Abstract
Background
It is well established that estrogens and other hormonal factors influence breast cancer susceptibility. We hypothesized that a woman’s total lifetime estrogen exposure accumulates changes in DNA methylation, detectable in the blood, which could be used in risk assessment for breast cancer.
Methods
An estimated lifetime estrogen exposure (ELEE) model was defined using epidemiological data from EPIC-Italy (n = 31,864). An epigenome-wide association study (EWAS) of ELEE was performed using existing Illumina HumanMethylation450K Beadchip (HM450K) methylation data obtained from EPIC-Italy blood DNA samples (n = 216). A methylation index (MI) of ELEE based on 31 CpG sites was developed using HM450K data from EPIC-Italy and the Generations Study and evaluated for association with breast cancer risk in an independent dataset from the Generations Study (n = 440 incident breast cancer cases matched to 440 healthy controls) using targeted bisulfite sequencing. Lastly, a meta-analysis was conducted including three additional cohorts, consisting of 1187 case-control pairs.
Results
We observed an estimated 5% increase in breast cancer risk per 1-year longer ELEE (OR = 1.05, 95% CI 1.04–1.07, P = 3 × 10−12) in EPIC-Italy. The EWAS identified 694 CpG sites associated with ELEE (FDR Q < 0.05). We report a DNA methylation index (MI) associated with breast cancer risk that is validated in the Generations Study targeted bisulfite sequencing data (ORQ4_vs_Q1 = 1.77, 95% CI 1.07–2.93, P = 0.027) and in the meta-analysis (ORQ4_vs_Q1 = 1.43, 95% CI 1.05–2.00, P = 0.024); however, the correlation between the MI and ELEE was not validated across study cohorts.
Conclusion
We have identified a blood DNA methylation signature associated with breast cancer risk in this study. Further investigation is required to confirm the interaction between estrogen exposure and DNA methylation in the blood.
Electronic supplementary material
The online version of this article (10.1186/s13148-019-0664-7) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, EWAS, Epigenetics, Breast cancer, Cancer risk, Estrogen exposure, Hormonal exposures, Biomarker
Background
Breast cancer is the most common women’s cancer, with an estimated 1.67 million cases diagnosed globally in 2012 [1]. Its crude incidence is rising due to an aging population and population-level changes in reproductive and lifestyle factors that affect breast cancer risk [2]. Up to 23% of breast cancer cases are considered preventable by lifestyle changes, such as maintaining a healthy weight and reducing alcohol consumption [3]. There is a need for improved risk assessment methods to target prevention and early detection to women at increased risk.
It is well established that estrogens play a role in breast cancer etiology, and women with higher circulating estrogen concentrations have an increased risk of breast cancer [4–6]. Several hormonal breast cancer risk factors contribute to a woman’s lifetime estrogen exposure. These include a younger age at menarche and an older age at menopause, which together define the reproductive span during which a woman is exposed to high levels of endogenous estrogens produced by the ovaries [7, 8]. The number of pregnancies is associated with a long-term decrease in both estrogen exposures and breast cancer risk [9, 10]. A small decrease in risk is seen for women who breastfeed for longer, and this decreases the total lifetime estrogen exposure [11, 12]. Exogenous hormones provided by oral contraceptives (OC) and hormone replacement therapy (HRT) increase risk during use, but risk returns to that for unexposed women 5–10 years after cessation [13–17]. Additionally, lifestyle risk factors for breast cancer such as higher postmenopausal body mass index (BMI), alcohol consumption, physical inactivity, and smoking can each affect circulating estrogen concentrations [4, 18–22].
Epigenetic mechanisms such as DNA methylation control gene expression and may be influenced by environmental and lifestyle exposures. Epigenome-wide association studies (EWAS) of blood DNA methylation for breast cancer risk measured have identified associations with global hypomethylation and several candidate genes, but these have generally not been replicated across studies [23–26]. Numerous large EWAS have identified epigenetic signatures for smoking [27], alcohol consumption [28], BMI [29], and aging [30, 31], and hypomethylation signatures associated with smoking can improve the prediction of lung cancer [32, 33]. We propose that an EWAS of breast cancer risk factors may identify CpG sites that could be used in risk prediction models [23]. We hypothesize that estrogen exposures over the lifetime give rise to accumulated changes in DNA methylation, detectable in the blood, which might add useful information to breast cancer risk prediction.
The aims of this study were to identify a DNA methylation signature reflecting a woman’s lifetime estrogen exposure and assess the signatures’ association with breast cancer risk. There is no standard model to estimate a woman’s total lifetime estrogen exposure, and numerous approaches have been used [34–39]. For example, the Pike model reflecting the “breast tissue aging” in relation to breast cancer risk includes a woman’s age at menarche, age at first full-term pregnancy, and age at menopause modeling changes over time [38]. In this study, we have used an estimated lifetime estrogen exposure (ELEE) model that reflects the reproductive span, comprising a woman’s time between age at menarche and age at menopause minus 1 year for each pregnancy and duration of breastfeeding, calculated at the time at recruitment. We performed an EWAS and identified 694 CpG sites associated with ELEE. We then developed a methylation signature of ELEE that showed association with breast cancer risk and was further validated in a large independent study cohort using targeted bisulfite sequencing, and a meta-analysis of three additional independent study cohorts.
Methods
Study cohorts
Data from two independent prospective cohort studies were used for the primary analysis: the Italian cohort from the European Prospective Investigation into Cancer and Nutrition study (EPIC-Italy) and the UK-based Generations Study. Questionnaire data and blood samples were collected at the time of study enrollment. EPIC-Italy included epidemiological questionnaire data from 32,059 women (dataset 1) and peripheral blood DNA methylation data measured using the HM450K array for 162 matched pairs of incident breast cancer cases and controls (dataset 2) [24]. The Generations Study cohort included HM450K peripheral blood DNA array data for a subset of 92 healthy women (dataset 3) [40] and 440 matched pairs of incident breast cancer cases and controls (independent from the HM450K dataset), who provided blood DNA samples used for targeted bisulfite sequencing (dataset 4). Inclusion criteria for incident breast cancer cases in the Generations Study, with blood samples taken prior to diagnosis, were the following: invasive ER-positive breast cancer with no previous history of (non-breast) cancer, white ethnicity, and completeness of epidemiological data. Controls were individually matched to cases on age at blood draw ± 5 years. Additional replication cohorts used in the meta-analysis included an additional 118 case-control pairs from EPIC-Italy (dataset 5), 435 case-control pairs from EPIC-IARC (dataset 6) [41], and 310 case-control pairs from the MCCS (dataset 7) [42]. Further information for these study cohorts is provided in Additional file 1: Supplementary material and methods.
EWAS of ELEE
Different ELEE models including a woman’s reproductive span (age at menopause minus age at menarche for postmenopausal women and age at recruitment minus age at menarche for premenopausal women), number of pregnancies, and breastfeeding duration were considered. The models were assessed for association with breast cancer risk in EPIC-Italy (n = 1193 cases and 30,671 controls, Additional file 2: Table S1) using age-adjusted Cox regression. The ELEE model selected for the EWAS included as many of these risk factors as possible without reducing the significance of the association with breast cancer for pre- and postmenopausal women. An EWAS with ELEE as the exposure and DNA methylation as the outcome was conducted for EPIC-Italy using a beta regression model on HM450K beta values. Subjects with missing information for ELEE were excluded (n = 87), as were cases with age at diagnosis < 50 (n = 28), to enrich for ER-positive disease, leaving 216 women for the EWAS (Additional file 2: Table S2). Potential confounders, known to influence methylation or estrogen levels, were adjusted for; these included age, BMI, alcohol consumption, and smoking duration, all reported at recruitment, as well as technical confounders including batch, position on batch, and white blood cell (WBC) composition [43]. Multiple testing was accounted for using the false discovery rate (FDR) Q values in R function “p.adjust.” Beta regression coefficients are not interpretable as methylation percentage changes. Therefore, to get interpretable estimates for the significantly associated CpG sites, i.e., percentage change in DNA methylation per unit longer ELEE, a linear mixed-effects regression model adjusted for the same variables, including random effects for batch and position on chip, was applied to beta values multiplied by 100.
