Abstract
Background
Prior studies showed that tumor glycolysis and tumor immune evasion are interdependent. However, a systematic investigation of the association between tumor glycolysis and tumor immunity in various cancers remains lacking.
Methods
Using the Cancer Genome Atlas (TCGA) datasets, we explored the association between glycolytic activity and immune signatures in 14 cancer types. We also explored the associations between glycolytic activity and tumor immunity associated genetic features, including PD-L1 expression, tumor mutation burden (TMB), and tumor aneuploidy. Moreover, we performed in vitro experiments to verify some findings from bioinformatics analysis. Furthermore, we explored the association between tumor glycolytic activity and immunotherapy response.
Findings
Glycolytic activity was likely correlated with active immune signatures in various cancers and highly glycolytic tumors presented an immune-stimulatory tumor microenvironment. Compared to TMB and aneuploidy, glycolytic activity was a stronger and more consistent predictor for immune signatures in diverse cancers. Both computational and experimental analyses showed that glycolysis could increase PD-L1 expression in tumor. Glycolytic activity had a strong correlation with apoptosis which was a strong positive predictor for immune signatures, suggesting that apoptosis could be an important medium connecting glycolytic activity with immune activity in cancer. Finally, highly glycolytic tumors exhibited a better immunotherapy response and a favorable survival in the immunotherapy setting.
Interpretation
Tumor glycolysis may increase tumor immunity in diverse cancers. Glycolytic activity enhances PD-L1 expression on tumor cells and thus promotes anti-PD-1/PD-L1 immunotherapy response. Thus, the tumor glycolytic activity could be a predictive biomarker for immunotherapy response in diverse cancers.
Fund
This work was supported by the China Pharmaceutical University (grant numbers 3150120001, 2632018YX01 to XW).
Keywords: Tumor glycolysis, Tumor immunity, Tumor immune microenvironment, PD-L1 expression, Tumor immunotherapy
Research in context section.
Evidence before this study
Dysregulated energy metabolism and immune evasion are two hallmarks of cancer. Prior studies showed that tumor glycolysis and tumor immune evasion are interdependent. Increased tumor glycolysis may impair immune elimination of tumor cells.
Added value of this study
Our study revealed a significant positive correlation between tumor glycolysis and tumor immunity in multiple cancer types, a finding different from previous observations. We found that tumor glycolysis could increase PD-L1 expression on tumor cells.
Implications of all the available evidence
Our findings implicate that the tumor glycolytic activity could be a predictive biomarker for immunotherapy response in diverse cancers.
Alt-text: Unlabelled Box
1. Introduction
Cancer immunotherapy has exhibited its effectiveness in treating various cancers [1,2]. Notably, the immune checkpoint blockade, such as targeting CTLA-4 (cytotoxic T-lymphocyte-associated protein 4), PD-1 (programmed cell death protein 1), and PD-L1 (programmed cell death 1 ligand), is being clinically used for therapy of diverse cancers [1]. Nevertheless, many cancer patients had limited or no response to current immunotherapeutic strategies [3]. To enhance antitumor immunotherapy response, the combination of immunotherapy with other therapeutic approaches, such as chemotherapy [4], radiotherapy [5], and targeted therapies [6,7], has been explored. Moreover, certain genetic or genomic biomarkers, such as PD-L1 expression [8], tumor mutation burden (TMB) [9], neoantigens [10], defective DNA mismatch repair (dMMR) or microsatellite instability (MSI) [11], and tumor aneuploidy [12], have been associated with cancer immunotherapy response.
Dysregulated energy metabolism and immune evasion are two hallmarks of cancer [13]. Cancer cells primarily utilize the glycolysis pathway for energy metabolism and reprogramme their microenvironment with enriched energy supply [14]. The weak expression of immunogenicity and the inhibition of cytotoxic T and NK cell activation often result to tumor immune evasion [15]. A number of studies have revealed that tumor metabolism and tumor immune evasion are interdependent [15,16]. The metabolic competition between immune cells and tumor cells may contribute to tumor immunosuppression [15]. Increased tumor glycolysis impairs immune elimination of tumor cells [16]. These prior studies provided interesting insights into the interplay between tumor metabolism and tumor immune evasion. However, a systematic exploration of the association between tumor metabolism and tumor immunity in various different cancer types is lacking. Also, how tumor metabolism affects tumor immunity and tumor immune microenvironment (TIM) remains incompletely understood.
With the rapid advances in next-generation sequencing (NGS) technologies, many large-scale cancer genomics datasets have been generated, e.g., the Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov) datasets, and numerous computational methods have been developed for investigating these datasets. In particular, many bioinformatics tools have been designed to explore genome-wide mRNA expression data, e.g., GSEA [17] for gene-set enrichment analysis, ESTIMATE [18] for evaluating the levels of tumor immune cell infiltration, ssGSEA [19,20] for single-sample gene-set enrichment analysis, and ABSOLUTE [21] for assessing tumor aneuploidy. In this study, using these bioinformatics tools, we explored the association between tumor glycolysis and immune signatures across 14 cancer types by analyzing the TCGA datasets. We also explored the association between tumor glycolysis and tumor immunity-associated molecular features, including PD-L1 expression, TMB, tumor aneuploidy, and apoptosis. Furthermore, we investigated the association between tumor glycolysis and tumor immunotherapy response using two cancer immunotherapy response datasets. Our study revealed a significant positive correlation between tumor glycolysis and tumor immunity across multiple cancer types, a phenomenon in contrast with previous observations [16]. Our findings implicate that the tumor glycolytic activity could be a predictive biomarker for immunotherapy response in diverse cancers.
2. Materials and methods
2.1. Materials
We downloaded the TCGA data from the genomic data commons data portal (https://portal.gdc.cancer.gov/). These data included RNA-Seq gene expression profiles, gene somatic mutations, somatic copy number alterations, protein expression profiles, and clinical data for 14 cancer types (Table 1). We obtained the gene sets that represented different immune signatures from several publications, including HLA [22], tumor-infiltrating lymphocytes (TILs) [23], immune cytolytic activity [24], and interferon (IFN) response [24]. The gene sets for glycolysis and other pathways were obtained from KEGG [25]. These gene sets are presented in the Supplementary Table S1. We downloaded the Hugo cohort immunotherapy response dataset [26] from the NCBI gene expression omnibus (https://www.ncbi.nlm.nih.gov/geo/, GSE78220), and the Nathanson cohort dataset [27] from the website http://www.hammerlab.org/melanoma-reanalysis/.
Table 1.
