Abstract
Background
Thyroid cancer is a type of endocrine cancers with rapidly increased incidence. Recent studies have indicated long non-coding RNAs (lncRNAs) played crucial roles in thyroid cancer tumorigenesis and progression. However, the roles of most lncRNAs in thyroid cancer were still unclear.
Material/Methods
We used TCGA (The Cancer Genome Atlas), GSE50901, GSE29265, and GSE33630 datasets to analyze the expression pattern of ZFAS1 (ZNFX1 antisense RNA 1). The correlation between ZFAS1 and clinicopathological features in thyroid cancer was analyzed. Cell proliferation and cell cycle assays were used to validate the roles of ZFAS1 in thyroid cancer cell lines. DAVID (the database for annotation, visualization and integrated discover) system was used to perform GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis. The starBase datasets and Cytoscape was used to perform ceRNA (competitive endogenous RNA) network.
Results
We demonstrated ZFAS1 was highly expressed in thyroid cancer compared to normal samples. Moreover, upregulation of ZFAS1 was positively correlated with clinicopathological features and poor prognosis in thyroid cancer. Functional validation showed knockdown of ZFAS1 suppressed cell proliferation and cell cycle in thyroid cancer cells. Bioinformatics analysis showed ZFAS1 was associated with translation, rRNA processing, intra-Golgi vesicle-mediated transport, ribosome, and ubiquitin-mediated proteolysis.
Conclusions
Our study suggested ZFAS1 could serve as a biomarker for thyroid cancer.
MeSH Keywords: Biological Markers; Cell Proliferation; Parathyroid Neoplasms; RNA, Long Noncoding
Background
Thyroid cancer is a type of endocrine cancers with rapidly increased incidence. However, the mechanism underlying thyroid cancer progression remains largely unclear. Long non-coding RNA (lncRNA) is a kind of non-coding RNA transcript longer than 200 bps. Recent studies have shown that lncRNAs plays crucial roles in thyroid cancer tumorigenesis and progression. For example, Huang et al. found that the lncRNA MALAT1 (metastasis associated lung adenocarcinoma transcript 1) was upregulated and promoted cell proliferation and invasion of thyroid cancer by increasing IQGAP1 (IQ motif containing GTPase activating protein 1) expression [1]. Several other lncRNAs, such as HIT000218960 [2], NR_036575.1 [3], and LOC100507661 [4], were also associated with thyroid cancer growth and metastasis. However, the roles of most lncRNAs in thyroid cancer are still unknown.
ZFAS1 (ZNFX1 antisense RNA 1) is a novel lncRNA identified to be dysregulated in human cancers, including breast cancer [5], gastric cancer [6], and colorectal cancer [7]. ZFAS1 was first described as a tumor suppressor in breast cancer. Of note, most publications have shown that ZFAS1 serves as an oncogene. In hepatocellular carcinoma, ZFAS1 promoted cancer metastasis by binding miR-150 to induce ZEB1 (zinc finger E-box binding homeobox 1), MMP14 (matrix metalloproteinase 14), and MMP16 expression [8]. In gastric cancer, ZFAS1 was found to be associated with proliferation, cell cycle, apoptosis, and epithelial-mesenchymal transition [9]. Similarly, it was shown that ZFAS1 could also promote cell growth and metastasis in breast cancer [5]. However, the expression pattern and function roles of ZFAS1 in thyroid cancer progression are still unclear.
Competitive endogenous RNA (ceRNA) hypothesis proposes that RNA modules, including lncRNAs, lncRNAs pseudogenes, and mRNAs could crosstalk with each other by competitively binding to microRNAs (miRNAs). Studies have suggested that the ceRNA mechanism plays crucial roles in cancer progression. For example, the BRAF pseudogene served as a ceRNA to induce lymphoma by regulating the BRAF and MAPK pathway. Wan et al. reported PTCH1-3′UTR promoted non-small cell lung cancer metastasis by activating miR-101-3p/SLC39A6. In thyroid cancer, H19 competitively binds to miR-17-5p to promote cancer progression by inducing YES1 expression.
In the present study, explored the expression of the lncRNA ZFAS1 in thyroid cancer tissues by analyzing TCGA dataset. TCGA is a public platform, which includes more than 30 types of human cancers with their clinicopathological information. TCGA platform was widely used to identify key regulators, mutations, and RNA splicing in human cancers. Furthermore, we performed bioinformatics analysis, such as co-expression, GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis, to reveal the potential roles of ZFAS1 in the development of thyroid cancer. Our study aimed to provide evidence to show ZFAS1 could act as an effective therapeutic strategy for thyroid cancer.