Laboratory analysis
HM450K array data generation has been described previously [24, 40–42]. For validation, targeted bisulfite sequencing was conducted in the Generations Study (n = 880) using the Fluidigm 48.48 Access Array. Forty-two CpG sites for validation were selected from the EWAS based on the magnitude of change in DNA methylation (> 0.1%) per 1-year longer ELEE (mixed-effects linear regression model coefficient) and statistical significance of association with ELEE (beta regression model P value < 7 × 10−5). To estimate WBC composition, five HM450K CpG probes that showed independent correlation with five different WBC types were included in the target panel (Additional file 3: Figure S1). The 880 Generations Study samples were sequenced on the Illumina MiSeq in 20 batches in four sequencing pools (6, 5, 5, and 4 batches). After quality control, two batches were rerun (batches 12 and 19) due to poor sequencing data, likely a result of decreased performance for the barcodes used for these batches. In the results, only the sequencing data from the new batches 12 and 19 are used, referring to sequencing pool 5. DNA methylation levels were extracted using Bismark [44] and analyzed in R version 3.3.2.
Statistical analysis
Quality control (QC) of the targeted sequencing data was conducted to exclude CpG sites (n = 42) with low coverage or with large difference in DNA methylation levels between duplicated pairs (n = 31 CpG sites passing quality control). A methylation index (MI) of ELEE was developed using the same 31 CpG sites in the HM450K data with complete information for ELEE from both EPIC-Italy (dataset 2, n = 237) and the Generations Study (dataset 3, n = 65, Additional file 2: Table S2). To develop the MI, ridge regression was conducted in a 10-fold cross-validation repeated 100 times, using the R package “glmnet” in “train” in the R package “caret” to estimate the following parameters: penalty coefficient (lambda) for ridge regression and regression coefficients for the model. The final MI model for predicted ELEE was calculated as a linear function, i.e., intercept plus the sum of the DNA methylation levels at the CpG sites included in the model weighted by their coefficient. The correlation between the MI and ELEE was evaluated in the Generations Study targeted sequencing data (dataset 4, pairs with coverage > 30 sequence reads for at least 10% of the remaining CpG sites) using the Pearson’s correlation coefficient. The association between the MI and risk of breast cancer was assessed using conditional logistic regression (R function “clogit” in the package “survival”) for matched case-control pairs in EPIC-Italy HM450K development data (dataset 2, n = 162 pairs), the Generations Study targeted sequencing validation data (dataset 4), and in each of the additional cohorts for the meta-analysis including EPIC-Italy (dataset 5, n = 118 pairs), EPIC-IARC (dataset 6, n = 420 pairs), and MCCS (dataset 7, n = 310 pairs). The meta-analysis of log odds ratios and standard errors was conducted using a weighted random-effects model, applying the restricted-maximum likelihood method (function “rma.uni” in the R package “metafor”). The Cochran’s Q statistics and I2 statistics were used to estimate heterogeneity between the studies; Q < 0.05 and I2 > 50% were defined as heterogeneous estimations. If the heterogeneity I2 was equal to 0, the meta-analysis behaved as a fixed-effects model. The ORs for all models were adjusted for baseline age, BMI, alcohol consumption, smoking duration, and estimated WBC composition in a multivariable model. Additionally, for each of the target CpG sites passing QC, the association with breast cancer risk was investigated using conditional logistic regression. Further details on methods and workflow are described in Additional file 1: Supplementary material and methods and Additional file 3: Figure S2.
Results
The ELEE is associated with breast cancer risk in EPIC-Italy
The estimated lifetime estrogen exposure (ELEE) was calculated as a woman’s reproductive span (age at menopause or, for premenopausal women, age at recruitment, minus age at menarche) minus 1 year per pregnancy and breastfeeding duration before recruitment in years. For EPIC-Italy (dataset 1, n = 1193 cases and 30,671 controls, Additional file 2: Table S1), a 1-year longer ELEE, ranging from 5 to 49, was associated with a 5% increase in breast cancer risk (hazard ratio (HR) = 1.05, 95% confidence interval (CI) 1.04–1.07, P = 3 × 10−12, Additional file 2: Table S3) and was associated with breast cancer risk for both pre- and postmenopausal women (HR = 1.09, 95% CI 1.03–1.15, P = 0.002, and HR = 1.03, 95% CI 1.01–1.05, P = 6 × 10−4, Additional file 2: Table S3).
EWAS identifies CpG sites associated with ELEE in EPIC-Italy
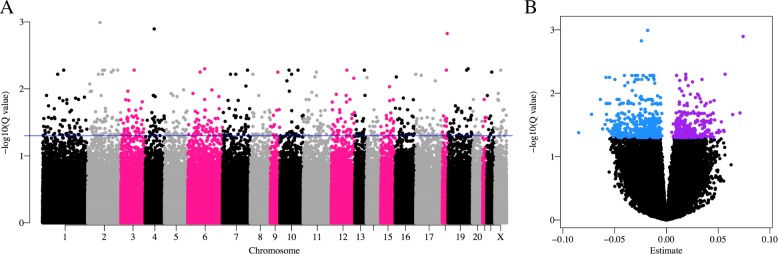
The EWAS of ELEE conducted in EPIC-Italy (dataset 2, n = 216, Additional file 2: Table S2) gave P values that were moderately inflated (Additional file 3: Figure S3), with a genomic inflation factor lambda of 1.13. After correction for multiple testing, the EWAS identified 694 CpG probes associated with ELEE (FDR Q < 0.05, Fig. 1a) with a mix of hypo- and hypermethylated CpG probes (Fig. 1b). Two sensitivity analyses were conducted firstly, including all cases and controls with complete information of ELEE (n = 237), and secondly for controls only (n = 119), and estimates from each analysis were highly correlated (r = 0.997 and r = 0.963, respectively, Additional file 3: Figure S4). All CpG probes were associated with ELEE (Q < 0.05) in the first analysis and 563 in the second analysis with controls only. Out of the 694 CpG probes associated with ELEE, CpG sites were selected for the targeted bisulfite sequencing. The selection was based on the largest magnitude of change in DNA methylation (> 0.1%) and statistical significance (P < 7 × 10−5). Furthermore, poorly performing assays were excluded before the sequencing in the Generations Study. A total of 42 CpG sites were included in the final target panel for targeted bisulfite sequencing in the Generations Study using the Fluidigm 48.48 Access Array (Table 1).
Fig. 1.
EWAS identifies CpG sites significantly associated with ELEE in EPIC-Italy. An EWAS with DNA methylation (HM450K beta values) as outcome and ELEE as exposure was conducted in EPIC-Italy (n = 216) using a beta regression model adjusted for age, BMI, alcohol consumption, and smoking duration (all variables reported at recruitment), and batch, position on batch, and WBC composition. a Manhattan plot of minus log10Q values for FDR-corrected P values from the EWAS of all 404,596 probes across the genome. Blue line indicates FDR Q value threshold of 0.05. b Volcano plot of the regression coefficients (estimates) from the beta regression model, showing significant hypomethylated (in blue) and hypermethylated probes (in purple)
Table 1.