The 14 cancer types analyzed.
| Cancer type | Full name | Sample size |
||
|---|---|---|---|---|
| Total | Highly glycolytic tumors | Lowly glycolytic tumors | ||
| BRCA | Breast invasive carcinoma | 1100 | 367 | 367 |
| DLBC | Lymphoid neoplasm difuse large B-cell lymphoma | 48 | 16 | 16 |
| GBM | Glioblastoma multiforme | 166 | 55 | 55 |
| KIRP | Kidney renal papillary cell carcinoma | 291 | 97 | 97 |
| LAML | Acute myeloid leukemia | 173 | 57 | 57 |
| LGG | Brain lower-grade glioma | 530 | 177 | 177 |
| LIHC | Liver hepatocellular carcinoma | 373 | 124 | 124 |
| LUAD | Lung adenocarcinoma | 517 | 172 | 172 |
| OV | Ovarian serous cystadenocarcinoma | 307 | 102 | 102 |
| PAAD | Pancreatic adeno-carcinoma | 179 | 60 | 60 |
| PCPG | Pheochromocytoma and paraganglioma | 184 | 61 | 61 |
| SARC | Sarcoma | 263 | 88 | 88 |
| SKCM | Skin cutaneous melanoma | 472 | 157 | 157 |
| UCEC | Uterine corpus endometrial carcinoma | 370 | 123 | 123 |
2.2. Glycolysis score, immune score, signature enrichment, biological and genomic features
2.2.1. Evaluation of tumor glycolysis score
We obtained the set of genes in the glycolysis pathway from KEGG [25], and applied the single-sample gene-set enrichment analysis (ssGSEA) [19,20] for the gene set to quantify the glycolytic activity (glycolysis score). We defined a tumor as highly glycolytic if its glycolysis score was in the upper third of all glycolysis scores in the same cancer type, and a tumor as lowly glycolytic if its glycolysis score was in the bottom third.
2.2.2. Evaluation of immune activity or immune signature enrichment in tumor
We used ESTIMATE [18] to assess tumor immune activity. ESTIMATE yields the immune score for each tumor sample that quantifies the immune activity (immune infiltration level) in that tumor sample based on its gene expression profiles. The single-sample gene-set enrichment analysis (ssGSEA) score [19,20] was utilized to quantify the enrichment levels of immune signatures in tumor. In evaluating the ratio between two different immune signatures, we used the base-2 log transformed value of the ratio between the mean expression levels of all marker genes in each immune signatures.
2.2.3. Gene-set enrichment analysis
We performed gene-set enrichment analysis of the gene expression profiles for each of the 14 cancer types by GSEA [17]. The KEGG [25] pathways significantly upregulated in highly glycolytic tumors and upregulated in lowly glycolytic tumors were identified using a FDR cutoff <0.1.
2.2.4. Evaluation of TMB, tumor aneuploidy, and tumor purity
For each tumor sample, we determined its TMB as the total count of somatic mutations (except silent mutations) detected in the tumor and used ABSOLUTE [21] to calculate its ploidy score representing the tumor aneuploidy, and evaluate its tumor purity.
2.3. In vitro experiments
2.3.1. Cell lines and cell culture
Human cancer cell lines MCF-7 (breast cancer), SJSA1 (osteosarcoma), and SK-OV-3 (ovarian carcinoma), and natural killer cells NK-92 were from the American Type Culture Collection (ATCC). MCF-7 and SJSA1 cells were incubated in Roswell Park Memorial Institute-1640 (RPMI-1640, GIBCO, USA) supplemented with 10% fetal bovine serum (FBS, GIBCO, USA). SK-OV-3 cells were cultured as a monolayer in McCoy's 5a (GIBCO, USA) medium supplemented with 10% FBS. All these cells were cultured in a humidified incubator at 37 °C and a 5% CO2 atmosphere, and were harvested in logarithmic growth phase.
2.3.2. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Tumor cells (MCF-7, SJSA1, and SK-OV-3) were harvested after treatment with 2-DG (5 mM, 48 h). The total RNA was isolated by Trizol (Invitrogen, USA) and was reversely transcribed into cDNA by the RevertAid First Strand cDNA Synthesis Kit (Termo Fisher, USA). Primers were diluted in nuclease-free water with the real-time PCR Master Mix (SYBR Green, TOYOBO Co., LTD, JAPAN). Relative copy number was determined as the fold-change of the gene of interest relative to β-actin. The qPCR was performed on an ABI 7500 FAST and Applied Biosystems StepOnePlus Real Time PCR machine. The PCR primer sequences were presented in Supplementary Table S2.
2.3.3. Co-culture of tumor cells with immune cells
A co-culture system was constructed by inserting the transwell chamber (Corning Inc., Corning, NY, USA) into a 6-well plate. Tumor cells (MCF-7, SJSA1, and SK-OV-3) were seeded on the 6-well plate at a density of 5 × 104 cells/well, and NK-92 cells were seeded on the membrane (polyethylene terephthalate, pore size, 0.4 μm) of the transwell chamber at a density of 5 × 104 cells/chamber. NK-92 and tumor cells were co-cultured in a humidified incubator at 37 °C and a 5% CO2 atmosphere for 48 h.
2.3.4. EdU proliferation assay
After the co-culture of tumor cells with immune cells for 48 h, an EdU (5-ethynyl-2′-deoxyuridine, Invitrogen, CA, USA) proliferation assay was performed to measure the proliferation ability of NK-92 cells. NK-92 cells were plated in 96-well plates at a density of 2 × 103 cells/well for 24 h. The cells were incubated with 10 μM EdU for 24 h at 37 °C before fixation, permeabilization, and EdU staining. The cell nuclei were stained with DAPI (Sigma) at a concentration of 1 μg/ml for 20 min. The proportion of the NK-92 cells incorporating EdU was determined with fluorescence microscopy. Each assay was performed in triplicate wells.
2.3.5. Western blotting
Tumor cells (MCF-7, SJSA1, and SK-OV-3) were harvested after treatment with 2-DG (5 mM, 48 h), and were washed twice with cold PBS. Next, the tumor cells were lysed in SDS buffer (1% SDS, 0.1 M Tris pH 7.4, 10% glycerol) supplemented with protease inhibitors. The protein concentration was determined by Bradford Protein Assay (Bio-rad). Samples were resolved by standard SDS-PAGE after normalization of the total protein content. After Western transfer, Nitrocellulose membranes (Millipore) were incubated with antibodies HK-2 (ab209847, Abcam) and PD-L1 (ab213254, Abcam). After incubation with the HRP-labeled secondary antibody (KGAA002–1, KeyGEN Biotech, China), proteins were visualized by enhanced chemiluminescence using a G: BOX chemiXR5 digital imaging system.
2.4. Statistical analysis, logistic regression and survival analysis
The experimental data were analyzed by Prism 5.0 software (GraphPad) and their mean ± SD were presented. The t-test P < .05 was utilized to determine the statistical significance. We calculated the correlation between two variables using the Spearman method. These variables included glycolytic activity, immune signature score, gene expression levels, TMB, aneuploidy, and pathway activity. The threshold of P < .05 (Spearman's correlation test) indicates the significance of correlation. To compare the contribution of different genomic features in the prediction of immune signature scores, we used logistic regression to construct a predictive model which contained three predictors, including glycolytic activity, TMB, and aneuploidy. The tumors with high (upper third) versus low (bottom third) immune signature scores were predicted.
Logistic regression was performed in R programming environment. The R function “glm” was used to fit the binary model. We specified the parameter “family” as “binomial” and other parameters as default in “glm”. The standardized regression coefficients (β values) were calculated using the function “lm.beta” in R package “QuantPsyc”.
We compared the survival (overall survival (OS) and disease-free survival (DFS)) of cancer patients separated by the median glycolysis score or the median expression level of specific genes. Kaplan-Meier curves were used to compare the survival time differences. The log-rank test P < .1 indicates the significance of survival time differences. The survival analyses were performed in both TCGA and the immunotherapy response datasets by R programming function “survfit” in “survival” package.
