Abstract
Background
HIV-1-specific CD8+ T cells are required for immune suppression of HIV-1 replication and elimination of the associated viral reservoirs. However, effective induction of functional HIV-1-specific CD8+ T cells from naïve cells remains problematic in the setting of human vaccine trials. In this study, we investigated priming of functional HIV-1-specific CD8+ T cells from naïve cells.
Methods
HIV-1-specific CD8+ T cells were primed from naïve T cells of HIV-1-seronegative individuals using TLR4 ligand LPS or STING ligand 3′3′-cGAMP in vitro. We established HIV-1-specific CD8+ T cell lines from primed T cells and then investigated functional properties of these cells.
Findings
HIV-1-specific CD8+ T cells primed with LPS failed to suppress HIV-1. In contrast, 3′3′-cGAMP effectively primed HIV-1-specific CD8+ T cells with strong ability to suppress HIV-1. 3′3′-cGAMP-primed T cells had higher expression levels of perforin and granzyme B than LPS-primed ones. The expression levels of granzyme B and perforin and viral suppression ability of 3′3′-cGAMP-primed T cells were positively correlated with the production level of type I IFN from PBMCs stimulated with 3′3′-cGAMP.
Interpretation
The present study demonstrates the potential of 3′3′-cGAMP to induce HIV-1-specific CD8+ T cells with strong effector function from naïve cells via a strong type I IFN production and suggests that this STING ligand may be useful for AIDS vaccine and cure treatment.
Keywords: HIV-1, CD8+ T cells, Priming, STING ligand, Naïve T cell
Research in context.
Evidence before this study
Latent reservoirs of HIV-1 have been the major obstacle for the eradication of HIV-1. Recent studies suggested that HIV-specific CD8+ T cells play an important role in purging HIV reservoirs. However, several T cell vaccine trials showed no protection against HIV-1 even though the vaccine induced HIV-1-specific CD8+ T cell responses, indicating that vaccine-induced CD8+ T cells did not have enough ability to prevent HIV-1 infection. These studies emphasize the importance of induction of high-functional CD8+ T cells from naïve cells for effective AIDS vaccine. Current therapeutic strategy “shock and kill” is based on reactivation of latent reservoirs (the shock) followed by eradication of the reservoirs (the kill) by immune effector such as CD8+ T cells. However, effective approaches to induce functional HIV-1-specific CD8+ T cells for eradication of latent reservoirs need to be developed. In mouse models, STING (Stimulator of IFN Gene) ligands showed a potent antitumor effect, which associated with an increase in the frequency of tumor-specific CD8+ T cells. However, there is no direct evidence that STING ligands have ability to induce functional HIV-1-specific CD8+ T cells from naïve cells.
Added value of this study
We show here that the STING ligand 3′3′-cGAMP enables the priming of HIV-1-specific CD8+ T cells from HIV-1-seronegative individual naïve cells. These cells present strong ability to suppress HIV-1 replication, which is associated with high-level production of type I IFN. These results indicated that 3′3′-cGAMP could promote the priming of HIV-1-specific CD8+ T cells with strong effector functions from naïve cells via a strong type I IFN production.
Implications of all the available evidence
This study demonstrates the potential application of the STING ligand 3′3′-cGAMP to induce highly-functional HIV-1-specific CD8+ T cells from naïve cells. The STING ligand 3′3′-cGAMP may be useful for AIDS vaccine development to prevent HIV-1 infection, and for cure strategy to eradicate latently-infected cells.
Alt-text: Unlabelled Box
1. Introduction
HIV-1-specific CD8+ T cells are key players in the control of HIV-1 infections and cure treatment [[1], [2], [3], [4], [5], [6]]. In recent years, their central role in purging HIV-1 reservoirs has also become obvious [7]. In an in vitro model of latency, expanded HIV-1-specific CD8+ T cells from ART-treated individuals were able to eliminate reactivated HIV-1-infected CD4+ T cells [8]. The induction of potent SIV-specific CD8+ T cells led to viral control and elimination of some SIV reservoirs in macaques vaccinated with a Rhesus CMV vector [9]. These studies have opened new therapeutic avenues where agents that reactivate latently-infected cells combine with immune interventions to induce the production of effective CD8+ T cells that can clear HIV-1 reservoirs in individuals on ART. Recent encouraging data show that the reduction in the viral reservoir upon treatment with TLR7 to reactive latently infected cells correlates with the magnitude of SIV-specific CD8+ T cell responses [10]. The induction of potent HIV-1-specific CD8+ T cell responses remains, therefore, a major objective to achieve a functional cure in the absence of treatment [11]. However, previous efforts to induce effective HIV-1-specific cellular immunity in human upon vaccination have failed [12,13], suggesting that the HIV-1-specific CD8+ T cells induced by the vaccines presented no benefit in preventing or controlling HIV-1 replication.
In recent years, several reports have emphasized the importance of functional or qualitative properties of CD8+ T cells for HIV-1 control [14,15]. In particular, a strong expression of T-bet, along with effector molecules such as perforin and granzyme B whose synthesis it promotes, were shown to correlate with anti-viral efficacy [16]. Recently, the induction during the early days following an HIV-1 infection of CD8+ T cells displaying a high level of T-bet and perforin showed a direct benefit on HIV-1 reservoir seeding in vivo, thus providing a strong rationale for designing interventions aimed at inducing these potent responses against HIV-1 [17].
The acquisition of such superior functional properties is thought to be crucially dependent on the priming of HIV-1-specific CD8+ T cells. Pathogen recognition receptor (PRR) ligands enhance the priming process to elicit more robust adaptive immune responses [[18], [19], [20], [21]]. Among PRR ligands, pathogen-derived factors such as lipopolysaccharides (LPS), which is a ligand for TLR4, have been recognized as potent adjuvants that enhance the survival, effector capability, and homing of antigen-stimulated T cells [22]. It is known that LPS can also prime melanoma-specific T cells from naïve T cells [23]. Agonists for the innate sensor STING (stimulator of IFN genes) are novel and highly promising immunomodulators that are currently tested in clinical trials to improve immune surveillance and control of tumors [24]. Cyclic dinucleotides such as 3′3′-cGAMP and 2′3′-cGAMP directly bind to the transmembrane molecule STING, thereby activating the TBK1-IRF3-dependent signaling pathway to induce the robust production of type-I IFNs [25]. Type I IFNs have been shown to enhance CD8+ T cell effector function ex vivo by increasing their killing ability [[26], [27], [28], [29]]. The link between type I IFN and HIV-1 infection have been intensively studied [30]. Type I IFN are reported to induce anti-HIV-1 effects by enhancing the expression of anti-viral genes such as APOBEC3G, thetherin, and SAM domain, suggesting that IFN-I responses are detrimental for viral replication and spread [31]. Moreover, administration of IFN-α to HIV-1-infected patients with Kaposi's sarcoma resulted in lower viral load and higher CD4/CD8 T cell ratio compared to placebo [32]. Several studies showed that IFN-α-treated patients had a less severe CD4 decline, lower HIV-1 load, fewer opportunistic infections, and slower disease progression with increased frequency of activated CD8 T cells [33]. Thus, previous studies imply that type I IFN also enhances HIV-1-specific T cell functions. However, it remains unclear whether STING ligands can be used as adjuvants to induce HIV antigen specific T cells. In humans, a recent study actually suggested a rather inhibitory effect of the STING pathway on adaptive immune responses [34]. Here we used an in vitro approach to prime HIV-1-specific CD8+ T cells from unfractionated peripheral blood mononuclear cells (PBMCs) derived from HIV-1-uninfected individuals. We investigated the ability of 3′3′-cGAMP to prime functional HIV-1-specific CD8+ T cells from naïve cells and compared it to that of LPS, which can elicit melanoma-specific T cells from naïve cells but does not induce type I IFN production [35].
2. Materials and methods
2.1. Subjects
Fifteen HLA-A*24:02+ HIV-1-seronegative individuals were recruited for this study, which was approved by the Ethical Committee of Kumamoto University, Japan. Written informed consent was obtained from all subjects according to the Declaration of Helsinki. Blood mononuclear cells (PBMCs) were separated from whole blood by the use of Ficoll-Paque PLUS.
2.2. Cell lines
C1R cells expressing HLA-A*24:02 (C1R-A*2402) were generated by transfecting C1R cells with HLA-A*24:02 genes as described previously [36]. These cells were cultured in RPMI 1640 medium (invitrogen) containing 5% fetal calf serum (FCS, R5) and 0.15 mg/ml hygromycin B.
