Abstract
Background
The heterotrimeric GTP-binding protein eIF2 forms a ternary complex with initiator methionyl-tRNA and recruits it to the 40S ribosomal subunit for start codon selection and thereby initiates protein synthesis. Mutations in EIF2S3, encoding the eIF2γ subunit, are associated with severe intellectual disability and microcephaly, usually as part of MEHMO syndrome.
Methods
Exome sequencing of the X chromosome was performed on three related males with normal head circumferences and mild learning difficulties, hypopituitarism (GH and TSH deficiencies), and an unusual form of glucose dysregulation. In situ hybridisation on human embryonic tissue, EIF2S3-knockdown studies in a human pancreatic cell line, and yeast assays on the mutated corresponding eIF2γ protein, were performed in this study.
Findings
We report a novel hemizygous EIF2S3 variant, p.Pro432Ser, in the three boys (heterozygous in their mothers). EIF2S3 expression was detectable in the developing pituitary gland and pancreatic islets of Langerhans. Cells lacking EIF2S3 had increased caspase activity/cell death. Impaired protein synthesis and relaxed start codon selection stringency was observed in mutated yeast.
Interpretation
Our data suggest that the p.Pro432Ser mutation impairs eIF2γ function leading to a relatively mild novel phenotype compared with previous EIF2S3 mutations. Our studies support a critical role for EIF2S3 in human hypothalamo-pituitary development and function, and glucose regulation, expanding the range of phenotypes associated with EIF2S3 mutations beyond classical MEHMO syndrome. Untreated hypoglycaemia in previous cases may have contributed to their more severe neurological impairment and seizures in association with impaired EIF2S3.
Fund
GOSH, MRF, BRC, MRC/Wellcome Trust and NIGMS funded this study.
Keywords: Hypopituitarism, Hypoglycaemia, Glucose dysregulation, Translation initiation, Protein synthesis, EIF2S3
Research in context.
Evidence before this study
A non-consanguineous white European pedigree with three affected male patients, presented with severe recurrent hypoglycaemia, short stature with GH and TSH deficiencies, an unusual form of glucose dysregulation, and mild learning difficulties. This combination of phenotypes has, to date, not been reported in the literature. There were no publications linking these phenotypes in any online journal listed on Pubmed or elsewhere.
Added value of this study
Exome sequencing of the X-chromosome revealed a novel missense variant c.1294C > T in EIF2S3 (p.Pro432Ser), encoding a subunit of the eukaryotic translation initiation factor 2, eIF2γ, in all three male patients and in the heterozygous mothers. Mutations in EIF2S3 have been described in patients with MEHMO syndrome, characterized by severe intellectual disability, microcephaly, short stature, epilepsy and accompanying midline and facial abnormalities. The functional and expression data in our study show a moderate but significant eIF2γ impairment, which relates to the milder phenotype in our three male patients.
Implications of all the available evidence
This milder loss of function compared with previous EIF2S3 mutations gives rise to a phenotype that is distinct from the classical spectrum of MEHMO syndrome. Untreated hypoglycaemia in the previously published cases may have contributed to their more severe impairment of neurodevelopment and seizures. We highlight that pancreatic and pituitary phenotypes appear to be associated with EIF2S3 mutations. Early identification of such patients with a rapid molecular diagnosis may lead to prevention of significant morbidity, and may be critical for the prevention of significant neurodevelopmental delay in these patients.
Alt-text: Unlabelled Box
1. Introduction
The eukaryotic translation initiation factor (eIF) 2 subunit 3 (EIF2S3) (NM_001415; Xp22.11) gene encodes the gamma (γ) subunit of the translation initiation factor eIF2; the largest of the three eIF2 subunits. In translation initiation, eIF2 binds GTP via eIF2γ, and the initiator methionyl-tRNA (Met-tRNAiMet), to form a ternary complex that scans mRNA for the AUG start codon [1]. Mutations in several translation factors, including eIF2 subunits, enhance translation initiation at near cognate codons such as AUU and UUG [[1], [2], [3]]. EIF2S3 is located within Xp21.1-p22.13, a region linked to a rare intellectual disability (ID) disorder designated as MEHMO syndrome (OMIM 300148) [4]. MEHMO syndrome exhibits phenotypic heterogeneity and is variably characterized by mental retardation, epileptic seizures, hypogonadism with hypogenitalism, microcephaly, and obesity. Life expectancy ranges from <1 year-adulthood and the condition is associated with significant morbidity and mortality.
An EIF2S3 missense substitution, p.Iso222Thr, was previously reported in a pedigree with three male children with MEHMO syndrome. Clinical features included moderate-to-severe intellectual disability (ID), microcephaly, short stature, epilepsy and facial dysmorphic features [5]. Each affected individual had unique additional features consisting of cleft lip/palate, behavioural problems, generalised seizures, post-pubertal microgenitalism and obesity. Two patients had growth hormone deficiency (GHD); however, no detailed data on pituitary function were presented, as the study focused on the neurological phenotype. Functional studies of the mutation in the corresponding residue in yeast eIF2γ revealed substantially impaired eIF2β binding to eIF2γ, and impaired translation start codon selection.
Further EIF2S3 mutations have been identified in patients with similar phenotypes. A missense substitution p.Iso259Met [6], and a frameshift p.Iso465Serfs*4 resulted in severe ID, microcephaly, GHD and epilepsy with additional features such as spastic quadriplegia, delayed puberty and genital abnormalities [6,7]. The patients also manifested hypoglycaemia, although the cause of this was unknown. A recent study reported the EIF2S3 p.Iso465Serfs*4 in three families with MEHMO syndrome, and a novel maternally inherited missense EIF2S3 variant, p.Ser108Arg, in an unrelated male patient with milder symptoms [7]. Therefore all reported mutations described in EIF2S3 have until now been identified in MEHMO patients with cardinal phenotypic features including significant ID and microcephaly. We now report a novel p.Pro432Ser missense EIF2S3 variant associated with partial loss of function in three boys with a milder phenotype including hypopituitarism and pancreatic dysfunction.
2. Materials and methods
2.1. DNA sequencing
The coding regions of the X-chromosome were sequenced in Pedigree 1 in the Department of Genetics, University Medical Center Utrecht, Netherlands, in collaboration with GOSgene, London UK. Next-generation sequencing of all protein coding sequences on the X chromosome (X-exome) was performed as previously described. Barcoded fragment libraries were pooled in equimolar ratios and enriched using multiplexed targeted genomic enrichment [8] with the Demo X-exome enrichment kit (Agilent Technologies, Santa Clara, CA, USA) and sequenced according to the SOLiD 3 Plus manual (Life Technologies, Carlsbad, CA, USA). Raw sequencing data were mapped against the GRCh37/hg18 reference genome using a custom bioinformatic pipeline based on the BWA algorithm. The percentage cut-off for the non-reference allele (NRA) of candidate single nucleotide variants and small indels was set to 15%. The cut-off for minimum sequence coverage was set to 10 reads. Filtering of variants that fit an X-linked dominant inheritance model was performed using non-stringent criteria for affected males being hemizygous for the candidate causal variant (>50% NRA). Subsequently, all common and rare polymorphisms present in Ensembl 65, ExAC, gnomAD or our in-house X-exome database of ~100 samples, were marked as known and not further considered for this study. The remaining variants were considered to be novel and thus fulfil the criteria for an ultra-rare disease. Protein prediction models were consulted, that use straightforward physical and comparative considerations reflecting pathogenicity of a variant. SIFT scores range from 0 (deleterious) to 1 (tolerated), whilst Polyphen-2 scores range from 0 (benign) to 1 (damaging) respectively.
2.2. PCR and direct sequencing analysis
PCR primers were designed using the Ensembl Genome Browser (http://www.ensembl.org/index.html), the UCSC genome browser (https://genome.ucsc.edu/) and the Primer3 input (http://bioinfo.ut.ee/primer3-0.4.0/). DNA was extracted from 103 patient blood samples from our cohort and the coding regions of EIF2S3 were amplified by PCR using exon flanking primers and the BIOTAQ™ DNA Polymerase kit (Bioline) on an Eppendorf Thermocycler. PCR products were treated with MicroClean reagent (Web Scientific). The precipitate was sequenced with forward and reverse respective primers using the BigDye® Terminator v1.1 Cycle Sequencing Kit (Life Technologies Ltd) on an Eppendorf Thermocycler. The samples were run on a 3730XL DNA Analyzer (Applied Biosystems/Hitachi, Japan). Detailed PCR and sequencing conditions and primer sequences are available upon request. Control databases ExAC and gnomAD were consulted.
