chase periods were confirmed by fluorescence microscopy and flow cytometry in both SD3-iH2B-GFP and SD2-iH2B-GFP lines grown as proliferating cultures on laminin-coated dishes (Fig. 1d, e; Fig. S1c; S3a, S3b). Distribution of flow cytometry peaks indicated that the iH2B-GFP reporter can track up to 9 cell divisions (Fig. S1d). Of note, we observed silencing of the reporter in a small population (<5%) of SD3-iH2B-GFP cells (Fig. 1e; denoted by “[s]”), in line with earlier reports on sporadic silencing at ASSVS1 locus [41]. On the other hand, no leaky (Dox-independent) expression of the iH2B-GFP reporter was observed, even after long -Dox chase periods (e.g. 20 days; Fig. 1d, e).
3.2. Quiescent cell populations in 3D GBM organoids
To characterize quiescent GBM cell populations, we utilized a 3D organoid culture approach in which GBM cells are expanded in floating Matrigel droplets [13]. GBM organoids mimic patient tumors, containing hypoxic gradients and cellular heterogeneity with both proliferative tumor bulk and slow-dividing stem-like cells [13]. We first generated 3D GBM organoids with the SD3-iH2B-GFP line, and tracked cell division using the paradigms of 2 week +Dox pulse followed by 2 or 4 week -Dox chase (Fig. 1f, g). During the chase period, we found decreasing number of H2B-GFP label-retaining (GFPhigh) qGBM cells as a result of cell divisions (Fig. 1h). Immunofluorescence for proliferation markers Ki67 and phospho-Vimentin showed absence of these markers in GFPhigh cells (Fig. 1i), confirming their relative quiescence. Quantification by flow cytometry revealed that the fraction of GFPhigh cells in GBM organoids was in the range of 2–5% after 2 week chase, with a substantial population of cells still labeled by intermediate levels of GFP, indicating ongoing proliferation (Fig. 1j). The fraction of GFPhigh cells decreased further to 0·2–1% after 4 week chase (Fig. 1j), indicating that qGBM cells continue to divide slowly between 2 and 4 week -Dox chase in GBM organoids. The SD2-iH2B-GFP line formed GBM organoids with a quiescent subpopulation in a similar manner (Fig. S3c, S3d), confirming general suitability of iH2B-GFP reporter and 3D GBM organoid approach for tracking quiescent GBM cell populations.
3.3. Quiescent GBM cells exhibit self-renewal capacity
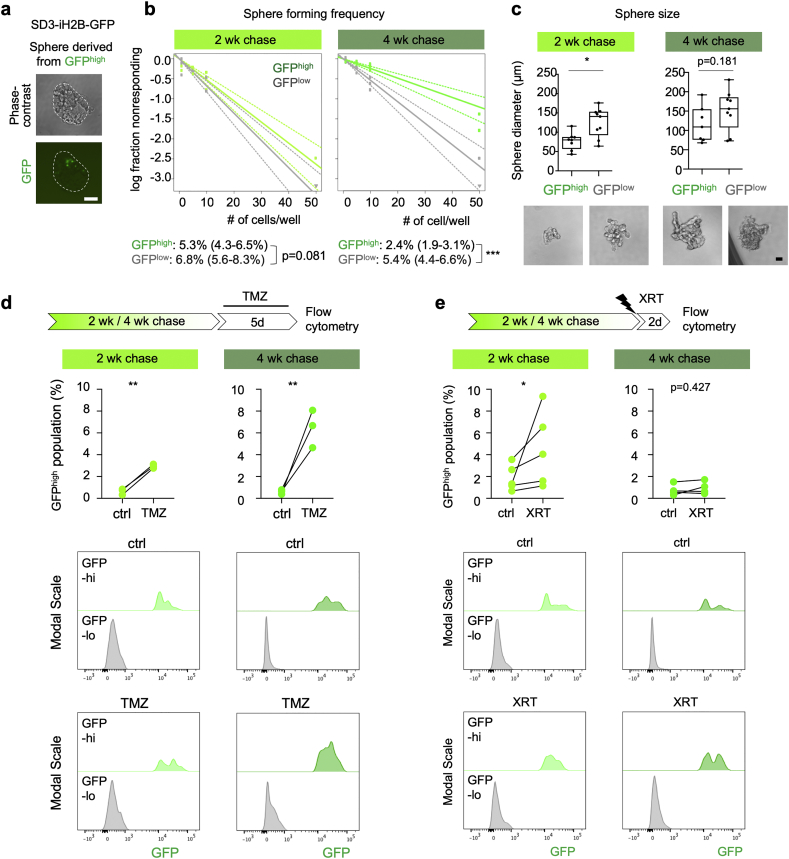
The quiescent population in GBM may contain either dormant stem cells that can be reawaken to enter cell cycle or terminally differentiated cells without the potential to spawn tumor re-expansion. To assess self-renewal capacity of qGBM cells, we separated GFPhigh and GFPlow populations from SD3-iH2B-GFP GBM organoids by FACS and compared sphere-forming potentials using a limiting dilution assay. Both GFPhigh and GFPlow populations formed gliomaspheres, indicating that the qGBM populations are not all terminally differentiated, but indeed harbor stem-like cells that can re-enter cell cycle under new environmental cues. Consistent with a high plasticity to switch back to a proliferative state, we observed that only a small fraction of GFPhigh cells retained the GFP label after 10 days in sphere culture condition, while a majority of GFPhigh cells diluted the H2B-GFP label after multiple rounds of cell division (Fig. 2a). The qGBM cells isolated after 2 week -Dox chase had an estimated effective sphere-forming rate of 5·3%, only slightly less than the 6·8% rate in pGBM cells (Fig. 2b). On the other hand, that rate dropped to 2·4% for qGBM cells isolated from 4 week chase organoids as compared to the 5·4% rate for pGBM (Fig. 2b). Sphere sizes were also measured as an indicator of proliferative rate, which revealed that the spheres derived from qGBM cells were on average smaller than those from pGBM (Fig. 2c), which may reflect the prior slow-dividing state of qGBM cells, thus the time needed to reawake from quiescent state. Similar results were obtained with GFPhigh (qGBM) cells from SD2-iH2B-GFP GBM organoids, with SD2 qGBM and pGBM populations displaying comparable sphere forming frequency and sphere size (Fig. S3e, S3f). Together, our finding that both qGBM and pGBM populations were able to spawn gliomaspheres indicates that quiescence does not simply equate stemness, but rather represents a specific cellular state.
Fig. 2.
Quiescent GBM cells exhibit self-renewal capacity and enhanced therapy resistance.
a) Phase-contrast and fluorescence images of a representative gliomasphere derived from GFPhigh cells after 10 day culture show that most previously GFPhigh cells have diluted their H2B-GFP label by cell divisions.
b) Limiting dilution sphere formation assay with GFPhigh and GFPlow cells. FACS-sorted cells from GBM organoids after 2 or 4 week -Dox chase were seeded in limiting dilutions in 96 well plates. Sphere forming frequency was quantified after 10 days by ELDA software. Results were generated from three independent experiments. Numbers in parenthesis represent 95% confidence interval. Statistical analysis was performed with ELDA software. ***, p < 0·001.
c) Sphere sizes and representative images of gliomaspheres derived from GFPhigh or GFPlow cells after 10 days of culture, with 10 cells seeded per well. Ten spheres per group were measured. Boxplots show 25th, 50th, and 75th percentiles of sphere diameters and whiskers represent minimum and maximum values. Statistical analysis was performed using Mann-Whitney test. *, p < 0·05.
d) Drug sensitivity assay for temozolomide (TMZ). Organoids after 2 or 4 week of –Dox chase were treated for 5 days with vehicle or 250 μM TMZ, and the fraction of live GFPhigh cells in organoids was quantified by flow cytometry. Quantification (middle panel) and flow cytometry histograms (bottom panels, y-axis normalized to modal scale) show that a higher proportion of surviving cells were GFPhigh in TMZ-treated organoids as compared to vehicle treatment (ctrl). Data were obtained from three independent experiments (10–12 pooled organoids per experiment). Significance was evaluated by binomial generalized linear mixed effect models. **, p < 0·01.
e) Radiation sensitivity assay. Organoids after 2 or 4 wk. chase were irradiated with one dose of 5 Gy, and 2 days later the fraction of live GFPhigh cells in organoids was quantified. Quantification (middle panel) and flow cytometry histogram (bottom panels) show a larger fraction of GFPhigh cells after XRT-treatment as compared to control (ctrl) in the 2 week chase paradigm, and a trend in the 4 week chase paradigm. Data represents five and four independent experiments for 2 week and 4 week chase, respectively. Significance was evaluated with binomial generalized linear mixed effect models. *, p < 0·05.
