Abstract
This revision of the classification of eukaryotes follows that of Adl et al., 2012 [J. Euk. Microbiol. 59(5)] and retains an emphasis on protists. Changes since have improved the resolution of many nodes in phylogenetic analyses. For some clades even families are being clearly resolved. As we had predicted, environmental sampling in the intervening years has massively increased the genetic information at hand. Consequently, we have discovered novel clades, exciting new genera and uncovered a massive species level diversity beyond the morphological species descriptions. Several clades known from environmental samples only have now found their home. Sampling soils, deeper marine waters and the deep sea will continue to fill us with surprises. The main changes in this revision are the confirmation that eukaryotes form at least two domains, the loss of monophyly in the Excavata, robust support for the Haptista and Cryptista. We provide suggested primer sets for DNA sequences from environmental samples that are effective for each clade. We have provided a guide to trophic functional guilds in an appendix, to facilitate the interpretation of environmental samples, and a standardized taxonomic guide for East Asian users.
Keywords: Algae, amoebae, biodiversity, ciliate, ecology, flagellate, fungus, microbiology, parasite, plankton, protozoa, systematics, taxonomy
THIS revision of the classification of eukaryotes updates that of the International Society of Protistologists (Adl et al. 2012). Since then, there has been a massive increase in DNA sequence information of phylogenetic relevance from environmental samples. We now have a much better sense of the undescribed biodiversity in our environment (De Vargas et al. 2015; Pawlowski et al. 2012). While significant, it still remains a partial estimation as several continents and soils in general are poorly sampled, and the deeper ocean is hard to reach. These new data clarified phylogenetic relationships and the new information is incorporated in this revision.
Systematics
We assembled the classification according to the principles outlined elsewhere, and we refer the reader to the introductions of both Adl et al. 2005 and 2012 for background information, and to Adl et al. 2007 for a discussion. Briefly, we adopted a hierarchical system without formal rank designations. The hierarchy is represented by indented paragraphs. The nomenclatural priority is given to the oldest name (and its authority) that correctly assembled genera or higher groups together into a clade, except where its composition was substantially modified. In these cases, we have used a newer term and its appropriate authorship. In cases where ranks were created to include a single lower rank, the higher ranks were eliminated as superfluous. In this scheme, monotypic taxa are represented by the genus only. Nested clades represent monophyletic lineages as best we know and para‐ or polyphyletic groups are so indicated.
This system of hierarchical nameless ranks, that ignores endings of clade names, has proved its utility in providing name stability as we reconstructed a new phylogenetic classification during the past 20 years. Clade names in this system do not change when their rank or composition changes, and it is only the authority for the name that changes when each clade description is adjusted (Cantino, 1998 Pleijel and Rouse, 2003). Where a new term is introduced in this classification, it is identified with “Adl et al. 2019” as the authority, or by the submitting author (e.g. Mann in Adl et al., 2019), and they are to be cited as emended in this publication. The descriptions provided are not intended to substitute for formal diagnoses. They are provided primarily for the student and general users to identify common morphological features, such as synapomorphies and apomorphies, within monophyletic lineages.
Figure 1.
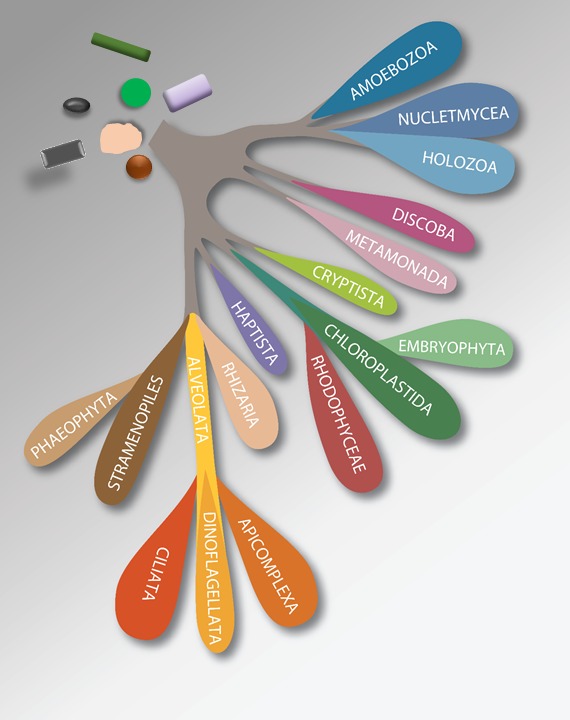
Overview of the diversity of protists among eukaryotes. Amoebozoa, Nucletmycea, and Holozoa together form the Amorphea; The Diaphoretickes includes Crypista, Chloroplastida and Embryophyta, Rhodophyceae, Haptista, Rhizaria, Alveolata (Apicomplexa, Dinoflagellata, Ciliata), Stramenopiles and Phaeophyta.
There are two novel components in this revision. First, we have provided trophic assignments for most taxa. This will prove useful in interpreting communities from environmental samples. Second, we informally suggest a phylum rank and classes in most clades to provide a point of reference in the classification hierarchy for the nonspecialist. This became possible, as there has been some stability at this level in the molecular phylogenetic reconstructions. It should be obvious that genera grouped into a clade then represent a family, and families into an order.
Nomenclature
This committee of the Society has had the responsibility of arbitrating nomenclature for protists in general. Historically, the task was simpler as most groups fell under one or the other of the two Codes of Nomenclature (algae and some other protists under the “International Code of Nomenclature for algae, fungi, and plants”, and protozoa under the “International Code of Zoological Nomenclature”), and few were described under both Codes. The Society was represented on the relevant committees. Notwithstanding that both Codes are incompatible, some have proposed to provide parallel classifications in each Code, while others proposed to adopt a modern unified code of nomenclature. Since the rearrangement of the classification along monophyletic lineages during the 1990s, many clades now include a mixture of taxa from both Codes. Several taxa, such as diatoms, are described in parallel under both Codes with different names. This situation created and perpetuates anomalies, such as the recent redescription of the dictyostellid amoebae with the botanical Code (Sheikh et al. 2018) for genera that are unarguably in Amoebozoa governed by the Zoological Code. Issues such as these have been thoroughly discussed in the past (Adl et al. 2007; Lahr et al. 2012). It has been the responsibility of this committee to discuss and arbitrate published phylogenetic hypotheses, proposals for new names and name changes. Underlying these discussions are principles of nomenclatural priority in the spirit of the codes of nomenclature.
A classification is unlike a phylogenetic tree in a publication, where the discovery of new clades, branches, or robust nodes ultimately leads to proposing new names. Newly named clades and nodes have their utility in phylogenetic analysis and discussion, but do not need to be formalized in the classification immediately. An overwhelming number of spent names have thus accumulated, with an increasing frequency over the past four decades, most of which are no longer—or never were—in common use. Many of these names were ephemeral, as their monophyly did not stand the test of (time) statistical analysis. The proliferation of these names reflects a methodological error practiced by some. That is to formalize names prematurely and try to reorganize classifications single‐handedly. As we argued before (Adl et al. 2012), this must be done with care, respecting nomenclatural priority, published as a proposal or a phylogenetic hypothesis first, to be verified by the community, and only eventually considered for change in the classification. The task of refereeing and classifying falls on Society committees representing communities of professionals. The very formal and slow process of voting to conserve or reject names through the tradition of the botanical code takes years as it has to proceed through committees and then approved by vote on the floor of the congress at 6‐year intervals. That is, however, too slow for the pace of changes today given the rate at which new information is becoming available.
In contrast to a phylogenetic tree, a classification system belongs to a community of users, and it is generated through discussions of the available evidence, for pragmatic purposes of teaching, curation, organizing data, archiving and communicating with a common language. It is a commonly agreed point of reference. It is not to be reimagined or re‐done at will by one individual. The Linnaean system that we have inherited has detailed codes of nomenclature that guide and regulate how living organisms are named, names changed and classified. The elaborate rules arise from disputes and mistakes made in the past, in part out of respect for each other's work. Instead of providing a long list of rejected and invalid names, we can specify that those not selected in this classification were considered nomina ambigua, nomina perplexa, nomina dubia, nomina nuda or did not have nomenclatural priority and are declared nomina rejicienda.
Another proposed classification of prokaryotes and eukaryotes was published recently (Ruggiero et al. 2015). This effort may be reasonable in their classification of the prokaryotes, but the eukaryote section does not pass standards of modern biology. Specifically, it is their refusal to use monophyly as a guiding principle, but to argue to retain “ancestral (paraphyletic) taxa when it seemed beneficial to do so” instead, even where monophyletic clades are already established. Their insistence on using a hodge‐podge of names that do not have nomenclatural priority, and that poorly describe the taxa included, further reduces its usefulness.
Classification
The super‐groups utilized since 2005 (Adl et al. 2005; Simpson and Roger 2004) are revised as follows (Fig. 1):
Eukaryotes now form two Domains called Amorphea and Diaphoretickes, with several additional clades that do not group into a third Domain.
In the Amorphea, the Opisthokonta, Breviatea and Apusomonadida now form a robust clade, as noted earlier (Adl et al. 2012), called Obazoa. Within the Opisthokonta, the Holozoa and Nucletmycea(/Holomycota) are robust clades with improved resolution of the basal sister lineages. In the Holozoa, the sponges and the other animals group together as the Metazoa (Porifera, Placozoa, Ctenophora, Cnidaria, Bilateria). In addition, a sister clade to the Amorphea comprising several genera was recently described as CRuMs (Brown et al. 2018).
There are two sister clades in Opisthokonta, the Holozoa and the Nucletmycea (/Holomycota). They share several characters, including synthesis of extracellular chitin in an exoskeleton, cyst/spore wall or cell wall of filamentous growth and hyphae; the extracellular digestion of substrates with osmotrophic absorption of nutrients; and other cell biosynthetic and metabolic pathways. Genera at the base of each clade are amoeboid and phagotrophic.
The Archaeplastida, Sar and several other clades remain a monophyletic clade under Diaphoretickes. The clade Cryptista comprising the cryptomonads, kathablepharids and Palpitomonas is well recognized and robust, although placement of its node within the Diaphoretickes remains problematic. In some but not all analyses, the clade appears inside the Archaeplastida. This position has always occurred from time to time in some phylogenies with weak support, but there is now stronger support for this association. We are not committed to their inclusion within the Archaeplastida but do note its likelihood. The inclusion of the Cryptista in the Archaeplastida would expand that group without affecting its defining criteria. Questioning the single origin of a plastid within the Archaeplastida is a rare minority opinion. Yet, the possibility of more than one plastid origin must not be ruled‐out until the cryptomonads are robustly positioned.
The new robust support for the Cryptista clade is accompanied by a similarly robust support for a clade comprising the Centroplasthelida and Haptophyta as the Haptista within the Diaphoretickes.
Nodes at the base of the Alveolata are better resolved with additional genera. The placeholder name Protalveolata is no longer required.
The Excavata comprise three clades: the Metamonada, the Discoba, and the Malawimonada. Their mutual relationships, as well as their relationships to other clades of eukaryotes, remain uncertain. We have dropped the supergroup Excavata in favour of the informal Excavates when referring to the “Discoba, Metamonada, Malawimonada”, as Incertae sedis in eukaryotes. The Excavates and several clades and genera fall outside of the two principal domains, but do not cluster together into a third domain.
This classification will serve as a primary starting reference for the taxonomic framework developed by UniEuk (unieuk.org; Berney et al. 2017), the Society supported, consensus‐driven, community‐based and expert‐driven international initiative to maintain a universal taxonomy for, at least, microbial eukaryotes. A specific aim of the UniEuk project is to apply one taxonomic framework to all genetic data in the International Nucleotide Sequence Database Collaboration (INSDC) repositories, which includes DDBJ (ddbj.nig.ac.jp), GenBank (ncbi.nlm.nih.gov) and ENA (ebi.ac.uk/ena) databases. The system's broad use and preservation will be ensured by a direct implementation of the UniEuk taxonomic framework into the ENA (European Nucleotide Archive) at EMBL‐EBI ( http://www.ebi.ac.uk/ena). The project will capture our collective knowledge on eukaryotic diversity, evolution, and ecology via three main modules (EukRef, EukBank and EukMap). EukRef (eukref.org; del Campo et al. 2018) uses a standardized, open‐source bioinformatics pipeline to generate homogenous, high‐quality curation of sequences (primarily 18S rDNA) available in INSDC databases. EukRef is fully operational; outputs include (on a lineage‐by‐lineage basis) taxonomically curated sequences, sequence alignments, phylogenetic trees and metadata. EukBank is a public repository of (primarily V4 18S rDNA) high‐throughput metabarcoding data sets, centralized at ENA, with standardized protocols for submitting data sets and metadata. EukMap (eukmap.unieuk.org) is an editable, user‐friendly representation of the UniEuk taxonomy in the form of a publicly navigable tree, where each node/taxon is associated with contextual data (taxonomic and ecological information, links to representative images, etc.). It will be operational by 2019 and will allow registered community members to directly interact with and inform the taxonomic framework, and to flag taxonomy issues requiring revision. As a whole, the UniEuk system will represent a community hub to centralize, standardize, and promote global knowledge on eukaryotic diversity, taxonomy and ecology.
Clarification of terms for trophic functional groups
Several terms were clarified to correct misuse of terminology in publications. In 2005, these were: eukaryote, prokaryote, algae, zoosporic fungi, protozoa, zooplankton, phytoplankton, cyst, spore and cilium. In 2012, they were related to the cytoskeleton and motility: lobopodia, lamellipodia, filopodia, granuloreticulopodia, reticulopodia, axopodia, centriole, centrosome, microtubular organizing centre (MTOC), basal body, kinetosome, kinetid and mastigont. In this revision, they pertain to trophic functional groups.
In addition to descriptions of morphology that accompany specimen, which is critical for understanding cell function and interpreting phylogenetic trees, improved descriptions of site and food preferences are required for an ecological interpretation of the role in the community and ecosystem. Often species lack sufficient description of the collection site or feeding habit.
To compare environmental DNA data sets, adequate metadata is necessary to select appropriate samples for comparison. The same issue exists when trying to re‐isolate a species or to verify the type specimen. Therefore, it is important that the environment and habitat is sufficiently described. Merely stating marine, terrestrial or soil is grossly inadequate. The soil, for example, is heterogeneous horizontally at the sub‐millimetre to regional scales. It is also stratified through the profile, and across the diameter of each ped. Whether a soil or aquatic sample, solution chemistry and site physical parameters contribute to define the niche space.
Because we care about nomenclature and the exact meaning of words and of names of things, especially species and their groupings into nodes and stems on phylogenetic trees, it is equally important to care for how we describe sampling sites and feeding habits. There are two parts to describing the feeding habit: what is eaten and how it is eaten.
Species that release enzymes extracellularly to digest substrates in their habitat, are generally called saprotrophic or lysotrophic, and contribute to the decomposition of organic matter. One incredible resource is FunGuild (Nguyen et al. 2016, ( https://github.com/UMNFuN/FUNGuild) to determine substrate utilization for saprotrophic fungi. Probably all eukaryotes are capable of osmotrophy, the acquisition of soluble nutrients through the cell membrane. For example, plants obtain their carbon for photosynthesis from the air, as well as some oxygen—however, they rely on osmotrophy through the roots to obtain all the other elements they need. Osmotrophy occurs through the ciliary pit, by pinocytosis, by diffusion, and by various membrane transport proteins. Some species have no alternative form of acquiring energy, are very poor at decomposing substrates and are strict osmotrophs relying on dissolved nutrients. Detritus eaters ingest particles derived from cells and tissues, decomposing organic matter, starch granules, plant or animal debris, or wood (microchip) fragments.
Species that eat other species are called consumers, and there are a variety of terms to describe the functional groups. Some acquire suspended particles in the solution and accumulate the particles by filtration into an oral region or cytostome (not filter‐feeders, as they do not feed on filters). The size of particles filtered out of the liquid depend on the current generated, and the structure of the feeding apparatus (Fenchel 1986), and it is a good idea to specify what size prey are ingested. The remaining consumers fall into two categories, the grazers and predators. Grazers, like a cow in a field of grasses, browse and ingest from surfaces covered with potential food items (e.g. an amoeba on a lawn of bacteria, or on soil particle surfaces). Predators pursue scarce prey according to optimal foraging theory, typically handle one prey at a time, and it is mathematically distinct (e.g. a Jakoba ingesting one bacterium). Species gather bacteria by filtration prior to phagocytosis, or directly by phagocytosis; it is best to specify “bacteria by filtration” or “bacteria by phagocytosis”. A popular term bacterivore has the unintended implication of voraciously devouring (voracitas L.) which is a false description of how many bacteria eaters acquire their prey, and an incomplete description. Use it, but be aware that some readers and reviewers will be more discriminating. In contrast, the more appropriate term –trophy (trophe Gr.), to eat for food and nourishment, sounds more awkward in English. For species that ingest unicellular protists by phagotrophy, the correct term is cytotrophy. Bacterium (Ehrenberg 1838) has been the word used to refer to a prokaryotic cell, while cell (Dutrochet 1824; Schleiden 1838; Schwann 1839) has been used since to refer to a eukaryotic cell. Mixotrophy refers to photosynthetic species that also ingest food by phagocytosis, and heterotrophs that retain prey plastids and symbionts.
There are two distinct mechanisms to feed on algal filaments (cellulosic cell wall) or fungal hyphae (chitinous cell wall). One mechanism is to slurp the filaments like noodles and ingest them, and the other is to penetrate through the cell wall. Those that puncture through phagocytose cytoplasm, and some species even penetrate inside to ingest cytoplasm along the tube or in the spore. It is best to distinguish between the cell wall material to digest and the mechanism of ingestion. Thus, we have mycotrophy or phycotrophy, by either swallowing (devoratis L.) or by penetrating (penetrando L.).
In microbial food webs, there are also consumers of consumers, typically by predation, that are equivalent above‐ground or in aquatic systems to carnivores (meat eaters), or other functional groups. Although 2° consumers, 3° consumers, and so on exist in microbial food webs, it is hardly correct to refer to carnivores in food webs where there is no meat.
Another poorly crafted term one encounters, albeit rarely, is eukaryovory. Although there are famous examples of eukaryovory (Saint‐Exupéry 1943), eukaryotes eating eukaryotes can include parasitism, as intracellular or extracellular parasites, on hosts that are protists or multicellular, with various grades of host specificity, and it is a poor substitute for cytotrophy.
We have summarized the higher level classification of eukaryotes in Table 1, with an estimate of the known number of genera, and providing informal phylum and class designations to help orient the student and users along the hierarchy, or nodes on a phylogenetic tree. The revised classification of eukaryotes is presented in Table 2, and genera that have not been studied enough to place in the classification are listed in Table 3 as incertae sedis Eukarya. Table 4 provides recommended primers for analysing DNA from environmental samples, noting that the choice of primers and depth of sequencing are important sources of variation between studies. Appendix S1 provides additional supporting literature that we considered important to understand the changes. Appendix S2 provides more detail about the trophic functional assignments across protists, by noting exceptions at the genus level. Appendix S3 provides a standardized guide to East Asian users for the new terminology.
Table 1.
Higher ranks of the eukaryotes suggesting the position of Linnaean ranks, and the number of known genera
| AMORPHEA |
|
| DIAPHORETICKES |
| Incertae sedis: Microhelliela maris, Ancoracysta twista, Rappemonads, Telonemia, Picozoa |
|
G = genus; F = family; O = order; C = class; P = phylum; K = kingdom.
The state of the classification in online databases are too poor to evaluate or work with this clade.
Table 2.
Classification of the higher ranks of the protists and multicellular organisms. The authority to whom the taxon name is attributed appears immediately after the taxon name. For purposes of nomenclature and stability of names in the classification, we have tried to retain the oldest term that correctly described the grouping, emended if necessary; in the square bracket following are inappropriate and incorrect names used in the literature, or that do not have nomenclatural priority. If the taxon name has been emended herein, the authority is indicated and the reference is to this manuscript (“emend. Adl et al. 2019”). Selected references to the literature since 2012 can be found in Appendix S1. Citations in the notes to this table can be found in the LITERATURE CITED. Named clades are monophyletic as best as we can determine; if paraphyly or polyphyly is suspected, it is indicated by P; robust clades recovered in phylogenetic analysis that do not have morphological diagnosis are indicated by R (ribo‐group); monotypic genera with only one described species are indicated by M; MTOC, microtubular organizing centre. * Denotes genera lacking DNA sequence information or known to require taxonomic revision
| AMORPHEA Adl et al. 2012 |
| The least inclusive clade containing Homo sapiens Linnaeus 1758, Neurospora crassa Shear and Dodge 1927 (both Opisthokonta), and Dictyostelium discoideum Raper 1935 (Amoebozoa). This is a node‐based definition in which all of the specifiers are extant; it is intended to apply to a crown clade; qualifying clause—the name does not apply if any of the following fall within the specified clade—Arabidopsis thaliana (Linnaeus) Heynhold 1842 (Archaeplastida), Tetrahymena thermophila Nanney and McCoy 1976 (Alveolata), Thalassiosira pseudonana Hasle and Hiemdal 1970 (Stramenopiles), Bigelowiella natans Moestrup and Sengco 2001 (Rhizaria), Euglena gracilis Klebs 1883 (Excavata) and Emiliania huxleyi (Lohmann) Hay and Mohler 1967 (Haptophyta). |
| Incertae sedis Amorphea: Obazoa Brown et al. 2013 (R) |
| Obazoa is a clade that is robustly recovered in phylogenetic trees and consists of the Opisthokonta and two other clades, Apusomonadida and Breviatea. It is the least inclusive clade containing Homo sapiens Linnaeus 1758 (Opisthokonta), Neurospora crassa Shear & Dodge 1927 (Opisthokonta), Pygsuia biforma Brown et al. 2013 (Breviatea) and Thecamonas trahens Larsen & Patterson 1990 (Apusomonadida). |
|
|
|
|
|
|
|
|
|
|
|
Table 3.
Genera incertae sedis in eukaryotes, with uncertain affiliation within protists
| Acinetactis |
| Actinastrum |
| Actinocoma |
| Actinolophus |
| Adinomonas |
| Aletium |
| Amphimonas |
| Amylophagus |
| Anaeramoeba |
| Aphelidiopsis |
| Asterocaelum |
| Asthmatos |
| Aurospora |
| Barbetia |
| Belaria |
| Belonocystis |
| Bertarellia |
| Bertramia |
| Bodopsis |
| Boekelovia |
| Branchipocola |
| Camptoptyche |
| Chalarodora |
| Cibdelia |
| Cichkovia |
| Cinetidomyxa |
| Cingula |
| Cladomonas |
| Clathrella |
| Codonoeca |
| Coelosporidium a |
| Copromonas |
| Cyanomastix |
| Cyclomonas |
| Cytamoeba |
| Dallingeria |
| Dictyomyxa |
| Dimastigamoeba |
| Dinemula |
| Dinoasteromonas |
| Diplocalium |
| Diplomita |
| Diplophysalis |
| Diploselmis |
| Dobellina |
| Ducelleria |
| Ectobiella |
| Elaeorhanis |
| Embryocola |
| Endamoeba |
| Endemosarca |
| Endobiella |
| Endomonas |
| Endospora |
| Enteromyxa |
| Eperythrocytozoon |
| Errera |
| Fromentella |
| Gymnococcus |
| Gymnophrydium |
| Haematotractidium |
| Hartmannina |
| Heliobodo |
| Heliomonas |
| Hermisenella |
| Heterogromia |
| Hillea |
| Hyalodaktylethra |
| Immanoplasma b |
| Isoselmis |
| Janickina |
| Kamera |
| Lagenidiopsis |
| Liegeosia |
| Luffisphaera c |
| Lymphocytozoon |
| Lymphosporidium |
| Macappella |
| Magosphaera |
| Malpighiella |
| Martineziella |
| Megamoebomyxa |
| Meringosphaera |
| Microcometes |
| Monochrysis |
| Monodus |
| Mononema |
| Myrmicisporidium |
| Naupliicola |
| Nephrodinium |
| Neurosporidium |
| Orbulinella |
| Ovicola |
| Palisporomonas |
| Pansporella |
| Paradinemula |
| Paraluffisphaera |
| Paramonas |
| Paraplasma |
| Parastasia |
| Parastasiella |
| Peliainia |
| Peltomonas |
| Petasaria |
| Phagodinium |
| Phanerobia |
| Phloxamoeba |
| Phyllomitus |
| Phyllomonas |
| Physcosporidium |
| Piridium |
| Pleuophrys |
| Pleuromastix |
| Protenterospora |
| Protomonas |
| Pseudoactiniscus |
| Pseudosporopsis |
| Rhizomonas |
| Rhynchodinium |
| Rigidomastix |
| Schewiakoffia |
| Sergentella |
| Serpentoplasma |
| Sphaerasuctans |
| Spongastericus |
| Spongocyclia |
| Stephanomonas |
| Strobilomonas |
| Tetradimorpha |
| Tetragonidium |
| Thaulirens |
| Topsentella |
| Toshiba |
| Trichonema |
| Urbanella |
Probably a junior synonym of Nephridiophaga, a Zygomycete.
Immanoplasma Neumann 1909 (see Kar 1990).
Belonocystis (Amoebozoa incertae sedis) and Luffisphaera maybe the same genus.
Table 4.