Material and Methods
TCGA dataset analysis
We used data from TCGA data portal and GSE50901 [10], GSE33630 [11,12], and GSE29265 to analysis the expression of ZFAS1 in thyroid cancer. GSE50901 was uploaded by Barros-Filho which contains 57 papillary thyroid carcinomas with a reference sample (pool of 9 adjacent normal thyroid tissue) and 4 papillary thyroid carcinomas against 4 matched adjacent normal thyroid tissues. GSE33630 and GSE29265 were contributed by Tomas: GSE33630 contains 11 anaplastic thyroid carcinomas, 49 papillary thyroid carcinomas, and 45 normal thyroids; GSE29265 contains 9 anaplastic thyroid carcinomas, together with 20 papillary thyroid carcinomas paired with their respective normal adjacent tissues.
TNM tumor classification system was developed and maintained by the Union for International Cancer Control. TNM is a notation system that describes the stage of a cancer, which originates from a solid tumor, using alphanumeric codes: T describes the size of the original (primary) tumor and whether it has invaded nearby tissue, N describes nearby (regional) lymph nodes that are involved, M describes distant metastasis (spread of cancer from one part of the body to another).
ZFAS1 mediated ceRNA networks
Competitive endogenous RNA (ceRNA) hypothesis proposes that RNA modules, including lncRNAs, lncRNAs pseudogenes, and mRNAs could crosstalk with each other by competitively binding to miRNAs. In this study, we constructed the ceRNA networks based on the ceRNA hypothesis. We used the Pearson correlation coefficient to determine the expression correlation. The ZFAS1-mRNA pairs which Pearson correlation coefficient are higher than 0.2 were chosen for analysis. ZFAS1 targeting miRNAs and mRNAs were analyzed by using starBase datasets.
Co-expression network construction and analysis
Co-expression network was constructed to explore the relationship between the lncRNAs and mRNAs. The Pearson correlation coefficient was calculated and the absolute value of the Pearson correlation coefficient ≥0.3 was picked out as meaningful. The co-expression network was illustrated using Cytoscape.
GO and KEGG pathway analysis
Molecule Annotation System (MAS, http://bioinfo.capitalbio.com/mas3/) is a web-based software toolkit for a whole data mining and function annotation solution to extract and analyze biological molecules relationships from public knowledgebase of biological molecules and signification. We used MAS3.0 system to perform GO and KEGG pathway analysis. A P value ≤0.05 was considered as significant.
Cell culture and cell transfection
CAL62 and SW579 were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS, Gibco) at 37°C in a humidified incubator with 5% CO2. On the day before transfection, the cells were harvested and seeded at 5×105 cells per well in a 6-well plate. Cells were transfected with small interfering RNA (siRNAs) using Lipofectamine 3000 (Invitrogen). siRNA for ZFAS1 (5′-CCCTGTGCTTTCATGAAAGTGAAGA-3′) and for NC (negative control) were purchased from BioTNT.
Real-time reverse transcription PCR (qRT-PCR) analysis
Total RNAs were extracted using Ultrapure RNA Kit (CWBIO, China) to extract RNAs. PrimeScript RT Reagent Kit (TaKaRA, China) was used for reverse transcript PCRs. The Ct values were normalized using β-actin as an internal control to estimate the differential expression of genes. Relative mRNA expression was calculated using the 2−ΔΔCt method. Each sample was run in triplicate to ensure quantitative accuracy. Primers for ZFAS1 were: forward, 5′ AAGCCACGTGCAGACATCTA 3′, reverse, 5′ CTACTTCCAACACCCGCATT 3′.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) assays were conducted to measure the cell proliferation. CCK-8 allows sensitive colorimetric assays for the determination of the number of viable cells in cell proliferation and cytotoxicity assays by using WST-8. Cell proliferation was determined at each indicated time points. The certain number of cells were seeded in 96-well plates and cultured in media containing 10% FBS. Absorbance was determined by a plate reader at a wavelength of 450 nm. The absorbance at 450 nm was selected as a reference. Triplicate wells were measured in each group to improve the accuracy.
Cell cycle assay
Flow cytometric analyses were performed as cell cycle assay to define the cycle distribution. Cells cultured in 10 cm dishes were trypsinized, harvested, then washed with phosphate-buffered saline (PBS) and fixed with 70% ethanol at 4°C for 2 hours. Cells were centrifuged and washed with PBS, then stained for DNA content in the propidium iodide/RNase I staining solution for 30 minutes in the dark. Cell cycle was analyzed using a flow cytometer.