3. Results
3.1. Tumor glycolytic activity likely positively correlate with immune signatures in various cancers
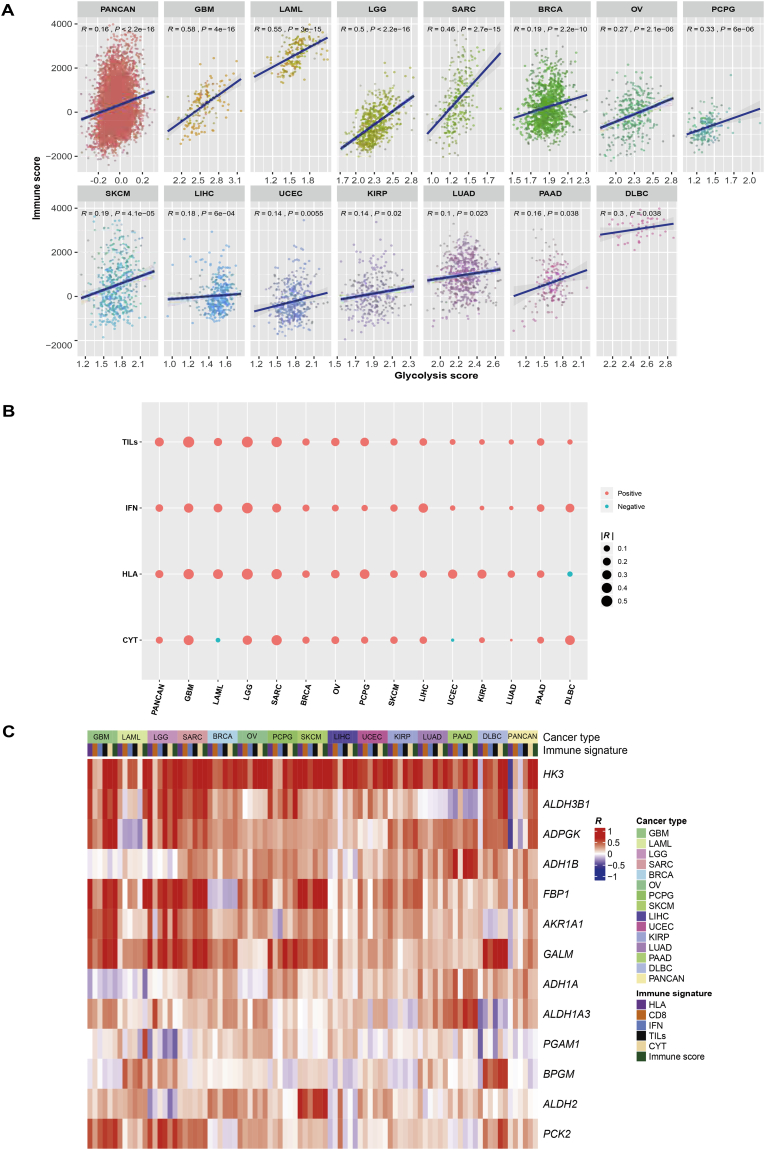
To explore the correlation between tumor glycolysis and tumor immunity, we analyzed cancer genomics data from 14 TCGA cancer types, including BRCA, DLBC, GBM, KIRP, LAML, LGG, LIHC, LUAD, OV, PAAD, PCPG, SARC, SKCM, and UCEC. Strikingly, we found a significant positive correlation between glycolytic activity (glycolysis score, quantified by the glycolysis pathway activity [25]) and immune activity (quantified by immune score [18]) both in pan-cancer and in the 14 individual cancer types (Spearman's correlation test, P < .05) (Fig. 1A). Moreover, tumor glycolysis positively correlated with diverse immune signatures in pan-cancer and multiple cancer types, including HLA expression, TILs infiltration, immune cytolytic activity, and IFN response (Fig. 1B). For example, glycolytic activity positively correlated with HLA expression both in pan-cancer and in 12 individual cancer types, and positively correlated with immune cytolytic activity both in pan-cancer and in 9 individual cancer types.
Fig. 1.
Tumor glycolytic activity tends to be positively correlated with immune signatures in diverse cancers. A. Glycolytic activity is positively correlated with immune score evaluated by ESTIMATE [18] in pan-cancer and 14 cancer types. The Spearman's correlation test P values and correlation coefficients (R) are shown. B. Glycolytic activity is positively correlated with diverse immune signatures in pan-cancer and 14 cancer types. TILs: tumor-infiltrating lymphocytes. IFN: interferon. CYT: immune cytolytic activity. R: Spearman's correlation coefficient. C. The expression levels of multiple glycolysis pathway genes are positively associated with immune signatures in pan-cancer and 14 cancer types. D. The glycolytic protein glucose-6-phosphate dehydrogenase (G6PD) has a significant positive expression correlation with immune score in pan-cancer and 9 cancer types. E. GSEA [17] identifies a number of immune-associated pathways which are upregulated in highly versus lowly glycolytic tumors in diverse cancers (FDR < 0.05). FDR: false discovery rate. F. The ratios between the expression levels of immune-stimulatory signatures and the expression levels of immune-inhibitory signatures are significantly higher in highly versus lowly glycolytic tumors in diverse cancers. M1: M1 macrophages. M2: M2 macrophages. *, P < .05. **, P < .01. ***, P < .001. It also applies to following figures.
Besides the glycolysis pathway (gene set), we found a number of glycolysis-related genes whose expression was positively associated with immune signatures in pan-cancer and individual cancer types (Fig. 1C). For example, the expression of ADPGK, AKR1A1, ALDH3B1, GALM, and HK3 was positively correlated with immune score as well as immune cytolytic activity both in pan-cancer and in at least 10 cancer types; the expression of ADPGK, AKR1A1, ALDH3B1, GALM, HK3, FBP1, and PCK2 was positively correlated with IFN response in at least 10 cancer types. Moreover, on the basis of the TCGA protein expression data, we found that the expression of glycolytic protein Glucose-6-phosphate dehydrogenase (G6PD) had a significant positive correlation with immune score in pan-cancer and in 9 cancer types (Fig. 1D).
We compared the gene expression profiles between highly glycolytic tumors and lowly glycolytic tumors, and identified the KEGG [25] pathways that were enriched in the respective groups in each of the 14 cancer types by GSEA [17]. Among the pathways upregulated in highly glycolytic tumors in >5 cancer types were those most associated with metabolism and immune signatures (Supplementary Table S3). The immune signature-associated pathways included antigen processing and presentation, autoimmune thyroid disease, intestinal immune network for IgA production, hematopoietic cell lineage, chemokine signaling, cytokine–cytokine receptor interaction, Toll-like receptor signaling, natural killer cell–mediated cytotoxicity, Fc gamma receptor–mediated phagocytosis, leukocyte transendothelial migration, B cell receptor signaling, NOD–like receptor signaling, and primary immunodeficiency (Fig. 1E, Supplementary Table S3). This result confirmed the elevated immune activity in highly glycolytic tumors.
Moreover, we observed a significant increase of the ratio between immune-stimulatory cells (CD8+ T cells, marker gene CD8A) and immune-inhibitory cells (CD4+ regulatory T cells, marker genes GPR1, FOXP3, CTLA4, IL32, C15orf53, and IL4) in highly versus lowly glycolytic tumors in 7 cancer types (Mann-Whitney U test, P < .05) (Fig. 1F). We also found a significant increase in the ratio between pro-inflammatory cytokines (marker genes IFNG, IL-1A, IL-1B, and IL-2) and anti-inflammatory cytokines (IL-4, IL-10, IL-11, and TGFB1) in 9 cancer types (Fig. 1F). A significant increase was also observed in the ratio between inflammation-inducing M1 macrophages (CD64, IDO, SOCS1, and CXCL10) and inflammation-inhibiting M2 macrophages (MRC1, TGM2, CD23, and CCL22) in 5 cancer types (Fig. 1F). These results suggest that highly glycolytic tumors tend to present an immune-stimulatory tumor microenvironment in diverse cancers.
