Abstract
Background
Genomic investigation of atypical adenomatous hyperplasia (AAH), the only known precursor lesion to lung adenocarcinomas (LUAD), presents challenges due to the low mutant cell fractions. This necessitates sensitive methods for detection of chromosomal aberrations to better study the role of critical alterations in early lung cancer pathogenesis and the progression from AAH to LUAD.
Methods
We applied a sensitive haplotype-based statistical technique to detect chromosomal alterations leading to allelic imbalance (AI) from genotype array profiling of 48 matched normal lung parenchyma, AAH and tumor tissues from 16 stage-I LUAD patients. To gain insights into shared developmental trajectories among tissues, we performed phylogenetic analyses and integrated our results with point mutation data, highlighting significantly-mutated driver genes in LUAD pathogenesis.
Findings
AI was detected in nine AAHs (56%). Six cases exhibited recurrent loss of 17p. AI and the enrichment of 17p events were predominantly identified in patients with smoking history. Among the nine AAH tissues with detected AI, seven exhibited evidence for shared chromosomal aberrations with matched LUAD specimens, including losses harboring tumor suppressors on 17p, 8p, 9p, 9q, 19p, and gains encompassing oncogenes on 8q, 12p and 1q.
Interpretation
Chromosomal aberrations, particularly 17p loss, appear to play critical roles early in AAH pathogenesis. Genomic instability in AAH, as well as truncal chromosomal aberrations shared with LUAD, provide evidence for mutation accumulation and are suggestive of a cancerized field contributing to the clonal selection and expansion of these premalignant lesions.
Fund
Supported in part by Cancer Prevention and Research Institute of Texas (CPRIT) grant RP150079 (PS and HK), NIH grant R01HG005859 (PS) and The University of Texas MD Anderson Cancer Center Core Support Grant.
Keywords: Atypical adenomatous hyperplasia, Lung adenocarcinoma, Preneoplasia, Pathogenesis, Allelic imbalance, Chromosomal instability
Research in context.
Evidence before this study
Atypical adenomatous hyperplasias (AAH), the only known precursor lesions to lung adenocarcinomas (LUAD), are challenging to identify and are often captured incidentally, thus making molecular interrogations of AAH limited. Due to the low cellular fractions of detectable somatic mutational processes within these premalignant tissues, little is known about its pathogenesis and progression to LUAD. Previously, mutations in known lung cancer drivers such as KRAS, BRAF and TP53 as well as gene expression and epigenetic modifications have been implicated in AAH development. Investigations of large chromosomal aberrations have been limited to microsatellite studies that have detected loss-of-heterozygosity in targeted chromosomal arms such as 9p, 9q, 16p and 17q.
Added value of this study
A genome-wide survey of chromosomal alterations that lead to allelic imbalance was performed using sensitive statistical techniques in premalignant AAH tissues and paired LUADs. Driver events comprising whole-chromosome or whole-arm mutations identified in this study, particularly the smoking-associated loss of 17p (TP53) in AAHs, further define the mutational landscape of a malignant transformation of these premalignant lesions. Additionally, the presence of shared, truncal events between matched AAH and LUAD further alludes to the early role of chromosomal imbalance in AAH pathogenesis as well as shared developmental trajectories among AAH and LUAD tissues.
Implications of all the available evidence
Additional clues from chromosomal aberrations in our present study aid our understanding of early mutational events in AAH pathogenesis, through a more comprehensive molecular characterization, allowing for a more complete picture of the genomics of lung cancer development. Chromosomal events identified in our study may contribute towards an improved screening for the early detection and possible prevention of premalignant disease, particularly in smokers or other individuals at elevated risk for lung cancer.
Alt-text: Unlabelled Box
1. Introduction
Lung adenocarcinoma (LUAD) is the most common histological subtype of lung cancer in both smokers and non-smokers [1]. In contrast to lung squamous cell carcinomas which is well characterized by histopathologically progressive lesions such as bronchial hyperplasias, metaplasias, dysplasias, and carcinomas in situ; the pathological sequence of LUAD development is poorly characterized [2]. Atypical adenomatous hyperplasia (AAH) is the only known precursor in the pathogenesis of LUAD. This premalignant lesion is challenging to identify and is, for the most part, captured incidentally [2]. Consequently, molecular interrogations of AAH have been few and our understanding of the pathobiology of this precursor lesion and the evolution of LUAD remains very limited.
Chromosomal instability is a hallmark of tumorigenesis and has been known to play a critical role in tumor initiation and progression [3]. Mechanisms leading to chromosomal instability include large chromosomal copy number alterations (e.g., whole-chromosome or whole-arm gain/loss) as well as regions of copy-neutral loss of heterozygosity (cnLOH) encompassing critical oncogenic drivers and tumor suppressor genes. Since these aberrations impact large portions of the genome, they play an important role in cancer development by paving the way for accumulating more mutations. Few reports have previously interrogated chromosomal alterations in AAH [4,5]. First, there is very limited tissue in AAH. Second, it is expected that genomic changes are present at low cellular fractions in preneoplastic tissue, or samples from these tissues include DNA from normal cells, thus compounding the challenge of rigorously identifying chromosomal alterations in a precursor lesion such as AAH. As such, few chromosomal aberrations have been previously identified in AAH. However, those documented include loss-of-heterozygosity (LOH) in 9p, 9q, 16p and 17q [4,5]. To date, the landscape of these chromosomal aberrations in AAH and their evolutionarily relationship with LUAD remain largely unexplored.
In this study, we interrogated matched AAH, LUAD and normal lung parenchyma (N) from 16 early-stage LUAD patients of East-Asian descent (n = 48 samples) using high-resolution single nucleotide polymorphism (SNP) arrays paired with sensitive haplotype-based techniques to detect genome-wide chromosomal aberrations present at low cellular fractions. We report the landscape of allelic imbalance (AI), comprising subtle copy number alterations (e.g., gain, loss) and cnLOH, in AAH and paired LUAD. Further, we integrated these AI events with published somatic single nucleotide variants (SNV) data from these samples within known cancer associated genes [6] to identify AAH lesions exhibiting multi-hit patterns of progression to matched LUADs. Findings from this comprehensive analysis of chromosomal instability provide a deeper understanding of the pathobiology of AAH and, thus, very early events in the pathogenesis of LUAD.
2. Material and methods
2.1. Clinical cohort
Matched normal lung parenchyma (N), atypical adenomatous hyperplasias (AAHs) and lung adenocarcinomas (LUADs) (n = 48 samples) were acquired from 16 patients with stage-I LUAD. Patients were evaluated at the Aichi Cancer Center (Nagoya, Japan) and Nagasaki University (Nagasaki, Japan) and were approved for study by institutional review boards. Informed consent was obtained from all patients recruited for the study. The diagnosis, specimen collection and slide preparation were carried out between 2011 and 2015, as described previously [6]. Specimens were obtained formalin-fixed and paraffin-embedded (FFPE) and assessed using hematoxylin and eosin/H&E staining. The AAH lesions were incidental and identified by radiological imaging. Normal lung was taken from resected areas and was confirmed histopathologically following H&E staining to be consistent with normal tissue devoid of preneoplastic or neoplastic cells. Tissues were pathologically examined following the World Health Organization guidelines on the classification of lung tumors [7]. Clinicopathological features of all patients are summarized in Table 1.
