Abstract
Next Generation Sequencing (NGS) combined with powerful bioinformatic approaches are revolutionising food microbiology. Whole genome sequencing (WGS) of single isolates allows the most detailed comparison possible hitherto of individual strains. The two principle approaches for strain discrimination, single nucleotide polymorphism (SNP) analysis and genomic multi-locus sequence typing (MLST) are showing concordant results for phylogenetic clustering and are complementary to each other. Metabarcoding and metagenomics, applied to total DNA isolated from either food materials or the production environment, allows the identification of complete microbial populations. Metagenomics identifies the entire gene content and when coupled to transcriptomics or proteomics, allows the identification of functional capacity and biochemical activity of microbial populations.
The focus of this review is on the recent use and future potential of NGS in food microbiology and on current challenges. Guidance is provided for new users, such as public health departments and the food industry, on the implementation of NGS and how to critically interpret results and place them in a broader context. The review aims to promote the broader application of NGS technologies within the food industry as well as highlight knowledge gaps and novel applications of NGS with the aim of driving future research and increasing food safety outputs from its wider use.
Keywords: Next generation sequencing, Whole genome sequencing, Metabarcoding, Metagenomics, Food safety and quality, Microbiology, Implementation, Data sharing
1. Introduction
In the last decade, next generation sequencing (NGS) has transformed from being solely a research tool to becoming routinely applied in many fields including diagnostics, outbreak investigations, antimicrobial resistance, forensics and food authenticity (Allard et al., 2017; Goodwin et al., 2016; Quainoo et al., 2017). The technology is developing at a rapid pace, with continuous improvement in quality and cost reduction ( The National Human Research Institute, 2017) and is having a major influence on food microbiology.
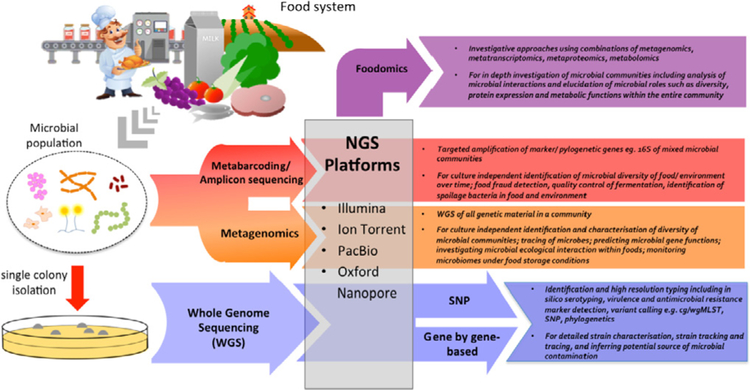
NGS in food microbiology is predominantly used in two ways: (i) determination of the whole genome sequence of a single cultured isolate (e.g. bacterial colony, a virus or any other organism) which is commonly referred to as “whole genome sequencing” (WGS) and (ii) “metagenomics”, where NGS is applied to a biological sample generating sequences of multiple (if not all) microorganisms in that sample. The high discriminatory power of WGS compared with traditional molecular typing tools is well established and WGS is gaining acceptance as a prospective surveillance tool for foodborne illness (Allard et al., 2016; Ashton et al., 2016; Jackson et al., 2016). WGS technology is increasingly replacing traditional microbial typing and characterisation techniques, providing faster and more precise answers.
The application of metagenomics for food safety and quality improvement is still in its infancy and offers exciting opportunities for predicting the presence or emergence of pathogens and spoilage microorganisms based on changes observed in entire microbial communities, as well as the potential to characterise unknown microbiota.
The focus of this review is on the recent use and future potential of NGS in food microbiology, also discussing current challenges in relation to all stakeholders involved. The review also aims to promote the use of NGS in the food industry while highlighting the knowledge gaps and future research needs to augment the value generated from the application of NGS technology to the users.
2. Description of technologies
Microbial genome sequencing has become main stream in the field of food microbiology due to the increasing affordability and improvements in the speed of sequencing and quality of the data. This is a consequence of the advancements in sequencing technologies collectively known as next generation sequencing. NGS encompasses both massively parallel and single-molecule sequencing which provide short and long sequencing reads respectively. Short-read sequencing is highly accurate and produces read lengths of 100–300 bp which are then assembled into incomplete or so called, draft genomes. Complete genomes cannot be generated from the short reads obtained in a single sequence run due to difficulties in assembling repetitive regions and large genomic rearrangements such as insertions, deletions and inversions. For many applications, including comparative genomics and phylogeny, this is not an issue but where complete genomes are required and for determining complex genomic regions, longer reads are necessary. Long-read sequencing produces reads from 10 to 50 Kb in length, but this is at the cost of higher error rates (Loman and Pallen, 2015). Currently, microbial DNA sequencing can be performed on a variety of platforms such as Illumina, Ion Torrent, PacBio and Nanopore. Table 1 provides a summary of these commonly used sequencing platforms whilst more detailed technology descriptions and comparisons are well described in a number of recent reviews including those of Deurenberg et al. (2017), Sekse et al. (2017) and Slatko et al. (2018).
Table 1.
Summary of commonly used Whole Genome Sequencing platforms.
| Platform | Sequencing technology | Read length | Output/run | Error rate | Example of use | Type of instrument and run time |
|---|---|---|---|---|---|---|
| Illumina | Sequencing by synthesis | Short reads 1 × 36bp – 2 × 300bp | 0.3–1000Gb | Low | Variant calling | Benchtop 2–29 h |
| Ion Torrent | Sequencing by synthesis | Short reads 200–400bp | 0.6–15Gb | Low | Variant calling | Benchtop 2–4 h |
| PacBio | Single molecule sequencing bysynthesis | Long reads Up to 60kb | 0.5–10Gb | High | De novo assembly of small bacterial genomes and large genome finishing | Large scale 0.5–4 h |
| Oxford Nanopore | Single molecule | Long reads Up to 100kb | 0.1–20Gb | High | Complete genome of isolates and metagenomics | Portable 1min-48 h |
2.1. Selection of technology
Which technology is used depends on what the sequencing data is to be used for and also on the throughput of sequencing. Maximising high throughput capabilities will result in low sequencing cost per sample. However, the number of samples sequenced in a single run is a function of the desired output and coverage and this varies depending on the application. For example, single nucleotide polymorphism (SNP) analysis of bacterial genomes can be performed with relatively low coverage meaning more DNA samples can be processed in a single sequencing run. In contrast, metagenomic analysis aiming to identify all microbial genes present in a sample needs far greater coverage and this limits the number of samples that can be included in a single run, usually increasing the sequencing cost per sample.
3. Whole genome sequencing of isolates
3.1. Current applications
WGS of microbial pathogens has been introduced into public health surveillance relatively rapidly compared with previous methodological advancements, with reports of its use from early adopters from around 2011 onwards (Koser et al., 2012; Lienau et al., 2011). Whilst initially used for the retrospective analyses of outbreaks of foodborne illnesses detected by typing technologies such as pulsed field gel electrophoresis (PFGE), WGS of microbial pathogens has now been introduced for prospective surveillance of bacterial foodborne pathogens in at least four countries: The United Kingdom, Denmark, France and The United States (Allard et al., 2016; Ashton et al., 2016; Jackson et al., 2016; Kvistholm Jensen et al., 2016; Moura et al., 2016). The year after WGS implementation for prospective assessment surveillance of listeriosis in the United States, more and smaller outbreaks were detected, outbreaks were detected earlier, the source of outbreaks was identified more often and the total number of outbreak related cases identified increased (Jackson et al., 2016). In the realm of public health, WGS is being introduced as a replacement technology, i.e. it will replace most current identification and characterisation methods in the microbiology laboratory such as serotyping, virulence profiling, antimicrobial resistance determination and previous molecular typing methods. In a public health setting replacing the plethora of traditional microbiological identification and typing methods with a single efficient analytical WGS workflow makes implementation cost-effective as well as providing public health with more accurate, actionable data than collected previously (Grant et al., 2018).
Following the lead of the public health sector, WGS is increasingly being considered for application in the food industry. This is not only due to the need to understand public health approaches but also because of the huge benefits and promises for improving food quality and safety afforded by this technology. A key and immediate benefit for the food industry is improved root cause analysis in a pathogen or spoilage contamination event. For example, WGS can help distinguish between new and recurrent introduction of an organism into the production environment. It can also be used for predicting traits such as virulence or antimicrobial resistance of a pathogen or the ability of a spoilage organism to break the preservation barriers of a product. Whilst, industry food safety testing does not demand the detailed microbial characterisation required by reference laboratories, WGS is being increasingly explored for tracking the source of microbial contamination (Rantsiou et al., 2017; Hoorde and Butler, 2018). As the cost of sequencing decreases with technology improvements it makes it more feasible for industry to consider incorporating its use.
3.2. The principles of WGS based tracking and tracing
Molecular subtyping methods have proved invaluable for tracking and tracing pathogens along the food chain, helping to identify sources of infection and the transmission route. (Gerner-Smidt et al., 2013). This includes when the source of infection due to the consequence of poor food handler practice as molecular typing can show that isolates from cases, the food handler or food service environment came from a common source. The additional information available through WGS greatly enhances our ability to determine the source of infection. Over time, bacteria accrue changes in their DNA and this can be used to measure their evolution. Whilst previous molecular subtyping methods detected sequence changes in a small portion of the microbial genome, WGS captures them across the entire genome and thus more accurately describes the genetic relatedness of strains. In tracking and tracing, the relatedness of bacterial sequences from outbreaks as well as the food production chain is assessed to determine if they could be part of the same transmission chain. However, as discussed in section 3.3 WGS data must be backed up by epidemiological evidence to prove and characterise a transmission chain.
Currently there are two main approaches to analysing genomic data to determine the relatedness between strains, namely SNP-based and the gene by gene-based approaches. Analysis of WGS data by either approach is a complex process in which multiple steps are combined to produce final results, such as SNP or allele matrices and phylogenetic trees (Timme et al., 2017). The large amount of data generated in WGS brings challenges for its analysis (Deurenberg et al., 2017; Wyres et al., 2014). This has led to multiple software solutions being developed, mainly through academic endeavours, which in general require specialist knowledge and expertise to deploy and run. However, more recently commercially developed software have become available, bringing a user-friendly interface, allowing non-bioinformatic experts, with the appropriate training in both bioinformatic software and final WGS result interpretation, to conduct analyses. The commercial software may be expensive but since limited bioinformatic expertise is needed, it may nevertheless be a more cost-efficient solution for many food industry users.
3.2.1. SNP approach
In the SNP-based approach, sequencing reads are aligned or mapped to a known sequenced reference genome, and the nucleotide differences in both coding and non-coding regions determined (Davis et al., 2015). For each isolate, every SNP relative to the reference genome is recorded and then used to quantify the genetic relatedness between strains. The selection of the reference genome is a critical step: the reference genome needs to be as completely sequenced, i.e. as contiguous as possible, and closely genetically related to the genomes being analysed (e.g. same serotype). A distantly related reference genome can result in an underestimation of the genetic relatedness of the isolates being investigated as it increases the likelihood of mismapping and decreases the regions that reads can be mapped to (Carriço et al., 2018; Schürch et al., 2018).
Variation in mobile genetic elements such as plasmids and prophage is, by definition, not restricted to vertical inheritance and therefore does not always reflect the true evolutionary history between strains and thus is not a reliable proxy for epidemiological relatedness. Repetitive regions such as prophage and insertion elements are often excluded implicitly due to ambiguous mapping (i.e. the sequencing reads can map to multiple places in the reference genome and are therefore ignored) or explicitly by masking regions of high SNP density. Despite such exclusions SNP analysis is usually performed using greater than 95% of the sequenced genome.
The number of SNP differences can vary depending on the reference strain, the reference mapping as well as the SNP calling method used (Pightling et al., 2015). There are several SNP analysis tools in the public domain which are under active development in addition to new ones coming online. This makes it challenging to compare them, particularly as no widely accepted guidelines or standards for selecting SNP analysis tools have been developed. Users are recommended to use previously validated SNP-based tools, such as those developed by the US Food and Drug Administration (FDA), Centers for Disease Control and Prevention (CDC), Public Health England (PHE) and Center for Genomic Epidemiology (CGE) that are available on Github and perform in-house verification, ideally using benchmarked data sets which are increasingly becoming available (Timme et al., 2017).
3.2.2. Gene by gene approach
Gene by gene analysis consists of assessing the variation in the coding regions i.e. the genes (or ‘loci’) of a bacterial genome (Maiden et al., 2013). In an extension to traditional 7-loci multi-locus sequence typing (MLST), the genes in either a defined core genome (cgMLST) or the whole genome (wgMLST), which includes the more variable accessory genes, are compared against a reference database of all known gene variants (alleles) for a particular species. Each gene or allele sequence is reduced to a number and genomes are compared based on the number of allele differences there are, comparable to the way the number of SNP differences are used. Since the reference is a database of loci and alleles from numerous strains, the analysis does not depend on the selection of a closely related reference strain for the precise assessment of the relatedness of genetically similar isolates. Often, prior to gene by gene analysis, sequencing reads are assembled, typically using the de novo based approach, into longer contiguous sequences (called contigs) which constitute a draft genome (i.e. one that still contains gaps). To assign an MLST type, the assembled short reads are compared using BLAST to a reference allele database (MLST scheme) holding all known allelic variants for each locus defined for a specific species. Variations, including SNPs, indels (insertions and deletions) and recombinations in the same gene are considered as a single allele difference. In some MLST pipelines, allele calling is completed with assembly-free allele calling whereby raw sequencing reads are mapped to alleles in a database. The choice of assembly or assembly-free allele calling usually depends upon whether a de novo assembly already exists or if reads have been mapped to a reference genome. A valuable evaluation of different MLST software for NGS sequencing data has been conducted by Page et al. (2017) using a validated dataset which provides information on accuracy, limitations and computational performance.
Traditional 7-gene MLST provides a broad phylogenetic relevant split of a species into sequence types (STs) and clonal complexes (CCs), whereas cgMLST provides highly detailed phylogenetically relevant information about the genetic relatedness of a species. wgMLST provides even more discrimination than cgMLST and this can be valuable for cluster investigations to discriminate between closely related isolates. However, because it includes sequence data possibly acquired by horizontal transfer, wgMLST analysis may not be as phylogenetically relevant when compared to cgMLST derived phylogeny. Thus, whilst genes on mobile elements are usually included in wgMLST they are often, as in SNP analysis, filtered out in the final analysis.
A public validated database with a shared nomenclature is recommended for comparisons, but ad hoc databases can also be created when a public reference is unavailable or insufficient. Examples of publicly available cgMLST schemes for common foodborne pathogens are provided in Table 2. There are currently no public cg/wg MLST schemes available for other foodborne bacteria, such as spoilage bacteria.
Table 2.
cgMLST and Genomic Reference databases for key food pathogens.
| Pathogen | DB location | Hosted by | Validation |
|---|---|---|---|
| Listeria monocytogenes | http://bigsdb.pasteur.fr/listeria/ | Institut Pasteur, FR | Moura et al. (2016) |
| Salmonella | https://enterobase.warwick.ac.uk/species/index/senterica | Warwick University, UK | – |
| Escherichia/Shigella | https://enterobase.warwick.ac.uk/species/index/ecoli | Warwick University, UK | – |
| Yersinia | https://enterobase.warwick.ac.uk/species/index/yersinia | Warwick University, UK | – |
| Campylobacter | https://pubmlst.org/campylobacter/ | University of Oxford, UK | – |
3.2.3. Phylogenetic analysis
The genetic variation detected by SNP or gene-by gene analysis can be used to infer phylogenetic relationships between bacterial isolates and this is usually displayed in the form a phylogenetic tree. The tree represents the calculated evolutionary model (obtained using different possible tree inference algorithms such as parsimony, maximum likelihood, and Bayesian or distance methods) of the isolates as a series of branches from the root or common ancestor. The isolates clustered together near the leaves of the tree are more closely related than other isolates elsewhere in the tree. The following references, Ajawatanawong (2017), Baldauf (2003), Hedge and Wilson (2016), and Yang and Rannala (2012) are recommended for a more in-depth review on the principles behind the construction and interpretation of phylogenetic trees.