Statistical analysis
Statistical comparisons between 2 groups were performed using t-test or Mann-Whitney U test according to the test condition. For more groups, one-way ANOVA followed by Newman-Keuls posthoc test was used. A value of P<0.05 was considered statistical significance.
Results
Long non-coding RNA ZFAS1 was upregulated in human thyroid cancer
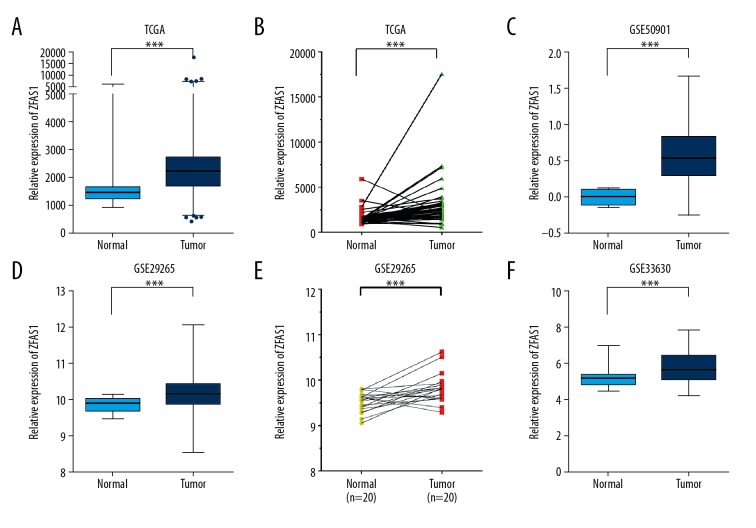
To determine the biological effect of ZFAS1, we analyzed TCGA dataset downloaded from cBioPortal [13,14] (http://www.cbioportal.org/), which included matched normal tissues (n=58) and thyroid cancer samples (n=510). It can be observed in Figure 1A and 1B that the level of ZFAS1 was significantly overexpressed in thyroid cancer tissues with respect to that in the non-cancerous tissues. Of note, this finding was further confirmed by 3 independent GEO datasets, including GSE50901 (Figure 1C), GSE29265 (Figure 1D, 1E), and GSE33630 (Figure 1F). These results suggested ZFAS1 expression was upregulated in human thyroid cancer.
Figure 1.
ZFAS1 was upregulated in human thyroid cancer. By analyzing TCGA (A, B), GSE50901 (C), GSE29265 (D, E), and GSE33630 (F) dataset, ZFAS1 was significantly overexpressed in thyroid cancer tissues compared with non-cancerous tissues. * P<0.05; *** P<0.001. ZFAS1 – ZNFX1 antisense RNA 1; TCGA – The Cancer Genome Atlas.
Upregulation of ZFAS1 was correlated with clinicopathological features in thyroid cancer
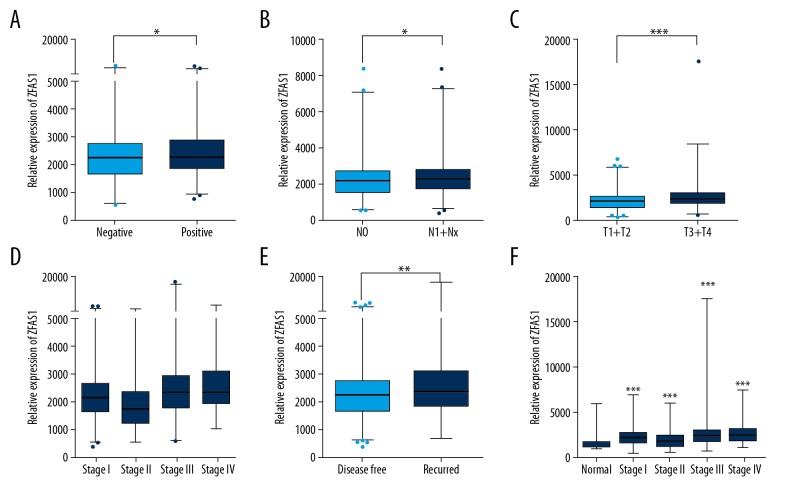
Next, we explored the association of ZFAS1 expression with clinicopathological features of thyroid cancer by analyzing the TCGA dataset. As shown in Figure 2, our analysis showed higher ZFAS1 expression was remarkably associated with lymph node metastasis (Figure 2A, P<0.05), N stage (Figure 2B, P<0.05), T stage (Figure 2C, P<0.001), and grade (Figure 2D). Interestingly, we observed ZFAS1 was upregulated in recurred thyroid cancer samples compared to recurrence-free thyroid cancer samples (Figure 2E, P<0.01). In order to explore whether ZFAS1 could serve as an early diagnosis, we compared the expression levels of ZFAS1 in all grade thyroid cancer samples to that in normal samples. Interestingly, we observed ZFAS1 was upregulated in earlier stages of thyroid cancer samples. These results suggested that ZFAS1 could serve as a new prognostic factor in thyroid cancer patients.