Table 1.
Clinicopathological features of the cohort.
| Case | Age | Sex | Smoking | Stage | Predominant tumor histology |
|---|---|---|---|---|---|
| 1 | 70 | M | Ever | IA | Papillary |
| 2 | 21 | F | Ever | IB | Acinar |
| 5 | 79 | F | Never | IA | Lepidic |
| 6 | 40 | M | Ever | IA | Lepidic |
| 7 | 72 | M | Never | IA | Papillary |
| 10 | 81 | F | Never | IA | AIS |
| 11 | 67 | M | Ever | IA | Lepidic |
| 13 | 79 | F | Ever | IA | MIA |
| 15 | 62 | M | Ever | IA | Lepidic |
| 16 | 64 | F | Never | IA | Papillary |
| 17 | 67 | M | Ever | IA | Acinar |
| 18 | 57 | F | Never | IA | Papillary |
| 19 | 71 | M | Ever | IB | Solid |
| 21 | 74 | F | Never | IA | Lepidic |
| 22 | 60 | M | Ever | IA | Lepidic |
| 23 | 65 | M | Ever | IB | Acinar |
AIS: Adenocarcinoma in situ.
MIA: Minimally invasive adenocarcinoma.
2.2. Genome-wide high-density array profiling
DNA was extracted using the AllPrep DNA FFPE Kit from Qiagen and suspended in AE buffer (DNA). Sample concentrations were measured on NanoDrop 1000 (Thermo Fisher Scientific) and DNA was quantified using the Quant-iT PicoGreen double-stranded DNA (dsDNA) kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The extracted DNA was then processed through the Infinium HD FFPE DNA Restoration protocol (Illumina Inc., San Diego, CA.) followed by SNP genotyping using the Illumina Infinium® Global Screening Array-24 v1.0 BeadChip array. Raw intensity files were analyzed with GenomeStudio Genotyping Module v2.0 (Illumina Inc., San Diego, CA.) to call genotypes, normalize and cluster data in order to obtain SNP metrics such as B-allele frequency (BAF) and log R ratio (LRR).
2.3. Identification of subtle genome-wide allelic imbalance at low cellular fractions
Allelic imbalance (AI) was inferred using hapLOH, to detect subtle patterns of BAFs at germline heterozygous markers consistent with a relative haplotype imbalance [8]. Using regions that exhibit deviations in their BAFs along with LRR intensities for markers within regions of AI, events were classified as gain, loss and copy-neutral loss of heterozygosity (cnLOH) as described previously [9]. Briefly, the event regions with LRR ≥ 0.05 were classified as gains while those with LRR ≤ −0.05 were classified as losses. Among the remaining calls, regions with BAF deviation of 0.1 or greater were classified as cnLOH. The event calls that were not classified into these three event types were annotated as subtle AI. Subsequently, detected AI events were specifically tested for statistical evidence of existence in other samples from the same individual, using a binomial test of similarities between the two sample-specific haplotypes in putative excess (derived from the sample BAFs) within each event region.
2.4. Intra-patient heterogeneity
For each patient, to assess shared as well as disparate patterns between the AAH and LUAD samples, we quantified proportions of the genome exhibiting AI in both samples (AAH and LUAD) as well as proportions of the genome harboring AAH or LUAD-specific AI. In addition, for shared AI events between matched AAH and LUAD, the regions exhibiting over-representation of opposite haplotypes were excluded since they might be suggestive of independent events or secondary events [10]. For each patient, markers profiled in the SNP genotyping array were annotated as either being in an event seen only in an AAH or LUAD sample or in a shared event seen in both tissues. Based on this, the proportion of markers within AI events in matched AAH and LUAD specimens were determined as shared events, while those specific to only one of the tissues were determined as the proportion of AAH-specific and LUAD-specific events. Phylogenetic trees were then constructed for each individual using shared and tissue-specific genomic AI proportions between the LUAD and AAH using the ape package in R.
2.5. Integration of single nucleotide mutations
All patients in this cohort (with the exception of patient 23) were profiled in a previous study for single nucleotide mutations (SNVs) in a sequencing panel of 409 known tumor associated genes [6]. Given the prevalence of EGFR mutations in this cohort [6], the samples exhibiting nonsynonymous mutations in this oncogene were assessed for patterns in their overall genomic AI burden. Further, the identified AI events in the present study as well as previously detected SNVs in known oncogenes and tumor suppressors [11,12] were assessed for patterns of two-hit mutations (AI and SNV) in AAH and LUAD as well as for patterns of shared-first hit (AI or SNV) between the AAH and LUAD of that individual with a LUAD-specific second hit.
3. Results
3.1. SNP array profiling of normal lung tissue, AAH, and early-stage LUAD
Atypical adenomatous hyperplasia, the only known precursor lesion to LUAD, is challenging to identify, presenting with minute amounts of tissue which further compounds the difficulty of detecting genomic alterations of low cellular fractions – expected conditions in such early, preneoplastic lesions. Thus, there is a dearth of knowledge of chromosomal alterations and instability in the pathogenesis of AAH. Using computational tools we developed based on haplotype information to model within-sample allele frequencies jointly [8], we sought to explore somatic genome-wide allelic imbalance events (AI) including copy number gains, losses and copy neutral loss-of-heterozygosity (cnLOH) that are present at low cellular fractions (e.g., as low as 3–5%) in AAHs and paired LUADs from 16 patients with early-stage disease (Table 1). Diagnosis and histopathologic determination of matched tissues (N, AAH and LUAD; n = 48) were conducted as described previously [6]. The cohort comprised 10 smokers and 6 nonsmokers (Table 1). Major clinicopathological variables are summarized in Table 1. The majority (n = 13) of LUADs in our study were stage IA with the remaining three cases determined to be IB (Table 1). Our cohort largely comprised invasive LUADs, with the exception of two cases that had a minimally invasive adenocarcinoma (MIA) and adenocarcinoma in situ (AIS) (Table 1). The predominant histological subtype of the LUADs in this cohort are also provided in Table 1. Since our primary focus involved AAHs, we treated the AIS/MIA specimens as matched tumor comparators in these patients. All 48 samples were profiled using SNP genotyping arrays and AI analysis was performed to identify megabase-scale chromosomal aberrations in normal lung parenchyma, AAH and LUAD.