2.3. Cell culture
A hybrid cell line (1.1B4 cells) formed by the electrofusion of a primary culture of human pancreatic islets with PANC-1, a human pancreatic ductal carcinoma cell line, was obtained from Public Health England (PHE). The cells were maintained in a humidified CO2 incubator at 37 °C in Roswell Park Memorial Institute (RPMI) medium 1640 containing l-glutamine (Life Technologies), supplemented with 10% fetal calf serum (FCS) and 5% penicillin/streptomycin (penstrep). The cells were washed in diluted Hanks Balanced Saline Solution (Gibco) (x1 HBSS), and trypsinised with trypsin (Gibco) diluted in x1 HBSS in line with manufacturer's instructions. Cells were passaged at >80% confluence.
2.4. Constructs containing shRNA cassettes
GFP-IRES-Puromycin-Zeomycin plasmids (pGIPZ) (11,744 bp), referred to in this study as Clone 1–4, contained small hairpin RNA (shRNA) cassettes targeting the EIF2S3 human gene, validated by the UCL Cancer Institute, and 100% matched with the expected hairpin sequence according to the Open Biosystems library database. An additional non-silencing (NS) pGIPZ plasmid containing a scrambled shRNA sequence was used as a control in transduction experiments. All five plasmids have a lentiviral (LV) backbone; long terminal repeats (LTR's) with modified U3 and a packaging signal. The pGIPZ plasmids also contained GFP and a puromycin-resistance cassette. This enabled the cells to be monitored for GFP expression and be puromycin-selected following transduction (the standard map of the pGIPZ plasmid is available upon request). A maxiprep of each plasmid was prepared and used in LV packaging and transduction assays.
2.5. Lentiviral packaging
LV packaging was carried out under appropriate containment by transient co-transfection of HEK293T cells (4.0 × 105 cells/well of a 6 well plate seeded 24 h prior transfection) for each pGIPZ plasmid in duplicate, with 500 ng of the pGIPZ vector constructs, 333 ng of packaging plasmid (pCMV-dR8.91), 333 ng of VSV-G envelope expressing plasmid (pMD2.G), Fugene HD (3.5 μl/well) transfecting agent (Promega) and opti-mem media (Thermo Fisher Scientific). Media was replaced after 24 h incubation at 37 °C, with subsequent harvesting and filtering of the vector 24 h later. Infectious titres were determined by limiting dilutions of LVs on HEK293T cells. Titres were comprised between 105 and 106 transducing units/ml.
2.6. Transduction of 1.1B4 cells using packaged LV vectors for stable gene knockdown
1.1B4 cells were seeded into 6-well plates and transduced at a multiplicity of infection (MOI) of 5, using the previously generated lentivirus under strict sterile and contained conditions. Cells were incubated at 37 °C for 72 h before media was removed. Fresh RPMI medium containing 10% FCS and 5% penstrep, also used on non-transduced cells, was supplemented with puromycin and added to the cells to positively select the transduced cells expressing shRNA. The cells were monitored and kept under puromycin selection for a further 10 days, trypsinised when confluent and expanded into T25cm2 flasks respectively. The cells were washed in x1 HBSS and visualised under an Olympus IX71 inverted fluorescence microscope for the presence of GFP until ready for lysing.
2.7. qPCR analysis
Cells were lysed using RNeasy lysis buffer (RLT) and RNA was extracted using the RNeasy MiniKit (Qiagen) including a DNase digestion step using the RNase-Free DNase Set (Qiagen). The High Capacity RNA-to-cDNA Kit (Applied Biosystems) was used to yield cDNA from each RNA population. Fast SYBR Green Master Mix (Life Technologies) was used in the qPCR reactions in this study. Intron flanking primers for qPCR were designed using the Universal Probe Library database (Roche) for the target gene (EIF2S3) and the three housekeeping genes (GAPDH, β-ACTIN and HPRT). The cDNA derived from the reverse transcription was diluted 1:5 in all qPCR reactions and nuclease-free water was used as the blank.
2.8. Apoptosis assay
1.1B4 cell populations were seeded into white 96-well sterile tissue culture treated microplates with clear bottoms (PerkinElmer, Cat: 6005181), at 10,000 cells/well in 200 μl of RPMI media, and incubated overnight at 37 °C. Cytokine mix (100 μl/well) containing 1 L-1β (50 U/ml), TNF-α (1000 U/ml), INF-γ (1000 U/ml) was added, diluted in media and incubated for 16 h at 37 °C, to induce caspase activity and thus increase apoptosis in cells. Caspase-Glo 3/7 reagent (Promega, Cat: G8090) was added at a 1:1 ratio of reagent:sample (100 μl) to each well and incubated for 1 h at room temperature before reading the luminescent signal generated on the luminometer. There were triplicate wells of each of the following for Clone 4 (the EIF2S3 knockdown cell line), non-transduced (NT) and NS cell populations: blank wells with no cells and media containing cytokine mix, wells containing cells with media without cytokine mix, and wells containing cells with cytokine mix.
2.9. In situ hybridisation on human embryonic sections
Digoxigenin (DIG) RNA probes were made using the purified pCMV-SPORT6 vector containing full-length human wild-type (WT) EIF2S3 cDNA (IMAGE ID: 4419438) (Source Bioscience). Human embryonic tissue sections were selected at Carnegie stage (CS) 16, 19, 20, 23 (equivalent to gestational age 5.5, 6, 7 and 8 weeks) respectively obtained from the Human Developmental Biology Resource (HDBR) tissue resource. Gene expression studies were performed by in situ hybridisation as previously described [9], to generate a human embryonic expression profile in the hypothalamo-pituitary (HP) region and the pancreas.
2.10. Yeast strains and plasmids
Yeast strain J515 (identical to J212 [10]; MATα leu2-3,-112 ura3-52 his3 gcd11Δ::KanMX p[GCD11, URA3]) was used for eIF2γ mutant analysis. Mutant and WT GCD11 constructs were introduced into J515 as the sole source of eIF2γ by plasmid shuffling [11]. Plasmids used in this study are listed and include p180 [GCN4-lacZ, URA3 12], p367 [HIS4(AUG)-lacZ, URA3 13], p391 [HIS4(UUG)-lacZ, URA3 13], pC2872 [His8-GCD11 (eIF2γ), LEU2 10], pC5856 [His8-gcd11-I318M in pC2872 [14]], pC5861 [His8-gcd11-S489 N-A491V in pC2872; this study] and pC5862 [His8-gcd11-S489 N-P490S-A491V in pC2872; this study].
2.11. β-galactosidase assays
For measurement of GCN4-lacZ, HIS4(UUG)-lacZ or HIS4(AUG)-lacZ expression, overnight yeast cultures were grown in synthetic complete (SC) medium and used to inoculate 25 ml of SC medium at OD600 = 0.25. For measurement of GCN4-lacZ, cultures were grown for 2 h (untreated or treated with 1 μg/ml sulfometuron methyl (SM), an inhibitor of leucine, isoleucine, and valine biosynthesis). Cultures continued growing for 6 h to OD600 ≤ 1 and β-galactosidase activities were determined as previously described [12,15]. For measurement of HIS4(UUG)-lacZ and HIS4(AUG)-lacZ expression, cultures were left untreated and grown for 6 h to OD600 ≤ 1, followed by measurement of β-galactosidase activities. Averages and standard deviations of β-galactosidase activities were calculated for three independent transformants. Statistical significance was calculated using an ANOVA test followed by a post hoc Tukey's test (P < 0.05).