Figure 2.
Overexpressed ZFAS1 is positively correlated with poor prognosis in thyroid cancer. (A–E). Overexpressed ZFAS1 was remarkably associated with lymph node metastasis (A, P<0.001), N stage (C, P<0.001), T stage (D, P<0.001), grade (E, P<0.001), and recurrence status (B, P<0.01). (F) ZFAS1 was remarkably upregulated in stage I, stage II, stage III, and stage IV thyroid cancer samples compared with normal samples. * P<0.05; ** P<0.01; *** P<0.001. ZFAS1 – ZNFX1 antisense RNA 1.
Knockdown of ZFAS1 inhibited cell proliferation and arrested cell cycle in thyroid cancer
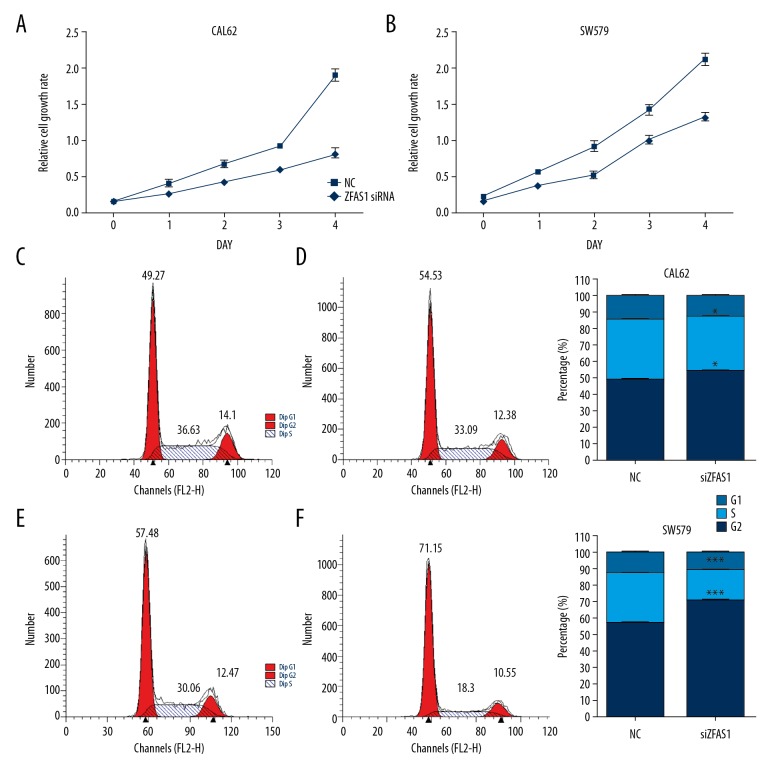
Furthermore, the CCK-8 assay was performed to evaluate the effect of ZFAS1 on cell proliferation. As shown in Figure 3A and 3B, we observed the proliferative viability of CAL62 and SW579 cells interfered with si-ZFAS1 was significantly suppressed. We also detect the percentage of thyroid cancer cells at G1, S, and G2 phases following ZFAS1 knockdown. From Figure 3C–3F, we found ZFAS1 knockdown significantly arrested cell cycle in SW579 and CAL62 cells by increasing G1 phase cells population and decreasing S and G2/M phase cells population.
Figure 3.
Function roles of ZFAS1 in thyroid cancer. (A, B) Knockdown of ZFAS1 suppressed proliferation of CAL62 and SW579 cells. (C–F) Knockdown of ZFAS1 significantly arrested cell cycle of CAL62 and SW579 cells by increasing the percentage of G1 phase cells and decreasing the percentage of S phase cells. Results are presented as the means ±SD of 3 independent experiments. Significance was defined as P<0.05 (* P<0.05; ** P<0.01; *** P<0.005; **** P<0.001). ZFAS1 – ZNFX1 antisense RNA 1; NC – negative control; SD – standard deviation.
