Abstract
Background
Melanoma has one of the fastest rising incidence rates of any cancer. It accounts for a small percentage of skin cancer cases but is responsible for the majority of skin cancer deaths. Early detection and treatment is key to improving survival; however, anxiety around missing early cases needs to be balanced against appropriate levels of referral and excision of benign lesions. Used in conjunction with clinical or dermoscopic suspicion of malignancy, or both, reflectance confocal microscopy (RCM) may reduce unnecessary excisions without missing melanoma cases.
Objectives
To determine the diagnostic accuracy of reflectance confocal microscopy for the detection of cutaneous invasive melanoma and atypical intraepidermal melanocytic variants in adults with any lesion suspicious for melanoma and lesions that are difficult to diagnose, and to compare its accuracy with that of dermoscopy.
Search methods
We undertook a comprehensive search of the following databases from inception up to August 2016: Cochrane Central Register of Controlled Trials; MEDLINE; Embase; and seven other databases. We studied reference lists and published systematic review articles.
Selection criteria
Studies of any design that evaluated RCM alone, or RCM in comparison to dermoscopy, in adults with lesions suspicious for melanoma or atypical intraepidermal melanocytic variants, compared with a reference standard of either histological confirmation or clinical follow‐up.
Data collection and analysis
Two review authors independently extracted all data using a standardised data extraction and quality assessment form (based on QUADAS‐2). We contacted authors of included studies where information related to the target condition or diagnostic threshold were missing. We estimated summary sensitivities and specificities per algorithm and threshold using the bivariate hierarchical model. To compare RCM with dermoscopy, we grouped studies by population (defined by difficulty of lesion diagnosis) and combined data using hierarchical summary receiver operating characteristic (SROC) methods. Analysis of studies allowing direct comparison between tests was undertaken. To facilitate interpretation of results, we computed values of specificity at the point on the SROC curve with 90% sensitivity as this value lies within the estimates for the majority of analyses. We investigated the impact of using a purposely developed RCM algorithm and in‐person test interpretation.
Main results
The search identified 18 publications reporting on 19 study cohorts with 2838 lesions (including 658 with melanoma), which provided 67 datasets for RCM and seven for dermoscopy. Studies were generally at high or unclear risk of bias across almost all domains and of high or unclear concern regarding applicability of the evidence. Selective participant recruitment, lack of blinding of the reference test to the RCM result, and differential verification were particularly problematic. Studies may not be representative of populations eligible for RCM, and test interpretation was often undertaken remotely from the patient and blinded to clinical information.
Meta‐analysis found RCM to be more accurate than dermoscopy in studies of participants with any lesion suspicious for melanoma and in participants with lesions that were more difficult to diagnose (equivocal lesion populations). Assuming a fixed sensitivity of 90% for both tests, specificities were 82% for RCM and 42% for dermoscopy for any lesion suspicious for melanoma (9 RCM datasets; 1452 lesions and 370 melanomas). For a hypothetical population of 1000 lesions at the median observed melanoma prevalence of 30%, this equated to a reduction in unnecessary excisions with RCM of 280 compared to dermoscopy, with 30 melanomas missed by both tests. For studies in equivocal lesions, specificities of 86% would be observed for RCM and 49% for dermoscopy (7 RCM datasets; 1177 lesions and 180 melanomas). At the median observed melanoma prevalence of 20%, this reduced unnecessary excisions by 296 with RCM compared with dermoscopy, with 20 melanomas missed by both tests. Across all populations, algorithms and thresholds assessed, the sensitivity and specificity of the Pellacani RCM score at a threshold of three or greater were estimated at 92% (95% confidence interval (CI) 87 to 95) for RCM and 72% (95% CI 62 to 81) for dermoscopy.
Authors' conclusions
RCM may have a potential role in clinical practice, particularly for the assessment of lesions that are difficult to diagnose using visual inspection and dermoscopy alone, where the evidence suggests that RCM may be both more sensitive and specific in comparison to dermoscopy. Given the paucity of data to allow comparison with dermoscopy, the results presented require further confirmation in prospective studies comparing RCM with dermoscopy in a real‐world setting in a representative population.
Keywords: Adult; Humans; Biopsy; Dermoscopy; Melanoma; Melanoma/diagnostic imaging; Melanoma/pathology; Microscopy, Confocal; Microscopy, Confocal/methods; Sensitivity and Specificity; Skin; Skin/pathology; Skin Neoplasms; Skin Neoplasms/diagnostic imaging; Skin Neoplasms/pathology
Plain language summary
What is the diagnostic accuracy of the imaging test reflectance confocal microscopy (RCM) for the detection of melanoma in adults?
What was the aim of the review?
The aim of this Cochrane Review was to find out how accurate reflectance confocal microscopy (RCM) was on its own and used in addition to dermoscopy compared to dermoscopy alone for diagnosing melanoma. Review authors in Cochrane included 18 publications to answer this question.
Why is improving the diagnosis of melanoma important?
Melanoma is one of the most dangerous forms of skin cancer. Not recognising a melanoma when it is present (called a false negative test result) delays surgery to remove it, risking cancer spreading to other parts in the body and possibly death. Diagnosing a skin lesion as a melanoma when it is not present (called a false positive result) may result in unnecessary surgery, further investigations, and patient anxiety.
What did the review study?
Microscopic techniques are used by skin cancer specialists to allow a more detailed, magnified examination of suspicious skin lesions than can be achieved using the naked eye alone. Currently, dermoscopy (a handheld device using natural light) can be used as part of the clinical examination of suspicious skin lesions. RCM is a new microscopic technique (a handheld device or static unit using infrared light) that can visualise deeper layers of the skin compared to dermoscopy. Both techniques are painless procedures, but RCM is more expensive, time consuming, and requires additional training. Dermoscopy can be used by general practitioners whereas RCM is likely to only be used by secondary care specialists in people who have been referred with a lesion suspicious for skin cancer. We sought to find out whether RCM should be used instead of, or in addition to, dermoscopy, to diagnose melanoma in any suspicious skin lesion or only in particularly difficult to diagnose skin lesions.
What were the main results of the review?
The review included 18 publications reporting data for 19 groups of participants with lesions suspected of melanoma. The main results were based on 16 of the 19 datasets (sets of information and results).
The review included nine datasets with 1452 lesions in people with any suspicious skin lesion, three of which compared RCM to dermoscopy. The results suggested that in 1000 lesions, of which 300 (30%) actually are melanoma:
‐ an estimated 396 would have an RCM result indicating melanoma was present, and of these, 126 (32%) would not be melanoma (false positive results); ‐ in the same group of 1000 lesions, dermoscopy would produce 406 false positive results, meaning RCM would avoid unnecessary surgery in 280 lesions compared to dermoscopy; ‐ of the 604 lesions with an RCM result indicating that melanoma was not present (and 324 lesions with a dermoscopy result indicating that melanoma was not present), 30 would actually be melanoma (false negative results). This equated to a false negative rate of 5% for RCM and 9% for dermoscopy.
The review also included seven datasets with 1177 lesions in people with particularly difficult to diagnose skin lesions, three of which compared RCM to dermoscopy. The results suggested that if skin specialists used RCM in a group of 1000 lesions, of which 200 (20%) were actually melanoma:
‐ an estimated 292 would have an RCM result indicating melanoma was present, and of these, 112 (38%) would not be melanoma (false positive results); ‐ in the same group of 1000 lesions, dermoscopy would produce 408 false positive results, meaning RCM would avoid unnecessary surgery in 296 lesions compared to dermoscopy; ‐ of the 708 lesions with an RCM result indicating that melanoma was not present (and 412 lesions with a dermoscopy result indicating that melanoma was not present), 20 would actually have melanoma (false negative results). This equates to a false negative rate of 3% for RCM and 5% for dermoscopy.
How reliable were the results of the studies of this review?
In all included studies, the diagnosis of melanoma was made by lesion biopsy (RCM/dermoscopy positive) (a biopsy involves taking a sample of body cells and examining them under a microscope), and the absence of melanoma was confirmed by biopsy or by follow‐up over time to make sure the skin lesion remained negative for melanoma (RCM/dermoscopy negative)*. This is likely to have been a reliable method for deciding whether people really had melanoma. Only a small number of studies compared the accuracy of dermoscopy and RCM. Most were conducted by specialist research teams with high levels of experience with RCM. Therefore, RCM may have appeared more accurate than it actually was. Participants in the nine studies of any suspicious lesion may have had very obvious disease compared to that seen in practice leading to a lower number of false positive results than would actually occur. It is not possible to recommend a definition of a positive RCM test that will reliably produce the results presented here due to differences between studies.
Who do the results of this review apply to?
Eleven studies were undertaken in Europe (61%), with the remainder undertaken in Oceania, North America, or more than one continent. Mean age ranged from 39 to 54.7 years. The percentage of people with melanoma ranged between 1.9% and 41.5% (a median (midpoint reading) of 19% for difficult to diagnose skin lesions and 32% for any suspicious lesion). The majority of studies only included people with certain types of skin lesion. In many studies, it was not clear what tests participants had received before RCM.
What are the implications of this review?
RCM appears to be an accurate test for identifying melanoma, and it may reduce the number of people receiving unnecessary surgery by up to three‐quarters compared to dermoscopy. There is considerable variation and uncertainty in results and in study conduct, reducing the reliability of findings. Use of RCM may be of most benefit in people with particularly difficult to diagnose lesions rather than people with any lesion suspicious for melanoma. Further research comparing RCM and dermoscopy in well described groups of people with difficult to diagnose skin lesions is needed.
How up‐to‐date is this review?
The review authors searched for and used studies published up to August 2016.
*In these studies, biopsy or clinical follow‐up were the reference standards (means of establishing final diagnoses).
Summary of findings
Summary of findings'. 'Summary of findings table.
| Question: | What was the diagnostic accuracy of reflectance confocal microscopy for the detection of cutaneous invasive melanoma and atypical intraepidermal melanocytic variants in adults? | |||||
| Population: | Adults with lesions suspicious for melanoma, including:
|
|||||
| Index test: | RCM | |||||
| Comparator test: | Dermoscopy | |||||
| Target condition: | Cutaneous invasive melanoma and atypical intraepidermal melanocytic variants | |||||
| Reference standard: | Histology with or without long‐term follow‐up | |||||
| Action: | If accurate, negative results of RCM will stop participants having unnecessary excision of skin lesions. | |||||
| Quantity of evidence | ||||||
| Number of cohorts | 19a | Total lesions with test results |
2838 | Total with melanoma |
658 | |
| Limitations | ||||||
| Risk of bias: | High risk for participant selection from exclusion of some difficult to diagnose types of lesion (8/20). High risk for the index test from data driven RCM threshold (4/20). High risk from inadequate reference standard (4/20) and unclear risk as it was not clear that the reference standard was interpreted blind to the RCM results in 18/20 studies. High risk from differential verification (6/20), timing of tests was not mentioned in 11/20. | |||||
| Applicability of evidence to question: | High concern from narrowly defined populations (12/20) and multiple lesions per participant (7/20). High concern for RCM applicability from blinded interpretation of images (10/20). Studies were dominated by 1 particularly expert research group (15/20). Little information was given concerning the expertise of the histopathologist. | |||||
| Findings: | All analyses were undertaken on subgroups of the studies | |||||
|
Test: RCM using RCM score algorithm at threshold ≥ 3 or likely ≥ 3 regardless of population | ||||||
| Datasets | Lesions | Melanomas | Sensitivity (95% CI) | Specificity (95% CI) | ||
| 6 | 1209 | 296 | 92% (87 to 95) | 72% (62 to 81) | ||
| Consistency: significant heterogeneity in specificity between studies. Includes both equivocal (4) and 'any suspicious lesion' (2) populations; both in‐person (3) and image based (3). | ||||||
| Numbers observed in a cohort of 1000 lesions being tested2 | ||||||
| Prevalence | True positive | False negative | False positive | True negative | ||
| (received necessary excision) | (did not receive required excision) | (inappropriately received excision) | (appropriately did not receive excision) | |||
| At prevalence 13% | 120 | 10 | 244 | 626 | ||
| At prevalence 23% | 212 | 18 | 216 | 554 | ||
| At prevalence 39% | 359 | 31 | 171 | 439 | ||
| Test: RCM versus dermoscopy:b any algorithm or threshold in 'any lesion suspicious for melanoma' populations [dermoscopy data denoted in brackets] | ||||||
| Datasets | Lesions | Melanomas |
Sensitivity (fixed) RCM [dermoscopy] |
Specificity RCM [dermoscopy] |
||
| 9 [3] | 1452 [451] | 370 [160] | 90% [90%] | 82% [42%] | ||
| Numbers observed in a cohort of 1000 lesions being testedb,c,d | ||||||
| Prevalence | True positive | False negative | False positive | True negative | ||
| (received necessary excision) | (did not receive required excision) | (inappropriately received excision) | (appropriately did not receive excision) | |||
| At prevalence 26% | 234 | 26 | 133 [429] ↓296 |
607 [311] ↑296 |
||
| At prevalence 30% | 270 | 30 | 126 [406] ↓280 |
574 [294] ↑280 |
||
| At prevalence 36% | 324 | 36 | 115 [371] ↓256 |
525 [269] ↑256 |
||
| Test: RCM versus dermoscopy: any algorithm or threshold in equivocal lesion populations [dermoscopy data denoted in brackets] | ||||||
| Datasets | Lesions | Melanomas |
Sensitivity (fixed) RCM [dermoscopy] |
Specificity RCM [dermoscopy] |
||
| 7 [3] | 1177 [645] | 180 [127] | 90% [90%] | 86% [49%] | ||
| Numbers observed in a cohort of 1000 lesions being testedc | ||||||
| Prevalence | True positive | False negative | False positive | True negative | ||
| (received necessary excision) | (did not receive required excision) | (inappropriately received excision) | (appropriately did not receive excision) | |||
| At prevalence 10% | 90 | 10 | 126 [459] ↓333 |
774 [441] ↑333 |
||
| At prevalence 20% | 180 | 20 | 112 [408] ↓296 |
688 [392] ↑296 |
||
| At prevalence 23% | 207 | 23 | 108 [393] ↓285 |
662 [377] ↑285 |
||
CI: confidence interval; RCM: reflectance confocal microscopy.
aThe denominator for the Limitations section was 20 because methodological quality was assessed separately for each of the 19 cohorts of lesions, and a further publication (Pellacani 2007a) reporting data for two of these cohorts (Guitera 2009b (Modena); Guitera 2009c (Sydney)) was also included and separately quality assessed, taking the total to 20. Pellacani 2007a was included only to allow analysis of additional algorithm thresholds and was not included in the main analyses.
b[ ] Dermoscopy data were denoted by square brackets throughout.
cThe numbers observed in a hypothetical cohort of lesions were estimated at the median and interquartile range in prevalence across the pooled datasets for each test.
dThe arrows ↓ ↑ indicated the change in number of false positive and true negative results as a result of RCM use.
Background
This review is one of a series of Cochrane Diagnostic Test Accuracy (DTA) Reviews on the diagnosis and staging of melanoma and keratinocyte skin cancers as part of the National Institute for Health Research (NIHR) Cochrane Systematic Reviews Programme. Appendix 1 shows the content and structure of the programme.
Target condition being diagnosed
Melanoma arises from uncontrolled proliferation of melanocytes, which are the epidermal cells that produce pigment or melanin. Melanoma can occur in any organ that contains melanocytes, including mucosal surfaces, the back of the eye, and lining around the spinal cord and brain, but most commonly arises in the skin. The incidence of melanoma rose to over 200,000 newly diagnosed cases worldwide in 2012 (Erdmann 2013; Ferlay 2015), with an estimated 55,000 deaths (Ferlay 2015). The highest incidence is observed in Australia with 13,134 new cases of melanoma of the skin in 2014 (ACIM 2017) and in New Zealand with 2341 registered cases in 2010 (HPA and MelNet NZ 2014). For 2014 in the USA, the predicted incidence was 73,870 per annum and the predicted number of deaths was 9940 (Siegel 2015). The highest rates in Europe are seen in north‐western Europe and the Scandinavian countries, with a highest incidence reported in Switzerland: 25.8 per 100,000 in 2012. Rates in England have tripled from 4.6 and 6.0 per 100,000 in men and women, respectively, in 1990, to 18.6 and 19.6 per 100,000 in 2012 (EUCAN 2012). In the UK, melanoma has one of the fastest rising incidence rates of any cancer, and has had the biggest projected increase in incidence between 2007 and 2030 (Mistry 2011). In the decade leading up to 2013, age standardised incidence increased by 46%, with 14,500 new cases in 2013 and 2459 deaths in 2014 (Cancer Research UK 2017a). Rates are higher in women than in men; however, the rate of incidence in men is increasing faster than in women (Arnold 2014).
Definitions: cutaneous melanoma refers to any skin lesion with malignant melanocytes present in the dermis, and includes superficial spreading, nodular, acral lentiginous, and lentigo maligna melanoma variants (Figure 1). Melanoma in situ refers to malignant melanocytes that are contained within the epidermis and have not yet invaded the dermis (i.e. intraepidermal), but are at risk of progression to melanoma if left untreated. Lentigo maligna, a subtype of melanoma in situ in chronically sun‐damaged skin, denotes another form of proliferation of abnormal melanocytes. Lentigo maligna can progress to invasive melanoma if its growth breaches the dermoepidermal junction during a vertical growth phase (when it becomes known as 'lentigo maligna melanoma'); however, its malignant transformation is both lower and slower than that of melanoma in situ (Kasprzak 2015). Melanoma in situ and lentigo maligna are both atypical intraepidermal melanocytic variants (also referred to as 'borderline evolving melanoma') (SEER). Melanoma is one of the most dangerous forms of skin cancer, with the potential to metastasise to other parts of the body via the lymphatic system and bloodstream. It accounts for only a small percentage of skin cancer cases but is responsible for up to 75% of skin cancer deaths (Boring 1994; Cancer Research UK 2017b).
1.
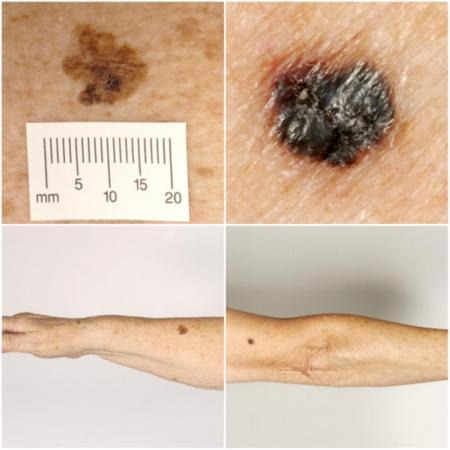
Sample photographs of superficial spreading melanoma (left) and nodular melanoma (right). Copyright © 2010 Dr Rubeta Matin: reproduced with permission.
In this diagnostic test accuracy (DTA) review we defined cutaneous invasive melanoma and atypical intraepidermal melanocytic variants as the primary target condition. We also examined accuracy for target conditions of cutaneous invasive melanoma alone, and any skin cancer or skin lesion with a high risk of progression to melanoma.
Prognosis: US data from 2007 to 2013 indicated five‐year survival of 98.5% for localised melanoma, dropping to 62.9% for those with regional spread (nodal disease) and 19.9% for disseminated disease (SEER 2017). Before the advent of targeted and immunotherapies, melanoma disseminated to distant sites and visceral organs was associated with median survival of six to nine months, a one‐year survival rate of 25%, and three‐year survival of 15% (Balch 2009; Korn 2008). Between 1975 and 2010, five‐year relative survival for melanoma in the US increased from 80% to 94%, with survival for localised disease estimated at 99%, regional disease at 70%, and distant disease at 18% in 2010 (Cho 2014). However, overall mortality rates showed little change, at 2.1 per 100,000 deaths in 1975 and 2.7 per 100,000 deaths in 2010 (Cho 2014). Increasing incidence in localised disease over the same period (from 5.7 to 21 per 100,000) suggested that much of the observed improvement in survival may have been due to earlier detection and heightened vigilance (Cho 2014); however, targeted therapies for stage IV melanoma (e.g. BRAF inhibitors) have improved survival expectation and immunotherapies are evolving such that long term survival is being documented (see below).
Treatment of melanoma
For primary melanoma, the mainstay of definitive treatment is wide local excision of the lesion, to remove both the tumour and any malignant cells that might have spread into the surrounding skin (Garbe 2016; Marsden 2010; NICE 2015a; SIGN 2017; Sladden 2009). Recommended surgical margins vary according to tumour thickness (Garbe 2016) and stage of disease at presentation (NICE 2015a).
Following histological confirmation of diagnosis, the lesion is staged according to the American Joint Committee on Cancer (AJCC) Staging System to guide treatment (Balch 2009). Stage 0 refers to melanoma in situ; stages I to II indicate localised melanoma; stage III occurs where there is regional metastasis; and stage IV indicates distant metastasis (Balch 2009). The main prognostic indicators can be divided into histological and clinical factors. Histologically, Breslow thickness is the single most important predictor of survival, as it is a quantitative measure of tumour invasion which correlates with the propensity for metastatic spread (Balch 2001). Microscopic ulceration, mitotic rate, microscopic satellites, regression, lymphovascular invasion, and nodular (rapidly growing) or amelanotic (lacking in melanin pigment) subtypes are also associated with worse prognosis (Moreau 2013; Shaikh 2012). Independent of tumour thickness, prognosis is worse in: older people; males; people with recurrent lesions; and in people with distant lymph node involvement (microscopic or macroscopic), widespread metastases, or both, at the time of primary presentation. There is debate regarding the prognostic effect from primary lesion site, with some evidence suggesting a worse prognosis for truncal lesions or lesions on the scalp or neck (Zemelman 2014).
In terms of local or regional interventions beyond wide local excision for primary lesions, completion lymphadenectomy (removal of all regional lymph nodes) is undertaken for people with clinically palpable lymph nodes and may be considered if micrometastatic disease is identified on sentinel lymph node biopsy (NICE 2015a), although no survival benefit has been shown to date for people undergoing sentinel node staging (Kyrgidis 2015; Morton 2014). Elective lymph node dissection (Eggermont 2007), adjuvant radiotherapy or adjuvant systemic treatments are not recommended for routine use in stage I, II, or III disease in the UK (NICE 2015a), and in many parts of Europe (Garbe 2016), other than interferon‐alpha (licensed by the US Food and drug Administration (FDA) and the European Medicines Agency (EMEA)) (Garbe 2016), which is effective for the treatment of high‐risk groups in terms of both disease‐free and overall survival in a Cochrane Review that found evidence for its effectiveness for disease‐free survival but not for overall survival (Mocellin 2013).
For stage IV melanoma, two distinct therapeutic approaches suggesting survival benefits in metastatic melanoma are available: targeting mutated signal transduction in the RAS‐RAF signalling pathway (e.g. BRAF‐inhibitors (Chapman 2012; Villanueva 2010) and MEK inhibitors (Dummer 2014; Larkin 2014), and immunomodulation (Chapman 2011; Hamid 2013; Hodi 2010)). Molecular targeted therapies recommended in the UK for unresectable or metastatic BRAF V600 mutation positive melanoma (around 45% of participants (Garbe 2016)) include the BRAF‐inhibitors dabrafenib (NICE 2014a), and vemurafenib (NICE 2012a), or trametinib (MEK inhibitor) in combination with dabrafenib (NICE 2016a). European guidelines recommend combinations of BRAF‐ and MEK‐inhibitors as standard treatment where indicated (Garbe 2016). Immunotherapy based approaches including ipilimumab (CTLA‐4 inhibitor) and PD‐1 inhibitors (nivolumab and pembrolizumab) have been approved in the US and Europe (Hodi 2010), and by the National Institute for Health and Care Excellence (NICE) in the UK both as single agents (NICE 2012b; NICE 2014b; NICE 2015b; NICE 2015c), and in combination (NICE 2016a; NICE 2016b). These have shown high response rates, and demonstrated the potential for a durable clinical response for the first time in the treatment of melanoma (Chapman 2011; Hamid 2013; Hodi 2010; Hodi 2016; Larkin 2015; Maio 2015; Sznol 2013).
A number of systemic therapies for stage IIIc and stage IV melanoma have been compared in a Cochrane Review (Pasquali 2018) and further NICE appraisals of new therapeutic agents, including binimetinib, talimogene laherparepvec, and temozolomide are underway (NICE 2018).
Index test(s)
Reflectance confocal microscopy (RCM), also known as confocal laser scanning microscopy or confocal microscopy, was first developed for skin imaging in the early 1990s (Rajadhyaksha 1995), and is emerging as a potential alternative or adjunct to dermoscopy for the diagnosis of skin cancer. It is a non‐invasive technology, which can be used to visualise horizontally sectioned images of the skin at a cellular lateral resolution of about 1 μm, in vivo to the depth of the upper dermis. The contrast for the monochrome images produced is achieved by the variation of the optical properties within the skin when illuminated by a near‐infrared light (830 nm) (see Figure 2). The greatest contrast is achieved from melanin, so that RCM is advocated as being particularly useful for assessing pigmented lesions.
2.
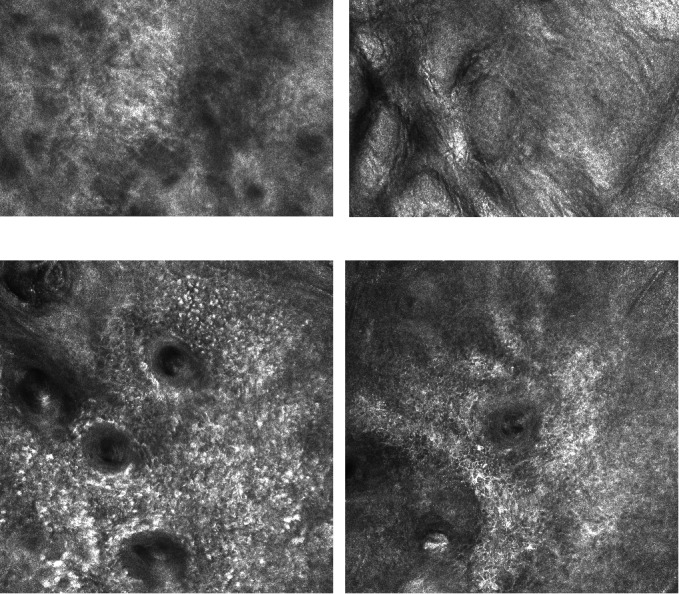
Reflectance confocal microscopy images of normal skin (top) and of lentigo maligna (bottom). Copyright © 2017 Dr Rakesh Patalay: reproduced with permission.
The Caliber I.D. VivaScope imaging systems are the only commercially available RCM devices (distributed by MAVIG in Europe). The Vivascope 1500 (and the previously available 1000 version) is a console based unit with an integrated dermoscope, whereas the Vivascope 3000 is a handheld device designed for superior ergonomics, allowing imaging of lesions inaccessible for the 1500 version (Figure 3). Imaging can be undertaken by clinicians or technicians following appropriate training (Edwards 2016). The length of time required for diagnosis has been estimated at 15 minutes for Vivascope 1500 (10 minutes of a technician's time for imaging and five minutes of a dermatologist's time for image interpretation) and 10 minutes for Vivascope 3000 (Edwards 2016). The company has estimated the mean cost per use of the 1500 system, including dermoscopy, as GBP 120 based on 2014 National Health Service (NHS) reference costs and an indicative price for Vivascope 1500 of GBP 95,224 (Edwards 2016).
3.
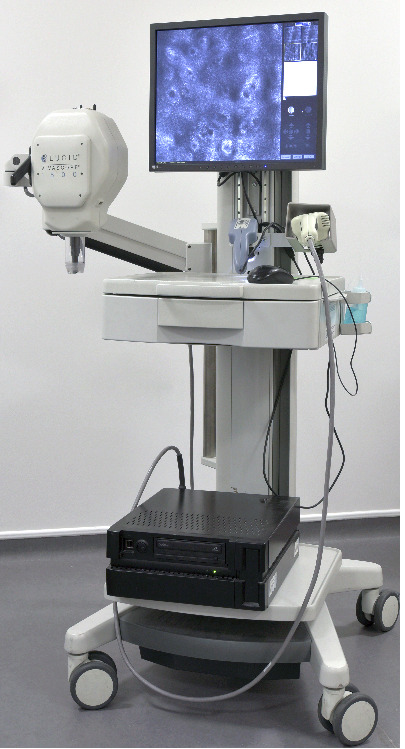
Caliber ID Vivascope 1500 with 3000 attachment. Copyright © 2017 Guy's & St Thomas' NHS Foundation Trust: reproduced with permission.
Various algorithms have been proposed for the interpretation of RCM images, relying on either numeric thresholds or qualitative indicators of test positivity according to the presence or absence of particular lesion characteristics. The lesion characteristics that are accepted as being associated with melanomas are: absence of the normal epidermis architecture, lack of delineation of the papillae (non‐edged papillae), irregular nests of atypical melanocytes, and the presence of large and highly refractile cells with prominent nuclei in higher epidermal layers (Edwards 2016; Pellacani 2007a).
Clinical pathway
The diagnosis of melanoma occurs in primary, secondary, and tertiary care settings by both generalist and specialist healthcare providers. People with concerns about a new or changing lesion will either present to their general practitioner or directly to a specialist in secondary care, which could include a dermatologist, plastic surgeon, general surgeon, or other specialist surgeon (such as an ear, nose, and throat (ENT) specialist or maxillofacial surgeon), or ophthalmologist (Figure 4). Current UK guidelines recommend that all suspicious pigmented lesions presenting in primary care should be assessed by taking a clinical history and visual inspection using the seven point checklist (MacKie 1990); lesions suspected to be melanoma should be referred for appropriate specialist assessment within two weeks (Chao 2013; Marsden 2010; NICE 2015d).
4.
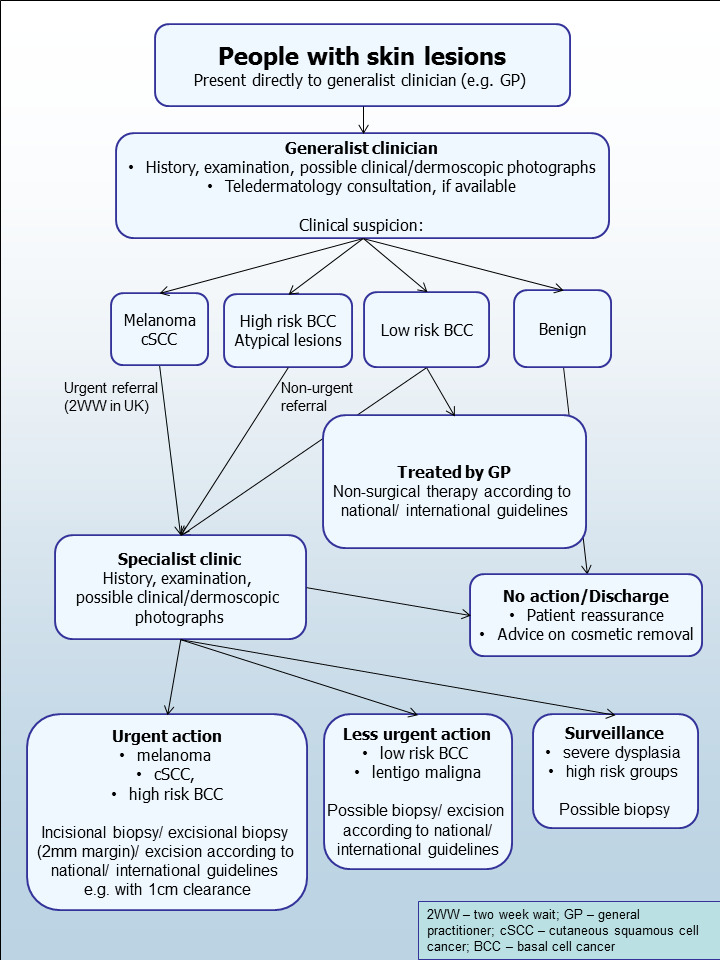
Current clinical pathway for people with skin lesions.
The specialist clinician will use history‐taking, visual inspection of the lesion (in comparison with other lesions on the skin), and usually dermoscopy to inform a clinical decision. If melanoma is suspected, then urgent excision is recommended. Other lesions such as suspected dysplastic naevi or premalignant lesions such as lentigo maligna may also be referred for a diagnostic biopsy, further surveillance, or reassurance and discharge. This is the point at which RCM is generally thought to have a role in patient management, most likely as an additional test to better identify people with lesions that can be monitored or reassured instead of being sent for urgent excision (Edwards 2016). RCM could also be considered as a primary diagnostic test (i.e. as a potential replacement for dermoscopy).
Prior test(s)
Fundamental to the diagnosis of skin cancer is clinical examination and history‐taking; however, a range of technologies have emerged to aid diagnosis to ideally reduce the number of excision biopsies. Dermoscopy in particular has become the most widely used tool for clinicians to try and obtain an accurate assessment of melanoma following visual inspection (Argenziano 1998; Argenziano 2012; Haenssle 2010; Kittler 2002).
Visual inspection of the skin is undertaken iteratively, using both implicit pattern recognition (non‐analytical reasoning) and more explicit 'rules' based on conscious analytical reasoning (Norman 2009), the balance of which will vary according to experience and familiarity with the diagnostic question. Various attempts have been made to formalise the "mental rules" involved in analytical pattern recognition for melanoma, ranging from a setting out of lesion characteristics that should be considered (Friedman 1985; Sober 1979), to formal scoring systems with explicit numerical thresholds. The seven point checklist, for example, assesses change in lesion size, shape, colour, inflammation, crusting or bleeding, sensory change, or diameter of 7 mm or greater (MacKie 1985; MacKie 1990). Other available tools include the ABCD(E) approach (Friedman 1985; Thomas 1998), and ugly duckling (Grob 1998).
Dermoscopy is a non‐invasive, in vivo technique that uses a hand‐held microscope and incident light (with or without oil immersion) to reveal subsurface images of the skin at increased magnification of ×10 to ×100 (Kittler 2011). Although widely used, the accuracy of dermoscopy largely depends on the experience and training of the examiner (Binder 1997; Kittler 2002; Kittler 2011). Pattern analysis (Pehamberger 1987; Steiner 1987) is thought to be the most specific and reliable technique to aid dermoscopy interpretation when used by specialists (Maley 2014); however, dermoscopic histological correlations have been established and diagnostic algorithms have been developed based on colour, aspect, pigmentation pattern, and skin vessels, including the ABCD rule for dermoscopy (Nachbar 1994; Stolz 1994), the Menzies approach (Menzies 1996), the seven point dermoscopy checklist (Annessi 2007; Argenziano 1998; Argenziano 2001), and the three point checklist (Gereli 2010).
The accuracy, and comparative accuracy, of visual inspection and dermoscopy and their associated scoring systems is summarised in a further review in this series (Dinnes 2018b).
Role of index test(s)
Used in conjunction with clinical or dermoscopic suspicion of malignancy (or both) in pigmented lesions, RCM is primarily advocated as a tool to reduce the number of unnecessary excisions (Ferrari 2015), especially in lesions that may be difficult to diagnose by clinical examination and dermoscopy alone (Guitera 2009a). RCM features have been shown to be strongly correlated with dermoscopic patterns (Pellacani 2014a). Moreover, small diameter melanomas (less than 5 mm diameter) may demonstrate specific dermoscopic and confocal features, such as marked cytological atypia and irregular nesting, which help to differentiate them from naevi (Pupelli 2013). One of the postulated advantages of RCM is its ability to differentiate seborrhoeic keratosis or non‐melanocytic lesions from a population of pigmented lesions.
Although the primary aim in diagnosing potentially life‐threatening conditions such as melanoma is to minimise false negative diagnoses (to avoid delay to diagnosis and even death), a test that can reduce false positive clinical diagnoses without missing true cases of disease has patient and resource benefits. False positive clinical diagnoses not only cause unnecessary morbidity from the biopsy, but also increase patient anxiety. Pigmented lesions are common so the resource implication for even a slight increase in the threshold to excise lesions in populations where melanoma rates are increasing will avoid a considerable healthcare burden to both patient and healthcare provider, as long as such lesions turn out to be harmless.
RCM is also being explored for its ability to differentiate lentigo maligna from actinic or seborrhoeic keratosis (de Carvalho 2015; Menge 2016). RCM could also develop a future role in guiding definitive therapeutic margins (Edwards 2016), and to evaluate response to topical chemotherapy for lentigo maligna; however, these uses are not under consideration in this review.
Alternative test(s)
A number of other tests are being reviewed as part of our series of Cochrane DTA reviews on the diagnosis of melanoma, including visual inspection and dermoscopy (Dinnes 2018a; Dinnes 2018b), teledermatology (Chuchu 2018a), mobile phone applications (Chuchu 2018b), computer‐aided diagnosis (CAD) techniques (Ferrante di Ruffano 2018b), optical coherence tomography (OCT) (Ferrante di Ruffano 2018a), and high‐frequency ultrasound (Dinnes 2018d).
OCT is an emerging optical imaging technology based on interferometry using a near infra‐red light source. It exploits differences in the refractive index in the skin to create vertically sectioned images in vivo, in real time. Vascular flow information can be extracted from the images, allowing neovascularisation to be visualised, which has potential for earlier diagnosis of melanoma (Kokolakis 2012; Themstrup 2015). High‐frequency ultrasound has shown good correlation with histology for measurement of melanoma thickness, but may also differentiate pigmented lesions, particularly for colour Doppler (Scotto di Santolo 2015). CAD or artificial intelligence based techniques process and manipulate lesion images using predefined algorithms to identify the features that discriminate malignant from benign lesions (Esteva 2017; Rajpara 2009). These techniques have been incorporated into commercially available handheld devices for ease of use in a clinic setting, including SIAscopy (Moncrieff 2002; Walter 2012), MelaFind (Hauschild 2014; Monheit 2011; Wells 2012), and the Nevisense Electrical Impedance Spectroscopy system (Malvehy 2014). However, CAD has most commonly been applied to digital dermoscopy images (Esteva 2017; Rajpara 2009).
Evidence permitting, the accuracy of available tests will be compared in an overview review, exploiting within‐study comparisons of tests and allowing the analysis and comparison of commonly used diagnostic strategies where tests may be used singly or in combination.
Other tests identified as potential candidates for review but for which we found no eligible studies included volatile organic compounds (including canine odour detection) (Abaffy 2010; Church 2001; D'Amico 2008; Gallagher 2008; Kwak 2013; Williams 1989), and gene expression analysis (Ferris 2012; Wachsman 2011).
We also considered and excluded a number of tests from the review including exfoliative cytology, which involves microscopic examination of a scraping taken from a skin lesion stained with Giemsa (Ruocco 2011); tests used in the context of monitoring people, such as total body photography of people with large numbers of typical or atypical naevi; and histopathological confirmation following lesion excision. Histopathological confirmation following lesion excision is the established reference standard for melanoma diagnosis and will be one of the standards against which the index tests are evaluated in these reviews.
Rationale
Our series of reviews of diagnostic tests used to assist clinical diagnosis of melanoma aims to identify the most accurate approaches to diagnosis and provide clinical and policy decision‐makers with the highest possible standard of evidence on which to base decisions. With increasing rates of melanoma incidence and the push towards the use of dermoscopy and other high resolution image analysis in primary care, the anxiety around missing early cases needs to be balanced to avoid referring too many people with benign lesions for a specialist opinion. It is questionable whether all skin cancers picked up by sophisticated techniques, even in specialist settings, help to reduce morbidity and mortality or whether newer technologies run the risk of increasing false positive diagnoses. It is also possible that use of some technologies (e.g. widespread use of dermoscopy in primary care with no training) could actually result in harm by missing melanomas if they are used as replacement technologies for traditional history‐taking and clinical examination of the entire skin. Many branches of medicine have noted the danger of such 'gizmo idolatry' amongst doctors (Leff 2008).
To date, the use of RCM has been limited by expense (in terms of both equipment and staff time) and the need for specialised training. Studies have demonstrated high sensitivity and specificity amongst experienced RCM users; however, in at least one study, the accuracy of the group on average was higher than that of any one individual observer (Farnetani 2015). A standardised system that is reproducible across users is therefore desirable. Ultimately it is thought that although RCM may augment diagnostic sensitivity when used in conjunction with clinical inspection and dermoscopy, its main contribution is an increase in specificity. However, the exact contribution of RCM as an adjunct to dermoscopy is not entirely clear (Edwards 2016; Stevenson 2013), and the number of RCM cases required to offset an unnecessary excision biopsy has not been assessed in a UK setting.
Although a set of billing codes for the USA have been agreed since January 2016 (Rajadhyaksha 2017), RCM is not recommended for routine use in the UK (Edwards 2016), Australia (Guitera 2017), or New Zealand (Sobarun 2015). Available systematic reviews are limited by currency (Stevenson 2013), and methods (Xiong 2016; e.g. failing to consider the nature of the target population, varying definitions of the target condition, and using an out of date meta‐analytic approach), or focus on selected studies considered to be more applicable to a UK setting (Edwards 2016). Furthermore, in a rapidly advancing field, there is a need for an up‐to‐date analysis of the accuracy of RCM in comparison to dermoscopy at different points in the clinical pathway.
As several reviews for each topic area followed the same methodology, generic protocols were prepared to avoid duplication of effort, one for diagnosis of melanoma (Dinnes 2015a), and one for diagnosis of keratinocyte skin cancers (Dinnes 2015b). The Background and Methods sections of this review therefore use some text that was originally published in the protocol concerning the evaluation of tests for the diagnosis of melanoma (Dinnes 2015a), and text that overlaps some of our other reviews (Dinnes 2018b). Table 2 provides a glossary of terms used.
1. Glossary of terms.
| Term | Definition |
| Amelanotic | Without melanin |
| Anti‐CTLA‐4 therapy system | Monoclonal antibody to CTLA‐4 (cytotoxic T‐lymphocyte‐associated protein 4); a protein that is involved in regulating the immune system |
| BRAF‐inhibitors | Therapeutic agents that inhibit the serine‐threonine protein kinase BRAF‐mutated metastatic melanoma |
| Driver mutations | Somatic gene mutations that are responsible for tumour progression |
| Elective lymph node dissection | Surgical removal of ≥ 1 lymph nodes in the absence of confirmed involvement with melanoma |
| Hybridised | The process of combining 2 biological molecules |
| Immune checkpoint targets | Signalling pathways that are inhibitory and switch off T cells in the immune system |
| Immunomodulation | Adjustment of the immune system in a person |
| Irregular nesting | Unbalanced asymmetrical arrangement of groups of melanocytes in the skin |
| Lymphovascular invasion | Tumour cells that have spread to involve the blood vessels and lymphatic vessels within the skin |
| MEK inhibitors | Drugs that inhibit the mitogen‐activated protein kinase enzymes that are often upregulated in melanoma |
| Microscopic satellites | Foci of melanoma observed histologically that are distinct from the original primary tumour |
| Mitotic rate | Microscopic evaluation of a number of cells actively dividing in a tumour |
| Mutated signal transduction | Activation of Ras proteins such that unintended and overactive signalling occurs and causes overgrowth of cells and higher rates of cell division |
| PD1 | Programmed cell death protein 1: a protein involved in downregulating the immune system |
| PD1‐L | Programmed cell death protein 1 receptor; expressed on T and B cells |
| Phenotypic risk | The various clinical/physical traits of a person determined by genetic and environmental factors that predispose people to melanoma |
| Prophylactic isolated limb perfusion | A medical procedure that directly delivers a drug through the bloodstream in a limb to the site affected by melanoma |
| Pseudopods | Temporary projections from cells that help cellular movement |
| RAS‐RAF signalling pathway | Family of proteins that serve as intermediary in transmitting extracellular signals from growth factor receptors that control cell growth, proliferation, and differentiation |
| RNA | Ribonucleic acid involved in coding, decoding, regulation, and expression of genes |
| Signal transduction | Occurs when extracellular signalling molecules activate a specific receptor, which then triggers cellular pathways |
| Spectroscopy | Study of the interaction between matter and electromagnetic radiation |
| Stratum corneum | Uppermost layer of the epidermis composed of dead keratinocytes (corneocytes) |
| Xeroderma pigmentosum | Autosomal recessive genetic disorder of DNA repair, resulting in an inability to repair damage caused by ultraviolet light leading to skin malignancies |
Objectives
To determine the diagnostic accuracy of reflectance confocal microscopy for the detection of cutaneous invasive melanoma and atypical intraepidermal melanocytic variants in adults with any lesion suspicious for melanoma and lesions that are difficult to diagnose, and to compare its accuracy with that of dermoscopy.
We estimated accuracy separately according to the point in the clinical pathway at which RCM was evaluated:
where it might have been used as an alternative to dermoscopy in participants with any lesion suspicious for melanoma;
where it might have been used in addition to dermoscopy in participants with equivocal lesions in whom a clear management decision could not be made following visual inspection and dermoscopy.
Secondary objectives
To determine the diagnostic accuracy of RCM in comparison to dermoscopy for the detection of:
cutaneous invasive melanoma alone;
any skin cancer (melanoma or other skin cancer) or skin lesion with a high risk of progression to melanoma.
We estimated accuracy separately according to the point in the clinical pathway at which RCM was evaluated:
where it might have been used in addition to current practice (which may or may not include dermoscopy) in participants with any lesion suspicious for melanoma;
where it might have been used as an addition to dermoscopy in participants with equivocal lesions in whom a clear management decision could not be made following visual inspection and dermoscopy.
For identifying cutaneous invasive melanoma and atypical intraepidermal melanocytic variants (the primary target condition):
to compare the accuracy of RCM to dermoscopy where both tests were evaluated in the same studies (direct test comparisons);
to determine the diagnostic accuracy of individual algorithms for RCM;
to determine the effect of observer experience.
Investigation of sources of heterogeneity
We aimed to consider a range of potential sources of heterogeneity for investigation across the series of reviews, as outlined in our generic protocol (Dinnes 2015a).
-
Population characteristics:
general versus higher risk populations;
participant population: primary/secondary/specialist unit;
lesion type: any pigmented; melanocytic;
inclusion of multiple lesions per participant;
ethnicity.
-
Index test characteristics:
in‐person versus remote image based RCM interpretations;
the nature and definition of criteria for test positivity;
observer experience with the index test.
-
Reference standard characteristics:
reference standard used;
whether histology‐reporting met pathology‐reporting guidelines;
use of excisional versus diagnostic biopsy;
whether two independent dermatopathologists reviewed histological diagnosis.
-
Study quality:
consecutive or random sample of participants recruited;
index test interpreted blinded to the reference standard result;
index test interpreted blinded to the result of any other index test;
presence of partial or differential verification bias (whereby only a sample of those subject to the index test were verified by the reference test or by the same reference test with selection dependent on the index test result);
use of an adequate reference standard;
overall risk of bias.
Methods
Criteria for considering studies for this review
Types of studies
We included test accuracy studies that allowed comparison of the result of the index test with that of a reference standard, including the following:
studies where all participants received a single index test and a reference standard;
studies where all participants received more than one index test and reference standard;
studies where participants were allocated (by any method) to receive different index tests or combinations of index tests and all received a reference standard (between‐person comparative (BPC) studies);
studies that recruited series of participants unselected by true disease status (referred to as case series for the purposes of this review);
diagnostic case‐control studies that separately recruited diseased and non‐diseased groups (see Rutjes 2005);
both prospective and retrospective studies; and
studies where previously acquired clinical or dermoscopic images were retrieved and prospectively interpreted for study purposes.
We excluded studies from which we could not extract 2×2 contingency data or if they included fewer than five melanoma cases.
Studies available only as conference abstracts were excluded; however, attempts were made to identify full papers for potentially relevant conference abstracts (Searching other resources).
Participants
We included studies in adults with pigmented skin lesions or lesions suspicious for melanoma.
We excluded studies that recruited only participants with malignant diagnoses and studies that compared test results in participants with malignancy compared with test results based on 'normal' skin as controls, due to the bias inherent in such comparisons (Rutjes 2006).
We excluded studies with more than 50% of participants aged 16 years and under.
Index tests
We included studies evaluating RCM alone, or RCM in comparison to dermoscopy.
We included all established algorithms or checklists to assist diagnosis. Studies developing new algorithms or methods of diagnosis (i.e. derivation studies) were included if they used a separate independent 'test set' of participants or images to evaluate the new approach. Studies that did not report data for a separate test set of participants or images were included only if the lesion characteristics investigated had previously been suggested as associated with melanoma and the study reported accuracy based on the presence or absence of particular combinations of characteristics. Studies using a statistical model to produce a data driven equation, or algorithm based on multiple diagnostic features, with no separate test set were excluded. Studies using cross‐validation approaches such as 'leave‐one‐out' cross‐validation were excluded (Efron 1983).
No exclusions were made according to test observer.
Target conditions
The primary target condition was defined as the detection of:
any form of invasive cutaneous melanoma, or
atypical intraepidermal melanocytic variants (i.e. including melanoma in situ, or lentigo maligna, which has a risk of progression to invasive melanoma).
Two additional definitions of the target condition were considered in secondary analyses, the detection of:
any form of invasive cutaneous melanoma alone, and
any skin cancer (melanoma or other skin cancer) or skin lesion with a high risk of progression to melanoma. This latter definition included other forms of skin cancer, such as basal cell carcinoma (BCC) and cutaneous squamous cell carcinoma (cSCC), as well as melanoma in situ, lentigo maligna, and lesions with severe melanocytic dysplasia.
The diagnosis of the keratinocyte skin cancers, BCC, and squamous cell carcinoma (SCC) as primary target conditions are the subject of a separate series of reviews (Dinnes 2015b).
Reference standards
The ideal reference standard was histopathological diagnosis of the excised lesion or biopsy sample in all eligible lesions. A qualified pathologist or dermatopathologist should have performed the histopathology. Ideally, reporting should have been standardised detailing a minimum dataset to include the histopathological features of the melanoma to determine the AJCC Staging System (e.g. Slater 2014). We did not apply the reporting standard as a necessary inclusion criterion, but extracted any pertinent information.
Partial verification (applying the reference test only to a subset of participants undergoing the index test) was of concern given that lesion excision or biopsy are unlikely to be carried out for all benign appearing lesions within a representative population sample. Therefore, we accepted clinical follow‐up of benign appearing lesions as an eligible reference standard, whilst recognising the risk of differential verification bias (as misclassification rates of histopathology and follow‐up will differ) in our quality assessment of studies.
Additional eligible reference standards included cancer registry follow‐up and 'expert opinion' with no histology or clinical follow‐up. Cancer registry follow‐up is considered less desirable than active clinical follow‐up, as follow‐up is not carried out within the control of the study investigators. Furthermore, if participant based analyses as opposed to lesion based analyses are presented, it may be difficult to determine whether the detection of a malignant lesion during follow‐up is the same lesion that originally tested negative on the index test.
All of the above were considered eligible reference standards with the following caveats:
all study participants with a final diagnosis of the target disorder must have a histological diagnosis, either subsequent to the application of the index test or after a period of clinical follow‐up, and
at least 50% of all participants with benign lesions must have had either a histological diagnosis or clinical follow‐up to confirm benignity.
Search methods for identification of studies
Electronic searches
The Information Specialist (SB) carried out a comprehensive search for published and unpublished studies. A single large literature search was conducted to cover all topics in the programme grant (see Appendix 1 for a summary of reviews included in the programme grant). This allowed for the screening of search results for potentially relevant papers for all reviews at the same time. A search combining disease related terms with terms related to the test names, using both text words and subject headings was formulated. The search strategy was designed to capture studies evaluating tests for the diagnosis or staging of skin cancer. As the majority of records were related to the searches for tests for staging of disease, a filter using terms related to cancer staging and to accuracy indices was applied to the staging test search, to try to eliminate irrelevant studies, for example, those using imaging tests to assess treatment effectiveness. A sample of 300 records that would be missed by applying this filter was screened and the filter adjusted to include potentially relevant studies. When piloted on MEDLINE, inclusion of the filter for the staging tests reduced the overall numbers by around 6000. The final search strategy, incorporating the filter, was subsequently applied to all bibliographic databases as listed below (Appendix 2). The final search result was cross‐checked against the list of studies included in five systematic reviews; our search identified all but one of the studies, and this study was not indexed on MEDLINE. The Information Specialist (SB) devised the search strategy, with input from the Information Specialist from Cochrane Skin. No additional limits were used.
We searched the following bibliographic databases to 29 August 2016 for relevant published studies:
MEDLINE via OVID (from 1946);
MEDLINE In‐Process & Other Non‐Indexed Citations via OVID; and
Embase via OVID (from 1980).
We searched the following bibliographic databases to 30 August 2016 for relevant published studies:
the Cochrane Central Register of Controlled Trials (CENTRAL; 2016, Issue 7) in the Cochrane Library;
the Cochrane Database of Systematic Reviews (CDSR; 2016, Issue 8) in the Cochrane Library;
Cochrane Database of Abstracts of Reviews of Effects (DARE; 2015, Issue 2);
CRD HTA (Health Technology Assessment) database, 2016, Issue 3;
CINAHL (Cumulative Index to Nursing and Allied Health Literature via EBSCO from 1960).
We searched the following databases for relevant unpublished studies using a strategy based on the MEDLINE search:
CPCI (Conference Proceedings Citation Index), via Web of Science™ (from 1990; searched 28 August 2016); and
SCI Science Citation Index Expanded™ via Web of Science™ (from 1900, using the 'Proceedings and Meetings Abstracts' Limit function; searched 29 August 2016).
We searched the following trials registers using the search terms 'melanoma', 'squamous cell', 'basal cell' and 'skin cancer' combined with 'diagnosis':
Zetoc (from 1993; searched 28 August 2016).
The US National Institutes of Health Ongoing Trials Register (www.clinicaltrials.gov); searched 29 August 2016.
NIHR Clinical Research Network Portfolio Database (www.nihr.ac.uk/research‐and‐impact/nihr‐clinical‐research‐network‐portfolio/); searched 29 August 2016.
The World Health Organization International Clinical Trials Registry Platform (apps.who.int/trialsearch/); searched 29 August 2016.
We aimed to identify all relevant studies regardless of language or publication status (published, unpublished, in press, or in progress). We applied no date limits.
Searching other resources
We screened relevant systematic reviews identified by the searches for their included primary studies, and included any missed by our searches. We checked the reference lists of all included papers, and subject experts within the author team have reviewed the final list of included studies. No electronic citation searching was conducted.
Data collection and analysis
Selection of studies
At least one review author (JDi or NC) screened titles and abstracts, with any queries discussed and resolved by consensus. A pilot screen of 539 MEDLINE references showed good agreement (89% with a kappa of 0.77) between screeners. We included primary test accuracy studies and test accuracy reviews (for scanning of reference lists) of any test used to investigate suspected melanoma, BCC, or cSCC at initial screening. Both a clinical review author (from one of a team of 12 clinician reviewers) and a methodologist review author (JDi or NC) independently applied inclusion criteria (Appendix 3) to all full text articles, and resolved disagreements by consensus or by a third party (JDe, CD, HW, and RM). We contacted authors of eligible studies when there were insufficient data to allow for the construction of 2×2 contingency tables.
Data extraction and management
One clinical (as detailed above) and one methodologist review author (JDi, NC, or LFR) independently extracted data concerning details of the study design, participants, index test(s) or test combinations, criteria for index test positivity, reference standards, and data required to complete a 2×2 diagnostic contingency table for each index test using a data extraction form piloted on five studies. Data were extracted at all available index test thresholds. We resolved disagreements by consensus or by a third party (JDe, CD, HW, and RM).
We contacted authors of included studies where information related to the target condition (in particular to allow the differentiation of invasive cancers from 'in situ' variants) or diagnostic thresholds were missing. We contacted authors of conference abstracts published from 2013 to 2015 to ask whether full data were available. If there was no full paper, conference abstracts were tagged and will be revisited during review updates.
Dealing with multiple publications and companion papers
Where we identified multiple reports of a primary study, we maximised yield of information by collating all available data. Where there were inconsistencies in reporting or overlapping study populations, we contacted study authors for clarification in the first instance. If this contact with authors was unsuccessful, we used the most complete and up‐to‐date data source where possible.
Assessment of methodological quality
We assessed risk of bias and applicability of included studies using the QUADAS‐2 checklist (Whiting 2011), tailored to the review topic (see Appendix 4). We piloted the modified QUADAS‐2 tool on five included full text articles. One clinical (as detailed above) and one methodologist review author (JDi, NC, or LFR) independently assessed quality for the remaining studies; we resolved disagreements by consensus or by a third party where necessary (JDe, CD, HW, and RM).
Statistical analysis and data synthesis
For the primary outcome of detection of invasive melanoma or atypical intraepidermal melanocytic variants, we conducted separate analyses according to the point in the clinical pathway that RCM was applied. Three groups of studies were formed:
RCM used as a replacement for dermoscopy in participants with lesions suspicious for melanoma, that is, no attempt to exclude those diagnosed as definite melanomas or as obviously benign on dermoscopy was described (denoted as studies in 'any lesion suspicious for melanoma' or 'any potential melanoma');
RCM used as an addition to dermoscopy in participants with equivocal lesions in whom a clear management decision could not be made following visual inspection and dermoscopy (denoted as studies in 'equivocal' lesions);
'Other' studies that did not fit into either of these categories.
Our unit of analysis was the lesion rather than the person. This is because in skin cancer initial treatment is directed to the lesion rather than systemically (thus it is important to be able to correctly identify cancerous lesions for each person), and it is the most common way in which the primary studies reported data. Although there was a theoretical possibility of correlations of test errors when the same people contributed data for multiple lesions, most studies included very few people with multiple lesions and any potential impact on findings was likely to be very small, particularly in comparison with other concerns regarding risk of bias and applicability. For each analysis, only one dataset was included per study to avoid multiple counting of lesions.
For each analysis undertaken, only one dataset was included per study to avoid over‐counting of lesions. Where multiple algorithms were assessed in an individual study, datasets were selected on the following preferential basis:
'no algorithm' reported; data presented for clinician's overall diagnosis or management decision;
pattern analysis or pattern recognition;
Pellacani's RCM score;
Segura algorithm;
presence of statistically significant lesion characteristics.
Where multiple thresholds per algorithm were reported, we included the standard or most commonly used threshold. If data for multiple observers were reported, data for the most experienced observer were used, and data for a single observer's diagnosis were used in preference to a consensus or mean across observers. If we were unable to choose a dataset based on the above 'rules,' we made a random selection of one dataset per study.
For each index test, algorithm, or checklist under consideration, we plotted estimates of sensitivity and specificity on coupled forest plots and in receiver operating characteristic (ROC) space. For tests that reported commonly used thresholds, we estimated summary operating points (summary sensitivities and specificities) with 95% confidence interval (CI) and prediction regions using the bivariate hierarchical model (Chu 2006; Reitsma 2005). Where inadequate data were available for the model to converge, we simplified the model, first by assuming no correlation between estimates of sensitivity and specificity and second by setting estimates of near zero variance terms to zero (Takwoingi 2015). Where all studies reported 100% sensitivity (or 100% specificity), we summed the numbers with disease (or no disease) across studies and used them to compute a binomial exact 95% CI. We assessed heterogeneity in estimates of sensitivity and specificity by inspection of the magnitude and statistical significance of the estimates of variance terms in the bivariate model.
We made comparisons between tests and in investigating heterogeneity by comparing summary receiver operator curves (SROC) using the hierarchical summary receiver operator curves (HSROC) model (Rutter 2001). This allowed incorporation of data at different thresholds and from different algorithms or checklists. We used an HSROC model that assumed a constant SROC shape between tests and subgroups, but allowed for differences in threshold and accuracy by addition of covariates. We assessed the significance of the differences between tests or subgroups by the likelihood ratio test assessing differences in both accuracy and threshold, and by a Wald test on the parameter estimate testing for differences in accuracy alone. We fitted simpler models when convergence was not achieved due to small numbers of studies, first assuming symmetric SROC curves (setting the shape term to zero), and then setting random‐effects variance estimates to zero.
We included data on the accuracy of dermoscopy, to allow comparisons of tests, only if reported in the studies of RCM due to the known substantial unexplained heterogeneity in all studies of the accuracy of dermoscopy (Dinnes 2018b). We made comparisons between dermoscopy results with RCM data from all RCM studies, and then only using RCM data from studies that also reported dermoscopy data for the same participants to enable a robust direct comparison (Takwoingi 2013).
We presented estimates of accuracy from HSROC models as diagnostic odds ratios (DOR; estimated where the SROC curve crossed the sensitivity = specificity line) with 95% CIs. We presented differences between tests and subgroups from HSROC analyses as relative DORs with 95% CIs. To facilitate interpretation in terms of rates of false positive and false negative diagnoses, we computed values of specificity at the point on the SROC curve with 90% sensitivity. We chose this value as it lay within the estimates for the majority of analyses. Results should only be considered as illustrative examples of possible specificities and differences in specificities that could be expected.
For computation of likely numbers of true positive, false positive, false negative, and true negative findings in the 'Summary of findings' tables, we applied these indicative values to lower quartiles, medians, and upper quartiles of the prevalence observed in the study groups.
We fitted the bivariate models using the meqrlogit command in STATA 13 and fitted HSROC models using the NLMIXED procedure in the SAS statistical software package (SAS 2012, version 9.3; SAS Institute, Cary, NC, USA) and the metadas macro (Takwoingi 2010).
Investigations of heterogeneity
We examined heterogeneity between studies by visually inspecting the forest plots of sensitivity and specificity and SROC plots. Where there was a sufficient number of studies identified, we performed meta‐regression by adding the potential source of heterogeneity as a covariate to a hierarchical model.
Sensitivity analyses
We performed no sensitivity analyses.
Assessment of reporting bias
Because of uncertainty about the determinants of publication bias for diagnostic accuracy studies and the inadequacy of tests for detecting funnel plot asymmetry (Deeks 2005), we performed no tests to detect publication bias.
Results
Results of the search
The review authors identified and screened 34,517 unique references for inclusion. Of these, we reviewed 1051 full text papers for eligibility for any one of the suite of reviews of tests to assist in the diagnosis of melanoma or keratinocyte skin cancer. Of the 1051 full text papers assessed, we excluded 848 from all reviews in our series (Figure 5 documents a PRISMA flow diagram of search and eligibility results). We tagged 85 studies as potentially eligible for the two RCM reviews; ultimately, we included 22 publications, 18 in this review and 10 in the review of RCM for the detection of keratinocyte skin cancers (six were included in both). Reasons for exclusion included: publications not being primary test accuracy studies (13 studies), lack of test accuracy data (12 studies), because they were derivation studies developing new algorithms or approaches to diagnosis without the use of separate training and test sets of data (eight studies), included ineligible populations (e.g. including only malignant lesions; six studies), did not assess eligible target conditions or did not adequately define the target condition (10 studies), inadequate sample size (15 studies), assessed the accuracy of individual RCM characteristics (five studies), or used ineligible reference standards (i.e. less than 50% of benign group with final diagnosis established by histology or follow‐up; three studies). A list of the 67 studies excluded from this review with reasons for exclusion is provided in the Characteristics of excluded studies table, with a list of all studies excluded from the full series of reviews available as a supplementary file (please contact skin.cochrane.org for a copy).
5.
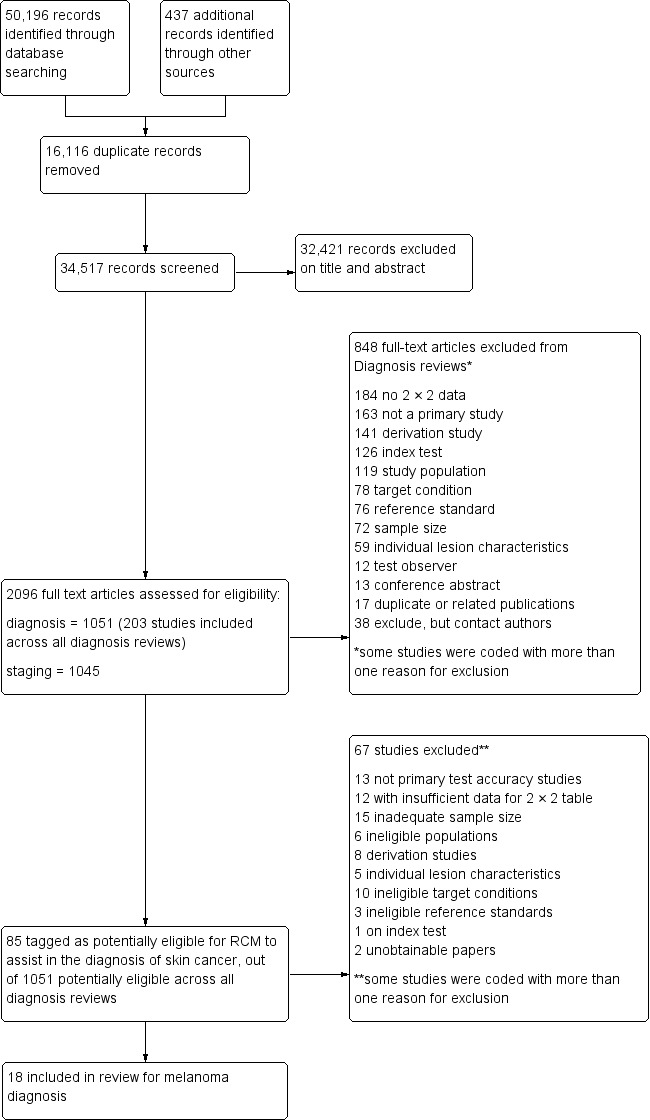
PRISMA flow diagram. RCM: reflectance confocal microscopy.
We contacted the corresponding authors of five studies and asked them to supply further information for this review. Two authors provided additional data in relation to Pupelli 2013 and Alarcon 2014a. Professor Pellacani further provided information on lesion overlap between several included studies that were coauthored by him (Guitera 2009b (Modena); Guitera 2009c (Sydney); Guitera 2012; Pellacani 2007a; Pellacani 2012; Pellacani 2014b (cons); Pellacani 2014c (doc)Ferrari 2015).
This review reported on 19 cohorts of participants with lesions suspected of melanoma, published in 18 study publications, and providing 67 datasets for RCM and seven datasets for dermoscopy. There were 2838 lesions, 658 with a diagnosis of melanoma. The total number of study participants could not be estimated due to lack of reporting in study publications. Two publications were split into two cohorts for the purposes of this review, one by Pellacani and colleagues (Pellacani 2014b (cons); Pellacani 2014c (doc)) and one by Guitera and colleagues (Guitera 2009b (Modena); Guitera 2009c (Sydney)). One of the 18 study publications (Pellacani 2007a) was based on a combined analysis of the two cohorts of lesions reported in the Guitera 2009a study and was included only to allow analysis of additional algorithm thresholds and was not included in the main analyses. A description of the various algorithms and thresholds used for diagnosis across the studies is provided in Appendix 5.
Methodological quality of included studies
The overall methodological quality of all included study cohorts is summarised in Figure 6 and Figure 7. The denominator for this section was 20 cohorts because of the inclusion of two reports for the same group of lesions (Guitera 2009a; Pellacani 2007a). Studies were generally at high or unclear risk of bias across all domains and of high or unclear concern regarding applicability of the evidence.
6.
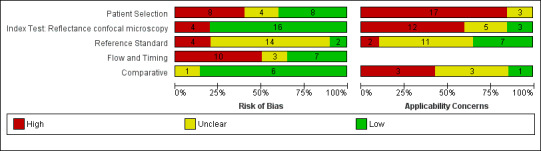
Risk of bias and applicability concerns graph: review authors' judgements about each domain presented as percentages across included studies.
7.
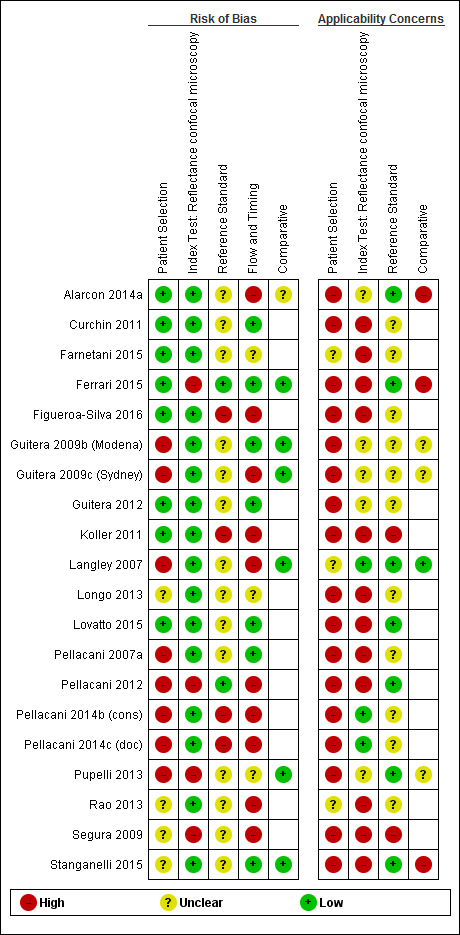
Risk of bias and applicability concerns summary: review authors' judgements about each domain for each included study.
Eight studies were at high risk of bias for participant selection due to inappropriate participant exclusions. Exclusions were variously made according to imaging failure, image quality, or particular lesion types such as lentigo maligna. Those at unclear risk of bias (four cohorts) did not clearly describe participant recruitment as random or consecutive. All cohorts were at high (17 cohorts) or unclear (three cohorts) concern regarding included participants and setting, due to restricted study populations (with 16 studies including only participants with melanocytic lesions, or even more narrowly defined populations such as nodular lesions) and inclusion of multiple lesions per participant (with seven including over 5% more lesions than participants and four not reporting the number of participants). Sixteen of the 20 cohorts included lesions selected for excision based on the clinical or dermoscopic diagnosis or selected retrospectively from histopathology databases; this was not considered of high concern regarding applicability for RCM studies as the primary role for RCM was to reduce unnecessary excisions.
Three‐quarters of cohorts were at low risk of bias in the index test domain; all studies reported blinding of RCM interpretation to the reference standard diagnosis and 16 reported prespecification of the diagnostic threshold. Over half of studies were at high concerns around the applicability of the index test, due to blinded interpretation of RCM images (fully blinded in six and providing only information on participant age and lesion site in four), lack of detail regarding the diagnostic threshold used (two studies), or interpretation by a non‐expert observer (one study). It was of note that 15 of the 20 cohorts were produced by, or in collaboration with, the same expert research team, led by Professor Pellacani, which may further reduce the generalisability of results.
Sixteen cohorts reported use of an acceptable reference standard, but only two clearly reported blinding of the reference standard to the RCM result. None of the cohorts reported blinding of histology to the referral diagnosis (based on clinical examination or dermoscopy), but this was not incorporated into the overall risk of bias for this domain. For the applicability of the reference standard, two reported using expert diagnosis for some lesions and 13 were unclear regarding histopathology interpretation by an experienced histopathologist or by a dermatopathologist.
Six cohorts did not use the same reference standard for all participants (differential verification), 11 were unclear on the interval between the application of the index test and excision for histology, and seven did not include all participants in the analysis primarily due to technical difficulties in imaging.
For the six cohorts comparing RCM with dermoscopy, three reported blinding between tests and three reported no blinding, but this did not contribute to the overall assessment of risk of bias. One study did not clearly report the interval between tests. The clinical applicability of the application of the tests was of high concern due to reporting of mean results for both tests (one study) and of unclear concern due to the image based nature of test interpretation (five studies).
Findings
Primary target condition: invasive melanoma and atypical intraepidermal melanocytic variants
In this section, we presented the results for studies of RCM versus dermoscopy for the primary target condition of invasive melanoma and atypical intraepidermal melanocytic variants (i.e. invasive malignant melanoma and melanoma in situ or lentigo maligna), according to the study population: studies in all those with 'any lesion suspicious for melanoma' versus those in participants with equivocal lesions. The studies used a number of different algorithms to assist RCM diagnosis; these are described in detail in Appendix 5.
Studies using reflectance confocal microscopy in any lesion suspicious for melanoma
The following section documents studies where RCM was used in all participants with lesions scheduled for excision. These populations included both clinically or dermoscopically obvious melanomas, along with some lesions that were likely to be benign, and a proportion of more difficult to diagnose (equivocal) lesions so that RCM was being evaluated as an addition to current practice (which may or may not have included dermoscopy).
Eight publications provided data for nine evaluations of RCM alone (Curchin 2011; Guitera 2009b (Modena); Guitera 2009c (Sydney); Guitera 2012; Koller 2011; Langley 2007; Pellacani 2014c (doc); Rao 2013; Segura 2009), three of which also included dermoscopy (Guitera 2009b (Modena); Guitera 2009c (Sydney); Langley 2007) (Table 3; Figure 8). All studies were case series (seven prospective in design and two unclear). Studies were undertaken in Europe (4; 44%), Oceania (2; 22%), North America (2; 22%), or in more than one continent (1; 11%). Four studies (44%) were undertaken in a secondary care setting, three (33) in specialist skin cancer units, and two (22%) in mixed secondary care and specialist units. Six cohorts reported inclusion of lesions scheduled for excision on the basis of clinical (Guitera 2012; Langley 2007), or dermoscopic (Guitera 2009b (Modena); Guitera 2009c (Sydney); Guitera 2012; Pellacani 2014c (doc)), suspicion of melanoma or due to lesions changes on follow‐up (Guitera 2009b (Modena); Guitera 2009c (Sydney); Langley 2007; Segura 2009). Two further cohorts included lesions scheduled for excision but did not describe any prior testing of participants (Curchin 2011; Rao 2013), and one provided no information as to lesion selection (Koller 2011). Two studies reported including any type of lesion (22%) (Curchin 2011; Rao 2013), three restricted to pigmented lesions only (33%) (Guitera 2012; Langley 2007; Pellacani 2014c (doc)), and four restricted to melanocytic (44%) lesions only (Guitera 2009b (Modena); Guitera 2009c (Sydney); Koller 2011; Segura 2009). Four studies (44%) excluded acral (Guitera 2009b (Modena); Guitera 2009c (Sydney); Guitera 2012) or awkwardly sited lesions (Langley 2007), and five (56%) reported excluding on RCM image quality (Guitera 2009b (Modena); Guitera 2009c (Sydney); Koller 2011; Langley 2007; Pellacani 2014c (doc)).
2. Summary characteristics for studies reporting reflectance confocal microscopy accuracy for the primary outcome.
| Characteristic | Any suspicious lesion | Equivocal |
| Number of publications | 8 | 7 |
| RCM datasets | 9 | 7 |
| Dermoscopy datasets | 3; 33.3% | 3; 42.9% |
| Study design | ||
| Prospective case series | 7; 77.8% | 3; 42.9% |
| Retrospective case series | – | 4; 57.1% |
| ‐ with prospective reinterpretation of images | – | 3; 75.0% |
| Case series (unclear data collection) | 2; 22.2% | – |
| Continent | ||
| Europe | 4; 44.4% | 7; 100.0% |
| North America | 2; 22.2% | – |
| Australasia | 2; 22.2% | – |
| Multicentre | 1; 11.1% | – |
| Setting | ||
| Secondary | 4; 44.4% | 3; 42.9% |
| Specialist clinic | 3; 33.3% | 4; 57.1% |
| Mixed | 2; 22.2% | – |
| Prior testing | ||
| Clinical examination | 1; 11.1% | – |
| Clinical examination or dermoscopy | 2; 22.2% | 2; 28.6% |
| Clinical examination and dermoscopy | 4; 44.4% | 3; 42.9% |
| Follow‐up of atypical lesions | – | 2; 28.6% |
| Selected for biopsy or excision | 2; 22.2% | – |
| Lesion characteristics | ||
| Any lesion (pigmented or nonpigmented) | 2; 22.2% | 1; 14.3% |
| Pigmented | 3; 33.3% | 1; 14.3% |
| Melanocytic only | 4; 44.4% | 5; 71.4% |
| Exclusion criteria | ||
| Excludes by site (acral/awkwardly sited) | 3; 33.3% | – |
| Excludes on image quality | 5; 55.6% | 3; 42.9% |
| Participant characteristics | ||
| Number of participants (median (range)) | 137 (42 to 195); 6 studies | 70 (62 to 264); 5 studies |
| Number of lesions (median (range)) | 131 (50 to 323) | 100 (60 to 308) |
| Lesion:participant ratio (median (range)) | 1.07 (1 to 1.19); 7 studies | 1.05 (1 to 1.22); 5 studies |
| Disease prevalence (mean (range)) | 27.6% (2.8% to 41.5%) | 18.2% (1.9 to 34.8%) |
| Melanoma in situ as % of disease positive | 25.0% (7.7% to 51.4%) | 28.6% (8.3 to 61.5%) |
| Vivascope | ||
| Vivascope 1000 | 2; 22.2% | – |
| Vivascope 1500 | 5; 55.6% | 7; 100.0% |
| Vivascope 1000 followed by 1500 | 2; 22.2% | – |
| RCM algorithms | ||
| No algorithm: observer diagnosis | 2; 22.2% | 1; 14.3% |
| No algorithm: selected lesion characteristics |
1; 11.1% | – |
| No algorithm: significant lesion characteristics |
– | 1; 14.3% |
| RCM score (including NR) | 5; 55.6% | 2; 28.6% |
| Segura algorithm | 1; 11.1% | 1; 14.3% |
| Pellcani 2‐step algorithm (including modified) | 0; 0.0% | 2; 28.6% |
| Diagnostic method | ||
| In‐person (real time interpretation) | 3; 33.3% | 1; 14.3% |
| Image based (remote interpretation) | 6; 66.7% | 6; 85.7% |
| RCM guided by dermoscopic image | ||
| Yes | 6; 66.7% | – |
| NR | 3; 33.3% | 7; 100.0% |
| Other test data available to observer | ||
| None | 2; 22.2% | 3; 42.9% |
| Lesion site, participant age or gender | 3; 33.3% | – |
| Dermoscopy image alone | 1; 11.1% | 2; 28.6% |
| Dermoscopy image plus participant age, site, or gender |
– | 1; 14.3% |
| In‐person (including dermoscopy) | 2; 22.2% | – |
| Unclear | 1; 11.1% | 1; 14.3% |
| Test interpretation | ||
| Number of observers (median (range)) | 1 (3 studies) 2 (4 studies) | 1 (4 studies) 3 (3 studies) |
| Single | 9; 100.0% | 5; 71.4% |
| Consensus of 3 | – | 1; 14.3% |
| Not reported | – | 1; 14.3% |
| Observer qualifications | ||
| Dermatologist | 4; 44.4% | 5; 71.4% |
| Not reported | 5; 55.6% | 2; 28.6% |
| Observer experience in practice | ||
| High | 3; 33.3% | 4; 57.1% |
| Not reported | 6; 66.7% | 3; 42.9% |
| Observer experience with RCM | ||
| High | 6; 66.7% | 5; 71.4% |
| Not reported | 3; 33.3% | 2; 28.6% |
| Reference Standard | ||
| Histology alone | 8; 88.9% | 6; 85.7% |
| Histology and clinical follow‐up | – | 1; 14.3% |
| Histology and expert diagnosis | 1; 11.1% | – |
NR: not reported; RCM: reflectance confocal microscopy.
8.