Recommended primers for environmental samples
| Supergroup or highest rank | Clades | Primer pair codes | Sequence length (bp) | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|---|---|---|
| Amorphea | Apusomonadida1 | 18S, EK‐42F & APU‐1R | 1,500‐2,200 | CTCAARGAYTAAGCCATGCA | CTTCCTTTGGTTAAAACAC |
| Amoebozoa | Tubulinea, Discosea, Variosea | 18S, RibA & RibB | Entire SSU molecule, variable | ACCTGGTTGATCCTDCCAGT | TGATCCATCTGCAGGTTCACCTAC |
| Tubulinea, Discosea, Variosea | 18S, RibA & S20R | ~1,800 | ACCTGGTTGATCCTDCCAGT | GACGGGCGGTGTGTACAA | |
| Nearly all clades | Cox‐I, LCO1490 & HCO2198 (modified Folmer primers) | ~660 | GGTCAACAAATCATAAAGATATTGG | TAAACTTCAGGGTGACCAAAAAATCA | |
| Arcellinida | 1st step, Euk 82F & Euk 1498 R, 2nd step, cloning | Variable | GAAACTGCGAATGGCTC | CYGCAGGTTCACCTA C | |
| Opisthokonta | Choanoflagellata2 | 18S, 42F & 1510R | ~1,750 | CTCAARGAYTAAGCCATGCA | CCTTCYGCAGGTTCACCTAC |
| Porifera3 | Demospongiae, Homoscleromorpha: | Cox‐I (Folmer primers), LCO1490 & HCO2198 | 658 | GGTCAACAAATCATAAAGATATTGG | TAAACTTCAGGGTGACCAAAAAATCA |
| 28S, C1 & D2, universal primers | 800–900 | ACCCGCTGAATTTAAGCAT | TCCGTGTTTCAAGACGGG | ||
| 28S D1–D2, Por28S‐15F & Por28S‐878R | 790–830 | GCGAGATCACCYGCTGAAT | CACTCCTTGGTCCGTGTTTC | ||
| 28S D3–D5, Por28S‐830F & Por28S‐1520R | 650‐660 | CATCCGACCCGTCTTGAA | GCTAGTTGATTCGGCAGGTG | ||
| 28S D3‐D5, NL4F & NL4R | 650–660 | GACCCGAAAGATGGTG AACTA | ACCTTGGAGACCTGA TGCG | ||
| Calcarea: | 28S C‐region, C2 & D2, or C2'modified & D2 | 430–470 | GAAAAGAACTTTGRARAGAGAGT or GAAA AGCACTTTGAAAAGAGA | TCCGTGTTTCAAGACGGG | |
| Hexactinellida: | 28S D3‐D5, NL4F & NL4R | 900–1,000 | GACCCGAAAGATGGTG AACTA | ACCTTGGAGACCTGATGCG | |
| 16S partial primers, 16S1fw & 16SH modified | 500 | TCGACTGTTTACCAAAAACATAGC | YRTAATTCAACATCGAGGTC | ||
| Fungi | Chytridiomycota4 , 5 , 6 , 7 | 18S, NS1 & NS4 | ~950–1,100 | ||
| PolySSU1 & PolySSU1R | Variable | TGATCCTTCYGCAGGTTCACC | |||
| 28S, LROR & LR5 | ~800‐950 | AACTAAGAACGGCCATGCAC | |||
| ITS1‐5.8S‐ITS2, ITS5 & ITS4 | ~614–987 | CCCGTGTTGAGTCAAATTAAGC | |||
| EF‐1a,* 983F & EF1aZ‐1R | ~1,150 | GAACGGCCATGCACCACCACC | |||
| EF‐1a‐like,** 983F & EFL‐RS2R | −~550–590 | GTTCTTGTGTTAATCTCAC | |||
| Fungi8 , 9 | ITS (See citation 9, Table S1) | Variable | |||
| 18S, AU2‐F & AU4‐R, & inner AUPH1 | TTTCGATGGTAGGATAGDGG | RTCTCACTAAGCCATTC, and inner AGAGCTMTCAATCTGTCAATCCT | ACTTCTGGRTGICCRAARAAYCA | ||
| Haptista | Haptophyta10 , 11 | 1F & 1528R (or EukA & EukB) | 1,800 | AACCTGGTTGATCCTGCCAGT | TGATCCTTCTGCAGGTTCACCTAC |
| 1,795 | ACCTGGTTGATCCTGCCAG | TGATCCTTCYGCAGGTTCAC | |||
| EukF & EukR | 830 | GGGTTCGATTCCGGAGAG | CCGTGTTGAGTCAAATT | ||
| TAReuk454FWD1 & TAReukREV3 | Variable | CCAGCA(G⁄C)C(C⁄T)GCGGTAATTCC | ACTTTCGTTCTTGATYRA | ||
| Hapto4 & Euk34r | 1,000 | ATGGCGAATGAAGCGGGC | GCATCGCCAGTTCTGCTTACC | ||
| LSU1 (Lhapto8 & Lhapto20R_bis) | 350‐400 | GGTATCGGAGAAGGTGAGAATCCT | TCAGACTCCTTGGTCCGTGTTTCT | ||
| Prym03‐3 & Hapto1R | 416 | GTAAATTGCCCGAATCCTG | CGAAACCAACAAAATAGCAC | ||
| 528FLong & PRYM01 + 7 | 399 | GCGGTAATTCCAGCTCCAA | GATCAGTGAAAACATCCCTGG | ||
| Pavlova‐V4F & 1528R | 904 | GTGAAATTCTTAGACCCACGGA | TGATCCTTCTGCAGGTTCACCTAC | ||
| 1F & Pavlova‐V4F2R | 593 | See 1F above | GTGAAATTCTTAGACCCACGGA | ||
| Pry421F & Pry1572R | 1,070 | AGCAGGCGCGTAAATTGCCCG | TCAACGYRCGCTGATGACA | ||
| Hap220F & Pav1702R | 1,400 | ACCGGTCTCCGGTTGCGTGC | TAGATGATAAGGTTTGGGTG | ||
| Centroplasthelida12 | Helio1979R | Variable | CACACTTACWAGGAYTTCCTCGTTSAAGACG | ||
| Cryptista13 , 14 | 18S‐0024F & 18S‐1757R | Variable | CTGGTTGATCCTGCCAGTAGT | CAGGTTCACCTACGGAAACCT | |
| 18S‐33F & 18S‐1768R | 1,700‐1,800 | CCT GCC AGT AGT CAT AYG CTT | TGA TCC TTC YGC AGG TTC ACC | ||
| Stramenopiles | Sar15 | 18S, SAR‐V3‐SSU F & R | 150 | TCGTCGGCAGCGTCAGATGTGTATAAGAGACA | ATGTGTATAAGAGACAGRACTACGAGCTTTTTAACTGC |
| Euglyphid16 | 1st step EuglySSUF & EuglyLSUR | Variable | GCGTACAGCTCATTATATCAGCA | GTTTGGCACCTTAACTCGCG | |
| 2nd step EuglySSUF & EuglySSUR | Variable | GCGTACAGCTCATTATATCAGCA | GCACCACCACCCATAGAATCWAGAAAGATC | ||
| Assulinidae17 | 1st step, COI: Eucox1F & Euglycox1R | Variable | GAYATGGCKTTNCCAAGATTAAA | AGCACCCATTGAHAAAACRTAATG | |
| 2nd step, Assucox 1F & Assucox 1R | Variable | AAYATGAGRGCYAGRGG | 5¢‐CGTAATGAAARTGWCCYACC | ||
| Amphitremida | 1st step, Euk 82F & Euk 1498 R, 2nd step, cloning | Variable | GAAACTGCGAATGGCTC | CYGCAGGTTCACCTAC | |
| Diatomea18 , 19 | rbcL, 646F& 998R | 379 | ATGCGTTGGAGAGARCGTTTC | GATCACCTTCTAATTTACCWACAACTG | |
| Diat_rbcL_708F (mixture of 3 primers) & two reverse primers R3_1 & R3_2 | 312 (amplicon 263) | 1: AGGTGAAGTAAAAGGTTCWTACTTAAA, and 2: AGGTGAAGTTAAAGGTTCWTAYTTAAA and 3: AGGTGAAACTAAAGGTTCWTACTTAAA | 1: CCTTCTAATTTACCWACWACTG, and 2: CCTTCTAATTTACCWACAACAG | ||
| Alveolata | Ciliophora20 | 18S V4, | Variable | ||
| Apicomplexa21 , 22 | 18S PF1 & R4 | 1,800 | GCGCTACCTGGTTGATCCTGCC | GATCCTTCTGCAGGTTCACCTAC | |
| 18S V4 TAReuk454FWD1 & TAReukREV3 | Variable | ACTTTCGTTCTTGAT(C⁄T)(A⁄G) | ACTTTCGTTCTTGAT(C⁄T)(A⁄G) | ||
| 18S V4, 346Fmix & 785R‐mix | Variable | CADCGACGGGTAACGGGGAATTA; CAGYGACGGGTAACGGGGAATTA; CAGYGACGGGTAACGGGGAATTA; CAGYGACGGGTAACGGGGAATTA | IIITATTCCATGCTGIAGTATTCA; IIITATTCCATGCTAAASTATTCA | ||
| Dinoflagellata**** , 23 , 24 | 18S V4, Next. For & Rev | Variable | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG[CCAGCASCYGCGGTAATTCC] | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG[ACTTTCGTTCTTGATYRATGA] | |
| Syndiniales | 18S V4, 528F & UnonMet | Variable | |||
| Rhizaria | Cercozoa | 18S V4, 3NDF & 1256R | ~500 | GGCAAGTCTGGTGCCAG | GCACCACCACCCAYAGAATCAAGAAAGAWCTTC |
| 18S V4***, 25F & 1256R | 1,200 | CATATGCTTGTCTCAAAGATTAAGCCA | GCACCACCACCCAYAGAATCAAGAAAGAWCTTC | ||
| Cyphoderiidae25 | Cox‐I, Eucox1F & Euglycox1R | Variable | GAYATGGCKTTNCCAAGATTAAA | AGCACCCATTGAHAAAACRTAATG | |
| Foraminifera26 | 18S, 14F1 & s17 | 300‐400 | AAGGGCACCACAAGAACGC | CGGTCACGTTCGTTGC | |
| Excavates | Fornicata27 | EukA & EukB* | Variable | ||
| Parabasalia | 16SI & 16S RR (or EukA & B) | Variable | TACTTGGTTGATCCTGCC | TCACCTACCGTTACCTTG | |
| ITS‐F & ITS‐R | Variable | TTCAGTTCAGCGGGTCTTCC | GTAGGTGAACCTGCCGTTGG | ||
| Jakobida | EukA ‐ EukB | Variable | |||
| Heterolobosea | ITS1‐5.8S & ITS2, JITS‐F & JITS‐R, (or EukA & EukB) | Variable | GTCTTCGTAGGTGAACCTGC | CCGCTTACTGATATGCTTAA | |
| Preaxostyla: Oxymonadida | Mon‐F & Mon‐R | Variable | GAAGTCATATGCTGTCTCAA, | TCACCTACGGAAACCTT | |
| Preaxostyla: Paratrimastigida, Trimastigida | EukA & EukB | 1,800–3,100 | CTGGTTGATCCTGCCAG | TGATCCTTCTGCAGGTTCACCTAC | |
| Euglenida Heterotrophs28 | See citation | Varies with the primer pairs | |||
| Euglenophyceae28 | See citation | Varies with the primer pair | |||
| Protist, general* , 27 | General Medlin primers | EukA‐F & EukB‐R | Variable | CTGGTTGATCCTGCCAG | TGATCCTTCTGCAGGTTCACCTAC |
| Protist general21 | General Stoeck primers | See citation | Variable |
Selective amplification of species, some clades missed.
Spizellomycetales.
Chytridiomycota except Spizellomycetales.
See citations for DINOREF in PR2 v.4.9.0.
Torruella, G., Moreira, D. & López‐García, P. 2017. Phylogenetic and ecological diversity of apusomonads, a lineage of deep‐branching eukaryotes Env. Microbiol. Rep. 9:113‐119. doi.org/10.1111/1758‐2229.12507.
Amaral‐Zettler, L.A., McCliment, E.A., Ducklow, H.W. & Huse, S.M. 2009. A Method for Studying Protistan Diversity Using Massively Parallel Sequencing of V9 Hypervariable Regions of Small‐Subunit Ribosomal RNA Genes. PLOS one 4 (12): https://doi.org/10.1371/journal.pone.0006372.
Morrow, C.C., Picton, B.E., Erpenbeck, D., Boury‐Esnault, N., Maggs, C.A. & Allcock, A.L. 2012. Congruence between nuclear and mitochondrial genes in Demospongiae: A new hypothesis for relationships within the G4 clade (Porifera: Demospongiae). Molecular Phylogenetics and Evolution 62: 174–190.
White, M.M., James, T.Y., O'Donnell, K., Cafaro, M.J., Tanabe, Y. & Sugiyama, J. 2006. Phylogeny of the Zygomycota based on nuclear ribosomal sequence data. Mycologia, 98(6): 872‐884. https://doi.org/10.1080/15572536.2006.11832617.
Simmons, D.R. 2011. Phylogeny of Powellomycetaceae fam. nov. and description of Geranomyces variabilis gen. et comb. Nov. Mycologia 103(6):1411‐1420.
Vilgalys, R. & Hester, M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriology 172(8):4238‐4246.
James, T.Y., Letcher, P.M., Longcore, J.E., Mozley‐Standridge, S.E., Porter, D., Powell, M.J., Griffith, G.W. & Vilgalys, R. 2006. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 98(6):860‐871.
Schoch, C.I., Seifert, K.A, Huhndorf, S., Robert, V., Spouge, J.L., Levesque, C.A., Chen, W. & Fungal Barcoding Consortium. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. PNAS 109 (16): 6241‐6246; https://doi.org/10.1073/pnas.1117018109.
Vandenkoornhuyse, P., Baldauf, S.L., Leyval, C., Straczek, J. &. Young, J.P.W. 2002. Extensive Fungal Diversity in Plant Roots Science 295 (5562): 2051 https://doi.org/10.1126/science.295.5562.2051.
Egge, E, Bittner, L, Andersen, T, Audic, S, de Vargas, C. 2013. 454 Pyrosequencing to Describe Microbial Eukaryotic Community Composition, Diversity and Relative Abundance: A Test for Marine Haptophytes. PLoS ONE 8(9): e74371. https://doi.org/10.1371/journal.pone.0074371.
Edvardsen, B., Egge, E.S. & Vaulot, D. 2016. Diversity and distribution of haptophytes revealed by environmental sequencing and metabarcoding – a review. Perspectives in Phycology 3 (2): 77–91.
Cavalier‐Smith, T. & von der Heyden, S. 2007. Molecular phylogeny, scale evolution and taxonomy of centrohelid heliozoan. Molecular Phylogenetics and Evolution 44(3): 1186‐1203. https://doi.org/10.1016/j.ympev.2007.04.019.
Kim, E., Simpson, A.G.B. & Graham, L.E. 2006. Evolutionary relationships of apusomonads inferred from taxon‐rich analyses of 6 nuclear encoded genes. Molec. Biol. Evol. 23(12):2455‐2466.
Kim, E. & Archibald, J. 2013. Ultrastructure and molecular phylogeny of the Cryptomonad Gonimonas avonlea sp. Nov. Protist 164(2):160‐182.
Sisson, C., Gulla‐Devaney, B., Katz, L.A. & Grattepanche, J‐D. 2018. Seed bank and seasonal patterns of eukaryotic SAR (Stramenopila, Alveolata, Rhizaria) clade in a New England vernal pool. J. Plankton Res. 00(00): 1–15. https://doi.org/10.1093/plankt/fby020.
Lara, E., Roussel‐Delif, L., Fournier, B., Wilkinson, D.M. & Mitchell, E.A.D. 2016. Soil microorganisms behave like macroscopic organisms: Patterns in the global distribution of soil euglyphid testate amoebae. Journal of Biogeography 43(3):520‐532.
Lara, E., Heger, T.J., Scheihing, R. & Mitchell, E.A.D. 2011. COI gene and ecological data suggest size‐dependent high dispersal and low intra‐specific diversity in free‐living terrestrial protists (Euglyphida: Assulina). Journal of Biogeography 38(4): 640‐650.
Kelly, M., Boonham, N., Juggins, S., Kille, P., Mann, D., Pass, D., Sapp, M., Sato, S. & Glover, R. 2018. A DNA based diatom metabarcoding approach for Water Framework Directive classification of rivers. SC140024/R, Environment Agency, Bristol. ISBN: 978‐1‐84911‐406‐6.
Vasselon, V., Rimet, F., Tapolczai, K. & Bouchez, A. 2017. Assessing ecological status with diatoms DNA metabarcoding: scaling up on a WFD monitoring network (Mayotte island, France). Ecol. Indicators 82: 1–12.
Lara, E., Berney, C., Harms, H. & Chatzinotas, A. 2007. Cultivation‐independent analysis reveals a shift in ciliate 18S rRNA gene diversity in a polycyclic aromatic hydrocarbon‐polluted soil. FEMS Microbiol. Ecol. 62:365‐373.
Stoeck, T., Bass, D., Nebel, Christen, R., Jones, M.C.M., Breiner, H‐W. & Richards, T. 2010. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19(1), 21‐31.
Janouškovec, J., Tikhonenkov, D.V., Burki, F., Howe, A.T., Kolisko, M., Mylnikov, A.P., & Keeling, P. 2015. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. PNAS 112(33), 10200–10207.
Piredda R., Tomasino, M.P., D'Erchia, A.M., Manzari, C., Pesole, G., Montresor, M., Kooistra, W.H.C F., Sarno, D. & Zingone, A. 2017. Diversity and temporal patterns of planktonic protist assemblages at a Mediterranean Long Term Ecological Research site FEMS Microbiology Ecology, 93(1), fiw200, doi.org/10.1093/femsec/fiw200.
Mordret, S., Piredda, R., Vaulot, D., Montresor, M., Kooistra, W.H.C.F. & Sarno. D. DINOREF: A curated dinoflagellate (Dinophyceae) reference database for the 18S rRNA gene. Mol Ecol Resour. 2018;1–14. https://doi.org/10.1111/1755-0998.12781.
Heger, T., Pawlowski, J., Lara, E., Leander, B.S., Todorov, M., Golemansky, V. & Mitchell, E.A.D. 2011. Comparing potential COI and SSU rDNA barcodes for assessing the diversity and phylogenetic relationships of cyphoderiid testate amoebae (Rhizaria: Euglyphida). Protist 162(1):131‐141.
Pawlowski, J. & Lecroq, B. 2010. Short rDNA barcodes for species identification in foraminifera. J. Eukaryot. Microbiol. 57(2):197‐205.
Medlin, L., Elwood, H.J., Stickel, S. & Sogin, M.L. 1988. The characterization of enzymatically amplified eukaryotic 16S‐like rRNA‐coding regions. Gene 71(2): 491‐499. https://doi.org/10.1016/0378-1119(88)90066-2.
Lax, G. & Simpson, AGB. 2013. Combining molecular data with classical morphology for uncultured phagotrophic euglenids (Excavata): a single cell approach. J. Eukaryot. Microbiol.. 60(6):615‐25. https://doi.org/10.1111/jeu.12068.
Appendices
Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes by Sina M. Adl, David Bass, Christopher E. Lane, Julius Lukeš, Conrad L. Schoch, Alexey Smirnov, Sabine Agatha, Cedric Berney, Matthew W. Brown, Fabien Burki, Paco Cárdenas, Ivan Čepička, Ludmila Chistyakova, Javier del Campo, Micah Dunthorn, Bente Edvardsen, Yana Eglit, Laure Guillou, Vladimír Hampl, Aaron A. Heiss, Mona Hoppenrath, Timothy Y. James, Anna Karnkowska, Sergey Karpov, Eunsoo Kim, Martin Kolisko, Alexander Kudryavtsev, Daniel J. G. Lahr, Enrique Lara, Line Le Gall, Denis H. Lynn, David G. Mann, Ramon Massana i Molera, Edward A. D. Mitchell, Christine Morrow, Jong Soo Park, Jan W. Pawlowski, Martha J. Powell, Daniel J. Richter, Sonja Rueckert, Lora Shadwick, Satoshi Shimano, Frederick W. Spiegel, Guifré Torruella i Cortes, Noha Youssef, Vasily Zlatogursky, Qianqian Zhang.
Appendix S1. Supplementary references.
Appendix S2. Functional group assignments.
Appendix S3. Translation guide for East Asian users.
Appendix S1. Selected Literature mostly since 2012.
AMORPHEA
Berney, C., Geisen, S., Van Wichelen, J., Nitsche, F., Vanormelingen, P., Bonkowski, M. & Bass, D. 2015. Expansion of the “reticulosphere”: diversity of novel branching and network‐forming amoebae helps to define Variosea (Amoebozoa). Protist, 166, 271–295.
Brown, M. W., Sharpe, S. C., Silberman, J. D., Heiss, A. A., Lang, B. F., Simpson, A. G. B. & Roger, A. J. 2013. Phylogenomics demonstrates that breviate flagellates are related to opisthokonts and apusomonads. Proc. R. Soc. Lond. B, 280:1769.
Cavalier‐Smith, T., Chao, E., E., & Lewis, R. 2016. 187‐Gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep‐branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution. Mol. Phylogenet. Evol., 99: 275–296.
Hamann, E., Gruber‐Vodicka, H., Kleiner, M., Tegetmeyer, H. E., Riedel, D., Littmann, S., Chen, J., Milucka, J., Viehweger, B., Becker, K. W., Dong, X., Stairs, C. W., Hinrichs, K.‐U., Brown, M. W., Roger, A. J. & Strous, M. 2016. Environmental Breviatea harbor mutualistic Acrobacter epibionts. Nature, 534:254–258.
Heiss, A. A., Lee, W. J., Ishida, K. & Simpson, A. G. B. 2015. Cultivation and characterisation of new species of apusomonads (the sister group to opisthokonts), including close relatives of Thecamonas (Chelonemonas n. gen.). J. Eukaryot. Microbiol., 62:637‐649.
Kang, S., Tice, A.K., Spiegel, F. W., Silberman, J. D., Pánek, T., Čepička, I., Kostka, M., Kosakyan, A., Alcântara, D. M., Roger, A. J., Shadwick, L. L., Smirnov, A., Kudryavstev, A., Lahr, D. J., & Brown, M. W. 2017. Between a pod and a hard test: the deep evolution of amoebae. Mol Biol Evol msx162. https://doi.org/10.1093/molbev/msx162.
Pánek, T, Zadrobílková, Walker, G., Brown, M. W., Gentekaki, E., Hroudrová, M., Kang, S. Roger, A. J., Tice, A. K., Vlček, Č., & Čepička, I. 2016. First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group. Molec. Phylogen. Evol., 98:41‐51.
Schaap, P., Winckler, T., Nelson, M., Alvarez‐Curto, E., Elgie, B., Hagiwara, H., Cavender, J., Milano‐Curto, A., Rozen, D. E., Dingermann, T., Mutzel, R. & Baldauf, S. 2006. Molecular phylogeny and evolution of morphology in the social amoebas. Science, 314: 661‐663.
Spiegel, F. W., Shadwick, L. L., Ndiritu, G. G., Brown, M. W., Aguilar, M., & Shadwick, J. D. L. 2017. Protosteloid Amoeboazoa (Protosteliids, Protosporangiida, Cavostellida, Schizoplasmodiida, Fractoviteliida, and sporcarpic members of Vanellida, Centramoebida, and Pellitida). In: Archibald, J. M.,Simpson, A. G. B., and Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-32669-6_12-1
Sheikh, S., MatsThulin Cavender, J.C., Escalante, R., Kawakami, S.I., Lado, C., Landolt, J.C., Nanjundiah, V., Queller, D.C., Strassmann, J.E., Spiegel, F.W., Stephenson, S.L., Vadell, S.M., & Baldauf, S.L. 2018. A new classification of the dictyostelids. Protist 169: 1‐28.
Wilkinson, D. M. & Mitchell, E. A. D. 2010. Testate amoebae and nutrient cycling with particular reference to soils. Geomicrobiol J., 27(6):520‐533. https://doi.org/10.1080/01490451 003702925.
Walker, G., Zadrobílková, E., Čepička., I. (2017) Archamoebae In: Archibald, J. M.,Simpson, A. G. B., & Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-28149-0_11
AMOEBOZOA
Berney, C., Geisen, S., Van Wichelen, J., Nitsche, F., Vanormelingen, P., Bonkowski, M. & Bass, D. 2015. Expansion of the “reticulosphere”: diversity of novel branching and network‐forming amoebae helps to define Variosea (Amoebozoa). Protist, 166, 271–295.
Cavalier‐Smith, T., Chao, E., E., & Lewis, R. 2016. 187‐Gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep‐branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution. Mol. Phylogenet. Evol., 99: 275–296.
Kang, S., Tice, A.K., Spiegel, F. W., Silberman, J. D., Pánek, T., Čepička, I., Kostka, M., Kosakyan, A., Alcântara, D. M., Roger, A. J., Shadwick, L. L., Smirnov, A., Kudryavstev, A., Lahr, D. J. & Brown, M. W. 2017. Between a pod and a hard test: the deep evolution of amoebae. Mol Biol Evol msx162. https://doi.org/10.1093/molbev/msx162.
Pánek, T, Zadrobílková, Walker, G., Brown, M. W., Gentekaki, E., Hroudrová, M., Kang, S. Roger, A. J., Tice, A. K., Vlček, Č., & Čepička, I. 2016. First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group. Molec. Phylogenet. Evol., 98:41‐51.
Schaap, P., Winckler, T., Nelson, M., Alvarez‐Curto, E., Elgie, B., Hagiwara, H., Cavender, J., Milano‐Curto, A., Rozen, D. E., Dingermann, T., Mutzel, R. & Baldauf, S. 2006. Molecular phylogeny and evolution of morphology in the social amoebas. Science, 314: 661‐663.
Spiegel, F. W., Shadwick, L. L., Ndiritu, G. G., Brown, M. W., Aguilar, M. & Shadwick, J. D. L. 2017. Protosteloid Amoeboazoa (Protosteliids, Protosporangiida, Cavostellida, Schizoplasmodiida, Fractoviteliida, and sporcarpic members of Vanellida, Centramoebida, and Pellitida). In: Archibald, J. M.,Simpson, A. G. B., and Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-32669-6_12-1
Sheikh, S., MatsThulin Cavender, J.C., Escalante, R., Kawakami, S.I., Lado, C., Landolt, J.C., Nanjundiah, V., Queller, D.C., Strassmann, J.E., Spiegel, F.W., Stephenson, S.L., Vadell, S.M. & Baldauf, S.L. 2018. A new classification of the dictyostelids. Protist 169: 1‐28.
Smirnov, A., V., Brown, S., 2004. Guide to the methods of study and identification of soil gymnamoebae. Protistology 3, 148–190.
Wilkinson, D. M. & Mitchell, E. A. D. 2010. Testate amoebae and nutrient cycling with particular reference to soils. Geomicrobiol J., 27(6):520‐533. https://doi.org/10.1080/01490451003702925.
Walker, G., Zadrobílková, E. & Čepička., I. 2017 Archamoebae In: Archibald, J. M.,Simpson, A. G. B. & Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-28149-0_11
OPISTHOKONTA
Holozoa
Grau‐Bové, X., Torruella, G., Donachie, S., Suga, H., Leonard, G., Richards, T.A. & Ruiz‐Trillo, I. 2017. Dynamics of genomic innovation in the unicellular ancestry of animals. eLife 6, e26036.
Hehenberger, E., Tikhonenkov, D.V., Kolisko, M., del Campo, J., Esaulov, A.S., Mylnikov, A.P., & Keeling, P.J., 2017. Novel predators reshape holozoan phylogeny and reveal the presence of a two‐ component signaling system in the ancestor of animals. Curr. Biol., 27: 2043‐2050.
Toruella, G., Mendoza, A., Grau Bauvé, X., Anto, M., Chaplin, M., del Campo, J., Eme, L., Perez Cordon, G., Whipps, C.M., Nichols, K.M., Paley, R., Roger, A.J., Sitja‐Bobadillas, A., Donachie, S., & Ruiz‐ Trillo, I. 2015. Phylogenomics Reveals Convergent Evolution of Lifestyles in Close Relatives of Animals and Fungi. Curr. Biol., 25: 2404‐2410. https://doi.org/10.1016/j.cub.2015.07.053
Choanoflagellata
Brunet, T. & King, N. 2017 The origin of animal multicellularity and cell differentiation. Developmental Cell, 43: 124‐140. https://doi.org/10.1016/j.devcel.2017.09.016
Budd, G.E. & Jensen, S. 2017. The origin of the animals and a “Savannah” hypothesis for early bilaterian evolution Biol. Rev. Camb Philos. Soc., 92: 446‐473.
Carr, M., Richter, D.J., Fozouni, P., Smith, T.J., Jeuck, A., Leadbeater, B.S.C. & Nitsche, F., 2017. A six‐gene phylogeny provides new insights into choanoflagellate evolution. Mol. Phylogenetics Evol., 107: 166–178.
Nitsche, F., Carr, M., Arndt, H. & Leadbeater, B. S. C. 2011. Higher level taxonomy and molecular phylogenetics of the Choanoflagellatea. J.Eukaryot. Microbiol., 58: 452‐462. https://doi.org/10.1111/j.1550‐ 7408.2011.00572.x
Richter, D.J. & Nitsche, F., 2017. Choanoflagellatea. In: Archibald, J.M., Simpson, A.G.B. & Slamovits, C.H. (Eds.) Handbook of the Protists. Springer International Publishing, pp. 1479‐1496.
Porifera
Cárdenas, P., Pérez, T. & Boury‐Esnault, N. 2012. Sponge Systematics facing new challenges. Adv.Marine Biol., 61: 79‐209.
Dohrmann, M., Kelley, C., Kelly, M., Pisera, A., Hooper, J. N. A. & Reiswig, H. M. 2017. An integrative systematic framework helps to reconstruct skeletal evolution of glass sponges (Porifera, Hexactinellida). Frontiers Zool., 14: 18.
Morrow, C. & Cárdenas, P. 2015. Proposal for a revised classification of the Demospongiae (Porifera). Frontiers Zool., 12: 1‐27.
Redmond, N. E., Morrow, C. C., Thacker, R. W., Diaz, M. C., Boury‐Esnault, N., Cárdenas, P., et al. 2013. Phylogeny and Systematics of Demospongiae in Light of New Small Subunit Ribosomal DNA (18S) Sequences. Integr. Comparat. Biol., 53: 388‐415.
Ruiz, C., Muricy, G., Lage, A., Domingos, C., Chenesseau, S. & Pérez, T. 2017. Descriptions of new sponge species and genus, including aspiculate Plakinidae, overturn the Homoscleromorpha classification. Zool. J. Linn. Soc., 179: 707‐724.
Voigt, O., Wülfing, E. & Wörheide, G. 2012. Molecular Phylogenetic Evaluation of Classification and Scenarios of Character Evolution in Calcareous Sponges (Porifera, Class Calcarea). PLoS ONE, 7, e33417.
Metazoa
Cannon, J.T., Vellutini, B.C., Smith, J., Ronquist, F., Jondelius, U. & Hejnol, A. 2016. Xenacoelomorpha is the sister group to Nephrozoa. Nature, 530: 89‐93.