Bioinformatics analysis of ZFAS1 in thyroid cancer
Co-expression analysis was widely used to explore the potential roles of lncRNAs in human diseases. Considering that ZFAS1 was a novel lncRNA in thyroid cancer, we constructed ZFAS1 mediated co-expression network to identify its potential mechanisms. The absolute value of the Pearson correlation coefficient ≥0.30 was selected as a cutoff to identified reliable ZFAS1-mRNA pairs. A total of 698 mRNAs were included in this network.
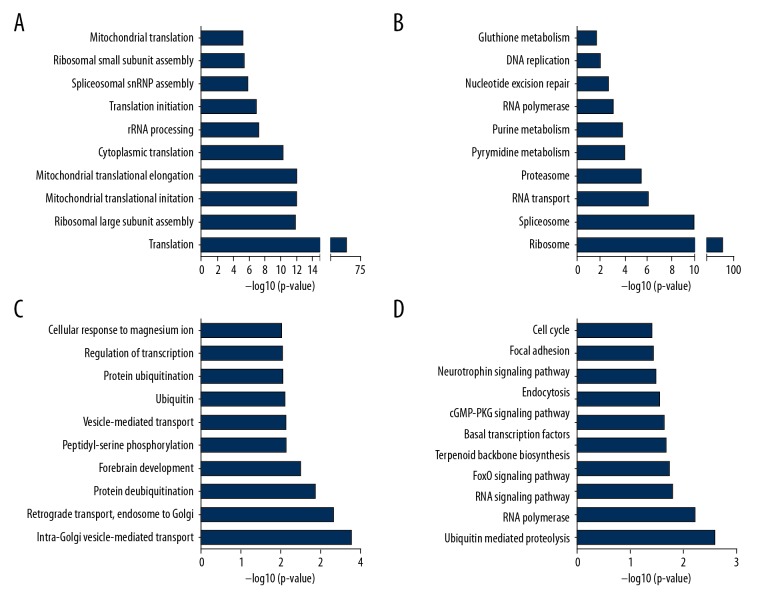
We next performed GO and KEGG pathway analysis based on the co-expressed genes of ZFAS1 by using the DAVID system (https://david.ncifcrf.gov/tools.jsp). GO analysis revealed that ZFAS1 positively co-expressing genes were involved in translation, rRNA processing, spliceosomal snRNP assembly regulation (Figure 4A), and ZFAS1 negatively co-expressing genes were involved in regulating intra-Golgi vesicle-mediated transport, retrograde transport, endosome to Golgi, protein deubiquitination, forebrain development, peptidyl-serine phosphorylation, vesicle-mediated transport, and ubiquitin (Figure 4C).
Figure 4.
GO and KEGG pathway analysis of co-expressed genes of ZFAS1. GO analysis of ZFAS1 positively co-expressing genes (A) and ZFAS1 negatively co-expressing genes (B). KEGG analysis of ZFAS1 positively co-expressing genes (C) and ZFAS1 negatively co-expressing genes (D). GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; ZFAS1 – ZNFX1 antisense RNA 1.
KEGG pathway analysis showed ZFAS1 was involved in regulating ribosome, spliceosome, RNA transport, proteasome, pyrimidine metabolism, purine metabolism, RNA polymerase, ubiquitin mediated proteolysis, RNA polymerase, FoxO signaling pathway, terpenoid backbone biosynthesis, basal transcription factors, cGMP-PKG signaling pathway, endocytosis, neurotrophin signaling pathway, focal adhesion, and cell cycle (Figure 4B, 4D).
