Abstract
The antioxidant (AOX) defense system is critical for combating whole-body oxidative stress, and the present study aimed to determine the consequences of a maternal high-fat (HF) diet on neonatal hepatic lipid accumulation, oxidative stress, the expression of AOX genes, as well as epigenetic histone modifications within Pon1, an AOX enzyme. Hepatic thiobarbituric acid reactive substances were significantly increased and nonesterified fatty acids decreased in offspring of HF-fed dams, while triglycerides increased in male but not female HF offspring when compared to controls (C). Pon1, Pon2, Pon3 and Sod2 were significantly increased in offspring of HF-fed dams when compared to C. However, the increase in Pon1 and Pon3 was only significant in male but not female offspring. When compared to C, the hepatic Pon1 promoter of male and female HF offspring had significantly more acetylated histone H4 as well as dimethylated histone H3 at lysine residue 4, which are both involved in transcriptional activation. Trimethylation of histone H3 at lysine residue 9, which is involved in transcriptional repression, was only associated with genes in females. Results from the present study reveal that a maternal HF diet affects hepatic metabolism in the neonate in a gender-specific manner, and these differences, in association with epigenetic modification of histones, may contribute to the known gender differences in oxidative balance.
Keywords: Developmental programming, Epigenetics, Oxidative stress, In uterine
1. Introduction
The intake of a high-fat (HF) diet during pregnancy impacts fetal development[1–3], and specifically, a recent study has demonstrated that markers of antioxidant (AOX) defense capacity were decreased in adult offspring of HF-fed dams [4]. Because the liver is infiltrated with byproducts of fatty acid oxidation and because altered handling of hepatic oxidation products is associated with liver damage and disease [5], the inappropriate programming of this crucial protective mechanism by a maternal HF diet may be deleterious for hepatic development and health. Additionally, because hepatic oxidative balance is often related to systemic antioxidant status [6–8], the altered production of these in the liver will likely have numerous consequences for all major organ systems.
The paraoxonase (PON) family of enzymes is critical for whole-system oxidative balance [9]. The liver is responsible for the synthesis of these enzymes that protect both itself as well as the rest of the body from prooxidant damage. Specifically, the PON1 enzyme, which has lactonase and esterase activities and is synthesized in the liver, has been shown to be a critical component of the AOX defense system, and its synthesis was altered in patients with hepatosteatosis [10]. Therefore, it is not surprising that modulations in PON1 enzyme activity in liver as well as serum have been correlated to liver disease and altered lipoprotein metabolism [11–14]. Furthermore, studies have suggested that gender differences in the response to oxidative stress may contribute to the differential susceptibility of males and females to developing chronic diseases [15–20], and the AOX defense system is likely responsible for these differences.
While the adult epigenome can be modified by various elements and toxins in the environment [21], studies have also shown that in utero exposures can result in significant epigenetic modifications, and these epigenetic marks can persist into adulthood to permanently control gene expression [22–24]. The covalent modification of histone tails – including acetylation (Ac) or methylation (Me) – represents a class of epigenetic changes that regulate transcriptional activity of genes by altering the state of the chromatin [25]. In primates, the chronic consumption of a maternal HF diet led to an increase in H3K14Ac and a trend of increase in H3K9Ac, H3K18Ac, H3K9Me2, trimethylated histone H3 at lysine residue 9 (H3K9Me3) and H3K27Me3, and these fetal hepatic tissues had an increase in triglycerides and nonalcoholic fatty liver disease [26]. Furthermore, a maternal HF diet in rats resulted in decreased association of acetylated histone H3 (H3Ac), dimethylated histone H3 at lysine residue 4 (H3K4Me2), H3K9Me3 and H3K27Me3 within the promoter of the hepatic gluconeogenic Pck1 gene, along with a decrease in H3K9Me3 and an increase in acetylated histone H4 (H4Ac) and H3K4Me2, and all of these modifications were associated with increased transcription and transcriptional rate of the Pck1 gene [27]. While studies have authenticated the importance of epigenetics in controlling the expression of AOX genes [28–30], few data are available showing the effect of gestational diet on this system.
A maternal HF diet in nonhuman primates was shown to increase hepatic oxidative stress in the offspring [31], and because the PON1 enzyme is synthesized in the liver and provides protection against system-wide oxidative damage [32], the current study aimed to determine its transcription, as well as the transcription of several AOX genes, in livers of both male and female offspring in response to a maternal HF diet. Furthermore, we wanted to investigate the fetal hepatic epigenome by focusing on Pon1 as a model for gender-specific histone modifications that occur in response to a maternal HF diet. In order to accomplish this, the present study utilized an obesity-resistant (OR) rat model without the concurrent development of gestational obesity. Our results demonstrate that the in utero exposure to an HF diet affects fetal hepatic oxidative balance and has a gender-specific effect on the expression of AOX genes in association with epigenetic modifications within the tested gene.
2. Methods
2.1. Animal and experimental design
Timed-pregnant female rats were obtained from Charles River (Wilmington, MA, USA) on embryonic day 2. The OR strain [Crl:OR(CD)] in the current experiment was developed from an outbred line of Crl:CD(SD) rats. This model does not become obese when fed HF diets. Dams were separated into two dietary treatments: five control (C; 64% carbohydrates, 20% protein, 16% fat), and five high fat (HF; 35% carbohydrates, 20% protein, 45% fat), where the HF diet had approximately 3.3, 3.17 and 1.8 times more saturated, monounsaturated and polyunsaturated fat, respectively, per kilocalorie consumed when compared to the C diet [27]. Pregnant animals were fed these diets ad libitum until embryonic day 20, when they were fasted overnight and underwent cesarean delivery to collect livers from offspring. All fetal liver tissue samples were immediately stored in liquid nitrogen and kept for further analysis. Fetal weights and gender were also recorded at this time.