3.2.4. Comparison between SNP and cg/wgMLST
The choice of which comparative genomic approach to use depends on the needs of the end-user and the epidemiological context. While either SNP or gene-by-gene approaches can be used to investigate a fixed number of strains associated with a particular contamination event, cgMLST might be more appropriate if multiple users need to systematically analyse every new isolate added to a common database, e.g., in an outbreak surveillance network, especially if the sequence information cannot be disclosed in the public domain. For investigating phylogeny, the use of either cgMLST or cgSNP may provide more robust analyses than wgMLST or wgSNP since it includes only regions of the genome present in all strains, however, the use of wgMLST or wgSNP can give higher resolution for strain discrimination. SNP and gene-by-gene approaches assess genetic variation in slightly different ways and should be viewed as being complementary and both used when one method alone does not provide a clear-cut answer or for stronger support for an association between isolates e.g. to confirm the source of an outbreak and support regulatory action. To date, both methods, have been shown to be equally discriminatory when calling strain relatedness and epidemiologically concordant for outbreak investigations (Chen et al., 2017; Cunningham et al., 2017; Katz et al., 2017). However, comparisons of the two approaches using WGS data from a wider range of foodborne pathogens in a variety of outbreak settings would be valuable and are in progress.
A major advantage of cg/wgMLST is that it can be standardized and harmonized by using an allele database with standardized allele calling and this approach is being adopted by PulseNet International (Nadon et al., 2017) to enable global strain comparisons for public health. The cg/wgMLST allele databases must be curated to maintain quality and, whilst most curation can be automated, manual curation by a subject matter expert in cg/wgMLST and microbiology is required if new alleles deviate from the quality thresholds defined for the automated curation.
An important difference between SNP and gene-by-gene approaches is the level of computational support required. SNP analysis has traditionally been performed using open source software requiring expert bioinformatic support, whereas cg/wg MLST has been implemented on both command-line open-source software and commercial solutions with user-friendly interfaces.
Maximal benefit from WGS of foodborne pathogens will be achieved if sequenced genomes are deposited in public databases in real time. Whilst there is general agreement on this principle, at present, not all agencies, organisations and companies are able to share their sequencing data. Raw sequencing data can be submitted to the international public archival resource the ‘Sequence Read Archive (SRA)’ either through the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/sra), the European Bioinformatics Institute (EBI) (www.ebi.ac.uk/ena) or the DNA Data Bank of Japan (DDBJ) (trace.ddbj.nig.ac.jp) with data shared between all three (Kodama et al., 2012). The NCBI pathogen detection website, which provides daily SNP based phylogenetic trees for all publicly available data, is also available to those able to make their pathogen sequence data public since it is a requirement from NCBI that the users submit their sequences to their public repository before their tools can be used. Users can upload their genomes and collect their results the following day using online web browsing tools. More considerations on data sharing are addressed in section 5.
A wide range of bioinformatic tools are available for analysing WGS data from bacterial isolates including those for the primary processing of raw data, e.g. for quality assessment, trimming and filtering of raw sequence data, and for secondary processing, such as sequence read assembly or alignment. There are also the tools for more detailed analysis of the data such as for species identification, marker gene detection, variant calling and phylogenetic analysis, amongst others. A selection of the more commonly used tools as well as bioinformatic suites containing such tools, are provided in Table 3.
Table 3.
Bioinformatic tools and pipelines for WGS analysis.
| Functionality | Name | Platform compatibility | Description | Link | Reference |
|---|---|---|---|---|---|
| Pre-processing of raw reads | Trimmomatic | Linux | Variety of useful trimming tasks for Illumina paired-end and single ended data (cut adapter and other Illumina-specific sequences, based on quality, …) | http://www.usadellab.org/cms/?page=trimmomatic | Bolger et al. (2014). |
| Quality control | FastQC | Linux | Quality control checks on raw sequence data with a modular set of analyses | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | None |
| checkM | Linux | Set of tools for assessing the quality of genomes recovered from isolates, single cells, or metagenomes (estimates of genome completeness and contamination, plots depicting key genomic characteristic, …) | http://ecogenomics.github.io/CheckM/ | Parks et al. (2015). | |
| Pre-processing of raw reads/Quality control | FaQCs | Linux | Combines several features including data quality visualization and trimming, filtering the PhiX control sequences, conversion of FASTQ formats, multi-threading. | https://github.com/LANL-Bioinformatics/FaQCs | Lo and Chain (2014). |
| De novo assembly | Velvet | Linux | De novo genomic assembler specially designed for short read sequencing technologies | https://www.ebi.ac.uk/∼zerbino/velvet/ | Zerbino and Birney (2008). |
| SPAdes | Linux/MacOS | Assembly toolkit containing various assembly pipelines which works with Illumina or IonTorrent reads and is capable of providing hybrid assemblies using PacBio, Oxford Nanopore and Sanger reads | http://cab.spbu.ru/software/spades/ | Bankevich et al. (2012). | |
| MIRA | Linux/MacOS | Whole genome shotgun and EST sequence assembler for Sanger, 454, Solexa (Illumina), IonTorrent data and PacBio | http://mira-assembler.sourceforge.net/docs/DefinitiveGuideToMIRA.html | Chevreux et al. (1999). | |
| HGA | Linux | Provide hierarchical genome assembly: de novo bacterial genome assembly using high coverage short sequencing reads | https://github.com/aalokaily/Hierarchical-Genome-Assembly-HGA | Chin et al. (2013). | |
| Canu | Linux | Fork of the Celera Assembler designed for high-noisesingle-molecule sequencing (such as the PacBio RSII or Oxford Nanopore MinION) | http://canu.readthedocs.io/en/latest/ | Koren et al. (2017). | |
| Reference Mapping | Burrows-Wheeler Aligner (BWA) | Linux | Align sequencing reads against a large reference genome and support Illumina, SOLiD, 454, Sanger reads, PacBio reads | http://bio-bwa.sourceforge.net/ | Li and Durbin (2009). |
| SMALT | Linux | Aligns DNA sequencing reads with a reference genome and support reads from Illumina, Roche-454, Ion Torrent, PacBio or ABI-Sanger | http://www.sanger.ac.uk/science/tools/smalt-0 | Ponstingl and Ning (2010). | |
| Bowtie2 | Windows/Linux/MacOS | Ultrafast and memory-efficient tool for aligning sequencing reads to long reference sequences. | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | Langmead and Salzberg (2012). | |
| Genome Viewer/Genome annotation | Prokka | Linux/MacOS | Tool to annotate bacterial, archaeal and viral genomes quickly and produce standards-compliant output files | https://github.com/tseemann/prokka | Seemann (2014). |
| NCBI prokaryotic genome annotation pipeline | Web-based | Designed to annotate bacterial and archaeal genomes (chromosomes and plasmids), including prediction of protein-coding genes, as well as other functional genome units such as structural RNAs, tRNAs, small RNAs, pseudogenes, control regions, direct and inverted repeats, insertion sequences, transposons and other mobile elements | https://www.ncbi.nlm.nih.gov/genome/annotation_prok/ | Angiuoli et al. (2008). | |
| RAST | Windows/Linux/MacOS | Fully-automated service for annotating complete or nearly complete bacterial and archaeal genomes, providing high quality genome annotations for these genomes across the whole phylogenetic tree | http://rast.nmpdr.org | Aziz et al. (2008). | |
| Variant/SNPcalling | SRST2 | Linux | Designed to take Illumina sequence data, a MLST database and/or a database of gene sequences (e.g. resistance genes, virulence genes, etc) and report the presence of STs and/or reference genes. | https://github.com/katholt/srst2 | Inouye et al. (2014). |
| VarScan2 | Windows/Linux/MacOS | Platform-independent mutation caller for targeted, exome, and whole-genome resequencing data generated on Illumina, SOLiD, Life/PGM, Roche/454, and similar instruments. | http://dkoboldt.github.io/varscan/ | Stead et al. (2013). | |
| BFCtools/SAMtools | Linux | Set of utilities that manipulate variant calls in the Variant Call Format (VCF) and its binary counterpart BCF | https://samtools.github.io/bcftools/ | Li (2011). | |
| kSNP | Linux/MacOS | SNP discovery and SNP annotation from whole genomes | https://sourceforge.net/projects/ksnp/files/ | Gardner et al.(2015). | |
| Mobile element detection | PhiSpy | Linux | Identify prophages in complete bacterial genome sequences | https://github.com/linsalrob/PhiSpy | Akhter et al. (2012). |
| PlasmidFinder | Web-based | Identify plasmids in total or partial sequenced isolates of bacteria | https://cge.cbs.dtu.dk/services/PlasmidFinder/ | Carattoli et al. (2014). | |
| Virulence/Resistome analysis | VirulenceFinder | Web-based | Identify virulence genes in total or partial sequenced isolates of bacteria | https://cge.cbs.dtu.dk/services/VirulenceFinder/ | Joensen et al. (2014). |
| VFDB | Web-based | Integrated and comprehensive online resource for curating information about virulence factors of bacterial pathogens | http://www.mgc.ac.cn/VFs/search_VFs.htm | Chen et al. (2016). | |
| MYKROBE PREDICTOR | Windows/Linux/MacOS | Analyse the whole genome of a bacterial sample and predict which drugs the infection is resistant to | http://www.mykrobe.com/products/predictor/ | Bradley et al. (2015). | |
| ResFinder | Web-based | Identify acquired antimicrobial resistance genes and/or find chromosomal mutations in total or partial sequenced isolates of bacteria | https://cge.cbs.dtu.dk/services/ResFinder/ | Zankari et al. (2012). | |
| Phylogenetic analysis | FastTree | Windows/Linux/MacOS | Infer approximately-maximum-likelihood phylogenetic trees from alignments of nucleotide or protein sequences. | http://www.microbesonline.org/fasttree/ | Price et al. (2010). |
| RAxML | Windows/Linux/MacOS | Programme for sequential and parallel Maximum Likelihood based inference of large phylogenetic trees. It can also be used for post-analyses of sets of phylogenetic trees, analyses of alignments and, evolutionary placement of short reads | https://sco.h-its.org/exelixis/web/software/raxml/index.html | Stamatakis (2014). | |
| PhyML | Web-based/Windows/Linux/MacOS | Phylogeny software based on the maximum-likelihood principle | http://www.atgc-montpellier.fr/phyml/ | Guindon et al. (2010). | |
| Visualization | Microreact system | Web-based | Phylogeographic analysis of SNP or MLST data | https://microreact.org/showcase | Argimón et al. (2016). |
| PHYLOViZ | Web-based/Windows/Linux | Epidemiological analysis and visualization of sequence (SNP and MLST) data | http://www.phyloviz.net/ | Nascimento et al. (2017). | |
| GenGIS | Windows/MacOS | Analysis of phylogenetic data and associated metadata on digital maps. | http://kiwi.cs.dal.ca/GenGIS/Main_Page | Parks et al. (2013). | |
| Bioinfonnatic suite/ pipeline | CLC Genomics Workbench | Windows/Linux/ MacOS | Analyse and visualize NGS data (resequencing, read mapping, de novo assembly, variant analysis, metagenomics, ...) | https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/ | None |
| BioNum erics | Windows | Quality control, assembly, reference mapping, SNP calling, wgMLST calling, phylogenetic tree, ... | http://www.applied-maths.com/bionumerics | None | |
| Ridom SeqSphere + | Windows | Quality control, assembly, reference mapping, SNP calling, cgMLST calling, phylogenetic tree, ... | http://www.ridom.de/seqsphere/ | Jünemann et al. (2013). | |
| Geneious | Windows/Linux/MacOS | Assembly, genome browser, SNP calling, phylogenetic tree, ... | http://www.geneious.com | Kearse et al. (2012). | |
| CFSAN SNP pipeline | Linux | Reference mapping, SNP calling | https://gitliub.com/CFSAN-Biostatistics/snp-pipeline | Davis et al. (2015). | |
| Lyve-SET SNp pipeline | Linux | Quality control, reference mapping, hqSNP calling, phylogenetic tree | https://gitliub.com/lskatz/lyve-SET | Katz et al. (2017). | |
| SNVPhyl (Single Nucleotide Variant PHYLogenomics) | Linux | Reference mapping, SNP calling, phylogenetic tree | https://snvphyl.readtliedocs.io/en/latest/ | Katz et al. (2017). | |
| Basepace | Cloud-computing platform | Quality control, assembly, reference mapping, SNP calling, cgMLST calling, plasmid, virulence, ...(over 70 bioinformatic tools) | https://emea.illumina.com/products/by-type/informatics-products/basespace-sequence-hub/apps.html | None | |
| Integrated Rapid Infectious Disease Analysis (IRIDA) platform | Linux | Data storage, management, assembly, reference mapping, SNP calling, phylogenetic tree | http://www.irida.ca/ | IRIDA (2017). |
3.3. Interpretation of results
The biological interpretation of the genetic relatedness of isolates using sequence data is often straightforward, provided all sequence quality control parameters are within the expected values and the genetic stability of the bacteria in question, e.g. their spontaneous mutation rates are known. In WGS analysis, the number of SNP/allele differences are used to construct phylogenetic trees providing information on the evolutionary history of the isolates. In a biological sense, a high sequence similarity by WGS analysis means that isolates share a recent common ancestor, and a low similarity means they do not (Pightling et al., 2018). It is a fundamental assumption in molecular epidemiology that phylogeny reflects epidemiological relatedness i.e. clinical isolates or clinical and food or environmental isolates that are phylogenetically closely related are likely to be epidemiologically or causally linked (Besser et al., 2018). Although this assumption is often true, it is not always so because of the complex or indirect connections that can occur at any point along the farm to fork continuum. Thus it is critical that epidemiological and food trace back evidence is used to support and facilitate the correct interpretation of WGS analysis. A key question to ask every time sequences are compared is: Does the phylogenetic result make epidemiological sense, i.e. does a sequence match between an isolate obtained from a food production plant/retailer/food service environment and a clinical isolate mean that the patient became infected by consuming food produced at that plant/retailer/food service? WGS analysis provides robust evidence that isolates are genetically related but it does not necessarily mean that a clinical case was infected directly from a food or a particular premise where WGS matched isolates were obtained. It is essential that epidemiological evidence is available to support the phylogenetic findings, determine the food vehicle, the original source of contamination, and mode of transmission.
Due to the inherent diversity of different bacterial species, different epidemiological contexts and different WGS analysis approaches, it is not possible, nor indeed wise, to define species-specific genetic cut off values at which strains are considered to be closely related (Pightling et al., 2018, Schürch et al., 2018). Some species or serotypes are more clonal than others, e.g., Salmonella ser. Enteritidis is highly clonal (Allard et al., 2013) whereas ser. Typhimurium is not. In addition, the environment a bacterial species exists in may also exert evolutionary pressure affecting mutation rate, and generation time (Deatherage et al., 2017). Thus, interpretation of the genetic relatedness of strains based on SNP/allele differences needs to be supported with expert knowledge of the particular pathogen including an understanding of its genetic diversity in the farm to fork environment and of the representativeness of the isolates under investigation (Besser et al., 2018; Schürch, 2018). WGS analysis of each foodborne outbreak scenario needs to be assessed independently with epidemiological and food chain investigations undertaken to provide as much information as possible for interpretation (Pightling et al., 2018, Schürch et al., 2018).
In general, if the sequences of two food pathogen isolates are highly related, for example within 0–20 SNP/allele differences, it is likely that the isolates share a recent common ancestor and probably originate from the same source (Wang et al., 2018). If such highly related isolates are cultured from different places in a food production plant, the most likely scenario is that the same strain has somehow spread within the production environment. Additional investigations are needed, however, to establish the actual transmission chain in order to mitigate the problem most efficiently.
If the sequences of two isolates are very different, for example > 50–100 SNPs/alleles different, in general, the isolates are deemed not to be related and it is not likely they come from the same source. Of course, such findings may still reflect a common underlying problem that requires investigation: multiple strains have been previously linked to outbreaks related to consumption of the same food product (‘polyclonal outbreaks’) and the presence of multiple strains in the food production environment may be indicative of general hygiene problems.
Isolates do not always fall within the above SNP/allele thresholds and thus can appear to lie between being highly related and unrelated. For example, isolates in a food processing plant may cluster separately from all other isolates in a database but still be 30 SNPs/alleles from each other. This indicates that the isolates share a common ancestor and may have evolved from a resident strain in the premises and potentially persistent (Elson et al., in publication). This can happen when microbial populations experience frequent reduction in numbers (i.e. by cleaning and disinfection), as random mutations can lead to diversification of the original resident strain. In addition, a factory environment offers several different environmental niches that enable isolates therein to undergo genetic drift, again causing strain diversification. Detection of isolates with this type of genetic variation, following cleaning and disinfection of a food premises, would indicate that the strain had not been eradicated by the cleaning/disinfection procedures employed or had constantly been re-introduced through independent events into the premises from external sources that supported conditions for strain diversification.
