Abstract
Background
Systemic treatment of advanced non-small cell lung cancer (NSCLC) has changed dramatically since the introduction of targeted therapies. The analysis of circulating tumor DNA (ctDNA) is a valuable approach to monitor the clonal evolution of tumors during treatment with EGFR-tyrosine kinase inhibitors (TKIs) and to detect resistance mutations.
Case presentation
A NSCLC patient with exon 19 deletion (ex19del) of EGFR was treated with osimertinib after multiple lines of treatment and obtained a partial response that lasted over 26 months. Blood was collected at each visit and ctDNA was extracted to monitor ex19del by digital droplet PCR. Within a few weeks from the beginning of osimertinib, ex19del disappeared from plasma but appeared again and steadily increased a few months later anticipating tumor progression. Interestingly, the change in ex19del was much more pronounced than other mutations, since T790M appeared 3 months after the increase of ex19del, and C797S was detectable a few weeks before clinical disease progression. Then the patient received cytotoxic chemotherapy, which was associated with a decrease in ex19del and disappearance of T790M and C797S; however, at disease progression, all EGFR mutations increased again in plasma together with MET amplification which was detected by NGS.
Conclusions
The measurement of ex19del changes in ctDNA is a simple and sensitive approach to monitor clinical outcome to osimertinib and, potentially, to other therapeutic interventions.
Keywords: Circulating tumor DNA, NSCLC, EGFR mutations, Treatment monitoring, EGFR-TKIs, Digital droplet PCR, NGS
Background
The presence of activating EGFR mutations, mainly ex19del, strongly predicts response to EGFR-TKIs; however, in 50–60% of these patients, resistance is acquired through the development of T790M, a second missense mutation of EGFR, which is indeed targeted by osimertinib [1]. Some patients retain EGFR oncogene addiction even after progression to osimertinib, as they may develop the C797S resistance mutation [2, 3].
The analysis of circulating tumor DNA (ctDNA) is a valuable approach to monitor the clonal evolution of tumors during treatment and to detect mutations capable of inducing resistance to EGFR-TKIs [4]. Even if the analysis of tumor tissue is required to select the appropriate treatment, it is indeed associated with several limitations, including invasiveness, inability to comprehensively capture tumor heterogeneity, and tissue availability for mutational testing. For these reasons, the analysis of EGFR mutations in ctDNA has recently emerged as a reliable, non-invasive alternative approach, showing high concordance with tissue molecular profile, with good sensitivity (> 65%) and high specificity (> 88%) [5].
Here, we report a case of ctDNA monitoring during osimertinib treatment and after disease progression, which provides evidence of the reliability of time-dependent changes in EGFR activating mutation to predict response to treatment.
Case presentation
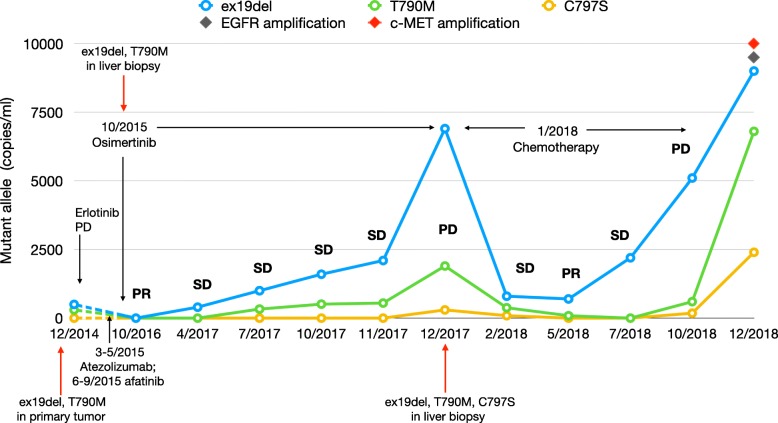
In October 2012, a 46-year-old woman was referred to our center for the presence of a large mass (50 × 70 mm) in the superior lobe of the left lung with homolateral pleural effusion. The patient was never smoker, without family history of cancer and without comorbidity. The cytological diagnosis was made using a CT-guided fine needle aspiration of the primary tumor and revealed an adenocarcinoma of the lung (TTF1+, CK7+) with the EGFR ex19del mutation. A PET-CT demonstrated the presence of liver and bone metastases and a nodule in the right breast, confirmed as a metastasis by fine needle aspiration. The patient received zoledronic acid 4 mg every 28 days and gefitinib 250 mg daily since November 2012 obtaining a partial response (PR). In August 2013, a disease progression (PD) was documented, with an increase in size of the primary tumor and size and number of liver metastases. A brain MRI revealed the presence of two cortical nodules, which were treated with stereotactic radiotherapy. The patient was enrolled in the IMPRESS trial and received 6 cycles of cisplatin and pemetrexed plus gefitinib obtaining again a PR that lasted until June 2014. Thereafter, a new lung metastasis