Laumer, C.E., Bekkouche, N., Kerbl, A., Goetz, F., Neves, R.C., Sørenson, M.V., Kristensen, R.M., Hejnol, A., Dunn, C.W., Giribet, G. & Worsaae, K. 2015. Spiralian phylogeny informs the evolution of microscopic lineages. Curr. Biol., 25: 2000‐2006.
Simion, P., Philippe, H., Baurain, D., Jager, M., Richter, D.J., Di Franco, A., Roure, B., Satoh, N., Queinnec, E., Ereskovsky, A., Lapebie, P., Corre, E., Delsuc, F., King, N., Wörheide, G. & Manuel, M. 2017. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol., 27: 958‐967.
Whelan, N.V., Kocot, K.M., Moroz, T.P., Mukherjee, K., Williams, P., Paulay, G., Moroz, L.L. & Halanych, K.M. 2017. Ctenophore relationships and their placement as the sister group to all other animals. Nat. Ecol. Evol., 1: 1737‐1746.
Holomycota
Bass, D., Czech, L., Williams, B.A.P., Berney, C., Dunthorn, M., Mahé, F., Torruella, G., Stentiford, G.D., Williams, T.A. 2018. Clarifying the Relationships between Microsporidia and Cryptomycota. J. Euk. Microbiol., 65: 773–782. https://doi.org/10.1111/jeu.12519
Jones, M. D. M., Richards, T. A., Hawksworth, D. L. & Bass, D. 2011. Validation and justification of the phylum name Cryptomycota. IMA Fungus, 2:173–175.
Karpov, S. A., Mamkaeva, M. A., Aleoshin, V. V., Nassonova, E., Lilje, O. & Gleason, F. H. 2014.Morphology, phylogeny, and ecology of the aphelids (Aphelidea, Opisthokonta) and proposal for the new superphylum Opisthosporidia.Front. Microbiol., 5:112.
Lara, E., Moriera, D. & Lopez‐Garcia, P. 2010. The environmental clade LKM11 and Rozella form the deepest branching clade of fungi. Protist, 161:116–121.
Liu, Y., Steenkamp, E.T., Brinkmann, H., Forget, L., Philippe, H. & Lang, F.B. 2009. Phylogenomic analyses predict sistergroup relationship of nucleariids and Fungi and paraphyly of zygomycetes with significant support. BMC Evol. Biol., 20099:272, https://doi.org/10.1186/1471-2148-9-272.
Fungi
Bauer, R., Garnica, S., Oberwinkler, F., Riess, K., Weiss, M. & Begerow, D. 2015. Entorrhizomycota: A New Fungal Phylum Reveals New Perspectives on the Evolution of Fungi. PLoS One, 10:e0128183.
Chang, Y., Wang, S. S., Sekimoto, S., Aerts, A. L., Choi, C., Clum, A., LaButti, K. M., Lindquist, E. A., Ngan, C. Y., Ohm, R. A., Salamov, A. A., Grigoriev, I. V., Spatafora, J. W. & Berbee, M. L. 2015. Phylogenomic Analyses Indicate that Early Fungi Evolved Digesting Cell Walls of Algal Ancestors of Land Plants. Genome Biol. . Evol., 7:1590‐1601.
Edlind, T. D., Li, J., Visvesvara, G. S., Vodkin, M. H., McLaughlin, G. L. & Katiyar, S. K. 1996. Phylogenetic analysis of beta‐tubulin sequences from amitochondrial protozoa. Mol. Phylogenet. Evol., 5:359‐67.
Hibbett, D. S., Binder, M., Bischoff, J. F., Blackwell, M., Cannon, P. F., Eriksson, O. E., Huhndorf, S., James, T., Kirk, P. M., Lücking, R., Lumbsch, T., Lutzoni, F., Matheny, P. B., Mclaughlin, D. J., Powell, M. J., Redhead, S., Schoch, C. L., Spatafora, J. W., Stalpers, J. A., Vilgalys, R., Aime, M. C., Aptroot, A., Bauer, R., Begerow, D., Benny, G. L., Castlebury, L. A., Crous, P. W., Dai, Y.‐C., Gams, W., Geiser, D. M., Griffith, G. W., Gueidan, C., Hawksworth, D. L., Hestmark, G., Hosaka, K., Humber, R. A., Hyde, K., Ironside, J. E., Kõljalg, U., Kurtzman, C. P., Larsson, K.‐H., Lichtwardt, R., Longcore, J., Miadlikowska, J., Miller, A., Moncalvo, J.‐M., Mozley‐Standridge, S., Oberwinkler, F., Parmasto, E., Reeb, V., Rogers, J. D., Roux, C., Ryvarden, L., Sampaio, J. P., Schüßler, A., Sugiyama, J., Thorn, R. G., Tibell, L., Untereiner, W. A., Walker, C., Wang, Z., Weir, A., Weiß, M., White, M. M., Winka, K., Yao, Y.‐J. & Zhang, N. 2007. A higher‐level phylogenetic classification of the Fungi. Mycological Res., 111:509‐547.
Hibbett, D. S., Blackwell, M., James, T. Y., Spatafora, J. W., Taylor, J. W. & Vilgalys, R. 2018. Phylogenetic taxon definitions for Fungi, Dikarya, Ascomycota and Basidiomycota. IMA Fungus, 9: 291–298.
James, T. Y., Kauff, F., Schoch, C. L., Matheny, P. B., Hofstetter, V., Cox, C. J., Celio, G., Gueidan, C., Fraker, E., Miadlikowska, J., Lumbsch, H. T., Rauhut, A., Reeb, V., Arnold, A. E., Amtoft, A., Stajich, J. E., Hosaka, K., Sung, G.‐H., Johnson, D., O'Rourke, B., Crockett, M., Binder, M., Curtis, J. M., Slot, J. C., Wang, Z., Wilson, A. W., Schuszler, A., Longcore, J. E., O'Donnell, K., Mozley‐Standridge, S., Porter, D., Letcher, P. M., Powell, M. J., Taylor, J. W., White, M. M., Griffith, G. W., Davies, D. R., Humber, R. A., Morton, J. B., Sugiyama, J., Rossman, A. Y., Rogers, J. D., Pfister, D. H., Hewitt, D., Hansen, K., Hambleton, S., Shoemaker, R. A., Kohlmeyer, J., Volkmann‐Kohlmeyer, B., Spotts, R. A., Serdani, M., Crous, P. W., Hughes, K. W., Matsuura, K., Langer, E., Langer, G., Untereiner, W. A., Lucking, R., Budel, B., Geiser, D. M., Aptroot, A., Diederich, P., Schmitt, I., Schultz, M., Yahr, R., Hibbett, D. S., Lutzoni, F., McLaughlin, D. J., Spatafora, J. W. & Vilgalys, R. 2006. Reconstructing the early evolution of Fungi using a six‐gene phylogeny. Nature, 443:818‐822.
Jones, M. D. M., Richards, T. A., Hawksworth, D. L. & Bass, D. 2011. Validation and justification of the phylum name Cryptomycota phyl. nov. IMA Fungus, 2:173‐175.
Jones, M. D. M., Forn, I., Gadelha, C., Egan, M. J., Bass, D., Massana, R. & Richards, T. A. 2011. Discovery of novel intermediate forms redefines the fungal tree of life. Nature, 474:200‐203.
Karpov, S. A., Mamkaeva, M. A., Aleoshin, V. V., Nassonova, E., Lilje, O. & Gleason, F. H. 2014. Morphology, phylogeny, and ecology of the aphelids (Aphelidea, Opisthokonta) and proposal for the new superphylum Opisthosporidia. Front. Microbiol., 5:112.
Keeling, P. J. 2003. Congruent evidence from alpha‐tubulin and beta‐tubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genet Biol, 38:298‐309.
Kirk, P. M., Cannon, P. F., Minter, D. W. & Stalpers, J. A. 2008. Ainsworth and Bisby's dictionary of the Fungi, 10th ed. CAB International., Wallingford , UK p. 771.
Lara, E., Moreira, D. & Lopez‐Garcia, P. 2010. The Environmental Clade LKM11 and Rozella Form the Deepest Branching Clade of Fungi. Protist, 161:116‐121.
Lee, S. C., Corradi, N., Doan, S., Dietrich, F. S., Keeling, P. J. & Heitman, J. 2010. Evolution of the sex‐related locus and genomic features shared in Microsporidia and Fungi. Plos ONE, 5:e10539.
Liu, Y., Steenkamp, E. T., Brinkmann, H., Forget, L., Philippe, H. & Lang, B. F. 2009. Phylogenomic analyses predict sistergroup relationship of nucleariids and fungi and paraphyly of zygomycetes with significant support. BMC Evol. Biol., 9:272.
Ren, R., Sun, Y., Zhao, Y., Geiser, D., Ma, H. & Zhou, X. 2016. Phylogenetic Resolution of Deep Eukaryotic and Fungal Relationships Using Highly Conserved Low‐Copy Nuclear Genes. Genome Biol. Evol., 8:2683‐701.
Spatafora, J. W., Chang, Y., Benny, G. L., Lazarus, K., Smith, M. E., Berbee, M. L., Bonito, G., Corradi, N., Grigoriev, I., Gryganskyi, A., James, T. Y., O'Donnell, K., Roberson, R. W., Taylor, T. N., Uehling, J., Vilgalys, R., White, M. M. & Stajich, J. E. 2016. A phylum‐level phylogenetic classification of zygomycete fungi based on genome‐scale data. Mycologia, 108:1028‐1046.
Tedersoo, L., Sanchez‐Ramirez, S., Koljalg, U., Bahram, M., Doring, M., D., S., May, T., Ryberg, M. & Abarenkov, K. 2018. High‐level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Diversity, 90:135‐159.
Chytridiomycota
Barr, D. J. S. 1980. An outline for the reclassification of the Chytridiales, and for a new order, the Spizellomycetales. Can. J. Bot., 58:2380–2394.
Karpov, S. A., Kobseva, A. A., Mamkaeva, M. A., Mamkaeva, K. A., Mikhailov, K. V.., Mirzaeva, G. S. & Aleoshin, V. V. 2014. Gromochytrium mamkaevae gen. & sp. nov. and two new orders: Gromochytriales and Mesochytriales (Chytridiomycetes). Personia, 32:115–126.
Letcher, P. M., Powell, M. J., Churchill, P. F. & Chambers, J. G. 2018. Ultrastructural and molecular phylogenetic delineation of a new order, the Rhizophydiales (Chytridiomycota). Mycol. Res., 110:898–915.
Letcher P. M., Powell, M. J., Barr, D. J. S., Churchill, P. F., Wakefield, W. S. & Picard, K. T. 2008. Rhizophlyctidales–a new order in Chytridiomycota. Mycol. Res., 112:1031–1048.
Longcore, J. E. & Simmons, D. R. 2012. The Polychytriales ord. nov. contains chitinophilic members of the rhizophlyctoid alliance. Mycologia, 104: 276–294.
Longcore, J. E., Simmons, D. R. & Letcher, P. M. 2016. Synchytrium microbalum sp. nov. is a saprobic species in a lineage of parasites. Fungal Biol., 120:1156–1164.
Mozley‐Standridge, S. E., Letcher, P. M., Longcore, J. E., Porter, D. & Simmons, D. R. 2009. Cladochytriales–a new order in the Chytridiomycota. Mycol. Res., 113:498–507.
Simmons, D. R., James, T. Y., Meyer, A. F. & Longcore, J. E. 2009. Lobulomycetales, a new order in the Chytridiomycota. Mycol. Res., 113:450–460.
Smith, D.S., Rocheleau, H., Chapados, J. T., Abbott, C., Ribero, S., Redhead, S. A., Lévesque, C. A., & De Boer, S. H., 2014. Phylogeny of the genus Synchytrium and the development of TaqMan PCR assay for sensitive detection of Synchytrium endobioticum in soil. Phytopath., 104:422–432.
Torruella, G., Grau‐Bové, X., Moreira, D., Karpov, S. A., Burns, J. A., Sebé‐Pedrós, A., Völcker E. & López‐García, P. 2019. Global transcriptome analysis of the aphelid Paraphelidium tribonemae supports the phagotrophic origin of fungi. Communications Biology. 1: 231, https://doi.org/10.1038/s42003-018-0235-z
Vélez, C. G., Letcher, P. M., Schultz, S., Powell, M. J. & Churchill, P. F. 2011. Molecular phylogenetic and zoospore ultrastructural analyses of Chytridium olla establish the limits of a monophyletic Chytridiales. Mycologia, 103:118–130.
DIAPHORETICKES ARCHAEPLASTIDA
Chloroplastida
Fawley, M.V., Yun, Y., & Qin, M. 2000. Phylogenetic analyses of 18S rDNA sequences reveal a new coccoid lineage of the Prasinophyceae (Chlorophyta). J. Phycol., 36:387‐393.
Fucikova, K., Leliaert, F., Cooper, E.D., Skaloud, P., D'Hondt, S., De Clerk, O., Gurgel, C.F.D., Lewis, L.A., Lewis, P.O., Lopez‐Bautista, J.M., Delwiche, C.F., Verbruggen, H. 2014. New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Frontiers Ecol. Evol., 2:67, https://doi.org/10.3389/fevo.2014.00063
Leliaert. F., Tronholm, A., Lemieux, C., Bhattacharya, D., Karol, K.G., Fredericq, S. 2016. Chloroplast phylogenomic analyses reveal the deepest‐branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci. Rep., 6:25367, https://doi.org/10.1038/srep25367
Lopes Dos Santos, A., Pollina, T., Gourvil, P., Corre, E., Marie, D., Garrido J.L., Rodríguez, F., Noël, M.H. & Vaulot, D., Eikrem W. 2017. Chloropicophyceae, a new class of picophytoplanktonic prasinophytes. Sci. Rep., 25:14019, https://doi.org/10.1038/s41598-017-12412-5
Nakayama, T., Marin, B., Kranz, H.D., Surek, B., Huss, V.A., Inouye, I. & Melkonian, M. 1998. The basal position of scaly green flagellates among the green algae (Chlorophyta) is revealed by analyses of nuclear‐encoded SSU rRNA sequences. Protist, 149:378‐380.
Rhodophyceae and Glaucophyta
Muñoz‐Gómez, S.A., Mejía‐Franco, F.G., Durnin, K., Colp, M., Grisdale, C.J., Archibald, J.M. & Slamovits C.H. 2017. The New Red Algal Subphylum Proteorhodophytina Comprises the Largest and Most Divergent Plastid Genomes Known. Curr. Biol., 27: 1677–1684.
Chong, J., Jackson, C., Kim, J. I., Yoon, H. S. & Reyes‐Prieto, A. 2014. Molecular markers from different genomic compartments reveal cryptic diversity within glaucophyte species. Mol. Phylogenet. Evol., 76:181–188.
Jackson, C., Clayden, S. & Reyes‐Prieto, A. 2015. The Glaucophyta: The blue‐green plants in a nutshell. Acta Soc. Bot. Pol., 84:149–165.
Jackson C.J., Reyes‐Prieto A. 2014. The Mitochondrial Genomes of the Glaucophytes Gloeochaete wittrockiana and Cyanoptyche gloeocystis: Multilocus Phylogenetics Suggests a Monophyletic Archaeplastida. Gen. Biol. Evol., 6: 2774–2785.
Price, D.C., Chan, C.X., Yoon, H.S., Yang, E.C., Qiu, H., Weber, A.P.M., Schwacke, R., Gross, J., Blouin, N.A., Lane, C., Reyes‐Prieto, A., Durnford, D.G., Neilson, J.A.D., Lang, B.F., Burger, G., Steiner, J.M., Loffelhardt, W., Meuser, J.E., Posewitz, M.C., Ball, S., Arias, M.C., Henrissat, B., Coutinho, P.M., Rensing, S.A., Symeonidi, A., Doddapaneni, H., Green, B.R., Rajah, V.D., Boore, J. & Bhattacharya, D. 2012. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science, 335:843–847.
Takahashi, T., Sato, M., Toyooka, K., Matsuzaki, R., Kawafune, K., Kawamura, M., Okuda, K. & Nozaki, H. 2014. Five Cyanophora (Cyanophorales, Glaucophyta) species delineated based on morphological and molecular data. J. Phycol., 50:1058–1069.
Takahashi, T., Nishida, T., Tuji, A., Saito, C., Matsuzaki, R., Sato, M., Toyooka, K., Yasuda, H. & Nozaki, H. 2016. Delineation of six species of the primitive algal genus Glaucocystis based on in situ ultrastructural characteristics. Sci. Rep., 6:29209.
Sar
Burki, F., Shalchian‐Tabrizi, K., Minge, M., Skjaeveland, A., Nikolaev, S. I., Jakobsen, K. S., & Pawlowski, J. 2007. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE, 2(8), e790. https://doi.org/10.1371/journal.pone.0000790
Hackett, J. D., Yoon, H. S., Li, S., Reyes‐Prieto, A., Rümmele, S. E., & Bhattacharya, D. 2007. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Molec. Biol. Evol., 24(8), 1702–1713. https://doi.org/10.1093/molbev/msm089
Rodriguez‐Ezpeleta, N., Brinkmann, H., Burger, G., Roger, A. J., Gray, M. W., Philippe, H., & Lang, B. F. 2007. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Current Biol., 17(16): 1420–1425. https://doi.org/10.1016/j.cub.2007.07.036
Stramenopiles
Aleoshin, V. V., A. P. Mylnikov, G. S. Mirzaeva, K. V. Mikhailov, and S. A. Karpov. 2016. Heterokont Predator Develorapax marinus gen. et sp. nov. ‐ A Model of the Ochrophyte Ancestor. Front. Microbiol., 7:1194.
Cavalier‐Smith, T. 2018. Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma, 255:297‐357.
Cavalier‐Smith, T., and J. M. Scoble. 2013. Phylogeny of Heterokonta: Incisomonas marina, a uniciliate gliding opalozoan related to Solenicola (Nanomonadea), and evidence that Actinophryida evolved from raphidophytes. Eur. J. Protistol., 49:328‐353.
Chang, F. H., J. Sutherland, and J. Bradford‐Grieve. 2017. Taxonomic revision of Dictyochales (Dictyochophyceae) based on morphological, ultrastructural, biochemical and molecular data. Phycol. Res. 65:235‐347.
Derelle, R., P. Lopez‐Garcia, H. Timpano, and D. Moreira. 2016. A Phylogenomic Framework to Study the Diversity and Evolution of Stramenopiles (=Heterokonts). Mol. Biol. Evol., 33:2890‐2898.
Lin, Y. C., T. Campbell, C. C. Chung, G. C. Gong, K. P. Chiang, and A. Z. Worden. 2012. Distribution patterns and phylogeny of marine stramenopiles in the north pacific ocean. Appl. Environ. Microbiol. 78:3387‐3399.
Massana, R., J. del Campo, M. E. Sieracki, S. Audic, and R. Logares. 2014. Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. ISME J ., 8:854‐866.
McCarthy, C. G. P., and D. A. Fitzpatrick. 2017. Phylogenomic Reconstruction of the Oomycete Phylogeny Derived from 37 Genomes. mSphere, 2.
Shiratori, T., T. Nakayama, and K. Ishida. 2015. A New Deep‐branching Stramenopile, Platysulcus tardus gen. nov., sp. nov. Protist, 166:337‐348.
Yang, E. C., G. H. Boo, H. J. Kim, S. M. Cho, S. M. Boo, R. A. Andersen, and H. S. Yoon. 2012. Supermatrix data highlight the phylogenetic relationships of photosynthetic stramenopiles. Protist, 163:217‐231.
Yubuki, N., T. Panek, A. Yabuki, I. Cepicka, K. Takishita, Y. Inagaki, and B. S. Leander. 2015. Morphological Identities of Two Different Marine Stramenopile Environmental Sequence Clades: Bicosoeca kenaiensis (Hilliard, 1971) and Cantina marsupialis (Larsen and Patterson, 1990) gen. nov., comb. nov. J. Eukaryot. Microbiol., 62:532‐542.
Diatomea
Ashworth, M. P., Nakov, T. & Theriot, E. C. 2013. Revisiting Ross and Sims (1971): toward a molecular phylogeny of the Biddulphiaceae and Eupodiscaceae (Bacillariophyceae). J. Phycol. 49:1207–1222.
Ichinomiya, M., Yoshikawa, S., Kamiya, M., Ohki, K., Takaichi, S. & Kuwata, A. 2011. Isolation and characterization of Parmales (Heterokonta/Heterokontophyta/Stramenopiles) from the Oyahio region, western North Pacific. J. Phycol. 47:144–151.
Ichinomiya, M., Lopes dos Santos, A., Gourvil, P., Yoshikawa, S., Kamiya, M., Ohki, K., Audic, S., Vargas, C. de, Noël, M.‐H., Vaulot, D. & Kuwata, A. 2016. Diversity and oceanic distribution of the Parmales (Bolidophyceae), a picoplanktonic group closely related to diatoms. ISME J. 10:2419–2434.
Mann, D.G., Crawford, R.M. & Round, F.E. 2017. Bacillariophyta. In: Handbook of the Protists (Archibald, J.M., Simpson, A.G.B. & Slamovits, C.H., eds), 62 pp. Springer, Cham. https://doi.org/10.1007/978-3-319-32669-6_29-1.
Medlin, L. K. 2016. Opinion: can coalescent models explain deep divergences in the diatoms and argue for the acceptance of paraphyletic taxa at all taxonomic hierarchies? Nova Hedwigia 102:107–128.
Medlin, L. K. & Kaczmarska, I. 2004. Evolution of the diatoms: V. Morphological and cytological support for the major clades and a taxonomic revision. Phycologia 43:245–270.
Nakov, T., Beaulieu, J. M. & Alverson, A. J. 2018. Accelerated diversification is related to life history and locomotion in hyperdiverse lineage of microbial eukaryotes (diatoms, Bacillariophta). New Phytol., https://doi.org/10.1111/nph.15137
Parks, M. B., Wickett, N. J. & Alverson, A. J. 2017. Signal, uncertainty, and conflict in phylogenomic data for a diverse lineage of microbial eukaryotes (diatoms, Bacillariophyta). Mol. Biol. Evol., 35:80–93.
Theriot, E. C., Ashworth, M. P., Nakov, T., Ruck, E. C. & Jansen, R. K. 2015. Dissecting signal and noise in diatom chloroplast protein encoding genes with phylogenetic information profiling. Mol. Phylogenet. Evol. 89:28–36.
Round, F. E., Crawford, R. M. & Mann, D. G. 1990. The diatoms. Biology and morphology of the genera. Cambridge: Cambridge University Press. 747 pp.
ALVEOLATA
Cumbo, V. R., Baird, A. H., Moore, R. B., Negri, A. P., Neilan, B. A., Salih, A., et al. 2013. Chromera velia is endosymbiotic in larvae of the reef corals Acropora digitifera and A. tenuis. Protist 164: 237–244.
Freeman, M. A., Fuss, J., Kristmundsson, A., Bjorbaekmo, M. F. M., Mangot, J. F., del Campo, J., Keeling, P. J., Shalchian‐Tabrizi, K. & Bass, D. 2017. X‐cells are globally distributed, genetically divergent fish parasites related to perkinsids and dinoflagellates. Curr. Biol., 27: 1645‐1651. https://doi.org/10.1016/j.cub.2017.04.045
Gile, G. H. & Slamovits, C. H. 2014 Transcriptomic analysis reveals evidence for a cryptic plastid in the colpodellid Voromonas pontica, a close relative of chromerids and apicomplexan parasites. PLoS ONE. 9: e96258. https://doi.org/10.1371/journal.pone.0096258
Mathur, V., del Campo, J., Kolisko, M. & Keeling, P. J. 2018. Global diversity and distribution of close relatives of apicomplexan parasites. Environ. Microbiol. 20: 2824–2833. https://doi.org/10.1111/1462-2920.14134
Oborník, M., Kručinská, J. & Esson, H. 2016. Life cycles of chromerids resemble those of colpodellids and apicomplexan parasites. Perspect. Phycol. 3: 21–27. https://doi.org/10.1127/pip/2016/0038
Oborník M., Lukeš J. 2015. The organellar genomes of Chromera and Vitrella, the phototrophic relatives of apicomplexan parasites. Annu. Rev. Microbiol. 69: 129‐144. https://doi.org/10.1146/annurev-micro-091014-104449
Okamoto, N., & Keeling, P. J. 2014. A comparative overview of the flagellar apparatus of dinoflagellate, perkinsids and colpodellids. Microorganisms, 2: 73–91. https://doi.org/10.3390/microorganisms2010073
Reñé, A., Alacid, E., Ferrera, I., & Garcés, E. 2017. Evolutionary trends of Perkinsozoa (Alveolata) characters based on observations of two new genera of parasitoids of dinoflagellates, Dinovorax gen. nov. and Snorkelia gen. nov. Front. Microbiol. 8. 1594. https://doi.org/10.3389/fmicb.2017.01594.
Tikhonenkov, D. V., Janouškovec, J., Mylnikov, A. P., Mikhailov, K. V., Simdyanov,T. G., Aleoshin, V. V. & Keeling, P. J. 2014. Description of Colponema vietnamicasp. n. and Acavomonas peruvianan. gen. n. sp., two new Alveolate phyla (Colponemidia nom. nov. and Acavomonidia nom. nov.) and their contributions to reconstructing the ancestral state of alveolates and eukaryotes. PLoS ONE, 9: e95467. https://doi.org/10.1371/journal.pone.0095467
Woo, Y.H., Ansari, H., Otto, T.D., Klinger, C.M., Kolisko, M., Michalek, J., et al. 2015. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. eLife. 4:1–41.
Yuan, C. L., Keeling, P. J., Krause, P. J., Horak, A., Bent, S., Rollend, L. & Hua, X. G. 2012. Colpodella spp.–like Parasite infection in woman, China. Emerg. Infect. Dis. 18: 125‐127. https://doi.org/10.3201/eid1801.110716.
Apicomplexa
Arisue, N. & Hashimoto, T. 2015. Phylogeny and evolution of apicoplasts and apicomplexan parasites. Parasitol. Int. 64: 254–259.
Cavalier‐Smith, T. 2018. Kingdom Chromista and its eight phyla: A new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma. 1: 297‐357. https://doi.org/10.1007/s00709-017-1147-3.
Cavalier‐Smith, T. 2014. Gregarine site‐heterogeneous 18S rDNA trees, revision of gregarine higher classification, and the evolutionary diversification of Sporozoa. Europ. J. Protistol. 50: 472‐495. https://doi.org/10.1016/j.ejop.2014.07.002.
Desportes, I. & Schrével, J. 2013. Treatise on Zoology ‐ Anatomy, Taxonomy, Biology. The Gregarines, Vol 1 & 2. Brill, Leiden.