Similarly, in outbreaks that are associated with a source that permits propagation of isolates, the sequence definition of the outbreak strain can be broader (up to 50 SNPs/allele differences or more). This is often seen, for example, in zoonotic outbreaks. This was the case in an outbreak in the US associated with exposure to small turtles, in which three Salmonella serotypes were involved, Poona, Pomona and Sandiego. The outbreak associated isolates of ser. Poona differed by up to 17 SNPs from each other and ser. Pomona isolates by up to 30 SNPs (https://www.cdc.gov/salmonella/small-turtles-03-12/epi.html). Similarly, 401 isolates associated with a multinational European outbreak of Salmonella Enteritidis 14b linked to eggs were shown to have a maximum of 23 SNPs between any genome (Dallman et al., 2016).
In outbreak investigations, it is critical and customary practice to gather supporting epidemiological evidence, such as patient interviews, confirming the consumption of the suspected food product, matching timelines, food trace backs and regulatory inspections, evidence of breakdown of food safety measures at the food producing plant, in addition to the phylogenetic information ascertained through WGS analysis of isolates to establish a causal relationship of a food product to an illness. Availability of such epidemiological evidence in addition to supporting WGS data can also link a food product to historical clinical cases (Schürch et al., 2018).
In conclusion, since biological relatedness, e.g. sequence similarity, imperfectly correlates with ecology/epidemiology, all available background data about the sources of the isolates and the reason for doing the comparison must be considered when interpreting sequence data. Sometimes additional descriptive data needs to be gathered to understand the sequence data. Therefore, for food safety and outbreak investigation purposes, sequence data alone cannot prove an epidemiological relationship between isolates.
3.4. The need for standardisation
To maximise the benefits from WGS, the data generated need to be accurate, reliable and globally comparable regardless of the sequencing platform, the bioinformatic approach and software used. Standardisation is the process whereby this is achieved and, whilst standards and guidance exist for human genetic sequencing, few have been available for microbial WGS. This is mainly because pathogen genomics is a rapidly developing field and comprises specialties, such as bioinformatics, which have not been subjected to microbiology laboratory standardisation procedures previously. However, many of the principles and quality practices developed for human sequencing are equally applicable to microbial WGS analysis (Gargis et al., 2016) and specific microbial WGS specific performance criteria and standards are becoming available (Kozyreva et al., 2017; Portmann et al., 2018). Just as with the microbial subtyping methods it is replacing, microbial WGS requires validation and verification and needs to be subject to all the quality assurance procedures that constitute a good laboratory quality management system. The WGS workflow consists of three components: sample preparation, sequencing and data analysis and the entire process, end to end, needs to be validated against existing typing methods e.g. PFGE or Multiple-Locus Variable number tandem repeat Analysis (MLVA), with a well-defined set of strains to ensure that the method works for the intended purpose by the end-user; this also facilitates the generation of interpretive guidelines for the consistent interpretation of results. Validation establishes performance specifications such as accuracy, precision, reproducibility, repeatability, sensitivity and specificity as well as discriminatory ability and epidemiological concordance. Quality control procedures are required for all components of the WGS process including sample DNA quality and quantity, sequence quality scores including depth of sequence coverage, read length and sequence quality, as well as the use of known positive and negative sample controls. As with other WGS components, the bioinformatic analysis process, once optimised, needs to be version controlled and any subsequent alterations will require some form of revalidation. Once the whole WGS process has been validated there needs to be regular independent assessment of its performance, i.e. verification, and this can be achieved through the use of internal quality controls, external quality controls and participation in proficiency tests.
Proficiency tests (PT) for microbial WGS analysis are being developed e.g. the Global Microbial Identifier (GMI) has been providing PTs for microbial WGS since 2015 (http://www.globalmicrobialidentifier.org/). Also, an end-user survey was published that provided information on capability, attitudes and practices of GMI community members (Moran-Gilad et al., 2015). This scheme provides bacterial strains for end to end testing, extracted DNA for sequencing and data analysis assessment and sequence data all from the same strain for bioinformatic analysis. Other quality initiatives include benchmarking activities in which well characterised sets of strains are available for evaluating the performance of bioinformatic pipelines. Recently, an outbreak benchmark dataset has been publicly released consisting of sequence data, sample metadata and corresponding known phylogenetic trees for L. monocytogenes, S. enterica ser. Bareilly, Escherichia coli, and Campylobacter jejuni and one simulated dataset (https://github.com/WGS-standards-and-analysis/Datasets), for laboratories to use to assess their bioinformatic tools and pipelines (Timme et al., 2017). Work has also been carried out under the EFSA funded Engage project (http://www.engage-europe.eu) to benchmark specific bioinformatic tools. A standard set of sequencing data has been used to evaluate different de novo assembly tools for predicting Salmonella serotypes as well as antimicrobial resistance gene profiling tools. The results of these benchmarking studies demonstrate that serotyping and predicting antimicrobial resistance in Salmonella using WGS data is a very feasible option.
3.5. Public health and regulatory actions based on WGS results
Increasingly, food regulators and public health scientists are monitoring sequence databases to identify indistinguishable isolates from patients, the food chain, and clustered clinical isolates which could indicate a foodborne outbreak. Such findings justify exploring the potential link between cases, and the food isolate(s). Using a WGS profile as part of the case definition in an outbreak investigation allows cases to be ruled in or out of the outbreak with a higher degree of resolution than previously possible. WGS evidence for isolates being the same strain is allowing cases to be attributable to outbreaks over longer time frames and to link cases from broader geographical areas than was possible with previous typing methods e.g. L. monocytogenes isolates from cases of listeriosis occurring over several years can be shown to be the same strain (Chen et al., 2017; Gillesberg Lassen et al., 2016; Kleta et al., 2017; Wilson et al., 2016); isolates of Salmonella Enteritidis from cases in different European countries have been demonstrated to be the same by SNP analysis and to have evolved from a common ancestor (Dallman et al., 2016). A more robust case definition gives increased power to subsequent epidemiological analyses such as case-control studies, as unrelated cases which may have been previously included as part of the outbreak, no longer confounds the analyses (Lienau et al., 2011). Sequence data from outbreak isolates can be compared to known sequence databases and may be found to match isolates associated with distinct geographical signals which may give indications to the possible original source of contamination and thus help to direct food chain and environmental investigations (Hoffmann et al., 2016).
The increased power of WGS analysis to demonstrate unequivocal genetic relatedness provides more robust evidence for public health action to be taken and may allow intervention at an earlier stage. However, as reiterated previously, epidemiological evidence is vital together with WGS evidence to ensure the appropriate public health and regulatory action is taken. Where WGS is being used routinely for public health surveillance of foodborne pathogens, a greater number of clusters or outbreaks are being detected, many of which would not have been detected by traditional typing methods (Franz et al., 2016). This obviously has resource implications for subsequent investigations and priorities on which outbreaks to focus on should be determined using a risk-based approach involving a variety of considerations, such as severity of illness, virulence of pathogen, infective dose, number of cases, time and geographical clustering of cases and likely exposure to the source in the future.
Outbreaks detected by WGS are investigated using similar approaches as used previously with cases being interviewed about their food exposures and case-control or case-case studies conducted as part of analytical epidemiological investigations to provide supporting evidence for the potential food source. Food authorities will conduct traceability investigations on implicated food products in order to confirm or refute links to the outbreak and if linked, to identify the root cause of the outbreak so that effective control measures can be implemented. In addition to the overwhelming evidence that WGS provides to outbreak investigations, it also provides support to prevent false positive association of a food to an outbreak. Where a pathogen has been identified in a food product or the food production environment, the isolate sequence can be compared with a database of human isolates to see if there are any matches. To date, PHE has been approached by the food industry on two separate occasions to compare pathogen WGS profiles with those from cases of human illness, on both occasions no matches were found (Grant, 2018; personal communication). However, regardless of whether any human illness is identified, the presence of the pathogen in a finished product (food) or critical food processing environment signifies a breakdown in preventative controls or hygienic conditions and may trigger investigation and/or compliance actions from the regulator.
3.6. Food safety management
Accurate source tracking during the investigation of a contamination event is one of the foremost applications of WGS in food safety management. Understanding if the pathogen or spoilage agent detected is the result of a sporadic contamination event or a recurrent one is essential to understanding the root cause of contamination and will facilitate the implementation or verification of control measures. This will allow industry to focus on priority areas for intervention either at the factory or at supplier level and enable effective monitoring to determine if the action has been successful. WGS can be used to improve supplier and raw material management and optimize efforts on environmental pathogen verification programmes. Improved root cause analysis will lead to better understanding of transmission routes and identification of new sources of contamination. The findings and resulting improvements in manufacturing and farming practices can then be shared with the entire food sector, not just the facility involved in the contamination event.
Besides direct matching of environmental isolates relative to a contamination, it is also possible for industry to compare isolates with entries in public databases as used by public health authorities and food regulators. Depending on the database used for comparison, valuable information can be gleaned such as the identification of potential novel sources (which may provide an indication on initial route of entry into the food production premises), geographic signals about possible origin of contamination and association with human illness. WGS can also lead to valuable insights to refine the ‘hazard identification’ step in microbial risk assessment process. Existing knowledge on organisms is most often gained by studying well characterised laboratory strains which may not necessarily truly represent the phenotypic diversity of the wider population. For example, Maury et al. (2016) recently identified additional novel virulence factors in L. monocytogenes by comparing genomes from clinical and food associated strains. Yahara et al. (2017) examined the impact of various stages of the poultry production chain on Campylobacter populations using WGS and Genome Wide Association Studies (GWAS). Disease-associated SNPs were distinct in ST-21 and ST-45 complexes and investigation of the function of genes containing associated elements demonstrated roles for formate metabolism, aerobic survival, oxidative respiration and nucleotide salvage, allowing potential links to be made between environmental robustness and virulence.
Many disciplines including predictive food microbiology and thermal processing are likely to benefit from the use of WGS data for phenotypic prediction. There are a range of web-based tools and publicly available databases for local use for this purpose, a selection of which are listed in Table 3. These tools identify the genes of interest by aligning draft genomes to a gene database. For example, the genome data obtained through the routine sequencing of every day isolates can be queried to predict traits such as the virulence profile, heat resistance, stress response, biofilm formation, resistance against antimicrobials and biocides by studying their phenotypic characteristics in parallel (Rantsiou et al., 2017). It is important to recognize that detailed genomic information does not necessarily translate into knowledge of gene expression.
Another area of use of WGS for risk assessment is for source attribution of sporadic foodborne illness, i.e. quantifying the relative contribution of different animal, environmental and food sources, including specific food commodity and production sources, to human illness (Pires et al., 2009). So far, the laboratory part of this activity has relied on phenotypic methods and older molecular subtyping methods by looking for characteristics that uniquely identify bacterial strains to any given source. However, recently, genomic data has been used to identify likely sources of infection. For instance, an analysis of 1810 genes comprising the pan-genome of 884 C. jejuni genomes identified 15 novel host-specific genetic markers that were used to attribute French and UK clinical isolates to chicken and ruminants, detecting a possible geographic difference in the relative importance of these sources (Thepault et al., 2017). In addition, gene by gene comparisons of C. jejuni have linked Finnish human disease isolates to temporally related chicken abattoir isolates (Kovanen et al., 2016). With the phylogenetic relevance of WGS, more reliable inferences about the common origin and therefore also the source of strains with similar WGS profiles can be made (Franz et al., 2016). However, to achieve this, new modelling approaches that can handle the huge amounts of sequence data must be developed. Once in place, this source attribution will become an extremely powerful tool identifying the areas of the food production that are associated with most human illnesses. This will help the food industry and others to prioritize food safety activities that are most likely to result in safer food and thereby also, reduce the burden of foodborne illness.
3.7. Industry implementation considerations
For industries and retailers with classically trained microbiologists and limited resources to spend, not only accuracy but also the practicality, simplicity and cost of a method are to be considered before implementing WGS. Ideally, a novel method would be cheaper or at least on par with those in current use. Simplicity means that in addition to sample handling, any software related solutions should be plug and play in both setup and utilization.
The most likely route for adoption by the industry is through an entry-level approach using cg/wg MLST with third party WGS or full 3rd party analysis. A number of commercial solutions are available and some have both cg/wg MLST and SNP analysis in their pipelines with an aim to identify primary clusters using MLST approach and SNP analysis to confirm the relatedness between isolates in a cluster. Key to enable adoption of WGS in routine application is simplification of the analysis and most important simplification of the finite reporting. The finite report of WGS typing analysis would ideally read: matching Yes/No/Maybe and analysis Success/Failed, which are parameters a non-skilled individual can interpret. The report should also include an explanation of the results describing caveats and reasoning behind the final interpretation.
A major consideration for industry adoption of WGS is that routine microbiological testing of foods doesn’t always require the detailed characterisation provided by sequencing and required by public health. Its adoption, use therefore will more likely to be on an as needs basis rather than a total replacement of existing methods. WGS is becoming more widely used by industry for tracking and tracing the origin of contamination; it is hoped that its success in this area, coupled with decreasing sequencing costs, will encourage its wider use.
3.8. Challenges to be addressed
Although WGS has revolutionized the molecular typing of pathogens, several scientific gaps and challenges exist that must be addressed to improve upon the interpretation of WGS data and enable widespread use of WGS in food safety management for the food industry including:
Further work on standardizing the end-to-end protocol to enable the global sharing and comparisons of WGS data.
Research to improve understanding of indistinguishable isolates from epidemiologically unrelated sources to strengthen the interpretation of WGS data.
Investigation into the role of environmental niches on the mutation rates of pathogens to support notions of relatedness. This would improve the interpretation of WGS data, specifically for developing guidance on SNP/allele cut off values and also for strains that may originate from different environments and support different growth rates but need to be considered in one investigation.
Exploration of the value of mobile genetic element (MGE) WGS analysis. In general, MGEs are excluded from WGS analysis although it is well known that these often contribute to virulence and antimicrobial resistance.
WGS of bacterial isolates is a disruptive technology in that it completely changes the way microbiology, in particular subtyping has traditionally been performed. This together with the significant analytical costs along with the knowledge and competency requirements are currently barriers for its wider use by industry.
4. Amplicon sequencing, metagenomics and metatranscriptomics
4.1. A definition of terms
Two approaches using NGS technologies are used to probe the species and functional diversity of microbial communities without bacterial culture: amplicon sequencing or metabarcoding, which involves the amplification and sequencing of specific marker gene families; and metagenomics, the random shotgun sequencing of the whole genomic content of communities.
It is important to differentiate between these two approaches that are sometimes erroneously combined under the term metagenomics (Forbes et al., 2017). We recommend using the term ‘metabarcoding’ when applying amplicon-based techniques and the term ‘metagenomics’ only when untargeted shotgun sequencing is applied. Both techniques eliminate the requirement for single colony isolation and have been highly successful for identifying and investigating uncultivable microorganisms (Cao et al., 2017; Forbes et al., 2017).
4.1.1. Amplicon-based (metabarcoding) microbial community profiling
This technology requires the isolation of DNA directly from samples that can include starter cultures, samples taken during production processing, the final food product and environmental samples. Extracted DNA undergoes targeted PCR amplification of phylogenetic marker genes; commonly the 16S rRNA gene for Archaea and Bacteria, the 18S rRNA gene for Eukaryotes (e.g. protists) and the internal transcribed spacer (ITS) of the ribosomal gene cluster sequences for fungal species. Massive parallel sequencing of these amplicons then generates an array of profiling information about the often-complex microbiota associated with food products. The sequencing data is then processed by dedicated bioinformatic pipelines (described in section 4.3 below) to structure and annotate this raw information into knowledge.
One of the benefits of the metabarcoding approach is the ability to follow the succession of microbial populations over time at various taxonomic levels. For example, oligotyping allows the differentiation of closely related microbial taxa using 16S rRNA gene sequence data (Eren et al., 2013). Compared to random shotgun sequencing (metagenomics), metabarcoding provides a cost-effective overview of the taxonomic composition of a sample and has already been applied to a variety of food products. The use of metabarcoding approaches to study the microbiology of fermented food production is well documented (Bokulich and Mills, 2012; Lusk et al., 2012; Parente et al., 2016; Warnecke and Hugenholtz, 2007) and has also been used for characterising the microbiota of food spoilage (de Boer et al., 2015). Just two examples include investigating the spoilage of dairy products by heat resistant spores of thermophilic bacilli (Zhao et al., 2013) and the proliferation of lactic acid bacteria in fresh cut lettuce, leading to acidification and loss of structure (Paillart et al., 2016). By surveying microbiota variations in fermented products during production, it may be possible to improve the production process by improving flavour or accelerate ripening, for example by adding novel strains at appropriate times or by changing environmental conditions to favour the development of specific microflora (Mayo et al., 2014). Metabarcoding approaches for the characterisation of microbial populations are currently commercially available through a range of companies.