2.2. Neonatal hepatic triglyceride (TG), nonesterified fatty acid (NEFA) and thiobarbituric acid reactive substances (TBARS) analysis
Frozen liver samples (100 mg) were ground using a mortar and pestle with liquid nitrogen and mixed with 0.3 ml saline (0.9% w/v NaCl) as previously reported [33]. Homogenized samples were quickly frozen in liquid nitrogen and kept in −70 °C. The samples were quickly thawed at 37 °C and diluted five times in saline to 1.5 ml. Twenty microliters of the diluted samples was incubated with 20 μl 1% deoxycholate at 37 °C for 5 min, and 10 μl of the samples was used to analyze either liver TG or NEFA. Hepatic TG was analyzed via the Thermo Infinity Triglycerides Liquid Stable Reagent (Thermo Fisher Scientific, Rockford, IL, USA) following company protocol and using a commercially available standard reference kit (Verichem Laboratories, Providence, RI). Hepatic NEFA concentration was determined using a commercially available kit (HR-2 Series, Wako Diagnostics, Richmond, VA, USA). TBARS assay (catalog no. 10009055, Cayman Chemical, Ann Arbor, MI, USA) was performed per company protocol to determine the concentration of malondialdehyde (MDA), a product of lipid peroxidation, within neonatal hepatic tissues. TG, NEFA and TBARS data are presented as the amount of TG, NEFA or MDA (in milligrams) per gram of liver.
2.3. RNA isolation and real-time polymerase chain reaction (RT-PCR) analysis
Livers from 10 offspring (5 male and 5 female) from each dietary group were randomly chosen for sampling, with all litters being represented. Total RNA was isolated using the GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich, St Louis, MO, USA), treated with DNase I to eliminate any DNA contamination and quantified using Nano Drop ND-1000 Spectophotometer while ensuring that the ratio of 260/280 was >1.9. cDNA was synthesized from 2 μg of RNA in a 20-μl reaction volume using the High Capacity cDNA Reverse Transcription Kit with random primers (Applied Biosystems, Foster City, CA, USA) and a thermal cycler (Applied Biosystems 2700, Foster City, CA, USA), with the following program: 25 °C for 10 min, 37 °C for 120 min, 85 °C for 5 s and a 4 °C hold. RT-PCR was performed using 25 ng cDNA as the template, SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), and 5 μmol/L of each forward and reverse primer (Table 1) in the 7300 Real-Time PCR System (Applied Biosystem, Foster City, CA, USA), with the following program: 95 °C for 10 min, 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s, 55 °C for 1 min and 95 °C for 15 s, with 40 cycles of steps 2 and 3. A serial dilution was used to create a standard curve for quantification, a dissociation curve was analyzed following each reaction, and a no-template control was included with all reactions to ensure that no additional products were synthesized during the PCR reaction. mRNA level of ribosomal protein L7a was utilized as the internal control.
Table 1.
Primer information
| Gene | Forward sequence and location | Reverse sequence and location | Transcript ID |
|---|---|---|---|
| Pon1 | |||
| mRNA & coding ChIP | AGTGAGGCCATCATTTCAGCC (+1149) | ATTCGTTGGTGAGCGGAGATC (+1221) | ENSRNOT00000011823 |
| Promoter ChIP | AGTTGTCAGAGCCACCAG (−35) | AGGATGGACAAGTGGAGG (+28) | ENSRNOT00000011823 |
| Pon2 | TAC GCC ACC AAT GAC CAC T (+584) | TGC CCA ACG TAG GTT CAA G (+658) | ENSRNOG00000009112 |
| Pon3 | TTTGCACCAGACAAGCCAG (+243) | TCCAGTGCTTGTGCCTTTG (+316) | ENSRNOG00000009096 |
| Acox1 | GCCTTTGTTGTCCCTATCCGT (+433) | ACCGATATCCCCGACAGTGAT (+504) | ENSRNOT00000042372 |
| Sod1 | CAGCGGATGAAGAGAGGCA (+239) | ACACATTGGCCACACCGTC (+310) | ENSRNOG00000002115 |
| Sod2 | GTTTGCAAGAAGTGAAGC (+738) | ACTACAAAACACCCACCA (+801) | ENSRNOG00000019048 |
| L7a | |||
| mRNA & Promoter ChIP | GAGGCCAAAAAGGTGGTCAATCC (+64) | CCTGCCCAATGCCGAAGTTCT (+127) | ENSRNOT00000006754 |
2.4. Chromatin immunoprecipitation (ChIP) analysis
The ChIP assay was previously reported in great detail and is therefore not described here [27]. As previously stated, livers from 10 offspring (5 male and 5 female) from each dietary group were randomly chosen for sampling and were the same samples as those used for mRNA analysis. Antibody information is listed in Table 2.
Table 2.