Scale bars: 50 μm (a, c).
3.4. Enhanced therapy resistance of quiescent GBM cells
A salient feature of GBM is tumor recurrence, driven by a therapy-resistant population with tumor initiating potential. We therefore tested whether qGBM cells display enhanced resistance relative to pGBM cells against temozolomide (TMZ), a DNA-alkylating agent that is the primary chemo drug for GBM, or against radiation therapy (XRT). For TMZ treatment, GBM organoids after 2 or 4 week -Dox chase (thus containing an established qGBM population) were cultured with or without TMZ for 5 days, and surviving cells were quantified by flow cytometry for GFP fluorescence levels. As expected, the TMZ treatment caused massive cell death in GBM organoids over the 5-day treatment, and changes in the relative proportion of GFPhigh population in the surviving cells would mostly reflect differential survival rates between qGBM and pGBM populations. Indeed, we found that the fraction of GFPhigh cells from 2 week chase organoids was <1% in the vehicle-treated group, but ~3% in the TMZ-treated group. As for the 4 week chase organoids, the fraction of GFPhigh cells was about 0·5% in vehicle-treated group, but 4–8% in TMZ treated group (Fig. 2d). Of note, TMZ treatment may slow down proliferation, leading to slight increase of GFP label retention; however, the close to 10-fold increase of the fraction of GFPhigh in TMZ-treated group in 4 week chase organoids favors the interpretation that qGBM cells exhibit higher TMZ resistance.
For XRT resistance studies, we irradiated GBM organoids with a single dose of 5 Gy and performed flow cytometry 2 days later. Similar to TMZ, the XRT treatment caused significant cell death in the organoids. We found that the fraction of GFPhigh cells was more than two-fold higher in organoids from the 2 week chase group after XRT as compared to control, and about 1·25-fold higher after XRT in organoids from the 4 week chase group (Fig. 2e). A second GBM cell line, SD2-iH2B-GFP, similarly demonstrated a trend for higher resistance of qGBM cells against TMZ and XRT (Fig. S3g, S3h).
3.5. Gene expression prolife of quiescent GBM cells reveals mesenchymal shift
To identify specific gene expression programs in qGBM cells, we performed next-generation RNA sequencing (RNA-Seq) of GFPhigh and GFPlow populations isolated by FACS from SD3-iH2B-GFP GBM organoids. Principal component analysis (PCA) of the transcriptome data showed that the replicates of each experimental group clustered closely to each other along axis PC1, indicating reproducibility of the experimental conditions (Fig. 3a). GFPhigh samples from both 2 and 4 week chase organoids clustered on one side of the PCA plot, opposite of GFPlow counterparts, indicating that qGBM cells express unique gene signatures distinct from pGBM cells (Fig. 3a). An example of RNA-Seq read coverage tracks for a differentially expressed gene (IGFBP3) between qGBM and pGBM cells is shown in Fig. 3b.
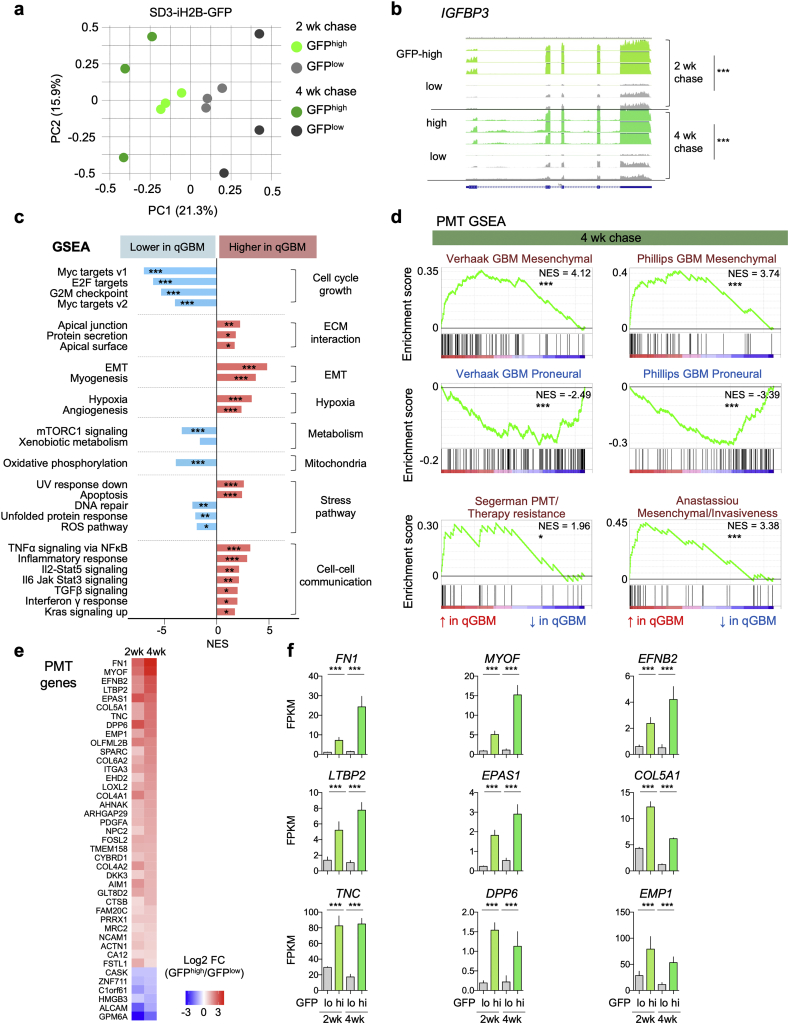
Fig. 3.
Quiescent GBM cells express unique gene signature with proneural-mesenchymal shift.
a) Principal component analysis (PCA) of RNA-Seq gene expression profiles from SD3 GFPhigh or GFPlow populations after 2 or 4 week -Dox chases. Each paradigm was replicated with three independent experimental samples.
b) RNA-Seq coverage tracks for differentially expressed gene IGFBP3 in GFPhigh and GFPlow populations after the indicated -Dox chases (track scales normalized by GAPDH expression). Statistical analysis was conducted with edgeR analysis of read counts, with Benjamini-Hochberg correction. *** indicates p < 0·001.
c) Gene set enrichment analysis (GSEA; Hallmark gene sets) of gene expression changes in SD3 GFPhigh vs. GFPlow populations (data from 4 week chase organoids shown). NES, normalized enrichment score. Significance calculated by GSEA for False discovery rate (FDR)-adjusted q-value. *, ** and *** indicate q < 0·05, q < 0·01 and q < 0·001, respectively.
d) GSEA results of the gene sets that are related to GBM transcriptional subtypes suggest proneural-mesenchymal transition (PMT) in SD3 GFPhigh cells (data from 4 week chase organoids shown). NES, normalized enrichment score. *, and *** indicate FDR-adjusted q < 0·05, and q < 0·001, respectively.
e) Heatmap of expression changes of PMT genes that are differentially expressed in SD3 GFPhigh cells relative to GFPlow cells from 2 wk and 4 wk chase organoids.
f) Absolute expression levels of the top differentially regulated PMT genes in SD3 GFPhigh or GFPlow populations in 2 and 4 week -Dox chase organoids. Data obtained from three independent experiments. Bars represent mean FPKM values and error bars represent standard error of the mean. Significance was evaluated using edgeR analysis of read counts, with Benjamini-Hochberg correction. *** indicates p < 0·001.
We next assessed global gene expression changes in qGBM relative to pGBM cells by Gene Set Enrichment Analysis (GSEA). The GSEA algorithm evaluates expression changes of all detectable genes for enrichment of specific gene sets [26]. We found that the top negatively enriched GSEA Hallmark gene sets in qGBM cells were associated with cell cycle and growth (e.g., MYC targets, E2F targets, and G2M checkpoint), and metabolic state (e.g., Oxidative Phosphorylation and mTORC1 signaling) (Fig. 3c; Fig. S4a). This is consistent with the slow-dividing nature and lower energy consumption state of qGBM cells. In addition, stress pathway gene sets (e.g. DNA repair and Unfolded protein response) were also downregulated in qGBM cells (Fig. 3c). The top positively enriched gene sets in qGBM relative to pGBM cells for both 2 wk. and 4 wk. -Dox chase were Epithelial mesenchymal transition (EMT), Myogenesis, Hypoxia, and several cell-cell communication pathways such as TGFβ, TNFα via NFκB, and IL6/JAK/Stat3 signaling (Fig. 3c; Fig. S4a). UV response down and Apoptosis gene sets were also significantly enriched in qGBM cells.