3.2. Genome-wide allelic imbalance in AAH
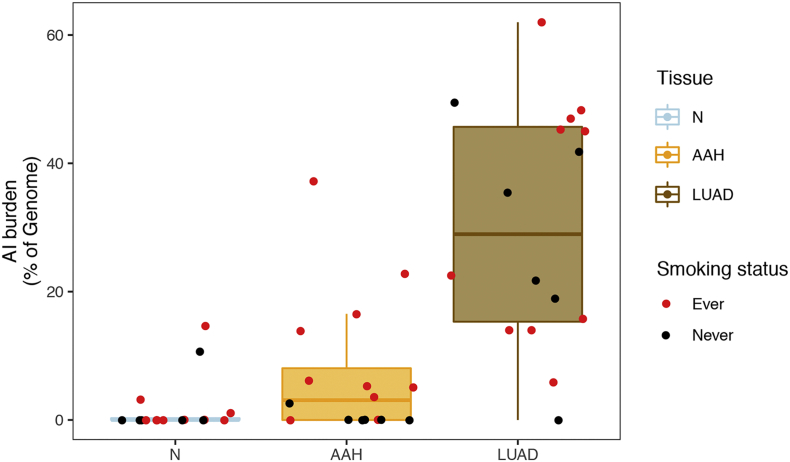
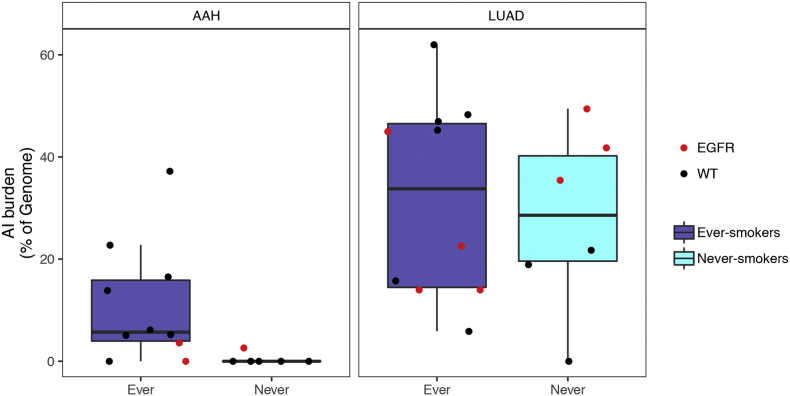
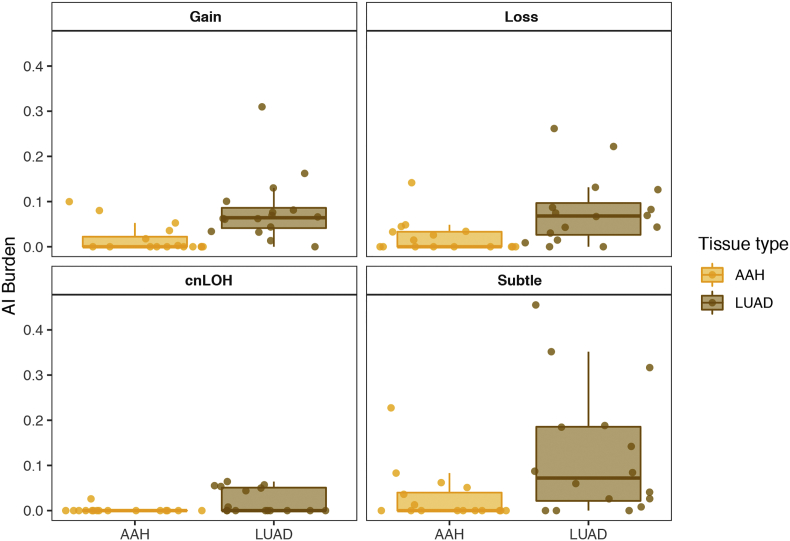
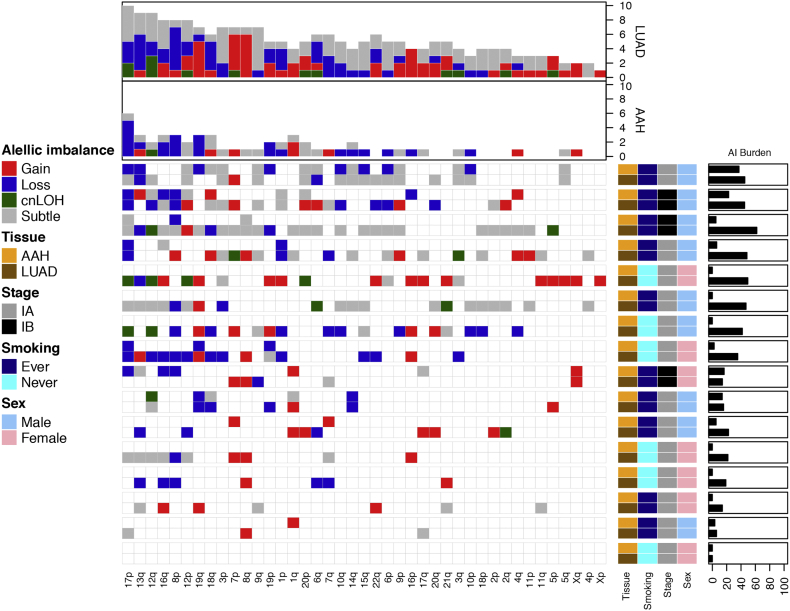
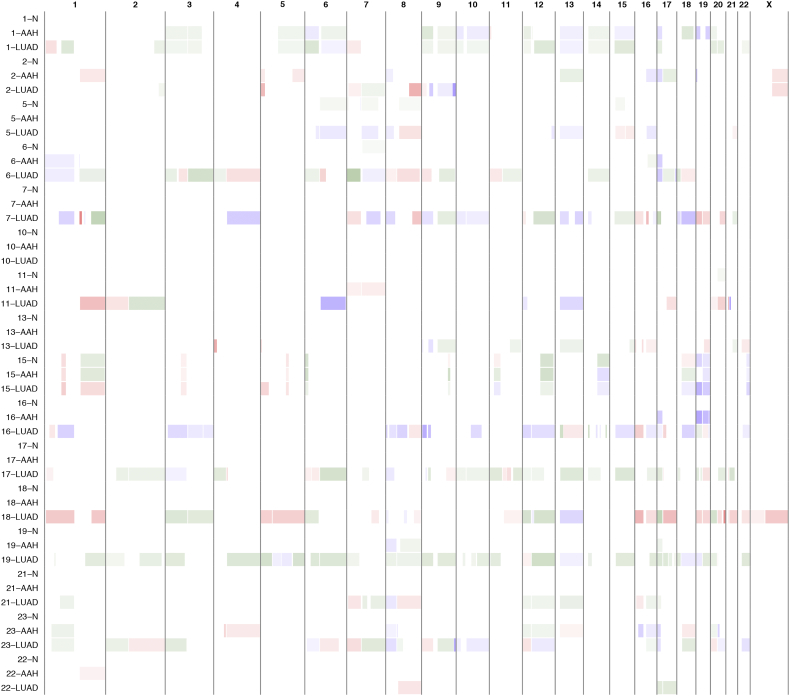
A haplotype-based computational framework, hapLOH, was used to infer subtle AI, including alterations present at lower cellular fractions [8]. Our cohort exhibited evidence for AI in nine AAHs (56%), 15 LUADs (94%) and four normal lung parenchyma tissues (25%) (Fig. 1). We identified 53 chromosome-arm AI events (≥ 50% of chromosomal arm) and 19 focal AI events (<50% of chromosomal arm) in AAHs; and 210 arm-level AI events and 97 focal events in LUADs (Supplementary Table 1). Overall, the detectable AI burden (defined as a percent of genome exhibiting AI) in AAHs was significantly lower than LUAD (Wilcoxon, P value = 0.0002; Fig. 1). While AAHs showed significantly higher AI burden in lifetime smokers compared to non-smokers (Wilcoxon, P value = 0.005), their matched LUADs showed similar distributions of AI burden between non-smokers and smokers (Fig. 1, Supplementary Fig. 1). Of note, the AI burdens of EGFR-mutant non-smoker LUADs were larger than those of smokers as well as non-smoker LUADs without the mutation (Supplementary Fig. 1, Supplementary Table 2). Higher overall genomic burden of chromosomal aberrations was observed across all event types, with cnLOH events being less common in both AAHs and LUADs compared to gains and losses (Supplementary Fig. 2). Recurrent allelic loss events in 17p harboring tumor suppressors TP53 (17p13) and PER1 (17p13), were the most frequently detected chromosomal aberrations in AAHs of our cohort (n = 6; Fig. 2). Additionally, five of these six cases with 17p loss events in AAHs were identified in patients with a history of tobacco use. Other recurrent AI events in AAHs included the following: gain of 1q, harboring oncogene ABL2 (1q25) and cell proliferation genes PARP1 (1q42) and PBX1 (1q23), gain of 18q harboring BCL2 (18q21), loss of 8p harboring tumor suppressor MTUS1 (8p22), loss of 16q encompassing CYLD (16q12), CDH1 (16q22), loss of 19p harboring KEAP1 (19p13), STK11 (19p13), SMARCA4 (19p13), and loss of 19q as well as mixed events on 13q (n = 3) (Fig. 2). The matched LUADs exhibited more complex patterns of allelic imbalance across the entire genome (Fig. 2). These tissues also showed frequent gains spanning known oncogenes including those on 8q (MYC: 8q24), 7p (EGFR: 7p11) and 2p (DNMT3A, ALK: 2p23); they showed loss or cnLOH events harboring tumor suppressors such as those on 12q (ARID2: 12q12, MLL2: 12q13), 3p (SETD2: 3p21, VHL: 3p25, FOXP1: 3p13), 9q (KLF4: 9q31, PTCH1:9q22, GNAQ: 9q21, TSC1: 9q34, ABL1, NOTCH1: 9q34), 18q (SMAD4: 18q21), and 6q (FOXO3: 6q21). Although the overall AI burden in AAHs was seemingly lower than in LUADs, four cases exhibited similar burdens across these matched tissues (Fig. 2). These cases also exhibited high sharing of specific AI events in AAHs and matched LUADs, including loss of chromosomal arms 17p (TP53, PER1: 17p13), 13q (RB1: 13q14), 19p (KEAP1, STK11, SMARCA4: 19p13), 19q and 9q (KLF4: 9q31, PTCH1:9q22, GNAQ: 9q21, TSC1: 9q34, ABL1, NOTCH1: 9q34). Of note, we identified AI events that exhibited patterns with opposite haplotypes in excess within the same event, between matched AAH and LUAD, signifying potentially independent events (Supplementary Table 3). In addition to chromosomal-arm AI events, we also identified subtle focal events in AAH of six patients that included 