appeared in the superior lobe of the left lung and the mammary nodule increased in dimensions. From June 2014 to December 2014 the patient received erlotinib 150 mg daily obtaining an initial stabilization of the disease (SD); however, within 6 months, she experienced again a PD with the increase of the mammary nodule and the appearance of a new bone metastasis in the sacrum. In December 2014, EGFR ex19del and T790M mutations were detectable in a new needle biopsy of the primary tumor; only at this time a digital PCR-based method was available for the analysis of circulating tumor DNA (ctDNA). Briefly, the method was optimized in order to recover a suitable amount of ctDNA for molecular analysis from 3 ml of plasma using the QIAmp Circulating Nucleic Acid Kit (Qiagen®, Valencia, CA). ctDNA was examined using the Prime PCR Probe Assay on a QX100™ Droplet Digital™ PCR System (BioRad®, Hercules, CA) for EGFR mutations (ex19del, T790M, and C797S) [6]. The ctDNA sample was considered as EGFR mutant when at least one droplet was above the fluorescence intensity threshold of 3000 and results were reported as copies/ml. The first plasma specimen was obtained in December 2014 and confirmed the presence of ex19del and T790M mutations (480 and 260 copies/ml, respectively; Fig. 1). The patient was treated with atezolizumab from March to May 2015 and received stereotactic radiotherapy on the lung primary tumor and on metastases of the left superior lobe, breast and bone (sacrum and D10). Due to PD, the patient was given afatinib 40 mg daily from June 2015 to September 2015, but she experienced a further PD with an increase in size of the primary tumor and lung nodules, the appearance of new multiple bilateral lung metastases, lymphangitic infiltration and liver metastases. At this time, the presence of ex19del and T790 M was again demonstrated in a biopsy of a liver metastasis. Since osimertinib was available, the patient was enrolled in the ASTRIS trial and given 80 mg daily starting from October 2015 with an immediate clinical benefit. At the first evaluation a PR was documented with disappearance of most of the lung nodules and lymphangitic infiltration, reduction of the primary tumor and of liver metastases as well. A disappearance of ex19del or T790M was demonstrated in ctDNA in a blood sample obtained in October 2016. However, in April 2017, ex19del appeared again (400 copies/ml) and in July 2017 it increased to 1000 copies/ml, while T790M was also detectable (330 copies/ml, Fig. 1). Then, ex19del and T790 M continued to increase to, respectively, 1600 and 510 copies/ml in October 2017, 2100 and 550 copies/ml in November 2017, and 6900 and 1900 copies/ml in December 2017.
Fig. 1.
Changes of EGFR mutations (copies/ml) in ctDNA of patient during treatments. PR: partial response; SD: stable disease; PD progressive disease. The numbers before the year indicate the months
A radiological progression was demonstrated with increase in size and number of liver metastases in December 2017. The patient underwent a new liver biopsy that confirmed the PD and the presence of ex19del and T790M, whereas the ctDNA showed also the appearance of C797S mutation (170 copies/ml), in addition to ex19del and T790M. The patient started chemotherapy with carboplatin and pemetrexed and in February 2018 she obtained an SD associated with a drop of ex19del (800 copies/ml), T790 M (380 copies/ml), and C797S (90 copies/ml) and then a PR in May 2018, with disappearance of C797S and reduction of ex19del (760 copies/ml) and T790M (90 copies/ml). In July 2018, however, ex19del strongly increased to 2200 copies/ml, even though T790M and C797S were undetectable. Finally, in October 2018, when a PD was documented, ex19del increased to 5100 copies/ml, while T790M and C797S appeared again in plasma with 600 and 180 copies/ml, respectively (Fig. 1). At this time a NextSeq 550 NGS platform (Illumina®, San Diego, CA) was available to analyse ctDNA by the AVENIO ctDNA Expanded Kit (Roche®, Pleasanton, CA). A plasma sample collected in December 2018 and analysed by NGS confirmed the presence of the ex19del, T790M and C797S and found, in addition, EGFR and c-MET amplifications, which were not present in tissue in the last re-biopsy of December 2017.