2.12. Patient phenotypes
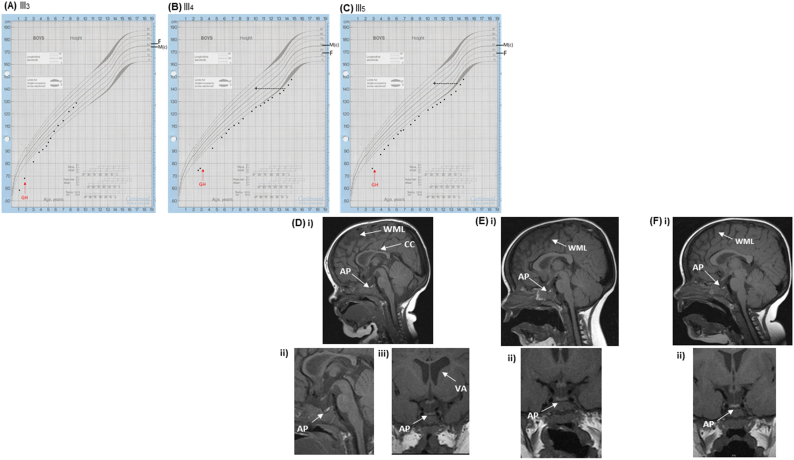
Three affected males, monozygotic (identical) twin brothers (III4, III5) and their maternal cousin (III3), born to a non-consanguineous white European pedigree, presented with severe recurrent hypoglycaemia, short stature with GH and TSH deficiencies, and an unique pancreatic phenotype (Table 1). They have an unusual form of glucose dysregulation which fluctuates between hyperinsulinaemic hypoglycaemia and post-prandial hyperglycaemia (Table 1). They were treated with rhGH (Fig. 1A-C), thyroxine and diazoxide together with chlorothiazide. The latter two medications were stopped as the hyperinsulinism resolved at 7 years of age; however, the twins manifested glucose dysregulation with post-prandial hyperglycaemia and variable fasting hypoglycaemia. Cortisol and prolactin concentrations are normal, and the twins have progressed through puberty. Endocrine data are presented in Table 1. Additional features in the three boys include intestinal lymphonodular hyperplasia with eosinophilic infiltration. Brain Magnetic Resonance Imaging (MRI) revealed variable white matter loss and a small anterior pituitary (AP) with a normal posterior pituitary (PP) and stalk in the three boys (Fig. 1D-F). In addition, hypoplasia of the corpus callosum and ventricular asymmetry were noted in patient III3 (Fig. 1D). Severe ID and microcephaly were not present in the boys, and all attend a mainstream school. The older twin brothers are maintaining educational parity with their peers, although they need some support.
Table 1.
Clinical data from the three affected males in Pedigree 1; III3, III4, III5. Patients presented with GH deficiency, and low IGF1 and IGFBP3 concentrations. Their cortisol and prolactin concentrations were normal. The twin brothers, III4 and III5, developed central hypothyroidism and were treated with thyroxine. Values following a gonadotrophin secretion test and a 3-day HCG test are shown. SDS, standard deviation score; NR, normal range; HCG, human chorionic gonadotrophin; SDS, standard deviation score; HC, head circumference.
| Patient | Birth weight kg (SDS) (gestation) | Age at presentation years | Height at presentation cm (SDS) | Peak GH to provocation μg/L | IGF1 ng/ml (NR) | IGFBP3 mg/L(NR) | Most recent morning cortisol nmol/L | FT4 (pre-treatment) pmol/L (NR) | TSH (pre-treatment) mU/L | PRL mU/L (NR) | Peak LH IU/L (GnRH at age 12 years) | Peak FSH IU/L (GnRH at age 12 years) | Peak testosterone to 3 day HCG nmol/L (age in years) | Most recent height SDS (age in years) | Glucose mmol/L (age in years) | Insulin mU/L (age in years) | HC SDS (age in years) | Diazoxide treatment (age in years) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| III3 | 2.1 (−2.8) (38/40) |
1.13 | 58.8 (−6.7) |
<0.1 on profile | <25 | <0.5 | 315 | 12.6 (12−22) Not treated |
5 | 360 (40–555) |
N/A | N/A | 0.992 (3) |
−0.30 (8.8) |
3.3 (0.25) |
5.9 (0.25) |
−2.2 (7.5) |
0.75–6.8 |
| III4 | 2.15 (−0.3) (34/40) |
2.2 | 71.5 (−4.4) |
1.1 | 9 (20–158) |
0.67 (1.2–3.7) |
183 | 11.4 (12–22) |
2.9 | 225 (40–555) |
6.3 | 4.0 | 1.99 (12) |
−2.07 (14.6) |
3.4 (2.2) |
6.8 (2.2) |
−1.06 (13.1) | 2.5–6.7 |
| III5 | 1.93 (−1.5) (34/40) |
2.2 | 69.5 (−5.2) |
0.7 | 10 (20–158) |
1.2 (1.2–3.7) |
241 | 11.3 (12–22) |
3.4 | 114 (40–555) |
8.1 | 3.4 | 3.64 (12) |
−2.05 (14.6) |
3.2 (2.2) |
4.9 (2.2) |
−1.38 (13.1) | 2.5–6.7 |
Fig. 1.
Growth charts from the three affected males from Pedigree 1. (A) III3, (B) III4, (C) III5. The labelled red arrows indicate when GH treatment was commenced in the three boys respectively. Delayed bone age in patients III3 and III4 is indicated by the cross with a dotted line to the chronological age when the measurement was taken. Parental heights are plotted respectively. GH, growth hormone; M(c), mother's corrected height; F, father's height. MRI scans from the three affected males, III3, III4, III5. D-F: The patients have a small AP with an otherwise structurally normal pituitary gland, indicated by the labelled white arrows respectively. (D-F) MRI images of affected patients. (D) (i-ii) Sagittal images of patient III3 showing generalised white matter loss, a small AP and a thin CC. (D) (iii) A coronal image of patient III3 showing a small AP and ventricular asymmetry with the right ventricle being larger than the left. (E) (i) A sagittal image of patient III4 showing generalised white matter loss and a small AP. (E) (ii) A coronal image of patient III4 showing a small AP. (F) (i) A sagittal image of patient III5 showing generalised white matter loss and a small AP. (F) (ii) A coronal image of patient III5 showing a small AP. CC, corpus callosum; AP, anterior pituitary; WML, white matter loss; VA, ventricular asymmetry. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.13. Patient III3
Patient III3 was the first born of non-identical twins via emergency caesarean at 38 weeks gestation [birth weight 2.1 kg (−2.8 SDS)]. He presented with poor feeding and hypoglycaemia 18 h after birth. He had congenital heart disease in the form of total anomalous pulmonary venous return at presentation, as well as severe gastro-oesophageal reflux requiring Nissen fundoplication. He was diagnosed with GHD and rhGH was commenced at 1.8 years of age. His thyroid function is currently normal. His gonadotrophin status is unknown as he is pre-pubertal. He had a blunted testosterone response to a 3-day HCG test; however, he was only 3 years of age when this test was performed (Table 1). He has global developmental delay needing 1:1 support in mainstream school, behavioural problems and mild hepatomegaly with normal liver function. His mother presented with secondary amenorrhoea. He is currently 8.8 years of age, with a weight of −0.2 SDS and height of −0.3 SDS, and continues to respond well to GH therapy. An oral glucose tolerance test (OGTT) performed at 8 years of age was normal with a blood glucose of 4.3 mmol/l at 2 h; however, he had symptomatic hypoglycaemia at 3 h with a blood glucose of 2.9 mmol/L and insulin of 8.6 mU/L, suggesting glucose dysregulation with hyperinsulinaemic hypoglycaemia. Continuous glucose monitoring showed blood glucose concentrations ranging from 3.1–10.3 mmol/l. Ophthalmological examination was normal.