2.3. In vitro priming of naïve HIV-1-specific CD8+ T cells
Naïve precursors specific for HLA-A*24:02-RF10 were primed in vitro by using an accelerated dendritic cell co-culture protocol [37,38]. On day 0, frozen-thawed PBMCs of HLA-A*24:02+ HIV-1-seronegative individuals were suspended at 5 × 106 cells/well in 24-well tissue culture plates containing AIM-V medium (Invitrogen) supplemented with Flt3L (50 ng/ml; R&D Systems). After 24 h, 500 μl of AIM-V medium supplemented with RF10 peptide (10 μM) and TLR4 ligand LPS (0.1 μg/ml; InvivoGen) or STING ligand 3′3′-cGAMP (10 μg/ml; InvivoGen) were added, and then FCS were further added to 10% by volume per well on day 2. On day 10, the cells were collected and then stained with RF10-tetramers.
2.4. Tetramer staining
HLA-A*24:02-RF10 peptide complexes (RF10-tetramer) were generated as previously described [39]. RF10-specific CD8+ T cells were stained with PE-conjugated RF10 tetramers at 37 °C for 30 min. After 2 washes with R5, the cells were stained with 7-AAD (BD Pharmingen) at room temperature for 10 min followed by incubation with APC-conjugated anti-CD8 mAb (DAKO, Denmark) at 4 °C for 30 min. Finally, the cells were washed twice with R5 and analyzed by using a FACS Cant II. Primed T cells unstained with tetramer were used as a negative control to determine gating of the tetramer+ population. The tetramer+ population was determined based on the gating of the negative control (<0% of tetramer+ cells).
2.5. Generation of HLA-A*24:02-restricted RF10-specific CD8+ T cell lines
On day 12 post priming, RF10-specific CD8+ T cells were stained with PE-conjugated RF10 tetramers, APC-conjugated anti-CD8 mAb, and 7-AAD; and sorted in U-bottomed 96-well microtiter plates (300–500 cells/well) using a FACS Aria (BD Biosciences). Each well contained 200 μl of cell mixture (1 × 106 irradiated allogeneic PBMCs from healthy donors and 1 × 105 irradiated C1R-A*24:02 cells prepulsed with the peptide at 100 nM in R10, containing 20 ng/ml of human rIL-2 (ProSpec), and 2.5% phytohemagglutinin soup). After 2 weeks culture, RF10-specific CD8+ T cells were used in functional assays after their purity was confirmed by flow cytometry analysis using tetramers. An HLA-A*24:02-restricted RF10-specific CTL clone (CTL52) from an HIV-1-infected individual was generated as previously described [40] and used as a positive control.
2.6. Intracellular staining of perforin, granzyme B, and T-bet
RF10-specific CD8+ T cell lines and the CTL52 CTL clone were stained with PE-conjugated RF10 tetramer, 7AAD, and APC-conjugated or Pacific blue-conjugated anti-CD8 mAb (BD Pharmingen), fixed with 4% paraformaldehyde solution, and then rendered permeable with 0.1% saponin buffer. Thereafter, the cells were stained with Alexa647-labeled anti-Granzyme B (BD Pharmingen) and Alexa488-labeled anti-Per (BD Pharmingen) or V450-labeled anti-T-bet (BD Horizon). Intracellular staining for T-bet was performed by using a Transcription Factor Buffer Set (BD Pharmingen) according to the manufacturer's instructions. All stained cells were analyzed on a FACS Canto II, on the same days. To clarify whether the CD8+ T cell lines had high expression level of granzyme B, perforin, and T-bet, we compared the expression level of these molecules in CD8+ T cell lines and in the clone, and calculated relative MFI ratio as: MFI for the CD8+ T cell lines/that for the clone.
2.7. HIV-1 replication suppression assay
The ability of RF10-specific CD8+ T cell lines to suppress HIV-1 replication was measured as described previously [40]. CD4+ T cells isolated from HLA-A*24:02+ healthy donor PBMCs were incubated with HIV-1 strain NL4–3-10F carrying the SF2 strain-derived Nef138–10 epitope sequence previously reported for an infectious proviral clone of HIV-1. After 5 h, the infected CD4+ T cells (Target, 2 × 104 cells) were co-cultured with RF10-specific CD8+ T cell lines or clone (Effector, 2 × 104, 2 × 103, and 0 cells) at an E:T ratio of 1:1, 0.1:1, 0:1. Culture supernatants were collected on day 5 after post-infection, and the concentration of p24 antigen in them was measured using an enzyme-linked immunosorbent assay kit (HIV-1 p24 Ag ELISA kit, ZeptoMetrix). The percent inhibition of HIV-1 replication was calculated as follows: % suppression = (1- concentration of p24 Ag in the supernatant of HIV-1-infected CD4+ T cells cultured with RF10-specific CD8+ T cells/concentration of p24 Ag in the supernatant of HIV-1-infected CD4+ T cells cultured without the T cells) × 100.
2.8. Cytokine production and polyfunctionality
RF10-specific CD8+ T cell lines were stimulated with C1R-A*2402 cells prepulsed with 100 nM RF10 peptide for 2 h at 37 °C. Brefeldin A (10 μg/ml, Sigma-aldrich) was added, and the cells were then incubated further for 4 h at 37 °C. Subsequently, they were stained with 7-AAD and Pacific blue-conjugated anti-CD8 mAb, fixed with 4% paraformaldehyde solution at 4 °C for 20 min, and then made permeable with 0.1% saponin buffer at 4 °C for 10 min. Thereafter, the cells were stained with FITC-conjugated anti-IFN-γ mAb (BD Pharmingen), APC-conjugated IL-2 mAbs (BD Pharmingen), PE-Cy7-conjugated TNF-α mAbs (BD Pharmingen), and APC-H7-conjugated MIP-1β mAbs (BD Pharmingen). Nonspecific production of cytokines was excluded by subtracting the data of the negative control, which was the same sample stimulated with C1R-A*2402 cells without the peptide and stained with the same mAbs. Poly-functionality was quantified as a standard index according to the formula: polyfunctionality index = F0 × 0/4 + F1 × 1/4 + F2 × 2/4 + F3 × 3/4 + F4 × 4/4, where Fi is the frequency of cells expressing i functions (i = 0, 1, 2, 3, or 4) [41].
2.9. Phenotypic analysis
RF10-specific CD8+ T cell lines were stained with APC-conjugated tetramers at 37 °C for 30 min. After 2 washes, the cells were stained with PE-Cy7-conjugated anti-CCR7 (BD Pharmingen) and 7-AAD at room temperature for 10 min followed by AmCyan-conjugated anti-CD8 (BD Biosciences), Pacific blue-conjugated anti-CD3 (BD Pharmingen), PE-conjugated anti-CD28 (BD Pharmingen), APC-eFluor® 780-labeled CD27 (Invitrogen), and FITC-conjugated anti-CD45RA (BD Pharmingen) at 4 °C for 30 min. The stained cells were analyzed by using a FACS Canto II.
2.10. Type I IFN and type II IFN measurements
Frozen-thawed PBMCs of HIV-1-seronegative individuals were suspended at 5 × 106 cells/well in 24-well tissue culture plates containing AIM-V medium supplemented with LPS (0.1 μg/ml) or 3′3′-cGAMP (10 μg/ml). After 24 h, supernatants were collected; and then the concentrations of IFN-α, −β, −ω, and –γ were measured by ELISA (Invitrogen).
2.11. Blocking of type I IFN in vitro priming
On day 0, frozen-thawed PBMCs of HLA-A*24:02+ HIV-1-seronegative individuals were suspended at 5 × 106 cells/well in 24-well tissue culture plates containing AIM-V medium (Invitrogen) supplemented with Flt3L (50 ng/ml; R&D Systems). For blocking experiment of type I IFN, mixture of antibodies (PBL Assay Science) directed against an human type I IFN receptor and several type I IFNs (IFN-α, −β, −ω, −κ, and ε) was added to the culture cells 30 min before stimulation of RF10 peptide (10 μM) and LPS or 3′3′-cGAMP (10 μg/ml; InvivoGen) at day 1. FCS was further added to 10% by volume per well on day 2. On day 10, the cells were collected and then stained with RF10-tetramers.