Discussion and conclusions
Molecular mechanisms underlying the acquired resistance to first- and second-generation EGFR-TKIs have been characterised and include MET amplification, T790M, BRAF and PIK3CA mutations, AXL overexpression and transformation to SCLC or squamous cell carcinoma [4, 7]. Moreover, mechanisms of acquired resistance to third generation EGFR-TKIs include L718Q or C797S EGFR mutations [8, 9]; in particular, the latter is found in about one third of cancers resistant to osimertinib [10]. The present patient received osimertinib after multiple lines of treatment and obtained a noteworthy PR that lasted over 26 months. Interestingly, clinical outcome was correlated with the number of ex19del ctDNA copies in plasma as it disappeared within a few weeks from the beginning of osimertinib and appeared again and steadily increased thereafter, anticipating tumor progression. The same pattern was seen during chemotherapy, with a decrease in ex19del during disease response and its relapse together with T790M and C797S at PD, suggesting that chemotherapy does not erase cellular clones bearing resistance mutations. Therefore, our data provide evidence that the changes in ex19del in ctDNA are more sensitive than T790M or C797S to monitor osimertinib response and resistance, as T790M appeared 3 months after the increase of activating mutation, and C797S was detectable just before PD. Thus, the efficacy of osimertinib is not simply predicted by T790M; accordingly, previous studies did not show a correlation between T790M levels and benefit of treatment [11–13].
The amount of T790M and C797S in ctDNA at disease progression to gefitinib and osimertinib was markedly lower than ex19del, despite their role in resistance to EGFR-TKIs. This finding is complex to explain due to the primary role of these mutations in driving tumor progression, when it should be expected a similar number of ex19del/T790M/C797S copies in ctDNA. However, a reasonable justification is the presence of multiple metastatic lesions bearing different resistance mutations as a result of clonal evolution.
The present evidence suggests that ex19del reflects tumor biology better than other EGFR molecular alterations, and can be used as a sensitive biomarker to monitor disease outcome. Amplification of EGFR has been documented in cells [14] and in patients resistant to osimertinib [15] and has been shown in one plasma sample of the present patient by NGS; this finding may explain, at least in part, the increase in ex19del copies in ctDNA at each disease progression. Therefore, further analysis should be done to better understand intratumor dynamic clonality as a result of multiple treatments administered to patients. In this respect, the non-invasive molecular analysis of ctDNA may allow a minimally invasive monitoring of disease and provide important information on cancer mutational landscape and emergence of druggable mutations, including c-MET as in the present patient.
In conclusion, this case report provides evidence of the significant activity of osimertinib in a T790M-mutated patient and of the involvement of C797S, EGFR and c-MET amplifications in drug resistance. The changes in ex19del activating mutation in ctDNA effectively predicted disease outcome and was easily detectable due to its larger amount with respect to T790M and C797S. Thus, ex19del measurement is a non-invasive approach to monitor clinical outcome and its increase suggests to perform additional molecular analysis to discover the druggable mutation responsible of drug resistance and adapt treatment strategy.
Acknowledgements
Not applicable.
Funding
This work was supported in part by a research funding granted to RD by Fondazione Cassa di Risparmio di Lucca, Italy and unrestricted research grant from AstraZeneca.
Availability of data and materials
The authors declare that all data supporting the findings of this study are available within the article. No datasets were generated or analysed during the current study.
Abbreviations
- CT
Computed tomography
- ctDNA
Circulating tumor DNA
- EGFR
Epidermal Growth Factor Receptor
- ex19del
Exon 19 deletion of EGFR
- NSCLC
Non-small cell lung cancer
- PD
Disease progression
- PET
Positron emission tomography
- PR
Partial response
- SD
Stabilization of disease
- TKIs
Tyrosine kinase inhibitors
Authors’ contributions
MDR, ER, CC, GP, SC, SV, AC, RD, IP made substantial contributions to conception and design. ER, MDR, SC, RD made substantial contributions to the laboratory analysis and interpretation of data; CC, GP, SV, AC, IP made substantial contributions to the acquisition of clinical data and their interpretation. ER, MDR, IP, RD have been involved in drafting the manuscript. CC, GP, SC, SV, AC, RD, IP have been involved in revising the paper critically for important intellectual content. ER, MDR, CC, GP, SC, SV, AC, RD, IP gave their final approval of the version to be published. Each author participated sufficiently in the work to take public responsibility for appropriate portions of the content; all the authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. RD is responsible for the financial support for the project leading to this publication.
Ethics approval and consent to participate
This case report is part of a translational study approved by the Ethics Committee of University Hospital (Pisa, Italy). A written consent has been obtained from the participant.
Consent for publication
A written consent has been obtained from the participant for her clinical details to be published in this study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marzia Del Re, Email: marzia.delre@gmail.com.
Eleonora Rofi, Email: eleonora0106@inwind.it.