Forest plot of tests: reflectance confocal microscopy (RCM) and dermoscopy data in any lesion suspicious for melanoma for detection of invasive melanoma (malignant melanoma (MM)) and atypical intraepidermal melanocytic variants (or melanoma in situ (MiS)). FN: false negative; FP: false positive; TN: true negative; TP: true positive.
The median sample size was 137 participants (range 42 to 195; reported in six studies) and 131 lesions (range 50 to 323). The median lesion:participant ratio was 1.07 (range 1 to 1.19) in seven studies (and not stated in Koller 2011 or Rao 2013). Five studies gave mean age and ranged from 41 to 53 years and mean percentage of men ranged from 39.9% to 54.3%. The mean prevalence of disease was 27.6% (range 2.8% to 41.5%). On average, melanoma in situ lesions made up 25% of the disease positive group, ranging from 7.7% to 51.4%. The spectrum in the disease negative groups also varied between studies with three studies including only benign melanocytic naevi (Koller 2011; Langley 2007; Segura 2009), three also including Spitz naevi (ranging from 3% (Guitera 2009c (Sydney)) to over 10% (Guitera 2009b (Modena); Guitera 2012)), and the three remaining studies including BCC, SCC, and seborrhoeic or actinic (or both) keratosis (Curchin 2011; Pellacani 2014c (doc); Rao 2013), amongst others (Pellacani 2014c (doc); Rao 2013).
Five studies used the Vivascope 1500 imaging system, two used Vivascope 1000, and the remaining two initially used the Vivascope 1000 and moved on to the Vivascope 1500 model during the course of the study. Six studies reported using dermoscopic images to help guide acquisition of RCM images. All studies reported diagnosis for a single observer rather than for a consensus of observers or mean value. Observers were dermatologists in four studies (44%), with three studies reporting observers to be expert or with high levels of experience in practice and six (67%) with high levels of experience with RCM. The remaining studies did not report these characteristics. Three studies undertook diagnosis in‐person with real time interpretation of RCM images. The remaining six studies undertook interpretation remotely based on RCM images alone (two studies); alongside the dermoscopic image of the same lesion (one study); or with information provided only on lesion site, participant age, or gender (three studies).
Eight studies made the reference standard diagnosis by histology alone (i.e. all lesions either excised or biopsied) and the remaining study based expert diagnosis opinion on "unequivocal clinical and conventional dermoscopic criteria" to establish the final diagnosis for 46% (31 participants) of the disease negative group (Koller 2011).
Reflectance confocal microscopy
The nine evaluations of RCM reported using Pellacani's RCM score for four datasets (50%). Curchin 2011 also applied the Guitera score for lesions suspected of lentigo maligna of the face (Guitera 2010). One study developed and applied the Segura algorithm (Segura 2009). The remaining studies reported test accuracy for selected RCM characteristics (Langley 2007), or for observer diagnosis of melanoma (Koller 2011; Rao 2013).
Estimates of sensitivities ranged from 63% to 100% and specificities from 57% to 95% (Figure 8). The low sensitivity of 63% in Koller 2011 appeared as an outlier, all other studies having values at or above 86%. Similarly, specificities were above 82% in all studies except Pellacani 2014c (doc) (57%), Guitera 2009b (Modena) (58%), and Guitera 2012 (62%). Guitera 2009b (Modena) and Guitera 2012 both had higher than expected percentages of Spitz nevi (19% for Guitera 2009b (Modena) and 11% for Guitera 2012), whereas Koller 2011 was one of two studies to use the Vivascope 1000 throughout. The lower specificity in Pellacani 2014c (doc) was more difficult to explain, but may have been related to the fact that all included lesions were considered to require excision based on dermoscopy alone, which may have affected the case‐mix of lesions in a way that we were not able to identify. Correctly identified BCC lesions were considered true negatives for the purposes of these calculations for Guitera 2012 and Rao 2013.
We pooled results across algorithms and thresholds as an SROC curve (Figure 9). Estimates of accuracy obtained from the curve suggested that the specificity of RCM would be 82% at a fixed threshold of 90% sensitivity (Table 4).
9.
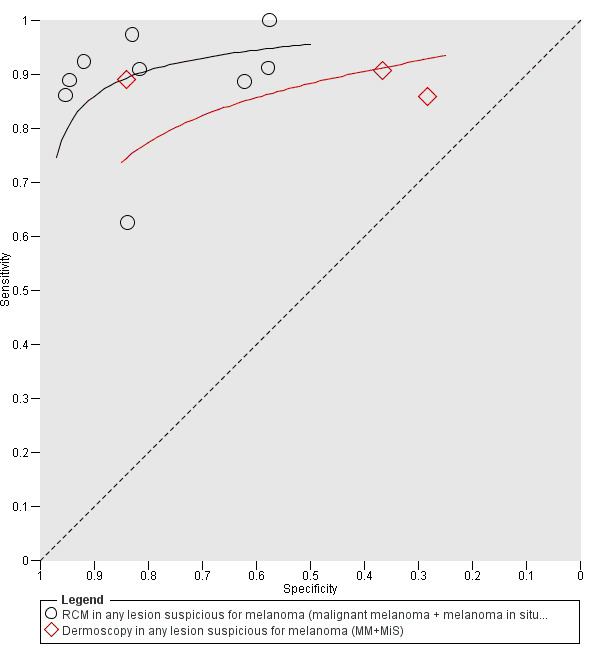
Summary receiver operating characteristic (ROC) comparing reflectance confocal microscopy (RCM) and dermoscopy in all lesions suspected of melanoma for detection of invasive melanoma or atypical intraepidermal melanocytic variants (malignant melanoma (MM) + melanoma in situ (MiS)).
3. Comparison of reflectance confocal microscopy with dermoscopy.
| Test | Studies | Participants |
DOR (95% CI) |
Specificity at 90% sensitivity |
Relative DOR (95% CI) |
P valuea (DOR) |
P valueb (HSROC models) |
| 'Any lesion suspicious for melanoma' studies (all studies) | |||||||
| RCM | 9 | 1452 | 57.5 (18.5 to 179.4) |
82% | 4.82 (2.16 to 10.8) |
0.0001 | < 0.001 |
| Dermoscopy | 3 | 451 | 14.4 (2.7 to 77.6) |
42% | |||
| 'Any lesion suspicious for melanoma' studies (direct comparisons) | |||||||
| RCM | 3 | 451 | 251.3 (5.7 to 11,050) |
93% | 4.96 (1.1 to 21.5) |
0.03 | < 0.001 |
| Dermoscopy | 3 | 451 | 50.6 (1.6 to 1634) |
41% | |||
| Equivocal lesion studies (all studies) | |||||||
| RCM | 7 | 1177 | 97.6 (30.3 to 313.8) |
86% | 20.1 (6.6 to 61.3) |
< 0.001 | < 0.001 |
| Dermoscopy | 3 | 645 | 3.0 (1.3 to 6.8) |
49% | |||
| Equivocal lesion studies (direct comparisons) | |||||||
| RCM | 3 | 645 | 154.5 (16.4 to 1457) |
94% | 22.1 (1.7 to 283.6) |
0.03 | < 0.001 |
| Dermoscopy | 3 | 645 | 7.0 (2.1 to 23.6) |
44% | |||
CI: confidence interval; DOR: diagnostic odds ratio; HSROC: hierarchical summary receiver operating characteristic curve; RCM: reflectance confocal microscopy.
aThe P value assessed whether the observed difference in DOR between RCM and dermoscopy was explicable by chance.
bThe P value was a global test assessing whether the observed differences in all HSROC parameters (accuracy and threshold) between RCM and dermoscopy was explicable by chance.
Comparison of reflectance confocal microscopy versus dermoscopy
The three evaluations of dermoscopy that were included in these RCM studies reported using pattern analysis to assist dermoscopy interpretation; two conducted them in‐person (Guitera 2009b (Modena); Langley 2007), and one was based on dermoscopic images with information on lesion site and participant age only (Guitera 2009c (Sydney)). Sensitivities for dermoscopy ranged from 86% to 91%; specificities ranged from 28% to 84% (Figure 8). The accuracy of dermoscopy was compared with the accuracy of RCM estimated from (a) all nine RCM studies (Figure 9) and estimated from direct comparisons in (b) with the subset of three studies that evaluated both RCM and dermoscopy (Figure 10). In both comparisons, the accuracy of RCM exceeded that of dermoscopy (Table 4). In (a), the DOR for RCM was 4.82 (95% CI 2.16 to 10.8; P = 0.0001) times that of the dermoscopy, in (b) it was 4.96 (95% CI 1.1 to 21.5; P = 0.03) times that of the dermoscopy. These effects corresponded to predicted differences in specificity of (a) 40% (82% versus 42%) and (b) 52% (93% versus 41%) at a fixed sensitivity of 90% (Table 4).
10.
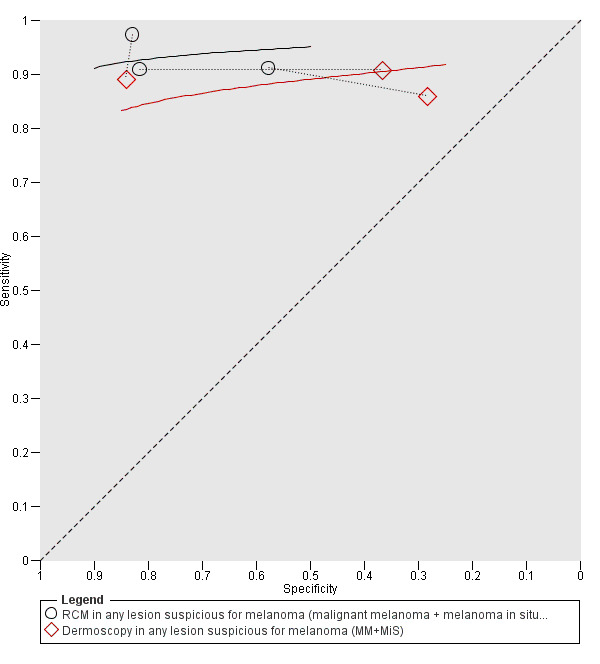
Summary receiver operating characteristic (ROC) of paired comparisons of reflectance confocal microscopy (RCM) and dermoscopy in all lesions suspected of melanoma for detection of invasive melanoma or atypical intraepidermal melanocytic variants (malignant melanoma (MM) + melanoma in situ (MiS)).
Equivocal lesion studies
We defined equivocal lesion studies as those in which RCM was used in participants with equivocal lesions in whom a clear management decision could not be made following visual inspection or dermoscopy (or both) (i.e. RCM was being evaluated as a potential addition to dermoscopy).
Seven publications provided data for seven evaluations of RCM alone (Alarcon 2014a; Farnetani 2015; Ferrari 2015; Lovatto 2015; Pellacani 2012; Pellacani 2014b (cons); Stanganelli 2015) and three of dermoscopy (Alarcon 2014a; Ferrari 2015; Stanganelli 2015) (Table 4; Figure 11). All studies were case series; three (43%) were prospective in design and four (57%) retrospective, three of which prospectively reinterpreted previously acquired RCM images. Studies were all undertaken in Europe (100%). Three studies (43%) were undertaken in a secondary care setting and four (57%) in specialist skin cancer units. All studies reported some degree of prior testing of participants, with two (29%) selecting lesions that were equivocal on either clinical examination or dermoscopy (Farnetani 2015; Pellacani 2012), three (43%) with all lesions equivocal on dermoscopy (Alarcon 2014a; Ferrari 2015; Pellacani 2014b (cons)), and two (29%) selecting lesions showing changes on digital follow‐up (Lovatto 2015; Stanganelli 2015). One study reported including any type of lesion (14%), one restricted to pigmented lesions only (14%), and five restricted to melanocytic (71%) lesions only. Three (43%) studies reported excluding lesions on RCM image quality.
11.
Forest plot of tests: reflectance confocal microscopy (RCM) and dermoscopy in equivocal lesion populations for detection of invasive melanoma (malignant melanoma (MM)) and atypical intraepidermal melanocytic variants (melanoma in situ (MiS). FN: false negative; FP: false positive; TN: true negative; TP: true positive.
The median sample size was 70 participants (range 62 to 264; reported in five studies) and 100 lesions (range 60 to 308), giving a median lesion:participant ratio of 1.05 (range 1 to 1.22). Five studies reported mean age, which ranged from 39 to 54.7 years and mean percentage of men was from 44.0% to 54.0%. The mean prevalence of the primary target condition of 18.2% (range 1.9% to 34.8%) was lower compared to the studies in any lesion suspicious for melanoma as would be expected in a group of more difficult to diagnose lesions. On average, melanoma in situ lesions made up 28.6% of the disease positive group, ranging from 8.3% to 61.5% (breakdown reported for four datasets). The spectrum in the disease negative groups also varied with four studies including only (Lovatto 2015) or primarily (Ferrari 2015; Pellacani 2012; Stanganelli 2015) benign naevi, although in one of these, non‐dysplastic naevi made up 41% of the disease negative group. Three included BCC and a range of other diagnoses including seborrhoeic or actinic keratosis (Alarcon 2014a; Farnetani 2015), or Spitz naevi (Pellacani 2014b (cons)).
All studies in this group used the Vivascope 1500 imaging system (100%); none reported the use of dermoscopic images to help guide acquisition of RCM images. Diagnosis was by a single observer in five (71%) studies, for a consensus of three observers in one study, and was not reported in the remaining study. Observers were qualified dermatologists in five studies (71%), and four studies reported observers to have high levels of experience in practice and five (71%) reported high levels of experience with (or training in) RCM. The remaining studies did not report these observer characteristics. One study undertook diagnosis in‐person with real time interpretation of RCM images; the remaining six studies undertook test interpretation remotely based on RCM images alone (three studies) or alongside the dermoscopic image of the same lesion (three studies), with information provided only on lesion site, participant age, or gender in one of these.
Six studies made the reference standard diagnosis by histology alone (i.e. all lesions either excised or biopsied) and in the remaining study, 227/308 lesions referred for RCM consultation underwent surveillance using sequential digital dermoscopy follow‐up and cancer registry searches for those lost to follow‐up; 28 lesions were excised during follow‐up and found to be benign (Pellacani 2014b (cons)).
Reflectance confocal microscopy
The seven evaluations of RCM reported using Pellacani's RCM score (Lovatto 2015), or use of the RCM score was assumed due to study authorship (Pellacani 2014b (cons)), the Segura algorithm (Alarcon 2014a), or the Pellacani two step algorithm for dysplastic naevi and melanoma (Pellacani 2012; Stanganelli 2015). The remaining studies reported test accuracy for the presence of statistically significant RCM characteristics (Ferrari 2015), or for observer diagnosis of melanoma (Farnetani 2015).
Estimates of sensitivities ranged from 80% to 100% and specificities from 67% to 95% (Figure 11). There were no obvious outliers or heterogeneity in sensitivities, and no consistent differences to potentially explain the observed heterogeneity in specificities. Correctly identified BCC lesions were considered true negatives for the purposes of these calculations for Farnetani 2015 and Pellacani 2014b (cons).
We pooled results across algorithms and thresholds as an SROC curve (Figure 12). Estimates of accuracy obtained from the curve suggested that specificity would be 86% at a fixed threshold of 90% sensitivity (Table 4). These values for specificity were higher than those observed in studies in any lesion suspicious for melanoma, reflecting the marginally higher values and lower variability of sensitivities in the equivocal lesion studies.
12.
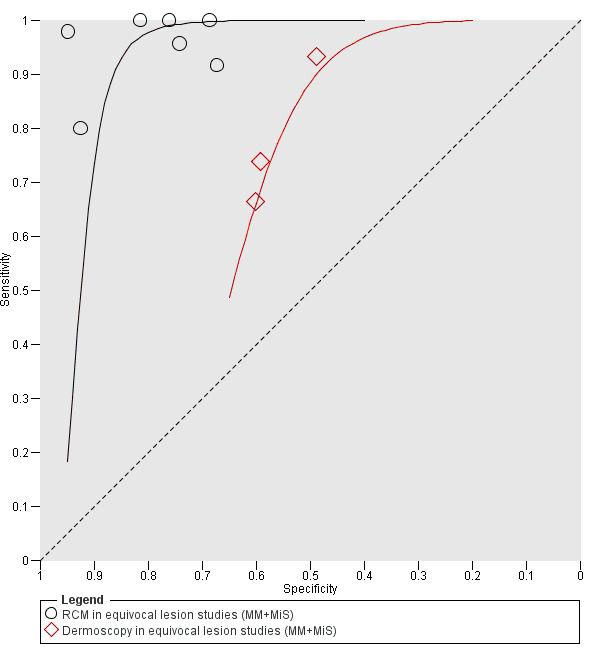
Summary receiver operating characteristic (ROC) comparing reflectance confocal microscopy (RCM) and dermoscopy in equivocal lesion populations for detection of invasive melanoma (malignant melanoma (MM)) and atypical intraepidermal melanocytic variants (or melanoma in situ (MiS)).
Comparison of reflectance confocal microscopy versus dermoscopy
The three evaluations of dermoscopy that were included in these RCM studies reported using the seven point checklist for dermoscopy (Ferrari 2015), or a revised version thereof (Stanganelli 2015), or did not report the approach to dermoscopy interpretation (Alarcon 2014a). All were image based diagnoses; two studies provided the RCM image with (Alarcon 2014a) or without (Ferrari 2015) additional participant or lesion information to assist diagnosis, and one providing a baseline dermoscopic image (Stanganelli 2015).
The accuracy of dermoscopy was compared with the accuracy of RCM estimated from (a) all seven RCM studies (Figure 12) and estimated from direct comparisons in (b) with the subset of three studies that evaluated both RCM and dermoscopy (Figure 13). The meta‐analytical model for the paired analysis (b) required assumptions of a symmetrical SROC curve and fixed effects for accuracy and threshold to obtain convergence.
13.
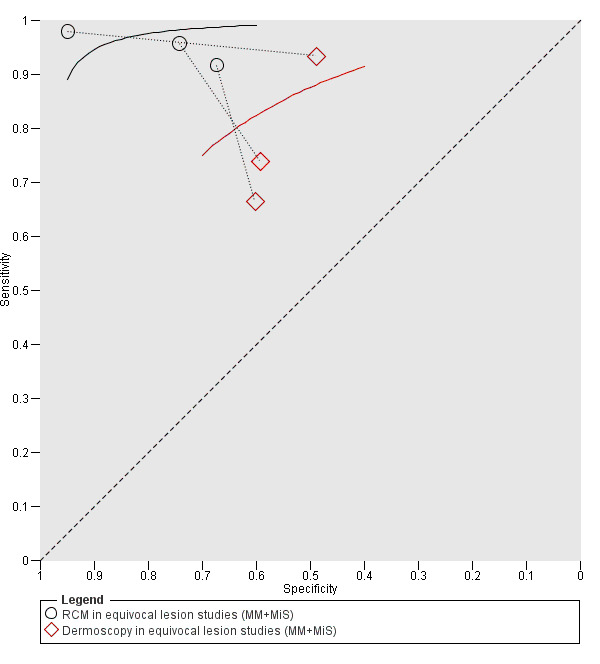
Summary receiver operating characteristic (ROC) for paired comparisons of reflectance confocal microscopy (RCM) and dermoscopy in equivocal lesion populations for detection of invasive melanoma (malignant melanoma (MM)) and atypical intraepidermal melanocytic variants (or melanoma in situ (MiS)).
It is notable that the accuracy of dermoscopy in these studies (DOR 3.0, 95% CI 1.3 to 6.8) was much lower than in those in any lesion suspicious for melanoma (DOR 14.4, 95% CI 2.7 to 77.6), as would be expected given that, by definition, these studies are those where diagnoses involving dermoscopy are equivocal (i.e. they include lesions to be excised because a clear diagnosis could not be reached on clinical examination or dermoscopy). In both comparisons, the accuracy of RCM exceeded that of dermoscopy (Table 4). In (a) the DOR for RCM was 20.1 (95% CI 6.6 to 61.3; P<0.001) times that for dermoscopy, in (b) it was 22.1 (95% CI 1.7 to 283.6; P = 0.03) times that of the dermoscopy. These effects corresponded to predicted differences in specificity of (a) 37% (86% versus 49%) and (b) 50% (94% versus 44%) at a fixed sensitivity of 90% (Table 4).
Analyses by algorithms used to assist reflectance confocal microscopy: all studies
The 18 included studies provided 25 datasets evaluating the accuracy of different algorithms or approaches to diagnosis with RCM at a number of different thresholds for test positivity for the detection of invasive melanoma or atypical intraepidermal melanocytic variants. A description of the various algorithms and thresholds is provided in Appendix 5. We excluded one dataset from Pellacani 2007a due to overlap in study population, algorithm, and threshold with a study by Guitera and colleagues (Guitera 2009b (Modena); Guitera 2009c (Sydney)).
Figure 14 provides forest plots of all algorithms for the detection of invasive melanoma or atypical intraepidermal melanocytic variants, with meta‐analytical estimates at each threshold presented in Table 5. We did not formally make any comparisons between the algorithms due to the small number of studies available evaluating each algorithm. Whilst the specificity of the computer assisted approach to analysis of RCM images (Koller 2011) appears to be much lower than any other algorithm, the ranges of values for different algorithms are largely comparable.
14.
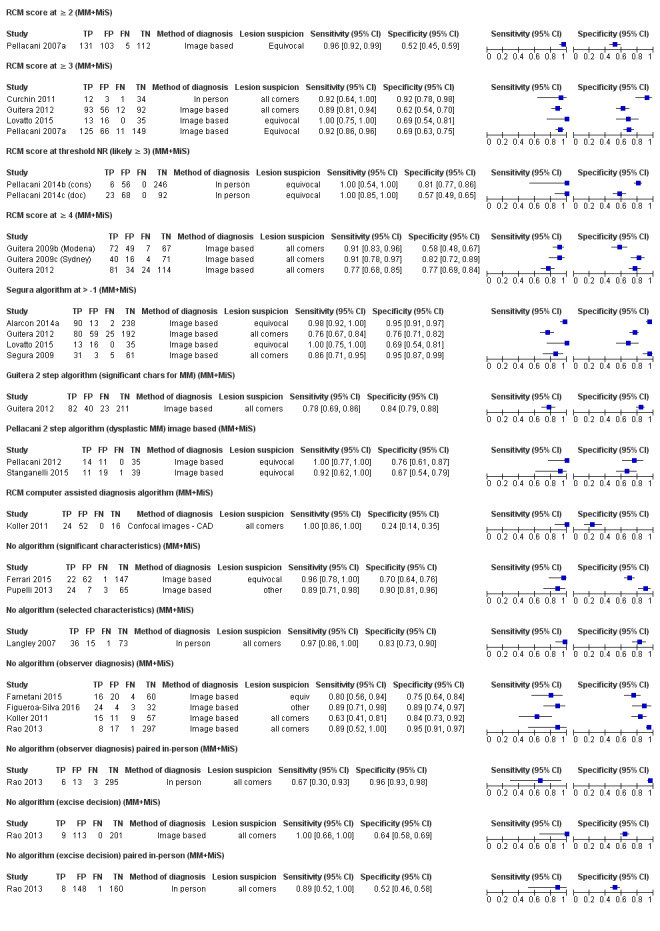
Forest plot: reflectance confocal microscopy (RCM) results by algorithm, threshold, and number of observers for diagnosis of invasive melanoma (malignant melanoma (MM)) and atypical intraepidermal melanocytic variants (or melanoma in situ (MiS)). FN: false negative; FP: false positive; TN: true negative; TP: true positive.
4. Pooled sensitivity and specificity for individual algorithms.
| Person/image |
Target condition Test |
Number of studies | Number of participants |
Pooled sensitivity (95% CI) |
Pooled specificity (95% CI) |
| Detection of invasive melanoma (MM) | |||||
| In‐person | RCM ≥ 3 | 1 | 50 | 1.00 (0.74 to 1.00) | 0.92 (0.79 to 0.98) |
| Image based | Segura > ‐1 | 1 | 100 | 0.96 (0.78 to 1.00) | 0.84 (0.74 to 0.92) |
| Image based | Guitera 2‐step (significant characteristics for MM) | 1 | 356 | 0.78 (0.65 to 0.89) | 0.86 (0.81 to 0.90) |
| Image based | No algorithm (observer diagnosis) | 1 | 63 | 0.88 (0.47 to 1.00) | 0.62 (0.48 to 0.75) |
| Image based | No algorithm (significant characteristics) | 1 | 140 | 1.00 (0.85 to 1.00) | 0.91 (0.85 to 0.96) |
| Image based | No algorithm (any threshold) | 2 | 203 | 0.98 (0.27 to 1.00) | 0.81 (0.52 to 0.94) |
| Detection of invasive melanoma or atypical intraepidermal melanocytic variants (MM+MiS) | |||||
| Image based | RCM ≥ 2 | 1 | 351 | 0.96 (0.92 to 0.99) | 0.52 (0.45 to 0.59) |
| Image based | RCM ≥ 3 | 3 | 668 | 0.91 (0.87 to 0.94)a | 0.67 (0.62 to 0.71)a |
| In‐person | RCM ≥ 3 | 1 | 50 | 0.92 (0.64 to 1.00) | 0.92 (0.78 to 0.98) |
| In‐person | RCM unstated but likely ≥ 3 | 2 | 366 | 1.00 (0.88 to 1.00)b | 0.62 (0.40 to 0.80) |
| Both | RCM ≥ 3 or likely ≥ 3 | 6 | 1209 | 0.92 (0.87 to 0.95) | 0.72 (0.62 to 0.81) |
| Image based | RCM ≥ 4 | 3 | 604 | 0.86 (0.76 to 0.92) | 0.73 (0.60 to 0.82) |
| Image based | Segura > ‐1 | 4 | 863 | 0.93 (0.76 to 0.98) | 0.88 (0.72 to 0.95) |
| Image based | Guitera 2‐step (significant characteristics) | 1 | 356 | 0.78 (0.69 to 0.86) | 0.84 (0.79 to 0.88) |
| Image based | Pellacani 2‐step | 2 | 130 | 0.96 (0.72 to 1.00) | 0.71 (0.61 to 0.79) |
| Image based | No algorithm (observer diagnosis) | 4 | 578 | 0.81 (0.65 to 0.91) | 0.88 (0.78 to 0.94) |
| In‐person | No algorithm (observer diagnosis) | 1 | 317 | 0.67 (0.30 to 0.93) | 0.96 (0.93 to 0.98) |
| Image based | No algorithm (significant characteristics) | 2 | 331 | 0.93 (0.78 to 0.98) | 0.81 (0.63 to 0.92) |
| In‐person | No algorithm (selected characteristics) | 1 | 125 | 0.97 (0.86 to 1.00) | 0.83 (0.73 to 0.90) |
| Image based | No algorithm (excise decision) | 1 | 323 | 1.00 (0.66 to 1.00) | 0.64 (0.58 to 0.69) |
| In‐person | No algorithm (excise decision) | 1 | 317 | 0.89 (0.52 to 1.00) | 0.52 (0.46 to 0.58) |
| Image based | RCM computer assisted | 1 | 92 | 1.00 (0.86 to 1.00) | 0.24 (0.14 to 0.35) |
| Detection of any skin cancer (melanoma or other skin cancer) or skin lesion with a high risk of progression to melanoma (any) | |||||
| In‐person | RCM ≥ or likely 3 | 3 | 541 | 0.98 (0.91 to 0.99)c | 0.75 (0.54 to 0.89)a |
| Image based | Segura > ‐1 | 1 | 356 | 0.80 (0.73 to 0.86) | 0.70 (0.63 to 0.77) |
| Image based | Pellacani two‐step | 1 | 60 | 0.89 (0.67 to 0.99) | 0.80 (0.65 to 0.91) |
| Image based | Guitera 2‐step (significant characteristics) | 1 | 356 | 0.92 (0.86 to 0.95) | 0.68 (0.61 to 0.75) |
| In‐person | No algorithm (observer diagnosis) | 1 | 317 | 0.78 (0.67 to 0.87) | 0.85 (0.80 to 0.89) |
| Image based | No algorithm (observer diagnosis) | 2 | 423 | 0.85 (0.77 to 0.90) | 0.87 (0.83 to 0.90) |
| In‐person | No algorithm (excise decision) | 1 | 317 | 0.85 (0.75 to 0.92) | 0.61 (0.55 to 0.68) |
| Image based | No algorithm (excise decision) | 1 | 323 | 0.90 (0.81 to 0.95) | 0.79 (0.73 to 0.84) |
CI: confidence interval; MiS: melanoma in situ (or lentigo maligna); MM: malignant melanoma; RCM: reflectance confocal microscopy.
aComputed without correlation between sensitivity and specificity.
bComputed using unstratified data as no false negatives.
cZero variance assumed for sensitivity random effect.
Pellacani's reflectance confocal microscopy score
Pellacani's RCM score was the most commonly evaluated formal algorithm for the detection of melanoma (eight studies; 10 datasets), with data reported at thresholds of two or greater, three or greater, and four or greater (Pellacani 2005; Pellacani 2007a). One study did not report the threshold used and contact with authors was unsuccessful (Pellacani 2014b (cons); Pellacani 2014c (doc)); as it cited one of the original Pellacani and colleagues papers (Pellacani 2007a), we assumed the recommended threshold of three or greater. The majority of datasets were image based, with only two studies providing data for in‐person evaluations (Curchin 2011; Pellacani 2014b (cons); Pellacani 2014c (doc)). The majority of datasets were from studies in any lesion suspicious for melanoma (Curchin 2011; Guitera 2009b (Modena); Guitera 2009c (Sydney); Guitera 2012; Pellacani 2007a; Pellacani 2014c (doc)), with only two from studies of equivocal lesions (Lovatto 2015; Pellacani 2014b (cons)). One study provided two datasets at different thresholds (Pellacani 2007a); hence, the total number of datasets was 10.
The pooled accuracy combining data from all six studies reporting (or assumed to be) at RCM score three or greater was a sensitivity of 92% (95% CI 87% to 95%) and specificity of 72% (95% CI 62% to 81%). Lower thresholds had higher sensitivity but lower specificity, higher thresholds had lower sensitivity but higher specificity (Table 5).
Segura score
Three studies evaluated the Segura algorithm, developed in Segura 2009 at the standard threshold of greater than ‐1 (Alarcon 2014a; Guitera 2012; Lovatto 2015). All datasets were image based; test interpretation was blinded to any further information in two (Lovatto 2015; Segura 2009), two provided the observer with participant age and lesion site (Alarcon 2014a; Guitera 2012), one of which also provided the dermoscopic image (Alarcon 2014a). Two studies were conducted in equivocal lesions (Alarcon 2014a; Lovatto 2015), and two in 'any lesion suspicious for melanoma' populations (Guitera 2012; Segura 2009). The pooled accuracy combining data from all four studies was a sensitivity of 92.6% (95% CI 76.2% to 98.0%) and specificity of 87.5% (95% CI 72.2% to 95.0%) (Table 5).
Other formally developed algorithms
Guitera 2012 reported a two step algorithm to first differentiate BCC from other lesions and then melanoma from the remaining lesions. In this single study, the melanoma component of the algorithm demonstrated a sensitivity of 78% (95% CI 69% to 86%) and specificity of 84% (95% CI 79% to 88%).
Pellacani 2012 also developed a two step algorithm, this time to differentiate dysplastic from non‐dysplastic lesions and then melanoma from dysplastic lesions. Stanganelli 2015 evaluated the same algorithm. Combined accuracy was a sensitivity of 96% (95% CI 72% to 100%) and specificity of 71% (95% CI 61% to 79%) (Table 5).
Finally, Koller 2011 reported a computer assisted approach to analysis of RCM images. This demonstrated a perfect sensitivity of 100% (95% CI 86% to 100%) but a very poor specificity of 24% (95% CI 14% to 35%) (Table 5).
'No algorithm' evaluations
Seven studies reported accuracy data for RCM without the use of a formally developed algorithm.
The three datasets reporting accuracy based on the presence of statistically significant characteristics (Ferrari 2015; Pupelli 2013), or selected lesion characteristics (Langley 2007), had sensitivities ranging from 89% to 97% and specificities from 70% to 90%.
The four datasets reporting accuracy for observer diagnosis of melanoma were all image based (all except one (Koller 2011) also providing the dermoscopic image to test interpreters), two conducted in 'any lesion suspicious for melanoma' populations (Koller 2011; Rao 2013), one in equivocal lesions (Farnetani 2015), and one in 'other' populations (Figueroa‐Silva 2016). The pooled accuracy of the four studies gave an estimated sensitivity of 81% (95% CI 65% to 91%) and specificity of 88% (95% CI 78% to 94%).
Rao 2013 provided a direct comparison of image based test interpretation by an experienced observer to in‐person real time diagnosis by a less experience observer. Sensitivity was lower for the in‐person evaluation (67%, 95% CI 30% to 93%) compared to image based (89%, 95% CI 52% to 100%), although CIs were wide and overlapping. Specificities were almost identical for the two approaches (96% versus 95%).
Overall, observed sensitivities appeared higher for studies reporting the use of a 'named' algorithm and were very similar (92%) between studies using the most widely used algorithms (Pellacani's RCM score and the Segura score). Summary specificity was higher for the Segura algorithm (87.5%) compared to the RCM score (72%), although it was used in fewer studies (Segura algorithm used four and RCM score used six), the number of lesions evaluated was higher (784 with Segura algorithm versus 420 with RCM score).
Investigations of heterogeneity
Results for formal investigations of heterogeneity are presented in Table 6, investigating the effects of use of any RCM scale versus no scale (Figure 15), in‐person versus image based (Figure 16), and whether RCM was used in all lesions or only equivocal lesions (Figure 17). Although RCM appeared to be more accurate when interpreted using a scale (relative DOR compared to studies not reporting use of a scale of 1.81, 95% CI 0.41 to 8.03), from in‐person studies (relative DOR in comparison to image based studies 4.77, 95% CI 0.56 to 40.8), and when used on equivocal lesions (relative DOR in comparison to 'any lesion suspicious for melanoma' populations 2.88, 95% CI 0.80 to 10.4), none of the differences reached statistical significance (Table 6).
5. Investigations of heterogeneity in reflectance confocal microscopy accuracy.
| Subgroup | Studies | Participants |
DOR (95% CI) |
Specificity at 90% sensitivity |
Relative DOR (95% CI) |
P value (DOR) |
P value (HSROC models) |
| Differences in participant pathway | |||||||
| Any lesion suspicious for melanoma | 9 | 1452 | 44.5 (19.8 to 99.9) |
81% | 2.88 (0.80 to 10.4) |
0.11 | 0.31 |
| Equivocal lesions | 7 | 1177 | 147.6 (37.2 to 585.7) |
94% | |||
| Differences in‐person and image based | |||||||
| Image based | 12 | 1963 | 54.1 (26.3 to 111.1) |
84% | 4.77 (0.56 to 40.8) |
0.15 | 0.13 |
| In‐person | 4 | 666 | 257.7 (28.7 to 2313) |
97% | |||
| Use of a scaling system | |||||||
| No scale used | 6 | 802 | 45.7 (16.2 to 128.6) |
83% | 1.81 (0.41 to 8.03) |
0.43 | 0.06 |
| Any scale used | 10 | 1663 | 82.8 (28.9 to 236.8) |
90% | |||
CI: confidence interval; DOR: diagnostic odds ratio; HSROC: hierarchical summary receiver operating characteristic.
15.
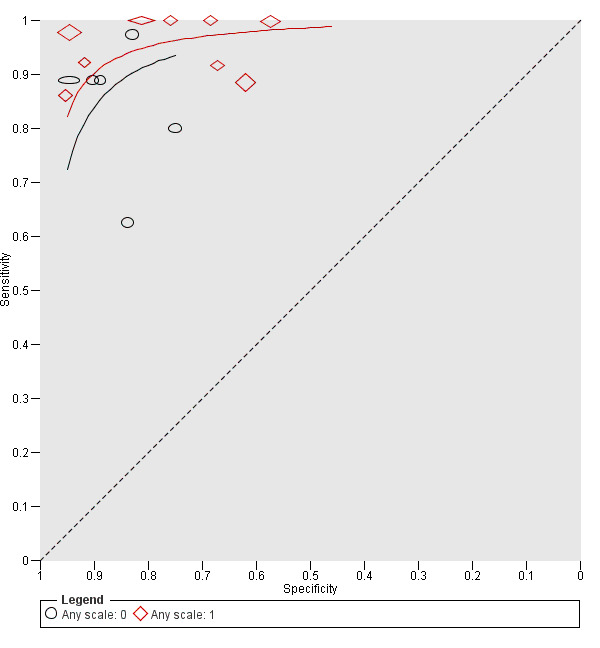
Summary receiver operating characteristic plot comparing studies which used and did not use an algorithm or scale to assist reflectance confocal microscopy (RCM) diagnosis (0 = none, 1 = tool used): outcome was detection of invasive melanoma (malignant melanoma (MM)) and atypical intraepidermal melanocytic variants (or melanoma in situ (MiS)).
16.
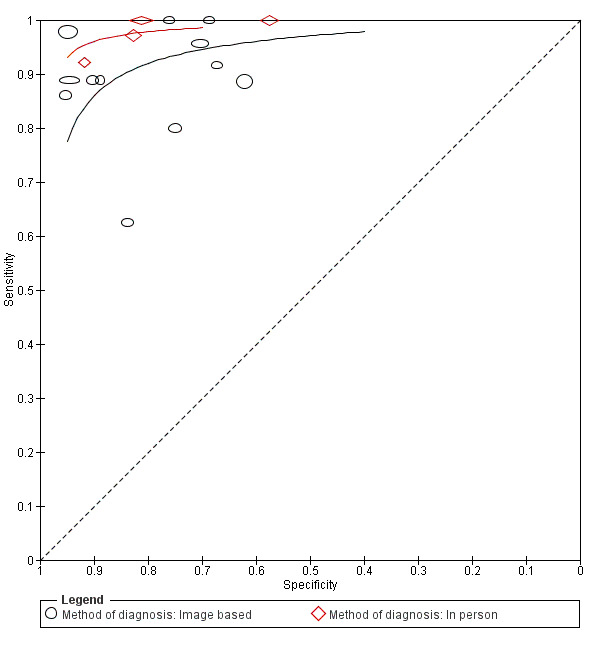
Summary receiver operating characteristic (ROC) plot: comparison of in‐person and Image based studies of reflectance confocal microscopy (RCM) for detection of invasive melanoma (malignant melanoma (MM)) and atypical intraepidermal melanocytic variants (or melanoma in situ (MiS)).
17.
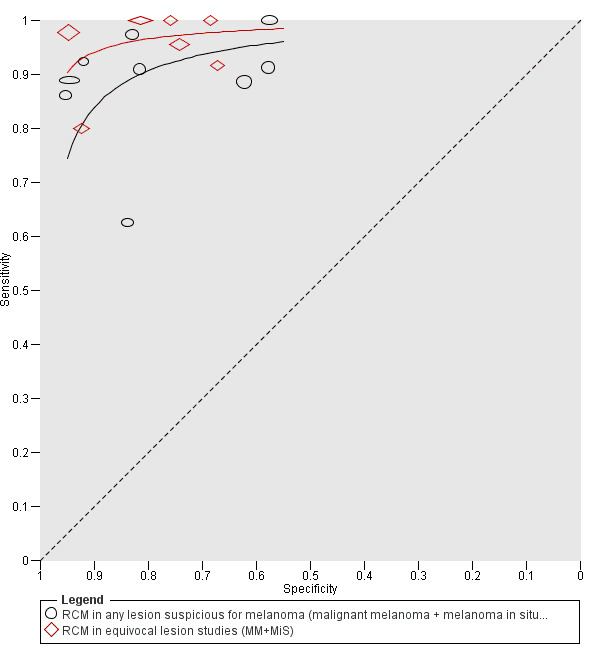
Summary receiver operating characteristic (ROC) plot comparing reflectance confocal microscopy (RCM) performance in studies of all lesions suspected of melanoma with those in participants with equivocal lesions (for detection of invasive melanoma (malignant melanoma (MM)) and atypical intraepidermal melanocytic variants (or melanoma in situ (MiS))).
The impact of observer experience on RCM accuracy is shown in Figure 18 for equivocal lesions and Figure 19 for 'any lesion suspicious for melanoma' populations. Overall, only three studies classified any observer as having low experience (Curchin 2011; Farnetani 2015; Rao 2013), too few to allow any conclusive analyses.
18.
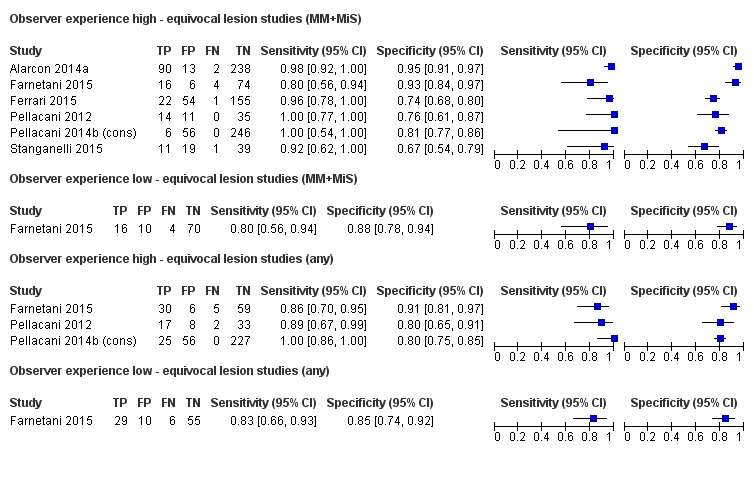
Forest plot: reflectance confocal microscopy (RCM) diagnosis in studies of participants with equivocal lesions by observer experience, for the detection of invasive melanoma (malignant melanoma (MM)), of invasive melanoma and atypical intraepidermal melanocytic variants (or melanoma in situ (MiS)) (MM+MiS), and of any potential skin cancer (melanoma or other skin cancer) or skin lesion with a high risk of progression to melanoma (any). FN: false negative; FP: false positive; TN: true negative; TP: true positive.
19.
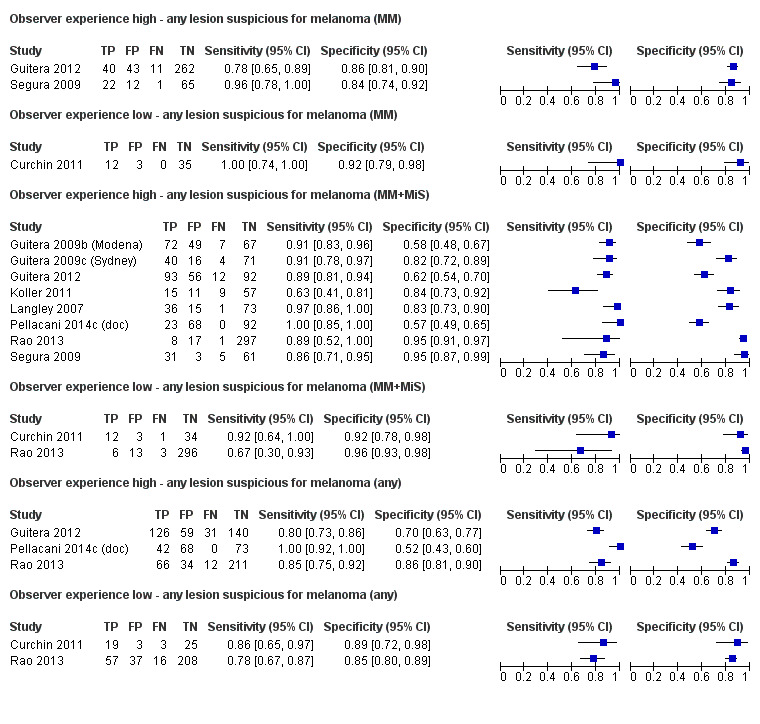
Forest plot: reflectance confocal microscopy (RCM) diagnosis in studies of all lesions suspected of melanoma by observer experience, for the detection of invasive melanoma (malignant melanoma (MM)), of invasive melanoma and atypical intraepidermal melanocytic variants (or melanoma in situ (MiS)) (MM+MiS), and of any potential skin cancer (melanoma or other skin cancer) or skin lesion with a high risk of progression to melanoma (any). FN: false negative; FP: false positive; TN: true negative; TP: true positive.
We were unable to undertake investigations of heterogeneity for other characteristics listed in the protocol due to lack of variation in characteristics, or absence of information in the study reports.
Target condition: invasive melanoma alone
In this section, we presented the results for studies of RCM for the target condition of invasive melanoma only; we identified no comparisons with dermoscopy for this target condition. All studies were conducted in 'any lesion suspicious for melanoma' populations (i.e. there was no attempt to exclude those diagnosed as definite melanomas or as obviously benign on dermoscopy).
Three study cohorts provided data for three evaluations of RCM (Curchin 2011; Guitera 2012; Segura 2009) (Figure 20). All studies were case series (two prospective in design and one unclear). Studies were undertaken in Europe (one), Oceania (one), or in more than one continent (one). Studies were in a secondary care setting (two) or a mixed secondary care specialist unit setting (one). Two studies reported including any type of lesion and one was restricted to melanocytic lesions only. One study excluded keratotic lesions. The sample size ranged from 42 to 330 participants and 50 to 356 lesions. The mean lesion:participant ratio was 1.11 (range 1.07 to 1.19). Mean age was given in two studies and ranged from 49.5 to 53 years; the percentage of men ranged from 39.9% to 53.4%. The mean prevalence of disease was 20.4% (range 14.3% to 24.0%). The percentage of melanoma in situ lesions in the disease negative group ranged from 2.6% to 17.7%.
20.
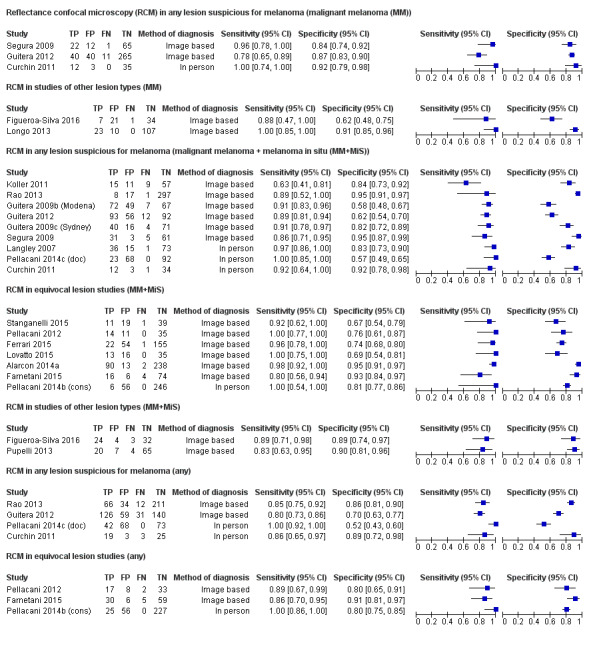
Forest plot of reflectance confocal microscopy (RCM) performance by study group and target condition definition (invasive melanoma alone (malignant melanoma (MM)), invasive melanoma and atypical intraepidermal melanocytic variants, or melanoma in situ (MiS) (MM+MiS), and for any skin cancer or skin lesion with a high risk of progression to melanoma (any)). FN: false negative; FP: false positive; TN: true negative; TP: true positive.
All studies used the Vivascope 1500 imaging system; two reporting the use of dermoscopic images to help the guide acquisition of RCM images. All studies reported diagnosis for a single observer, though only one clearly reported that this was by an experienced dermatologist. One reported in‐person real‐time interpretation of RCM images and two reported RCM interpretation remotely from the participant (one was blinded to any other information, and one supplied details of participant age and lesion site). All studies made the reference standard diagnosis by histology alone.
Segura 2009 developed and applied a new algorithm (denoted the Segura algorithm) to a set of melanocytic lesions at a threshold of greater than ‐1 (excluded BCCs and benign non‐melanocytic lesions); sensitivity was 96% (95% CI 78% to 100%) and specificity was 84% (95% CI 74% to 92%) (Figure 21).
21.
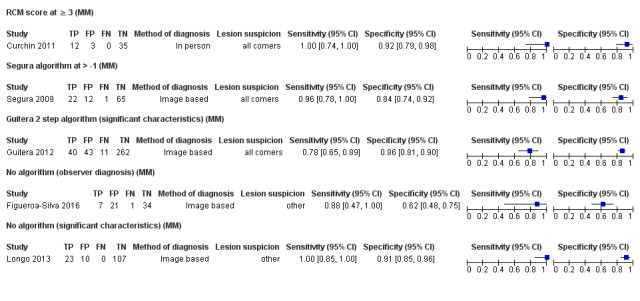
Forest plot: reflectance confocal microscopy (RCM) results by algorithm and threshold for diagnosis of invasive melanoma (MM). FN: false negative; FP: false positive; TN: true negative; TP: true positive.
Curchin 2011 used Pellacani's RCM score at a threshold of three or greater and also applied the Guitera score for lesions suspected of lentigo maligna of the face (Guitera 2010); sensitivity was 100% (95% CI 74% to 100%) and specificity was 92% (95% CI 79% to 98%).
Guitera 2012 reported the melanoma component of a two step algorithm to have a sensitivity of 78% (95% CI 65% to 89%) and specificity of 86% (95% CI 81% to 90%). Correctly identified melanoma in situ, BCC, and SCC were considered true negative results for the purposes of these calculations.
There were insufficient data to make any overall summary of test accuracy for this target condition.
Target condition: any skin cancer (melanoma or other skin cancer) or skin lesion with a high risk of progression to melanoma
In this section, we presented the results for studies of RCM versus dermoscopy for the target condition of any skin cancer (melanoma or other skin cancer) or skin lesion with a high risk of progression to melanoma (i.e. any invasive skin cancer, melanoma in situ or lentigo maligna and lesions with severe dysplasia), according to the study population: studies in any lesion suspicious for melanoma versus those in participants with equivocal lesions.
Studies in any lesion suspicious for melanoma
Four study cohorts provided data for four RCM evaluations (Curchin 2011; Guitera 2012; Pellacani 2014c (doc); Rao 2013) (Figure 20). All studies were case series (two prospective in design and two unclear). Studies were undertaken in Europe (one), North America (one), Oceania (one), or in more than one continent (one). Studies were undertaken in a secondary care setting (two) or a mixed secondary care specialist unit setting (two). Three studies included all lesion types and Pellacani 2014c (doc) included pigmented lesions only. The sample size ranged from 42 to 330 participants (reported in three studies) and 50 to 356 lesions. The lesion:participant ratio ranged from 1.07 to 1.19. Mean age was given in three studies and ranged from 41 to 53 years; the percentage of men was ranged from 44% to 54% (three studies). The mean prevalence of disease was 33.8% (range 22.9% to 44.1%). The percentage of invasive melanoma or melanoma in situ lesions in the disease positive group was 12% (Rao 2013), 46% (Pellacani 2014c (doc), 59% (Curchin 2011), and 63% (Guitera 2012); invasive SCCs were included as disease positive for both Rao 2013 (5% of disease positive group) and Guitera 2012 (54% of disease positive group) but could not be differentiated from seborrhoeic keratoses in Curchin 2011 and were therefore included in the disease negative group (six lesions; or 21% of disease negative group). Pellacani 2014c (doc) did not report including any cSCCs.
