Summary
Aims
To explore the prognostic and clinicopathological features of glioma with Paxillin (PXN) expression based on a large number of samples.
Methods
RNA sequencing data of 325 glioma samples from Chinese Glioma Genome Atlas (CGGA) database were obtained as discovery set. Three additional datasets were further obtained as validation sets. The protein expression pattern of PXN in glioma was measured by IHC. Kaplan–Meier survival and multivariate Cox analysis were used to estimate the survival distributions. Moreover, the functional annotation of PXN was also analyzed.
Results
In the discovery set, PXN overexpression was significantly associated with high‐grade glioma as well as the higher mortality in survival analysis (log‐rank test, P < 0.01). The results of the other validation datasets showed similar findings. PXN also served as an independent prognostic biomarker in glioblastoma patients. Functional assays showed that PXN contributed to glioma cell proliferation and invasion.
Conclusion
PXN plays as an oncogene in glioma progression and suggests a new potential biotarget for therapy.
Keywords: Glioma, Oncogene, Prognosis, PXN
Introduction
Glioblastoma multiforme (GBM), the most common and malignant type of glioma, remains one of the most lethal cancer in center nervous system (CNS) 1, 2. Despite the advances in combination of resection, chemotherapy, and radiotherapy, the median survival of GBM patients is only about 1 year 3. The major clinical challenge for GBM therapy is that tumor cells deeply infiltrate into adjacent normal brain tissues and/or migrate to the contralateral cerebral hemisphere at early stage and escape from surgical removal and are relatively resistant to radiotherapy as well as chemotherapy, resulting in tumor recurrence 4, 5. GBM invasion is the combination of specific proteases that enable the remodeling of extracellular matrix (ECM) component and the extracellular integrin interactions 6, which are mediated by numerous regulators, including PXN.
PXN, a phosphorylated adaptor protein, plays a critical role in cytoskeletal remodeling and focal adhesion process by recruiting diverse cytoskeleton and signaling proteins into a complex and coordinating the transmission of downstream targets to regulate cell migration and invasion 7, 8, 9. PXN has been reported to be dysregulated and acts as an oncogene in malignancy progression of many human cancers 10, 11, 12, 13, 14, 15, 16, 17, 18. However, only limited studies showed that PXN was present in some glioma cell lines 6, 19, and little was known about the expression pattern of PXN in glioma samples, as well as the clinical features and prognostic value of PXN in glioma. In this study, we evaluated the expression pattern, prognostic value, and biological associations of PXN from CGGA RNA sequencing expression profiling of glioma samples and validated in three other independent datasets. We further demonstrated the functional roles of PXN in glioma cell lines. All these results indicate PXN is a novel prognostic biomarker with potential anti‐invasion therapeutic implications in GBM.
Methods
PXN Expression Analysis in Datasets
RNA sequencing expression data and clinical information of 325 glioma samples (109 grade II, 72 grade III, and 144 grade IV samples) from Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn) were obtained as the discovery set. The Cancer Genome Atlas (TCGA) RNA sequencing database (http://cancergenome.nih.gov), the whole‐genome mRNA expression microarray data of Repository for Molecular Brain Neoplasia Data (REMBRANDT) (http://caintegrator-info.nci.nih.gov/REMBRANDT), and GSE16011 data (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16011) were obtained as validation sets. CGGA and TCGA RNA sequencing data were log2‐transformed. In the four datasets, only samples with definite WHO classification were included for expression analysis, and only GBM samples with survival data were used for survival analysis (two samples diagnosed as GBM but defined as WHO I grade in REMBRANDT dataset were excluded).
Immunohistochemistry (IHC)
Immunohistochemical staining with streptavidin–biotin immunoperoxidase assay was performed on an independent group of 41 formalin fixed, paraffin‐embedded glioma tissues (15 grade II, 12 grade III, 14 grade IV) from CGGA database to detect PXN expression using rabbit anti‐human PXN (Abcam, Cambridge, MA, USA, 1:500) primary antibody as previously carried out 20. The staining score = staining intensity × proportion of positively stained tumor cells. High PXN expression was defined as a staining score >4, while low expression was defined as a staining score ≤4. Written informed consents were all obtained from the patients, and the study was approved by the ethics committee of the hospitals involved.