11p gain encompassing HRAS and IGF2 (11p15), 5q gain spanning RAD50 (5q31), FGFR4 and NSD1 (5q35), 19p loss comprising STK11 (19p13), 3p amplification at FOXP1 (3p13), 11p gain encompassing the oncogene WT1 (11p13), 17q loss harboring NF1 (17q11) and 4q gain covering KIT (4q12) (Supplementary Table 1, Supplementary Fig. 3). Finally, AI detected in the four normal lung parenchyma tissues included three patients with smoking history (Fig. 1) and exhibited large chromosomal loss events on 19p and 19q, gain of 18q as well as several subtle, yet, large events on 1q, 6q, 7q, 8q, and 20q (Supplementary Table 1, Supplementary Fig. 3). Three of these cases exhibited events that were shared with matched LUAD specimens and the remaining case showed events shared with both AAH and LUAD tissues (Supplementary Table 1, Supplementary Fig. 3).
Fig. 1.
Chromosomal allelic imbalance burden in normal, AAH and LUAD tissues. Regions with subtle chromosomal allelic imbalance (AI) were identified in the normal (N), AAH and matched LUAD tissues using genome-wide genotype array profiling as described in the Methods section. AI burdens, defined as a percent of the genome, are represented by box plots for each tissue type (N, AAH and LUAD). The burden for each patient is shown as a point overlaid on the boxplots. The points are colored red if the patient had a smoking history and black if the patient was a non-smoker. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Supplementary Fig. 1.
Chromosomal allelic imbalance burden in AAH and LUAD based on tobacco history. Identification of allelic imbalance (AI) and estimation of genomic AI burden was performed as described in the Methods section. AI burdens, defined as a percent of the genome, are represented by box plots for each tissue type (AAH and LUAD) based on tobacco history (never-smoker and ever-smoker). The burdens for each individual case are overlaid as points on the boxplot. Specifically, samples exhibiting EGFR mutations are shown as red dots.
Supplementary Fig. 2.
Chromosomal allelic imbalance burden in AAH and LUAD for each event type. Regions with chromosomal allelic imbalance (AI) were identified in AAH and matched LUADs and classified into gain, loss and copy-neutral loss of heterozygosity (cnLOH), from genome-wide genotype array profiling as described in the Methods section. The remaining unclassifiable events are shown as “subtle”. AI burdens, defined as a percent of the genome aberrant, are represented by tissue-level box plots (AAH and LUAD) for each event category. The burdens for each individual case are overlaid as points on the boxplot.
Fig. 2.
Genome-wide chromosomal arm allelic imbalance events in matched AAH and LUAD. We identified 53 subtle chromosomal arm events in AAHs and 210 chromosomal arm events in matched LUADs across 16 stage-I LUAD patients. The distribution and type of these events are shown, with rows representing individual patients and columns representing chromosome arms. Each individual row is further divided to show profiles of matched AAH and LUAD from that individual. The events are annotated as gain (red), loss (blue) or copy-neutral loss of heterozygosity (cnLOH, green) while unclassifiable events are annotated as subtle (gray). The overall burden across all chromosomal arms is shown in the bar plots at the top, while allelic imbalance burdens in each sample are shown on the right. Patients are also annotated to denote their clinicopathological features. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Supplementary Fig. 3.