As gliomas are not epithelial tumors, the process of EMT is not directly applicable to GBM. A related scenario has been described as proneural-mesenchymal transition (PMT), i.e. a shift from a proneural GBM subtype towards a mesenchymal GBM subtype [28,29,42,43]. Notably, PMT has also been associated with increased therapy resistance and slower cell cycling for GBM cells [29,42]. Indeed, when we performed GSEA for gene sets of PMT-related signatures based on transcriptome of qGBM cells from SD3-iH2B-GFP organoids, we found that mesenchymal subtype signatures were positively enriched, while proneural subtype gene signatures were negatively enriched (Fig. 3d; Fig. S4b). In addition, other mesenchymal features that are associated with PMT, such as multitherapy resistance [29] and invasiveness [30] were also positively enriched in qGBM cells. We further analyzed PMT genes that were significantly regulated in qGBM cells from both 2 and 4 wk. chase paradigms, and filtered for those above minimal threshold for fold changes and absolute expression levels, and we found that a majority of these PMT genes was upregulated in qGBM cells (Fig. 3e, f).
3.6. Differential expression of ECM-associated genes in quiescent GBM cells
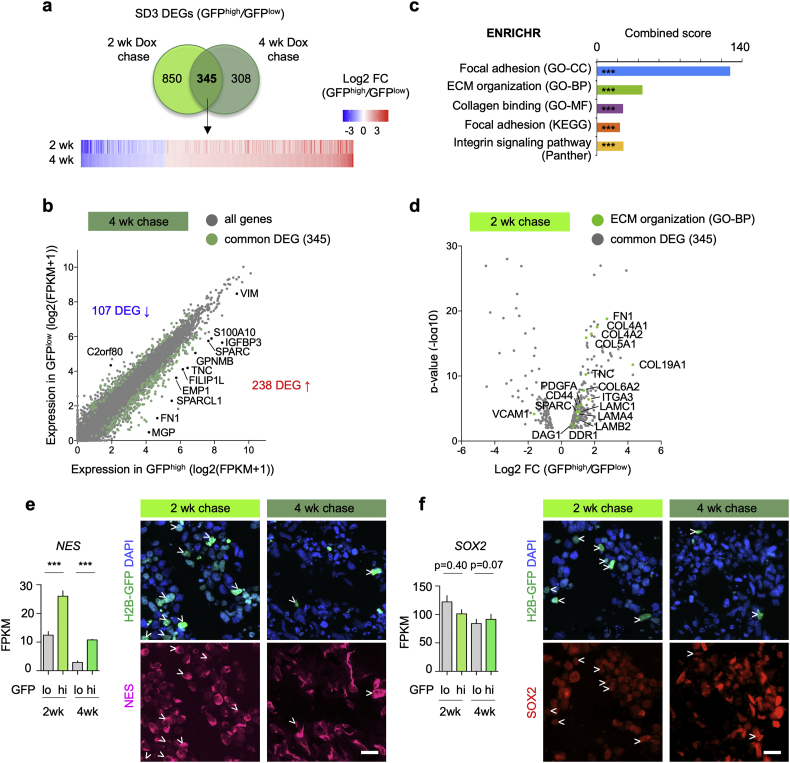
We next analyzed differentially expressed genes (DEGs) between qGBM and pGBM in SD3-iH2B-GFP organoids, and detected 1195 DEGs after 2 week chase and 653 DEGs after 4 week chase (Fig. 4a). Intersection of the two sets of DEGs revealed 345 common DEGs (Fig. 4a; Table S1), which we further focused on for in depth analyses. Among these common DEGs, 238 were upregulated in qGBM relative to pGBM cells, and 107 downregulated (Fig. 4b). We performed gene ontology (GO) and pathway analysis of the common DEGs with the ENRICHR platform [25], which revealed a predominant enrichment for genes ontologies and pathways that are associated with ECM components and interactions, e.g., Focal adhesion, ECM organization, Collagen binding, and Integrin signaling (Fig. 4c; Fig. S5a). Notably, these genes were mostly upregulated in qGBM cells, e.g. FN1, tenascinC (TNC), several collagens (COL12A1, COL4A2, COL4A2, COL5A1, etc.), laminins (LAMC1, LAMA4), and ECM receptor integrin α3 (Fig. 4d; Fig. S5b). These findings suggest that quiescent GBM cells may be actively engaged in modifying their own niche ECM microenvironment.
Fig. 4.
qGBM cells engage ECM interaction.
a) Venn diagram illustrates overlap of differentially expressed genes (DEGs; significance cut-off of adjusted p < 0·01; filtered for protein-coding genes) in GFPhigh relative to GFPlow cells from SD3-iH2B-GFP GBM organoids at 2 and 4 week -Dox chase stages. Data were obtained from three independent experiments for each paradigm. Bottom, heatmap depicts fold-changes of transcription of 345 common DEG at 2 and 4 week -Dox chase stages.
b) Dot plot of genes by absolute expression levels in GFPhigh (x-axis) and GFPlow cells (y-axis; 4 week chase paradigm). DEGs shared by GFPhigh cells at 2 and 4 wk chase are marked by green dots. About twice as many DEGs were upregulated than downregulated in GFPhigh cells. Selected DEGs are labeled.
c) ENRICHR gene ontology (GO) and pathway analysis of common DEGs of SD3 qGBM cells reveals highest enrichment for ontologies/pathways associated with ECM interaction and ECM components (CC, Cellular Component; BP, Biological Pathway; MF, Molecular Function). Top result of each category is shown. X-axis indicates combined score as calculated by ENRICHR (adjusted p-value multiplied by z-score). *** indicates p < 0·001.
d) Volcano plot showing significantly regulated genes of GO term ECM organization (marked by green dots) among common DEGs (2 week chase organoid).
e, f) Gene expression graphs and immunofluorescence images of 3D GBM organoids for neural stem cell markers Nestin and SOX2. While Nestin (e) was upregulated in qGBM (GFPhigh) cells (arrowheads), SOX2 (f) was not significantly changed. Data were combined from three independent experiments per paradigm. Statistical analysis was performed using edgeR analysis of read counts, with Benjamini-Hochberg correction. *** indicates p < 0·001.
Scale bars: 20 μm (e, f). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We next surveyed expression changes of established stem cell markers in qGBM cells. Interestingly, the neural stem cell marker Nestin was consistently upregulated in GFPhigh cells in SD3-iH2B-GFP organoids at both 2 and 4 week chase stages. However, the expression of other stem cell marker such as SOX2, OLIG2, OCT4, PAX6, SOX9, CDKN1C (Kip2), NR2E1 (TLX), NANOG, or CD133 was not significantly changed in GFPhigh relative to GFPlow cells (Fig. 4e, f; Fig. S6). Similarly, no enrichment for the glioma stem cell markers A2B5, SSEA-1, or Integrin 6 was detected in qGBM cells (data not shown). These results indicate that quiescence in GBM does not simply equate stemness, but rather represents a distinct cellular feature.
We next assessed protein expression levels of ECM-related DEGs in GBM organoids by immunofluorescence (IF) (Fig. 5). We detected high IF intensity for fibronectin (FN1), tenascin C (TNC), and collagen IV (COLIV) adjacent to GFPhigh nuclei in GBM organoids at both stages of -Dox chase, although IF signals were sometimes also detected in areas further away from GFPhigh nuclei, which may reflect deposited matrix proteins by earlier generations of slow-dividing GBM cells. We also examined ECM receptor CD44 and one of its ligands, SPP1 (also known as Osteopontin), both of which showed elevated protein expression levels in cells with GFPhigh nuclei in both 2 and 4 week chase organoids (Fig. 5). Interestingly, CD44 and SPP1 have been shown to promote the GBM stem cell compartment [44].