Gene Ontology (GO) Analysis and Gene Set Enrichment Analysis (GSEA) of PXN‐Associated Genes
The Pearson correlation analysis of PXN and other genes in whole‐genome gene expression profile was performed in CGGA dataset. To detect the biological processes that correlate with PXN expression in glioma, PXN positively correlated genes (r > 0.4, P < 0.01) were analyzed by DAVID (http://david.abcc.ncifcrf.gov/home.jsp) and GSEA.
Cell Culture and Transfection
Cell culture and transfection were carried out as previously reported 20. PXN small interference RNA (siRNA) and negative control (NC) RNA was constructed by GenePharma Co., Inc. (Shanghai, China). PXN siRNA sense sequence is 5′‐GCAGCAACCUUUCUGAACUTT‐3′, and NC sense sequence is 5′‐UUCUCCGAACGUGUCACGUTT‐3′. PXN over‐expressing vectors and NC vectors were constructed by GeneChem Co., Inc. (Shanghai, China).
Clonogenic and Invasion Assays
The Clonogenic and transwell invasion assays were performed as previously reported 20, 21.
Western Blot
Western blot was applied as previously carried out 22. Immunoblot analysis was performed with rabbit anti‐PXN polyclonal antibody (Abcam, 1:1500) followed by HRP‐conjugated goat anti‐rabbit secondary antibodies. Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) was used for loading control.
Statistical Analysis
Statistical analysis was performed using SPSS 16.0 (Armonk, NY, USA), R 3.2.1 and GraphPad Prism 5.0 statistical software (La Jolla, CA, USA). Descriptive statistics were shown as mean ± standard deviation (SD). Student's t‐test, chi‐square test, and one‐way ANOVA test were used to analyze the significance of differences. Overall survival time (OS) was calculated from the date of histological diagnosis until death or the last follow‐up. Kaplan–Meier survival analysis was used to estimate the survival distributions, and the log‐rank test was used to assess the statistical significance between stratified survival groups. Patients with lower than median expression level of PXN were defined as low expression, while patients with higher than or equal to the median value were considered as high expression. Univariate and multivariate Cox regression analysis including gender, age, extent of resection, Karnofsky performance status (KPS), isocitrate dehydrogenase1 (IDH1) mutation status, O‐6‐methylguanine‐DNA methyltransferase (MGMT) methylation status, temozolomide (TMZ) treatment, and radiotherapy were used to assess prognostic value of PXN in GBM. A two‐sided P value <0.05 was considered statistically significant.
Results
PXN Expression is Associated with Glioma Grade and Shows a Subtype Preference
Firstly, we screened the differentially expressed genes from the CGGA dataset and found that PXN expression was positively correlated with tumor grade (one‐way ANOVA, P < 0.01). These results were validated in TCGA, REMBRANDT, and GSE16011 databases (Figure 1A–D). To analyze PXN expression pattern in the different molecular subtypes of glioma, we annotated the four datasets using TCGA classification systems by PAM 23, 24. Based on TCGA subtype classification system, the mesenchymal subtype had the highest PXN expression while the neural subtype showed the lowest, in the four datasets (Figure 1E–H). Additionally, IDH1 wild‐type glioma samples showed higher PXN expression values than those with mutant IDH1 (Figure 1I–K). These results indicated that PXN had a mesenchymal subtype and IDH1 wild‐type preference.
Figure 1.
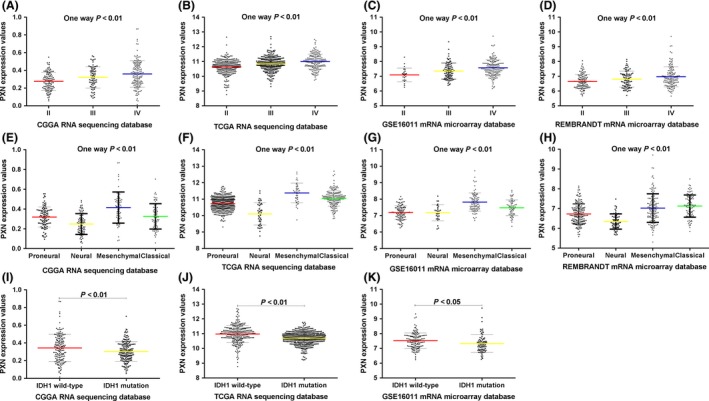
PXN RNA expression pattern in CGGA and other validation datasets. (A–D) The expression values of PXN are positively correlated with tumor grade. (E–H) PXN expression shows mesenchymal subtype preference according to TCGA classification system. (I–K) Patients with wild‐type IDH1 have higher PXN expression than those with mutant IDH1. A single spot is the PXN expression value of an individual patient. Lines in the middle are the mean expression value. Error bars represent standard deviation (SD).