2.12. TCR binding avidity
RF10-specific CD8+ T cell lines were stained with various concentrations of PE-conjugated RF10 tetramers (0, 0.1, 1, 10, 100 nM) at 37 °C for 30 min. After 2 washes with R5, the cells were stained with 7-AAD (BD Pharmingen) at room temperature for 10 min followed by incubation with APC-conjugated anti-CD8 mAb (DAKO, Denmark) at 4 °C for 30 min. TCR avidity between LPS-primed T cell lines and 3′3′-cGAMP T cell lines was compared in terms of TCR binding index. TCR binding index was calculated as follow: (EC50 of primed T cell line/EC50 of control clone CTL52).
2.13. TCR clonotype analysis
RF10-specific CD8+ T cell lines established from HIV-1-seronegative individuals were stained with PE-conjugated tetramers, APC-conjugated anti-CD8 mAb, and 7-AAD; and then the tetramer+ CD8+ 7-AAD− cells were sorted by using a FACS Aria. The sorted single cells were plated into each well of a 96-well plate (total 48 wells per cell line). For samples from sorted single RF10-specific CD8+ T cells, unbiased identification of TCR αβ chain usage was assessed as previously described [42]. Using this method, PCR success rates of 47% and 62% on average were obtained for TCRα and TCRβ chains respectively [42]. All PCR amplifications of full-length TCRα/β chains were sequenced (8–22 cells were sequenced per line). TCR gene usage was determined with reference to the ImMunoGeneTics (IMGT) database [43].
2.14. Statistical analysis
Univariate statistical analyses were performed using Prism software (GraphPad). Groups were compared using a paired t-test or Mann-Whitney U tests. Correlations were determined using the Spearman rank test. P values <.05 were considered significant.
3. Results
3.1. In vitro priming of HIV-1-specific CD8+ T cells from naïve cells
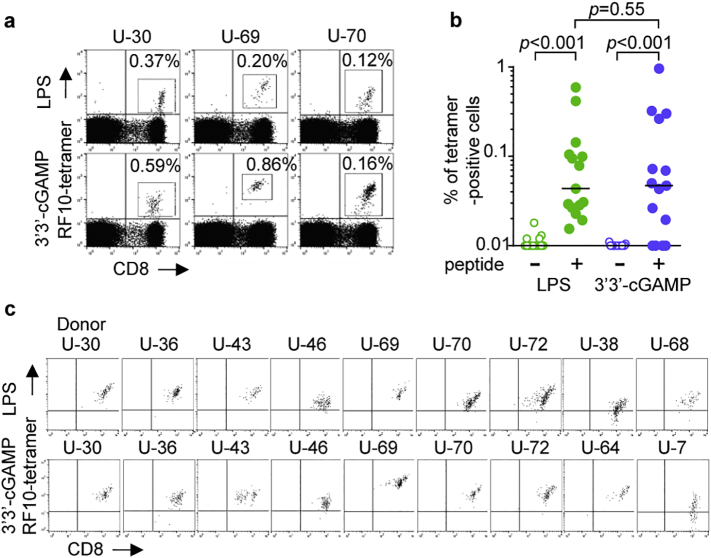
To investigate in vitro priming of naïve CD8+ T cells specific for immunodominant HLA-A*24:02-restricted HIV-1 epitope Nef RF10 (RYPLTFGWCF) [40,44], we recruited 15 HLA-A*24:02+ HIV-1-seronegative individuals. PBMCs from these individuals were stimulated with Flt3 ligand for 24 h followed by stimulation with RF10 peptide and TLR4 ligand LPS or STING ligand 3′3′-cGAMP. Both of these ligands yielded successful in vitro priming of RF10-specific CD8+ T cells (Fig. 1a). RF10-specific CD8+ T cells were measured by using HLA-A*24:02 tetramer with RF10 peptide. No significant difference was found in the frequency of RF10-tetramer+ CD8+ T cells between LPS-primed and 3′3′-cGAMP-primed CD8+ T cells (Fig. 1b), suggesting that there was no difference between these 2 ligands in terms of the magnitude of RF10-specific CD8+ T cell expansion upon priming. Due to the low frequency of RF10-specific CD8+ T cells among total primed T cells, it was difficult to analyze functional properties of primed T cells with these ligands. Therefore, we sought to establish RF10-specific T cell lines for further functional analysis. For this purpose, primed RF10-tetramer+ CD8+ T cells were sorted and cultured for two additional weeks. We successfully established RF10-specific CD8+ T cell lines primed with LPS and/or 3′3′-cGAMP from 11 HLA-A*24:02+ HIV-1-seronegative individuals for further analysis. Both LPS-primed T cell lines and 3′3′-cGAMP-primed ones were established from 7 individuals, whereas either one was obtained from 4 individuals (Fig. 1c). The binding ability of the tetramer varied among these T cell lines (Fig. 1c), suggesting that these T cell lines expressed different TCRs or the same TCR with different affinity. We used these T cell lines for functional assays.
Fig. 1.
Priming of HIV-1-specific CD8+ T cells from naïve cells of HLA-A*24:02+ HIV-1-seronegative individuals.
RF10-specific CD8+ T cells from 15 HLA-A*24:02+ HIV-1-seronegative individuals were primed with LPS or 3′3′-cGAMP. (a) Representative results of tetramer staining of HLA-A*24:02-restricted RF10-specific CD8+ T cells primed with LPS or 3′3′-cGAMP. The frequency of the tetramer+ cells among the CD8+ T cell population is indicated. (b) Summarized results for the frequency of RF10-specific CD8+ T cells. Horizontal bars indicate median values. Statistical analysis was conducted by use of the non-parametric Mann Whitney test. (c) Tetramer staining of RF10-specific CD8+ T cell lines established from 11 HLA-A*24:02+ HIV-1-seronegative individuals. Both LPS-primed and 3′3′-cGAMP-primed RF10-specific CD8+ T cell lines were established from 7 individuals, while either one was obtained from 4 individuals. These T cell lines were stained with HLA-A*24:02-RF10 tetramers.
3.2. Ability of 3′3′cGAMP-primed RF10-specific CD8+ T cell lines to suppress HIV-1 replication
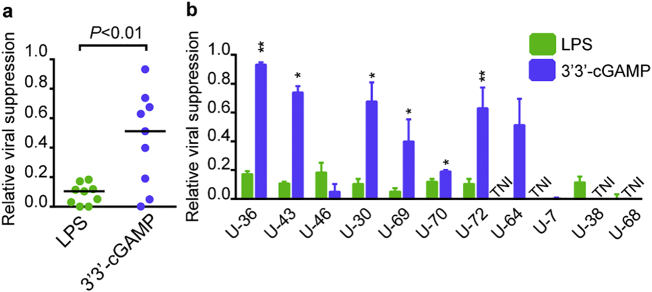
To investigate the efficacy of RF10-specific CD8+ T cells primed from naïve cells to suppress replication of HIV-1, we measured the ability of LPS-primed RF10-specific T cell lines or 3′3′cGAMP-primed ones to suppress HIV-1 replication in primary CD4+ T cells infected with NL4-3, and compared to that of HLA-A*24:02-restricted RF10-specific CTL clone (CTL52) presenting a strong HIV suppression capacity [40,44]. Despite successful priming, all LPS-primed RF10-specific T cell lines presented a poor HIV suppression capacity (Supplementary Fig. 1). In contrast, 3′3′-cGAMP-primed RF10-specific CD8+ T suppressed the replication of HIV-1 (Supplementary Fig. 1). 3′3′cGAMP-primed RF10-specific T cell lines exhibited a significantly higher viral suppression ratio than LPS-primed ones (Fig. 2a). Five out of the nine 3′3′cGAMP-primed T cell lines showed a > 0.5 ratio, whereas all of the nine LPS-primed ones showed a < 0.2 ratio (Fig. 2a). The former T cell lines showed higher viral suppression capacity than the latter T ones from the same individual in six out of seven cases (Fig. 2b). Taken together, these results suggest that 3′3′-cGAMP effectively primed RF10-specific T cells with a much stronger viral suppression ability than LPS-primed ones.
Fig. 2.
Ability of HIV-1-specific CD8+ T cells primed from naïve cells to suppress HIV-1 replication.
(a) Ability of RF10-specific T cell lines primed with LPS or 3′3′-cGAMP to suppress the replication of HIV-1. NL4–3-infected CD4+ T cells from an HLA-A*24:02+ donor were co-cultured with T cell lines at different E:T ratios. Relative viral suppression ratio was calculated as follows: percent inhibition of primed T cell lines/that of control clone. The results at the E:T ratio of 1:1 are shown. (b) Comparison of relative viral suppression ability between LPS-primed T cell lines and 3′3′cGAMP-primed ones from each individual. Data are shown as the means and SD of triplicate assays. Statistical analysis was conducted by use of the non-parametric Mann Whitney test (a), a paired t-test (b). Horizontal bars indicate median values (a). *P < .05, **P < .01. TNI, T cells were not induced.