List of the 42 target CpG sites included in the targeted bisulfite sequencing
| HM450K probe | Chr | Position | Nearest gene | Distance to gene | EWAS of ELEE in EPIC-Italyb | |||
|---|---|---|---|---|---|---|---|---|
| Estimate | SE | P value | Q value | |||||
| cg01893629 | chr12 | 34494825 | ALG10 | 313588 | − 0.14 | 0.03 | 8.15E−05 | 4.96E−02 |
| cg08254089a | chr20 | 36933189 | BPI | 0 | − 0.28 | 0.08 | 7.04E−05 | 4.78E−02 |
| cg21590238 | chr12 | 121454837 | C12orf43 | 536 | − 0.29 | 0.08 | 2.67E−06 | 1.44E−02 |
| cg21153102 | chr15 | 41252147 | CHAC1 | 3429 | − 0.43 | 0.14 | 6.16E−05 | 4.58E−02 |
| cg03340215a | chr15 | 83315615 | CPEB1 | 0 | − 0.31 | 0.08 | 4.78E−05 | 4.21E−02 |
| cg06968859 | chr2 | 80724209 | CTNNA2 | 0 | − 0.26 | 0.05 | 2.52E−09 | 1.02E−03 |
| cg12105860a | chr12 | 31742801 | DENND5B | 0 | 0.14 | 0.03 | 1.58E−06 | 1.21E−02 |
| cg16840364a | chr4 | 84539569 | GPAT3 | 12541 | − 0.14 | 0.06 | 4.77E−05 | 4.21E−02 |
| cg08835688 | chr7 | 50849931 | GRB10 | 0 | − 0.14 | 0.03 | 4.50E−05 | 4.11E−02 |
| cg08349826 | chr16 | 10346403 | GRIN2A | 69791 | − 0.11 | 0.04 | 4.19E−05 | 4.02E−02 |
| cg23681866 | chr6 | 29895175 | HLA-J | 0 | − 1.33 | 0.31 | 2.17E−05 | 3.13E−02 |
| cg22968966 | chr16 | 22959875 | HS3ST2 | 32215 | − 0.40 | 0.12 | 7.95E−05 | 4.92E−02 |
| cg15127563 | chr2 | 231729487 | ITM2C | 132 | 0.36 | 0.09 | 1.77E−05 | 2.95E−02 |
| cg20020161 | chr2 | 231732669 | ITM2C | 0 | − 0.14 | 0.04 | 4.49E−05 | 4.11E−02 |
| cg22097768 | chr17 | 61615913 | KCNH6 | 0 | − 0.20 | 0.05 | 4.88E−05 | 4.25E−02 |
| cg17969123 | chr19 | 18745971 | KLHL26 | 1865 | − 0.15 | 0.04 | 3.18E−05 | 3.65E−02 |
| cg05422360 | chrX | 75648455 | MAGEE1 | 0 | − 0.44 | 0.12 | 1.72E−07 | 5.26E−03 |
| cg01768446 | chr16 | 89982419 | MC1R | 1866 | − 0.13 | 0.03 | 4.57E−05 | 4.13E−02 |
| cg25372296 | chr1 | 98510328 | MIR137HG | 0 | 0.33 | 0.09 | 3.83E−06 | 1.65E−02 |
| cg04519403a | chr5 | 79298951 | MTX3 | 11862 | − 0.24 | 0.05 | 7.51E−06 | 2.20E−02 |
| cg12091786 | chr20 | 61877942 | NKAIN4 | 0 | − 0.37 | 0.10 | 2.07E−05 | 3.06E−02 |
| cg25279613a | chr7 | 24956523 | OSBPL3 | 0 | 0.20 | 0.06 | 4.95E−06 | 1.96E−02 |
| cg24536703 | chr11 | 77183438 | PAK1 | 0 | − 0.32 | 0.09 | 2.71E−05 | 3.43E−02 |
| cg24036523 | chr14 | 73712256 | PAPLN | 0 | − 0.45 | 0.11 | 3.05E−05 | 3.65E−02 |
| cg16720405 | chr3 | 122790178 | PDIA5 | 0 | − 0.17 | 0.05 | 5.17E−05 | 4.33E−02 |
| cg13674411 | chr1 | 204232677 | PLEKHA6 | 0 | 0.11 | 0.03 | 3.50E−05 | 3.76E−02 |
| cg20684174 | chr11 | 7541255 | PPFIBP2 | 0 | − 0.13 | 0.03 | 1.23E−05 | 2.67E−02 |
| cg01430588a | chr17 | 56769767 | RAD51C | 194 | − 0.32 | 0.09 | 6.04E−05 | 4.51E−02 |
| cg22273487 | chr20 | 32580931 | RALY | 525 | 0.19 | 0.05 | 1.60E−05 | 2.84E−02 |
| cg22343083 | chr8 | 54786401 | RGS20 | 0 | − 0.30 | 0.07 | 1.92E−06 | 1.26E−02 |
| cg22758104 | chr17 | 50465 | RPH3AL | 11713 | − 0.23 | 0.06 | 4.62E−07 | 6.07E−03 |
| cg16733643a | chr1 | 41575522 | SCMH1 | 0 | − 0.42 | 0.11 | 5.00E−05 | 4.25E−02 |
| cg17588491 | chr22 | 25198892 | SGSM1 | 3242 | − 0.17 | 0.04 | 4.41E−05 | 4.06E−02 |
| cg13971030 | chr11 | 35366721 | SLC1A2 | 0 | − 0.49 | 0.14 | 9.19E−06 | 2.41E−02 |
| cg17567562a | chr3 | 47687980 | SMARCC1 | 0 | − 0.48 | 0.11 | 1.18E−05 | 2.66E−02 |
| cg10298859 | chr13 | 112883993 | SPACA7 | 146656 | − 0.20 | 0.04 | 2.26E−07 | 5.26E−03 |
| cg19216791 | chr19 | 5568216 | TINCR | 210 | − 0.25 | 0.07 | 4.13E−05 | 4.02E−02 |
| cg25936380 | chr2 | 120981591 | TMEM185B | 606 | − 0.29 | 0.07 | 2.22E−07 | 5.26E−03 |
| cg01824466a | chr8 | 95959531 | TP53INP1 | 0 | − 0.23 | 0.08 | 8.08E−05 | 4.96E−02 |
| cg26657235 | chr6 | 150378972 | ULBP3 | 4367 | − 0.12 | 0.03 | 1.32E−05 | 2.76E−02 |
| cg08551047 | chr15 | 91473569 | UNC45A | 0 | − 0.58 | 0.14 | 2.87E−06 | 1.46E−02 |
| cg20394620a | chrX | 48541924 | WAS | 260 | − 0.20 | 0.05 | 3.02E−06 | 1.46E−02 |
SE standard error
aMarked probes did not pass quality control in the targeted sequencing data and were not included in the analysis of the methylation index
bResult from the EWAS of ELEE in EPIC-Italy (n = 216, dataset 2). The estimates correspond to regression coefficients from a mixed-effects linear regression model (percentage change in DNA methylation per unit longer ELEE), and P values from the beta regression model, which have been corrected for multiple testing using FDR (Q values)
Quality control of targeted sequencing data prior to model development
Targeted bisulfite sequencing of 42 target regions using the Fluidigm 48.48 Access Array was conducted for 880 samples (440 matched case-control pairs) from the Generations Study (dataset 4). The targeted sequencing data was of high quality: average sequencing depth per CpG site per sample was 1740, > 97% of the reads were assigned to a sample (i.e., had a barcode sequence), and > 99% of the paired reads were aligned to target regions (Additional file 2: Table S4). Additionally, a high correlation in DNA methylation values was observed between batches, but with some variability in some of the assays (mean r = 0.88). Eleven CpG sites were excluded from the analysis due either to low coverage across batches or to high variation in DNA methylation levels between duplicated pairs. Out of the 880 samples, subjects were excluded due to incorrect case-control status (1 pair) or low coverage in > 10% remaining target CpG sites (n = 100 matched pairs where at least one sample of the pair had low coverage), leaving a total of 678 samples (339 matched case-control pairs, Table 2) for the analyses. In the 328 matched case-control pairs in the Generations Study with coverage > 30 for all five WBC CpG sites, no difference (P > 0.05) in DNA methylation levels between cases and controls were observed (Additional file 3: Figure S1).
Table 2.