3.7. TGFβ and HIF1A are potential upstream regulators of GBM quiescence
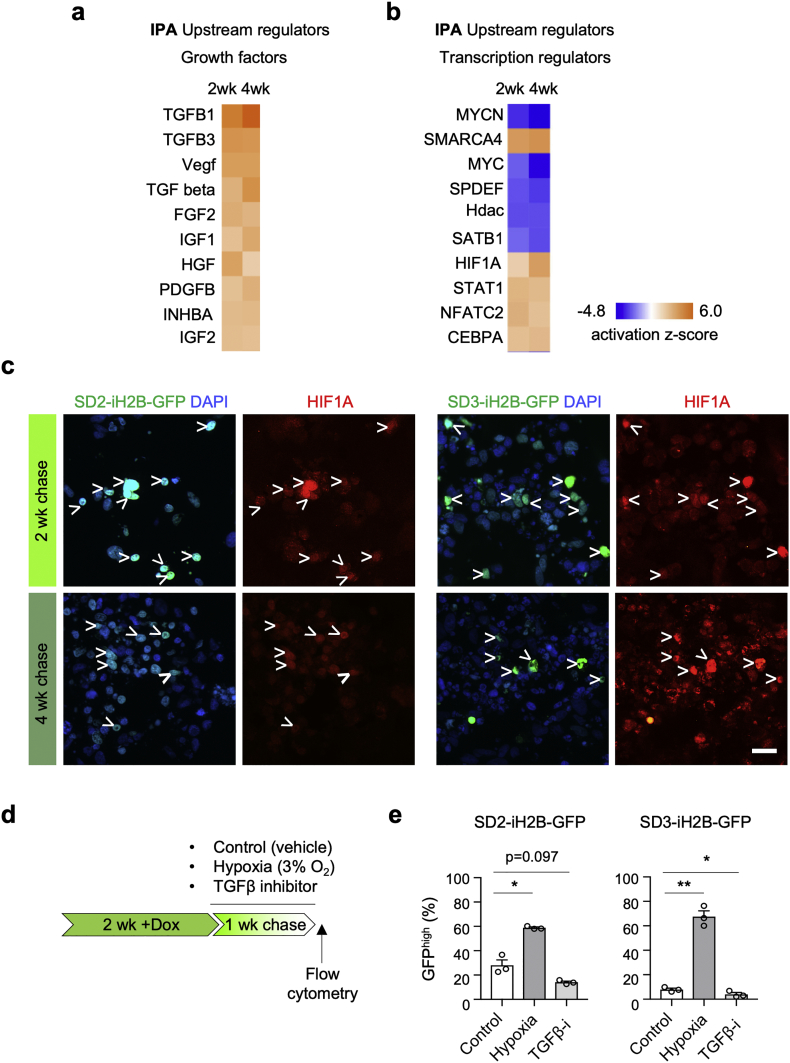
To identify factors that regulate DEG expression in qGBM cells, we applied Ingenuity Pathway Analysis (IPA) for Upstream Regulators to the transcriptome data of SD3 qGBM cells. Among the category Growth factors, several TGFβ family members were identified as top candidates (Fig. 6a), consistent with earlier reports that TGFβ can promote tumor dormancy [45] and induce an EMT-like gene program [46]. Among the category Transcriptional regulators, inactivation of MYCN and MYC were detected as top ranked candidates (Fig. 6b), echoing the GSEA result of downregulation of MYC targets (see Fig. 3c). Another transcription regulator with high activation score was the chromatin remodeler SMARCA4 (also known as Brg1), which has been implicated in EMT related processes [47]. In addition, hypoxia induced factor HIF1A had a high activation z-score in qGBM cells (Fig. 6b), which again echoes the GSEA result of an enriched Hypoxia gene set in qGBM cells. Moreover, the presence of immunoreactivity for HIF1A in SD2- and SD3-iH2B-GFP organoids confirmed hypoxia signaling in qGBM cells (Fig. 6c).
Fig. 6.
Functional assays confirm role of TGFβ and hypoxia in promoting GBM cell quiescence.
a, b) Upstream regulator analysis of common DEGs in SD3 qGBM cells with Ingenuity pathway analysis (IPA), filtered for growth factors (a) or transcriptional regulators (b). TGFβ family members were top candidate upstream growth factors for quiescence gene program. The terms Vegf, Tgf beta, and Hdac denote gene families with multiple members. MYC and MYCN were top transcriptional regulators with negative activation z-score.
c) Immunofluorescence staining for hypoxia-induced factor 1α (HIF1A) demonstrates hypoxia signaling in quiescent GBM cells (H2B-GFP+; arrowheads) of SD2- and SD3-iH2B-GFP organoids.
d) Experimental paradigm for functional intervention on GBM organoids during one week chase period.
e) Quantification of GFPhigh cells in SD2- or SD3-iH2B-GFP organoids after 1 week chase revealed that hypoxia (3% O2) increased ratio of qGBM cells. In contrast, treatment with TGFβ inhibitor SB-431542 (TGFβ-i) reduced ratio of qGBM cells. Data were combined from three independent experiments (10–12 pooled organoids per experiment). Significance was evaluated by paired t-test. * and ** indicate p < 0·05 and p < 0·01, respectively.
Scale bar: 20 μm (c).
3.8. Functional assays for validation of upstream regulators of quiescence
We took advantage of the GBM organoid system to functionally test TGFβ and hypoxia pathways as potential upstream regulators of GBM cell quiescence. Organoids from both SD2- and SD3-iH2B-GFP cells were pulsed with Dox for 2 weeks, followed by -Dox chase for one week under either hypoxic culture condition (inside a gas-controlled chamber) or in the presence of a TGFβ inhibitor (Fig. 6d). We found that hypoxia indeed increased the fraction of quiescent GFPhigh cells in organoids as compared to normoxic conditions in both lines (Fig. 6e). Conversely, the fraction of qGBM cells in organoids expanded in the presence of a TGFβ inhibitor was significantly reduced as compared to vehicle-treated controls (Fig. 6e). Together, these results support hypoxia and TGFβ signaling as niche factors promoting GBM quiescence.
3.9. NanoString gene expression analysis verifies quiescence gene signatures in diverse GBM subtypes
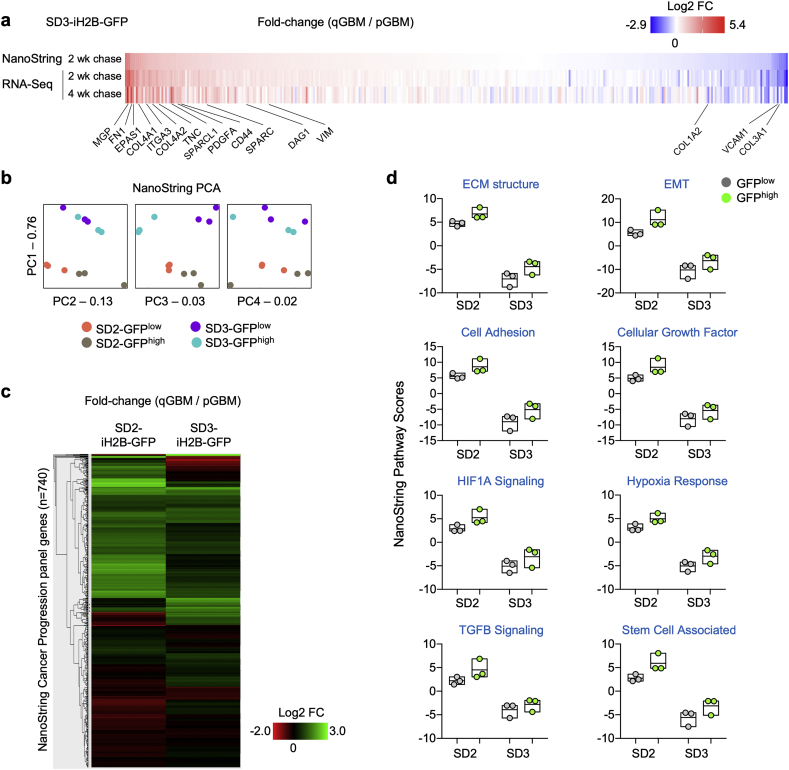
For comparative evaluation of gene expression programs in qGBM cells derived from different GBM molecular subtypes, we performed NanoString gene expression analysis of qGBM vs. pGBM cells isolated from both SD3-iH2B-GFP (proneural subtype) and SD2-iH2B-GFP (mesenchymal subtype) organoids after 2 weeks -Dox chase, utilizing the NanoString Pan-Cancer Progression panel platform that measures 740 genes involved in cancer progression. We first compared NanoString and RNA-Seq data of SD3 qGBM cells, which verified faithfulness of the NanoString platform (Fig. 7a). We then evaluated NanoString data by principal component analysis (PCA), which showed that SD2 and SD3 samples were clearly different from each other, as expected for different GBM subtypes (Fig. 7b). For both lines, NanoString data showed that the GFPhigh (qGBM) samples were separated from the GFPlow (pGBM) samples, indicating unique quiescence gene signatures (Fig. 7b). We next clustered gene expression changes in SD2 and SD3 qGBM cells relative to pGBM cells by heatmap analysis, which showed that majority of gene changes occurred in a common direction for both lines with some exceptions, which are expected for different GBM molecular subtypes (Fig. 7c).
Fig. 7.
