Abstract
Drosophila segmentation is regulated by a complex network of transcription factors that include products of the pair-rule genes (PRGs). PRGs are expressed in early embryos in the primorida of alternate segmental units, establishing the repeated, segmental body plan of the fly. Despite detailed analysis of the regulatory logic among segmentation genes, the relationship between these genes and the morphological formation of segments is still poorly understood, since regulation of transcription factor expression is not sufficient to explain how segments actually form and are maintained. Cell surface proteins containing Leucine rich repeats (LRR) play a variety of roles in development, and those expressed in segmental patterns likely impact segment morphogenesis. Here we explore the relationships between the PRG network and segmentally expressed LRR-encoding (sLRR) genes. We examined expression of Toll2, Toll6, Toll7, Toll8 and tartan (trn) in wild type or PRG mutant embryos. Expression of each sLRR-encoding gene is dynamic, but each has a unique register along the anterior-posterior axis. The registers for different sLRRs are off-set from one another resulting in a continually changing set of overlapping expression patterns among the sLRR-encoding genes themselves and between the sLRR-encoding genes and the PRGs. Accordingly, each sLRR-encoding gene is regulated by a unique combination of PRGs. These findings suggest that one role of the PRG network is to promote segmentation by establishing a cell surface code: each row of cells in the two-segment-wide primordia expresses a unique combination of sLRRs, thereby translating regulatory information from the PRGs to direct segment morphogenesis.
Keywords: Toll family, tartan, pair rule genes, segmentation
Introduction
Segmentation can be broken down into three steps: segment specification, differentiation or morphogenesis of segments, and maintenance of segments once they are formed. The process of segmentation has been extensively studied in Drosophila melanogaster where a cascade of regulatory genes specifies formation of embryonic segments (Lawrence and Johnston, 1989; Nusslein-Volhard and Wieschaus, 1980). This hierarchy is initiated by a set of maternal genes that define the anterior-posterior (AP) axis. Gap genes then define broad regions within the embryo, and, together with maternal genes, control expression of the pair rule genes (PRGs). PRGs are expressed in the primordia of alternating segment-wide units, typically in a pattern of 7 stripes at the cellular blastoderm stage (PR-stripes). The PRGs are essential for formation of segments, evidenced by the finding that in embryos homozygous for mutations in any of the nine PRGs, alternate segmental units are missing. The last set of segmentation genes, the segment polarity genes (SPGs), define anterior and posterior compartments within each segment and are typically expressed in 14-stripe patterns (segmental stripes). Definition of segments is completed very early, by the end of the cellular blastoderm stage of development, when PR-stripes peak (Chan and Gehring, 1971; Lohs-Schardin et al., 1979). The PRGs and SPGs encode regulatory proteins, primarily transcription factors, whose cross-regulatory interactions have been well-studied. Although these regulatory interactions have been documented in some detail, regulatory proteins themselves cannot direct segment formation. Rather, these regulators must control downstream target genes that encode products more directly involved in segment morphogenesis.
Genes that encode cell surface proteins are good candidates for playing direct roles in segment morphogenesis. Subsequent to segment establishment, the embryo undergoes two dramatic morphogenetic movements, germband extension and germband retraction. During these movements, cells specified to be part of a particular segment remain associated and retain their segmental identity (Chan and Gehring, 1971; Gergen et al., 1986; Gilbert, 2010; Grosshans and Wieschaus, 2000; Irvine and Wieschaus, 1994; Lohs-Schardin et al., 1979; Wieschaus et al., 1991). Several groups previously showed that genes encoding cell surface proteins belonging to the Toll family are expressed in patterns suggestive of PR-regulation. Specifically, Drosophila Toll-2, Toll-6, Toll-7 and Toll-8 were shown to be expressed in early embryos in PR-like stripes (Eldon et al., 1994; Kambris et al., 2002). The Toll-2 stripes lie posterior to those of even-skipped (eve), and Toll-8 stripes overlap those of eve (Kambris et al., 2002). By germband extension, Toll-2 and Toll-8 stripes have doubled in the trunk, with Toll-2 overlapping and possibly extending posterior to wingless (wg) and Toll-8 lying posterior to wg stripes and potentially partially overlapping Toll-2 (Eldon et al., 1994; Kambris et al., 2002). Toll-6 is expressed in stripes late in cellular blastoderm, with secondary stripes evident by early germband extension (Kambris et al., 2002). Toll-7 is expressed during germband extension in 14 stripes that overlap those of engrailed (en) (Kambris et al., 2002).
Tartan (Trn), like the segmentally expressed Tolls, contains leucine rich repeats (LRR) in its extracellular domain (Figure 1A), is apically localized, and has been shown to function as an adhesion molecule in a variety of contexts (Artero et al., 2003; Mao et al., 2008; Milan et al., 2005; Milán et al., 2001). Thus, Trn may act with the segmentally expressed Toll family members to translate the spatial information provided by the PRGs into actions that produce or maintain actual segments. trn is expressed in a PR-like pattern of eight stripes, seven of which overlap with the PRG fushi tarazu (ftz). These seven stripes were lost in a ftz mutant background (Chang et al., 1993; Field et al., 2016). Regulation of trn by Ftz, and its partner Ftz-F1, appears to be direct, as a trn-enhancer was identified that directs reporter gene expression in a ftz-dependent seven-stripe pattern (Field et al., 2016).
Figure 1.
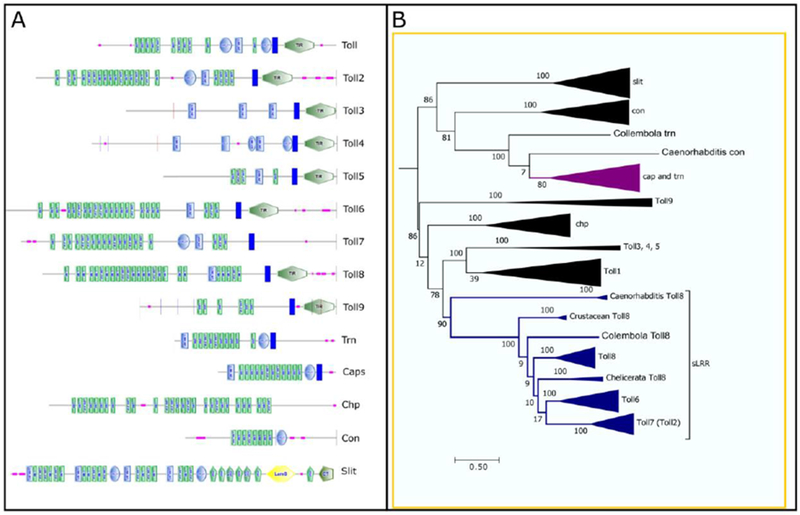
Trn shares LRR repeats with the Toll family proteins. A) Diagrams of Drosophila sLRR proteins domains. Diagrams modified from those produced by the SMART program. Most Tolls have large Leucine Rich Repeat regions (LRR, green rectangles), C- (blue ovals) or N- (light blue rectangles) type cysteine rich flanking regions, a transmembrane domain (TM, dark blue rectangles) and an internal Toll/Interleukin-1 Receptor (TIR, green hexagons) domain. Pink squares represent regions of low complexity. Tartan (Trn) and Capricious (Caps) share two of these features, the LRR region and the TM domain, but lack the TIR domain. Chaoptin (Chp), Connectin (Con) and Slit all contain LRR domains and are involved in cell adhesion, but lack the TM and TIR domains. Slit also contains a laminin G domain (LamG, yellow hexagon), an EGF domain (GFP, vertical green pentagons), and a cystein knot-like domain (CT, horizontal green pentagon). B) Simplified phylogenetic tree showing the relationship between Trn and Toll family proteins. For the full tree see Supplemental Figure 1. Orthologs were identified by reciprocal blast of protein sequences, aligned with Clustal Omega and placed in a midpoint rooted RaxML phylogenetic tree using Topali. The clade containing Trn is purple. The clades containing the Tolls that are expressed in a segmental pattern are blue. Numbers at the junctions are bootstrap values from 100 iterations which provide statistical support for the nodes. The size of the triangle is proportional to the number of orthologs within the clade.