PXN Protein was Measured in an Independent Group of Glioma Samples by IHC
We further detected the protein level of PXN in an independent group of 41 glioma patients from CGGA database by IHC. Similar to PXN RNA expression pattern, PXN protein was low expressed in WHO grade II glioma samples (11 low expressions, four high expressions), comparing with WHO III (four low expressions, eight high expressions) and WHO IV (three low expressions, 11 high expressions) groups (P < 0.013, chi‐square test) (Figure 2A,B). Thus, PXN expression was elevated according to the malignancy of glioma in protein level.
Figure 2.
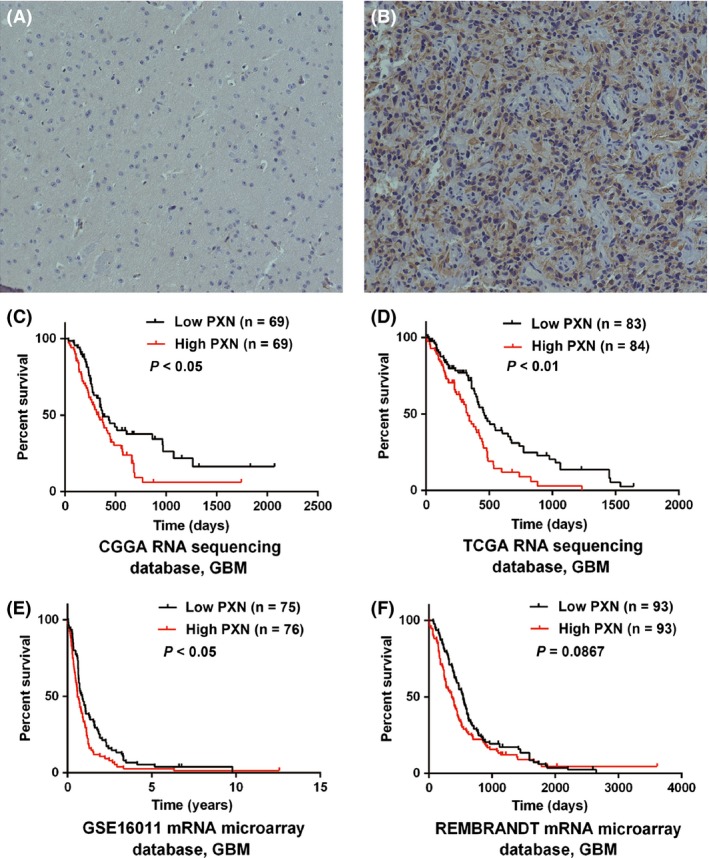
PXN protein expression pattern and prognostic value in GBM patients. (A) WHO II grade sample, showed low expression of PXN in cytoplasm (magnification: ×200); (B) WHO IV grade sample, showed high expression of PXN (magnification: ×200). (C–F) Except for GBM patients in REMBRANDT dataset, according to PXN expression value, patients with GBM could be divided into two groups with significantly different prognosis in three datasets, respectively. High group, patients with higher PXN expression than or equal to the median one. Low group, patients with lower PXN level than the median one.
High PXN Expression Confers a Poor Prognosis in GBM Patients
The association of PXN expression with prognosis of GBM patients was investigated through Kaplan–Meier survival curve analysis with a log‐rank test of 138 GBM patients in CGGA dataset. Patients with high PXN expression had a significantly worse overall survival time in contrast to those with low PXN expression (P < 0.05) (Figure 2C). Furthermore, similar results were also observed in TCGA and GSE16011 validation sets (P < 0.05, P < 0.01, respectively) (Figure 2D,E). Although it was not statistically significant in REMBRANDT dataset, overall survival days in high PXN group still showed a decreased trend compared with low group (median OS: high group 360 days, low group 540 days, P = 0.0867) (Figure 2F).