Gene expression analysis of qGBM cells from different GBM subtype confirms mesenchymal shift in quiescent GBM cells of 3D organoids.
a) Heatmaps of gene expression changes of qGBM relative to pGBM cells in 3D organoids as measured by NanoString and RNA-Seq verified reliability of Nanostring platform as compared to RNA-Seq approach to measure gene expression changes. Selected common DEGs are labeled.
b) Principal component analysis (PCA) of NanoString gene expression profiles from SD2- and SD3-iH2B-GFP GFPhigh or GFPlow populations after 2 or 4 week -Dox chases (three independent experiments per paradigm).
c) Heatmap of gene expression changes of qGBM relative to pGBM cells in SD2 and SD3 organoids shows largely common patterns, with some differences between the two cell lines (note: SD2 cells are classified as mesenchymal GBM subtype, SD3 cells as proneural GBM subtype).
d) NanoString pathway score analysis of gene expression changes in SD2 and SD3 qGBM relative to pGBM cells reveals a common pattern of increased pathway scores for pathways associated with ECM, EMT, hypoxia, and TGFβ signaling. Each data point represents an independent experiment with 10–12 pooled organoids.
NanoString Pathway Score analysis was carried out for functional annotation of gene expression changes in both SD2 and SD3 qGBM relative to pGBM cells (Fig. 7d). In general, pathway score values were higher for SD2 cells (both qGBM and pGBM populations) than for SD3 cells, reflecting the underlying differences between mesenchymal and proneural GBM subtypes. Notably, for both SD2 and SD3 lines, pathway scores for ECM structure, EMT, and Cell Adhesion were increased in the same direction in qGBM relative to pGBM counterparts, indicating that qGBM cells undergo a general shift towards increased mesenchymal features in both GBM subtypes (Fig. 7d). Importantly, mirroring the results from RNA-Seq analysis of SD3 qGBM cells, NanoString pathway scores for HIF1A signaling, hypoxia and TGFβ pathways were also higher in qGBM than pGBM cells for both SD2 and SD3 lines, supporting them as potential niche factors for GBM quiescence (Fig. 7d).
3.10. Identification of qGBM gene signatures in TCGA GBM patients
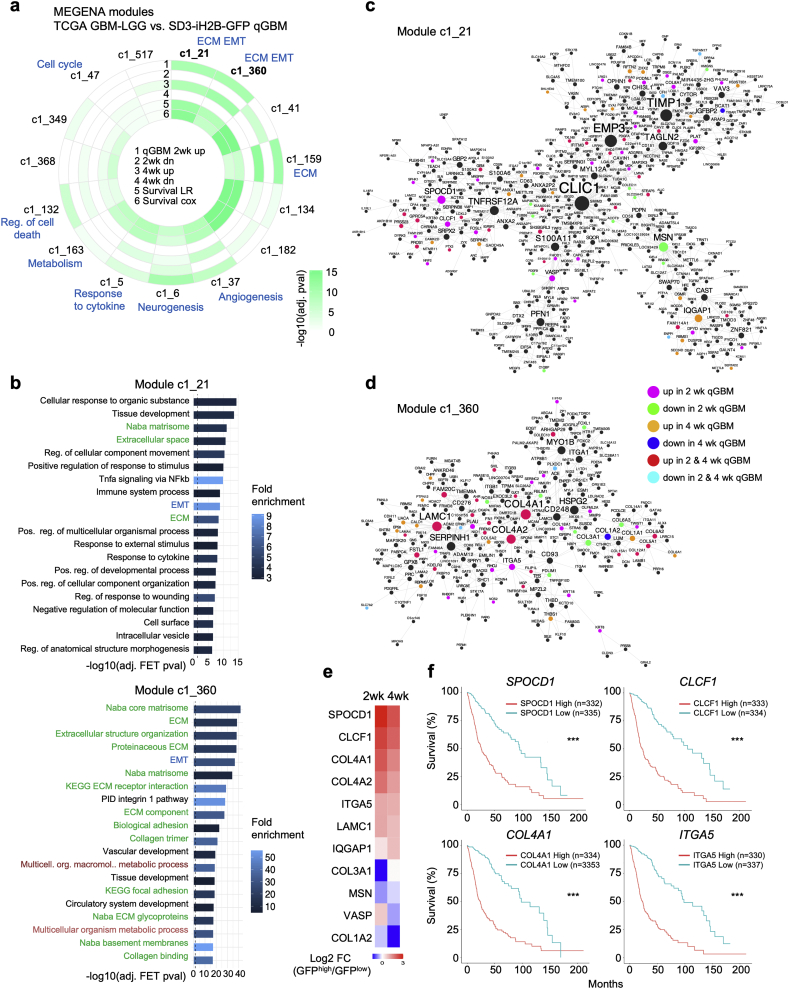
To understand how the transcriptional profile that we have identified in quiescent GBM cells using the organoid approach relates to gene expression networks in glioma patients, we applied co-expression network analysis on the NIH The Cancer Genome Atlas (TCGA) glioma database to identify gene modules that are enriched for qGBM gene signatures. To this end, we first surveyed the TCGA gene expression data for GBM and low-grade glioma patients (n = 666) and constructed a MEGENA co-expression network [31] to identify modules of co-regulated genes and their hubs. We found several gene modules in gliomas that showed not only enrichment for quiescence DEGs at 2 or 4 week -Dox chase stages, but also significant association with patient survival (Fig. 8a; Table S2). The two highest ranked modules, c1_21 and c1_360, were significantly enriched for EMT and ECM related GO terms and pathways (Fig. 8b). Several additional gene modules were also enriched for ECM and matrisome functions, as well as functions related to metabolism and regulation of cell death (Fig. 8a, b; Table S3). These results largely mirror the functional categories identified in our GSEA and ENRICHR GO analysis of qGBM DEGs, thus demonstrating clinical relevance of our quiescence gene profiles from the GBM organoid approach.
Fig. 8.
Analysis of TCGA glioma patient data for gene co-expression modules that are enriched for qGBM signatures and associated with survival.
a) MEGENA gene co-expression modules from TCGA glioma patient data that show enrichment for genes up or downregulated in qGBM cells at 2 or 4 weeks chase. Modules are ranked by their enrichment for qGBM genes and association with patient survival. Tracks, starting from outermost: enrichment for genes up- or downregulated at 2 (tracks 1 and 2, respectively) or 4 weeks (tracks 3 and 4, respectively); log-rank (track 5) and Cox regression (track 6) p-values of association of module first principal component (PC1) with survival in TCGA patient cohort. Selected enriched functions of the modules are indicated in blue.
b) Functional annotation terms (MSigDB, WikiPathways) that are enriched in the two highest ranked modules c1_21 and c1_360. Top 20 significant terms shown (Fisher's Exact Test for overrepresentation, Benjamini-Hochberg-adjusted p ≤ 0·05). Up to 20 top enriched terms in all modules that are significantly enriched for qGBM signature are provided in Supplemental Table S3. Green text font highlights ECM associated functions, blue EMT program, and dark red metabolism-associated functions.
c, d) Network visualization of gene connectivity and hub genes of modules c1_21 (c) and c1_360 (d). Node size is proportional to number of neighboring genes. Colour legend indicates if a network gene is also a DEG in qGBM at 2 or 4 week -Dox chase stages.
e) Heatmap depicting expression changes of hub genes identified by network analysis (c, d) that are differentially expressed in qGBM relative to pGBM cells. Colour key represents log2 fold change (FC).
f) Kaplan-Meier survival graphs of glioma patients stratified by high or low expression of the indicated hub genes (TCGA GBM-LGG dataset; split by median gene expression; web platform GlioVis (http://gliovis.bioinfo.cnio.es/)). Horizontal axis indicates duration (months) since initial diagnosis. Statistical analysis was carried out by log-rank test. *** indicates p < 0·001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We next identified network hubs of the top ranked modules c1_21 and c1_360 (Fig. 8c, d), which contained several genes previously not connected with a function in glioma (Table S4). Many of these hub genes showed differential expression in qGBM cells (Fig. 8e). Moreover, there was significant association of the expression levels of these hub genes with survival in TCGA GBM-LGG patients (comprised of grade II-IV glioma), including multiple ECM-related genes (COL4A1, ITGA5), as well as transcription factors (e.g. SPOCD1) and cytokines (e.g. CLCF1) (Fig. 8f).