Flegontov, P. Michálek, J., Tomčala, A., Janouškovec, J., Jirků, M., Lai, D. H., Hajdůšková, E., Otto, T. D., Keeling, P. J., Pain, A., Oborník, M. & Lukeš, J. 2015. Divergent mitochondrial respiratory chains in phototrophic relatives of apicomplexan parasites. Mol. Biol. Evol. 32: 1115‐1131. https://doi.org/10.1093/molbev/msv021
Ghazy, A. A., Abdel‐Shafy, S. & Shaapan, R. M. 2015. Cryptosporidiosis in animals and man: 1. taxonomic classification, life cycle, epidemiology and zoonotic importance. Asian J. Epidemiol. 8: 48‐63. https//doi.org/10.3923/aje.2015.48.63
Heintzelman, M. B. 2015. Gliding motility in apicomplexan parasites. Semin. Cell. Dev. Biol. 46:135–142. https://doi.org/10.1016/j.semcdb.2015.09.020
Iritani, D., Wakeman, K. & Leander, B.S. 2018. Molecular phylogenetic positions of two new marine gregarines (Apicomplexa) from the intestines of Lumbrineris inflata (Polychaeta) show patterns of co‐ evolution: Paralecudina anankea nov. sp. and Lecudina caspera nov. sp. J. Eukaryot. Microbiol. 65:211‐219. https://doi.org/10.1111/jeu.12462
Janouškovec, J., Tikhonenkov, D. V., Burki, F., Howe, A. T., Kolisko, M., Mylnikov, A. P., et al. 2015. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc. Natl. Acad. Sci. USA 112: 10200–10207. https://doi.org/10.1111/1462-2920.14134
Janouškovec, J. Horák, A., Barrot, K. L., Rohwer, F. L. & Keeling, P. J. 2012. Global analysis of plastid diversity reveals new apicomplexan‐related lineages associated with coral reefs. Curr. Biol. 22: R518–9. https://doi.org/10.1016/j.cub.2012.04.047
Karadjian, G., Chavatte, J.‐M., & Landau, I. 2015. Systematic revision of the adeleid haemogregarines, with creation of Bartazoon n. g., reassignment of Hepatozoon argantis Garnham, 1954 to Hemolivia, and molecular data on Hemolivia stellata. Parasite. 22: 31. https://doi.org/10.1051/parasite/2015031
Megía‐Palma, R., Martínez, J., Nasri, I., Cuervo, J. J., Martín, J., Acevedo, I., Belliure, J., Ortega, J., García‐Roa, R., Selmi, S. & Merino, S. 2016. Phylogenetic relationships of Isospora, Lankesterella, and Caryospora species (Apicomplexa: Eimeriidae) infecting lizards. Org. Divers. Evol. 16: 275‐288. https://doi.org/10.1007/s13127-015-0253-3
Muñoz‐Gómez S. A. & Slamovits C. H. 2018. Chapter Three ‐ Plastid Genomes in the Myzozoa. In: Chaw, S.‐M. & Jansen, R. K. Adv. Bot. Res. 85: 55‐94. https://doi.org/10.1016/bs.abr.2017.11.015.
Ogedengbe, M. E., El‐Sherry, S., Ogedengbe, J. D., Chapman, H. D. & Barta, J. R. 2018. Phylogenies based on combined mitochondrial and nuclear sequences conflict with morphologically defined genera in the eimeriid coccidia (Apicomplexa). Int. J. Parasitol. 48: 59‐69. https://doi.org/10.1016/j.ijpara.2017.07.008.
Ogedengbe, J. D., Ogedengbe, M. E., Hafeez, M. A. & Barta, J. R. 2015. Molecular phylogenetics of eimeriid coccidia (Eimeriidae, Eimeriorina, Apicomplexa, Alveolata): A preliminary multi‐gene and multi‐ genome approach. Parasitol. Res. 114: 4149‐4160. https://doi.org/10.1007/s00436-015-4646-1
Rueckert, S. & Horák, A. 2017. Archigregarines of the English Channel revisited: New molecular data on Selenidium species including early described and new species and the uncertainties of phylogenetic relationships. PLoS ONE 12: e0187430. https://doi.org/10.1371/journal.pone.0187430
Rueckert, S., Wakeman, K.C. & Leander, B.S. 2013. Discovery of a diverse clade of gregarine apicomplexans (Apicomplexa: Eugregarinorida) from Pacific eunicid and onuphid polychaetes, including descriptions of Paralecudina n. gen., Trichotokara japonica n. sp., and T. eunicae n. sp. J. Eukaryot. Microbiol. 60:121‐136. https://doi.org/10.1111/jeu.12015
Ryan, U., Fayer, R. & Xiao, L. (2014) Cryptosporidium species in humans and animals: Current understanding and research needs. Parasitol., 141: 1667‐1685. https://doi.org/10.1017/s0031182014001085
Schrével, J., Valigurová, A., Prensier, G., Chambouvet, A., Florent, I. & Guillou, L. 2016. Ultrastructure of Selenidium pendula, the type species of archigregarines, and phylogenetic relations to other marine Apicomplexa. Protist, 167: 339‐368. https://doi.org/10.1016/j.protis.2016.06.001.
Seeber, F. & Steinfelder, S. 2016. Recent advances in understanding apicomplexan parasites. F1000Research. 5:1369. https://doi.org/10.12688/f1000research.7924.1
Simdyanov, T. G., Paskerova, G. G., Valigurová, A., Diakin, A., Kováčiková, M., Schrével, J., Guillou, L., Dobrovolskij, A. A. & Aleoshin, V. V. (2018) First ultrastructural and molecular phylogenetic evidence from the blastogregarines, an early branching lineage of plesiomorphic Apicomplexa. Protist, 169: 697–726. https://doi.org/10.1016/j.protis.2018.04.006.
Votýpka, J., Modrý, D., Oborník, M., Šlapeta, J. & Lukeš, J. 2017. Apicomplexa. In: Archibald, J. M.,Simpson, A. G. B., and Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-32669-6_12-1.
Wakeman, K.C., Heintzelman, M.B. & Leander, B.S. 2014. Comparative ultrastructure and molecular phylogeny of Selenidium melongena n. sp. and S. terebellae Ray 1930 demonstrate niche partitioning in marine gregarine parasites (apicomplexa). Protist. 165: 493‐511. https://doi.org/10.1016/j.protis.2014.05.007
Wakeman, K., Reimer, J.D., Jenke‐Kodama, H. & Leander, B.S. 2014. Molecular phylogeny and ultrastructure of Caliculium glossobalani n. gen. et sp. (Apicomplexa) from a Pacific Glossobalanus minutus (Hemichordata) confounds the relationships between marine and terrestrial gregarines. J. Eukaryot. Microbiol. 61:343‐353.
Ciliophora
Antipa, G.A., Dolan, J.R., Lynn, D.H., Obolkina, L.A. & Strüder‐Kypke, M.C. 2016. Molecular phylogeny and evolutionary relationships between the ciliate genera Peniculistoma and Mytilophilus (Peniculistomatidae, Pleuronematida). J. Eukaryot. Microbiol., 63:642‐650.
Bourland, W., Rotterová, J. & Čepička, I. 2017. Morphologic and molecular characterization of seven species of the remarkably diverse and widely distributed metopid genus Urostomides Jankowski, 1964 (Armophorea, Ciliophora). Eur. J. Protistol., 61:194‐232.
Bourland, W.A., Hampikian, G. & Vďačný, P. 2012. Morphology and phylogeny of a new woodruffiid ciliate, Etoschophrya inornata sp. n. (Ciliophora, Colpodea, Platyophryida), with an account on evolution of platyophryids. Zool. Scr., 41:400‐416.
Chen, X., Ma, H.‐G., Al‐Rasheidm, K. A. S. & Miao, M. 2015. Molecular data suggests the ciliate Mesodinium (Protista: Ciliophora) might represent an undescribed taxon at class level. Zool. Syst., 40:31‐40.
Dunthorn, M., Otto, J., Berger, S. A., Stamatakis, A., Mahé, F., Romac, S., de Vargas, C., Audic, S., BioMarKs Consortium, Stock, A., Kauff, F. & Stoeck, T. 2014. Placing environmental next‐generation sequencing amplicons from microbial eukaryotes into a phylogenetic context. Mol. Biol. Evol., 31:993‐1009.
Fan, X., Pan, H., Li, L., Jiang, J., Al‐Rasheid, K.A.S. & Gu, F. 2014. Phylogeny of the poorly known ciliates, Microthoracida, a systematically confused taxon (Ciliophora), with morphological reports of three species. J. Eukaryot. Microbiol., 61:227‐237.
Feng, J.‐M., Jiang, C.‐Q., Warren, A., Tian, M., Cheng, J., Liu, G.‐L., Xiong, J. & Miao, W. 2015. Phylogenomic analyses reveal subclass Scuticociliatia as the sister group of subclass Hymenostomatia within class Oligohymenophorea. Mol. Phylogenet. Evol., 90:104‐111.
Fernandes, N.M., Vizzoni, V.F., Borges, B.d.N., Soares, C.A.G., da Silva‐Neto, I.D. & Paiva, T.d.S. 2018. Molecular phylogeny and comparative morphology indicate that odontostomatids (Alveolata, Ciliophora) form a distinct class‐level taxon related to Armophorea. Mol. Phylogenet. Evol., 126:382‐389.
Foissner, W., Stoeck, T., Agatha, S. & Dunthorn, M. 2011. Intraclass evolution and classification of the Colpodea (Ciliophora). J. Eukaryot. Microbiol., 58:397‐415.
Foissner, W., Bourland, W.A., Wolf, K.W., Stoeck, T. & Dunthorn, M. 2014. New SSU‐rDNA sequences for eleven colpodeans (Ciliophora, Colpodea) and description of Apocyrtolophosis nov. gen. Eur. J. Protistol., 50:40‐46.
Gao, F., Katz, L.A. & Song, W. 2012. Insights into the phylogenetic and taxonomy of philasterid ciliates (Protozoa, Ciliophora, Scuticociliatia) based on analyses of multiple molecular markers. Mol. Phylogenet. Evol., 64:308‐317.
Gao, F., Katz, L.A. & Song, W. 2013. Multigene‐based analyses on evolutionary phylogeny of two controversial ciliate orders: Pleuronematida and Loxocephalida (Protista, Ciliophora, Oligohymenophorea). Mol. Phylogenet. Evol., 68:55‐63.
Gao, F., Warren, A., Zhang, Q., Gong, J., Miao, M., Sun, P., Xu, D., Huang, J., Yi, Z. & Song, W. 2016. The all‐data‐based evolutionary hypothesis of ciliated protists with a revised classification of the Phylum Ciliophora (Eukaryota, Alveolata). Sci. Rep., 6:24874.
Gentekaki, E., Kolisko, M., Gong, Y. & Lynn, D. 2017. Phylogenomics solves a long‐standing evolutionary puzzle in the ciliate world: the subclass Peritrichia is monophyletic. Mol. Phylogenet. Evol., 106:1‐5.
Gentekaki, E., Kolisko, M., Boscaro, V., Bright, K. J., Dini, F., Di Giuseppe, G., Gong, Y., Miceli, C., Modeo, L., Molestina, R.E., Petroni, G., Pucciarelli, S., Roger, A.J., Strom, S.L. & Lynn, D.H. 2014. Large‐ scale phylogenomic analysis reveals the phylogenetic position of the problematic taxon Protocruzia and unravels the deep phylogenetic affinities of the ciliate lineages. Mol. Phylogenet. Evol., 78:36‐42.
Gong, J., Stoeck, T., Yi, Z., Miao, M., Zhang, Q., Roberts, D. M., Warren, A. & Song, W. 2009. Small subunit rRNA phylogenies show that the class Nassophorea is not monophyletic (Phylum Ciliophora). J. Eukaryot. Microbiol., 56:339‐347.
Johnson, M. D., Tengs, T., Oldach, D. W., Delwiche, C. F. & Stoecker, D. K. 2004. Highly divergent SSU rRNA genes found in the marine ciliates Myrionecta rubra and Mesodinium pulex. Protist, 155:347‐359.
Kittelmann, S., Devente, S.R., Kirk, M.R., Seedorf, H., Dehority, B.A. & Janssen, P.H. 2015. Phylogeny of intestinal ciliates, including Charonina ventriculi, and comparison of microscopy and 18S rRNA gene pyrosequencing for rumen ciliate community structure analysis. Appl. Environ. Microbiol., 81:2433‐2444.
Liu, A., Yi, Z., Lin, X., Hu, X., Al‐Farraj, S. A. & Al‐Rasheid, K. A. S. 2015. Molecular phylogenetic lineage of Plagiopogon and Askenasia (Protozoa, Ciliophora) revealed by their gene sequences. J. Ocean Univ. China, 14:724‐730.
Lynn, D. H. 2008. The ciliated protozoa: characterization, classification, and guide to the literature, 3rd edition. Springer, Dordrecht.
Lynn, D.H., Kolisko, M. & Bourland, W. 2018. Phylogenomic analysis of Nassula variabilis n. sp., Furgasonia blochmanni, and Pseudomicrothorax dubius confirms a nassophorean clade. Protist, 169:180‐189.
Orsi, W., Edgcomb, V., Faria, J., Foissner, W., Fowle, W. H., Hohman, T., Suarez, P., Taylor, C., Taylor, G. T., Vd'ačný, P. & Epstein, S. 2012. Class Cariacotrichea, a novel ciliate taxon from the anoxic Cariaco Basin, Venezuela. Int. J. Syst. Evol. Microbiol., 62:1425‐1433.
Pan, H., Stoeck, T. 2017. Redescription of the halophile ciliate, Blepharisma halophilum Ruinen, 1938 (Ciliophora, Heterotrichea, Heterotrichida) shows that the genus Blepharisma is non‐monophyletic. Eur. J. Protistol., 61:20‐28.
Santoferrara, L. F., Alder, V.V. & McManus, G.B. 2017. Phylogeny, classification and diversity of Choreotrichia and Oligotrichia (Ciliophora, Spirotrichea). Mol. Phylogenet. Evol., 112:12‐22.
Sauvadet, A.‐L., Lynn, D.H., Roussel, E. G., Le Panse, S., Bigeard, E., Schrével, J. &, Guillou, L. 2017. Redescription and phylogenetic analyses of Durchoniella spp. (Ciliophora, Astomatida) associated with the polychaete Cirriformia tentaculata (Montagu, 1808). Eur. J. Protistol., 61:265‐277.
Shin, M. K., Hwang, U. W., Kim, W., Wright, A.‐D. G., Krawczyk, C. & Lynn, D. H. 2000. Phylogenetic position of the ciliates Phacodinium (Order Phacodiniida) and Protocruzia (Subclass Protocruziidia) and systematics of the spirotrich ciliates examined by small subunit ribosomal RNA gene sequences. Europ. J. Protistol., 36:293‐302.
Vďačný, P. & Rataj, M. 2017. Evaluation of systematic position of helicoprorodontids and chaeneids (Ciliophora, Litostomatea): an attempt to break long branches in 18S rRNA gene phylogenies. J. Eukaryot. Microbiol., 64:608‐621.
Wang, P., Wang, Y., Wang, C., Zhang, T., Al‐Farraj, S.A. & Gao, F. 2017. Further consideration on the phylogeny of the Ciliophora: analyses using both mitochondrial and nuclear data with focus on the extremely confused class Phyllopharyngea. Mol. Phylogenet. Evol., 112:96‐106.
Xu, Y., Shao, C., Miao, M. & Song, W. 2013. Redescription of Parasonderia vestita (Kahl, 1928) comb. nov. (Ciliophora, Plagiopylida), with notes on its phylogeny based on SSU rRNA gene. Eur. J. Protistol., 49:106‐113.
Yan, Y., Xu, Y., Al‐Farraj, S.A., Al‐Rasheid, K.A.S. & Song, W. 2016. Morphology and phylogeny of three trachelocercids (Protozoa, Ciliophora, Karyorelictea), with description of two new species and insight into the evolution of the family Trachelocercidae. Zool. J. Linn. Soc.‐Lond., 177:306‐319.
Zhan, Z., Xu, K. & Dunthorn, M. 2013. Evaluating molecular support for and against the monophyly of the Peritrichia and phylogenetic relationships within the Mobilida (Ciliophora, Oligohymenophorea) Zool. Scripta, 42:213‐226.
Zhan, Z., Xu, K., Warren, A. & Gong, Y. 2009. Reconsideration of phylogenetic elationships of the subclass Peritrichia (Ciliophora, Oligohymenophorea) based on small subunit ribosomal RNA gene sequences, with the establishment of a new subclass Mobilia Kahl, 1933. J. Eukaryot. Microbiol., 56:552‐558.
Zhang, Q., Fan, X., Clamp, J.C., Al‐Rasheid, K.A.S. & Song, W. 2010. Description of Paratetrahymena parawassi n. sp. using morphological and molecular evidence and a phylogenetic analysis of Paratetrahymena and other taxonomically ambiguous genera in the order Loxocephalida (Ciliophora, Oligohymenophorea). J. Eukaryot. Microbiol., 57:483‐493.
Zhang, Q., Miao, M., Strüder‐Kypke, M.C., Al‐Rasheid, K.A.S., Al‐Farraj, S.A. & Song, W. 2011. Molecular evolution of Cinetochilum and Sathrophilus (Protozoa, Ciliophora, Oligohymenophorea), two genera of ciliates with morphological affinities to scuticociliates. Zool. Scr., 40:317‐325.
Zhang, Q., Yi, Z., Fan, X., Warren, A., Gong, J. & Song, W. 2014. Further insights into the phylogeny of two ciliate classes Nassophorea and Prostomatea (Protista, Ciliophora). Mol. Phylogenet. Evol., 70:162‐170.
Zhang, X., Ji, D., Zhang, Q. & Li, C. 2015. Description and phylogeny of a new prostomatid, Metacystis similis nov. spec. (Protista, Ciliophora) from the East China Sea. Zootaxa, 4033:584‐592.
Zhao, X., Miao, M., Chen, X., Ma, H. & Al‐Rasheid, K.A.S. 2014. A phylogenetic reconsideration of suctorian ciliates (Protista, Ciliophora, Phyllopharyngea) based on small subunit rRNA gene sequences. Zool. Scr., 43:206‐216.
Dinoflagellata
Bachvaroff, T. R., Gornik, S. G., Concepcion, G. T., Waller, R. F., Mendez, G. S., Lippmeier, J. C. & Delwiche, C. F. 2014. Dinoflagellate phylogeny revisited: using ribosomal proteins to resolve deep branching dinoflagellate clades. Mol. Phylogen. Evol., 70: 314‐322.
Boutrup, P. V., Moestrup, Ø., Tillmann, U. & Daugbjerg, N. 2016. Katodinium glaucum (Dinophyceae) revisited: proposal of new genus, family and order based on ultrastructure and phylogeny. Phycologia, 55(2): 147‐164.
Hoppenrath, M. 2017. Dinoflagellate taxonomy ‐ a review and proposal of a revised classification. Mar. Biodiv., 47: 381‐403.
Janouskovec, J., Gavelis, G. S., Burki, F., Dinh, D., Bachvaroff, T. R., Gornik, S. G., Bright, K. J., Imanian, B., Strom, S. L., Delwiche, C. F., Waller, R. F., Fensome, R. A., Leander, B. S., Rohwer, F. L. & Saldarriaga, J.F. 2017. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. PNAS, USA, 114: E171‐E180.
Orr, R. J. S., Murray, S. A., Stüken, A., Rhodes, L. & Jakobsen, K. S. 2012. When naked became armored: an eight‐gene phylogeny reveals monophyletic origin of theca in dinoflagellates. PLoS ONE, 7(11): e50004.
Takano, Y., Yamaguchi, H., Inouye, I., Moestrup, Ø. & Horiguchi, T. 2014. Phylogeny of five species of Nusuttodinium gen. nov. (Dinophyceae), a genus of unarmoured kleptoplastidic dinoflagellates. Protist, 165(6): 759‐778.
RHIZARIA
Bass, D., Tikhonenkov, D.V., Foster, R., Dyal, P., Janouskovec, J., Keeling, P.J., Gardner, M., Neuhauser, S., Hartikainen, H., Mylnikov, A.P., & Berney, C. 2018. Rhizarian ‘Novel Clade 10’ revealed as abundant and diverse planktonic and terrestrial flagellates, including Aquavolon n. gen. J. Eukaryot. Microbiol. 65: 828–842. https://doi.org/10.1111/jeu.12524.
Cavalier‐Smith, T., Chao, E.E., & Lewis, R. 2018. Multigene phylogeny and cell evolution of chromist infrakingdom Rhizaria: contrasting cell organisation of sister phyla Cercozoa and Retaria. Protoplasma, 255: 1517‐1574. https://doi.org/10.1007/s00709-018-1241-1.
Krabberød, A.K., Orr, R.J., Bråte, J., Kristensen, T., Bjørklund, K.R., & Shalchian‐Tabrizi, K. 2017. Single cell transcriptomics, mega‐phylogeny, and the genetic basis of morphological innovations in Rhizaria. Mol. Biol. Evol. 34:1557‐1573. https://doi.org/10.1093/molbev/msx075.
Sierra, R., Cañas‐Duarte, S.J., Burki, F., Schwelm, A., Fogelqvist, J., Dixelius, C., González‐García, L.N., Gile, G.H., Slamovits, C.H., Klopp, C., Restrepo, S., Arzul, I., & Pawlowski, J. 2016. Evolutionary origins of Rhizarian parasites. Mol. Biol. Evol. 33:980‐983. https://doi.org/10.1093/molbev/msv340.
Cercozoa
Bass, D., Silberman, J.D., Brown, M.W., Pearce, R.A., Tice, A.T., Jousset, A., Geisen, S., & Hartikainen, H. 2016. Coprophilic amoebae and flagellates, including Guttulinopsis, Rosculus and Helkesimastix, characterise a divergent and diverse rhizarian radiation and contribute to a large diversity of faecal‐associated protists. Env. Microbiol. 18:1604‐1619. https://doi.org/10.1111/1462-2920.13235.
Bass, D., Yabuki, A., Santini, S., Romac, S., & Berney, C. 2012. Reticulamoeba is a long‐branched Granofilosean (Cercozoa) that is missing from sequence databases. PLoS One 7:e49090. https://doi.org/10.1371/journal.pone.0049090.
Bugge Harder, C., Rønn, R., Brejnrod, A., Bass, D., Al‐Soud, W.A. & Ekelund, F. 2016. Local diversity of heathland Cercozoa explored by in‐depth sequencing. ISME J. 10: 2488‐97.
Cavalier‐Smith, T. & Oates, B. (2012). Ultrastructure of Allapsa vibrans and the Body Plan of Glissomonadida (Cercozoa). Protist, 163: 165–187.
Cavalier‐Smith, T. & Karpov, S.A. 2012. Paracercomonas Kinetid Ultrastructure, Origins of the Body Plan of Cercomonadida, and Cytoskeleton Evolution in Cercozoa. Protist, 163, 47–75.
Chatelain, A.P., Meisterfeld, R., Roussel‐Delif, L., & Lara, E. 2013. Sphenoderiidae (fam. nov.), a new clade of euglyphid testate amoebae characterized by small, round scales surrounding the aperture. Protist 164:782‐792. https://doi.org/10.1016/j.protis.2013.08.001.
Dumack, K., Baumann, C. & Bonkowski, M. 2016. A Bowl with Marbles: Revision of the Thecate Amoeba Genus Lecythium (Chlamydophryidae, Tectofilosida, Cercozoa, Rhizaria) Including a Description of Four New Species and an Identification Key. Protist, 167: 440–459.
Dumack, K., Schuster, J., Bass, D., & Bonkowski, M. 2016. A novel lineage of ‘naked filose amoebae’; Kraken carinae gen. nov. sp. nov. (Cercozoa) with a remarkable locomotion by disassembly of its cell body. Protist 167:268‐278. https://doi.org/10.1016/j.protis.2016.04.002.
Dumack, K., Flues, S., Hermanns, K. & Bonkowski, M. 2017. Rhogostomidae (Cercozoa) from soils, roots and plant leaves (Arabidopsis thaliana): Description of Rhogostoma epiphylla sp. nov. and R. cylindrica sp. nov. Europ. J. Protistol., 60, 76–86.
Dumack, K., Mausbach, P., Hegmann, M. & Bonkowski, M. 2017. Polyphyly in the Thecate Amoeba Genus Lecythium (Chlamydophryidae, Tectofilosida, Cercozoa), Redescription of its Type Species L. hyalinum, Description of L. jennyae sp. nov. and the Establishment of Fisculla gen. nov. and Fiscullidae fam. nov. Protist, 168: 294–310.
Dumack, K., Mylnikov, A.P. & Bonkowski, M. 2017. Evolutionary Relationship of the Scale‐Bearing Kraken (incertae sedis, Monadofilosa, Cercozoa, Rhizaria): Combining Ultrastructure Data and a Two‐ Gene Phylogeny. Protist, 168: 362–373.
Dumack, K., Öztoprak, H., Rüger, J. & Bonkowski, M. 2017. Shedding Light on the Polyphyletic Thecate Amoeba Genus Plagiophrys: Transition of Some of its Species to Rhizaspis (Tectofilosida, Thecofilosea, Cercozoa) and the Establishment of Sacciforma gen. nov. and Rhogostomidae fam. nov. (Cryomonadida, Thecofilosea, Cercozoa). Protist, 168: 92–108.
Dumack, K., Bonkowski, M., Clauss, S. & Völcker, E. 2018. Phylogeny and redescription of the testate amoeba Diaphoropodon archeri (Chlamydophryidae, Thecofilosea, Cercozoa), De Saedeleer 1934, and annotations on the polyphyly of testate amoebae with agglutinated tests in the Cercozoa. J. Eukaryot. Microbiol. [Epub ahead of print] https://doi.org/10.1111/jeu.12474.
Dumack, K., Siemensma, F. & Bonkowski, M. 2018. Rediscovery of the testate amoeba genus Penardeugenia (Thaumatomonadida, Imbricatea). Protist, 169:29‐42. https://doi.org/10.1016/j.protis.2017.12.002.
Hess, S. & Melkonian, M. 2013. The mystery of clade X: Orciraptor gen. nov. and Viridiraptor gen. nov. are highly specialised, algivorous amoeboflagellates (Glissomonadida, Cercozoa). Protist, 164:706‐747. https://doi.org/10.1016/j.protis.2013.07.003.
Kosakyan, A., Gomaa, F., Lara, E. & Lahr, D.J.G. 2016. Current and future perspectives on the systematics, taxonomy and nomenclature of testate amoebae. Eur. J. Protistol., 55:105‐117. https://doi.org/10.1016/j.ejop.2016.02.001.
Lee, W.J. & Park, J.S. 2016. Placement of the unclassified Cyranomonas australis Lee 2002 within a novel clade of Cercozoa. Eur. J. Protistol., 56:60‐66. https://doi.org/10.1016/j.ejop.2016.06.004.
Ngo, C.N., Braithwaite, K.S., Bass, D., Young, A.J. & Croft, B.J. 2018. Phytocercomonas venanatans, a new species of Cercozoa associated with chlorotic streak of sugarcane. Phytopathology, 108:479‐486. https://doi.org/10.1094/phyto-07-17-0237-r.
Nicholls, K.H. 2012. Zoelucasa sablensis n. gen. et n. sp. (Cercozoa, Incertae sedis), a new scale‐ covered flagellate from marine sandy shores. Acta Protozoologica, 51(2): 113‐117.
Scoble, J.M. & Cavalier‐Smith, T. 2014 Scale evolution, sequence phylogeny, and taxonomy of thaumatomonad Cercozoa: 11 new species and new genera Scutellomonas, Cowlomonas, Thaumatospina and Ovaloplaca. Europ. J. Protistol., 50: 270–313.
Schuler, G.A., Tice, A.K., Pearce, R.A., Foreman, E., Stone, J., Gammill, S., Wilson, J.D., Reading, C., Silberman, J.D., Brown, M.W., Phylogeny and Classification of Novel Diversity in Sainouroidea (Cercozoa, Rhizaria) Sheds Light on a Highly Diverse and Divergent Clade. Protist https://doi.org/10.1016/j.protis.2018.08.002
Shiratori, T. & Ishida, K‐I. 2016. Trachyrhizium urniformis n. g., n. sp., a Novel Marine Filose Thecate Amoeba Related to a Cercozoan Environmental Clade (Novel Clade 4) J. Euk. Microbiol., 63:722–731.
Shiratori, T., Yabuki, A. & Ishida, K‐I. 2012. Esquamula lacrimiformis n. g., n. sp., a New Member of Thaumatomonads that Lacks Siliceous Scales. J. Eukaryot. Microbiol., 59(6): 527–536.
Shiratori, T., Yokoyama, A. & Ishida, K‐I. 2014. Phylogeny, Ultrastructure, and Flagellar Apparatus of a New Marimonad Flagellate Abollifer globosa sp. nov. (Imbricatea, Cercozoa). Protist, 165: 808–824.
Shiratori, T., Fujita, S., Shimizu, T., Nakayama, T. & Ishida, K. 2017. Viridiuvalis adhaerens gen. et sp. nov., a novel colony‐forming chlorarachniophyte. J. Plant Res., 130:999‐1012. https://doi.org/10.1007/s10265-017-0961-1.
Yabuki, A. & Ishida, K. 2018. An orphan protist Quadricilia rotundata finally finds its phylogenetic home in Cercozoa. J. Eukaryot. Microbiol. [Epub ahead of print] https://doi.org/10.1111/jeu.12502.
Yabuki, A. & Ishida, K‐I. 2011. Mataza hastifera n. g., n. sp.: A Possible New Lineage in the Thecofilosea (Cercozoa). J. Eukaryot. Microbiol., 58(2): 94–102.