PXN is an Independent Prognostic Biomarker for GBM Patients
To further determine the prognostic value of PXN in GBM patients, a univariate Cox regression analysis was employed. As shown in Table 1, high PXN expression was shown to be a risk factor for GBM patients [P = 0.017, HR = 1.677, 95% confidence interval (CI): 1.099–2.558]. Additionally, some other factors including age, MGMT methylation status, TMZ treatment, KPS score, extent of resection, and radiotherapy were all significantly associated with the overall survival time of GBM patients. Next, we performed multivariate Cox proportional hazards analysis incorporating PXN expression, age, MGMT methylation state, TMZ treatment, KPS score, extent of resection, and radiotherapy. The analysis revealed that PXN was an independent prognostic factor for the overall survival of GBM patients. Taken together, these results imply that PXN might be a novel prognostic biomarker for GBM patients.
Table 1.
Cox proportional hazard regression analyses of PXN expression and clinicopathological factors affecting overall survival of GBM patients
| Variable | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P Value | |
| Gender (male vs. female) | 1.227 | 0.795–1.893 | 0.355 | |||
| PXN (high vs. low) | 1.677 | 1.099–2.558 | 0.017 | 1.939 | 1.112–3.38 | 0.020 |
| IDH1 (mutation vs. wild type) | 0.638 | 0.380–1.072 | 0.089 | |||
| MGMT (methylation vs. no) | 0.564 | 0.364–0.872 | 0.010 | 0.628 | 0.379–1.038 | 0.070 |
| Age (≥60 vs. <60) | 1.723 | 1.041–2.850 | 0.034 | 1.278 | 0.693–2.356 | 0.432 |
| TMZ (received vs. no) | 0.336 | 0.214–0.528 | <0.01 | 0.493 | 0.287–0.847 | 0.010 |
| Radiotherapy (received vs. no) | 0.412 | 0.259–0.654 | <0.01 | 0.368 | 0.218–0.621 | <0.01 |
| Increasing KPS | 0.967 | 0.952–0.982 | <0.01 | 0.969 | 0.947–0.991 | 0.006 |
| Extent of resection (subtotal vs. total) | 1.645 | 1.090–2.482 | 0.018 | 1.562 | 0.922–2.646 | 0.097 |
HR, hazard ratio.
PXN is Tightly Correlated with Cell Apoptosis, Proliferation, and Adhesion
To investigate biological process associated with PXN expression in glioma, Pearson correlation analysis between PXN expression and other genes in whole‐genome RNA sequencing data was performed in 325 CGGA glioma samples. And the 436 significantly positively correlated genes (r > 0.4, P < 0.01) were used for GO analysis. The top GO terms indicated that PXN was significantly associated with gene sets related to cell adhesion, proliferation, motion, and migration (Figure 3A). The KEGG pathway analysis denoted focal adhesion (Figure 3B). And the GSEA analysis showed the similar results (Figure 3C,D). These analyses indicate that PXN might have an essential role on glioma cell proliferation and invasion.
Figure 3.
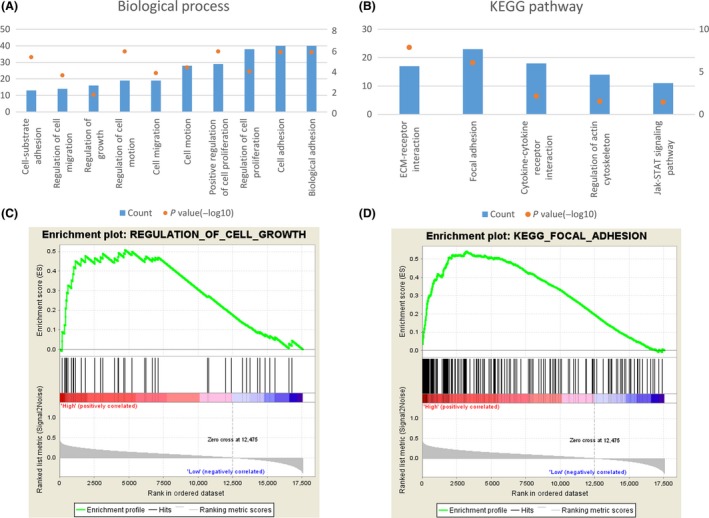
(A) GO analysis of the biological processes of PXN positively correlated genes in CGGA dataset. (B) GO analysis of the KEGG pathway of PXN‐associated genes. The left Y‐axis indicates gene counts, and the right Y‐axis indicates the adjusted (−log10) P values. (C) GSEA analysis of PXN‐associated gene sets and representative enrichment plots. (D) KEGG pathway enriched in high PXN group of glioma patients analyzed by GSEA. NES, normalized enrichment score.