Table of characteristics for case-control pairs in EPIC-Italy and the Generations Study
| EPIC-Italy | The Generations Study | |||||
|---|---|---|---|---|---|---|
| Cases (n = 162) | Controls (n = 162) | Cases (n = 339) | Controls (n = 339) | P b | ||
| Age | Mean (st.dev.), years | 52.9 (7.2) | 53.0 (7.1) | 53.9 (10.3) | 54.1 (10.4) | 0.054 |
| Time to diagnosis | Mean (st.dev.), years | 5.3 (4.4) | NA | 4.0 (2.4) | NA | 0.022 |
| Menopausal status | n (%) | 0.690 | ||||
| Premenopausal | 52 (32.1%) | 49 (30.2%) | 135 (39.8%) | 127 (37.5%) | ||
| Postmenopausal | 85 (52.5%) | 87 (53.7%) | 204 (60.2%) | 212 (62.5%) | ||
| Age at menarche | Mean (st.dev.), years | 12.7 (1.4) | 12.7 (1.7) | 12.7 (1.4) | 12.7 (1.5) | 0.914 |
| Age at menopause | Mean (st.dev.), years | 50.2 (3.7) | 49.1 (3.8) | 50.3 (4.3) | 50.1 (4.5) | 0.126 |
| Number of pregnancies | Mean (st.dev.) | 1.6 (1.1) | 1.7 (1.0) | 1.9 (1.1) | 1.9 (1.2) | 4 × 10 −4 |
| Ever breastfed | n (%) | 103 (63.6%) | 112 (69.1%) | 265 (78.2%) | 274 (80.8%) | 0.179 |
| Breastfeeding duration | Mean (st.dev.), years | 0.7 (0.6) | 0.8 (0.7) | 0.9 (1.0) | 0.8 (0.9) | 2 × 10 −5 |
| BMI | Mean (st.dev.), kg/m2 | 25.8 (4.1) | 25.3 (4.3) | 25.7 (4.3) | 25.2 (4.3) | 0.626 |
| Alcohol consumptiona | Mean (st.dev.) | 5.5 (7.0) | 7.4 (9.9) | 15.8 (16.7) | 14.6 (15.5) | 2 × 10 −26 |
| Smoking status | n (%) | 8 × 10 −10 | ||||
| Smoker | 31 (19.1%) | 36 (22.2%) | 25 (7.4%) | 23 (6.8%) | ||
| Former | 23 (14.2%) | 41 (25.3%) | 98 (28.9%) | 86 (25.4%) | ||
| Never | 106 (65.4%) | 85 (52.5%) | 216 (63.7%) | 230 (67.9%) | ||
| Smoking duration | Mean (st.dev.), years | 8.1 (12.6) | 10.6 (13.4) | 4.4 (9.0) | 3.9 (9.0) | 4 × 10 −10 |
| OC ever | n (%) | 59 (36.4 %) | 67 (41.4 %) | 253 (74.6%) | 256 (75.5%) | < 2 × 10 −16 |
| OC duration | Mean (st.dev.), years | 4.1 (5.1) | 5.6 (5.8) | 8.1 (6.4) | 8.8 (6.8) | 4 × 10 −9 |
| HRT ever | n (%) | 23 (14.2 %) | 28 (17.3 %) | 239 (70.5%) | 241 (71.1%) | 2 × 10 −7 |
| HRT duration | Mean (st.dev.), years | 2.8 (2.7) | 2.6 (3.4) | 6.7 (5.7) | 5.4 (4.3) | 5 × 10 −8 |
aAlcohol consumption in the Generations Study reported in average units per week, converted to average gram per day by multiplying with 8 (1 unit = 8 g alcohol) and dividing by 7
bP values indicate differences between all subjects from EPIC-Italy and all subjects from the Generations Study: t test for continuous variables, chi-squared test for categorical variables
The MI is associated with breast cancer risk
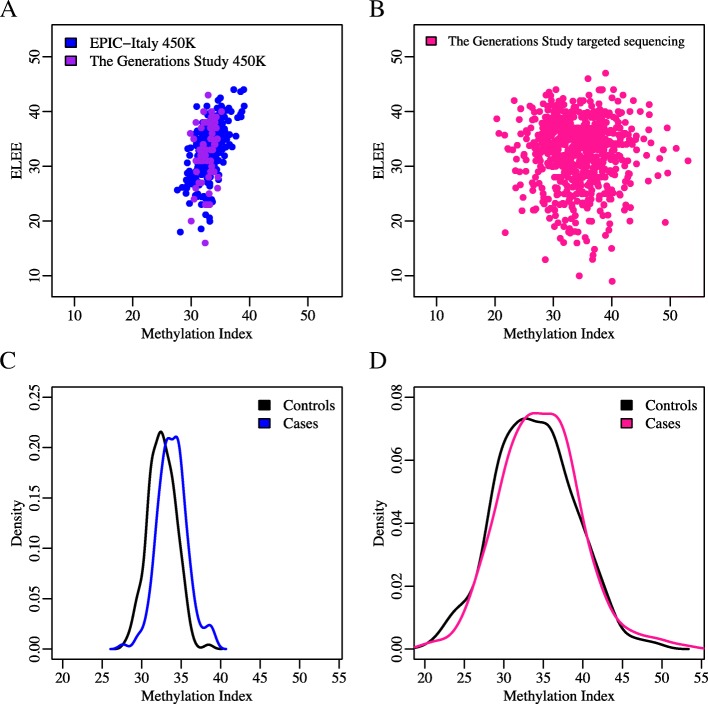
A methylation index (MI) to predict ELEE was developed in HM450K data from EPIC-Italy (dataset 2, n = 237) and the Generations Study (dataset 3, n = 65, Additional file 2: Table S2) using ridge regression on the 31 target CpG sites passing QC in the targeted bisulfite sequencing data. As expected, the MI correlated with ELEE in the development data; a high correlation was observed in EPIC-Italy (r = 0.60, P = 6 × 10−25) and moderately correlated in the Generations Study HM450K data (r = 0.27, P = 0.027, Fig. 2a). The correlation between the MI and ELEE, however, was not replicated in the Generations Study targeted sequencing data (n = 678, r = − 0.04, P = 0.340, Fig. 2b). We also observed no association between the ELEE and breast cancer risk in the Generations Study (n = 339 matched case-control pairs, age-adjusted odds ratio (OR) = 1.01, 95% CI 0.98–1.04, P = 0.562), in contrast to EPIC-Italy subjects from the HM450K dataset (n = 162 matched case-control pairs, age-adjusted OR = 1.10, 95% CI 1.03–1.17, P = 0.007). The correlations between the MI and ELEE were similar for pre- and postmenopausal women in the Generations Study, but stronger for postmenopausal women (r = 0.72, P = 2 × 10−22) than for premenopausal women (r = 0.53, P = 7 × 10−7) in EPIC-Italy (test for heterogeneity between pre- and postmenopausal women in EPIC-Italy: Q = 0.79, I2 = 0 (no heterogeneity); Generations Study: Q = 0.42, I2 = 0 (no heterogeneity)).
Fig. 2.