ChIP antibody information
| Antibody | Species | Source | Catalog number |
|---|---|---|---|
| IgG | Rabbit | Santa Cruz | sc-2027 |
| H3Ac | Rabbit | Millipore | 06-599 |
| H4Ac | Rabbit | Upstate | 06-866 |
| H3K4Me2 | Rabbit | Millipore | 07-030 |
| H3K9Me3 | Rabbit | Upstate | 07-442 |
2.5. Statistical analysis
Results are reported as means±S.E.M. Neonatal liver TG, TBARS and gene expression data were analyzed using two-way analysis of variance (ANOVA) with interactions (SAS software, Cary, NC, USA) with diet and gender as main effects. When an interaction was found between factors (P<.05), the post hoc Tukey test was used to identify specific differences within diet or within gender. For hepatic ChIP analysis in offspring (n=10, 5 male and 5 female), each gender was analyzed individually to determine the effect of diet on each antibody using Student's t test, while the effect of antibody between IgG and H3K9Me3 was determined using two-way ANOVA with antibody and gender as main effects.
2.6. Statement of ethics
We certify that all applicable institutional and governmental regulations regarding the ethical use of animals were followed during this research (University of Illinois Institutional Animal Care and Use Committee approval #09112).
3. Results
3.1. Maternal gestational characteristics
The following has been previously reported [27]; therefore, the data are not shown here. Briefly, gestational food intake and body weight did not differ between C and HF dams. Dams fed the HF diet gained significantly more weight than C, possibly because the offspring of these dams were heavier. However, when fetal and placental weights were subtracted from this weight to determine actual mass gained during gestation, the two groups did not differ.
3.2. Offspring observations
As previously reported [27], there were no differences in litter size between the two dietary groups, and at the time of cesarean delivery on gestational day 21, the offspring of dams fed the HF diet were significantly heavier when compared to the offspring of dams that were fed the C diet during gestation. At birth, the offspring of HF-fed dams had significantly higher (P<.05) hepatic TBARS content when compared to the offspring of C dams, and there was no effect of gender or an interaction between diet and gender (Table 3). There was no effect of diet or gender on hepatic TG content in offspring at birth; however, there was a significant interaction between the factors (P<.05), which was due to the fact that while male offspring had a significant (P<.05) increase in hepatic TG in response to a maternal HF diet, female offspring did not respond to diet (Table 3). Hepatic NEFA content was significantly lower (P<.05) in offspring of HF-fed dams when compared to C, and there was no effect of gender or interaction between diet and gender (Table 3). Acox1 mRNA content was measured as a marker of fatty acid oxidation and was significantly higher in female than in male offspring (P<.05), and there was a significant interaction between diet and gender, which, similarly to hepatic TG content, was due to the fact that while male offspring had a significant (P<.05) increase in hepatic Acox1 in response to a maternal HF diet, female offspring did not respond to diet (Table 3).
Table 3.
Neonatal hepatic components
| Variable | Diet and gender |
P value |
|||||
|---|---|---|---|---|---|---|---|
| C |
HF |
D | G | D×G | |||
| Male | Female | Male | Female | ||||
| TBARS (per g tissue) | 0.20±0.02 | 0.19±0.03 | 0.31±0.04 | 0.28±0.05 | <.05 | NS | NS |
| TG (mg per g liver) | 0.61±0.05 | 0.96±0.13 | 1.22±0.23 * | 0.82±0.07 | NS | NS | <.05 |
| NEFA (mg per g liver) | 0.47±0.04 | 0.65±0.12 | 0.34±0.03 | 0.36±0.05 | <.05 | NS | NS |
| Acox1 (mRNA to L7a) | 0.90±0.06 | 2.54±0.32# | 1.62±0.10 * | 1.97±0.31 | NS | <.05 | <.05 |
P<.05 comparing the within-gender (G) effect of diet (D).
P<.05 comparing the within-diet (D) effect of gender (G).
3.3. Gene expression
Hepatic Pon1 mRNA content was significantly higher (P<.005) in offspring of HF-fed dams when compared to C offspring, and there was a significant (P<.05) interaction between the factors, which was due to the fact that male HF offspring had significantly (P<.005) higher Pon1 expression than C offspring, while females did not respond to diet. Additionally, HF female offspring had significantly (P<.05) lower Pon1 expression than HF males (Table 4). Offspring of HF-fed dams had significantly higher (P<.05) Pon2 mRNA content, with no effect of gender and no interaction between diet and gender (Table 4). Similar to Pon1 expression, hepatic Pon3 mRNA content was significantly higher (P<.005) in offspring of HF-fed dams when compared to C offspring, and there was a significant (P<.05) interaction between the factors, which was due to the fact that male HF offspring had significantly (P<.005) higher Pon3 expression than C offspring, while females did not respond to diet. Additionally, C female offspring had significantly (P<.05) higher Pon3 expression than C males (Table 4). There was no effect of diet or gender, or an interaction between the two factors on hepatic Sod1 expression (Table 4). However, Sod2 mRNA was significantly (P<.05) higher in female than male offspring and significantly (P<.005) higher in offspring of HF-fed dams when compared to C offspring (Table 4).
Table 4.
Neonatal hepatic antioxidant gene expression (normalized to L7a)
| mRNA (to L7a) |
Diet and gender |
P value |
|||||
|---|---|---|---|---|---|---|---|
| C |
HF |
D | G | D×G | |||
| Male | Female | Male | Female | ||||
| Pon1 | 3.06±0.29 | 3.83±0.76 | 6.89±0.52 * | 4.91±0.45# | <.005 | NS | <.05 |
| Pon2 | 1.01±0.12 | 0.98±0.19 | 1.35±0.10 | 1.52±0.09 | <.05 | NS | NS |
| Pon3 | 1.57±0.04 | 2.07±0.18# | 2.46±0.09 * | 2.25±0.06 | <.005 | NS | <.05 |
| Sod1 | 1.12±0.06 | 1.18±0.09 | 1.07±0.20 | 1.24±0.14 | NS | NS | NS |
| Sod2 | 0.48±0.06 | 0.72±0.09# | 0.82±0.03 * | 0.94±0.07 | <.005 | <.05 | NS |
P<.005 comparing the within-gender (G) effect of diet (D).