PXN Promotes Glioma Cell Proliferation and Invasion
To explore the functional role of PXN in glioma, we first detected PXN protein levels in H4, LN229, U87, U251 glioma cell lines, as shown in Figure 4A; H4 had the lowest expression level while U87 had the highest. Then, PXN overexpressing vector was employed to elevate PXN expression in H4 cells (Figure 4B), and PXN small interference RNA (siRNA) was used to silence PXN expression in U87 cells (Figure 4C). Invasion and clonogenic assays were performed to evaluate the function of PXN in glioma cell lines, and the gain‐of‐function assay in H4 cells indicated that ectopic of PXN promoted cell invasion and proliferation (Figure 4D,E), while loss‐of‐function assay in U87 cells showed that antagonism of PXN expression suppressed the invasive ability of cells, as well as the formation of clones (Figure 4F,G).
Figure 4.
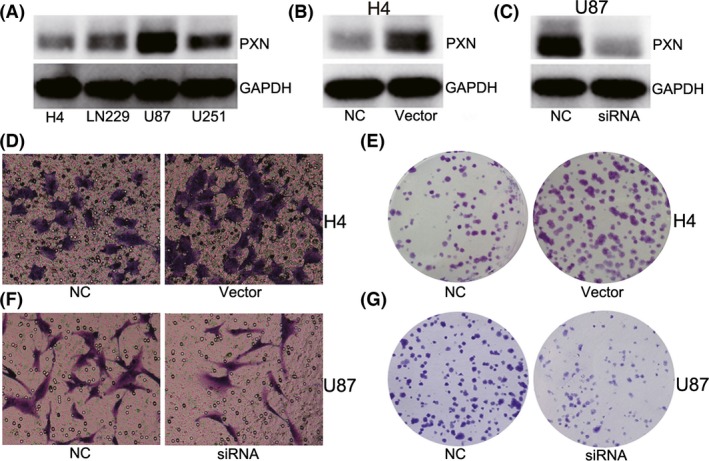
PXN plays as an oncogene in glioma cell lines. (A) PXN protein level was evaluated in four glioma cell lines. (B) PXN protein was up‐regulated by PXN‐overexpression vectors in H4 cells. (C) PXN protein was decreased by PXN siRNA in U87 cells. (D and F) Clonogenic and transwell assay indicates that ectopic of PXN expression promoted cell invasion and proliferation. (E and G) Reduction in PXN expression suppressed cell invasion and proliferation. NC means negative control group, and siRNA means PXN small interfering RNA treatment group.
Discussion
GBM contributes to about 50% of gliomas 25, which has an inevitable recurrence for its highly invasion behavior that tumor cells diffusely infiltrating into the adjacent normal brain tissues during the early stage of disease. Tumor migration and invasion is a complex and multistep process and contains the surrounding ECM component remodeling, morphological polarization, membrane extension, the formation of cell substratum attachments, contractile force and traction, and the release of attachments 26. During this process, cell attachments to substratum play crucial role, which is mediated by coordinating signals from ECM components 27, integrin family 28, focal adhesion‐linked molecules, and the actin cytoskeleton 29.
Among these regulators, we focused on PXN, a focal adhesion‐associated protein 30, which functions as an adaptor protein through interactions with integrin family numbers, growth factors, and cell surface receptors to regulate downstream gene expression, matrix organization, cell proliferation, motility, and migration 30, 31, 32, 33. Consistently, in this study, we found that PXN expression was positively correlated with tumor grade in mRNA profiling of four independent datasets, as well as PXN protein expression difference which was measured by IHC from an independent validation set of samples. The above results indicate both PXN RNA and protein were associated with tumor malignancy, which has been reported in other cancers 10, 12, 14, 15, 16, 17.