4.1.2. Metagenomic microbiome profiling
Metagenomics generates sequencing information from the genetic material in a sample, permits identification of individual strains and can allow the prediction of functions encoded by microbial communities. This approach has already permitted measurement of population diversity levels in situ (Baker et al., 2006; Venter et al., 2004) and the determination of gene families specific to or enriched in a habitat (Tyson et al., 2004).
Metagenomics is also being explored for the detection, identification and characterisation of pathogens in food (Aw et al., 2016; Leonard et al., 2015, 2016) and in the food chain environment (Yang et al., 2016). Whilst low detection limits have been reported for bacterial pathogens spiked into foods this follows several hours of culture-based enrichment coupled with high sequencing depth to ensure capture of the genomic diversity within the sample (Sekse et al., 2017). However, metagenomics provides an opportunity to survey the diversity and the dynamic abundance of microorganisms within a sample in a less biased manner than metabarcoding and is being used to improve culture-based enrichment methods (Forbes et al., 2017). Shotgun metagenomics can provide a valuable, rapid view of the presence of genetic markers specifying species, serotype, virulence and AMR genes etc. although, at present, these markers usually cannot be assigned to specific bacterial genomes due to the complexity of the metagenomic data (Leonard et al., 2016; Yang et al., 2016). Future metagenomic and metabarcoding bioinformatic developments are likely to make this, and the ability to investigate phylogeny, possible (Ottesen et al., 2016; Truong et al., 2017).
4.2. Meta-omics for microbiome functional characterisation
The field of environmental omics (or meta-omics) has drastically expanded our knowledge about microbial communities (Waldor et al., 2015), prompting a paradigm shift in which the complete microbial community is considered rather than single species. The importance of ecological interactions among microorganisms is now recognized and needs to be included in a global framework to further develop models of the function of community eco-systems (Raes and Bork, 2008). Metagenomics alone is a powerful approach for characterising microbial communities but holds even greater potential when combined with other complementary “omics” technologies such as the measurement of mRNA expression (meta-transcriptomics), detection and categorisation of proteins (proteomics) and metabolite concentration (metabolomics) (Warnecke and Hugenholtz, 2007). The term “foodomics” has been coined to refer to the application of ‘omics technologies in food processing, nutrition and food safety (Cifuentes, 2009). In particular, the combination of metagenomics and metaproteomics holds great potential for the survey of food production, assessing food safety, authenticity and quality (Josic et al., 2017). It is possible to use mass spectrometry (MS)-based proteomic methods to evaluate protein abundance and partitioning of metabolic functions within natural microbial communities (Ram et al., 2005). Undoubtedly, the translation of ‘omics technologies to food microbiology will have an important impact in the food industry (Brown et al., 2017; Walsh et al., 2017). Noteworthy, computational biology advances enabling the description of environmental genomes and their expression in situ have accompanied these new technologies (Segata et al., 2013).
4.3. Computational tools for microbiome characterisation
Most barcoding bioinformatic pipelines start by the cleaning and quality-filtering of 16S rRNA gene or other conserved target amplicons, before their clustering in Operational Taxonomic Units (OTUs), typically at 97% similarity (Konstantinidis and Tiedje, 2005). Pipelines such as mothur (Schloss et al., 2009) and QIIME 2 (http://qiime.org/; Caporaso et al., 2010) perform the entire analysis from raw sequences to OTUs abundance matrices. OTU delineation is useful to detect distinct lineages, to estimate diversity and assess microbial community structure. Nonetheless, this approach is far from perfect and suffers from the fact that a single sequence identity cut-off is inappropriate to delineate true taxonomic lineages such as the species or genus levels, since it overestimates the evolutionary similarity, underestimates the number of substitutions compared to a multiple alignment and does not consider the variability of the 16S rRNA gene or other conserved targets across the tree or network of life (Nguyen et al., 2016).
An attractive alternative to the delineation of OTUs are oligotyping approaches. They take advantage of the ever-increasing quality of reads, do not rely on any clustering algorithm or sequence identity thresholds to identify OTUs and enable analysis of the diversity of closely related but distinct bacterial organisms usually grouped into OTUs (Eren et al., 2013). Two oligotyping implementations are currently available, a supervised ‘oligotyping’ (Eren et al., 2014) and an unsupervised one ‘MED’ (Eren et al., 2015). Another promising approach aims at correcting sequencing errors to enable resolving the fine-scale variation of 16S rRNA reads. The DADA2 package extends the Divisive Amplicon Denoising Algorithm (DADA), a model-based approach for correcting amplicon errors without constructing OTUs (Rosen et al., 2012), which appears to surpass the current state of the art algorithms including QIIME, mothur and MED (Callahan et al., 2016).
Co-occurrence and correlation analyses applied to metabarcoding and metagenomics data (Table 4) are increasingly being used for the prediction of species interactions and the analyses of microbial community structures (Faust and Raes, 2012). A variety of tools are currently available to reconstruct ecological networks and network analyses are revealing unexpected keystone species involved in key ecosystem functions at the global level (Guidi et al., 2016).
Table 4.
Bioinformatic pipelines for metabarcoding, meta-omics analyses and ecological network inference.
| Functionality | Name | Description | Link | Reference |
|---|---|---|---|---|
| Metabarcoding pipeline | QDME2 | Complete metabarcoding workflow: from raw reads to abundance tables | https://qiime2.org/ | Caporaso et al. (2010). |
| MOTHUR | Complete metabarcoding workflow: from raw reads to abundance tables | https://www.mothur.org/ | Schloss et al. (2009). | |
| Oligotyping | Computational method to identify subtle variations among 16S Ribosomal RNA gene sequences | http://merenlab.org/software/oligotyping/ | Eren et al. (2014). | |
| DADA2 | From raw reads to amplicon sequence variant abundance table | https://github.com/benjjneb/dada2 | Callahan et al. (2016). | |
| Meta-omics pipeline | MG-RAST | Complete metagenomic workflow: from raw reads to functional annotations | http://metagenomics.anl.gov/ | Meyer et al. (2008). |
| MOCAT2 | Complete metagenomic workflow: from raw reads to functional annotations | http://mocatembl.de/ | Kultima et al. (2016). | |
| ANvro | Omics data analysis and visualization platform | http://merenlab.org/software/anvio/ | Eren et al. (2015b) | |
| IMP | Complete metagenomic and metatranscriptomic integrative workflow | http://r31ab.uni.lu/web/imp/ | Narayanasamy et al. (2016) | |
| Network inference | Co Net | Ensemble correlation-based network inference | http://psbweb05.psb.ugent.be/conet/ | Faust et al. (2012). |
| sparCC | Correlation-based network inference | https://bitbucket.org/yonatanf/sparcc | Friedman and Aim (2012). | |
| SPIEC-EASI | Inference of graphical models of species association from genomics data | https://github.com/zdkl23/SpiecEasi | Kurtz et al. (2015). | |
| eLSA | Inference of time-dependent associations in time series datasets | https://bitbucket.org/charade/elsa | Xia et al. (2011). |
These tools are very useful to predict microbial interactions and capture the structure of microbial ecosystems but their predictions are very difficult to validate due to the lack of known and validated species interactions in the environment. In addition, predictions of these tools vary widely in sensitivity and precision (Weiss et al., 2016).
Various pipelines for the pre-processing, assembly, clustering and analyses are available for genomic/metatranscriptomic bioinformatic analyses (Table 4), such as MOCAT2 (Kultima et al., 2016), MetAMOS (Treangen et al., 2013) and IMP (Narayanasamy et al., 2016) as standalone frameworks and MG-RAST (Wilke et al., 2016) and Anvi’o (Eren et al., 2015b) as web-based platforms. For the functional annotations of meta-omics data, the most commonly used databases remain KEGG (Kanehisa et al., 2017), COG (Huerta-Cepas et al., 2016) and Pfam (Finn et al., 2016) for functional classifications. Last but not least, bioinformatic platforms implementing complete workflows such as Galaxy (Afgan et al., 2016; Bornich et al., 2016) and EDGE (Li et al., 2017) allow development and deployment of customized pipelines tailored to the needs of the biologists. In-depth bioinformatic expertise will be required to use these tools and to interpret the results obtained, though customization options and the availability of commercial solutions aim to simplify these steps and make it more accessible to the microbiologists.
4.4. Applications of metagenomics in food safety
The absence of a well-curated and high-quality standard database of genomic sequence for pathogenic, probiotic, and functional microbes is a significant hindrance to the implementation of metagenomic-based methods for food safety management (Weimer et al., 2016). Groups such as the Consortium for Sequencing the Food Supply Chain (CSFSC), founded by IBM and Mars Incorporated, are putting efforts into collecting genome information on pathogenic bacteria across the food supply chain, as well as characterising and quantifying the microbiome before and after processing to use genomic and metagenomic data to assure food safety, authenticity and traceability (IBM, 2015; Mars, 2015; Weimer et al., 2016; Welser, 2015). DNA and RNA sequence information collected from food samples by the CSFSC will be used to describe a microbial baseline representing normal microbe communities, which can be applied to track the source of contamination and for food authentication (IBM, 2015; Mars, 2015). Using data from CSFSC’s research, IBM is developing a scalable web-based bioinformatic workbench, the Metagenomics Computation and Analytics Workbench (MCAW), designed to analyse metagenomic and metatranscriptomic sequence data for assessing microbiological hazards and for food authentication in the supply chain. It also provides a service for the storage and management of raw genomic sequences and analysis results (Edlund et al., 2016). The work done to date within the CSFSC and its related MCAW bioinformatic tool offer a model of high-quality genomic and metagenomic database collection, as well as a bioinformatic workbench that can eventually apply NGS to food safety. Similar approaches are being applied by smaller service providers, who are aiming to use NGS to characterise pathogens in food ingredients and products. These combined studies and efforts will potentially bring about a new perspective on microbiological risk assessment and a basis for mitigation strategies as well as related implications for current food safety management norms.
4.5. Issues and challenges
The evaluation of the complete functional repertoire of a microbial population remains difficult due to the incomplete nature of the functional annotation of individual genes or proteins in public databases. As an example, a recent global ocean reference gene catalogue has been annotated at roughly 50% using the eggNOG orthologous genes database (Huerta-Cepas et al., 2016) and only at roughly 30% using the KEGG metabolic pathways database (Kanehisa and Goto, 2000). In recent years, detailed functional categories present in the KEGG (Kanehisa et al., 2017) and SEED (Aziz et al., 2012; Overbeek et al., 2005) databases have been used to annotate and compare genomes and metagenomes using the KEGG Automatic Annotation Server (KAAS) (Moriya et al., 2007), Metagenomics Rapid Annotation using Subsystem Technology (Wilke et al., 2016), and Metagenome Analyzer systems (Huson et al., 2007, 2016). However, these functional categories often remain broad and do not allow the distinguishing of metabolic and physiological features. New tools are required to characterise potential physiological and metabolic pathways (De Filippo et al., 2012) such as the MAPLE system (Takami et al., 2016) which uses KEGG module annotations and permits the estimation of functional abundance and indicates the working probability of the KEGG module based on completion ratio results.
As with traditional microbiological methods, sampling is an extremely important first step in collecting relevant microbiological information from the food processing environment and final products (International Commission on Microbiological Specifications for Foods and Christian and Roberts, 1986; Ni et al., 2013). The diversity in types of samples will be reflected in variations in cell densities, cell viability and the presence of biofilms. Unfortunately, the large variety of matrices in food production does not allow for a one-size fits all solution. Therefore, process and product specific sampling schemes need to be designed. Misinterpretation of results, especially in samples containing low number of microbial cells, can be caused due to the contamination that may originate from reagents used for DNA extraction (Biesbroek et al., 2012). DNA from dead cells may also give a false impression of the microbial load in a food product or processing environment. Preculturing may be used for enrichment of viable cells. However, this must consider microorganisms that require specific growth conditions such as higher temperature, oxygen availability and/or specific nutritional factors (Zhao et al., 2013) and growth requirements for every microorganism are not known). In the case of metatranscriptomic analysis, pre-culturing is of course undesirable, as it would affect the physiological state of the cells. In addition, samples need to be processed as quickly as possible for RNA extraction, stored at −80 °C or fixed using solutions such as RNALater. This is crucial to get an accurate picture of the microbial activity in a sample.
Nucleic acid extraction methods undoubtedly affect the nature as well as the quality and quantity of DNA/RNA obtained from the microorganisms present in a sample, and thus they influence the experimental results. It is essential to keep this in mind during data interpretation and highlights the need to use extraction methods that are optimal for a given study or know what biases the nucleic extraction method may introduce (Bag et al., 2016; Klenner et al., 2017; Cottier et al., 2018; Panek et al., 2018; Vaidya et al., 2018). The matrix from which DNA or RNA is purified for metagenomic/metatranscriptomic analysis also requires special attention. In the case of DNA isolation, the product often contains plant or animal nucleic acid that would also yield sequence information, thereby diluting relevant microbial sequence information. To overcome this there are protocols for removing non-microbial DNA (Feehery et al., 2013, Gosiewski et al., 2014, 2017). The matrix contents may also interfere with performance of molecular analysis as it may inhibit the required biochemical reactions (de Boer et al., 2015). A potential approach to eliminate matrix components is to retrieve microbes by differential centrifugation and filtration from aqueous solutions. Biofilms are sometimes highly rigid making these complex microbial communities difficult to homogenize (Corcoll et al., 2017). Options to open-up these communities include enzyme treatment combined with strong shear forces such as sonication and bead beating.
The issue of metagenomic approaches to detect and characterise specific strains and traits in clinical specimens without the need for using culture is becoming pressing in public health as clinical laboratories are increasingly moving away from culturing bacterial pathogens to detecting them directly in specimens by PCR or enzyme immunoassays (Marder et al., 2017). Metabarcoding after amplification of a single or a few conserved genes may be used to detect different species in a specimen but will fail to detect pathotypes within a species that includes commensals, e.g., E. coli which includes the verocytotoxin producing (Shiga toxin producing, VTEC/STEC), enteroaggregative (EAEC), enteropathogenic (EPEC), entero-invasive (EIEC) pathotypes and Shigella, and less virulent variants of pathogenic species, e.g. non-O1, non-O139 serotypes of Vibrio cholerae. This problem could be solved by targeting genes that encode the virulence factors associated with these pathotypes or serotypes but while this might be feasible with serotype encoding genes, it is often not feasible with virulence associated genes that are commonly present on mobile genetic elements, e.g. plasmids and phages, as it might be impossible to determine which, of multiple bacteria in the specimen, they belong to. This is an active area of current research (Spencer et al., 2015).
Traditional metabarcoding usually does not provide sufficient resolution to differentiate between different isolates or between samples. This is needed for source tracking similar to WGS of cultured isolates. One solution to this problem is to use a similar and potentially compatible approach to wgMLST for analysing sequences of cultured isolates. As many loci as possible (up to a few thousand) are selected from the wgMLST schemes for amplification and sequencing directly from the specimen. This approach is currently being tested for detection and subtyping of Salmonella with the goal of designing a culture independent detection and subtyping system that approximates the resolution of the wgMLST scheme (CDC unpublished).
Metagenomic shotgun sequencing is also being pursued for simultaneous detection and subtyping of pathogens without culture. It has worked in retrospective studies of specimens from outbreaks where the pathogen involved had already been identified by culture (Huang et al., 2017; Loman et al., 2013). However, without prior knowledge of the pathogen, a number of issues need to be resolved such as the aforementioned linking of genes on mobile genetic elements to the strains they belong to. Recent developments in single cell sequencing look promising in addressing this issue for both metabarcoding and metagenomics (Lan et al., 2017; Spencer et al., 2015).
In addition to the issues discussed here, critical improvements in the sequencing technologies and bioinformatics are needed before metabarcoding or shotgun metagenomics can be implemented cost-effectively for diagnostics and subtyping of foodborne pathogens in support of public health and food safety. However, the rapid progress of developments in NGS is likely to herald the demise of bacterial culture as one of the principle methods in food microbiology.