Endomyxa
Berney, C., Romac, S., Mahé, F., Santini, S., Siano, R. & Bass, D. 2013. Vampires in the oceans: predatory cercozoan amoebae in marine habitats. ISME J., 7:2387‐2399. https://doi.org/10.1038/ismej.2013.116.
Gong, Y., Patterson, D.J., Li, Y., Hu, Z., Sommerfeld, M., Chen, Y. & Hu, Q. 2015 Vernalophrys algivore gen. nov., sp. nov. (Rhizaria: Cercozoa: Vampyrellida), a new algal predator isolated from outdoor mass culture of Scenedesmus dimorphus. Appl. Environ. Microbiol., 81:3900 –3913. https://doi.org/10.1128/aem.00160-15.
Hartikainen, H., Stentiford, G.D., Bateman, K.S., Berney, C., Feist, S.W., Longshaw, M., Okamura, B., Stone, D., Ward, G., Wood, C. & Bass, D. 2014. Mikrocytids are a broadly distributed and divergent radiation of parasites in aquatic invertebrates. Curr. Biol., 24:807‐812. https://doi.org/10.1016/j.cub.2014.02.033.
Hartikainen, H., Ashford, O.S., Berney, C., Okamura, B., Feist, S.W., Baker‐Austin, C., Stentiford, G.D. & Bass, D. 2014. Lineage‐specific molecular probing reveals novel diversity and ecological partitioning of haplosporidians. ISME J., 8: 177‐86.
Hess, S. 2017. Description of Hyalodiscus flabellus sp. nov. (Vampyrellida, Rhizaria) and identification of its bacterial endosymbiont, “Candidatus Megaira polyxenophila” (Rickettsiales, Alphaproteobacteria). Protist, 168:109‐133. https://doi.org/10.1016/j.protis.2016.11.003.
Hess, S., Sausen, N. & Melkonian, M. 2012. Shedding Light on Vampires: The Phylogeny of Vampyrellid Amoebae Revisited. PLoS ONE, 7(2): e31165. https://doi.org/10.1371/journal.pone.0031165
Neuhauser, S., Kirchmair, M., Bulman, S. & Bass, D. 2014. Cross‐kingdom host shifts of phytomyxid parasites. BMC Evol. Biol., 14:33. https://doi.org/10.1186/1471-2148-14-33.
Ward, G.M., Bennett, M., Bateman, K., Stentiford, G.D., Kerr, R., Feist, S.W., Williams, S.T., Berney, C. & Bass, D. 2016. A new phylogeny and environmental DNA insight into paramyxids: an increasingly important but enigmatic clade of protistan parasites of marine invertebrates. Int. J. Parasitol., 46:605‐619. https://doi.org/10.1016/j.ijpara.2016.04.010.
Ward, G.M., Neuhauser, S., Groben, R., Ciaghi, S., Berney, C., Romac, S. & Bass, D. 2018. Environmental sequencing fills the gap between parasitic haplosporidians and free‐living giant amoebae. J. Eukaryot. Microbiol., 65: 574–586. https://doi.org/10.1111/jeu.12501.
Retaria
Biard, T., Pillet, L., Decelle, J., Poirier, C., Suzuki, N. & Not, F. 2015. Towards an integrative morpho‐molecular classification of the Collodaria (Polycystinea, Radiolaria). Protist, 166:374‐388. https://doi.org/10.1016/j.protis.2015.05.002.
Decelle, J., Martin, P., Paborstava, K., Pond, D.W., Tarling, G., Mahé, F., de Vargas, C., Lampitt, R. & Not, F. 2013. Diversity, ecology and biogeochemistry of cyst‐forming Acantharia (Radiolaria) in the oceans. PLoS ONE, 8:e53598. https://doi.org/10.1371/journal.pone.0053598.
Gooday, A.J., Holzmann, M., Caulle, C., Goineau, A., Kamenskaya, O., Weber, A.A.T. & Pawlowski, J. 2017. Giant protists (xenophyophores, Foraminifera) are exceptionally diverse in parts of the abyssal eastern Pacific licensed for polymetallic nodule exploration. Biological Conservation, 207:106‐116. https://doi.org/10.1016/j.biocon.2017.01.006.
Holzmann, M. & Pawlowski, J. 2017. An updated classification of rotaliid foraminifera based on ribosomal DNA phylogeny. Mar. Micropal., 132:18‐34. https://doi.org/10.1016/j.marmicro.2017.04.002.
Pawlowski, J., Holzmann, M. & Tyszka, J. 2013. New supraordinal classification of Foraminifera: Molecules meet morphology. Mar. Micropal., 100:1‐10. https://doi.org/10.1016/j.marmicro.2013.04.002.
Siemensma, F., Apothéloz‐Perret‐Gentil, L., Holzmann, M., Clauss, S., Völcker, E. & Pawlowski, J. 2017. Taxonomic revision of freshwater foraminifera with the description of two new agglutinated species and genera. Eur. J. Protistol., 60:28‐44. https://doi.org/10.1016/j.ejop.2017.05.006.
Cryptista
Burki, F., Okamoto, N., Pombert, J.‐F. & Keeling, P. J. 2012. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc. Roy. Soc. B: Biol. Sci., 279(1736): 2246–2254. https://doi.org/10.1098/rspb.2011.2301
Yabuki, A., Kamikawa, R., Ishikawa, S. A., Kolisko, M., Kim, E., Tanabe, A. S., et al. 2014. Palpitomonas bilix represents a basal cryptist lineage: insight into the character evolution in Cryptista. Sci. Reports, 4: 4641. https://doi.org/10.1038/srep04641
Cavalier‐Smith, T., Chao, E. E. & Lewis, R. 2015. Multiple origins of Heliozoa from flagellate ancestors: New cryptist subphylum Corbihelia, superclass Corbistoma, and monophyly of Haptista, Cryptista, Hacrobia and Chromista. Mol. Phylogenet. Evol., 93: 331–362. https://doi.org/10.1016/j.ympev.2015.07.004
Haptista
Edvardsen, B., Egge, E.S. & Vaulot, D. 2016. Diversity and distribution of haptophytes revealed by environmental sequencing and metabarcoding – a review. Perspectives in Phycology, 3, Issue 2, p. 77–91.
Burki, F., Kaplan, M., Tikhonenkov, D. V., Zlatogursky, V., Minh, B. Q., Radaykina, L. V., et al. 2016. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc. Royal Soc. B: Biol. Sci., 283(1823). https://doi.org/10.1098/rspb.2015.2802 .
Cavalier‐Smith, T., Chao, E. E. & Lewis, R. 2015. Multiple origins of Heliozoa from flagellate ancestors: New cryptist subphylum Corbihelia, superclass Corbistoma, and monophyly of Haptista, Cryptista, Hacrobia and Chromista. Mol. Phylogenet. Evol., 93, 331–362. https://doi.org/10.1016/j.ympev.2015.07.004.
Egge, E.S., Bittner, L., Andersen, T., Audic, S., de Vargas, C. & Edvardsen, B. 2013. 454 Pyrosequencing to Describe Microbial Eukaryotic Community Composition, Diversity and Relative Abundance: A Test for Marine Haptophytes. PLOS ONE, 8 e74371.
Centroplasthelida
Bass, D., Chao, E. E., Nikolaev, S., Yabuki, A., Ishida, K. I., Berney, C., Pakzad, U., Wylezich, C. & Cavalier‐Smith, T. 2009. Phylogeny of novel naked filose and reticulose Cercozoa: Granofilosea cl. n. and Proteomyxidea revised. Protist, 160: 75‐109.
Cavalier‐Smith, T. & Chao, E. E. 2012. Oxnerella micra sp. n.(Oxnerellidae fam. n.), a tiny naked centrohelid, and the diversity and evolution of heliozoa. Protist, 163: 574‐601.
Cavalier‐Smith, T. & Scoble J. M. 2013. Phylogeny of Heterokonta: Incisomonas marina, a uniciliate gliding opalozoan related to Solenicola (Nanomonadea), and evidence that Actinophryida evolved from raphidophytes. Europ. J. Protistol., 49: 328‐353.
Febvre‐Chevalier, C., & Febvre, J. 1984. Axonemal microtubule pattern of Cienkowskya mereschkovskyi and a revision of heliozoan taxonomy. Origins of life, 13: 315‐338.
Krabberød, A. K., Orr, R. J., Bråte, J., Kristensen, T., Bjørklund, K. R. & Shalchian‐Tabrizi, K. 2017. Single Cell Transcriptomics, Mega‐Phylogeny, and the Genetic Basis of Morphological Innovations in Rhizaria. Mol. Biol. Evol., 34; 1557‐1573.
Mikrjukov, K. A. 1996. Revision of Genera and Species Composition of Lower Centroheliozoa. II. Family Raphidiophryidae n. fam. Archiv. Protistenkd., 147: 205‐212.
Mikrjukov, K. A. 2000. Taxonomy and phylogeny of heliozoa. I. The order Desmothoracida Hertwig et Lesser, 1874. Acta Protozool., 39; 81‐97.
Mikrjukov, K. A. 2002. Centrohelid heliozoans (Centroheliozoa). KMK Scientific, Moscow. Sekiguchi, H., Kawachi, M., Nakayama, T. & Inouye I. 2003. A taxonomic re‐evaluation of the Pedinellales (Dictyochophyceae), based on morphological, behavioural and molecular data. Phycologia, 42: 165–182.
Shishkin, Y., Drachko, D., Klimov, V. I. & Zlatogursky, V. V. 2018. Yogsothoth knorrus gen. n., sp. n. and Y. carteri sp. n. (Yogsothothidae fam. n., Haptista, Centroplasthelida), with notes on Evolution and Systematics of Centrohelids. Protist, https://doi.org/10.1016/j.protis.2018.06.003.
Zlatogursky, V. V. 2016. There and back again: parallel evolution of cell coverings in centrohelid heliozoans. Protist, 167; 51‐66.
Zlatogursky, V. V., Drachko, D., Klimov, V. I. & Shishkin, Y. 2018. On the phylogenetic position of the genus Raphidocystis (Haptista: Centroplasthelida) with notes on the dimorphism in centrohelid life cycle. Europ. J. Protistol., 64: 82‐90.
Excavates
Bennett, M. S. & Triemer, R. E. 2012 A new method for obtaining nuclear gene sequences from field samples and taxonomic revisions of photosynthetic euglenoids Lepocinclis (Euglena) helicoideus and Lepocinclis (Phacus) horridus (Euglenophyta). J. Phycol., 48: 254‐60.
Bennett, M. S., Triemer R. E. 2014 The genus Cyclidiopsis: an obituary. J. Eukaryot. Microbiol., 61:166‐72.
Bennett, M. S., Wiegert, K. E. & Triemer, R.E. 2014 Characterization of Euglenaformis gen. nov. and the chloroplast genome of Euglenaformis [Euglena] proxima (Euglenophyta). Phycologia, 53: 66–73.
Breglia, S. A., Yubuki, N. & Leander, B. S., 2013 Ultrastructure and molecular phylogenetic position of Heteronema scaphurum: a eukaryovorous euglenid with a cytoproct. J. Eukaryot. Microbiol., 60: 107‐20.
Cavalier‐Smith, T. 2013 Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa. Eur. J. Protistol., 49: 115‐178.
Cavalier‐Smith, T. 2016 Higher classification and phylogeny of Euglenozoa. Eur. J. Protistol., 56: 250‐276.
Cavalier‐Smith, T., Chao, E. E. & Vickerman, K. 2016 New phagotrophic euglenoid species (new genus Decastava; Scytomonas saepesedens; Entosiphon oblongum), Hsp90 introns, and putative euglenoid Hsp90 pre‐mRNA insertional editing. Eur. J. Protistol., 56: 147‐170.
Čepička, I., Dolan, M. F. & Gile, H. F. 2017. Parabasalia. In: Archibald, J. M.,Simpson, A. G. B., and Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-32669-6_12-1.
Čepička, I., Hampl, V. & Kulda, J. 2010 Critical taxonomic revision of parabasalids with description of one new genus and three new species. Protist, 161: 400‐433.
Chan, Y.‐F., Moestrup, Ø. & Chang, J. 2013 On Keelungia pulex nov. gen. et nov. sp., a heterotrophic euglenoid flagellate that lacks pellicular plates (Euglenophyceae, Euglenida). Eur. J. Protistol., 49: 15‐31.
Gibson, W. 2017 Kinetoplastea. In: Archibald, J. M.,Simpson, A. G. B. & Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-32669-6_12-1.
Hampl, V. 2017 Preaxostyla. In: Archibald, J. M., Simpson,G. B., and Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-32669-6_12-1
Hanousková, P., Táborský, P. & Čepička, I. 2018 Dactylomonas gen. nov., a novel lineage of heterolobosean flagellates with unique ultrastructure, closely related to the amoeba Selenaion koniopes Park, de Jonckheere & Simpson, 2012. J. Eukaryot. Microbiol., in press.
Heiss, A. A., Kolisko, M., Ekelund, F., Brown, M. W., Roger, A. J. & Simpson, A. G. B. 2018. Combined morphological and phylogenomic re‐examination of malawimonads, a critical taxon for inferring the evolutionary history of eukaryotes. R. Soc. Open Sci., 5:171707. https://doi.org/10.1098/rsos.171707.
Kamikawa, R., Kolisko, M., Nishimura, Y., Yabuki, A., Brown, M. W., Ishikawa, S. A., Ishida, K., Roger, A. J., Hashimoto, T. & Inagaki, Y. 2014 Gene content evolution in discobid mitochondria deduced from the phylogenetic position and complete mitochondrial genome of Tsukubamonas globosa. Genome Biol. Evol., 6: 306‐315.
Karnkowska, A., Bennett, M. S., Watza, D., Kim, J. I., Zakryś, B. & Triemer, R. E. 2015 Phylogenetic relationships and morphological character evolution of photosynthetic euglenids (Excavata) inferred from taxon‐rich analyses of five genes. J. Eukaryot. Microbiol., 62: 362‐73.
Kaufer, A., Ellis, J., Stark, D. & Barratt, J. 2017 The evolution of trypanosomatid taxonomy. Parasites & Vectors, 10: 287.
Kostygov, A.Y. & Yurchenko, V. 2017 Revised classification of the subfamily Leishmaniinae (Trypanosomatidae). Folia Parasitol., 64: 020.
Kulda, J., Nohýnková, E. & Čepička, I. 2017 Retortamonadida (with notes on Carpediemonas‐like organisms and Caviomonadidae). In: Archibald, J. M.,Simpson, A. G. B., and Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-32669-6_12-1.
Lax, G. & Simpson, A. G. B. 2013 Combining molecular data with classical morphology for uncultured phagotrophic euglenids (Excavata): a single‐cell approach. J. Eukaryot. Microbiol., 60: 615‐25.
Leander, B. S., Lax, G., Karnkowska, A. & Simpson, A. G. B. 2017 Euglenida. In: Archibald, J. M.,Simpson, A. G. B., and Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-32669-6_12-1
Lee, W. J. & Simpson, A. G. 2014 Ultrastructure and molecular phylogenetic position of Neometanema parovale sp. nov. (Neometanema gen. nov.), a marine phagotrophic euglenid with skidding motility. Protist, 165: 452‐72.
Lee, W. J. & Simpson, A. G. 2014 Morphological and molecular characterisation of Notosolenus urceolatus Larsen and Patterson 1990, a member of an understudied deep‐branching euglenid group (petalomonads). J. Eukaryot. Microbiol., 61: 463‐79.
Lukeš, J., Butenko, A., Hashimi, H., Maslov, D.A., Votýpka, J. & Yurchenko, V. 2018 Trypanosomatids are much more than just trypanosomes: clues from the expanded family tree. Trends Parasitol., 34: 466‐480.
Pánek, T., Simpson, A. G. B., Brown, M. W. & Dyer, B. D. 2017 Heterolobosea. In: Archibald, J. M.,Simpson, A. G. B., and Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-32669-6_12-1.
Pánek T., Táborský P., Pachiadaki, M. G., Hroudová, M., Vlček, Č., Edgcomb, V. P. & Čepička, I. 2017 Combined culture‐based and culture‐independent approaches provide insights into diversity of jakobids, an extremely plesiomorphic eukaryotic lineage. Front. Microbiol., 6: 1288.
Okamoto, N., Gawryluk, R.M.R., del Campo, J., Strassert, J.F.H., Lukeš, J., Richards, T.A., Worden, A.Z., Santoro, A.E. & Keeling, P.J. 2018 A revised taxonomy of diplonemids including the Eupelagonemidae n. fam. and a type species, Eupelagonema oceanica n. gen. & sp. J. Euk. Microbiol., (in press)
Radek, R., Strassert, J. F., Krüger, J., Meuser, K., Scheffrahn, R. H. & Brune, A. 2014 Phylogeny and ultrastructure of Oxymonas jouteli, a rostellum‐free species, and Opisthomitus longiflagellatus sp. nov., oxymonadid flagellates from the gut of Neotermes jouteli. Protist, 165: 384‐99.
Simpson, A. G. B. 2017. Jakobida. In: Archibald, J. M.,Simpson, A. G. B., & Slamovits, C., eds. Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.) Springer Reference Works (e‐book) https://doi.org/10.1007/978-3-319-32669-6_12-1.
Strassert, J. F. H., Tikhonenkov, D. V., Pombert, J. F., Kolisko, M., Tai, V., Mylnikov, A. P. & Keeling, P. J. 2016. Moramonas marocensis gen. nov., sp. nov.: a jakobid flagellate isolated from desert soil with a bacteria‐like, but bloated mitochondrial genome. Open Biol., 6: 150239.
Tashyreva, D., Prokopchuk, G., Yabuki, A., Kaur, B., Faktorová, D., Votýpka, J., Kusaka, C., Fujikura, K., Shiratori, T., Ishida, K.‐I., Horák, A. & Lukeš, J. 2018. Phylogeny and morphology of new diplonemids from Japan. Protist, 169: 158‐179.
Treitli, S. C., Kotyk, M., Yubuki, N., Jirounková, E., Vlasáková, J., Smejkalová, P., Šípek, P., Čepička, I. & Hampl, V. 2018. Molecular and morphological diversity of the oxymonad genera Monocercomonoides and Blattamonas gen. nov. Protist. 169:744–783. https://doi.org/10.1016/j.protis.2018.06.005
Yamaguchi, A., Yubuki, N. & Leander, B. S. 2012. Morphostasis in a novel eukaryote illuminates the evolutionary transition from phagotrophy to phototrophy: description of Rapaza viridis n. gen. et sp. (Euglenozoa, Euglenida). BMC Evol. Biol., 12: 29.
Yubuki, N., Zadrobílková, E. & Čepička, I. 2017. Ultrastructure and molecular phylogeny of Iotanema spirale gen. nov. et sp. nov., a new lineage of endobiotic Fornicata with strikingly simplified ultrastructure. J. Eukaryot. Microbiol., 64: 422‐433.
Zhang, Q., Táborský, P., Silberman, J. D., Pánek, T., Čepička, I. & Simpson, A. G. B. 2015. Marine isolates of Trimastix marina form a plesiomorphic deep‐branching lineage within Preaxostyla, separate from other known trimastigids (Paratrimastix n. gen.). Protist, 166: 468‐91.
Incertae sedis Eukarya
Foissner, I. & Foissner, W. (1993): Revision of the family Spironemidae Doflein (Protista, Hemimastigophora), with description of two new species, Spironema terricola n. sp. and Stereonema geiseri n. g., n. sp. J. Eukaryot. Microbiol., 40:422–438.
Glücksman, E., Snell, E. A. & Cavalier‐Smith, T. (2013) Phylogeny and evolution of Planomonadida (Sulcozoa): eight new species and new genera Fabomonas and Nutomonas. Europ. J. Protistol., 49:179–200.
Hausmann, K., Weitere, M., Wolf, M. & Arndt, H. 2002. Meteora sporadica gen. et sp. nov. (Protista incertae sedis) — an extraordinary free‐living protist from the Mediterranean deep sea. Europ. J. Protistol., 38:171–177.
Janouškovec, J., Tikhonenkov, D. V., Burki, F., Howe, A. T., Rohwer, F. L., Mylnikov, A. P. & Keeling, P. J. (2017). A New Lineage of Eukaryotes Illuminates Early Mitochondrial Genome Reduction. Curr. Biol., 27(23), 3717–3724.e5. https://doi.org/10.1016/j.cub.2017.10.051
CRuMs
Brown, M. W., Heiss, A. A., Kamikawa, R., Inagaki, Y., Yabuki, A., Tice A. K., Shiratori, T., Ishida, K., Hashimoto, T., Simpson, A. G. B. & Roger, A. J. 2018. Phylogenomics places orphan protistan lineages in a novel eukaryotic super‐group. Genome Biol. Evol., 10:427–433.
Brugerolle, G. 2006. Description of a new freshwater heterotrophic flagellate Sulcomonas lacustris affiliated to the collodictyonids. Acta Protozool., 45:175–182.
Brugerolle, G., Bricheux, G., Philippe, H. & Coffe, G. 2002. Collodictyon triciliatum and Diphylleia rotans ( = Aulacomonas submarina) form a new family of flagellates (Collodictyonidae) with tubular mitochondrial cristae that is phylogenetically distant from other flagellate groups. Protist, 153:59–70.
Glücksman, E., Snell, E. A., Berney, C., Chao, E. E., Bass, D. & Cavalier‐Smith, T. 2011. The novel marine gliding zooflagellate genus Mantamonas (Mantamonadida ord. n.: Apusozoa). Protist, 162:207–221.
Yabuki, A., Ishida, K. & Cavalier‐Smith, T. 2013. Rigifila ramosa n. gen., n. sp. a filose apusozoan with a distinctive pellicle, is related to Micronuclearia. Protist, 164:75–88.
Appendix S2. Trophic functional groups across protist diversity.
Trophic functional group assignments facilitate interpretation of community structure and food web assembly from DNA sequence information identification of “operational taxonomic unit” (OTU). Here we tried to provide details of trends in eating habits of non‐parasitic protists. Many genera have never been cultured so their feeding preferences are unknown. For definition of terms, please refer to the main text, page 6.
Across protists, in most cases it is safe to assume that species within a genus are most likely to have the same trophic function, with rare exceptions. However, it is not the case in some taxa, where there can be considerable variation between species within a genus. That is the case, for example, in Dinoflagellata, Arcellinida, and Euglyphida.
In the Amoebozoa, genera can be assumed to ingest bacteria by phagocytosis with the following exceptions: Amphizonella (Corycida) and Dermamoeba also ingest Cyanobacteria; the Euamoebida genera Amoeba, Cashia, Chaos, Polychaos, Trichamoeba and Deuteramoeba are also cytotrophic; amoebae of the genera Polychaos and Pseudothecamoeba ingest diatoms. Some species or genera have a preference for heterotrophic bacteria over of photosynthetic ones, or even discriminate against Cyanobacteria such as Copromyxa and Copromyxella. In Protosteliida, Protostelium spp. prefer yeasts but some will also eat bacteria, and several can eat coccoid green algae, except P. nocturnum which only eats bacteria; they can be cannibalistic in culture as can Cavosteliida and some Protosporangiidae. In Schizoplasmodiidae, all do best in culture on a mixture of yeast and bacteria, though some will survive on bacteria alone; when they eat yeasts, they puncture the cell wall and ingest only the protoplast. Multicilia (Holomastigida) is cytotrophic on other amoebae. Dictyostelia are primarily bacterivorous but they will eat yeasts in culture, and can also be cannibalistic in culture; zygotes are cannibalistic exclusively. Myxogastria ciliated stage is bacterivorous, their plasmodia are omnivorous and often cannibalistic when they first form. The Entamoebae are mostly commensal or parasitic and can eat bacteria by phagotrophy. A few strains of Sappinia and Acanthamoeba can be parasitic. Some Archamoebae, Tricholimax, Thecamoeba, Korotnevella, Gocevia, Paragocevia, Leptomyxa and Mayorella species are omnivores. There are some diatom specialists as in Difflugia and Phryganella. The ability to feed on yeast might be more widespread than reported. Similarly, in their habitat some soil amoebae probably ingest small Cercozoa such as Glissomonads. In the Arcellinida, at least species of Nebela and Cryptodifflugia are known to prey on nematodes. The tendency to cannibalism amongst some amoebae may be due to crowding in culture.
In Arcellinida certain species of Cucurbitella, Difflugia, Heleopera, Hyalosphenia and Netzelia form symbiosis with Trebouxiophyta species and can be considered mixotrophic.
Free‐living basal genera of Holozoa and Nucletmycea are amoeboid, phagotrophic on bacteria. Parasitic ones, including Microsporidia, are intracellular parasites. The Choanoflagellata feed on bacteria by filtration through the collar. The Fungi are saprotrophic on external substrate, obtaining their nutrient by osmotrophy through a chitinous cell wall. Lichenous fungi are capable of photosynthesis. Mycorrhizal fungi receive most or all of their organic molecules from the host plant. Many Fungi are parasitic (see table 2). For details of substrate preference the reader is referred to FUNGuild (Nguyen et al., 2016, https://github.com/UMNFuN/FUNGuild).
In the Archaeplastida, ignoring the Embryophyta, all Chloroplastida are photosynthetic with chl a, b, with the exception of Helicosporidium (insect gut parasite), Polytomella, Polytoma, Prototheca which have 2° lost photosynthesis and are strictly osmotrophic. Among the Rhodophyceae, some Cyanidiales are facultative heterotrophs. Amongs the red algae certain species in several genera have lost photosynthesis and are parasites of other red algae. For example: Gracilariophila oryzoides, Harveyella mirabilis, Choreocolax polysiphoniae, Janczewskia gardneri, Faucheocolax attenuata, Bostrychiocolax australis, Dawsoniocolax bostrychiae, Coccotylus hartzii, Epulo multipedes (see Blouin and Lane, 2012, Table S1). Nearly all of the parasitic genera end with the term “colax” to indicate the lifestyle. Most parasites are sister species with the hosts, and many of these genera need to be merged with the genus of their hosts, as they were described separately. This is in progress but obviously correcting the taxonomy is a slow process.
The Cryptista ingest bacteria prey by phagocytosis, and the photosynthetic ones are mixotrophic. In Haptophyta, Coccolithales are autotrophic, while genera without scales can be mixotrophic or heterotrophic on bacteria, with or without haptonema contributing to bacteria capture. The Centroplasthelida are typically cytotrophic.
In the Apicomplexa, all species are historically treated as parasites. Recent observation suggest some may be endobionts as commensals or at least not as parasites, especially among some invertebrate hosts. However, there are no free‐living species known. The presence of a relic plastid genome in the Apicomplexa, and apicoplast in some, provides useful metabolic targets for drug treatment development. In contrast, both Dinoflagellata and Ciliophora provide a wide diversity of trophic functional groups.
The Dinoflagellata functional assignments vary between closely related genera, and are shown in Appendix 2, Table 2.1.
The Ciliophora are to be assumed to ingest bacteria by filtration except as noted below. Some genera of Karyorelictea (eg. Kentrophoridae) can be considered grazers as they live on sand surfaces covered in bacteria, and often harbour symbiotic bacteria. We discriminate between surface‐feeding and water‐column filtration feeding. Surface‐feeding ciliophores spend most of their time gathering food particles from surfaces rather than by swimming in a volume of water. Thus clearing rates are calculated by surface area rather than volume of liquid. Most ciliophores (with exceptions noted in table 2.2) ingest bacteria, and many also ingest small protists (labelled omnivorous), while others can ingest larger protists by predation. Many omnivores can also ingest some detritus if the cytostome permits. In Ciliophora one should not assume that a small cell or a small cytostome implies a bacteria diet. There are parasites, histophagous species, endocommensals, and epibionts. The Stichotrichida contain species that can ingest larger protists and even small ciliophores for example in some species of Oxytrichidae. The Litostomatea are redoubtable predatory cytotrophs, and some can ingest prey larger than themselves. Entodiniomorphida are often (or typically) commensals in rumen. Among the Colpodea a small number of genera are cytotrophic, for example Bursaria (and maybe other Bursariomorphida). In the Nassulida, Nassula is phycotrophic by swallowing and can ingest cyanobacteria filaments. In the Prorodontida several genera, eg. Coleps, are attracted to dead or wounded invertebrates to the leaking tissues. The Astomatia can be found as symbionts in Oligochaetae gut. Some Tetrahymenida genera are parasitic or histophagous. The Suctoria (Phyllopharyngea) use specialised tentacles to capture and manipulate prey, typically other ciliophores or small invertebrates; they are cytotrophic predators and 3° consumers as predators of invertebrates. The Rhynchodia (Phyllopharyngea) are parasites of bivalves. Among the Scuticociliatia the Philasterida contain cytotrophic genera, for example Uronema. See Appendix table 2.2 for more details.