P<.05 comparing the within-diet (D) effect of gender (G).
3.4. Histone modifications related to transcription activation
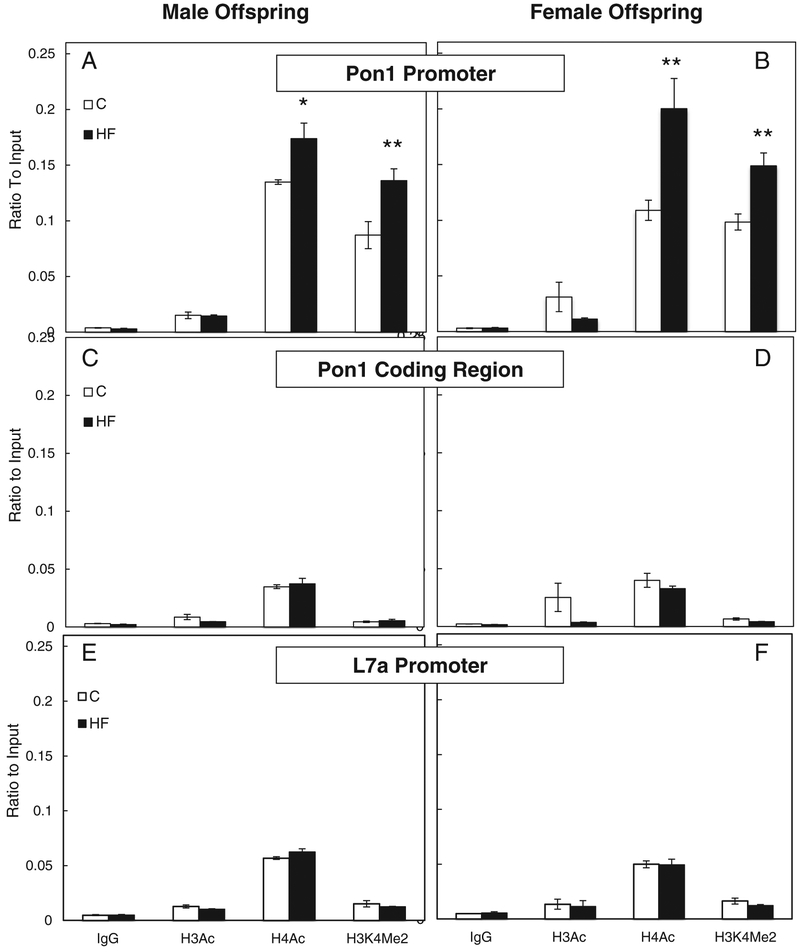
The ChIP assay was performed to investigate the association of modified histones to Pon1 gene expression. Normal rabbit IgG antibody was used as the negative control, indicating nonspecific binding of protein–DNA. Proteins were considered negative for binding if the resulting value was equal to or less than the IgG value. At the Pon1 promoter, H3Ac was not significantly different between C and HF offspring in males (Fig. 1A) but slightly decreased in females (P=.11, Fig. 1B). At the coding region, H3Ac was also not significantly different between C and HF offspring in males (Fig. 1C) but slightly decreased in females (P=.11, Fig. 1D). At the promoter, H4Ac was significantly higher in both male (P<.05, Fig. 1A) and female (P<.01, Fig. 1B) HF offspring when compared to C offspring; whereas there was no significant change in H4Ac within the Pon1 coding region in either males (Fig. 1C) or females (Fig. 1D). At the promoter, H3K4Me2 was also significantly higher in HF male (P<.01, Fig. 1A) and female (P<.01, Fig. 1B) offspring when compared to C, and there was no significant change in H3K4Me2 within the Pon1 coding region in either male (Fig. 1C) or female (Fig. 1D) offspring. The L7a ribosomal protein gene was used as a control to normalize all mRNA data because its expression was not affected by either diet or gender, and histone modifications within the L7a promoter are also presented to demonstrate any histone modifications occurring within the promoter of a gene whose expression was not affected by diet or gender. No histone modifications corresponding to transcription activation were observed in response to diet within the L7a promoter in male (Fig. 1E) or female (Fig. 1F) offspring.
Fig. 1.
Gender-specific hepatic Pon1 gene-activating histone modifications. Histone modifications within the hepatic Pon1 promoter region [male (A) and female (B)] and coding region [male (C) and female (D)], as well as the L7a promoter region [male (E) and female (F)] in offspring of dams fed the C or HF diet during gestation. Values are means±S.E.M., n=10. *Different from C, P<.05 within each gender and modification. **Different from C, P<.01 within each gender and modification.