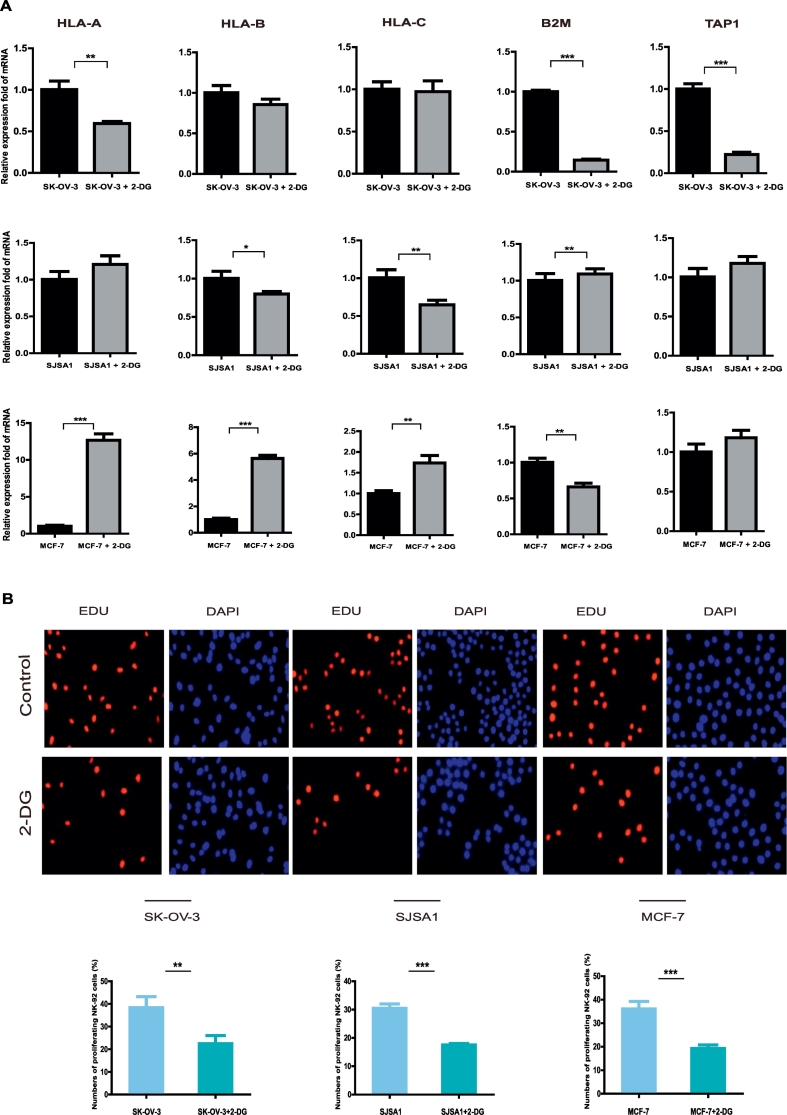
To experimentally verify the findings from bioinformatics analysis, we used the glycolysis inhibitor 2-deoxy-d-glucose (2-DG) to treat three cancer cell lines, including MCF-7 (breast cancer), SJSA1 (osteosarcoma), and SK-OV-3 (ovarian carcinoma), and compared the expression levels of MHC class I genes (HLA-A, HLA—B, HLA—C, B2M, and TAP1) between the pre- and post-treated cell lines. We observed a significant reduction of the HLA-A, B2M, and TAP1 expression levels in post-treated SK-OV-3 (Student's t-test, P < .05) and the HLA-B and HLA-C expression levels also decreased in post-treated SK-OV-3 although the differences were not statistically significant (P > .05) (Fig. 2A). The HLA-B and HLA-C expression levels were significantly lower in post-treated SJSA1, whereas B2M had higher expression levels in post-treated SJSA1 (Fig. 2A). For MCF7, we observed a significant decrease of B2M expression and a significant increase of HLA-A, HLA—B, and HLA-C expression in post-treated cells (Fig. 2A). These results suggest that inhibition of glycolysis tends to downregulate HLA expression in cancer although this action is cellular context dependent. Furthermore, we used the EdU proliferation assay to compare the proliferation potential between NK92 cells co-cultured with the cancer cell lines and those co-cultured with the cancer cell lines treated with 2-DG for 24 h. We observed a significant reduction of the proliferation potential of the NK92 cells co-cultured with the post-treated cancer cells, and this result was consistent across all three cell lines (Fig. 2B). This experimental result suggests that elevated glycolytic activity may result in increased tumor immunity, and therefore verified the finding from bioinformatics analysis.
Fig. 2.
In vitro experiments demonstrate that tumor glycolytic activity may promote tumor immune activity. A. The expression levels of MHC Class I genes are altered when tumor glycolysis is inhibited in SK-OV-3 (ovarian carcinoma), SJSA1 (osteosarcoma), and MCF-7 (breast cancer) cell lines by 2-deoxy-d-glucose (2-DG), evident by real-time quantity PCR. B. The proliferation potential of the NK92 cells co-cultured with tumor cells is significantly reduced when glycolysis is inhibited in the tumor cells, evident by EdU proliferation assay.
Collectively, these computational and experimental analyses demonstrated that glycolytic activity tended to be positively associated with immune activity in diverse cancers.
3.2. Using tumor glycolytic activity to predict immune signatures in cancers
To explore the predictability of tumor glycolytic activity for tumor immunity, we used logistic regression to evaluate the contribution of tumor glycolytic activity in predicting two immune signatures (immune score and immune cytolytic activity). We defined two groups of cancer samples based on immune signature scores (upper third versus bottom third) in the logistic regression model, and explored the relative contribution of glycolytic activity, TMB, and aneuploidy in predicting immune signatures, considering that both TMB [9] and tumor aneuploidy [12] have been demonstrated to be significantly associated with immune signatures.