The MI is associated with breast cancer risk. The MI was developed in combined HM450K data from EPIC-Italy (dataset 2, n = 237) and the Generations Study (dataset 3, n = 65) using ridge regression. The correlation between the MI and ELEE and the association between the MI and breast cancer risk were evaluated. a The correlations between the MI and ELEE in the development of HM450K data were as follows: r = 0.60 and P = 6 × 10−25 for EPIC-Italy and r = 0.27 and P = 0.027 for the Generations Study b The MI and ELEE were not correlated in the Generations Study targeted sequencing data (r =− 0.04, P = 0.340). c Density plot of the MI values in controls and cases in EPIC-Italy HM450K data. The MI was significantly associated with breast cancer risk in EPIC-Italy (n = 162 pairs, OR = 1.51, 95% CI 1.26–1.82, P = 1 × 10−5). d Density plot of the MI values in controls and cases in the Generations Study targeted sequencing data. The MI was significantly associated with breast cancer risk in the Generations Study (n = 339 pairs, OR = 1.04, 95% CI 1.01–1.08, P = 0.022). ORs were adjusted for age, BMI, alcohol consumption, and smoking duration (all variables reported at recruitment) and WBC composition
The association between the MI and breast cancer risk was tested using matched case-control pairs from EPIC-Italy HM450K data and the Generations Study targeted sequencing data (n = 162 and 339 pairs respectively, Table 2). In a multivariable model, each unit increase in the MI, ranging from 27.6 to 39.1, was associated with a 51% increase in breast cancer risk in EPIC-Italy (OR = 1.51, 95% CI 1.28–1.82, P = 1 × 10−5, Fig. 2c). The association was validated in the Generations Study targeted sequencing data: A comparable analysis of the Generations Study data gave an estimated 4% increase in risk per unit increase in MI, which ranged from 20.3 to 53.1 (OR = 1.04, 95% CI 1.01–1.08, P = 0.022, Fig. 2d). The OR estimate was greater for postmenopausal women in both EPIC-Italy and the Generations Study (OR = 1.91, 95% CI 1.29–2.82, P = 0.001 and OR = 1.07, 95% CI 1.02–1.12, P = 0.006, respectively) than for premenopausal women (OR = 1.61, 95% CI 1.17–2.22, P = 0.004 and OR = 1.01, 95% CI 0.94–1.09, P = 0.713, respectively). However, the test for heterogeneity between pre- and postmenopausal women in EPIC-Italy, Q = 0.19 and I2 = 43, and Generations Study, Q = 0.24 and I2 = 27, suggested no significant heterogeneity.
Sensitivity analysis on the model development was also conducted using controls only in the combined EPIC-Italy and the Generations Study HM450K data (datasets 2 and 3, n = 184). The control-only MI model showed a very similar association with breast cancer risk as previously when tested on the case-control pairs from the Generations Study (n = 339 pairs, OR = 1.04 per unit increase in MI, 95% CI 1.01–1.08, P = 0.012).
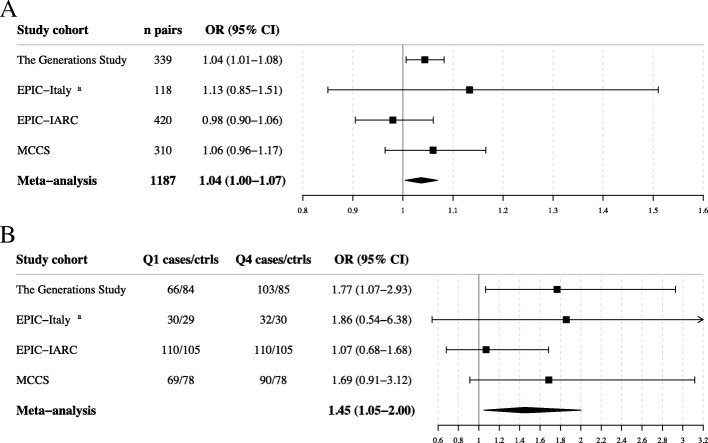
Meta-analysis of the association between MI and breast cancer risk
The association between the MI and breast cancer risk was examined in a meta-analysis, excluding the discovery data from EPIC-Italy. The analysis included 2374 women (1187 matched case-control pairs) from four prospective study cohorts, the Generations Study targeted sequencing data, additional subjects from EPIC-Italy, EPIC-IARC, and MCCS, with mean time to diagnosis of 4.0, 8.5, 7.5, and 7.9 years, respectively (Additional file 2: Table S7). All estimates were adjusted for baseline age, BMI, smoking duration, alcohol consumption, and WBC composition. The combined meta-analysis for MI as a continuous variable showed low heterogeneity across study cohorts (Q = 0.45, I2 = 0%) and an association with breast cancer risk, with 4% increase in risk per one unit increase in the MI (OR = 1.04, 95% CI 1.00–1.07, P = 0.024, Fig. 3a). Women in the highest quartile compared with the lowest quartile of MI had higher breast cancer risk with a combined OR of 1.45 (OR = 1.45, 95% CI 1.05–2.00, P = 0.024, Fig. 3b) and low heterogeneity (Q = 0.44, I2 = 16%). There was no significant association between the MI and breast cancer risk between pairs with a shorter time to diagnosis (less than median) in the combined meta-analysis (OR = 1.03, 95% CI 0.98–1.08, P = 0.241), but there was a significant association in pairs with time to diagnosis above the median (OR = 1.05, 95% CI 1.01–1.10, P = 0.021) (Additional file 2: Table S8). Lastly, the MI did not correlate with the ELEE in any of these four study cohorts (Additional file 2: Table S9).
Fig. 3.
Meta-analysis of the association between MI and breast cancer risk. The association between MI and risk for breast cancer, as a continuous variable (a) or as a categorical variable (b), was estimated in the four studies included in the meta-analysis using conditional logistic regression adjusted for age, BMI, alcohol consumption, and smoking duration (all variables reported at recruitment) and WBC composition. The log odds ratios were combined in a meta-analysis using restricted-maximum likelihood model. The square boxes represent the odds ratios (ORs) and the lines the 95% confidence intervals (CIs). aEPIC-Italy corresponds to the new EPIC-Italy samples, not included in the development of the MI
To explore non-linearity in the association between the MI and breast cancer risk, the MI was stratified into quartiles defined by the distribution in controls in each cohort. A higher breast cancer risk was observed for women in the highest quartile compared with those in the lowest quartile in EPIC-Italy (ORQ4_vs_Q1 = 5.45, 95% CI 2.17–13.67, P = 3 × 10−4) and in the Generations Study (ORQ4_vs_Q1 = 1.77, 95% CI 1.07–2.93, P = 0.027, Additional file 2: Table S5), but not in three additional cohorts. In the meta-analysis, excluding the EPIC-Italy development data, increased breast cancer risk was observed for women in the highest quartile compared with the lowest quartile (OR = 1.45, 95% CI 1.05–2.00, P = 0.024) and modest heterogeneity (Q = 0.44; I2 = 16%, Additional file 2: Table S5).
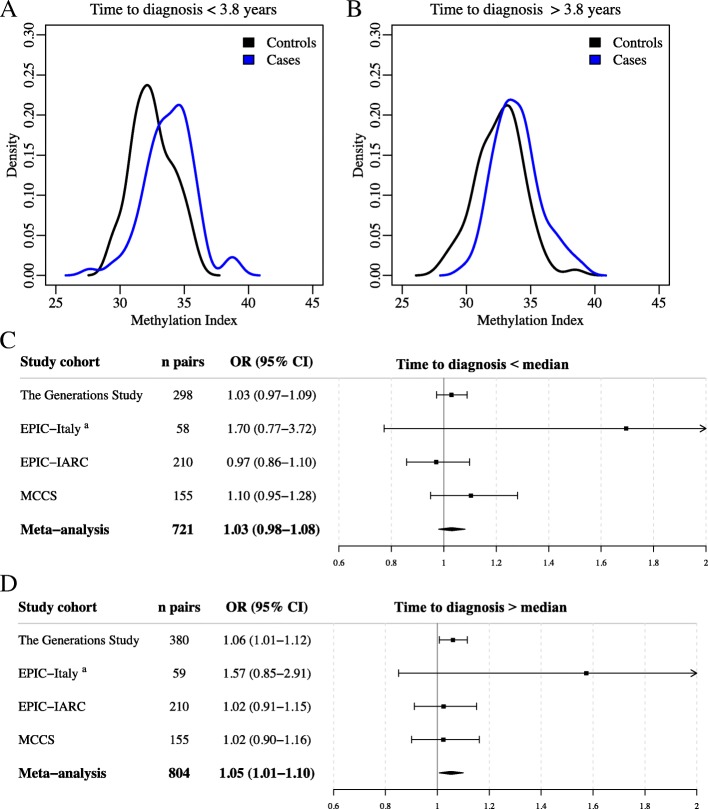
Reverse causation would be indicated if the association between MI and breast cancer risk was higher in cases with a short time to diagnosis. To explore this, we investigated the association between the MI and breast cancer risk stratified by median time to diagnosis in cases. In the EPIC-Italy development data, and in the meta-analysis, the association between the MI and breast cancer risk appeared to be stronger with longer time to diagnosis. In EPIC-Italy, the association between the MI and breast cancer risk was significant for both groups but with a higher OR for pairs with a longer time to diagnosis (n = 81 pairs in both groups, OR = 1.47, 95% CI 1.12–1.93, P = 0.005 vs OR = 1.84, 95% CI 1.30–2.61, P = 0.001; Fig. 4a, b). In the meta-analysis, for pairs with shorter time to diagnosis, there was no significant association with breast cancer risk (n = 721 pairs, OR = 1.03, 95% CI 0.98–1.08, P = 0.241, Fig. 4c); however, the MI was associated with breast cancer risk for pairs with a time to diagnosis above the median (n = 804 pairs, OR = 1.05, 95% CI 1.01–1.10, P = 0.021, Fig. 4d). Therefore, the data do not support reverse causation as a mechanism for the association with DNA methylation.