Functional roles for these cell surface proteins remained elusive for some time, because deletion of any single Toll family gene failed to cause segmentation defects (Yagi et al., 2010, W.R.A. and L.P., unpublished). However, RNAi-mediated knock-down indicated that Toll-2, Toll-6 and Toll-8 have roles in germband extension (Pare et al., 2014). While knockdown of any single gene did not affect elongation, double and triple gene knock-downs reduced axis elongation. Similarly, in the flour beetle Tribolium castaneum, Toll-7 and Toll-10 are expressed in striped patterns, and simultaneous knock-down of both genes reduced mediolateral intercalation during germband extension (Benton et al., 2016). The defects in germband extension seen for knockdown of Drosophila Tolls are reminiscent of defects seen in eve and runt (run) mutants, consistent with these genes regulating Toll gene expression (Butler et al., 2009; Irvine and Wieschaus, 1994; Pare et al., 2014).
In this paper, we determined the phylogenetic relationships between trn and the segmentally expressed Toll genes, referred to here collectively as ‘segmentally expressed Leucine-Rich Repeat (sLRR)’-encoding genes. We examined the expression patterns of these sLRR-encoding genes in wild type and segmentation gene mutant backgrounds throughout early embryogenesis and developed a model for the dynamic positioning of these genes relative to one another and the PRGs during early embryogenesis. Expression of each sLRR-encoding gene was affected by multiple PRGs. However, regulatory interactions are complex: the complement of PRG regulators, and the direction of regulation by PRGs were unique for each sLRR, suggesting that each is regulated by different elements of the PRG network.
Materials and Methods
Drosophila genetics
Drosophila mutant strains used in this study: ftz9h39/TM3Ser, actin-GFP; h22/TM3Ser, actin-GFP; run2/FM7, actin-GFP; prd12/CyO, actin-GFP; odd5/CyO, actin-GFP; eve1/CyO, actin-GFP; slpΔ34/CyO, actin-GFP; and en54/CyO, actin-GFP. Each allele has been characterized as a null or a strong hypomorphic allele, except for eve1. Null alleles of eve produce a denticle lawn phenotype rather than a PR phenotype. At 25°C eve1 produces a PR phenotype (Frasch et al., 1988). Initially, we used actin-GFP balancers to identify homozygous mutant embryos, expecting that the GFP marker would be visible throughout embryogenesis, such that homozygous mutant embryos would be identified by the lack of expression. However, we found that reliable identification of the mutant embryos was difficult. Thereafter, we examined expression of genes of interest in embryos at cellular blastoderm, gastrulation and germband extension stages for each genotype, without double staining for a marker gene. Because both parents were heterozygous for each PRG mutation, 25% of the embryos would be expected to be homozygous mutant. A PRG mutation was considered to have a strong effect on a particular sLRR when expression differed from wild type in at least 20% of the embryos. If expression was altered in fewer than 10% of the embryos, the PRG was considered to have no effect. Those PRG mutations that affected expression in 10 - 15% of embryos are described as having a weak effect; between 15 and 20% affected are described as having a moderate effect.
Phylogenetic analysis
Orthologs were identified by blasting the protein sequence (blastp) of the Drosophila gene of interest (Trn, Cap, Chp, Con, Slit, Toll1, Toll2/18w, Toll3/MstProx, Toll4, Toll5/Tehao, Toll6, Toll7, Toll8/Tollo, or Toll9) against the non-redundant protein sequences (nr) collection of Caenorhabditis, Chelicerata, Myriapoda, Crustacea, Collembola, Orthoptera, Isoptera, Hemiptera, Hymenoptera, Coleoptera, Lepidoptera, Siphonaptera, and Mecoptera in NCBI. The protein sequence of the top 3-4 “hits” (excluding sequences of low-quality proteins, partial sequences and additional isoforms of a single gene) was blasted back against the Drosophila melanogaster non-redundant protein sequences (nr) collection in NCBI. If the reciprocal blast identified the original Drosophila gene as the first hit, the putative ortholog was retained for use in the phylogenetic analysis. Sequences for all orthologs of a particular gene were aligned in Clustal Omega (Li et al., 2015), then imported into Aliview (Larsson, 2014) for evaluation. Sequences that were at least 100 amino acids shorter than the majority of the orthologs were presumed to be partial sequences and were discarded from the pool. A master set of sequences for all orthologs of all genes was generated by combining the subsets of individual gene sequences. The master set was aligned in Clustal Omega, then imported into Topali (Milne et al., 2009) for phylogenetic analysis using the Maximum Likelihood algorithm RaxML, WAG model with 100 bootstraps (Stamatakis and Swiss Federal Institute of Technology Lausanne, 2018). Additional formatting of trees was performed in MEGA7 (Tamura et al., 2013) or Dendroscope3 (Huson and Scornavacca, 2012).
Drosophila gene expression analysis
Drosophila embryo in situ hybridization with digoxygenin-, biotin- or fluorescein-labeled probes was carried out using modifications of the protocols in Tautz and Pfeifle 1989 (Tautz and Pfeifle, 1989) and Kosman et al. 2004 (Kosman et al., 2004). Details are provided in supplemental material. Reagents for TSA detection were: Sigma goat anti-biotin HRP conjugated (1:1000 dilution), Roche mouse anti-digoxygenin (1:250 dilution), Life Technologies goat anti-mouse IgG HRP conjugated (1:100 dilution), ThermoFisher AlexaFlour 488 tyramide reagent, and ThermoFisher AlexaFluor 555 tyramide reagent. Reagents used for colorometric detection were: Roche anti-digoxygenin AP conjugated Fab fragments (1:2000 dilution), Roche anti-fluorescein AP conjugated Fab fragments (1:200 dilution), MP Biomedicals mouse anti-beta-galactosidase biotin conjugated (1/1000), NBT (Roche 11383213001) -BCIP (Roche 11383221001) -INT -BCIP (Roche 11681460001), ABC (vector labs Vectastain Elite ABC-HRP Kit) and DAB (SigmaFAST 3,3-Diaminobensidine tablets).
Results
As mentioned above, cell surface proteins likely play direct roles in morphogenesis in the Drosophila embryo. The Toll family genes have been examined in this light (Pare et al. 2014), but Trn encodes another cell surface protein expressed in PR-like stripes, which could function similarly to or together with Tolls. Trn shares LRR and transmembrane domains with the Toll family but lacks a Toll/Il-1 Receptor (TIR) domain (Fig. 1A). To assess whether Trn is related to the Tolls, we constructed a phylogenetic tree comparing Toll1-9, trn, and capricious (caps) which is closely related to trn (Shishido et al., 1998). We included three additional divergent LRR protein-encoding genes that mediate cell adhesive interactions in Drosophila: Choaptin (Chp) and Connectin (Con) are LRR proteins that are membrane associated via a phosphatidylinositol link rather than a transmembrane domain (Krantz and Zipursky, 1990; Nose et al., 1992; Van Vactor et al., 1988), while Slit is a secreted LRR protein (Rothberg et al., 1988; Rothberg et al., 1990). Orthologs for each gene were identified from up to three species within the following groups: Caenorhabditis, Chelicerata, Myriapoda, Crustacea, Collembola, Orthoptera, Isoptera, Hemiptera, Hymenoptera, Coleoptera, Lepidoptera, Siphonaptera, Mecoptera. We used Drosophila protein sequences as queries for blastp searches of the non-redundant protein sequences database for each order. The protein sequences of candidate orthologs were then used for reciprocal blastp searches of the Drosophila non-redundant protein sequences database. Only candidates that returned the original Drosophila gene were considered as orthologs.