3.3. TCR avidity and clonotype of HIV-1-specific CD8+ T cells primed from naïve cells
We next aimed at better defining the determining factors of HIV-1 suppressive capacity in 3′3′-cGAMP-primed RF10-specific CD8+ T cells. Since TCR avidity is an important determinant of the capacity for CD8+ T cells to suppress HIV-1 [45], we compared TCR avidity of the T cell lines primed with the two ligands, based on the measure of TCR binding index (Supplementary Fig. 2a). There was no significant difference in this index between LPS-primed RF10-specific T cell lines and 3′3′-cGAMP-primed ones (Fig. 3a). We also observed no correlation between viral suppression ability and TCR avidity in LPS-primed T cell lines and 3′3′-cGAMP-primed ones (Fig. 3b). These results suggest that TCR avidity was not a critical factor for viral suppression ability of primed T cell lines but that other factors may have critically contributed to strong viral suppression ability in this setting.
Fig. 3.
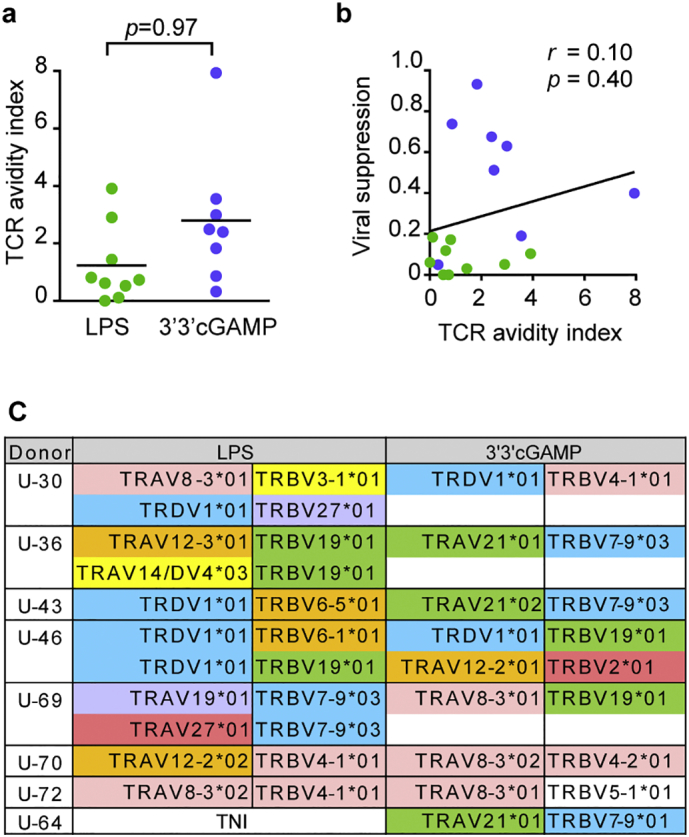
TCR avidity and TCR clonotype of RF10-specific CD8+ T cell lines from naïve cells.
(a) The mean fluorescence intensity (MFI) of RF10-specific CD8+ T cell lines stained with tetramers (0, 0.1, 1, 10, and 100 nM) was measured by flow cytometry. TCR avidity was compared between LPS-primed T cell lines and 3′3′cGAMP-primed ones based on TCR binding indexes: EC50 of primed T cell line/EC50 of the control clone. (b) Correlation between relative viral suppression ratio and TCR avidity index for LPS-primed T cell lines (green) and 3′3′cGAMP-primed ones (blue). Each dot represents a T cell line established from each individual. Statistical analysis was conducted by use of the non-parametric Mann Whitney test (a) or Spearman correlations test (b). (c) Clonotypic analysis of RF10-specific CD8+ T cell lines established from naïve T cells. LPS-primed T cell lines and 3′3′cGAMP-primed ones were sorted at the single-cell level, and then TCRαβ-chain sequences were analyzed. TNI, T cells were not induced. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Despite TCR repertoire diversity and flexibility, HIV-specific CD8+ T cell populations are frequently characterized by specific clonotypic immunodominance or even public TCRs in HIV-1-infected individuals [[44], [45], [46], [47], [48], [49]]. For example, Nef RF10-specific CD8+ T cells having TRVD1*01/TRBV28*01 clonotype were predominantly elicited among chronically HIV-1-infected HLA-A*24:02+ Japanese individuals [44,46]. These findings suggest that a particular TCR clonotype may also be recruited among RF10-specific CD8+ T cells primed with LPS or 3′3′-cGAMP from naïve T cells. We therefore analyzed the TCR clonotype of LPS-primed RF10-specific CD8+ T cell lines and 3′3′-cGAMP-primed ones at the single-cell level to clarify whether these 2 different ligands selected the same T cell clones or not. Except for 1 donor (U-46), different TCR clonotypes were found between LPS-primed RF10-specific CD8+ T cells and 3′3′-cGAMP-primed ones established from the same donors (Fig. 3c and Supplementary Fig. 2b). This TCR repertoire diversity highlights the random selection of antigen-specific clonotypes among a polyclonal naïve T cell population upon priming, indicating that 3′3′-cGAMP did not preferentially select T cells expressing high-functioning TCRs.
3.4. Differentiation status and cytokine production of HIV-1-specific CD8+T cells primed from naïve cells
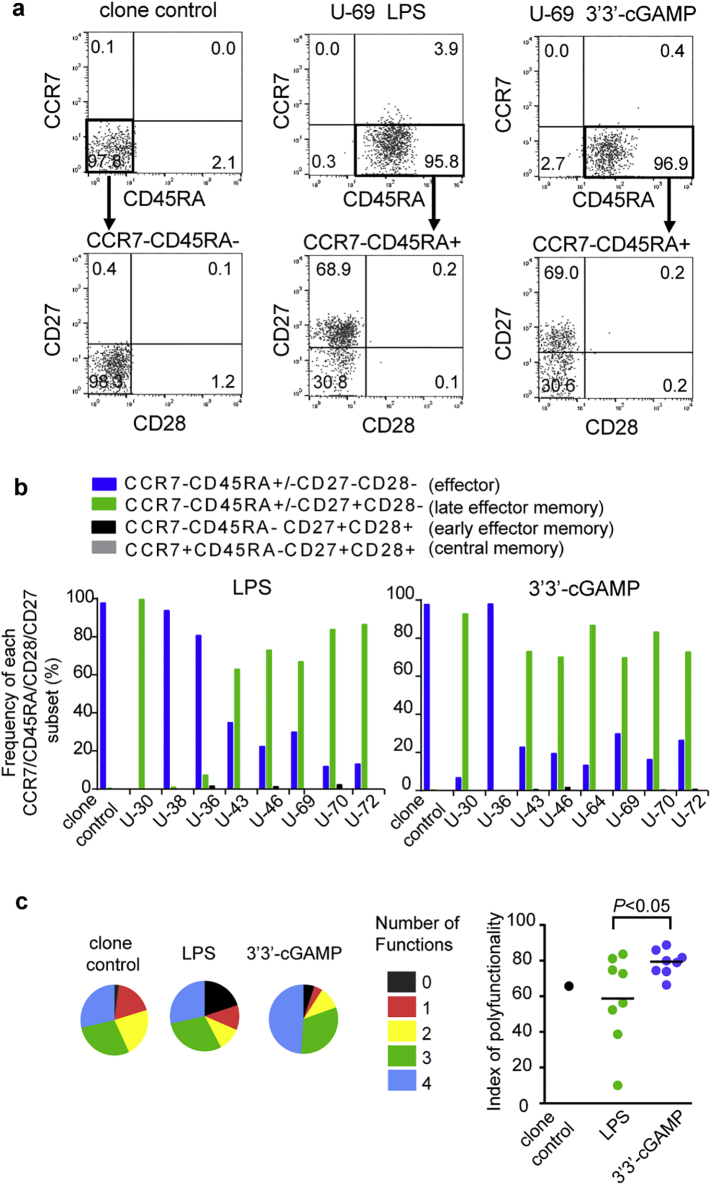
We analyzed differentiation status of HIV-1-specific T cells primed with 3′3′-cGAMP or LPS. Human CD8+ T cells are classified into five major populations based on their expression of four cell-surface markers: CD27highCD28+CD45RA+CCR7+ (naive), CD27+CD28+CD45RA−CCR7+ (central memory), CD27+CD28+CD45RA−CCR7− (early effector memory), CD27lowCD28−CD45RA+/-CCR7− (late effector memory), and CD27−CD28−CD45RA+/-CCR7− (effector) [50]. To investigate the differentiation stage of the primed T cell lines, we analyzed their phenotype by using these four markers (Fig. 4a). The control CTL clone expressed the CD27−CD28−CD45RA−CCR7− effector phenotype. In contrast, both LPS-primed T cell lines and 3′3′cGAMP-primed ones presented the late effector memory phenotype (Fig. 4a and b), although some of the lines showed the effector phenotype. These findings indicate that T cell lines primed with these ligands had differentiated to a similar stage.