4.6. Validation and benchmarking
As with any new technology undergoing rapid development, end-to-end validation and standardization of NGS is challenging. However, the need for validation, benchmarking and standardization are crucial to define guidelines and best practices for application in food safety and quality management.
Despite the availability of various laboratory protocols and many dedicated tools for the analysis of amplicon and metagenomic sequencing data, their validation is often limited due to the complex nature of environmental or food samples. The variety of protocols and software solutions for NGS applications continues to expand, which makes validation and standardization a hurdle for specific applications. However, several comparative studies have been carried out to test the performance and benchmark various methods and tools at the different steps of a meta-omics survey; namely the sample preparation (Lewandowska et al., 2017), the DNA/RNA extraction (Knudsen et al., 2016; Yuan et al., 2012), the library preparation (Jones et al., 2015; Schirmer et al., 2015), the sequencing platform used (Tremblay et al., 2015) and the bioinformatic approach applied (Siegwald et al., 2017). Nevertheless, standardization in the field is still in its infancy and the comparison and validation of these protocols and tools are essential to gain meaningful information and to make intra- and inter laboratory exchange of information effective (Costea et al., 2017).
With respect to bioinformatic analyses, state of the art pipelines exist that include crucial steps such as adaptor removal, matrix genome sequence removal (meat, vegetables, fruit etc.), low-quality read filtering, contig assembly and finally perform searches against regularly updated databases (Olson et al., 2017; Schlaberg et al., 2017). Singer et al. (2016) reported the use of a defined mock community with complete reference genomes for the benchmarking and validation of metagenomic sequencing and a public resource has recently been created for microbiome bioinformatic benchmarking (Bokulich et al., 2016; Singer et al., 2016). The importance of validation and benchmarking is often overlooked but is essential for a sound interpretation of the data in the context of food safety (e.g. pathogen identification).
The current stage of validation and standardisation with respect to strain detection as well as the assignation of virulence and resistance markers to specific species or strains is more advanced in WGS compared to metagenomics. This can easily be explained by the inherent differences between both approaches: WGS enables easy access to genomes one at a time, at low throughput, while metagenomics is adapted to assess fragmented genomes of complex samples at a high throughput. Nevertheless, new bioinformatic approaches are now enabling the identification of conspecific (i.e. belonging to the same species) strains from metagenomic sequence data (Luo et al., 2015; Zolfo et al., 2017), although these approaches often rely on complete genome information available in public databases.
5. Considerations and challenges related to data sharing
The food industry is truly global, producing and trading items around the world. Processed goods and raw commodities are transported between continents and undergo a variety of investigations by exporting, as well as importing, countries. This results in data generation at several stages and in different countries by different organizations and companies. In this context, NGS is increasingly being applied, as outlined in detail in the preceding sections. It is widely acknowledged that maximal benefit from NGS will be fully realised through the global sharing of sequence data together with an agreed minimal set of descriptive metadata (FAO, 2016). Industry will benefit if their isolates are included in scientific analyses that ultimately leads to a deeper understanding of global microbial diversity, ecology and distribution of organisms. Public health will benefit both from enhanced outbreak detection and resolution but also because industry will proactively implement more effective prevention and control measures based on NGS intelligence.
Currently, industry is concerned that safeguards do not exist to protect companies from regulatory actions, as well as for protecting the company’s reputation and brand equity and this is forcing companies to limit sharing to a legal minimum even though the benefits of data sharing are readily recognized. Thus, to encourage sharing, risk needs to be reduced whilst benefits enhanced, and value demonstrated (FAO, 2016). Some of the key aspects to be addressed to encourage data sharing are described in the following sections.
5.1. Correct data interpretation
WGS data amenable to gross misinterpretation at the hands of poorly-trained personnel can pose serious risks to the food industry, especially in the age of social media. Mechanisms to prevent and tackle these concerns must be addressed for industry to engage with an open data model (FAO, 2016; Taboada et al., 2017). This was highlighted recently by the Technical University of Denmark, where a preliminary analysis reported the presence of monkey DNA in burgers. Following further analysis, this was shown to be cattle DNA (Sep 2016 http://www.food.dtu.dk/english/news/2016/08/mapping-foods-dna-can-reveal-fraud?id=800739d1-f72d-4c57-bab1-4376e0a87bc7). Database limitations and short reads used for data comparison were identified as reasons for the erroneous interpretation of the sequence results, highlighting the critical importance of specialised knowledge for analysing and interpreting WGS data.
Furthermore, particularly within the field of microbial metagenomics, standards for data interpretation are not available or agreed upon, and this can lead to conflicting reporting of the same results (Clooney et al., 2016). This applies not only to the different approaches and data analysis methods, but also when the same approach is used but the conclusions differ.
5.2. Legal clarity/due diligence
In the majority of WGS source tracking investigations, sequence data from closely related strains are included in the analysis to precisely understand the relatedness of the isolates being studied. This is usually achieved by querying the sequences of interest against a public sequence database comprising strains isolated from multiple sources. This can potentially result in the clustering of a food/environmental isolate being analysed with a clinical isolate. The situation becomes complicated when a link is found between a historical patient and a recent in-house isolate and vice-versa with regards to the subsequent steps which must be taken by the food processor from a due diligence perspective. In the USA, all foodborne pathogens obtained through surveillance and inspection are sequenced and the sequences are uploaded into the public domain where they will reside for the life of the database. Matches to any isolates that share a recent common ancestor may cause further investigations by the federal government. In most cases, no regulatory actions will occur without supplemental information, either regarding food exposure or unhygienic observations within the farm to fork continuum. The regulatory response depends on what is found during inspection and how the industry responds according to long term existing practices to inspection and regulation. WGS is just the newest subtyping tool being applied but fundamentally regulatory decision-making and actions are largely unchanged. WGS helps regulators to recognize potential problems earlier because of the higher precision of the technology leading to a more rapid response to improve food safety and public health. Regulators are interested in when a company became aware of a contamination issue and what was done to alleviate this, and prevent recurrence. However, uptake of NGS technologies by industry will also enable industry to investigate potential hygiene or contamination issues in their premises more thoroughly, facilitating root cause analysis and provide them with the opportunity to be far more proactive in tackling such contamination issues (Amini, 2017; FAO, 2016b). Routine use of WGS will mean that food companies are far more aware of what is going on in their production environments and be more pre-emptive in preventing foodborne illness rather than just reacting to it.
5.3. Data ownership
There are concerns that the use of publicly available WGS data could result in trade barriers and even lead to local legal actions due to countries operating within different legal frameworks. Thus, there is a strong desire to establish and agree on a global, harmonised legal framework to facilitate open sharing (FAO, 2016). Potential solutions to some of the issues could be agreed defined delays in data sharing or even a ‘grace period’ without legal consequences in order to promote active data sharing. Considerable effort in terms of cooperation and coordination in this area is required to achieve the aim open sharing of WGS data. It is important for industry to develop mechanisms to both share and protect sensitive information so it can contribute to WGS databases more comfortably.
6. Future prospects for improving food safety
The food industry will increasingly adopt NGS technologies for a wide variety of food microbiological investigations and Fig. 1 summarizes the four different approaches that might be taken depending on the requirement, the available resources and the interest and experience of each company.
Fig. 1.
Summary of potential NGS use by the food industry.
6.1. Whole genome sequencing
One of the key applications of WGS in the food industry will be to understand the root cause of a contamination event in so it can be addressed swiftly. The entire end to end WGS process needs to be convenient, rapid and affordable for WGS to be widely used routinely. The further development of easy-to-use bioinformatic pipelines and the harmonization of analysis methods will help to facilitate this. WGS needs to be adopted not as an add-on to existing microbiological characterisation techniques but as a replacement for existing identification and typing methods in order for the cost benefit to be realised.
Industry will greatly benefit if phenotypic characteristics such as growth and inactivation profiles can be predicted based on analysis of the genome. However, because phenotypic responses are often also controlled at the transcriptional and post-transcriptional level, multi-omics approaches will play a key role for pathogen characterisation in the future. Furthermore, data generated from WGS and metagenomics are likely to be integrated with predictive microbiology for greater control of food safety and quality along the food chain. In the future, genomic databases may be linked to websites dealing with predictive microbiology such as ComBase (http://www.combase.cc/index.php/en/).
Maximal food safety benefit from WGS depends on data sharing and it is anticipated that industry will develop a mechanism to both share and protect sensitive information, so it can contribute to the WGS databases more comfortably. The further development of easy to use bioinformatic pipelines and the harmonization of the methods are also required.
6.2. Metagenomic analysis
Metagenomic tools can improve understanding of the microbial ecology of food processing lines. Within a microbial community, interactions between pathogens and the associated microbiome may indicate the existence of a specific pathogen species or impact its colonization. Variations in environmental factors such as pH, salt concentration, and water activity, caused by processing and handling treatments may lead to corresponding changes in the microbial community (Weimer et al., 2016). Food producers will be able to either validate or improve current microbial hazard management using the metagenomic approach to monitor the occurrence and abundance of microbes and genes in the microbial community of food processing lines.
For microbial spoilage risk management, it is important to monitor changes in the microbial community during storage to plan appropriate processing, treatment and storage conditions for food products (Ercolini, 2013). Metagenomic tools can help anticipate microbial spoilage by studying changes in the diversity or proportion of spoilage associated microbes in the microbiota of food products (Ercolini et al., 2011; Kable et al., 2016), as well as monitoring the behaviour of starter/spoilage-associated populations in cultured food (Masoud et al., 2012). These tools have allowed researchers to develop understanding of defects with unknown origin and to develop strategies to eliminate those defects such as those affecting meat and seafood (Chaillou et al., 2015), sausage meat (Hultman et al., 2015), Chinese rice wine (Hong et al., 2016) and continental cheeses (Quigley et al., 2016). The information gleaned from these applications has been used to select starter cultures used to produce fermented foods with more consistent quality (Galimberti et al., 2015), to identify biomarkers for ripeness and quality, and to optimize environmental conditions during production of cheeses (Wolfe et al., 2014), by driving formation of microbial communities to produce foods with desired properties. Applications of metagenomic studies have revealed that difference in soil microbiota have an impact on the flavours of wines produced in different geographic regions (Zarraonaindia et al., 2015).
Metagenomic and metatranscriptomic approaches also have great potential in becoming valuable options for detecting food authenticity and integrity by precisely describing the microbial community of a specific food product. Traditional DNA barcoding methodologies based on (PCR) and Sanger sequencing are limited by their low-throughput nature and the need for high DNA purity and concentration of food samples (Shokralla et al., 2014). These limitations are being addressed by high-throughput NGS technologies including metagenomic approaches, which provide more information on the microbial community populations and biological ingredients of a food product, as well as allowing culture-independent testing. Metagenome prediction software has also been used to understand the impact of modified atmospheres on metabolic pathways, to aid the design of preservation systems (Ferrocino and Cocolin, 2017). These metagenomic approaches, when combined with other ‘omics technologies such as proteomics and metabolomics have the potential to link particular species in a community with functional characteristics, such as flavour production or production of harmful metabolites such as biogenic amines in rice wine (Liu et al., 2016).
There are challenges regarding utilization of the metagenome for the food industry including the detection of DNA originating from dead microbes as well as low sensitivity of detection compared with culture based methods as well as the relatively high costs and further developments in these areas are being pursued.
6.3. The impact of NGS application on food trade and food industry
NGS application in food safety management is likely to become a game changer for global food trade. While the main players continue to push for NGS technologies for global food safety management, there is also an urgent need to close the technological gap between the less-advanced food producing countries to facilitate global food trade. Developing countries have significant concerns over the possible imbalance of trade opportunities, since they might not be able to provide the same level of WGS-based data as others (FAO, 2016). Obstacles to using WGS include lack of infrastructure e.g. basic utilities and/or internet access and the need to develop a skilled trained workforce both at regulatory and food industry level to perform and interpret WGS data.
It is important that international efforts to facilitate the transition from old technologies to NGS globally continue to offer opportunities to these countries, in terms of technology and training, knowledge exchange, restructuring of the food safety system within the country and also by improving the local food industry in the country. The emergence of NGS technology could be a turning point to bridge the gap between less-advanced food producing countries and the developed nations.
Finally, the ultimate extension of the impact of NGS will be a reduction in food industry costs. The cost of generating bacterial genomic sequences is still decreasing rapidly and within the next few years it is expected that the cost of applying NGS technology will easily out-compete the cost of microbiological culture and physiological examination. This cost reduction is additional to the transformational food industry benefits that this new technology is set to deliver.
Complete list of members of the Expert Group
| Dr Kathie Grant – Chair | Public Health England | UK |
| Dr Balamurugan Jagadeesan – Vice-Chair | Nestlé Research Center, Nestec Ltd | CH |
| Prof. Frank Aarestrup | Technical University of Denmark (DTU) | DK |
| Dr Marc Allard | Food and Drug Administration (FDA) | US |
| Dr Samuel Chaffron | CNRS and University of Nantes | FR |
| Dr Lay Ching Chai* | University of Malaya | MY |
| Dr John Chapman | Unilever | NL |
| Dr Peter Gerner-Smidt* | Centers for Disease Control and Prevention (CDC) | US |
| Prof. Dag Harmsen | Munster University Hospital | DE |
| Dr Mitsuru Katase** | Fuji Oil Co., Ltd. | JP |
| Dr Bon Kimura* | Tokyo University of Marine Science & Technology | JP |
| Mr Sebastien Leuillet | Institut Merieux (Merieux NutriSciences) | FR |
| Dr Peter McClure | Mondelez International | UK |
| Dr Trevor Phister | PepsiCo International | UK |
| Dr Masami Takeuchi | Food and Agricultural Organisation (FAO) | IT |
| Dr Silin Tang | Mars Global Food Safety Center | CN |
| Dr Jos van der Vossen | The Netherlands Organisation for Applied Scientific Research (TNO) | NL |
| Dr Anett Winkler | Cargill | BE |
| Dr Yinghua Xiao | Aria Foods | DK |
The participation of these experts was supported by ILSI Japan, ILSI North America or ILSI Southeast Asia Region.
This company is a member of ILSI Japan.
Acknowledgements
The authors would like to thank Ms Lilou van Lieshout, Dr Belén Márquez-García and Dr Tobias Recker, Scientific Project Managers at ILSI Europe who facilitated scientific meetings and coordinated the overall project management and administrative tasks related to the completion of this work. The authors would like to thank Prof. Frank Aarestrup, Prof. Dag Harmsen, Dr Robèr Kempermann, Dr Trevor Phister and Dr Masami Takeuchi, for their contributions and suggestions when developing the publication. The authors would like to thank ILSI Europe’s Microbiological Food Safety Task Force, the ILSI North America Food Microbiology Committee, ILSI Japan Next Generation Sequencing Project, and ILSI Southeast Asia Region for their support as specified in the funding sources section
Funding sources
This work was conducted by an expert group brought together by the European branch of the International Life Sciences Institute (ILSI Europe), with support from the ILSI North America Food Microbiology Committee, ILSI Japan Next Generation Sequencing Project, and ILSI Southeast Asia Region. The expert group and this publication were coordinated by the ILSI Europe Microbiological Food Safety Task Force. Industry members of this task force are listed on the ILSI Europe website at http://ilsi.eu/task-forces/food-safety/microbiological-food-safety/. Experts were not paid for the time spent on this work; however, the non-industry members within the expert group were offered support for travel and accommodation cost from the ILSI Europe Microbiological Food Safety Task Force, the ILSI North America Food Microbiology Committee, ILSI Japan, and ILSI Southeast Asia Region to attend meetings to discuss the manuscript. The ILSI Europe Microbiological Food Safety Task Force offered to all non-industry members a small compensatory sum (honoraria) with the option to decline. The research reported is the result of a scientific evaluation in line with ILSI Europe’s framework to provide a precompetitive setting for public–private partnership. The opinions expressed herein and the conclusions of this publication are those of the authors and do not necessarily represent the views of ILSI Europe nor those of its member companies. For further information about ILSI Europe, please email info@ilsieurope.be or call +32 2771 00 14.
Footnotes
About ILSI Europe
ILSI Europe fosters collaboration among the best scientists from industry, academia and the public sector to provide evidence-based scientific solutions and to pave the way forward in nutrition, food safety, consumer trust and sustainability. To deliver science of the highest quality and integrity, scientists collaborate and share their unique expertise in expert groups, workshops, symposia and resulting publications. ILSI Europe’s activities are mainly funded by its member companies. In addition, ILSI Europe receives funding from the European Union-funded projects they partner with and from projects initiated by Member States’ national authorities.