2.14. Patients III4 and III5
Identical twin brothers were born via caesarean section at 34 weeks gestation; patient III4 had a birth weight of 2.15 kg (−0.3 SDS), and patient III5 had a birth weight of 1.93 kg (−1.5 SDS). Initial presentation with gastrointestinal symptoms and failure to thrive led to a diagnosis of intestinal lymphonodular hyperplasia and eosinophilic infiltration. Both patients presented with hypoglycaemic seizures at 2 years of age, and a diagnosis of GHD was made. Both boys had a microphallus increasing in size following commencement of GH treatment at 3.2 years of age. The boys also developed central hypothyroidism at 2 years of age, and were treated with thyroxine. Patient III4 had small undescended testes, however they descended spontaneously by 2 years of age. His twin brother, III5, had normal descended testes on initial examination. The twins had feeding difficulties and poor weight gain, possibly partly due to their dairy-free diet. By 4 years of age, the brothers had delayed speech development with mild conductive hearing loss, and behavioural and mild learning difficulties were present by the age of 6 years. A standard clinical microarray performed in both boys was normal. Behavioural difficulties were more severe in III5; however, patient III4 often had episodes of twitching, possibly related to hypoglycaemia. Patient III4 also had hepatomegaly, myopia and a squint. Both brothers underwent a tonsillectomy and adenoidectomy at 4 years of age due to recurrent tonsillitis and upper airway obstruction. At 10 years of age, the brothers had impaired glucose tolerance upon testing; the glucose concentrations fluctuated between hypoglycaemia and hyperglycaemia (Table 2). Subsequently, the HbA1c was elevated in both boys [HbA1C 47 mmol/mol (N20-42), 2 h peak glucose on OGTT 13 mmol/L with insulin 33.2 mU/L in III4; HbA1C 43 mmol/mol (N20-42), 2 h peak glucose on OGTT 13.5 mmol/L with insulin 30.5 mU/L in III5]. Both boys had evidence of persisting hyperinsulinaemic hypoglycaemia (fasting glucose of 2.9 mmol/L with insulin of 2.5 mU/L in III5, blood glucose 5 h post-oral glucose load 2.7 mmol/L with insulin 10.9 mU/L in III4) at the age of 13.6 years.
Table 2.
Glucose tolerance test in patient III4, off Diazoxide. A prolonged oral glucose tolerance test was performed on patient III4 at the age of 10 years, off Diazoxide treatment. At 0 min the patient had a low basal glucose with detectable insulin. Glucose increased over time and after 2 h (120 mins) the patient had a high blood glucose. Glucose then decreased and by 5 h (300 mins) the patient was hypoglycaemic with a detectable insulin.
| Time (mins) | −30 | 0 | 30 | 60 | 90 | 120 | 150 | 180 | 210 | 240 | 270 | 300 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose (mmol/L) | 3.6 | 3.2 | 10.2 | 12.1 | 8.8 | 8.4 | 7.8 | 6.2 | 5.5 | 5.5 | 4.0 | 2.7 |
| Insulin (mU/L) | 5.6 | 4.9 | 21.2 | 21.3 | 19.5 | 22 | 22 | 20.5 | 19.4 | 16.1 | 10.7 | 5.5 |
Gonadotrophin secretion tested at 12 years of age was normal (Table 1). Patient III4 had a blunted testosterone response to a 3-day HCG test, while his brother III5 had a borderline testosterone response (Table 1). Nevertheless, both twin brothers have progressed through puberty spontaneously (the most recent pubertal examination at 14.6 years showed Genitalia (G) stage 4 with testicular volumes of 12 and 25 mls in III4 and G4 with testicular volumes of 25 and 20 mls in III5). Both brothers are growing well on rhGH [heights of 147.7 cm (−2.07 SDS) and 147.8 cm (−2.05 SDS) for III4 and III5 respectively, with delayed bone ages at a chronological age of 14.1 years (Fig. 1B-C]. The BMI SDS were + 1.48 for III4 and 0.57 for III5 respectively.
Their mother had osteoporosis due to secondary amenorrhoea, with menarche at 13 years and cessation of periods at 16 years of age, after which she received oestrogen supplementation (CycloProgynova). In later life she required a hysterectomy. An MRI of the brain was normal in the mother. Maternal height was 162.9 cm and their father had a height of 169 cm (mid-parental height: -0.34 SDS).
3. Results
3.1. Mutational analysis
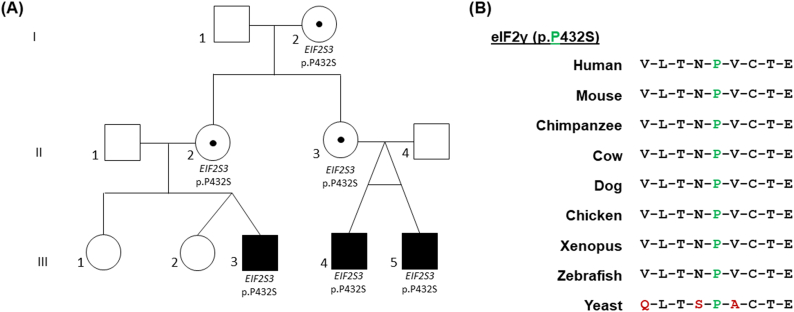
The segregation of the disease phenotype in affected male individuals only, was suggestive of an X-linked mode of inheritance. Consequently, sequencing of the X chromosome in Pedigree 1 revealed a novel hemizygous variant in EIF2S3 (ENST00000253039.8): ChrX_24091319 C/T, c.1294C > T, p.Pro432Ser (GRCh37) in all 3 affected males, inherited from their heterozygous mothers (sisters). This variant is located at a highly conserved residue in the C-terminal domain of eIF2γ across multiple species (Fig. 2B). Protein prediction models predict this variant to be deleterious (Polyphen2 = 0.971, SIFT = 0). The EIF2S3 c.1294C > T, p.Pro432Ser variant was absent from control databases including the Exome Aggregation Consortium (ExAC) (www.exac.broadinstitute.org) and the Genome Aggregation Database 2018 (gnomAD) (www.gnomad.broadinstitute.org). This was the only potential pathogenic variant identified and was considered to be the most likely genetic cause of the disease in the patients.
Fig. 2.
(A) Pedigree 1 harbouring the EIF2S3 (p.Pro432Ser) variant. This pedigree consists of three affected individuals that are hemizygous for the EIF2S3 (p.Pro432Ser) variant, represented by the black shaded squares labelled with ‘EIF2S3 p.Pro432Ser’. Patients III4 and III5 are monozygotic twins. The circles containing a dot highlight the females that carry the variant in heterozygous form. Unshaded squares/circles represent males and females that were negative for the variant respectively. The roman numerals on the left of the image depict the generation within the pedigree. The numbers ‘1’ – ‘5’ distinguish between each individual within that generation, which are referred to in the text. (B) Conservation of the substituted eIF2γ residue. Conservation of the P432 amino acid (highlighted in green) that is substituted in Pedigree 1. Amino acids that differ from the human WT protein sequence are highlighted in red (only in the yeast sequence). The surrounding protein sequence in eIF2γ is very highly conserved across multiple species. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. EIF2S3 expression analysis using in situ hybridisation in human embryos reveals expression in the developing endocrine organs
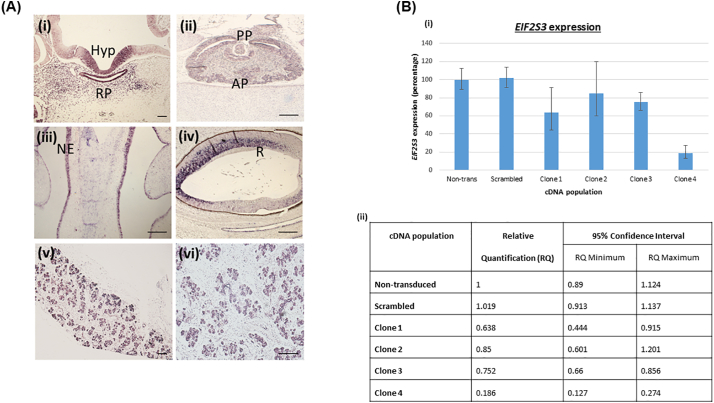
EIF2S3 mRNA transcripts were visualised in the hypothalamus and Rathke's pouch, both the AP and PP, the progenitor cells of the nasal epithelium, and the retina of the eye at both CS20 and 23 of human development respectively (Fig. 3A(i-iv)). The EIF2S3 transcripts were also strongly detected in the pancreas of a 13-week old human fetus (Fig. 3A v, vi). The EIF2S3 DIG-labelled sense control probe had negative staining.