Fig. 4.
Differentiation phenotype and cytokine production of RF10-specific CD8+ T cells primed with LPS or 3′3′-cGAMP.
(a) The differentiation stage of RF10-specific CD8+ T cell lines primed with LPS or 3′3′-cGAMP and of the control CTL clone was analyzed by using the following 4 cell-surface markers: CD27, CD28, CD45RA, and CCR7. Representative flow cytometric results of the RF10-specific CD8+ T cells. RF10 tetramer+ CD8+ T cells were gated and analyzed for CD45RA and CCR7 expression. The CCR7−CD45RA− or CCD7−CD45RA+ subset was further divided into subpopulations based on the expression of CD27 and CD28. (b) Summarized results showing the frequency of each of the CCD7 CD45RA CD27 CD28 subsets among the RF10 tetramer+ CD8+ T cell population. (c) Ability of RF10-specific CD8+ T cell lines to produce multiple cytokines or a chemokine. Responses of LPS-primed or 3′3′-cGAMP-primed T cell lines to C1R-A*24:02 cells pulsed with RF10 peptide were analyzed by measuring the production levels of IFN-γ, IL-2, TNF-α, and MIP-1β. Responses are grouped into 0–4 functions and are summarized in the pie chart (left panel). Poly-functional ability of T cell lines was compared in terms of poly-functionality index (right panel). The method for the calculation of the poly-functionality index was shown in Materials and Methods. Statistical analysis was conducted by use of the non-parametric Mann Whitney test. Horizontal bars indicate median values.
We next investigated the ability of these T cell lines to produce different cytokines or chemokines (IFN-γ, IL-2, TNF-α and MIP-1β). The RF10-specific CTL clone 52 established from an HIV-1-infected HLA-A*24:02+ individual was used as control for further analysis, since this clone had a strong ability to suppress HIV-1 replication [40,44]. Both LPS-primed T cell lines and 3′3′cGAMP-primed ones had the ability to produce all of these cytokines and chemokine in response to RF10 peptide, although no difference was found in their production between these cells (Supplementary Fig. 3). However, overall 3′3′-cGAMP-primed T cell lines showed significantly a higher polyfunctionality index than LPS-primed ones (Fig. 4c and Supplemental Table 1), suggesting that 3′3′-cGAMP primed more polyfunctional CD8+ T cells than did LPS.
3.5. Expression of cytolytic effector molecules in HIV-1-specific CD8+ T cells primed with 3′3′-cGAMP
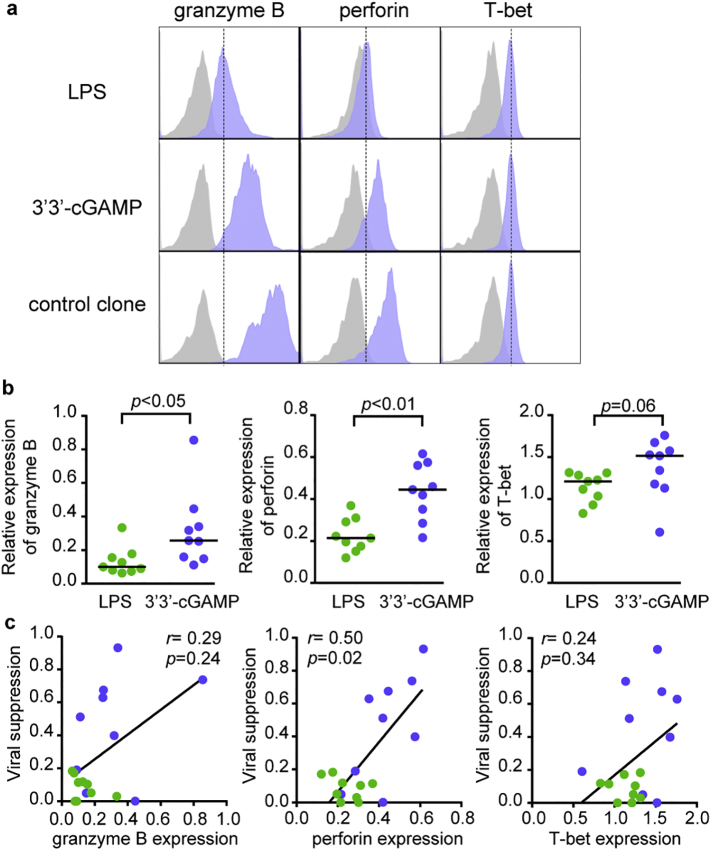
To investigate the expression level of cytolytic effector molecules in T cells primed with 3′3′-cGAMP or LPS, we measured intracellular granzyme B and perforin as well as T-bet in the RF10-specific CD8+ T cell lines (Fig. 5a). 3′3′-cGAMP-primed RF10-specific T cell lines displayed higher expression levels of granzyme B (P < .05), perforin (P < .01), and T-bet (P = .06) than LPS-primed ones (Fig. 5b and Supplementary Fig. 4). Thus, LPS-primed T cell lines had not been fully differentiated into effector T cells expressing sufficient amounts of granzyme B and perforin. We found a significant positive correlation between viral suppression ability and perforin expression in the primed T cell lines (Fig. 5c). There was also a clear trend when considering 3′3′-cGAMP-primed CD8+ T cells only (Supplementary Fig. 5). The expression level of perforin appears therefore as a key factor for viral suppression capacity of 3′3′-cGAMP-primed RF10-specific T cells. This is in line with previous results on HCMV or EBV-specific T cells [50]. Overall, 3′3′-cGAMP promoted the induction of HIV-1 suppressive CD8+ T cells, not through the selection of naïve T cells with high TCR avidity, but by enhancing intrinsic functional properties of primed T cells.
Fig. 5.
Expression of intracellular cytolytic effector molecules in RF10-specific CD8+ T cells primed from naïve cells.