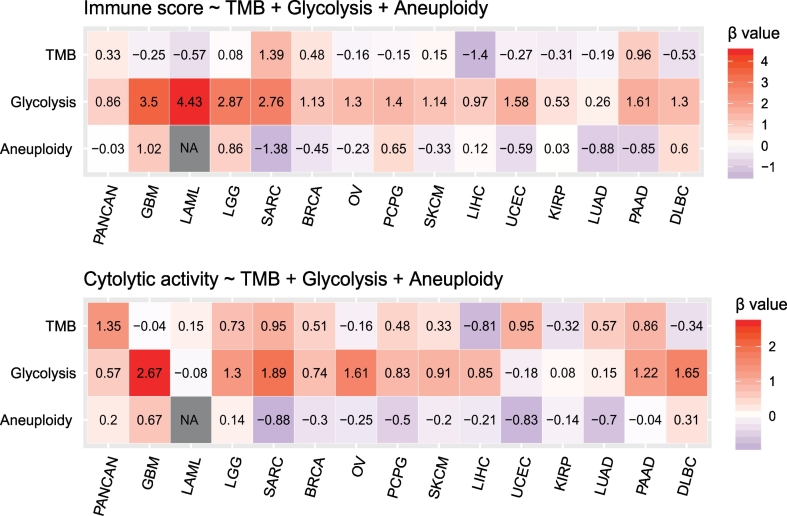
In the pan-cancer analysis, both glycolytic activity (β coefficient: β = 0.86, P < 2 × 10−16) and TMB (β = 0.33, P = .004) significantly predicted the immune score and aneuploidy is not significant in predicting the immune score (β = −0.03, P = .77). For the immune cytolytic activity, glycolytic activity (β = 0.57, P = 1.19 × 10−9), TMB (β = 1.35, P = 4.05 × 10−9) and aneuploidy (β = 0.20, P = .04) all represented positive predictors (Fig. 3). In the analysis of 14 individual cancer types, glycolytic activity was a positive predictor of the immune score in 11 cancer types, and TMB represented a positive predictor in 3 cancer types and aneuploidy a negative predictor in 3 cancer types. Moreover, glycolytic activity represented a positive predictor of the immune cytolytic activity in 8 cancer types, as compared to TMB being a positive predictor in 3 cancer types and aneuploidy a negative predictor in 2 cancer types (Fig. 3). These results suggest that glycolytic activity is a stronger predictor than TMB and aneuploidy for immune signatures.
Fig. 3.
Logistic regression analysis shows that glycolytic activity is a stronger predictor than TMB and aneuploidy for immune signatures in pan-cancer and diverse individual cancer types. The tumors with high (upper third) versus low (bottom third) immune signature scores are predicted. Two immune signatures (immune score calculated by ESTIMATE [18] and immune cytolytic activity) are predicted by glycolytic activity, TMB, and aneuploidy. TMB (tumor mutation burden) is defined as the total somatic mutation count in tumor. Tumor aneuploidy is evaluated by ABSOLUTE [21]. β coefficients for each predictor in pan-cancer or individual cancer types are shown.
In fact, with immune score, tumor glycolysis had a significant positive correlation in all 14 cancer types, TMB exhibited a positive correlation in 2 cancer types and a negative correlation in 3 cancer types, and aneuploidy had a negative correlation in 4 cancer types and a positive correlation in 1 cancer type (Spearman's correlation test, P < .05) (Supplementary Table S4). In addition, with the immune cytolytic activity, tumor glycolysis had a significant positive correlation in 9 cancer types, TMB had a positive correlation in 4 caner types, and aneuploidy had a negative correlation in 5 caner types (Supplementary Table S4). These results again suggest that tumor glycolytic activity may contribute to the alterations of tumor immune activity in a stronger and more consistent manner compared to TMB and aneuploidy.
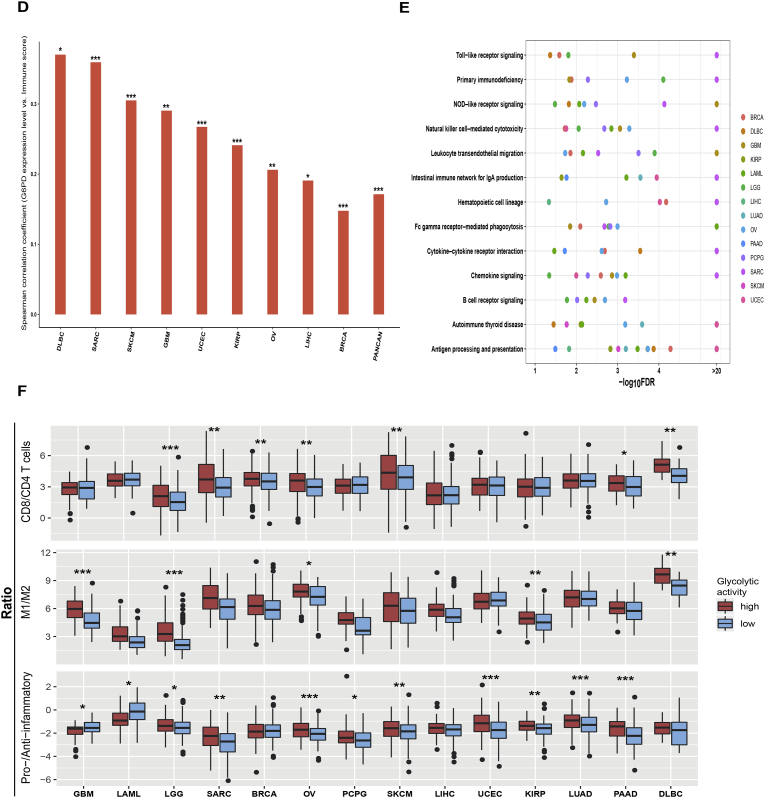
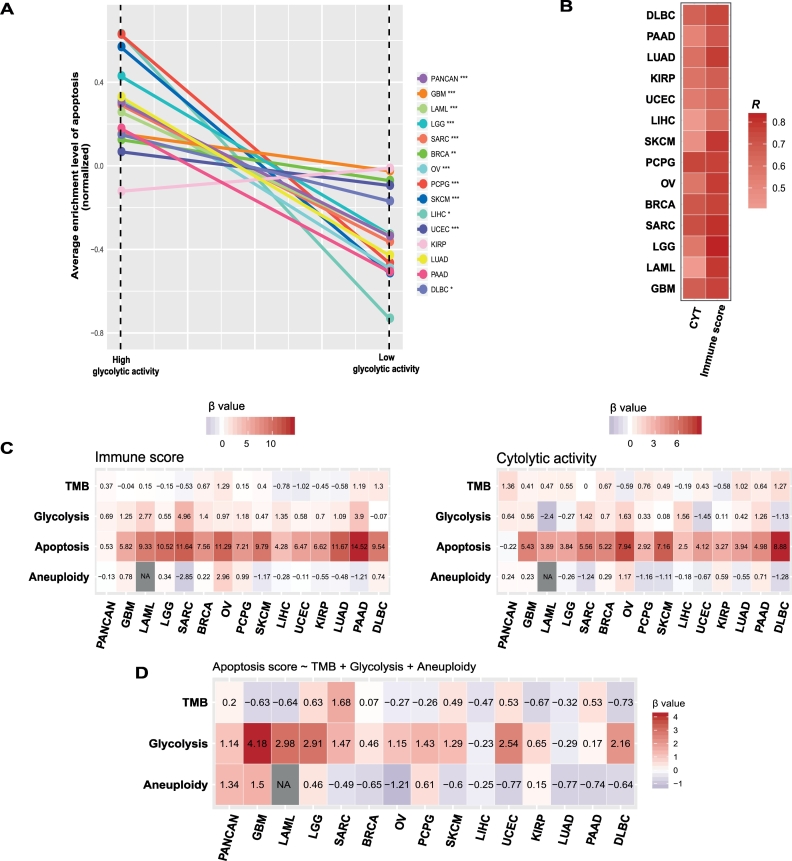
Besides plentiful metabolism and immune-associated pathways (Supplementary Table S3), the apoptosis pathway was more enriched in highly versus lowly glycolytic tumors in 11 cancer types (Mann-Whitney U test, P < .05) (Fig. 4A). Interestingly, the activity of apoptosis was significantly associated with immune score and immune cytolytic activity in all 14 cancer types consistently in a positive direction (Fig. 4B). When we added apoptosis activity into the logistic regression model to predict the immune signatures (immune score and immune cytolytic activity), we observed a significant reduction of the contribution (β value) from glycolytic activity in predicting the immune signatures in pan-cancer and many individual cancer types (Fig. 4C). Moreover, glycolytic activity was significant as a positive predictor in predicting apoptosis activity in pan-cancer and most individual cancer types (Fig. 4D), and apoptosis activity was a significant positive predictor for immune signatures in pan-cancer and 14 individual cancers (Fig. 4C). Altogether, these data suggest that apoptosis may play a key role in connecting tumor glycolysis with tumor immunity in these cancers.