Fig. 3.
(A) Human EIF2S3 expression in the hypothalamo-pituitary axis, eye and pancreas in the developing human embryo. In situ hybridization using the antisense probe against the human EIF2S3 mRNA transcript (hEIF2S3) on human sections. Expression is representative of two embryos. The scale bars represent 200 μM in (i-v), and 100 μM in (vi). (i - iii) Transverse sections of the brain, (i) mRNA EIF2S3 transcripts were localised to the ventral hypothalamus and Rathke's pouch (CS20). (ii) EIF2S3 expression was seen in the AP and PP although not throughout the whole pituitary tissue (CS23). (iii) Defined EIF2S3 transcripts were seen in progenitor cells in the nasal epithelium (CS23). (iv) Sagittal section of the eye. EIF2S3 transcripts were localised in the retina (CS23). (v-vi) Sagittal sections from a pancreas obtained from a 13 week old fetus, showing expression of the EIF2S3 transcript in the Islets of Langerhans. Abbreviations: Hyp, hypothalamus; RP, Rathke's pouch; AP, anterior pituitary; PP, posterior pituitary; NE, nasal epithelium; R, retina. (B) EIF2S3 qPCR expression analysis. (i) Histogram showing the relative quantification (RQ) of EIF2S3 expression (shown as a percentage), against GAPDH, β-ACTIN and HPRT in cDNA derived from transduced 1.1B4 cells, compared to non-transduced cells. This non-transduced control population is thus termed as having 100% EIF2S3 expression. There are five different cDNA populations derived from cells transduced with different shRNA cassette-containing constructs: Non-silencing Scrambled, Clone 1, Clone 2, Clone 3, Clone 4. Error bars depict the lowest percentage value on the minus and the highest percentage value on the plus error bar respectively, generated by StepOne Real-Time PCR System software (ThermoFisher Scientific). (ii) Table showing qPCR Relative Quantification (RQ) values for EIF2S3 expression against GAPDH, β-ACTIN and HPRT compared to non-transduced cDNA, with a 95% confidence interval range: minimum RQ and maximum RQ, for all five cDNA populations.
3.3. Cohort screening
Upon identification of the EIF2S3 (p.Pro432Ser) variant in Pedigree 1, 103 patients with variable congenital hypopituitarism phenotypes, +/− structural midline brain defects on MRI, were screened for EIF2S3 variants; isolated GHD (n = 16), SOD (n = 37), hypogonadotropic hypogonadism/Kallmann syndrome (n = 4), and multiple pituitary hormone deficiencies with variable endocrine deficits and no other midline defects (n = 46). No further variants were identified in these patients. However, there were no other patients in this cohort that had glucose dysregulation similar to that observed in Pedigree 1.
4. Functional analysis
4.1. Generation of an EIF2S3 knockdown human pancreatic β cell line
A stable EIF2S3 knockdown (KD) cell line was established in a human hybrid pancreatic β cell line, 1.1B4 cells, using (LV) expressing target-specific short hairpin RNA (shRNA) and green fluorescent protein (GFP). Seventy-two hours after transduction the cells fluoresced green, indicating successful transduction with shRNA-encoding LV. The qPCR expression analysis showed that the most efficient shRNA construct (Clone 4) knocked down EIF2S3 gene expression by 82%, with a relative quantification (RQ) of 0.186 and a 95% confidence interval (CI) ranging between 0.127 and 0.274 when compared to non-transduced cells (RQ: 1, CI: 0.89–1.124) (Fig. 3B).
4.2. Apoptosis assays in the EIF2S3 knockdown cell line demonstrate higher caspase activity and cell death
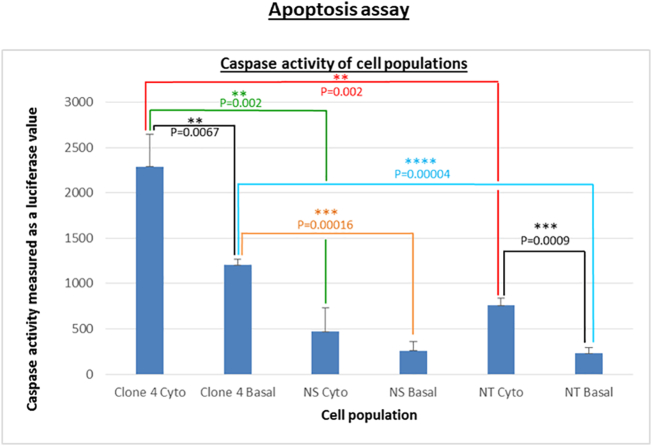
Following transduction of 1.1B4 cells using shRNA Clone 4, cell viability appeared to be reduced on visual observation. Significantly higher caspase activity was observed in both the KD (Clone 4) and NT control cell populations after the addition of cytokines, compared to basal [Clone 4 (2288 ± 358.31 versus (vs) 1203 ± 63.52 p = 0.0067) and NT (758 ± 84.06 vs 230 ± 59.76 p = 0.0009)] (Fig. 4). Though the NS shRNA transduced control cell population did not reach a statistically significant difference in caspase activity after the addition of cytokines, it followed a similar trend to the other cell populations. Consistent with the essential role of the translational apparatus in cell viability, there was significantly higher caspase activity in the clone 4 EIF2S3 KD cells compared to the two control populations, both basally [NS 1203 ± 63.52 vs 258 ± 100.49, p = 0.00016 and NT 1203 ± 63.52 vs 230 ± 59.76, p = 0.00004] and after the addition of cytokines [NS 2288 ± 358.31 vs 468 ± 261.10, p = 0.002 and NT 2288 ± 358.31 vs 758 ± 84.06, p = 0.002] respectively (Fig. 4).
Fig. 4.
Apoptosis assay comparing caspase activity in EIF2S3 KD cells compared to controls, with and without cytokine treatment. Results from an apoptosis assay shown as the mean ± SD of 3 independent experiments with each assay performed in triplicate, on NT 1.1B4 cells, NS shRNA transduced 1.1B4 cells and EIF2S3 KD (Clone 4) 1.1B4 cells. The statistical test used when analysing data significance was an unpaired 2-tailed t-test. Caspase activity is measured as luciferase values on a luminometer. Caspase activity is significantly higher after cytokine treatment compared to the basal caspase activity in Clone 4 (2288 ± 358.31 versus (vs) 1203 ± 63.52, p = 0.0067) and NT (758 ± 84.06 vs 230 ± 59.76 p = 0.0009) cell populations respectively. Basal caspase activity is significantly higher in Clone 4 compared with NS (1203 ± 63.52 vs 258 ± 100.49, p = 0.00016; displayed in orange) and NT (1203 ± 63.52 vs 230 ± 59.76 p = 0.00004; displayed in blue) respectively. Clone 4 cells treated with cytokine mix have a significantly higher caspase activity compared with NS (2288 ± 358.31 vs 468 ± 261.10 p = 0.002; displayed in green) and NT (2288 ± 358.31 vs 758 ± 84.06 p = 0.002; displayed in red) cytokine-treated cell populations respectively. The error bars represent the standard deviation (SD) of the mean. NS, scrambled non-silencing; NT, non-transduced. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
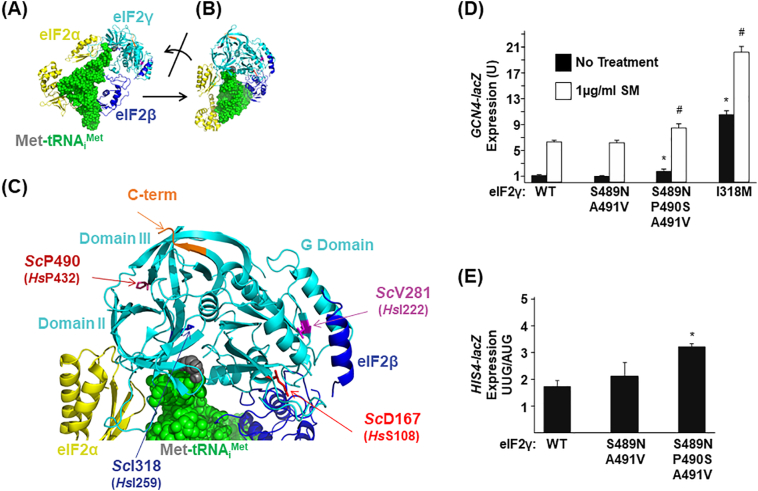
4.3. Altered translational control and start codon selection stringency in EIF2S3 mutant yeast
As there are no protein structure data available for the human eIF2 complex or the isolated human eIF2γ protein, the yeast (Saccharomyces cerevisiae) structure presents the best current model of the eIF2 complex [16]. The eIF2α docks on domain II of eIF2γ and the acceptor arm of Met-tRNAiMet binds into a groove between the G domain and domains II and III of eIF2γ (Fig. 5A-C). The yeast P490 residue (homologous to human Pro432) lies on a β-strand within domain III of eIF2γ. To test if the Pro432Ser mutation impairs eIF2 function, the mutation was introduced into the yeast GCD11 gene encoding eIF2γ [17]. As the residues flanking the mutated Pro residue differ between yeast and human, a locally humanized version of yeast eIF2γ was generated by replacing Ser489 and Ala491 with Asn and Val, respectively, to mimic human eIF2γ (Fig. 2B). When expressed in yeast cells, the humanized eIF2γ-S489 N-A491V mutant and the patient eIF2γ-S489 N-P490S-A491V mutant derivative had no significant effect on yeast cell growth.