As shown in Figure 1B, and in more detail in Supplemental Figure 1, the orthologs of each gene form well defined clades with high confidence levels. capricious and tartan orthologs are so closely related that they cannot be separated into individual clades. The sLRR-encoding genes most closely related to trn and caps are slit and connectin. All the Tolls and chaoptin form a clade separate from that containing trn, caps, slit and con.
The Tolls that are expressed in segmental patterns (Toll2, 6, 7 and 8) form a clade of their own separate from other Tolls and chaoptin. Within this clade, Toll8 orthologs show the most divergence from one another. The Toll6 orthologs all fall within one clade, and most potential orthologs of Dmel-Toll2 return Dmel-Toll7 when blasted against Drosophila genes. Since Toll2 and Toll7 are closely linked on chromosome 2, this suggests they are the results of a recent duplication in lineages leading to Drosophila.
trn and Toll2, Toll6, Toll7 and Toll8 are expressed in non-overlapping segmental patterns
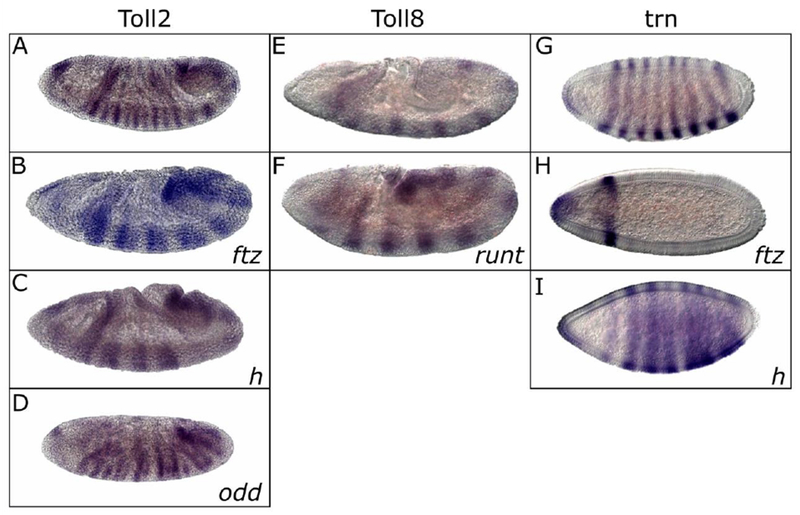
The expression of the sLRR-encoding genes was compared to that of segmentation genes expressed in early embryos by double in situ hybridization (Fig. 2). Expression of each sLRR-encoding gene overlapped exactly with that of one segmentation gene. Specifically, at the cellular blastoderm stage, the seven anterior Toll2 stripes overlapped with those of the PRG run (Fig. 2A-C). The anterior 7 Toll8 stripes at cellular blastoderm overlapped with the seven stripes of the PRG hairy (h) (Fig. 2D-F). trn was detected in eight stripes at the cellular blastoderm stage, with stripes 2-8 overlapping the seven ftz stripes (Fig. 2G-I). Two sLRR-encoding genes were not detectable at early cellular blastoderm. Toll6 expression was first reliably detectable at late cellular blastoderm. The number of Toll6 stripes changes rapidly, but Toll6 stripes overlapped sloppy paired (slp1) stripes (Fig. 2J-L). Finally, Toll7 was detected during germband extension in a pattern of 14 stripes that overlapped ectodermal en stripes (Fig. 2M-O). Thus, each sLRR-encoding gene was expressed in a distinct spatio-temporal domain, overlapping the pattern of one segmentation gene.
Figure 2.
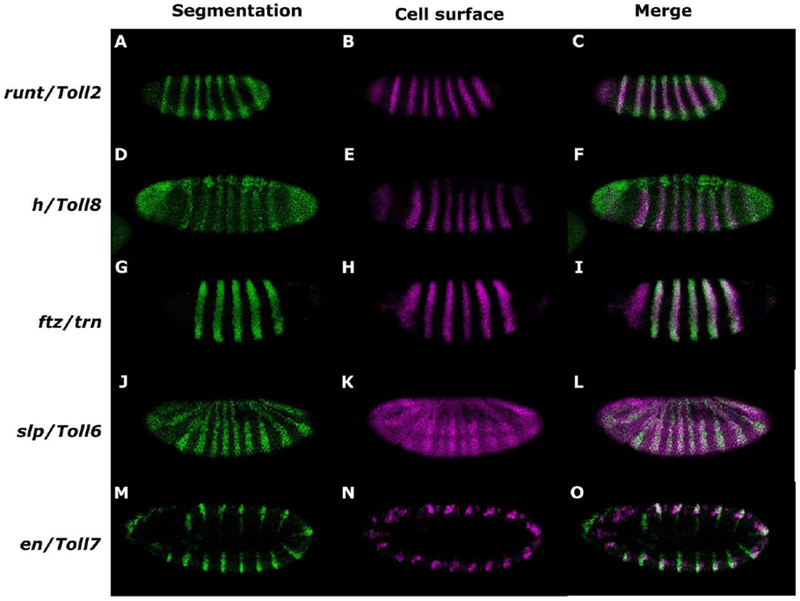
Expression of each sLRR encoding gene overlaps with a segmentation gene in early Drosophila embryos. Expression patterns were monitored in w1118 embryos by in situ hybridization using digoxigenin- or fluorescein-labeled probes, as indicated. In each row, the first panel shows the pattern of the segmentation gene, the second shows expression of the sLRR-encoding gene and the final shows the merge. (A, D, G, J, M) segmentation gene expression. (B, E, H, K, N) sLRR-encoding gene expression. (C, F, I, L, O) merged images. Expression of run (A, C) overlaps with Toll2 (B, C). h (D, F) overlaps Toll8 (E, F). ftz (G, I) overlaps trn (H, I). slp (J, L) overlaps Toll6 (K, L), and en (M, O) overlaps Toll7 (N, O). Embryos are oriented anterior, left; dorsal, top.
sLRR genes are expressed dynamically during Drosophila embryogenesis
Although a static pattern of sLRR expression is implied by single snapshot-views of expression such as that shown in Figure 2, the sLRR-encoding genes are expressed in dynamic patterns that change rapidly during early embryogenesis. We and others (Chang et al., 1993; Eldon et al., 1994; Kambris et al., 2002) monitored expression of the sLRR-encoding genes through cellular blastoderm, gastrulation and germband extension stages (Fig. 3, Supp. Fig. 2). Toll2 expression was detected in a pattern of 8 stripes at the cellular blastoderm stage with an additional anterior stripe that does not reach the dorsal side and a region of expression near the anterior end of the embryo (Fig. 3A). The cephalic furrow formed between the partial stripe and the first full stripe. The 5 most anterior full stripes remained a constant 2-3 cells wide from the cellular blastoderm stage through germband extension, while the two most posterior stripes were initially 4-5 cells wide, but narrowed to 2-3 cells before germband extension. Secondary stripes began to appear during gastrulation or early germband extension (Fig. 3B). Initially 1 cell wide, they quickly widened to 2-3 cells like the primary stripes. During germband extension (Figure 3C), 14 Toll2 stripes were detected in a segmentally repeating pattern.
Figure 3.