Also in the Alveolates, the colpodellids, Colponema, Acavomonas, and Oxyrrhis are cytotrophic predators, while the Perkinsidae are parasitic as trophozoites.
Stramenopiles in the Ochrophyta are almost all photosynthetic. The Actinophryidae are cytotrophic. The Opalinata, Bicosoecida, and Placidida are phagotrophic on bacteria, with the exception of the Proteromonada and Blastocystis (osmotrophic) which are intestinal commensals or parasites. The Hyphochytriales and Peronosporomycetes are saprotrophs, but the latter contains numerous plant parasites. The Pirsoniales are parasites on diatoms.
Rhizaria. The Cercomonadida, Paracercomonadida, and Glissomonadida can be assumed to be phagotrophic predators on bacteria, with the notable exception of the Viridiraptoridae which are phycotrophic (mycotrophy not known) by penetration. The Vampyrellida are mycotrophic or phycotrophic. The Euglyphida show some variation, although most are phagotrophic on bacteria. Observations indicate that the larger the cell the more likely it is to be cytotrophic; in fact the larger species cannot survive on bacteria alone. Many Hyalospheniidae need to prey at least partly on Euglyphida and use the scales to build their own test. Symbiosis occurs between Trebouxiophyta (photosynthetic) with several genera of Rhizaria. This implicates at least some species of Placocista (Euglyphida), Amphitrema and Archerella (Labyrinthulomycetes). Chlorarachnea are photosynthetic but some species are mixotrophic on bacteria. In the Endomyxa, the Phytomyxea are plant root parasites, while the Ascetosporea are parasites of invertebrates.
The Foraminifera are primarily bacteria eaters, but can ingest detritus, other protists, and larger forms can prey on small invertebrates.
Among the Discoba (Excavates) Heterolobosea graze bacteria by phagocytosis, except Stephanopogon and Creneis which are cytotrophic. Some strains of Naegleria fowleri can cause primary amoebic meningoencephalitis, penetrating through the nose. The Jakobida and Tsukubomonas are predatory on bacteria which are ingested by phagocytosis. The Diplonema are cytotrophic by phagocytosis and have an important role in aphotic marine waters; some species are parasitic or symbiotic with animals. Among the Euglenida (Discoba) the Aphagea (Rhabdomonas, Gyropaigne, Menoidium, Parmidium and Rhabdospira), Astasia and Distigma are osmotrophic; Euglenophyceae (except Rapaza) are photosynthetic with chl a, b in plastids, or secondarily osmotrophic, and Rapaza is cytotrophic; the Peranemids, Anisonemids, Pleotiids, and Petalomonadida are phagotrophic. Certain clades graze on bacteria by phagotrophy or are cytotrophic predators. Among the Kinetoplastea (Discoba) all except the Trypanosomatida are predatory phagotrophs on bacteria. The Trypanosomatida are animal blood and tissue parasites, except Phytomonas which lives in plant latex vessels, and all are transferred by insect vectors.
The Metamonada (Fornicata, Parabasalia, and Preaxostyla Excavates) are predatory or grazers on bacteria, ingested by phagocytosis. The Preaxostyla are endosymbiont detritivores that phagocytose wood microchips, and acquire many nutrients by osmotrophy from syntrophic bacteria; some genera may also ingest bacteria by phagocytosis. Giardia (Diplomonadida) are osmotrophic gut parasites. Several genera have species that can be parasitic in humans and other animals.
Human parasites. For protists that are human parasites, see Adl and Matheson (2019) in ASM‐Manual of Clinical Microbiology chapter 132 (Taxonomy and Classification of Human Eukaryotic Parasites), 12th edn.
Definitions. We further provide descriptions and clarifications for symbisois, microbiome, holobiont, intra‐ cellular and extracellular locations, that relate to interactions between organisms.
Symbiosis is a sustained relationship between at least two individuals (same or different species), living in direct contact or close enough one to the other, during part or the whole live cycle of the two partners. This interaction is transmitted either vertically (from one generation to the other) or horizontally (acquired de novo at each generation). Symbiosis is the antonym of transitory interactions (predatory). A symbiosis can be beneficial for both partners (mutualism) or at the other extreme, deleterious for one of these partners (parasite or parasitoid). In general, the larger species of this association is called the host, the smallest one is the symbiont. Examples of symbiosis: photosymbiosis (corals and Symbiodinium, kleptoplast), mycorrhizae, lichens, parasitism and hyperparasitism (parasite of parasite). Some specialist predators may also be considered as symbionts (example: Helcion living exclusively on laminarian). A virus is a parasitic symbiont.
The microbiome is a microbial community living in, on or at the close vicinity of an individual. Examples: gut community, biosphere of phytoplanktonic cells, microbes living on the skin/cuticule of metazoan, in the rhizopshere, microbes living in specialized tissues, or part of the cell. Unicellular symbionts are part of the microbiome of their host.
Holobiont. Ecoevolutionary concept considering an individual together with its microbiome as the true unit on which evolutionary processes will act.
Intracellular (endocellular) versus extracellular. Intracellular symbionts are those where the parasite cytoplasm and nucleus enter inside the host and reside inside the host cell. In some extracellular symbionts, part of the cytoplasm (including the mitochondrion) may penetrate inside the host cell, but never the parasite nucleus. Endobionts penetrate the host tissue between cells, or inside a matrix, but do not reside inside host cells. Mycorrhizae are symbiotic endobionts.
Appendix 2, Table 2.1. Trophic Functional Assignments for Dinoflagellates where known
L = photosynthetic, M = mixotrophic (photosynthesis and phagotrophy), B = phagotrophic on bacteria, C = cytotrophic (phagotrophic on protists), H = heterotrophic. In general, few dinoflagellates nutritional strategy is really investigated, documented, and understood. For most of the taxa we can only indirectly judge and it is more or less a guess. Some genera are difficult to assign because some species are mixotrophic, and for other species we do not know whether they feed in addition to phototrophy. In Amphidinium s.s. for example we have pure heterotrophic but also photosynthetic and mixotrophic species. For these genera there is species level variation in trophic functions. Heterotrophic taxa are the species that have to feed on something, because they are not photosynthetic but we do not know what they feed on (only food bodies or vacuoles are visible but the content cannot be identified and the feeding process has not been recorded).
| •Alveolata |
| ••Dinoflagellata |
| •••Noctilucales |
| Abedinium H , Cachonodinium H , Craspedotella H , Cymbodinium H , Kofoidinium C , Leptodiscus H , Noctiluca M C , Petalodinium H , Pomatodinium H , Scaphodinium H , Spatulodinium M H. |
| •••Dinophyceae |
| ••••Gymnodiniphycidae |
| •••••Gymnodinium |
| Barrufeta M C , Dissodinium L M C , Erythropsidinium C , Greuetodinium C?, Gymnodinium L M , Lepidodinium L , Nematodinium M C , Nusuttodinium M C , Pellucidodinium C , Polykrikos C M , Proterythropsis C , Warnowia C. |
| •••••Amphidinium L M C. |
| ••••• Gyrodinium C. |
| •••••Kareniaceae |
| Brachidinium M , Karenia M B C , Karlodinium M B C , Takayama M C. |
| •••••Ceratoperidiniaceae |
| Ceratoperidinium L, Kirithra L. |
| •••••Torodiniales |
| Kapelodinium C , Torodinium M |
| •••••Levanderina |
| Levanderina M C. |
| •••••Margalefidinium |
| Margalefidinium M C |
| •••••Cochlodinium |
| Cochlodinium strangulatum M? |
| •••••Ptychodiscales |
| Achradina B , Amphitolus, Balechina H C?, Ptychodiscus L , Sclerodinium. |
| •••••Borghiellaceae. |
| Baldinia M C , Borghiella L. |
| •••••Tovelliaceae. |
| Bernardinium, Esopotrodinium M C , Jadwigia L , Tovellia L M. |
| •••••Suessiaceae |
| Ansanella L , Asulcocephalium L , Biecheleria M C , Biecheleriopsis L M , Leiocephalium L , Pelagodinium L , Polarella L , Prosoaulax M C , Protodinium, Symbiodinium L M B , Yihiella L M. |
| ••••Peridiniphycidae |
| •••••Gonyaulacales |
| Alexandrium L M C , Amylax L M C , Ceratium L M C , Ceratocorys L , Coolia L , Fukuyoa L , Fragilidium |
| L M C , Gambierdiscus L M , Goniodoma L , Gonyaulax L M C , Lingulodinium L M C , Ostreopsis L M, |
| Pentaplacodinium L , Peridiniella L , Protoceratium L M, Pyrocystis L , Pyrodinium L , Pyrophacus L , Tripos L M C. |
| •••••Peridiniales |
| Amphidiniopsis H C?, Archaeperidinium C , Blastodinium M, Diplopelta C , Diplopsalis C , Diplopsalopsis C , Herdmania H C?, Niea C , Oblea C , Palatinus L , Parvodinium L , Peridinium L , Peridiniopsis L C , Preperidinium C , Protoperidinium C , Qia C , Vulcanodinium L. |
| •••••Thoracosphaeraceae |
| Aduncodinium C , Amyloodinium, Apocalathium, Blastodinium, Chimonodinium, Cryptoperidiniopsis M C , Duboscquodinium C , Ensiculifera L , Leonella, Luciella C , Naiadinium L M?, Paulsenella C , Pentapharsodinium L , Pfiesteria M C , Scrippsiella L M C , Stoeckeria C , Theleodinium L M?, Thoracosphaera L , Tintinnophagus C , Tyrannodinium C. |
| •••••Podolampadaceae |
| Blepharocysta C?, Gaarderiella, Heterobractum, Lissodinium, Mysticella, Podolampas M C. |
| •••••Kryptoperidiniaceae |
| Blixaea L M, Durinskia L M, Galeidinium L M, Kryptoperidinium L M, Unruhdinium L. |
| •••••Heterocapsaceae |
| Heterocapsa L M C. |
| •••••Amphidomataceae |
| Amphidoma L , Azadinium L. |
| •••••Oxytoxaceae. |
| Corythodinium M C?, Oxytoxum L M? |
| •••••Centrodiniaceae |
| Centrodinium L |
| ••••Dinophysales |
| Amphisolenia L M, Citharistes M B , Dinofurcula H , Dinophysis M C , Histioneis M B , Latifascia H , Metadinophysis L , Metaphalacroma H , Ornithocercus M B , Oxyphysis C , Parahistioneis M B , Phalacroma (M) C , Pseudophalacroma H , Sinophysis H M? B , Triposolenia M. |
| ••••Prorocentrales |
| Mesoporus L , Prorocentrum L M C. |
| •••Incertae sedis Dinoflagellata: |
| e.g., Adenoides L , Akashiwo M C , Amphidiniella L M, Ankistrodinium C , Apicoporus M C , Bispinodinium L M, Bysmatrum L M C , Cabra H C?, Cladopyxis, Crypthecodinium C , Cucumeridinium H , Dactylodinium L M, Dicroerisma H , Gloeodinium, Grammatodinium L M, Gynogonadinium, Gyrodiniellum C , Halostylodinium L , Heterodinium L?, Moestrupia L , Paragymnodinium M C , Phytodinium, Pileidinium L , Plagiodinium L , Planodinium H C?, Pseudadenoides L , Pseudothecadinium L , Pyramidodinium L , Roscoffia H C?, Rhinodinium H C?, Sabulodinium H , Sphaerodinium L , Spiniferodinium L M, Testudodinium L M, Thecadinium L M, Thecadiniopsis L , Togula L. |
| •••Incertae sedis Dinoflagellata [Blastodiniales Chatton 1906, no longer valid]. Amyloodinium*, Apodinium*, Cachonella, Crepidoodinium M, Haplozoon, Oodinium M, Piscinodinium M, Protoodinium M . *ectoparasites |
Appendix 2, Table 2.2. Trophic functional assignments in CILIOPPHORES
All genera feed on bacteria by filtration (B) except where noted. Omnivorous genera (O) ingest small protists in addition to bacteria. Those that are labelled cytrophic (C) need to ingest protists for growth, in addition to bacteria that are co‐ingested. Many omnivores can occasionally ingest detritus (D). Those that feed on fungi (F) do so by digesting a small hole, while those that feed on other filaments (f) (Cyanobacteria and algae) do so by swallowing. Predators (P) are distinguished from those that obtain their food particles by filtration (the default state in ciliophores) or by grazing where noted. Histophagous genera (P*) are distinguished from parasites (X) and commensals (*). Osmotrophic (S) genera feed extensively, or exclusively, by pinocytosis and other membrane transport mechanisms.
| •Alveolata |
| ••Ciliophora |
| •••Postodesmatophora |
| ••••Karyorelictea |
| •••••Kentrophoridae(Kentrophoros) B |
| •••••Loxodida |
| ••••••Cryptopharyngidae (Cryptopharynx) O C |
| ••••••Loxodidae (Loxodes) O C |
| •••••Geleiidae (Geleia) O C |
| ••••Heterotrichea |
| •••••Blepharismidae (Blepharisma) B |
| •••••Climacostomidae (Climacostomum) O |
| •••••Condylostomatidae (Chattonidium, Condylostoma) B |
| •••••Fabreidae (Fabrea) B |
| •••••Gruberiidae (Gruberia) B |
| •••••Coliphorina |
| ••••••Folliculinidae (Folliculina) B |
| ••••••Maristentoridae (Maristentor) O |
| •••••Peritromidae (Peritromus) B |
| •••••Spirostomidae (Anigsteinia, Spirostomum) B |
| •••••Stentoridae (Stentor) O or C or P |
| •••Intramacronucleata |
| ••••SAL |
| Incertae sedis SAL: Mesodiniidae (Mesodinium) Incertae sedis SAL: Phacodinium |
| ••••• Spirotrichea |
| ••••••Euplotia |
| •••••••Euplotida |
| ••••••••Aspidiscidae (Aspidisca) O |
| ••••••••Certesiidae (Certesia) O |
| ••••••••Euplotidae (Euplotes) O |
| ••••••••Gastrocirrhidae (Gastrocirrhus) O |
| ••••••••Uronychidae (Diophrys, Uronychia) O |
| •••••••Discocephalida |
| ••••••••Discocephalidae(Discocephalus, Prodiscocephalus, Paradiscocephalus) O |
| ••••••••Pseudoamphisiellidae (Leptoamphisiella, Pseudoamphisiella) O |
| ••••••Perilemmaphora |
| •••••••Hypotrichia |
| ••••••••Stichotrichida |
| •••••••••Amphisiellidae (Amphisiella, Bistichella) O |
| •••••••••Atractosidae (Atractos) O |
| •••••••••Epiclintidae (Epiclintes) O |
| •••••••••Gonostomatidae (Cotterillia, Gonostomum) O |
| •••••••••Halteriidae (Halteria, Meseres) O |
| •••••••••Holostichidae (Holosticha, Uncinata) O |
| •••••••••Kahliellidae (Deviata, Kahliella) O |
| ••••••••• Keronidae (Kerona) O |
| •••••••••Oxytrichidae (Cyrtohymena, Gastrostyla, Oxytricha, Stylonychia) O |
| •••••••••Parabirojimidae (Parabirojimia, Tunicothrix) O |
| •••••••••Plagiotomidae (Plagiotoma) O |
| •••••••••Psammomitridae (Psammomitra) O |
| •••••••••Pseudoamphisiellidae (Pseudoamphisiella) O |
| •••••••••Psilotrichidae (Psilotricha, Urospinula) O |
| •••••••••Schmidingerotrichidae (Schmidingerothrix) O |
| •••••••••Spirofilidae (Spirofilopsis, Strongylidium) O |
| •••••••••Trachelostylidae (Trachelostyla) O |
| •••••••••Uroleptidae (Paruroleptus, Uroleptus) O |
| ••••••••Urostylida |
| •••••••••Bergeriellidae (Bergeriella, Neourostylopsis) O |
| •••••••••Hemicycliostylidae (Hemicycliostyla, Australothrix) O |
| •••••••••Pseudokeronopsidae (Apoholosticha, Pseudokeronopsis) O |
| •••••••••Pseudourostylidae (Pseudourostyla) O |
| •••••••••Urostylidae (Bakuella, Diaxonella, Urostyla) O C |
| •••••••Oligotrichea |
| ••••••••Oligotrichida |
| •••••••••Cyrtostrombidiidae (Cyrtostrombidium) O |
| •••••••••Pelagostrombidiidae (Pelagostrombidium) O |
| •••••••••Strombidiidae (Strombidium) O |
| •••••••••Tontoniidae (Laboea, Tontonia) O |
| ••••••••Choreotrichida |
| •••••••••Strobilidiina |
| ••••••••••Leegaardiellidae (Leegaardiella) O |
| ••••••••••Lohmanniellidae (Lohmanniella) O |
| ••••••••••Strobilidiidae (Strobilidium) O |
| ••••••••••Strombidinopsidae (Strombidinopsis) O |
| •••••••••Tintinnina (particle size ingested is determined by the lorica diameter) |
| ••••••••••Ascampbelliellidae (Ascampbelliella) O |
| ••••••••••Cyttarocylididae (Cyttarocylis) O |
| ••••••••••Dictyocystidae (Dictyocysta) O |
| ••••••••••Epiplocylididae (Epiplocylis) O |
| ••••••••••Eutintinnidae (Dartintinnus, Eutintinnus) O |
| ••••••••••Favellidae (Favella) O |
| ••••••••••Nolaclusiliidae (Nolaclusilis) O |
| ••••••••••Petalotrichidae (Petalotricha) O |
| ••••••••••Ptychocylididae (Cymatocylis, Ptychocylis) O |
| ••••••••••Rhabdonellidae (Metacylis, Rhabdonella, Schmidingerella O |
| ••••••••••Stenosemellidae (Stenosemella) O |
| ••••••••••Tintinnidae (Amphorellopsis, Salpingacantha, Salpingella, Tintinnus) O |
| ••••••••••Tintinnidiidae (Tintinnidium) O |
| ••••••••••Undellidae (Undella) O |
| ••••••••••Xystonellidae (Dadayiella, Parafavella, Xystonella) O |
| •••••• Licnophoridae (Licnophora, Prolicnophora) O |
| ••••••Kiitrichidae (Caryotricha, Kiitricha) O |
| •••••Lamellicorticata |
| ••••••Armophorea |
| Incertae sedis Armophorea: Mylestomatidae (Mylestoma) O |
| •••••••Metopida anaerobic‐aerotolerant |
| ••••••••Metopidae (Metopus) O |
| ••••••••Apometopidae (Cirranter, Urostomides) O |
| ••••••••Tropidoatractidae (Palmarella, Tropidoatractus) O |
| •••••••Clevelandellida anaerobic, gut * |
| ••••••••Clevelandellidae (Clevelandella) O |
| ••••••••Inferostomatidae (Inferostoma) O |
| ••••••••Neonyctotheridae (Neonyctotherus) O |
| ••••••••Nyctotheridae (Nyctotherus) O |
| ••••••••Sicuophoridae (Sicuophora) O |
| •••••••Caenomorphidae (Caenomorha) O |
| •••••••Odontostomatida |
| ••••••••Discomorphellidae (Discomorphella) B |
| ••••••••Epalxellidae (Epalxella, Saprodinium) B |
| •••••• Litostomatea |
| •••••••Rhynchostomatia |
| ••••••••Dileptida |
| •••••••••Dileptidae (Apodileptus, Dileptus, Pseudomonilicaryon) P |
| •••••••••Dimacrocaryonidae (Dimacrocaryon, Monomacrocaryon, Rimaleptus) P |
| ••••••••Tracheliidae (Trachelius) O P |
| •••••••Haptoria |
| ••••••••Lacrymariidae (Lacrymaria) P |
| ••••••••Haptorida |
| •••••••••Enchelyodonidae (Enchelyodon, Fuscheria) P |
| •••••••••Homalozoonidae (Homalozoon) P |
| •••••••••Pleuroplitidae (Pleuroplites) P |
| ••••••••Didiniidae (Didinium, Monodinium) P |
| ••••••••Pleurostomatida |
| •••••••••Amphileptidae (Amphileptus) P |
| •••••••••Litonotidae (Litonotus) P |
| •••••••••Kentrophyllidae (Kentrophyllum, Epiphyllum) P |
| ••••••••Spathidiida |
| •••••••••Acropisthiidae (Acropisthium, Chaenea) P |
| •••••••••Actinobolinidae (Actinobolina) P |
| •••••••••Apertospathulidae (Apertospathula) P |
| •••••••••Enchelyidae (Enchelys) P |
| •••••••••Pseudoholophryidae (Pseudoholophrya) P |
| •••••••••Spathidiidae (Spathidium) P |
| •••••••••Trachelophyllidae (Trachelophyllum) P |
| •••••••Helicoprorodontidae (Helicoprorodon) P |
| •••••••Trichostomatia (anaerobic endosymbiont in vertebrates) |
| ••••••••Vestibuliferida (anaerobic endosymbiont in fish) |
| •••••••••Balantidiidae (Balantidium, some X, Neobalantidium) B |
| •••••••••Buetschliidae (Buetschlia) B |
| •••••••••Paraisotrichidae (Paraisotrichia) B |
| •••••••••Protocaviellidae (Protocaviella) B |
| •••••••••Protohalliidae (Protohallia) B |
| •••••••••Pycnotrichidae (Pycnothrix, perhaps Buxtonella) B |
| •••••••Isoendo |
| ••••••••Isotrichidae (Dasytricha, Isotricha) (anaerobic gut * in ungulate ruminants) B |
| ••••••••Entodiniomorphida |
| •••••••••Blepharocorythina (anaerobic gut * in mammals) |
| ••••••••••Blepharocorythidae (Blepharocorys) B |
| ••••••••••Parentodiniidae (Parentodinium) B |
| ••••••••••Pseudoentodiniidae (Pseudoentodinium) B |
| •••••••••Entodiniomorphina |
| ••••••••••Cycloposthiidae (Cycloposthium) B |
| ••••••••••Gilchristidae (Gilchristia) B |
| ••••••••••Ophryoscolecidae (Entodinium, Ophryoscolex, Polyplastron) B |
| ••••••••••Polydiniellidae (Polydiniella) B |
| ••••••••••Rhinozetidae (Rhinozeta) B |
| ••••••••••Spirodiniidae (Spirodinium) B |
| ••••••••••Telamodiniidae (Telamodinium) B |
| ••••••••••Troglodytellidae (Troglodytella) B |
| ••••••••Macropodiniida (anaerobic gut * in fore‐stomach of marsupial macropodids and vombatids) |
| ••••••••••Amylovoracidae (Amylovorax) B |
| ••••••••••Macropodiniidae (Macropodinium) B |
| ••••••••••Polycostidae (Polycosta) B |
| ••••CONTHREEP |
| Incertae sedis CON‐threeP: Askenasia, Cyclotrichiidae, Paraspathidium, |
| Pseudotrachelocercidae, Discotrichidae. B |
| ••••• Phyllopharyngea |
| ••••••Synhymeniida f, cyanobacteria |
| •••••••Nassulopsidae (Nassulopsis) f |
| •••••••Orthodonellidae (Orthodonella, Zosterodasys) f |
| •••••••Scaphidiodontidae (Chilodontopsis, Scaphidiodon) f |
| •••••••Synhymeniidae (Synhymenia) f |
| ••••••Subkinetalia |
| •••••••Cyrtophoria |
| ••••••••Chlamydodontida |
| •••••••••Chilodonellidae (Chilodonella) O C |
| •••••••••Chitonellidae (Chitonella) O C |
| •••••••••Chlamydodontidae (Chlamydodon) O C |
| •••••••••Gastronautidae (Gastronauta) O C |
| •••••••••Kryoprorodontidae (Gymnozoum) O C |
| •••••••••Lynchellidae (Chlamydonella, Lynchella) O C |
| ••••••••Dysteriida |
| •••••••••Dysteriidae (Dysteria, Trochilia) O C |
| •••••••••Hartmannulidae Hartmannula) O C |
| •••••••••Kyaroikeidae (Kyaroikeus) O C |
| •••••••••Plesiotrichopidae (Plesiotrichopus) O C |
| •••••••••Chonotrichia |
| ••••••••••Exogemmida ? |
| •••••••••••Chilodochonidae (Chilodochona) |
| •••••••••••Filichonidae (Filichona) |
| •••••••••••Helichonidae (Heliochona) |
| •••••••••••Lobochonidae (Lobochona) |
| •••••••••••Phyllochonidae (Phyllochona) |
| •••••••••••Spirochonidae (Spirochona) |
| ••••••••••Cryptogemmida ? |
| •••••••••••Actinichonidae (Actinichona) |
| •••••••••••Echinichonidae (Echinichona) |
| •••••••••••Inversochonidae (Inversochona) |
| •••••••••••Isochonidae (Isochona) |
| •••••••••••Isochonopsidae (Isochonopsis) |
| •••••••••••Stylochonidae (Stylochona) |
| •••••••Rhynchodia |
| ••••••••Hypocomidae (Hypocoma) S, X in marine invertebrates |
| ••••••••Rhynchodida |
| •••••••••Ancistrocomidae (Ancistrocoma) C or X in marine invertebrates |
| •••••••••Sphenophryidae (Sphenophrya) C or X in marine invertebrates |
| •••••••Suctoria (P larger species can capture small invertebrates) |
| ••••••••Exogenida |
| •••••••••Allantosomatidae (Allantosoma) C P |
| •••••••••Dentacinetidae (Dentacineta) C P |
| •••••••••Dendrosomididae (Dendrosomides) C P |
| •••••••••Ephelotidae (Ephelota) C P |
| •••••••••Lecanophryidae (Lecanophrya) C P |
| •••••••••Metacinetidae (Metacineta) C P |
| •••••••••Manuelophryidae (Manuelophrya) C P |
| •••••••••Ophryodendridae (Ophryodendron) C P |
| •••••••••Paracinetidae (Paracineta) C P |
| •••••••••Phalacrocleptidae (Phalacrocleptes) |
| •••••••••Podophryidae (Podophrya) C P |
| •••••••••Praethecacinetidae (Praethecacineta) C P |
| •••••••••Rhabdophryidae (Rhabdophrya) C P |
| •••••••••Severonidae (Severonis) C P |
| •••••••••Spelaeophryidae (Spelaeophrya) C P |
| •••••••••Tachyblastonidae (Tachyblaston) C P |
| •••••••••Thecacinetidae (Thecacineta) C P |
| ••••••••Endogenida |
| •••••••••Acinetidae (Acineta) C P |
| •••••••••Acinetopsidae (Acinetopsis) C P |
| •••••••••Choanophryidae (Choanophrya) C P |
| •••••••••Corynophryidae (Corynophrya) C P |
| •••••••••Dactylostomatidae (Dactylostoma) C P |
| •••••••••Dendrosomatidae (Dendrosoma) C P |
| •••••••••Endosphaeridae (Endosphaera) C P |
| •••••••••Erastophryidae (Erastophrya) C P |
| •••••••••Pseudogemmidae (Pseudogemma) C P |
| •••••••••Rhynchetidae (Rhyncheta) C P |