Expression levels of granzyme B, perforin, and T-bet in RF10-specific CD8+ T cell lines primed with LPS or 3′3′-cGAMP. The expression level of each molecule in LPS-primed T cell lines (n = 9) or 3′3′-cGAMP-primed ones (n = 9) was measured by intracellular staining with analysis by flow cytometry. (a) Representative staining data of HLA-A*24:02-restricted RF10-specific CD8+ T cell lines primed with LPS or 3′3′-cGAMP in the same individual. (b) Comparison of relative expression of granzyme B, perforin, and T-bet between LPS-primed T cell lines and 3′3′cGAMP-primed ones in terms of relative MFI ratio. Relative MFI ratio was calculated as MFI for primed T cell lines/that for the control clone. (c) Correlation between relative viral suppression ratio and relative expression of granzyme B, perforin, and T-bet in LPS-primed T cell lines (green) and 3′3′cGAMP-priemd ones (blue). Each dot represents a T cell line from each individual. Statistical analysis was conducted by use of the non-parametric Mann Whitney test (b) or Spearman correlations test (c). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Correlation between IFN production and effector functions of 3′3′-cGAMP-primed CD8+ T cells
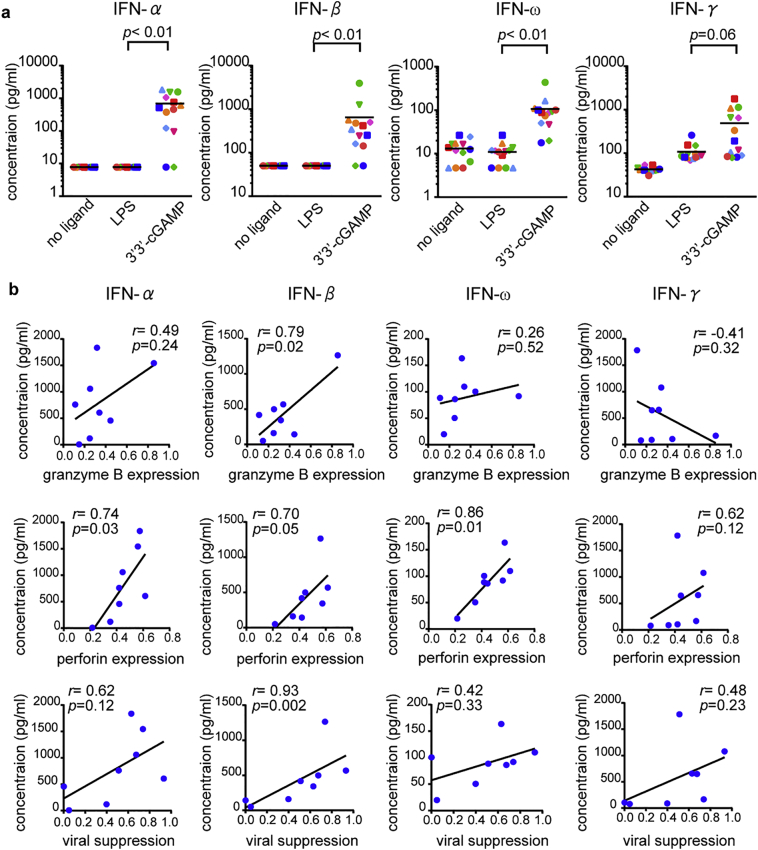
To get mechanistic insights underlying effective priming of HIV-1-specific CD8+ T cells with strong effector functions in the presence of 3′3′-cGAMP, we investigated the production of IFN from HIV-1-seronegative individuals PBMCs. We stimulated PBMCs from 13 HIV-1-seronegative individuals with LPS or 3′3′-cGAMP, and then measured the production levels of type I IFN (IFN-α, −β, and -ω) and type II IFN (IFN-γ). 3′3-’cGAMP induced significantly higher level of IFN production than LPS, except in the case of IFN-γ production (Fig. 6a). These results indicate that a stronger type I IFN production was triggered by 3′3′-cGAMP than by LPS during T-cell priming. To investigate the effect of type I IFN production on the effector function of 3′3′-primed HIV-specific CD8+ T cells, we further examined the correlation between IFN production induced by 3′3′-cGAMP and the expression level of cytolytic effector molecules or viral suppression ability of 3′3′-cGAMP-primed RF10-specific T cell lines established from 8 individuals. The expression level of perforin in 3′3′-GAMP-primed T cells correlated positively with the production of all type I IFNs, and the one of granzyme B was significantly correlated with IFN-β production levels (Fig. 6b). Viral suppression capacity was also significantly correlated with IFN-β and showed a trend for a positive correlation with IFN-α (Fig. 6b).
Fig. 6.
Correlation between type I IFN production and effector functions of RF10-specific CD8+ T cells primed with 3′3′-cGAMP.
(a) Induction of type I IFN and type II IFN production from PBMCs by LPS or 3′3′-cGAMP. PBMCs from 13 HIV-1-seronegative individuals were stimulated with LPS or 3′3′-cGAMP for 24 h, and then the production levels of IFN-α, −β, −ω, and -γ were measured by using ELISA. (b) Correlation between type I and type II IFN production by 3′3′-cGAMP and relative expression of granzyme B (upper), perforin (middle), or relative viral suppression ratio (lower) of 3′3′cGAMP-primed T cells. Each dot represents each individual whose T cell lines had been analyzed for the expression of perforin and granzame B as well as their ability to suppress viral replication. Statistical analysis was conducted by use of the non-parametric Mann Whitney test (a) or Spearman correlations test (b).
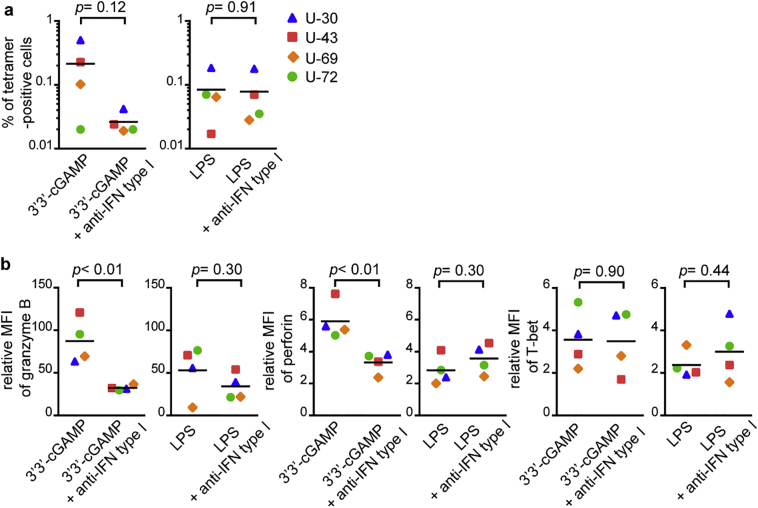
To clarify the effect of type I IFN on induction of functional T cells, we further investigated the blocking effect of anti-type I IFN blocking antibodies on the priming of functional T cells. The presence of type I IFN blocking antibodies during priming yielded a trend towards lower frequency of the 3′3′-cGAMP-primed T cells, whereas there was no effect of these antibodies on LPS-primed ones (Fig. 7a). This suggests that type I IFNs play a role in the induction of the specific T cells from naïve cells. The expression levels of granzyme B and perforin in 3′3′-cGAMP-primed HIV-1-specific T cell lines were significantly decreased in the presence of anti-type I IFN blocking antibodies, whereas levels in LPS-primed cell lines were not affected (Fig. 7b). These results support the idea that type I IFN production induced by 3′3′-cGAMP is a key factor for induction of functional T cells from naïve T cells. Type I IFN has been reported to promote the expression level of cytolytic molecules and cytotoxic effector function in CD8+ T cells [[26], [27], [28], [29]]. Given these reports and the present findings, we propose that high-level of type I IFN production induced by 3′3′-cGAMP is the mechanism by which the effector functions of HIV-1-specific CD8+ T cells primed from naïve cells was enhanced.
Fig. 7.
Effect of type I IFN blocking on induction of functional RF10-specific CD8+ T cells primed with 3′3′-cGAMP.
RF10-specific CD8+ T cells from 4 HLA-A*24:02+ HIV-1-seronegative individuals were primed with LPS or 3′3′-cGAMP in the presence or absence of anti-IFN type I blocking antibodies. Blocking effect of anti-type I IFN antibodies on the frequency of primed-RF10-specific CD8+ T cells (a), and expression of granzyme B, perforin, and T-bet in primed T cell lines (b). Expression levels of granzyme B, perforin, and T-bet were compared in terms of relative MFI. Relative MFI was calculated as MFI for T cell lines/that for negative control, being respective unstained samples. Horizontal bars indicate median values. Statistical analysis was conducted by use of the non-parametric Mann Whitney test.
4. Discussion
We demonstrated that STING ligand 3′3′-cGMAP elicited functional HIV-1-specific CD8+ T cells expressing high levels of perforin and granzyme B, although TLR4 ligand LPS did not do so. STING ligands are known to promote cross-presentation, which is key for cross-priming and effective induction of CD8+ T cells [51]; and they also generate particularly strong type I IFN responses [52] that lead directly to up-regulated expression of T-bet and subsequently to perforin and granzyme B production in primed CD8+ T cells [26,53]. Indeed, type I IFN signaling drives the acquisition of the cytolytic function of CD8+ T cells [[27], [28], [29]]. A mouse model study showed that IFN-α and -β provided signal-III directly to CD8+ T cells via a STAT4-dependent pathway to develop cytolytic function and production of IFN-γ [53]. The present study demonstrated that 3′3′-cGAMP produced a much higher level of type I IFN than did LPS and that the production level of this IFN was correlated with the expression levels of perforin and granzyme B in 3′3′-cGAMP-primed T cells. These results suggest that robust type I IFN production induced by 3′3′cGAMP promoted the priming of HIV-1-specific CD8+ T cells with strong cytolytic effector function. Therefore, rather than acting on signal I between T cells and APCs (i.e., TCR-pMHC signaling), STING ligands exploited signal III (i.e., stimulatory cytokines) in order to boost intrinsic cellular factors such as perforin in T cells, thus enhancing their cytolytic functions.