3.5. Histone modifications related to transcription repression
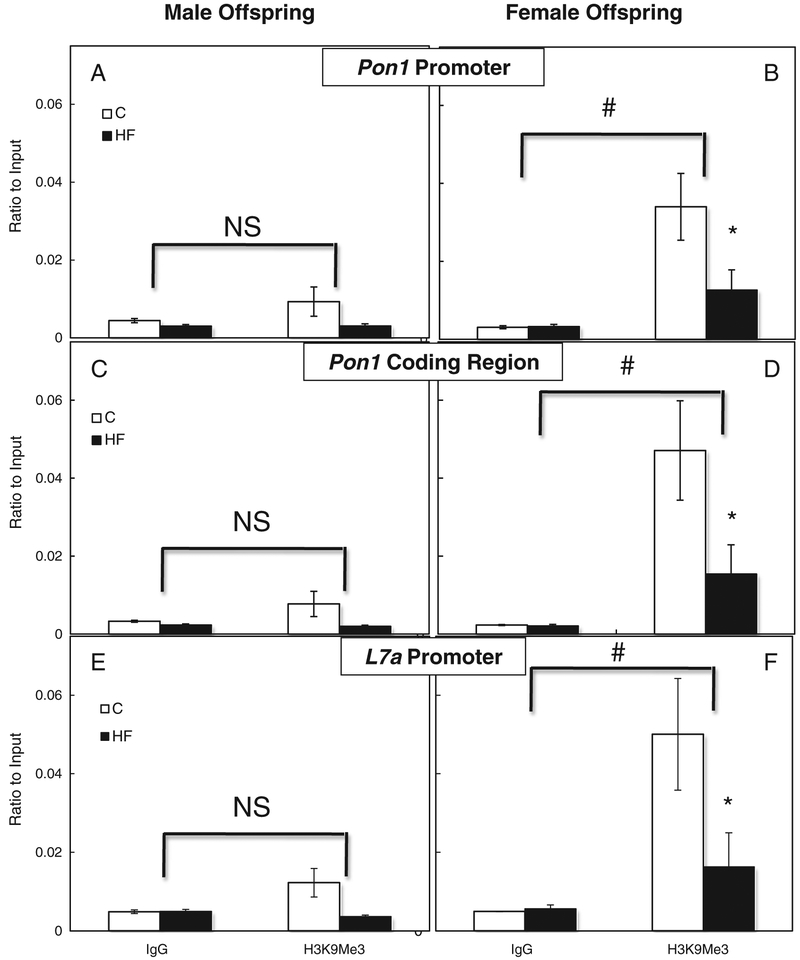
Methylation of lysine 9 of histone H3 (H3K9Me3) is a marker of condensed, inactive chromatin. There was no significant difference between H3K9Me3 and IgG in male offspring at the promoter (Fig. 2A) and coding region (Fig. 2C) of the Pon1 gene and at the promoter (Fig. 2E) of the L7a gene. This indicates that lysine 9 on histone H3 was not methylated in male offspring. However, in female offspring, the differences between H3K9Me3 and IgG at the promoter (Fig. 2B) and coding region (Fig. 2D) of the Pon1 gene and at the promoter (Fig. 2F) of the L7a gene were all significant (P<.05). This indicates that histone H3 was modified by methylation of lysine 9 in female offspring to induce a more inactive chromatin state. Although H3K9Me3 was significantly reduced in livers of HF female offspring when compared to C in a non-gene-specific manner (Fig. 2B, D, F), it was still significantly higher than nonspecific IgG binding, indicating a generally condensed chromatin state in females.
Fig. 2.
Gender-specific hepatic Pon1 H3K9Me3 association. Comparison between IgG and H3K9Me3 binding as well as diet effect within the hepatic Pon1 promoter region [male (A) and female (B)] and coding region [male (C) and female (D)] as well as the L7a promoter region [male (E) and female (F)] in offspring of dams fed the C or HF diet during gestation. Values are means±S.E.M., n=10 (5 male and 5 female). #Different from IgG within each gender, P<.01. *Different from C within each gender and modification, P<.05.
4. Discussion
Maternal nutrition is a key regulator of fetal growth and development, and our study is the first, to our knowledge, to suggest that a maternal HF diet, independent of gestational obesity, increases fetal hepatic oxidative stress, and the AOX system that responds to this stimulus appears to be regulated epigenetically. In rodents, these phenotypic gender differences have been shown to also exist in adulthood [34,35], and our findings provide the first potential mechanistic explanation for these differences. It is likely that the early hit of oxidative stress and the premature activation of the AOX system may influence the efficacy of this system in the future, thereby regulating the adult offspring's response to stress events.
Previous reports in rats have suggested that males may be more susceptible to the deleterious effects of a maternal HF diet, which include their tendency to have increased oxidative stress when compared with females [36]. While both males and females in the present study had increased hepatic TBARS, a marker of lipid peroxidation, the differential genetic response may be an indicator of innate gender differences in the mechanisms that govern both the induction and response to oxidative stress. Both male and female offspring had decreased hepatic NEFA content, which may result from an increase in hepatic fatty acid oxidation. We previously reported that gluconeogenesis was increased in livers of these neonates [27], which agrees with the current data, since an increase in gluconeogenesis and fatty acid oxidation typically occur concurrently. However, while fatty acids were decreased in both genders in response to a maternal HF diet, Acox1, which encodes the enzyme responsible for the regulatory step of palmitoyl fatty acid oxidation, was approximately twofold increased in male but not female offspring in response to maternal HF feeding. In males, hepatic TGs were also significantly increased, but this was not observed in females. An increase in TG synthesis occurs in response to excess NEFA availability, suggesting that males accumulate and potentially uptake more lipids in response to a maternal HF diet when compared to their basal state than do females, which may also explain the increase of Acox1 expression as an attempt to deal with this excess. Therefore, it appears that the differential regulation of the AOX system occurs in direct response to the inherent differences in lipid metabolism between males and females exposed to a maternal HF diet.