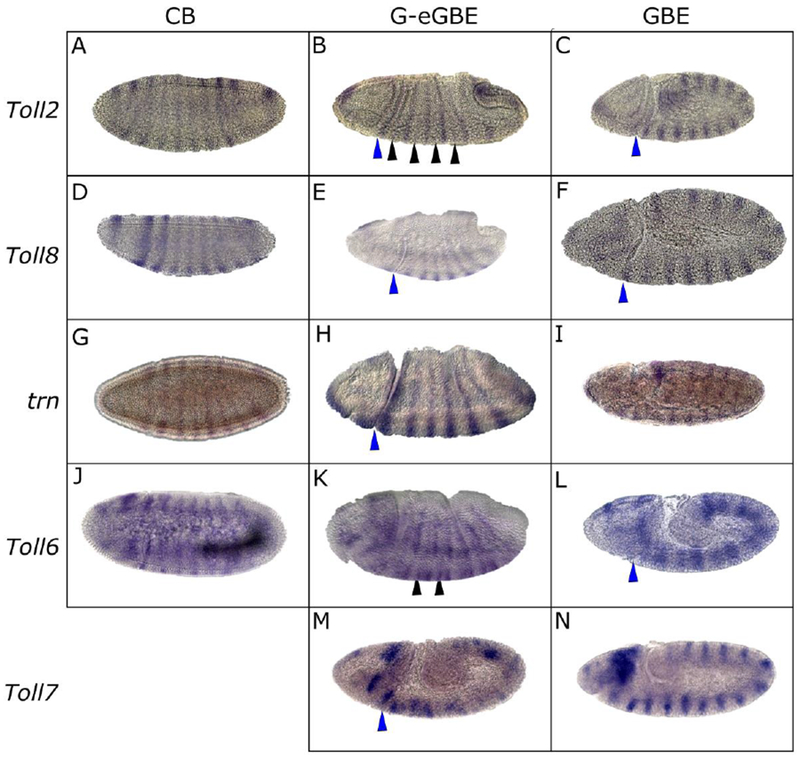
sLRR-encoding gene expression evolves continuously throughout early embryogenesis. Expression patterns were monitored in w1118 embryos by in situ hybridization using digoxigenin labeled probes. Toll2 (A-C); Toll8 (D-F); trn (G-I); Toll6 (J-L); Toll7 (M-N). In each row, embryos are ordered in increasing age from cellular blastoderm (CB), through gastrulation (G) or early germband extension (eGBE), to fully extended germband (GBE) stages. Embryos oriented anterior, left; dorsal, top. Black arrowheads indicate secondary stripes. Blue arrowheads indicate the cephalic furrow.
Toll8 expression first appeared at cellular blastoderm (Fig. 3D). Near the anterior of the embryo, there was a wide stripe (4-5 cells), then a gap of about 12 cells. Posterior to the gap, there was a set of 8 stripes, with the first stripe being slightly wider (4-5 cells) than the others (3-4 cells). The most anterior stripe faded during gastrulation while the set of 8 stripes was maintained, and the first stripe in the set narrowed to 3-4 cells (Fig. 3E). The cephalic furrow formed just posterior to first stripe in this set of 8 stripes. Weak secondary Toll8 stripes appeared between the primary stripes during mid to late germband extension. By the end of germband extension, 14 Toll8 stripes were apparent in the trunk, each approximately 2 cells wide (Fig. 3F), along with patches of stain in the head.
As described previously, trn was initially detected as a set of 8 stripes approximately 3 cells wide at the cellular blastoderm stage (Fig. 3G). These stripes were maintained during gastrulation (Fig.3H) and early germband extension. The cephalic furrow formed between the first and second stripe. Late in germband extension, epidermal trn was very briefly and weakly expressed in a segmental pattern of stripes 1-2 cells wide, with additional expression in neuronal tissue (Fig. 3I).
The first set of Toll6 stripes consists of 7 full stripes posterior to a single partial stripe that does not reach the ventral side (Fig. 3J). The stripes were 2-3 cells wide and appeared late in cellular blastoderm or early in gastrulation with the most anterior stripe appearing first. At gastrulation, the cephalic furrow formed on top of the most anterior stripe, and slightly narrower secondary stripes were detected between the primary stripes (Fig. 3K). When the germband was fully extended, a set of 14 Toll6 stripes that had narrowed to single cell width were detected (Fig. 3L). In addition, there were patches of expression in the head region throughout this period of development.
Toll7 expression was first detected at early germband extension, when a set of 14 ‘messy’ stripes appear. As the germband extended, the stripes became more refined until they were a single cell wide (Fig. 3M, N). In addition, there were patches of expression in the head region. In a variable percent of embryos (0-30% depending upon the sample), a broad band of expression was observed at the internal edge of the Toll7 stripes, which is partially visible in the embryos shown in Fig. 3N, 6J.
Figure 6.
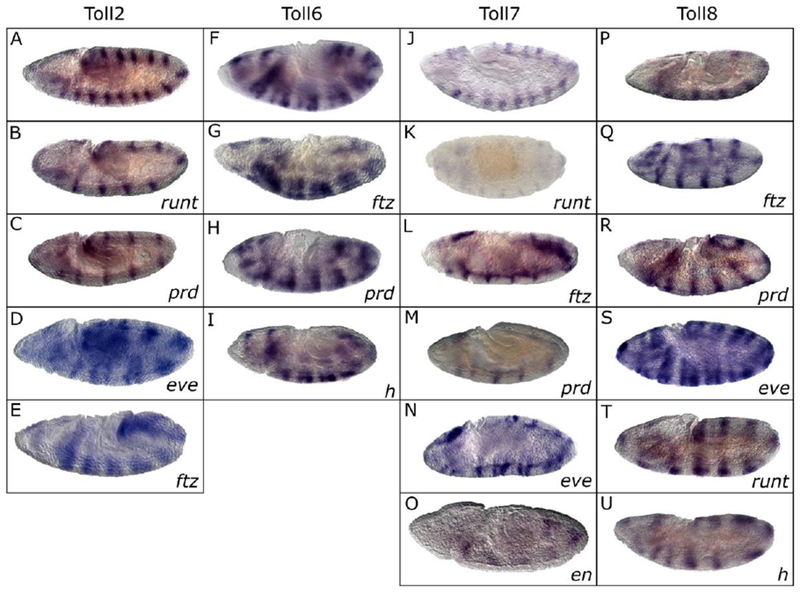
Several PRG mutations alter expression of each sLRR at the germband extended stage. Expression patterns were monitored by in situ hybridization using digoxigenin labeled probes to the sLRR encoding gene listed at the top of each column on embryos homozygous for a PRG mutation as indicated in the lower right corner of each panel. The top panel in each column (A,F,J, P) shows the wild-type pattern for comparison. (A-E) Toll2 in run (B), prd (C), eve (D) and ftz (E); (F-I) Toll6 in ftz (G), prd (H) and h (I); (J-O) Toll7 in run (K), ftz (L), prd (M), eve (N) and en (O); (P-U) Toll8 in ftz (Q), prd (R), eve (S) run (T) and h (U). Embryos are oriented anterior, left; dorsal, top.
In sum, Toll 2, 6, and 8 were expressed in striped patterns from cellular blastoderm through germband extension which shifted from double to single segment periodicity. The secondary stripes appeared during gastrulation for Toll2 and Toll6. Toll8 secondary stripes appeared in the middle of germband extension, trn was expressed in alternate segment primordia from cellular blastoderm to mid germband extension and shifted to single segment periodicity for a very short period at the end of germband extension. Toll7 was expressed only during germband extension and with single segment periodicity.