Fig. 5.
The p.P490S mutation in yeast eIF2γ, corresponding to the human eIF2γ-Pro432Ser mutation, impairs translation initiation. (A) Ribbon and sphere representation of Saccharomyces cerevisiae eIF2–GDPNP–Met-tRNAiMet ternary complex (PDB code 3JAP [16] using PyMOL software (Schrodinger)). Components are coloured as follows: tRNAiMet, green spheres; Met, grey spheres; eIF2α, yellow ribbons; eIF2β, blue ribbons; and eIF2γ, cyan ribbons. (B) Rotation of the eIF2–GDPNP–Met-tRNAiMet ternary complex to highlight the site of the EIF2S3 patient mutation identified in this study, ScP490 (HsP432, burgundy). (C) Magnification of the eIF2–GDPNP–Met-tRNAiMet ternary complex from panel B. The three domains of eIF2γ are indicated, and the sites of five mutations identified in patients with MEHMO syndrome or its variants are highlighted in stick representation including ScP490 (HsP432, burgundy), a C-terminal frameshift mutation (C-term, orange), ScV281 (HsI222, magenta), ScD167 (HsS108, red), and ScI318 (HsI259, blue). (D-E) A GCN4-lacZ reporter (D) or HIS4(AUG)-lacZ and HIS4(UUG)-lacZ reporters (E) were introduced into yeast strains containing the indicated WT or mutant forms of eIF2γ. Means and standard deviations of β-galactosidase activities were calculated for three independent transformants. The asterisk (*) and hashtag (#) indicate a significant difference from WT eIF2γ in the No Treatment and SM treatment conditions, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A more sensitive assay to examine eIF2 function in yeast relies on translational control of the GCN4 gene [18]. Conditions that lower eIF2 ternary complex levels, like phosphorylation of eIF2α or mutations that impair eIF2 function [[1], [2], [3],10,19], elevate GCN4 expression. A GCN4-lacZ reporter was introduced into isogenic strains expressing WT eIF2γ, humanized eIF2γ-S489 N,A491V, the humanized mutant eIF2γ-S489 N,P490S,A491V or eIF2γ-I318M corresponding to the published eIF2γ-Iso259Met mutation [6]. As shown in Fig. 5D, GCN4-lacZ expression was low in cells expressing WT eIF2γ or humanized eIF2γ-S489 N,A491V. The p.P490S mutation resulted in a modest, but statistically significant, ~1.7-fold increase in GCN-lacZ expression, while the I318M mutation [7] conferred nearly an 11-fold increase in GCN4-lacZ expression. Treatment with SM to activate eIF2α phosphorylation and lower eIF2 ternary complex levels [20], resulted in a ~6-fold increase in GCN4-lacZ expression in cells expressing WT eIF2γ, while GCN4-lacZ expression was de-repressed nearly 9-fold in cells expressing the eIF2γ-S489 N,P490S,A491V mutant and over 20-fold in cells expressing the eIF2γ-I318M mutant (Fig. 5D).
Mutations in yeast eIF2 subunits that impair Met-tRNAiMet binding, enhance GTP hydrolysis, or disrupt complex integrity have been found to relax the stringency of translation start site selection and enhance initiation at differing codons [7,8]. To characterise mutations in this context, a set of HIS4-lacZ reporter constructs with an AUG or UUG start codon were introduced into strains expressing eIF2γ WT or mutant. In eIF2γ WT, the HIS4(UUG)-lacZ reporter was expressed at ~1.6% the level observed with the canonical HIS4(AUG)-lacZ reporter (Fig. 5E) consistent with previous reports [3,13]. A similar low level of UUG initiation was observed in cells expressing the humanized eIF2γ-S489 N,A491V mutant (Fig. 5E, middle bar). Introduction of the P490S mutation, however, increased HIS4(UUG)-lacZ expression to 3% of the value obtained with the AUG-initiated reporter (Fig. 5E, third bar).
5. Discussion
We have identified a novel EIF2S3 mutation in children manifesting a phenotype characterized by mild learning difficulties, hypopituitarism with GH and TSH deficiencies, and an unusual form of glucose dysregulation with early hypoglycaemia and later hyperglycaemia [9]. The ‘probability of loss-of-function (LoF) intolerance’ (pLI) value for EIF2S3 is 0.92, (calculated from controls in ExAC. A pLI ≥ 0.9 value is an extremely LoF intolerant gene). The observed number of missense variants from population genetic data is significantly lower than expected with a Z score of 3.81 (ExAC). Positive Z scores indicate intolerance to variation, thus indicating that EIF2S3 is highly intolerant to protein coding variation. The p.Pro432Ser variant alters a highly conserved residue in the eIF2γ C-terminal domain (Fig. 2B) and segregates in an X-linked recessive manner in three generations of Pedigree 1 (Fig. 2A), having been inherited from the heterozygous maternal grandmother.
Murine Eif2s3 expression in the pancreas, hypothalamus and pituitary (Mouse Genome Informatics), the abundant human EIF2S3 expression in the postnatal brain and endocrine tissues, and high eIF2γ protein expression in the postnatal brain and pancreas (Human Protein Atlas) have previously been established. Prominent EIF2S3 expression in the hypothalamus, Rathke's pouch, AP, PP, progenitor cells of the nasal epithelium, and retina of the eye (Fig. 3A(i-iv)) at CS20 and 23 has been established in this study. At CS23, the retina is in the process of differentiating into different cell types. The expression appears to be most prominent in the developing inner nuclear layer, in a region consistent with developing ganglion cells (Fig. 3A(iv)). However, ophthalmological examination has revealed no retinal changes in the three boys. We further examined expression in the human pancreas, due to the previously documented expression of murine Eif2s3 in the pancreas (MGI), the presence of an unusual pancreatic phenotype in the patients in Pedigree 1, and the early onset diabetes in previously published cases. Expression was most prominent in β cell progenitors within the Islets of Langerhans in the pancreatic tissue of a 13-week old fetus (Fig. 3A(v-vi)). These data provide a human embryonic EIF2S3 gene expression profile in a developmental context.