Chromosomal arm and focal allelic imbalance events in matched normal lung parenchyma, AAH and LUAD. The genomic locations of the identified chromosomal allelic imbalance events were plotted for all 48 samples of matched normal lung parenchyma, AAH and LUAD from 16 patients. Allelic imbalance regions are first classified as gains (red) or losses (blue) as described in the Methods section, the intensity of which is based on the log R ratio of the event. The remaining events are annotated in green as subtle and copy-neutral loss of heterozygosity (cnLOH) events, intensity of which is based on B-allele frequency deviation for the event region, with darker shaded regions representing increased evidence for cnLOH.
3.3. Genomic evolution processes in AAH and/to LUAD
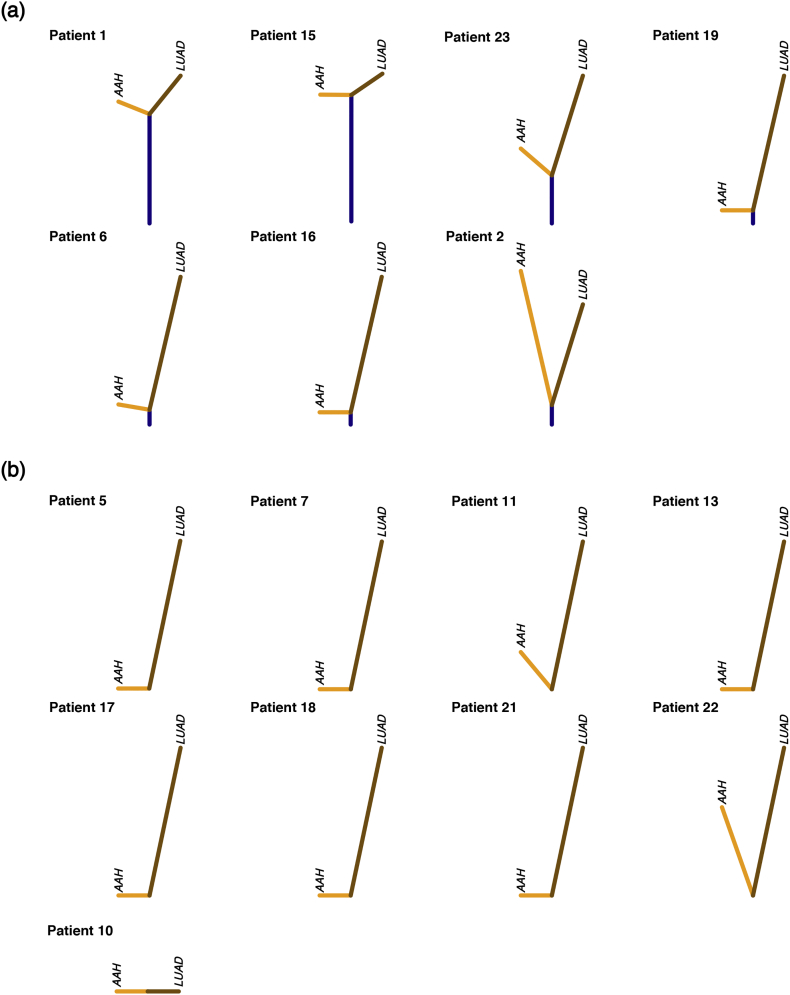
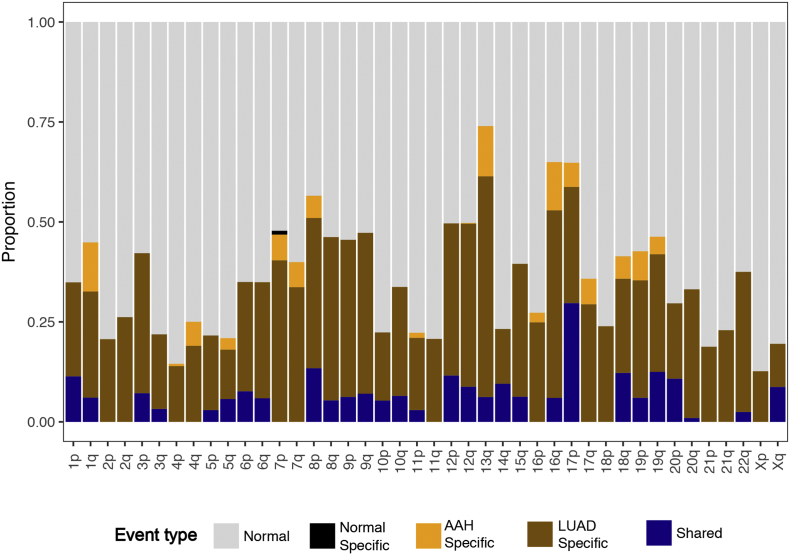
We next used the identified chromosomal-arm and focal AI events of matched AAH and LUAD tissues for all patients to construct phylogenetic trees depicting the genomic evolution of these tissues. We identified seven patients exhibiting regions of shared AI events between matched AAH and LUAD forming trunks of phylogenetic trees (Fig. 3a). The length of the trunk, and therefore the extent of shared events between matched AAH and LUAD varied across patients. Truncal events included chromosomal arms that spanned known lung cancer associated genes such as loss or cnLOH events harboring tumor suppressors such as 17p (TP53, PER1: 17p13), 8p (MTUS1: 8p22), 9p (CDKN2A: 9p21), 9q (KLF4: 9q31, PTCH1: 9q22, GNAQ: 9q21, and ABL1, NOTCH1, TSC1: 9q34), 19p (KEAP1, STK11, SMARCA4: 19p13), as well as gains of chromosomal arms encompassing oncogenes such as 8q (MYC: 8q24), 12p (KRAS: 12p12), 1q (ABL2: 1q25). Patient 1 showed the largest percentage of shared AI events (36.9%) that included subtle events on chromosomal arms 3p, 5q, 6p, 6q, 9p, 9q, 12p and 17p, LUAD-specific events such as on 1p, 7p and 12q and AAH-specific events on 18q, 19p and 19q. Patients 15 and 23 also exhibited shared AI events between AAH and LUAD (15.6% and 16.0% respectively) that included chromosomal arms 1q, 11p, 18q and 19p in patient 15 and 1p, 12p, 12q, 16q, 17p, 18q, 20p and 20q in patient 23. While patient 15 showed similar overall AI burdens in both AAH and LUAD tissues, patient 23 showed an overall higher AI burden in LUAD compared to its AAH with LUAD-specific AI events including 1p, 2q, 3p, 7p, 9p, 9q and AAH-specific AI events on 4q and 13q. Patient 6 and 19 exhibited shared AI events in a small proportion of the genome (5.2% and 6.1% respectively) followed by patients 2 (2.3%), and 16 (3.2%) showing much lesser sharing between matched AAH and LUADs. Further, among all seven cases with evidence for shared AI between matched AAH and LUAD, a majority were identified as smokers (6 of 7) with only one case identified as a non-smoker (patient 16). In the remaining cases, the AAH and LUAD showed distinct and independent AI profiles (Fig. 3b). These cases exhibited private somatic AI events unique to AAH or LUAD such as those on 1q, 7p, 7q, 13q and 16q. The distribution of shared AI events as well as AAH-specific and LUAD-specific AI events across the genome is shown in Supplementary Fig. 4.