About ILSI North America
ILSI North America is a public, non-profit foundation that provides a forum to advance understanding of scientific issues related to the nutritional quality and safety of the food supply by sponsoring research programs, educational seminars and workshops, and publications. ILSI North America receives support primarily from its industry membership.
About ILSI Japan
ILSI Japan plays a role in worldwide activities of ILSI, and positively consults on the specific issues in Japan. Based on the latest and sound science, ILSI Japan carries out projects to resolve and disseminate scientific issues in the fields of health, nutrition, food safety, and the environment while ensuring the international harmonization. The purpose of these projects is to contribute to better nutrition, improved health, food safety and the environment for the Japanese and people all over the world.
About ILSI Southeast Asia Region
ILSI Southeast Asia Region seeks to achieve ILSI’s mission by fostering collaboration among experts from academia, government, and industry, to provide a balance of perspectives and ensure that the scientific outcomes of their activities are useful at the national, regional and international levels for public health improvement. Through its Nutrition, Food Safety and Sustainability Science Clusters, ILSI Southeast Asia Region initiates and coordinates scientific and community programs, research, and capacity building training through sharing and dissemination of the latest scientific knowledge and information in Southeast Asia, Australia and New Zealand. In Southeast Asia, these activities span across the 10 ASEAN countries of Brunei, Cambodia, Indonesia, Laos, Malaysia, Myanmar, Philippines, Thailand, Singapore and Vietnam.
References
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J, 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44, W3–W10. 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajawatanawong P, 2017. Molecular phylogenetics: concepts for a newcomer. Adv. Biochem. Eng. Biotechnol 160, 185–196. 10.1007/10_2016_49. [DOI] [PubMed] [Google Scholar]
- Akhter S, Aziz RK, Edwards RA, 2012. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 40(16), e126. doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard MW, Luo Y, Strain E, Pettengill J, Timme R, Wang C, Li C, Keys CE, Zheng J, Stones R, Wilson MR, Musser SM, Brown EW, 2013. On the evolutionary history, population genetics and diversity among isolates of Salmonella Enteritidis PFGE pattern JEGX01.0004. PLoS One 8(1), e55254. doi: 10.1371/journal.pone.0055254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard MW, Strain E, Melka D, Bunning K, Musser SM, Brown EW, Timme R, 2016. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J. Clin. Microbiol 54 (8), 1975–1983. 10.1128/JCM.00081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard MW, Bell R, Ferreira CM, Gonzalez-Escalona N, Hoffmann M, Muruvanda T, Ottesen A, Ramachandran P, Reed E, Sharma S, Stevens E, Timme R, Zheng J, Brown EW, 2017. Genomics of foodborne pathogens for microbial food safety. Curr. Opin. Biotechnol 49, 224–229. 10.1016/j.copbio.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Amini S, 2017. NGS in Food Safety: seeing what was not possible before. Food Safety Tech Sept 20 2017. (https://foodsafetytech.com/feature_article/ngs-food-safety-seeing-never-possible/).
- Angiuoli SV, Gussman A, Klimke W, Cochrane G, Field D, Garrity G, Kodira CD, Kyrpides N, Madupu R, Markowitz V, Tatusova T, Thomson N, White O, 2008. Towards an online repository of Standard Operating Procedures (SOPs) for (meta) genomic annotation. OMICS 12 (2), 137–141. 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argimón S, Abudahab K, Goater RJ, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MT, Yeats CA, Grundmann H, Spratt BG, Aanensen DM, 2016. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom 2(11), e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton PM, Nair S, Peters TM, Bale JA, Powell DG, Painset A, Tewolde R, Schaefer U, Jenkins C, Dallman TJ, de Pinna EM, Grant KA, Salmonella Whole Genome Sequencing Implementation Group., 2016. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 4, e1752. doi: 10.7717/peerj.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw TG, Wengardt S, Rose RB, 2016. Metagenomic analysis of viruses associated with field-grown and retail lettuce identifies human and animal viruses. Int. J. Food Microbiol 233, 50–56. 10.1016/j.ijfoodmicro.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O, 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Devoid S, Disz T, Edwards RA, Henry CS, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Stevens RL, Vonstein V, Xia F, 2012. SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS One 7(10), e48053. doi: 10.1371/journal.pone.0048053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bag S, Saha B, Mehta O, Anbumani D, Kumar N, Dayal M, 2016. An improved method for high quality metagenomics DNA extraction from human and environmental samples. Sci. Rep 31 (6), 26775 https://doi.org/10.0.4.14/srep26775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Tyson GW, Webb RI, Flanagan J, Hugenholtz P, Allen EE, Banfield JF, 2006. Lineages of acidophilic archaea revealed by community genomic analysis. Science 314, 1933–1935. 10.1126/science.1132690. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, 2003. The deep roots of eukaryotes. Science 300 (5626), 1703–1706. 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA, 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol 19 (5), 455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E, 2018. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin. Microbiol. Infect 24, 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesbroek G, Sanders EAM, Roeselers G, Wang X, Caspers MPM, Trzciński K, et al. (2012) Deep sequencing analyses of low density microbial communities: working at the boundary of accurate microbiota detection. PloS One 7(3): e32942. doi.org/10.1371/journal.pone.0032942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Rideout JR, Mercurio WG, Shiffer A, Wolfe B, Maurice CF, Dutton RJ, Turnbaugh PJ, Knight R, Caporaso JG, 2016. mockrobiota: a public resource for microbiome bioinformatics benchmarking. mSystems 1(5), e00062–16. doi: 10.1128/mSystems.00062-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Mills DA, 2012. Next-generation approaches to the microbial ecology of food fermentations. BMB Rep 45, 377–389. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 (15), 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornich C, Grytten I, Hovig E, Paulsen J, Cech M, Sandve GK, 2016. Galaxy Portal: interacting with the galaxy platform through mobile devices. Bioinformatics 32, 1743–1745. 10.1093/bioinformatics/btw042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, Earle S, Pankhurst LJ, Anson L, de Cesare M, Piazza P, Votintseva AA, Golubchik T, Wilson DJ, Wyllie DH, Diel R, Niemann S, Feuerriegel S, Kohl TA, Ismail N, Omar SV, Smith EG, Buck D, McVean G, Walker AS, Peto TE, Crook DW, Iqbal Z, 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun 6, 10063 10.1038/ncomms10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EW, Gonzalez-Escalona N, Stones R, Timme R, Allard MW, 2017. The rise of genomics and the promise of whole genome sequencing for understanding microbial foodborne pathogens. In: Gurtler JB, Doyle MP, Kornacki JL (Eds.), Foodborne Pathogens: Virulence Factors and Host Susceptibility Springer International Publishing, London: ), pp. 333–351. [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP, 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Fanning S, Proos S, Jordan K, Srikumar S, 2017. A review on the applications of next generation sequencing technologies as applied to food-related microbiome studies. Front. Microbiol 8, 1829 10.3389/fmicb.2017.01829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R, 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H, 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother 58 (7), 3895–3903. 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriço JA, Rossi M, Moran-Gilad J, Van Domselaar G, Ramirez M, 2018. A primer on microbial bioinformatics for nonbioinformaticians. Clin. Microbiol. Infect 24 (4), 342–349. 10.1016/j.cmi.2017.12.015. [DOI] [PubMed] [Google Scholar]
- Chaillou S, Chaulot-Talmon A, Caekebeke H, Cardinal M, Christieans S, Denis C, Desmonts MH, Dousset X, Feurer C, Hamon E, Joffraud JJ, La Carbona S, Leroi F, Leroy S, Lorre S, Macé S, Pilet MF, Prévost H, Rivollier M, Roux D, Talon R, Zagorec M, Champomier-Vergès MC, 2015. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J 9 (5), 1105–1118. 10.1038/ismej.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zheng D, Liu B, Yang J, Jin Q, 2016. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res 44 (D1), D694–D697. 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Luo Y, Carleton H, Timme R, Melka D, Muruvanda T, 2017. Whole genome and core genome multilocus sequence typing and single nucleotide polymorphism analyses of Listeria monocytogenes associated with an outbreak linked to cheese, United States, 2013. Appl. Environ. Microbiol 83(15), e00633–17. doi: 10.1128/AEM.00633-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S, 1999. Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB) 99, 45–56. [Google Scholar]
- Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J, 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10 (6), 563–569. 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- Cifuentes A, 2009. Food analysis and foodomics. J. Chromatogr. A 1216 (43), 7109 10.1016/j.chroma.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Clooney AG, Fouhy F, Sleator RD, O’Driscoll A, Stanton C, Cotter PD, Claesson MJ, 2016. Comparing apples and oranges?: next generation sequencing and its impact on microbiome analysis. PloS One 11(2), e01480281–16. doi: 10.1371/journal.pone.0148028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoll N, Österlund T, Sinclair L, Eiler A, Kristiansson E, Backhaus T, et al. , 2017. Comparison of four DNA extraction methods for comprehensive assessment of 16S rRNA bacterial diversity in marine biofilms using high-throughput sequencing. FEMS Microbiol. Lett 364 10.1093/femsle/fnx139.fnx139. [DOI] [PubMed] [Google Scholar]
- Costea PI, Zeller G, Sunagawa S, Pelletier E, Alberti A, Levenez F, Tramontano M, Driessen M, Hercog R, Jung FE, Kultima JR, Hayward MR, Coelho LP, Allen-Vercoe E, Bertrand L, Blaut M, Brown JRM, Carton T, Cools-Portier S, Daigneault M, Derrien M, Druesne A, de Vos WM, Finlay BB, Flint HJ, Guarner F, Hattori M, Heilig H, Luna RA, van Hylckama Vlieg J, Junick J, Klymiuk I, Langella P, Le Chatelier E, Mai V, Manichanh C, Martin JC, Mery C, Morita H, O’Toole PW, Orvain C, Patil KR, Penders J, Persson S, Pons N, Popova M, Salonen A, Saulnier D, Scott KP, Singh B, Slezak K, Veiga P, Versalovic J, Zhao L, Zoetendal EG, Ehrlich SD, Dore J, Bork P, 2017. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol 35 (11), 1069–1076. 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- Cottier F, Srinivasan KG, Yurieva M, Liao W, Poidinger M, Zolezzi F, 2018. Advantages of meta-total RNA sequencing (MeTRS) over shotgun metagenomics and amplicon-based sequencing in the profiling of complex microbial communities. npj Biofilms Microbiomes 4 (1), 2 10.1038/s41522-017-0046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham SA, Chia N, Jeraldo PR, Quest DJ, Johnson JA, Boxrud DJ, 2017. Comparison of two whole-genome sequencing methods for analysis of three methicillin-resistant Staphylococcus aureus outbreaks. J. Clin. Microbiol 55 (6), 1946–1953. 10.1128/JCM.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallman T, Inns T, Jombart T, Ashton P, Loman N, Chatt C, Messelhaeusser U, Rabsch W, Simon S, Nikisins S, Bernard H, le Hello S, Jourdan da-Silva N, Kornschober C, Mossong J, Hawkey P, de Pinna E, Grant K, Cleary P, 2016. Phylogenetic structure of European Salmonella Enteritidis outbreak correlates with national and international egg distribution network. Microb. Genom 2(8), e000070. doi: 10.1099/mgen.0.000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Pettengill JB, Luo Y, Payne J, Shpuntoff A, Rand H, Strain E, 2015. CFSAN SNP Pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Comput. Sci 1, e20. doi: 10.7717/peerj-cs.20. [DOI] [Google Scholar]
- Deatherage DE, Kepner JL, Bennett AF, Lenski RE, Barrick JE, 2017. Specificity of genome evolution in experimental populations of < em > Escherichia coli < / em > evolved at different temperatures. Proc. Natl. Acad. Sci. Unit. States Am. 7;114(10):E1904 LP-E1912. doi: 10.1073/pnas.1616132114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Caspers M, Sanders JW, Kemperman R, WIjman J, Lommerse G, Roeselers G, Montijn R, Abee T, Kort R, 2015. Amplicon sequencing for the quantification of spoilage microbiota in complex foods including bacterial spores. Microbiome 3, 30 10.1186/s40168-015-0096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Ramazzotti M, Fontana P, Cavalieri D, 2012. Bioinformatic approaches for functional annotation and pathway inference in metagenomics data. Briefings Bioinf 13 (6), 696–710. 10.1093/bib/bbs070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, Garcia-Cobos S, Kooistra-Smid AM, Raangs EC, Rosema S, Veloo AC, Zhou K, Friedrich AW, Rossen JW, 2017. Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol 243, 16–24. 10.1016/j.jbiotec.2016.12.022. [DOI] [PubMed] [Google Scholar]
- Edlund SB, Beck KL, Haiminen N, Parida LP, Storey DB, Weimer BC, Kaufman JH, Chamliss DD, 2016. Design of the MCAW compute service for food safety bioinformatics. IBM J. Res. Dev 60 (5–6), 7580716 10.1147/JRD.2016.2584798. [DOI] [Google Scholar]
- Elson R, Awofisayo-Okuyelu A, Greener T, Swift C, Painset A, Amar C, Newton A, Aird H, Swindlehurst M, Elviss N, Foster K, Dallman TJ, Ruggles R, Grant K Utility of WGS to describe the persistence and evolution of L. monocytogenes strains within crabmeat processing environments linked to outbreaks. J. Food Protect. (Accepted for publication). [DOI] [PubMed]
- Ercolini D, Ferrocino I, Nasi A, Ndagijimana M, Vernocchi P, La Storia A, 2011. Monitoring of microbial metabolites and bacterial diversity in beef stored under different packaging conditions. Appl. Environ. Microbiol 77 (20), 7372–7381. 10.1128/AEM.05521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercolini D, 2013. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol 79 (10), 3148–3155. 10.1128/AEM.00256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML, 2013. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol 4, 1111–1119. 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Borisy GG, Huse SM, Mark Welch JL, 2014. Oligotyping analysis of the human oral microbiome. Proc. Natl. Acad. Sci. U. S. A 111, E2875–E287E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML, 2015a. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9, 968–979. 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO, 2015b. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO, 2016. Applications of Whole Genome Sequencing in Food Safety Management http://www.fao.org/3/a-i5619e.pdf.
- Faust K, Raes J, 2012. Microbial interactions: from networks to models. Nat. Rev. Microbiol 10 (8), 538–550. 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C, 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol 8(7), e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feehery GR, Yigit E, Oyola SO, Langhorst BW, Schmidt VT, Stewart FJ, 2013. A method for selectively enriching microbial DNA from contaminating vertebrate host DNA. PLoS One 28;8(10):e76096–e76096. doi: 10.1371/journal.pone.0076096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrocino I, Cocolin L, 2017. Current perspectives in food-based studies exploiting multi-omics approaches. Curr. Opin. Food Sci 13, 10–15. 10.1016/j.cofs.2017.01.002. [DOI] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A, 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44 (D1), D279–D285. 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes JD, Knox NC, Ronholm J, Pagotto F, Reimer A, 2017. Metagenomics: the next culture-independent game changer. Front. Microbiol 8, 1069 10.3389/fmicb.2017.01069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz E, Gras LM, Dallman T, 2016. Significance of whole genome sequencing for surveillance, source attribution and microbial risk assessment of foodborne pathogens. Curr. Opin. Food Sci 8, 74–79. 10.1016/j.cofs.2016.04.004. [DOI] [Google Scholar]
- Friedman J, Alm EJ, 2012. Inferring correlation networks from genomic survey data. PLoS Comput. Biol 8(9), e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimberti A, Bruno A, Mezzasalma V, De Mattia F, Bruni I, Labra M, 2015. Emerging DNA-based technologies to characterize food ecosystems. Food Res. Int 69, 424–433. 10.1016/j.foodres.2015.01.017. [DOI] [Google Scholar]
- Gardner SN, Slezak T, Hall BG, 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31 (17), 2877–2878. 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- Gargis AS, Kalman L, Lubin IM, 2016. Assuring the quality of next-generation sequencing in clinical microbiology and public health laboratories. J. Clin. Microbiol 54 (12), 2857–2865. 10.1128/JCM.00949-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner-Smidt P, Hyytia-Trees E, Barrett T, 2013. In: Doyle M, Buchanan R (Eds.), Molecular Source Tracking and Molecular Subtyping in Food Microbiology: Fundamentals and Frontiers, fourth ed. ASM Press, Washington DC: ), pp. 1059–1077. 10.1128/9781555818463.ch43. [DOI] [Google Scholar]
- Gillesberg Lassen S, Ethelberg S, Björkman JT, Jensen T, Sørensen G, Kvistholm Jensen A, Sørensen G, Kvistholm Jensen A, Müller L, Nielsen EM, Mølbak K, 2016. Two listeria outbreaks caused by smoked fish consumption-using whole-genome sequencing for outbreak investigations. Clin. Microbiol. Infect 22 (7), 620–624. 10.1016/j.cmi.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, McCombine WR, 2016. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet 17, 333–351. 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosiewski T, Ludwig-Galezowska AH, Huminska K, Sroka-Oleksiak A, Radkowski P, Salamon D, 2017. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method - the observation of DNAemia. Eur. J. Clin. Microbiol. Infect. Dis 36 (2), 329–336. 10.1007/s10096-016-2805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosiewski T, Jurkiewicz-Badacz D, Sroka A, Brzychczy-Włoch M, Bulanda M, 2014. A novel, nested, multiplex, real-time PCR for detection of bacteria and fungi in blood. BMC Microbiol 14 (1), 144 10.1186/1471-2180-14-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant K, Jenkins C, Arnold C, Green J, Zambon M, 2018. Implementing Pathogen Genomics: a Case Study www.gov.uk/government/publications/implementing-pathogen-genomics-a-case-study.