The 1.1B4 human hybrid pancreatic cells [[21], [22], [23]] were used to perform a stable knockdown of the EIF2S3 gene, using an integrative LV vector expressing EIF2S3-targeting shRNA to generate an EIF2S3 KD cell line. The cells transduced with LV vector encoding EIF2S3-targeting shRNA failed to survive for as long as control cell populations (Fig. 4). Furthermore, the EIF2S3 KD cell line had significantly higher basal and cytokine-stimulated caspase activities compared to control cells (Fig. 4), suggesting increased apoptosis. The impaired cell survival and increased caspase activities in our EIF2S3 KD human cell line in this study is consistent with the essential role of eIF2γ in initiating protein synthesis within the cell [24].
eIF2α phosphorylation inhibits global protein synthesis under conditions of endoplasmic reticulum (ER) stress [25]. In vivo, eIF2α phosphorylation is essential for the pancreas and liver to provide glucose homeostasis. Defects in eIF2-mediated translational control in the pancreas can cause a β-cell deficiency and contribute to diabetes. Mice with a homozygous mutation p.Ser51Ala that abolishes phosphorylation of eIF2α died within 18 h of birth due to prolonged hypoglycaemia associated with reduced glycogen stores and defective gluconeogenesis [26]. Homozygous mutant embryos and neonates displayed a reduction in insulin content 50% and 35% that of WT mice respectively; neonates had 20% of WT serum insulin concentrations, and both had pancreatic β cell deficiency [26]. A subtle reduction in eIF2α phosphorylation in heterozygous p.Ser51Ala mice leads to an increase in β cell death in some treated cells [27] and type 2 diabetes when the animals are stressed by a physiologically relevant high fat diet (HFD). Delayed folding and/or misfolding of proinsulin in the HFD-fed mutant islets was associated with reduced production of mature insulin [28]. These studies are consistent with the glucose dysregulation observed in Pedigree 1, and suggest that eIF2γ may also contribute to glucose homeostasis in humans.
The yeast assays conducted in this study show a modest increase in GCN4-lacZ expression in the p.Pro432Ser yeast homologue (p.Pro490Ser), compared to the robust increase in the yeast model of the human p.Iso259Met mutation (p.Iso318Met) [14] (Fig. 5D) associated with a more severe phenotype [6], indicating a defect in ternary complex formation or delivery of Met-tRNAiMet to the ribosome. These modest impacts on GCN4-lacZ expression, and the ~2-fold increase in non-AUG initiation seen with p.Pro490Ser (Fig. 5E) suggesting a relaxed start site selection stringency, are comparable to previously reported yeast assays on the human MEHMO syndrome mutations p.Iso222Thr and Iso465Serfs*4 [5,7]. Cumulatively, our results demonstrate that the human p.Pro432Ser eIF2γ modestly impairs eIF2 function and may cause disease by altering the fidelity of translation start site selection.
The patients in our study exhibited hypoketotic hypofattyacidaemic hyperinsulinaemic hypoglycaemia. The pathogenesis underlying the hypoglycaemia observed in previous patients was not established [6]. The C-terminal location of the variants and the phenotypes encompassing GHD and hypoglycaemia described by Moortgat et al. support the pathogenicity of EIF2S3 (p.Pro432Ser) in our phenotypically similar patients. Previous reports describing EIF2S3 mutations have focussed on ID with microcephaly [[5], [6], [7]]; however, two previously published patients carrying the EIF2S3 frameshift p.Iso465Serfs*4 [7] were re-evaluated for the presence of a pancreatic phenotype. Neonatal hypoglycaemia, early onset insulin-dependent diabetes, and variable hypopituitarism have since been noted in some of these patients [29]. However, the cause of the neonatal hypoglycaemia has not, to date, been established. Our studies clearly show impaired glucose dysregulation with initial hypoglycaemia due to hyperinsulinism, followed by an unusual form of diabetes that occurs in the second decade of life and is not insulin-dependent, unlike the early onset diabetes reported in two previously published cases [6,29]. The affected males in Pedigree 1 in our study differ from all previously described cases in having a much milder neurodevelopmental phenotype, with the twin boys attending a mainstream school. Our patients do not have microcephaly, epilepsy, or obesity which are prominent phenotypes in previous patients diagnosed with MEHMO syndrome [5] (Table 3). Patients III3, III4 and III5 from Pedigree 1 had variable generalised white matter loss in the brain, findings which are consistent with previously published reports, where patients with EIF2S3 mutations manifested a global reduction in white matter on MRI [6]. However, this may be attributable to hypoketotic hypoglycaemia at birth. Intriguingly, white matter loss is a prominent feature of Vanishing White Matter (VWM) disease, in which patients have mutations in eIF2B [30,31], the guanine-nucleotide exchange factor for eIF2. Thus, mutations impairing eIF2 and eIF2B may display some phenotypic overlap [9].
Table 3.
Clinical phenotypes of male patients with EIF2S3 mutations from four separate studies. Clinical phenotypes from previous reports by Borck et al 5, Moortgat et al 6, Skopkova et al 7, Stanik J et al 29, and from this current study, respectively. Not all unique additional features listed under each study were present in every patient described; each male within the study had various combinations of these features.
| Borck et al. 2012 Mol Cell | Moortgat et al. 2016 AJMG | Skopkova et al. 2017 Hum Mutat, Stanik J et al. 2018 Physiol Res | This study – Pedigree 1 |
|---|---|---|---|
| EIF2S3, p.Iso222Thr in the highly conserved GTP-binding (G) domain | EIF2S3, p.Iso259Met and p.Iso465Serfs*4 in two unrelated pedigrees in the C-terminal domain | EIF2S3, p.Iso465Serfs*4 and pSer108Arg in four unrelated pedigrees | EIF2S3, p.Pro432Ser in the C-terminal domain |
Three males: 2 brothers and maternal uncle
|
Three males: 2 brothers, 1 unrelated male
|
Four unrelated male patients:
|
Three males: 2 brothers and maternal male cousin
|
The three unrelated males with severe MEHMO syndrome only:
| |||
| Unique additional features: Cleft lip/palate
|
Unique additional features: Spastic quadriplegia
|
Unique additional features:
|
Unique additional features:
|
We have shown, for the first time to our knowledge, that the hypoglycaemia observed in our pedigree, and likely those previously published, is associated with hyperinsulinism (Table 3), and may then be followed by the evolution of non-autoimmune diabetes, as shown in a previously published murine model [32]. The significantly higher apoptosis in the EIF2S3 KD human cell line, the human HP and pancreatic expression profile, the yeast assays, and the segregation of the EIF2S3 (p.Pro432Ser) variant with the hypopituitarism and glucose dysregulation phenotype in Pedigree 1, all suggest that the p.Pro432Ser mutation moderately, but significantly, impairs eIF2γ function and leads to a relatively mild phenotype affecting human HP and pancreatic function [9]. This milder loss of function compared with previous EIF2S3 mutations, gives rise to a phenotype that is distinct from the classical spectrum of MEHMO syndrome.
Variable effects on brain development are clearly associated with mutations in EIF2S3. It is worth considering that untreated hypoketotic hypoglycaemia in the previously published cases may have contributed to their more severe impairment of neurodevelopment and seizures. The insights provided by our study reporting both pancreatic and pituitary phenotypes associated with EIF2S3 mutations may be critical for the prevention of significant neurodevelopmental delay in these patients. Early identification of such patients with a rapid molecular diagnosis may lead to prevention of significant morbidity in these patients. The identification of further patients with EIF2S3 mutations will allow more genotype-phenotype correlation studies that will shed further light on the role of eIF2γ in brain, HP, and pancreatic development and function in humans.
Corresponding author statement
I, Professor Mehul T Dattani, confirm that I had full access to all the data in the study and had final responsibility for the decision to submit for publication as the corresponding author.
Declaration of interests
All authors express no conflict of interest and have nothing to disclose.
Ethics statement
The appropriate ethical approval for the genetics and human embryo tissue expression studies have been made prior to this project taking place. The patients/patient guardians gave full consent to all clinical and genetic studies carried out on their blood/DNA.
Author contributions
LC. Gregory: Performed all of the cell culture, the knockdown studies, apoptosis assays, the expression analysis on human embryonic tissue, and wrote the manuscript.
CB. Ferriera, W. Qasim: Packaged the lentiviral vector and transduced cells to generate the knockdown cell line, and provided the secure and licensed laboratory for lentiviral work to take place.
SK. Young-Baird, TE. Dever: The yeast assays.
M. Harakolova, G. van Haaften, HJ. Williams, GOSgene: Conducted the exome sequencing analysis, filtering and interpretation of exome data.