Fig. 3.
Phylogenetic reconstruction of truncal, AAH-specific and LUAD-specific chromosomal aberrations. Matched AAH and LUAD specimens from individual patients were assessed for patterns of shared as well as tissue-specific allelic imbalance events and phylogenetic rooted trees were constructed as described in the Methods section. Cases exhibiting any evidence for shared events are shown in (a) and remaining cases are shown in (b). Vertical distances in each tree are scaled to the proportion of shared as well as tissue-specific events. Shared events, thereby trunks of the trees, are shown in dark blue; while tissue-specific events are shown separately for AAH (orange) and LUAD (brown). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Supplementary Fig. 4.
Distribution of shared and tissue-specific allelic imbalance events across chromosomal arms. The identified allelic imbalance (AI) events across all patients were averaged and assessed for chromosomal patterns of shared as well as tissue-specific events. A stacked bar plot representing the proportion of normal region (gray), shared AI (blue), AAH-specific AI (orange), LUAD-specific AI (brown) and normal tissue-specific AI (black) for each chromosome arm is shown.
3.4. Somatic multi-hit progression of AAH to LUAD
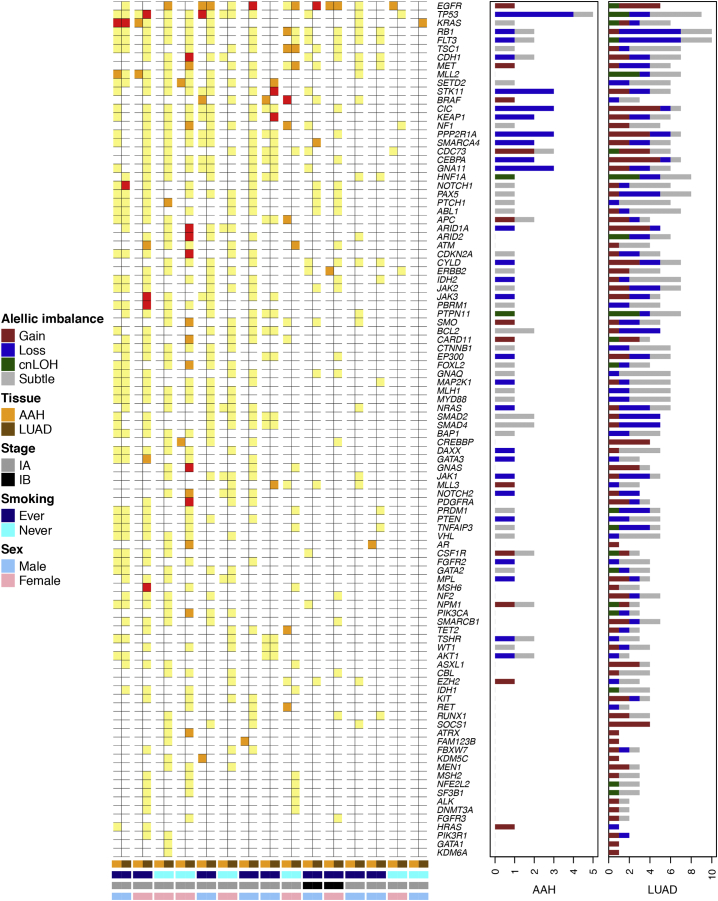
We integrated our previous analysis of single nucleotide mutations (SNVs) within this cohort [6] to identify cancer driver genes [11,12] exhibiting somatic multi-hit mutational processes (i.e. mutation and a chromosomal-arm or focal AI events encompassing the mutated gene). While the LUADs exhibited somatic-two hit events in known cancer associated genes such as EGFR, TP53, KRAS, CDH1, JAK3, ARID1A, ARID2, CDKN2A, GNAS and MSH6, we identified only two AAH cases with such patterns. One case exhibited a KRAS mutation and a 12p gain, that was shared between its AAH and LUAD tissues and another case with an AAH-specific BRAF/7p gain event (Supplementary Table 4, Supplementary Fig. 5). We also identified an additional two cases that exhibited a single shared AI event (i.e. present in AAH and LUAD) with a LUAD-specific second mutation hit (SNV) such as subtle AI on 9q/NOTCH1 and 17p/TP53 (Supplementary Table 4, Supplementary Fig. 5).
Supplementary Fig. 5.
Progressive and somatic two-hit processes in matched AAH and LUADs. Known cancer driver genes [[11], [12]] within regions of allelic imbalance or with single nucleotide mutations (SNVs) [6] were examined. The figure depicts genes exhibiting somatic “two-hits” (both SNVs and AI; red) in AAHs and LUADs as well as those exhibiting a first shared hit (either SNV: orange or AI: yellow) in the AAH accompanied by a second tumor-specific hit in the matched LUAD. For samples with allelic imbalance, the event types are shown as bar plots on the right, accompanying each gene, in both the AAH and LUAD specimens.
4. Discussion
There is a lack of understanding of the molecular aberrations, such as acquired megabase-scale chromosomal alterations, that lead to genomic instability in AAHs, the only known precursor lesion to the largest subtype of non-small cell lung cancer, LUAD. In our study, we sought to address this gap in knowledge by profiling matched normal lung parenchyma, AAH and LUAD specimens from 16 stage-I patients (n = 48) to characterize the genomic landscape of large acquired chromosomal allelic imbalance in AAHs, specifically a subset of progressive events that are shared with matched LUADs.
The challenges in physical acquisition of incidental AAHs, in addition to the low mutant cell fractions that are typical for these samples, have limited the molecular characterization of these premalignant lesions to date. Statistical approaches to discover even large chromosomal alterations in these samples may be limited to mutant cell fractions exceeding 15% with standard SNP array technology. Here we applied hapLOH, a sensitive, haplotype-based method [8] that offers resolution at 5% mutant fraction. From our results, the frequency of detectable allelic imbalance events in these samples may at first appear high. However, aspects of our analysis, study design, and findings in other nonmalignant tissues serve to contextualize these findings. First, the increased power from our statistical approach captures a critical region of the within-sample mutation frequency spectrum, specifically, mutations in a small proportion of cells, consistent with their involvement in early stage of development and the heterogeneous nature of the tissues. Second, the use of SNP arrays, instead of whole-exome sequencing, allows for more power to detect copy number changes, particularly those leading to AI, due to more comprehensive genomic coverage (more heterozygous markers queried) and more consistent evaluations of total intensities (cf. read depths). Third, we applied additional statistical testing; when we detected an event in a tissue, we specifically looked at that same region in other matched tissues. Finally, rates of “half” for premalignant lesions or field cancerization samples demonstrating detectable AI have been observed in the lung [9] and colon [13].