All studies used the Vivascope 1500 imaging system and reported the use of dermoscopic images to help the guide acquisition of RCM images. All studies reported diagnosis for a single observer, though only one clearly reported that this was by an experienced dermatologist. Two reported in‐person real time interpretation of RCM images (Curchin 2011; Pellacani 2014c (doc)), and two reported RCM interpretation remotely from the participant (Guitera 2012 supplied details of participant age and lesion site and Rao 2013 presented the dermoscopic image to aid interpretation). All studies made the reference standard diagnosis by histology alone.
Curchin 2011 and Pellacani 2014c (doc) used Pellacani's RCM score at a threshold of three or greater, Guitera 2012 reported data for their new two step algorithm for detection of BCC or melanoma, and Rao 2013 reported observer diagnosis. Estimates of sensitivities ranged from 85% to 100% and specificities from 52% to 89%.
Equivocal lesion studies
Three studies provided data for three evaluations of RCM alone (Farnetani 2015; Pellacani 2012; Pellacani 2014b (cons)) (Figure 20). All studies were case series (two prospective in design and one retrospective with prospective reinterpretation of images). All studies were undertaken in Europe (Italy). Two studies were undertaken in a secondary care setting and one in a specialist clinic. One study recruited any lesion type, one restricted to pigmented lesions and one to melanocytic lesions only. The sample size ranged from 62 to 252 participants (reported in two studies) and 60 to 308 lesions (one study). The lesion:participant ratios where reported were 1.03 (Pellacani 2012) and 1.22 (Pellacani 2014b (cons)). Mean age was given in two studies and ranged from 41 to 47.7 years; the percentage of male participants ranged from 44% to 52% (2 studies). The mean prevalence of disease was 24.9% (range 8.1% to 35.0%). The mean percentage of invasive melanoma or melanoma in situ lesions in the disease positive group was 51.6% (range 24% to 73.6%).
All studies used the Vivascope 1500 imaging system and reported diagnosis for a single observer. Observers were described as dermatologists in two studies. One reported in‐person real time interpretation of RCM images (Pellacani 2014b (cons)) and two reported RCM interpretation remotely from the participant (one blinded (Pellacani 2012) and one appeared to supply dermoscopic image to aid interpretation (Farnetani 2015), although this was not well reported). The reference standard diagnosis was made by histology alone in two studies and was supplemented by clinical and cancer registry follow‐up in the other (Pellacani 2014b (cons)).
One study developed a two step algorithm to differentiate dysplastic from non‐dysplastic lesions and then melanomas from dysplastic lesions (Pellacani 2012), one reported the observers overall diagnosis (Farnetani 2015), and one did not report the algorithm used but was assumed to have used Pellacani's RCM score, based on study authorship (Pellacani 2014b (cons)). Estimates of sensitivities ranged from 86% to 100% and specificities from 80% to 91%.
Analyses by algorithms used to assist reflectance confocal microscopy: all studies
The six included studies provided 11 datasets evaluating the accuracy of different algorithms or approaches to diagnosis with RCM at different thresholds for test positivity for the detection of any skin cancer or skin lesion with a high risk of progression to melanoma. A description of the various algorithms and thresholds used for diagnosis across the studies is provided in Appendix 5. We excluded Pellacani 2007a due to an overlap in study population, algorithm, and threshold with a study by Guitera and colleagues (Guitera 2009b (Modena); Guitera 2009c (Sydney)).
Figure 22 provides forest plots of all algorithms and thresholds, with meta‐analytical estimates at each threshold presented in Table 5. We did not formally make any comparisons between the algorithms due to the small number of studies available evaluating each algorithm.
22.
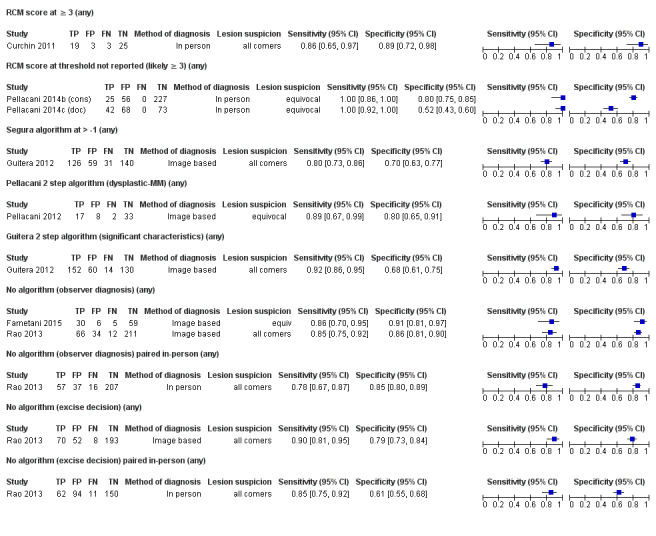
Forest plot: reflectance confocal microscopy (RCM) results by algorithm and threshold for diagnosis of any skin cancer or skin lesion with a high risk of progression to melanoma (any). FN: false negative; FP: false positive; TN: true negative; TP: true positive.
Pellacani's reflectance confocal microscopy score
Three datasets evaluated Pellacani's RCM score, one reported results at a threshold of three or greater (Curchin 2011 in an 'any potential melanoma' population), and two that did not report the threshold used but the recommended threshold of three or greater was assumed, as discussed above (from a single study in equivocal lesions (Pellacani 2014b (cons); Pellacani 2014c (doc)). All three datasets were from in‐person evaluations of RCM, one with interpretation by an RCM novice (Curchin 2011), and the other two by members of a 'confocal unit' assumed to be expert in RCM use.
RCM sensitivities were relatively high in all studies (86% or greater), with lower specificities in the two studies of dermoscopically equivocal lesions (Pellacani 2014b (cons); Pellacani 2014c (doc)). The pooled accuracy combining data from all three datasets reporting (or assumed to be) at RCM score three or greater was a sensitivity of 98% (95% CI 91% to 99%) and specificity of 75% (95% CI 54% to 89%) (Table 5).
Other formally developed algorithms
A single dataset each evaluated three other algorithms for detection of any skin cancer (melanoma or other skin cancer) or skin lesion with a high risk of progression to melanoma. Guitera 2012 evaluated both the Segura algorithm and Guitera two step algorithm, and Pellacani 2012 evaluated the Pellacani two step algorithm.
All studies used image based evaluations of RCM, one study conducted in equivocal lesions was blinded to any further information (Pellacani 2012), and the other (in an any potential melanoma population) provided details of participant age and lesion site only (Guitera 2012).
Guitera 2012 reported a sensitivity of 92% and specificity of 68% for their two step algorithm (with nine SCCs considered as disease positive) compared to a sensitivity of 80% and specificity of 70% for the Segura algorithm (the nine SCCs could only be considered disease negative for this calculation due to lack of disaggregated data, however, classing the nine SCCs as disease negative for the Guitera two‐step algorithm made very little difference to the estimate of sensitivity and specificity). Pellacani 2012 estimated sensitivity of 89% and specificity of 80% for their two‐step algorithm.
'No algorithm' evaluations
Two studies reported accuracy data for observer diagnosis with RCM without the use of a formally developed algorithm by different observers and at more than one threshold.
Farnetani 2015 reported data for in‐person diagnosis of malignancy by nine different observers with varying levels of experience. Rao 2013 provided a comparison of image based test interpretation by an experienced observer to in‐person real time diagnosis by a less experienced observer in an 'any lesion suspicious for melanoma' population for the diagnosis of any malignancy and for the decision to excise a lesion. There was a slight discrepancy in the number of lesions between in‐person (318 lesions) and image based (323 lesions) interpretations. For the diagnosis of any malignancy, the sensitivity was 78% (95% CI 67% to 87%) for in‐person diagnosis and 85% (95% CI 75% to 92%) for image based; specificities were 85% (95% CI 80% to 89%) for in‐person diagnosis and 86% (95% CI 81% to 90%) for imaged based. For the decision to excise a lesion, the sensitivity was 85% (95% CI 75% to 92%) for in‐person diagnosis and 90% (95% CI 81% to 95%) for image based; specificities were 61% (95% CI 55% to 68%) for in‐person diagnosis and 79% (95% CI 73% to 84%) for image based.
For the diagnosis of malignancy by an experienced observer, the pooled data across the two studies (using the image based data from Rao 2013) gave an estimated sensitivity of 85% (95% CI 77% to 90%) and specificity of 87% (95% CI 83% to 90%).
Evaluations of reflectance confocal microscopy in other study populations
We identified three evaluations of RCM in other study populations.
Pupelli 2013 selected confocal images of 24 melanomas of less than 5 mm diameter and three histologically confirmed small‐diameter naevi controls per melanoma (72 lesions) that were excised within the same time frame. Images were interpreted alongside the dermoscopic image plus information on participant age and site. The presence of three statistically significant lesion characteristics led to an estimated sensitivity of 83% (95% CI 63% to 95%) and specificity of 90% (95% CI 81% to 96%) for the detection of invasive melanoma and atypical intraepidermal melanocytic variants.
Figueroa‐Silva 2016 included a series of 63 images of pigmented lesions with a clear‐cut 'dermoscopy island,' defined as "a well‐circumscribed area showing a uniform dermoscopic pattern, different from the rest of the lesion." A single observer assessed the RCM images (blinded to all other information apart from dermoscopy) for the presence or absence of a number of lesion characteristics and provided an overall diagnosis. For the detection of invasive melanoma and atypical intraepidermal melanocytic variants the estimated sensitivity was 89% (95% CI 71% to 98%) and the estimated specificity was 89% (95% CI 74% to 97%). For the detection of invasive melanoma only (seven lesions) and considering the 19 melanoma in situ lesions as disease negative, the sensitivity was 88% and specificity was 62%.
Longo 2013 examined the use of RCM for the diagnosis of nodular melanoma. A series of images of clinically nodular lesions (defined as cutaneous palpable or superficial seated lesions) were interpreted by a single dermatologist blinded to all other information. For the diagnosis of invasive nodular lesions, the sensitivity was 100% and specificity was 91%.
Discussion
Summary of main results
Studies evaluated a range of study populations using a number of different algorithms. Sensitivity was generally high across studies and target conditions, but there was considerable heterogeneity in specificity. Studies were generally at high or unclear risk of bias across almost all domains and of high or unclear concern regarding applicability of the evidence, limiting the strength of conclusion that can be drawn. Table 1 presents key results for the primary target condition of cutaneous invasive melanoma or atypical intraepidermal melanocytic variants.
Across all algorithms and thresholds assessed, the Pellacani RCM score at a threshold of three or greater had the largest number of datasets for any one threshold; sensitivity was estimated at 92% and specificity at 72%. RCM accuracy was similar between 'any lesion suspicious for melanoma' studies and equivocal lesion studies, with sensitivities consistently around or above 90% but with much greater variation in specificities. In comparison to dermoscopy, RCM was more accurate in both participant groups (i.e. groups with all lesions suspected of melanoma, and in equivocal lesion populations). Due to differences in the algorithms and thresholds used between studies, analysis required use of an SROC curve, and to aid interpretation we quoted 'typical' summary results assuming a fixed sensitivity of 90% for both tests. Table 1 translated these estimates to a hypothetical cohort of 1000 lesions.
For 'any lesion suspicious for melanoma' studies, specificities were 82% for RCM and 42% for dermoscopy at a sensitivity of 90% for both tests. At disease prevalences of 26%, 30%, and 36%, using RCM as an alternative to dermoscopy would reduce false positives (or number of excisions that would be performed) by 296, 280, and 256 per 1000 compared with dermoscopy alone. Both tests would miss 26, 30 and 36 melanomas at each respective prevalence of melanoma.
For equivocal lesion studies, specificities were 86% for RCM and 49% for dermoscopy at a sensitivity of 90% for both tests. At disease prevalences of 10%, 20%, and 23%, using RCM in addition to dermoscopy would reduce the number of excisions by 333, 296, and 285 per 1000. Both tests would miss 10, 20, and 23 melanomas at each respective prevalence of melanoma. Investigations of heterogeneity were limited due to paucity of data but suggested higher RCM accuracy in equivocal lesions and from in‐person evaluations of RCM images.
Strengths and weaknesses of the review
The strengths of this review included an in‐depth and comprehensive electronic literature search, systematic review methods including double extraction of papers by both clinicians and methodologists, and contact with authors to allow study inclusion or clarify data. A clear analysis structure according to patient pathway was adopted to allow test accuracy in different study populations to be estimated and a detailed and replicable analysis of methodological quality was undertaken.
The main concerns for the review were a result of the poor reporting of primary studies, in particular forcing some assumptions to be made to allow studies to be split by pathway and in separating studies by the different definitions of the target condition. In terms of the separation of studies by pathway, although some assumptions were made, it emerged that the studies in each group could almost have been separated by disease prevalence, with higher rates in the 'any lesion suspicious for melanoma' group (ranging from 26% to 42% with one outlier at 3%), as would be expected in studies that included more obvious melanomas, and lower rates in the equivocal lesion group (ranging from 2% to 27%), again as would be expected with more clinically difficult lesions.
Three of the 16 cohorts included in our primary analyses for detection of invasive melanoma or atypical intraepidermal melanocytic variants did not provide clear identification of the target condition. The inclusion of melanoma in situ lesions as disease positive was eventually discerned from the text of two papers (Alarcon 2014a; Ferrari 2015), and was assumed for the third study (Farnetani 2015). Where studies included other invasive skin cancers in the study population, we attempted to class any that were correctly identified as true negative results as opposed to false positives, on the basis that removal of any skin cancer in the attempt to identify melanomas would not be a negative consequence of the test. This relied on studies providing a disaggregation of test results according to final lesion classification and was not always possible, particularly when invasive SCCs were not separated from 'in situ' lesions such as Bowen's disease.
Finally, observer expertise is key for any diagnostic process based on visual inspection, with both non‐analytical pattern recognition (implicit identification) and analytical pattern recognition (using more explicit 'rules' based on conscious analytical reasoning) employed to varying extents between clinicians, according to factors such as experience and familiarity with the diagnostic question (Norman 2009). Notably, research in this field has been dominated by a single expert group and results obtained from a more typical range of specialists in different countries, healthcare systems, and settings are needed. A lack of clear reporting of observer training and experience in RCM made analysis difficult.
Given these limitations, our results should be considered as exploratory rather than conclusive. However, our results are generally in accord with those of other recently published systematic reviews (Edwards 2016; Xiong 2016), one of which was conducted as part of a technology assessment report for NICE (Edwards 2016), despite differences in methodological approaches. Xiong 2016 did not consider varying definitions of the target condition in their primary analysis but pooled all studies regardless of detection of melanoma, BCC, or SCC (our examination of RCM for the diagnosis of keratinocyte skin cancers was reported in a separate systematic review in our series (Dinnes 2018c)). In a secondary analysis, eight studies with melanoma as the 'focus' were pooled, producing estimates of sensitivity of 92.7% (95% CI 90.0% to 94.9%) and specificity of 78.3% (95% CI 0.76% to 0.81%) (Xiong 2016). There was no consideration to differences in participant populations, we excluded two studies from our review (Gerger 2005; Guitera 2010), and two of the included studies reported on the same set of lesions (Guitera 2009a; Pellacani 2007a).
The Edwards 2016 review did not conduct a meta‐analysis, instead selecting studies considered to be more applicable to a UK setting. Using studies with 'optimistic' accuracy data (sensitivity 97% and specificity 94% in Alarcon 2014a) and with 'less favourable' (sensitivity 100% and specificity 51% in Pellacani 2014a) accuracy, deterministic incremental cost‐effectiveness ratios (ICER) for RCM in comparison to 'usual practice' were estimated for participants with dermoscopically equivocal lesions (assuming that two‐thirds of lesions would be excised and the remainder monitored). Resulting quality‐adjusted life years ranged GBP 8877 using 'optimistic' data to GBP 19,095 (Edwards 2016). The report concluded that data were lacking to allow generalisability to a UK setting.
Applicability of findings to the review question
The data included in this review were generally applicable to the clinical setting. Most of the studies used the current version of the only commercially available RCM system, the Vivascope 1500. Narrow definitions of the eligible study populations and lack of clarity regarding the patient pathway and any prior testing may restrict applicability, and the use of remote image based diagnosis largely by RCM experts further restrict the transferability of results to a clinical setting.
Authors' conclusions
Implications for practice.
Reflectance confocal microscopy (RCM) may have a potential role in clinical practice, particularly for the assessment of melanocytic lesions identified as equivocal following visual inspection and dermoscopy, where the evidence suggests that RCM may be both more sensitive and specific in comparison to dermoscopy. Given the paucity of data to allow comparison with dermoscopy, we presented illustrative data assuming that both tests are similarly sensitive. On this basis, for all lesions suspicious for melanoma, with RCM essentially used as a replacement for dermoscopy, the number of inappropriate excisions could potentially be reduced by up to two‐thirds. Given the additional expense and training required for RCM, the evidence for improved accuracy is insufficient to support its widespread use in a general population of people with lesions suspicious for melanoma. For an equivocal lesion population, the evidence for equivalent sensitivity between RCM and dermoscopy is more tenuous (with RCM likely to be the more sensitive test given that this group of lesions has already been identified as equivocal on dermoscopy), but even assuming equivalent sensitivity, inappropriate excisions could be reduced by as much as three‐quarters. If superior sensitivity of RCM could be demonstrated for this group, considerable patient benefit could be gained in terms of fewer missed melanomas and reduced morbidity. Digital monitoring in those considered negative on RCM could further reduce harms from any missed cases; however, resource implications and patient impact from such a policy would have to be taken into account.
Implications for research.
Further prospective evaluation of RCM in a standard healthcare setting with a clearly defined and representative population of participants with dermoscopically equivocal lesions and with RCM results interpreted in a usual practice setting by observers representative of those who would normally interpret images is appropriate to confirm the suggested increase in accuracy over dermoscopy. A multicentre approach would allow confirmation that results are replicable across centres and that the technology can be implemented across a health service. Prospective recruitment of consecutive series of participants, with test interpretation blinded to the reference standard diagnosis and with prespecified and clearly defined diagnostic thresholds for determining test positivity are easily achieved. Systematic follow‐up of non‐excised lesions avoids over‐reliance on a histological reference standard and allows results to be more generalisable to routine practice. A standardised approach to diagnosis, and clear identification of the level of training and experience required to achieve good results is also required. Any future research study needs to be clear about the diagnostic pathway followed by study participants prior to study enrolment, and should conform to the updated Standards for Reporting of Diagnostic Accuracy (STARD) guideline (Bossuyt 2015).
What's new
| Date | Event | Description |
|---|---|---|
| 18 December 2018 | Amended | Affiliations, Disclaimer and Sources of support updated. |
Acknowledgements
Members of the Cochrane Skin Cancer Diagnostic Test Accuracy Group include:
the full project team (Susan Bayliss, Naomi Chuchu, Clare Davenport, Jonathan Deeks, Jac Dinnes, Kathie Godfrey, Rubeta Matin, Colette O'Sullivan, Yemisi Takwoingi, Hywel Williams);
our 12 clinical reviewers (Rachel Abbott, Ben Aldridge, Oliver Bassett, Sue Ann Chan, Alana Durack, Monica Fawzy, Abha Gulati, Jacqui Moreau, Lopa Patel, Daniel Saleh, David Thompson, Kai Yuen Wong) and two methodologists (Lavinia Ferrante di Ruffano, Louise Johnston) who assisted with full text screening, data extraction, and quality assessment across the entire suite of reviews of diagnosis and staging and skin cancer,
and all members of our Advisory Group (Jonathan Bowling, Seau Tak Cheung, Colin Fleming, Matthew Gardiner, Abhilash Jain, Susan O'Connell , Pat Lawton, John Lear, Mariska Leeflang, Richard Motley, Paul Nathan, Julia Newton‐Bishop, Miranda Payne, Rachael Robinson, Simon Rodwell, Julia Schofield, Neil Shroff, Hamid Tehrani, Zoe Traill, Fiona Walter, Angela Webster).
Cochrane Skin editorial base wishes to thank Michael Bigby, who was the Dermatology Editor for this review; the clinical referees, Emma Craythorne and a referee who wishes to remain anonymous; and the consumer referee, Brian Stafford. We also wish to thank the Cochrane DTA editorial base and colleagues, as well as Anne Lawson, who copy‐edited this review.
Appendices
Appendix 1. Current content and structure of the Programme Grant
| LIST OF REVIEWS | Number of studies | |
| Diagnosis of melanoma | ||
| 1 | Visual inspection | 49 |
| 2 | Dermoscopy +/‐ visual inspection | 104 |
| 3 | Teledermatology | 22 |
| 4 | Smartphone applications | 2 |
| 5a | Computer‐assisted diagnosis – dermoscopy‐based techniques | 42 |
| 5b | Computer‐assisted diagnosis – spectroscopy‐based techniques | Review amalgamated into 5a |
| 6 | Reflectance confocal microscopy | 18 |
| 7 | High‐frequency ultrasound | 5 |
| Diagnosis of keratinocyte skin cancer (BCC and cSCC) | ||
| 8 | Visual inspection +/‐ Dermoscopy | 24 |
| 5c | Computer‐assisted diagnosis – dermoscopy‐based techniques | Review amalgamated into 5a |
| 5d | Computer‐assisted diagnosis – spectroscopy‐based techniques | Review amalgamated into 5a |
| 9 | Optical coherence tomography | 5 |
| 10 | Reflectance confocal microscopy | 10 |
| 11 | Exfoliative cytology | 9 |
| Staging of melanoma | ||
| 12 | Imaging tests (ultrasound, CT, MRI, PET‐CT) | 38 |
| 13 | Sentinel lymph node biopsy | 160 |
| Staging of cSCC | ||
| Imaging tests review | Review dropped; only one study identified | |
| 13 | Sentinel lymph node biopsy | Review amalgamated into 13 above (n = 15 studies) |
Appendix 2. Final search strategies
Melanoma search strategies to August 2016
Database: Ovid MEDLINE(R) 1946 to August week 3 2016
Search strategy:
1 exp melanoma/
2 exp skin cancer/
3 exp basal cell carcinoma/
4 basalioma$1.ti,ab.
5 ((basal cell or skin) adj2 (cancer$1 or carcinoma$1 or mass or masses or tumour$1 or tumor$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or lesion$1 or malignan$ or nodule$1)).ti,ab.
6 (pigmented adj2 (lesion$1 or mole$ or nevus or nevi or naevus or naevi or skin)).ti,ab.
7 (melanom$1 or nonmelanoma$1 or non‐melanoma$1 or melanocyt$ or non‐melanocyt$ or nonmelanocyt$ or keratinocyt$).ti,ab.
8 nmsc.ti,ab.
9 (squamous cell adj2 (cancer$1 or carcinoma$1 or mass or masses or tumor$1 or tumour$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or epithelial or lesion$1 or malignan$ or nodule$1) adj2 (skin or epiderm$ or cutaneous)).ti,ab.
10 (BCC or CSCC or NMSC).ti,ab.
11 keratinocy$.ti,ab.
12 Keratinocytes/
13 or/1‐12
14 dermoscop$.ti,ab.
15 dermatoscop$.ti,ab.
16 photomicrograph$.ti,ab.
17 exp epiluminescence microscopy/
18 (epiluminescence adj2 microscop$).ti,ab.
19 (confocal adj2 microscop$).ti,ab.
20 (incident light adj2 microscop$).ti,ab.
21 (surface adj2 microscop$).ti,ab.
22 (visual adj (inspect$ or examin$)).ti,ab.
23 ((clinical or physical) adj examin$).ti,ab.
24 3 point.ti,ab.
25 three point.ti,ab.
26 pattern analys$.ti,ab.
27 ABCD$.ti,ab.
28 menzies.ti,ab.
29 7 point.ti,ab.
30 seven point.ti,ab.
31 (digital adj2 (dermoscop$ or dermatoscop$)).ti,ab.
32 artificial intelligence.ti,ab.
33 AI.ti,ab.
34 computer assisted.ti,ab.
35 computer aided.ti,ab.
36 neural network$.ti,ab.
37 exp diagnosis, computer‐assisted/
38 MoleMax.ti,ab.
39 image process$.ti,ab.
40 automatic classif$.ti,ab.
41 image analysis.ti,ab.
42 SIAscop$.ti,ab.
43 Aura.ti,ab.
44 (optical adj2 scan$).ti,ab.
45 MelaFind.ti,ab.
46 SIMSYS.ti,ab.
47 MoleMate.ti,ab.
48 SolarScan.ti,ab.
49 VivaScope.ti,ab.
50 (high adj3 ultraso$).ti,ab.
51 (canine adj2 detect$).ti,ab.
52 ((mobile or cell or cellular or smart) adj ((phone$1 adj2 app$1) or application$1)).ti,ab.
53 smartphone$.ti,ab.
54 (DermoScan or SkinVision or DermLink or SpotCheck).ti,ab.
55 Mole Detective.ti,ab.
56 Spot Check.ti,ab.
57 (mole$1 adj2 map$).ti,ab.
58 (total adj2 body).ti,ab.
59 exfoliative cytolog$.ti,ab.
60 digital analys$.ti,ab.
61 (image$1 adj3 software).ti,ab.
62 (teledermatolog$ or tele‐dermatolog$ or telederm or tele‐derm or teledermoscop$ or tele‐dermoscop$ or teledermatoscop$ or tele‐dermatoscop$).ti,ab.
63 (optical coherence adj (technolog$ or tomog$)).ti,ab.
64 (computer adj2 diagnos$).ti,ab.
65 exp sentinel lymph node biopsy/
66 (sentinel adj2 node).ti,ab.
67 nevisense.mp. or HFUS.ti,ab.
68 electrical impedance spectroscopy.ti,ab.
69 history taking.ti,ab.
70 patient history.ti,ab.
71 (naked eye adj (exam$ or assess$)).ti,ab.
72 (skin adj exam$).ti,ab.
73 physical examination/
74 ugly duckling.mp. or UD.ti,ab.
75 ((physician$ or clinical or physical) adj (exam$ or triage or recog$)).ti,ab.
76 ABCDE.mp. or VOC.ti,ab.
77 clinical accuracy.ti,ab.
78 Family Practice/ or Physicians, Family/ or clinical competence/
79 (confocal adj2 microscop$).ti,ab.
80 diagnostic algorithm$1.ti,ab.
81 checklist$.ti,ab.
82 virtual imag$1.ti,ab.
83 volatile organic compound$1.ti,ab.
84 dog$1.ti,ab.
85 gene expression analy$.ti,ab.
86 reflex transmission imag$.ti,ab.
87 thermal imaging.ti,ab.
88 elastography.ti,ab.
89 or/14‐88
90 (CT or PET).ti,ab.
91 PET‐CT.ti,ab.
92 (FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical$).ti,ab.
93 exp Deoxyglucose/
94 deoxy‐glucose.ti,ab.
95 deoxyglucose.ti,ab.
96 CATSCAN.ti,ab.
97 exp Tomography, Emission‐Computed/
98 exp Tomography, X‐ray computed/
99 positron emission tomograph$.ti,ab.
100 exp magnetic resonance imaging/
101 (MRI or fMRI or NMRI or scintigraph$).ti,ab.
102 exp echography/
103 Doppler echography.ti,ab.
104 sonograph$.ti,ab.
105 ultraso$.ti,ab.
106 doppler.ti,ab.
107 magnetic resonance imag$.ti,ab.
108 or/90‐107
109 (stage$ or staging or metasta$ or recurrence or sensitivity or specificity or false negative$ or thickness$).ti,ab.
110 "Sensitivity and Specificity"/
111 exp cancer staging/
112 or/109‐111
113 108 and 112
114 89 or 113
115 13 and 114
Database: Ovid MEDLINE(R) In‐Process & Other Non‐Indexed Citations 29 August 2016
Search strategy:
1 basalioma$1.ti,ab.
2 ((basal cell or skin) adj2 (cancer$1 or carcinoma$1 or mass or masses or tumour$1 or tumor$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or lesion$1 or malignan$ or nodule$1)).ti,ab.
3 (pigmented adj2 (lesion$1 or mole$ or nevus or nevi or naevus or naevi or skin)).ti,ab.
4 (melanom$1 or nonmelanoma$1 or non‐melanoma$1 or melanocyt$ or non‐melanocyt$ or nonmelanocyt$ or keratinocyt$).ti,ab.
5 nmsc.ti,ab.
6 (squamous cell adj2 (cancer$1 or carcinoma$1 or mass or masses or tumor$1 or tumour$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or epithelial or lesion$1 or malignan$ or nodule$1) adj2 (skin or epiderm$ or cutaneous)).ti,ab.
7 (BCC or CSCC or NMSC).ti,ab.
8 keratinocy$.ti,ab.
9 or/1‐8
10 dermoscop$.ti,ab.
11 dermatoscop$.ti,ab.
12 photomicrograph$.ti,ab.
13 (epiluminescence adj2 microscop$).ti,ab.
14 (confocal adj2 microscop$).ti,ab.
15 (incident light adj2 microscop$).ti,ab.
16 (surface adj2 microscop$).ti,ab.
17 (visual adj (inspect$ or examin$)).ti,ab.
18 ((clinical or physical) adj examin$).ti,ab.
19 3 point.ti,ab.
20 three point.ti,ab.
21 pattern analys$.ti,ab.
22 ABCD$.ti,ab.
23 menzies.ti,ab.
24 7 point.ti,ab.
25 seven point.ti,ab.
26 (digital adj2 (dermoscop$ or dermatoscop$)).ti,ab.
27 artificial intelligence.ti,ab.
28 AI.ti,ab.
29 computer assisted.ti,ab.
30 computer aided.ti,ab.
31 neural network$.ti,ab.
32 MoleMax.ti,ab.
33 image process$.ti,ab.
34 automatic classif$.ti,ab.
35 image analysis.ti,ab.
36 SIAscop$.ti,ab.
37 Aura.ti,ab.
38 (optical adj2 scan$).ti,ab.
39 MelaFind.ti,ab.
40 SIMSYS.ti,ab.
41 MoleMate.ti,ab.
42 SolarScan.ti,ab.
43 VivaScope.ti,ab.
44 (high adj3 ultraso$).ti,ab.
45 (canine adj2 detect$).ti,ab.
46 ((mobile or cell or cellular or smart) adj ((phone$1 adj2 app$1) or application$1)).ti,ab.
47 smartphone$.ti,ab.
48 (DermoScan or SkinVision or DermLink or SpotCheck).ti,ab.
49 Mole Detective.ti,ab.
50 Spot Check.ti,ab.
51 (mole$1 adj2 map$).ti,ab.
52 (total adj2 body).ti,ab.
53 exfoliative cytolog$.ti,ab.
54 digital analys$.ti,ab.
55 (image$1 adj3 software).ti,ab.
56 (teledermatolog$ or tele‐dermatolog$ or telederm or tele‐derm or teledermoscop$ or tele‐dermoscop$ or teledermatoscop$ or tele‐dermatoscop$).ti,ab.
57 (optical coherence adj (technolog$ or tomog$)).ti,ab.
58 (computer adj2 diagnos$).ti,ab.
59 (sentinel adj2 node).ti,ab.
60 nevisense.mp. or HFUS.ti,ab.
61 electrical impedance spectroscopy.ti,ab.
62 history taking.ti,ab.
63 patient history.ti,ab.
64 (naked eye adj (exam$ or assess$)).ti,ab.
65 (skin adj exam$).ti,ab.
66 ugly duckling.mp. or UD.ti,ab.
67 ((physician$ or clinical or physical) adj (exam$ or triage or recog$)).ti,ab.
68 ABCDE.mp. or VOC.ti,ab.
69 clinical accuracy.ti,ab.
70 (Family adj (Practice or Physicians)).ti,ab.
71 (confocal adj2 microscop$).ti,ab.
72 clinical competence.ti,ab.
73 diagnostic algorithm$1.ti,ab.
74 checklist$.ti,ab.
75 virtual imag$1.ti,ab.
76 volatile organic compound$1.ti,ab.
77 dog$1.ti,ab.
78 gene expression analy$.ti,ab.
79 reflex transmission imag$.ti,ab.
80 thermal imaging.ti,ab.
81 elastography.ti,ab.
82 or/10‐81
83 (CT or PET).ti,ab.
84 PET‐CT.ti,ab.
85 (FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical$).ti,ab.
86 deoxy‐glucose.ti,ab.
87 deoxyglucose.ti,ab.
88 CATSCAN.ti,ab.
89 positron emission tomograph$.ti,ab.
90 (MRI or fMRI or NMRI or scintigraph$).ti,ab.
91 Doppler echography.ti,ab.
92 sonograph$.ti,ab.
93 ultraso$.ti,ab.
94 doppler.ti,ab.
95 magnetic resonance imag$.ti,ab.
96 or/83‐95
97 (stage$ or staging or metasta$ or recurrence or sensitivity or specificity or false negative$ or thickness$).ti,ab.
98 96 and 97
99 82 or 98
100 9 and 99
Database: Embase 1974 to 29 August 2016
Search strategy:
1 *melanoma/
2 *skin cancer/
3 *basal cell carcinoma/
4 basalioma$.ti,ab.
5 ((basal cell or skin) adj2 (cancer$1 or carcinoma$1 or mass or masses or tumour$1 or tumor$1 or neoplasm$ or adenoma$ or epithelioma$ or lesion$ or malignan$ or nodule$)).ti,ab.
6 (pigmented adj2 (lesion$1 or mole$ or nevus or nevi or naevus or naevi or skin)).ti,ab.
7 (melanom$1 or nonmelanoma$1 or non‐melanoma$1 or melanocyt$ or non‐melanocyt$ or nonmelanocyt$ or keratinocyt$).ti,ab.
8 nmsc.ti,ab.
9 (squamous cell adj2 (cancer$1 or carcinoma$1 or mass or tumor$1 or tumour$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or epithelial or lesion$1 or malignan$ or nodule$1) adj2 (skin or epiderm$ or cutaneous)).ti,ab.
10 (BCC or cscc).mp. or NMSC.ti,ab.
11 keratinocyte.ti,ab.
12 keratinocy$.ti,ab.
13 or/1‐12
14 dermoscop$.ti,ab.
15 dermatoscop$.ti,ab.
16 photomicrograph$.ti,ab.
17 *epiluminescence microscopy/
18 (epiluminescence adj2 microscop$).ti,ab.
19 (confocal adj2 microscop$).ti,ab.
20 (incident light adj2 microscop$).ti,ab.
21 (surface adj2 microscop$).ti,ab.
22 (visual adj (inspect$ or examin$)).ti,ab.
23 ((clinical or physical) adj examin$).ti,ab.
24 3 point.ti,ab.
25 three point.ti,ab.
26 pattern analys$.ti,ab.
27 ABCD$.ti,ab.
28 menzies.ti,ab.
29 7 point.ti,ab.
30 seven point.ti,ab.
31 (digital adj2 (dermoscop$ or dermatoscop$)).ti,ab.
32 artificial intelligence.ti,ab.
33 AI.ti,ab.
34 computer assisted.ti,ab.
35 computer aided.ti,ab.
36 neural network$.ti,ab.
37 MoleMax.ti,ab.
38 exp diagnosis, computer‐assisted/
39 image process$.ti,ab.
40 automatic classif$.ti,ab.
41 image analysis.ti,ab.
42 SIAscop$.ti,ab.
43 (optical adj2 scan$).ti,ab.
44 Aura.ti,ab.
45 MelaFind.ti,ab.
46 SIMSYS.ti,ab.
47 MoleMate.ti,ab.
48 SolarScan.ti,ab.
49 VivaScope.ti,ab.
50 confocal microscop$.ti,ab.
51 (high adj3 ultraso$).ti,ab.
52 (canine adj2 detect$).ti,ab.
53 ((mobile or cell$ or cellular or smart) adj ((phone$1 adj2 app$1) or application$1)).ti,ab.
54 smartphone$.ti,ab.
55 (DermoScan or SkinVision or DermLink or SpotCheck).ti,ab.
56 Spot Check.ti,ab.
57 Mole Detective.ti,ab.
58 (mole$1 adj2 map$).ti,ab.
59 (total adj2 body).ti,ab.
60 exfoliative cytolog$.ti,ab.
61 digital analys$.ti,ab.
62 (image$1 adj3 software).ti,ab.
63 (optical coherence adj (technolog$ or tomog$)).ti,ab.
64 (teledermatolog$ or tele‐dermatolog$ or telederm or tele‐derm or teledermoscop$ or tele‐dermoscop$ or teledermatoscop$).mp. or tele‐dermatoscop$.ti,ab.
65 (computer adj2 diagnos$).ti,ab.
66 *sentinel lymph node biopsy/
67 (sentinel adj2 node).ti,ab.
68 nevisense.ti,ab.
69 HFUS.ti,ab.
70 electrical impedance spectroscopy.ti,ab.
71 history taking.ti,ab.
72 patient history.ti,ab.
73 (naked eye adj (exam$ or assess$)).ti,ab.
74 (skin adj exam$).ti,ab.
75 *physical examination/
76 ugly duckling.ti,ab.
77 UD sign$.ti,ab.
78 ((physician$ or clinical or physical) adj (exam$ or recog$ or triage)).ti,ab.
79 ABCDE.ti,ab.
80 clinical accuracy.ti,ab.
81 *general practice/
82 (confocal adj2 microscop$).ti,ab.
83 clinical competence/
84 diagnostic algorithm$.ti,ab.
85 checklist$1.ti,ab.
86 virtual image$1.ti,ab.
87 volatile organic compound$1.ti,ab.
88 VOC.ti,ab.
89 dog$1.ti,ab.
90 gene expression analys$.ti,ab.
91 reflex transmission imaging.ti,ab.
92 thermal imaging.ti,ab.
93 elastography.ti,ab.
94 dog$1.ti,ab.
95 gene expression analys$.ti,ab.
96 reflex transmission imaging.ti,ab.
97 thermal imaging.ti,ab.
98 elastography.ti,ab.
99 or/14‐93
100 PET‐CT.ti,ab.
101 (CT or PET).ti,ab.
102 (FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical$).ti,ab.
103 exp Deoxyglucose/
104 CATSCAN.ti,ab.
105 deoxyglucose.ti,ab.
106 deoxy‐glucose.ti,ab.
107 *positron emission tomography/
108 *computer assisted tomography/
109 positron emission tomograph$.ti,ab.
110 *nuclear magnetic resonance imaging/
111 (MRI or fMRI or NMRI or scintigraph$).ti,ab.
112 *echography/
113 Doppler.ti,ab.
114 sonograph$.ti,ab.
115 ultraso$.ti,ab.
116 magnetic resonance imag$.ti,ab.
117 or/100‐116
118 (stage$ or staging or metasta$ or recurrence or sensitivity or specificity or false negative$ or thickness$).ti,ab.
119 "Sensitivity and Specificity"/
120 *cancer staging/
121 or/118‐120
122 117 and 121
123 99 or 122
124 13 and 123
Database: Cochrane Library (Wiley) 2016 searched 30 August 2016 CDSR Issue 8 of 12 2016 CENTRAL Issue 7 of 12 2016 HTA Issue 3 of 4 July 2016 DARE Issue 3 of 4 2015
Search strategy:
#1 melanoma* or nonmelanoma* or non‐melanoma* or melanocyt* or non‐melanocyt* or nonmelanocyt* or keratinocyte*
#2 MeSH descriptor: [Melanoma] explode all trees
#3 "skin cancer*"
#4 MeSH descriptor: [Skin Neoplasms] explode all trees
#5 skin near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*)
#6 nmsc
#7 "squamous cell" near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*) near/2 (skin or epiderm* or cutaneous)
#8 "basal cell" near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*)
#9 pigmented near/2 (lesion* or nevus or mole* or naevi or naevus or nevi or skin)
#10 #1 or #2 or #3 or #4 or #5 or #6 or #7 or #8 or #9
#11 dermoscop*
#12 dermatoscop*
#13 Photomicrograph*
#14 MeSH descriptor: [Dermoscopy] explode all trees
#15 confocal near/2 microscop*
#16 epiluminescence near/2 microscop*
#17 incident next light near/2 microscop*