The PON1 enzyme is synthesized in the liver and acts as a potent antioxidant in circulation, especially in atherosclerotic plaques, where lipid peroxidation is increased. Polymorphisms in the human PON1 gene are associated with an increase in various diseases, including diabetes, heart disease and hyperlipidemia [37–40]. Gender differences in the activity of the PON enzymes in response to HF feeding have been previously demonstrated. A study in adult animals fed an HF (cafeteria diet, 64% fat) diet showed that males and females had gender-specific responses in PON1 serum activity, with no response in the male and a significant decrease by HF feeding in females, bringing the level of PON1 activity to that of males [41]. However, there was no effect of the HF diet on the expression of Pon1 in the liver in either males or females. In adults, PON1's AOX function is most often associated with HDL in circulation where it is thought to contribute to the prevention of atherosclerosis development [42]. However, in the neonate, circulating HDL has been shown to be quite low [43], suggesting that the increase in hepatic Pon1 gene expression in the current study was likely a local response to oxidative stress and an appropriate marker of enzyme activity. In addition to Pon1, we observed that the expression of the other two PONs (Pon2 and Pon3) as well as Sod2 (MnSOD) was also elevated in response to a maternal HF diet, but only Pon3 followed Pon1's expression pattern – being increased in males but not females in response to a maternal HF diet.
Nonalcoholic fatty liver disease (NAFLD) and cirrhosis are typically accompanied by increased hepatic oxidative stress [44], and NAFLD has been associated with decreased AOX capacity and a decrease in the expression of AOX genes [45]. Our previous study showed that adult offspring of HF-fed mothers had decreased hepatic AOX gene expression, which is generally though to signal decreased antioxidant defense capacity [4]. These differences were observed in offspring consuming a control postweaning diet, suggesting that the in utero environment has the potential to program adult phenotypes, regardless of the postnatal diet. Because the current study utilized OR rats to isolate the effects of fat intake from those of maternal adiposity typically observed in animal models of diet-induced obesity, it is possible that this current study and our previous study using Sprague–Dawley rats actually model different pathologies. However, it is also possible that the overexpression of the AOX system early on increases the basal expression of these genes and disrupts the feedback mechanisms that regulate their transcription in adulthood. Further studies will be critical for determining the exact role that maternal diet has in the oxidative balance in offspring and whether altered PON expression in livers of neonates corresponds to its decreased activity in the plasma of adult animals.
Pon1 was used in the present study as a model for studying histone modifications in a gene that had a gender-specific pattern of transcription in response to a maternal HF diet. Additionally, unlike the other genes studied, the PON1 enzyme is a known extrahepatic AOX, so its expression in liver impacts systemic AOX balance. As previously discussed, the essential role of PON1 during AOX defense has been confirmed; the regulation of the PON1 gene remains poorly understood. Our histone modification analysis of the Pon1 promoter and coding region presents a novel mechanism for the in utero activation of this gene by a maternal HF diet. Both the male and female promoters showed a histone code that correlates to the active transcription of genes. The acetylation of histone H4 and the dimethylation of histone H3 at lysine residue 4 are associated with active transcription, and these were both significantly increased by a maternal HF diet in the Pon1 promoter of male and female offspring. However, the Pon1 gene was only significantly increased by a maternal HF diet in male livers but not female, which may be due to the presence of H3K9Me3, a known potent inhibitor of transcription, within the female but not male promoter as well as coding regions. We observed a similar pattern of H3K9Me3 modification within the L7a promoter, indicating a female-specific modification. However, unlike the promoter and coding regions of Pon1, there were no transcription activation-associated modifications present within L7a in response to diet. These complex histone interactions and patterns are likely responsible for differences in the expression of these genes [46]. Future in-depth analyses of gene-wide histone modifications, as well as studies of other epigenetic mechanisms, will be essential for determining the precise regulation of Pon1, as well as of other AOX genes in response to maternal HF diet.
In the current study, we focused on the epigenetic regulation of the Pon1 gene to demonstrate the consequence of a maternal HF diet for the histone code of a critical AOX gene. However, we observed that other AOX genes were also increased in the livers of offspring in response to a maternal HF diet, so it is reasonable to presume that these genes are also regulated at the epigenetic level in response to the in utero environment, and in some of these genes, the changes may be gender specific. More studies are needed to fully unravel the role of epigenetics in the regulation of the AOX system, but our preliminary results suggest that the in utero environment is a potent regulator of AOX genes, which can potentially result in the dysregulation of the AOX system in adult offspring. These outcomes could result in altered handling of prooxidants in adult offspring whose mothers consumed an HF diet and could potentially explain the systemic differences often observed between the male and female response to oxidative stress.
Acknowledgments
This project was supported by the USDA Cooperative State Research, Education and Extension Service, Hatch project number # ILLU-698-374 and ILLU-698-324W1005, and an NIEHS training grant (T32ES007326).