3.11. Expression of ECM associated genes correlates with glioma grade and patient survival
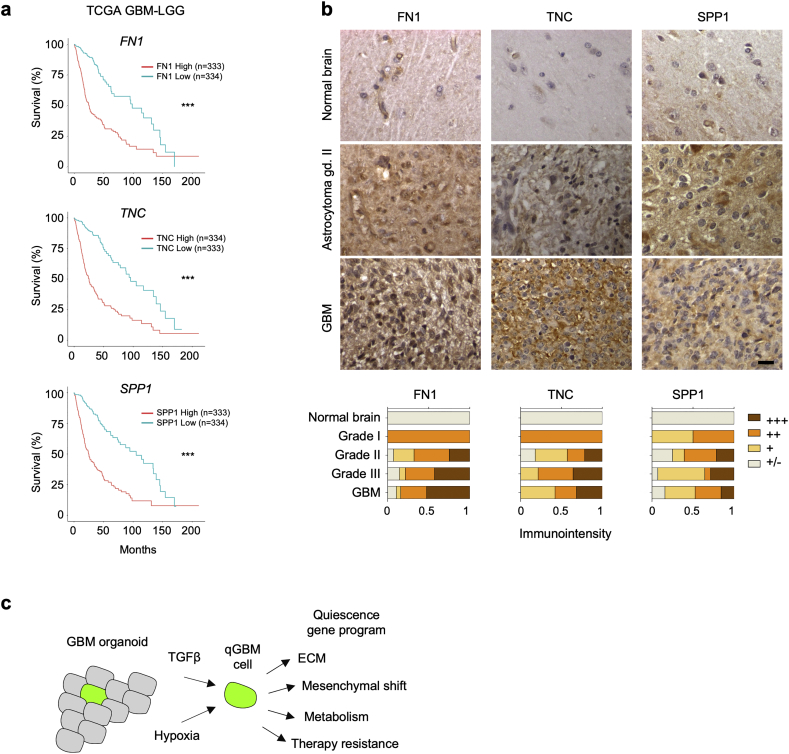
To further examine the clinical significance of qGBM DEGs that are associated with ECM, we analyzed patient data and tissue microarrays, focusing on SPP1, FN1, and TNC. We surveyed the TCGA GBM-LGG patient data set and conducted survival analysis using the GlioVis platform [34], which indicated that high expression of these genes correlated with shorter patient survival (Fig. 9a). Immunohistochemistry of glioma tissue microarrays showed that increased protein expression levels of these DEGs correlated with higher tumor grades (Fig. 9b). This finding was also corroborated by the mRNA expression levels for these genes in the TCGA data, which showed increased expression of FN1, TNC, and SPP1 in relation to glioma histology and grade (Fig. S7).
Fig. 9.
Expression of ECM-associated quiescence genes correlates with glioma grade and patient survival.
a) Kaplan-Meier survival graphs of glioma patients stratified by high or low expression of selected ECM-associated DEG of qGBM cells (TCGA GBM-LGG dataset; split by median gene expression; web platform GlioVis (http://gliovis.bioinfo.cnio.es/)). Horizontal axis indicates duration (months) since initial diagnosis. Statistical analysis was carried out by log-rank test. *** indicates p < 0·001.
b) Immunohistochemistry images of glioma samples on tissue microarray for ECM components show increased protein expression of SPP1, FN1, and TNC in high grade gliomas. Bottom panels show quantification of staining intensities in different samples (arbitrary scale).
c) Model of quiescence gene programs and niche factors for quiescent GBM cells, derived from our in vitro GBM organoid studies.
Scale bar: 20 μm (b).
4. Discussion
Our approach of utilizing an iH2B-GFP reporter in a 3D GBM organoid culture model supports the hypothesis that GBM can harbor a quiescent population with self-renewal potential and enhanced therapy resistance. Gene expression profiling revealed unique gene signatures in qGBM cells that are distinct from proliferative counterparts. Transcriptional changes in qGBM cells not only align with reduced proliferation and lower energy need, but also facilitate physiological adaptions to enhance stress-coping capabilities and engage a shift towards mesenchymal features, including expression of ECM-associated proteins of the microenvironment. Moreover, bioinformatic analyses and functional in vitro assays suggest that hypoxia and TGFβ signaling act as niche factors that promote GBM quiescence (Fig. 9c).
One important concept emerging from our studies is that tumor cell quiescence does not simply equate self-renewal capacity or stemness. First, in sphere formation assays, qGBM cells did not exhibit higher stem cell frequency than pGBM counterparts; in fact, qGBM cells isolated from 4 week chase GBM organoids displayed a slightly lower sphere-forming capacity than pGBM cells. One caveat is that freshly isolated qGBM cells need time to transition from quiescence to active cycling, which is a prerequisite for sphere formation, thus the sphere formation assay might under-estimate the self-renewal capacity of qGBM cells. In congruence, the average size of spheres derived from qGBM was smaller than from pGBM, which indicates reduced proliferation rate for qGBM cells. Similar results have been reported in two previous studies that used dye labeling (e.g., CFSE) for tracking of quiescent, label retaining cells (LRCs) in gliomasphere cultures: one study described reduction of sphere-forming capacity of LRCs [9] and another study reduction in both sphere-forming capacity and proliferative rate of LRCs [6]. Thus, our study with genetic iH2B-GFP reporter and 3D GBM organoids supports and extends these earlier findings. It is worth mentioning that a third dye labeling study reported increased sphere forming capacity and higher proliferation rate for LRCs [8]. These divergent findings may reflect inter- and intratumoral heterogeneity of GBM. Moreover, we found that qGBM cells did not display increased expression of stem cell marker genes, including SOX2, OLIG2, and CD133, which parallels findings by Liu and colleagues that SOX2 is more associated with proliferating stem cells [48]. Similarly, one of the earlier dye retention studies also showed that expression levels of SOX2 were not increased in quiescent GBM cells [8]. Taken together, our data further clarify that quiescence is a separate cellular/metabolic state distinct from stemness. In fact, as both qGBM and pGBM cells exhibit sphere-forming capacity, the stem cell compartment of GBM most likely consists of both a proliferative and a quiescent subpopulation. Our study therefore brings attention to the quiescent subpopulation in the stem cell compartment in order to tackle GBM recurrence. Future in vivo studies will further compare tumor initiating capacity of qGBM and pGBM cells, particularly after treatment.
What are unique features of qGBM cells that could contribute to tumor recurrence? The previous dye retention studies with gliomaspheres revealed that quiescent GBM populations display elevated therapy resistance and tumor forming capacity in xeno-transplants [6,8,9]. The results from our study echo these findings and further define “quiescence gene programs” that shed light on specific features that may underlie malignant potency qGBM cells. Although the proneural and mesenchymal GBM cell lines in our study had different baseline score levels for pathways associated with mesenchymal features, reflecting underlying differences of GBM subtypes [49], both GBM cell lines displayed a similar direction of shift towards increased mesenchymal features in qGBM relative to pGBM cells. In the case of the proneural GBM cell line SD3, qGBM cells clearly engaged proneural-mesenchymal transition (PMT), which has been linked with elevated malignancy and therapy resistance [29,42]. In the case of the mesenchymal line SD2, qGBM cells also showed increased pathway scores for mesenchymal features. It will be interesting to examine in future studies whether qGBM cells of classical molecular subtype also display a mesenchymal shift.
How do qGBM cells acquire and maintain quiescence? As part of the PMT of SD3 qGBM cells, we found that a number of genes encoding ECM components was upregulated, e.g. genes encoding fibronectin, collagens, tenascin C, and ECM receptors such as integrins and CD44. CD44 is a receptor for ECM component hyaluronic acid (HA) and secreted protein osteopontin (SPP1), and it has been proposed as a functional GBM stem cell marker [44,50] and to promote GBM invasion [51]. Our data suggest that qGBM cells may modify their own microenvironment by actively organizing ECM, a model that implies reciprocal interactions between qGBM cells and their microenvironment, in line with the model that microenvironmental niches determine the distinct physiology of GBM cells [52]. Indeed, for adult neural stem cells, the ECM plays an active role as site for integration of niche signals [53]. ECM organization may also be an intrinsic aspect for qGBM cells undergoing a mesenchymal shift, similar to the concept of EMT in epithelial cancers [54]. For instance, tenascin C has been shown to be a highly expressed ECM protein in malignant brain tumors and functions to decrease tumor cell proliferation while promoting invasion [55]. Our results extend this notion by linking tenascin C and other previously less well-characterized ECM components to GBM quiescence. Evidently, expression of ECM components by quiescent GBM cells may also modulate interactions with stromal cells, e.g. tumor-associated macrophages and endothelial cells, and future studies with in vivo paradigms will shed light on the influence of ECM on GBM-stromal interactions.
It will be also interesting to investigate in future studies whether qGBM cells are more migratory than pGBM. Earlier studies on mesenchymal transition-like processes in GBM have demonstrated that EMT-associated factors promote invasiveness of GBM [56]. It is thus conceivable that qGBM cells with mesenchymal transition-like features disseminate widely in GBM. The organoid culture model is not ideal to directly address this question, and future orthotopic in vivo transplant studies will be needed to gauge migratory behavior of quiescent cells in the complex microenvironment of the host brain.