#18 surface near/2 microscop*
#19 "visual inspect*"
#20 "visual exam*"
#21 (clinical or physical) next (exam*)
#22 "3 point"
#23 "three point"
#24 "pattern analys*"
#25 ABDC
#26 menzies
#27 "7 point"
#28 "seven point"
#29 digital near/2 (dermoscop* or dermatoscop*)
#30 "artificial intelligence"
#31 "AI"
#32 "computer assisted"
#33 "computer aided"
#34 AI
#35 "neural network*"
#36 MoleMax
#37 "computer diagnosis"
#38 "image process*"
#39 "automatic classif*"
#40 SIAscope
#41 "image analysis"
#42 "optical near/2 scan*"
#43 Aura
#44 MelaFind
#45 SIMSYS
#46 MoleMate
#47 SolarScan
#48 Vivascope
#49 "confocal microscopy"
#50 high near/3 ultraso*
#51 canine near/2 detect*
#52 Mole* near/2 map*
#53 total near/2 body
#54 mobile* or smart near/2 phone*
#55 cell next phone*
#56 smartphone*
#57 "mitotic index"
#58 DermoScan or SkinVision or DermLink or SpotCheck
#59 "Mole Detective"
#60 "Spot Check"
#61 mole* near/2 map*
#62 total near/2 body
#63 "exfoliative cytolog*"
#64 "digital analys*"
#65 image near/3 software
#66 teledermatolog* or tele‐dermatolog* or telederm or tele‐derm or teledermoscop* or tele‐dermoscop* or teledermatoscop* or tele‐dermatolog*
#67 "optical coherence" next (technolog* or tomog*)
#68 computer near/2 diagnos*
#69 sentinel near/2 node*
#70 #11 or #12 or #13 or #14 or #15 or #16 or #17 or #18 or #19 or #20 or #21 or #22 or #23 or #24 or #25 or #26 or #27 or #28 or #29 or #30 or #31 or #32 or #33 or #34 or #35 or #36 or #37 or #38 or #39 or #40 or #41 or #42 or #43 or #44 or #45 or #46 or #47 or #48 or #49 or #50 or #51 or #52 or #53 or #54 or #55 or #56 or #57 or #58 or #59 or #60 or #61 or #62 or #63 or #64 or #65 or #66 or #67 or #68 or #69
#71 ultraso*
#72 sonograph*
#73 MeSH descriptor: [Ultrasonography] explode all trees
#74 Doppler
#75 CT or PET or PET‐CT
#76 "CAT SCAN" or "CATSCAN"
#77 MeSH descriptor: [Positron‐Emission Tomography] explode all trees
#78 MeSH descriptor: [Tomography, X‐Ray Computed] explode all trees
#79 MRI
#80 MeSH descriptor: [Magnetic Resonance Imaging] explode all trees
#81 MRI or fMRI or NMRI or scintigraph*
#82 "magnetic resonance imag*"
#83 MeSH descriptor: [Deoxyglucose] explode all trees
#84 deoxyglucose or deoxy‐glucose
#85 "positron emission tomograph*"
#86 #71 or #72 or #73 or #74 or #75 or #76 or #77 or #78 or #79 or #80 or #81 or #82 or #83 or #84 or #85
#87 stage* or staging or metasta* or recurrence or sensitivity or specificity or "false negative*" or thickness*
#88 MeSH descriptor: [Neoplasm Staging] explode all trees
#89 #87 or #88
#90 #89 and #86
#91 #70 or #90
#92 #10 and #91
#93 BCC or CSCC or NMCS
#94 keratinocy*
#95 #93 or #94
#96 #10 or #95
#97 nevisense
#98 HFUS
#99 "electrical impedance spectroscopy"
#100 "history taking"
#101 "patient history"
#102 naked next eye near/1 (exam* or assess*)
#103 skin next exam*
#104 "ugly duckling" or (UD sign*)
#105 MeSH descriptor: [Physical Examination] explode all trees
#106 (physician* or clinical or physical) near/1 (exam* or recog* or triage*)
#107 ABCDE
#108 "clinical accuracy"
#109 MeSH descriptor: [General Practice] explode all trees
#110 confocal near microscop*
#111 "diagnostic algorithm*"
#112 MeSH descriptor: [Clinical Competence] explode all trees
#113 checklist*
#114 "virtual image*"
#115 "volatile organic compound*"
#116 dog or dogs
#117 VOC
#118 "gene expression analys*"
#119 "reflex transmission imaging"
#120 "thermal imaging"
#121 elastography
#122 #97 or #98 or #99 or #100 or #101 or #102 or #103 or #104 or #105 or #106 or #107 or #108 or #109 or #110 or #111 or #112 or #113 or #114 or #115 or #116 or #117 or #118 or #119 or #120 or #121
#123 #70 or #122
#124 #96 and #123
#125 #96 and #90
#126 #125 or #124
#127 #10 and #126
Database : CINAHL Plus (EBSCO) 1937 to 30 August 2016
Search strategy:
S1 (MH "Melanoma") OR (MH "Nevi and Melanomas+")
S2 (MH "Skin Neoplasms+")
S3 (MH "Carcinoma, Basal Cell+")
S4 basalioma*
S5 (basal cell) N2 (cancer* or carcinoma* or mass or masses or tumor* or tumour* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*)
S6 (pigmented) N2 (lesion* or mole* or nevus or nevi or naevus or naevi or skin)
S7 melanom* or nonmelanoma* or non‐melanoma* or melanocyt* or non‐melanocyt* or nonmelanocyt*
S8 nmsc
S9 TX BCC or cscc or NMSC
S10 (MH "Keratinocytes")
S11 keratinocyt*
S12 S1 OR S2 OR S3 OR S4 OR S5 OR S6 OR S7 OR S8 OR S9 OR S10 OR S11
S13 dermoscop* or dermatoscop* or photomicrograph* or (3 point) or (three point) or ABCD* or menzies or (7 point) or (seven point) or AI or Molemax or SIASCOP* or Aura or MelaFind or SIMSYS or MoleMate or SolarScan or smartphone* or DermoScan or SkinVision or DermLink or SpotCheck
S14 (epiluminescence or confocal or incident or surface) N2 (microscop*)
S15 visual N1 (inspect* or examin*)
S16 (clinical or physical) N1 (examin*)
S17 pattern analys*
S18 (digital) N2 (dermoscop* or dermatoscop*)
S19 (artificial intelligence)
S20 (computer) N2 (assisted or aided)
S21 (neural network*)
S22 (MH "Diagnosis, Computer Assisted+")
S23 (image process*)
S24 (automatic classif*)
S25 (image analysis)
S26 SIAScop*
S27 (optical) N2 (scan*)
S28 (high) N3 (ultraso*)
S29 elastography
S30 (mobile or cell or cellular or smart) N2 (phone*) N2 (app or application*)
S31 (mole*) N2 (map*)
S32 total N2 body
S33 exfoliative cytolog*
S34 digital analys*
S35 image N3 software
S36 teledermatolog* or tele‐dermatolog* or telederm or tele‐derm or teledermoscop* or tele‐dermoscop* or teledermatoscop* or tele‐dermatoscop* teledermatolog* or tele‐dermatolog* or telederm or tele‐derm or teledermoscop*
S37 (optical coherence) N1 (technolog* or tomog*)
S38 computer N2 diagnos*
S39 sentinel N2 node
S40 (MH "Sentinel Lymph Node Biopsy")
S41 nevisense or HFUS or checklist* or VOC or dog*
S42 electrical impedance spectroscopy
S43 history taking
S44 "Patient history"
S45 naked eye
S46 skin exam*
S47 physical exam*
S48 ugly duckling
S49 UD sign*
S50 (physician* or clinical or physical) N1 (exam*)
S51 clinical accuracy
S52 general practice
S53 (physician* or clinical or physical) N1 (recog* or triage)
S54 confocal microscop*
S55 clinical competence
S56 diagnostic algorithm*
S57 checklist*
S58 virtual image*
S59 volatile organic compound*
S60 gene expression analys*
S61 reflex transmission imag*
S62 thermal imaging
S63 S13 or S14 or S15 OR S16 OR S17 OR S18 OR S19 OR S20 OR S21 OR S22 OR S23 OR S24 OR S25 OR S26 OR S27 OR S28 OR S29 OR S30 OR S31 OR S32 OR S33 OR S34 OR S35 OR S36 OR S37 OR S38 OR S39 OR S40 OR S41 OR S42 OR S43 OR S44 OR S45 OR S46 OR S47 OR S48 OR S49 OR S50 OR S51 OR S52 OR S53 OR S54 OR S55 OR S56 OR S57 OR S58 OR S59 OR S60 OR S61 OR S62
S64 CT or PET
S65 PET‐CT
S66 FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical*
S67 (MH "Deoxyglucose+")
S68 deoxy‐glucose or deoxyglucose
S69 CATSCAN
S70 CAT‐SCAN
S71 (MH "Deoxyglucose+")
S72 (MH "Tomography, Emission‐Computed+")
S73 (MH "Tomography, X‐Ray Computed")
S74 positron emission tomograph*
S75 (MH "Magnetic Resonance Imaging+")
S76 MRI or fMRI or NMRI or scintigraph*
S77 echography
S78 doppler
S79 sonograph*
S80 ultraso*
S81 magnetic resonance imag*
S82 S64 OR S65 OR S66 OR S67 OR S68 OR S69 OR S70 OR S71 OR S72 OR S73 OR S74 OR S75 OR S76 OR S77 OR S78 OR S79 OR S80 OR S81
S83 stage* or staging or metasta* or recurrence or sensitivity or specificity or (false negative*) or thickness
S84 (MH "Neoplasm Staging")
S85 S83 OR S84
S86 S82 AND S85
S87 S63 OR S86
S88 S12 AND S87
Database: Science Citation Index SCI Expanded (Web of Science) 1900 to 30 August 2016
Conference Proceedings Citation Index (Web of Science) 1900 to 1 September 2016
Search strategy:
#1 (melanom* or nonmelanom* or non‐melanoma* or melanocyt* or non‐melanocyt* or nonmelanocyt* or keratinocyt*)
#2 (basalioma*)
#3 ((skin) near/2 (cancer* or carcinoma or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*))
#4 ((basal) near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*))
#5 ((pigmented) near/2 (lesion* or mole* or nevus or nevi or naevus or naevi or skin))
#6 (nmsc or BCC or NMSC or keratinocy*)
#7 ((squamous cell (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*))
#8 (skin or epiderm* or cutaneous)
#9 #8 AND #7
#10 #9 OR #6 OR #5 OR #4 OR #3 OR #2 OR #1
#11 ((dermoscop* or dermatoscop* or photomicrograph* or epiluminescence or confocal or "incident light" or "surface microscop*" or "visual inspect*" or "physical exam*" or 3 point or three point or pattern analy* or ABCDE or menzies or 7 point or seven point or dermoscop* or dermatoscop* or AI or artificial or computer aided or computer assisted or neural network* or Molemax or image process* or automatic classif* or image analysis or siascope or optical scan* or Aura or melafind or simsys or molemate or solarscan or vivascope or confocal microscop* or high ultraso* or canine detect* or cellphone* or mobile* or phone* or smartphone or dermoscan or skinvision or dermlink or spotcheck or spot check or mole detective or mole map* or total body or exfoliative psychology or digital or image software or optical coherence or teledermatology or telederm* or teledermoscop* or teledermatoscop* or computer diagnos* or sentinel))
#12 ((nevisense or HFUS or impedance spectroscopy or history taking or patient history or naked eye or skin exam* or physical exam* or ugly duckling or UD sign* or physician* exam* or physical exam* or ABCDE or clinical accuracy or general practice or confocal microscop* or clinical competence or diagnostic algorithm* or checklist* or virtual image* or volatile organic or VOC or dog* or gene expression or reflex transmission or thermal imag* or elastography))
#13 #11 or #12
#14 ((PET or CT or FDG or deoxyglucose or deoxy‐glucose or fluorodeoxy* or radiopharma* or CATSCAN or positron emission or computer assisted or nuclear magnetic or MRI or FMRI or NMRI or scintigraph* or echograph* or Doppler or sonograph* or ultraso* or magnetic reson*))
#15 ((stage* or staging or metast* or recurrence or sensitivity or specificity or false negative* or thickness*))
#16 #14 AND #15
#17 #16 OR #13
#18 #10 AND #17
Refined by: DOCUMENT TYPES: (MEETING ABSTRACT OR PROCEEDINGS PAPER)
Appendix 3. Full text inclusion criteria
| Criterion | Inclusion | Exclusion |
| Study design |
For diagnostic and staging reviews
|
|
| Target condition |
|
|
| Population |
For diagnostic reviews
For staging reviews
|
|
| Index tests |
For diagnosis
For staging
Any test combination and in any order Any test positivity threshold Any variation in testing procedure (e.g. radioisotope used) |
|
| Reference standard |
For diagnostic studies
For studies of imaging tests for staging
For studies of SLNB accuracy for staging
|
For diagnostic studies
|
| BCC: basal cell carcinoma; cSCC: cutaneous squamous cell carcinoma; CT: computed tomography; FNAC: fine needle aspiration cytology; LND: lymph node dissection; MRI: magnetic resonance imaging; PET: positron emission tomography; PET‐CT: positron emission tomography computed tomography; RCT: randomised controlled trial; SCC: squamous cell carcinoma; SLN+: positive sentinel lymph node; SLn: negative sentinel lymph node; SLNB: sentinel lymph node biopsy. | ||
Appendix 4. Quality assessment (based on QUADAS‐2)
The QUADAS‐2 checklist was tailored to the review topic as follows below (Whiting 2011).
Participant selection domain (1)
Selective recruitment of study participants can be a key influence on test accuracy. In general terms, all participants eligible to undergo a test should be included in a study, allowing for the intended use of that test within the context of the study. We considered studies that separately sampled malignant and benign lesions to have used a case‐control design; and those that supplemented a series of suspicious lesions with additional malignant or benign lesions to be at unclear risk of bias
In terms of exclusions, we considered studies that excluded particular lesion types (e.g. lentigo maligna), particular lesion sites, or that excluded lesions on the basis of image quality or lack of observer agreement (e.g. on histopathology) to be at high risk of bias.
In judging the applicability of participant populations to the review question, we considered restriction to particular lesion populations, such as melanocytic, nodular, high risk, or restrictions by size to be of high concern for applicability.
Given that diagnosis of skin cancer is primarily lesion‐based, there is the potential for study participants with multiple lesions to contribute disproportionately to estimates of test accuracy, especially if they are at particular risk of having skin cancer. We considered studies that included a high number of lesions in relation to the number of study to be less representative than studies conducted in a more general population participants (i.e. if the difference between the number of included lesions and number of included participants was greater than 5%).
Index test domain (2)
Given the potential for subjective differences in test interpretation for melanoma, the interpretation of the index test blinded to the result of the reference standard is a key means of reducing bias. For prospective studies and retrospective studies that used the original index test interpretation, the diagnosis will by nature be interpreted and recorded before the result of the reference standard is known; however, studies using previously acquired images could be particularly susceptible to information bias. For these studies to be at low risk of bias, we required a clear indication that observers were unaware of the reference standard diagnosis at time of test interpretation. An item was also added to assess the presence of blinding between interpretations of different algorithms, however this item was not included in the overall assessment of risk of bias.
Prespecification of the index test threshold was considered present if the study clearly reported that the threshold used was not data driven, i.e. was not based on study results. Studies that did not clearly describe the threshold used but that required clinicians to record a diagnosis or management decision for a lesion were considered to be unclear on this criterion. Studies reporting accuracy for multiple numeric thresholds, where receiver operating characteristic (ROC) analysis was used to select the threshold, or that reported accuracy for the presence of independently significant lesion characteristics with no separate test set of lesions were considered at high risk of bias.
In terms of applicability of the index test to the review question, we required the test to be applied and interpreted as it would be in a clinical practice setting, i.e. in‐person or face‐to‐face with the patient, and by a single observer as opposed to a consensus decision or mean across multiple observers. Image based studies were considered to be high concern, although reflectance confocal microscopy (RCM) image interpretations where the observer was also supplied with a clinical or dermoscopic image of the lesion along with some participant characteristics were considered 'unclear.'
Despite the often subjective nature of test interpretation, it is also important for study authors to outline the particular lesion characteristics that were considered to be indicative for melanoma, particularly where established algorithms or checklists were not used. Studies were considered of low concern if the threshold used was established in a prior study or sufficient threshold details were presented to allow replication.
The experience of the examiner will also impact on the applicability of study results. We required studies to describe the test interpreter as 'experienced' or 'expert' in RCM to have low concern about applicability.
Reference standard domain (3)
In an ideal study, consecutively recruited participants should all undergo incisional or excisional biopsy of the skin lesion regardless of level of clinical suspicion of melanoma. In reality, both partial and differential verification bias are likely. Partial verification bias may occur where histology is the only reference standard used, and only those participants with a certain degree of suspicion of malignancy based on the result of the index test undergo verification, the others either being excluded from the study or defined as being disease negative without further assessment or follow‐up, as discussed above.
Differential verification bias will be present where other reference standards are used in addition to histological verification of suspicious lesions. A typical example of verification bias in skin cancer occurs when investigators do not biopsy people with benign appearing lesions but instead follow them up for a period of time to determine whether any malignancy subsequently develops (these would be false negatives on the index test). We defined an 'adequate' reference standard as: all disease positive people having a histological reference standard either at the time of application of the index test or after a period of clinical follow‐up; and at least 80% of disease negative participants have received a histological diagnosis, with up to 20% undergoing at least three months' follow‐up of benign appearing lesions.
A further challenge is the potential for incorporation bias, i.e. where the result of the index test is used to help determine the reference standard diagnosis. It is normal practice for the clinical diagnosis (usually by visual inspection or dermoscopy) to be included on pathology request forms and for the histopathologist to use this diagnosis to help with the pathology interpretation. Although inclusion of such clinical information on the histopathology request form is theoretically a form of incorporation bias, blinded interpretation of the histopathology reference standard is not normal practice, and enforcement of such conditions would significantly limit the generalisability of the study results. For studies evaluating RCM, this item was divided into two questions, first whether the reference standard was blinded to the index test result (RCM), and second whether it was blinded to the clinical diagnosis. Only the response to the first part (i.e. blinding to RCM) was included in our overall assessment of risk of bias for the reference standard domain.
In judging the applicability of the reference standard to our review question, scored studies as high concern around applicability if they used expert diagnosis (with no follow‐up) as a reference standard in any participant, or did not report histology interpretation by a dermatopathologist.
Flow and timing domain (4)
In the ideal study, the diagnosis based on the index test and reference standard should be made consecutively or as near to each other in time as possible to avoid changes in lesion over time. For lesions with a histological reference standard, we defined a one‐month period as an appropriate interval between application of the index test and the reference standard. For studies using clinical follow‐up, a minimum three‐month follow‐up period has been defined as at low risk of bias for detecting false negatives. This interval was chosen based on a study showing that most false negative melanomas will be diagnosed within three months of the initial negative index test although a small number will be diagnosed up to 12 months subsequently (Altamura 2008).
In assessing whether all participants were included in the analysis, we considered studies at high risk of bias if participants were excluded following recruitment.
Comparative domain
A comparative domain was added to the QUADAS‐2 checklist for studies comparing the accuracy of RCM and dermoscopy. Items were included to assess the presence blinding of interpretation between tests, and to specify a maximum of one‐month interval between application of index tests, as intervals greater than these may be accompanied by changes in tumour characteristics. As it would not be normal practice for RCM to be interpreted blinded to the clinical or dermoscopic diagnosis, the scoring of this item did not contribute to our overall assessment of risk of bias. We also considered whether both tests were applied and interpreted in a clinically applicable manner.
The following tables use text that was originally published in the QUADAS‐2 tool by Whiting and colleagues (Whiting 2011).
| Item | Response (delete as required) |
| Participant selection (1) risk of bias | |
| 1) Was a consecutive or random sample of participants or images enrolled? |
Yes: if paper stated consecutive or random No: if paper described other method of sampling Unclear: if participant sampling not described |
| 2) Was a case‐control design avoided? |
Yes: if consecutive or random or case‐control design clearly not used No: if study described as case‐control or described sampling specific numbers of participants with particular diagnoses Unclear: if not described |
3) Did the study avoid inappropriate exclusions, e.g.
|
Yes: if inappropriate exclusions were avoided No: if lesions were excluded that might have affected test accuracy, e.g. 'difficult to diagnose' lesions, or where disagreement between evaluators was observed Unclear: if not clearly reported but there was suspicion that difficult to diagnose lesions may have been excluded |
4) For between‐person comparative studies only (i.e. allocating different tests to different study participants):
|
For A)
For B)
For C)
|
| Could the selection of participants have introduced bias? For non‐comparative and within‐person comparative studies
For between‐person comparative studies
|
For non‐comparative and within‐person comparative studies
For between‐person comparative studies
|
| Participant selection (1) concerns regarding applicability | |
1) Were the included participants and chosen study setting appropriate to answer the review question, i.e. were the study results generalisable?
|
A) For studies that contributed to the analysis of participants with a primary presentation of a skin lesion (i.e. test naive) Yes: if participants included in the study appeared to be generally representative of those who might present in a usual practice setting No: if study participants appeared to be unrepresentative of usual practice, e.g. in terms of severity of disease, demographic features, presence of differential diagnosis or comorbidity, setting of the study, and previous testing protocols Unclear: if insufficient details were provided to determine the generalisability of study participants B) For studies that contributed to the analysis of referred participants (i.e. who have already undergone some form of testing) Yes: if study participants appeared to be representative of those who might be referred for further investigation. If the study focused only on those with equivocal lesions, for example, we would suggest that this was not representative of the wider referred population No: if study participants appeared to be unrepresentative of usual practice, e.g. if a particularly high proportion of participants have been self‐referred or referred for cosmetic reasons. Other factors to consider include severity of disease, demographic features, presence of differential diagnosis or comorbidity, setting of the study, and previous testing protocols Unclear: if insufficient details were provided to determine the generalisability of study participants |
| 2) Did the study avoid including participants with multiple lesions? |
Yes: if the difference between the number of included lesions and number of included participants was less than 5% No: if the difference between the number of included lesions and number of included participants was greater than 5% Unclear: if it is not possible to assess |
Is there concern that the included participants do not match the review question?
|
|
| Index test (2) risk of bias (to be completed per test evaluated) | |
| 1) Was the index test or testing strategy result interpreted without knowledge of the results of the reference standard? |
Yes: if index test described as interpreted without knowledge of reference standard result or, for prospective studies, if index test was always conducted and interpreted prior to the reference standard No: if index test described as interpreted in knowledge of reference standard result Unclear: if index test blinding was not described |
| 2) Was the diagnostic threshold at which the test was considered positive (i.e. melanoma present) prespecified? |
Yes: if threshold was prespecified (i.e. prior to analysing study results) No: if threshold was not prespecified Unclear: if not possible to tell whether or not diagnostic threshold was prespecified |
| 3) For within‐person comparisons of index tests or testing strategies (i.e. > 1 index test applied per participant): was each index test result interpreted without knowledge of the results of other index tests or testing strategies? |
Yes: if all index tests were described as interpreted without knowledge of the results of the others No: if the index tests were described as interpreted in the knowledge of the results of the others Unclear: if it was not possible to tell whether knowledge of other index tests could have influenced test interpretation N/A: if only 1 index test was evaluated |
| Could the conduct or interpretation of the index test have introduced bias? For non‐comparative and between‐person comparison studies
For within‐person comparative studies
|
For non‐comparative and between‐person comparison studies
For within‐person comparative studies
|
| Index test (2) concern about applicability | |
| 1) Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? e.g. previously evaluated/established
|
Yes: if a previously evaluated/established tool to aid diagnosis of melanoma was used or if the diagnostic threshold used was established in a previously published study No: if an unfamiliar/new tool to aid diagnosis of melanoma was used, if no particular algorithm was used, or if the objective threshold reported was chosen based on results in the current study Unclear: if insufficient information was reported |
| 2) Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? Study results can only be reproduced if the diagnostic threshold was described in sufficient detail. This item applies equally to studies using pattern recognition and those using checklists or algorithms to aid test interpretation |
Yes: if the criteria for diagnosis of melanoma were reported in sufficient detail to allow replication No: if the criteria for diagnosis of melanoma were not reported in sufficient detail to allow replication Unclear: if some but not sufficient information on criteria for diagnosis to allow replication were provided |
| 3) Was the test interpretation carried out by an experienced examiner? |
Yes: if the test was interpreted by ≥ 1 speciality accredited dermatologists, or by examiners of any clinical background with special interest in dermatology and with any formal training in the use of the test No: if the test was not interpreted by an experienced examiner (see above) Unclear: if the experience of the examiner was not reported in sufficient detail to judge or if examiners were described as 'expert' with no further detail given N/A: if system based diagnosis, i.e. no observer interpretation |
Is there concern that the index test, its conduct, or interpretation differed from the review question?
|
|
| Reference standard (3) risk of bias | |
| 1) Is the reference standard likely to correctly classify the target condition? A) Disease positive: ≥ 1 of the following:
B) Disease negative: ≥ 1 of the following:
|
A) Disease positive Yes: if all participants with a final diagnosis of melanoma underwent 1 of the listed reference standards No: if a final diagnosis of melanoma for any participant was reached without histopathology Unclear: if the method of final diagnosis was not reported for any participant with a final diagnosis of melanoma or if the length of clinical follow‐up used was not clear or if a clinical follow‐up reference standard was reported in combination with a participant‐based analysis and it was not possible to determine whether the detection of a malignant lesion during follow‐up was the same lesion that originally tested negative on the index test B) Disease negative Yes: if ≥ 80% of benign diagnoses were reached by histology and up to 20% were reached by clinical follow‐up for a minimum of 3 months following the index test No: if > 20% of benign diagnoses were reached by clinical follow‐up for ≤ 3 months following the index test or if clinical follow‐up period was < 3 months Unclear: if the method of final diagnosis was not reported for any participant with benign or non‐melanoma diagnosis |
| 2) Were the reference standard results interpreted without knowledge of the results of the index test? Please score this item for all studies even though histopathology interpretation is usually conducted with knowledge of the clinical diagnosis (from visual inspection or dermoscopy or both). We will deal with this by not including the response to this item in the 'Risk of bias' assessment for these tests. For reviews of all other tests, this item will be retained |
Yes: if the reference standard diagnosis was reached blinded to the index test result No: if the reference standard diagnosis was reached with knowledge of the index test result Unclear: if blinded reference test interpretation was not clearly reported |
| Could the reference standard, its conduct, or its interpretation have introduced bias? For visual inspection/dermoscopy evaluations
For all other tests
|
For visual inspection/dermoscopy evaluations
For all other tests
|
| Reference standard (3) concern about applicability | |
| 1) Are index test results presented separately for each component of the target condition (i.e. separate results presented for those with invasive melanoma, melanoma in situ, lentigo maligna, severe dysplasia, BCC, and cSCC)? |
Yes: if index test results for each component of the target condition could be disaggregated No: if index test results for the different components of the target condition could not be disaggregated Unclear: if not clearly reported |
| 2) Expert opinion (with no histological confirmation) was not used as a reference standard 'Expert opinion' means diagnosis based on the standard clinical examination, with no histology or lesion follow‐up ***do not complete this item for teledermatology studies |
Yes: if expert opinion was not used as a reference standard for any participant No: if expert opinion was used as a reference standard for any participant Unclear: if not clearly reported |
| 3) Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? |
Yes: if histology interpretation was reported to be carried out by an experienced histopathologist or dermatopathologist No: if histology interpretation was reported to be carried out by a less experienced histopathologist Unclear: if the experience/qualifications of the pathologist were not reported |
Is there concern that the target condition as defined by the reference standard does not match the review question?
***For teledermatology studies only
|
***For teledermatology studies only
|
| Flow and timing (4): risk of bias | |
| 1) Was there an appropriate interval between index test and reference standard? A) For histopathological reference standard, was the interval between index test and reference standard ≤ 1 month? B) If the reference standard included clinical follow‐up of borderline/benign appearing lesions, was there ≥ 3 months' follow‐up following application of index test(s)? |
A) Yes: if study reported ≤ 1 month between index and reference standard No: if study reported > 1 month between index and reference standard Unclear: if study did not report interval between index and reference standard B) Yes: if study reported ≥ 3 months' follow‐up No: if study reported < 3 months' follow‐up Unclear: if study did not report the length of clinical follow‐up |
| 2) Did all participants receive the same reference standard? |
Yes: if all participants underwent the same reference standard No: if > 1 reference standard was used Unclear: if not clearly reported |
| 3) Were all participants included in the analysis? |
Yes: if all participants were included in the analysis No: if some participants were excluded from the analysis Unclear: if not clearly reported |
| 4) For within‐person comparisons of index tests Was the interval between application of index tests ≤ 1 month? |
Yes: if study reported ≤ 1 month between index tests No: if study reported > 1 month between index tests Unclear: if study did not report the interval between index tests |
| Could the participant flow have introduced bias? For non‐comparative and between‐person comparison studies
For within‐person comparative studies
|
For non‐comparative and between‐person comparison studies
For within‐person comparative studies
|
| BCC: basal cell carcinoma; cSCC: cutaneous squamous cell carcinoma. | |
Appendix 5. Details of reflectance confocal microscopy algorithms and diagnostic thresholds for diagnosis
| RCM algorithms (based on analysis of training set) | |||||
| RCM score (Pellacani 2005; Pellacani 2007a) | Segura score: 2 step to identify melanocytic first and then melanomas (Segura 2009) | Guitera 2 step method for BCC and MM (Guitera 2012) | Pellacani 2 step method for dysplastic lesions and then MM (Pellacani 2012) | ||
| Used in: Curchin 2011 (with LM score) Guitera 2009a Guitera 2012 Lovatto 2015 Pellacani 2007a Pellacani 2014a |
Used in: Alarcon 2014a Guitera 2012 Lovatto 2015 Segura 2009 |
– | Used in: Pellacani 2012 Stanganelli 2015 |
||
| RCM score ≥ 2, ≥ 3, ≥ 4 Presence of 2 major features (each scoring 2):
Presence of 4 minor features (each scored 1)
|
Cut‐off of > ‐1 = 'most probable melanoma' Within melanocytic lesions 2 protective features associated with benign lesions (score ‐1 each)
2 risk features associated with melanoma (score +1 each)
Lesions were assigned a value from ‐2 to 2 according to the presence or absence of these factors |
Correctly identified as MM or BCC (based on independently significant features as id from training set) Melanoma:
|
2 step algorithm (≥ 3 characteristics present, 2 at step 1 and 1 at step 2) Step 1: identified dysplastic naevus Presence of cytologic atypia (≥ 1 present)
and Presence of architectural atypia (≥ 1 present)
Step 2: identified melanoma from dysplastic naevus (≥ 1 characteristic present)
|
||
| RCM 'no algorithm'(selected lesion characteristics, independently significant characteristics identified, or 'observer diagnosis') | |||||
| Langley 2007 (based on Langley 2001) | Ferrari 2015 |
Koller 2011 (MM) Rao 2013 (MM/BCC/SCC) Farnetani 2015 (MM and BCC) |
|||
| ≥ 1 characteristic present (selected from prior study) Any 1 of:
|
Independently significant features (these 4 features are referenced to Pellacani 2012 as 'melanoma clues') For featureless lesions (score 0‐2 on dermoscopy 7PCL), presence of ≥ 1 of:
For equivocal lesions (score 3‐4 on dermoscopy 7PCL), presence of ≥ 1 of:
≥ 1 characteristic present for each |
Observer diagnosis Koller 2011 'diagnoses based on 'expert experience' Rao 2013 (MM/BCC/SCC) Observers gave diagnosis and excise decision (no further details) Farnetani 2015 (MM and BCC) Evaluators completed a 'pattern description' (presence/absence of a number of RCM features) and gave an overall diagnosis of malignant (melanoma or BCC) or benign |
|||
| RCM 'no algorithm'(developed for specific study populations) | |||||
|
Longo 2013 Nodular lesions |
Pupelli 2013 ≤ 5 mm melanocytic lesions |
Figueroa‐Silva 2016 'Thin' MM with dermoscopic island |
|||
Independently significant features:
|
Independently significant features:
|
Overall diagnosis reported; features assessed included:
|
|||
| 7PCL: 7 point checklist; BCC: basal cell carcinoma; DEJ: dermoepidermal junction; LM: lentigo meligna; MM: malignant melanoma; RCM: reflectance confocal microscopy; SCC: squamous cell carcinoma. | |||||
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
1. Test.
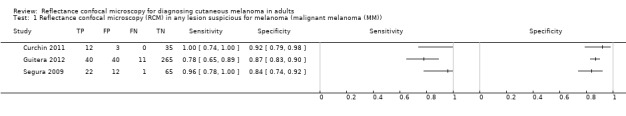
Reflectance confocal microscopy (RCM) in any lesion suspicious for melanoma (malignant melanoma (MM)).
2. Test.
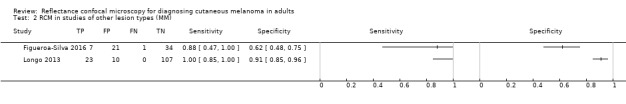
RCM in studies of other lesion types (MM).
3. Test.
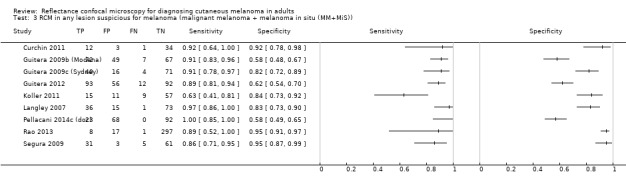
RCM in any lesion suspicious for melanoma (malignant melanoma + melanoma in situ (MM+MiS)).
4. Test.
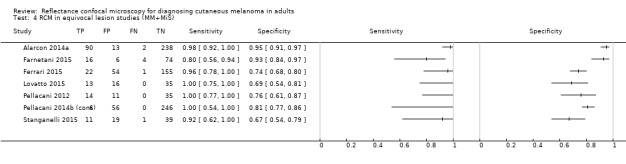
RCM in equivocal lesion studies (MM+MiS).
5. Test.
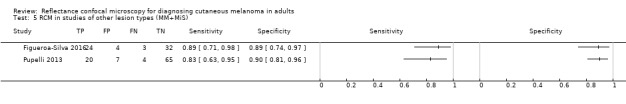
RCM in studies of other lesion types (MM+MiS).
6. Test.
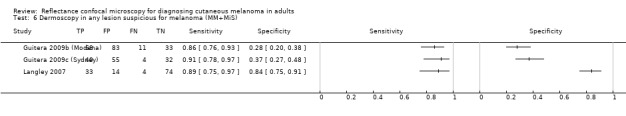
Dermoscopy in any lesion suspicious for melanoma (MM+MiS).
7. Test.
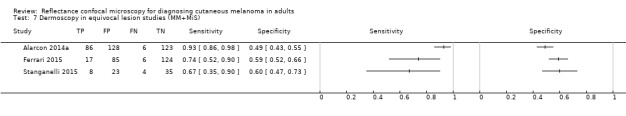
Dermoscopy in equivocal lesion studies (MM+MiS).
8. Test.