Fig. 4.
Time to diagnosis and the association between the MI and breast cancer risk. Matched case-control pairs were stratified on median time to diagnosis in EPIC-Italy HM450K data (dataset 2) and in the four study cohorts included in the meta-analysis. The association between the MI and breast cancer risk was analyzed in the two groups. a The MI was significantly associated with breast cancer risk in EPIC-Italy pairs with a shorter time to diagnosis (n = 81 pairs, OR = 1.47, 95% CI 1.12–1.93, P = 0.005). b The MI was significantly associated with breast cancer risk in EPIC-Italy pairs with a longer time to diagnosis (n = 81 pairs, OR = 1.84, 95% CI 1.30–2.61, P = 0.001). c The combined meta-analysis including pairs with shorter time to diagnosis showed no significant association between the MI and breast cancer risk (OR = 1.03, 95% CI 0.98–1.08, P = 0.241). d The combined meta-analysis including pairs with shorter time to diagnosis showed no significant association between the MI and breast cancer risk (OR = 1.05, 95% CI 1.01–1.10, P = 0.021). The log odds ratios were combined in the meta-analyses using restricted-maximum likelihood model. ORs were adjusted for age, BMI, alcohol consumption, and smoking duration (all variables reported at recruitment) and WBC composition
Individual associations between each of the 31 target CpG sites and breast cancer risk were further examined in the Generations Study (dataset 4). Four CpG sites located nearest the genes CTNNA2, GRB10, RPH3AL, and TINCR showed individual associations with breast cancer risk in a multivariable model (P < 0.05, Additional file 2: Table S6) and were also associated with breast cancer risk in matched case-control pairs in EPIC-Italy (dataset 2).
Discussion
In this study, we have performed an EWAS of lifetime estrogen exposure using the HM450K array and identified 694 CpG sites (FDR Q < 0.05) associated with ELEE in the EPIC-Italy study cohort. In addition to this, we have conducted a validation step in a case-control study nested within a large independent cohort, the Generations Study, using targeted bisulfite sequencing. We have developed a methylation index (MI) to predict ELEE using DNA methylation levels at 31 CpG sites and tested the MI for association with breast cancer risk. Although the MI did not correlate with ELEE in the validation cohorts, it was associated with breast cancer risk. Women in the highest quartile of the MI in the Generations Study had 77% higher risk for breast cancer compared with women in the lowest quartile (Additional file 2: Table S5). In the meta-analysis, including three other independent datasets, the highest quartile had a 45% higher risk compared with the lowest quartile (Fig. 4).
There are several potential explanations why the correlation between the MI and ELEE was only seen in the discovery EPIC-Italy dataset and not in the additional validation cohorts. Firstly, it is possible that the observed association between MI and ELEE in EPIC-Italy is a false positive and that the MI was over fitted in this study cohort. It may also be possible that the measured methylation index could capture the biological effects of unknown confounders that are not included in the calculated ELEE model. Alternatively, population-specific differences between the cohorts (or between sub-cohorts of EPIC-Italy) cannot be excluded. For example, there are more smokers in the EPIC studies compared with the Generations Study and MCCS, which considerably affects DNA methylation. Also, there are different patterns in breastfeeding, number of pregnancies, and OC and HRT use across the studies, which may all affect the total lifetime estrogen exposure and DNA methylation. With the current evidence, we conclude that the methylation index developed does not directly predict ELEE.
We observed no evidence for reverse causation with a higher association between the MI and breast cancer risk for cases with a longer time to diagnosis (Fig. 4). In the meta-analysis, the association with breast cancer risk was only significant for the cases with the time to diagnosis greater than the median in the continuous MI model (OR = 1.05, 95% CI 1.01–1.10, P = 0.021, Additional file 2: Table S8). More work is needed using longitudinal studies to understand the dynamics of this MI over time.
In this study, we used an ELEE model based on a woman’s age at recruitment (premenopausal women) or age at menopause (postmenopausal women), age at menarche, number of pregnancies, and breastfeeding duration. The model does not include all the variables that can affect estrogen exposure, for example, menstrual cycle regularity, long-term pregnancies with miscarriage or abortion as outcome, and current use of HRT or OC. Hormonal risk factors are difficult to assess comprehensively via questionnaires; for example, the short-term outcome of being pregnant is increased estrogen levels and breast cancer risk, but in the long term, the estrogen levels and risk are reduced compared with nulliparous women [9, 10, 45]. Subtracting 1 year for each pregnancy instead of 9 months for each pregnancy did not materially change the results and might reflect the lifetime estrogen exposure better because there is a delay of 3 months on average before ovulatory cycling resumes. OC and HRT use is associated with elevated risk in current users, but it is not known how it will contribute to the lifetime estrogen exposure or DNA methylation. For this reason, we have not included OC and HRT use in our ELEE model. Other hormonal exposures accompanied by estrogens, such as progesterones, were not considered in this study. We acknowledge that the ELEE model is not a perfect model for cumulative estrogen exposure; however, it was hypothesized that if DNA methylation signature could be identified as an intermediate biological phenotype for the exposure, this might be more accurate measurement than questionnaire-based estimates.
We acknowledge the small sample size of the discovery EWAS in EPIC-Italy (n = 216) and potential false positive hits due to artefactual inflation of test statistics. We attempted to reduce the likelihood of false positive associations by correcting for multiple testing and restricting the MI signature to CpGs showing the largest changes in DNA methylation. Replication and validation are important steps to identify valid DNA methylation biomarkers. For validation of the MI and breast cancer risk, we used an independent method, targeted bisulfite sequencing, on a case-control study nested within a large independent study cohort (the Generations Study, n = 880) and a meta-analysis across four independent study cohorts (n = 2374). Another limitation that we identified was that not all 31 CpG sites are present on the updated Illumina HumanMethylation EPIC (850K) array, which precludes the possibility of including 850K studies in this analysis without changing the model.
Breast cancer risk assessment needs further improvement to be able to identify women at low or high risk of developing breast cancer that would warrant a preventive intervention. It remains to be explored if epigenetic signatures, in combination with other existing risk models, polygenic risk scores and breast density measurements, will improve breast cancer risk prediction and stratification. Furthermore, blood sampling is an accessible and less invasive method that is relatively easy to include into population screening. Targeted prevention approaches, including chemoprevention or lifestyle changes, for high-risk women might reduce the breast cancer incidence rate.
Conclusion
In this study, a DNA methylation signature in blood associated with breast cancer risk was identified. However, the methylation signature, although developed from ELEE associations in the EPIC-Italy cohort, was not associated with lifetime estrogen exposure in the subsequent cohorts analyzed. Further investigation is required to confirm the interaction between estrogen exposure and DNA methylation in blood, and how epigenetic signatures might improve risk assessment models.