Abbreviations:
- H3Ac
acetylated histone H3
- H4Ac
acetylated histone H4
- AOX
antioxidant
- H3K4Me2
dimethylation of histone H3 at lysine residue 4
- H3K9Me3
trimethylation of histone H3 at lysine residue 9
- HF
high fat
- PON
paraoxonase
- SOD
superoxide dismutase
References
- [1].Aaltonen J, Ojala T, Laitinen K, Poussa T, Ozanne S, Isolauri E. Impact of maternal diet during pregnancy and breastfeeding on infant metabolic programming: a prospective randomized controlled study. Eur J Clin Nutr 2011;65:10–9. [DOI] [PubMed] [Google Scholar]
- [2].Bayol SA, Simbi BH, Stickland NC. A maternal cafeteria diet during gestation and lactation promotes adiposity and impairs skeletal muscle development and metabolism in rat offspring at weaning. J Physiol 2005;567:951–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bruce KD, Cagampang FR, Argenton M, Zhang J, Ethirajan PL, Burdge GC, et al. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology 2009;50:1796–808. [DOI] [PubMed] [Google Scholar]
- [4].Zhang X, Strakovsky RS, Zhou D, Zhang Y, Pan YX. A maternal high-fat diet represses the expression of antioxidant defense genes and induces the cellular senescence pathway in the liver of male offspring rats. J Nutr 2011;141:1254–9. [DOI] [PubMed] [Google Scholar]
- [5].Jaeschke H Reactive oxygen and mechanisms of inflammatory liver injury: present concepts. J Gastroenterol Hepatol 2011;26(Suppl 1):173–9. [DOI] [PubMed] [Google Scholar]
- [6].Franco R, Schoneveld OJ, Pappa A, Panayiotidis MI. The central role of glutathione in the pathophysiology of human diseases. Arch Physiol Biochem 2007;113:234–58. [DOI] [PubMed] [Google Scholar]
- [7].Marra M, Sordelli IM, Lombardi A, Lamberti M, Tarantino L, Giudice A, et al. Molecular targets and oxidative stress biomarkers in hepatocellular carcinoma: an overview. J Transl Med 2011;9:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shen H, Li M, Wang B, Lai IK, Robertson LW, Ludewig G. Dietary antioxidants (selenium and N-acetylcysteine) modulate paraoxonase 1 (PON1) in PCB 126-exposed rats. Environ Sci Pollut Res Int 2013. [Epub ahead of print; http://dx.doi.org.10.1007/S11356-013-1690-1]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Precourt LP, Amre D, Denis MC, Lavoie JC, Delvin E, Seidman E, et al. The three-gene paraoxonase family: physiologic roles, actions and regulation. Atherosclerosis 2011;214:20–36. [DOI] [PubMed] [Google Scholar]
- [10].Atamer A, Bilici A, Yenice N, Selek S, Ilhan N, Atamer Y. The importance of paraoxonase 1 activity, nitric oxide and lipid peroxidation in hepatosteatosis. J Int Med Res 2008;36:771–6. [DOI] [PubMed] [Google Scholar]
- [11].Prakash M, Shetty JK, Tripathy S, Verma M, Vasudev S, Bhandary PV. Serum paraoxonase in alcohol abusers associated with alcoholic liver disease. Clin Chim Acta 2007;378:232–4. [DOI] [PubMed] [Google Scholar]
- [12].Kumon Y, Nakauchi Y, Suehiro T, Shiinoki T, Tanimoto N, Inoue M, et al. Proinflammatory cytokines but not acute phase serum amyloid A or C-reactive protein, downregulate paraoxonase 1 (PON1) expression by HepG2 cells. Amyloid 2002;9:160–4. [DOI] [PubMed] [Google Scholar]
- [13].Feingold KR, Memon RA, Moser AH, Grunfeld C. Paraoxonase activity in the serum and hepatic mRNA levels decrease during the acute phase response. Atherosclerosis 1998;139:307–15. [DOI] [PubMed] [Google Scholar]
- [14].Aviram M, Rosenblat M, Bisgaier CL, Newton RS, Primo-Parmo SL, La Du BN. Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions. A possible peroxidative role for paraoxonase. J Clin Invest 1998;101: 1581–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bahado-Singh RO, Schenone M, Cordoba M, Shieh WS, Maulik D, Kruger M, et al. Male gender significantly increases risk of oxidative stress related congenital anomalies in the non-diabetic population. J Matern Fetal Neonatal Med 2011;24:687–91. [DOI] [PubMed] [Google Scholar]
- [16].Hirakawa Y, Masuda Y, Kuzuya M, Iguchi A, Kimata T, Uemura K. Impact of gender on in-hospital mortality of patients with acute myocardial infarction undergoing percutaneous coronary intervention: an evaluation of the TAMIS-II data. Intern Med 2007;46:363–6. [DOI] [PubMed] [Google Scholar]
- [17].Legato MJ. Gender-specific medicine: the view from Salzburg. Gend Med 2004;1: 61–3. [DOI] [PubMed] [Google Scholar]
- [18].Liang Q, Sheng Y,Jiang P,Ji L, Xia Y, Min Y, et al. The gender-dependent difference of liver GSH antioxidant system in mice and its influence on isoline-induced liver injury. Toxicology 2011;280:61–9. [DOI] [PubMed] [Google Scholar]
- [19].Malorni W, Straface E, Matarrese P, Ascione B, Coinu R, Canu S, et al. Redox state and gender differences in vascular smooth muscle cells. FEBS Lett 2008;582:635–42. [DOI] [PubMed] [Google Scholar]
- [20].Golab F, Kadkhodaee M, Xu J, Soleimani M. Male susceptibility to hepatic damage in acute uremia in rats. Urology 2011;78:232.e1–232.e6. [DOI] [PubMed] [Google Scholar]
- [21].Vandegehuchte MB, Janssen CR. Epigenetics and its implications for ecotoxicology. Ecotoxicology 2011;20:607–24. [DOI] [PubMed] [Google Scholar]