| •••••••••Solenophryidae (Solenophrya) C P |
| •••••••••Tokophryidae (Tokophrya) C P |
| •••••••••Trichophryidae (Trichophrya) C P |
| ••••••••Evaginogenida |
| •••••••••Cometodendridae (Cometodendron) C P |
| •••••••••Cyathodiniidae (Cyathodinium) C P |
| •••••••••Dendrocometidae (Dendrocometes) C P |
| •••••••••Discophryidae (Discophrya) C P |
| •••••••••Enchelyomorphidae (Enchelyomorpha) C P |
| •••••••••Heliophryidae (Heliophrya) C P |
| •••••••••Periacinetidae (Periacineta) C P |
| •••••••••Prodiscophryidae (Prodiscophrya) C P |
| •••••••••Rhynchophryidae (Rhynchophrya) C P |
| •••••••••Stylocometidae (Stylocometes) C P |
| •••••••••Trypanococcidae (Trypanococcus) C P |
| ••••• Colpodea |
| ••••••Bursariomorphida |
| •••••••Bryometopidae (Bryometopus, Thylakidium) P |
| •••••••Bursaridiidae (Bursaridium, Paracondylostoma) P |
| •••••••Bursariidae (Bursaria) P |
| ••••••Colpodida (smaller species <20 µm are probably ingesting primarily bacteria) |
| Incertae sedis Colpodida: Bardeliellidae, Hausmanniellidae, Ilsiellidae, Marynidae, Pseudochlamydonellidae. O |
| •••••••Bryophryina |
| ••••••••Bryophryidae (Bryophrya) O |
| ••••••••Sandmanniellidae (Sandmanniella) O |
| •••••••Colpodina |
| ••••••••Colpodidae (Colpoda) O |
| ••••••••Grandoriidae (Grandoria) O |
| ••••••••Tillinidae (Tillina) O |
| ••••••• Grossglockneriidae (Grossglockneria, Pseudoplatyophrya) F |
| ••••••Cyrtolophosidida |
| •••••••Cyrtolophosididae (Cyrtolophosis) O |
| •••••••Kreyellidae (Kreyella) O |
| ••••••Platyophryida (C ingest small green algae) |
| •••••••Ottowphryidae (Ottowphrya, Platyophryides) C |
| •••••••Platyophryidae (Platyophrya) C |
| •••••••Sagittariidae (Sagittaria) C |
| •••••••Sorogenidae (Sorogena) C |
| •••••••Woodruffiidae (Etoschophrya, Rostrophrya, Woodruffia) C |
| ••••• Nassophorea |
| ••••••Colpodidiidae (Colpodidium) B |
| ••••••Nassulida (f, cyanobacteria) |
| •••••••Furgasoniidae (Furgasonia, Wolfkosia) f |
| •••••••Nassulidae (Nassula, Obertrumia) f |
| ••••••Microthoracida |
| •••••••Microthoracidae (Drepanomonas, Microthorax) f |
| •••••••Leptopharyngidae (Pseudomicrothorax, Leptopharynx B) f |
| ••••• Prostomatea |
| •••••• Apsiktratidae (Apsikrata) O |
| ••••••Prorodontida |
| •••••••Balanionidae (Balanion) C D |
| •••••••Cryptocaryonidae (Cryptocaryon) C D |
| •••••••Colepidae (Coleps, Plagiopogon) C D |
| •••••••Holophryidae (Holophrya) C D |
| •••••••Lagynidae (Lagynus) C D |
| •••••••Metacystidae (Metacystis, Vasicola) C D |
| •••••••Placidae (Placus) C D |
| •••••••Plagiocampidae (Plagiocampa) C D |
| •••••••Prorodontidae (Prorodon) C D |
| •••••••Urotrichidae (Urotricha) C D |
| ••••• Plagiopylea |
| ••••••Plagiopylida |
| •••••••Epalxellidae (Epalxella) B |
| •••••••Plagiopylidae (Plagiopyla) B |
| •••••••Sonderiidae (Sonderia) B |
| •••••••Trimyemidae (Trimyema) B |
| ••••• Oligohymenophorea |
| ••••••Apostomatia |
| •••••••Apostomatida |
| ••••••••Colliniidae (Collinia, Metacollinia) S |
| ••••••••Cyrtocaryidae (Cyrtocaryum) S |
| ••••••••Foettingeriidae (Foettingeria) S |
| ••••••••Pseudocolliniidae (Fusiforma, Pseudocollinia) S |
| •••••••Astomatophorida |
| ••••••••Opalinopsidae (Opalinopsis) S |
| •••••••Pilisuctorida |
| ••••••••Conidophryidae (Conidophrys) S |
| ••••••Astomatia |
| •••••••Anoplophryidae 910 (Anoplophrya) S |
| •••••••Buetschliellidae (Buetschliella) S |
| •••••••Clausilocolidae (Clausilocola) S |
| •••••••Contophryidae (Contophyra) S |
| •••••••Haptophryidae (Haptophrya) S |
| •••••••Hoplitophryidae (Hoplitophrya) S |
| •••••••Intoshellinidae (Intoshellina) S |
| •••••••Maupasellidae (Maupasella) S |
| •••••••Radiophryidae (Radiophrya) S |
| ••••••Hymenostomatia |
| •••••••Tetrahymenida (verify each species life cycle, macrostome stage is C) |
| Incertae sedis Tetrahymenida: Trichospiridae (Trichospira) B P* |
| ••••••••Curimostomatidae (Curimostoma) B P* |
| ••••••••Glaucomidae (Glaucoma) B P* |
| ••••••••Spirozonidae (Spirozona) B P* |
| ••••••••Tetrahymenidae (Tetrahymena) B P* |
| ••••••••Turaniellidae (Colpidium, Dexiostoma, Turaniella) B P* |
| •••••••Ophryoglenida |
| ••••••••Ichthyophthiriidae (Ichthyophthirius) P* P |
| ••••••••Ophryoglenidae (Ophryoglena) P* P |
| ••••••Peniculia |
| •••••••Peniculida |
| ••••••••Clathrostomatidae (Clathrostoma) O |
| ••••••••Frontoniidae (Disematostoma, Frontonia) O |
| ••••••••Lembadionidae (Lembadion) O |
| ••••••••Maritujidae (Marituja) O |
| ••••••••Neobursaridiidae (Neobursaridium) O |
| ••••••••Parameciidae (Paramecium) O |
| ••••••••Paranassulidae (Paranassula) O |
| ••••••••Stokesiidae (Stokesia) O |
| •••••••Urocentridae (Urocentrum) O |
| ••••••Peritrichia |
| •••••••Sessilida |
| ••••••••Astylozoidae (Astylozoon, Hastatella) O |
| ••••••••Ellobiophryidae (Ellobiophrya) O |
| ••••••••Epistylididae (Epistylis) O |
| ••••••••Lagenophryidae (Lagenophrys) O |
| ••••••••Operculariidae (Opercularia) O |
| ••••••••Rovinjellidae (Rovinjella) O |
| ••••••••Scyphidiidae Kahl, 133 (Scyphidia) O |
| ••••••••Termitophryidae (Termitophrya) O |
| ••••••••Usconophryidae (Usconophrys) O |
| ••••••••Vaginicolidae (Cothurnia, Pyxicola, Thuricola, Vaginicola) O |
| ••••••••Vorticellidae (Carchesium, Vorticella) O |
| ••••••••Zoothamniidae (Haplocaulus, Zoothamnium) O |
| •••••••Mobilida |
| ••••••••Polycyclidae (Polycycla) O |
| ••••••••Trichodinidae (Trichodina) O |
| ••••••••Trichodinopsidae (Trichodinopsis) O |
| ••••••••Urceolariidae (Leiotrocha, Urceolaria) O |
| ••••••Scuticociliatia |
| •••••••Philasterida |
| ••••••••Cohnilembidae (Cohnilembus) O |
| ••••••••Cryptochilidae (Cryptochilum.) O |
| ••••••••Entodiscidae (Entodiscus) O |
| ••••••••Entorhipidiidae (Entorhipidium) O |
| ••••••••Orchitophryidae (Orchitophrya) O |
| ••••••••Paralembidae (Anophrys, Paralembus) O |
| ••••••••Parauronematidae (Parauronema) O |
| ••••••••Philasteridae (Kahlilembus, Philaster) O |
| ••••••••Pseudocohnilembidae (Pseudocohnilembus) O |
| ••••••••Schizocaryidae (Schizocaryum) O |
| ••••••••Thigmophryidae (Thigmophrya) O |
| ••••••••Thyrophylacidae (Thyrophylax) O |
| ••••••••Uronematidae (Uronema) C |
| ••••••••Urozonidae (Urozona) O |
| •••••••Pleuronematida |
| ••••••••Calyptotrichidae (Calyptotricha) O |
| ••••••••Conchophthiridae (Conchophthirus) O |
| ••••••••Ctedoctematidae (Ctedoctema) O |
| •••••••• Cyclidiidae (Cristigera, Cyclidium) O |
| ••••••••Dragescoidae (Dragescoa) O |
| ••••••••Eurystomatellidae (Eurystomatella) O |
| ••••••••Histiobalantiidae (Histiobalantium) O |
| ••••••••Peniculistomatidae (Peniculistoma) O |
| ••••••••Pleuronematidae (Pleuronema) O |
| ••••••••Thigmocomidae (Thigmocoma) O |
| •••••••Thigmotrichida |
| ••••••••Ancistridae (Ancistrum) B |
| ••••••••Hemispeiridae (Hemispeira) B |
| ••••••••Hysterocinetidae (Hysterocineta) B |
| ••••••••Paraptychostomidae (Paraptychostomum) B |
| •••••••Loxocephalida |
| ••••••••Cinetochilidae (Cinetochilum, Sathrophilus) B |
| ••••••••Loxocephalidae (Cardiostomatella, Dexiotricha, Loxocephalus) B |
Appendix 3. Table 3.1.
Protist names and common names for East Asian translations
| Higher level ranks and supergroups | Phylum | Important sub‐divisions in Chinese characters | Common name translation |
|---|---|---|---|
| Amoebozoa | Tubulinea | Corycida (皮殼葉状根足綱) 皮壳管状根足纲 | Leathery‐shell amoebae |
| Echinamoebida (多針葉状根足綱) 多针管状根足纲 | Amoebae with spiny pseudopodia | ||
| Elardia (三組葉状根足綱) 三组管状根足纲 | Amoeboid group including three groups (Euamoebida, Leptomyxida, Arcellinida) | ||
| Arcellinida (有殼葉状變形綱) 有壳管状根足纲 | Testate amoebae partially enclosed in a simple shell | ||
| Evosea | Variosea (多圓錐型 根足綱) 多圆锥型根足纲 | Amoebae with many conical‐shaped pseudopodia | |
| Eumycetozoa (眞菌根足綱) 真粘菌根足纲 | Genuine (mushroom) slime mold amoebae | ||
| Cutosea (眞皮根足綱) 真皮根足纲 | Amoebae with a discrete skin | ||
| Archamoebea (原始根足綱) 古变形纲 | Ancient (first) amoebae | ||
| Discosea | Stygamoebida (細片根足綱) 细片根足纲 | Amoebae with pseudopodia resembling tooth‐pick or splinters | |
| Centramoebia (中心體根足綱) 中心体根足纲 | Centrosome‐bearing amoebae | ||
| Incertae sedis Holozoa | Filasterea (星狀絲足綱) 星状丝足纲 | Rounded amoebae with pseudopodia like fingers | |
| Ichtyosporea (孢子型魚病根足綱) 孢子型鱼病根足纲 | Fish pathogenic amoebae forming a spore | ||
| Choanoflagellata (立襟鞭毛綱) 领鞭毛纲 | Flagellates with a collar‐like ring | ||
| Porifera | Demospongiae (普通海綿綱) 普通海绵纲 | Common sponges | |
| Homoslceromorpha (同骨海綿綱) 同骨海绵纲 | Sponge with undifferentiated cytoskeleton | ||
| Calcarea (石灰海綿綱) 石灰海绵纲 | Chalk sponges | ||
| Hexactinellida (玻璃海綿綱) 玻璃海绵纲 | Glass sponges | ||
| Nucletmycea | Opisthosporidia | Aphelidea (藻寄生性根足綱) 藻寄生性根足纲 | Amoeboid endobiotic parasitoids of algae |
| Microsporidia (原始寄生性擬菌綱) 原始寄生拟菌纲 | Primitive fungus‐like parasite | ||
| Blastocladiales(厚壁囊菌綱) 厚壁囊菌纲 | Fungi having a thick‐walled resting spore | ||
| Neocallimastigaceae (新多鞭毛菌綱) 新美鞭菌纲 | New fungi having many/pretty flagella | ||
| Chytridiomycota (壺狀菌綱) 壶菌纲 | Fungi resembling a broken‐cracked egg | ||
| Mucoromycota (粘液菌綱) 粘液菌纲 | Mucoid or sugar fungi | ||
| Zoopagomycota (動物生菌綱) 捕虫霉纲 | Fungi growing on animals, Fungi able to catch bugs | ||
| Taphrinomycotina (外囊菌綱) 外囊菌纲 | Fungi having an outer ascus | ||
| Saccharomycetales (酵母菌綱) 酵母菌纲 | Fungi associated with fermentation | ||
| Pezizomycotina (周鉢菌綱) 盘菌纲 | (Bowl‐shaped fungi), plate‐shaped fungi | ||
| Agaricomycotina (擔子菌綱) 担子菌纲 | Fungi forming a sterigma | ||
| Pucciniomycotina (銹菌綱) 柄锈菌纲 | Fungi with stem and is associated with rust disease | ||
| Ustilaginomycotina (黑穗菌綱) 黑穗菌纲 | Fungi associated with smut disease | ||
| Wallemiomycotina (無子實體菌綱) 无子实体菌纲 | Fungi without palisade of basidia | ||
| Rhodophyceae | Proteorhodophytina (原始紅藻綱) 原始红藻纲 | Primitive red algae | |
| Eurhodophytina (眞正紅藻綱) 真红藻纲 | Genuine red alage, which have two phases or three phases | ||
| Chloroplastida | Chlorophyta (綠藻綱) 绿藻纲 | Green algae | |
| Charophyta (輪藻綱) 轮藻纲 | Wheel‐shaped green algae | ||
| Bigyra | Nanomonadea (矮小鞭毛綱) 微小鞭毛纲 | Tiny brown flagellates | |
| Opalinata (皮下共生鞭毛綱) 蛙片虫纲 | Animal endobionts; Flagellates mostly found in frog, with a flat shape like a slice | ||
| Placidida (小突起鞭毛綱) 小突起鞭毛纲 | Flagellate having a papilla | ||
| Bicosoecida (毫髮鞭毛綱) 毫发鞭毛纲 | Flagellates having tiny flagellar hairs | ||
| Labyrinthulomycetes (網形鞭毛綱) 网丝鞭毛纲 | Gliding flagellates producing a network of filaments | ||
| Pseudophyllomitidae (非附着鞭毛綱) 无附着鞭毛纲 | Flagellates lacking the two adhering flagella | ||
| Gyrista | Developea (後固着鞭毛綱) 后附鞭毛纲 | Flagellates with adhering posterior flagellum | |
| Hyphochytriales (菌絲體鞭毛綱) 菌丝体鞭毛纲 | Flagellates having hypha‐like structures | ||
| Peronosporomycetes (植物病原體擬菌綱) 植病拟菌纲 | Fungus‐like plant pathogens | ||
| Pirsoniales (硅藻寄生綱) 硅藻寄生纲 | Diatom parasites | ||
| Actinophryidae (太陽綱) 太阳纲 | Sunlight (or star light)‐shaped protozoa | ||
| Chrysophyceae (黃褐藻綱) 金藻纲 | Golden brown algae | ||
| Eustigmatales (眞眼點褐藻綱) 真眼点褐藻纲 | Brown algae with a big eye spot | ||
| Phaeophyceae (大褐藻綱) 褐藻纲 | The large brown algae | ||
| Phaeothamniophyceae (黃赤藻綱) 褐枝藻纲 | Yellow‐red algae with branches | ||
| Raphidophyceae (針鞭毛藻綱) 针胞藻纲 | Algae with needle shaped flagellum | ||
| Xanthophyceae (黃綠藻綱) 黄绿藻纲 | Yellow‐green algae | ||
| Bolidomonas (迅游泳藻綱) 迅游藻纲 | Diatom‐like algae with rapid swimming | ||
| Diatomeae | Diatomeae (硅褐藻綱) 硅褐藻纲 | Brown algae like glass box with lid | |
| Dictyochophyceae (硅質鞭毛藻綱) 硅鞭藻纲 | Algae producing a siliceous skeleton | ||
| Pelagophyceae (浮生褐藻綱) 浮生褐藻纲 | Filamentous brown algae living in the sea; brown algae living a planktonic habitat | ||
| Pinguiophyceae (脂褐藻綱) 脂褐藻纲 | Brown algae containing (a high concentration of) fatty acids | ||
| Incertae sedis Alveolata | Colpodellida (捕食性鞭毛綱) 捕食鞭毛纲 | Predatory flagellates | |
| Dinoflagellata | Syndiniales (共生性渦鞭毛藻綱) 共生涡鞭藻纲 | Parasitic dinoflagellates, symbiotic dinoflagellates | |
| Noctilucales (夜光藻綱) 夜光藻纲 | Algae with bioluminescence | ||
| Dinophyceae (渦鞭毛藻綱) 涡鞭藻纲 | Algae with spiraling motility, algal flagellates with a spiral or girdle groove | ||
| Apicomplexa | Aconoidasida (無圓錐頂端複合體綱) 无圆锥体顶复纲 | Apicomplexa lacking a conoid | |
| Conoidasida (圓錐頂端複合體綱) 圆锥体顶复纲 | Apicomplexa possessing a conoid | ||
| Ciliophora | Karyorelictea (核殘跡纖毛綱) 核残迹纤毛纲 | Ciliophores having relict of parents’ macronulei | |
| Heterotrichea (異毛纖毛綱) 异毛纤毛纲 | Ciliophores with different length of flagella | ||
| Spirotrichea (型 旋纖毛綱) 旋唇纤毛纲 | Ciliophores with spiraling adoral zone of membranelles | ||
| Armophorea (鬪帽纖毛綱) 盔帽纤毛纲 | Ciliophores having the appearance of military helmets | ||
| Litostomatea (裂口纖毛綱) 裂口纤毛纲 | Ciliophores with a cytostome with oral dome | ||
| Phyllopharyngea (葉咽纖毛綱) 叶咽纤毛纲 | Ciliophores with a leaf‐shaped cytopharynx | ||
| Colpodea (腎形纖毛綱) 肾形纤毛纲 | Ciliophores with a kidney shape | ||
| Prostomatea (前口纖毛綱) 前口纤毛纲 | Ciliophores with cytostome at the anterior pole | ||
| Plagiopylea (斜毛纖毛綱) 斜毛纤毛纲 | Ciliophores with oblique slit flagella | ||
| Oligohymenophorea (貧膜纖毛綱) 寡膜纤毛纲 | Ciliophores with a small paroral membrane | ||
| Nassophorea (篮口纖毛綱) 篮口纤毛纲 | Ciliophores showing a basket‐shaped oral structure | ||
| Cercozoa | Silicofilosea (硅質絲狀根足綱) 硅质丝足纲 | Filose amoebae covered by siliceous or glass scales, vase‐shaped shell | |
| Foraminifera | Monothalamea (單房室有孔綱) 单房室有孔纲 | Foraminiferans with single chamber test | |
| Tubothalamea (管狀有孔綱) 管状有孔纲 | Foraminiferans with tubular chamber test | ||
| Lagenida (單層有孔綱) 瓶状有孔纲 | Foraminiferans with monolamellar test | ||
| Radiolaria | Acantharia (放射棘綱) 等辐骨纲 | Protozoa with axopods and filopodia; protist with radiated spicula of same length | |
| Polycystinea (多孔囊綱) 多孔纲 | Protozoa (with a sac) covered by many pores | ||
| Metamonada | Fornicata (拱門形纖維綱) 拱形纤维纲 | Protozoa with an arched B‐fiber | |
| Parabasalia (副基体綱) 副基体纲 | Protozoa with one or more parabasal apparatus | ||
| Preaxostyla (二重纖維綱) 二重纤维纲 | Protozoa with I‐fiber with double‐cross matrix | ||
| Discoba | Jakobida (單背翼綱) 单背翼纲 | Protozoa with a single dorsal vane in the posterior flagellum | |
| Heterolobosea (噴出形根足綱) 异叶足纲 | Amoebae with eruptive pseudopodia; amoebae with differentiated leaf‐shaped pseudopodia | ||
| Euglenozoa | Kinetoplastid (運動核質鞭毛類) 动质体纲 | Protozoa with kinetoplast | |
| Euglenid (軟豆鞭毛類) 软豆鞭毛类 | Yellow‐green flagellates | ||
| Haptophyta | Prymnesiophyceae (碳酸鑛物化藻綱) 碳酸质鳞片藻纲 | Algae with CO2‐mineralized scales | |
| Coccolithophorid (圓石藻類) 颗石藻类 | Haptophyte with calcareous (chalk) scales | ||
| Centroplasthelida | Pterocystida (無外骨格鞭毛綱) 无外骨骼鞭毛纲 | Protozoa without any exoskeletal elements | |
| Panacanthocystida (石鱗鞭毛綱) 石鳞鞭毛纲 | Protozoa with siliceous scales or with organic spicules | ||
| Cryptista | Cryptophyceae (隱鞭毛藻型) 隐藻纲 | New algae with prominent ejectisomes; cryptic algae |
Acknowledgments
After the first author, D. Bass, C.E Lane, J. Lukeš, C. L. Schoch and A. Smirnov have contributed equally and are to be considered second authors; subsequent authors are listed alphabetically and are to be considered third authors.
We were saddened and hurt by the untimely loss of two dear colleagues, D.H. Lynn and J. Clamp, both ciliatologists.
Research support was provided as follows: SMA by NSERC 249889‐2007; DB by NERC NE/H009426/1 and NE/H000887/1; MWB by NSF 1456054; FB by a Fellowship from Science for Life Laboratory and VR/2017‐04563; PC by EU‐Horizon 2020 research and innovation program through the SponGES project 679849 (This document reflects only the authors’ view and the Executive Agency for Small and Medium‐sized Enterprises (EASME) is not responsible for any use that may be made of the information it contains); IC by CSF 18‐18699S; BE by RCN TaxMArc 268286/GMR; LG by ANR HAPAR (ANR‐14‐CE02‐0007); VH MK JL by ERDF; MEYS with ERC 771592 CZ 1.05/1.1.00/02.0109 BIOCEV; SK by RSF 16‐14‐10302; MK by CSF GA18‐28103S; CEL by NSF 1541510 and NIH‐AI124092; EL by CAM: 2017‐T1/AMB‐5210; and by grant 2017‐T1/AMB‐5210 from the program "Atracción de talentos" from the Consejería de Educación, Juventud y Deporte, Comunidad de Madrid; JL by ERC CZ LL1601 and OPVVV 16_019/0000759; MP by NSF DEB‐1455611; DJR by the Beatriu de Pinós postdoctoral programme of the Government of Catalonia's Secretariat for Universities and Research of the Ministry of Economy and Knowledge; CLS by the intramural research program of the National Library of Medicine, National Institutes of Health; AS by RSF 17‐14‐01391 and RFBR 16‐04‐01454 NY by NSF DEB 1557102; VZ by RFBR 16‐34‐60102 mol‐a‐dk; UniEuk and EukRef by the Gordon and Betty Moore Foundation.
We thank numerous colleagues who were consulted ad hoc throughout this process. In addition, we specifically thank Alexander Ereskovsky (CNRS, Station marine d'Endoume, Marseille, France) for help with the sponges; and Iñaki Ruiz‐Trillo (ICREA ‐ Institut de Biologia Evolutiva, CSIC‐Universitat Pompeu Fabra, Barcelona, Catalonia, Spain) with the Holozoa; David S. Hibbett (Biology Department, Clark University, Worcester, MA USA, and Radcliffe Institute for Advanced Study, Harvard University, Cambridge, MA) with the Holomycota; Isabelle Florent (Institut de Systématique, Évolution, Biodiversité, Muséum National d'Histoire Naturelle, Sorbonne Universités, Paris, France) with Apicomplexa; Shauna Murray (Climate Change Cluster, University of Technology Sydney, Australia), Albert Reñé (Dept. Biologia Marina i Oceanografia, Institut de Ciències del Mar, CMIMA (CSIC), Barcelona, Spain) and Nicolas Chomérat (IFREMER, ODE/UL/LER Bretagne Occidentale, Concarneau, France) for dinoflagellate primers and barcoding; Urban Tillmann (Alfred Wegener Institut, Helmholz‐Zentrum für Polar‐ und Meeresforschung, Bremerhaven, Germany) and Per Juel Hansen (Marine Biological Section, Dept. of Biology, University of Copenhagen, Denmark) for the dinoflagellate literature and functional assignments; William Bourland (Biology, Boise State University) for discussions on ciliates; Alastair Simpson (Dalhousie University) for discussions on higher level ranking and structure; Angela Mele (Philadelphia) for the cover art.
Footnotes
The ability of a unicellular organism or a cell type in a multicellular organism to actively change the conformation of the entire cell body by extending and retracting pseudopodia; pseudopodia are used for cell movement over the substratum and/or for feeding.
Monolayer of scales covering the cell adhering to the substratum from the dorsal surface; the ventral surface of the cell remains free. Known in amoebae of the genus Cochliopodium.
Layer of fibrous material covering the cell, adhering to the substratum from the dorsal surface; the ventral surface remains free. Known in amoebae of the genera Gocevia, Paragocevia and Ovalopodium.
Single amoeboid cell differentiates into a usually stalked, subaerial structure that supports one to many propagules termed spores. As defined here, this kind of sporocarp has only ever been observed in Amoebozoa and is potentially synapomorphic for Amoebozoa. Should this prove the case, non‐sporocarpic amoebozoans are the products of reductive evolution.
Amoebae aggregate into a multicellular mass that develops into a multicellular, subaerial fruiting body consisting of either distinct stalk cells and spores or non‐differentiated encysted cells (usually also called spores). Sorocarpic development is found in two lineages of amoebozoans, the Dictyostelia (Eumycetozoa) and in Copromyxa (Tubulinea).
The species Schoutedamoeba minuta described by Van Vichelen et al. (2016) has a hartmannelid morphology (monopodial cells with pronounced frontal hyaline cap) but in SSU tree it shows affinities with Variosea, although with no support. More robust data are necessary to clarify its position among Amoebozoa.
The taxon name Stereomyxa ramosa is used in Tekle et al. (2016) and Tekle and Wood (2017) for an isolate that is a distinct genus named Dracoamoeba (see Tice et al. 2016). To date, no molecular data on a true Stereomyxa are available and thus it remains incertae sedis.
The name Unda is used in Tekle et al. (2016) as well as in Tekle and Wood (2017) for an isolate of Vannella as noted in Cavalier‐Smith et al. (2016) and Kang et al. (2017).
Variable cell projections, smooth in outline, with rounded tips, which participate in the relocation of the main cytoplasmic mass of the cell and include both the granuloplasm and the hyaloplasm (sensu Smirnov 2008).
Schaudinn (1899) reported a complex life cycle in Trichosphaerium (Corycida) that included biciliated stages, which undergo copulation; no further confirmation of this observation has been obtained.
Strain numbers and source data for these isolates are provided by Kang et al. (2017).
The genus Atrichosa by Cavalier‐Smith et al. 2016 is considered here a junior synonym of Trichosphaerium until the opposite is shown. The position of the genera Penardochlamys, Microcorycia, Zonomyxa and Parmulina, which were listed by Meisterfeld (2002) under “Microcoryciidae” is not clear; by their morphological characters they may belong to this lineage as well but this requires demonstration by molecular data.
The SSU rRNA sequence of Trigonopyxis arcula AY848967 is almost identical to Bullinularia indica AY848970, and represents probably a contamination.
This species typically positioned as sister clade of both Sphaerothecina and Difflugina in beta‐tubulin (Lahr et al. 2011), SSU rRNA (Lara et al. 2008) and multigene phylogenies (Lahr et al. 2013), so it is listed here as a separate lineage.
In the most complete version spores from a sporocarp germinate as ciliated‐amoebae, cells that are reversibly amoeboid or ciliated then go on to develop into an obligate amoeboid stage that cannot produce cilia, with the obligate amoeba differentiating into one or more sporocarps. However, some members are always ciliated or obligate amoebae. The kinetid structures of swimming stages are diagrammed in Spiegel et al. (2017), Mikryukov and Mylnikov (1998), Hibberd (1983), and Pánek et al. (2016).
Recent papers with relatively broad taxon sampling, based on SSU phylogeny (Berney et al. 2015) and multigene phylogeny (Kang et al. 2017) suggest grouping of some variosean genera into higher rank clades. However, SSU phylogeny shows little or no statistical support for many of these groupings, while many important variosean taxa are not yet represented in the multigene trees. Therefore, we prefer to be cautious about including certain higher level taxa proposed in these studies at this time. Hence, we list most variosean clades under the similarly high level regardless of the traditional ranks until more robust groups are established.