Construction of ZFAS1-mediated ceRNA network in thyroid cancer
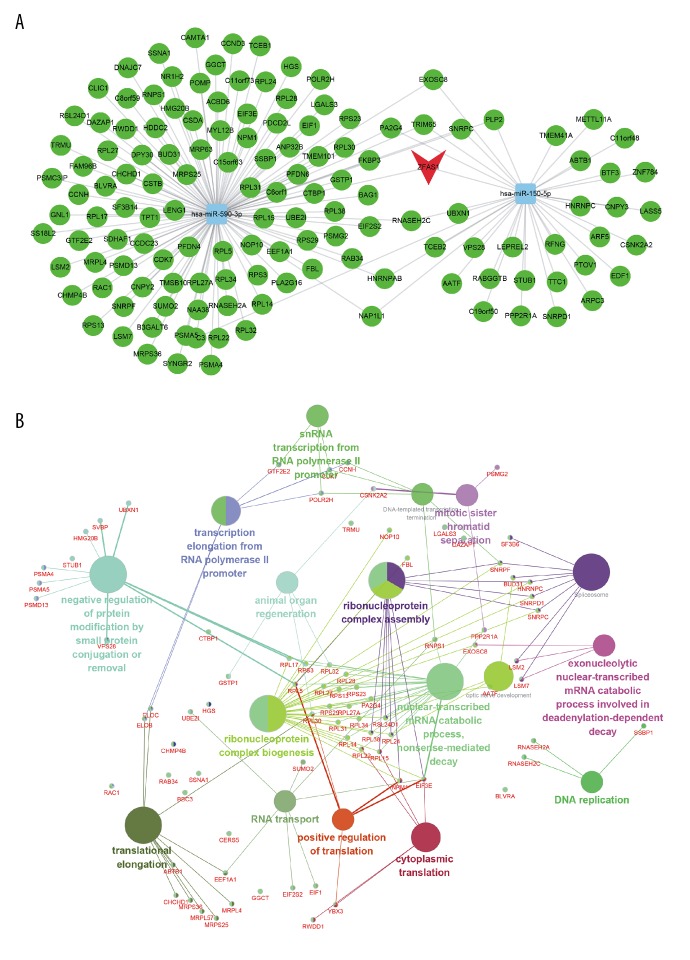
Previous studies have suggested that lncRNAs could serve as competing endogenous RNAs in cancer cell. Here, we also constructed ZFAS1-related ceRNA networks in thyroid cancer. We first predicted ZFAS1 targeting miRNAs and mRNAs by using starBase datasets. Next, we constructed ZFAS1-related ceRNA networks based on positive related ZFAS1-target (Pearson score ≥0.45) pairs. As shown in Figure 5A, a ZFAS1-related ceRNA network was constructed for thyroid cancer. Two miRNAs (hsa-miR-150-5p and hsa-miR-590-3p) and 137 mRNAs were included in this ceRNA network.
Figure 5.
The ZFAS1-mediated ceRNA network (A) and the potential roles of the ceRNA network (B). ZFAS1 – ZNFX1 antisense RNA 1; ceRNA – competitive endogenous RNA.
Furthermore, we used Cytoscape ClueGo plug-in to explore the potential roles involved in ZFAS1-mediated ceRNA network. We found ZFAS1-mediated ceRNA network was associated with in regulating DNA replication, transcription, mitotic sister chromatid separation, and translation (Figure 5B).
Discussion
LncRNAs have been implicated to be an important regulator in thyroid cancer. A series of lncRNAs, such as H19 [15], CNALPTC1 [16], AFAP1-AS1 [17], GAS8-AS1 [18], and TUG1 [19], have been associated with cell proliferation, migration, and invasion regulation. A recent study showed lncRNAs, such as PTCSC3 [20], could also affect drug resistance in thyroid cancer. Of note, a few studies indicated that lncRNAs was associated with cancer progression and could serve as biomarkers for thyroid cancer. For example, lncRNA SPRY4-IT1 was observed to be overexpressed and correlated with poor prognosis in thyroid cancer [21]. In the present study, we demonstrated for the first time that ZFAS1 was overexpressed in thyroid cancer tissues and associated with associated with age, lymph node metastasis, and histological type in thyroid cancer. In order to explore whether ZFAS1 could play a role in early diagnosis, we compared the expression level of ZFAS1 in all grades of thyroid cancer samples to that in normal samples. Interestingly, we observed ZFAS1 was upregulated in the earlier stage thyroid cancer samples. Importantly, higher ZFAS1 expression levels was significantly associated with shorter recurrence-free survival time in thyroid cancer. These results suggested ZFAS1 could serve as a biomarker for thyroid cancer.
ZFAS1 has been found to play crucial roles in the progression of many different human cancers. ZFAS1 was identified as a novel biomarker for the diagnosis of hepatocellular carcinoma [22], prostate cancer [23], non-small cell lung cancer [24], and glioma [25]. Although Askarian et al. [26] first reported that ZFAS1 serves as a tumor suppressor, most reports have identified ZFAS1 as an oncogenic gene in human cancers. Xie et al. found lncRNA ZFAS1 promoted cell proliferation and invasion in colorectal cancer [27]. However, the roles of ZFAS1 in thyroid cancer remained unknown. Here, we performed cellular experiments to explore its roles and found ZFAS1 knockdown suppressed the proliferative ability by inducing cell cycle arrest. Inducing the cell cycle arrest is a well-known anti-cancer mechanism. Dysregulation of cell cycle progression subverts the dynamic balance of cell proliferation and cell death, thereby leading to cancer development. Emerging studies suggest that lncRNAs are involved in the regulation of cell cycle progression. For instance, GAS5 regulated cell cycle arrest in human neuroblastoma. And Linc00152 was found to be involved in cell cycle arrest in gastric cancer. Here, we found knockdown of ZFAS1 significantly increased the percentage of G1 phase cells but decreased the percentage of S phase cells. Our results were consistent with previous findings that ZFAS1 played oncogenic roles in human cancers.