- Guidi L, Chaffron S, Bittner L, Eveillard D, Larhlimi A, Roux S, Darzi Y, Audic S, Berline L, Brum J, Coelho LP, Espinoza JCI, Malviya S, Sunagawa S, Dimier C, Kandels-Lewis S, Picheral M, Poulain J, Searson S, Tara Oceans coordinators, Stemmann, L., Not F, Hingamp P, Speich S, Follows M, Karp-Boss L, Boss E, Ogata H, Pesant S, Weissenbach J, Wincker P, Acinas SG, Bork P, de Vargas C, Iudicone D, Sullivan MB, Raes J, Karsenti E, Bowler C, Gorsky G, 2016. Plankton networks driving carbon export in the oligotrophic ocean. Nature 532 (7600), 465–470. 10.1038/nature16942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O, 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol 59 (3), 307–321. 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hedge J, Wilson DJ, 2016. Practical approaches for detecting selection in microbial genomes. PLoS Comput. Biol 12(2), e1004739. doi : 10.1371/journal.pcbi.1004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Luo Y, Monday SR, Gonzalez-Escalona N, Ottesen AR, Muruvanda T, Wang C, Kastanis G, Keys C, Janies D, Senturk IF, Catalyurek UV, Wang H, Hammack TS, Wolfgang WJ, Schoonmaker-Bopp D, Chu A, Myers R, Haendiges J, Evans PS, Meng J, Strain EA, Allard MW, Brown EW, 2016. Tracing origins of the Salmonella Bareilly strain causing a food-borne outbreak in the United States. J. Infect. Dis 213 (4), 502–508. 10.1093/infdis/jiv297. [DOI] [PubMed] [Google Scholar]
- Hong X, Chen J, Liu L, Wu H, Tan H, Xie G, Xu Q, Zou H, Yu W, Wang L, Qin N, 2016. Metagenomic sequencing reveals the relationship between microbiota composition and quality of Chinese Rice Wine. Sci. Rep 6, 26621 10.1038/srep26621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AD, Luo C, Pena-Gonzalez A, Weigand MR, Tarr CL, Konstantinidis KT, 2017. Metagenomics of two severe foodborne outbreaks provides diagnostic signatures and signs of coinfection not attainable by traditional methods. Appl. Environ. Microbiol 83(3), pii: e02577–16. doi: 10.1128/AEM.02577-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P, 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44 (D1), D286–D293. 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman J, Rahkila R, Ali J, Rousu J, Björkroth KJ, 2015. Meat processing plant microbiome and contamination patterns of cold-tolerant bacteria causing food safety and spoilage risks in the manufacture of vacuum-packaged cooked sausages. Appl. Environ. Microbiol 81 (20), 7088–7097. 10.1128/AEM.02228-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC, 2007. MEGAN analysis of metagenomic data. Genome Res 17 (3), 377–386. 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S, Ruscheweyh HJ, Tappu R, 2016. MEGAN Community Edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol 12(6), e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IBM., 2015. Consortium for Sequencing the Food Supply Chain: IBM Research and Mars Tackle Global Health with Food Safety Partnership, [online] Available: http://www.research.ibm.com/client-programs/foodsafety/.
- Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE, 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6 (11), 90 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Commission on Microbiological Specifications for Foods & Christian, J. H. B & Roberts TA, 1986, Microorganisms in Foods. 2, Sampling for Microbiological Analysis: Principles and Specific Applications/International Commission on Microbiological Specifications for Foods (ICMSF) of the International Union of Microbiological Societies, second ed., Blackwell Scientific Publications, Oxford, England: xiii:293. [Google Scholar]
- IRIDA, 2017. IRIDA – Integrated Rapid Infectious Disease Analysis Project Available at: http://irida.ca.
- Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, Carleton H, Katz LS, Stroika S, Gould LH, Mody RK, Silk BJ, Beal J, Chen Y, Timme R, Doyle M, Fields A, Wise M, Tillman G, Defibaugh-Chavez S, Kucerova Z, Sabol A, Roache K, Trees E, Simmons M, Wasilenko J, Kubota K, Pouseele H, Klimke W, Besser J, Brown E, Allard M, Gerner-Smidt P, 2016. Implementation of nationwide real-time whole-genome sequencing to enhance Listeriosis outbreak detection and investigation. Clin. Infect. Dis 63, 380–386. 10.1093/cid/ciw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM, 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol 52 (5), 1501–1510. 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MB, Highlander SK, Anderson EL, Li W, Dayrit M, Klitgord N, Fabani MM, Seguritan V, Green J, Pride DT, Yooseph S, Biggs W, Nelson KE, Venter JC, 2015. Library preparation methodology can influence genomic and functional predictions in human microbiome research. Proc. Natl. Acad. Sci. U. S. A 112 (45), 14024–14029. 10.1073/pnas.1519288112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josic D, Persuric Z, Resetar D, Martinovic T, Saftic L, Kraljevic-Pavelic S, 2017. Use of foodomics for control of food processing and assessing of food safety. Adv. Food Nutr. Res 81, 187–229. 10.1016/bs.afnr.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D, 2013. Updating benchtop sequencing performance comparison. Nat. Biotechnol 31 (4), 294–296. 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- Kable ME, Srisengfa Y, Laird M, Zaragoza J, Mcleod J, Heidenreich J, Marco ML, 2016. The core and seasonal microbiota of raw bovine milk in tanker trucks and the impact of transfer to a milk processing facility. mBio 7(4), pii: e00836–16. doi: 10.1128/mBio.00836-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28 (1), 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K, 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45 (D1), D353–D361. 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LS, Griswold T, Williams-Newkirk AJ, Wagner D, Petkau A, Sieffert C, Van Domselaar G, Deng X, Carleton HA, 2017. A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front. Microbiol 8, 375 10.3389/fmicb.2017.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A, 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28 (12), 1647–1649. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenner J, Kohl C, Dabrowski PW, Nitsche A, 2017. Comparing viral metagenomics extraction methods. Curr. Issues Mol. Biol 24, 59–70. 10.21775/cimb.024.059. [DOI] [PubMed] [Google Scholar]
- Kleta S, Hammerl JA, Dieckmann R, Malorny B, Borowiak M, Halbedel S, Prager R, Trost E, Flieger A, Wilking H, Vygen-Bonnet S, Busch U, Messelhäußer U, Horlacher S, Schönberger K, Lohr D, Aichinger E, Luber P, Hensel A, Al Dahouk S, 2017. Molecular tracing to find source of protracted invasive Listeriosis outbreak, Southern Germany, 2012–2016. Emerg. Infect. Dis 23 (10), 1680–1683. 10.3201/eid2310.161623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen BE, Bergmark L, Munk P, Lukjancenko O, Priemé A, Aarestrup FM, Pamp SJ, 2016. Impact of sample type and DNA isolation procedure on genomic inference of microbiome composition. mSystems, 1(5). pii: e00095–16. doi: 10.1128/mSystems.00095-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Shumway M, Leinonen R, 2012. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res 40 (Database issue), D54–D56. 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM, 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A 102 (7), 2567–2572. 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM, 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27 (5), 722–736. 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koser CU, Ellington MJ, Cartwright EJP, Gillespie SH, Brown NM, Farrington M, Holden MT, Dougan G, Bentley SD, Parkhill J, Peacock SJ, 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 8(8), e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovanen S, Kivisto R, Llarena A-K, Zhang J, Kärkkäinen U-M, Tuuminen T, Uksila J, Hakkinen M, Rossi M, Hänninen M-L, 2016. Tracing isolates from domestic human Campylobacter jejuni infections to chicken slaughter batches and swimming water using whole-genome multilocus sequence typing. Int. J. Food Microbiol 226, 53–60. 10.1016/j.ijfoodmicro.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Kozyreva VK, Truong CL, Greninger AL, Crandall J, Mukhopadhyay R, Chaturvedi V, 2017. Validation and implementation of clinical laboratory improvements act-compliant whole-genome sequencing in the public health microbiology laboratory. J. Clin. Microbiol 55 (8), 2502–2520. 10.1128/JCM.00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultima JR, Coelho LP, Forslund K, Huerta-Cepas J, Li SS, Driessen M, Voigt AY, Zeller G, Sunagawa S, Bork P, 2016. MOCAT2: a metagenomic assembly, annotation and profiling framework. Bioinformatics 32 (16), 2520–2523. 10.1093/bioinformatics/btw183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA, 2015. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol 11(5), e1004226. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvistholm Jensen A, Nielsen EM, Bjorkman JT, Jensen T, Muller L, Persson S, Bjerager G, Perge A, Krause TG, Kiil K, Sørensen G, Andersen JK, Mølbak K, Ethelberg S, 2016. Whole-genome sequencing used to investigate a nationwide outbreak of listeriosis caused by ready-to-eat delicatessen meat, Denmark, 2014. Clin. Infect. Dis 63, 64–70. 10.1093/cid/ciw192. [DOI] [PubMed] [Google Scholar]
- Lan F, Demaree B, Ahmed N, Abate A, 2017. SiC-Seq: single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat. Biotechnol 35 (7), 640–646. 10.1038/nbt.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL, 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard SR, Mammel MK, Lacher DW, Elkins CA, 2015. Application of metagenomic sequencing to food safety: detection of Shiga Toxin-producing Escherichia coli on fresh bagged spinach. Appl. Environ. Microbiol 81 (23), 8183–8191. 10.1128/AEM.02601-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard SR, Mammel MK, Lacher DW, Elkins CA, 2016. Strain-level discrimination of shiga toxin-producing Escherichia coli in spinach using metagenomic sequencing. PLoS One 11(12), e0167870. doi: 10.1371/journal.pone.0167870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska DW, Zagordi O, Geissberger FD, Kufner V, Schmutz S, Böni J, Metzner KJ, Trkola A, Huber M, 2017. Optimization and validation of sample preparation for metagenomic sequencing of viruses in clinical samples. Microbiome 5 (1), 94 10.1186/s40168-017-0317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R, 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 (14), 1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27 (21), 2987–2993. 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PE, Lo CC, Anderson JJ, Davenport KW, Bishop-Lilly KA, Xu Y, Ahmed S, Feng S, Mokashi VP, Chain PS, 2017. Enabling the democratization of the genomics revolution with a fully integrated web-based bioinformatics platform. Nucleic Acids Res 45 (1), 67–80. 10.1093/nar/gkw1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienau EK, Strain E, Wang C, Zheng J, Ottesen AR, Keys CE, Hammack TS, Musser SM, Brown EW, Allard MW, Cao G, Meng J, Stones R, 2011. Identification of a salmonellosis outbreak by means of molecular sequencing. N. Engl. J. Med 364 (10), 981–982. 10.1056/NEJMc1100443. [DOI] [PubMed] [Google Scholar]
- Liu SP, Yu JX, Wei XL, Ji ZW, Zhou ZL, Meng XY, Mao J, 2016. Sequencing-based screening of functional microorganism to decrease the formation of biogenic amines in Chinese rice wine. Food Contr 64, 98–104. 10.1016/j.foodcont.2015.12.013. [DOI] [Google Scholar]
- Loman NJ, Constantinidou C, Christner M, Rohde H, Chan JZ, Quick J, Weir JC, Quince C, Smith GP, Betley JR, Aepfelbacher M, Pallen MJ, 2013. A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. J. Am. Med. Assoc 309 (14), 1502–1510. 10.1001/jama.2013.3231. [DOI] [PubMed] [Google Scholar]
- Loman NJ, Pallen MJ, 2015. Twenty years of bacterial genome sequencing. Nat. Rev. Microbiol 13(12), 787–794. doi: 10.1038/nrmicro3565. [DOI] [PubMed] [Google Scholar]
- Lo CC, Chain PS, 2014. Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinf 15, 366 10.1186/s12859-014-0366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Knight R, Siljander H, Knip M, Xavier RJ, Gevers D, 2015. ConStrains identifies microbial strains in metagenomic datasets. Nat. Biotechnol 33, 1045–1052. doi: 10.1038/nbt.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk TS, Ottesen AR, White JR, Allard MW, Brown EW, Kase JA, 2012. Characterization of microflora in Latin-style cheeses by next-generation sequencing technology. BMC Microbiol 12, 254 10.1186/1471-2180-12-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND, 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol 11 (10), 728–736. 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder EP, Cieslak PR, Cronquist AB, Dunn J, Lathrop S, Rabatsky-Her T, Ryan P, Smith K, Tobin-D’Angelo M, Vugia DJ, Zansky S, Holt KG, Wolpert BJ, Lynch M, Tauxe R, Geissler AL, 2017. Incidence and trends of infections with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance -foodborne diseases active surveillance network, 10 U.S. Sites, 2013–2016. MMWR Morb. Mortal. Wkly. Rep 66 (15), 397–403. 10.15585/mmwr.mm6615a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars., 2015. IBM Research and Mars, Inc. Launch Pioneering Effort to Drive Advances in Global Food Safety, [Online] Available: http://www.mars.com/nordics/en/press-center/press-list/news-releases.aspx?SiteId=94&Id=6369.