The genus Planoprotostelium is subsumed into Protostelium (Shadwick et al. 2017).
This is the only group revealed in the paper by Lahr et al. (2011) which we suggest to apply because it is fully supported in a phylogenomic study by Kang et al. (2017) and combines several monotypic lineages; many of them group with each other in SSU trees as well.
Coenonia was seen only once by Van Tieghem (1884) who described but never illustrated it. On the basis of his description, it seems reasonable to conclude it was a dictyostelid.
Many taxa in need of revision because of rampant paraphyly.
The species Mycamoeba gemmipara in Blandenier et al. (2017) groups with Dermamoeba; however. the phylogenetic analysis in this paper uses a limited number of taxa and does not show Dermamoebida as a clade, so we list this genus as a separate branch until it position is better resolved.
Two known species of this genus—J. chaetognathi (Grassi 1881) and J. pigmentifera (Grassi 1881) are not triangular but monopodial in locomotion which may be a consequence of their parasitic life style. No molecular data on this genus are available, so it is left among the Dactylopodida provisionally, basing on the presence of a kinetoplastid intracellular symbiont (PLO), which appeares to have originated only once in the evolution of paramoebids (Sibbald et al. 2017).
This genus may be a junior synonym of Vannella, being a life form of some Vannella species.
These genera group together in some SSU phylogenetic trees, but usually they appear as separate branches in phylogenetic studies. In the phylogenomic study of Kang et al. (2017) they do not form a clade, but this is the only region of the tree that is not fully supported. Taking into account the superficial similarity of the ultrastructure, including the unique shape of the mitochondrial cristae, in these two genera we suggest keeping this assemblage as a potential branch in the tree unless the opposite is proven with increased taxon sampling.
Teretosporea Torruella et al. 2015 (R) is a monophyletic clade consisting of at least Corallochytrium and Ichthyosporea; Pluriformea Hehenberger et al. 2017 (R) is a monophyletic clade consisting of at least Corallochytrium and Syssomonas. These are two competing phylogenetic hypotheses.
Choanomonada was an unfortunate error in spelling that was corrected in an erratum, Journal of Eukaryotic Microbiology 60 (3): 321, published online March 11, 2013.
The clade that comprises the Metazoa and Choanoflagellata is called Choanozoa Brunet and King 2017 [Choanozoa Cavalier‐Smith et al. 1991].
The clade comprising the Porifera, Trichoplax, Cnidaria, Ctenophora, and Bilateria is called Metazoa, Metazoa Haeckel 1874.
Although monotypic, genetic diversity of isolates indicate there are probably multiple genera, and it is probably a sister clade to Cnidaria (Schierwater and DeSalle 2018; and Srivastava et al. 2008).
Nucletmycea and Holomycota were proposed at about the same period and they are to be considered synonymous, but Nucletmycea has nomenclatural priority having been published first. Since, Holomycota has come into more usage in favour of the symmetry with Holozoa.
There is no agreement on the definition, thus placement, of a clade named Fungi. Variations differ from the historical understanding of Fungi that included social amoebae, now dispersed across protists, and several other clades also dispersed across protists.
Holomycota is the sister lineage to Holozoa and is comprised of Fungi and the group Rotosphaerida containing nucleariids. In this classification, we include Opisthosporidia (Karpov et al. 2014), comprising the endoparasitic lineages Rozellida (Cryptomycota) Aphelida and Microsporidia, in the Fungi, noting that this is an ongoing point of contention. New genomic and transcriptomic data provide modest to poor support for the monophyly of an Opisthosporidia clade (Toruella et al. 2018), but multiple molecular studies nonetheless support the placement of all three lineages at the base of the Fungi (Torruella et al. 2019; Karpov et al. 2017a, 2017b; Letcher et al. 2017; Lopez‐Escardo et al. 2018). A major point of contention has been the fact that Opisthosporidia all have a vegetative phase that is unbounded by a cell wall, unlike most Fungi, and that Aphelida and Rozella have an intracellular (parasitic) phagotrophic feeding mode (Karpov et al. 2014; Powell et al. 2017). On the other hand, all of the Opisthosporidia share with other Fungi a chitinous cell wall at some stage in their life cycle. Recent analysis of an aphelid transcriptome (the lineage that has most retained ancestral characters within opisthosporidians) suggests a genomic composition and metabolism more aligned with the free‐living chytrid fungi with degradative enzymes, than other Opisthosporidia (Toruella et al. 2019). But an alternative interpretation places emphasis on Opisthosporida cell biology which is more similar to other Opisthokonta than to Fungi, because of the phagotrophic nature of their nutrition. Ultimately, additional cell biology and genomic sequencing is needed for these poorly known organisms. Because there is no synapomorphy for the Fungi regardless of whether the Opisthosporidia are included (Richards et al. 2017), determining the constituency of the Fungi will rely on which traits are deemed most relevant (presence of chitinous cell wall or osmoheterotrophy) as both traits have been convergently derived across the eukaryote tree. A similar debate is ongoing about the distinction between Rozellida (Cryptomycota) and Microsporidia (Bass et al. 2018) as some rozellids have a spore structure very similar to Microsporidia but a genomic content less reduced than core microsporidia, and more like other Fungi. The stance taken in this classification is to follow the majority opinion of the mycological community to include Opisthosporidia in the Fungi, noting that this consensus may change with the discovery of new taxa and improved phylogenies.
The placement of Aphelidea in the Opisthosporidia is unstable and may change.
This revision reflects numerous advances in the phylogeny of the diatoms over the last decade. Due to our poor taxon sampling outside of the Mediophyceae and pennate diatoms, and the known and anticipated diversity of all diatoms, many clades appear at a high classification level (and the higher level classification is rather flat). Nomenclature follows the botanical code (ICN). Some of the basal nodes will probably become better resolved in the future, and would permit additional subdivisions. The genera are provided as examples only, and are far from complete lists.
The ordering of the major taxa follows the arrangement of most SSU rRNA phylogenies and recent phylogenomic analyses (e.g. Gentekaki et al., 2014). The current classification contains some polyphyletic or paraphyletic taxa marked by (P) which require further investigations (morphologic and/or genetic). Families are arranged alphabetically, unless phylogeny places a family at a higher hierarchical rank.
A node including a probably monophyletic clade composed of Sainouridae and Thecofilosea is proposed, called Helkesida Cavalier‐Smith 2018 (R).
Ancestral condition probably a gliding heterotrophic cell with two cilia with divergent kinetosomes, and relatively rigid pellicle that helped to define a distinct ventral groove from which filose pseudopods extended, but without the dense extracellular theca of Thecofilosea or internal silica skeleton of Ebriacea and Phaeodaria. Includes Cercozoa with often imbricate silica scales and their closest nonscaly relatives.
Centrohelida Kühn was introduced to include both centrohelids and gymnosphaerids; Febvre‐Chavalier and Febvre (1984) were the first who recognized and named this taxon.
One more genus name—Heteroraphidiophrys, mentioned in Mikrjukov (2002) was never formally introduced and needs to be avoided; the organisms designated needs to be re‐isolated, carefully studied and provided with formal description.
Genera Pseudoraphidiophrys, Pseudoraphidocystis, Pterocystis, Raineriophrys altogether obviously unify a group of closely related organisms which need a careful taxonomic revision involving morphological and molecular data. The genus name Echinocystis sometimes used instead of Raineriophrys is multiply preoccupied and should be avoided.
For more detailed description of the location and nomenclature of SSU rRNA gene expansions in centrohelids see Shishkin et al., 2018.
Separation of this genus from Heterophrys requires further justification.
Cryptista has been used since 1989 with a variety of definitions, and varying in composition and rank. Unless the authority and date is specified, it is uncertain which meaning, or rank, of Cryptista is meant. However, the term has uses in phylogenetic trees and it has been somewhat adopted in the literature, although without precision over its meaning. For nomenclatural stability Cryptista is defined here according to its latest use in phylogenetic trees.
Presently, there is no phylogenetic taxonomy for phagotrophic euglenids as a whole. Traditional systems based on, for example, the presence or absence of conspicuous feeding apparatuses, are artificial. Some possibly monophyletic taxa have been proposed that would cover restricted subsets of the phagotrophic euglenids: for example, the Ploeotiida, including Ploeotia, Entosiphon, Lentomonas, and presumably Keelungia; and the Petalomonadida, including Petalomonas, Notosolenus and Calycimonas, and presumably also taxa such as Sphenomonas. The limited taxon sampling for molecular sequence data is a significant impediment, especially as many traditional genera are probably polyphyletic.
Literature Cited
- Adl, M. S. , Leander, B. S. , Simpson, A. G. B. , Archibald, J. M. , Anderson, O. R. , Barta, J. R. , Bass, D. , Bowser, S. S. , Brugerolle, G. , Farmer, M. A. , Karpov, S. , Kolisko, M. , Lane, C. E. , Lodge, J. , Lynn, D. H. , Mann, D. G. , Meisterfeld, R. , Mendoza, L. , Moestrup, Ø. , Mozley‐Standridge, S. E. , Smirnov, A. V. & Spiegel, F. W. 2007. Diversity, nomenclature and taxonomy of protists. Syst. Biol., 56(4):684–689. [DOI] [PubMed] [Google Scholar]
- Adl, M. S. , Simpson, A. G. B. , Farmer, M. A. , Andersen, R. A. , Anderson, O. R. , Barta, J. R. , Bowser, S. S. , Brugerolle, G. , Fensome, R. A. , Fredericq, S. , James, T. Y. , Karpov, S. , Kugrens, P. , Krug, J. , Lane, C. E. , Lewis, L. A. , Lodge, J. , Lynn, D. H. , Mann, D. G. , McCourt, R. M. , Mendoza, L. , Moestrup, Ø. , Mozley‐Standridge, S. E. , Nerad, T. A. , Shearer, C. A. , Smirnov, A. V. , Spiegel, F. W. & Taylor, F. J. R. 2005. The new classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol., 52(5):399–451. [DOI] [PubMed] [Google Scholar]
- Adl, S. M. , Simpson, A. G. , Lane, C. E. , Lukeš, J. , Bass, D. , Bowser, S. S. , Brown, M. W. , Burki, F. , Dunthorn, M. , Hampl, V. , Heiss, A. , Hoppenrath, M. , Lara, E. , Le Gall, L. , Lynn, D. H. , McManus, H. , Mitchell, E. A. D. , Mozley‐Stanridge, S. E. , Parfrey, L. W. , Pawlowski, J. , Rueckert, S. , Shadwick, L. , Schoch, C. , Smirnov, A. & Spiegel, F. W. 2012. The revised classification of eukaryotes. J. Eukaryot. Microbiol., 59:429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, D. , Czech, L. , Williams, B. A. P. , Berney, C. , Dunthorn, M. , Mahé, F. , Torruella, G. , Stentiford, G. D. & Williams, T. A. 2018. Clarifying the relationships between microsporidia and cryptomycota. J. Euk. Microbiol., 65(6):773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney, C. , Ciuprina, A. , Bender, S. , Brodie, J. , Edgcomb, V. , Kim, E. , Rajan, J. , Wegener Parfrey, L. , Adl, S. , Audic, S. , Bass, D. , Caron, D. A. , Cochrane, G. , Czech, L. , Dunthorn, M. , Geisen, S. , Oliver Glöckner, F. , Mahé, F. , Quast, C. , Kaye, J. Z. , Simpson, A. G. B. , Stamatakis, A. , del Campo, J. , Yilmaz, P. & de Vargas, C. 2017. UniEuk: time to speak a common language in protistology! J. Eukaryot. Microbiol., 64:407–411. 10.1111/jeu.12414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney, C. , Geisen, S. , Van Wichelen, J. , Nitsche, F. , Vanormelingen, P. , Bonkowski, M. & Bass, D. 2015. Expansion of the “reticulosphere”: diversity of novel branching and network‐forming amoebae helps to define Variosea (Amoebozoa). Protist, 166:271–295. [DOI] [PubMed] [Google Scholar]
- Blandenier, Q. , Seppey, C. V. W. , Singer, D. , Vlimant, M. , Simon, A. , Duckert, C. & Lara, E. 2017. Mycamoeba gemmipara nov. gen., nov. sp., the first cultured member of the environmental Dermamoebidae Clade LKM74 and its unusual life cycle. J. Eukaryot. Microbiol., 64:257–265. [DOI] [PubMed] [Google Scholar]
- Brown, M. W. , Heiss, A. A. , Kamikawa, R. , Inagaki, Y. , Yabuki, A. , Tice, A. K. , Shiratori, T. , Ishida, K. , Hashimoto, T. , Simpson, A. G. B. & Roger, A. J. 2018. Phylogenomics places orphan protistan lineages in a novel eukaryotic super‐group. Genome Biol. Evol., 10:427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campo, J. , Kolisko, M. , Boscaro, V. , Santoferrara, L. F. , Massana, R. , Guillou, L. , Simpson, A. G. B. , Berney, C. , de Vargas, C. , Brown, M. , Keeling, P. & Parfrey, L. W. 2018. EukRef: phylogenetic curation of ribosomal RNA to enhance understanding of eukaryotic diversity and distribution. PLOS Biol. 2018;16:e2005849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantino, P. D. 1998. Binomials, hyphenated uninomials, and phylogenetic nomenclature. Taxon 47:425–429. [Google Scholar]
- Cavalier‐Smith, T. , Chao, E. E. & Lewis, R. 2016. 187‐Gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep‐branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution. Mol. Phylogenet. Evol., 99:275–296. [DOI] [PubMed] [Google Scholar]
- De Vargas, C. , Audic, S. , Henry, N. , Decelle, J. , Mahé, F. , Logares, R. , Lara, E. , Berney, Ć. , Le Bescot, N. , Probert, I. , Carmichael, M. , Poulain, J. , Romac, S. , Colin, S. , Aury, J.‐M. , Bittner, L. , Chaffron, S. , Dunthorn, M. , Engelen, S. , Flegontova, O. , Guidi, L. , Horák, A. , Jaillon, O. , Lima‐Mendez, G. , Lukeš, J. , Malviya, S. , Morard, R. , Mulot, M. , Scalco, E. , Siano, R. , Vincent, F. , Zingone, A. , Dimier, C. , Picheral, M. , Searson, S. , Kandels‐Lewis, S. , Acinas, S. G. , Bork, P. , Bowler, C. , Gorsky, G. , Grimsley, N. , Hingamp, P. , Iudicone, D. , Not, F. , Ogata, H. , Pesant, S. , Raes, J. , Sieracki, M. E. , Speich, S. , Stemmann, L. , Sunagawa, S. , Weissenbach, J. , Wincker, P. , Karsenti, E. , Boss, E. , Follows, M. , Karp‐Boss, L. , Krzic, U. , Reynaud, E. G. , Sardet, C. , Sullivan, M. B. & Velayoudon, D. 2015. Eukaryotic plankton diversity in the sunlit ocean. Science, 348(6237):1261605 10.1126/science.1261605 [DOI] [PubMed] [Google Scholar]
- Dutrochet, R. J. H. 1824. Recherches anatomiques et physiologiques sur la structure intime des animaux et des végétaux. Baillière, Paris. [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg, C. G. 1838. Über das Massenverhältniss der jetzt lebenden Kiesel‐Infusorien und über ein neues Infusorien‐Conglomerat als Polierschiefer von Jastraba in Ungarn. Königlich Akademie der Wissenschaften zu Berlin, Abhandlungen; Vol. 1: p. 109–135, pl.1‐2. [Google Scholar]
- Fenchel, T. 1986. Protozoan filter feeding. Prog. Protistol., 1:65–114. Eds. Corliss, J.O. & Patterson, D.J. Biopress Ltd. [Google Scholar]
- Grassi, B. 1881. Intorno ai chetognati. Reale Ist. Lombardo Sci. Lett., 2:185–224. [Google Scholar]
- Hibberd, D. J. 1983. Ultrastructure of the colonial colouless flagellates Phalansterium digitatum Stein (Phalansteriida ord. nov.) and Spongomonas uvella (Spongomonadida ord. nov.). Protistologica, 19:523–535. [Google Scholar]
- Karpov, S. A. , Torruella, G. , Moreira, D. , Mamkaeva, M. A. & López‐García, P. 2017a. Molec. Phylogeny of Paraphelidium letcheri sp. nov. (Aphelida, Opisthosporidia) J. Euk. Micro. 64(5): 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov, S. A. , Tcvetkova, V. S. , Mamkaeva, M. A. , Torruella, G. , Timpano, H. , Moreira, D. , Mamanazarova, K. S. & López‐García, P. 2017b. Morphological and genetic diversity of opisthosporidia: New aphelid Paraphelidium tribonemae gen. et sp. nov. J. Euk. Micro. 64(2):204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S. , Tice, A. K. , Spiegel, F. W. , Silberman, J. D. , Pánek, T. , Čepička, I. , Kostka, M. , Kosakyan, A. , Alcântara, D. M. , Roger, A. J. , Shadwick, L. L. , Smirnov, A. , Kudryavstev, A. , Lahr, D. J. & Brown, M. W. 2017. Between a pod and a hard test: the deep evolution of amoebae. Mol. Biol. Evol., 34:2258–2270. 10.1093/molbev/msx162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar, D. 1990. A pirhemocyton‐like parasite of the blenny, Blennius pholis L. (Teleostei: Blennidae) and its relationship to immune‐plasma Neumann 1909. Int. J. Parasitol., 3:235e241. [DOI] [PubMed] [Google Scholar]
- Lahr, D. J. G. , Grant, J. R. & Katz, L. A. 2013. Multigene phylogenetic reconstruction of the Tubulinea (Amoebozoa) corroborates four of the six major lineages, while additionally revealing that shell composition does not predict phylogeny in the Arcellinida. Protist, 164:323–339. [DOI] [PubMed] [Google Scholar]
- Lahr, D. J. G. , Grant, J. , Nguyen, T. , Lin, J. H. & Katz, L. A. 2011. Comprehensive phylogenetic reconstruction of Amoebozoa based on concatenated analyses of SSU‐rDNA and actin genes. PLoS ONE, 6:e22780 10.1371/journal.pone.0022780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr, J. G. , Lara, E. & Mitchell, E. A. D. 2012. Time to regulate microbial eukaryote nomenclature. Biol. J. Linn. Soc., 107(3):469–476. 10.1111/j.1095-8312.2012.01962.x. [DOI] [Google Scholar]
- Lara, E. , Heger, T. J. , Ekelund, F. , Lamentowicz, M. & Mitchell, E. A. D. 2008. Ribosomal RNA genes challenge the monophyly of the Hyalospheniidae (Amoebozoa: Arcellinida). Protist, 159:165–176. [DOI] [PubMed] [Google Scholar]
- Letcher, P. M. , Longcore, J. E. , Quandt, C. A. , Leite, D. D. S. , James, T. Y. & Powell, M. J. 2017. Morphological, molecular, and ultrastructural characterization of Rozella rhizoclosmatii, a new species in Cryptomycota. Fungal Biol., 121(1):1–10. [DOI] [PubMed] [Google Scholar]
- López‐Escardó, D. , López‐García, P. , Moreira, D. , Ruiz‐Trillo, I. & Torruella, G. 2018. Parvularia atlantis gen. et sp. nov., a Nucleariid Filose Amoeba (Holomycota, Opisthokonta). J. of Euk. Microbiol. 65(2): 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisterfeld, R. 2002. Order Arcellinida Kent 1880 In: Lee J. J., Leedale G. F. & Bradbury P. (ed.), An Illustrated Guide to the Protozoa, 2nd ed Society of Protozoologists, Lawrence, KS: (Year 2000). p. 827–860. [Google Scholar]
- Mikryukov, K. A. & Mylnikov, A. P. 1998. The fine structure of a carnivorous multiflagellar protist, Multicilia marina Cienkowski, 1881 (Flagellata incertae sedis). Eur. J. Protistol., 34:391–401. [Google Scholar]
- Nguyen, N. H. , Song, Z. , Bates, S. T. , Branco, S. , Tedersoo, L. , Menke, J. , Schilling, J. S. & Kennedy, P. G. 2016. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol., 20:241–248. [Google Scholar]
- Pánek, T. , Zadrobílková, E. , Walker, G. , Brown, M. W. , Gentekaki, E. , Hroudrová, M. , Kang, S. , Roger, A. J. , Tice, A. K. , Vlček, Č. & Čepička, I. 2016. First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group. Mol. Phylogenet. Evol., 98:41–51. [DOI] [PubMed] [Google Scholar]
- Pawlowski, J. , Audic, S. , Adl, S. , Bass, D. , Belbahri, L. , Berney, C. , Bowser, S. S. , Cepicka, I. , Decelle, J. , Dunthorn, M. , Fiore‐Donno, A. M. , Gile, G. H. , Holzmann, M. , Jahn, R. , Jirků, M. , Keeling, P. J. , Kostka, M. , Kudryavtsev, A. , Lara, E. , Lukeš, J. , Mann, D. G. , Mitchell, E. A. D. , Nitsche, F. , Romeralo, M. , Saunders, G. W. , Simpson, A. G. B. , Smirnov, A. V. , Spouge, J. L. , Stern, R. F. , Stoeck, T. , Zimmermann, J. & Schindel, D. 2012. CBOL protist working group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol., 10(11):e1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, M. J. , Letcher, P. M. & James, T. Y. 2017. Ultrastructural characterization of the host–parasite interface between Allomyces anomalus (Blastocladiomycota) and Rozella allomycis (Cryptomycota). Fungal Biol. 121(6–7): 561–572. [DOI] [PubMed] [Google Scholar]
- Pleijel, F. , and Rouse G. W.. 2003. Ceci n'est pas une pipe: Clades and phylogenetic nomenclature. J. Zool. Syst. Evol. Res. 41:162–174. [Google Scholar]
- Quandt, C. A. , Beaudet, D. , Corsaro, D. , Walochnik, J. , Michel, R. , Corradi, N. , & James, T. Y. 2017. The genome of an intranuclear parasite, Paramicrosporidium saccamoebae, reveals alternative adaptations to obligate intracellular parasitism. Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, T. A. , Leonard, G. & Wideman, J. G. 2017. What defines the “Kingdom” Fungi? Microbiol. Spectrum 01 Jun 2017, 5(3) DOI: 10.1128/microbiolspec.FUNK-0044-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero, M. A. , Dennis, P. G. , Orrell, T. M. , Bailly, N. , Bourgoin, T. , Brusca, R. C. , Cavalier‐Smith, T. , Guiry, M. D. & Kirk, P. 2015. A higher level classification of all living organisms. PLoS ONE, 10(6):e0130114 10.1371/journal.pone.0119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint‐Exupéry, A. 1943. Le petit prince. Reynal & Hitchcock, New York, NY. [Google Scholar]
- Schaudinn, F. 1899. Untersuchungen uber den Generations wechsel von Trichosphaerium sieboldi. Schn. Abh. K. Preuss. Akad. Wiss., Berlin, (Suppl):1–93. [Google Scholar]
- Schierwater, B. & DeSalle, R. 2018. Placozoa. Curr. Biol., 28:R97–R98. [DOI] [PubMed] [Google Scholar]
- Schleiden, M. J. 1838. Beiträge zur phytogenesis. Veit et Comp, Berlin. [Google Scholar]
- Schwann, T. 1839. Mikroskopische Untersuchungen über die Uebereinstimmung in der Struktur und dem Wachsthum der Thiere und Pflanzen, Berlin. [PubMed] [Google Scholar]
- Shadwick, J. D. L. , Silberman, J. D. & Spiegel, F. W. 2017. Variation in the SSUrDNA of the genus Protostelium leads to a new phylogenetic understanding of the genus and of the species concept for Protostelium mycophaga (Protosteliida, Amoebozoa). J. Eukaryot. Microbiol., 65:331–344. 10.1111/jeu.12476 [DOI] [PubMed] [Google Scholar]
- Sheikh, S. , MatsThulin Cavender, J. C. , Escalante, R. , Kawakami, S. I. , Lado, C. , Landolt, J. C. , Nanjundiah, V. , Queller, D. C. , Strassmann, J. E. , Spiegel, F. W. , Stephenson, S. L. , Vadell, S. M. & Baldauf, S. L. 2018. A new classification of the dictyostelids. Protist, 169:1–28. [DOI] [PubMed] [Google Scholar]
- Sibbald, S. J. , Cenci, U. , Colp, M. , Eglit, Y. , O'Kelly, C. J. & Archibald, J. M. 2017. Diversity and evolution of Paramoeba spp. and their kinetoplastid endosymbionts. J. Eukaryot. Microbiol., 64:598–607. 10.1111/jeu.12394 [DOI] [PubMed] [Google Scholar]
- Simpson, A. G. B. & Roger, A. J. 2004. The real “kingdoms” of eukaryotes. Curr. Biol., 14:R693–R896. 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Smirnov, A. 2008. Amoebas, Lobose In: Schaechter M. (ed.), Encyclopedia of Microbiology. Elsevier Science Publishers, Oxford: p. 558–577. [Google Scholar]
- Spiegel, F. W. , Shadwick, L. L. , Ndiritu, G. G. , Brown, M. W. , Aguilar, M. & Shadwick, J. D. L. 2017. Protosteloid Amoeboazoa (Protosteliids, Protosporangiida, Cavostellida, Schizoplasmodiida, Fractoviteliida, and sporcarpic members of Vanellida, Centramoebida, and Pellitida) In: Archibald J. M., Simpson A. G. B. & Slamovits C. (ed.), Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.). Springer; Reference Works (e‐book). 10.1007/978-3-319-32669-6_12-1 [DOI] [Google Scholar]
- Srivastava, M. , Begovic, E. , Chapman, J. , Putnam, N. H. , Hellsten, U. , Kawashima, T. , Kuo, A. , Mitros, T. , Salamov, A. , Carpenter, M. L. , Signorovitch, A. Y. , Moreno, M. A. , Kamm, K. , Grimwood, J. , Schmutz, J. , Shapiro, H. , Grigoriev, I. V. , Buss, L. W. , Schierwater, B. , Dellaporta, S. L. & Rokhsar, D. S. 2008. The Trichoplax genome and the nature of placozoans. Nature, 454(7207):955–960. 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Tekle, Y. I. , Anderson, O. R. , Katz, L. A. , Maurer‐Alcala, X. X. , Romero, M. A. & Molestina, R. 2016. Phylogenomics of ‘Discosea’: A new molecular phylogenetic perspective on Amoebozoa with flat body forms. Mol. Phylogenet. Evol., 99:144–154. [DOI] [PubMed] [Google Scholar]
- Tekle, Y. I. & Wood, F. C. 2017. Longamoebia is not monophyletic: phylogenomic and cytoskeleton analyses provide novel and well‐resolved relationships of amoebozoan subclades. Mol. Phylogenet. Evol., 114:249–260. 10.1016/j.ympev.2017.06.019 [DOI] [PubMed] [Google Scholar]
- Tice, A. K. , Shadwick, L. L. , Fiore‐Donno, A. M. , Geisen, S. , Kang, S. , Schuler, G. A. , Spiegel, F. W. , Wilkinson, K. A. , Bonkowski, M. , Dumack, K. , Lahr, D. J. G. , Voelcker, E. , Clauβ, S. , Zhang, J. & Brown, M. W. 2016. Expansion of the molecular and morphological diversity of Acanthamoebidae (Centramoebida, Amoebozoa) and identification of a novel life cycle type within the group. Biol. Direct, 11:69 10.1186/s13062-016-0171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torruella, G. , Grau‐Bové, X. , Moreira, D. , Karpov, S. A. , & Burns, J. A. , Sebé‐Pedrós, A. , Völcker, E. & López‐García, P. 2019. Global transcriptome analysis of the aphelid Paraphelidium tribonemae supports the phagotrophic origin of fungi. Communications Biology. In press. 10.1038/s42003-018-0235-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tieghem, M. P. 1884. Cœnonia, genre nouveau de Myxomycètes a plasmode agrégé. Bull. Soc. Bot. France, 31:303–306. 10.1080/00378941.1884.10828252. [DOI] [Google Scholar]
- Van Vichelen, J. , D'Hondt, S. , Claeys, M. , Vyverman, W. , Berney, C. , Bass, D. & Vanormelingen, P. 2016. A hotspot of amoebae diversity: 8 new naked amoebae associated with the planktonic bloom‐forming cyanobacterium Microcystis . Acta Protozool., 55:61–87. [Google Scholar]