Considering the importance of TCR avidity for CD8+ T cell effector functions and efficacy [45], we speculated that the STING ligand might have promoted the selection of HIV-1-specific T cell clonotypes having high-avidity TCRs. However, this was not the case, as there was no significant difference in TCR avidity between 3′3′-cGAMP-primed RF10-specific T cells and LPS-primed ones, as well as no direct correlation between TCR avidity and HIV-1 suppressive capacity. Clonotypic analyses at the single-cell level showed that a variety of TCR clonotypes were recruited within HIV-1-specific CD8+ T cell population primed under different adjuvant conditions from the same or different donors, indicating the random selection of antigen-specific clonotypes among a polyclonal naïve T cell population upon priming. Of note, 1 clonotype (TRAV1*01/TRBV19*01) found in a donor (U-46) primed T cell population, was also previously identified in a RF10-specific CD8+ T cell population from a chronically HIV-1-infected patient [44,46], illustrating the connection between in vitro and in vivo priming of HIV-1-specific CD8+ T cells.
We also showed that although seemingly polyfunctional HIV-1-specific CD8+ T cells could be successfully primed with LPS, they actually lacked important functional attributes of efficacy such as perforin expression and the ability to suppress HIV-1 replication. This is reminiscent of the immunogenicity but poor efficacy of previous T cell-based HIV-1 vaccines in clinical trials [12,13] and implies that further developments are needed to induce more effective CD8+ T cells able to impact positively vaccination outcomes. Our findings highlight the potential of 3′3′-cGAMP to induce potent CD8+ T cell immunity against HIV-1. STING ligands may therefore present an interest in HIV vaccine development.
Although encouraging, the present work represents only a preliminary step towards the potential use of cGAMP as an adjuvant to induce effective HIV-1 specific CD8+ T cells, as multiple issues need to be solved before envisaging clinical trials. First, the present study is in vitro, and may not reflect the induction of T-cells in vivo, where the APCs and environment in lymphoid tissue differ. Second, our study focuses primarily on the induction of T cells with immediate effector functions, while the goal of vaccination is generally on inducing long-lived cells with strong regenerative capacity. Further studies are therefore needed to address these issues, as well as the homing of primed HIV-1-specific T cells to lymphoid tissues in vivo, or the selection of HIV-1 protective epitopes. Our previous studies demonstrated that CD8+ T cells specific for conserved Gag and Pol regions have strong ability to suppress HIV-1 replication in vivo and in vitro [[54], [55], [56]], suggesting that these T cells may have ability to eradicate the latent viral reservoir. STING ligands may therefore be incorporated into nanoparticles such as amphiphiles or polymers as delivery agents, together with conserved and protective HIV-1 epitopes, in order to generate effective anti-HIV-1 immunity in lymphoid tissues. In association with molecules able to reactivate the latent reservoir, the immediate immuno-stimulatory potential of cGAMP may be crucial to eradicate efficiently latently HIV-1 infected cells towards a functional cure of HIV-1+ patients [57].
To conclude, we present here a reliable in vitro approach to prime functional human CD8+ T cells specific for an HIV-1 antigen. STING ligands appear to have a strong ability to prime HIV-1-specific CD8+ T cells with strong effector functions enhanced by high-level production of type-I IFN. Our findings represent an important methodological advance, enabling for the first time the study of HIV-1-specific CD8+ T cell priming outside the complex setting of a vaccine clinical trial towards the functional cure and potentially the prevention of HIV-1 infection. The present work paves the way for the use of potent adjuvants such as STING ligands in the fight against HIV-1.
Funding
This work was supported by a grant-in-aid (18fk0410021h0001) for AIDS Research from Japan Agency for Medical Research and Development (AMED), by a grant-in-aid (16K19159) for Young Scientists, and by an AIDS International Collaborative Research Grant from the Ministry of Education, Science, Sports, and Culture of Japan. Funding sources had no involvement in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Declaration of interests
The authors have no financial conflicts of interest.
Author's contributions
N.K performed experiments, analyzed data, and wrote the manuscript. X.S. generated the virus and purified HLA-peptide monomer. T.A. purified HLA-peptide monomer. A.L. contributed to perform experiment on T-cell priming. T.Y. contributed to prepare reagents. V.A. designed the study, analyzed data and wrote the manuscript. M.T. designed the study, supervised all experiments, and wrote the manuscript. All authors revised and edited the manuscript.
Acknowledgements
We thank Sachiko Sakai and Sachie Kawazoe for their secretarial assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.03.078.
Appendix A. Supplementary data
Supplementary material
References
- 1.Borrow P., Lewicki H., Hahn B.H., Shaw G.M., Oldstone M.B. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koup R.A., Safrit J.T., Cao Y., Andrews C.A., McLeod G., Borkowsky W. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogg G.S., Jin X., Bonhoeffer S., Dunbar P.R., Nowak M.A., Monard S. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 4.Appay V., Douek D.C., Price D.A. CD8+ T cell efficacy in vaccination and disease. Nat Med. 2008;14:623–628. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- 5.Jones R.B., Walker B.D. HIV-specific CD8+ T cells and HIV eradication. J Clin Invest. 2016;126:455–463. doi: 10.1172/JCI80566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiepiela P., Ngumbela K., Thobakgale C., Ramduth D., Honeyborne I., Moodley E. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 7.Deeks S.G.H.I.V. Shock and kill. Nature. 2012;487:439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 8.Shan L., Deng K., Shroff N.S., Durand C.M., Rabi S.A., Yang H.C. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity. 2012;36:491–501. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen S.G., Piatak M., Ventura A.B., Hughes C.M., Gilbride R.M., Ford J.C. Immune clearance of highly pathogenic SIV infection. Nature. 2013;502:100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borducchi E.N., Cabral C., Stephenson K.E., Liu J., Abbink P., Ng'ang'a D. Ad26/MVA therapeutic vaccination with TLR7 stimulation in SIV-infected rhesus monkeys. Nature. 2016;540:284–287. doi: 10.1038/nature20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun T.W., Justement J.S., Moir S., Hallahan C.W., Ehler L.A., Liu S. Suppression of HIV replication in the resting CD4+ T cell reservoir by autologous CD8+ T cells: implications for the development of therapeutic strategies. Proc Natl Acad Sci U S A. 2001;98:253–258. doi: 10.1073/pnas.98.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchbinder S.P., Mehrotra D.V., Duerr A., Fitzgerald D.W., Mogg R., Li D. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the step study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammer S.M., Sobieszczyk M.E., Janes H., Karuna S.T., Mulligan M.J., Grove D. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med. 2013;369:2083–2092. doi: 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almeida J.R., Price D.A., Papagno L., Arkoub Z.A., Sauce D., Bornstein E. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Migueles S.A., Laborico A.C., Shupert W.L., Sabbaghian M.S., Rabin R., Hallahan C.W. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 16.Hersperger A.R., Martin J.N., Shin L.Y., Sheth P.M., Kovacs C.M., Cosma G.L. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood. 2011;117:3799–3808. doi: 10.1182/blood-2010-12-322727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takata H., Buranapraditkun S., Kessing C., Fletcher J.L., Muir R., Tardif V. Delayed differentiation of potent effector CD8. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aag1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutjahr A., Tiraby G., Perouzel E., Verrier B., Paul S. Triggering intracellular receptors for vaccine adjuvantation. Trends Immunol. 2016;37:716. doi: 10.1016/j.it.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Smyth K., Garcia K., Sun Z., Tuo W., Xiao Z. TLR agonists are highly effective at eliciting functional memory CTLs of effector memory phenotype in peptide immunization. Int Immunopharmacol. 2013;15:67–72. doi: 10.1016/j.intimp.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warger T., Osterloh P., Rechtsteiner G., Fassbender M., Heib V., Schmid B. Synergistic activation of dendritic cells by combined toll-like receptor ligation induces superior CTL responses in vivo. Blood. 2006;108:544–550. doi: 10.1182/blood-2005-10-4015. [DOI] [PubMed] [Google Scholar]