Fig. 4.
Associations among tumor glycolysis, apoptosis, and tumor immune activity. A. The apoptosis pathway is more enriched in highly versus lowly glycolytic tumors in pan-cancer and diverse individual cancer types (Mann-Whitney U test, P < .05). B. Apoptosis activity is positively associated immune signatures in all 14 cancer types. R: Spearman's correlation coefficient. C. Logistic regression analysis shows that apoptosis activity is a strong positive predictor for immune signatures in all 14 cancer types. D. Glycolytic activity is a significant positive predictor for apoptosis activity in pan-cancer and most of the 14 cancer types. β coefficients (β value) for each predictor in pan-cancer or individual cancer types are shown.
3.3. Tumor glycolytic activity positively correlates with PD-L1 expression and with immunotherapy response
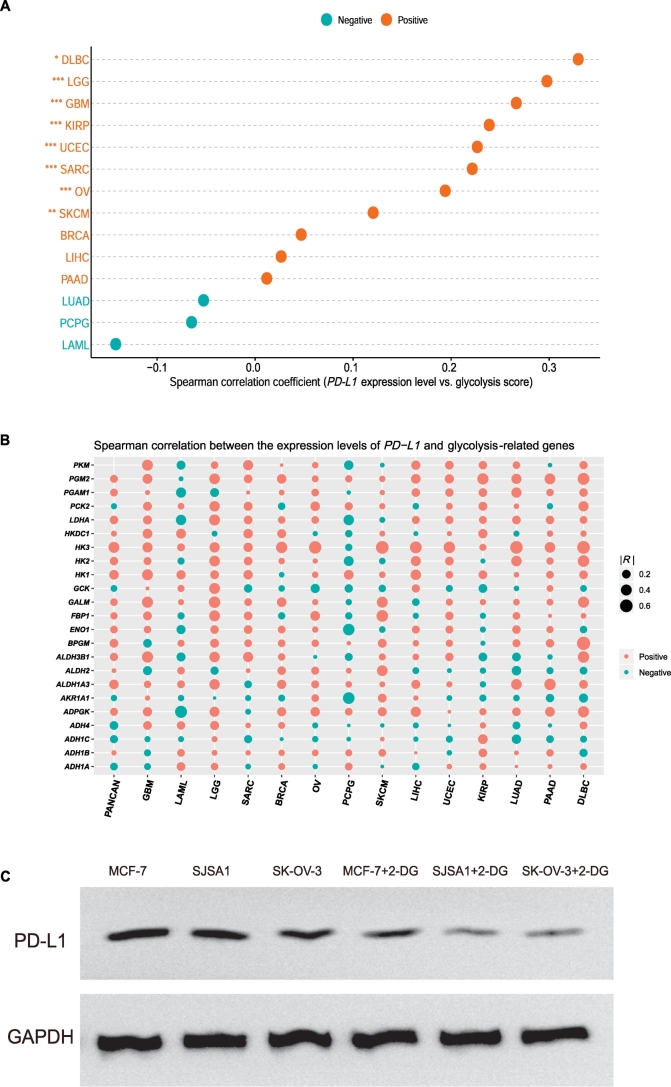
We examined the correlation of PD-L1 expression with glycolytic activity in the 14 cancer types, and found that the expression levels of PD-L1 significantly positively correlated with glycolytic activity in 8 cancer types (DLBC, GBM, LGG, OV, KIRP, SARC, SKCM, and UCEC) (Spearman's correlation test, P < .05) (Fig. 5A). Moreover, PD-L1 expression showed a significant positive correlation with the expression of numerous glycolysis-related genes in pan-cancer and many individual cancers, e.g., HK3, LDHA, and PGM2 (Fig. 5B). Furthermore, in vitro experiments confirmed that PD-L1 expression was significantly decreased when tumor glycolysis was inhibited in all three cell lines (MCF-7, SJSA-1, and SK-OV-3) tested (Fig. 5C). This finding suggests that the PD-1/PD-L1 blockade therapy could be more effective against highly glycolytic tumors, since the PD-L1 expression is a biomarker for the active response to PD-1/PD-L1 pathway inhibition [8]. Interestingly, a previous study showed that PD-L1 could promote tumor glycolysis [15]. Collectively, these findings suggest a mutual positive regulation relationship between tumor PD-L1 expression and tumor glycolysis.
Fig. 5.
Tumor glycolysis promotes PD-L1 expression on tumor cells in diverse cancers. A. The PD-L1 expression levels are positively correlated with glycolytic activity in various cancer types. Spearman's correlation test P values and correlation coefficients are shown. B. The PD-L1 expression levels are positively correlated with the expression levels of numerous glycolysis-related genes in pan-cancer and various cancer types. R: Spearman's correlation coefficient. C. In vitro experiments demonstrate that PD-L1 expression is significantly reduced in MCF-7 (breast cancer), SJSA1 (osteosarcoma), and SK-OV-3 (ovarian carcinoma) when tumor glycolysis is inhibited in these cell lines by 2-deoxy-d-glucose (2-DG), evident by Western blotting.
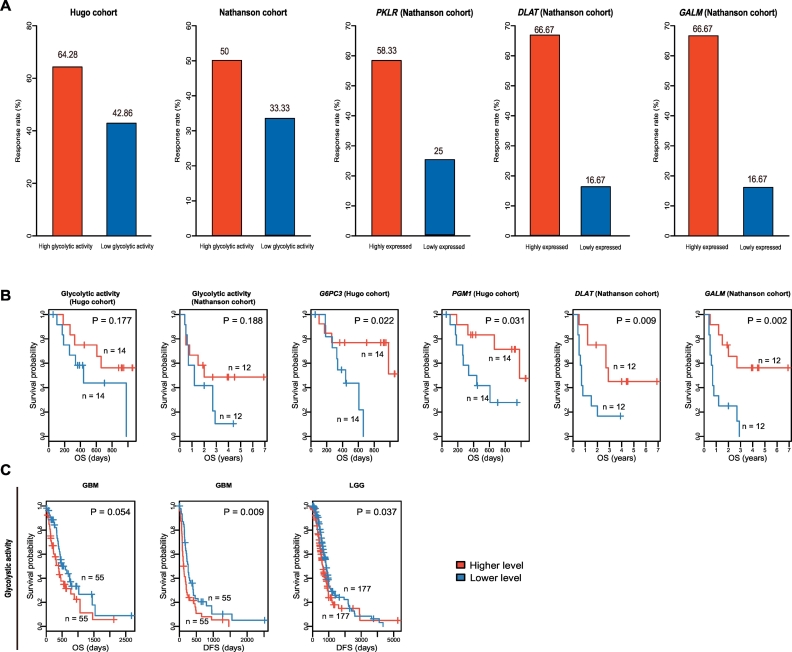
We investigated the correlation between tumor glycolytic activity and tumor immunotherapy response (PD-1/PD-L1 or CTLA-4 inhibition) using two cancer (melanoma) immunotherapy response-associated datasets (Hugo cohort [26] and Nathanson cohort [27]). First, on the basis of the glycolysis pathway activity, we found that the immunotherapy response rate was higher in higher versus lower glycolysis pathway activity tumors in both cohorts (Fig. 6A). Second, based on the expression of PKLR, a key gene instructing for producing pyruvate kinase involved in the final step of the glycolytic pathway [28], we found that the immunotherapy response rate was higher in tumors more highly expressing PKLR than those more lowly expressing PKLR in Nathanson cohort (Fisher's exact test, P = .21, OR = 3.94) (Fig. 6A). Moreover, we found 9 of the 12 tumors highly expressing PKLR being the tumors with higher glycolysis pathway activity, and 9 of the 12 tumors lowly expressing PKLR being the tumors with lower glycolysis pathway activity. It indicates that the PKLR expression levels significantly correlate with the glycolysis score in a positive direction, and thus may reflect the glycolysis pathway activity. In the same cohort, the expression of two glycolysis-associated genes DLAT and GALM had a significant positive correlation with the immunotherapy response (Fisher's exact test, P = .036 and OR = 8.91 for both genes) (Fig. 6A). These results indicate that tumor glycolytic activity tends to be positively associated with immunotherapy response.