Unique expression for each sLRR-encoding gene
To examine how relative expression of the sLRR-encoding genes changes over the course of development, we first examined expression of Toll2 relative to each of the others (Figure 4; see Figure 7 for summary). At cellular blastoderm, Toll6 stripe 1 was evident as a full stripe but other stripes were just beginning to emerge, allowing us to determine stripe register. The 9 Toll2 stripes were positioned adjacent and posterior to Toll6 stripes (Fig. 4 A, B; Supp. Fig. 3 K, L). During gastrulation, Toll2 secondary stripes formed between the Toll6 stripes (Figure 4 C, D; Supp. Fig. 3 M-P). As the germband elongated, the Toll6 stripes narrowed from the original width of 2-3 cells to 1-2 cells (Fig. 4 E, F; Supp. Fig. 3 Q, R).
Figure 4.
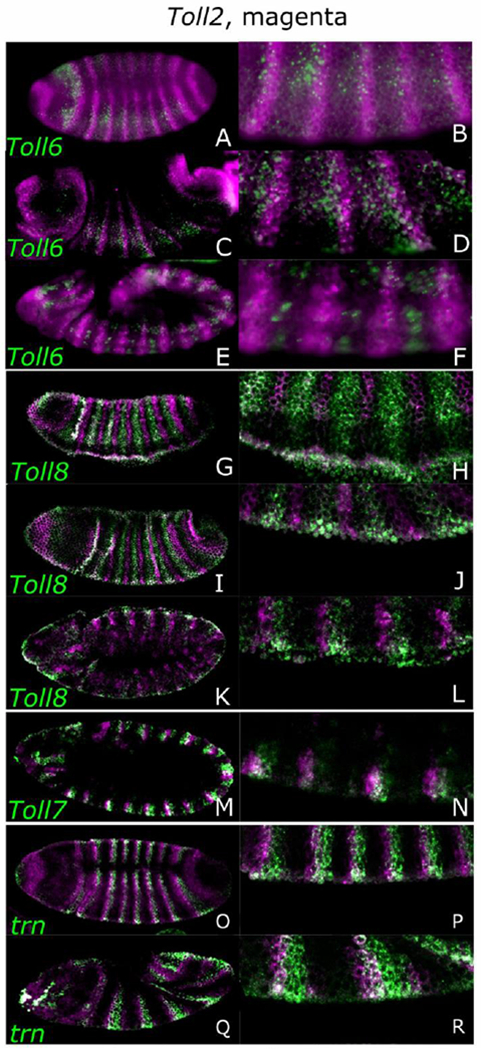
sLRR relationships change during development. Expression patterns were monitored in w1118 embryos by in situ hybridization using digoxigenin- or biotin-labeled probes, as indicated. Embryos are oriented anterior, left; dorsal, top. Images to the right of each panel are a higher magnification view of the embryo to the left. Toll2, expression (magenta) is compared to that of a second sLRR-encoding gene (green), as indicated in the lower left corner, at different stages of embryogenesis: Toll6 at cellular blastoderm (A, B), gastrulation (C, D) and germband extension (E, F), Toll8 at cellular blastoderm (G, H), gastrulation (I, J) and germband extension (K, L), Toll7 at germband extension (M, N) and trn at CB (O, P), early germband extension (Q, R).
Figure 7.
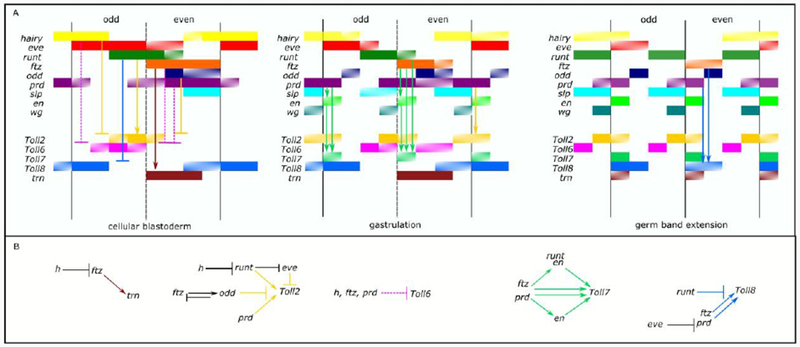
The relationships between the PRGs and the sLRR genes are complex and suggest that multiple PRGs regulate each sLRR gene. A) Schematic representation of PRG and sLRR-encoding gene expression. The diagrams, using the format of Clark and Akam (Clark and Akam, 2016), represent snapshots of a continuously evolving pattern at cellular blastoderm, gastrulation and germ-band extension. Gene expression is indicated by a colored bar to the right of the gene name: hairy (yellow), eve (red), run (dark green), ftz (orange), odd-skipped (dark blue), paired (purple), sloppy-paired (light blue), engrailed (light green), wingless (blue-green), Toll2 (gold), Toll6 (pink), Toll7 (medium green), Toll8 (medium blue), trn (brown). Patterned bars represent rapidly changing expression. The black vertical lines represent the eventual position of the parasegment boundaries. Arrows or T-bars represent possible activation or repression respectively. Solid arrows or T-bars indicate possible direct regulations. Interactions affecting the primary Toll stripes are shown in the cellular blastoderm panel while those affecting the secondary Toll stripes are diagramed in the gastrulation or germband extended panel. Arrow color matches that of the Toll gene affected. B) Regulatory relationships between PRGs and sLRR genes. The diagrams show proposed regulatory interactions between PRGs and sLRR-encoding genes. PRGs that activate primary sLRR stripes are located above the sLRRs. PRGs that activate secondary sLRR stripes are located below the sLRRs. PRGs that repress sLRR stripes are on the same level as the sLRR. Previously described regulatory relationships are shown as black arrows or bars.
Toll2 and Toll8 stripes never overlapped (Fig. 4 G-L). Initially they alternated with small or no gaps (Fig. 4 G, H), but during germband elongation, the Toll8 stripes narrowed, generating gaps between the anterior edge of the Toll2 stripes and the posterior edge of the Toll8 stripes (Fig. 4 K, L). The relationship between Toll7 and Toll2 remained stable, with Toll2 stripes overlapping and extending anterior to the Toll7 stripes (Fig. 4 M, N, Supp. Fig. 3A, B). Finally, Toll2 stripes were positioned adjacent and anterior to trn stripes through cellular blastoderm and gastrulation stages of development (Fig. 4 O-R). Colorimetric double in situs are shown in Supplemental Figure 3 with better resolution for the Toll2-Toll6 and Toll2-Toll7 double in situ hybridizations and additional combinations of sLRR-encoding genes. In sum, the sLRR-encoding genes are expressed in evolving patterns. While each is expressed in a unique domain, expression is partially overlapping for several (Fig. 7A), similar to the situation for the PRGs.
Mutations in segmentation genes have specific effects on the expression sLRR-encoding genes
The sLRR-encoding genes are expressed in overlapping segmental patterns that closely match those of segmentation genes. To determine whether mutations in these genes affect expression of the sLRR-encoding genes, we examined expression of each sLRR-encoding gene in PRG or en mutants. All sLRR-encoding genes showed altered expression in multiple PRG mutant backgrounds suggesting that the network of sLRR-encoding genes responds to the PRG network. Each sLRR-encoding gene was affected by a different set of PRGs, but run, eve, ftz, and paired (prd) had strong effects on all of the sLRR-encoding genes during at least one stage of embryogenesis.
Changes in expression at gastrulation were observed in some PRG mutant backgrounds. Toll2 expression was altered in ftz, h and a small fraction of odd skipped (odd) mutant embryos (Fig. 5B-D), with changes evident earliest at gastrulation. In all three mutant backgrounds, fusion and/or uneven spacing of the stripes, resulting in doublets of stripes with a wider gap between the doublets, was observed. Changes in Toll8 expression were not observed at gastrulation in h, ftz, odd or prd mutant backgrounds, but Toll8 stripes were fused or expanded in run mutant embryos (Fig. 5F). The only effects seen at cellular blastoderm were on trn. At cellular blastoderm, trn stripes 2-8 were lost in ftz mutant embryos (Fig. 5H) and all trn stripes expanded throughout the trunk region in h mutants (Fig. 5I). No effect of PRG mutant background was seen for Toll6 or Toll7 at these developmental stages.