Carla Cappelli, Email: carlacappelli1@gmail.com.
Gianfranco Puppo, Email: g.puppo@hotmail.it.
Stefania Crucitta, Email: stefania.crucitta@gmail.com.
Simona Valeggi, Email: simona10.74@gmail.com.
Antonio Chella, Email: anto.kell@tiscali.it.
Romano Danesi, Phone: +39050992632, Email: romano.danesi@unipi.it.
Iacopo Petrini, Email: iacopo.petrini@unipi.it.
References
- 1.Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WS, et al. Osimertinib or platinum-Pemetrexed in EGFR T790M-positive lung Cancer. N Engl J Med. 2017;376(7):629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, Lai Z, Markovets A, Vivancos A, Kuang Y, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21(6):560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, Feeney N, Sholl LM, Dahlberg SE, Redig AJ, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4(11):1527–34. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Re M, Vasile E, Falcone A, Danesi R, Petrini I. Molecular analysis of cell-free circulating DNA for the diagnosis of somatic mutations associated with resistance to tyrosine kinase inhibitors in non-small-cell lung cancer. Expert Rev Mol Diagn. 2014;14(4):453–468. doi: 10.1586/14737159.2014.908120. [DOI] [PubMed] [Google Scholar]
- 5.Passiglia F, Rizzo S, Maio MD, Galvano A, Badalamenti G, Listi A, Gulotta L, Castiglia M, Bazan V, Russo A, et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: a systematic review and meta-analysis. Sci Rep. 2018;8(1):13379. doi: 10.1038/s41598-018-30780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Re M, Tiseo M, Bordi P, D'Incecco A, Camerini A, Petrini I, Lucchesi M, Inno A, Spada D, Vasile E, et al. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget. 2017;8(8):13611–13619. doi: 10.18632/oncotarget.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim SM, Syn NL, Cho BC, Soo RA. Acquired resistance to EGFR targeted therapy in non-small cell lung cancer: mechanisms and therapeutic strategies. Cancer Treat Rev. 2018;65:1–10. doi: 10.1016/j.ctrv.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Bersanelli M, Minari R, Bordi P, Gnetti L, Bozzetti C, Squadrilli A, Lagrasta CA, Bottarelli L, Osipova G, Capelletto E, et al. L718Q mutation as new mechanism of acquired resistance to AZD9291 in EGFR-mutated NSCLC. J Thorac Oncol. 2016;11(10):e121–e123. doi: 10.1016/j.jtho.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 9.Planchard D, Loriot Y, Andre F, Gobert A, Auger N, Lacroix L, Soria JC. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Ann Oncol. 2015;26(10):2073–2078. doi: 10.1093/annonc/mdv319. [DOI] [PubMed] [Google Scholar]
- 10.Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, Kurtz DM, Stehr H, Scherer F, Karlovich CA, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung Cancer. N Engl J Med. 2018;378(2):113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 12.Remon J, Caramella C, Jovelet C, Lacroix L, Lawson A, Smalley S, Howarth K, Gale D, Green E, Plagnol V, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol. 2017;28(4):784–790. doi: 10.1093/annonc/mdx017. [DOI] [PubMed] [Google Scholar]
- 13.Del Re M, Bordi P, Petrini I, Rofi E, Mazzoni F, Belluomini L, Vasile E, Restante G, Di Costanzo F, Falcone A, et al. Patients with NSCLC may display a low ratio of p.T790M vs. activating EGFR mutations in plasma at disease progression: implications for personalised treatment. Oncotarget. 2017;8(49):86056–86065. doi: 10.18632/oncotarget.20947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakatani K, Yamaoka T, Ohba M, Fujita KI, Arata S, Kusumoto S, Taki-Takemoto I, Kamei D, Iwai S, Tsurutani J, et al. KRAS and EGFR amplifications mediate resistance to rociletinib and osimertinib in acquired afatinib-resistant NSCLC harboring exon 19 deletion/T790M in EGFR. Mol Cancer Ther. 2018;18(1):112–26. doi: 10.1158/1535-7163.MCT-18-0591. [DOI] [PubMed] [Google Scholar]
- 15.Knebel FH, Bettoni F, Shimada AK, Cruz M, Alessi JV, Negrao MV, Reis LFL, Katz A, Camargo AA. Sequential liquid biopsies reveal dynamic alterations of EGFR driver mutations and indicate EGFR amplification as a new mechanism of resistance to osimertinib in NSCLC. Lung Cancer. 2017;108:238–241. doi: 10.1016/j.lungcan.2017.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article. No datasets were generated or analysed during the current study.