Fig. 6.
Tumor glycolytic activity is correlated with immunotherapy response. A. High glycolytic activity is associated with increased immunotherapy response rate in two melanoma cohorts (Hugo cohort [26] and Nathanson cohort [27]). B. Kaplan-Meier survival curves show that the glycolysis score and the expression of the glycolysis-associated genes are positively correlated with survival prognosis in the immunotherapeutic cohorts. C. Kaplan-Meier survival curves show that the glycolysis score is negatively correlated with survival prognosis in multiple TCGA cohorts without immunotherapy. OS: overall survival. DFS: disease-free survival. The log-rank test P values are shown in the survival curves.
Furthermore, we found that the glycolysis score had a positive correlation with survival prognosis in both cohorts (Fig. 6B). Moreover, the higher expression levels of numerous glycolysis-associated genes were significantly associated with a better survival in the immunotherapeutic cohorts (Fig. 6B and Supplementary Fig. S1A). These genes included G6PC3, PGM1, DLAT, GALM, HK3, FBP1, ALDH3B1, and ACSS2. However, when we analyzed the TCGA datasets in which the tumor patients were not treated with immunotherapy, we found that the glycolysis score had a significant negative correlation with survival prognosis in GBM and LGG (Fig. 6C). Moreover, the upregulation of these glycolysis-associated genes were associated with a poor prognosis in multiple cancer types including melanoma (Supplementary Fig. S1B). The discrepant correlations of glycolytic activity with survival between the patients treated with immunotherapy and the patients in the absence of such therapy could be attributed to the higher immunotherapy response rate in highly glycolytic tumors relative to lowly glycolytic tumors. This can be justified by that the expression of DLAT and GALM was significantly positively correlated with immunotherapy response and with survival in Nathanson cohort.
4. Discussion
In this study, we explored the correlation between tumor glycolytic activity and tumor immune activity in 14 cancer types. Our data showed that tumor glycolytic activity tended to be positively associated with diverse immune signatures in these cancers. Interestingly, among the three cancer types in which glycolytic activity and immune score had the highest correlation (correlation coefficient ≥ 0.5), two were brain-associated tumors (GBM and LGG). This suggests a particularly significant positive correlation between glycolysis and immune/inflammation activity in brain tumors. This outstanding correlation may be associated with the high glycolysis energy metabolism in brain astrocytes [29] which has a correlation with both GBM and LGG, whereas the related mechanism remains to be elucidated.
It should be noted that the tumor glycolysis score may not thoroughly reflect the tumor itself glycolytic activity considering the possibly incomplete tumor purity. However, when we analyzed high-purity tumors and low-purity tumors, we observed similar results that glycolytic activity positively correlated with diverse immune signatures in pan-cancer and multiple cancer types (Supplementary Table S5). Furthermore, we added the tumor purity as a predictor into the logistic regression model with the glycolytic activity, TMB, and aneuploidy predictors for predicting immune signatures. We obtained similar results that glycolytic activity was a positive predictor for immune signatures in multiple individual cancer types (Supplementary Fig. S2). As expected, tumor purity is a strong negative predictor for immune signatures in pan-cancer and multiple individual cancer types.
In addition, we compared the expression levels of the 67 glycolysis pathway genes between highly glycolytic tumors and lowly glycolytic tumors. We found that 43 (64%) genes were significantly upregulated in highly glycolytic tumors in at least half of the 14 cancer types (Student's t-test, FDR < 0.1) (Supplementary Table S6). Notably, many key genes for glycolytic reactions were upregulated in highly glycolytic tumors in diverse cancers. For example, HK3, which encodes hexokinase that catalyzes the first reaction in glycolysis (conversion of d-glucose into glucose-6-phosphate), had significantly higher expression levels in highly glycolytic tumors than in lowly glycolytic tumors in all 14 cancer types. GPI, which encodes the enzyme phosphoglucose isomerase involved in the second reaction in glycolysis (conversion of glucose-6-phosphate to fructose-6-phosphate), had significantly higher expression levels in highly glycolytic tumors in 12 cancer types. PKM, which encodes a pyruvate kinase involved in the final step of glycolysis (conversion of phosphoenolpyruvate into pyruvate), was upregulated in highly glycolytic tumors in 12 cancer types. These data suggest that our classification of highly and lowly glycolytic tumors using the gene-set score is reasonable.
Moreover, in comparison with TMB and aneuploidy, glycolytic activity exhibited a stronger and more consistent correlation with immune signatures in these cancers. Prior studies have revealed that certain genomic features, such as TMB and neoantigens [30], are associated with active antitumor immune signatures. We examined the correlation between glycolytic activity and TMB in the 14 cancer types, and found that glycolytic activity had a significant positive correlation with TMB in 2 cancer types (UCEC and KIRP) and had a significant negative correlation with TMB in 2 cancer types (LUAD and LAML) (Spearman's correlation test, P < .05) (Supplementary Table S7). This suggests that TMB may not be the key factor explaining the significant correlation between glycolytic activity and immune signatures in these cancers. In addition, we examined the correlation between glycolytic activity and tumor aneuploidy which correlates with reduced antitumor immune signatures [12]. A significant negative correlation was observed between them in 4 cancer types (LIHC, OV, LUAD, and GBM) (Supplementary Table S7). This data suggests that aneuploidy is not a necessary factor explaining the relationship between glycolytic activity and immune signatures, although it may be connected with the relationship in a few cancer types.