Dermoscopy in studies of other lesion types (MM+MiS).
9. Test.
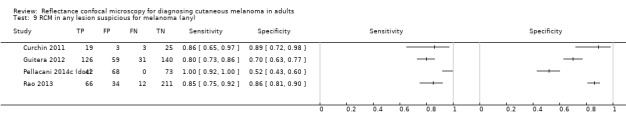
RCM in any lesion suspicious for melanoma (any).
10. Test.
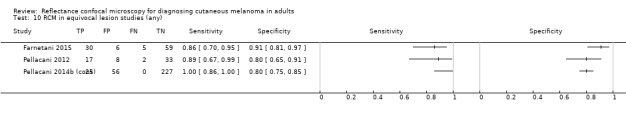
RCM in equivocal lesion studies (any).
11. Test.

RCM score at ≥ 3 (MM).
12. Test.

Segura algorithm at > ‐1 (MM).
13. Test.

Guitera 2 step algorithm (significant characteristics) (MM).
14. Test.

No algorithm (observer diagnosis) (MM).
15. Test.

No algorithm (significant characteristics) (MM).
16. Test.

RCM score at ≥ 2 (MM+MiS).
17. Test.
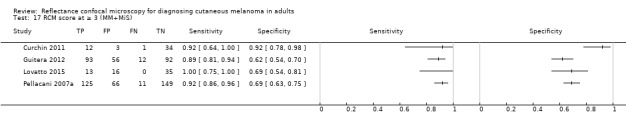
RCM score at ≥ 3 (MM+MiS).
18. Test.
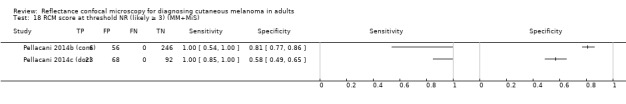
RCM score at threshold NR (likely ≥ 3) (MM+MiS).
19. Test.
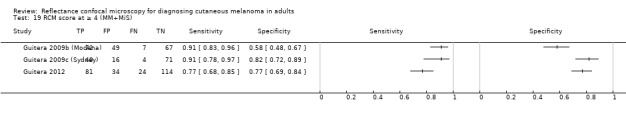
RCM score at ≥ 4 (MM+MiS).
20. Test.
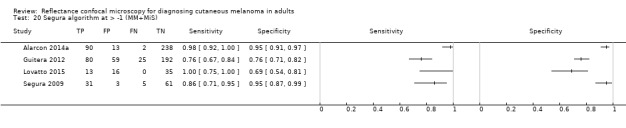
Segura algorithm at > ‐1 (MM+MiS).
21. Test.

Guitera 2 step algorithm (significant chars for MM) (MM+MiS).
22. Test.
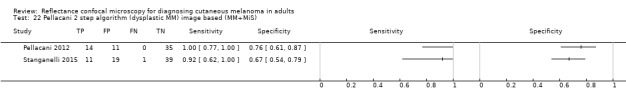
Pellacani 2 step algorithm (dysplastic MM) image based (MM+MiS).
23. Test.

RCM computer assisted diagnosis algorithm (MM+MiS).
24. Test.
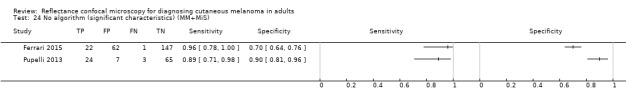
No algorithm (significant characteristics) (MM+MiS).
25. Test.

No algorithm (selected characteristics) (MM+MiS).
26. Test.
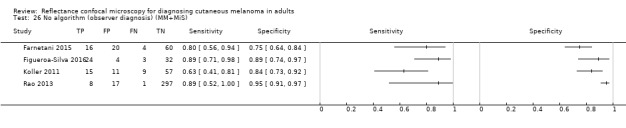
No algorithm (observer diagnosis) (MM+MiS).
27. Test.

No algorithm (observer diagnosis) paired in‐person (MM+MiS).
28. Test.

No algorithm (excise decision) (MM+MiS).
29. Test.

No algorithm (excise decision) paired in‐person (MM+MiS).
30. Test.

RCM score at ≥ 3 (any).
31. Test.
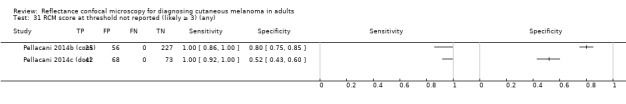
RCM score at threshold not reported (likely ≥ 3) (any).
32. Test.

Segura algorithm at > ‐1 (any).
33. Test.

Pellacani 2 step algorithm (dysplastic‐MM) (any).
34. Test.

Guitera 2 step algorithm (significant characteristics) (any).
35. Test.
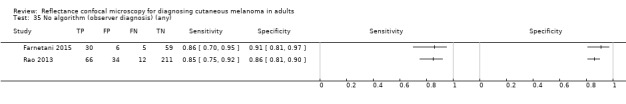
No algorithm (observer diagnosis) (any).
36. Test.

No algorithm (observer diagnosis) paired in‐person (any).
37. Test.

No algorithm (excise decision) (any).
38. Test.

No algorithm (excise decision) paired in‐person (any).
39. Test.
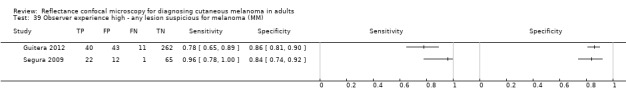
Observer experience high ‐ any lesion suspicious for melanoma (MM).
40. Test.

Observer experience low ‐ any lesion suspicious for melanoma (MM).
41. Test.

MM1 observer experience high other.
42. Test.

MM1 observer experience not reported other.
43. Test.
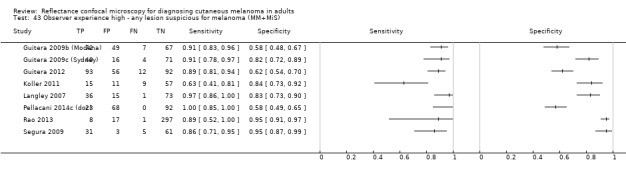
Observer experience high ‐ any lesion suspicious for melanoma (MM+MiS).
44. Test.
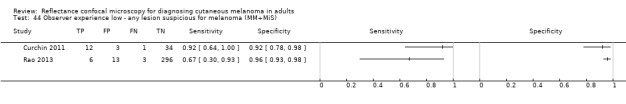
Observer experience low ‐ any lesion suspicious for melanoma (MM+MiS).
45. Test.
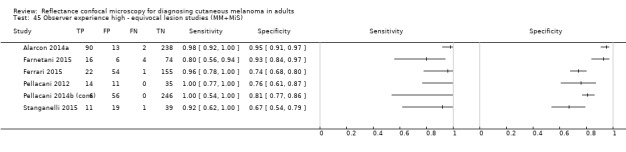
Observer experience high ‐ equivocal lesion studies (MM+MiS).
46. Test.

Observer experience low ‐ equivocal lesion studies (MM+MiS).
47. Test.

Observer experience not reported ‐ equivocal lesion studies (MM+MiS).
48. Test.
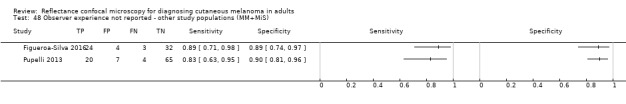
Observer experience not reported ‐ other study populations (MM+MiS).
49. Test.
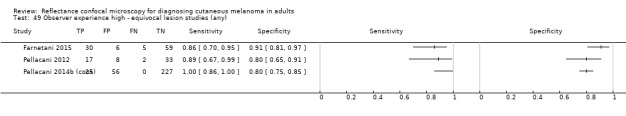
Observer experience high ‐ equivocal lesion studies (any).
50. Test.