Most previous studies of AAHs have used microsatellite and fluorescent in situ hybridization techniques to target a limited number of chromosomal loci for loss of heterozygosity (LOH) predominantly on 3p, 9p, 9q, 16p, 17q, and 17p [4,5,14]. Our findings not only corroborate these previously described LOH events but also provide better resolution of genome-wide gain, loss and cnLOH, including previously undocumented aberrations such as those on chromosomes 1, 7, 8, 12 and 19. Chromosomal aberrations such as loss and cnLOH of arms 9p, 12q, 17p, 19p and 19q and gain of 1q, 8q, 18q, 7p and 7q in AAHs of our cohort have been shown in previous studies of chromosomal changes in non-small cell lung cancer (NSCLC), including EGFR-mutant LUADs, of Asian patients, that form a major subset of our cohort [[15], [16], [17], [18], [19], [20], [21]]. Further that these changes are not only shared with NSCLCs but exhibit reduced overall proportions in AAHs compared to matched LUADs are consistent with the morphological changes in these lesions and might suggest their role in malignant transformation of these premalignant lesions. Chromosomal aberrations identified in our study have also been previously described in premalignant lesions of other tumor sites [22,23]. For example, the most common event in AAHs of our cohort, 17p loss, has been previously described as an early event, preceding mutations in TP53, and a predictor of neoplastic progression in Barrett's esophagus, a premalignant lesion which predisposes to esophageal adenocarcinoma [[22], [23], [24], [25]]. Another study described the importance of loss events such as 17p, 8p and 13q in addition to early LOH events of 3p and 9p in conferring increased relative risk of malignant transformation in oral premalignant lesions [26]. Further, the higher incidence of 17p loss events observed here compared to previous studies [5], particularly in smokers, might be attributable to the East Asian origin of this cohort. These findings implicate a role of chromosomal imbalances early in the development and progression of these preneoplastic lesions.
Genomic instability has been shown to play a critical role in field carcinogenesis thereby providing an environment for accumulating more mutations necessary for a normal tissue transitioning to pre-invasive and invasive phenotypes [27]. Shared and potentially truncal chromosomal imbalance events revealed in our phylogenetic analysis of matched AAH and LUAD tissues suggest mechanisms of clonal selection in development of AAH from normal lung parenchyma as well as their progression to LUAD. Further, that these AI events were also accompanied by LUAD-specific secondary mutation hits in genes established as critical drivers of AAH and LUAD pathogenesis [6,12,28], such as 12p gain/KRAS, 9q subtle/NOTCH1 and 17p subtle/TP53, are suggestive of a multi-step mutational accumulation involved in AAH and LUAD pathogenesis. Additionally allelic imbalance identified in AAHs in our study including loss of 1p, 8p, 17p, 19p and 19q, and gain of 1q, 7p and 8q, as well as subtle events on 13q, were previously reported in multiple tumor-adjacent and distant airway epithelia from early-stage NSCLC patients [9]. These findings provide evidence for a premalignant lesion arising from the field of cancerization and suggest a key role of accumulating chromosomal imbalance-driven genomic instability and selection early in AAH and NSCLC pathogenesis.
Novel findings from our study describe the importance of subtle chromosomal aberrations and genome instability in the development of AAH. A recognized challenge with the analysis of non-malignant or premalignant tissues is the low proportion of cells harboring a mutation (mutant cell fraction), which is expected for such tissues. We overcome this challenge by modeling the “B allele” frequency data jointly, via comparisons to germline haplotype information, facilitating detection of AI; however, categorization of AI into specific mutation types still remains a challenge, since the data for categorization (log R ratios) are inherently noisy. Additionally, our cohort was limited in size to further compare and contrast different pathological subtypes of LUAD. Future and larger studies are warranted to identify patterns of changing chromosomal aberrations along the development of AAH to LUAD as well as to assess the prognostic value of these aberrations in predicting their progression to invasive LUAD, such as what is observed in oral cancers [29,30] and lower grade gliomas [31]. Longitudinal studies of genomic instability in the progression of normal to a cancerized field resulting in premalignant and malignant phenotypes can further help delineate the evolutionary dynamics of AAH and LUAD pathogenesis. Findings from such studies will be crucial for improved screening, early detection and possible prevention of premalignant disease, particularly in smokers and other high risk individuals.
The following are the supplementary data related to this article.
Supplementary material
Authors' contributions
S.Sivakumar, F.A.San Lucas, I.I.Wistuba, P.Scheet and H.Kadara conceived and designed the study. J.Fujimoto, J.Fukuoka and Y.Yatabe provided specimens and clinical databases. S.Sivakumar, F.A.San Lucas, T.L.McDowell, W.Lang, J.Fujimoto, N.Kallsen, S.Peyton, G.E.Davies and E.A.Ehli performed experiments. S.Sivakumar, F.A.San Lucas, Y.A.Jakubek, T.L.McDowell, P.Scheet and H.Kadara analyzed the data. S.Sivakumar, F.A.San Lucas, J.Zhang, P.A.Futreal, J.Fowler, E.T.Hawk., I.I.Wistuba, P.Scheet and H.Kadara interpreted the results. S.Sivakumar, F.A.San Lucas, P.Scheet and H.Kadara wrote the paper. All authors approved the final version of the manuscript.
Declaration of interests
Ms. Sivakumar has nothing to disclose.
Dr. San Lucas has nothing to disclose.
Dr. Jakubek has nothing to disclose.
Ms. McDowell has nothing to disclose.
Ms. Lang has nothing to disclose.
Mr. Kallsen has nothing to disclose.
Ms. Peyton has nothing to disclose.
Dr. Davies has nothing to disclose.
Dr. Fukuoka has nothing to disclose.
Dr. Yatabe has nothing to disclose.
Dr. Zhang has nothing to disclose.
Dr. Futreal has nothing to disclose.
Dr. Fowler has nothing to disclose.
Dr. Fujimoto has nothing to disclose.
Dr. Ehli has nothing to disclose.
Dr. Hawk has nothing to disclose.
Dr. Wistuba reports grants and personal fees from Genentech/Roche, grants and personal fees from Medimmune/AstraZeneca, grants and personal fees from Bristol-Myers Squibb, personal fees from Pfizer, grants and personal fees from HTG Molecular, grants and personal fees from Asuragen, grants and personal fees from Merck, personal fees from GlaxoSmithKline, personal fees from MSD, grants from DepArray, grants from Adaptimmune, grants from EMD Serono, grants from Takeda, grants from Amgen, grants from Karus, grants from Johnson & Johnson, grants from Bayer, grants from 4D, grants from Novartis, grants from Perkin-Elmer/Akoya, outside the submitted work.
Dr. Kadara has nothing to disclose.
Dr. Scheet has nothing to disclose.