- 21.Kawai T., Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 22.McAleer J.P., Rossi R.J., Vella A.T. Lipopolysaccharide potentiates effector T cell accumulation into nonlymphoid tissues through TRIF. J Immunol. 2009;182:5322–5330. doi: 10.4049/jimmunol.0803616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubsky P., Saito H., Leogier M., Dantin C., Connolly J.E., Banchereau J. IL-15-induced human DC efficiently prime melanoma-specific naive CD8+ T cells to differentiate into CTL. Eur J Immunol. 2007;37:1678–1690. doi: 10.1002/eji.200636329. [DOI] [PubMed] [Google Scholar]
- 24.Corrales L., McWhirter S.M., Dubensky T.W., Gajewski T.F. The host STING pathway at the interface of cancer and immunity. J Clin Invest. 2016;126:2404–2411. doi: 10.1172/JCI86892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubensky T.W., Kanne D.B., Leong M.L. Rationale, progress and development of vaccines utilizing STING-activating cyclic dinucleotide adjuvants. Ther Adv Vaccines. 2013;1:131–143. doi: 10.1177/2051013613501988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hervas-Stubbs S., Riezu-Boj J.I., Gonzalez I., Mancheño U., Dubrot J., Azpilicueta A. Effects of IFN-α as a signal-3 cytokine on human naïve and antigen-experienced CD8(+) T cells. Eur J Immunol. 2010;40:3389–3402. doi: 10.1002/eji.201040664. [DOI] [PubMed] [Google Scholar]
- 27.Kolumam G.A., Thomas S., Thompson L.J., Sprent J., Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med. 2005;202:637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiesel M., Crouse J., Bedenikovic G., Sutherland A., Joller N., Oxenius A. Type-I IFN drives the differentiation of short-lived effector CD8+ T cells in vivo. Eur J Immunol. 2012;42:320–329. doi: 10.1002/eji.201142091. [DOI] [PubMed] [Google Scholar]
- 29.Moseman E.A., Wu T., de la Torre J.C., Schwartzberg P.L., McGavern D.B. Type I interferon suppresses virus-specific B cell responses by modulating CD8. Sci Immunol. 2016;1 doi: 10.1126/sciimmunol.aah3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bosinger S.E., Utay N.S. Type I interferon: understanding its role in HIV pathogenesis and therapy. Curr HIV/AIDS Rep. 2015;12:41–53. doi: 10.1007/s11904-014-0244-6. [DOI] [PubMed] [Google Scholar]
- 31.Soper A., Kimura I., Nagaoka S., Konno Y., Yamamoto K., Koyanagi Y. Type I interferon responses by HIV-1 infection: association with disease progression and control. Front Immunol. 2018;8:1823. doi: 10.3389/fimmu.2017.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane H.C., Kovacs J.A., Feinberg J., Herpin B., Davey V., Walker R. Anti-retroviral effects of interferon-alpha in AIDS-associated Kaposi's sarcoma. Lancet. 1988;2:1218–1222. doi: 10.1016/s0140-6736(88)90811-2. [DOI] [PubMed] [Google Scholar]
- 33.Asmuth D.M., Murphy R.L., Rosenkranz S.L., Lertora J.J., Kottilil S., Cramer Y. Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon Alfa-2a in HIV-1-monoinfected participants: a phase II clinical trial. J Infect Dis. 2010;201:1686–1696. doi: 10.1086/652420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerboni S., Jeremiah N., Gentili M., Gehrmann U., Conrad C., Stolzenberg M.C. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J Exp Med. 2017;214:1769–1785. doi: 10.1084/jem.20161674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longhi M.P., Trumpfheller C., Idoyaga J., Caskey M., Matos I., Kluger C. Et.al, dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J Exp Med. 2009;206:1589–1602. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikeda-Moore Y., Tomiyama H., Miwa K., Oka S., Iwamoto A., Kaneko Y. Identification and characterization of multiple HLA-A24-restricted HIV-1 CTL epitopes: strong epitopes are derived from V regions of HIV-1. J Immunol. 1997;159:6242–6252. [PubMed] [Google Scholar]
- 37.Lissina A., Briceño O., Afonso G., Larsen M., Gostick E., Price D.A. Priming of qualitatively superior human effector CD8+ T cells using TLR8 ligand combined with FLT3 ligand. J Immunol. 2016;196:256–263. doi: 10.4049/jimmunol.1501140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinuzzi E., Afonso G., Gagnerault M.C., Naselli G., Mittag D., Combadière B. acDCs enhance human antigen-specific T-cell responses. Blood. 2011;118:2128–2137. doi: 10.1182/blood-2010-12-326231. [DOI] [PubMed] [Google Scholar]
- 39.Altman J.D., Moss P.A., Goulder P.J., Barouch D.H., McHeyzer-Williams M.G., Bell J.I. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 40.Fujiwara M., Tanuma J., Koizumi H., Kawashima Y., Honda K., Mastuoka-Aizawa S. Different abilities of escape mutant-specific cytotoxic T cells to suppress replication of escape mutant and wild-type human immunodeficiency virus type 1 in new hosts. J Virol. 2008;82:138–147. doi: 10.1128/JVI.01452-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsen M., Sauce D., Arnaud L., Fastenackels S., Appay V., Gorochov G. Evaluating cellular polyfunctionality with a novel polyfunctionality index. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun X., Saito M., Sato Y., Chikata T., Naruto T., Ozawa T. Unbiased analysis of TCRα/β chains at the single-cell level in human CD8+ T-cell subsets. PLoS One. 2012;7:e40386. doi: 10.1371/journal.pone.0040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lefranc M.P., Giudicelli V., Duroux P., Jabado-Michaloud J., Folch G., Aouinti S. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015;43(Database issue):D413–D422. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun X., Fujiwara M., Shi Y., Kuse N., Gatanaga H., Appay V. Superimposed epitopes restricted by the same HLA molecule drive distinct HIV-specific CD8+ T cell repertoires. J Immunol. 2014;193:77–84. doi: 10.4049/jimmunol.1400375. [DOI] [PubMed] [Google Scholar]
- 45.Lissina A., Chakrabarti L.A., Takiguchi M., Appay V. TCR clonotypes: molecular determinants of T-cell efficacy against HIV. Curr Opin Virol. 2016;16:77–85. doi: 10.1016/j.coviro.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Sun X., Shi Y., Akahoshi T., Fujiwara M., Gatanaga H., Schönbach C. Effects of a single escape mutation on T cell and HIV-1 co-adaptation. Cell Rep. 2016;15:2279–2291. doi: 10.1016/j.celrep.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 47.Motozono C., Kuse N., Sun X., Rizkallah P.J., Fuller A., Oka S. Molecular basis of a dominant T cell response to an HIV reverse transcriptase 8-mer epitope presented by the protective allele HLA-B*51:01. J Immunol. 2014;192:3428–3434. doi: 10.4049/jimmunol.1302667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ladell K., Hashimoto M., Iglesias M.C., Wilmann P.G., McLaren J.E., Gras S. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity. 2013;38:425–436. doi: 10.1016/j.immuni.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 49.Iglesias M.C., Almeida J.R., Fastenackels S., van Bockel D.J., Hashimoto M., Venturi V. Escape from highly effective public CD8+ Tcell clonotypes by HIV. Blood. 2011;118:2138–2149. doi: 10.1182/blood-2011-01-328781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takata H., Takiguchi M. Three memory subsets of human CD8+ T cells differently expressing three cytolytic effector molecules. J Immunol. 2006;177:4330–4340. doi: 10.4049/jimmunol.177.7.4330. [DOI] [PubMed] [Google Scholar]
- 51.Lirussi D., Ebensen T., Schulze K., Trittel S., Duran V., Liebich I. Type I IFN and not TNF, is essential for cyclic di-nucleotide-elicited CTL by a cytosolic cross-presentation pathway. EBioMedicine. 2017;22:100–111. doi: 10.1016/j.ebiom.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crouse J., Kalinke U., Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol. 2015;15:231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 53.Curtsinger J.M., Valenzuela J.O., Agarwal P., Lins D., Mescher M.F. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J Immunol. 2005;174:4465–4469. doi: 10.4049/jimmunol.174.8.4465. [DOI] [PubMed] [Google Scholar]
- 54.Murakoshi H., Akahoshi T., Koyanagi M., Chikata T., Naruto T., Maruyama R. Clinical control of HIV-1 by cytotoxic T cells specific for multiple conserved epitopes. J Virol. 2015;89:5330–5339. doi: 10.1128/JVI.00020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ondondo B., Murakoshi H., Clutton G., Abdul-Jawad S., Wee E.G., Gatanaga H. Novel conserved-region T-cell mosaic vaccine with high global HIV-1 coverage is recognized by protective responses in untreated infection. Mol Ther. 2016;24:832–842. doi: 10.1038/mt.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murakoshi H., Zou C., Kuse N., Akahoshi T., Chikata T., Gatanaga H. CD8+ T cells specific for conserved, cross-reactive gag epitopes with strong ability to suppress HIV-1 replication. Retrovirology. 2018;15:46. doi: 10.1186/s12977-018-0429-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trautmann L. Kill: boosting HIV-specific immune responses. Curr Opin HIV AIDS. 2016;11:409–416. doi: 10.1097/COH.0000000000000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material