Observer experience low ‐ equivocal lesion studies (any).
51. Test.
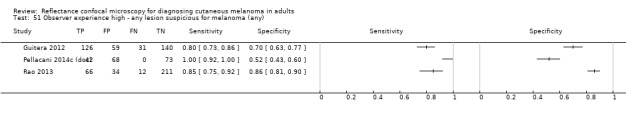
Observer experience high ‐ any lesion suspicious for melanoma (any).
52. Test.
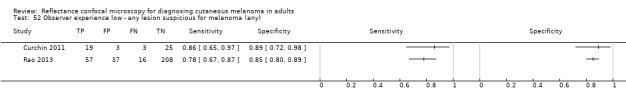
Observer experience low ‐ any lesion suspicious for melanoma (any).
53. Test.
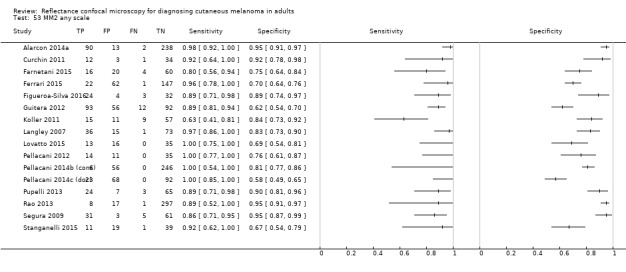
MM2 any scale.
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Alarcon 2014a.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: prospective Recruitment: 1 June 2011 to 30 May 2012 Country: Spain |
||
| Patient characteristics and setting |
Inclusion criteria: dermoscopically equivocal pigmented lesions, assumed to be melanocytic, seen at Melanoma Unit Setting: specialist unit (skin cancer/pigmented lesions clinic); Melanoma Unit of the Hospital Clinic of Barcelona Prior testing: dermatoscopic suspicion in all cases Setting for prior testing: specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: non‐melanocytic appearance and lesions referred for immediate excision or scheduled for digital follow‐up based on dermoscopy. Sample size (participants): number eligible: unclear; number included: unclear Sample size (lesions): number eligible: 343; number included: 264 Participant characteristics: Age: mean: unknown; median: 54.7; range: 8‐89 (for 264 excised) years Gender: 136 (51.5% of 264 excised) men Fitzpatrick phototype: type I to II: 42 (46%) of melanoma; type III to IV: 50 (54%) of melanoma Lesion characteristics: pigmented: 100% Lesion site: head/neck: 73 (27.7%); trunk: 135 (51.1%); limbs: 49 (18.6%); acral: 7 (7%). Thickness/depth: ≤ 1 mm: 86/92 melanoma; 6 > 1 mm |
||
| Index tests |
Dermoscopy: no algorithm used Method of diagnosis: dermoscopic images Prior test data: clinical examination or case notes site and age (or both); dermoscopy and RCM interpretation appeared to have been conducted by same observer with no indication of blinding. Diagnostic threshold: NR; no details Diagnosis based on: single observer Number of examiners: 1 of 3 Observer qualifications: dermatologist Experience in practice: not described Experience with index test: high experience/'expert' users; 3 dermatologists with expertise in RCM Any other details: all lesions imaged with a digital camera (Canon PowerShot G10; Canon, Tokyo, Japan) and a high resolution dermatoscope (DermLite Photo; 3Gen LLC, Dana Point, CA, USA) RCM: Segura algorithm used. RCM: Vivascope 1500 Method of diagnosis: confocal images (remote) Prior test data: lesion site or participant age only (or both); dermoscopy and RCM interpretation appeared to have been conducted by same observer with no indication of blinding. Diagnostic threshold: > ‐1 (presence of 2 protective criteria in the basal layer with a score of ‐1 was considered (i) edged papillae and (ii) presence of typical cells in the basal layer; and the presence of 2 risk criteria with a score of 1 was also considered: (i) presence of round pagetoid cells in upper layers of the epidermis; and (ii) presence of the nucleated cells found within the dermal papillae. A threshold score > ‐1 was used to obtain a diagnosis of melanoma). Diagnosis based on: single observer; 1 of 3 examiners Observer qualifications: dermatologist Experience in practice: high experience or 'expert' Experience with index test: high experience/'expert' users Other details: in vivo confocal microscopy performed with a commercially available reflectance confocal microscope (Vivascope 1500; Caliber Imaging and Diagnostics, Rochester, NY, USA), which uses a near‐infrared laser at a wavelength of 830 nm with a maximum power of 35 mW. |
||
| Target condition and reference standard(s) |
Type of reference standard: histology and clinical follow‐up of 1 year Details: histology of excision: 264; follow‐up: 79 Target condition (final diagnoses): melanoma (in situ and invasive, or NR): 92; BCC: 12; benign naevus: 107; other (including SK and AK: 53); plus 79 followed up with no histological classification |
||
| Flow and timing |
Excluded participants: following the use of dermoscopy, 343 lesions classified as equivocal would eventually have been excised. After the addition of RCM, 264/343 (77%) lesions judged as suggestive of malignancy according to the criteria followed in the study, and therefore were excised. The 79 lesions without criteria of malignancy upon RCM examination were scheduled for clinical or digital follow‐up; these were not included in accuracy calculations by the authors but data provided to allow their inclusion. Time interval between index test(s) and reference standard: histology undertaken on the same day as RCM. Unclear time gap from dermoscopy. |
||
| Comparative | Time interval between index test(s): not specified but appeared consecutive | ||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Unclear | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| High | |||
| DOMAIN 5: Comparative | |||
| Was each index test result interpreted without knowledge of the results of other index tests or testing strategies? | No | ||
| Was the interval between application of the index tests less than one month? | Unclear | ||
| Were all tests applied and interpreted in a clinically applicable manner? | No | ||
| Unclear | High | ||
Curchin 2011.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: prospective Period of data collection: January 2010 to May 2010 Country: Australia |
||
| Patient characteristics and setting |
Inclusion criteria: consecutive participants from dermatology department's minor excision booking list Setting: secondary (general dermatology) Prior testing: selected for excision (no further details) Setting for prior testing: unspecified Exclusion criteria: none reported Sample size (participants): number included: 42 Sample size (lesions): number included: 50 Participant characteristics: none reported Lesion characteristics: none reported |
||
| Index tests |
RCM: Vivascope 1500 RCM score: used RCM score and LM score for suspected lentigo maligna of the face (Guitera 2010) Method of diagnosis: in‐person Prior test data: dermoscopy "dermoscopic and RCM images were aligned over the top of each other so that correlation between the two could be made." Diagnostic threshold: RCM score: ≥ 3; LM score for suspected lentigo maligna of the face (Guitera 2010); threshold NR Diagnosis based on: single observer Number of examiners: possibly 1 Observer qualifications: NR Experience in practice: not described Experience with index test: low experience/novice users; analysis performed by a novice to RCM analysis after completing an RCM analysis course in Modena, Italy. Other details: macroscopic images obtained using a 14.7 megapixel digital camera (Canon Power Shot G10, Canon, Tokyo, Japan). A dermoscopic image taken using the dermoscopic camera attached to the Vivascope 1500 RCM System. RCM images were then captured using the Vivascope 1500 (Lucid Inc., Rochester, NY, USA). |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: number participants/lesions: disease positive: 21; disease negative: 29 Target condition (final diagnoses) melanoma (invasive): 12; melanoma (in situ): 1; BCC: 9; cSCC: 6 (included SK or AK, or both); 'benign' diagnoses: 23 |
||
| Flow and timing | Time interval to reference test: participants asked to come to the clinic (for imaging) 1 hour prior to their scheduled surgery. | ||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | Unclear | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Low | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Yes | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | No | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| Low | |||
Farnetani 2015.
| Study characteristics | |||
| Patient sampling |
Study design: case series of consecutively and retrospectively selected participants by an expert dermoscopist for a web based interobserver reliability study Data collection: retrospective image selection/prospective interpretation Period of data collection: NR Country: Italy (lesion image acquisition); observers located in the US (3), Europe (4), Australia (1), and Israel (1) |
||
| Patient characteristics and setting |
Inclusion criteria: diagnostically equivocal lesions excised due to clinical or dermoscopic suspicion of melanoma, where a specific clinical and dermoscopic diagnosis could not be rendered with certainty. Lesions selected by an expert dermoscopist blinded to final diagnosis Setting: secondary (general dermatology). All included RCM images were collected at the Department of Dermatology of the University of Modena and Reggio Emilia (Modena, Italy) Prior testing: clinical or dermatoscopic suspicion, or both Setting for prior testing: secondary (general dermatology) Exclusion criteria: poor quality index test image; no additional selection criteria considered in case selection Sample size (participants): number included: NR Sample size (lesions): number included: 100 Participant characteristics: none reported Lesion characteristics: none reported |
||
| Index tests |
RCM: no algorithm. Vivascope 1500 Method of diagnosis: confocal images (remote) 3 RCM mosaic images presented per lesion Prior test data: dermoscopy. "Each case for evaluation had a high‐resolution dermoscopic image obtained with a dermoscopic lens that was attached to a digital camera;" "No additional clinical information (eg, age and melanoma or lesion history) was provided to evaluators." Diagnostic threshold: evaluators completed a 'pattern description' (presence/absence of a number of RCM features) and gave an overall diagnosis of malignant (melanoma or BCC) or benign; no specific threshold used. Derivation aspect to study: discriminant analysis used to identified RCM features independently associated with malignancy, melanoma, and BCC. 3/6 discriminatory RCM features were more frequently observed in melanoma: the presence of pagetoid cells, the presence of atypical cells at the DEJ, and irregular epidermal architecture; 3/6 discriminatory RCM features were more frequently observed in BCCs: basaloid cord‐like structures, presence of ulceration, and a specific DEJ pattern. Accuracy was not estimated for combinations of these particular features. Diagnosis based on: results presented for each of 9 observers and for majority diagnosis; i.e. consensual diagnosis by ≥ 5/9 evaluators. Also presented mean across 9 observers and across 6 more experienced and 3 less experienced observers. Number of examiners: 9. 15 were invited to participate, 9 agreed. Between 15 June 2010 and 24 August 2010, participants were asked to evaluate 10 cases per week for 10 consecutive weeks. Observer qualifications: dermatologist Experience in practice: not described Experience with index test: low experience/novice users: 3 with < 3 years' RCM experience; high experience/'expert' users: 6 with ≥ 3 years' RCM experience |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: none reported Number of participants/lesions: disease positive: 35; disease negative: 65 Target condition (final diagnoses): melanoma (in situ and invasive, or NR): 20; BCC: 15; SK: 7; melanocytic nevi: 55; actinic keratoses: 3 |
||
| Flow and timing |
Excluded participants: excised lesions only included Time interval to reference test: NR Time interval between index test(s): N/A |
||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | Yes | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| Unclear | |||
Ferrari 2015.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: retrospective image selection/prospective interpretation Period of data collection: 2010 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: melanocytic lesions with equivocal clinical or dermoscopic features (or both) that underwent excision and had a complete set of dermoscopy and RCM images with histopathology report. Only dermoscopically featureless (retrospectively scoring 0‐2 on 7 point checklist) or equivocal lesions (those scoring 3‐4 on dermoscopy 7 point checklist) included in RCM evaluation. Setting: secondary (general dermatology) Prior testing: clinical or dermatoscopic suspicion (or both) Setting for prior testing: secondary (general dermatology) Exclusion criteria: non‐melanocytic appearance; unequivocal appearance 90 'positive‐clear cut' lesions (scoring ≥ 5 on 7 point checklist) were excluded from RCM evaluation; poor quality index test image "Only lesions with high quality dermoscopic images, a complete set of confocal images and histopathology report available were included in the study;" other characteristic: incomplete histopathology report. Sample size (participants): number included: NR Sample size (lesions): number eligible: 322; number included: 322* for dermoscopy; 232 for RCM (*232 for each test included in this review) Participant characteristics: none reported Lesion characteristics: thickness/depth: overall: mean 1.05 (SD 16) mm, range 0‐10 mm (70 melanomas); those scoring 0‐2 on 7 point checklist: mean 0.18 (SD 0.42) mm; range 0‐0.94 mm) (6 melanomas); those scoring 3‐4 on 7 point checklist: mean 0.36 (SD 0.42) mm, range 0‐1.4 mm (17 melanomas) |
||
| Index tests |
Dermoscopy: 7 point checklist Method of diagnosis: dermoscopic images Prior test data: lesion site and age provided; dermoscopy and RCM interpretation appeared to have been conducted by same observer with no indication of blinding Diagnostic threshold: all thresholds reported. Data extracted using standard threshold ≥ 3 Diagnosis based on: single observer Number of examiners: 1 of 3 Observer qualifications: dermatologist. All images interpreted independently by 1 of the 3 dermatologists with expertise in RCM. Experience in practice: not described Experience with index test: high experience/'expert' users; 3 dermatologists with expertise in RCM Any other details: all lesions imaged with a digital camera (Canon PowerShot G10; Canon, Tokyo, Japan) and a high resolution dermatoscope (DermLite Photo; 3Gen LLC, Dana Point, CA, USA). RCM: no algorithm (presence of significant characteristics); criteria taken from Pellacani (Two step); 4 features described as 'melanoma clues,' referenced to Pellacani 2012. Final criteria tested on data and only those predictive used. RCM: Vivascope 1500 Method of diagnosis: confocal images (remote) Prior test data: dermoscopy "Dermoscopic and confocal microscopic images were evaluated, in blind from histological diagnosis, by a dermatologist trained in dermoscopy and RCM." Diagnostic threshold: 2×2 data for chosen qualitative threshold For featureless lesions (score 0‐2 on dermoscopy 7PCL), presence of ≥ 1 of:
For equivocal lesions (score 3‐4 on dermoscopy 7PCL), presence of ≥ 1 of:
Derivation aspect to study: previously published RCM parameters demonstrated useful for melanoma detection were selected. Evaluated confocal features were:
Selection of characteristics indicative of skin cancer: logistic regression Characteristics selected: as above Diagnosis based on: consensus (3 observers) Number of examiners: 3 Observer qualifications: dermatologists Experience in practice: high/expert Experience with index test: high/expert Other details: confocal imaging was performed with near‐infrared reflectance mode confocal laser scanning microscope (Vivascope 1500; MAVIG GmbH, Munich, Germany). |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: not further described Number participants/lesions: 232 out of originally selected 322; disease positive: 23; disease negative: 209 Target condition (final diagnoses): melanoma (in situ or invasive): 23; benign naevus: 195; Spitz naevus: 14 |
||
| Flow and timing |
Excluded participants: 90 'positive‐clear cut' lesions scoring ≥ 5 excluded from RCM evaluation Time interval to reference test: images taken 'before excision' |
||
| Comparative | Dermoscopy and RCM interpretation appeared to have been conducted by same observer with no indication of blinding | ||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Low | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| High | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| Low | |||
| DOMAIN 5: Comparative | |||
| Was each index test result interpreted without knowledge of the results of other index tests or testing strategies? | Unclear | ||
| Was the interval between application of the index tests less than one month? | Yes | ||
| Were all tests applied and interpreted in a clinically applicable manner? | No | ||
| Low | High | ||
Figueroa‐Silva 2016.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: retrospective image selection/prospective interpretation Period of data collection: January 2010 to February 2015 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: all pigmented lesions with a clear‐cut DI and available dermoscopic and RCM images were included Setting: secondary (general dermatology) Prior testing: dermatoscopic suspicion in all cases Setting for prior testing: secondary (general dermatology) Exclusion criteria: none reported Sample size (participants): number eligible: NR; number included: 61 Sample size (lesions): number eligible: 1964 pigmented lesions; number included: 63 Participant characteristics: Mean age: 44.1 (SD 14.8) years Gender: 43% men Lesion characteristics: lesion site: trunk: 37 lesions (60%) |
||
| Index tests |
RCM: no algorithm (observer diagnosis); Vivascope 1500 Method of diagnosis: confocal images (remote) Prior test data: dermoscopic image provided but blinded to histology and clinical information Diagnostic threshold: melanoma or not based on pattern analysis. RCM mosaics were evaluated for the presence/absence of: cobblestone pattern, pagetoid cells, architecture type (ringed, meshwork or clod prevalent pattern at DEJ, regular/irregular), and atypical cells at the DEJ. All RCM criteria were evaluated on both the DI and the rest of the lesion. Diagnosis based on: 1 observer. 1 investigator reviewed all RCM images and rendered a diagnosis; 2 other investigators separately reviewed the dermoscopic images according to 4 DI patterns. Observer qualifications: NR; probably dermatologist given setting Experience in practice: not described Experience with index test: not described Other details: dermoscopic images previously collected using a digital camera (Canon Powershot; Canon, New York, NY, USA) equipped with a contact, non‐polarised dermatoscope (DermLite Photo 3Gen, San Juan Capistrano, CA, USA) using a 20‐fold magnification. RCM images acquired with a near‐infrared, reflectance mode, confocal microscope (Vivascope 1500 MAVIG GmbH, Munich, Germany). Minimum of 3 mosaics were obtained per lesion at 3 different skin levels (superficial epidermal layers, DEJ, and papillary dermis) as described elsewhere (Debarbieux 2013). |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis plus follow‐up Details: histology (not further described): disease positive: 27; disease negative: 19 Clinical follow‐up plus histology of suspicious lesions: lesions were followed up on the basis of original RCM interpretation, i.e. at time of participant presentation. All lesions would have been excised on basis of dermoscopy alone; length of follow‐up: ≥ 1 year (mean 22 months); number participants: 17 lesions (47% of all disease negative) Target condition (final diagnoses): melanoma (invasive): 8; melanoma (in situ): 19; benign naevus: 36 |
||
| Flow and timing |
Excluded participants: NR Time interval to reference test: NR Time interval between index test(s): N/A |
||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| High | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | Yes | ||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| High | |||
Guitera 2009b (Modena).
| Study characteristics | |||
| Patient sampling |
Study design: case series (random sample of 50% of benign lesions included to increase the prevalence of melanoma) Data collection: prospective Period of data collection: September 2004 to August 2007 Country: Italy and Australia Note: data were separated by country for the purposes of this review due to mixed use of imaged based and in‐person dermoscopy interpretation according to country. Guitera 2009b (Modena) reported data for lesions recruited in Modena, Italy. *The dataset also overlapped Pellacani 2007a, which reported data for RCM only but at alternative RCM score thresholds. |
||
| Patient characteristics and setting |
Inclusion criteria: melanocytic lesions suspicious of melanoma based on dermatoscopic diagnostic criteria or lesion change; only 50% of observed benign lesions included Setting: secondary (general dermatology) Prior testing: clinical or dermatoscopic suspicion (or both) or changes on digital monitoring Setting for prior testing: secondary (general dermatology) Exclusion criteria: lesions on soles/palms excluded; lentigo maligna excluded; lesions used in previous assessments or RCM model development Sample size (participants): number included: 195 Sample size (lesions): number eligible: 195; number included 195 Participant characteristics: Median age: 42 (7‐88) years; IQR 32 to 59 years Gender: 54.3% men Lesion characteristics: pigmented: 92%; non‐pigmented: 8% (included amelanotic lesions or those with tan, light grey, or pale blue pigment only); lesion thickness/depth: median 0.65 mm (IQR25 to IQR75: 0.23 to 1.01) |
||
| Index tests |
Dermoscopy: pattern analysis Method of diagnosis: in‐person; dermoscopy diagnosis made at time of first consultation, prior to RCM Prior test data: clinical examination or case notes (or both) Diagnostic threshold: NR Diagnosis based on: 1 observer Observer qualifications: dermatologist (described as Modena expert based in Dermatology Department) Experience in practice: high experience/'expert' Experience with index test: high experience/'expert' users; 'expert'; no further details Other details: hand‐held dermoscope (Delta 10, Heine, Herrsching, Germany) RCM: Pellacani RCM score; Vivascope 1000 and Vivascope 1500, Lucid Inc., Henrietta, NY, USA Method of diagnosis: confocal images (remote) Prior test data: lesion site or participant age (or both) provided. Confocal images from Modena were scored by an expert (located in Sydney) retrospectively and blinded to dermoscopy and pathological diagnosis, but not to information of site and age. Diagnostic threshold: 6 diagnostic features scored: non‐edged papillae and cytological atypia at the dermal‐epidermal junction scored 2 each; round pagetoid cells intraepidermally, widespread pagetoid infiltration in the epidermis, nucleated cells found within the dermal papillae, and cerebriform nests in the dermis scored 1 each. Total score > 3 indicated MM. Diagnosis based on: 1 observer Observer qualifications: NR; presumed dermatologist based on study setting and expert nature of observers Experience in practice: NR; based on study setting Experience with index test: NR, but both observers coauthored studies developing RCM Other details: some differences between Vivascope 1000 and Vivascope 1500 exist. "The former is a more cumbersome instrument, as 4 mm images required laborious reprocessing. Furthermore, single capture images were slightly smaller in size, however, showing a similar quality with respect to the Vivascope 1500." |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: none provided; disease positive: 79; disease negative: 116 Target condition (final diagnoses): melanoma (invasive): 61; melanoma (in situ): 18; benign naevus: 94; Spitz naevus 22 |
||
| Flow and timing |
Excluded participants: none reported Time interval to reference test: imaged prior to biopsy Time interval between index test(s): imaged prior to biopsy |
||
| Comparative | Confocal images from Modena were scored in Sydney retrospectively and blinded to dermoscopy but not age and lesion site | ||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| High | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Unclear | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Unclear | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| Low | |||
| DOMAIN 5: Comparative | |||
| Was each index test result interpreted without knowledge of the results of other index tests or testing strategies? | Yes | ||
| Was the interval between application of the index tests less than one month? | Yes | ||
| Were all tests applied and interpreted in a clinically applicable manner? | Unclear | ||
| Low | Unclear | ||
Guitera 2009c (Sydney).
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: prospective Period of data collection: September 2004 to August 2007 Country: Italy and Australia Note: data were separated by country for the purposes of this review due to mixed use of imaged based and in‐person dermoscopy interpretation according to country. Guitera 2009c (Sydney) reported data for lesions recruited in Sydney, Australia *The dataset also overlapped Pellacani 2007a, which reported data for RCM only but at alternative RCM score thresholds. |
||
| Patient characteristics and setting |
Inclusion criteria: melanocytic lesions suspicious of melanoma based on dermatoscopic diagnostic criteria or lesion change Setting: specialist unit (skin cancer/pigmented lesions clinic); Australia Melanoma Diagnostic centre Prior testing: clinical or dermatoscopic suspicion (or both) or changes on digital monitoring Setting for prior testing: specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: lesions on soles/palms excluded; lentigo maligna excluded; lesions used in previous assessments or RCM model development; 25/156 lesions rejected for poor quality dermoscopy image Sample size (participants): number included: 156 Sample size (lesions): number eligible: 156; number included: 131 Participant characteristics: Median age: 52 (range 19‐90) years; IQR 40 to 63 years Gender: 59% men Lesion characteristics: pigmented: 75%; non‐pigmented: 25% (included amelanotic lesions or those with tan, light grey, or pale blue pigment only); lesion thickness/depth: median 0.40 mm (IQR 0 to 0.84) |
||
| Index tests |
Dermoscopy: pattern analysis Method of diagnosis: dermoscopic images Prior test data: lesion site and age available Diagnostic threshold: NR Diagnosis based on: 1 observer; dermoscopy diagnosis of Sydney lesions was made retrospectively on the images in a random order, blinded to RCM and pathological diagnosis but not to information of site and age, by a Modena expert using pattern analysis (Pehamberger 1993). Observer qualifications: dermatologist; not clearly reported, but described as Modena expert based in Dermatology Department Experience in practice: high experience or 'expert' Experience with index test: high experience/'expert' users; 'expert': no further details Other details: high resolution digital oil immersion dermoscopy camera (Sentry, Polartechnics Ltd, Sydney, NSW, Australia) RCM: Pellacani RCM score; Vivascope 1000 and Vivascope 1500, Lucid Inc., Henrietta, NY, USA Method of diagnosis: confocal images (remote) Prior test data: lesion site or participant age only (or both). Confocal images from Sydney were scored by an expert (located in Modena), retrospectively and blinded to dermoscopy and pathological diagnosis, but not to information of site and age. Diagnostic threshold: 6 diagnostic features scored: non‐edged papillae and cytological atypia at the dermal‐epidermal junction scored 2 each; round pagetoid cells intraepidermally, widespread pagetoid infiltration in the epidermis, nucleated cells found within the dermal papillae, and cerebriform nests in the dermis scored 1 each. Total score > 3 indicated MM. Diagnosis based on: 1 observer Observer qualifications: NR; dermatologist assumed based on study setting and expert nature of observers Experience in practice: NR; based on study setting Experience with index test: NR, but both observers coauthored studies developing RCM Other details: some differences between Vivascope 1000 and Vivascope 1500 exist. "The former is a more cumbersome instrument, as 4 mm images required laborious reprocessing. Furthermore, single capture images were slightly smaller in size, however, showing a similar quality with respect to the Vivascope 1500." |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: none provided; disease positive: 44; disease negative: 87 Target condition (final diagnoses): melanoma (invasive): 28; melanoma (in situ): 16; benign naevus: 84; Spitz naevus 3 |
||
| Flow and timing |
Excluded participants: 25/156 lesions rejected for poor quality dermoscopy image, blinded to the diagnostician Time interval to reference test: NR Time interval between index test(s): NR |
||
| Comparative | Confocal images from Sydney were scored by an expert (located in Modena), retrospectively and blinded to dermoscopy and pathological diagnosis, but not to information of site and age. | ||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| High | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Unclear | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Unclear | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| High | |||
| DOMAIN 5: Comparative | |||
| Was each index test result interpreted without knowledge of the results of other index tests or testing strategies? | Yes | ||
| Was the interval between application of the index tests less than one month? | Yes | ||
| Were all tests applied and interpreted in a clinically applicable manner? | Unclear | ||
| Low | Unclear | ||
Guitera 2012.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: NR Period of data collection: NR Country: Australia and Italy Test set derived: randomly split into training and test sets |
||
| Patient characteristics and setting |
Inclusion criteria: consecutive participants presenting or found with suspicious lesions, including all macules of the face and neck suspicious for lentigo maligna, and which would be subjected to biopsy or excision to rule out an epithelial tumour or an MM following conventional clinical and dermoscopy diagnosis and with lesion location amenable to RCM; described as predominantly melanocytic or suspicious for BCC. Setting: mixed, lesions recruited from Modena (general dermatology) and Sydney (skin cancer/pigmented lesions clinic) Prior testing: clinical or dermatoscopic suspicion (or both) Exclusion criteria: location/site of lesion keratotic, sole, and palm lesions excluded Sample size (participants): number eligible: 663 Sample size (lesions): number eligible: 710; number included: 356 in test set, 253 melanocytic Participant characteristics: Median age (full sample): 53 (IQR 39 to 66) years, range: 6‐90 years Gender: 354 (53.4% of full sample) men Lesion characteristics: NR |
||
| Index tests |
RCM: RCM score and Segura algorithm; also derived own independently significant features for MM and BCC; Vivascope 1500 Method of diagnosis: confocal images (remote) Prior test data: lesion site or participant age (or both); "RCM features were described by two expert observers, blinded from any clinical information, dermoscopy, and clinical aspects, but not for the location and age of the patient." Diagnostic threshold: Pellacani RCM score (Pellacani 2007a): > 3 and > 2; Segura (Segura 2009) "calculated with a threshold of zero;" own new 2 step model identified 7 independently significant features for MM (assume presence of any one indicated T+): cerebriform nests, atypical cobblestone pattern with small nucleated cells in the epidermis, marked cytological atypia, pagetoid cells, disarranged epidermal layer with no honey comb, large interpapillae spaces filled with honeycomb, dense nest. 8 independently significant features for BCC: polarised in the honeycomb, linear telangiectasia‐like horizontal vessels, basaloid cord or nodule, epidermal shadow, convoluted glomerular‐like vessels, non‐visible papillae, cerebriform nests, disarray of the epidermal layer Derivation aspect to study: lesion characteristics assessed a series of 48 features, corresponding to previous observations (Pellacani 2007a; Guitera 2009a), and new descriptors were considered at 3 different depth levels. Descriptions and definitions provided. Selection of characteristics indicative of skin cancer by multivariate discriminant analysis performed on the training set. Diagnosis based on: single observer (n = 1 of 2) Observer qualifications: dermatologist Experience in practice: high experience or 'expert' Experience with index test: high experience/'expert' users |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: not further described; full sample; disease positive: 335; disease negative: 375 Target condition (final diagnoses): test set only. Melanoma (in situ and invasive, or NR): 105; BCC: 52; cSCC: 9; benign naevus: 132; Spitz naevus: 16; AK: 8; benign macule of the face: 31; dermatofibroma: 3 |
||
| Flow and timing |
Excluded participants: none Imaged prior to biopsy |
||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | Unclear | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Low | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Unclear | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Low | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| Low | |||
Koller 2011.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: prospective (retrospective image selection/prospective interpretation for training set) Period of data collection: July 2007 to June 2008 Country: Austria Training set lesions were evaluated retrospectively (also reported in Gerger 2005). |
||
| Patient characteristics and setting |
Inclusion criteria: melanocytic skin lesions recruited from Department of Dermatology; lesions were not selected according to presence or absence of particular RCM features. Setting: secondary (general dermatology) Prior testing: unclear; some assessment conducted as only melanocytic lesions included Setting for prior testing: secondary (general dermatology) Exclusion criteria: non‐melanocytic appearance Sample size (participants): number included: NR Sample size (lesions): number included: 92 (test set only) Participant characteristics: none reported Lesion characteristics: ulcerated: 1/24 melanomas; mean thickness/depth: 0.75 mm (SD 1.06; range in situ to 3.7 mm) |
||
| Index tests |
RCM: no algorithm overall diagnosis and development of new CAD model using training set of lesions; Vivascope 1000 Method of diagnosis: confocal images (remote) Prior test data: no further information used observer was blinded with regard to the clinical or histopathological diagnosis Diagnostic threshold: human observer: diagnosis based on 'expert experience;' RCM characteristics NR; CAD interpretation: 30.47% (set for sensitivity of 100%) Derivation aspect to study: lesion characteristics assessed: in each RCM image, a set of 39 analysis parameters were measured. Selection of characteristics indicative of skin cancer classification: procedure was performed by the CART (Classification and Regression Trees) analysis software from Salford Systems (San Diego, CA, USA). Characteristics selected: N/A Diagnosis based on: 1 observer Observer qualifications: independent clinical dermatologist interpreted RCM images but image acquisition not described in detail. Experience in practice: not described Experience with index test: high experience/'expert' users |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis plus expert diagnosis Details: histology (not further described): disease positive: 24; disease negative: 37. Expert opinion based on unequivocal clinical and conventional dermoscopic criteria: disease positive: 0; disease negative: 31 Target condition (final diagnoses): melanoma invasive: 18; melanoma in situ: 6; benign naevus: 68 |
||
| Flow and timing | Suspicious lesions were excised after clinical, dermoscopic, and confocal examination and subjected to standard histopathological assessment. | ||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Low | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | No | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| High | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| High | |||
Langley 2007.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: prospective Period of data collection: February 2002 to May 2005 Country: Canada |
||
| Patient characteristics and setting |
Inclusion criteria: participants with suspicious pigmented lesions scheduled for biopsy due to clinical suspicion of malignancy determined by clinical appearance or a history of change in the lesion. Setting: specialist unit (skin cancer/pigmented lesions clinic), Division of Dermatology PLC and the Plastic Surgery Clinics Prior testing: clinical suspicion of malignancy without dermatoscopic suspicion Setting for prior testing: specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: non‐pigmented; poor quality index test image; lesion site or previous diagnostic biopsy Sample size (participants): number eligible: 127; number included: 125 Sample size (lesions): number eligible: 127; number included: 125 Participant characteristics: Mean age: 44.2 (range 16 to 84) years Lesion characteristics: median thickness 0.62 (range 0.20 to 7.92) mm |
||
| Index tests |
Dermoscopy: pattern analysis Method of diagnosis: in‐person Prior test data: clinical examination or case notes (or both) Diagnostic threshold: qualitative pattern analysis; no further details Diagnosis based on: 1 observer Observer qualifications: NR; likely dermatologist Experience in practice: not described Experience with index test: not described Any other details: dermoscopy performed and diagnosis rendered using the pattern analysis method. Clinical photograph obtained with a Nikon D1X digital camera, and with a Nikon F401s camera with a 60 mm lens with dermatophot attachment RCM: no algorithm; selected characteristics based on the criteria described in authors' initial series (Langley 2001); Vivascope 1000 Method of diagnosis: in‐person Prior test data: "Clinical, dermoscopic and confocal examinations were conducted sequentially by a single reviewer (R.L.)." Diagnostic threshold: any 1 of: epidermal disarray with loss of the normal honeycomb pattern; a grainy image; pagetoid cells in the epidermis; complex branching dendrites or dendritic cells; atypical and pleomorphic refractile cells, and the presence of bright, highly refractile particles. Diagnosis based on: 1 observer with experience in CSLM performed the imaging and examined all images in real time Observer qualifications: dermatologist Experience in practice: not described Experience with index test: high experience/'expert' users |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: definitive diagnosis was made by a dermatopathologist; disease positive: 37; disease negative: 88 Target condition (final diagnoses): melanoma (invasive): 22; melanoma (in situ): 15; benign naevus: 88 |
||
| Flow and timing |
Excluded participants: 2 excluded from the database due to technical difficulties with the imaging. Interval: when CSLM imaging was complete, the lesions were removed by excisional biopsy. |
||
| Comparative | Clinical, dermoscopic, and confocal examinations conducted sequentially by a single reviewer | ||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included participants and chosen study setting appropriate? | Unclear | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| High | Unclear | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Yes | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Yes | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Low | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| High | |||
| DOMAIN 5: Comparative | |||
| Was each index test result interpreted without knowledge of the results of other index tests or testing strategies? | No | ||
| Was the interval between application of the index tests less than one month? | Yes | ||
| Were all tests applied and interpreted in a clinically applicable manner? | Yes | ||
| Low | Low | ||
Longo 2013.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: retrospective image selection/prospective interpretation Period of data collection: NR Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: clinically nodular lesions (defined as cutaneous palpable/superficial seated lesions and not subcutaneous ones) that underwent excision Setting: Department of Dermatology, University of Modena and Reggio Emilia and Dermatology and Skin Cancer Unit, Arcispedale S. Maria Nuova IRCCS, Reggio, Italy Prior testing: selected for excision (no further detail) Exclusion criteria: none reported Sample size (participants): number included: 140 Sample size (lesions): number included: 140 Participant characteristics: Mean age: 50 (SD 19.7) years Gender: 45.7% men Lesion characteristics: 'most' lesions on the trunk; dermatofibroma mainly located on extremities; mean thickness 16 (SD 1.82) mm; 23 'pure' nodular melanomas |
||
| Index tests |
RCM: no algorithm (correct diagnosis of each histological category); also identified independently significant features (could not include data for MM as does not give breakdown of nodular melanoma and melanoma metastases; no response to author contacted); Vivascope model NR; likely Vivascope 1500 given publication date Method of diagnosis: confocal images (remote) Prior test data: no further information used; blinded to dermoscopic image Diagnostic threshold: 'RCM pattern analysis' (referenced to Longo 2011; Pellacani 2007a; amongst others) Multivariate analysis identified 3 positive independent significant features for MM plus melanoma in situ (widespread pagetoid distribution (graded as focal, localised, widespread); many atypical cells; cerebriform nests) and 4 positive independent significant features for BCC (tumour islands (dark silhouettes or tightly packed basaloid islands); cauliflower architecture; bright filaments within the tumour islands; and presence of bright collagen). Derivation aspect to study: each of 36 criteria were also scored for presence or absence. MM: epidermis: honeycombed or cobblestone pattern; disarray of epidermis; Pagetoid spread; Pagetoid cell shape; Pagetoid cell distribution DEJ: non‐specific architecture; cytological atypia (moderate, severe); dermis: sheet‐like structures; dermal nesting; prominent vascularity (enlarged vessels covering > 50% of the lesion surface); inflammatory infiltrate covering > 50% of the lesion surface. BCC: epidermis: honeycombed or cobblestone pattern; disarray of epidermis; ulceration or erosions; DEJ: cauliflower architecture; non‐specific architecture; dermis: dark silhouettes; tightly packed cells; bright filaments within tumour islands; prominent vascularity (enlarged vessels covering more than 50% of the lesion surface); inflammatory infiltrate covering > 50% of the lesion surface. SCC: epidermis: honeycombed or cobblestone pattern; disarray of epidermis; ulceration or erosions; scales; keratin inclusion/plugs; dermis: prominent vascularity ( enlarged vessels covering > 50% of the lesion surface); inflammatory infiltrate (covering > 50% of the lesion surface). Selection of characteristics indicative of skin cancer: univariate and then multivariate discriminant analysis was also performed to identify independently significant RCM criteria for MM plus melanoma in situ vs all other diagnoses, BCC vs all other diagnoses, SCC vs all other diagnoses. Diagnosis based on: 1 observer Observer qualifications: dermatologist Experience in practice: 5 years' experience in RCM and therefore presumably in practice Experience with index test: 5 years' experience in RCM |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: disease positive: 23 nodular melanoma; disease negative: 117 Target condition (final diagnoses): melanoma (invasive): 23 nodular; BCC: 28; cSCC: 6; other malignant: 9 melanoma metastases; benign naevus: 25 (14 compound, 8 intradermal, 3 blue naevi); 7 Spitz naevi; SK: 14; 5 vascular and 6 other benign lesions |
||
| Flow and timing |
Excluded participants: 8 not evaluable and 3 'non‐specific' RCM results reported (appeared to be excluded from derivation of independently significant characteristics) Not evaluable: lesions where all the 3 levels (epidermis, DEJ, and upper dermis) were not explorable for any reason that hampered the collection of quality images or the exploration of DEJ/superficial dermis. Non‐specific: lesions where a diagnosis could not be formulated, despite the possibility of exploring all 3 levels, because of the impossibility of recognising diagnostic features with enough confidence. |
||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| Unclear | |||
Lovatto 2015.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: retrospective image selection/prospective interpretation Period of data collection: January 2006 to January 2009 Country: Spain |
||
| Patient characteristics and setting |
Inclusion criteria: consecutive high risk participants with atypical melanocytic lesions excised because of change following sequential digital dermoscopy follow‐up. Required to have ≥ 2 of the following characteristics: > 100 melanocytic naevi; high number of atypical melanocytic lesions under dermoscopy; personal or familial history of melanoma; or predisposing genetic mutations for melanoma (i.e. CDKN2A mutation‐carriers, xeroderma pigmentosum). Setting: specialist unit (skin cancer/pigmented lesions clinic) Prior testing: changes on digital monitoring follow‐up with total body photography and digital dermoscopy Setting for prior testing: specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: unequivocal appearance/diagnosis Sample size (participants): number included: 51 Sample size (lesions): number included: 64 Participant characteristics: Mean age: 42 (SD 11.7) years; range 25‐69 years. Gender: 47% men; Other details: history of melanoma/skin cancer: 25%; family history of melanoma: 24%; genetic predisposition: 4% (CDKN2A mutation); 20% with both personal and familial history of melanoma. Fitzpatrick phototype I to II: 71%; type III to IV: 29% Lesion characteristics: mean total dermoscopy score at follow‐up (i.e. on excision): 5.44 for naevus group and 5.55 for melanoma group. 5 Lesions at ≤ 1 mm thickness (3 with Breslow 0.5 mm, and 1 each at 0.6 mm and 0.7 mm) |
||
| Index tests |
RCM: RCM score; Segura algorithm; Vivascope 1500 Method of diagnosis: confocal images (remote) Prior test data: no further information used; blinded to dermoscopy and histopathologic diagnosis Diagnostic threshold: Segura algorithm (Segura 2009) included 4 diagnostic features: 2 protective criteria in the basal layer with a score of ‐1 these were (i) edged papillae and (ii) typical cells in the basal layer; and 2 risk criteria with a score of +1, these were (i) roundish pagetoid cells in upper layers of the epidermis and (ii) nucleated cells within the dermal papillae. Melanoma must be considered when the total score is ≥ 0. RCM score (Pellacani 2007a): 2 major criteria scoring 2 points; these were (i) presence of cytologic atypia and (ii) non‐edged papillae at basal layer and 4 minor criteria scoring 1 point; these were (i) presence of roundish cells in superficial layers spreading upward in a pagetoid fashion, (ii) pagetoid cells widespread throughout the lesion, (iii) cerebriform clusters in the papillary dermis, and (iv) nucleated cells within dermal papilla. Score ≥ 3. Diagnosis based on: unclear; could be consensus or mean (n = 3 observers) Observer qualifications: NR; likely dermatologists (based in Dermatology Department) Experience in practice: not described Experience with index test: not described Other details: Vivascope 1500 (Lucid Inc., Henrietta, NY, USA) |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: consensus of 3 skilled histopathologists with experience in the field of melanocytic skin lesions; reviewed at the dermatopathology conference; disease positive: 13; disease negative: 51 Target condition (final diagnoses): melanoma (invasive): 5; melanoma (in situ): 8; benign naevus: 51 (melanocytic naevus with variable degree of atypia) |
||
| Flow and timing |
Excluded participants: NR Time interval to reference test: consecutive; images taken 'before excision' Time interval between index test(s): N/A |
||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Low | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | Yes | ||
| Low | |||
Pellacani 2007a.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: prospective Period of data collection: NR Country: Italy and Australia Test set derived: for multivariate analysis, the study sample was randomly divided into a training set and a test set, each comprising 50% of the lesions. Data included relate to full sample. *The dataset also overlapped Guitera 2009a, which reported data for dermoscopy as well as RCM; have only included Pellacani 2007a data related to alternative RCM score thresholds; not included in primary analysis. |
||
| Patient characteristics and setting |
Inclusion criteria: all melanocytic lesions excised to exclude melanoma, based upon dermoscopy, sequential digital monitoring, or history of change in standard clinical practice, were included. Setting: secondary (general dermatology) Italy; specialist unit (skin cancer/pigmented lesions clinic) Australia Prior testing: clinical or dermatoscopic suspicion (or both); changes on digital monitoring Exclusion criteria: lesions of palms and soles were not included; non‐melanocytic appearance; lesions excised for cosmetic reasons or solely due to a participant request; lentigo maligna excluded Sample size (participants): number included: 332 Sample size (lesions): number included: 351; 156 from Australia and 195 from Italy Participant characteristics: Median age: 47.7 (IQR 35.9 to 60.4) years Gender: 52% men Lesion site: head/neck: 15; trunk: 68; abdomen and chest: 135 on the back; upper limbs/shoulder: 50; lower limbs/hip: 83. ≤ 1 mm thickness: 66% (62/136); 1.01‐2.00 mm: 25% (23); 2.01‐4.00 mm: 9% (8); median thickness 0.49 mm (IQR 0 to 0.89) |
||
| Index tests |
RCM: RCM score. Also identified features independently correlated with malignancy by means of discriminant analysis on the training set, unable to include as only AUC presented; Vivascope 1000s and Vivascope 1500s Method of diagnosis: confocal images (remote) Prior test data: lesion site or participant age only (or both) Diagnostic threshold: data presented for all RCM scores from ≥ 1 to ≥ 8; data extracted for ≥ 2, ≥ 3, and ≥ 4 (included here only for ≥ 2, ≥ 3). Diagnosis based on: 2 single observers; 1 from the University of Modena evaluated the Sydney cases, and 1 from Sydney evaluated the Modena cases Observer qualifications: dermatologist Experience in practice: not described Experience with index test: high experience/'expert' users Other details: Vivascope 1000s and Vivascope 1500s, Lucid Inc., Henrietta, NY, USA |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: not further described; disease positive: 136; disease negative: 215 Target condition (final diagnoses): melanoma (invasive): 94; melanoma (in situ): 42; benign naevus: 215 |
||
| Flow and timing | RCM images acquired before biopsy | ||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| Low | |||
Pellacani 2012.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: prospective Period of data collection: 1 January 2008 to 31 March 2008 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: melanocytic lesions with equivocal clinical or dermatoscopic features (or both) Setting: secondary (general dermatology) not mentioned in text, just in author institution details Prior testing: clinical or dermatoscopic suspicion (or both) Setting for prior testing: secondary (general dermatology) (it was inferred that the participant was evaluated in the same unit) Exclusion criteria: non‐melanocytic appearance; unequivocal appearance/diagnosis; disagreement between evaluators on tumour histological classification Sample size (participants): number included: 62 Sample size (lesions): number eligible: 64; number included: 60 Participant characteristics: none reported Lesion characteristics: none reported |
||
| Index tests |
RCM: Pellacani (Two step) own new algorithm based on evaluation of a list of previously published parameters and some new descriptors (cites Pellacani 2009a; Pellacani 2009b; Scope 2007); Vivascope 1500 Method of diagnosis: confocal images (remote) Prior test data: no further information used "blinded from any clinical, dermatoscopic, or histopathologic information" Diagnostic threshold: ≥ 3 chars present, 2 at step 1 and 1 at step 2. Step 1: to identify dysplastic naevus: presence of cytologic atypia (≥ 1 present) including: round pagetoid cells, atypical cells at DEJ. Presence of architectural atypia (≥ 1 present) including: irregular junctional nests, short interconnections between junctional nests, non‐homogenous cellularity within junctional nests. Step 2: to identify melanoma from dysplastic naevus (≥ 1 present): widespread (≥ 50% of lesional area) round pagetoid cells, widespread (≥ 50% of lesional area) atypical cells at the DEJ, and non‐edged papillae (≥ 10% of the lesional area) Derivation aspect to study: lesions were evaluated for a list of previously published parameters and for some new descriptors specifically introduced for this study. Selection of characteristics indicative of skin cancer: for multivariate analysis, binary logistic regression was performed for the identification of the independently significant features in distinguishing among non‐dysplastic nevi, dysplastic nevi, and MM. Stepwise forward selection and goodness‐of‐fit statistics were used to select the features and determine whether the model adequately described the data. P < 0.01 was considered significant for the correlation tests, whereas a P < 0.05 was used for the other statistical tests. Diagnosis based on: 1 observer Observer qualifications: dermatologist Experience in practice: high experience or 'expert' Experience with index test: high experience/'expert' users Other details: RCM used a low‐power 830 nm laser beam that generated horizontal sections of the skin of 1.0 μm lateral resolution up to approximately 200 μm in depth. A minimum of 3 mosaics, with a maximum area of 8 × 8 mm were obtained per lesion, 1 in the superficial epidermis (stratum granulosum/spinosum), 1 at the DEJ, and 1 in the papillary dermis. |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: disease positive: 14; disease negative: 46 Target condition (final diagnoses): melanoma (invasive): 10; melanoma in situ: 4. Benign: severe dysplasia: 5; 7 showed mild dysplasia, 15 moderate; 19 non‐dysplastic nevi |