C. Gaston-Massuet: Supervised and provided consumables/laboratory space for the expression studies.
RA. Rahman: Provided helpful discussion and insight into the pancreatic phenotype and assisted with the pancreatic cell work.
D. Kelberman: Supervised the study.
P. Shah, MT. Dattani: Consultant paediatricians for the patients and provided all of the clinical details and tests for the patients in this study.
S. Camper, ICAF. Robinson: Provided helpful insight and discussion, expertise and advice throughout the project.
All co-authors expressed their views and assisted in writing the manuscript.
Acknowledgements and funding sources
We thank Edwin Cuppen and Isaac J Nijman for their work on data analysis and the supervision of the next generation sequencing performed in Utrecht, NL. Great Ormond Street Hospital (GOSH) charity and the Medical Research Foundation (MRF) (grant# 535963) funded this study. The human embryonic and fetal material was provided by the Joint MRC/Wellcome Trust (grant# MR/R006237/1) Human Developmental Biology Resource (http://hdbr.org). The analysis performed by GOSgene in this study is in part supported by the National Institute for Health Research (NIHR), GOSH and Biomedical Research Centre (BRC). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. The yeast assays performed in this study were supported by the Intramural Research Program of the NIH, NICHD. S.K.Y.-B. was supported by a Postdoctoral Research Associate (PRAT) fellowship from the National Institute of General Medical Sciences (NIGMS), award number: 1Fi2GM123961. The funders did not have any role in study design, data collection, data analysis, interpretation or in the writing of the report.
References
- 1.Hinnebusch A.G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev. 2011;75(3):434–467. doi: 10.1128/MMBR.00008-11. (first page of table of contents) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang H.K., Yoon H., Hannig E.M., Donahue T.F. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11(18):2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorris D.R., Erickson F.L., Hannig E.M. Mutations in GCD11, the structural gene for eIF-2 gamma in yeast, alter translational regulation of GCN4 and the selection of the start site for protein synthesis. EMBO J. 1995;14(10):2239–2249. doi: 10.1002/j.1460-2075.1995.tb07218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinmuller R., Steinberger D., Muller U. MEHMO (mental retardation, epileptic seizures, hypogonadism and -genitalism, microcephaly, obesity), a novel syndrome: assignment of disease locus to xp21.1-p22.13. Eur J Hum Genet. 1998;6(3):201–206. doi: 10.1038/sj.ejhg.5200180. [DOI] [PubMed] [Google Scholar]
- 5.Borck G., Shin B.S., Stiller B., Mimouni-Bloch A., Thiele H., Kim J.R. eIF2gamma mutation that disrupts eIF2 complex integrity links intellectual disability to impaired translation initiation. Mol Cell. 2012;48(4):641–646. doi: 10.1016/j.molcel.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moortgat S., Desir J., Benoit V., Boulanger S., Pendeville H., Nassogne M.C. Two novel EIF2S3 mutations associated with syndromic intellectual disability with severe microcephaly, growth retardation, and epilepsy. Am J Med Genet A. 2016;170(11):2927–2933. doi: 10.1002/ajmg.a.37792. [DOI] [PubMed] [Google Scholar]
- 7.Skopkova M., Hennig F., Shin B.S., Turner C.E., Stanikova D., Brennerova K. EIF2S3 mutations associated with severe X-linked intellectual disability syndrome MEHMO. Hum Mutat. 2017;38(4):409–425. doi: 10.1002/humu.23170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harakalova M., Mokry M., Hrdlickova B., Renkens I., Duran K., van Roekel H. Multiplexed array-based and in-solution genomic enrichment for flexible and cost-effective targeted next-generation sequencing. Nat Protoc. 2011;6(12):1870–1886. doi: 10.1038/nprot.2011.396. [DOI] [PubMed] [Google Scholar]
- 9.Gregory L.C. 2016. Investigation of new candidate genes in a cohort of patients with familial congenital hypopituitarism and associated disorders. [Google Scholar]
- 10.Alone P.V., Cao C., Dever T.E. Translation initiation factor 2gamma mutant alters start codon selection independent of Met-tRNA binding. Mol Cell Biol. 2008;28(22):6877–6888. doi: 10.1128/MCB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boeke J.D., Trueheart J., Natsoulis G., Fink G.R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 12.Hinnebusch A.G. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5(9):2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donahue T.F., Cigan A.M. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol. 1988;8(7):2955–2963. doi: 10.1128/mcb.8.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young-Baird S.K., Shin B.S., Dever T.E. MEHMO syndrome mutation EIF2S3-I259M impairs initiator met-tRNAiMet binding to eukaryotic translation initiation factor eIF2. Nucleic Acids Res. 2018;47(2):855–867. doi: 10.1093/nar/gky1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moehle C.M., Hinnebusch A.G. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11(5):2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llacer J.L., Hussain T., Marler L., Aitken C.E., Thakur A., Lorsch J.R. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell. 2015;59(3):399–412. doi: 10.1016/j.molcel.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannig E.M., Cigan A.M., Freeman B.A., Kinzy T.G. GCD11, a negative regulator of GCN4 expression, encodes the gamma subunit of eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13(1):506–520. doi: 10.1128/mcb.13.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 19.Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 20.Wek S.A., Zhu S., Wek R.C. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15(8):4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasu S., McClenaghan N.H., McCluskey J.T., Flatt P.R. Cellular responses of novel human pancreatic beta-cell line, 1.1B4 to hyperglycemia. Islets. 2013;5(4):170–177. doi: 10.4161/isl.26184. [DOI] [PubMed] [Google Scholar]
- 22.McCluskey J.T., Hamid M., Guo-Parke H., McClenaghan N.H., Gomis R., Flatt P.R. Development and functional characterization of insulin-releasing human pancreatic beta cell lines produced by electrofusion. J Biol Chem. 2011;286(25):21982–21992. doi: 10.1074/jbc.M111.226795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo-Parke H., McCluskey J.T., Kelly C., Hamid M., McClenaghan N.H., Flatt P.R. Configuration of electrofusion-derived human insulin-secreting cell line as pseudoislets enhances functionality and therapeutic utility. J Endocrinol. 2012;214(3):257–265. doi: 10.1530/JOE-12-0188. [DOI] [PubMed] [Google Scholar]
- 24.Yatime L., Mechulam Y., Blanquet S., Schmitt E. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc Natl Acad Sci U S A. 2007;104(47):18445–18450. doi: 10.1073/pnas.0706784104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7(6):1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 27.Chambers K.T., Unverferth J.A., Weber S.M., Wek R.C., Urano F., Corbett J.A. The role of nitric oxide and the unfolded protein response in cytokine-induced beta-cell death. Diabetes. 2008;57(1):124–132. doi: 10.2337/db07-0944. [DOI] [PubMed] [Google Scholar]
- 28.Scheuner D., Vander Mierde D., Song B., Flamez D., Creemers J.W., Tsukamoto K. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11(7):757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 29.Stanik J., Skopkova M., Stanikova D., Brennerova K., Barak L., Ticha L. Neonatal hypoglycemia, early-onset diabetes and hypopituitarism due to the mutation in EIF2S3 gene causing MEHMO syndrome. Physiol Res. 2018;67(2):331–337. doi: 10.33549/physiolres.933689. [DOI] [PubMed] [Google Scholar]
- 30.Schiffmann R., Fogli A., van der Knaap M.S., Boespflug-Tanguy O. Childhood Ataxia with central nervous system Hypomyelination/Vanishing white matter. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., editors. GeneReviews((R)). Seattle (WA) University of Washington, Seattle University of Washington, Seattle GeneReviews is a registered trademark of the University of Washington; Seattle: 1993. All rights reserved. [PubMed] [Google Scholar]
- 31.Li W., Wang X., Van Der Knaap M.S., Proud C.G. Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol Cell Biol. 2004;24(8):3295–3306. doi: 10.1128/MCB.24.8.3295-3306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ladiges W.C., Knoblaugh S.E., Morton J.F., Korth M.J., Sopher B.L., Baskin C.R. Pancreatic beta-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54(4):1074–1081. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]