Besides, PXN can combine with several oncogene to mislead the normal adhesion and growth factor signaling pathways which results in uncontrolled cell proliferation, migration, and invasion 32, 34, 35, 36. Additionally, phosphorylation of PXN can activate ERK signaling pathway, which further increases the association of focal adhesion kinase (FAK) with PXN to induce cell migration and invasion 13, 37, 38, 39. In this study, the biological roles of PXN in gliomas were evaluated by in silico and in vitro studies, where a positive correlation of PXN expression with a series of tumor oncogenes, driving cell proliferation, mitosis, and invasion, including TGF‐ß1, MAPK, and PLK family members, was observed. Functional assays showed that reduction of PXN expression significantly suppressed U87MG cells proliferation and invasion, while ectopic PXN expression promoted H4 cells proliferation and invasion. All these results are in concordance with the previous reports that PXN is an oncogene in tumor progression. PXN was reported to be regulated by TGF‐ß1, which further promoted malignant astrocytoma cell spreading, attachment, and adhesion 19. And phosphorylated PXN might contribute to TMZ treatment resistant [40].
PXN has been found to be an oncogene and with prognostic value in various malignant human carcinomas 10, 11, 12, 13, 14, 15, 16, 17, 41, 42, 43, 44. Similarly in this study, we found that GBM patients with higher expression of PXN had a worse survival than those with lower PXN expression. It remained to be an independent prognostic marker for GBM patients in multivariate Cox regression analysis.
The relationship between clinical features and PXN expression was not yet clear, and a better understanding of the expression pattern and functional role of PXN in glioma progressive process may provide better diagnosis, prognosis of glioma patient, as well as the development of personalized therapeutics to improve the clinical outcomes.
Conclusion
In conclusion, our study showed that the expression level of PXN increased with the ascending grade of glioma. PXN was preferentially expressed in IDH1 wild‐type patients, mesenchymal subtype of TCGA classification. Moreover, it conferred a worse overall survival and could act as an independent prognostic biomarker in GBM patients. Finally, we demonstrated that PXN played as an oncogene in GBM cells. All of these results suggest that PXN is a novel biomarker and could be a potential target for GBM treatment.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by 1. National Natural Science Foundation of China (Grant No. 81302183, 81502606, 81502494) 2. Natural Science Foundation of Jiangsu Province (Grant No. BK20141099) 3. Beijing Science and Technology Plan (No. Z131100006113018, Z141100000214009) 4. The National Key Research and Development Plan (No. 2016YFC0902500) 5. Capital Medical Development Research Fund (2016‐1‐1072).
The first two authors contributed equally to this work.
Contributor Information
Zheng‐Xiang Yang, Email: wuxiyangz@126.com.
Tao Jiang, Email: taojiang1964@163.com.
References
- 1. Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: Diagnostic, prognostic, and predictive markers. Lancet Neurol 2010;9:717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang‐Xuan K, Delattre JY. Primary brain tumours in adults. Lancet 2012;379:1984–1996. [DOI] [PubMed] [Google Scholar]
- 3. Okumus NO, Gursel B, Meydan D, Ozdemir O, Odabas E, Gonullu G. Prognostic significance of concomitant radiotherapy in newly diagnosed glioblastoma multiforme: A multivariate analysis of 116 patients. Ann Saudi Med 2012;32:250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drappatz J, Norden AD, Wen PY. Therapeutic strategies for inhibiting invasion in glioblastoma. Expert Rev Neurother 2009;9:519–534. [DOI] [PubMed] [Google Scholar]
- 5. Demuth T, Berens ME. Molecular mechanisms of glioma cell migration and invasion. J Neurooncol 2004;70:217–228. [DOI] [PubMed] [Google Scholar]