||
| Flow and timing |
Excluded participants: 4 lesions were excluded as dermatopathologists could not agree on pathology (in 2 cases discordance was for MM vs dysplastic naevus diagnosis, and in the other 2 between dysplastic and non‐dysplastic naevus) Time interval to reference test: before excision, all lesions were recorded by means of digital dermatoscopy and RCM |
||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| High | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| High | |||
Pellacani 2014b (cons).
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: prospective Period of data collection: January 2010 to December 2010 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: participants requesting a mole check or with suspicion of melanoma who were referred to PLC and who were then found to have atypical lesions on dermoscopy. Those in whom diagnosis could not be determined on dermoscopy were referred for an 'outcome decision' (consultation group). Participants were referred on the basis of both urgent access (melanoma suspected in a single lesion by an expert or other dermatologist) and scheduled access (referred for dermoscopy and total body examination). Setting: specialist unit (skin cancer/pigmented lesions clinic) Prior testing: dermatoscopic suspicion in all cases. All participants underwent dermoscopy in the PLC; those with dermoscopically atypical lesions were referred for RCM, either to document a lesion already selected for excision (documentation group, reported in Pellacani 2014c (doc)) or for an 'outcome decision' (consultation group), i.e. diagnosis could not be determined on dermoscopy. Setting for prior testing: specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: clinically or dermatoscopically (or both) clear‐cut epithelial tumours were not enrolled; poor quality index test image. In 9 cases, RCM could not be performed (5 RCM documentation and 4 RCM consultation) due to the presence of artefacts, hyperkeratosis, ulcerations, or a combination of these, impeding imaging. Sample size (participants): number eligible: 1005 examined with dermoscopy; number included: 252 referred for RCM consultation Sample size (lesions): number eligible: NR; number included: 308 for RCM documentation Participant characteristics: Median age: 41.7 (IQR 31.9 to 52.1) Gender: for all 1005 referred participants: 443 (44%) men Other details: consultation group only: history of melanoma/skin cancer: 23 (7%); family history of melanoma 30 (10%). Fitzpatrick phototype I to II: 150 (49%); type III to IV: 116 (38%) Lesion characteristics: lesion site (full sample) head/neck: 9%; trunk: 59%; upper limbs/shoulder: 12%; lower limbs/hip: 20% |
||
| Index tests |
RCM: RCM score; Vivascope 1500 Method of diagnosis: in‐person Prior test data: participants were "referred to confocal unit;" confocal reader was blinded to the participant pathway and aware that lesions were dermoscopically atypical for 'RCM documentation' or for 'RCM consultation.' Diagnostic threshold: NR; Pellacani 2005 cited Diagnosis based on: 1 observer Observer qualifications: dermatologist Experience in practice: not described Experience with index test: not described but 'confocal unit' described Other details: dermatoscopy examinations were conducted using the Dermlite HR (3Gen LLC, San Juan Capistrano, CA, USA). Lesions that were scheduled for digital monitoring were also acquired by means of FotoFinder (TeachScreen GmbH, Bad Birnbach, Germany) using 20‐fold magnification. |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis plus follow‐up and cancer registry follow‐up Histology (not further described): 81 (consultation group) (overall dataset 292 excised (see Pellacani 2014b (cons))) Clinical follow‐up: 227, 28 of which were subsequently excised (including above) because of observed dermatoscopic changes (all benign). Most non‐excised lesions (178/199 (89.4%)) were followed up for 1 year; the others were lost at 1‐year follow‐up Cancer registry follow‐up: those lost to clinical follow‐up were checked on the tumour registry; no melanomas were diagnosed in participants scheduled for follow‐up after baseline examinations. Target condition (final diagnoses): melanoma (invasive): 13; melanoma (in situ): 9; BCC: 19; melanoma metastasis: 1; Clark naevus: 71; Spitz naevus: 5; solar lentigo, SK, or lichen planus‐like keratosis: 0; other benign: 207 (8 with histological diagnosis (25 Clark naevi, 2 Spitz naevi, and 1 benign non‐melanocytic lesion) and 199 benign on follow‐up) |
||
| Flow and timing | Excluded participants: 9 excluded due to RCM failure | ||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included participants and chosen study setting appropriate? | Yes | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Yes | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| High | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | Yes | ||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| High | |||
Pellacani 2014c (doc).
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: prospective Period of data collection: January 2010 to December 2010 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: participants requesting a mole check or with suspicion of melanoma who were referred to PLC and who were then found to have atypical lesions on dermoscopy. Those in whom excision was required on dermoscopy were referred for RCM documentation (documentation group). Participants were referred on the basis of both urgent access (melanoma suspected in a single lesion by an expert or other dermatologist) and scheduled access (referred for dermoscopy and total body examination). Setting: specialist unit (skin cancer/pigmented lesions clinic) Prior testing: dermatoscopic suspicion in all cases. All participants underwent dermoscopy in the PLC; those with dermoscopically atypical lesions were referred for RCM, either to document a lesion already selected for excision (documentation group, as reported here) or for an 'outcome decision' (consultation group, reported in Pellacani 2014b (cons)), i.e. diagnosis could not be determined on dermoscopy. Setting for prior testing: specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: clinically or dermatoscopically (or both) clear‐cut epithelial tumours were not enrolled; poor quality index test image. In 9 cases, RCM could not be performed (5 RCM documentation and 4 RCM consultation) due to the presence of artefacts, hyperkeratosis, ulcerations, or a combination of these, impeding imaging. Sample size (participants): number eligible: 1005 examined with dermoscopy; number included: 171 referred for RCM documentation Sample size (lesions): number eligible: NR; number included: 183 for RCM documentation Participant characteristics: Median age: 41.2 (IQR 35 to 63) years Gender: for all 1005 referred participants: 443 (44%) men Other details: history of melanoma/skin cancer: 8 (5%); family history of melanoma: 13 (8%). Fitzpatrick phototype I to II: 99 (58%); type III to IV: 72 (42%) Lesion characteristics: lesion site (full sample): head/neck: 9%; trunk: 59%; upper limbs/shoulder: 12%; lower limbs/hip: 20% |
||
| Index tests |
RCM: RCM score; Vivascope 1500 Method of diagnosis: in‐person Prior test data: participants were "referred to confocal unit;" confocal reader was blinded to the participant pathway and aware that lesions were dermoscopically atypical for 'RCM documentation' or for 'RCM consultation.' Diagnostic threshold: NR; Pellacani 2005 cited. Diagnosis based on: 1 observer Observer qualifications: dermatologist Experience in practice: not described Experience with index test: not described but 'confocal unit' described Other details: dermatoscopy examinations were conducted using the Dermlite HR (3Gen LLC, San Juan Capistrano, CA, USA). Lesions that were scheduled for digital monitoring were also acquired by means of FotoFinder (TeachScreen GmbH, Bad Birnbach, Germany) using 20‐fold magnification. |
||
| Target condition and reference standard(s) |
Type of reference standard: histology alone for documentation group; 227 from consultation group were referred for follow‐up (see Pellacani 2014b (cons)) Target condition (final diagnoses): melanoma (invasive): 13; melanoma (in situ): 9; BCC: 19; melanoma metastasis: 1; Clark naevus: 121; Spitz naevus: 8; solar lentigo, SK, or lichen planus‐like keratosis: 7; other benign: 5 (1 haemosiderotic dermatofibroma, 1 xanthogranuloma, 1 viral wart, and 2 non‐specific inflammatory dermatoses) |
||
| Flow and timing | Excluded participants: 9 excluded due to RCM failure | ||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included participants and chosen study setting appropriate? | Yes | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Yes | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| High | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | Yes | ||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| High | |||
Pupelli 2013.
| Study characteristics | |||
| Patient sampling |
Study design: case control Data collection: retrospective Period of data collection: 2007‐2011 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: melanomas < 5 mm consecutively excised; plus 3 histologically confirmed small‐diameter naevi per included melanoma Setting: specialist unit (skin cancer/pigmented lesions clinic) (from author's institution) Prior testing: selected for excision (no further detail), All had undergone dermoscopy and RCM to be included Setting for prior testing: specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: lesion size > 5 mm excluded; disagreement between evaluators on tumour histological classification Sample size (participants): number included: 96 Sample size (lesions): number included: 96 Participant characteristics: Mean age: MM: 48 (IQR 17 to 77) years; naevi: 41 (IQR 6 to 82) years Gender: MM 54% men; naevi: 58% men Lesion characteristics: lesion site: trunk: 62% naevi; lower limbs/hip: 46% melanomas. Mean thickness 0.37 (SD 0.44 mm); melanoma diameter in situ MM: 10 < 1 mm, 3 ≥ 1 mm |
||
| Index tests |
Dermoscopy: 7 point checklist Method of diagnosis: dermoscopic images Prior test data: body site and age provided; RCM images may also have been available at time of image interpretation Diagnostic threshold: ≥ 3 Diagnosis based on: likely single observer (number NR) Observer qualifications: NR, likely dermatologist Experience in practice: not described Experience with index test: not described Any other details: dermoscopic images were acquired by means of a polarised dermatoscope (DermLite FOTO; 3Gen Inc., San Juan Capistrano, CA, USA). RCM: no algorithm independently significant features; Vivascope 1500 Method of diagnosis: confocal images (remote) Prior test data: lesion site or participant age only (or both). "Dermoscopic and confocal microscopic images were evaluated ‐ in blind from histological diagnosis, but not from the body site or the age of the patient." Diagnostic threshold: appeared to be ≥ 1 characteristic present. 3 characteristics were identified as independently significant (presence of ≥ 5 pagetoid cells per mm2, tangled lines within the epidermis, and atypical roundish cells at the DEJ). Sensitivity and specificity to allow 2×2 estimation were obtained from authors. Derivation aspect to study: lesion characteristics assessed. RCM parameters as published previously (all described). Selection of characteristics indicative of skin cancer multivariate analysis (logistic regression) Diagnosis based on: likely single observer (number NR) Observer qualifications: NR, likely dermatologist Experience in practice: not described Experience with index test: not described Other details: confocal imaging was performed with a near‐infrared reflectance‐mode confocal laser scanning microscope (Vivascope 1500; Lucid Inc., Rochester, NY, USA). The instrument and acquisition methods have been described elsewhere. |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: disease positive: 24; disease negative: 72 Target condition (final diagnoses): melanoma (invasive): 13; melanoma (in situ): 11; benign naevus: 65 (29 junctional, 19 compound, intradermal, 8 blue, 4 lentigo simplex); Spitz naevus: 7 |
||
| Flow and timing |
Excluded participants: NR Time interval to reference test: NR Time interval between index test(s): NR |
||
| Comparative | "Dermoscopic and confocal microscopic images were evaluated ‐ in blind from histological diagnosis, but not from the body site or the age of the patient." | ||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| High | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Unclear | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| High | Unclear | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Unclear | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Low | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| Unclear | |||
| DOMAIN 5: Comparative | |||
| Was each index test result interpreted without knowledge of the results of other index tests or testing strategies? | No | ||
| Was the interval between application of the index tests less than one month? | Yes | ||
| Were all tests applied and interpreted in a clinically applicable manner? | Unclear | ||
| Low | Unclear | ||
Rao 2013.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: NR; appeared to be prospective but not explicitly stated Period of data collection: June 2010 to September 2011 Country: US |
||
| Patient characteristics and setting |
Inclusion criteria: lesions removed for cosmetic or medical reasons (no further details; 'teleconsultation setting') Setting: secondary (general dermatology); private (based on author institutions) Prior testing: NR; unclear whether selection for excision was based on clinical assessment alone or including dermoscopy. Setting for prior testing: unspecified Exclusion criteria: 6 cases excluded due to "insufficient information" Sample size (participants): NR Sample size (lesions): number eligible: 340; number included: 334. 318/334 reported for reader 1; 323/334 reported for reader 2 Participant characteristics: none reported Lesion characteristics: none reported |
||
| Index tests |
RCM: no algorithm; overall observer diagnosis; Vivascope 1500 Method of diagnosis: in‐person US (reader 1; less experienced); confocal images (remote) Modena, Italy; reader 2 (more experienced) (*data used for primary analysis and QUADAS scoring) Prior test data: clinical examination or case notes (or both) and dermoscopy; "diagnosis was based on the dermoscopic image and confocal microscopy evaluation before excision." Diagnostic threshold: NR. Observers gave diagnosis and excise decision (no further details) Diagnosis based on: 2 single observers Observer qualifications: NR presume dermatologists Experience in practice: not described Experience with index test: low experience/novice users. Reader 1 (US) had 1 year of experience at the beginning of the study. High experience/'expert' users. Reader 2 (Italy) had > 9 years of experience with RCM. Other details: images were sent via Vivanet (CaliberID, Rochester, NY, USA), a Health Insurance Portability and Accountability Act‐compliant server. |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: not further described; disease positive: 78; disease negative: 256 Target condition (final diagnoses): melanoma (invasive): 8; melanoma (in situ): 1; BCC: 27; cSCC: 42; benign nevi: 176; SK: 22; AK: 24; other: 23 |
||
| Flow and timing | Excluded participants: 6 described as excluded because of insufficient information. Of the 334 participants included, reader 1 provided diagnoses for 318 of them and reader 2 provided diagnoses for 323. | ||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included participants and chosen study setting appropriate? | Unclear | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | Unclear | ||
| Were sufficient details of diagnostic thresholds provided? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| High | |||
Segura 2009.
| Study characteristics | |||
| Patient sampling |
Study design: case series. Authors separately describes recruitment of 'possibly malignant' and clinically/dermoscopically benign but seemed to be from same overall population Data collection: prospective Period of data collection: November 2005 to June 2006 Country: Spain |
||
| Patient characteristics and setting |
Inclusion criteria: all participants attending dermatology department or melanoma unit with a lesion suggestive of malignancy (study participation did not affect the clinical decision or the excision schedule) and participants with lesions known to be clinically and dermatoscopically benign; only melanocytic included in 2×2 Setting: mixed (Dermatology Department and the Melanoma Unit of the Hospital Clinic, Barcelona, Spain) Prior testing: clinical or dermatoscopic suspicion (or both) Exclusion criteria: none reported Sample size (participants): number eligible: 143 Sample size (lesions): number eligible: 154; number included: 100 (melanocytic only) Participant characteristics: Mean age: 49.45 years Gender: 39.9% men Other details: 13 had personal or family history of melanoma; 'most' described as having dysplastic mole syndrome and 'most' with dermatoscopic changes recorded during follow‐up examinations Lesion characteristics: lesion site: head: 34 (22%); trunk: 82 (53%); lower limbs/hip: 22 (14%); upper limbs: 14 (9%); neck: 2 (1.3%) |
||
| Index tests |
RCM: Segura own new algorithm; Vivascope 1500 Method of diagnosis: confocal images (remote) Prior test data: no further information used "stored confocal images were evaluated afterward, without regard to clinical or dermatoscopic data." Diagnostic threshold: cut‐off of > ‐1 = 'most probable melanoma.' Within melanocytic lesions 2 protective features associated with benign lesions (score ‐1 each); typical basal cells and edged papillae 2 risk features associated with melanoma (score +1 each); roundish pagetoid cells and atypical dermal nucleated cells. Lesions were assigned a value from ‐2 to 2 according to the presence or absence of these factors. Derivation aspect to study: lesion characteristics assessed: superficial layer: honeycombed pattern, cobblestone pattern, epidermal disarray, pagetoid cells. DEJ: visible dermal papilla, typical basal cells, marked atypia basal cells, cells in sheet like structures, junctional clusters. Papillary dermis: dermal nests, nucleated dermal cells, plump bright cells, bright hyper reflecting spots, enlarged dermal vessels. Selection of characteristics indicative of skin cancer: multivariate analysis using logistic regression to develop an algorithm in which benign (protective) features given a value of ‐1 and malignant (risk) features a value of +1 Diagnosis based on: single observer (number NR) Observer qualifications: NR. 2 observers described for the interobserver reproducibility study (120 images) but this appeared separate to RCM interpretations used for the accuracy study. Experience in practice: not described Experience with index test: not described Other details: study participation (RCM) did not affect the clinical decision or the excision schedule. Study aimed to develop a 2‐step process, first to differentiate melanocytic from non‐melanocytic lesions, second to differentiate malignant from benign within the melanocytic group. The first step was not extracted but note that relatively poor accuracy was observed (sensitivity for detection of ML 59%, specificity 96.7%) |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis plus expert diagnosis Details: both diagnostic and therapeutic excisions performed Number participants/lesions: 139 in total; including 92 melanocytic lesions; disease positive: melanocytic: 36 melanomas; non‐melanocytic: 27 BCC: disease negative: melanocytic: 56; non‐melanocytic: 20 Expert opinion: of the 154 included lesions, 15 clinically and dermatoscopically benign did not undergo excision: 8 were melanocytic benign nevi and 7 were non‐melanocytic Target condition (final diagnoses): melanoma (invasive): 23; melanoma (in situ): 13; BCC: 0 (27 BCC in non‐melanocytic lesion group; not included in 2×2); benign naevus: 64 (32 dysplastic, 20 common, 7 congenital, 2 blue, 2 Reed, and 1 Meyerson nevi); 27 benign NML not included in 2×2 (8 SK, 5 solar lentigines, 4 benign lichenoid keratoses, 4 vascular lesions, 3 actinic keratoses, 2 dermatofibromas, and 1 sebaceous hyperplasia) |
||
| Flow and timing | Excluded participants: all (54) non‐melanocytic | ||
| Comparative | |||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | No | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| High | |||
Stanganelli 2015.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: retrospective Period of data collection: July 2010 to July 2012 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: lesions excised at the Skin Cancer Unit on the basis of clinical or dermoscopic changes (or both) at follow‐up suggesting a malignancy and with available dermoscopy, RCM, and histological images and reports. Setting: specialist unit (skin cancer/pigmented lesions clinic) Prior testing: lesions showing clinical or dermoscopic changes on follow‐up Setting for prior testing: specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: unequivocal appearance/diagnosis Sample size (participants): number included: 70 Sample size (lesions): number included: 70 Participant characteristics: Mean age: women: 39 years, men: 40 years Gender: 38 (54%) men Other details: history of melanoma/skin cancer: 26 (37%). Total naevus counts: 27 (39%) > 50 melanocytic naevi: 33 (47%) 10‐50 naevi; and 10 (14%) < 10 naevi. Fitzpatrick phototype type I to II 19 (27%); type III to IV 50 (73%) Lesion characteristics: lesion site: head/neck: 5; trunk: 56; upper limbs/shoulder: 1; lower limbs/hip: 8. Median thickness 0.4 (range 0.2‐1) mm. Mean diameter at baseline: 8 (range 2‐22) mm; mean at follow‐up: 9 (range 3‐24) mm |
||
| Index tests |
Dermoscopy: revised 7 point checklist Method of diagnosis: dermoscopic images. Appears to be image based comparison of follow‐up images with baseline images to determine criteria indicating significant change Prior test data: baseline and follow‐up dermoscopic images were compared to detect structural or chromatic changes or the development of new dermoscopic features indicative of melanoma Diagnostic threshold: a score of 'no change' was assigned if all variables remained constant, with a tolerance of major axis change of 2 mm (Beer 2011; Terushkin 2012); 'minor change' if there was only symmetrical change in structural or chromatic pattern; 'moderate change' if either structural or chromatic changes were asymmetrical, but there were no melanoma‐specific criteria; and 'major change' if there were asymmetrical structural and chromatic changes, or the appearance of melanoma‐specific criteria. Diagnosis based on: unclear; NR for dermoscopy Observer qualifications: NR, likely dermatologists (RCM images in same study were evaluated jointly by 3 expert dermatologists who had no knowledge of the clinical, dermoscopic, or histopathology information) Experience in practice: not described Experience with index test: not described RCM: Pellacani (two step algorithm). Methods cited RCM score (Pellacani 2005); also refers to weighting according to extent and distribution for differential diagnosis with dysplastic naevus (Pellacani 2012). From discussion: "We were able to distinguish benign and malignant lesions accurately using a previously proposed algorithm for differentiating dysplastic naevus and melanoma that considers the extent and distribution of RCM parameters" (Pellacani 2012). Vivascope 1500 Method of diagnosis: confocal images (remote) Prior test data: no further information used observers "had no knowledge of the clinical, dermoscopic or histopathology information, and reached a consensus or majority opinion." Diagnostic threshold: NR in detail. "Each lesion was classified considering the main melanoma features (Pellacani 2005) and weighted according to extent and distribution for differential diagnosis with dysplastic naevus (Pellacani 2012)." Diagnosis based on: consensus 3 expert dermatologists Observer qualifications: dermatologist Experience in practice: high experience or 'expert' Experience with index test: high experience/'expert' users Other details: RCM images were obtained with a Vivascope 1500 (Lucid Inc., MAVIG GmbH, Munich, Germany) using an 830 nm laser at a maximum power of 20 mW. Methods and acquisition settings have been described previously (Pellacani 2007a). RCM images of 0.5 × 0.5 mm were acquired with a lateral resolution of 1 lm and an axial resolution of 3‐5 lm and assembled into composite images that covered 4‐8 mm2 mosaics (Pellacani 2009a). Composite images were obtained at 3 different depths, corresponding to the stratum granulosum/spinosum, the DEJ, and the papillary dermis. |
||
| Target condition and reference standard(s) |
Type of reference standard: histological diagnosis alone Details: not further described; disease positive: 12; disease negative: 58 Target condition (final diagnoses): melanoma (invasive): 11; melanoma (in situ): 1; 55 melanocytic naevi (79%) and 3 non‐melanocytic lesions (4%) |
||
| Flow and timing | RCM imaging performed before surgical excision | ||
| Comparative | (RCM) observers "had no knowledge of the clinical, dermoscopic or histopathology information, and reached a consensus or majority opinion." | ||
| Notes | – | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included participants and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Reflectance confocal microscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| For studies reporting the accuracy of multiple diagnostic thresholds, was each threshold or algorithm interpreted without knowledge of the results of the others? | |||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were sufficient details of diagnostic thresholds provided? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Was the minimum clinical follow‐up after application of index test(s) adequate? | |||
| If more than one algorithm evaluated for the same test, was the interval between application of the different algorithms 1 month or less? | |||
| Low | |||
| DOMAIN 5: Comparative | |||
| Was each index test result interpreted without knowledge of the results of other index tests or testing strategies? | Yes | ||
| Was the interval between application of the index tests less than one month? | Yes | ||
| Were all tests applied and interpreted in a clinically applicable manner? | No | ||
| Low | High | ||
7PCL: 7 point check list; AK: actinic keratosis; AUC: area under the curve; BCC: basal cell carcinoma; CAD: computer assisted diagnosis; CSLM: confocal scanning laser microscope; cSCC: cutaneous squamous cell carcinoma; DEJ: dermoepidermal junction; DI: dermoscopy island; IQR: interquartile range; LM: lentigo meligna; MM: malignant melanoma; N/A: not available/applicable; NR: not reported; PLC: pigmented lesion clinic; RCM: reflectance confocal microscopy; SD: standard deviation; SK: seborrhoeic keratosis.
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Agero 2006 | Excluded on sample size Only 5 lesions |
| Ahlgrimm‐Siess 2010 | Excluded on study population and sample size. 2 cases of BCC |
| Ahlgrimm‐Siess 2011 | Excluded on study population and sample size. 2 cases of SCC |
| Alarcon 2014b | Excluded on sample size |
| Amjadi 2011 | Excluded on study population; included only BCC (82)/SCC (48) and 8 AK/SK lesions; primary aim appeared to be to differentiate BCC and SCC despite describing inclusion of clinically difficult to diagnose non‐pigmented lesions |
| Bassoli 2012 | Excluded on target condition Aimed to identify criteria for specific diagnosis of LPLK using in vivo RCM |
| Benati 2015 | Excluded on individual lesion characteristics |
| Braga 2009 | Excluded on sample size Case reports |
| Carrera 2015 | Excluded as not a primary study |
| Castro 2015 | Excluded on target condition; eligible for keratinocyte review only |
| de Carvalho 2015 | Excluded on individual lesion characteristics Excluded on 2×2 data |
| de Carvalho 2016 | Excluded on target condition and sample size |
| Edwards 2016 | Excluded, not a primary study Systematic review |
| Eichert 2010 | Review/comment paper |
| Gareau 2009 | Excluded on study population Only BCC cases |
| Gerger 2005 | Excluded on reference standard Only 1/3 of disease negative group had adequate reference test Excluded duplicate or related publication; data reported as training set in Koller 2011 (#860) |
| Gerger 2006 | Excluded on reference standard Only 30/120 benign were excised (30/90 benign nevi and 0/30 SK) |
| Gerger 2008a | Excluded on reference standard All MMs were excised plus 14/50 benign; remainder diagnosed on clinical/dermoscopic criteria |
| Gerger 2008b | Excluded on reference standard; included 70 melanocytic lesions ‐ 20 MM (all histologically verified); 70 benign naevi (28% histologically verified, and the rest diagnosed with dermoscopy only) |
| Giambrone 2015 | Excluded on target condition Excluded but contact authors ‐ no information provided on the target condition, text describes malignant vs benign cutaneous lesions. Authors contacted 8 May 2017 |
| Gill 2014 | Excluded as derivation study; looking for correlation with histological features Excluded on 2×2 data; looked at correlation between RCM features and histological features; not test accuracy Excluded duplicate or related publication; same lesions reportedly included in Pellacani 2012 |
| Gonzalez 2002 | Excluded on study population. BCC only |
| Gonzalez 2013 | Excluded, not a primary study |
| Guida 2015 | Excluded, not a primary study Systematic review |
| Guitera 2010 | Excluded on target condition; only looking at LM and not LMM |
| Guitera 2013 | Excluded on study population; LM and LMM only Excluded on target condition; data only available for LM Excluded on 2×2 data |
| Haenssle 2006 | Excluded on index test; surveillance study estimating accuracy of different approaches to follow‐up |
| Hennessy 2010 | Excluded on 2×2 data |
| Hoogedoorn 2014 | Excluded conference abstract |
| Hoogedoorn 2015 | Excluded on sample size |
| Humphrey 2006 | Excluded on study population Excluded as derivation study; assessed lesion vascularity |
| Incel 2015 | Excluded on 2×2 data; excluded but contacted authors. Sensitivity and specificity given in Table 3 but not clear how the disease negative groups are comprised (i.e. BCC vs what? The 37 benign or some other definition?) and not clear what threshold was used |
| Kadouch 2015a | Systematic review |
| Kadouch 2015b | Excluded, not a primary study Clinical trial protocol |
| Kose 2014 | Excluded, not a test accuracy study Excluded on 2×2 data |
| Langley 2001 | Excluded on 2×2 Excluded but contacted authors; contact authors for RCM 2×2 data can only get 2×2 for clinical diagnosis |
| Langley 2006 | Excluded on sample size |
| Losi 2014 | Excluded if individual lesion characteristics Excluded on 2×2 data |
| Maier 2013 | Excluded on study population; all study participants had final diagnosis of melanoma |
| Malvehy 2012 | Excluded, not a primary study; review article |
| Menge 2016 | Excluded on target population; included participants with primary possible recurrent or previously treated lesions (or both) and did not disaggregate results. Also included multiple lesions per participant (63 'sites' from 17 participants; unclear how many of the 39 LM positive on histology had melanoma) |
| Miller 2011 | Excluded on target condition and on 2×2 data; not an accuracy study |
| Nobre 2011 | Excluded on sample size; case report |
| Nori 2004 | Excluded on target condition; eligible for keratinocyte review only |
| Pellacani 2005 | Excluded if derivation study; used leave one out |
| Pellacani 2007b | Excluded if individual lesion characteristics; looked at blue hue not overall diagnosis Excluded if derivation study |
| Pellacani 2008 | Excluded on 2×2 data; no accuracy data provided in the study, looked at correlation of RCM features to dermoscopy and histology |
| Pellacani 2009a | Excluded on 2×2 data; study tested concordance of terminology used in RCM, not accuracy. |
| Peppelman 2013 | Excluded on study population; only presented data for subtypes of BCC Excluded on 2×2 data; did not give accuracy data |
| Peppelman 2015 | Excluded as derivation study Excluded on 2×2 data; no data for overall accuracy |
| Peppelman 2016 | Excluded, not a primary study; RCT protocol |
| Puig 2012 | Excluded on sample size; case report |
| Reggiani 2015 | Excluded, not a primary study; systematic review |
| Rishpon 2009 | Excluded on sample size; only 3 invasive SCC Excluded if derivation study RCM characteristics for SCC |
| Röwert‐Huber 2007 | Review/comment paper |
| Salerni 2011 | Excluded on sample size; < 5 cases |
| Scope 2009 | Excluded on sample size |
| Scope 2014 | Excluded, not a primary study; editorial paper |
| Soyer 2013 | Excluded, not a primary study; comment on a primary study (Longo 2013) |
| Steiner 1992 | Excluded on sample size; only 2 melanomas |
| Stephens 2013 | Excluded on sample size |
| Stevenson 2013 | Excluded, not a primary study; systematic review of RCM |
| Tannous 2009 | Excluded on sample size; only 2 MMs |
| Willard 2011 | Excluded on sample size; case study |
| Witkowski 2016 | Excluded on target condition; eligible for keratinocyte review only |
| Xiong 2016 | Excluded, not a primary study, systematic review of RCM |
| Yelamos 2016 | Excluded, not a primary study |
AK: actinic keratosis; BCC: basal cell carcinoma; LM: lentigo meligna; LPLK: lichen planus‐like keratosis; MM: malignant melanoma; RCM: reflectance confocal microscopy; SCC: squamous cell carcinoma; SK: seborrhoeic keratosis.
Characteristics of studies awaiting classification [ordered by study ID]
Borsari 2016.
| Study characteristics | |||
| Patient sampling | Not yet assessed | ||
| Patient characteristics and setting | Not yet assessed | ||
| Index tests | Not yet assessed | ||
| Target condition and reference standard(s) | Not yet assessed | ||
| Flow and timing | Not yet assessed | ||
| Comparative | Not yet assessed | ||
| Notes | Published October 2016; after search dates | ||
Guitera 2016.
| Study characteristics | |||
| Patient sampling | Not yet assessed | ||
| Patient characteristics and setting | Not yet assessed | ||
| Index tests | Not yet assessed | ||
| Target condition and reference standard(s) | Not yet assessed | ||
| Flow and timing | Not yet assessed | ||
| Comparative | Not yet assessed | ||
| Notes | Published October 2016; after search dates | ||
Jain 2017.
| Study characteristics | |||
| Patient sampling | Not yet assessed | ||
| Patient characteristics and setting | Not yet assessed | ||
| Index tests | Not yet assessed | ||
| Target condition and reference standard(s) | Not yet assessed | ||
| Flow and timing | Not yet assessed | ||
| Comparative | Not yet assessed | ||
| Notes | Published March 2017; conference abstract only | ||
Ludzik 2016.
| Study characteristics | |||
| Patient sampling | Not yet assessed | ||
| Patient characteristics and setting | Not yet assessed | ||
| Index tests | Not yet assessed | ||
| Target condition and reference standard(s) | Not yet assessed | ||
| Flow and timing | Not yet assessed | ||
| Comparative | Not yet assessed | ||
| Notes | Published September 2016; after search dates | ||
Differences between protocol and review
Inclusion criteria amended to remove inclusion of participants "at high risk of developing melanoma, including those with a family history or previous history of melanoma skin cancer, atypical or dysplastic naevus syndrome, or genetic cancer syndromes" as these are not a target population for RCM use.
Primary objectives and primary target condition changed from detection of invasive melanoma alone to the detection of cutaneous invasive melanoma and atypical intraepidermal melanocytic variants, as the latter is more clinically relevant to the practicing clinician. The detection of the target condition of invasive melanoma alone has instead been included as a secondary objective.
For the primary objective, study populations that could not be clearly identified as either 'any lesion suspicious for melanoma ' or 'equivocal lesions' were considered separately as 'other lesion' studies.
We amended the text to clarify that studies available only as conference abstracts would be excluded from the review unless full papers could be identified; studies available only as conference abstracts do not allow a comprehensive assessment of study methods or methodological quality.
Secondary objectives tailored to the individual test, with three objectives added: to compare the accuracy of RCM to dermoscopy where both tests have been evaluated in the same studies; to determine the diagnostic accuracy of individual algorithms for RCM; and to determine the effect of observer experience. Heterogeneity investigations were limited by the data available.
Studies using cross‐validation, such as 'leave‐one‐out' cross‐validation were excluded rather than included as these methods are not sufficiently robust and are likely to produce unrealistic estimates of test accuracy. To improve clarity of methods, this text from the protocol, "We will include studies developing new algorithms or methods of diagnosis (i.e. derivation studies) if they use a separate independent 'test set' of participants or images to evaluate the new approach. We will also include studies using other forms of cross validation, such as 'leave‐one‐out' cross‐validation (Efron 1983). We will note for future reference (but not extract) any data on the accuracy of lesion characteristics individually, e.g. the presence or absence of a pigment network or detection of asymmetry" has been replaced with "We included all established algorithms or checklists to assist diagnosis. Studies developing new algorithms or methods of diagnosis (i.e. derivation studies) were included if they used a separate independent 'test set' of participants or images to evaluate the new approach. Studies that did not report data for a separate test set of participants or images were included only if the lesion characteristics investigated had previously been suggested as associated with melanoma and the study reported accuracy based on the presence or absence of particular combinations of characteristics. Studies using a statistical model to produce a data driven equation, or algorithm based on multiple diagnostic features, with no separate test set were excluded. Studies using cross‐validation approaches such as 'leave‐one‐out' cross‐validation were excluded (Efron 1983)."
We proposed to supplement the database searches by searching the annual meetings of appropriate organisations (e.g. British Association of Dermatologists Annual Meeting, American Academy of Dermatology Annual Meeting, European Academy of Dermatology and Venereology Meeting, Society for Melanoma Research Congress, World Congress of Dermatology, European Association of Dermato Oncology); however, due to volume of evidence retrieved from database searches and time restrictions we were unable to do this.
For quality assessment, the QUADAS‐2 tool was further tailored according to the review topic. In terms of analysis, restriction to analysis of per participant data was not performed due to lack of data. Sensitivity analyses were not performed as planned due to lack of data.
Contributions of authors
JD was the contact person with the editorial base. JD co‐ordinated contributions from the coauthors and wrote the final draft of the review. SEB conducted the literature searches. JD, NC, DS, and LP screened papers against eligibility criteria. JD and NC obtained data on ongoing and unpublished studies. JD, NC, DS, and LP appraised the quality of papers. JD, NC, DS, and LP extracted data for the review and sought additional information about papers. JD entered data into Review Manager 5. JD and JJD analysed and interpreted data. JD, JJD, NC, YT, and CD worked on the methods sections. JD, DS, RP, RNM, and HCW drafted the clinical sections of the background and responded to the clinical comments of the referees. JD, JJD, CD, and YT responded to the methodology and statistics comments of the referees. KG was the consumer coauthor and checked the review for readability and clarity, as well as ensuring outcomes were relevant to consumers. JD was the guarantor of the update.
Disclaimer
This project presents independent research supported by the National Institute for Health Research, via Cochrane Infrastructure funding to the Cochrane Skin Group and Cochrane Programme Grant funding, and the NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham. The views and opinions expressed therein are those of the authors and do not necessarily reflect those of the Systematic Reviews Programme, NIHR, NHS or the Department of Health and Social Care.
Sources of support
Internal sources
No sources of support supplied
External sources
-
NIHR Systematic Review Programme, UK.
This project was funded by an NIHR Cochrane Systematic Reviews Programme Grant (13/89/15)
-
The National Institute for Health Research (NIHR), UK.
The NIHR, UK, is the largest single funder of the Cochrane Skin Group
NIHR clinical fellowship, UK.
-
NIHR Birmingham Biomedical Research Centre, UK.
JD, JJD and YT receive support from the NIHR Birmingham Biomedical Research Centre
Declarations of interest
JD: nothing to declare. JJD: nothing to declare. DS: nothing to declare. NC: nothing to declare. SEB: nothing to declare. LP: nothing to declare. CD: nothing to declare. YT: nothing to declare. KG: nothing to declare. RNM: "my institution received a grant for a Barco NV commercially sponsored study to evaluate digital dermoscopy in the skin cancer clinic. My institution also received Oxfordshire Health Services Research Charitable Funds for carrying out a study of feasibility of using the Skin Cancer Quality of Life Impact Tool (SCQOLIT) in non melanoma skin cancer. I have received royalties for the Oxford Handbook of Medical Dermatology (Oxford University Press). I have received payment from Public Health England for the "Be Clear on Cancer" skin cancer report. I have no conflicts of interest to declare that directly relate to the publication of this work." RP: nothing to declare. HCW: I am director of the NIHR Health Technology Assessment (HTA) Programme. HTA is part of the NIHR, which also supports the NIHR systematic reviews programme from which this work is funded.
Edited (no change to conclusions)
References
References to studies included in this review
Alarcon 2014a {published data only}
- Alarcon I, Carrera C, Palou J, Alos L, Malvehy J, Puig S. Impact of in vivo reflectance confocal microscopy on the number needed to treat melanoma in doubtful lesions. British Journal of Dermatology 2014;170(4):802‐8. [ER4:17941078; PUBMED: 24124911] [DOI] [PMC free article] [PubMed] [Google Scholar]
Curchin 2011 {published data only}
- Curchin CE, Wurm EM, Lambie DLj, Longo C, Pellacani G, Soyer HP. First experiences using reflectance confocal microscopy on equivocal skin lesions in Queensland. Australasian Journal of Dermatology 2011;52(2):89‐97. [ER4:15465900; PUBMED: 21605091] [DOI] [PubMed] [Google Scholar]
Farnetani 2015 {published data only}
- Farnetani F, Scope A, Braun RP, Gonzalez S, Guitera P, Malvehy J, et al. Skin cancer diagnosis with reflectance confocal microscopy: reproducibility of feature recognition and accuracy of diagnosis. JAMA Dermatology 2015;151(10):1075‐80. [ER4:25233569; PUBMED: 25993262] [DOI] [PubMed] [Google Scholar]
Ferrari 2015 {published data only}
- Ferrari B, Pupelli G, Farnetani F, Carvalho NT, Longo C, Reggiani C, et al. Dermoscopic difficult lesions: an objective evaluation of reflectance confocal microscopy impact for accurate diagnosis. Journal of the European Academy of Dermatology and Venereology 2015;29(6):1135‐40. [DOI: 10.1111/jdv.12769; ER4:20569458; PUBMED: 25303304] [DOI] [PubMed] [Google Scholar]
Figueroa‐Silva 2016 {published data only}
- Figueroa‐Silva O, Cinotti E, Almeida Silva T, Moscarella E, Lallas A, Ciardo S, et al. Diagnostic accuracy of reflectance confocal microscopy for lesions typified by dermoscopic island. Journal of the European Academy of Dermatology and Venereology : JEADV 2016;30(9):1594‐8. [ER4:25012335; PUBMED: 27109574] [DOI] [PubMed] [Google Scholar]
Guitera 2009b (Modena) {published data only}
- Guitera P, Pellacani G, Longo C, Seidenari S, Avramidis M, Menzies SW. In vivo reflectance confocal microscopy enhances secondary evaluation of melanocytic lesions. Journal of Investigative Dermatology 2009;129(1):131‐8. [ER4:15465945; PUBMED: 18633444] [DOI] [PubMed] [Google Scholar]
Guitera 2009c (Sydney) {published data only}
- Guitera P, Pellacani G, Longo C, Seidenari S, Avramidis M, Menzies SW. In vivo reflectance confocal microscopy enhances secondary evaluation of melanocytic lesions. Journal of Investigative Dermatology 2009;129(1):131‐8. [PUBMED: 18633444] [DOI] [PubMed] [Google Scholar]
Guitera 2012 {published data only}
- Guitera P, Menzies SW, Longo C, Cesinaro AM, Scolyer RA, Pellacani G. In vivo confocal microscopy for diagnosis of melanoma and basal cell carcinoma using a two‐step method: analysis of 710 consecutive clinically equivocal cases. Journal of Investigative Dermatology 2012;132(10):2386‐94. [ER4:15465942; PUBMED: 22718115] [DOI] [PubMed] [Google Scholar]
Koller 2011 {published data only}
- Koller S, Wiltgen M, Ahlgrimm‐Siess V, Weger W, Hofmann‐Wellenhof R, Richtig E, et al. In vivo reflectance confocal microscopy: automated diagnostic image analysis of melanocytic skin tumours. Journal of the European Academy of Dermatology and Venereology : JEADV 2011;25(5):554‐8. [ER4:15465979; PUBMED: 20735518] [DOI] [PubMed] [Google Scholar]
Langley 2007 {published data only}
- Langley RG, Walsh N, Sutherland AE, Propperova I, Delaney L, Morris SF, et al. The diagnostic accuracy of in vivo confocal scanning laser microscopy compared to dermoscopy of benign and malignant melanocytic lesions: a prospective study. Dermatology 2007;215(4):365‐72. [ER4:15465985; PUBMED: 17912001] [DOI] [PubMed] [Google Scholar]
Longo 2013 {published data only}
- Longo C, Farnetani F, Ciardo S, Cesinaro AM, Moscarella E, Ponti G, et al. Is confocal microscopy a valuable tool in diagnosing nodular lesions? A study of 140 cases. British Journal of Dermatology 2013;169(1):58‐67. [ER4:15465992; PUBMED: 23374159] [DOI] [PubMed] [Google Scholar]
Lovatto 2015 {published data only}
- Lovatto L, Carrera C, Salerni G, Alos L, Malvehy J, Puig S. In vivo reflectance confocal microscopy of equivocal melanocytic lesions detected by digital dermoscopy follow‐up. Journal of the European Academy of Dermatology and Venereology : JEADV 2015;29(10):1918‐25. [ER4:25012311; PUBMED: 25752663] [DOI] [PubMed] [Google Scholar]
Pellacani 2007a {published data only}
- Pellacani G, Guitera P, Longo C, Avramidis M, Seidenari S, Menzies S. The impact of in vivo reflectance confocal microscopy for the diagnostic accuracy of melanoma and equivocal melanocytic lesions. Journal of Investigative Dermatology 2007;127(12):2759‐65. [ER4:15466047; PUBMED: 17657243] [DOI] [PubMed] [Google Scholar]
Pellacani 2012 {published data only}
- Pellacani G, Farnetani F, Gonzalez S, Longo C, Cesinaro AM, Casari A, et al. In vivo confocal microscopy for detection and grading of dysplastic nevi: a pilot study. Journal of the American Academy of Dermatology 2012;66(3):e109‐21. [ER4:15466043; PUBMED: 21742408] [DOI] [PubMed] [Google Scholar]
Pellacani 2014b (cons) {published data only}
- Pellacani G, Pepe P, Casari A, Longo C. Reflectance confocal microscopy as a second‐level examination in skin oncology improves diagnostic accuracy and saves unnecessary excisions: a longitudinal prospective study. British Journal of Dermatology 2014;171(5):1044‐51. [PUBMED: 24891083] [DOI] [PubMed] [Google Scholar]
Pellacani 2014c (doc) {published data only}
- Pellacani G, Pepe P, Casari A, Longo C. Reflectance confocal microscopy as a second‐level examination in skin oncology improves diagnostic accuracy and saves unnecessary excisions: a longitudinal prospective study. British Journal of Dermatology 2014;171(5):1044‐51. [ER4:20569486; PUBMED: 24891083] [DOI] [PubMed] [Google Scholar]
Pupelli 2013 {published data only}
- Pupelli G, Longo C, Veneziano L, Cesinaro AM, Ferrara G, Piana S, et al. Small‐diameter melanocytic lesions: morphological analysis by means of in vivo confocal microscopy. British Journal of Dermatology 2013;168(5):1027‐33. [ER4:15466070; PUBMED: 23301553] [DOI] [PubMed] [Google Scholar]
Rao 2013 {published data only}
- Rao BK, Mateus R, Wassef C, Pellacani G. In vivo confocal microscopy in clinical practice: comparison of bedside diagnostic accuracy of a trained physician and distant diagnosis of an expert reader. Journal of the American Academy of Dermatology 2013;69(6):e295‐300. [ER4:15466076; PUBMED: 24035553] [DOI] [PubMed] [Google Scholar]
Segura 2009 {published data only}
- Segura S, Puig S, Carrera C, Palou J, Malvehy J. Development of a two‐step method for the diagnosis of melanoma by reflectance confocal microscopy. Journal of the American Academy of Dermatology 2009;61(2):216‐29. [ER4:20569494; PUBMED: 19406506] [DOI] [PubMed] [Google Scholar]
Stanganelli 2015 {published data only}
- Stanganelli I, Longo C, Mazzoni L, Magi S, Medri M, Lanzanova G, et al. Integration of reflectance confocal microscopy in sequential dermoscopy follow‐up improves melanoma detection accuracy. British Journal of Dermatology 2015;172(2):365‐71. [ER4:20569496; PUBMED: 25154446] [DOI] [PubMed] [Google Scholar]
References to studies excluded from this review
Agero 2006 {published data only}
- Agero AL, Busam KJ, Benvenuto‐Andrade C, Scope A, Gill M, Marghoob AA, et al. Reflectance confocal microscopy of pigmented basal cell carcinoma. Journal of the American Academy of Dermatology 2006;54(4):638‐43. [PUBMED: 16546585] [DOI] [PubMed] [Google Scholar]
Ahlgrimm‐Siess 2010 {published data only}
- Ahlgrimm‐Siess V, Cao T, Oliviero M, Hofmann‐Wellenhof R, Rabinovitz HS, Scope A. The vasculature of nonmelanocytic skin tumors in reflectance confocal microscopy: vascular features of basal cell carcinoma. Archives of Dermatology 2010;146(3):354. [DOI] [PubMed] [Google Scholar]
Ahlgrimm‐Siess 2011 {published data only}
- Ahlgrimm‐Siess V, Cao T, Oliviero M, Hofmann‐Wellenhof R, Rabinovitz HS, Scope A. The vasculature of nonmelanocytic skin tumors in reflectance confocal microscopy: vascular features of squamous cell carcinoma in situ. Archives of Dermatology 2011;147(2):264. [DOI] [PubMed] [Google Scholar]
Alarcon 2014b {published data only}
- Alarcon I, Carrera C, Turegano P, Malvehy J, Puig S. Basal cell carcinoma with spontaneous regression: added value of reflectance confocal microscopy when the dermoscopic diagnosis is uncertain. Journal of the American Academy of Dermatology 2014;71(1):e7‐9. [PUBMED: 24947714] [DOI] [PubMed] [Google Scholar]
Amjadi 2011 {published data only}
- Amjadi M, Coventry BJ, Greenwood JE. Reflectance confocal microscopy in the diagnosis of non‐melanoma skin cancer and benign lesions versus normal skin: a blinded prospective trial. Internet Journal of Plastic Surgery 2011;7(2):1‐6. [Google Scholar]
Bassoli 2012 {published data only}
- Bassoli S, Rabinovitz HS, Pellacani G, Porges L, Oliviero MC, Braun RP, et al. Reflectance confocal microscopy criteria of lichen planus‐like keratosis. Journal of the European Academy of Dermatology and Venereology : JEADV 2012;26(5):578‐90. [PUBMED: 21605173] [DOI] [PubMed] [Google Scholar]
Benati 2015 {published data only}
- Benati E, Argenziano G, Kyrgidis A, Moscarella E, Ciardo S, Bassoli S, et al. Melanoma and naevi with a globular pattern: confocal microscopy as an aid for diagnostic differentiation. British Journal of Dermatology 2015;173(5):1232‐8. [PUBMED: 26212145] [DOI] [PubMed] [Google Scholar]
Braga 2009 {published data only}
- Braga JC, Scope A, Klaz I, Mecca P, Gonzalez S, Rabinovitz H, et al. The significance of reflectance confocal microscopy in the assessment of solitary pink skin lesions. Journal of the American Academy of Dermatology 2009;61(2):230‐41. [PUBMED: 19398144] [DOI] [PubMed] [Google Scholar]
Carrera 2015 {published data only}
- Carrera C. High‐risk melanoma patients: can unnecessary naevi biopsies be avoided?. British Journal of Dermatology 2015;172(2):313‐5. [PUBMED: 25660675] [DOI] [PubMed] [Google Scholar]
Castro 2015 {published data only}
- Castro RP, Stephens A, Fraga‐Braghiroli NA, Oliviero MC, Rezze GG, Rabinovitz H, et al. Accuracy of in vivo confocal microscopy for diagnosis of basal cell carcinoma: a comparative study between handheld and wide‐probe confocal imaging. Journal of the European Academy of Dermatology and Venereology 2015;29(6):1164‐9. [DOI: 10.1111/jdv.12780; ER4:20569441] [DOI] [PubMed] [Google Scholar]
de Carvalho 2015 {published data only}