- Masoud W, Vogensen FK, Lillevang S, Abu AW, Sørensen SJ, Jakobsen M, 2012. The fate of indigenous microbiota, starter cultures, Escherichia coli, Listeria innocua and Staphylococcus aureus in Danish raw milk and cheeses determined by pyrosequencing and quantitative real time (qRT)-PCR. Int. J. Food Microbiol 153 (1–2), 192–202. 10.1016/j.ijfoodmicro.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Maury MM, Tsai YH, Charlier C, Touchon M, Chenal-Francisque V, Leclercq A, Criscuolo A, Gaultier C, Roussel S, Brisabois A, Disson O, Rocha EPC, Brisse S, Lecuit M, 2016. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet 48, 308–313. 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo B, Rachid CT, Alegría A, Leite AM, Peixoto RS, Delgado S, 2014. Impact of next generation sequencing techniques in food microbiology. Curr. Genom 15 (4), 293–309. 10.2174/1389202915666140616233211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA, 2008. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinf 9, 386 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Gilad J, Sintchenko V, Pedersen SK, Wolfgang WJ, Pettengill J, Strain E, Hendriksen RS, 2015. Proficiency testing for bacterial whole genome sequencing: an end-user survey of current capabilities, requirements and priorities. BMC Infect. Dis 15, 174 10.1186/s12879-015-0902-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M, 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35, W182–W185. 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, Tarr C, Björkman JT, Dallman T, Reimer A, Enouf V, Larsonneur E, Carleton H, Bracq-Dieye H, Katz LS, Jones L, Touchon M, Tourdjman M, Walker M, Stroika S, Cantinelli T, Chenal-Francisque V, Kucerova Z, Rocha EP, Nadon C, Grant K, Nielsen EM, Pot B, Gerner-Smidt P, Lecuit M, Brisse S, 2016. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol 2, 16185 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadon C, Van Valle I, Gerner-Smidt P, Campos J, Chinen I, Concepcion-Acevedo J, Gilpin B, Smith AM, Man Kam K, Perez E, Trees E, Kubota K, Takkinen J, Nielsen EM, Carleton H, FWD-NEXT Expert Panel, 2017. PulseNet International: vision for the implementation of whole genome sequencing (WGS) for global foodborne disease surveillance. Euro Surveill. 22(23), pii 30544. doi: 10.2807/1560-7917.ES.2017.22.23.30544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanasamy S, Jarosz Y, Muller EE, Heintz-Buschart A, Herold M, Kaysen A, Laczny CC, Pinel N, May P, Wilmes P, 2016. IMP: a pipeline for reproducible reference-independent integrated metagenomic and metatranscriptomic analyses. Genome Biol 17 (1), 260 10.1186/s13059-016-1116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, Vaz C, 2017. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 33 (1), 128–129. 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- Nguyen NP, Warnow T, Pop M, White B, 2016. A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. NPJ Biofilms Microbiomes 2, 16004 10.1038/npjbiofilms.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Yan Q, Yu Y, 2013. How much metagenomic sequencing is enough to achieve a given goal? Sci. Rep. 11;3:1968. doi : 10.0.4.14/srep01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ND, Treangen TJ, Hill CM, Cepeda-Espinoza V, Ghurye J, Koren S, Pop M, 2017. Metagenomic assembly through the lens of validation: recent advances in assessing and improving the quality of genomes assembled from metagenomes. Brief. Bioinform. bbx09 10.1093/bib/bbx098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen A, Ramachandran P, Reed E, White JR, Hasan N, Subramanian P, Ryan G, Jarvis K, Grim C, Daquiqan N, Hanes D, Allard M, Colwell R, Brown E, Chen Y, 2016. Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol 16 (1), 275 10.1186/s12866-016-0894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HV, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V, 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33 (17), 5691–5702. 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Alikhan N-F, Carleton HA, Seemann T, Keane JA, Katz LS, 2017. Comparison of multi-locus sequence typing software for next generation sequencing data. Microb. Genom 10.1099/mgen.0.000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillart MJM, van der Vossen JMBM, Levin E, Lommen E, Otma EC, Snels JCMA, Woltering EJ, 2016. Bacterial population dynamics and sensorial quality loss in modified atmosphere packed fresh-cut iceberg lettuce. Postharvest Biol. Technol 124, 91–99. 10.1016/j.postharvbio.2016.10.008. [DOI] [Google Scholar]
- Panek M, Čipčić Paljetak H, Barešić A, Perić M, Matijašić M, Lojkić I, 2018. Methodology challenges in studying human gut microbiota – effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci. Rep 8 (1), 5143 10.1038/s41598-018-23296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente E, Cocolin L, De Filippis F, Zotta T, Ferrocino I, O’sullivan O, Neviani E, De Angelis M, Cotter PD, Ercolini D, 2016. FoodMicrobionet: a database for the visualisation and exploration of food bacterial communities based on network analysis. Int. J. Food Microbiol 219, 28–37. 10.1016/j.ijfoodmicro.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Parks DH, Mankowski T, Zangooei S, Porter MS, Armanini DG, Baird DJ, Langille MG, Beiko RG, 2013. GenGIS 2: geospatial analysis of traditional and genetic biodiversity, with new gradient algorithms and an extensible plugin framework. PLoS One 8(7), e69885. doi: 10.1371/journal.pone.0069885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW, 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25 (7), 1043–1055. 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pightling AW, Petronella N, Pagotto F, 2015. Choice of reference-guided sequence assembler and SNP caller for analysis of Listeria monocytogenes short-read sequence data greatly influences rates of error. BMC Res. Notes 8 (1), 748 10.1186/s13104-015-1689-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pightling AW, Pettengill JB, Luo Y, Baugher JD, Rand H, Strain E, 2018. Interpreting whole-genome sequence analyses of foodborne bacteria for regulatory applications and outbreak investigations. Front. Microbiol 9, 1482 10.3389/fmicb.2018.01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires SM, Evers EG, van Pelt W, Ayers T, Scallan E, Angulo FJ, Havelaar A, Hald T, Med-Vet-Net Workpackage 28 Working Group, 2009. Attributing the human disease burden of foodborne infections to specific sources. Foodb. Pathog. Dis 6 (4), 417–424. 10.1089/fpd.2008.0208. [DOI] [PubMed] [Google Scholar]
- Ponstingl H, Ning Z, 2010. SMALT – a New Mapper for DNA Sequencing Reads. F1000 Posters
- Portmann AC, Fournier C, Gimonet J, Ngom-Bru C, Barretto C, Baert L, 2018. A validation approach of an end-to-end whole genome sequencing workflow for source tracking of Listeria monocytogenes and Salmonella enterica. Front. Microbiol 14 (9), 446 10.3389/fmicb.2018.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP, 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5(3), e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quainoo S, Coolen JPM, van Hijum SAFT, Huynen MA, Melchers WJG, van Schaik W, Wertheim HFL, 2017. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin. Microbiol. Rev 30, 1015–1063. 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley L, O’Sullivan DJ, Daly D, O’Sullivan O, Burdikova Z, Vana R, Beresford TP, Ross RP, Fitzgerald GF, McSweeney PL, Giblin L, Sheehan JJ, Cotter PD, 2016. Thermus and the pink discoloration defect in cheese. mSystems 1(3), pii e00023–16. doi: 10.1128/mSystems.00023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Bork P, 2008. Molecular eco-systems biology: towards an understanding of community function. Nat. Rev. Microbiol 6 (9), 693–699. 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- Ram RJ, Verberkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC 2nd, Hettich RL, Banfield JF, 2005. Community proteomics of a natural microbial biofilm. Science 308(5730), 1915–1920. doi: 10.1126/science.1109070. [DOI] [PubMed] [Google Scholar]
- Rantsiou K, Kathariou S, Winkler A, Skandamis P, Saint-Cyr MJ, Rouzeau-Szynalski K, Amézquita A, 2017. Next generation microbiological risk assessment: opportunities of whole genome sequencing (WGS) for foodborne pathogen surveillance, source tracking and risk assessment. Int. J. Food Microbiol (in press). doi: 10.1016/j.ijfoodmicro.2017.11.007. [DOI] [PubMed] [Google Scholar]
- Rosen MJ, Callahan BJ, Fisher DS, Holmes SP, 2012. Denoising PCR-amplified metagenome data. BMC Bioinf 13, 283 10.1186/1471-2105-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Ijaz UZ, D’Amore R, Hall N, Sloan WT, Quince C, 2015. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 43(6), e37. doi: 10.1093/nar/gku1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G, Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology, Microbiology Resource Committee of the College of American Pathologists, 2017. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch. Pathol. Lab Med 141 (6), 776–786. 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF, 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol 75 (23), 7537–7541. 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV, 2018. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin. Microbiol. Infect 24 (4), 350–354. 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- Seemann T, 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 (14), 2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Segata N, Boernigen D, Tickle TL, Morgan XC, Garrett WS, Huttenhower C, 2013. Computational meta’omics for microbial community studies. Mol. Syst. Biol 9, 666 10.1038/msb.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekse C, Holst-Jensen A, Dobrindt U, Johannessen GS, Li W, Spilsberg B, Shi J, 2017. High throughput sequencing for detection of foodborne pathogens. Front. Microbiol 8, 2029 10.3389/fmicb.2017.02029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokralla S, Gibson JF, Nikbakht H, Janzen DH, Hallwachs W, Hajibabaei M, 2014. Next-generation DNA barcoding: using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens. Mol. Ecol. Resour 14 (5), 892–901. 10.1111/1755-0998.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegwald L, Touzet H, Lemoine Y, Hot D, Audebert C, Caboche S, 2017. Assessment of common and emerging bioinformatics pipelines for targeted metagenomics. PLoS One 12(1), e0169563. doi: 10.1371/journal.pone.0169563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer E, Andreopoulos B, Bowers RM, Lee J, Deshpande S, Chiniquy J, Ciobanu D, Klenk HP, Zane M, Daum C, Clum A, Cheng JF, Copeland A, Woyke T, 2016. Next generation sequencing data of a defined microbial mock community. Sci. Data 3, 160081 10.1038/sdata.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatko BE, Garner AF, Ausubel FM, 2018. Overview of next-generation sequencing technologies. Curr. Protoc. Mol. Biol 122, e59 10.1002/cpmb.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SJ, Tamminen MV, Preheim SP, Guo MT, Briggs AW, Brito IL, A Weitz, D., Pitkänen LK, Vigneault F, Juhani Virta MP, Alm EJ, 2015. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J 10 (2), 427–436. 10.1038/ismej.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead LF, Sutton KM, Taylor GR, Quirke P, Rabbitts P, 2013. Accurately identifying low-allelic fraction variants in single samples with next-generation sequencing: applications in tumor subclone resolution. Hum. Mutat 34 (10), 1432–1438. 10.1002/humu.22365. [DOI] [PubMed] [Google Scholar]
- Taboada EN, Graham MR, Carriço JA, Van Domselaar G, 2017. Food safety in the age of next generation sequencing, bioinformatics, and open data access. Front. Microbiol 8, 909 10.3389/fmicb.2017.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami H, Taniguchi T, Arai W, Takemoto K, Moriya Y, Goto S, 2016. An automated system for evaluation of the potential functionome: MAPLE version 2.1.0. DNA Res 23 (5), 467–475. 10.1093/dnares/dsw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The National Human Research Institute, 2017. https://www.genome.gov/images/content/costpergenome_2017.jpg.
- Thepault A, Méric G, Rivoal K, Pascoe B, Mageiros L, Touzain F, Rose V, Béven V, Chemaly, Sheppard S 2017. Genome-wide identification of host-segregating epidemiological markers for source attribution in Campylobacter jejuni. Appl. Environ. Microbiol, 83 (7), e03085–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Rand H, Shumway M, Trees EK, Simmons M, Agarwala R, Davis S, Tillman GE, Defibaugh-Chavez S, Carleton HA, Klimke WA, Katz LS, 2017. Benchmark datasets for phylogenomic pipeline validation, applications for foodborne pathogen surveillance. PeerJ 5, e3893. doi: 10.7717/peerj.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Koren S, Sommer DD, Liu B, Astrovskaya I, Ondov B, Darling AE, Phillippy AM, Pop M, 2013. MetAMOS: a modular and open source metagenomic assembly and analysis pipeline. Genome Biol 14 (1), R2 10.1186/gb-2013-14-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay J, Singh K, Fern A, Kirton ES, He S, Woyke T, Lee J, Chen F, Dangl JL, Tringe SG, 2015. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol 6, 771 10.3389/fmicb.2015.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N, 2017. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27 (4), 626–638. 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF, 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428 (6978), 37–43. 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Vaidya JD, van den Bogert B, Edwards JE, Boekhorst J, van Gastelen S, Saccenti E, 2018. The effect of DNA extraction methods on observed microbial communities from fibrous and liquid rumen fractions of dairy cows. Front. Microbiol 92 10.3389/fmicb.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoorde K, Butler F, 2018. Use of next‐generation sequencing in microbial risk assessment. EFSA Journal 16 (S1). 10.2903/j.efsa.2018.e16086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO, 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304 (5667), 66–74. 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Waldor MK, Tyson G, Borenstein E, Ochman H, Moeller A, Finlay BB, Kong HH, Gordon JI, Nelson KE, Dabbagh K, Smith H, 2015. Where next for microbiome research? PLoS Biol. 13(1), e1002050. doi: 10.1371/journal.pbio.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh AM, Crispie F, Claesson MJ, Cotter PD, 2017. Translating omics to food microbiology. Annu. Rev. Food Sci. Technol 8, 113–134. 10.1146/annurev-food-030216-025729. [DOI] [PubMed] [Google Scholar]
- Wang Yu, Pettengill James B., Pightling Arthur, Timme Ruth, Allard Marc, Strain Errol, Rand Hugh, 2018. J. Food Protect 10.4315/0362-028X.JFP-18-093. (in press). [DOI] [PubMed] [Google Scholar]
- Warnecke F, Hugenholtz P, 2007. Building on basic metagenomics with complementary technologies. Genome Biol 8 (12), 231 10.1186/gb-2007-8-12-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer BC, Storey DB, Elkins CA, Baker RC, Markwell P, Chambliss DD, Edlund SB, Kaufman JH, 2016. Defining the food microbiome for authentication, safety, and process management. IBM J. Res. Dev, 60(5–6), 1:1–1:13. doi: 10.1147/JRD.2016.2582598. [DOI] [Google Scholar]
- Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y, Xia LC, Xu ZZ, Ursell L, Alm EJ, Birmingham A, Cram JA, Fuhrman JA, Raes J, Sun F, Zhou J, Knight R, 2016. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J 10 (7), 1669–1681. 10.1038/ismej.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welser J, 2015. Sequ encing the Food Supply Chain: How a New Consortium Will Improve Food Safety Forbes BrandVoice®; [online] Available: http://www.forbes.com/sites/ibm/2015/01/29/sequencing-the-food-supply-chain-how-a-new-consortium-will-improve-food-safety/” \h. [Google Scholar]
- Wilke A, Bischof J, Gerlach W, Glass E, Harrison T, Keegan KP, Paczian T, Trimble WL, Bagchi S, Grama A, Chaterji S, Meyer F, 2016. The MG-RAST metagenomics database and portal in 2015. Nucleic Acids Res 44 (D1), D590–D594. 10.1093/nar/gkv1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MR, Brown E, Keys C, Strain E, Luo Y, Muruvanda T, Grim C, Jean-Gilles Beaubrun J, Jarvis K, Ewing L, Gopinath G, Hanes D, Allard MW, Musser S, 2016. Whole genome DNA sequence analysis of Salmonella subspecies enterica serotype Tennessee obtained from related peanut butter foodborne outbreaks. PloS One 11(6), e0146929. doi: 10.1371/journal.pone.0146929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe BE, Button JE, Santarelli M, Dutton RJ, 2014. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 158, 422–433. 10.1016/j.cell.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres KL, Conway TC, Garg S, Queiroz C, Reumann M, Holt K, Rusu LI, 2014. WGS analysis and interpretation in clinical and public health microbiology laboratories: what are the requirements and how do existing tools compare? Pathogens 3 (2), 437–458. 10.3390/pathogens3020437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia LC, Steele JA, Cram JA, Cardon ZG, Simmons SL, Vallino JJ, Fuhrman JA, Sun F, 2011. Extended local similarity analysis (eLSA) of microbial community and other time series data with replicates. BMC Syst. Biol 5 (Suppl. 2), S15 10.1186/1752-0509-5-S2-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara K, Méric G, Taylor A, de Vries SPW, Murray S, Pascoe B, Mageiros L, Torralbo A, Vidal A, Ridley A, Komukai S, McCarthy N, Harris D, Bray JE, Jolley KA, Maiden M, Bentley SD, Parkhill J, Bayliss CD, Grant A, Maskell D, Didelot X, Kelly DJ, Sheppard SK, 2017. Genome-wide association of functional traits linked with Campylobacter jejuni survival from farm to fork. Environ. Microbiol 19 (1), 361–380. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B, 2012. Molecular phylogenetics: principles and practice. Nat. Rev. Genet 13, 303–314. 10.1038/nrg3186. [DOI] [PubMed] [Google Scholar]
- Yang X, Noyes NR, Doster E, Martin JN, Linke LM, Magnuson RJ, Yang H, Geornaras I, Woerner DR, Jones KL, Ruiz J, Boucher C, Morley PS, Belk KE, 2016. Use of metagenomic shotgun sequencing technology to detect foodborne pathogens within the microbiome of the beef production chain. Appl. Environ. Microbiol 82, 2433–2443. 10.1128/AEM.00078-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ, 2012. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One 7(3), e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV, 2012. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother 67 (11), 2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S, Bokulich NA,, Mills DA, Martin G, Taghavi S, van der Lelie D, Gilbert JA, 2015. The soil microbiome influences grapevine-associated microbiota. mBio 6(2), pii e02527–14. doi: 10.1128/mBio.02527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E, 2008. Velvet: algorithms for the novo short read assembly using the Bruijn graphs. Genome Res 18 (5), 821–829. 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Caspers MPM, Metselaar KI, de Boer P, Roeselers G, Moezelaar R, Nierop Groot M, Montijn RC, Abee T, Kort R, 2013. Abiotic and microbiotic factors controlling biofilm formation by thermophilic sporeformers. Appl. Environ. Microbiol 79 (18), 5652–5660. 10.1128/AEM.00949-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfo M, Tett A, Jousson O, Donati C, Segata N, 2017. MetaMLST: multi-locus strain-level bacterial typing from metagenomic samples. Nucleic Acids Res. 45(2), e7. doi: 10.1093/nar/gkw837. [DOI] [PMC free article] [PubMed] [Google Scholar]