The mechanisms involved in ZFAS1 regulating cancer progression have been identified by several groups. ZFAS1 was found to act as a ceRNA, which could sponge miR-329 [28], miR-150 [29], and miR-484 [30] to promote their targets. In addition, Xu et al. found that ZFAS1 could block Wnt/β-catenin signaling in gastric cancer proliferation and metastasis [31]. Nie et al. found that ZFAS1 could epigenetically repress KLF2 and NKD2 expression in gastric cancer. In the present study, we performed a co-expression analysis for ZFAS1 to explore potential roles of lncRNAs; such methods are widely applied in various human diseases [32]. We observed ZFAS1 were significantly associated with translation, rRNA processing, intra-Golgi vesicle-mediated transport, retrograde transport, endosome to Golgi, protein deubiquitination, ribosome, spliceosome, RNA transport, ubiquitin mediated proteolysis, RNA polymerase, and FoxO signaling pathway. Moreover, we constructed a ZFAS1-mediated ceRNA network in thyroid cancer that included 2 miRNAs (hsa-miR-150-5p and hsa-miR-590-3p) and 137 mRNAs. Bioinformatics analysis revealed this ceRNA network was involved in regulating DNA replication, transcription, mitotic sister chromatid separation, and translation.
Conclusions
Our study demonstrated that ZFAS1 was highly expressed and correlated with poor prognosis in thyroid cancer. Knockdown of ZFAS1 was observed to suppress cell proliferation and cell cycle. Bioinformatics analysis showed ZFAS1 was associated with translation, rRNA processing, intra-Golgi vesicle-mediated transport, ribosome, and ubiquitin mediated proteolysis. Our study suggested ZFAS1 could serve as a biomarker for thyroid cancer.
Footnotes
Source of support: Departmental sources
Conflicts of interest
None.
References
- 1.Huang JK, Ma L, Song WH, et al. MALAT1 promotes the proliferation and invasion of thyroid cancer cells via regulating the expression of IQGAP1. Biomed Pharmacother. 2016;83:1–7. doi: 10.1016/j.biopha.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 2.Li T, Yang XD, Ye CX, et al. Long noncoding RNA HIT000218960 promotes papillary thyroid cancer oncogenesis and tumor progression by upregulating the expression of high mobility group AT-hook 2 (HMGA2) gene. Cell Cycle. 2017;16:224–31. doi: 10.1080/15384101.2016.1261768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun W, Lan X, Wang Z, et al. Overexpression of long non-coding RNA NR_036575.1 contributes to the proliferation and migration of papillary thyroid cancer. Med Oncol. 2016;33:102. doi: 10.1007/s12032-016-0816-y. [DOI] [PubMed] [Google Scholar]
- 4.Kim D, Lee WK, Jeong S, et al. Upregulation of long noncoding RNA LOC100507661 promotes tumor aggressiveness in thyroid cancer. Mol Cell Endocrinol. 2016;431:36–45. doi: 10.1016/j.mce.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–91. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie F, Yu X, Huang M, et al. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget. 2017;8:38227–38. doi: 10.18632/oncotarget.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorenoor N, Faltejskova-Vychytilova P, Hombach S, et al. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget. 2016;7:622–37. doi: 10.18632/oncotarget.5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T, Xie J, Shen C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75:3181–91. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 9.Zhou H, Wang F, Chen H, et al. Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging (Albany, NY) 2016;8:2023–38. doi: 10.18632/aging.101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barros-Filho MC, Marchi FA, Pinto CA, et al. High diagnostic accuracy based on CLDN10, HMGA2, and LAMB3 transcripts in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2015;100:E890–99. doi: 10.1210/jc.2014-4053. [DOI] [PubMed] [Google Scholar]
- 11.Tomas G, Tarabichi M, Gacquer D, et al. A general method to derive robust organ-specific gene expression-based differentiation indices: Application to thyroid cancer diagnostic. Oncogene. 2012;31:4490–98. doi: 10.1038/onc.2011.626. [DOI] [PubMed] [Google Scholar]