- Carvalho N, Farnetani F, Ciardo S, Ruini C, Witkowski AM, Longo C, et al. Reflectance confocal microscopy correlates of dermoscopic patterns of facial lesions help to discriminate lentigo maligna from pigmented nonmelanocytic macules. British Journal of Dermatology 2015;173(1):128‐33. [PUBMED: 25413382] [DOI] [PubMed] [Google Scholar]
de Carvalho 2016 {published data only}
- Carvalho N, Guida S, Abraham LS, Cesinaro AM, Farnetani F, Bonamonte D, et al. Pink melanocytic and non‐melanocytic lesions: how reflectance confocal microscopy can help in differential diagnosis. Journal of the European Academy of Dermatology and Venereology : JEADV 2016;30(6):1026‐9. [PUBMED: 25753043] [DOI] [PubMed] [Google Scholar]
Edwards 2016 {published data only}
- Edwards SJ, Mavranezouli I, Osei‐Assibey G, Marceniuk G, Wakefield V, Karner C. VivaScope 1500 and 3000 systems for detecting and monitoring skin lesions: a systematic review and economic evaluation. Health Technology Assessment (Winchester, England) 2016;20(58):1‐260. [PUBMED: 27483991] [DOI] [PMC free article] [PubMed] [Google Scholar]
Eichert 2010 {published data only}
- Eichert S, Mohrle M, Breuninger H, Rocken M, Garbe C, Bauer J. Diagnosis of cutaneous tumors with in vivo confocal laser scanning microscopy. Journal der Deutschen Dermatologischen Gesellschaft 2010;8(6):400‐10. [DOI] [PubMed] [Google Scholar]
Gareau 2009 {published data only}
- Gareau DS, Karen JK, Dusza SW, Tudisco M, Nehal KS, Rajadhyaksha M. Sensitivity and specificity for detecting basal cell carcinomas in Mohs excisions with confocal fluorescence mosaicing microscopy. Journal of Biomedical Optics 2009;14(3):034012. [PUBMED: 19566305] [DOI] [PMC free article] [PubMed] [Google Scholar]
Gerger 2005 {published data only}
- Gerger A, Koller S, Kern T, Massone C, Steiger K, Richtig E, et al. Diagnostic applicability of in vivo confocal laser scanning microscopy in melanocytic skin tumors. Journal of Investigative Dermatology 2005;124(3):493‐8. [PUBMED: 15737188] [DOI] [PubMed] [Google Scholar]
Gerger 2006 {published data only}
- Gerger A, Kerl H, Samonigg H, Langsenlehner U, Krippl P, Smolle J. Sensitivity and specificity of confocal laser scanning microscopy for in vivo diagnosis of malignant skin tumors. Journal of Investigative Dermatology 2006;126(Suppl 3):s114. [DOI] [PubMed] [Google Scholar]
Gerger 2008a {published data only}
- Gerger A, Wiltgen M, Langsenlehner U, Richtig E, Horn M, Weger W, et al. Diagnostic image analysis of malignant melanoma in in vivo confocal laser‐scanning microscopy: a preliminary study. Skin Research & Technology 2008;14(3):359‐63. [PUBMED: 19159384] [DOI] [PubMed] [Google Scholar]
Gerger 2008b {published data only}
- Gerger A, Hofmann‐Wellenhof R, Langsenlehner U, Richtig E, Koller S, Weger W, et al. In vivo confocal laser scanning microscopy of melanocytic skin tumours: diagnostic applicability using unselected tumour images. British Journal of Dermatology 2008;158(2):329‐33. [DOI] [PubMed] [Google Scholar]
Giambrone 2015 {published data only}
- Giambrone D, Alamgir M, Masud A, Bronsnick T, Rao B. The diagnostic accuracy of in vivo confocal microscopy in clinical practice. Journal of the American Academy of Dermatology 2015;73(2):317‐9. [PUBMED: 26183976] [DOI] [PubMed] [Google Scholar]
Gill 2014 {published data only}
- Gill M, Longo C, Farnetani F, Cesinaro AM, Gonzalez S, Pellacani G. Non‐invasive in vivo dermatopathology: identification of reflectance confocal microscopic correlates to specific histological features seen in melanocytic neoplasms. Journal of the European Academy of Dermatology and Venereology : JEADV 2014;28(8):1069‐78. [PUBMED: 24147614] [DOI] [PubMed] [Google Scholar]
Gonzalez 2002 {published data only}
- Gonzalez S, Tannous Z. Real‐time in vivo confocal reflectance microscopy of basal cell carcinoma. Journal of the American Academy of Dermatology 2002;47(6):869‐74. [PUBMED: 12451371] [DOI] [PubMed] [Google Scholar]
Gonzalez 2013 {published data only}
- Gonzalez S. Should reflectance confocal microscopy be the gold standard for basal cell carcinoma diagnosis?. Imaging in Medicine 2013;5(4):299‐301. [DOI: 10.2217/IIM.13.36] [DOI] [Google Scholar]
Guida 2015 {published data only}
- Guida S, Longo C, Casari A, Ciardo S, Manfredini M, Reggiani C, et al. Update on the use of confocal microscopy in melanoma and non‐melanoma skin cancer. Giornale Italiano di Dermatologia e Venereologia 2015;150(5):547‐63. [PUBMED: 26140397] [PubMed] [Google Scholar]
Guitera 2010 {published data only}
- Guitera P, Pellacani G, Crotty KA, Scolyer RA, Li LX, Bassoli S, et al. The impact of in vivo reflectance confocal microscopy on the diagnostic accuracy of lentigo maligna and equivocal pigmented and nonpigmented macules of the face. Journal of Investigative Dermatology 2010;130(8):2080‐91. [PUBMED: 20393481] [DOI] [PubMed] [Google Scholar]
Guitera 2013 {published data only}
- Guitera P, Moloney FJ, Menzies SW, Stretch JR, Quinn MJ, Hong A, et al. Improving management and patient care in lentigo maligna by mapping with in vivo confocal microscopy. JAMA Dermatology 2013;149(6):692‐8. [PUBMED: 23553208] [DOI] [PubMed] [Google Scholar]
Haenssle 2006 {published data only}
- Haenssle HA, Krueger U, Vente C, Thoms KM, Bertsch HP, Zutt M, et al. Results from an observational trial: digital epiluminescence microscopy follow‐up of atypical nevi increases the sensitivity and the chance of success of conventional dermoscopy in detecting melanoma. Journal of Investigative Dermatology 2006;126(5):980‐5. [PUBMED: 16514414] [DOI] [PubMed] [Google Scholar]
Hennessy 2010 {published data only}
- Hennessy R, Jacques S, Pellacani G, Gareau D. Clinical feasibility of rapid confocal melanoma feature detection. Proceedings of SPIE. 2 March 2010; Vol. 7548. [DOI: 10.1117/12.842824] [DOI]
Hoogedoorn 2014 {published data only}
- Hoogedoorn L, Peppelman M, Erp PEJ, Gerritsen MJP. The use of in vivo reflectance confocal microscopy in clinical practice: prospective differentiation of difficult to distinguish nodular basal cell carcinomas and intradermal nevi. Nederlands Tijdschrift voor Dermatologie en Venereologie 2014;24(1):48. [EMBASE: 71623025] [Google Scholar]
Hoogedoorn 2015 {published data only}
- Hoogedoorn L, Peppelman M, Blokx WA, Erp PE, Gerritsen MJ. Prospective differentiation of clinically difficult to distinguish nodular basal cell carcinomas and intradermal nevi by non‐invasive reflectance confocal microscopy: a case series study. Journal of the European Academy of Dermatology and Venereology : JEADV 2015;29(2):330‐6. [PUBMED: 24841762] [DOI] [PubMed] [Google Scholar]
Humphrey 2006 {published data only}
- Humphrey S, Walsh NM, Delaney L, Propperova I, Langley RG. Prognostic significance of vascularity in cutaneous melanoma: pilot study using in vivo confocal scanning laser microscopy. Journal of Cutaneous Medicine and Surgery 2006;10(3):122‐7. [PUBMED: 17241587] [DOI] [PubMed] [Google Scholar]
Incel 2015 {published data only}
- Incel P, Gurel MS, Erdemir AV. Vascular patterns of nonpigmented tumoral skin lesions: confocal perspectives. Skin Research and Technology 2015;21(3):333‐9. [DOI] [PubMed] [Google Scholar]
Kadouch 2015a {published data only}
- Kadouch DJ, Schram ME, Leeflang MM, Limpens J, Spuls PI, Rie MA. In vivo confocal microscopy of basal cell carcinoma: a systematic review of diagnostic accuracy. Journal of the European Academy of Dermatology and Venereology : JEADV 2015;29(10):1890‐7. [PUBMED: 26290493] [DOI] [PubMed] [Google Scholar]
Kadouch 2015b {published data only}
- Kadouch DJ, Wolkerstorfer A, Elshot Y, Zupan‐Kajcovski B, Crijns MB, Starink MV, et al. Treatment of basal cell carcinoma using a one‐stop‐shop with reflectance confocal microscopy: study design and protocol of a randomized controlled multicenter trial. JMIR Research Protocols 2015;4(3):e109. [PUBMED: 26362616] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kose 2014 {published data only}
- Kose K, Cordova M, Duffy M, Flores ES, Brooks DH, Rajadhyaksha M. Video‐mosaicing of reflectance confocal images for examination of extended areas of skin in vivo. British Journal of Dermatology 2014;171(5):1239‐41. [PUBMED: 24720744] [DOI] [PMC free article] [PubMed] [Google Scholar]
Langley 2001 {published data only}
- Langley RG, Rajadhyaksha M, Dwyer PJ, Sober AJ, Flotte TJ, Anderson RR. Confocal scanning laser microscopy of benign and malignant melanocytic skin lesions in vivo. Journal of the American Academy of Dermatology 2001;45(3):365‐76. [PUBMED: 11511832] [DOI] [PubMed] [Google Scholar]
Langley 2006 {published data only}
- Langley RG, Burton E, Walsh N, Propperova I, Murray SJ. In vivo confocal scanning laser microscopy of benign lentigines: comparison to conventional histology and in vivo characteristics of lentigo maligna. Journal of the American Academy of Dermatology 2006;55(1):88‐97. [PUBMED: 16781299] [DOI] [PubMed] [Google Scholar]
Losi 2014 {published data only}
- Losi A, Longo C, Cesinaro AM, Benati E, Witkowski A, Guitera P, et al. Hyporeflective pagetoid cells: a new clue for amelanotic melanoma diagnosis by reflectance confocal microscopy. British Journal of Dermatology 2014;171(1):48‐54. [PUBMED: 24329036] [DOI] [PubMed] [Google Scholar]
Maier 2013 {published data only}
- Maier T, Sattler EC, Braun‐Falco M, Korting HC, Ruzicka T, Berking C. Reflectance confocal microscopy in the diagnosis of partially and completely amelanotic melanoma: report on seven cases. Journal of the European Academy of Dermatology and Venereology : JEADV 2013;27(1):e42‐52. [PUBMED: 22324783] [DOI] [PubMed] [Google Scholar]
Malvehy 2012 {published data only}
- Malvehy J, Hanke‐Martinez M, Costa J, Salerni G, Carrera C, Puig S. Semiology and pattern analysis in nonmelanocytic lesions. Reflectance Confocal Microscopy for Skin Diseases. Berlin, Heidelberg: Springer, 2012:239‐52. [DOI: 10.1007/978-3-642-21997-9_18] [DOI] [Google Scholar]
Menge 2016 {published data only}
- Menge TD, Hibler BP, Cordova MA, Nehal KS, Rossi AM. Concordance of handheld reflectance confocal microscopy (RCM) with histopathology in the diagnosis of lentigo maligna (LM): a prospective study. Journal of the American Academy of Dermatology 2016;74(6):1114‐20. [PUBMED: 26826051] [DOI] [PubMed] [Google Scholar]
Miller 2011 {published data only}
- Miller JH, Chrisler WB, Wang X, Sowa MB. Confocal microscopy for modeling electron microbeam irradiation of skin. Radiation and Environmental Biophysics 2011;50(3):365‐9. [PUBMED: 21604000] [DOI] [PubMed] [Google Scholar]
Nobre 2011 {published data only}
- Nobre Moura F, Dalle S, Depaepe L, Durupt F, Balme B, Thomas L. Melanoma: early diagnosis using in vivo reflectance confocal microscopy. Clinical and Experimental Dermatology 2011;36(2):209‐11. [PUBMED: 20659120] [DOI] [PubMed] [Google Scholar]
Nori 2004 {published data only}
- Nori S, Rius‐Diaz F, Cuevas J, Goldgeier M, Jaen P, Torres Abel, et al. Sensitivity and specificity of reflectance‐mode confocal microscopy for in vivo diagnosis of basal cell carcinoma: a multicenter study. Journal of the American Academy of Dermatology 2004;51(6):923‐30. [ER4:15466027; PUBMED: 15583584] [DOI] [PubMed] [Google Scholar]
Pellacani 2005 {published data only}
- Pellacani G, Cesinaro AM, Seidenari S. Reflectance‐mode confocal microscopy of pigmented skin lesions ‐ improvement in melanoma diagnostic specificity. Journal of the American Academy of Dermatology 2005;53(6):979‐85. [PUBMED: 16310058] [DOI] [PubMed] [Google Scholar]
Pellacani 2007b {published data only}
- Pellacani G, Bassoli S, Longo C, Cesinaro AM, Seidenari S. Diving into the blue: in vivo microscopic characterization of the dermoscopic blue hue. Journal of the American Academy of Dermatology 2007;57(1):96‐104. [PUBMED: 17485141] [DOI] [PubMed] [Google Scholar]
Pellacani 2008 {published data only}
- Pellacani G, Longo C, Malvehy J, Puig S, Carrera C, Segura S, et al. In vivo confocal microscopic and histopathologic correlations of dermoscopic features in 202 melanocytic lesions. Archives of Dermatology 2008;144(12):1597‐608. [PUBMED: 19075142] [DOI] [PubMed] [Google Scholar]
Pellacani 2009a {published data only}
- Pellacani G, Vinceti M, Bassoli S, Braun R, Gonzalez S, Guitera P, et al. Reflectance confocal microscopy and features of melanocytic lesions: an internet‐based study of the reproducibility of terminology. Archives of Dermatology 2009;145(10):1137‐43. [PUBMED: 19841401] [DOI] [PubMed] [Google Scholar]
Peppelman 2013 {published data only}
- Peppelman M, Wolberink EA, Blokx WA, Kerkhof PC, Erp PE, Gerritsen MJ. In vivo diagnosis of basal cell carcinoma subtype by reflectance confocal microscopy. Dermatology 2013;227(3):255‐62. [PUBMED: 24158236] [DOI] [PubMed] [Google Scholar]
Peppelman 2015 {published data only}
- Peppelman M, Nguyen KP, Hoogedoorn L, Erp PE, Gerritsen MJ. Reflectance confocal microscopy: non‐invasive distinction between actinic keratosis and squamous cell carcinoma. Journal of the European Academy of Dermatology and Venereology 2015;29(7):1302‐9. [PUBMED: 25357235] [DOI] [PubMed] [Google Scholar]
Peppelman 2016 {published data only}
- Peppelman M, Nguyen KP, Alkemade HA, Maessen‐Visch B, Hendriks JC, Erp PE, et al. Diagnosis of basal cell carcinoma by reflectance confocal microscopy: study design and protocol of a randomized controlled multicenter trial. JMIR Research Protocols 2016;5(2):e114. [PUBMED: 27363577] [DOI] [PMC free article] [PubMed] [Google Scholar]
Puig 2012 {published data only}
- Puig S, Giacomo TB, Serra D, Cabrini F, Alos L, Palou J, et al. Reflectance confocal microscopy of blue nevus. European Journal of Dermatology 2012;22(4):552‐3. [PUBMED: 22735078] [DOI] [PubMed] [Google Scholar]
Reggiani 2015 {published data only}
- Reggiani C, Manfredini M, Mandel VD, Farnetani F, Ciardo S, Bassoli S, et al. Update on non‐invasive imaging techniques in early diagnosis of non‐melanoma skin cancer. Giornale Italiano di Dermatologia e Venereologia 2015;150(4):393‐405. [PUBMED: 26184797] [PubMed] [Google Scholar]
Rishpon 2009 {published data only}
- Rishpon A, Kim N, Scope A, Porges L, Oliviero MC, Braun RP, et al. Reflectance confocal microscopy criteria for squamous cell carcinomas and actinic keratoses. Archives of Dermatology 2009;145(7):766‐72. [PUBMED: 19620557] [DOI] [PubMed] [Google Scholar]
Röwert‐Huber 2007 {published data only}
- Röwert‐Huber J, Patel MJ, Forschner T, Ulrich C, Eberle J, Kerl H, et al. Actinic keratosis is an early in situ squamous cell carcinoma: a proposal for reclassification. British Journal of Dermatology 2007;156(Suppl 3):8‐12. [PUBMED: 17488400] [DOI] [PubMed] [Google Scholar]
Salerni 2011 {published data only}
- Salerni G, Lovatto L, Carrera C, Palou J, Alos L, Puig‐Butille JA, et al. Correlation among dermoscopy, confocal reflectance microscopy, and histologic features of melanoma and basal cell carcinoma collision tumor. Dermatologic Surgery 2011;37(2):275‐9. [PUBMED: 21281387] [DOI] [PubMed] [Google Scholar]
Scope 2009 {published data only}
- Scope A, Mecca PS, Marghoob AA. skINsight lessons in reflectance confocal microscopy: rapid diagnosis of pigmented basal cell carcinoma. Archives of Dermatology 2009;145(1):106‐7. [PUBMED: 19153366] [DOI] [PubMed] [Google Scholar]
Scope 2014 {published data only}
- Scope A, Longo C. Recognizing the benefits and pitfalls of reflectance confocal microscopy in melanoma diagnosis. Dermatology Practical & Conceptual 2014;4(3):67‐71. [DOI: 10.5826/dpc.0403a13] [DOI] [PMC free article] [PubMed] [Google Scholar]
Soyer 2013 {published data only}
- Soyer HP, Prow TW. Reflectance confocal microscopy in the diagnosis of nodular skin lesions. British Journal of Dermatology 2013;169(1):4. [PUBMED: 23834114] [DOI] [PubMed] [Google Scholar]
Steiner 1992 {published data only}
- Steiner A, Pehamberger H, Binder M, Wolff K. Pigmented Spitz nevi: improvement of the diagnostic accuracy by epiluminescence microscopy. Journal of the American Academy of Dermatology 1992;27(5 Pt 1):697‐701. [PUBMED: 1430390] [DOI] [PubMed] [Google Scholar]
Stephens 2013 {published data only}
- Stephens A, Fraga‐Braghiroli N, Oliviero M, Rabinovitz H, Scope A. Spoke wheel‐like structures in superficial basal cell carcinoma: a correlation between dermoscopy, histopathology, and reflective confocal microscopy. Journal of the American Academy of Dermatology 2013;69(5):e219‐21. [PUBMED: 24124839] [DOI] [PubMed] [Google Scholar]
Stevenson 2013 {published data only}
- Stevenson AD, Mickan S, Mallett S, Ayya M. Systematic review of diagnostic accuracy of reflectance confocal microscopy for melanoma diagnosis in patients with clinically equivocal skin lesions. Dermatology Practical & Conceptual 2013;3(4):19‐27. [DOI: 10.5826/dpc.0304a05] [DOI] [PMC free article] [PubMed] [Google Scholar]
Tannous 2009 {published data only}
- Tannous Z, Al‐Arashi M, Shah S, Yaroslavsky AN. Delineating melanoma using multimodal polarized light imaging. Lasers in Surgery and Medicine 2009;41(1):10‐6. [PUBMED: 19143015] [DOI] [PubMed] [Google Scholar]
Willard 2011 {published data only}
- Willard K, Warschaw KE, Swanson DL. Use of reflectance confocal microscopy to differentiate hidrocystoma from basal cell carcinoma. Dermatologic Surgery 2011;37(3):392‐4. [PUBMED: 21314800] [DOI] [PubMed] [Google Scholar]
Witkowski 2016 {published data only}
- Witkowski AM, Ludzik J, DeCarvalho N, Ciardo S, Longo C, DiNardo A, et al. Non‐invasive diagnosis of pink basal cell carcinoma: how much can we rely on dermoscopy and reflectance confocal microscopy?. Skin Research and Technology 2016;22(2):230‐7. [ER4:25012281; PUBMED: 26338448] [DOI] [PubMed] [Google Scholar]
Xiong 2016 {published data only}
- Xiong YD, Ma S, Li X, Zhong X, Duan C, Chen Q. A meta‐analysis of reflectance confocal microscopy for the diagnosis of malignant skin tumours. Journal of the European Academy of Dermatology and Venereology : JEADV 2016;30(8):1295‐302. [PUBMED: 27230832] [DOI] [PubMed] [Google Scholar]
Yelamos 2016 {published data only}
- Yelamos O, Nehal KS. Integrating clinical information, dermoscopy and reflectance confocal microscopy to improve the diagnostic accuracy and confidence of amelanotic and lightly pigmented melanomas. British Journal of Dermatology 2016;175(6):1147‐8. [PUBMED: 27996145] [DOI] [PubMed] [Google Scholar]
References to studies awaiting assessment
Borsari 2016 {published data only}
- Borsari S, Pampena R, Lallas A, Kyrgidis A, Moscarella E, Benati E, et al. Clinical indications for use of reflectance confocal microscopy for skin cancer diagnosis. JAMA Dermatology 2016;152(10):1093‐8. [PUBMED: 27580185] [DOI] [PubMed] [Google Scholar]
Guitera 2016 {published data only}
- Guitera P, Menzies SW, Argenziano G, Longo C, Losi A, Drummond M, et al. Dermoscopy and in vivo confocal microscopy are complementary techniques for diagnosis of difficult amelanotic and light‐coloured skin lesions. British Journal of Dermatology 2016;175(6):1311‐9. [PUBMED: 27177158 ] [DOI] [PubMed] [Google Scholar]
Jain 2017 {published data only}
- Jain M, Pulijal SV, Rajadhyaksha M. The bedside diagnostic accuracy of a novice reflectance confocal microscopy reader for skin cancer detection in vivo in real‐time: understanding challenges and potential pitfalls. Proceedings of SPIE. 24 March 2017; Vol. 10060. [DOI: 10.1117/12.2255685] [DOI]
Ludzik 2016 {published data only}
- Ludzik J, Witkowski AM, Roterman‐Konieczna I, Bassoli S, Farnetani F, Pellacani G. Improving diagnostic accuracy of dermoscopically equivocal pink cutaneous lesions with reflectance confocal microscopy in telemedicine settings: double reader concordance evaluation of 316 cases. PloS One 2016;11(9):e0162495. [PUBMED: 27606812] [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional references
Abaffy 2010
- Abaffy T, Duncan R, Riemer DD, Tietje O, Elgart G, Milikowski C, et al. Differential volatile signatures from skin, naevi and melanoma: a novel approach to detect a pathological process. PloS One 2010;5(11):e13813. [PUBMED: 21079799] [DOI] [PMC free article] [PubMed] [Google Scholar]
Altamura 2008
- Altamura D, Avramidis M, Menzies SW. Assessment of the optimal interval for and sensitivity of short‐term sequential digital dermoscopy monitoring for the diagnosis of melanoma. Archives of Dermatology 2008;144(4):502‐6. [PUBMED: 18427044] [DOI] [PubMed] [Google Scholar]
Annessi 2007
- Annessi G, Bono R, Sampogna F, Faraggiana T, Abeni D. Sensitivity, specificity, and diagnostic accuracy of three dermoscopic algorithmic methods in the diagnosis of doubtful melanocytic lesions: the importance of light brown structureless areas in differentiating atypical melanocytic nevi from thin melanomas. Journal of the American Academy of Dermatology 2007;56(5):759‐67. [PUBMED: 17316894] [DOI] [PubMed] [Google Scholar]
Argenziano 1998
- Argenziano G, Fabbrocini G, Carli P, Giorgi V, Sammarco E, Delfino M. Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions. Comparison of the ABCD rule of dermatoscopy and a new 7‐point checklist based on pattern analysis. Archives of Dermatology 1998;134(12):1563‐70. [PUBMED: 9875194] [DOI] [PubMed] [Google Scholar]
Argenziano 2001
- Argenziano G, Soyer HP. Dermoscopy of pigmented skin lesions ‐ a valuable tool for early diagnosis of melanoma. Lancet Oncology 2001;2(7):443‐9. [PUBMED: 11905739] [DOI] [PubMed] [Google Scholar]
Argenziano 2012
- Argenziano G, Albertini G, Castagnetti F, Pace B, Lernia V, Longo C, et al. Early diagnosis of melanoma: what is the impact of dermoscopy?. Dermatologic Therapy 2012;25(5):403‐9. [PUBMED: 23046019] [DOI] [PubMed] [Google Scholar]
Arnold 2014
- Arnold M, Holterhues C, Hollestein LM, Coebergh JW, Nijsten T, Pukkala E, et al. Trends in incidence and predictions of cutaneous melanoma across Europe up to 2015. Journal of the European Academy of Dermatology and Venereology : JEADV 2014;28(9):1170‐8. [PUBMED: 23962170] [DOI] [PubMed] [Google Scholar]
Balch 2001
- Balch CM, Soong SJ, Gershenwald JE, Thompson JF, Reintgen DS, Cascinelli N, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. Journal of Clinical Oncology 2001;19(16):3622‐34. [PUBMED: 11504744] [DOI] [PubMed] [Google Scholar]
Balch 2009
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. Journal of Clinical Oncology 2009;27(36):6199‐206. [PUBMED: 19917835] [DOI] [PMC free article] [PubMed] [Google Scholar]
Beer 2011
- Beer J, Xu L, Tschandl P, Kittler H. Growth rate of melanoma in vivo and correlation with dermatoscopic and dermatopathologic findings. Dermatology Practical and Conceptual 2011;1(1):59‐67. [PUBMED: 24396722] [DOI] [PMC free article] [PubMed] [Google Scholar]
Binder 1997
- Binder M, Schwarz M, Steiner A, Kittler H, Muellner M, Wolff K, et al. Epiluminescence microscopy of small pigmented skin lesions: short‐term formal training improves the diagnostic performance of dermatologists. Journal of the American Academy of Dermatology 1997;36(2 Pt 1):197‐202. [PUBMED: 9039168] [DOI] [PubMed] [Google Scholar]
Boring 1994
- Boring CC, Squires TS, Tong T, Montgomery S. Cancer statistics, 1994. CA: a Cancer Journal for Clinicians 1994;44(1):7‐26. [PUBMED: 8281473] [DOI] [PubMed] [Google Scholar]
Bossuyt 2015
- Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527. [DOI: 10.1136/bmj.h5527; PUBMED: 26511519] [DOI] [PMC free article] [PubMed] [Google Scholar]
Cancer Research UK 2017a
- Cancer Research UK. Skin cancer incidence statistics. www.cancerresearchuk.org/health‐professional/cancer‐statistics/statistics‐by‐cancer‐type/skin‐cancer/incidence (accessed prior to 1 November 2017).
Cancer Research UK 2017b
- Cancer Research UK. Skin cancer statistics. www.cancerresearchuk.org/health‐professional/cancer‐statistics/statistics‐by‐cancer‐type/skin‐cancer#heading‐One (accessed prior to 19 July 2017).
Chao 2013
- Chao D, London Cancer (North and East). Guidelines for cutaneous malignant melanoma management August 2013. www.londoncancer.org/media/76373/london‐cancer‐melanoma‐guidelines‐2013‐v1.0.pdf. London: London Cancer North and East Alliance, (accessed 25 February 2015).
Chapman 2011
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. New England Journal of Medicine 2011;364(26):2507‐16. [PUBMED: 21639808] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chapman 2012
- Chapman PB, Hauschild A, Robert C, Larkin J, Haanen JB, Ribas A, et al. Updated overall survival (OS) results for BRIM‐3, a phase III randomized, open‐label, multicenter trial comparing BRAF inhibitor vemurafenib (vem) with dacarbazine (DTIC) in previously untreated patients with BRAFV600E‐mutated melanoma. Journal of Clinical Oncology 2012;30(15 Suppl 1):8502. [EMBASE: 71004853] [Google Scholar]
Cho 2014
- Cho H, Mariotto AB, Schwartz LM, Luo J, Woloshin S. When do changes in cancer survival mean progress? The insight from population incidence and mortality. Journal of the National Cancer Institute. Monographs 2014;2014(49):187‐97. [PUBMED: 25417232] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chu 2006
- Chu H, Cole SR. Bivariate meta‐analysis for sensitivity and specificity with sparse data: a generalized linear mixed model approach (comment). Journal of Clinical Epidemiology 2006;59(12):1331‐2. [PUBMED: 17098577] [DOI] [PubMed] [Google Scholar]
Chuchu 2018a
- Chuchu N, Dinnes J, Takwoingi Y, Matin RN, Bayliss SE, Davenport C, et al. Teledermatology for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013193] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chuchu 2018b
- Chuchu N, Takwoingi Y, Dinnes J, Matin RN, Bassett O, Moreau JF, et al. Smartphone applications for triaging adults with skin lesions that are suspicious for melanoma. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013192] [DOI] [PMC free article] [PubMed] [Google Scholar]
Church 2001
- Church J, Williams H. Another sniffer dog for the clinic?. Lancet 2001;358(9285):930. [PUBMED: 11575380] [DOI] [PubMed] [Google Scholar]
D'Amico 2008
- D'Amico A, Bono R, Pennazza G, Santonico M, Mantini G, Bernabei M, et al. Identification of melanoma with a gas sensor array. Skin Research and Technology 2008;14(2):226‐36. [PUBMED: 18412567] [DOI] [PubMed] [Google Scholar]
Debarbieux 2013
- Debarbieux S, Depaepe L, Poulalhon N, Balme B, Dalle S, Thomas L. Reflectance confocal microscopy accurately discriminates between benign and malignant melanocytic lesions exhibiting a 'dermoscopic island'. Journal of the European Academy of Dermatology and Venereology 2013;27(2):159‐65. [PUBMED: 22486883] [DOI] [PubMed] [Google Scholar]
Deeks 2005
- Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. Journal of Clinical Epidemiology 2005;58(9):882‐93. [PUBMED: 16085191] [DOI] [PubMed] [Google Scholar]
Dinnes 2018a
- Dinnes J, Deeks JJ, Grainge MJ, Chuchu N, Ferrante di Ruffano L, Matin RN, et al. Visual inspection for diagnosing cutaneous melanoma in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013194] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018b
- Dinnes J, Deeks JJ, Chuchu N, Ferrante di Ruffano L, Matin RN, Thomson DR, et al. Dermoscopy, with and without visual inspection, for diagnosing melanoma in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD011902.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018c
- Dinnes J, Deeks JJ, Chuchu N, Saleh D, Bayliss SE, Takwoingi Y, et al. Reflectance confocal microscopy for diagnosing keratinocyte skin cancers in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013191] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018d
- Dinnes J, Bamber J, Chuchu N, Bayliss SE, Takwoingi Y, Davenport C, et al. High‐frequency ultrasound for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013188] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dummer 2014
- Dummer R, Arenberger P, Ascierto PA, Groot JW, Hallmeyer S, Lotem M, et al. 1130TiP‐NEMO: a phase 3 trial of binimetinib (MEK162) versus dacarbazine in patients with advanced NRAS‐mutant melanoma who are untreated or have progressed after any number of immunotherapy regimens. Annals of Oncology 2014;25(Suppl 4):iv392. [PUBMED: 28171154] [Google Scholar]
Efron 1983
- Efron B. Estimating the error rate of a prediction rule: improvement on cross‐validation. Journal of the American Statistical Association 1983;78(382):316‐31. [DOI: 10.1080/01621459.1983.10477973] [DOI] [Google Scholar]
Eggermont 2007
- Eggermont AM, Gore M. Randomized adjuvant therapy trials in melanoma: surgical and systemic. Seminars in Oncology 2007;34(6):509‐15. [PUBMED: 18083374] [DOI] [PubMed] [Google Scholar]
Erdmann 2013
- Erdmann F, Lortet‐Tieulent J, Schuz J, Zeeb H, Greinert R, Breitbart EW, et al. International trends in the incidence of malignant melanoma 1953‐2008 ‐ are recent generations at higher or lower risk?. International Journal of Cancer 2013;132(2):385‐400. [PUBMED: 22532371] [DOI] [PubMed] [Google Scholar]
Esteva 2017
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature 2017;542(7639):115‐8. [PUBMED: 28117445] [DOI] [PMC free article] [PubMed] [Google Scholar]
EUCAN 2012
- EUCAN, International Agency for Research on Cancer. Malignant melanoma of skin: estimated incidence, mortality & prevalence for both sexes, 2012. eco.iarc.fr/eucan/Cancer.aspx?Cancer=20. International Agency for Research on Cancer, (accessed 29 July 2015).
Ferlay 2015
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer 2015;136(5):E359‐86. [PUBMED: 25220842] [DOI] [PubMed] [Google Scholar]
Ferrante di Ruffano 2018a
- Ferrante di Ruffano L, Dinnes J, Deeks JJ, Chuchu N, Bayliss SE, Davenport C, et al. Optical coherence tomography for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013189] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ferrante di Ruffano 2018b
- Ferrante di Ruffano L, Takwoingi Y, Dinnes J, Chuchu N, Bayliss SE, Davenport C, et al. Computer‐assisted diagnosis techniques (dermoscopy and spectroscopy‐based) for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013186] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ferris 2012
- Ferris LK, Harris RJ. New diagnostic aids for melanoma. Dermatologic Clinics 2012;30(3):535‐45. [PUBMED: 22800557] [DOI] [PMC free article] [PubMed] [Google Scholar]
Friedman 1985
- Friedman RJ, Rigel DS, Kopf AW. Early detection of malignant melanoma: the role of physician examination and self‐examination of the skin. CA: a Cancer Journal for Clinicians 1985;35(3):130‐51. [PUBMED: 3921200] [DOI] [PubMed] [Google Scholar]
Gallagher 2008
- Gallagher M, Wysocki CJ, Leyden JJ, Spielman AI, Sun X, Preti G. Analyses of volatile organic compounds from human skin. British Journal of Dermatology 2008;159(4):780‐91. [PUBMED: 18637798] [DOI] [PMC free article] [PubMed] [Google Scholar]
Garbe 2016
- Garbe C, Peris K, Hauschild A, Saiag P, Middleton M, Bastholt L, et al. Diagnosis and treatment of melanoma. European consensus‐based interdisciplinary guideline ‐ update 2016. European Journal of Cancer 2016;63:201‐17. [PUBMED: 27367293] [DOI] [PubMed] [Google Scholar]
Gereli 2010
- Gereli MC, Onsun N, Atilganoglu U, Demirkesen C. Comparison of two dermoscopic techniques in the diagnosis of clinically atypical pigmented skin lesions and melanoma: seven‐point and three‐point checklists. International Journal of Dermatology 2010;49(1):33‐8. [PUBMED: 20465608] [DOI] [PubMed] [Google Scholar]
Grob 1998
- Grob JJ, Bonerandi JJ. The 'ugly duckling' sign: identification of the common characteristics of nevi in an individual as a basis for melanoma screening. Archives of Dermatology 1998;134(1):103‐4. [PUBMED: 9449921] [DOI] [PubMed] [Google Scholar]
Guitera 2009a
- Guitera P, Pellacani G, Longo C, Seidenari S, Avramidis M, Menzies SW. In vivo reflectance confocal microscopy enhances secondary evaluation of melanocytic lesions. Journal of Investigative Dermatology 2009;129(1):131‐8. [PUBMED: 18633444] [DOI] [PubMed] [Google Scholar]
Guitera 2017
- Guitera P, Menzies S, Chamberlain A, Soyer P, Cancer Council Australia Melanoma Guidelines Working Party. What is the role of confocal microscopy in melanoma diagnosis? Clinical practice guidelines for the diagnosis and management of melanoma. wiki.cancer.org.au/australiawiki/index.php?oldid=158690 (accessed 18 May 2017).
Haenssle 2010
- Haenssle HA, Korpas B, Hansen‐Hagge C, Buhl T, Kaune KM, Rosenberger A, et al. Seven‐point checklist for dermatoscopy: performance during 10 years of prospective surveillance of patients at increased melanoma risk. Journal of the American Academy of Dermatology 2010;62(5):785‐93. [PUBMED: 20226567] [DOI] [PubMed] [Google Scholar]
Hamid 2013
- Hamid O, Sosman JA, Lawrence DP, Sullivan RJ, Ibrahim N, Kluger HM, et al. Clinical activity, safety, and biomarkers of MPDL3280A, an engineered PD‐L1 antibody in patients with locally advanced or metastatic melanoma (mM). Journal of Clinical Oncology 2013;31(15 Suppl 1):9010. [EMBASE: 71099860] [Google Scholar]
Hauschild 2014
- Hauschild A, Chen SC, Weichenthal M, Blum A, King HC, Goldsmith J, et al. To excise or not: impact of MelaFind on German dermatologists' decisions to biopsy atypical lesions. Journal der Deutschen Dermatologischen Gesellschaft 2014;12(7):606‐14. [PUBMED: 24944011] [DOI] [PubMed] [Google Scholar]
Hodi 2010
- Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. New England Journal of Medicine 2010;363(8):711‐23. [PUBMED: 20525992] [DOI] [PMC free article] [PubMed] [Google Scholar]
Hodi 2016
- Hodi FS, Chesney J, Pavlick AC, Robert C, Grossmann KF, McDermott DF, et al. Combined nivolumab and ipilimumab versus ipilimumab alone in patients with advanced melanoma: 2‐year overall survival outcomes in a multicentre, randomised, controlled, phase 2 trial. Lancet Oncology 2016;17(11):1558‐68. [DOI: 10.1016/S1470-2045(16)30366-7; PUBMED: 27622997] [DOI] [PMC free article] [PubMed] [Google Scholar]
HPA and MelNet NZ 2014
- Health Promotion Agency and the Melanoma Network of New Zealand (MelNet). New Zealand Skin Cancer Primary Prevention and Early Detection Strategy 2014 to 2017. www.sunsmart.org.nz//sites/default/files/documents/NZ%20Skin%20Cancer%20PrimaryPrevention%20and%20EarlyDetection%20Strategy%202014%20to%202017%20FINAL%20VERSION%20%23406761.pdf. Cancer Society of New Zealand, (accessed 29 May 2018).
Kasprzak 2015
- Kasprzak JM, Xu YG. Diagnosis and management of lentigo maligna: a review. Drugs in Context 2015;4:212281. [PUBMED: 26082796] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kittler 2002
- Kittler H, Pehamberger H, Wolff K, Binder M. Diagnostic accuracy of dermoscopy. Lancet Oncology 2002;3(3):159‐65. [PUBMED: 11902502] [DOI] [PubMed] [Google Scholar]
Kittler 2011
- Kittler H, Rosendahl C, Cameron A, Tschandl P. Dermatoscopy. An Algorithmic Method Based on Pattern Analysis. Vienna (Austria): Facultas WUV, 2011. [ISBN‐10: 3708907175] [Google Scholar]
Kokolakis 2012
- Kokolakis A, Zacharakis G, Krasagakis K, Lasithiotakis K, Favicchio R, Spiliopoulos G, et al. Prehistological evaluation of benign and malignant pigmented skin lesions with optical computed tomography. Journal of Biomedical Optics 2012;17(6):066004. [PUBMED: 22734760] [DOI] [PubMed] [Google Scholar]
Korn 2008
- Korn EL, Liu PY, Lee SJ, Chapman JA, Niedzwiecki D, Suman VJ, et al. Meta‐analysis of phase II cooperative group trials in metastatic stage IV melanoma to determine progression‐free and overall survival benchmarks for future phase II trials. Journal of Clinical Oncology 2008;26(4):527‐34. [PUBMED: 18235113] [DOI] [PubMed] [Google Scholar]
Kwak 2013
- Kwak J, Gallagher M, Ozdener MH, Wysocki CJ, Goldsmith BR, Isamah A, et al. Volatile biomarkers from human melanoma cells. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences 2013;931:90‐6. [PUBMED: 23770738] [DOI] [PubMed] [Google Scholar]
Kyrgidis 2015
- Kyrgidis A, Tzellos T, Mocellin S, Apalla Z, Lallas A, Pilati P, et al. Sentinel lymph node biopsy followed by lymph node dissection for localised primary cutaneous melanoma. Cochrane Database of Systematic Reviews 2015, Issue 5. [DOI: 10.1002/14651858.CD010307.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Larkin 2014
- Larkin J, Ascierto PA, Dréno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF‐mutated melanoma. New England Journal of Medicine 2014;371(20):1867‐76. [PUBMED: 25265494] [DOI] [PubMed] [Google Scholar]
Larkin 2015
- Larkin J, Lao CD, Urba WJ, McDermott DF, Horak C, Jiang J, et al. Efficacy and safety of nivolumab in patients with BRAF V600 mutant and BRAF wild‐type advanced melanoma: a pooled analysis of 4 clinical trials. JAMA Oncology 2015;1(4):433‐40. [PUBMED: 26181250] [DOI] [PubMed] [Google Scholar]
Leff 2008
- Leff B, Finucane TE. Gizmo idolatry. JAMA 2008;299(15):1830‐2. [PUBMED: 18413879] [DOI] [PubMed] [Google Scholar]
Longo 2011
- Longo C, Rito C, Beretti F, Cesinaro AM, Piñeiro‐Maceira J, Seidenari S, et al. De novo melanoma and melanoma arising from pre‐existing nevus: in vivo morphologic differences as evaluated by confocal microscopy. Journal of the American Academy of Dermatology 2011;65(3):604‐14. [PUBMED: 21715047] [DOI] [PubMed] [Google Scholar]
MacKie 1985
- MacKie RM, English J, Aitchison TC, Fitzsimons CP, Wilson P. The number and distribution of benign pigmented moles (melanocytic naevi) in a healthy British population. British Journal of Dermatology 1985;113(2):167‐74. [PUBMED: 4027184] [DOI] [PubMed] [Google Scholar]
MacKie 1990
- MacKie RM. Clinical recognition of early invasive malignant melanoma. BMJ 1990;301(6759):1005‐6. [PUBMED: 2249043] [DOI] [PMC free article] [PubMed] [Google Scholar]
Maio 2015
- Maio M, Grob JJ, Aamdal S, Bondarenko I, Robert C, Thomas L, et al. Five‐year survival rates for treatment‐naive patients with advanced melanoma who received ipilimumab plus dacarbazine in a phase III trial. Journal of Clinical Oncology 2015;33(10):1191‐6. [PUBMED: 25713437] [DOI] [PMC free article] [PubMed] [Google Scholar]
Maley 2014
- Maley A, Rhodes AR. Cutaneous melanoma: preoperative tumor diameter in a general dermatology outpatient setting. Dermatologic Surgery 2014;40(4):446‐54. [PUBMED: 24479783] [DOI] [PubMed] [Google Scholar]
Malvehy 2014
- Malvehy J, Hauschild A, Curiel‐Lewandrowski C, Mohr P, Hofmann‐Wellenhof R, Motley R, et al. Clinical performance of the Nevisense system in cutaneous melanoma detection: an international, multicentre, prospective and blinded clinical trial on efficacy and safety. British Journal of Dermatology 2014;171(5):1099‐107. [PUBMED: 24841846] [DOI] [PMC free article] [PubMed] [Google Scholar]
Marsden 2010
- Marsden JR, Newton‐Bishop JA, Burrows L, Cook M, Corrie PG, Cox NH, et al. BAD guidelines: revised UK guidelines for the management of cutaneous melanoma 2010. British Journal of Dermatology 2010;163(2):238‐56. [PUBMED: 20608932] [DOI] [PubMed] [Google Scholar]
Menzies 1996
- Menzies SW, Ingvar C, Crotty KA, McCarthy WH. Frequency and morphologic characteristics of invasive melanomas lacking specific surface microscopic features. Archives of Dermatology 1996;132(10):1178‐82. [PUBMED: 8859028] [PubMed] [Google Scholar]
Mistry 2011
- Mistry M, Parkin DM, Ahmad AS, Sasieni P. Cancer incidence in the United Kingdom: projections to the year 2030. British Journal of Cancer 2011;105(11):1795‐803. [PUBMED: 22033277] [DOI] [PMC free article] [PubMed] [Google Scholar]
Mocellin 2013
- Mocellin S, Lens MB, Pasquali S, Pilati P, Chiarion Sileni V. Interferon alpha for the adjuvant treatment of cutaneous melanoma. Cochrane Database of Systematic Reviews 2013, Issue 6. [DOI: 10.1002/14651858.CD008955.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Moncrieff 2002
- Moncrieff M, Cotton S, Claridge E, Hall P. Spectrophotometric intracutaneous analysis: a new technique for imaging pigmented skin lesions. British Journal of Dermatology 2002;146(3):448‐57. [PUBMED: 11952545] [DOI] [PubMed] [Google Scholar]
Monheit 2011
- Monheit G, Cognetta AB, Ferris L, Rabinovitz H, Gross K, Martini M, et al. The performance of MelaFind: a prospective multicenter study. Archives of Dermatology 2011;147(2):188‐94. [PUBMED: 20956633] [DOI] [PubMed] [Google Scholar]
Moreau 2013
Morton 2014
- Morton DL, Thompson JF, Cochran AJ, Mozzillo N, Nieweg OE, Roses DF, et al. Final trial report of sentinel‐node biopsy versus nodal observation in melanoma. New England Journal of Medicine 2014;370(7):599‐609. [PUBMED: 24521106] [DOI] [PMC free article] [PubMed] [Google Scholar]
Nachbar 1994
- Nachbar F, Stolz W, Merkle T, Cognetta AB, Vogt T, Landthaler M, et al. The ABCD rule of dermatoscopy. High prospective value in the diagnosis of doubtful melanocytic skin lesions. Journal of the American Academy of Dermatology 1994;30(4):551‐9. [PUBMED: 8157780] [DOI] [PubMed] [Google Scholar]
NICE 2012a
- National Institute for Health and Care Excellence. Vemurafenib for treating locally advanced or metastatic BRAF V600 mutation‐positive malignant melanoma. www.nice.org.uk/guidance/ta269. London: National Institute of Health and Care Excellence,, (accessed prior to 19 July 2017).
NICE 2012b
- National Institute for Health and Care Excellence. Ipilimumab for previously treated advanced (unresectable or metastatic) melanoma. www.nice.org.uk/guidance/ta268. London: National Institute of Health and Care Excellence, (accessed prior to 19 July 2017).
NICE 2014a
- National Institute for Health and Care Excellence. Dabrafenib for treating unresectable or metastatic BRAF V600 mutation‐positive melanoma. www.nice.org.uk/guidance/ta321. London: National Institute of Health and Care Excellence, (accessed prior to 19 July 2017).
NICE 2014b
- National Institute for Health and Care Excellence. Ipilimumab for previously untreated advanced (unresectable or metastatic) melanoma. www.nice.org.uk/guidance/ta319. London: National Institute of Health and Care Excellence, (accessed prior to 19 July 2017).
NICE 2015a
- National Institute for Health and Care Excellence. Melanoma: assessment and management. www.nice.org.uk/guidance/ng14. London: National Institute for Health and Care Excellence, (accessed prior to 19 July 2017).
NICE 2015b
- National Institute for Health and Care Excellence. Pembrolizumab for advanced melanoma not previously treated with ipilimumab. www.nice.org.uk/guidance/ta366. London: National Institute of Health and Care Excellence,, (accessed prior to 19 July 2017).
NICE 2015c
- National Institute for Health and Care Excellence. Pembrolizumab for treating advanced melanoma after disease progression with ipilimumab. www.nice.org.uk/guidance/ta357. London: National Institute of Health and Care Excellence, (accessed prior to 19 July 2017).
NICE 2015d
- National Institute for Health and Care Excellence. Suspected cancer: recognition and referral. www.nice.org.uk/guidance/ng12. London: National Institute for Health and Care Excellence, (accessed prior to 19 July 2017).
NICE 2016a
- National Institute for Health and Care Excellence. Trametinib in combination with dabrafenib for treating unresectable or metastatic melanoma. www.nice.org.uk/guidance/ta396. London: National Institute of Health and Care Excellence, (accessed prior to 19 July 2017).
NICE 2016b
- National Institute for Health and Care Excellence. Nivolumab in combination with ipilimumab for treating advanced melanoma. www.nice.org.uk/guidance/ta400. London: National Institute of Health and Care Excellence, (accessed prior to 19 July 2017).
NICE 2018
- National Institute for Health and Care Excellence. Skin Cancer. www.nice.org.uk/guidance/conditions‐and‐diseases/cancer/skin‐cancer#panel‐in‐development (accessed 05 October 2018).
Norman 2009
- Norman G, Barraclough K, Dolovich L, Price D. Iterative diagnosis. BMJ 2009;339:b3490. [PUBMED: 19773326] [DOI] [PubMed] [Google Scholar]
Pasquali 2018
- Pasquali S, Hadjinicolaou AV, Chiarion Sileni V, Rossi CR, Mocellin S. Systemic treatments for metastatic cutaneous melanoma. Cochrane Database of Systematic Reviews 2018, Issue 2. [DOI: 10.1002/14651858.CD011123; CD011123] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pehamberger 1987
- Pehamberger H, Steiner A, Wolff K. In vivo epiluminescence microscopy of pigmented skin lesions. I. Pattern analysis of pigmented skin lesions. Journal of the American Academy of Dermatology 1987;17(4):571‐83. [PUBMED: 3668002] [DOI] [PubMed] [Google Scholar]
Pehamberger 1993
- Pehamberger H, Binder M, Steiner A, Wolff K. In vivo epiluminescence microscopy: improvement of early diagnosis of melanoma. Journal of Investigative Dermatology 1993;100(3):356S‐62S. [PUBMED: 8440924] [DOI] [PubMed] [Google Scholar]
Pellacani 2009b
- Pellacani G, Scope A, Ferrari B, Pupelli G, Bassoli S, Longo C, et al. New insights into nevogenesis: in vivo characterization and follow‐up of melanocytic nevi by reflectance confocal microscopy. Journal of the American Academy of Dermatology 2009;61(6):1001‐13. [PUBMED: 19833408] [DOI] [PubMed] [Google Scholar]
Pellacani 2014a
- Pellacani G, Pepe P, Casari A, Longo C. Reflectance confocal microscopy as a second‐level examination in skin oncology improves diagnostic accuracy and saves unnecessary excisions: a longitudinal prospective study. British Journal of Dermatology 2014;171(5):1044‐51. [PUBMED: 24891083] [DOI] [PubMed] [Google Scholar]
Rajadhyaksha 1995
- Rajadhyaksha M, Grossman M, Esterowitz D, Webb RH, Anderson RR. In vivo confocal scanning laser microscopy of human skin: melanin provides strong contrast. Journal of Investigative Dermatology 1995;104(6):946‐52. [PUBMED: 7769264] [DOI] [PubMed] [Google Scholar]
Rajadhyaksha 2017
- Rajadhyaksha M, Marghoob A, Rossi A, Halpern AC, Nehal KS. Reflectance confocal microscopy of skin in vivo: from bench to bedside. Lasers in Surgery and Medicine 2017;49(1):7‐19. [PUBMED: 27785781] [DOI] [PMC free article] [PubMed] [Google Scholar]
Rajpara 2009
- Rajpara SM, Botello AP, Townend J, Ormerod AD. Systematic review of dermoscopy and digital dermoscopy/artificial intelligence for the diagnosis of melanoma. British Journal of Dermatology 2009;161(3):591‐604. [PUBMED: 19302072] [DOI] [PubMed] [Google Scholar]
Reitsma 2005
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. Journal of Clinical Epidemiology 2005;58(10):982‐90. [PUBMED: 16168343] [DOI] [PubMed] [Google Scholar]
Ruocco 2011
- Ruocco E, Brunetti G, Vecchio M, Ruocco V. The practical use of cytology for diagnosis in dermatology. Journal of the European Academy of Dermatology & Venereology 2011;25(2):125‐9. [PUBMED: 20553359] [DOI] [PubMed] [Google Scholar]
Rutjes 2005
- Rutjes AW, Reitsma JB, Vandenbroucke JP, Glas AS, Bossuyt PM. Case‐control and two‐gate designs in diagnostic accuracy studies. Clinical Chemistry 2005;51(8):1335‐41. [PUBMED: 15961549] [DOI] [PubMed] [Google Scholar]
Rutjes 2006
- Rutjes AW, Reitsma JB, Nisio M, Smidt N, Rijn JC, Bossuyt PM. Evidence of bias and variation in diagnostic accuracy studies. CMAJ 2006;174(4):469‐76. [PUBMED: 16477057] [DOI] [PMC free article] [PubMed] [Google Scholar]
Rutter 2001
- Rutter CM, Gatsonis CA. A hierarchical regression approach to meta‐analysis of diagnostic test accuracy evaluations. Statistics in Medicine 2001;20(19):2865‐84. [PUBMED: 11568945] [DOI] [PubMed] [Google Scholar]
SAS 2012 [Computer program]
- SAS Institute Inc.. SAS 2012. Version 9.3. Cary (NC): SAS Institute Inc., 2012.
Scope 2007
- Scope A, Benvenuto‐Andrade C, Agero AL, Malvehy J, Puig S, Rajadhyaksha M, et al. In vivo reflectance confocal microscopy imaging of melanocytic skin lesions: consensus terminology glossary and illustrative images. Journal of the American Academy of Dermatology 2007;57(4):644‐58. [PUBMED: 17630045] [DOI] [PubMed] [Google Scholar]
Scotto di Santolo 2015
SEER 2017
- SEER. Cancer stat facts: melanoma of the skin (the Surveillance, Epidemiology, and End Results (SEER) Program). www.seer.cancer.gov/statfacts/html/melan.html (accessed prior to 01 November 2017).
Shaikh 2012
- Shaikh WR, Xiong M, Weinstock MA. The contribution of nodular subtype to melanoma mortality in the United States, 1978 to 2007. Archives of Dermatology 2012;148(1):30‐6. [PUBMED: 21931016] [DOI] [PubMed] [Google Scholar]
Siegel 2015
- Siegel R, Miller K, Jemal A. Cancer statistics, 2015. CA: a Cancer Journal for Clinicians 2015;65(1):5‐29. [PUBMED: 25559415] [DOI] [PubMed] [Google Scholar]
SIGN 2017
- Scottish Intercollegiate Guidelines Network. Cutaneous melanoma. www.sign.ac.uk/sign‐146‐melanoma.html. Scotland: SIGN, (accessed prior to 19 July 2017).
Sladden 2009
- Sladden MJ, Balch C, Barzilai DA, Berg D, Freiman A, Handiside T, et al. Surgical excision margins for primary cutaneous melanoma. Cochrane Database of Systematic Reviews 2009, Issue 10. [DOI: 10.1002/14651858.CD004835.pub2] [DOI] [PubMed] [Google Scholar]
Slater 2014
- Slater D, Walsh M. Standards and datasets for reporting cancers: dataset for the histological reporting of primary cutaneous malignant melanoma and regional lymph nodes, May 2014. www.rcpath.org/Resources/RCPath/Migrated%20Resources/Documents/G/G125_DatasetMaligMelanoma_May14.pdf. London: Royal College of Pathologists, (accessed 29 July 2015).
Sobarun 2015
- Sobarun P. Reflectance confocal microscopy. www.dermnetnz.org/topics/reflectance‐confocal‐microscopy/ (accessed 18 May 2017).
Sober 1979
- Sober AJ, Fitzpatrick TB, Mihm MC, Wise TG, Pearson BJ, Clark WH, et al. Early recognition of cutaneous melanoma. JAMA 1979;242(25):2795‐9. [PUBMED: 501893] [PubMed] [Google Scholar]
Steiner 1987
- Steiner A, Pehamberger H, Wolff K. In vivo epiluminescence microscopy of pigmented skin lesions. II. Diagnosis of small pigmented skin lesions and early detection of malignant melanoma. Journal of the American Academy of Dermatology 1987;17(4):584‐91. [PUBMED: 3668003] [DOI] [PubMed] [Google Scholar]
Stolz 1994
- Stolz W, Riemann A, Cognetta AB, Pillet L, Abmayer W, Holzel D, et al. ABCD rule of dermatoscopy: a new practical method for early recognition of malignant melanoma. European Journal of Dermatology 1994;4(7):521‐7. [EMBASE: 24349113] [Google Scholar]
Sznol 2013
- Sznol M, Chen L. Antagonist antibodies to PD‐1 and B7‐H1 (PD‐L1) in the treatment of advanced human cancer. Clinical Cancer Research 2013;19(5):1021‐34. [PUBMED: 23460533] [DOI] [PMC free article] [PubMed] [Google Scholar]
Takwoingi 2010
- Takwoingi Y, Deeks J. MetaDAS: a SAS macro for meta‐analysis of diagnostic accuracy studies. User Guide Version 1.3. 2010. www.methods.cochrane.org/sites/methods.cochrane.org.sdt/files/public/uploads/MetaDAS%20Readme%20v1.3%20May%202012.pdf (accessed prior to 17 July 2017).
Takwoingi 2013
- Takwoingi Y, Leeflang MM, Deeks JJ. Empirical evidence of the importance of comparative studies of diagnostic test accuracy. Annals of Internal Medicine 2013;158(7):544‐54. [PUBMED: 23546566] [DOI] [PubMed] [Google Scholar]
Takwoingi 2015
- Takwoingi Y, Guo B, Riley RD, Deeks JJ. Performance of methods for meta‐analysis of diagnostic test accuracy with few studies or sparse data. Statistical Methods in Medical Research 2015;24:1‐19. [DOI: 10.1177/0962280215592269] [DOI] [PMC free article] [PubMed] [Google Scholar]
Terushkin 2012
- Terushkin V, Dusza SW, Scope A, Argenziano G, Bahadoran P, Cowell L, et al. Changes observed in slow‐growing melanomas during long‐term dermoscopic monitoring. British Journal of Dermatology 2012;166(6):1213‐20. [DOI: 10.1111/j.1365-2133.2012.10846.x] [DOI] [PubMed] [Google Scholar]
Themstrup 2015
- Themstrup L, Jemec GB. Optical coherence tomography and its role for delineating the thickness of keratinocyte dysplasia and neoplasia. Current Problems in Dermatology 2015;46:95‐100. [PUBMED: 25561212] [DOI] [PubMed] [Google Scholar]
Thomas 1998
- Thomas L, Tranchand P, Berard F, Secchi T, Colin C, Moulin G. Semiological value of ABCDE criteria in the diagnosis of cutaneous pigmented tumors. Dermatology 1998;197(1):11‐7. [PUBMED: 9693179] [DOI] [PubMed] [Google Scholar]
Villanueva 2010
- Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga‐Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF‐1R/PI3K. Cancer Cell 2010;18(6):683‐95. [PUBMED: 21156289] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wachsman 2011
- Wachsman W, Morhenn V, Palmer T, Walls L, Hata T, Zalla J, et al. Noninvasive genomic detection of melanoma. British Journal of Dermatology 2011;164(4):797‐806. [PUBMED: 21294715] [DOI] [PMC free article] [PubMed] [Google Scholar]
Walter 2012
- Walter FM, Morris HC, Humphrys E, Hall PN, Prevost AT, Burrows N, et al. Effect of adding a diagnostic aid to best practice to manage suspicious pigmented lesions in primary care: randomised controlled trial. BMJ 2012;345:e4110. [PUBMED: 22763392] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wells 2012
- Wells R, Gutkowicz‐Krusin D, Veledar E, Toledano A, Chen SC. Comparison of diagnostic and management sensitivity to melanoma between dermatologists and MelaFind: a pilot study. Archives of Dermatology 2012;148(9):1083‐4. [PUBMED: 22986873] [DOI] [PubMed] [Google Scholar]
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529‐36. [PUBMED: 22007046] [DOI] [PubMed] [Google Scholar]
Williams 1989
- Williams H, Pembroke A. Sniffer dogs in the melanoma clinic?. Lancet 1989;1(8640):734. [PUBMED: 2564551] [DOI] [PubMed] [Google Scholar]
Zemelman 2014
- Zemelman VB, Valenzuela CY, Sazunic I, Araya I. Malignant melanoma in Chile: different site distribution between private and state patients. Biological Research 2014;47(1):34. [PUBMED: 25204018] [DOI] [PMC free article] [PubMed] [Google Scholar]
References to other published versions of this review
Dinnes 2015a
- Dinnes J, Matin RN, Moreau JF, Patel L, Chan SA, Wong KY, et al. Tests to assist in the diagnosis of cutaneous melanoma in adults: a generic protocol. Cochrane Database of Systematic Reviews 2015, Issue 10. [DOI: 10.1002/14651858.CD011902] [DOI] [Google Scholar]
Dinnes 2015b
- Dinnes J, Wong KY, Gulati A, Chuchu N, Leonardi‐Bee J, Bayliss SE, et al. Tests to assist in the diagnosis of keratinocyte skin cancers in adults: a generic protocol. Cochrane Database of Systematic Reviews 2015, Issue 10. [DOI: 10.1002/14651858.CD011901] [DOI] [Google Scholar]