- [22].Perera F, Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reprod Toxicol 2011;31:363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vanhees K, Coort S, Ruijters EJ, Godschalk RW, van Schooten FJ, Barjesteh van Waalwijk van Doorn-Khosrovani S. Epigenetics: prenatal exposure to genistein leaves a permanent signature on the hematopoietic lineage. FASEB J 2011;25:797–807. [DOI] [PubMed] [Google Scholar]
- [24].Hanley B, Dijane J, Fewtrell M, Grynberg A, Hummel S, Junien C, et al. Metabolic imprinting, programming and epigenetics – a review of present priorities and future opportunities. Br J Nutr 2010;104(Suppl 1):S1–S25. [DOI] [PubMed] [Google Scholar]
- [25].Esteller M Epigenetics in cancer. N Engl J Med 2008;358:1148–59. [DOI] [PubMed] [Google Scholar]
- [26].Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, et al. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol 2008;41: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Strakovsky RS, Zhang X, Zhou D, Pan YX. Gestational high fat diet programs hepatic phosphoenolpyruvate carboxykinase (Pck) expression and histone modification in neonatal offspring rats. J Physiol 2011;589:2707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Donkena KV, Young CY, Tindall DJ. Oxidative stress and DNA methylation in prostate cancer. Obstet Gynecol Int 2010;2010:302051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lawless MW, O'Byrne KJ, Gray SG. Oxidative stress induced lung cancer and COPD: opportunities for epigenetic therapy. J Cell Mol Med 2009;13:2800–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Soriano FX, Papadia S, Bell KF, Hardingham GE. Role of histone acetylation in the activity-dependent regulation of sulfiredoxin and sestrin 2. Epigenetics 2009;4: 152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest 2009;119:323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Furlong CE, Suzuki SM, Stevens RC, Marsillach J, Richter RJ, Jarvik GP, et al. Human PON1, a biomarker of risk of disease and exposure. Chem Biol Interact 2010;187: 355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Miao B, Zondlo S, Gibbs S, Cromley D, Hosagrahara VP, Kirchgessner TG, et al. Raising HDL cholesterol without inducing hepatic steatosis and hypertriglyceridemia by a selective LXR modulator. J Lipid Res 2004;45:1410–7. [DOI] [PubMed] [Google Scholar]
- [34].Gustavsson C, Soga T, Wahlstrom E, Vesterlund M, Azimi A, Norstedt G, et al. Sex-dependent hepatic transcripts and metabolites in the development of glucose intolerance and insulin resistance in Zucker diabetic fatty rats. J Mol Endocrinol 2011;47:129–43. [DOI] [PubMed] [Google Scholar]
- [35].Gomez-Perez Y, Gianotti M, Llado I, Proenza AM. Sex-dependent effects of high-fat-diet feeding on rat pancreas oxidative stress. Pancreas 2011;40: 682–8. [DOI] [PubMed] [Google Scholar]
- [36].Bayol SA, Simbi BH, Fowkes RC, Stickland NC. A maternal “junk food” diet in pregnancy and lactation promotes nonalcoholic Fatty liver disease in rat offspring. Endocrinology 2010;151:1451–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Leviev I, Poirier O, Nicaud V, Evans A, Kee F, Arveiler D, et al. High expressor paraoxonase PON1 gene promoter polymorphisms are associated with reduced risk of vascular disease in younger coronary patients. Atherosclerosis 2002;161: 463–7. [DOI] [PubMed] [Google Scholar]
- [38].Leviev I, Righetti A, James RW. Paraoxonase promoter polymorphism T(−107)C and relative paraoxonase deficiency as determinants of risk of coronary artery disease. J Mol Med 2001;79:457–63. [DOI] [PubMed] [Google Scholar]
- [39].Suehiro T, Nakamura T, Inoue M, Shiinoki T, Ikeda Y, Kumon Y, et al. A polymorphism upstream from the human paraoxonase (PON1) gene and its association with PON1 expression. Atherosclerosis 2000;150:295–8. [DOI] [PubMed] [Google Scholar]
- [40].Gaidukov L, Rosenblat M, Aviram M, Tawfik DS. The 192R/Q polymorphs of serum paraoxonase PON1 differ in HDL binding, lipolactonase stimulation, and cholesterol efflux. J Lipid Res 2006;47:2492–502. [DOI] [PubMed] [Google Scholar]
- [41].Thomas-Moya E, Gomez-Perez Y, Fiol M, Gianotti M, Llado I, Proenza AM. Gender related differences in paraoxonase 1 response to high-fat diet-induced oxidative stress. Obesity (Silver Spring) 2008;16:2232–8. [DOI] [PubMed] [Google Scholar]
- [42].Soran H, Younis NN, Charlton-Menys V, Durrington P. Variation in paraoxonase-1 activity and atherosclerosis. Curr Opin Lipidol 2009;20:265–74. [DOI] [PubMed] [Google Scholar]
- [43].Johansson MB. Lipoproteins and lipids in fetal, neonatal and adult rat serum. Biol Neonate 1983;44:278–86. [DOI] [PubMed] [Google Scholar]
- [44].Ha HL, Shin HJ, Feitelson MA, Yu DY. Oxidative stress and antioxidants in hepatic pathogenesis. World J Gastroenterol 2010;16:6035–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Videla LA. Oxidative stress signaling underlying liver disease and hepatoprotective mechanisms. World J Hepatol 2009;1:72–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Murr R Interplay between different epigenetic modifications and mechanisms. Adv Genet 2010;70:101–41. [DOI] [PubMed] [Google Scholar]