Additional files
Supplementary material and methods. (DOCX 65 kb)
Supplementary Tables S1 to S9. (DOCX 118 kb)
Supplementary Figures S1–S4. (DOCX 2232 kb)
Acknowledgements
The authors acknowledge infrastructure support from the Imperial Experimental Cancer Medicine Centre, Cancer Research UK Imperial Centre, the National Institute for Health Research Imperial Biomedical Research Center, and the Ovarian Cancer Action Research Centre. The Generations Study is funded by Breast Cancer Now and the Institute of Cancer Research. The Institute of Cancer Research acknowledges funding to the National Institute for Health Research Biomedical Research Centre. We thank Breast Cancer Now for funding this work as part of Programme Funding to the Breast Cancer Now Toby Robins Research Centre. The authors would like to thank the study participants, study staff, and the doctors, nurses, and other healthcare staff and data providers who have contributed to all cohorts. EPIC Italy was supported by the Italian Association for Research on Cancer (IARC). The Melbourne Collaborative Cohort Study (MCCS) cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further augmented by Australian National Health and Medical Research Council grants 209057, 396414, and 1074383 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry and the Australian Institute of Health and Welfare, including the National Death Index and the Australian Cancer Database. The MCCS methylation work was supported by the National Health and Medical Research Council (grant number 1011618) and the Victorian Breast Cancer Research Consortium. The work performed by the Epigenetics Group at IARC was supported by grants from the Institut National du Cancer (INCa, France), the European Commission (EC) Seventh Framework Programme (FP7) Translational Cancer Research (TRANSCAN) Framework, the Fondation ARC pour la Recherche sur le Cancer (France), and la Ligue Nationale (Française) Contre le Cancer to ZH. The authors acknowledge the postdoctoral fellowship support (to SA) from the International Agency for Research on Cancer, partially supported by the EC FP7 Marie Curie Actions–People–Co-funding of regional, national, and international programmes (COFUND).
Funding
This work was funded by a Breast Cancer Now PhD scholarship (2014NovPhD403).
Availability of data and materials
The original EPIC-Italy 450K dataset supporting the results of this article is available in the Gene Expression Omnibus (GEO) repository, accession GSE51057. The targeted bisulfite sequencing datasets generated during and/or analysed during the current study are not publicly available due to ethical considerations but are available from the corresponding author on reasonable request.
Abbreviations
- BMI
Body mass index
- ELEE
Estimated lifetime estrogen exposure
- EPIC
The European Prospective Investigation into Cancer and Nutrition
- EWAS
Epigenome-wide association study
- FDR
False discovery rate
- HRT
Hormone replacement therapy
- IARC
International Agency for Research on Cancer
- MCCS
Melbourne Collaborative Cohort Study
- MI
Methylation index
- OC
Oral contraceptives
- QC
Quality control
Authors’ contributions
This study was conceived, designed, and interpreted by JMF, PV, and AJ. AJ performed all laboratory experiments and statistical analyses. Cohort study data from EPIC-Italy was provided by DP, GM, SG, CA, RT, MCG, FF, CS, SP, AM, and SP. Cohort study data from the Generations Study was provided by MJ, MS, NO, KT, NJ, OF, and AS. Data and analyses for MCCS for the meta-analysis was provided and conducted by VP, LB, P-AD, MCS, GGG, DRE, RLM, and GS. Data and analyses for EPIC-IARC for the meta-analysis was provided and conducted by SA, CC, VC, IR, and ZH. The manuscript was drafted by JMF and AJ, several authors contributed to the manuscript writing (OF, AS, MJ, MS, P-AD, GGG, and RLM), and all authors reviewed and approved the final manuscript.
Ethics approval and consent to participate
All study participants signed informed consent forms, and each cohort was approved by the national ethical review boards: The Generations Study: South Thames Multicentre Research Ethics Committee (reference MREC 03/01/014), and EPIC-Italy and EPIC-IARC: The ethics committees of the Human Genetics Foundation (HuGeF) and IARC respectively. MCCS: The MCCS study protocol was approved by the Cancer Council Victoria’s Human Research Ethics.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.International Agency for Research on Cancer. EPIC study. 2018. http://epic.iarc.fr. Accessed: September 2018.
- 2.Eccles SA, Aboagye EO, Ali S, Anderson AS, Armes J, Berditchevski F, et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast cancer research: BCR. 2013;15(5):R92. [DOI] [PMC free article] [PubMed]
- 3.Cancer Research UK. Breast cancer statistics 2018. https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/breast-cancer. Accessed: November 2018.
- 4.Endogenous Hormones Breast Cancer Collaborative Group Circulating sex hormones and breast cancer risk factors in postmenopausal women: reanalysis of 13 studies. Br J Cancer. 2011;105(5):709–722. doi: 10.1038/bjc.2011.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endogenous Hormones Breast Cancer Collaborative Group Sex hormones and risk of breast cancer in premenopausal women: a collaborative reanalysis of individual participant data from seven prospective studies. Lancet Oncol. 2013;14(10):1009–1019. doi: 10.1016/S1470-2045(13)70301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoemaker MJ, Folkerd EJ, Jones ME, Rae M, Allen S, Ashworth A, et al. Combined effects of endogenous sex hormone levels and mammographic density on postmenopausal breast cancer risk: results from the Breakthrough Generations Study. Br J Cancer. 2014;110(7):1898–1907. doi: 10.1038/bjc.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collaborative Group on Hormonal Factors in Breast Cancer Menarche, menopause, and breast cancer risk: individual participant meta-analysis, including 118 964 women with breast cancer from 117 epidemiological studies. Lancet Oncol. 2012;13(11):1141–1151. doi: 10.1016/S1470-2045(12)70425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horn J, Asvold BO, Opdahl S, Tretli S, Vatten LJ. Reproductive factors and the risk of breast cancer in old age: a Norwegian cohort study. Breast cancer research and treatment. 2013;139(1):237–243. doi: 10.1007/s10549-013-2531-0. [DOI] [PubMed] [Google Scholar]
- 9.Ewertz M, Duffy SW, Adami HO, Kvale G, Lund E, Meirik O, et al. Age at first birth, parity and risk of breast cancer: a meta-analysis of 8 studies from the Nordic countries. International journal of cancer. 1990;46(4):597–603. doi: 10.1002/ijc.2910460408. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein L, Pike MC, Ross RK, Judd HL, Brown JB, Henderson BE. Estrogen and sex hormone-binding globulin levels in nulliparous and parous women. Journal of the National Cancer Institute. 1985;74(4):741–745. [PubMed] [Google Scholar]
- 11.Collaborative Group on Hormonal Factors in Breast Cancer Breast cancer and breastfeeding: collaborative reanalysis of individual data from 47 epidemiological studies in 30 countries, including 50302 women with breast cancer and 96973 women without the disease. Lancet (London, England) 2002;360(9328):187–195. doi: 10.1016/S0140-6736(02)09454-0. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury R, Sinha B, Sankar MJ, Taneja S, Bhandari N, Rollins N, et al. Breastfeeding and maternal health outcomes: a systematic review and meta-analysis. Acta Paediatr. 2015;104(467):96–113. doi: 10.1111/apa.13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collaborative Group on Hormonal Factors in Breast Cancer Breast cancer and hormone replacement therapy: collaborative reanalysis of data from 51 epidemiological studies of 52,705 women with breast cancer and 108,411 women without breast cancer. Lancet. 1997;350(9084):1047–1059. doi: 10.1016/S0140-6736(97)08233-0. [DOI] [PubMed] [Google Scholar]