Contributor Information
Humam Kadara, Email: hkadara@mdanderson.org.
Paul Scheet, Email: pscheet@alum.wustl.edu.
References
- 1.Herbst R.S., Heymach J.V., Lippman S.M. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadara H., Scheet P., Wistuba I.I., Spira A.E. Early events in the molecular pathogenesis of lung cancer. Cancer Prev Res. 2016;9:518–527. doi: 10.1158/1940-6207.CAPR-15-0400. [DOI] [PubMed] [Google Scholar]
- 3.Negrini S., Gorgoulis V.G., Halazonetis T.D. Genomic instability — an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 4.Takamochi K., Ogura T., Suzuki K., Kawasaki H., Kurashima Y., Yokose T. Loss of heterozygosity on chromosomes 9q and 16p in atypical adenomatous hyperplasia concomitant with adenocarcinoma of the lung. Am J Pathol. 2001;159:1941–1948. doi: 10.1016/S0002-9440(10)63041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wistuba I.I., Gazdar A.F. Lung cancer preneoplasia. Annu Rev Pathol. 2006;1:331–348. doi: 10.1146/annurev.pathol.1.110304.100103. [DOI] [PubMed] [Google Scholar]
- 6.Sivakumar S., Anthony San Lucas F., McDowell T.L., Lang W., Xu L., Fujimoto J. Genomic landscape of atypical adenomatous hyperplasia reveals divergent modes to lung adenocarcinoma. Cancer Res. 2017;77:6119–6130. doi: 10.1158/0008-5472.CAN-17-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travis W.D., Brambilla E., Nicholson A.G., Yatabe Y., Austin J.H.M., Beasley M.B. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 8.Vattathil S., Scheet P. Haplotype-based profiling of subtle allelic imbalance with SNP arrays. Genome Res. 2013;23:152–158. doi: 10.1101/gr.141374.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakubek Y., Lang W., Vattathil S., Garcia M., Xu L., Huang L. Genomic landscape established by allelic imbalance in the cancerization field of a normal appearing airway. Cancer Res. 2016;76:3676–3683. doi: 10.1158/0008-5472.CAN-15-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakubek Y.A., San Lucas F.A., Scheet P. Directional allelic imbalance profiling and visualization from multi-sample data with RECUR. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borras E., San Lucas F.A., Chang K., Zhou R., Masand G., Fowler J. Genomic landscape of colorectal mucosa and adenomas. Cancer Prev Res. 2016;9:417–427. doi: 10.1158/1940-6207.CAPR-16-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitaguchi S., Takeshima Y., Nishisaka T., Inai K. Proliferative activity, p53 expression and loss of heterozygosity on 3p, 9p and 17p in atypical adenomatous hyperplasia of the lung. Hiroshima J Med Sci. 1998;47:17–25. [PubMed] [Google Scholar]
- 15.Balsara B.R., Testa J.R. Chromosomal imbalances in human lung cancer. Oncogene. 2002;21:6877. doi: 10.1038/sj.onc.1205836. [DOI] [PubMed] [Google Scholar]
- 16.Nahar R., Zhai W., Zhang T., Takano A., Khng A.J., Lee Y.Y. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun. 2018;9:216. doi: 10.1038/s41467-017-02584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo F.-Y., Chang J.-W., Chang I.-S., Chen Y.-J., Hsu H.-S., Huang S.-F.K. The database of chromosome imbalance regions and genes resided in lung cancer from Asian and Caucasian identified by array-comparative genomic hybridization. BMC Cancer. 2012;12:235. doi: 10.1186/1471-2407-12-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinayanuwattikun C., Le Calvez-Kelm F., Abedi-Ardekani B., Zaridze D., Mukeria A., Voegele C. Elucidating genomic characteristics of lung cancer progression from in situ to invasive adenocarcinoma. Sci Rep. 2016;6:31628. doi: 10.1038/srep31628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding X.-J., Liu M.-X., Ao L., Liang Y.-R., Cao Y. Frequent loss of heterozygosity on chromosome 12q in non-small-cell lung carcinomas. Virchows Arch. 2011;458:561–569. doi: 10.1007/s00428-011-1042-9. [DOI] [PubMed] [Google Scholar]
- 20.de Bruin E.C., McGranahan N., Mitter R., Salm M., Wedge D.C., Yates L. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weir B.A., Woo M.S., Getz G., Perner S., Ding L., Beroukhim R. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett M.T., Sanchez C.A., Prevo L.J., Wong D.J., Galipeau P.C., Paulson T.G. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet. 1999;22:106–109. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X., Galipeau P.C., Sanchez C.A., Blount P.L., Maley C.C., Arnaudo J. Single nucleotide polymorphism-based genome-wide chromosome copy change, loss of heterozygosity, and aneuploidy in Barrett's Esophagus neoplastic progression. Cancer Prev Res. 2008;1:413–423. doi: 10.1158/1940-6207.CAPR-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid B.J., Prevo L.J., Galipeau P.C., Sanchez C.A., Longton G., Levine D.S. Predictors of progression in Barrett's esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol. 2001;96:2839–2848. doi: 10.1111/j.1572-0241.2001.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolan K., Morris A.I., Gosney J.R., Field J.K., Sutton R. Loss of heterozygosity on chromosome 17p predicts neoplastic progression in Barrett's esophagus. J Gastroenterol Hepatol. 2003;18:683–689. doi: 10.1046/j.1440-1746.2003.03048.x. [DOI] [PubMed] [Google Scholar]
- 26.Rosin M.P., Cheng X., Poh C., Lam W.L., Huang Y., Lovas J. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin Cancer Res. 2000;6:357–362. [PubMed] [Google Scholar]
- 27.Abbosh C., Venkatesan S., Janes S.M., Fitzgerald R.C., Swanton C. Evolutionary dynamics in pre-invasive neoplasia. Curr Opin Syst Biol. 2017;2:1–8. doi: 10.1016/j.coisb.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izumchenko E., Chang X., Brait M., Fertig E., Kagohara L.T., Bedi A. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat Commun. 2015;6:8258. doi: 10.1038/ncomms9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bremmer J.F., Brakenhoff R.H., Broeckaert M.A.M., Beliën J.A.M., Leemans C.R., Bloemena E. Prognostic value of DNA ploidy status in patients with oral leukoplakia. Oral Oncol. 2011;47:956–960. doi: 10.1016/j.oraloncology.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 30.Siebers T.J.H., Bergshoeff V.E., Otte-Höller I., Kremer B., Speel E.J.M., van der Laak J.A.W.M. Chromosome instability predicts the progression of premalignant oral lesions. Oral Oncol. 2013;49:1121–1128. doi: 10.1016/j.oraloncology.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Roy D.M., Walsh L.A., Desrichard A., Huse J.T., Wu W., Gao J. Integrated genomics for pinpointing survival loci within arm-level somatic copy number alterations. Cancer Cell. 2016;29:737–750. doi: 10.1016/j.ccell.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material