- 12.Dom G, Tarabichi M, Unger K, et al. A gene expression signature distinguishes normal tissues of sporadic and radiation-induced papillary thyroid carcinomas. Br J Cancer. 2012;107:994–1000. doi: 10.1038/bjc.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, Yang J, Zhu X, et al. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283:2326–39. doi: 10.1111/febs.13741. [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Zhou L, Wang H, et al. Long noncoding RNA CNALPTC1 promotes cell proliferation and migration of papillary thyroid cancer via sponging miR-30 family. Am J Cancer Res. 2018;8:192–206. [PMC free article] [PubMed] [Google Scholar]
- 17.Dai W, Tian Y, Jiang B, Chen W. Down-regulation of long non-coding RNA AFAP1-AS1 inhibits tumor growth, promotes apoptosis and decreases metastasis in thyroid cancer. Biomed Pharmacother. 2018;99:191–97. doi: 10.1016/j.biopha.2017.12.105. [DOI] [PubMed] [Google Scholar]
- 18.Pan W, Zhou L, Ge M, et al. Whole exome sequencing identifies lncRNA GAS8-AS1 and LPAR4 as novel papillary thyroid carcinoma driver alternations. Hum Mol Genet. 2016;25:1875–84. doi: 10.1093/hmg/ddw056. [DOI] [PubMed] [Google Scholar]
- 19.Lei H, Gao Y, Xu X. LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta Biochim Biophys Sin (Shanghai) 2017;49:588–97. doi: 10.1093/abbs/gmx047. [DOI] [PubMed] [Google Scholar]
- 20.Fan M, Li X, Jiang W, et al. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med. 2013;5:1143–46. doi: 10.3892/etm.2013.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Sun Z, Li S, et al. LncRNA SPRY4-IT was concerned with the poor prognosis and contributed to the progression of thyroid cancer. Cancer Gene Ther. 2018;25:39–46. doi: 10.1038/s41417-017-0003-0. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Xie J, Shen C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75:3181–91. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Yang C, Xie S, Cheung E. Long non-coding RNA GAS5 and ZFAS1 are prognostic markers involved in translation targeted by miR-940 in prostate cancer. Oncotarget. 2018;9:1048–62. doi: 10.18632/oncotarget.23254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian FM, Meng FQ, Wang XB. Overexpression of long-noncoding RNA ZFAS1 decreases survival in human NSCLC patients. Eur Rev Med Pharmacol Sci. 2016;20:5126–31. [PubMed] [Google Scholar]
- 25.Gao K, Ji Z, She K, et al. Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and promotes glioma cell progression by activation of the Notch signaling pathway. Biomed Pharmacother. 2017;87:555–60. doi: 10.1016/j.biopha.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–91. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie S, Ge Q, Wang X, et al. Long non-coding RNA ZFAS1 sponges miR-484 to promote cell proliferation and invasion in colorectal cancer. Cell Cycle. 2018;17:154–61. doi: 10.1080/15384101.2017.1407895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JS, Liu QH, Cheng XH, et al. The long noncoding RNA ZFAS1 facilitates bladder cancer tumorigenesis by sponging miR-329. Biomed Pharmacother. 2018;103:174–81. doi: 10.1016/j.biopha.2018.04.031. [DOI] [PubMed] [Google Scholar]
- 29.Xia B, Hou Y, Chen H, et al. Long non-coding RNA ZFAS1 interacts with miR-150-5p to regulate Sp1 expression and ovarian cancer cell malignancy. Oncotarget. 2017;8:19534–46. doi: 10.18632/oncotarget.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie S, Ge Q, Wang X, et al. Long non-coding RNA ZFAS1 sponges miR-484 to promote cell proliferation and invasion in colorectal cancer. Cell Cycle. 2018;17:154–61. doi: 10.1080/15384101.2017.1407895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu W, He L, Li Y, et al. Silencing of lncRNA ZFAS1 inhibits malignancies by blocking Wnt/beta-catenin signaling in gastric cancer cells. Biosci Biotechnol Biochem. 2018;82:456–65. doi: 10.1080/09168451.2018.1431518. [DOI] [PubMed] [Google Scholar]
- 32.Nie F, Yu X, Huang M, et al. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget. 2017;8:38227–38. doi: 10.18632/oncotarget.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]