- 6. Belot N, Rorive S, Doyen I, et al. Molecular characterization of cell substratum attachments in human glial tumors relates to prognostic features. Glia 2001;36:375–390. [DOI] [PubMed] [Google Scholar]
- 7. Huang C, Rajfur Z, Borchers C, Schaller MD, Jacobson K. JNK phosphorylates paxillin and regulates cell migration. Nature 2003;424:219–223. [DOI] [PubMed] [Google Scholar]
- 8. Brown MC, Turner CE. Paxillin: Adapting to change. Physiol Rev 2004;84:1315–1339. [DOI] [PubMed] [Google Scholar]
- 9. Deakin NO, Turner CE. Distinct roles for paxillin and Hic‐5 in regulating breast cancer cell morphology, invasion, and metastasis. Mol Biol Cell 2011;22:327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li L, Wang J, Gao L, Gong L. Expression of paxillin in laryngeal squamous cell carcinoma and its prognostic value. Int J Clin Exp Pathol 2015;8:9232–9239. [PMC free article] [PubMed] [Google Scholar]
- 11. Sen A, De Castro I, Defranco DB, et al. Paxillin mediates extranuclear and intranuclear signaling in prostate cancer proliferation. J Clin Invest 2012;122:2469–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu DW, Chuang CY, Lin WL, Sung WW, Cheng YW, Lee H. Paxillin promotes tumor progression and predicts survival and relapse in oral cavity squamous cell carcinoma by microRNA‐218 targeting. Carcinogenesis 2014;35:1823–1829. [DOI] [PubMed] [Google Scholar]
- 13. Huang CC, Wu DW, Lin PL, Lee H. Paxillin promotes colorectal tumor invasion and poor patient outcomes via ERK‐mediated stabilization of Bcl‐2 protein by phosphorylation at Serine 87. Oncotarget 2015;6:8698–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen DL, Wang ZQ, Ren C, et al. Abnormal expression of paxillin correlates with tumor progression and poor survival in patients with gastric cancer. J Transl Med 2013;11:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Short SM, Yoder BJ, Tarr SM, et al. The expression of the cytoskeletal focal adhesion protein paxillin in breast cancer correlates with HER2 overexpression and may help predict response to chemotherapy: A retrospective immunohistochemical study. Breast J 2007;13:130–139. [DOI] [PubMed] [Google Scholar]
- 16. Jagadeeswaran R, Surawska H, Krishnaswamy S, et al. Paxillin is a target for somatic mutations in lung cancer: Implications for cell growth and invasion. Cancer Res 2008;68:132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim G, Davidson B, Henning R, et al. Adhesion molecule protein signature in ovarian cancer effusions is prognostic of patient outcome. Cancer 2012;118:1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao Y, Zhang X, Guda K, et al. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc Natl Acad Sci U S A 2010;107:2592–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han X, Stewart JE Jr, Bellis SL, et al. TGF‐beta1 up‐regulates paxillin protein expression in malignant astrocytoma cells: Requirement for a fibronectin substrate. Oncogene 2001;20:7976–7986. [DOI] [PubMed] [Google Scholar]
- 20. Pu Y, Li S, Zhang C, Bao Z, Yang Z, Sun L. High expression of CXCR3 is an independent prognostic factor in glioblastoma patients that promotes an invasive phenotype. J Neurooncol 2015;122:43–51. [DOI] [PubMed] [Google Scholar]
- 21. Wu Z, Sun L, Wang H, et al. MiR‐328 expression is decreased in high‐grade gliomas and is associated with worse survival in primary glioblastoma. PLoS ONE 2012;7:e47270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun L, Yan W, Wang Y, et al. MicroRNA‐10b induces glioma cell invasion by modulating MMP‐14 and uPAR expression via HOXD10. Brain Res 2011;1389:9–18. [DOI] [PubMed] [Google Scholar]
- 23. Yan W, Zhang W, You G, et al. Molecular classification of gliomas based on whole genome gene expression: A systematic report of 225 samples from the Chinese Glioma Cooperative Group. Neuro Oncol 2012;14:1432–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Visser O, Ardanaz E, Botta L, et al. Survival of adults with primary malignant brain tumours in Europe; Results of the EUROCARE‐5 study. Eur J Cancer 2015;51:2231–2241. [DOI] [PubMed] [Google Scholar]