Which upstream pathways could induce quiescence and activate the mesenchymal transition-like gene programs in qGBM cells? In respect to growth factors, our bioinformatic analyses and functional assays point towards TGFβ signaling as a potential inducer of quiescence. This is in agreement with previous studies demonstrating that TGFβ drives dormancy of disseminated tumor cells, as, for instance, in epithelial carcinomas [57]. Interestingly, a recent study on quiescence in squamous cell carcinoma, which also used a H2B-GFP label retention strategy, identified TGFβ/Smad2 signaling as a key pathway for maintaining quiescence and acquiring high chemotherapy resistance [58]. For GBM, TGFβ has been shown to promote self-renewal potential and tumorigenicity of GBM stem cells [59]. TGFβ can also be an inducer of an EMT-like shift in GBM cells, leading to increased invasion [60]. Another intriguing relation is between hypoxia and GBM quiescence. In the original description of the GBM organoid model, it was shown that cells in inner areas of the organoid are in hypoxic state [13]. We also observed that the majority of qGBM cells was located in central areas of organoids. Furthermore, hypoxia pathway genes were highly enriched in qGBM cells, while HIF1A was predicted as an upstream regulator. When GBM organoids were cultured in hypoxic conditions, the relative fraction of qGBM cells increased. Consistently, hypoxia can directly induce mesenchymal transition [61] and a hypoxia-induced gene program can promote the GBM stem cell compartment [62,63]. Our data thus supports a link of TGFβ, hypoxia, quiescence, mesenchymal shift, and stemness in GBM. It should be noted that our organoid culture studies did not provide information regarding the sources and niche effects of TGFβ and hypoxia in GBM, which requires future in vivo studies.
In terms of candidate transcriptional regulators of quiescence, we identified several chromatin remodelers. For instance, SMARCA4 (also known as Brg1) is a the ATPase subunit of the BAF chromatin remodeling complex [64]. The SMARCA4 gene itself was not differentially regulated in qGBM cells, but regulation of SMARCA4 activity may occur by interaction with other proteins; for instance, recruitment of SMARCA4 by the transcription factor ZEB1 has been shown during EMT of epithelial cancers [47].
Our gene co-expression network analysis of TCGA glioma patients revealed gene modules that are not only enriched for quiescence gene signature, but also associated with patient survival. We also identified hub genes in the modules that were not previously connected to a function in glioma. Future studies are worthwhile to examine their importance in promoting glioma malignancy; it is noteworthy that the survival association of these hub genes was determined, as an exploratory effort, using the TCGA GBM-LGG patient data, which comprises both low and high grade gliomas; however, survival analyses of these genes using only GBM (grade IV, IDH wild-type) patients did not reveal significant associations (data not shown). One possible scenario is that the link of these genes to patient survival may be partly due to their increased expression in GBM vs. LGG, and not strictly attributable to cellular quiescence. Another likely scenario is that these hub genes might exert an important role in a subpopulation of GBM cells, thus further analysis on single-cell level or in the context of niche microenvironment will be needed.
Our transcriptomic analysis also revealed differential down-regulation of apoptosis, DNA repair, and stress pathways in qGBM cells relative to pGBM counterparts. This may explain how qGBM exhibit higher therapy resistance against both the DNA alkylating agent TMZ and radiation therapy. Upon treatment, proliferative cells activate DNA repair and ER stress pathways in an attempt to repair, but failing to do so may trigger apoptosis. In contrast, qGBM cells with downregulated stress pathways may avoid apoptosis.
In summary, our studies on quiescence in GBM organoid cultures not only support the presence of a quiescent population in GBM that displays self-renewal capacity and high therapy resistance, but also go a step further in defining quiescence gene programs that may shed light on: i) how qGBM cells acquire and maintain quiescence (through ECM organization and interaction with niche factors such as TGFβ and hypoxia), ii) how qGBM cells cope with stress induced by chemo and radiation therapy (through regulation of DNA repair/ER stress pathway), and iii) how qGBM cells gain malignant potency (by engaging PMT or a mesenchymal shift). Our findings provide a starting point for developing combinatorial therapeutic strategies that target not only the proliferative GBM tumor bulk but also the quiescent population of GBM.
The following are the supplementary data related to this article.
List of DEG in qGBM.xlsx
Lists of differentially expressed genes (DEGs) between GFPhigh (quiescent) and GFPlow (proliferative) cells in organoids of GBM cell line SD3. DEGs from 2 and 4 week -Dox chase organoids are shown (filtered for protein coding genes). Also listed are the common DEGs of both chase paradigms.
Genes in top modules.xlsx
List of genes comprising the top 15 MEGENA co-expression modules that were enriched for qGBM DEGs.
Functional annotation of top modules.xlsx
Functional annotations for MEGENA co-expression modules that are significantly enriched for qGBM DEGs.
Hub genes of c1_21 c1_360 Pubmed sorted.xlsx
List of top hub genes for the modules c1_21 and c1_360 showing the number of Pubmed citations for a given query term (e.g., ‘glioma.pubmed’ means a query of the gene name and “glioma”), as well as status of a gene as significant qGBM DEG (0=false, 1=true).
Supplementary Figures S1–S6
Gene set matrix file (.gmx) with gene sets that were used to analyze proneural-mesenchymal transition (PMT) by gene set enrichment analysis (GSEA)
Funding
This project was supported by National Institutes of Health grants NIH-NINDS R21NS085466 (to H.Z. and R.H.F.), R01NS092735 (to R.H.F.), R01NS107462 (to H.Z. and R.H.F.), and NCI Cancer Center support grant P30CA196521 (Tisch Cancer Institute). M.K. and C.C.F. were supported by DFG grants FR2938/7–1 and CRC 1123 (Z2) to C.C.F.
Declaration of interests
R.H. Friedel and H. Zou report grants from the National Institutes of Health and M. Kluge and C.C. Friedel report grants from the Deutsche Forschungsgemeinschaft during the conduct of the study. The authors declare no other relationships/conditions/circumstances that present a potential conflict of interest.
Author contributions
R.T., H.Z., and R.H.F. designed the study and wrote the manuscript, R.T., Y.H., J.T.-G., N.M.T., and R.H.F. performed experiments, N.D. contributed preliminary data, M.K., C.C.F., I.K., Y.W., and B.Z. conducted bioinformatic analyses, and J. L. conducted statistical analyses.
Acknowledgements
We thank Dirk Hockemeyer (UC Berkeley) for the pAAVS1-Puro-H2BGFP plasmid. The pAAVS1-Neo-M2rtTA plasmid was a gift from Rudolph Jaenisch (Addgene plasmid #60843), and the pX330 plasmid was a gift from Feng Zhang (Addgene #42230). We thank the Mount Sinai qPCR core for assistance with the NanoString system and the flow cytometry core and Gerardo Ferrer for support for flow cytometry analysis, and Aarthi Ramakrishnan for help with RNA-Seq data management. We also wish to acknowledge the support of the Biostatistics Shared Resource Facility, Tisch Cancer Institute.
Contributor Information
Hongyan Zou, Email: hongyan.zou@mssm.edu.
Roland H. Friedel, Email: roland.friedel@mssm.edu.
References
- 1.Kleinhues P., Barnholtz-Sloan J., Ohgaki H. World Cancer report 2014. International Agency for Research on Cancer; Lyon: 2014. Tumours of the nervous system. [Google Scholar]
- 2.Ostrom Q.T., Gittleman H., Xu J., Kromer C., Wolinsky Y., Kruchko C. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009-2013. Neuro Oncol. 2016;18(suppl_5):v1–v75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huse J.T., Holland E.C. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 4.Chen J., McKay R.M., Parada L.F. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149(1):36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visvader J.E. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 6.Deleyrolle L.P., Harding A., Cato K., Siebzehnrubl F.A., Rahman M., Azari H. Evidence for label-retaining tumour-initiating cells in human glioblastoma. Brain. 2011;134(Pt 5):1331–1343. doi: 10.1093/brain/awr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J., Li Y., Yu T.S., McKay R.M., Burns D.K., Kernie S.G. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richichi C., Brescia P., Alberizzi V., Fornasari L., Pelicci G. Marker-independent method for isolating slow-dividing cancer stem cells in human glioblastoma. Neoplasia. 2013;15(7):840–847. doi: 10.1593/neo.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campos B., Gal Z., Baader A., Schneider T., Sliwinski C., Gassel K. Aberrant self-renewal and quiescence contribute to the aggressiveness of glioblastoma. J Pathol. 2014;234(1):23–33. doi: 10.1002/path.4366. [DOI] [PubMed] [Google Scholar]
- 10.Kanda T., Sullivan K.F., Wahl G.M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8(7):377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 11.Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W.E., Rendl M. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs E., Horsley V. Ferreting out stem cells from their niches. Nat Cell Biol. 2011;13(5):513–518. doi: 10.1038/ncb0511-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubert C.G., Rivera M., Spangler L.C., Wu Q., Mack S.C., Prager B.C., Couce M., McLendon R.E., Sloan A.E., Rich J.N. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76(8):2465–2477. doi: 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le A.P., Huang Y., Pingle S.C., Kesari S., Wang H., Yong R.L. Plexin-B2 promotes invasive growth of malignant glioma. Oncotarget. 2015;6(9):7293–7304. doi: 10.18632/oncotarget.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeKelver R.C., Choi V.M., Moehle E.A., Paschon D.E., Hockemeyer D., Meijsing S.H. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20(8):1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1–2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Andrews S. Fastqc: A Quality Control Tool for High Throughput Sequence Data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc Available online at.
