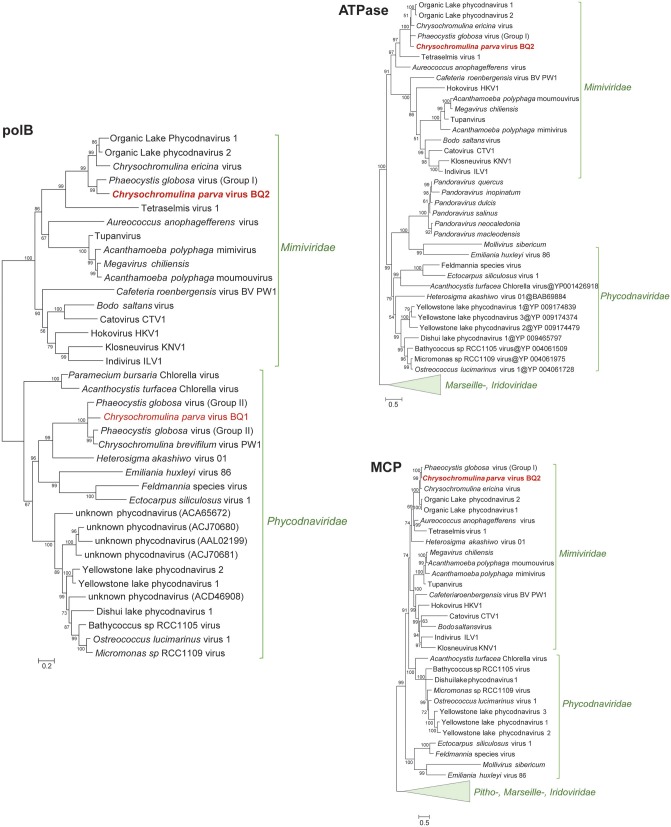
In the original article, there was a mistake in Figure 2 as published. In the figure, the tree branch closest to Chrysochromulina parva virus BQ2 labeled as “Phaeocystis globosa virus (group II)” should have been labeled “Phaeocystis globosa virus (group I).” Similarly, the two branches labeled as “Phaeocystis globosa virus (group I)” and “Phaeocystis globosa virus” adjacent to Chrysochromulina parva virus BQ1 should have both been labeled as “Phaeocystis globosa virus (group II).”
Figure 2.
Approximately maximum-likelihood phylogenetic trees of B-family DNA polymerase (polB), A32-like virion packaging ATPase (ATPase), and the major capsid protein (MCP). The polB tree is constructed on protein fragments corresponding to PCR amplicons reported in Mirza et al. (2015). When more than one MCP gene was present in a genome (Mimiviridae), a paralog closest to Phycodnaviridae was chosen for the MCP tree. Node support (aLRT-SH statistic) >50% are shown. Accession numbers for the polB sequences are provided in Supplementary Table 1.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
References
- Mirza S. F., Staniewski M. A., Short C. M., Long A. M., Chaban Y. V., Short S. M. (2015). Isolation and characterization of a virus infecting the freshwater algae Chrysochromulina parva. Virology 486, 105–115. 10.1016/j.virol.2015.09.005 [DOI] [PubMed] [Google Scholar]