Figure 5.
A few PRG mutations alter early expression of Toll2, Toll8 and trn. Expression patterns were monitored during the cellular blastoderm or gastrulation stage of development in PRG mutant embryos by in situ hybridization using digoxigenin labeled probes. sLRR-encoding gene indicated at the top of each column; PRG mutation indicated in the lower right corner of each panel. The top panel (A, E, G) in each column shows the wild type pattern for comparison. (A-D) Toll2 in ftz (B), h (C) or odd (D) (E-F) Toll8 in run (F) (G-I) trn in ftz (H) and h (I). Embryos are oriented anterior, left; dorsal, top.
Changes in sLRR expression patterns were more common in PRG mutant embryos during germband extension (Figure 6). All sLRR-encoding genes are expressed in a 13-14-stripe pattern in wild type embryos at this stage (Fig. 6 A, F, J, P; Fig. 3I for trn). Alternate Toll2 stripes were absent in run and, prd, mutant embryos (Fig. 6B, C). The primary stripes were missing in run mutant embryos, while the secondary stripes were missing in prd mutant embryos. Embryos mutant for eve and ftz had fused Toll2 stripes (Fig. 6D, E). In a moderate fraction of odd mutant embryos, Toll2 stripes were unevenly spaced, as they were in younger odd mutants (data not shown). Although Toll2 stripes were unevenly spaced in many h mutant embryos at gastrulation, only a small fraction of the h mutant embryos were affected at germband extension. Those that were affected were missing alternate stripes (data not shown).
The Toll6 pattern was altered in most ftz, prd and h mutant embryos. Toll6 stripes appeared to be fused in embryos mutant for ftz (Fig 6G), prd (Fig. 6H), or h (Fig. 6I). The Toll7 pattern was altered in a moderate fraction of run, ftz, prd, and eve mutant embryos, with even numbered stripes missing in ftz and run mutant embryos and odd numbered stripes missing in prd mutant backgrounds (Fig. 6K-M). eve mutant embryos had unevenly spaced Toll7 stripes (Fig. 6N) while in a moderate number of en mutant embryos, all stripes were missing (Fig. 6O).
Secondary Toll8 stripes were missing in ftz mutants (Fig. 6Q). Since it was somewhat difficult to determine which stripes were missing in the prd mutants (6R), prd mutant embryos and their heterozygous siblings were double stained for En and Toll8. During early germband extension, all the En stripes were present, but only the primary Toll8 stripes were clearly visible. The secondary stripes were very weak or absent. In prd mutant embryos at a similar stage, the remaining En stripes also showed strong Toll8 staining (data not shown). This suggests that the remaining Toll8 stripes are the primary stripes and that the Toll8 stripes lost in prd mutants are the secondary stripes. In eve mutants, Toll8 stripes were unevenly spaced (Fig. 6S), while in run mutants, Toll8 stripes were fused, as they were at earlier stages (Fig. 5F compared to Fig. 6T). In h mutant embryos, Toll8 stripes appear expanded or fused (Fig. 6U). Since there is only fleeting expression of trn in stripes during germband extension, it was difficult to discern alterations in the trn pattern at this stage in PRG mutant embryos.
Discussion
Genes regulating segmentation have been studied intensively over many years, and a complex network of regulatory interactions among them has been described in Drosophila (Reviewed in (Jaynes and Fujioka, 2004; Peel et al., 2005; Schroeder et al., 2011; Wieschaus and Nüsslein-Volhard, 2016). However, the downstream targets of this network responsible for the mechanical implementation of PRG network directives are not well understood. As cell surface proteins capable of forming heterodimers, the sLRRs are good candidates for these functional PRG-target genes involved in defining and/or maintaining cellular interactions within and between segments. Although PRG expression patterns are often described in relatively simple terms (e.g., ftz is expressed in seven stripes at blastoderm), they actually change rapidly and continuously (Clark and Akam, 2016). As shown in Figure 7A, expression of both the PRGs and the sLRR-encoding genes changes significantly and quickly during the early stages of Drosophila embryogenesis, and spatial relationships between different pairs of genes shift over time. Because expression of each sLRR-encoding gene overlaps expression of more than one PRG at various points during development, each sLRR-encoding gene has the potential to be controlled by more than one PRG. Consistent with this, expression of each sLRR-encoding gene was altered in embryos carrying mutations in different PRGs (Figures 5 and 6).
Considering our observations here, along with data from the extensive literature documenting expression patterns of and regulatory interactions among PRGs (i.e., PR-gene cross-regulation) summarized in Table S1, we propose a model for regulatory interactions between PRGs and the sLRRs (Fig. 7B). Note that the experiments presented in this manuscript do not indicate whether interactions between regulators and their targets are direct or indirect; however, they provide a framework for testing such interactions in the future. The following logic was used to establish this model: If a set of sLRR stripes is lost in a particular PRG mutant background, and the PRG expression pattern overlaps with the sLRR stripes that are lost, the simplest explanation is that the PRG activates that sLRR. This scenario is observed in the following cases: run and Toll2 primary stripes (Fig. 6B); prd and Toll2 secondary stripes (Fig. 6C); en and Toll7 (Fig. 6O); run and Toll7 even stripes (Fig. 6K); ftz and Toll8 secondary stripes (Fig. 6Q); ftz and trn stripes 2-8 (Fig. 5H).
If we observed that a PRG was expressed in the regions between sLRR stripes, and loss of the PRG caused expansion of the sLRR stripes, the simplest explanation would be that the PRG acts as a repressor of that particular Toll gene. This pattern fits the expansion of Toll8 in run mutant embryos (Fig. 5F, 6T); Toll2 in h and odd mutant embryos (Fig. 5B, C and 6E); and trn in h mutant embryos (Fig. 5I). In the case of the fusion or doublets of Toll2, 7 and 8 in an eve mutant background (Fig. 6D, N and S), it seems likely that this is due to the effects of the eve mutation on the width of the odd-numbered parasegments. As clearly described in the model of Jaynes and Fujioka (2004), partial loss of Eve causes the odd-numbered parasegments to narrow (Coulter and Wieschause, 1988; DiNardo and O’Farrell, 1987; Fujioka et al., 1995; Jaynes and Fujioka, 2004), which would bring the Toll stripes closer together, explaining the doublets we observed.
If we observed an effect on an sLRR from a PRG that only partially or transiently overlapped the sLRR, we checked to see if activation or repression during that period would be reasonable in light of the observed phenotype. For example, although h, ftz and prd partially overlap the Toll6 primary stripes, loss of any of these PRG causes fusion/doublets of Toll6 (Fig. 6G-I) suggesting loss of repression. One possibility is that these PRGs act as repressors but expression of these PRGs is normally low enough in the overlap region to allow expression of Toll6. When the PRG is missing, Toll6 spreads from the normal region into adjacent cells. Similarly, prd expression overlaps part of the regions covered by the Toll8 primary and secondary stripes (Fig. 7A). Since the secondary stripes of Toll8 are missing in prd mutant embryos (Figure 6R and data not shown) prd probably acts with ftz to activate the secondary stripes. In this model, the Toll8 primary stripes lack an activator. It may be that Toll8 is activated by a gene that is expressed throughout the early embryo (eg. opa or ftz-f1), and the stripes are produced by repression of Toll8 by runt (Fig. 7A). Alternatively, there may be a specific activator that was not included in this study.