We found that numerous HLA genes were upregulated in highly glycolytic tumors compared to lowly glycolytic tumors via bioinformatics analysis (Supplementary Fig. S3). This result was verified by in vitro experiments to a certain degree (Fig. 2A). It suggests that tumor glycolysis may promote tumor immunogenicity in diverse cancers. PD-L1 expression on tumor cells indicates an antitumor immunosuppression mechanism in tumor [31]. Our computational and experimental results showed that tumor glycolysis increased PD-L1 expression in diverse cancers, suggesting that highly glycolytic tumors are more likely to inhibit antitumor immunity compared to lowly glycolytic tumors. This is consistent with a recent study showing that increased tumor glycolysis suppressed antitumor immunity [16]. This finding could explain why highly glycolytic tumor patients showed no a significantly better prognosis than lowly glycolytic tumor patients in these cancer types analyzed, even though the former had higher levels of antitumor immune cell infiltration than the latter. However, in the anti-PD-1/PD-L1 immunotherapy setting, highly glycolytic tumors tended to exhibit better survival than lowly glycolytic tumors. This suggests that highly glycolytic tumors are more likely to respond to anti-PD-1/PD-L1 therapy than lowly glycolytic tumors. The more active response to anti-PD-1/PD-L1 therapy in highly glycolytic tumors may be due to the elevated PD-L1 expression on these tumors. Of course, the association between tumor glycolysis and immunotherapy response revealed in this study needs to be validated in larger cohorts.
There are several limitations in the present study. First, only three tumor cell lines were used for in vitro experiments. Second, all experiments were in vitro that would overlook the effect of tumor microenvironment on tumor immunity. To overcome these limitations, more cell lines for in vitro experiments and further in vivo experiments are necessary for confirming the present findings. That would be a priority for our future study.
The following are the supplementary data related to this article.
Correlate tumor glycolytic activity with survival prognosis in cancers with immunotherapy and cancers without immunotherapy. A. The expression of glycolysis-associated genes are positively associated with survival prognosis in the immunotherapeutic cohorts (log-rank test, P < .05). B. The expression of numerous glycolysis-associated genes are negatively associated with survival prognosis in multiple TCGA cohorts without immunotherapy (log-rank test, P < .1). OS: overall survival. DFS: disease-free survival. Kaplan-Meier curves are used to compare the survival time differences, and the log-rank test P values are shown in the survival curves.
Logistic regression analysis shows that glycolytic activity is a positive predictor for immune signatures in multiple cancer types when the tumor purity is added as a predictor. The tumors with high (upper third) versus low (bottom third) immune signature scores are predicted. Two immune signatures (immune score calculated by ESTIMATE [18] and immune cytolytic activity) are predicted by glycolytic activity, aneuploidy, TMB, and purity. TMB (tumor mutation burden) is defined as the total somatic mutation count in tumor. Tumor aneuploidy is evaluated by ABSOLUTE [21]. β coefficients for each predictor in pan-cancer or individual cancer types are shown.
Numerous HLA genes are upregulated in highly glycolytic tumors versus lowly glycolytic tumors (Student's t-test, FDR < 0.05). FC: fold change.
Supplementary material
Funding sources
This work was supported by the China Pharmaceutical University (grant numbers 3150120001, 2632018YX01 to XW).
Conflicts of interest
The authors declare that they have no competing interests.
Authors' contributions
ZJ performed data analyses and helped prepare for and wrote the manuscript. ZL performed data analyses and experiments, and wrote the manuscript. ML performed data analyses and helped prepare for the manuscript. CC performed data analyses. XW conceived the research, designed analysis strategies, performed data analyses, and wrote the manuscript. All the authors read and approved the final manuscript.
Acknowledgments
Not applicable.
References
- 1.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359(6382):1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.June C.H. CAR T cell immunotherapy for human cancer. Science. 2018;359(6382):1361–1365. doi: 10.1126/science.aar6711. [DOI] [PubMed] [Google Scholar]
- 3.Braun D.A., Burke K.P., Van Allen E.M. Genomic approaches to understanding response and resistance to immunotherapy. Clin Cancer Res. 2016;22(23):5642–5650. doi: 10.1158/1078-0432.CCR-16-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y. Combination of DC/CIK adoptive T cell immunotherapy with chemotherapy in advanced non-small-cell lung cancer (NSCLC) patients: a prospective patients' preference-based study (PPPS) Clin Transl Oncol. 2018 doi: 10.1007/s12094-018-1968-3. [DOI] [PubMed] [Google Scholar]
- 5.Kang J., Demaria S., Formenti S. Current clinical trials testing the combination of immunotherapy with radiotherapy. J Immunother Cancer. 2016;4:51. doi: 10.1186/s40425-016-0156-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moya-Horno I. Combination of immunotherapy with targeted therapies in advanced non-small cell lung cancer (NSCLC) Ther Adv Med Oncol. 2018;10 doi: 10.1177/1758834017745012. (p. 1758834017745012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goel S. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548(7668):471–475. doi: 10.1038/nature23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel S.P., Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 9.Goodman A.M. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yarchoan M. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17(4):209–222. doi: 10.1038/nrc.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao Y., Freeman G.J. The microsatellite instable subset of colorectal cancer is a particularly good candidate for checkpoint blockade immunotherapy. Cancer Discov. 2015;5(1):16–18. doi: 10.1158/2159-8290.CD-14-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davoli T. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;(6322):355. doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Warburg O., Wind F., Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang C.H. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162(6):1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cascone T. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 2018;27(5):977–987. doi: 10.1016/j.cmet.2018.02.024. (e4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshihara K. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbie D.A. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter S.L. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z. A comprehensive immunologic portrait of triple-negative breast cancer. Transl Oncol. 2018;11(2):311–329. doi: 10.1016/j.tranon.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massink M.P.G. Genomic profiling of CHEK2*1100delC-mutated breast carcinomas. BMC Cancer. 2015;15:877. doi: 10.1186/s12885-015-1880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rooney M.S. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugo W. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nathanson T. Somatic mutations and neoepitope homology in melanomas treated with CTLA-4 blockade. Cancer Immunol Res. 2017;5(1):84–91. doi: 10.1158/2326-6066.CIR-16-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christofk H.R. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 29.Magistretti P.J., Allaman I. A cellular perspective on brain energy metabolism and functional imaging. Neuron. 2015;86(4):883–901. doi: 10.1016/j.neuron.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 30.Rizvi N.A. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juneja V.R. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med. 2017;214(4):895–904. doi: 10.1084/jem.20160801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlate tumor glycolytic activity with survival prognosis in cancers with immunotherapy and cancers without immunotherapy. A. The expression of glycolysis-associated genes are positively associated with survival prognosis in the immunotherapeutic cohorts (log-rank test, P < .05). B. The expression of numerous glycolysis-associated genes are negatively associated with survival prognosis in multiple TCGA cohorts without immunotherapy (log-rank test, P < .1). OS: overall survival. DFS: disease-free survival. Kaplan-Meier curves are used to compare the survival time differences, and the log-rank test P values are shown in the survival curves.
Logistic regression analysis shows that glycolytic activity is a positive predictor for immune signatures in multiple cancer types when the tumor purity is added as a predictor. The tumors with high (upper third) versus low (bottom third) immune signature scores are predicted. Two immune signatures (immune score calculated by ESTIMATE [18] and immune cytolytic activity) are predicted by glycolytic activity, aneuploidy, TMB, and purity. TMB (tumor mutation burden) is defined as the total somatic mutation count in tumor. Tumor aneuploidy is evaluated by ABSOLUTE [21]. β coefficients for each predictor in pan-cancer or individual cancer types are shown.
Numerous HLA genes are upregulated in highly glycolytic tumors versus lowly glycolytic tumors (Student's t-test, FDR < 0.05). FC: fold change.
Supplementary material