- 19.Bonfert T., Kirner E., Csaba G., Zimmer R., Friedel C.C. ContextMap 2: fast and accurate context-based RNA-seq mapping. BMC Bioinformatics. 2015;16:122. doi: 10.1186/s12859-015-0557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 22.Robinson M.D., McCarthy D.J., Smyth G.K. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- 24.Kluge M., Friedel C.C. Watchdog - a workflow management system for the distributed analysis of large-scale experimental data. BMC Bioinformatics. 2018;19(1):97. doi: 10.1186/s12859-018-2107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Segerman A., Niklasson M., Haglund C., Bergström T., Jarvius M., Xie Y. Clonal variation in drug and radiation response among glioma-initiating cells is linked to proneural-mesenchymal transition. Cell Rep. 2016;17(11):2994–3009. doi: 10.1016/j.celrep.2016.11.056. [DOI] [PubMed] [Google Scholar]
- 30.Anastassiou D., Rumjantseva V., Cheng W., Huang J., Canoll P.D., Yamashiro D.J. Human cancer cells express slug-based epithelial-mesenchymal transition gene expression signature obtained in vivo. BMC Cancer. 2011;11:529. doi: 10.1186/1471-2407-11-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song W.M., Zhang B. Multiscale embedded gene co-expression network analysis. PLoS Comput Biol. 2015;11(11) doi: 10.1371/journal.pcbi.1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slenter D.N., Kutmon M., Hanspers K., Riutta A., Windsor J., Nunes N. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46(D1):D661–D667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowman R.L., Wang Q., Carro A., Verhaak R.G., Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19(1):139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bender R., Lange S. Adjusting for multiple testing--when and how? J Clin Epidemiol. 2001;54(4):343–349. doi: 10.1016/s0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- 36.Verkuilen J., Smithson M. Mixed and mixture regression models for continuous bounded responses using the Beta distribution. J Edu Behav Stat. 2012;37(1):82–113. [Google Scholar]
- 37.Ferrari S., Cribari-Neto F. Beta regression for modelling rates and proportions. J Appl Stat. 2004;31(7):799–815. [Google Scholar]
- 38.Pollard S.M., Yoshikawa K., Clarke I.D., Danovi D., Stricker S., Russell R. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 39.Galli R., Binda E., Orfanelli U., Cipelletti B., Gritti A., De Vitis S. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 40.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 41.Ordovás L., Boon R., Pistoni M., Chen Y., Wolfs E., Guo W. Efficient recombinase-mediated cassette exchange in hPSCs to study the hepatocyte lineage reveals AAVS1 locus-mediated transgene inhibition. Stem Cell Rep. 2015;5(5):918–931. doi: 10.1016/j.stemcr.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhat K.P.L., Balasubramaniyan V., Vaillant B., Ezhilarasan R., Hummelink K., Hollingsworth F. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halliday J., Helmy K., Pattwell S.S., Pitter K.L., LaPlant Q., Ozawa T. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci U S A. 2014;111(14):5248–5253. doi: 10.1073/pnas.1321014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pietras A., Katz A.M., Ekström E.J., Wee B., Halliday J.J., Pitter K.L. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14(3):357–369. doi: 10.1016/j.stem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sosa M.S., Bragado P., Aguirre-Ghiso J.A. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14(9):611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindley L.E., Briegel K.J. Molecular characterization of TGFbeta-induced epithelial-mesenchymal transition in normal finite lifespan human mammary epithelial cells. Biochem Biophys Res Commun. 2010;399(4):659–664. doi: 10.1016/j.bbrc.2010.07.138. [DOI] [PubMed] [Google Scholar]
- 47.Sánchez-Tilló E., Lázaro A., Torrent R., Cuatrecasas M., Vaquero E.C., Castells A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29(24):3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 48.Zhu Z., Khan M.A., Weiler M., Blaes J., Jestaedt L., Geibert M. Targeting self-renewal in high-grade brain tumors leads to loss of brain tumor stem cells and prolonged survival. Cell Stem Cell. 2014;15(2):185–198. doi: 10.1016/j.stem.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q., Hu B., Hu X., Kim H., Squatrito M., Scarpace L. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32(1):42–56. doi: 10.1016/j.ccell.2017.06.003. [e6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anido J., Sáez-Borderías A., Gonzàlez-Juncà A., Rodón L., Folch G., Carmona M.A. TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18(6):655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 51.Kim Y., Kumar S. CD44-mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility. Mol Cancer Res. 2014;12(10):1416–1429. doi: 10.1158/1541-7786.MCR-13-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hambardzumyan D., Bergers G. Glioblastoma: defining tumor niches. Trends Cancer. 2015;1(4):252–265. doi: 10.1016/j.trecan.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fidoamore A., Cristiano L., Antonosante A., d'Angelo M., Di Giacomo E., Astarita C. Glioblastoma stem cells microenvironment: the paracrine roles of the niche in drug and radioresistance. Stem Cells Int. 2016;2016 doi: 10.1155/2016/6809105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 55.Xia S., Lal B., Tung B., Wang S., Goodwin C.R., Laterra J. Tumor microenvironment tenascin-C promotes glioblastoma invasion and negatively regulates tumor proliferation. Neuro Oncol. 2016;18(4):507–517. doi: 10.1093/neuonc/nov171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siebzehnrubl F.A., Silver D.J., Tugertimur B., Deleyrolle L.P., Siebzehnrubl D., Sarkisian M.R. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5(8):1196–1212. doi: 10.1002/emmm.201302827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bragado P., Estrada Y., Parikh F., Krause S., Capobianco C., Farina H.G. TGF-β2 dictates disseminated tumour cell fate in target organs through TGF-β-RIII and p38α/β signalling. Nat Cell Biol. 2013;15(11):1351–1361. doi: 10.1038/ncb2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown J.A., Yonekubo Y., Hanson N., Sastre-Perona A., Basin A., Rytlewski J.A. TGF-β-induced quiescence mediates Chemoresistance of tumor-propagating cells in squamous cell carcinoma. Cell Stem Cell. 2017;21(5):650–664. doi: 10.1016/j.stem.2017.10.001. [e8] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peñuelas S., Anido J., Prieto-Sánchez R.M., Folch G., Barba I., Cuartas I. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15(4):315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 60.Joseph J.V., Conroy S., Tomar T., Eggens-Meijer E., Bhat K., Copray S. TGF-β is an inducer of ZEB1-dependent mesenchymal transdifferentiation in glioblastoma that is associated with tumor invasion. Cell Death Dis. 2014;5:e1443. doi: 10.1038/cddis.2014.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joseph J.V., Conroy S., Pavlov K., Sontakke P., Tomar T., Eggens-Meijer E. Hypoxia enhances migration and invasion in glioblastoma by promoting a mesenchymal shift mediated by the HIF1α-ZEB1 axis. Cancer Lett. 2015;359(1):107–116. doi: 10.1016/j.canlet.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Li Z., Bao S., Wu Q., Wang H., Eyler C., Sathornsumetee S. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soeda A., Park M., Lee D., Mintz A., Androutsellis-Theotokis A., McKay R.D. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene. 2009;28(45):3949–3959. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- 64.Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6(6):1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of DEG in qGBM.xlsx
Lists of differentially expressed genes (DEGs) between GFPhigh (quiescent) and GFPlow (proliferative) cells in organoids of GBM cell line SD3. DEGs from 2 and 4 week -Dox chase organoids are shown (filtered for protein coding genes). Also listed are the common DEGs of both chase paradigms.
Genes in top modules.xlsx
List of genes comprising the top 15 MEGENA co-expression modules that were enriched for qGBM DEGs.
Functional annotation of top modules.xlsx
Functional annotations for MEGENA co-expression modules that are significantly enriched for qGBM DEGs.
Hub genes of c1_21 c1_360 Pubmed sorted.xlsx
List of top hub genes for the modules c1_21 and c1_360 showing the number of Pubmed citations for a given query term (e.g., ‘glioma.pubmed’ means a query of the gene name and “glioma”), as well as status of a gene as significant qGBM DEG (0=false, 1=true).
Supplementary Figures S1–S6
Gene set matrix file (.gmx) with gene sets that were used to analyze proneural-mesenchymal transition (PMT) by gene set enrichment analysis (GSEA)