For some regulatory interactions, alternative hypotheses can be proposed by considering known relationships among PRGs. First, since ftz activates trn, it is likely that the expansion of trn expression in h mutant embryos (Figure 5I) is due to the expansion of ftz in h mutants (Ingham and Gergen, 1988). Second, the expansion of Toll2 in h mutant embryos (Fig. 5C) would be due to the expansion of runt which occurs in h mutant embryos (Ingham and Gergen, 1988; Jiménez et al., 1996). Third, the loss of Toll2 in runt mutant embryos would be explained by persistence of the proposed Toll2 repressor eve, since runt would normally repress eve in the posterior of odd-numbered parasegments (Ingham and Gergen, 1988; Jiménez et al., 1996; Manoukian and Krause, 1993).
Three cases fit none of the categories mentioned above. The expansion of Toll2 in ftz mutant embryos (Fig. 5B, 6E) suggests that ftz acts as a repressor of Toll2. However, ftz expression overlaps the posterior of the even Toll2 stripes through much of early embryogenesis, making this simple explanation unlikely. The expansion of Toll2 in ftz mutant embryos is probably due to loss of odd in ftz mutants, (Jaynes and Fujioka, 2004; Nasiadka and Krause, 1999), with odd acting as a Toll2 repressor. ftz and prd regulate alternate en stripes (DiNardo and O’Farrell, 1987; Fujioka et al., 1996; Howard and Ingham, 1986; Ish-Horowicz et al., 1989), and all three genes impact Toll7 expression (Fig. 6L, M and O), suggesting that ftz and prd could exert their effects on Toll7 via en. However, we propose that the effects of ftz and prd on Toll7 are only partially indirect, since mutations in both ftz and prd have more consistent, strong effects on even and odd Toll7 stripes, respectively, than does en (Fig. 6L, M, O). It should also be noted that the Toll7 and en expression patterns do not completely overlap since en expression extends beyond the ventral ectoderm (Kornberg, 1981a, b) while Toll7 expression does not (Kambris et al., 2002), Fig. 2N, 4M). Thus, it seems likely that although en likely regulates part of the Toll7 expression pattern there are additional factors regulating Toll7. The effect of h on Toll8 (Fig 6U) remains difficult to interpret.
These results extend those of Pare et al. (2014), who examined the effect of eve and run on three Toll family genes. Our study made use of strong hypomorphs of eve and run (Frasch et al., 1987; Lifschytz and Falk, 1968; Torres and Sánchez, 1992) while Pare et al. (2014) used eve and run null mutants (Duffy and Gergen, 1991; Nusslein-Volhard and Wieschaus, 1980). This led to some differences in expression patterns observed but these differences are consistent with the use of these different types of alleles. For example, we observed Toll2 doublets in eve mutants (Fig. 5f, 6t), while Pare et al. observed a broad expansion of Toll2 stripes in an eve mutant background. In one case, our results were inconsistent with Pare et al. (2014): they reported complete loss of Toll8 stripes in eve mutants while we found doublets. One possible explanation is that, given that eve represses run (Fujioka et al., 1995; Manoukian and Krause, 1992) a null mutation in eve would allow the run domain to expand more widely than a weaker eve allele causing complete repression of the Toll8 stripes.
Overall, our data suggest that regulatory information from the PRGs establishes a code of cell surface proteins that mark cells with unique identities. Specifically, the repeating units comprised of one odd- and one even-numbered parasegment primoridia are generally thought to consist of 8 rows of cells at the blastoderm stage. Combinatorial action of the PRGs directs expression of the sLRRs such that each row expresses a unique combination of sLRR genes at gastrulation (Supp. Fig. 4). Specifically, in these repeating units, cell 1 is marked by the expression of Toll2,7 and 8; cell2 expresses only Toll8; cell 3 expressed Toll6; cell 4, Toll2 and 6; cell 5, Toll 2,7,8, and trn; cell 6, Toll 6,8, trn; cell 7, Toll 6, trn; cell 8, only Toll 2. By the end of germband extension, a segmentally repeating matching pattern emerges that matches that seen in the odd PS at gastrulation (Supp. Fig. 4). Thus, the alternate segment periodicity and combinatorial action of the PRGs directs the establishment of sets of cells along the anterior-posterior axis in double-width segmental units, each with a unique cellular identity.
Supplementary Material
Highlights.
Tartan and related Toll proteins form a subset of the Leucine Rich Repeat family of cell surface proteins that is distinguished by expression in segmentally repeating pattern during Drosophila development (sLRR proteins).
sLRR-encoding genes have dynamic expression patterns, with each expressed in a unique register along the anterior-posterior axis, marking each cell in the double segment primorida with a unique cell surface code.
The sLRR-encoding gene are regulated by the pair-rule gene network, linking determination to differentiation during segmentation.
Acknowledgements
Thanks to Judy Wexler and Alys Jarvela for helpful comments on the manuscript. We thank reviewer 1 for the detailed comments and helpful suggestions that significantly improved the manuscript. Fluorescent images were performed at the University of Maryland imaging core facility. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. Funding: This work was supported by National Institutes of Health grant GM113230 to LP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Artero R, Furlong EE, Beckett K, Scott MP, Baylies M, 2003. Notch and Ras signaling pathway effector genes expressed in fusioncompetent and founder cells during Drosophila myogenesis. 6257–6272. [DOI] [PubMed]
- Benton MA, Pechmann M, Frey N, Stappert D, Conrads KH, Chen YT, Stamataki E, Pavlopoulos A, Roth S, 2016. Toll Genes Have an Ancestral Role in Axis Elongation. Curr Biol 26, 1609–1615. [DOI] [PubMed] [Google Scholar]
- Butler LC, Blanchard GB, Kabla AJ, Lawrence NJ, Welchman DP, Mahadevan L, Adams RJ, Sanson B, 2009. Cell shape changes indicate a role for extrinsic tensile forces in Drosophila germ-band extension. Nat Cell Biol 11, 859–864. [DOI] [PubMed] [Google Scholar]
- Chan LN, Gehring W, 1971. Determination of blastoderm cells in Drosophila melanogaster. Proc Natl Acad Sci U S A 68, 2217–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Price BD, Bockheim S, Boedigheimer MJ, Smith R, Laughon A, 1993. Molecular and genetic characterization of the Drosophila tartan gene. Dev Biol 160, 315–332. [DOI] [PubMed] [Google Scholar]
- Clark E, Akam M, 2016. Odd-paired controls frequency doubling in Drosophila segmentation by altering the pair-rule gene regulatory network. [DOI] [PMC free article] [PubMed]
- Coulter DE, Wieschause, 1988. Gene activities and segmental patterning in Drosophila: analysis of odd-skipped and pair-rule double mutants. Genes and Development 2, 1812–1823. [DOI] [PubMed] [Google Scholar]
- DiNardo S, O’Farrell PH, 1987. Establishment and refinement of segmental pattern in the Drosophila embryo: spatial control of engrailed expression by pair-rule genes., 1212–1225. [DOI] [PubMed]
- Duffy JB, Gergen JP, 1991. The Drosophila segmentation gene runt acts as a position-specific numerator element necessary for the uniform expression of the sex-determining gene Sex-lethal. Genes Dev 5, 2176–2187. [DOI] [PubMed] [Google Scholar]
- Eldon E, Kooyer S, D’Evelyn D, Duman M, Lawinger P, Botas J, Bellen H, 1994. The Drosophila 18 wheeler is required for morphogenesis and has striking similarities to Toll. Development 120, 885–899. [DOI] [PubMed] [Google Scholar]
- Field A, Xiang J, Anderson WR, Graham P, Pick L, 2016. Activation of Ftz-F1-Responsive Genes through Ftz/Ftz-F1 Dependent Enhancers. PLoS ONE 11, 0163128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M, 1987. Characterization and localization of the even-skipped protein of Drosophila. Embo j 6, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Warrior R, Tugwood J, Levine M, 1988. Molecular analysis of even-skipped mutants in Drosophila development. Genes Dev 2, 1824–1838. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB, Goto T, 1995. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development 121, 4371–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Miskiewicz P, Raj L, Gulledge AA, Weir M, Goto T, 1996. Drosophila Paired regulates late even-skipped expression through a composite binding site for the paired domain and the homeodomain. 2697–2707. [DOI] [PubMed]
- Gergen JP, Coulter D, Wieschaus EF, 1986. Segmental pattern and blastoderm cell identities. Gametogenesis and the Early Embryo, 195–220. [Google Scholar]
- Gilbert SF, 2010. Developmental Biology, Ninth Edition Sinauer Associates, Sunderland, MA. [Google Scholar]
- Grosshans J, Wieschaus E, 2000. A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell 101, 523–531. [DOI] [PubMed] [Google Scholar]
- Howard K, Ingham P, 1986. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell 44, 949–957. [DOI] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C, 2012. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Oxford Journals, Systemic Biology, pp. 1061–1067. [DOI] [PubMed] [Google Scholar]
- Ingham P, Gergen P, 1988. Interactions between the pair-rule genes runt, hairy, even-skipped and fushi tarazu and the establishment of periodic pattern in the Drosophila embryo. 51–60.