- 14.Million Women Study Collaborators Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet (London, England) 2003;362(9382):419–427. doi: 10.1016/S0140-6736(03)14065-2. [DOI] [PubMed] [Google Scholar]
- 15.Chlebowski RT, Rohan TE, Manson JE, Aragaki AK, Kaunitz A, Stefanick ML, et al. Breast cancer after use of estrogen plus progestin and estrogen alone: analyses of data from 2 women’s health initiative randomized clinical trials. JAMA oncology. 2015;1(3):296–305. doi: 10.1001/jamaoncol.2015.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlton BM, Rich-Edwards JW, Colditz GA, Missmer SA, Rosner BA, Hankinson SE, et al. Oral contraceptive use and mortality after 36 years of follow-up in the Nurses’ Health Study: prospective cohort study. BMJ (Clinical research ed) 2014;349:g6356. doi: 10.1136/bmj.g6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gierisch JM, Coeytaux RR, Urrutia RP, Havrilesky LJ, Moorman PG, Lowery WJ, et al. Oral contraceptive use and risk of breast, cervical, colorectal, and endometrial cancers: a systematic review. Cancer Epidemiol Biomarkers Prev. 2013;22(11):1931–1943. doi: 10.1158/1055-9965.EPI-13-0298. [DOI] [PubMed] [Google Scholar]
- 18.Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet (London, England) 2008;371(9612):569–578. doi: 10.1016/S0140-6736(08)60269-X. [DOI] [PubMed] [Google Scholar]
- 19.Endogenous Hormones Breast Cancer Collaborative Group Body mass index, serum sex hormones, and breast cancer risk in postmenopausal women. Journal of the National Cancer Institute. 2003;95(16):1218–1226. doi: 10.1093/jnci/djg022. [DOI] [PubMed] [Google Scholar]
- 20.Romieu I, Scoccianti C, Chajes V, de Batlle J, Biessy C, Dossus L, et al. Alcohol intake and breast cancer in the European prospective investigation into cancer and nutrition. International journal of cancer. 2015;137(8):1921–1930. doi: 10.1002/ijc.29469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collaborative Group on Hormonal Factors in Breast Cancer Alcohol, tobacco and breast cancer--collaborative reanalysis of individual data from 53 epidemiological studies, including 58,515 women with breast cancer and 95,067 women without the disease. Br J Cancer. 2002;87(11):1234–1245. doi: 10.1038/sj.bjc.6600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahmann PH, Friedenreich C, Schuit AJ, Salvini S, Allen NE, Key TJ, et al. Physical activity and breast cancer risk: the European Prospective Investigation into Cancer and Nutrition. Cancer Epidemiol Biomarkers Prev. 2007;16(1):36–42. doi: 10.1158/1055-9965.EPI-06-0582. [DOI] [PubMed] [Google Scholar]
- 23.Johansson A, Flanagan JM. Epigenome-wide association studies for breast cancer risk and risk factors. Trends in cancer research. 2017;12:19–28. [PMC free article] [PubMed] [Google Scholar]
- 24.van Veldhoven K, Polidoro S, Baglietto L, Severi G, Sacerdote C, Panico S, et al. Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis. Clinical epigenetics. 2015;7:67. doi: 10.1186/s13148-015-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan K, Garcia-Closas M, Orr N, Fletcher O, Jones M, Ashworth A, et al. Intragenic ATM methylation in peripheral blood DNA as a biomarker of breast cancer risk. Cancer research. 2012;72(9):2304–2313. doi: 10.1158/0008-5472.CAN-11-3157. [DOI] [PubMed] [Google Scholar]
- 26.Joo JE, Dowty JG, Milne RL, Wong EM, Dugue PA, English D, et al. Heritable DNA methylation marks associated with susceptibility to breast cancer. Nature communications. 2018;9(1):867. doi: 10.1038/s41467-018-03058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shenker NS, Ueland PM, Polidoro S, van Veldhoven K, Ricceri F, Brown R, et al. DNA methylation as a long-term biomarker of exposure to tobacco smoke. Epidemiology. 2013;24(5):712–716. doi: 10.1097/EDE.0b013e31829d5cb3. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Marioni RE, Hedman AK, Pfeiffer L, Tsai PC, Reynolds LM, et al. A DNA methylation biomarker of alcohol consumption. Molecular psychiatry. 2016. [DOI] [PMC free article] [PubMed]
- 29.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horvath S. DNA methylation age of human tissues and cell types. Genome biology. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Schottker B, Ordonez-Mena J, Holleczek B, Yang R, Burwinkel B, et al. F2RL3 methylation, lung cancer incidence and mortality. International journal of cancer. 2015;137(7):1739–1748. doi: 10.1002/ijc.29537. [DOI] [PubMed] [Google Scholar]
- 33.Baglietto L, Ponzi E, Haycock P, Hodge A, Bianca Assumma M, Jung CH, et al. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. International journal of cancer. 2017;140(1):50–61. doi: 10.1002/ijc.30431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fox M, Berzuini C, Knapp LA. Cumulative estrogen exposure, number of menstrual cycles, and Alzheimer’s risk in a cohort of British women. Psychoneuroendocrinology. 2013;38(12):2973–2982. doi: 10.1016/j.psyneuen.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Atsma F, van der Schouw YT, Grobbee DE, Kors JA, Bartelink ML. Lifetime endogenous estrogen exposure and electrocardiographic frontal T axis changes in postmenopausal women. Maturitas. 2009;63(4):347–351. doi: 10.1016/j.maturitas.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Hesson J. Cumulative estrogen exposure and prospective memory in older women. Brain and cognition. 2012;80(1):89–95. doi: 10.1016/j.bandc.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Jansen SC, Temme EH, Schouten EG. Lifetime estrogen exposure versus age at menopause as mortality predictor. Maturitas. 2002;43(2):105–112. doi: 10.1016/S0378-5122(02)00183-4. [DOI] [PubMed] [Google Scholar]
- 38.Pike MC, Krailo MD, Henderson BE, Casagrande JT, Hoel DG. ‘Hormonal’ risk factors, ‘breast tissue age’ and the age-incidence of breast cancer. Nature. 1983;303(5920):767–770. doi: 10.1038/303767a0. [DOI] [PubMed] [Google Scholar]
- 39.Smith CA, McCleary CA, Murdock GA, Wilshire TW, Buckwalter DK, Bretsky P, et al. Lifelong estrogen exposure and cognitive performance in elderly women. Brain and cognition. 1999;39(3):203–218. doi: 10.1006/brcg.1999.1078. [DOI] [PubMed] [Google Scholar]
- 40.Flanagan JM, Brook MN, Orr N, Tomczyk K, Coulson P, Fletcher O, et al. Temporal stability and determinants of white blood cell DNA methylation in the breakthrough generations study. Cancer Epidemiol Biomarkers Prev. 2015;24(1):221–229. doi: 10.1158/1055-9965.EPI-14-0767. [DOI] [PubMed] [Google Scholar]
- 41.Ambatipudi S, Horvath S, Perrier F, Cuenin C, Hernandez-Vargas H, Le Calvez-Kelm F, et al. DNA methylome analysis identifies accelerated epigenetic ageing associated with postmenopausal breast cancer susceptibility. European journal of cancer (Oxford, England : 1990) 2017;75:299–307. doi: 10.1016/j.ejca.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Severi G, Southey MC, English DR, Jung CH, Lonie A, McLean C, et al. Epigenome-wide methylation in DNA from peripheral blood as a marker of risk for breast cancer. Breast cancer research and treatment. 2014;148(3):665–673. doi: 10.1007/s10549-014-3209-y. [DOI] [PubMed] [Google Scholar]
- 43.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics (Oxford, England) 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersson TM, Johansson AL, Fredriksson I, Lambe M. Cancer during pregnancy and the postpartum period: a population-based study. Cancer. 2015;121(12):2072–2077. doi: 10.1002/cncr.29325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material and methods. (DOCX 65 kb)
Supplementary Tables S1 to S9. (DOCX 118 kb)
Supplementary Figures S1–S4. (DOCX 2232 kb)
Data Availability Statement
The original EPIC-Italy 450K dataset supporting the results of this article is available in the Gene Expression Omnibus (GEO) repository, accession GSE51057. The targeted bisulfite sequencing datasets generated during and/or analysed during the current study are not publicly available due to ethical considerations but are available from the corresponding author on reasonable request.