- 26. Lauffenburger DA, Horwitz AF. Cell migration: A physically integrated molecular process. Cell 1996;84:359–369. [DOI] [PubMed] [Google Scholar]
- 27. Gladson CL. The extracellular matrix of gliomas: Modulation of cell function. J Neuropathol Exp Neurol 1999;58:1029–1040. [DOI] [PubMed] [Google Scholar]
- 28. Calderwood DA, Shattil SJ, Ginsberg MH. Integrins and actin filaments: Reciprocal regulation of cell adhesion and signaling. J Biol Chem 2000;275:22607–22610. [DOI] [PubMed] [Google Scholar]
- 29. Taylor KA, Taylor DW, Schachat F. Isoforms of alpha‐actinin from cardiac, smooth, and skeletal muscle form polar arrays of actin filaments. J Cell Biol 2000;149:635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schaller MD. Paxillin: A focal adhesion‐associated adaptor protein. Oncogene 2001;20:6459–6472. [DOI] [PubMed] [Google Scholar]
- 31. Deramaudt TB, Dujardin D, Noulet F, et al. Altering FAK‐paxillin interactions reduces adhesion, migration and invasion processes. PLoS ONE 2014;9:e92059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Turner CE. Paxillin interactions. J Cell Sci 2000;113(Pt 23):4139–4140. [DOI] [PubMed] [Google Scholar]
- 33. Turner CE. Paxillin and focal adhesion signalling. Nat Cell Biol 2000;2:E231–E236. [DOI] [PubMed] [Google Scholar]
- 34. Das K, Bohl J, Vande SB. Pol, Identification of a second transforming function in bovine papillomavirus type 1 E6 and the role of E6 interactions with paxillin, E6BP, and E6AP. J Virol 2000;74:812–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nikolopoulos SN, Turner CE. Molecular dissection of actopaxin‐integrin‐linked kinase‐Paxillin interactions and their role in subcellular localization. J Biol Chem 2002;277:1568–1575. [DOI] [PubMed] [Google Scholar]
- 36. Ma PC, Kijima T, Maulik G, et al. c‐MET mutational analysis in small cell lung cancer: Novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res 2003;63:6272–6281. [PubMed] [Google Scholar]
- 37. Liu ZX, Yu CF, Nickel C, Thomas S, Cantley LG. Hepatocyte growth factor induces ERK‐dependent paxillin phosphorylation and regulates paxillin‐focal adhesion kinase association. J Biol Chem 2002;277:10452–10458. [DOI] [PubMed] [Google Scholar]
- 38. Wu DW, Wu TC, Wu JY, et al. Phosphorylation of paxillin confers cisplatin resistance in non‐small cell lung cancer via activating ERK‐mediated Bcl‐2 expression. Oncogene 2014;33:4385–4395. [DOI] [PubMed] [Google Scholar]
- 39. Ishibe S, Joly D, Liu ZX, Cantley LG. Paxillin serves as an ERK‐regulated scaffold for coordinating FAK and Rac activation in epithelial morphogenesis. Mol Cell 2004;16:257–267. [DOI] [PubMed] [Google Scholar]
- 40. Ueno H, Tomiyama A, Yamaguchi H, et al. Augmentation of invadopodia formation in temozolomide‐resistant or adopted glioma is regulated by c‐Jun terminal kinase‐paxillin axis. Biochem Biophys Res Commun 2015;468:240–247. [DOI] [PubMed] [Google Scholar]
- 41. Liu Z, Yang Z, Jiang S, et al. Paxillin and carbonic anhydrase IX are prognostic markers in gallbladder squamous cell/adenosquamous carcinomas and adenocarcinomas. Histopathology 2014;64:921–934. [DOI] [PubMed] [Google Scholar]
- 42. Panousis D, Patsouris E, Lagoudianakis E, et al. The value of TOP2A, EZH2 and paxillin expression as markers of aggressive breast cancer: Relationship with other prognostic factors. Eur J Gynaecol Oncol 2011;32:156–159. [PubMed] [Google Scholar]
- 43. Panousis D, Xepapadakis G, Lagoudianakis E, et al. Prognostic value of EZH2, paxillin expression and DNA ploidy of breast adenocarcinoma: Correlation to pathologic predictors. J BUON 2013;18:879–885. [PubMed] [Google Scholar]
- 44. Wu DW, Cheng YW, Wang J, Chen CY, Lee H. Paxillin predicts survival and relapse in non‐small cell lung cancer by microRNA‐218 targeting. Cancer Res 2010;70:10392–10401. [DOI] [PubMed] [Google Scholar]