- Irvine KD, Wieschaus E, 1994. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development 120, 827–841. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D, Pinchin SM, Ingham PW, Gyurkovics HG, 1989. Autocatalytic ftz activation and metameric instability induced by ectopic ftz expression. Cell 57, 223–232. [DOI] [PubMed] [Google Scholar]
- Jaynes JB, Fujioka M, 2004. Drawing lines in the sand: even-skipped et al and parasegment boundaries. Developmental Biology 269, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez G, Pinchin SM, Ish-Horowicz D, 1996. In vivo interactions of the Drosophila Hairy and Runt transcriptional repressors with target promoters. EMBO J 15, 7088–7098. [PMC free article] [PubMed] [Google Scholar]
- Kambris Z, Hoffmann JA, Imler JL, Capovilla M, 2002. Tissue and stage-specific expression of the Tolls in Drosophila embryos. Gene Expr Patterns 2, 311–317. [DOI] [PubMed] [Google Scholar]
- Kornberg T, 1981a. Compartments in the abdomen of Drosophila and the role of the engrailed locus. Developmental Biology 86, 363–372. [DOI] [PubMed] [Google Scholar]
- Kornberg T, 1981b. Engrailed: a gene controlling compartment and segment formation in Drosophila. Proc. Natl. Acad. Sci. USA 78, 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E, 2004. Multiplex Detection of RNA Expression in Drosophila Embryos. 846. [DOI] [PubMed]
- Krantz DE, Zipursky SL, 1990. Drosophila chaoptin, a member of the leucine-rich repeat family, is a photoreceptor cell-specific adhesion molecule. EMBO J 9, 1969–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A, 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets, Bioinformatics, pp. 3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P, 1989. Pattern formation in the Drosophila embryo: allocation of cells to parasegments by even-skipped and fushi tarazu. Development 105, 761–767. [DOI] [PubMed] [Google Scholar]
- Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R, 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43, W580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschytz E, Falk R, 1968. Fine structure analysis of a chromosome segment in Drosophila melanogaster. Analysis of x-ray-induced lethals. Mutat Res 6, 235–244. [DOI] [PubMed] [Google Scholar]
- Lohs-Schardin M, Cremer C, Nusslein-Volhard C, 1979. A fate map for the larval epidermis of Drosophila melanogaster: localized cuticle defects following irradiation of the blastoderm with an ultraviolet laser microbeam. Dev Biol 73, 239–255. [DOI] [PubMed] [Google Scholar]
- Manoukian AS, Krause HM, 1992. Concentration-dependent activities of the even-skipped protein in Drosophila embryos. Genes Dev 6, 1740–1751. [DOI] [PubMed] [Google Scholar]
- Manoukian AS, Krause HM, 1993. Control of segmental asymmetry in Drosophila embryos. Development 118, 785–796. [DOI] [PubMed] [Google Scholar]
- Mao Y, Kerr M, Freeman M, 2008. Modulation of Drosophila Retinal Epithelial Integrity by the Adhesion Proteins Capricious and Tartan, PLoS ONE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan M, Perez L, Cohen SM, 2005. Boundary formation in the Drosophila wing: functional dissection of Capricious and Tartan. Dev Dyn 233, 804–810. [DOI] [PubMed] [Google Scholar]
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F, 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops, Bioinformatics, pp. 126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miláxn M, Weihe U, Pérez L, Cohen SM, cohen@embl-heidelberg.de, 2001. The LRR Proteins Capricious and Tartan Mediate Cell Interactions during DV Boundary Formation in the Drosophila Wing. Cell 106, 785–794. [DOI] [PubMed] [Google Scholar]
- Nasiadka A, Krause HM, 1999. Kinetic analysis of segmentation gene interactions in Drosophila embryos. Development 126, 1515–1526. [DOI] [PubMed] [Google Scholar]
- Nose A, Mahajan VB, Goodman CS, 1992. Connectin: A homophilic cell adhesion molecule expressed on a subset of muscles and the motoneurons that innervate them in Drosophila. Cell 70, 553–567. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E, 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801. [DOI] [PubMed] [Google Scholar]
- Pare AC, Vichas A, Fincher CT, Mirman Z, Farrell DL, Mainieri A, Zallen JA, 2014. A positional Toll receptor code directs convergent extension in Drosophila. Nature 515, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel AD, Chipman AD, Akam M, 2005. Arthropod Segmentation: beyond the Drosophila paradigm. Nature Reviews Genetics 6, 905–916. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Hartley DA, Walther Z, Artavanis-Tsakonas S, 1988. slit: An EGF-homologous locus of D. melanogaster involved in the development of the embryonic central nervous system. Cell 55, 1047–1059. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S, 1990. slit: an extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes Dev 4, 2169–2187. [DOI] [PubMed] [Google Scholar]
- Schroeder MD, Greer C, Gaul U, 2011. How to make stripes: deciphering the transition from non-periodic to periodic patterns in Drosophila segmentation. 3067–3078. [DOI] [PMC free article] [PubMed]
- Shishido E, Takeichi M, Nose A, 1998. Drosophila Synapse Formation: Regulation by Transmembrane Protein with Leu-Rich Repeats, CAPRICIOUS. Science, 2118–2121. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Swiss Federal Institute of Technology Lausanne, S.o.C.a.C.S.P.M., STATION 14, CH-1015 Lausanne, Switzerland, 2018. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0, Mol Biol Evol, pp. 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C, 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback | SpringerLink. Chromosoma 2, 81–85. [DOI] [PubMed] [Google Scholar]
- Torres M, Sánchez L, 1992. The segmentation gene runt is needed to activate Sex-lethal, a gene that controls sex determination and dosage compensation in Drosophila. Genet Res 59, 189–198. [DOI] [PubMed] [Google Scholar]
- Van Vactor D, Krantz DE, Reinke R, Lawrence Zipursky S, 1988. Analysis of mutants in chaoptin, a photoreceptor cell-specific glycoprotein in Drosophila, reveals its role in cellular morphogenesis. Cell 52, 281–290. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C, 2016. The Heidelberg Screen for Pattern Mutants of Drosophila: A Personal Account. Annu Rev Cell Dev Biol 32, 1–46. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Sweeton D, Costa M, 1991. Convergence and extension during germband elongation in Drosophila embryos, in: Keller R (Ed.), Gastrulation. Plenum Press, New York. [Google Scholar]
- Yagi Y, Nishida Y, Ip YT, 2010. Functional analysis of Toll-related genes in Drosophila. Dev Growth Differ 52, 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


